
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 08 March 2021
Sec. Genomic Medicine
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.535123
This article is part of the Research TopicPersonal Genomes: Accessing, Sharing, and InterpretationView all 13 articles
Although best practices have emerged on how to analyse and interpret personal genomes, the utility of whole genome screening remains underdeveloped. A large amount of information can be gathered from various types of analyses via whole genome sequencing including pathogenicity screening, genetic risk scoring, fitness, nutrition, and pharmacogenomic analysis. We recognize different levels of confidence when assessing the validity of genetic markers and apply rigorous standards for evaluation of phenotype associations. We illustrate the application of this approach on a family of five. By applying analyses of whole genomes from different methodological perspectives, we are able to build a more comprehensive picture to assist decision making in preventative healthcare and well-being management. Our interpretation and reporting outputs provide input for a clinician to develop a healthcare plan for the individual, based on genetic and other healthcare data.
A great deal of literature has been generated over the past decade defining best practices for clinical interpretation of personal genomes (Nykamp et al., 2017; Biesecker et al., 2018; Brandt et al., 2019; Machini et al., 2019). Some additional approaches involve the simultaneous analysis of parents and child, for example in the case of pediatric diagnosis for children with rare diseases (Wright et al., 2018). Other studies have used family genomes to assign the precise chromosomal position of variants (Roach et al., 2011). To our knowledge, however, the use of genome analysis for screening and disease prevention remains underdeveloped. To address this shortcoming, our current study sheds light on two areas. First, we provide a comprehensive whole genome analysis of pathogenicity screening, genetic risk, pharmacogenomic, fitness, and nutrition trait analysis. Second, we discuss the joint interpretation of these results within the perspective of a family of five for whom we have deep phenotypic knowledge, allowing us to find “true positive” predictions based on the family observations.
In the past, we have performed assessment of personal genome analysis for the same set of family individuals using direct to consumer data and crowdsourcing methods (Glusman et al., 2012; Corpas et al., 2015). We were limited by the amount of data available at the time (DNA chip or exome) as well as a lack of reference data sources and analysis platforms to help with the interpretation that have appeared in recent years [e.g., gnomAD (Karczewski et al., 2019), ClinVar (Landrum et al., 2014)]. In this new iteration, we perform whole genome sequencing analysis for the same family of five and expand from our previous research to encompass a more comprehensive set of analyses and individual genomic data following published standard practice for interpretation of results as much as possible. Following standard practice is not always possible given that authoritative guidelines for interpretation of variants (Richards et al., 2015) are mostly applied to pathogenicity screening, rather than preventative healthcare using personal genomes. We were indeed able to perform a pathogenicity screening for the five members of the family quintet. For four genomes we also report genetic risk scores for 49 phenotypes using published Genome Wide Association Study (GWAS) markers (see Supplementary Table 1). We make a distinction between our genetic marker score notation and polygenic risk scores in the literature (Khera et al., 2018; Georgi et al., 2019; Meisner et al., 2019; Palmer, 2019; Torkamani and Topol, 2019) as we only use markers reported above a certain threshold of probability (as defined by GWAS studies).
To date, genome analysis of pathogenicity screening, genetic risk scoring for cardiovascular disease and some pharmacogenomics characterization has been performed by the MedSeq project for 100 individuals (Machini et al., 2019). Compared to this study, we offer novel perspectives on several fronts: (1) Our genetic risk analysis encompasses mental, metabolic, and autoimmune diseases, in addition to only cardiovascular being done in the MedSeq Project. (2) We include a systematic curation of known nutrition and fitness markers following newly developed guidelines to evaluate the scientific validity of gene x lifestyle interventions (Grimaldi et al., 2017). (3) Our deep phenotype and clinical knowledge of analyzed participants, helps us interpret and report results in a familial context within a wellness and prevention point of view. In addition, this work provides a proof-of-principle approach about an application of genetic risk scores within a family-oriented preventative healthcare and well-being case, recognizing that we are studying only one family and therefore this represents only an illustration of our proposed methodology for comprehensive whole genome analysis. Whenever possible, we use established guidelines from the American College for Medical Genetics and Genomics (ACMG), Food and Drug Administration (FDA), the Clinical Pharmacogenetics Implementation Consortium (CPIC), and other specialized organizations. We also discuss how different whole genome analysis methods can be integrated into more actionable outcomes for the individual and his or her relatives.
This project builds on prior work (Glusman et al., 2012; Corpas et al., 2015). We started as an open source project in 2010 using the data available from direct to consumer providers. As the project evolved and exome sequencing was performed, a consent form was created and signed for the collection of samples, analysis, and publishing of results. This form identified participants as voluntary donors of their genetic data to the public domain and educated participants, making them aware of the potential discomforts and risks that doing this research might bring.
Here we base our analysis on the whole genome rather than the exome. To facilitate this work, further collection of samples has been performed in order to sequence and analyse whole personal genomes for this family. All participants underwent a new consent process and signed a consent form accepting the terms and conditions of this work as well as the potential consequences of performing such analysis. When developing the consent framework, we drew on the Personal Genome Project UK (PGP-UK Consortium, 2018) as an example of a rigorous approach to informed consent. As a result, the consent process developed for this work included the following elements: (a) participants underwent extensive training on the risks of genetic analysis including the risks of publishing personal genetic data; (b) participants completed an exam to demonstrate their comprehension of the risks and protocols associated with participating in genetic analysis which may be published and (c) participants were judged truly capable of giving informed consent. Consent forms were signed by all family individuals or their next to kin (in the case of a deceased member). This ethical framework has been independently assessed and approved by the Ethics Committee of Universidad Internacional de La Rioja (code PI:029/2020).
We selected this family dataset for two reasons: (1) We have performed and published in the past decade two studies describing state of the art personal genomics analysis for a family of related individuals using array chip data and Illumina exome data (Glusman et al., 2012; Corpas et al., 2015). (2) The accumulated genetic studies and follow up of the disease and lifestyle history of the family through their continuous research have afforded us a deep knowledge of their phenotypes and disease history. Figure 1 shows the family pedigree. In it we have individuals PT00010A (Aunt), who is the sister of PT00008A (Mother). PT00007A (Father) is Mother's spouse and both have two children (PT00009A and PT00002A; Daughter and Son). From here onwards, and for simplicity, we refer to family members as (Aunt, Father, Mother, Daughter, Son). All individuals of the family except Aunt had their DNA sequenced from saliva, whereas Aunt's DNA was sequenced from hair (see Supplementary Materials for details). This is because at the time of sample collection Aunt was already deceased (see next section for phenotypic details). Thirty-six hairs were retrieved from a personal comb only she used and her DNA extracted from hair roots using a different protocol described in the Supplementary Materials.
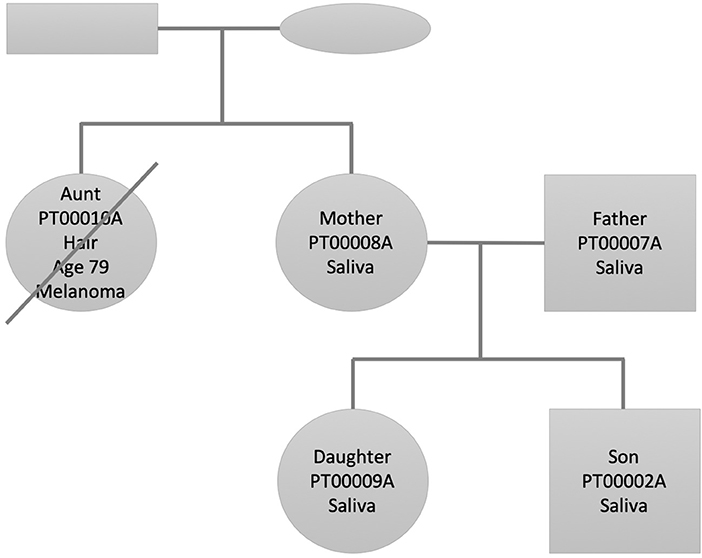
Figure 1. Family pedigree showing the relationship, gender (square: male, circle: female), and sample used for whole genome sequencing (saliva/hair). The crossed circle indicates a deceased individual.
When we analyzed the variant output of all samples, we benchmarked against Fabric Genomics Clinical Grade Scoring Rules (http://help.fabricgenomics.com/hc/en-us/articles/206433937-Appendix-4-Clinical-Grade-Scoring-Rules; accessed 7/January/2020), where Clinical Grade is a measure of a variant file's overall quality and fitness for clinical interpretation. The hair sample failed the criteria for clinical-grade coverage, genotype quality, homozygous/heterozygous ratio, and transition/transversion ratio (Table 1).
We performed a further analysis of quality of variants by counting those that pass the default standard filters of quality for interpretation given our analysis software (Table 2; see Supplementary Materials). For Aunt, we eliminated all variants below the threshold of QUAL < 20. The performance of the variant count and the level of coverage was sufficient to include Aunt in pathogenicity screening, but not sufficient for participation in the rest of the analysis.
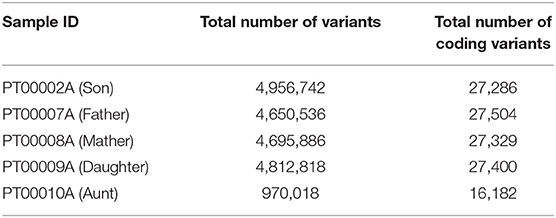
Table 2. The total number of variants for all saliva samples and the total number of coding variants for each family member.
We conducted research into the family disease and lifestyle history. This research consisted of face-to-face interviews with all family members, during which they were asked about past illnesses, hospitalizations, reasons of death for past relatives and any ongoing condition that they think might related to the phenotypes and traits we analyse in this study. At the time of our last interview (October 2020), Mother and Father are in their mid-eighties, a similar age Aunt would be, had she not passed due to metastasised melanoma at age 79. Daughter is in her late fifties and Son mid-forties. All members of the family have been diagnosed obese or overweight at some point in their adulthood years. Childhood obesity was present in both Son and Daughter. Mother had a benign breast tumor removed in her early forties. She has also suffered from a history of low blood pressure and was diagnosed with chronic inflammation of her colon in her sixties, suffering from lower abdominal pain ever since. Father has a history of high blood pressure and heart problems. He has recently been diagnosed with atrial fibrillation. He displays difficulty breathing at moderate exertion levels and has been taking anticoagulants to prevent thromboembolism as a consequence of his atrial fibrillation, with some episodes of adverse drug reactions to warfarin. He is suspected to be lactose intolerant. In addition to her metastasised melanoma, before Aunt's passing she suffered from several episodes of venous thromboembolism, treated with anticoagulants (warfarin). There is no history in the family of diabetes or Parkinson's disease, although the father of both Mother and Aunt was diagnosed with Alzheimer's disease in his mid-eighties. Apart from Aunt's melanoma, there is no history of any other malignancy known to the family, no major mental health episodes or alcohol dependence diagnosed to date. All family members except Mother reported being light smokers for a period of their lives, all having quit more than a decade ago except daughter who still smokes several cigarettes a day.
All single nucleotide variants and indels were filtered according to three different gene panels: (1) genes present in the OMIM morbid list (Amberger et al., 2015), (2) ACMG 59 genes (Kalia et al., 2017), and (3) a Hereditary Cancer panel of 52 genes (https://info.fabricgenomics.com/ace; accessed 10/February/2020). All three panels required pathogenic or likely pathogenic alleles matching ClinVar (Landrum et al., 2014) evidence. The variant prioritization was based on their ClinVar evidence, their frequency in gnomAD (Karczewski et al., 2019), the 1000 Genomes Project (The 1000 Genomes Project Consortium, 2015) and their predicted variant effect (i.e., loss of function, non-synonymous or other). Our selection of frequency threshold is based on the gnomAD database (https://gnomad.broadinstitute.org/faq) criteria of common variant sites, defined as frequency >0.01. For each of the variants that passed the filtering, we classified them following the guidelines proposed by the ACMG (Richards et al., 2015) into 5 categories; from most to least pathogenic these categories are: pathogenic, likely pathogenic, uncertain significance, likely benign, benign. Relevant scientific literature as well as a number of algorithms were also used to assess each prioritized variant [i.e., OMICIA (Coonrod et al., 2013), VAAST (Hu et al., 2013), VVP (Flygare et al., 2018), and CADD (Rentzsch et al., 2019)].
Genetic risk scores, also called genetic predisposition scores, aim to quantify the cumulative effects of a number of variants affecting multiple genes, which may individually confer only small risk susceptibility. Genetic risk scores are not diagnostic, as a high-risk score does not necessarily mean that a person will develop a condition, and a low score does not mean that they will not develop it. Nevertheless, genetic risk scores may be pointers for further exploration when looking for potential preventative interventions, particularly for multigenic conditions like diabetes type 2, hypertension or many mental illnesses. They can be useful when other independent sources of risk information are also concordant [e.g., genotype/phenotype additional knowledge (Fahed et al., 2020), family history, imaging data]. A database of 4,688 published GWAS SNPs was generated encompassing 49 common diseases (we call these common diseases “phenotypes” from now onwards; Supplementary Table 1), their risk alleles and weighted contributions (odds ratio or beta scores). These phenotypes were selected according to GWAS Catalog criteria (https://www.ebi.ac.uk/gwas) as having studies including a primary GWAS analysis, defined as array-based genotyping and analysis of 100,000+ pre-QC SNPs selected to tag variation across the genome and without regard to gene content. Individual SNP-trait associations were collected with a statistical significance (SNP-trait p-value <1.0 x 10−5) in the overall (initial GWAS and replication) population. To create genetic risk scores, each collected SNP marker was required to possess (a) the risk allele and (b) the measurement or effect size that this risk allele confers to the individual that carries this mutation. A genetic risk score was calculated as the sum of the weights of all the phenotype's risk alleles observed in the individual divided by the total number of alleles reported for that phenotype. We used the final (Phase 3) dataset of the 1000 Genomes Project containing data for 2,504 individuals from 26 populations to calculate their genetic risk scores for each of the 49 phenotypes. The 1000 Genomes Project individuals became our background distribution of genetic risks against which to measure how far from the mean each of the family participant lies. We required that the identified GWAS SNPs are also present in the 1000 Genomes Project since individuals from the 1000 Genomes Project were used as a background population to which compare the participant's score. In order to control for potential differences in results due to the ethnic diversity of the background population, we also performed the analysis using a background population of only the 503 European (CEU) participants in the 1000 Genomes Project, given that all family members are of European origin.
We plotted the genetic risk score of each family member to establish whether he or she lies on the higher tail of the distribution of scores in relation to the calculated risk scores of 1000 Genomes Project individuals. In order to evaluate whether a member of the family had a reportable genetic risk, we applied a two standard deviations (2SD) threshold from the mean genetic risk score of the background 1000 Genomes individual distribution for a particular phenotype (equivalent to the top 5 percentile normal distribution of a predicted risk). We use a threshold of 2SD to give confidence that results are not attributable to chance. Furthermore, scores from both 1000 Genome individuals and family members are calculated independently. Multiple testing correction is not performed since the objective here is to identify family members in the extreme risk tail of the 1000 Genomes background distribution of calculated scores. For completeness, we also noted those phenotypes for which a greater than one standard deviation (1SD) from the mean background genetic risk is reached by the tested family individual.
We analyzed three well-known genes influencing pharmacogenomic responses, all of them forming part of the Cytochrome P450 family: CYP2D6, CYP2C9, CYP2C19. In order to extract relevant pharmacogenomic data, we rely on Food and Drug Administration (FDA) and European Medicines Agency (EMA) guidance sourced via the PharmGKB database (https://www.pharmgkb.org). We also take guidance from the Clinical Pharmacogenetics Implementation Consortium (CPIC; https://cpicpgx.org), the Association for Molecular Pathology and College of American Pathologists (https://www.amp.org), and the American College of Medical Genetics and Genomics (https://www.acmg.net). In order to perform the genotyping of pharmacological genes we allow the extraction of non-variant positions if required.
The testing of specific positions within a gene provides an accurate representation of the metaboliser status of an individual for that particular gene. For instance, if we were to assume that an individual has a nucleotide change 1846G>A in CYP2D6, this polymorphism is determinant for allele *4. If there are no other variants, it is presumed that the other allele this person harbors is a wild haplotype, being denoted as *4/*1 [see (Nofziger et al., 2020) for more detail]. Once both haplotypes for an individual are identified, a lookup table is referenced where the pharmacological effect of the observed haplotypes are indexed.
Pharmacological analysis also depends on whether analyzed genes have their copy number altered. We performed a consensus-based algorithm prediction using the short-read sequencing data for Father, Mother, Daughter and Son (Supplementary Materials). This analysis did not yield significant evidence for presence of copy number alterations in all Cytochrome P450 genes analyzed here.
Besides pathogenicity screening, genetic risk scoring, and pharmacogenomics, there is further useful information that can be extracted from whole genomes using genotyping. In particular, we identify two areas of interest that provide further information about a person's genetic load: fitness and nutrition. We recognize that these areas of genetic analysis are less developed than pathogenicity screening, and so we add rigor to the analysis by first evaluating the quality of supporting evidence before testing for the presence of variants in the family. To evaluate the scientific validity and evidence for genotype-based dietary or fitness advice, we first performed a literature search to identify an initial list of potential genetic markers, and then adopted the proposed recommendations of Grimaldi et al. (2017) for specific gene x interactions and their relation to a health outcome. This framework allows us to establish levels of confidence for each of the SNPs or groups of SNPs we analyse for fitness and nutrition, according to a set of peer-reviewed guidelines. These guidelines differentiate four levels of scientific evidence assessment:
• “Convincing”: gene x interaction is based on at least 3 studies with high subject numbers, showing the relation and mechanistic knowledge.
• “Probable” is based on several studies showing the relation and/or some mechanistic understanding.
• “Possible”: based on a few studies showing the relation.
• “Not demonstrated” is any level of evidence below the established above.
The levels of assessment above rely on the following criteria:
• “Study quality rating”: either A, B, C or D, based on whether a study is (a) interventional or observational; (b) prospective or retrospective, (c) whether it is randomized, placebo controlled and blinded; (d) the number of subjects with effect alleles (where possible); (e) the effect magnitude; (f) P-values, false discovery rate and multiple testing and; (g) replications in other populations and meta-analyses.
• “Type of gene x interaction”: direct phenotype, intermediate phenotype, or indirect phenotype.
• “Nature of genetic variant”: causal, in linkage disequilibrium with functional variant or associated but unknown function.
• “Biological plausibility,” rated as high, medium, low, or unknown, based on our critical assessment of current understanding of the physiological effect of identified SNPs.
Our initial selection and classification of genotyping markers for fitness and nutrition are shown in Tables 3, 4. We then assess each marker according to the above criteria of scientific evidence, carrying forwards for analysis in the family participants those classified as convincing (fitness n = 2; nutrition n = 13) and probable (fitness n = 5; nutrition n = 1).
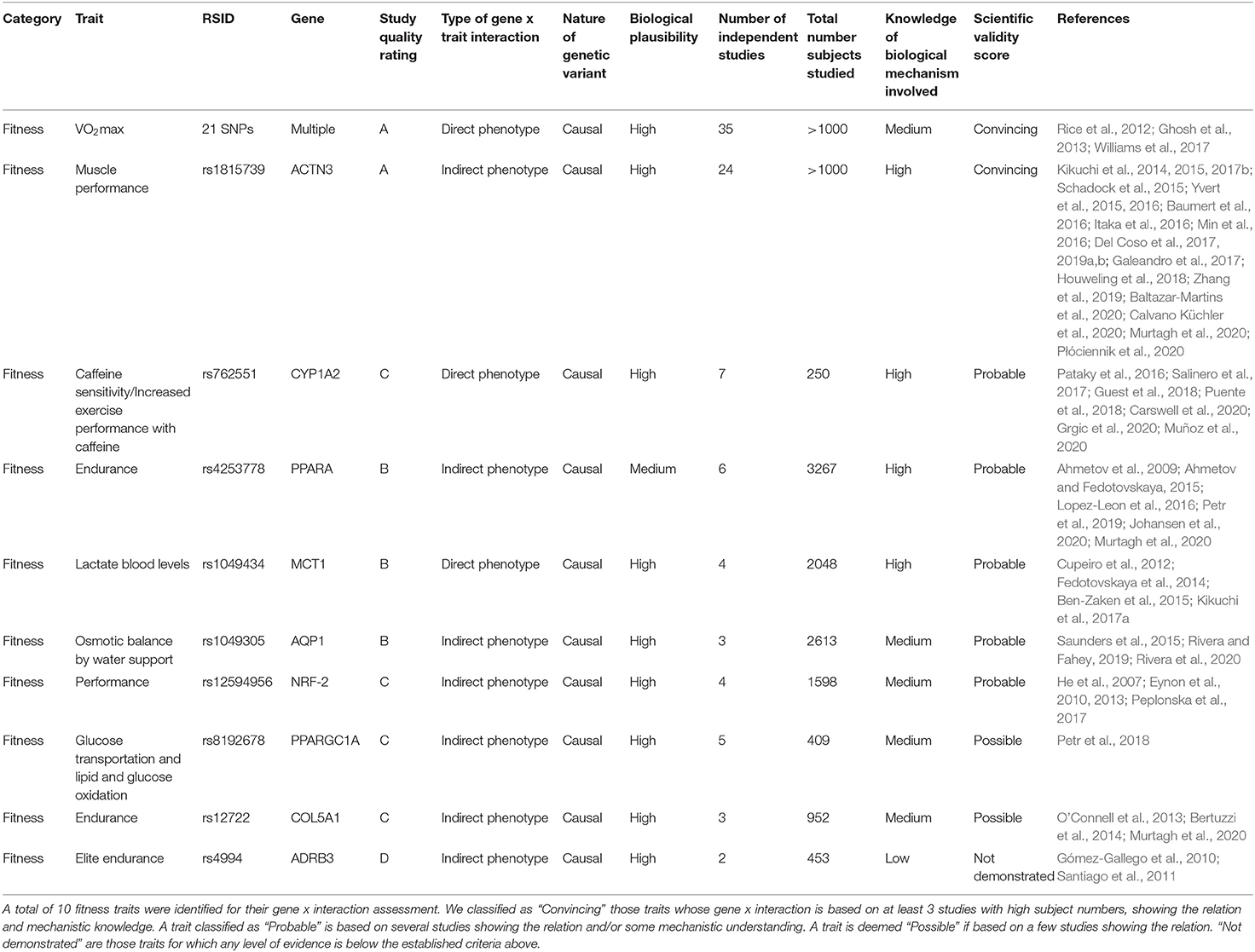
Table 3. Summary of fitness trait analysis candidates assessed according to the scientific validity score as proposed by Grimaldi et al. (2017).
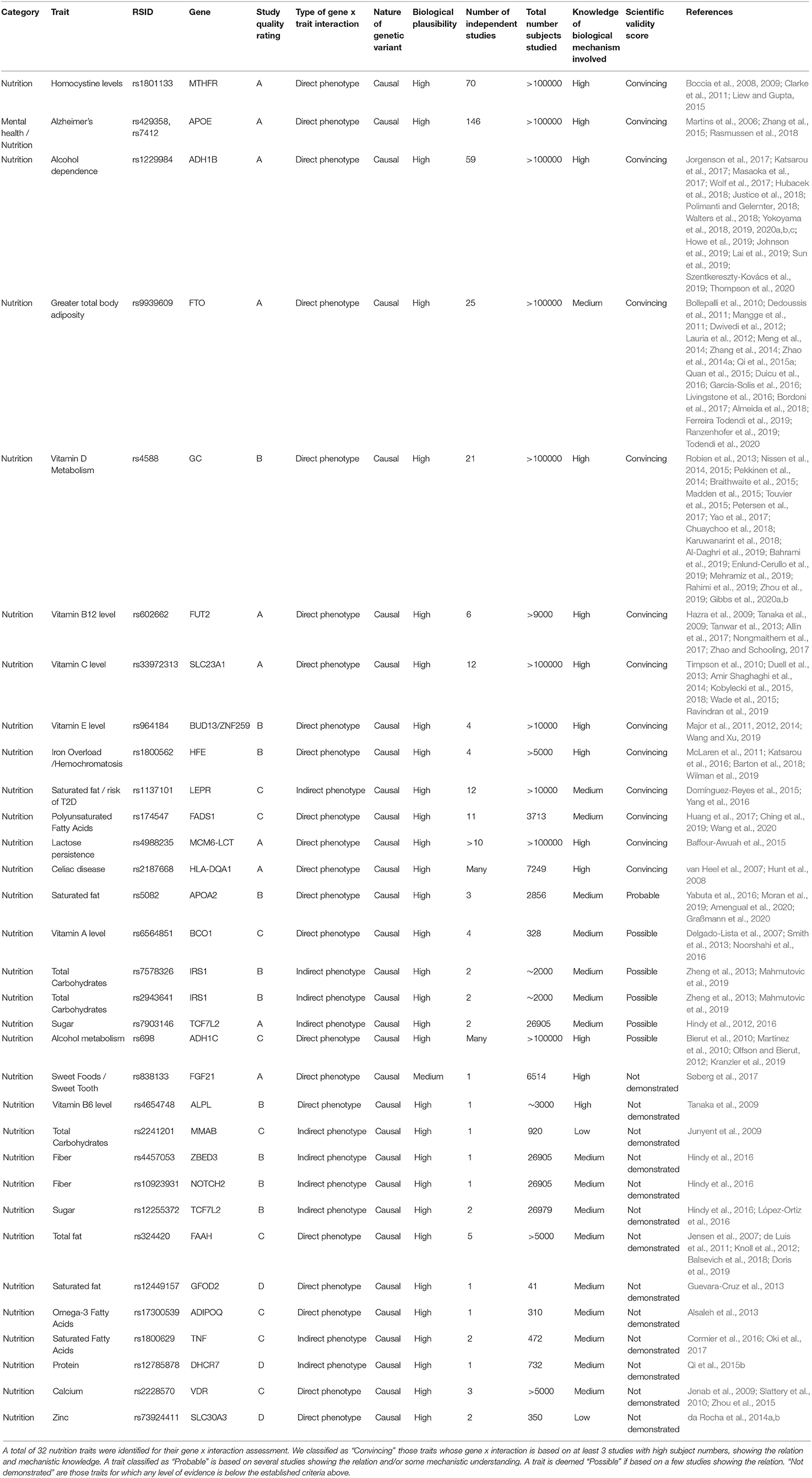
Table 4. Summary of nutrition trait analysis candidates assessed according to the scientific validity score as proposed by Grimaldi et al. (2017).
Having made the selection of relevant SNPs according to the above framework, we proceed to analyse the family. The trait analysis is performed as follows. First, a list of all the positions of the SNPs to be tested is created. All those positions are queried in the VCF files for each of the family members and all observed alleles are then recorded. The observed alleles are then interpreted via lookup tables collected from the scientific literature.
An exception to the above approach concerns the phenotype susceptibility to VO2max trainability, where we use a specific study. To calculate the VO2Max trainability genetic score, we follow the methodology outlined in Bouchard et al. (2011), that identifies SNPs associated with improvements in VO2Max. This study provides a panel of 21 SNPs that accounted for 49% of the variance in VO2Max trainability.
Figure 2 shows a summary of the pedigree and filtered variants found listed within each individual. For Son, when searching for pathogenic or likely pathogenic mutations within a panel of 4,100 OMIM morbid genes, we found that only two mutations passed our prioritization and filtering criteria (see Methods section). No other mutations passed the threshold criteria within the ACMG59 and Hereditary Cancer panels. The first mutation is a heterozygous missense change of C → T; c.200C>T; p.Thr67Ile within the CTH gene. This change has been associated to cystathioninuria, a disorder observed in 1 out of 20,000 individuals (ORPHANET, Pavan et al., 2017). However, the ExAC (Lek et al., 2016) frequency in non-Finnish European is (~1 in 100), so much higher than the prevalence of the disorder. We also observe that this missense variant is not excessively constrained: its missense z-score is −0.127428 (excessively constrained genes are those with a missense z-score > 3.09, corresponding to a p-value < 0.001). Multiple lines of computational evidence suggest no impact on the gene. Our current assessment is that this variant is benign according to the ACMG scoring and inferred classification and is therefore not considered any further. The second mutation for Son corresponds to chr11:111764842 (rs1805076) producing C → T; c.269G>A; p.Gly90Asp in PPP2R1B. This gene encodes a regulatory subunit of protein phosphatase 2. Protein phosphatase 2 is one of the four major Ser/Thr phosphatases, and it is implicated in the negative control of cell growth and division. While ClinVar evidence suggests a matching allele to cause lung cancer, the computational and other sources of evidence are inconclusive, hence we infer this variant to be of uncertain significance (VUS).
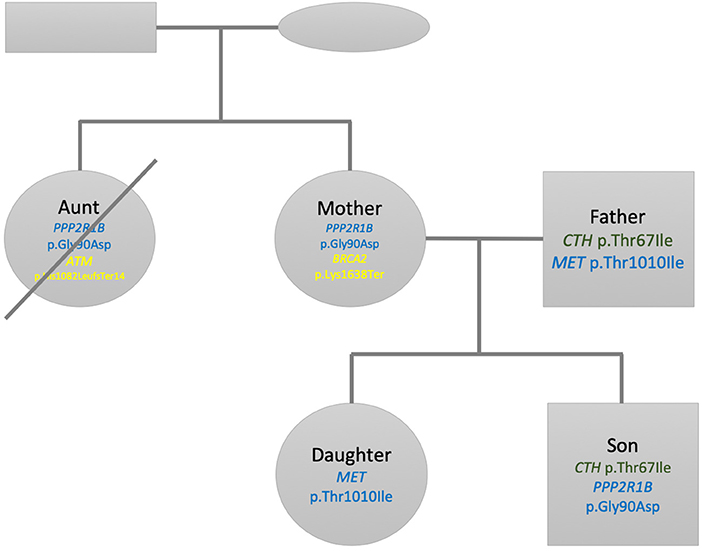
Figure 2. Family pedigree showing the relationship, gender (square: male, circle: female), and variants found listed within each individual. Green variants are inferred as benign, blue are variants of unknown significance and yellow pathogenic variants according to ACMG scoring. The crossed circle indicates a deceased individual.
For Father two variants pass the filters. The first variant is selected from the 4100 genes OMIM panel and corresponds to the heterozygous missense change of C → T; c.200C>T; p.Thr67Ile within the CTH gene, which is the same one Son has. We set the same classification as above and conclude it to be a benign variant as well. The second variant is located in the MET gene, part or our Hereditary Cancer panel, on chr7:116771936 (rs56391007) and produces C → T; c.3029C>T; p.Thr1010Ile. This gene encodes a member of the receptor tyrosine kinase family of proteins and the product of the proto-oncogene MET. The MET gene is associated with autosomal dominant hereditary papillary renal cell carcinoma (Takahashi et al., 2002). According to ORPHANET (Pavan et al., 2017), the prevalence of this cancer is less than 1 in 1,500,000, while the allele frequency of this variant is 1 in ~89 in non-Finnish European, higher than expected for the disorder. There are seven pathogenic and 15 likely pathogenic ClinVar missense variants in this gene while there are two benign and 25 likely benign ClinVar missense variants in this gene. Evidence thus indicates that missense variants are not a common mechanism of disease. In addition, there are multiple sources that point to this mutation being both pathogenic and likely benign. We conclude that this variant is of uncertain significance.
The first selected mutation for Mother comes from the OMIM disease gene panel and is the same as the one previously reported for Son corresponding to chr11:111764842 (rs1805076) producing C → T; c.269G>A; p.Gly90Asp in PPP2R1B. We apply the same criteria as above, inferring this variant effect being of uncertain significance. The second variant selected for interpretation in Mother corresponds to a heterozygous stop gained mutation in chr13:32339267 (rs886040553) producing the change A → T; c.4912A>T; p.Lys1638Ter in the BRCA2 gene. The gene is included both in the ACMG 59 and our Hereditary Cancer gene panel. The impact of this mutation is stop-gained in a splice site. ClinVar evidence contains several entries in its classification history all matching the allele and all pathogenic. This variant is absent from gnomAD or the 1000 genomes project. We thus classify this entry as pathogenic according to the ACMG scoring and inferred classification and as associated to hereditary breast and ovarian cancer syndrome. However, since the gene is autosomal recessive, both alleles need to be affected in order to cause the disorder.
For Daughter we did not find any mutation in the panels OMIM disease genes and ACMG 59 after our filtering criteria. One mutation passes our criteria for the Hereditary Cancer panel, corresponding to the variant located in chr7:116771936 (rs56391007) producing C → T; c.3029C>T; p.Thr1010Ile, located in the MET gene, identified for Father as well. As above, we conclude that this variant is of uncertain significance.
We prioritize the filtered variants from Aunt's variant file. After performing the selection of variants above the VCF threshold quality of 20, we identify three variants. Among these three variants, we decided to discard a homozygous variant corresponding to chr1:25563142 (rs121908326) in the LDLRAP1 gene, as this one did not qualify to have a sufficient genotype quality (minimum acceptable threshold = 40). Among the remaining two variants, the first one corresponds to chr11:111764842 (rs1805076), producing C → T; c.269G>A; p.Gly90Asp in PPP2R1B, already observed in Son and Mother. We apply the same criteria as above, inferring this variant effect being of uncertain significance. The remaining heterozygous variant, chr11:108272812 (rs587776549) in the ATM gene, produces a frameshift mutation CATC → CTGATc.3245_3247delinsTGAT p.His1082LeufsTer14. The ATM protein plays a critical role assisting cells in recognizing damaged or broken DNA strands, enabling them to repair broken strands and work on maintenance of the stability of the cell's genetic information. The mutation identified here has been described as pathogenic, involved in ataxia-telangiectasia syndrome and has also been linked to a hereditary cancer predisposition (Laake et al., 2000). Mutations to the ATM gene have a 20% to 30% lifetime risk of lymphoid, gastric, breast, central nervous system and skin, including melanoma (Choi et al., 2016). We conclude this variant as being pathogenic.
Given the limitations of the VCF file produced for Aunt, for the reminder of the results section, we are only able to perform further analyses for Father, Mother, Daughter and Son; analyses which include genetic risk scores, pharmacogenomics and nutrition/fitness traits.
From our initial list of 49 GWAS phenotypes (Supplementary Table 1), we identified members of the family who have a risk score (or predisposition) of one or two standard deviations (SD) from the average risk score of the 1000 genomes population for the same condition. Table 5 shows the phenotypes whose genetic score from the initial list is more than 2SD (yellow) and those with more than 1SD (green). The GWAS studies from which the genetic risk SNPs originated are sourced in the “Reference Studies” column.
First, we find that no one phenotype in yellow (>2SD) occurs in isolation. There is either another member of the family in yellow or green (>1SD). This occurs for the predicted higher risks of ulcerative colitis and nicotine withdrawal symptoms. Second, we observe that related phenotypes with high risk overlap (although not always) in the same family individual. For instance, ulcerative colitis is a subtype of inflammatory bowel disease (Ronald et al., 2006; Wu et al., 2009a; Uhlig and Muise, 2017). We find that Mother and Daughter have risks overlapping both phenotypes whereas Son only ulcerative colitis. Father has a risk for three of the four phenotypes for the mental health category. For the disorders where we observe this overlapping phenotype risk, there is scientific literature (Ronald et al., 2006; Wu et al., 2009a; Uhlig and Muise, 2017) supporting this pattern. Third, there are high risk phenotypes in a parent also observed in their offspring. For instance, ulcerative colitis is not present in Father but it is predicted high risk in Mother together with both children displaying inherited risk.
If we describe results according to categories, for general health, we thus find that ulcerative colitis constitutes a phenotype where the risk of the condition is shared among several family members (Mother, Daughter, Son; Supplementary Figures 1A,B). The high risk for ulcerative colitis also overlaps with the higher than average (>1SD) risk of inflammatory bowel disease in two same family members (Mother, Daughter). For cancer there is a more elevated than normal (>1SD) predicted risk of breast cancer among Mother and Daughter, with some isolated moderately high (>1SD) predicted risks of bladder carcinoma for Daughter, glaucoma for Son and prostate cancer for Father. For mental health, Father has higher than normal (>1SD) predicted risk on bipolar disorder, depression, and posttraumatic disorder. Mental health diseases share similar markers thus influencing the greater number of potentially deleterious yet related phenotypes observed in Father. For dependence and withdrawal symptoms phenotypes we find that the paternal line has a higher than average alcohol (>1SD) dependence predicted risk whereas the maternal line passes on to Son a high predicted risk (>2SD) of nicotine withdrawal symptoms. Phenotypes in green (>1SD) that are not shared with other family members are not considered any further. Graphical representation of the ulcerative colitis results using both the whole 1000 Genomes background population (2,504 individuals) and only the Europeans (503) may be found in Supplementary Figure 1.
We analyzed the metaboliser status of three cytochrome P450 genes (CYP2C9, CYP2C19, CYP2D6) affecting pharmacological responses in Father, Mother, Daughter, and Son. We also look at some pharmacology-related SNPs in additional genes.
CYP2C9 is responsible for the metabolic clearance of up to 15–20% of all drugs undergoing Phase 1 metabolisation, including warfarin, phenytoin, and oral hypoglycaemics (source: Get to Know an Enzyme: CYP2C91). Some of the more potent CYP2C9 inhibitors include amiodarone, fluorouracil, metronidazole, and sulphaphenazole. Dangerous drug-drug interaction can arise when an inhibitor is added to a therapeutic regime that includes drugs with a low therapeutic index, such as s-warfarin. Inducers, such as rifampicin, can substantially increase CYP2C9 activity (source: Get to Know an Enzyme: CYP2C9). For CYP2C9, Father, Mother and Son have a predicted metaboliser status of intermediate (*1/*2). For Daughter, the predicted metaboliser status for CYP2C9 is poor (*2/*2).
Warfarin is an anticoagulant used in the prevention and treatment of venous thrombosis, pulmonary embolism, and the complications associated with atrial fibrillation and/or cardiac valve replacement (Dean, 2018). Warfarin metabolism is influenced by genetic polymorphisms in CYP2C9 and VKORC1 (Biss et al., 2012). Carriers of the common allelic variants (*2 or *3) of the CYP2C9 are associated with a lower warfarin dose requirement accompanied by a greater tendency to experience haemorrhagic complications. In addition, adults with VKORC1 (rs9923231) CC alleles require higher warfarin doses than TC or TT. Based on these alleles, we found that Son and Father have a *1/*2 CYP2C9 variant and a TT for rs9923231. Mother has a *1/*2 CYP2C9 variant and a CT for rs9923231. Daughter has a *2/*2 CYP2C9 variant and a TT for rs9923231. This makes Son, Father and Mother intermediate metabolisers and Daughter a poor metaboliser of warfarin.
CYP2C19 is a liver enzyme that acts on at least 10% of drugs in current clinical use (source: Genetics Home Reference2; see references), most notably the antiplatelet treatment clopidogrel (Plavix) but also drugs that treat pain associated with ulcers, such as omeprazole, antiseizure drugs such as mephenytoin, the antimalarial proguanil, and the anxiolytic diazepam. For this gene we found Son, Mother and Daughter to be predicted normal metabolisers (*1/*1) whereas Father is predicted an intermediate metaboliser (*17/*4A).
For CYP2D6, the final cytochrome we analyse here, we are able to estimate the metabolism and elimination of approximately 25% of clinically used drugs including the opiate codeine (Wang et al., 2009). CYP2D6 is highly polymorphic in the human population, with marked inter-racial variation observed. Individuals are identified as ultra-rapid (UM), extensive (EM), intermediate (IM) or poor metaboliser (PM), according to the number of functional alleles.
For members of this family we find that there is considerable variation in the alleles detected. For Son and Father, we find them to be predicted extensive metabolisers (*2/*41 and *1/*2, respectively). Mother has the following star alleles *10/*2/*41/*4 [activity score: 0.5–1 (Gaedigk et al., 2018)] which make her predicted range between an intermediate and extensive metaboliser. Daughter has *10/*4/*20, which makes her a poor or intermediate predicted metaboliser (activity score 0–0.5).
rs12979860 is a SNP near the IL28B gene, encoding interferon-lambda-3 (IFN-lambda-3). This SNP influences hepatitis C treatment-induced viral clearance. It is associated with an approximately twofold change in response to pegylated interferon-alpha (PEG-IFN-alpha) plus ribavirin (RBV) treatment, both among patients of European ancestry (p = 1.06 x 10e-25). Research indicates that the virus was eradicated in ~80% of CC patients, compared to only about 25% of those with TT, while CT response was intermediate (Elkader and Sproule, 2005). We found that Son and Father carry a CC genotype, whereas Mother and Daughter carry a CT genotype.
First, we performed a filtering of the SNPs associated with fitness traits in order to determine which of them should be applied to our family cohort. The full results of our filtering can be found in Table 3. From a total of 10 markers initially selected for genotyping, we classified two as “Convincing” (VO2max, and rs1815739 for ACTN3), five as “Probable,” two as “Possible” and one “Not demonstrated.” As an example of the application of this framework, in Table 3, the best studied SNP marker is rs1815739 for ACTN3. We identified 24 studies for rs1815739 in fitness, most of which suggested a significant decrease in muscle performance by the effect allele (also known as X allele). Based on these studies, we classify the biological plausibility of this marker as high. Our scientific evidence assessment for rs1815739 is “Convincing.”. For “increased performance with caffeine,” we assess existing scientific evidence as “Probable” because despite finding 7 studies, the total number of participants summed by all seven studies is only 250. Within those, there is also one study not showing significant differences in performance with coffee intervention (Pataky et al., 2016; Salinero et al., 2017; Guest et al., 2018; Puente et al., 2018; Carswell et al., 2020; Grgic et al., 2020; Muñoz et al., 2020).
For family trait analysis, we only apply those markers that are either classified as “Convincing” or “Probable”. In the next section we describe in detail our selected fitness analysis results.
Table 6 summarizes the fitness traits analyzed for 4 family members. Concerning VO2Max trainability, training response markers within the 21 SNP panel show Son scoring 13/21 favorable alleles, Father and Mother 16/21 favorable alleles and Daughter scores 15/21 favorable alleles. This contrasts with ≥19 of these alleles associated with elite athletes (Bouchard et al., 2011; Rice et al., 2012; Ghosh et al., 2013; Williams et al., 2017).
The ACTN3 R577X (rs1815739) C>T base substitution results in the transformation of an arginine amino acid (R) to a premature stop codon (X). X allele homozygotes are deficient in the alpha-actinin-3 protein, which is associated with a lower fast-twitch fiber percentage and potentially increased injury risk (Yang et al., 2003; Massidda et al., 2019). We found that Father, Mother and Daughter have a CT genotype (XR); whereas Son, harbors a homozygote X allele genotype (XX).
A polymorphism in the CYP1A2 gene (rs762551; AA genotype) has been associated with improved exercise performance when combined with caffeine intake, with no effect for those with the AC genotype and diminished performance in those with the CC genotype (Guest et al., 2018). We found that most family members (Son, Father, and Daughter) had an AA genotype for this SNP, whereas Mother had a CA genotype.
The role of the peroxisome proliferator activated receptor alpha (PPARA) gene intron 7 G/C polymorphism (rs4253778) is also tested in the family. Athletes with high ability in endurance sports have a higher frequency of the G allele (Lopez-Leon et al., 2016). We found that Son and Mother did not have any of the G allele, whereas Father and Daughter had a G allele each.
For the MCT1 gene's rs1049434, we find all family members to have the TT genotype, associated with lower lactate levels (Cupeiro et al., 2012; Fedotovskaya et al., 2014; Ben-Zaken et al., 2015; Kikuchi et al., 2017a). For the AQP1 gene, which is associated with osmotic balance and fluid loss when exercising, possession of the C allele has been associated with faster cardiorespiratory endurance (Rivera and Fahey, 2019). For this gene, we found C (favorable) alleles in Son, Father, and Daughter, while no C alleles were found in Mother. Finally, for rs12594956 in NRF-2, we find that the genotypes observed (CA/CC) in all family members are not associated with the effect allele (He et al., 2007; Eynon et al., 2010, 2013; Peplonska et al., 2017).
Our analysis includes markers involved in the metabolism of main components of diet: carbohydrates, fats, and proteins. We also look at metabolization of essential nutritional components such as vitamins, minerals, and specific dietary substances like lactose, whose metabolism is strongly linked to a genetic marker according to our suggested framework. Table 7 provides a summary of the nutrition markers explained in this section.
We performed a filtering of the SNPs associated with nutrition traits to select SNPS to be applied to our family cohort. The full results of our filtering can be found in Table 5. From a total of 32 markers initially selected for analysis, we classified 13 as “Convincing,” one as “Probable,” five as “Possible”, and 12 “Not demonstrated.” An example of convincing scientific evidence for nutrition interventions in Table 4 includes MTHFR (rs1801133). This SNP is said to affect homocysteine concentrations, which are influenced by dietary folate (Boccia et al., 2008, 2009; Clarke et al., 2011; Liew and Gupta, 2015). A large number of studies (n = 70) have been performed to date about this interaction, including randomized trials. We evaluate this interaction as having a high biological plausibility. An example of nutrition marker we classify as possible is BCO1 (rs6564851). According to our research (Table 4), there are 4 studies with a number of total subjects analyzed of 328 (Yabuta et al., 2016; Moran et al., 2019; Amengual et al., 2020; Graßmann et al., 2020). Our judgement of the underlying knowledge of the biological mechanism involved is medium and there are some cases where a potential intervention may not have the desired effect. We do not include this marker in our subsequent analyses.
Same as in the fitness category, for family trait analysis we only apply those markers that are either classified as “Convincing” or “Probable.” In the next section we describe in detail our selected nutrition trait analysis results.
The B vitamins contribute to DNA synthesis and methylation, with homocysteine as a by-product of their metabolism associated with coronary heart disease, stroke, and neurological disease (Tanaka et al., 2009). “A” alleles in the rs1801133 SNP within the MTHFR gene have been associated with higher homocysteine levels and reduced folic acid processing (Tanaka et al., 2009). We note that Father and Mother have one A allele whereas Daughter has the two A (“detrimental”) alleles. Son has the two G alleles genotype. Next, the presence of the A allele in rs602662 SNP in FUT2, has been associated with higher B12 concentrations (Tanaka et al., 2009). We found the presence of an A allele in Father and Daughter, and no A allele presence in the other individuals. With regards to circulating concentrations of vitamin C (L-ascorbic acid), a variation at rs33972313 (SLC23A1 gene) has been associated with a reduction in circulating concentrations of L-ascorbic acid (Timpson et al., 2010). None of the family members have the predicted detrimental allele. With regards to vitamin D, rs4588 was genotyped. Son was found homozygous for the major allele (GG) and the rest of the family heterozygous for the minor allele (GT). The effect allele for higher a-tocopherol concentration in plasma (G) is found in both alleles in Mother (GG) and one allele in Son and Daughter (Major et al., 2011, 2012, 2014; Wang and Xu, 2019).
With regards to dietary fat, we analyse a number of SNPs in genes involved in nutrition: FTO, APOA2, FADS1, and LEPR. With regards to rs9939609 FTO variant alleles (homozygous = AA and heterozygous = AT), both Son and Daughter are heterozygous for the risk allele and Father is homozygous for the risk allele. Each additional copy of the rs9939609 A allele has been associated with a BMI increase of a mean of 0.10 Z-score units, equivalent to ~0.4 kg/m2 (Sonestedt et al., 2009; Tanofsky-Kraff et al., 2009; Zhao et al., 2014b). For the observed genotypes in APOA2 and FADS1, there is no associated effect (Yabuta et al., 2016; Huang et al., 2017; Ching et al., 2019; Moran et al., 2019; Amengual et al., 2020; Graßmann et al., 2020; Wang et al., 2020). For LEPR, a study found that rs1137101 AG and GG carriers with a high fat total intake had 3.0 times higher risk of obesity and 4.1 times higher risk of high cholesterol levels than those with a low intake of total fat (Domínguez-Reyes et al., 2015). All family members are carriers of the risk allele (G) of rs1137101.
The HFE protein interacts with other proteins on the cell surface to detect the amount of iron in the body (Katsarou et al., 2019). For rs1800562, a SNP in HFE, an A allele was not observed in any of the individuals analyzed here. This A allele causes ~85% of all cases of hemochromatosis (Katsarou et al., 2019). The rs2187668 SNP's CC alleles in all family members have not been associated with Celiac disease (van Heel et al., 2007; Hunt et al., 2008). MCM6-LCT regulates lactose persistence. According to a recent study (Mattar et al., 2012), both genotypes of rs4988235 GA and AA were associated with the lactase-persistence phenotype, indicating that the presence of one single lactase-persistence allele in the heterozygous state has a dominant effect, rendering the person a lactose digester, whereas the genotype CC, when the lactase-persistence allele T is absent, is consistent with lactose maldigestion. Father's genotype was found to be CC associated with an increased likelihood of being lactose intolerant.
Our main objective is to provide insight into the current development status of personal genomics, using whole genome sequencing, illustrated by a use case of a family of five. To that end, we provide pathogenicity screening, genetic risk scoring, pharmacogenomics and fitness and nutrition trait analysis of the family. This approach is tailored for the situation where knowledge of the disease and lifestyle history of the family is used to “validate” some of the findings. A main limitation of this approach is the post-hoc reasoning that only allows to find true positive predictions based on the family observations. In contrast, those risks and phenotypes that are not reflected in the family so far can neither be confirmed nor rejected as it is unclear whether those predictions are “wrong” or whether the conditions have not had their time of onset yet. In addition, there are other limitations stemming from the different methodologies and resources used for analysis and interpretation, which we summarize in Table 8.

Table 8. List of known limitations of the methodologies we have performed for our analysis and the countermeasures we have adopted to contain them.
For instance, short read whole genome sequencing provides a limited capacity for detecting copy number and structural variants, which are particularly relevant for Pharmacogenomic analysis. To mitigate this shortcoming, we run prediction algorithms (Supplementary Materials) and find no significant prediction of copy number changes in pharmacologically important genes.
For pathogenicity screening, current standards and literature focus on genes (e.g., American College of Medical Genetics and Genomics), and therefore pathogenicity screening does not typically cover intergenic regions. Knowledge bases used for variant annotation may contain inconsistent or incomplete information, and therefore we only report variants where there is consensus among both literature, database and bioinformatic algorithm prediction, within a set of established guidelines. Moreover, while the field of genetics is evolving constantly, it is also a well-known limitation that many variants are currently classified as unknown significance. We do not report variants of unknown significance, but ensure that we use databases that remain current so that we can deploy the latest variant research in the analysis.
Concerning trait analysis in fitness and nutrition, we set a framework for selection and validation of fitness and nutrition markers to mitigate the limitations specific to phenotypes in these areas (smaller study sizes, weaker phenotype – genotype relationships). Application of this framework results in a reduction in the number of markers we were able to test in our family members. Although this filtering has restricted the number of resulting inferences, it has increased the robustness of the analysis.
Finally, the genetic risk analysis we provide here has not been tested in an independent population, and as such serves as an illustration of a potential approach and a template for further work.
When screening for pathogenicity we find that Son and Father have C → T; c.200C>T; p.Thr67Ile within the CTH gene. Father and Daughter share the mutation C → T; c.3029C>T; p.Thr1010Ile, located in the MET gene. Son and Mother share C → T; c.269G>A; p.Gly90Asp in PPP2R1B. All of these mutations are not deemed reportable due to the unknown significance nature of the inferences. The reportable variant is the A → T; c.4912A>T; p.Lys1638Ter in the BRCA2 gene for Mother in a recessive context. Mother had a benign breast tumor removed in her forties but it was never analyzed. Therefore, it is not possible to ascertain whether her BRCA2 gene mutation had any role in her benign tumor formation. Fortunately, this mutation is heterozygous and Father does not carry a known pathogenic mutation in this gene. Both children did not inherit Mother's pathogenic BRCA2 mutation and therefore are unable to pass it on to their offspring.
Aunt passed away in 2013, aged 79, due to a metastasised melanoma. For this participant, we transform our screening into a quasi-diagnostic setting given that we would like to identify a potential genetic cause for her demise. We were able to retrieve 36 hairs 4 years after her death from one of her combs. The DNA was carefully handled (see Supplementary Materials). We were able to assess pathogenicity among those variants that passed our strict quality filters. A heterozygous frameshift mutation, chr11:108272812 (rs587776549) in the ATM gene was identified. Recently, the Pan-Cancer Analysis of Whole Genomes Consortium confirmed that many cancer driver mutations are two-hit inactivation events (ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium, 2020), with 17% of patients having rare germline protein-truncating variants (PTVs) in cancer-predisposition genes, DNA-damage response genes and somatic driver genes. Biallelic inactivation due to somatic alteration on top of a germline PTV was observed in 4.5% of patients overall, with 81% of these affecting known cancer-predisposition genes (such as ATM). We thus hypothesize that the loss of function of one copy of the ATM gene could have contributed to her melanoma. Although Aunt's genome only provides information about her germline genetics and not the actual somatic mutations that led to the disease that ended her life, a more targeted cancer therapy (than the general chemotherapy she was administrated with) targeting defects in the DNA repair caused by ATM was already available while she was still alive (Kelley et al., 2014) and was never used.
We observe there is a conserved family risk of ulcerative colitis, running in Son, Mother, and Daughter. Ulcerative colitis is a long-term condition that results in inflammation and ulcers of the colon and rectum. It has also been found that both Mother and Daughter have a >1SD risk of inflammatory bowel disease, of which ulcerative colitis is a type. The primary symptoms of active disease are abdominal pain and diarrhea mixed with blood. Mother has reported suffering from a recurrent abdominal pain associated with inflammation of her colon. Her symptoms have appeared intermittently but are more recurrent in older age, affecting her quality of life. Given that ulcerative colitis begins most commonly between the ages of 15 and 25 with a second peak of onset in the 6th decade of life, Mother's reported symptoms are concordant with her ulcerative colitis / bowel disease susceptibility. We also note that according to Sen and Stark (2019), CYP2D6*4 polymorphisms may be risk factors for ulcerative colitis. Both Mother and Daughter display CYP2D6*4.
We have noted that for warfarin, the genotyping analysis has shown that members of the family are either intermediate or poor metabolisers. According to FDA guidance (Dean, 2012), Daughter requires 20% of the standard initial recommended dose and would take a more prolonged time to achieve the maximum anticoagulant effect. Son, Father, and Mother require a 60% of the standard initial recommended dose. This information is particularly relevant to Father, who was recently diagnosed with atrial fibrillation. Atrial fibrillation is a heart condition that causes an irregular and often abnormally fast heart rate. People with atrial fibrillation who have a high or moderate risk of having a stroke are usually prescribed warfarin. This was the case of Father, who was recommended to take warfarin to stop the risk of blood clotting. It has been reported by Father, that as soon as he started taking warfarin, he began to experience sores in legs, changes in the skin color, and severe pain in his lower half of the body. We note that his predicted response to warfarin is concordant with warfarin sensitivity (Vu and Gooderham, 2017). Hence, knowledge of this genetic predisposition would have been helpful to the clinician when making an initial prescription.
We also note that for both Mother and Daughter their predicted metaboliser status for CYP2D6 is either intermediate or poor. This has important implications in the specific dosage required by these individuals to receive the appropriate effects for pain relievers such as codeine and tramadol (Smith et al., 2019). So far there is some anecdotal evidence that Mother and Daughter are not able to cope well opiates, but nothing that was confirmed medically. Of note, the three most susceptible individuals to ulcerative colitis, Mother, Son, and Daughter are predicted to be normal metabolisers of drugs that treat pain associated with ulcers, such as omeprazole (Dickinson, 1994).
We performed an investigation of the literature to identify candidate fitness gene x interactions and their relation to a health outcome (see Methods section). Several limitations were noted throughout these studies, including the robustness of significance for identified variants, small sample sizes, limited cohorts focused primarily on Caucasian populations, and minimal baseline data (Williams et al., 2017). These factors are combined with differences in exercise training programs, diet and other environmental gene expression mediators between studies. As a result, we are able to classify as “Convincing” (2 of 10 candidates) or “Probable” (5 of 10). Overall, we found that fitness studies were made with a smaller sample size compared to nutrition. For instance, ACTN3's rs1815739, one of the most studied fitness-related SNPs, there are >1000 participants studied in total, which would put this SNP among the lowest sample sizes if included in nutrition markers, where we found seven markers with >100,000 study participants.
For all family members, the predicted genetic VO2max trainability was predicted average or less than average, contrasting with their lower predicted levels of blood lactate accumulation. With the exception of Mother, the family harbor variants in AQP1 alleles associated with endurance and fluid balance. Their genotype also predicts a predisposition to improved exercise performance if done with caffeine [with the exception of Mother; (Guest et al., 2018)]. Son is unique in the family in having the XX genotype for rs1815739 in ACTN3. Deficiency in α-actinin-3 can be accompanied by higher body fatness, lower muscle strength and higher muscle flexibility and range of motion (Yang et al., 2003; Massidda et al., 2019). A study suggested that recreational marathon runners who have the ACTN3 XX genotype could benefit from personalized strength training to improve their performance more than their counterparts with other ACTN3 genotypes (Del Coso et al., 2019b).
Compared to fitness studies, we found a greater number of candidate nutrition phenotypes passed our filtering (14 out of 32 initially selected phenotypes; Table 5). Larger sample sizes and a greater number of studies with concordant results were the main reasons for a larger number of nutrition phenotypes passing our filtering. As with fitness, we choose to analyse those traits whose scientific validity score is convincing or probable and report those that are likely to display pointers for further action or deemed reportable given the family disease and lifestyle history.
Congruent with the general lower likelihood of predicted alcohol dependence by rs1229984 in ADH1B, there is no history of alcohol addiction in the family. For all members of the family except Mother, there is a predicted increase in total body adiposity as suggested by FTO rs9939609. With regards to vitamin-related traits, all family members with the exception of Son are predicted to be less likely to respond to vitamin D supplements. For vitamin B12 levels, Mother and Son are predicted to harbor lower B12 levels and higher for Father and Daughter. All family members are predicted lower serum L-ascorbic acid.
For homocysteine levels, we found that all family members except Son are predicted to be higher. Reduction of plasma homocysteine levels has been observed with supplementation of vitamin B12 and folic acid (Boccia et al., 2008; Clarke et al., 2011; Liew and Gupta, 2015). Several other studies have observed associations between lower circulating vitamin B12 levels and adverse metabolic health profiles, with insulin resistance, cardiovascular disorders, and adiposity as important features (Hazra et al., 2009; Tanwar et al., 2013; Allin et al., 2017; Nongmaithem et al., 2017; Zhao and Schooling, 2017).
With regards to lactose consumption, we were able to confirm that Father, suspected to be lactose intolerant, has the lactose intolerant genotype.
When performing a screening study in an individual, for some variants which can confer significant disease risk, it is important to report not just positive findings, but also negative ones if there is family history of the disease. The ApoE2, E3, and E4 isoforms, which are encoded by the ε2, ε3, and ε4 alleles of the APOE gene, respectively, differ from one another at amino acid residues 112 and/or 158. There is a significant association between the ε4 allele of APOE and Alzheimer's disease. APOE ε4 increases the risk of Alzheimer's disease and lowers the age of disease onset in a gene-dose-dependent manner (Liu et al., 2013). A small proportion of apo ε2 homozygotes, develop type III hyperlipoproteinemia, a highly atherogenic form disorder of lipoprotein metabolism characterized by the accumulation of remnant particles derived from the incomplete catabolism of triglyceride-rich lipoproteins (März et al., 2000). All the family members whose genomes were analyzed for this study exhibit the wild type ε3/ε3, meaning that no association to Alzheimer's disease is conferred. This is also further supported by analysis we performed for family members using genetic risks of Alzheimer's disease (Supplementary Table 1). The fact that there is history of Alzheimer's disease in the maternal line, makes it interesting to ascertain whether genetic risk for this disease is present in the family. It was thus of special interest for the family to research this trait, with the positive outcome that all family members display the less risky ε3/ε3 alleles. We were also in search of negative findings for classified pathogenic mutations that fall within any of the ACMG 59 genes and were able to find only one positive finding for Mother in BRCA2. Our variant analysis did not find any other mutation within the 59 genes. As always, the fact that no mutation was found does not necessarily mean a particular disease might not develop.
Part of the novelty of the present study revolves around the integration of genetic screening, genetic risk scores and trait analysis. A further layer of integration is constituted by the familial context our participant dataset provides. As stated in Methods, each family individual is tested independently for each of the genetic screening panels, genetic score phenotypes and trait analysis. Although the overlap between each of these methods can only be partial, we now explore the degree of consistency and support that each of the results conveys.
For obesity, the family history indicates a persistent tendency toward this phenotype. At the level of genotyping, the rs9939609 FTO marker analysis, shown to be the most contributing to obesity (Sonestedt et al., 2009; Tanofsky-Kraff et al., 2009; Zhao et al., 2014b), yields all genotyped individuals except Mother to carry the risk allele. We acknowledge that the specific contribution of this SNP can only be small. When integrating this genotypic result with GWAS-based genetic risk score, we observe that the obesity risk slightly increased (>1SD) for Father and Son.
Both analysis of specific SNPs in the APOE gene (see Table 7), and genetic risk scores do not suggest an increased risk for all family members of Alzheimer's disease. This is also congruent with the observed disease history of the analyzed family, where both parents are highly advanced in years of age (mid-eighties) and no signs for the disease have been observed yet. This does not rule out the possibility that any member of the family could develop Alzheimer's disease at any point in the future. It does rule out, however, both parents having developed early onset Alzheimer's disease. For alcohol dependency, there has not been observed any tendency of addictive behavior in the family. The rs1229984 ADH1B marker supports this phenotype. However, the >1SD predicted genetic risk for alcohol dependency in Father, Daughter and Son does not. A way to reconciling this result is that the genetic risk is moderately higher than average and therefore it does not strongly rule out the possibility of a false positive or random fluctuation, since the observed genetic risk for alcohol dependence may be the result of fluctuations in the score that are not significant. Another integration of different analysis sources is the pathogenic heterozygous variant for BRCA2 p.Lys1638Ter observed in Mother and her >1SD genetic risk observed for breast cancer. An interpretation of this finding is that she only carries one defective allele for the gene, increasing her risk but not high enough to make it to our >2SD average score threshold.
With regards to integrating the results of similar (yet independent) tests performed in different individuals of the family, we note the coincidence of phenotypic history of irritable colon of Mother with her >2SD increased risk of ulcerative colitis. As mentioned earlier, this >2SD phenotype risk is also observed in Son and not so strongly in Daughter (>1SD), suggesting a pointer for preventative action on the part of Son.
Results for members of the family were communicated either in person or via phone call. For Son, his results have had an impact in his training exercise program, which has a lot more stretching and warming up, with less emphasis on speed and more on building up his endurance. The predicted ulcerative colitis risk for three members of the family was communicated to Mother, who is already displaying some symptoms of the disease, and to Son, who is already taking steps to bring these results forward to his general practitioner as part of his future health management plans. Father's possible explanation of his adverse reaction to warfarin has also been discussed and he has currently discontinued taking the medicine, having discussed it first with his cardiologist. The communication of Aunt's result of her mutation in the ATM gene has not led to any concrete actions by her partner.
The family has been exposed to genetic testing for a decade (Corpas, 2012), and as such were generally comfortable knowing results of genetic analysis. Even so, attention was paid in particular to make sure they were aware of the ramifications of knowing their results of tests related to more serious and harder to treat conditions (such as ulcerative colitis).
As was the case when the same family was analyzed with direct-to-consumer genotyping methods (Corpas, 2012), the tendency to discuss “whose genome is best” is a recurrent pattern that could affect other families when communicating genetic test results. We stress the importance of discussing such results with qualified professionals such as genetic counselors.
Compared to the communication of the results in 2012, we also note the change in attitude toward sharing of personal genomic data. Individuals are less keen to share their genetic data now, arguing that their perceptions regarding the privacy of their data have been changed by their increased awareness of the importance of protecting individual's personal data.
By looking at the genome from various methodological angles and applying distinct analytical frameworks as appropriate, we were able to build a “genetic story” of each individual. We built this story in part through having whole genomes as the basis for the analysis. The approach is applied here for a family, but we believe it is also valid for individuals. Our most notable findings for the family were around susceptibility to ulcerative colitis, and in the areas of fitness, nutrition, and pharmacogenomics.
Concerning ulcerative colitis, when analyzing genetic risk scores, we noted that the recurrent intestinal pain Mother has been affected from for years is concordant with her substantially increased risk of suffering from ulcerative colitis. Moreover, this high risk is predicted in three out of the five members of the family, two of them overlapping with increased risk of inflammatory bowel disease, ulcerative colitis being one type of this disease. We report this susceptibility to ulcerative colitis/inflammatory bowel disease as a potential lead for preventative intervention in at least one family member (Son) who is currently asymptomatic.
We observed some associations for fitness and nutrition variants which passed our quality control framework and as such we believe are valuable for relevant nutritional and exercise science specialists to help the family in making plans in those areas.
We were also able to hypothesize a genetic contribution to the development of melanoma leading to the passing of Aunt. A pathogenic heterozygous germline mutation was reported in her ATM gene. This gene has been described as being involved in DNA repair and the information gathered here could have been exploited for targeted cancer therapy if caught on time.
Concordance between an adverse reaction to warfarin and a prediction for low dosing requirement was observed in Father, which he has already acted on, in consultation with his cardiologist. There were also informative results for Mother and Daughter regarding their likely metaboliser status for certain drugs. While not relevant to them at the moment, this information could be shared with their physician in the event that these drugs become necessary in the future, with the hope of reducing trial and error in prescribing and so cutting down the possibility of adverse reactions.
We believe that, taken together, these results represent relevant information which the family can use, when working with the appropriate healthcare professionals, to proactively promote their health and well-being. Any one element of the analysis would not allow this genetic “story” to be compellingly told, but when all them are put together, the narrative becomes more actionable, increasing the applicability of whole genome screening to pre-emptive healthcare and well-being management.
The sources of genetic markers used in this study are included in Table 3 and Table 4. The sources for genetic risk score creation are included in Supplementary Table 1. The gene panel utilised for cancer screening is in the Supplementary materials. The family genome variation data for this manuscript is not publicly available because they were not consented for open access. Request to access the family genome variation data should be directed to Manuel Corpas (bS5jb3JwYXNAY3BtLm9ubA==).
We confirm that written informed consent was obtained from the individuals or appropriate next to kin individual for the publication of any potentially identifiable images or data included in this article.
MC and EL conceived the experiments and performed the analysis. MC wrote the paper with contributions from all authors. All authors read and approved the paper.
At the time of writing, MC, KM, AM, and EL are associated with Cambridge Precision Medicine Limited. VM is a full-time employee of Fabric Genomics.
We are grateful to Mahsa Shabani for helpful guidance on the development of an ethical framework for this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.535123/full#supplementary-material
1. ^Get to Know an Enzyme: CYP2C9 Pharmacy Times. Available at: https://www.pharmacytimes.com/publications/issue/2008/2008-03/2008-03-8462 [accessed October 15, 2019].
2. ^Genetics Home Reference CYP2C19 gene. Genetics Home Reference. Available at: https://ghr.nlm.nih.gov/gene/CYP2C19 [accessed October 15, 2019].
Ahmetov, I. I., and Fedotovskaya, O. N. (2015). Current progress in sports genomics. Adv. Clin. Chem. 70, 247–314. doi: 10.1016/bs.acc.2015.03.003
Ahmetov, I. I., Williams, A. G., Popov, D. V., Lyubaeva, E. V., Hakimullina, A. M., Fedotovskaya, O. N., et al. (2009). The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum. Genet. 126, 751–761. doi: 10.1007/s00439-009-0728-4
Al-Daghri, N. M., Mohammed, A. K., Bukhari, I., Rikli, M., Abdi, S., Ansari, M. G. A., et al. (2019). Efficacy of vitamin D supplementation according to vitamin D-binding protein polymorphisms. Nutrition 63–64, 148–154. doi: 10.1016/j.nut.2019.02.003
Allin, K. H., Friedrich, N., Pietzner, M., Grarup, N., Thuesen, B. H., Linneberg, A., et al. (2017). Genetic determinants of serum vitamin B12 and their relation to body mass index. Eur. J. Epidemiol. 32, 125–134. doi: 10.1007/s10654-016-0215-x
Almeida, S. M., Furtado, J. M., Mascarenhas, P., Ferraz, M. E., Ferreira, J. C., Monteiro, M. P., et al. (2018). Association between LEPR, FTO, MC4R, and PPARG-2 polymorphisms with obesity traits and metabolic phenotypes in school-aged children. Endocrine 60, 466–478. doi: 10.1007/s12020-018-1587-3
Alsaleh, A., Crepostnaia, D., Maniou, Z., Lewis, F. J., Hall, W. L., Sanders, T. A. B., et al. (2013). Adiponectin gene variant interacts with fish oil supplementation to influence serum adiponectin in older individuals. J. Nutr. 143, 1021–1027. doi: 10.3945/jn.112.172585
Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F., and Hamosh, A. (2015). OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43, D789–D798. doi: 10.1093/nar/gku1205
Amengual, J., Coronel, J., Marques, C., Aradillas-García, C., Morales, J. M. V., Andrade, F. C. D., et al. (2020). β-Carotene oxygenase 1 activity modulates circulating cholesterol concentrations in mice and humans. J. Nutr. 150, 2023–2030. doi: 10.1093/jn/nxaa143
Amir Shaghaghi, M., Bernstein, C. N., Serrano León, A., El-Gabalawy, H., and Eck, P. (2014). Polymorphisms in the sodium-dependent ascorbate transporter gene SLC23A1 are associated with susceptibility to Crohn disease. Am. J. Clin. Nutr. 99, 378–383. doi: 10.3945/ajcn.113.068015
Baffour-Awuah, N. Y., Fleet, S., Montgomery, R. K., Baker, S. S., Butler, J. L., Campbell, C., et al. (2015). Functional significance of single nucleotide polymorphisms in the lactase gene in diverse US patients and evidence for a novel lactase persistence allele at−13909 in those of European ancestry. J. Pediatr. Gastroenterol. Nutr. 60, 182–191. doi: 10.1097/MPG.0000000000000595
Bahrami, A., Khayyatzadeh, S. S., Jaberi, N., Tayefi, M., Mohammadi, F., Ferns, G. A., et al. (2019). Common polymorphisms in genes related to vitamin D metabolism affect the response of cognitive abilities to vitamin D supplementation. J. Mol. Neurosci. 69, 150–156. doi: 10.1007/s12031-019-01344-6
Balsevich, G., Sticht, M., Bowles, N. P., Singh, A., Lee, T. T. Y., Li, Z., et al. (2018). Role for fatty acid amide hydrolase (FAAH) in the leptin-mediated effects on feeding and energy balance. Proc. Natl. Acad. Sci. U.S.A. 115, 7605–7610. doi: 10.1073/pnas.1802251115
Baltazar-Martins, G., Gutiérrez-Hellín, J., Aguilar-Navarro, M., Ruiz-Moreno, C., Moreno-Pérez, V., López-Samanes, Á., et al. (2020). Effect of genotype on sports performance, exercise-induced muscle damage, and injury epidemiology. Sports 8:99. doi: 10.3390/sports8070099
Barton, J. C., McLaren, C. E., Chen, W.-P., Ramm, G. A., Anderson, G. J., Powell, L. W., et al. (2018). Cirrhosis in hemochromatosis: independent risk factors in 368 HFE p.C282Y homozygotes. Ann. Hepatol. 17, 871–879. doi: 10.5604/01.3001.0012.3169
Baumert, P., Lake, M. J., Stewart, C. E., Drust, B., and Erskine, R. M. (2016). Genetic variation and exercise-induced muscle damage: implications for athletic performance, injury and ageing. Eur. J. Appl. Physiol. 116, 1595–1625. doi: 10.1007/s00421-016-3411-1
Ben-Zaken, S., Eliakim, A., Nemet, D., Rabinovich, M., Kassem, E., and Meckel, Y. (2015). Differences in MCT1 A1470T polymorphism prevalence between runners and swimmers. Scand. J. Med. Sci. Sports 25, 365–371. doi: 10.1111/sms.12226
Berndt, S. I., Gustafsson, S., Mägi, R., Ganna, A., Wheeler, E., Feitosa, M. F., et al. (2013). Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 45, 501–512. doi: 10.1038/ng.2606
Bertuzzi, R., Pasqua, L. A., Bueno, S., Lima-Silva, A. E., Matsuda, M., Marquezini, M., et al. (2014). Is the COL5A1 rs12722 gene polymorphism associated with running economy? PLoS ONE 9:e106581. doi: 10.1371/journal.pone.0106581
Bierut, L. J., Agrawal, A., Bucholz, K. K., Doheny, K. F., Laurie, C., Pugh, E., et al. (2010). A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. U.S.A. 107, 5082–5087. doi: 10.1073/pnas.0911109107
Biesecker, L. G., the ClinGen Sequence Variant Interpretation Working Group, and Harrison, S. M. (2018). The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet. Med. 20, 1687–1688. doi: 10.1038/gim.2018.42
Biss, T. T., Avery, P. J., Brandão, L. R., Chalmers, E. A., Williams, M. D., Grainger, J. D., et al. (2012). VKORC1 and CYP2C9 genotype and patient characteristics explain a large proportion of the variability in warfarin dose requirement among children. Blood 119, 868–873. doi: 10.1182/blood-2011-08-372722
Boccia, S., Boffetta, P., Brennan, P., Ricciardi, G., Gianfagna, F., Matsuo, K., et al. (2009). Meta-analyses of the methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and risk of head and neck and lung cancer. Cancer Lett. 273, 55–61. doi: 10.1016/j.canlet.2008.07.026
Boccia, S., Hung, R., Ricciardi, G., Gianfagna, F., Ebert, M. P. A., Fang, J.-Y., et al. (2008). Meta- and pooled analyses of the methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and gastric cancer risk: a huge-GSEC review. Am. J. Epidemiol. 167, 505–516. doi: 10.1093/aje/kwm344
Bollepalli, S., Dolan, L. M., Deka, R., and Martin, L. J. (2010). Association of FTO gene variants with adiposity in African-American adolescents. Obesity 18, 1959–1963. doi: 10.1038/oby.2010.82
Bordoni, L., Marchegiani, F., Piangerelli, M., Napolioni, V., and Gabbianelli, R. (2017). Obesity-related genetic polymorphisms and adiposity indices in a young Italian population. IUBMB Life 69, 98–105. doi: 10.1002/iub.1596
Bouchard, C., Sarzynski, M. A., Rice, T. K., Kraus, W. E., Church, T. S., Sung, Y. J., et al. (2011). Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. J. Appl. Physiol. 110, 1160–1170. doi: 10.1152/japplphysiol.00973.2010
Braithwaite, V. S., Jones, K. S., Schoenmakers, I., Silver, M., Prentice, A., and Hennig, B. J. (2015). Vitamin D binding protein genotype is associated with plasma 25OHD concentration in West African children. Bone 74, 166–170. doi: 10.1016/j.bone.2014.12.068
Brandt, T., Sack, L. M., Arjona, D., Tan, D., Mei, H., Cui, H., et al. (2019). Adapting ACMG/AMP sequence variant classification guidelines for single-gene copy number variants. Genet. Med. 22, 336–344. doi: 10.1038/s41436-019-0655-2
Calvano Küchler, E., Arid, J., Palinkas, M., Ayumi Omori, M., de Lara, R. M., Napolitano Gonçalves, L. M., et al. (2020). Genetic polymorphisms in contribute to the etiology of bruxism in children. J. Clin. Pediatr. Dent. 44, 180–184. doi: 10.17796/1053-4625-44.3.8
Carswell, A. T., Howland, K., Martinez-Gonzalez, B., Baron, P., and Davison, G. (2020). The effect of caffeine on cognitive performance is influenced by CYP1A2 but not ADORA2A genotype, yet neither genotype affects exercise performance in healthy adults. Eur. J. Appl. Physiol. 120, 1495–1508. doi: 10.1007/s00421-020-04384-8
Ching, Y. K., Chin, Y. S., Appukutty, M., Ramanchadran, V., Yu, C. Y., Ang, G. Y., et al. (2019). Interaction of dietary linoleic acid and α-linolenic acids with rs174547 in gene on metabolic syndrome components among vegetarians. Nutrients 11:1686. doi: 10.3390/nu11071686
Choi, M., Kipps, T., and Kurzrock, R. (2016). ATM mutations in cancer: therapeutic implications. Mol. Cancer Ther. 15, 1781–1791. doi: 10.1158/1535-7163.MCT-15-0945
Choquet, H., Paylakhi, S., Kneeland, S. C., Thai, K. K., Hoffmann, T. J., Yin, J., et al. (2018). A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 9, 2278. doi: 10.1038/s41467-018-04555-4
Chuaychoo, B., Tungtrongchitr, R., Kriengsinyos, W., Tuntipopipat, S., On-Nom, N., and Chupeerach, C. (2018). Correlation of vitamin D binding protein gene polymorphism and protein levels in chronic obstructive pulmonary disease compared with non-chronic obstructive pulmonary disease subjects. Per. Med. 15, 371–379. doi: 10.2217/pme-2018-0005
Cichon, S., Mühleisen, T. W., Degenhardt, F. A., Mattheisen, M., Miró, X., Strohmaier, J., et al. (2011). Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am. J. Hum. Genet. 88, 372–381. doi: 10.1016/j.ajhg.2011.01.017
Clarke, R., Halsey, J., Bennett, D., and Lewington, S. (2011). Homocysteine and vascular disease: review of published results of the homocysteine-lowering trials. J. Inherit. Metab. Dis. 34, 83–91. doi: 10.1007/s10545-010-9235-y
Coonrod, E. M., Margraf, R. L., Russell, A., Voelkerding, K. V., and Reese, M. G. (2013). Clinical analysis of genome next-generation sequencing data using the Omicia platform. Expert Rev. Mol. Diagn. 13, 529–540. doi: 10.1586/14737159.2013.811907
Cormier, H., Rudkowska, I., Lemieux, S., Couture, P., and Vohl, M.-C. (2016). Expression and Sequence Variants of Inflammatory Genes; Effects on Plasma Inflammation Biomarkers Following a 6-Week Supplementation with Fish Oil. Int. J. Mol. Sci. 17:375. doi: 10.3390/ijms17030375
Cornelis, M. C., Kacprowski, T., Menni, C., Gustafsson, S., Pivin, E., Adamski, J., et al. (2016). Genome-wide association study of caffeine metabolites provides new insights to caffeine metabolism and dietary caffeine-consumption behavior. Hum. Mol. Genet. 25, 5472–5482. doi: 10.1093/hmg/ddw334
Corpas, M. (2012). A family experience of personal genomics. J. Genet. Counsel. 21, 386–391. doi: 10.1007/s10897-011-9473-7
Corpas, M., Valdivia-Granda, W., Torres, N., Greshake, B., Coletta, A., Knaus, A., et al. (2015). Crowdsourced direct-to-consumer genomic analysis of a family quartet. BMC Genomics 16:910. doi: 10.1186/s12864-015-1973-7
Cupeiro, R., González-Lamuño, D., Amigo, T., Peinado, A. B., Ruiz, J. R., Ortega, F. B., et al. (2012). Influence of the MCT1-T1470A polymorphism (rs1049434) on blood lactate accumulation during different circuit weight trainings in men and women. J. Sci. Med. Sport 15, 541–547. doi: 10.1016/j.jsams.2012.03.009
da Rocha, T. J., Blehm, C. J., Bamberg, D. P., Fonseca, T. L. R., Tisser, L. A., de Oliveira Junior, A. A., et al. (2014a). The effects of interactions between selenium and zinc serum concentration and SEP15 and SLC30A3 gene polymorphisms on memory scores in a population of mature and elderly adults. Genes Nutr. 9:377. doi: 10.1007/s12263-013-0377-z
da Rocha, T. J., Korb, C., Schuch, J. B., Bamberg, D. P., de Andrade, F. M., and Fiegenbaum, M. (2014b). SLC30A3 and SEP15 gene polymorphisms influence the serum concentrations of zinc and selenium in mature adults. Nutr. Res. 34, 742–748. doi: 10.1016/j.nutres.2014.08.009
de Luis, D. A., Gonzalez Sagrado, M., Aller, R., Izaola, O., and Conde, R. (2011). Effects of C358A missense polymorphism of the endocannabinoid degrading enzyme fatty acid amide hydrolase on weight loss after a hypocaloric diet. Metabolism 60, 730–734. doi: 10.1016/j.metabol.2010.07.007
Dean, L. (2012). “Warfarin therapy and genotype,” in Medical Genetics Summaries, eds V. M. Pratt, S. A. Scott, M. Pirmohamed, B. Esquivel, M. S. Kane, B. L. Kattman et al. [Bethesda, MD: National Center for Biotechnology Information (US)].
Dean, L. (2018). “Warfarin therapy and VKORC1 and CYP genotype,” in Medical Genetics Summaries [Internet], eds V. M. Pratt, S. A. Scott, M. Pirmohamed, B. Esquivel, M. S. Kane, B. L. Kattman et al. [Bethesda, MD: National Center for Biotechnology Information (US)].
Dedoussis, G. V. Z., Yannakoulia, M., Timpson, N. J., Manios, Y., Kanoni, S., Scott, R. A., et al. (2011). Does a short breastfeeding period protect from FTO-induced adiposity in children? Int. J. Pediatr. Obes. 6, e326–e335. doi: 10.3109/17477166.2010.490269
Del Coso, J., Hiam, D., Houweling, P., Pérez, L. M., Eynon, N., and Lucía, A. (2019a). More than a “speed gene”: ACTN3 R577X genotype, trainability, muscle damage, and the risk for injuries. Eur. J. Appl. Physiol. 119, 49–60. doi: 10.1007/s00421-018-4010-0
Del Coso, J., Moreno, V., Gutiérrez-Hellín, J., Baltazar-Martins, G., Ruíz-Moreno, C., Aguilar-Navarro, M., et al. (2019b). R577X genotype and exercise phenotypes in recreational marathon runners. Genes 10:413. doi: 10.3390/genes10060413
Del Coso, J., Salinero, J. J., Lara, B., Gallo-Salazar, C., Areces, F., Puente, C., et al. (2017). ACTN3 X-allele carriers had greater levels of muscle damage during a half-ironman. Eur. J. Appl. Physiol. 117, 151–158. doi: 10.1007/s00421-016-3507-7
Delgado-Lista, J., Perez-Jimenez, F., Tanaka, T., Perez-Martinez, P., Jimenez-Gomez, Y., Marin, C., et al. (2007). An apolipoprotein A-II polymorphism (-265T/C, rs5082) regulates postprandial response to a saturated fat overload in healthy men. J. Nutr. 137, 2024–2028. doi: 10.1093/jn/137.9.2024
Dickinson, J. B. (1994). Is omeprazole helpful in inflammatory bowel disease? J. Clin. Gastroenterol. 18, 317–319. doi: 10.1097/00004836-199406000-00012
Domínguez-Reyes, T., Astudillo-López, C. C., Salgado-Goytia, L., Muñoz-Valle, J. F., Salgado-Bernabé, A. B., Guzmán-Guzmán, I. P., et al. (2015). Interaction of dietary fat intake with APOA2, APOA5 and LEPR polymorphisms and its relationship with obesity and dyslipidemia in young subjects. Lipids Health Dis. 14:106. doi: 10.1186/s12944-015-0112-4
Doris, J. M., Millar, S. A., Idris, I., and O'Sullivan, S. E. (2019). Genetic polymorphisms of the endocannabinoid system in obesity and diabetes. Diabetes Obes. Metab. 21, 382–387. doi: 10.1111/dom.13504
Duell, E. J., Lujan-Barroso, L., Llivina, C., Muñoz, X., Jenab, M., Boutron-Ruault, M.-C., et al. (2013). Vitamin C transporter gene (SLC23A1 and SLC23A2) polymorphisms, plasma vitamin C levels, and gastric cancer risk in the EPIC cohort. Genes Nutr. 8, 549–560. doi: 10.1007/s12263-013-0346-6
Duicu, C., Mărginean, C. O., Voidăzan, S., Tripon, F., and Bănescu, C. (2016). FTO rs 9939609 SNP is associated with adiponectin and leptin levels and the risk of obesity in a cohort of romanian children population. Medicine 95:e3709. doi: 10.1097/MD.0000000000003709
Dwivedi, O. P., Tabassum, R., Chauhan, G., Ghosh, S., Marwaha, R. K., Tandon, N., et al. (2012). Common variants of FTO are associated with childhood obesity in a cross-sectional study of 3,126 urban Indian children. PLoS ONE 7:e47772. doi: 10.1371/journal.pone.0047772
Elkader, A., and Sproule, B. (2005). Buprenorphine: clinical pharmacokinetics in the treatment of opioid dependence. Clin. Pharmacokinet. 44, 661–680. doi: 10.2165/00003088-200544070-00001
Enlund-Cerullo, M., Koljonen, L., Holmlund-Suila, E., Hauta-Alus, H., Rosendahl, J., Valkama, S., et al. (2019). Genetic variation of the vitamin D binding protein affects vitamin D status and response to supplementation in infants. J. Clin. Endocrinol. Metab. 104, 5483–5498. doi: 10.1210/jc.2019-00630
Eynon, N., Alves, A. J., Sagiv, M., Yamin, C., Sagiv, M., and Meckel, Y. (2010). Interaction between SNPs in the NRF2 gene and elite endurance performance. Physiol. Genomics 41, 78–81. doi: 10.1152/physiolgenomics.00199.2009
Eynon, N., Ruiz, J. R., Bishop, D. J., Santiago, C., Gómez-Gallego, F., Lucia, A., et al. (2013). The rs12594956 polymorphism in the NRF-2 gene is associated with top-level Spanish athlete's performance status. J. Sci. Med. Sport 16, 135–139. doi: 10.1016/j.jsams.2012.05.004
Fahed, A. C., Wang, M., Homburger, J. R., Patel, A. P., Bick, A. G., Neben, C. L., et al. (2020). Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun. 11:3635. doi: 10.1038/s41467-020-17374-3
Fedotovskaya, O. N., Mustafina, L. J., Popov, D. V., Vinogradova, O. L., and Ahmetov, I. I. (2014). A common polymorphism of the MCT1 gene and athletic performance. Int. J. Sports Physiol. Perform. 9, 173–180. doi: 10.1123/ijspp.2013-0026
Ferreira Todendi, P., de Moura Valim, A. R., Klinger, E., Reuter, C. P., Molina, S., Martínez, J. A., et al. (2019). The role of the genetic variants IRX3 rs3751723 and FTO rs9939609 in the obesity phenotypes of children and adolescents. Obes. Res. Clin. Pract. 13, 137–142. doi: 10.1016/j.orcp.2019.01.005
Ferreira, M. A. R., O'Donovan, M. C., Meng, Y. A., Jones, I. R., Ruderfer, D. M., Jones, L., et al. (2008). Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 40, 1056–1058. doi: 10.1038/ng.209
Figueroa, J. D., Ye, Y., Siddiq, A., Garcia-Closas, M., Chatterjee, N., Prokunina-Olsson, L., et al. (2014). Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum. Mol. Genet. 23, 1387–1398. doi: 10.1093/hmg/ddt519
Flygare, S., Hernandez, E. J., Phan, L., Moore, B., Li, M., Fejes, A., et al. (2018). The VAAST Variant Prioritizer (VVP): ultrafast, easy to use whole genome variant prioritization tool. BMC Bioinformatics 19:57. doi: 10.1186/s12859-018-2056-y
Gaedigk, A., Dinh, J. C., Jeong, H., Prasad, B., and Leeder, J. S. (2018). Ten years' experience with the CYP2D6 activity score: a perspective on future investigations to improve clinical predictions for precision therapeutics. J. Pers. Med. 8:15. doi: 10.3390/jpm8020015
Galeandro, V., Notarnicola, A., Bianco, A., Tafuri, S., Russo, L., Pesce, V., et al. (2017). ACTN3/ACE genotypes and mitochondrial genome in professional soccer players performance. J. Biol. Regul. Homeost. Agents 31, 207–213.
García-Solís, P., Reyes-Bastidas, M., Flores, K., García, O. P., Rosado, J. L., Méndez-Villa, L., et al. (2016). Fat mass obesity-associated (FTO) (rs9939609) and melanocortin 4 receptor (MC4R) (rs17782313) SNP are positively associated with obesity and blood pressure in Mexican school-aged children. Br. J. Nutr. 116, 1834–1840. doi: 10.1017/S0007114516003779
Gelernter, J., Kranzler, H. R., Sherva, R., Almasy, L., Koesterer, R., Smith, A. H., et al. (2014). Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol. Psychiatry 19, 41–49. doi: 10.1038/mp.2013.145
Georgi, B., Mielke, J., Chaffin, M., Khera, A. V., Gelis, L., Mundl, H., et al. (2019). Leveraging human genetics to estimate clinical risk reductions achievable by inhibiting factor XI. Stroke 50, 3004–3012. doi: 10.1161/STROKEAHA.119.026545
Ghosh, S., Vivar, J. C., Sarzynski, M. A., Sung, Y. J., Timmons, J. A., Bouchard, C., et al. (2013). Integrative pathway analysis of a genome-wide association study of (V)O(2max) response to exercise training. J. Appl. Physiol. 115, 1343–1359. doi: 10.1152/japplphysiol.01487.2012
Gibbs, D. C., Bostick, R. M., McCullough, M. L., Um, C. Y., Flanders, W. D., Jenab, M., et al. (2020a). Association of prediagnostic vitamin D status with mortality among colorectal cancer patients differs by common, inherited vitamin D-binding protein isoforms. Int. J. Cancer 147, 2725–2734. doi: 10.1002/ijc.33043
Gibbs, D. C., Song, M., McCullough, M. L., Um, C. Y., Bostick, R. M., Wu, K., et al. (2020b). Association of circulating vitamin D with colorectal cancer depends on vitamin D-binding protein isoforms: a pooled, nested, case-control study. JNCI Cancer Spectr. 4:kz083. doi: 10.1093/jncics/pkz083
Glusman, G., Cariaso, M., Jimenez, R., Swan, D., Greshake, B., Bhak, J., et al. (2012). Low budget analysis of Direct-To-Consumer genomic testing familial data. F1000Res. 1:3. doi: 10.12688/f1000research.1-3.v1
Gómez-Gallego, F., Ruiz, J. R., Buxens, A., Altmäe, S., Artieda, M., Santiago, C., et al. (2010). Are elite endurance athletes genetically predisposed to lower disease risk? Physiol. Genomics 41, 82–90. doi: 10.1152/physiolgenomics.00183.2009
Graßmann, S., Pivovarova-Ramich, O., Henze, A., Raila, J., Ampem Amoako, Y., King Nyamekye, R., et al. (2020). SNP rs6564851 in the BCO1 gene is associated with varying provitamin a plasma concentrations but not with retinol concentrations among adolescents from rural Ghana. Nutrients 12:1786. doi: 10.3390/nu12061786
Grgic, J., Pickering, C., Bishop, D. J., Schoenfeld, B. J., Mikulic, P., and Pedisic, Z. (2020). CYP1A2 genotype and acute effects of caffeine on resistance exercise, jumping, and sprinting performance. J. Int. Soc. Sports Nutr. 17:21. doi: 10.1186/s12970-020-00349-6
Grimaldi, K. A., van Ommen, B., Ordovas, J. M., Parnell, L. D., Mathers, J. C., Bendik, I., et al. (2017). Proposed guidelines to evaluate scientific validity and evidence for genotype-based dietary advice. Genes Nutr. 12:35. doi: 10.1186/s12263-017-0584-0
Guest, N., Corey, P., Vescovi, J., and El-Sohemy, A. (2018). Caffeine, CYP1A2 genotype, and endurance performance in athletes. Med. Sci. Sports Exerc. 50, 1570–1578. doi: 10.1249/MSS.0000000000001596
Guevara-Cruz, M., Lai, C.-Q., Richardson, K., Parnell, L. D., Lee, Y.-C., Tovar, A. R., et al. (2013). Effect of a GFOD2 variant on responses in total and LDL cholesterol in Mexican subjects with hypercholesterolemia after soy protein and soluble fiber supplementation. Gene 532, 211–215. doi: 10.1016/j.gene.2013.09.055
Hällfors, J., Palviainen, T., Surakka, I., Gupta, R., Buchwald, J., Raevuori, A., et al. (2019). Genome-wide association study in Finnish twins highlights the connection between nicotine addiction and neurotrophin signaling pathway. Addict. Biol. 24, 549–561. doi: 10.1111/adb.12618
Hazra, A., Kraft, P., Lazarus, R., Chen, C., Chanock, S. J., Jacques, P., et al. (2009). Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum. Mol. Genet. 18, 4677–4687. doi: 10.1093/hmg/ddp428
He, Z., Hu, Y., Feng, L., Lu, Y., Liu, G., Xi, Y., et al. (2007). NRF2 genotype improves endurance capacity in response to training. Int. J. Sports Med. 28, 717–721. doi: 10.1055/s-2007-964913
Hindy, G., Mollet, I. G., Rukh, G., Ericson, U., and Orho-Melander, M. (2016). Several type 2 diabetes-associated variants in genes annotated to WNT signaling interact with dietary fiber in relation to incidence of type 2 diabetes. Genes Nutr. 11:6. doi: 10.1186/s12263-016-0524-4
Hindy, G., Sonestedt, E., Ericson, U., Jing, X.-J., Zhou, Y., Hansson, O., et al. (2012). Role of TCF7L2 risk variant and dietary fibre intake on incident type 2 diabetes. Diabetologia 55, 2646–2654. doi: 10.1007/s00125-012-2634-x
Hou, L., Bergen, S. E., Akula, N., Song, J., Hultman, C. M., Landén, M., et al. (2016). Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum. Mol. Genet. 25, 3383–3394. doi: 10.1093/hmg/ddw181
Houweling, P. J., Papadimitriou, I. D., Seto, J. T., Pérez, L. M., Coso, J. D., North, K. N., et al. (2018). Is evolutionary loss our gain? The role of ACTN3 p.Arg577Ter (R577X) genotype in athletic performance, ageing, and disease. Hum. Mutat. 39, 1774–1787. doi: 10.1002/humu.23663
Howard, D. M., Adams, M. J., Shirali, M., Clarke, T.-K., Marioni, R. E., Davies, G., et al. (2018). Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 9:1470. doi: 10.1038/s41467-018-03819-3
Howe, L. J., Lawson, D. J., Davies, N. M., St Pourcain, B., Lewis, S. J., Davey Smith, G., et al. (2019). Genetic evidence for assortative mating on alcohol consumption in the UK Biobank. Nat. Commun. 10:5039. doi: 10.1038/s41467-019-12424-x
Hu, H., Huff, C. D., Moore, B., Flygare, S., Reese, M. G., and Yandell, M. (2013). VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet. Epidemiol. 37, 622–634. doi: 10.1002/gepi.21743
Huang, M.-C., Chang, W.-T., Chang, H.-Y., Chung, H.-F., Chen, F.-P., Huang, Y.-F., et al. (2017). FADS gene polymorphisms, fatty acid desaturase activities, and HDL-C in type 2 diabetes. Int. J. Environ. Res. Public Health 14:572. doi: 10.3390/ijerph14060572
Hubacek, J. A., Jirsa, M., Bobak, M., Pelclova, D., and Zakharov, S. (2018). Aldehyde dehydrogenase 2 polymorphism affects the outcome of methanol poisoning in exposed humans. Clin. Genet. 94, 445–449. doi: 10.1111/cge.13411
Hunt, K. A., Zhernakova, A., Turner, G., Heap, G. A. R., Franke, L., Bruinenberg, M., et al. (2008). Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 40, 395–402. doi: 10.1038/ng.102
ICGC/TCGA and Pan-Cancer Analysis of Whole Genomes Consortium (2020). Pan-cancer analysis of whole genomes. Nature 578, 82–93. doi: 10.1038/s41586-020-1969-6
Ikeda, M., Takahashi, A., Kamatani, Y., Okahisa, Y., Kunugi, H., Mori, N., et al. (2018). A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry 23, 639–647. doi: 10.1038/mp.2016.259
Itaka, T., Agemizu, K., Aruga, S., and Machida, S. (2016). G allele of the IGF2 ApaI polymorphism is associated with judo status. J. Strength Cond. Res. 30, 2043–2048. doi: 10.1519/JSC.0000000000001300
Jenab, M., McKay, J., Bueno-de-Mesquita, H. B., van Duijnhoven, F. J. B., Ferrari, P., Slimani, N., et al. (2009). Vitamin D receptor and calcium sensing receptor polymorphisms and the risk of colorectal cancer in European populations. Cancer Epidemiol. Biomarkers Prev. 18, 2485–2491. doi: 10.1158/1055-9965.EPI-09-0319
Jensen, D. P., Andreasen, C. H., Andersen, M. K., Hansen, L., Eiberg, H., Borch-Johnsen, K., et al. (2007). The functional Pro129Thr variant of the FAAH gene is not associated with various fat accumulation phenotypes in a population-based cohort of 5,801 whites. J. Mol. Med. 85, 445–449. doi: 10.1007/s00109-006-0139-0
Johansen, J.-M., Goleva-Fjellet, S., Sunde, A., Gjerløw, L. E., Skeimo, L. A., Freberg, B. I., et al. (2020). No change - no gain; the effect of age, sex, selected genes and training on physiological and performance adaptations in cross-country skiing. Front. Physiol. 11:581339. doi: 10.3389/fphys.2020.581339
Johnson, E. C., St Pierre, C. L., Meyers, J. L., Aliev, F., McCutcheon, V. V., Lai, D., et al. (2019). The genetic relationship between alcohol consumption and aspects of problem drinking in an ascertained sample. Alcohol. Clin. Exp. Res. 43, 1113–1125. doi: 10.1111/acer.14064
Jorgenson, E., Thai, K. K., Hoffmann, T. J., Sakoda, L. C., Kvale, M. N., Banda, Y., et al. (2017). Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol. Psychiatry 22, 1359–1367. doi: 10.1038/mp.2017.101
Junyent, M., Parnell, L. D., Lai, C.-Q., Lee, Y.-C., Smith, C. E., Arnett, D. K., et al. (2009). Novel variants at KCTD10, MVK, and MMAB genes interact with dietary carbohydrates to modulate HDL-cholesterol concentrations in the Genetics of Lipid Lowering Drugs and Diet Network Study. Am. J. Clin. Nutr. 90, 686–694. doi: 10.3945/ajcn.2009.27738
Justice, A. C., Smith, R. V., Tate, J. P., McGinnis, K., Xu, K., Becker, W. C., et al. (2018). AUDIT-C and ICD codes as phenotypes for harmful alcohol use: association with ADH1B polymorphisms in two US populations. Addiction 113, 2214–2224. doi: 10.1111/add.14374
Kalia, S. S., Adelman, K., Bale, S. J., Chung, W. K., Eng, C., Evans, J. P., et al. (2017). Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 19, 249–255. doi: 10.1038/gim.2016.190
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alföldi, J., Wang, Q., et al. (2019). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. doi: 10.1038/s41586-020-2308-7
Karuwanarint, P., Phonrat, B., Tungtrongchitr, A., Suriyaprom, K., Chuengsamarn, S., Schweigert, F. J., et al. (2018). Vitamin D-binding protein and its polymorphisms as a predictor for metabolic syndrome. Biomark. Med. 12, 465–473. doi: 10.2217/bmm-2018-0029
Katsarou, M.-S., Karakonstantis, K., Demertzis, N., Vourakis, E., Skarpathioti, A., Nosyrev, A. E., et al. (2017). Effect of single-nucleotide polymorphisms in ADH1B, ADH4, ADH1C, OPRM1, DRD2, BDNF, and ALDH2 genes on alcohol dependence in a Caucasian population. Pharmacol. Res. Perspect. 5:e00326. doi: 10.1002/prp2.326
Katsarou, M.-S., Latsi, R., Papasavva, M., Demertzis, N., Kalogridis, T., Tsatsakis, A. M., et al. (2016). Population-based analysis of the frequency of HFE gene polymorphisms: Correlation with the susceptibility to develop hereditary hemochromatosis. Mol. Med. Rep. 14, 630–636. doi: 10.3892/mmr.2016.5317
Katsarou, M.-S., Papasavva, M., Latsi, R., and Drakoulis, N. (2019). Hemochromatosis: hereditary hemochromatosis and HFE gene. Vitam. Horm. 110, 201–222. doi: 10.1016/bs.vh.2019.01.010
Kelley, M. R., Logsdon, D., and Fishel, M. L. (2014). Targeting DNA repair pathways for cancer treatment: what's new? Future Oncol. 10, 1215–1237. doi: 10.2217/fon.14.60
Khera, A. V., Chaffin, M., Aragam, K. G., Haas, M. E., Roselli, C., Choi, S. H., et al. (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224. doi: 10.1038/s41588-018-0183-z
Kiemeney, L. A., Sulem, P., Besenbacher, S., Vermeulen, S. H., Sigurdsson, A., Thorleifsson, G., et al. (2010). A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat. Genet. 42, 415–419. doi: 10.1038/ng.558
Kiemeney, L. A., Thorlacius, S., Sulem, P., Geller, F., Aben, K. K. H., Stacey, S. N., et al. (2008). Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 40, 1307–1312. doi: 10.1038/ng.229
Kikuchi, N., Fuku, N., Matsumoto, R., Matsumoto, S., Murakami, H., Miyachi, M., et al. (2017a). The association between MCT1 T1470A polymorphism and power-oriented athletic performance. Int. J. Sports Med. 38, 76–80. doi: 10.1055/s-0042-117113
Kikuchi, N., Nakazato, K., Min, S.-K., Ueda, D., and Igawa, S. (2014). The ACTN3 R577X polymorphism is associated with muscle power in male Japanese athletes. J. Strength Cond. Res. 28, 1783–1789. doi: 10.1519/JSC.0000000000000338
Kikuchi, N., Yoshida, S., Min, S.-K., Lee, K., Sakamaki-Sunaga, M., Okamoto, T., et al. (2015). The ACTN3 R577X genotype is associated with muscle function in a Japanese population. Appl. Physiol. Nutr. Metab. 40, 316–322. doi: 10.1139/apnm-2014-0346
Kikuchi, N., Zempo, H., Fuku, N., Murakami, H., Sakamaki-Sunaga, M., Okamoto, T., et al. (2017b). Association between ACTN3 R577X polymorphism and trunk flexibility in 2 different cohorts. Int. J. Sports Med. 38, 402–406. doi: 10.1055/s-0042-118649
Knoll, N., Volckmar, A.-L., Pütter, C., Scherag, A., Kleber, M., Hebebrand, J., et al. (2012). The fatty acid amide hydrolase (FAAH) gene variant rs324420 AA/AC is not associated with weight loss in a 1-year lifestyle intervention for obese children and adolescents. Horm. Metab. Res. 44, 75–77. doi: 10.1055/s-0031-1291306
Kobylecki, C. J., Afzal, S., Davey Smith, G., and Nordestgaard, B. G. (2015). Genetically high plasma vitamin C, intake of fruit and vegetables, and risk of ischemic heart disease and all-cause mortality: a Mendelian randomization study. Am. J. Clin. Nutr. 101, 1135–1143. doi: 10.3945/ajcn.114.104497
Kobylecki, C. J., Afzal, S., and Nordestgaard, B. G. (2018). Genetically high plasma vitamin C and urate: a Mendelian randomization study in 106 147 individuals from the general population. Rheumatology 57, 1769–1776. doi: 10.1093/rheumatology/key171
Kranzler, H. R., Zhou, H., Kember, R. L., Vickers Smith, R., Justice, A. C., Damrauer, S., et al. (2019). Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 10:1499. doi: 10.1038/s41467-019-09480-8
Laake, K., Jansen, L., Hahnemann, J. M., Brondum-Nielsen, K., Lönnqvist, T., Kääriäinen, H., et al. (2000). Characterization of ATM mutations in 41 Nordic families with ataxia telangiectasia. Hum. Mutat. 16, 232–246. doi: 10.1002/1098-1004(200009)16:3<232::AID-HUMU6>3.0.CO;2-L
Lai, D., Wetherill, L., Bertelsen, S., Carey, C. E., Kamarajan, C., Kapoor, M., et al. (2019). Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav. 18:e12579. doi: 10.1111/gbb.12579
Landrum, M. J., Lee, J. M., Riley, G. R., Jang, W., Rubinstein, W. S., Church, D. M., et al. (2014). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985. doi: 10.1093/nar/gkt1113
Lauria, F., Siani, A., Bammann, K., Foraita, R., Huybrechts, I., Iacoviello, L., et al. (2012). Prospective analysis of the association of a common variant of FTO (rs9939609) with adiposity in children: results of the IDEFICS study. PLoS ONE 7:e48876. doi: 10.1371/journal.pone.0048876
Lek, M., Karczewski, K. J., Minikel, E. V., Samocha, K. E., Banks, E., Fennell, T., et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. doi: 10.1038/nature19057
Liew, S.-C., and Gupta, E. D. (2015). Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: epidemiology, metabolism and the associated diseases. Eur. J. Med. Genet. 58, 1–10. doi: 10.1016/j.ejmg.2014.10.004
Liu, C.-C., Liu, C.-C., Kanekiyo, T., Xu, H., and Bu, G. (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118. doi: 10.1038/nrneurol.2012.263
Liu, J. Z., van Sommeren, S., Huang, H., Ng, S. C., Alberts, R., Takahashi, A., et al. (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986. doi: 10.1038/ng.3359
Livingstone, K. M., Celis-Morales, C., Papandonatos, G. D., Erar, B., Florez, J. C., Jablonski, K. A., et al. (2016). FTO genotype and weight loss: systematic review and meta-analysis of 9563 individual participant data from eight randomised controlled trials. BMJ 354:i4707. doi: 10.1136/bmj.i4707
Lopez-Leon, S., Tuvblad, C., and Forero, D. A. (2016). Sports genetics: the PPARA gene and athletes' high ability in endurance sports. A systematic review and meta-analysis. Biol. Sport 33, 3–6. doi: 10.5604/20831862.1180170
López-Ortiz, M. M., Garay-Sevilla, M. E., Tejero, M. E., and Perez-Luque, E. L. (2016). Analysis of the interaction between transcription factor 7-like 2 genetic variants with nopal and wholegrain fibre intake: effects on anthropometric and metabolic characteristics in type 2 diabetes patients. Br. J. Nutr. 116, 969–978. doi: 10.1017/S0007114516002798
Machini, K., Ceyhan-Birsoy, O., Azzariti, D. R., Sharma, H., Rossetti, P., Mahanta, L., et al. (2019). Analyzing and reanalyzing the genome: findings from the MedSeq Project. Am. J. Hum. Genet. 105, 177–188. doi: 10.1016/j.ajhg.2019.05.017
Madden, K., Feldman, H. A., Chun, R. F., Smith, E. M., Sullivan, R. M., Agan, A. A., et al. (2015). Critically ill children have low vitamin D-binding protein, influencing bioavailability of vitamin D. Ann. Am. Thorac. Soc. 12, 1654–1661. doi: 10.1513/AnnalsATS.201503-160OC
Mahmutovic, L., Bego, T., Sterner, M., Gremsperger, G., Ahlqvist, E., Velija Asimi, Z., et al. (2019). Association of IRS1 genetic variants with glucose control and insulin resistance in type 2 diabetic patients from Bosnia and Herzegovina. Drug Metab Pers Ther 34. doi: 10.1515/dmpt-2018-0031
Major, J. M., Yu, K., Chung, C. C., Weinstein, S. J., Yeager, M., Wheeler, W., et al. (2012). Genome-wide association study identifies three common variants associated with serologic response to vitamin E supplementation in men. J. Nutr. 142, 866–871. doi: 10.3945/jn.111.156349
Major, J. M., Yu, K., Weinstein, S. J., Berndt, S. I., Hyland, P. L., Yeager, M., et al. (2014). Genetic variants reflecting higher vitamin e status in men are associated with reduced risk of prostate cancer. J. Nutr. 144, 729–733. doi: 10.3945/jn.113.189928
Major, J. M., Yu, K., Wheeler, W., Zhang, H., Cornelis, M. C., Wright, M. E., et al. (2011). Genome-wide association study identifies common variants associated with circulating vitamin E levels. Hum. Mol. Genet. 20, 3876–3883. doi: 10.1093/hmg/ddr296
Mangge, H., Renner, W., Almer, G., Weghuber, D., Möller, R., and Horejsi, R. (2011). Rs9939609 variant of the fat mass and obesity-associated gene and trunk obesity in adolescents. J. Obes. 2011:186368. doi: 10.1155/2011/186368
Martínez, C., Galván, S., Garcia-Martin, E., Ramos, M. I., Gutiérrez-Martín, Y., and Agúndez, J. A. G. (2010). Variability in ethanol biodisposition in whites is modulated by polymorphisms in the ADH1B and ADH1C genes. Hepatology 51, 491–500. doi: 10.1002/hep.23341
Martins, I. J., Hone, E., Foster, J. K., Sünram-Lea, S. I., Gnjec, A., Fuller, S. J., et al. (2006). Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer's disease and cardiovascular disease. Mol. Psychiatry 11, 721–736. doi: 10.1038/sj.mp.4001854
März, W., Nauck, M. S., Fisher, E., Hoffmann, M. M., and Wieland, H. (2000). “The molecular mechanisms of inherited hypercholesterolemia,” in From Molecule to Men, eds M. Zehender, H. Just, and G. Breithardt (Heidelberg: Steinkopff), 151–169. doi: 10.1007/978-3-642-57724-6_13
Masaoka, H., Ito, H., Gallus, S., Watanabe, M., Yokomizo, A., Eto, M., et al. (2017). Combination of ALDH2 and ADH1B polymorphisms is associated with smoking initiation: a large-scale cross-sectional study in a Japanese population. Drug Alcohol Depend. 173, 85–91. doi: 10.1016/j.drugalcdep.2016.12.015
Massidda, M., Voisin, S., Culigioni, C., Piras, F., Cugia, P., Yan, X., et al. (2019). ACTN3 R577X polymorphism is associated with the incidence and severity of injuries in professional football players. Clin. J. Sport Med. 29, 57–61. doi: 10.1097/JSM.0000000000000487
Matsuda, K., Takahashi, A., Middlebrooks, C. D., Obara, W., Nasu, Y., Inoue, K., et al. (2015). Genome-wide association study identified SNP on 15q24 associated with bladder cancer risk in Japanese population. Hum. Mol. Genet. 24, 1177–1184. doi: 10.1093/hmg/ddu512
Mattar, R., de Campos Mazo, D. F., and Carrilho, F. J. (2012). Lactose intolerance: diagnosis, genetic, and clinical factors. Clin. Exp. Gastroenterol. 5, 113–121. doi: 10.2147/CEG.S32368
Mbarek, H., Milaneschi, Y., Fedko, I. O., Hottenga, J.-J., de Moor, M. H. M., Jansen, R., et al. (2015). The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168, 739–748. doi: 10.1002/ajmg.b.32379
McLaren, C. E., Garner, C. P., Constantine, C. C., McLachlan, S., Vulpe, C. D., Snively, B. M., et al. (2011). Genome-wide association study identifies genetic loci associated with iron deficiency. PLoS ONE 6:e17390. doi: 10.1371/journal.pone.0017390
Mehramiz, M., Khayyatzadeh, S. S., Esmaily, H., Ghasemi, F., Sadeghi-Ardekani, K., Tayefi, M., et al. (2019). Associations of vitamin D binding protein variants with the vitamin D-induced increase in serum 25-hydroxyvitamin D. Clin. Nutr. 29, 59–64. doi: 10.1016/j.clnesp.2018.12.005
Meisner, A., Kundu, P., and Chatterjee, N. (2019). Case-only analysis of gene-environment interactions using polygenic risk scores. Am. J. Epidemiol. 188, 2013–2020. doi: 10.1093/aje/kwz175
Meng, X.-R., Song, J.-Y., Ma, J., Liu, F.-H., Shang, X.-R., Guo, X.-J., et al. (2014). Association study of childhood obesity with eight genetic variants recently identified by genome-wide association studies. Pediatr. Res. 76, 310–315. doi: 10.1038/pr.2014.88
Min, S.-K., Lim, S.-T., and Kim, C.-S. (2016). Association of ACTN3 polymorphisms with BMD, and physical fitness of elderly women. J. Phys. Ther. Sci. 28, 2731–2736. doi: 10.1589/jpts.28.2731
Moran, N. E., Thomas-Ahner, J. M., Fleming, J. L., McElroy, J. P., Mehl, R., Grainger, E. M., et al. (2019). Single nucleotide polymorphisms in β-carotene oxygenase 1 are associated with plasma lycopene responses to a tomato-soy juice intervention in men with prostate cancer. J. Nutr. 149, 381–397. doi: 10.1093/jn/nxy304
Mühleisen, T. W., Leber, M., Schulze, T. G., Strohmaier, J., Degenhardt, F., Treutlein, J., et al. (2014). Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun. 5:3339. doi: 10.1038/ncomms4339
Muñoz, A., López-Samanes, Á., Aguilar-Navarro, M., Varillas-Delgado, D., Rivilla-García, J., Moreno-Pérez, V., et al. (2020). Effects of and genotypes on the ergogenic response to caffeine in professional handball players. Genes 11:933. doi: 10.3390/genes11080933
Murtagh, C. F., Brownlee, T. E., Rienzi, E., Roquero, S., Moreno, S., Huertas, G., et al. (2020). The genetic profile of elite youth soccer players and its association with power and speed depends on maturity status. PLoS ONE 15:e0234458. doi: 10.1371/journal.pone.0234458
Nievergelt, C. M., Maihofer, A. X., Mustapic, M., Yurgil, K. A., Schork, N. J., Miller, M. W., et al. (2015). Genomic predictors of combat stress vulnerability and resilience in U.S. Marines: a genome-wide association study across multiple ancestries implicates PRTFDC1 as a potential PTSD gene. Psychoneuroendocrinology 51, 459–471. doi: 10.1016/j.psyneuen.2014.10.017
Nissen, J., Rasmussen, L. B., Ravn-Haren, G., Andersen, E. W., Hansen, B., Andersen, R., et al. (2014). Common variants in CYP2R1 and GC genes predict vitamin D concentrations in healthy Danish children and adults. PLoS ONE 9:e89907. doi: 10.1371/journal.pone.0089907
Nissen, J., Vogel, U., Ravn-Haren, G., Andersen, E. W., Madsen, K. H., Nex,ø, B. A., et al. (2015). Common variants in CYP2R1 and GC genes are both determinants of serum 25-hydroxyvitamin D concentrations after UVB irradiation and after consumption of vitamin D3-fortified bread and milk during winter in Denmark. Am. J. Clin. Nutr. 101, 218–227. doi: 10.3945/ajcn.114.092148
Nofziger, C., Turner, A. J., Sangkuhl, K., Whirl-Carrillo, M., Agúndez, J. A. G., Black, J. L., et al. (2020). PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 107, 154–170. doi: 10.1002/cpt.1643
Nongmaithem, S. S., Joglekar, C. V., Krishnaveni, G. V., Sahariah, S. A., Ahmad, M., Ramachandran, S., et al. (2017). GWAS identifies population-specific new regulatory variants in FUT6 associated with plasma B12 concentrations in Indians. Hum. Mol. Genet. 26, 2551–2564. doi: 10.1093/hmg/ddx071
Noorshahi, N., Sotoudeh, G., Djalali, M., Eshraghian, M. R., Keramatipour, M., Basiri, M. G., et al. (2016). APOA II genotypes frequency and their interaction with saturated fatty acids consumption on lipid profile of patients with type 2 diabetes. Clin. Nutr. 35, 907–911. doi: 10.1016/j.clnu.2015.06.008
Nykamp, K., The Invitae Clinical Genomics Group, Anderson, M., Powers, M., Garcia, J., Herrera, B., et al. (2017). Sherloc: a comprehensive refinement of the ACMG–AMP variant classification criteria. Genet. Med. 19, 1105–1117. doi: 10.1038/gim.2017.37
O'Connell, K., Posthumus, M., Schwellnus, M. P., and Collins, M. (2013). Collagen genes and exercise-associated muscle cramping. Clin. J. Sport Med. 23, 64–69. doi: 10.1097/JSM.0b013e3182686aa7
Oki, E., Norde, M. N., Carioca, A. A. F., Souza, J. M. P., Castro, I. A., Marchioni, D. M. L., et al. (2017). Polymorphisms of the TNF-α gene interact with plasma fatty acids on inflammatory biomarker profile: a population-based, cross-sectional study in São Paulo, Brazil. Br. J. Nutr. 117, 1663–1673. doi: 10.1017/S0007114517001416
Olfson, E., and Bierut, L. J. (2012). Convergence of genome-wide association and candidate gene studies for alcoholism. Alcohol. Clin. Exp. Res. 36, 2086–2094. doi: 10.1111/j.1530-0277.2012.01843.x
Palmer, J. R. (2019). Polygenic risk scores for breast cancer risk prediction: lessons learned and future opportunities. J. Natl. Cancer Inst. 112, 555–556. doi: 10.1093/jnci/djz176
Pataky, M. W., Womack, C. J., Saunders, M. J., Goffe, J. L., D'Lugos, A. C., El-Sohemy, A., et al. (2016). Caffeine and 3-km cycling performance: effects of mouth rinsing, genotype, and time of day. Scand. J. Med. Sci. Sports 26, 613–619. doi: 10.1111/sms.12501
Pavan, S., Rommel, K., Mateo Marquina, M. E., Höhn, S., Lanneau, V., and Rath, A. (2017). Clinical practice guidelines for rare diseases: the orphanet database. PLoS ONE 12:e0170365. doi: 10.1371/journal.pone.0170365
Pekkinen, M., Saarnio, E., Viljakainen, H. T., Kokkonen, E., Jakobsen, J., Cashman, K., et al. (2014). Vitamin D binding protein genotype is associated with serum 25-hydroxyvitamin D and PTH concentrations, as well as bone health in children and adolescents in Finland. PLoS ONE 9:e87292. doi: 10.1371/journal.pone.0087292
Peplonska, B., Adamczyk, J. G., Siewierski, M., Safranow, K., Maruszak, A., Sozanski, H., et al. (2017). Genetic variants associated with physical and mental characteristics of the elite athletes in the Polish population. Scand. J. Med. Sci. Sports 27, 788–800. doi: 10.1111/sms.12687
Petersen, R. A., Larsen, L. H., Damsgaard, C. T., Sørensen, L. B., Hjorth, M. F., Andersen, R., et al. (2017). Common genetic variants are associated with lower serum 25-hydroxyvitamin D concentrations across the year among children at northern latitudes. Br. J. Nutr. 117, 829–838. doi: 10.1017/S0007114517000538
Petr, M., Maciejewska-Skrendo, A., Zajac, A., Chycki, J., and Stastny, P. (2019). Association of elite sports status with gene variants of peroxisome proliferator activated receptors and their transcriptional coactivator. Int. J. Mol. Sci. 21:162. doi: 10.3390/ijms21010162
Petr, M., Stastny, P., Zajac, A., Tufano, J. J., and Maciejewska-Skrendo, A. (2018). The role of peroxisome proliferator-activated receptors and their transcriptional coactivators gene variations in human trainability: a systematic review. Int. J. Mol. Sci. 19:472. doi: 10.3390/ijms19051472
PGP-UK Consortium (2018). Personal Genome Project UK (PGP-UK): a research and citizen science hybrid project in support of personalized medicine. BMC Med. Genomics 11:108. doi: 10.1186/s12920-018-0423-1
Pirastu, N., Joshi, P. K., de Vries, P. S., Cornelis, M. C., McKeigue, P. M., Keum, N., et al. (2017). GWAS for male-pattern baldness identifies 71 susceptibility loci explaining 38% of the risk. Nat. Commun. 8:1584. doi: 10.1038/s41467-017-01490-8
Płóciennik, Ł. A., Zaucha, J., Zaucha, J. M., Łukaszuk, K., Józwicki, M., Płóciennik, M., et al. (2020). Detection of epistasis between ACTN3 and SNAP-25 with an insight towards gymnastic aptitude identification. PLoS ONE 15:e0237808. doi: 10.1371/journal.pone.0237808
Polimanti, R., and Gelernter, J. (2018). ADH1B: From alcoholism, natural selection, and cancer to the human phenome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 177, 113–125. doi: 10.1002/ajmg.b.32523
Psychiatric, G. W. A. S., and Consortium Bipolar Disorder Working Group (2011). Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983. doi: 10.1038/ng.943
Puente, C., Abián-Vicén, J., Del Coso, J., Lara, B., and Salinero, J. J. (2018). The CYP1A2−163C>A polymorphism does not alter the effects of caffeine on basketball performance. PLoS ONE 13:e0195943. doi: 10.1371/journal.pone.0195943
Qi, Q., Downer, M. K., Kilpeläinen, T. O., Taal, H. R., Barton, S. J., Ntalla, I., et al. (2015a). Dietary intake, FTO genetic variants, and adiposity: a combined analysis of over 16,000 children and adolescents. Diabetes 64, 2467–2476. doi: 10.2337/db14-1629
Qi, Q., Zheng, Y., Huang, T., Rood, J., Bray, G. A., Sacks, F. M., et al. (2015b). Vitamin D metabolism-related genetic variants, dietary protein intake and improvement of insulin resistance in a 2 year weight-loss trial: POUNDS Lost. Diabetologia 58, 2791–2799. doi: 10.1007/s00125-015-3750-1
Quan, L.-L., Wang, H., Tian, Y., Mu, X., Zhang, Y., and Tao, K. (2015). Association of fat-mass and obesity-associated gene FTO rs9939609 polymorphism with the risk of obesity among children and adolescents: a meta-analysis. Eur. Rev. Med. Pharmacol. Sci. 19, 614–623.
Rafnar, T., Sulem, P., Thorleifsson, G., Vermeulen, S. H., Helgason, H., Saemundsdottir, J., et al. (2014). Genome-wide association study yields variants at 20p12.2 that associate with urinary bladder cancer. Hum. Mol. Genet. 23, 5545–5557. doi: 10.1093/hmg/ddu264
Rafnar, T., Vermeulen, S. H., Sulem, P., Thorleifsson, G., Aben, K. K., Witjes, J. A., et al. (2011). European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum. Mol. Genet. 20, 4268–4281. doi: 10.1093/hmg/ddr303
Rahimi, M. H., Mollahosseini, M., Mirzababaei, A., Yekaninejad, M. S., Maghbooli, Z., and Mirzaei, K. (2019). Interactions between vitamin D binding protein variants and major dietary patterns on the odds of metabolic syndrome and its components in apparently healthy adults. Diabetol. Metab. Syndr. 11:28. doi: 10.1186/s13098-019-0422-1
Ramírez, J., van Duijvenboden, S., Ntalla, I., Mifsud, B., Warren, H. R., Tzanis, E., et al. (2018). Thirty loci identified for heart rate response to exercise and recovery implicate autonomic nervous system. Nat. Commun. 9:1947. doi: 10.1038/s41467-018-04148-1
Ranzenhofer, L. M., Mayer, L. E. S., Davis, H. A., Mielke-Maday, H. K., McInerney, H., Korn, R., et al. (2019). The FTO gene and measured food intake in 5- to 10-year-old children without obesity. Obesity 27, 1023–1029. doi: 10.1002/oby.22464
Rasmussen, K. L., Tybjærg-Hansen, A., Nordestgaard, B. G., and Frikke-Schmidt, R. (2018). Absolute 10-year risk of dementia by age, sex and genotype: a population-based cohort study. CMAJ 190, E1033–E1041. doi: 10.1503/cmaj.180066
Ravindran, R. D., Sundaresan, P., Krishnan, T., Vashist, P., Maraini, G., Saravanan, V., et al. (2019). Genetic variants in a sodium-dependent vitamin C transporter gene and age-related cataract. Br. J. Ophthalmol. 103, 1223–1227. doi: 10.1136/bjophthalmol-2018-312257
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J., and Kircher, M. (2019). CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894. doi: 10.1093/nar/gky1016
Rice, T. K., Sarzynski, M. A., Sung, Y. J., Argyropoulos, G., Stütz, A. M., Teran-Garcia, M., et al. (2012). Fine mapping of a QTL on chromosome 13 for submaximal exercise capacity training response: the HERITAGE Family Study. Eur. J. Appl. Physiol. 112, 2969–2978. doi: 10.1007/s00421-011-2274-8
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424. doi: 10.1038/gim.2015.30
Rivera, M. A., and Fahey, T. D. (2019). Association between aquaporin-1 and endurance performance: a systematic review. Sports Med Open 5:40. doi: 10.1186/s40798-019-0213-0
Rivera, M. A., Fahey, T. D., López-Taylor, J. R., and Martínez, J. L. (2020). The association of aquaporin-1 gene with marathon running performance level: a confirmatory study conducted in male hispanic marathon runners. Sports Med. Open 6:16. doi: 10.1186/s40798-020-00243-0
Roach, J. C., Glusman, G., Hubley, R., Montsaroff, S. Z., Holloway, A. K., Mauldin, D. E., et al. (2011). Chromosomal haplotypes by genetic phasing of human families. Am. J. Hum. Genet. 89, 382–397. doi: 10.1016/j.ajhg.2011.07.023
Robien, K., Butler, L. M., Wang, R., Beckman, K. B., Walek, D., Koh, W.-P., et al. (2013). Genetic and environmental predictors of serum 25-hydroxyvitamin D concentrations among middle-aged and elderly Chinese in Singapore. Br. J. Nutr. 109, 493–502. doi: 10.1017/S0007114512001675
Ronald, A., Happ,é, F., Price, T. S., Baron-Cohen, S., and Plomin, R. (2006). Phenotypic and genetic overlap between autistic traits at the extremes of the general population. J. Am. Acad. Child Adolesc. Psychiatry 45, 1206–1214. doi: 10.1097/01.chi.0000230165.54117.41
Rothman, N., Garcia-Closas, M., Chatterjee, N., Malats, N., Wu, X., Figueroa, J. D., et al. (2010). A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 42, 978–984. doi: 10.1038/ng.687
Salinero, J. J., Lara, B., Ruiz-Vicente, D., Areces, F., Puente-Torres, C., Gallo-Salazar, C., et al. (2017). CYP1A2 genotype variations do not modify the benefits and drawbacks of caffeine during exercise: a pilot study. Nutrients 9:269. doi: 10.3390/nu9030269
Santiago, C., Ruiz, J. R., Buxens, A., Artieda, M., Arteta, D., González-Freire, M., et al. (2011). Trp64Arg polymorphism in ADRB3 gene is associated with elite endurance performance. Br. J. Sports Med. 45, 147–149. doi: 10.1136/bjsm.2009.061366
Saunders, C. J., Posthumus, M., O'Connell, K., September, A. V., and Collins, M. (2015). A variant within the AQP1 3'-untranslated region is associated with running performance, but not weight changes, during an Ironman Triathlon. J. Sports Sci. 33, 1342–1348. doi: 10.1080/02640414.2014.989535
Schadock, I., Schneider, A., Silva, E. D., Buchweitz, M. R. D., Correa, M. N., Pesquero, J. B., et al. (2015). Simple method to genotype the ACTN3 r577x polymorphism. Genet. Test. Mol. Biomark. 19, 253–257. doi: 10.1089/gtmb.2014.0299
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. doi: 10.1038/nature13595
Schumacher, F. R., Al Olama, A. A., Berndt, S. I., Benlloch, S., Ahmed, M., Saunders, E. J., et al. (2018). Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 50, 928–936. doi: 10.1038/s41588-018-0142-8
Sen, A., and Stark, H. (2019). Role of cytochrome P450 polymorphisms and functions in development of ulcerative colitis. World J. Gastroenterol. 25, 2846–2862. doi: 10.3748/wjg.v25.i23.2846
Slattery, M. L., Wolff, R. K., Herrick, J. S., Caan, B. J., and Samowitz, W. (2010). Calcium, vitamin D, VDR genotypes, and epigenetic and genetic changes in rectal tumors. Nutr. Cancer 62, 436–442. doi: 10.1080/01635580903441204
Smith, C. E., Tucker, K. L., Arnett, D. K., Noel, S. E., Corella, D., Borecki, I. B., et al. (2013). Apolipoprotein A2 polymorphism interacts with intakes of dairy foods to influence body weight in 2 U.S. populations. J. Nutr. 143, 1865–1871. doi: 10.3945/jn.113.179051
Smith, D. M., Weitzel, K. W., Elsey, A. R., Langaee, T., Gong, Y., Wake, D. T., et al. (2019). CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet. Med. 21, 1842–1850. doi: 10.1038/s41436-018-0431-8
Smith, E. N., Bloss, C. S., Badner, J. A., Barrett, T., Belmonte, P. L., Berrettini, W., et al. (2009). Genome-wide association study of bipolar disorder in European American and African American individuals. Mol. Psychiatry 14, 755–763. doi: 10.1038/mp.2009.43
Søberg, S., Sandholt, C. H., Jespersen, N. Z., Toft, U., Madsen, A. L., von Holstein-Rathlou, S., et al. (2017). FGF21 is a sugar-induced hormone associated with sweet intake and preference in humans. Cell Metab. 25, 1045–1053.e6. doi: 10.1016/j.cmet.2017.04.009
Sonestedt, E., Roos, C., Gullberg, B., Ericson, U., Wirfält, E., and Orho-Melander, M. (2009). Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am. J. Clin. Nutr. 90, 1418–1425. doi: 10.3945/ajcn.2009.27958
Stein, M. B., Chen, C.-Y., Ursano, R. J., Cai, T., Gelernter, J., Heeringa, S. G., et al. (2016). Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of US army soldiers. JAMA Psychiatry 73, 695–704. doi: 10.1001/jamapsychiatry.2016.0350
Sun, Y., Chang, S., Wang, F., Sun, H., Ni, Z., Yue, W., et al. (2019). Genome-wide association study of alcohol dependence in male Han Chinese and cross-ethnic polygenic risk score comparison. Transl. Psychiatry 9, 249. doi: 10.1038/s41398-019-0586-3
Szentkereszty-Kovács, Z., Fiatal, S., Szegedi, A., Kovács, D., Janka, E., Herszényi, K., et al. (2019). The prevalence of ADH1B and OPRM1 alleles predisposing for alcohol consumption are increased in the Hungarian psoriasis population. Arch. Dermatol. Res. 311, 435–442. doi: 10.1007/s00403-019-01915-y
Takahashi, M., Kahnoski, R., Gross, D., Nicol, D., and Teh, B. T. (2002). Familial adult renal neoplasia. J. Med. Genet. 39, 1–5. doi: 10.1136/jmg.39.1.1
Tanaka, T., Scheet, P., Giusti, B., Bandinelli, S., Piras, M. G., Usala, G., et al. (2009). Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet. 84, 477–482. doi: 10.1016/j.ajhg.2009.02.011
Tanofsky-Kraff, M., Han, J. C., Anandalingam, K., Shomaker, L. B., Columbo, K. M., Wolkoff, L. E., et al. (2009). The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am. J. Clin. Nutr. 90, 1483–1488. doi: 10.3945/ajcn.2009.28439
Tanwar, V. S., Chand, M. P., Kumar, J., Garg, G., Seth, S., Karthikeyan, G., et al. (2013). Common variant in FUT2 gene is associated with levels of vitamin B(12) in Indian population. Gene 515, 224–228. doi: 10.1016/j.gene.2012.11.021
The 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68–74. doi: 10.1038/nature15393
Thompson, A., Cook, J., Choquet, H., Jorgenson, E., Yin, J., Kinnunen, T., et al. (2020). Functional validity, role, and implications of heavy alcohol consumption genetic loci. Sci. Adv. 6:eaay5034. doi: 10.1126/sciadv.aay5034
Timpson, N. J., Forouhi, N. G., Brion, M.-J., Harbord, R. M., Cook, D. G., Johnson, P., et al. (2010). Genetic variation at the SLC23A1 locus is associated with circulating concentrations of L-ascorbic acid (vitamin C): evidence from 5 independent studies with >15,000 participants. Am. J. Clin. Nutr. 92, 375–382. doi: 10.3945/ajcn.2010.29438
Todendi, P. F., Martínez, J. A., Reuter, C. P., Klinger, E. I., Fiegenbaum, M., and Rosane de Moura Valim, A. (2020). Influence of FTO (Fat mass and obesity) gene and parental obesity on Brazilian children and adolescents adiposity. J. Pediatr. Endocrinol. Metab. 33, 975–982. doi: 10.1515/jpem-2019-0594
Torkamani, A., and Topol, E. (2019). Polygenic risk scores expand to obesity. Cell 177, 518–520. doi: 10.1016/j.cell.2019.03.051
Touvier, M., Deschasaux, M., Montourcy, M., Sutton, A., Charnaux, N., Kesse-Guyot, E., et al. (2015). Determinants of vitamin D status in Caucasian adults: influence of sun exposure, dietary intake, sociodemographic, lifestyle, anthropometric, and genetic factors. J. Invest. Dermatol. 135, 378–388. doi: 10.1038/jid.2014.400
Uhlig, H. H., and Muise, A. M. (2017). Clinical genomics in inflammatory bowel disease. Trends Genet. 33, 629–641. doi: 10.1016/j.tig.2017.06.008
van Heel, D. A., Franke, L., Hunt, K. A., Gwilliam, R., Zhernakova, A., Inouye, M., et al. (2007). A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 39, 827–829. doi: 10.1038/ng2058
Vu, T. T., and Gooderham, M. (2017). Adverse drug reactions and cutaneous manifestations associated with anticoagulation. J. Cutan. Med. Surg. 21, 540–550. doi: 10.1177/1203475417716364
Wade, K. H., Forouhi, N. G., Cook, D. G., Johnson, P., McConnachie, A., Morris, R. W., et al. (2015). Variation in the SLC23A1 gene does not influence cardiometabolic outcomes to the extent expected given its association with L-ascorbic acid. Am. J. Clin. Nutr. 101, 202–209. doi: 10.3945/ajcn.114.092981
Walters, R. K., Polimanti, R., Johnson, E. C., McClintick, J. N., Adams, M. J., Adkins, A. E., et al. (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 21, 1656–1669. doi: 10.1038/s41593-018-0275-1
Wang, B., Yang, L.-P., Zhang, X.-Z., Huang, S.-Q., Bartlam, M., and Zhou, S.-F. (2009). New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab. Rev. 41, 573–643. doi: 10.1080/03602530903118729
Wang, M., Li, Z., Chu, H., Lv, Q., Ye, D., Ding, Q., et al. (2016). Genome-wide association study of bladder cancer in a chinese cohort reveals a new susceptibility locus at 5q12.3. Cancer Res. 76, 3277–3284. doi: 10.1158/0008-5472.CAN-15-2564
Wang, T., and Xu, L. (2019). Circulating vitamin E levels and risk of coronary artery disease and myocardial infarction: a mendelian randomization study. Nutrients 11:153. doi: 10.3390/nu11092153
Wang, Y., Tang, Y., Ji, Y., Xu, W., Ullah, N., Yu, H., et al. (2020). Association between rs174547 and levels of long-chain polyunsaturated fatty acids: a meta-analysis. Br. J. Nutr. doi: 10.1017/S0007114520005103. [Epub ahead of print].
Willer, C. J., Schmidt, E. M., Sengupta, S., Peloso, G. M., Gustafsson, S., Kanoni, S., et al. (2013). Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283. doi: 10.1038/ng.2797
Williams, C. J., Williams, M. G., Eynon, N., Ashton, K. J., Little, J. P., Wisloff, U., et al. (2017). Genes to predict VO trainability: a systematic review. BMC Genomics 18:831. doi: 10.1186/s12864-017-4192-6
Wilman, H. R., Parisinos, C. A., Atabaki-Pasdar, N., Kelly, M., Thomas, E. L., Neubauer, S., et al. (2019). Genetic studies of abdominal MRI data identify genes regulating hepcidin as major determinants of liver iron concentration. J. Hepatol. 71, 594–602. doi: 10.1016/j.jhep.2019.05.032
Wolf, J. M., Simon, D., Béria, J. U., Tietzmann, D. C., Stein, A. T., and Lunge, V. R. (2017). Analysis of the association of nonsynonymous polymorphisms in ADH genes with hazardous drinking in HIV-1-positive individuals. Alcohol. Clin. Exp. Res. 41, 1866–1874. doi: 10.1111/acer.13486
Wright, C. F., FitzPatrick, D. R., and Firth, H. V. (2018). Paediatric genomics: diagnosing rare disease in children. Nat. Rev. Genet. 19, 253–268. doi: 10.1038/nrg.2017.116
Wu, X., Liu, Q., and Jiang, R. (2009a). Align human interactome with phenome to identify causative genes and networks underlying disease families. Bioinformatics 25, 98–104. doi: 10.1093/bioinformatics/btn593
Wu, X., Ye, Y., Kiemeney, L. A., Sulem, P., Rafnar, T., Matullo, G., et al. (2009b). Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 41, 991–995. doi: 10.1038/ng.421
Yabuta, S., Urata, M., Wai Kun, R. Y., Masaki, M., and Shidoji, Y. (2016). Common SNP rs6564851 in the BCO1 gene affects the circulating levels of β-carotene and the daily intake of carotenoids in healthy Japanese women. PLoS ONE 11:e0168857. doi: 10.1371/journal.pone.0168857
Yang, M. M., Wang, J., Fan, J. J., Ng, T. K., Sun, D. J., Guo, X., et al. (2016). Variations in the obesity gene “LEPR” contribute to risk of type 2 diabetes mellitus: evidence from a meta-analysis. J Diabetes Res 2016:5412084. doi: 10.1155/2016/5412084
Yang, N., MacArthur, D. G., Gulbin, J. P., Hahn, A. G., Beggs, A. H., Easteal, S., et al. (2003). ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 73, 627–631. doi: 10.1086/377590
Yao, P., Sun, L., Lu, L., Ding, H., Chen, X., Tang, L., et al. (2017). Effects of genetic and nongenetic factors on total and bioavailable 25(OH)D responses to vitamin D supplementation. J. Clin. Endocrinol. Metab. 102, 100–110. doi: 10.1210/jc.2016-2930
Yokoyama, A., Omori, T., and Yokoyama, T. (2020a). Risk factors for esophageal iodine-unstained lesions and changing trends among Japanese alcohol-dependent men (2003-2018). Cancer Sci. 112, 734–743. doi: 10.1111/cas.14753
Yokoyama, A., Taniki, N., Hara, S., Haysashi, E., Nakamoto, N., Mizukami, T., et al. (2018). Slow-metabolizing ADH1B and inactive heterozygous ALDH2 increase vulnerability to fatty liver in Japanese men with alcohol dependence. J. Gastroenterol. 53, 660–669. doi: 10.1007/s00535-017-1402-6
Yokoyama, A., Taniki, N., Nakamoto, N., Tomita, K., Hara, S., Mizukami, T., et al. (2020b). Associations among liver disease, serum lipid profile, body mass index, ketonuria, meal skipping, and the alcohol dehydrogenase-1B and aldehyde dehydrogenase-2 genotypes in Japanese men with alcohol dependence. Hepatol. Res. 50, 565–577. doi: 10.1111/hepr.13475
Yokoyama, A., Yokoyama, T., Matsui, T., Mizukami, T., Kimura, M., Matsushita, S., et al. (2020c). Impacts of interactions between ADH1B and ALDH2 genotypes on alcohol flushing, alcohol reeking on the day after drinking, and age distribution in Japanese alcohol-dependent men. Pharmacogenet. Genomics 30, 54–60. doi: 10.1097/FPC.0000000000000395
Yokoyama, A., Yokoyama, T., Omori, T., Maesato, H., Takimura, T., Iwahara, C., et al. (2019). Endoscopic screening using esophageal iodine staining and genotypes of ADH1B and ALDH2 in Japanese alcohol-dependent women. PLoS ONE 14:e0210546. doi: 10.1371/journal.pone.0210546
Yvert, T., Miyamoto-Mikami, E., Murakami, H., Miyachi, M., Kawahara, T., and Fuku, N. (2016). Lack of replication of associations between multiple genetic polymorphisms and endurance athlete status in Japanese population. Physiol. Rep. 4:e13003. doi: 10.14814/phy2.13003
Yvert, T., Santiago, C., Santana-Sosa, E., Verde, Z., Gómez-Gallego, F., López-Mojares, L. M., et al. (2015). Physical-capacity-related genetic polymorphisms in children with cystic fibrosis. Pediatr. Exerc. Sci. 27, 102–112. doi: 10.1123/pes.2014-0050
Zhang, M., Zhao, X., Cheng, H., Wang, L., Xi, B., Shen, Y., et al. (2014). Age- and sex-dependent association between FTO rs9939609 and obesity-related traits in Chinese children and adolescents. PLoS ONE 9:e97545. doi: 10.1371/journal.pone.0097545
Zhang, Q., Cao, Y., Chen, J., Shen, J., Ke, D., Wang, X., et al. (2019). ACTN3 is associated with children's physical fitness in Han Chinese. Mol. Genet. Genomics 294, 47–56. doi: 10.1007/s00438-018-1485-7
Zhang, Y., Tang, H.-Q., Peng, W.-J., Zhang, B.-B., and Liu, M. (2015). Meta-analysis for the association of apolipoprotein E ε2/ε3/ε4 polymorphism with coronary heart disease. Chin. Med. J. 128, 1391–1398. doi: 10.4103/0366-6999.156803
Zhao, J. V., and Schooling, C. M. (2017). Homocysteine-reducing B vitamins and ischemic heart disease: a separate-sample Mendelian randomization analysis. Eur. J. Clin. Nutr. 71, 267–273. doi: 10.1038/ejcn.2016.246
Zhao, X., Xi, B., Shen, Y., Wu, L., Hou, D., Cheng, H., et al. (2014a). An obesity genetic risk score is associated with metabolic syndrome in Chinese children. Gene 535, 299–302. doi: 10.1016/j.gene.2013.11.006
Zhao, X., Yang, Y., Sun, B.-F., Zhao, Y.-L., and Yang, Y.-G. (2014b). FTO and obesity: mechanisms of association. Curr. Diab. Rep. 14:486. doi: 10.1007/s11892-014-0486-0
Zheng, J.-S., Arnett, D. K., Parnell, L. D., Smith, C. E., Li, D., Borecki, I. B., et al. (2013). Modulation by dietary fat and carbohydrate of IRS1 association with type 2 diabetes traits in two populations of different ancestries. Diabetes Care 36, 2621–2627. doi: 10.2337/dc12-2607
Zhou, J.-C., Zhu, Y., Gong, C., Liang, X., Zhou, X., Xu, Y., et al. (2019). The haplotype of the vitamin D binding protein is a risk factor for a low plasma 25-hydroxyvitamin D concentration in a Han Chinese population. Nutr. Metab. 16:5. doi: 10.1186/s12986-019-0332-0
Keywords: whole genome sequencing, personal genomics, interpretation, precision medicine, genetic risk score, pharmacogenomics, nutrigenomics
Citation: Corpas M, Megy K, Mistry V, Metastasio A and Lehmann E (2021) Whole Genome Interpretation for a Family of Five. Front. Genet. 12:535123. doi: 10.3389/fgene.2021.535123
Received: 14 February 2020; Accepted: 15 February 2021;
Published: 08 March 2021.
Edited by:
Mehdi Pirooznia, National Heart, Lung, and Blood Institute (NHLBI), United StatesReviewed by:
Bastian Greshake Tzovaras, Centre de Recherches Interdisciplinaires (CRI), FranceCopyright © 2021 Corpas, Megy, Mistry, Metastasio and Lehmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manuel Corpas, bS5jb3JwYXNAY3BtLm9ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.