- 1International Institute of Tropical Agriculture, Ibadan, Nigeria
- 2International Institute of Tropical Agriculture, Nairobi, Kenya
- 3CGIAR Excellence in Breeding Platform, El Batan, Mexico
- 4National Root Crops Research Institute, Umudike, Nigeria
- 5Department of Global Development, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, United States
Cassava is crucial for food security of millions of people in sub-Saharan Africa. The crop has great potential to contribute to African development and is increasing its income-earning potential for small-scale farmers and related value chains on the continent. Therefore, it is critical to increase cassava production, as well as its quality attributes. Technological innovations offer great potential to drive this envisioned change. This paper highlights genomic tools and resources available in cassava. The paper also provides a glimpse of how these resources have been used to screen and understand the pattern of cassava genetic diversity on the continent. Here, we reviewed the approaches currently used for phenotyping cassava traits, highlighting the methodologies used to link genotypic and phenotypic information, dissect the genetics architecture of key cassava traits, and identify quantitative trait loci/markers significantly associated with those traits. Additionally, we examined how knowledge acquired is utilized to contribute to crop improvement. We explored major approaches applied in the field of molecular breeding for cassava, their promises, and limitations. We also examined the role of national agricultural research systems as key partners for sustainable cassava production.
Introduction
The agricultural sector is key to economic growth in Africa. The recent report on the Global Hunger Index indicates that over half of the world’s food-insecure people live in Africa (FSIN, 2019). Sustainable agricultural production is imperative to curb food insecurity, reduce poverty, and impact the livelihood of smallholder farmers (Ojijo et al., 2016; Donkor et al., 2017). Cassava is among the six commodities defined by the African Heads of States as strategic crops for the continent, given its significant contribution to the livelihoods of African farmers and its potential for transforming African economies (Feleke et al., 2016).
Cassava (Manihot esculenta Crantz) is a root crop grown throughout the tropics by more than 800 million people (Nassar and Ortiz, 2010). It can grow with minimal inputs under marginal soil conditions and in regions prone to drought. Though mainly cultivated for its starchy roots, nutrient-dense cassava leaves are also consumed as vegetables in many regions of Africa (Spencer and Ezedinma, 2017). Due to its long harvest window, cassava roots are used as a food reserve during periods of food shortage or during the lean season before harvest of other crops. Although its cultivation has traditionally been associated with subsistence farming, the crop is gradually becoming an industrial crop, which is processed into different products, including bread, pasta, and couscous-like products (Bechoff et al., 2018; Mtunguja et al., 2019). Apart from the food industry, cassava starch is used for textiles, the paper industry, in the manufacture of plywood and veneer adhesives, glucose and dextrin syrups (Tonukari et al., 2015; Spencer and Ezedinma, 2017; Waisundara, 2018).
Sub-Saharan Africa accounts for 61.1% of the world’s cassava production (FAOSTAT, 2020). Although some increase have been achieved recently, mainly due to expansion in crop area, cassava average productivity (9.1 t/ha) on the continent is well below the average cassava fresh root yield (21.5 t/ha) recorded in Asia (Spencer and Ezedinma, 2017; FAOSTAT, 2020). In order to satisfy the forecasted increase in demand for cassava food and non-food products, and to harness the enormous potential offered by the crop, cassava production in sub-Saharan Africa must be increased (Khandare and Choomsook, 2019; Otekunrin and Sawicka, 2019; FAOSTAT, 2020). Some of the constraints inherent to its production include pests (Kalyebi et al., 2018; Koros et al., 2018) and diseases caused by bacteria (Fanou et al., 2018) and viruses (Patil et al., 2015; Alicai et al., 2019) leading to significant yield losses. Another drawback is the cyanogenic glucosides, which upon hydrolysis, produces toxic hydrogen cyanide (Akinpelu et al., 2011). Cassava is an energy-dense food, mainly composed of starch with low levels of protein and other nutrients important for a balanced diet. Populations that consume cassava as a staple are at high risk of protein, vitamin A, zinc, and/or iron deficiency (Gegios et al., 2010; Stephenson et al., 2010). These challenges guide the breeding objectives: (a) high yield in terms of dry matter per unit land area; (b) resistance to diseases, such as cassava mosaic disease (CMD), cassava brown streak disease (CBSD), and cassava bacteria blight (CBB), and pests such as cassava green mites (CGM) and whiteflies; (c) improved starch quality and quantity; (d) low hydrogen cyanide potential; (e) improved nutritional value or biofortification; (f) adaption to a wide range of environments; (g) improved plant type for mechanization; and (h) end user characteristics, including processing, cooking, and organoleptic properties (Teeken et al., 2020).
Conventional breeding has been efficient in providing a continuous supply of improved cultivars that have resulted in a dramatic increase in yield of most major crops (Prohens, 2011). Conventional cassava breeding is based on phenotype-based recurrent selection, which relies on the production of full-sib and/or half-sib progenies followed by successive clonal selection stages, including single row trials, preliminary, advanced, and uniform yield trials (Ceballos et al., 2016). Many cassava varieties have been developed and released through conventional breeding (Malik et al., 2020). Breeding cassava is a challenging task due to the heterozygous genetic make-up of the crop. The development of improved varieties is time consuming due to its long breeding cycle (12 months). New tools and technologies have the potential to improve the efficiency of conventional breeding, especially when several traits are being selected at the same time. Modernization of breeding programs, through the application of innovative tools, is vital for more efficient agriculture, especially in the context of climate change, shrinking resources, land scarcity, and increased food demand. Biotechnology and new genomic approaches have the potential to enhance genetic gain, speed up the development of better cultivars, and impact the livelihoods of smallholder farmers.
We highlighted the genomic resources available in cassava and their potential applications. We also reviewed the state of knowledge of the phenotyping technologies currently available and their shortcomings. Additionally, we explored the potential and implications for integration of technological innovations in cassava genetics and breeding. With farmers being the ultimate beneficiaries, we examined how to ensure regular, sustainable high level of cassava production in sub-Saharan Africa; thereby, contributing to food security challenges and improved livelihoods through income generation.
Genomic Resources and Their Use
Genomic resources for cassava have increased substantially in recent years. Thousands of simple sequence repeats (SSR) markers have been developed from expressed sequence tags and enriched genomic DNA libraries (reviewed in Ferguson et al., 2011). The cassava chromosome-scale reference genome and the re-sequencing of diverse accessions, has identified a large number of sequence polymorphisms. Single nucleotide polymorphisms (SNPs), methylation polymorphism, Insertions-Deletions (Indels), and structural variants have been identified (Sakurai et al., 2013; Wang et al., 2014; Xia et al., 2014; Bredeson et al., 2016). A cassava haplotype map harboring 25.9 million SNPs and 19 million indels has been developed (Ramu et al., 2017). High-throughput genotyping platforms (e.g., GoldenGate assay) have allowed researchers to simultaneously interrogate tens of thousands of SNPs at a reduced cost (Ferguson et al., 2012). The development of low cost genotyping technologies based on multiplex sequencing platforms, such as genotyping by sequencing, has enabled rapid and accurate high-density fingerprinting using SNP markers (Elshire et al., 2011; ICGMC, 2015; Rabbi et al., 2015; Kamanda et al., 2020)5). These resources provide valuable tools that have contributed to genetic research and molecular breeding of cassava.
Genetic Diversity
Genetic diversity is of paramount importance in crop improvement through breeding. The extent and nature of genetic variation existing within cassava landraces and cultivars from selected African countries have been assessed using molecular markers (Kawuki et al., 2009; Kamanda et al., 2020). These studies have revealed genetic differentiation between African and South American germplasm, as well as differentiation between cassava landraces within Africa. Those from East, South, and Central Africa are somewhat differentiated from those from West Africa (Kawuki et al., 2009; Ferguson et al., 2019; Adjebeng-Danquah et al., 2020). The narrow genetic base of African cassava breeding lines is attributable to intense selection pressure for CMD resistance with recurrent selection using few parents and the clonal nature of the crop (Turyagyenda et al., 2012; Wolfe et al., 2016; Ferguson et al., 2019). Slightly higher genetic diversity has been reported in landraces in comparison to elite accessions (Ferguson et al., 2019), which is typical of most crops. It is likely that the genepool of pre-breeding germplasm will become more diverse as breeders begin to incorporate variation that responds better to consumer preferences.
Genetic Redundancy and Breeding Impact
Fingerprinting of cassava accessions using molecular markers has many applications. Genetic redundancy represents a challenge to efficiently manage and optimize the conservation of genetic resources in genebanks and breeders’ collections. SNP markers have been used to confirm that particular accessions are not identical, and others are possible duplicates (Ferguson et al., 2012). They have also been used to assess the adoption of improved varieties (Turyagyenda et al., 2012; Rabbi et al., 2015; Wossen et al., 2017). Molecular markers have an important role to play as farmers frequently give different names to the same cultivar or landrace. Thus, they become difficult to identify, particularly as cassava varieties are not easy to distinguish morphologically. This enables the correct assessment of adoption rates, which in turn, influences breeding priorities and agricultural policies (Kretzschmar et al., 2018). Molecular markers have also been used to assess the integrity of putative mapping populations, select true-cross progeny, validate crosses, and guide parental selection (Rabbi et al., 2012; ICGMC, 2015; Masumba et al., 2017).
Phenotyping of Key Traits
Phenotyping is important to support crop breeding. The acquisition of phenotype data remains a bottleneck hindering cassava genetic studies and full-deployment of genomics-assisted breeding. Several approaches have been used to phenotype breeding lines and germplasm collections for nutrition (carotenoids, cyanogenic potential), yield and yield components (dry matter content), quality (starch physiochemical and functional properties, texture, and pasting properties), biotic stresses (disease resistance), and root system architecture. In the following section, we detail the current phenotyping strategies used in cassava.
Carotenoids
Breeding cassava roots with enhanced levels of provitamin A carotenoids is a high priority in some breeding programs. The color of the cassava storage root parenchyma has been correlated with the total carotenoids content; thus, used to evaluate carotenoids content (Iglesias et al., 1997; Afonso et al., 2017). A challenge has been to efficiently distinguish the subtle differences within each color group, as this is difficult by eye. To overcome this limitation, quantification of total carotenoids content by ultraviolet-visible spectrophotometry, as well as identification and quantification of β-carotene and its isomers by high-performance liquid chromatography (HPLC), have been employed (Carvalho et al., 2012; Belalcazar et al., 2016). These approaches are accurate, but have many drawbacks, including cost, time-needed for analysis, labor-intensive methods, and requirements for laboratory infrastructure and trained technical staff, which is not always available to breeding programs in Africa (Udoh et al., 2017). Alternative portable devices, such as ICheckTM carotene, have been proven useful for rapid field evaluation and could be valuable in remote areas with no laboratory facilities or electricity (Esuma et al., 2016; Jaramillo et al., 2018). Color instruments designed to quantify the Commission International de l’Eclairage (CIELAB) color parameters have also been successfully used to evaluate carotenoids content in cassava root samples in an efficient way (Afonso et al., 2017). Near-infrared spectroscopy (NIRS) is another promising approach that has been explored for carotenoids quantification with demonstrated high prediction accuracy (Sánchez et al., 2014; Ikeogu et al., 2019).
Cyanogenic Potential
Cassava contains naturally occurring, but potentially toxic compounds called cyanogenic glycosides, which release hydrogen cyanide (HCN) on hydrolysis. This can be highly toxic to humans and animals if not removed through processing. It is important that the levels of cyanogenic glycosides are measured. Several approaches have been used to quantify cyanide potential, including the titration method (Moriasi et al., 2017; Iliya and Madumelu, 2019), the alkaline picrate method (Fukushima et al., 2016; Moriasi et al., 2017), and the metal-based chemosensors (Tivana et al., 2014). These approaches involved multi-step reactions and necessitate trained personnel. The picrate method, although easy to use, is very slow (minimum 12 h) and the chemicals used are hazardous. Recently, it was shown that NIRS could efficiently be used to distinguish roots with high or low cyanogenic potential (Sánchez et al., 2014).
Dry Matter Content
Storage root dry matter content (DMC) reflects the proportion of useable fresh root yield. DMC is commonly measured using either specific gravity through suspension of a root sample in water and air, or the oven-drying method, which has been the most widely used method. This is where a representative root sample is weighted wet and then oven dried to constant weight (Fukuda et al., 2010; Teye et al., 2011). The oven-drying method is tedious when working with a large number of samples. Likewise, it is difficult to implement this approach where the source of electricity is unreliable (Teye et al., 2011). Both oven-drying and specific gravity could be substituted by NIRS, which has been shown to predict DMC with a high degree of accuracy (Belalcazar et al., 2016).
Starch Physiochemical and Functional Properties
Physiochemical properties among cassava cultivars determine root quality attributes important for processing and consumption. The main constituent of cassava storage roots is starch, which is composed of amylose and amylopectin. Both of these play a crucial role in retrogradation, gelling, pasting, crystallinity, gelatinization temperature, viscosity, texture, cooking, eating, and processing quality of cassava (Ayetigbo et al., 2018). Amylose content (AC) is the most important factor influencing cooking and textural quality. The AC in cassava has been estimated using iodine colorimetry (Sandoval-Aldana and Fernandez, 2013; Boonpo and Kungwankunakorn, 2017) or Megazyme amylose/amylopectin assay kit (Chisenga et al., 2019). Iodine colorimetry is prone to inter-laboratory variability due to the complexity of the procedure and relies on the development of a suitable curve of known amylose to-amylopectin ratios. The swelling power of starch determines its specific functional properties when utilized in food products (Noranizan et al., 2010). Swelling power and solubility patterns of cassava flour have been determined using the Leach (1959) and Kainuma et al. (1967) method, respectively (Chisenga et al., 2019; Ma’Aruf and Abdul, 2020). Starch gelatinization properties (onset, peak and conclusion gelatinization temperature, and enthalpy) and retrogradation have been determined using differential scanning calorimetry (DSC). DSC can be run at a rate of four samples per hour (Thirathumthavorn and Trisuth, 2008; Tappiban et al., 2020). Crystallinity measurement requires the use of an x-ray diffractometer, a piece of complex and expensive equipment (Chatpapamon et al., 2019). In sweet potato, it was shown that NIRS could predict most physiochemical and thermal properties of starch with acceptable precision (Lu et al., 2006a,b). NIRS was sufficiently accurate for the determination of total starch and amylose in barley in the study of Ping et al. (2013). Meanwhile, Cozzolino et al. (2013) demonstrated that swelling properties and water solubility could be determined in whole grain barley using NIRS spectroscopy. NIRS technology could potentially be used to predict functional and physiochemical properties of cassava or cassava-based products.
Texture and Pasting Properties
Texture is a critical factor for consumer acceptance of cassava. Sensory analysis has been used for characterizing cassava and cassava-based product texture properties. The sensory descriptors assessed included texture, appearance, odor, taste, masticability (Akely et al., 2016; Adinsi et al., 2019). The cost associated with training and maintaining a descriptive panel combined with the low throughput of sensory evaluations has prompted the development of less costly and less time-consuming approaches. Instrumental methods using a texture analyzer that mimics mastication have been used to evaluate and/or predict the texture of raw, cooked, and processed cassava products. The parameters measured include hardness, springiness, adhesiveness, gel strength, mouldability, elasticity, smoothness, appearance, thickness, and general acceptability (Rodríguez-Sandoval et al., 2008; Maieves et al., 2012; Rosales-soto et al., 2016; An et al., 2019; Ma’Aruf and Abdul, 2020). Pasting properties of cassava products are key determinants of quality. Rapid Visco Analyzer (RVA) has been used to evaluate pasting properties of cassava accessions, and the parameters estimated include peak viscosity, setback viscosity, final viscosity, pasting temperature, and time to reach peak viscosity. Using RVA, less than five samples can be processed in 1 h (Rosales-soto et al., 2016; Chatpapamon et al., 2019). Although not yet reported for cassava, NIRS has been shown to adequately predict texture and pasting properties of rice (Meullenet et al., 2002; Chueamchaitrakun et al., 2011) and sweet potato (Lu et al., 2006a,b). Therefore, the ability of NIRS to predict cassava pasting properties should be explored.
Biotic Constraints
Different purposes required different discrimination methods for quantification of disease incidence and/or severity. For example, for breeding purposes, a visual scale of 1–5 from disease-free to highly diseased may be sufficient, where breeders will only retain those cultivars with a score lower than two. However, this is subjective and dependent upon personal perceptions. On the other hand, QTL mapping or other applications may require greater resolution or more accuracy (Garcia-Oliveira et al., 2020). Accurate evaluation and novel approaches for cassava disease detection are needed to efficiently assess disease severity (Chiang et al., 2016). Root necrosis indexes for CBSD evaluation, which account for the root sample size or their economic value, have been proposed to replace traditional based evaluation (Kawuki et al., 2019). Image analysis has similarly been used (Garcia-Oliveira et al., 2020; Nakatumba-Nabende et al., 2020). Analysis of field images combined with various algorithms, including K mean clustering algorithms (Anderson et al., 2015), artificial neutral network (Abdullakasim et al., 2011), and more recently, machine learning techniques and convolutional neutral network (Owomugisha and Mwebaze, 2016; Sambasivam and Opiyo, 2020) have provided a more accurate and objective assessment of disease severity and incidence. The smart phone-based diagnostic system (NURU-AI) is being developed to support remote diagnosis by smallholder farmers in Africa for real-time prediction of the state of cassava health (Owomugisha and Mwebaze, 2016; Ramcharan et al., 2017, 2019).
Root System Architecture
Uniformity in size and shape of cassava roots is an important breeding objective. Routine assessment of storage root size and shape have relied on visual scores that are both subjective, time consuming, labor-intensive and destructive. Implementation of non-invasive approaches include image analysis of roots to provide unbiased quantitative data on important root traits. High throughput three-dimensional imaging and non-destructive methods such as ground penetrating radar (GPR) have recently been developed and used for quantifying cassava storage roots and their parameters for estimation of size and shape under field condition (Delgado et al., 2017, 2019). Kengkanna et al. (2019) developed cassava shovelomics to evaluate root architecture traits. More recently, Yonis et al. (2020), demonstrated the feasibility of root phenotyping using image capture and analysis. Routine implementation of these new phenotyping solutions offers new opportunities for cassava breeders to efficiently and precisely select and release cultivars with root architecture that are favorable for harvesting, processing, as well as selection for early bulking characteristics. The development of robust phenotyping technologies for large-scale phenotyping with increased precision at reduced cost will enable efficient screening of larger populations. Precise phenotyping is invaluable for carrying out downstream analysis, including characterization of genetic factors that contribute to phenotypic variation.
Connecting Genotypic and Phenotypic Information and Discovery of Useful Genes and/or Quantitative Trait Loci
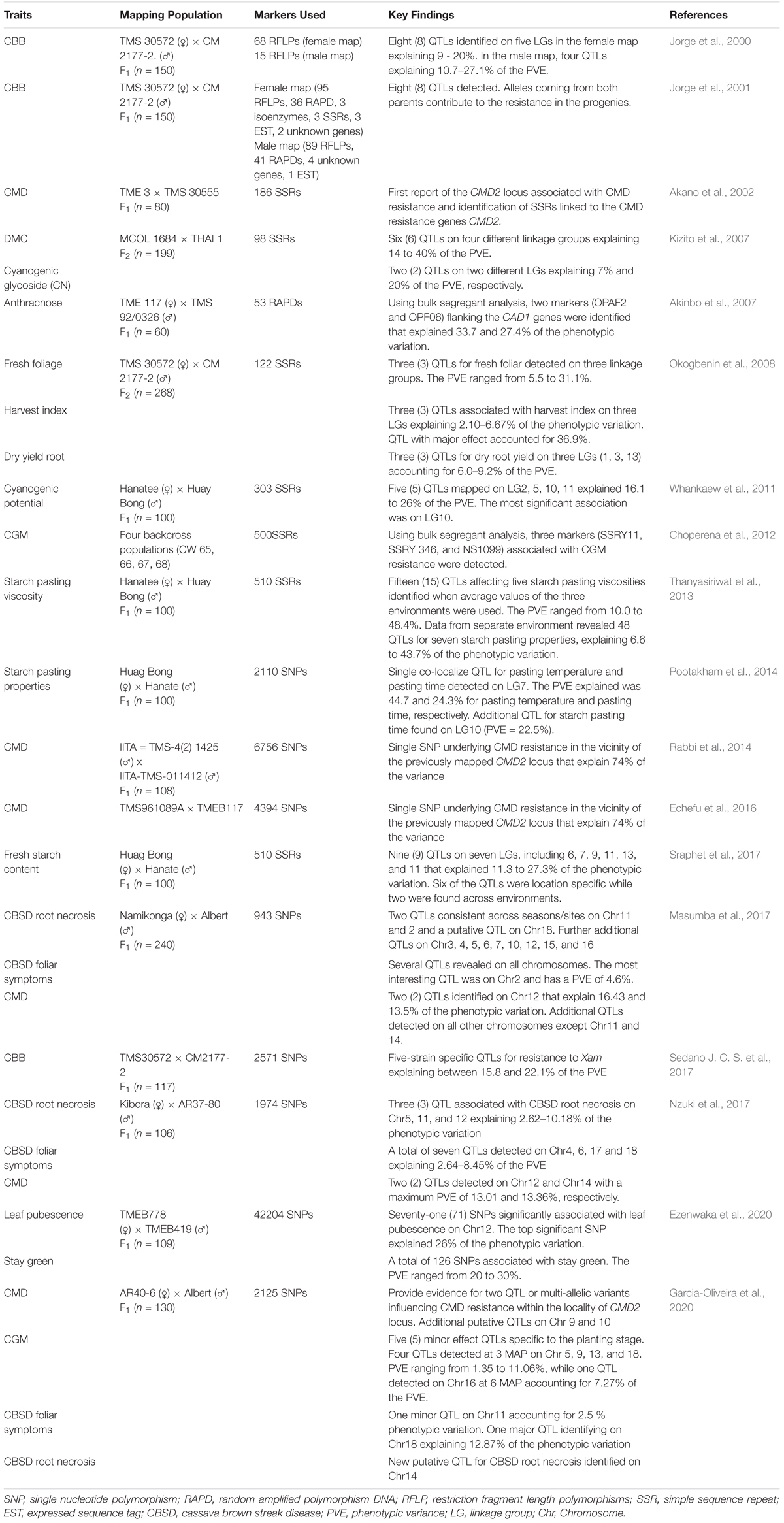
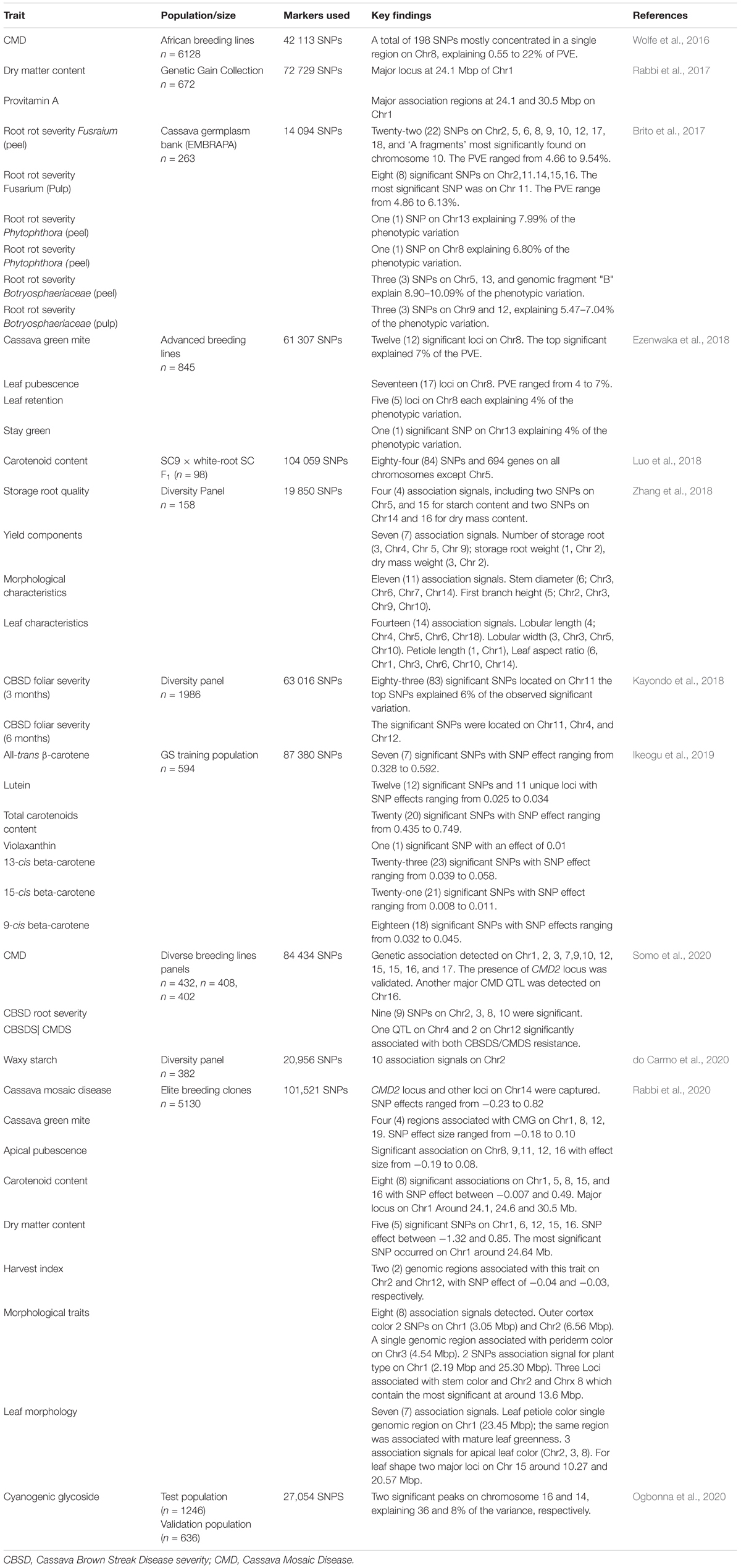
Genotype variations have been linked to corresponding differences in phenotypes and the genetic basis of the phenotypic variability evaluated to identify genes and/or quantitative trait loci (QTLs) associated with the traits of interest. Two approaches have been used for genotype-phenotype association: (1) classical QTL mapping using experimental populations derived from bi-parental crosses with contrasting phenotypes (Collard et al., 2005); and (2) genome-wide association studies (GWAS) mapping that use germplasm collections and incorporate historical events that have occurred during domestication of the crop. Classical QTL analysis in cassava has been made possible with the development of numerous genetic linkage maps (Table 1) and use of statistical approaches that have resulted in several QTLs identified for economically important traits (Table 2). With the decreasing cost of DNA sequencing, new genotyping technologies and their improved accuracy, GWAS is being increasingly used for genetic analysis of traits. GWAS has enabled the exploration of allelic diversity that exists in natural populations, the discovery of beneficial alleles in several crops. Marker-trait associations in cassava have focused on a few important traits of interest.
Cassava Mosaic Disease
The dominant gene CMD2 underlying CMD resistance was discovered in the farmer-preferred Nigerian landrace TME 3 (Akano et al., 2002). A new linked QTL underlying CMD resistance named CMD3 that explained 11% of the phenotypic variance (PVE) was later identified 36 cM away of CMD2 gene by Okogbenin et al. (2012). The qualitative nature of CMD resistance was confirmed in subsequent studies, and a single locus with a large effect in the vicinity of the previously mapped CMD2 locus was uncovered (Rabbi et al., 2014; Echefu et al., 2016; Table 2). The first GWAS mapping study in cassava conducted by Wolfe et al. (2016) identified 198 significant SNPs associated with CMD severity on 14 chromosomes. The significant SNPs were mostly concentrated in a single region on chromosome 8 (based on Manihot esculenta v5 genome assembly corresponding to chromosome 12 on v5.1 and v6.1) that account for 30 to 60% of variation in genetic resistance. Additional regions with small effects, including one on chromosome 9 that co-located with the CMD1 resistance were also reported (Wolfe et al., 2016; Table 3). The study of Wolfe et al. (2016) substantiated bi-parental mapping studies (Akano et al., 2002; Okogbenin et al., 2012) reporting the single major gene, CMD2, determining CMD resistance and a second QTL, CMD3, closely linked to CMD2. A key outcome of the Wolfe et al. (2016) study was the lack of other major-effect loci. Likewise, significant interactions between the significant SNPs were disclosed. Nzuki et al. (2017) found two QTLs associated with CMD on chromosomes 12 and 14. The percentage of variation explained (PVE) by these QTLs were 13.01 and 13.36%, respectively. The QTL on chromosome 12 was confirmed as the QTL linked to the CMD2 locus. Masumba et al. (2017) detected two highly significant CMD resistance QTLs on chromosome 12 defining the CMD2 locus, as well as additional putative QTLs on other chromosomes. Similar observations were made by Garcia-Oliveira et al. (2020) who found two closely linked loci on chromosome 12 and additional QTLs on chromosomes 9 and 10. Further dissection of the major QTL on chromosome 12 (Manihot esculenta v5.1 and 6.1 genome assembly) revealed the presence of two possible epistatic loci and/or multiple resistance alleles, which may account for the difference between moderate and strong disease resistance in the germplasm (Masumba et al., 2017; Nzuki et al., 2017; Garcia-Oliveira et al., 2020; Table 2). Gaps in the pseudochromosome 12 region containing the CMD2 loci caused by highly repetitive DNA might explain the two separate loci (Kuon et al., 2019). Somo et al. (2020) identified QTLs associated with CMD severity across all chromosomes, except on chromosomes 4, 5, 6, 8, 11, 13, and 18. They validated the presence of the previously reported CMD2 locus and detected another major QTL on chromosome 16. Nzuki et al. (2017) and Rabbi et al. (2020) confirmed the role of CMD2 loci as the major gene for CMD resistance and reported two additional loci on chromosome 14 (Table 3).
Cassava Brown Streak Disease
Quantitative trait loci associated with CBSD root necrosis were identified on chromosomes 5, 11, 12, and 15 by Nzuki et al. (2017). The detected QTLs explained up to 10.18% of the PVE. Masumba et al. (2017) reported two consistent QTLs linked to resistance to CBSD-induced root necrosis on chromosomes 2 and 11, as well as a putative QTL on chromosome 18. Further additional putative QTLs were detected on other chromosomes (3, 4, 5, 6, 7, 10, 12, 15, and 16). In addition to QTLs found on chromosome 8 and 18, new putative QTL for CBSD root necrosis was identified on chromosome 14 (Garcia-Oliveira et al., 2020). Seven QTLs associated with CBSD foliar symptoms were found on chromosomes 4, 6, 15, 17, and 18. The most significant QTL explained 8.45% of the PVE (Nzuki et al., 2017). The study of Masumba et al. (2017) revealed several QTLs on all chromosomes linked with CBSD foliar symptoms, the most interesting of which was found on chromosome 2 and explained 4.6% of the PVE. Garcia-Oliveira et al. (2020) identified one major QTL on chromosome 18 that explained 12.87% of the phenotypic variation, but at a slightly different position. These same authors also reported a minor QTL on chromosome 11 (Table 2). The GWAS approach was used by Kayondo et al. (2018) to unravel the genetic architecture of CBSD. The polygenic control mechanism of resistance for CBSD and its instability across the environment was highlighted. Eighty-three (83) loci associated with foliar symptoms at 3 months after planting (MAP) were identified on chromosome 11. The top SNPs explained 6% of the phenotypic variance. Significant SNPs were identified on chromosomes 11, 4, and 12 for foliar severity score at 6 MAP. Recently, Somo et al. (2020) using a diverse panel of breeding lines, identified QTLs conferring resistance to CBSD on chromosomes 9 and 11 that accounted for 9 and 5% of PVE, respectively. Nine markers representing four loci on chromosomes 2, 3, 8, and 10 associated with resistance to CBSD for root necrosis were also reported by these authors (Table 2). Putative regions on chromosomes 11 and 15 were shown to be associated with both CBSD foliar and root necrosis symptoms (Nzuki et al., 2017), suggesting that CBSD root necrosis and CBSD foliar symptoms are most likely to be influenced to some extent by the same gene(s) or by closely linked genes at this locus. However, most of the QTLs reported have been associated with either root necrosis or foliar symptoms, supporting the notion that resistance to foliar and root symptoms of CBSD are largely under different genetic control (Masumba et al., 2017; Garcia-Oliveira et al., 2020). The detected QTLs were not consistent across studies. Different mapping populations were used in the case of experimental populations. There might be some variations in CBSD response. Furthermore, QTLs tend to be population specific in bi-parental populations. Likewise, different environmental conditions, population size (106–1986 samples), and the subjectivity of the scoring system used for data collection could have affected QTL detection and localization. The use of different versions of the cassava genome assembly makes it challenging to compare some of these results. Therefore, these QTLs should be mapped onto the most recent version of the cassava reference genome.
Cassava Green Mite and Related Traits
Nzuki et al. (2017) detected on chromosomes 5 and 10 QTLs associated with cassava green mite (CGM) with maximum PVE of 10.56 and 10.08%, respectively. Recently, 95 SNP markers significantly associated with CGM resistance were reported by Ezenwaka et al. (2020). The significant markers concentrated in a single region of the left arm side on chromosome 12. The variance explained by the significant markers ranged from 18 to 31%. Garcia-Oliveira et al. (2020) detected five QTL for CGM resistance at 3 and 6 MAP, all with minor effect on chromosomes 5, 9, 13, and 18. While the QTLs on chromosome 9 was found in all four environment tested, the QTL on chromosome 8 co-localized with previously reported marker for CGM resistance (Table 2). The first GWAS to identify SNPs linked to CGM and CGM-related traits was performed by Ezenwaka et al. (2018) who found 35 significant SNP markers, including 12, 17, 5, and 1 associated with CGM, leaf pubescence, leaf retention, and stay green, respectively. All the significant markers were found on chromosome 8, except the SNP associated with stay green, which was identified on chromosome 13. Some of the significant SNP markers on chromosome 8 reported by these authors were also detected in the recent study of Rabbi et al. (2020) who identified other putative loci on chromosomes 1, 12, and 9. Association analysis of CGM-related traits, apical pubescence identified significant loci on five chromosomes, two of which co-located in the same regions underlying resistance to CGM on chromosomes 8 and 12 (Rabbi et al., 2020; Table 3).
Cassava Bacterial Blight
Two QTLs associated with cassava bacterial blight (CBB) caused by Xanthomonas axonopodis pv. manihotis (Xam) were reported on linkage group (LG) 4 and LG8 that explained 12.6 and 10.9% of the field resistance to CBB, respectively (Sedano J. S. et al., 2017). Another study conducted by Sedano J. C. S. et al. (2017) found five strain-specific QTLs conferring resistance to Xam that explained 15.8 and 22.1% of the phenotypic variance. Three of the associated QTLs were found to be effective against Xam318 strain and explained 17.3 to 18.8% of the phenotypic variance, while two were detected for Xam681 and accounted for 15.8–22.1% of the phenotypic variance (Table 2).
Cassava Root Rot
The complex nature of cassava root rot disease (CRR) was highlighted by Brito et al. (2017), who identified 38 significant SNPs associated with CRR. Of these, 8 and 22 were related to the severity of dry root rot in the pulp and peel, respectively, while the other eight were associated with soft root rot and black root rot (Table 3).
Carotenoids and Storage Root Color
Three QTLs that control the content of carotenoids and four QTLs linked to the color of cassava roots pulp were identified by Morillo et al. (2013) (Table 2). Root yellowness resulting from carotenoid accumulation elucidated through GWAS has revealed major association regions that govern this trait around 24.1 and 30.5 Mbp of chromosome 1 (Rabbi et al., 2017). More recently, using a larger diversity panel, Rabbi et al. (2020) confirmed these previous findings. They found five new genomic regions associated with carotenoid content on chromosomes 5, 8, 15, and 16. Luo et al. (2018) identified 84 SNPs distributed in all chromosomes, except chromosome 5, associated with carotenoid traits. Ikeogu et al. (2019) identified 42 unique SNPs significantly associated with variation in total carotenoid content on chromosomes 1, 2, 4, 13, 14 and 15. Additional regions for variation in the total carotenoid content, as well as the individual carotenoids, were uncovered. Some regions associated with more than a single carotenoid were identified, suggesting the possibility of pleiotropic effects (Table 3). The level of phenotypic variability, the SNP frequencies and distributions, and the difference in sample size (98–5130 samples) between the panels used for GWAS might explain the differences in QTLs detected. The wider coverage of diversity could increase the detection of true novel associations, while small sample size could lead to some spurious associations.
Cyanogenic Glucosides
Kizito et al. (2007) reported two QTLs (SSRY105, SSRY42) on two different linkage groups controlling cyanogenic glucosides. The two QTLs explained 7% and 20% of phenotypic variation, respectively. Also, both QTLs showed additive effects. Five QTLs associated with cyanogenic potential were identified across four linkage groups, including LG2, 5, 10, and 11 (Whankaew et al., 2011). The percentage of phenotypic variance explained from all detected QTLs ranged from 15.9 to 26.0% (Table 2). One (SSRY42) of the two QTLs reported by Kizito et al. (2007) was found in this latest study. More recently, Ogbonna et al. (2020) reported two genomic regions, in chromosomes 16 and 14, associated with hydrogen cyanide potential in cassava. The most significant marker was found on chromosome 16. Chromosomes 16 and 14 tagged SNPs explained 36 and 8% of phenotypic variance, respectively (Table 3). The significant region on chromosome 14 coincides with previously reported cyanide associated QTL reported by Kizito et al. (2007).
Dry Matter Content
Six QTLs detected on four different LGs were reported to control DMC using a bi-parental mapping population. Individual QTL explained 14 to 40% of the variance. It was shown that additive, dominance, and overdominance effects play a role in the expression of this trait (Kizito et al., 2007; Table 2). Using genome-wide association mapping, DMC was found to be associated with a major locus occurring on a 24.1 Mbp region of chromosome 1 (Rabbi et al., 2017). This locus was confirmed by the recent study of Rabbi et al. (2020), who reported another major locus on chromosome 6 as well as three additional loci on chromosomes 12, 15, and 16 (Table 3).
Starch and Starch Quality
The study of Thanyasiriwat et al. (2013) revealed the complex inheritance of starch pasting properties. Using average values of three environments, these authors reported 15 QTLs on LG1, 4, 6, 7, 8, 10 12, 13, 14, 16, and 18 affecting five starch pasting viscosities (peak viscosity, hot paste viscosity, cool paste viscosity, set back, and pasting temperature). The detected QTLs explained 10.0 to 48.4% of the phenotypic variance. Based on analysis of each environment, 48 QTLs significantly associated with seven starch pasting viscosities (peak viscosity, hot paste viscosity, break down, cool paste viscosity, setback, pasting time, and pasting temperature) were detected on all LGs except LG2, 4, and 7. The PVE ranged from 6.6 to 43.7. Thanyasiriwat et al. (2013) also reported two major QTLs on LG1 and LG6 for pasting temperature, which accounted for more than 70% of the phenotypic variation. Pootakham et al. (2014) identified a single co-localized QTL controlling pasting temperature and pasting time on LG7. The major QTL explaining 44.7 and 24.3% of the phenotypic variance. These authors reported additional QTL associated with starch pasting time on LG 10 that accounted for 22.5% of the phenotypic variance. A total of nine QTLs controlling fresh starch content was identified on seven linkage groups (LG4, 6, 7, 9, 11, 13, and 16). Among these, six QTLs were location-specific, and three QTLs were detected across three environments. The percentage of phenotypic variance explained by the QTLs ranged from 11.3 to 27.3% of the phenotypic variation (Sraphet et al., 2017; Table 2). Using GWAS, 10 SNP associated with waxy phenotypes were identified on chromosome 2 that co-located in genic regions that included five known genes and five genes of unknown function (do Carmo et al., 2020; Table 3).
Agronomy and Physiology Traits
Eight QTLs associated with fresh root yield were identified on seven linkage groups (LG1, 2, 6, 9,12,13, and 16) from a bi-parental mapping population, of which two QTLs on linkage group 16 were found across two environments. These QTLs explained 12.9 to 40% of phenotypic variation (Sraphet et al., 2017; Table 2). Zhang et al. (2018) reported 36 loci related to 11 agronomic traits, including leaf characteristics, morphological characteristics, yield components, and root quality that were identified by GWAS analyses. They found seven SNPs associated with yield components that explained about 14.95% of the phenotypic variance on average. Morphological characteristics exhibited 11 association signals and explained 12.23 to 20.86% of the phenotypic variance. A total of 14 SNPs were identified from leaf characteristics, which explained 11.9 to 22.6% of the phenotypic variance. Genomic regions associated with harvest index were uncovered on chromosomes 2, 3, 4, 6, 8, 9, 12, 14, and 15. Two association signals for outer cortex color were found on chromosomes 1 (3.05 Mbp) and 2 (6.56 Mb). A single genomic region on chromosome 3 (4.54 Mbp) has been linked to periderm color. Two association signals were found for plant types on chromosome 1 (2.19 and 25.30 Mbp). Five loci associated with stem color variation were reported on chromosomes 2 and 8. The most significant loci were around 13.6 Mbp on chromosome 8. A single genomic region at around 23.45 Mbp on chromosome 1 was found concomitantly associated with leaf petiole color and mature leaf greenness. Variation in leaf color was found to be associated with three loci occurring on chromosomes 2, 3, and 8. At around 10.27 and 20.57 Mbp on chromosome 15, two loci associated were identified (Rabbi et al., 2020; Table 3). Seventy-one (71) markers significantly associated with leaf pubescence were identified by Ezenwaka et al. (2020) on chromosome 12. The variance explained by these significant markers ranged from 18 to 26%. The same study of Ezenwaka et al. (2020) reported 126 SNP markers associated with stay green on chromosome 12, and the variance explained by the detected QTLs ranged from 20 to 30% (Table 2).
Numerous favorable alleles, functional loci or regions linked to traits of interest have been identified through marker-trait associations and their phenotypic contribution identified, giving a glimpse to the genetics underlying phenotype variation. However, the designation of linkage groups/chromosomes are sometimes different, making it challenging to align QTL detected in different studies (Garcia-Oliveira et al., 2020). Synchronization of cassava QTL information and the development of a cassava QTL repository is needed. A cassava QTL database that contains QTL information systematically aligned to the cassava reference genome, as well as information about germplasm and genetic material used in the QTL studies, will be a useful resource for both cassava geneticists and breeders (Ni et al., 2009; Yonemaru et al., 2010; Said et al., 2015). A benefit envisioned from the identification of molecular markers tightly linked to traits of interest is their deployment in breeding as an indirect selection method to accelerate the rate of genetic gain.
QTL Utilization in Cassava Breeding
Numerous QTL controlling a wide variety of traits have been identified to be utilized in marker-assisted selection (MAS) known as forward selection (Tables 2, 3). MAS is a technique of indirect selection of traits. Its implementation is still challenging in many breeding programs (Chukwu et al., 2019; Cobb et al., 2019). Although panels of molecular markers closely linked to key cassava traits have been identified, successful applied experiences of MAS in cassava breeding are limited (Table 4). Markers tightly associated with CMD2 have been effectively used to identify genotypes bearing the CMD2 gene in West African germplasm, introgression of CMD2 genes into various cassava germplasm, and selection of parental lines for planned crosses. This has enabled breeders to focus on fewer genotypes at an early stage of the breeding program, saving the breeder time and labor cost (Blair et al., 2007; Okogbenin et al., 2012; do Carmo et al., 2015). MAS is appropriate for moderate to highly inherited traits that are controlled by a few major genes and not sufficiently predictive for quantitatively inherited traits controlled by many genes at different loci, each contribution to a small effect of the phenotypic expression (Singh et al., 2019). The time and investment required to develop reliable markers have also been a limiting factor for MAS implementation in cassava breeding programs in SSA. Likewise, the use of convenient phenotyping proxies do not always translate into meaningful selection targets for the breeding programs (Cobb et al., 2019). Future endeavors should prioritize cassava traits for marker development using a stage-gate system (SGS) to manage trait development. This will require the development of well-defined cassava product profiles, a set of targeted attributes the new variety should meet (Ragot et al., 2018). The establishment of a SGS will ensure accountability, transparency, and data-driven advancement decisions. This strategy is currently being implemented with the support of Excellence in Breeding (EiB)1. MAS should be considered only when proven more efficient than phenotypic selection to justify the expenses of development and deployment of the locus in a breeding program. Likewise, marker development for the traits retained should be feasible in a short to medium term to avoid waste of resources (Collard and Mackill, 2008). The International Institute of Tropical Agriculture (IITA) and partners have designed and converted molecular markers for several major gene traits, including provitamin A, CGM, DMC, to low-plex-high-throughput marker assay2. The reliability and utility of these markers are currently being evaluated in different genetic backgrounds and across different environments. A KASP assay has recently been employed to develop and validated diagnostic markers for HCN content (Ogbonna et al., 2020). The use of shared genotyping services could substantially reduce the genotyping costs, enabling the screening of a larger number of accessions at the seedling stage. Development of novel breeding strategies or models that not only capture additive effects, but also account for dominance and epistatic interactions, could contribute to the introgression of the QTLs into desired varieties. Likewise, the model should account for QTL-environment interactions for an effective MAS scheme (Singh et al., 2019). Recent genomic innovations have prompt for the search of new tools for population improvement.
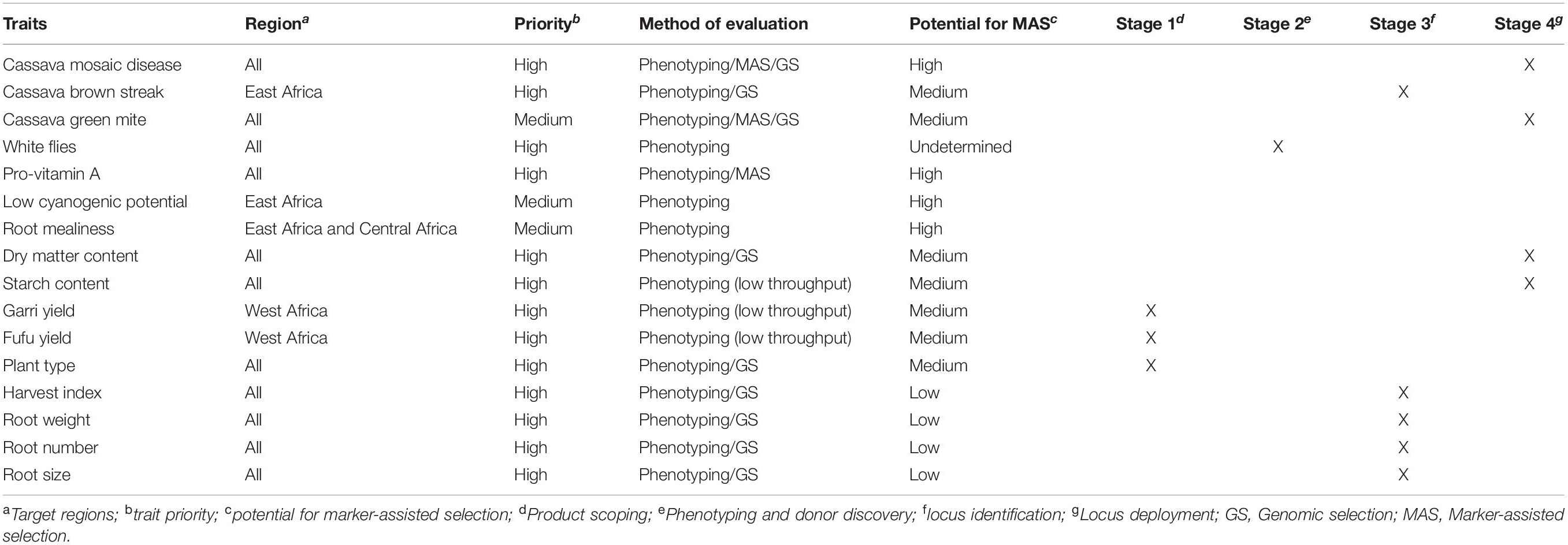
Table 4. Key cassava traits, target regions, potential for GS and/or MAS and the current stage of trait development.
Genomic Selection and It’s Potential in Cassava Breeding
Genomic selection (GS), also referred to as genomic prediction, can complement MAS; with MAS being used for highly heritable traits controlled by one or a few markers, and GS being used for more quantitative traits. GS relies on predicting the genomic estimated breeding values (GEBV) of an individual using a trait specific model built by simultaneously fitting information provided by thousands of molecular markers spread throughout the genome (Meuwissen et al., 2001). Fitting all markers simultaneously allows a substantial fraction of trait heritability missed by QTLs or association mapping to be captured. These are likely to be small effect alleles (Resende et al., 2012). The approach was first championed in dairy cattle (Jonas and de Koning, 2013). There are great expectations for the use of GS in cassava breeding. Since the first GS studies in cassava conducted by de Oliveira et al. (2012) and Ly et al. (2013), highlighting the potential of GS, several studies have been published (Wolfe et al., 2017; Ozimati et al., 2018; de Andrade et al., 2019; Yonis et al., 2020). GS has been utilized in cassava breeding to increase resistance against CMD and CBSD in cassava populations (Wolfe et al., 2016; Kayondo et al., 2018; Ozimati et al., 2018). Ozimati et al. (2018) highlighted the potential of GS as a pre-emptive breeding strategy. Torres et al. (2019) predicted good progress in selecting clones for traits such as fresh root yield (FRY), dry matter content (DMC), fresh shoot yield (FSY), harvest index (HI), dry yield (DY) using GS. Yonis et al. (2020) assessed genomic prediction for root size and shape based on root traits extracted from digital images. Greater predictive ability has been reported for DMC, CMD, and, to a lesser extent, HI compared to other traits (Wolfe et al., 2016; de Andrade et al., 2019). This success has been attributed to their high to moderate heritability, large-effect QTLs and low genotype × environment interaction (Torres et al., 2019; Yonis et al., 2020). GS is especially attractive for complex traits controlled by many QTLs with low heritability that are difficult or expensive to assess or are measured late in the breeding cycle (de Oliveira et al., 2012). It could drastically reduce the breeding cycle by choosing new parents based on GEBV rather than actual phenotypes and limiting the size and number of field experiments. However, refined strategies should be adopted for complex traits. Non-additive genetic variation prevails for low heritability cassava traits and should be accounted for. Therefore, models that capture non-additive effects should be applied (Wolfe et al., 2016). GS, like any other approaches, is facing various challenges. The GS models predict poorly across populations, and consequently, the strategy requires continuous re-calibration with every breeding cycle (Wolfe et al., 2017). Allele frequency changes, introgression of new alleles, SNP-QTL linkage disequilibrium association, lack of relatedness between germplasm and population structure, as well as genotype-by-environment interaction effects, compromise the efficiency of GS. To overcome some of these limitations, dual-purpose population development and variety development pipelines have been applied to ensure training data are closely related to the new clones and for continuously updating the training model (Santantonio et al., 2020). GS could be more robust by integrating biological knowledge. The inclusion of QTL markers associated with the trait of interest could increase the robustness of genetic evaluation (Ozimati et al., 2018; Lan et al., 2020). Other variables affecting the precision of the prediction model include the size of the training population, the number of markers used in the model, the trait genetic architecture, and heritability (de Oliveira et al., 2012; Ly et al., 2013; Wolfe et al., 2017; Somo et al., 2020). Poor predictions have been reported across breeding programs limiting the prospect of sharing data from different locations, breeding programs, and countries (Wolfe et al., 2017; Somo et al., 2020). An international project funded by UK Foreign, Commonwealth and Development Office (FCDO) and the Bill and Melinda Gates Foundation (Gates Foundation) named the Next Generation Cassava Breeding Project3 is currently using this approach in four African research institutes, including the National Crop Resources Research Institute (NaCRRI) in Uganda, the National Root Crops Research Institute (NRCRI) in Nigeria, the Tanzania Agriculture Research Institute (TARI) in Tanzania, and IITA in Nigeria. A multi-trait genomic selection strategy is being applied using program-based selection indices to efficiently improve quantitative traits simultaneously. The selection is based on yield, DMC, virus resistance to CMD and CBSD, and good plant architecture. Outstanding clones are currently advanced toward the variety release pipeline. The adoption of GS has guided the mating design, enabling rapid pyramiding of favorable allele combinations and development of progeny with the improved allelic combinations (de Oliveira et al., 2012). Moreover, GS is being used to increase genetic gain by decreasing the breeding cycle time, increasing selection accuracy, and increasing selection intensity in early generations. In order to identify a more effective breeding strategy and further improve breeding efficiency, computational simulations are currently being performed with the support of EIB4 to: (1) compare different selection methods; (2) define optimal crossing strategies; and (3) determine appropriate recycling times. The data intensive nature of GS and widespread use of this approach across more breeding programs requires development of shared computational infrastructures and analysis pipelines.
Development of Computational Tools for a Greater Impact
Breeding modernization will necessitate the development of robust statistical tools and innovative analytical pipelines and platforms to accommodate the enormous quantity of data, which is being generated. Due to its clonal propagation method, there is a narrow timeline between harvesting and the next cassava planting season. This necessitates quick decision making and effective data management. Information systems are required to track samples, store genomic and phenotypic information, merge data, and conduct analysis to guide decision making (Santantonio et al., 2020). It is from this perspective that an open access repository, such as Cassavabase5, has been developed to centralize information access to cassava research data and support cassava breeding programs (Fernandez-Pozo et al., 2015). Cassavabase contains phenotypic, pedigree, and genomics data generated over 30 years by cassava programs in Africa, Asia, and South America. Cassavabase also encompasses computational tools to facilitate analyses.
Genome Editing Advances and Prospects
Genome-editing is becoming a popular molecular tool for functional genomics, as well as crop improvement. Clustered regularly interspaced palindromic repeats (CRISPR/CRISPR-associated protein 9 (Cas9)-mediated genome-editing has rapidly become the most popular genome engineering approach due to its simplicity, efficiency, specificity, multiplexing and ease to adapt. Most of the CRISPR/Cas9-based genome-editing is reported for seed crops; however, recently, it is also established for clonally propagated crops such as potato, banana, and cassava (Butler et al., 2016; Odipio et al., 2017; Kaur et al., 2018; Naim et al., 2018; Tripathi et al., 2019; Ntui et al., 2020). The accessibility of the genetic transformation system and reference genomes for cassava made it possible to realize the potential of CRISPR-based genome-editing for basic and applied research to improve economically important cassava traits. The CRISPR/Cas9-based genome-editing system was demonstrated by knocking out the Phytoene desaturase (MePDS) gene in two cultivars of cassava, TMS60444 and TME204 (Odipio et al., 2017). Genome-editing was further applied for developing a cassava variety with resistance against two species of Ipomovirus, cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV), causing CBSD (Gomez et al., 2019). The targeted mutations in the translation initiation factor 4E (eIF4E) isoforms nCBP-1 and nCBP-2 in the edited cassava variety showed a reduction in disease severity and virus accumulation in the storage tuberous roots upon glasshouse challenge of edited cassava lines with CBSV. Even though the mutations in the nCBP-1 and nCBP-2 genes conferred enhanced resistance to CBSD, complete resistance was not obtained. This suggests that total resistance to CBSD can be developed by stacking the genome-editing approach of disrupting eIF4E isoforms with other resistance strategies such as RNAi (Gomez et al., 2019). Later, researchers have attempted to apply this technology to develop resistance against a geminivirus, African cassava mosaic virus (ACMV) (Mehta et al., 2019). However, the edited cassava plants did not show significant resistance against ACMV in greenhouse inoculation experiments. It might be due to the evolution of editing-resistant geminiviruses in genome-edited cassava. CRISPR/Cas9 based genome-editing can be coupled to genetic improvements in cassava for traits such as starch improvement and early flowering. Cassava roots normally produce large quantities of starch, having high amylose levels, a crystallizable component that is more soluble in water. However, the starch with low amylose levels, known as “waxy starch,” is preferred for food processing and other industrial uses. Bull et al. (2018) reported the application of CRISPR/Cas9 for manipulating starch biosynthesis and improving the starch quality in the storage. They generated edited cassava plants with mutations in two genes: protein targeting to starch (PTST1) or granule bound starch synthase (GBSS) involved in amylose biosynthesis, leading to reduction or elimination of amylose content. This, in turn, can improve the quality of starch cassava roots for commercial use. The authors also demonstrated accelerated breeding by transferring the Arabidopsis FLOWERING LOCUS T gene in the genome-edited events of cassava for early flowering. Genome-editing can multiplex the traits, and researchers can develop cassava varieties with the waxy starch and early flowering. Despite still being in its infancy, genome-editing offers promising prospects for cassava improvement and could shorten the breeding process. Novel plant varieties could be directly used for crop production or as pre-breeding materials (Xu et al., 2019). Technological developments like this should be followed by their adoption to increase productivity.
Current Status of Technology Applications in Sub-Saharan Africa
Effective implementation of technological development is key to enhancing the productivity and profitability of cassava on the continent. Genetic studies conducted by national institutes or academia in sub-Saharan Africa have been mainly focused on germplasm characterization, genetic diversity assessment, varietal identification, linkage mapping, and classical QTL mapping (Fregene et al., 1997; Kawuki et al., 2009; Rabbi et al., 2015; Adjebeng-Danquah et al., 2020). Recently, collaborative projects involving international institutions, NARS, along with substantial funding from the donor communities, have contributed to the increased number of genome-wide association studies. Several gene(s)/QTLs underlying key cassava traits have been identified and trait-linked markers, a pre-requisite for MAS have been developed (Ogbonna et al., 2020; Rabbi et al., 2020). The effective implementation of MAS hampered by economic obstacles is currently being addressed with the support of the EIB platform2, who seeks to mainstream the use of genotyping data. The platform through subsidies from the Gates Foundation offers small breeding programs access to high quality genotyping services, including low-density (Kompetitive allele specific PCR (KASP)) and mid-density (DArTAg) genotyping to foster the progressive integration of MAS into NARS crop breeding programs. The EIB low-density genotyping service is being used by IITA and NARS partners in Uganda, Tanzania, and Nigeria for quality control, identification of cassava accessions with the desired alleles, and validation of trait-markers. Genomic selection-based pilot projects are ongoing in Nigeria, Uganda, and Tanzania and the appealing perspectives offered by this approach, including shortening of the breeding cycle and speed-up of variety development have been highlighted (Wolfe et al., 2017; Kayondo et al., 2018; Somo et al., 2020). The mid-density genotyping, DArTAg, a target genotyping provided by Diversity Arrays is being used as an alternative to GBS for genomic selection applications. Although traditional methodologies are still widely used for trait phenotyping, integration of high throughput phenotyping is on course. NIRS spectroscopy is being explored by few NARS programs to predict some key cassava traits (i.e., carotenoids and DMC) and promising results have been reported (Ikeogu et al., 2017, 2019). NIRS evaluation and optimization for other traits, including gari, fufu, starch, and root mealiness is ongoing. In collaboration with IDS GeoRadar6, IITA is testing a prototype commercial GPR for routine cassava root phenotyping. The unmanned aerial vehicle (UAV) is currently used at IITA by the Cassava Source-Sink Project to quantify aboveground plant growth (Sonnewald et al., 2020). Image recognition has been evaluated for high accuracy disease detection and mobile, as well as web-based tools developed for cassava disease scoring and monitoring7. An application programming interface has been implemented within Cassavabase in order to process cassava root images. Breeders can upload cassava root necrosis sectional image captured during harvest and the result will be returned back to Cassavabase. IITA and NARS partners have digitized all processes; data are uploaded, processed, and accessed within Cassavabase to minimize human errors. CRISPR/Cas9-based genome-editing is mainly driven by IITA, in partnership with a handful of national research organizations. The difficulty in acquiring laboratory supplies, as well as the need for constant and sufficient level of funding, constitute a bottleneck for the implementation of gene-editing technology by NARS. Likewise, the legal framework and appropriate regulatory structures needed to guide the use of this technology are still lacking in several African countries, hampering the movement of plants from laboratory to the field (Tripathi et al., 2020). Furthermore, biosafety considerations remain a public concern. Winning consumer’s acceptance and trust is key to the effective implementation of this new breeding technique and to harness its full potential. Few cassava breeding programs have benefited and/or explored these recent technological developments. For greater impact, the knowledge and technologies outlined here should be disseminated and transferred to other cassava breeding programs on the continent.
Technology Transfer and Capacity Development for More Sustainable Cassava Production
Sustained research and innovation capacity is imperative for agricultural transformation in Africa (Ojijo et al., 2016). Most African national agricultural research systems (NARS) do not have well-funded cassava breeding programs with sufficient technical critical mass. Many programs routinely evaluate and multiply clones imported from larger breeding programs (i.e., from IITA or CIAT). Although most NARS would maintain certain capacity to develop and release varieties adapted to local agro-ecologies and local preferences, their breeding capacities need to be strengthened and modernized to be effective. Many of the technological innovations are new to NARS and access to equipment, reagents, and skilled personnel is challenging (Tester and Langridge, 2010). It is with this in mind that an initiative such as the Cassava Community of Practice and Partnership (CoPP) has been established through the NextGen Cassava Project to serve as a platform to disseminate and facilitate the transfer of proven tools, methods, technologies, and products. The CoPP provides the initial technical backstopping, fosters collaboration, facilitates connectivity, and creates opportunities for peer-to-peer learning between members. This will enable the establishment of best practices and procedures and implementation of recent technological innovations by NARS, increase breeding efficiencies, and create a broader and deeper impact. Successful integration and application of innovative technologies necessitate technical expertise. Scaling up the research capabilities of NARS, through training, will hone their skills and knowledge (Bull et al., 2011). The CoPP concept is not new. A successful example is the sweet potato breeding Community of Practice, which has strengthened national capacities leading to the development and released of several user-preferred varieties with impact on the quality of life of small farmers (SASHA, 2019). The cassava CoPP will need to evolve into a cohesive network with clear accountabilities and expectations. NARS and CGIAR will need to work together in a coordinated breeding network for economies of scale. Government involvement will be needed to sustain the gain achieved by the breeders.
Conclusion and Perspectives
It takes several years to develop improved cassava varieties. Genomic resources have facilitated the evaluation of local and regional genetic diversity and profiling of breeding materials. New phenotyping approaches are substituting traditional trait evaluation and have been used for marker-trait associations, enabling identification of numerous QTLs for key traits. Although notable signs of progress have been achieved, especially for less complex traits, genetic architecture is still not fully understood. For quantitatively inherited traits, phenotypic plasticity and the difficult phenotypic evaluation complicates matters further. There is a need to scale up multi-environment evaluation. The development of advanced high throughput and accurate cassava phenotyping approaches is imperative. Translating those results obtained into practical breeding methodologies and coherent biological knowledge is needed. QTL results should be exploited, and validated trait-markers developed. Priorities will have to be set to ensure return on investment. Therefore, establishment of a formal advancement system with well-defined metrics is needed. Genetic gain should be routinely monitored. Strategies need to be constantly revised as priorities evolve, and new challenges emerge. MAS and GS are complementary breeding approaches that should be used in tandem. More attention should be given to quality traits, which have received less attention; whereas quality traits influence varietal adoption and product utilization. Key quality traits need to be defined and translated into biochemical parameters. Advanced technologies, such as genome-editing, could play a prominent role in cassava improvement. However, further investigation will be required to ensure maximum benefits. It will be important that the new technologies and tools developed are used by NARS. Therefore, regional networks, shared expertise, and service will be of prime importance. Last, but not the least, the support and involvement of national and regional governments is crucial for sustainable cassava production on the continent.
Author Contributions
EN conceived this review and drafted the manuscript. CE conceived the idea and edited the manuscript. LT drafted a section of the manuscript. MF, IR, EH, KSI, PK, and CE provided critical edits. All authors contributed to the article and approved the submitted version.
Funding
The authors thank the UK’s Foreign, Commonwealth and Development Office (FCDO) and the Bill and Melinda Gates Foundation (Grant INV-007637, http://www.gatesfoundation.org) for their financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ https://excellenceinbreeding.org/blog/applying-stage-gates-better-manage-public-breeding-programs
- ^ https://excellenceinbreeding.org/module3/kasp
- ^ NextGen cassava, https://www.nextgencassava.org
- ^ https://excellenceinbreeding.org/module2
- ^ https://www.cassavabase.org/
- ^ https://idsgeoradar.com/
- ^ http://www.mcrops.org/
References
Abdullakasim, W., Powbunthorn, K., Unartngam, J., and Takigawa, T. (2011). An images analysis technique for recognition of brown leaf spot disease in cassava. Tarım Makinaları Bilim. Derg. 7, 165–169.
Adinsi, L., Akissoé, N., Escobar, A., Prin, L., Kougblenou, N., Dufour, D., et al. (2019). Sensory and physicochemical profiling of traditional and enriched gari in Benin. Food Sci. Nutr. 00, 1–11. doi: 10.1002/fsn3.1201
Adjebeng-Danquah, J., Manu-Aduening, J., Asante, I. K., Agyare, R. Y., Gracen, V., and Offei, S. K. (2020). Genetic diversity and population structure analysis of Ghanaian and exotic cassava accessions using simple sequence repeat (SSR) markers. Heliyon 6:e03154. doi: 10.1016/j.heliyon.2019.e03154
Afonso, T., Moresco, R., Uarrota, V. G., Navarro, B. B., da Nunes, E. C. N., Maraschin, M., et al. (2017). UV-Vis and CIELAB based chemometric characterization of Manihot esculenta carotenoid contents∗. J. Integr. Bioinform. 14:20170056. doi: 10.1515/jib-2017-0056
Akano, A. O., Dixon, A. G. O., Mba, C., Barrera, E., and Fregene, M. (2002). Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor. Appl. Genet. 105, 521–525. doi: 10.1007/s00122-002-0891-7
Akely, P. M. T., Djina, Y., Konan, B. R., Irie, K., Kouame, L. P., and Amani, N. G. (2016). Study of Varietal Influence Post Conservation on Biochemical and Sensory Qualities of Attiéké and Boiled Cassava (Manihot esculenta Crantz). Agric. Sci. 7, 127–136. doi: 10.4236/as.2016.73012
Akinbo, O., Gedil, M., Ekpo, E. J. A., Oladele, J., and Dixon, A. G. O. (2007). Detection of RAPD markers-linked to resistance to cassava anthracnose disease. African J. Biotechnol. 6, 677–682. doi: 10.5897/AJB2007.000-2068
Akinpelu, A. O., Amamgbo, L. E. F., Olojede, A. O., and Oyekale, A. S. (2011). Health implications of cassava production and consumption. J. Agric. Soc. Res. 11, 118–125.
Alicai, T., Szyniszewska, A. M., Omongo, C. A., Abidrabo, P., Okao-Okuja, G., Baguma, Y., et al. (2019). Expansion of the cassava brown streak pandemic in Uganda revealed by annual field survey data for 2004 to 2017. Sci. Data 6:327. doi: 10.1038/s41597-019-0334-9
An, O., Uj, U., and Re, K. (2019). Physicochemical properties of cassava processing residue flour and sensory evaluation of fufu prepared from it. J. Nutraceuticals Food Sci. 4, 1–6.
Anderson, B., Eghan, M. J., Asare-Bediako, E., and Buah-Bassuah, P. K. (2015). Optical imaging method for determining symtoms severity of cassava mosaic disease. Appl. Phys. Res. 7:34.
Ayetigbo, O., Latif, S., Abass, A., and Müller, J. (2018). Comparing characteristics of root, flour and starch of biofortified yellow-flesh and white-flesh cassava variants, and sustainability considerations: A review. Sustain 10:3089. doi: 10.3390/su10093089
Bechoff, A., Tomlins, K., Fliedel, G., Becerra Lopez-lavalle, L. A., Westby, A., Hershey, C., et al. (2018). Cassava traits and end-user preference: Relating traits to consumer liking, sensory perception, and genetics. Crit. Rev. Food Sci. Nutr. 58, 547–567. doi: 10.1080/10408398.2016.1202888
Belalcazar, J., Dufour, D., Andersson, M. S., Pizarro, M., Luna, J., Londoño, L., et al. (2016). High-throughput phenotyping and improvements in breeding cassava for increased carotenoids in the roots. Crop Sci. 56, 2916–2925. doi: 10.2135/cropsci2015.11.0701
Blair, M. W., Fregene, M. A., Beebe, S. E., and Ceballos, H. (2007). “Marker-assisted selection in common beans and cassava,” in Marker-assisted selection: current status and future perspectives in crops, livestock, forestry and fish, eds E. Guimarães, J. Ruane, B. Scherf, A. Sonnino, and J. Dargie (Rome: FAO), 81–115.
Boonpo, S., and Kungwankunakorn, S. (2017). Study on amylose iodine complex from cassava starch by colorimetric method. J. Adv. Agric. Technol. 4, 345–349. doi: 10.18178/joaat.4.4.345-349
Bredeson, J. V., Lyons, J. B., Prochnik, S. E., Wu, G. A., Ha, C. M., Edsinger-Gonzales, E., et al. (2016). Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 34, 562–571. doi: 10.1038/nbt.3535
Brito, A. C., Oliveira, S. A. S., and Oliveira, E. J. (2017). Genome-wide association study for resistance to cassava root rot. J. Agric. Sci. 155, 1424–1441. doi: 10.1017/S0021859617000612
Bull, S. E., Ndunguru, J., Gruissem, W., Beeching, J. R., and Vanderschuren, H. (2011). Cassava: Constraints to production and the transfer of biotechnology to African laboratories. Plant Cell Rep. 30, 779–787. doi: 10.1007/s00299-010-0986-6
Bull, S. E., Seung, D., Chanez, C., Mehta, D., Kuon, J. E., Truernit, E., et al. (2018). Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci. Adv. 4:eaat6086. doi: 10.1126/sciadv.aat6086
Butler, N. M., Baltes, N. J., Voytas, D. F., and Douches, D. S. (2016). Geminivirus-mediated genome editing in potato (Solanum tuberosum l.) using sequence-specific nucleases. Front. Plant Sci. 7:1045. doi: 10.3389/fpls.2016.01045
Carvalho, L. M. J., Oliveira, A. R. G., Godoy, R. L. O., Pacheco, S., Nutti, M. R., De Carvalho, J. L. V., et al. (2012). Retention of total carotenoid and β-carotene in yellow sweet cassava (Manihot esculenta Crantz) after domestic cooking. Food Nutr. Res. 56:15788. doi: 10.3402/fnr.v56i0.15788
Ceballos, H., Pérez, J. C., Barandica, O. J., Lenis, J. I., Morante, N., Calle, F., et al. (2016). Cassava breeding I: The value of breeding value. Front. Plant Sci. 7:1227. doi: 10.3389/fpls.2016.01227
Chatpapamon, C., Wandee, Y., Uttapap, D., Puttanlek, C., and Rungsardthong, V. (2019). Pasting properties of cassava starch modified by heat-moisture treatment under acidic and alkaline pH environments. Carbohydr. Polym. 215, 338–347. doi: 10.1016/j.carbpol.2019.03.089
Chiang, K. S., Bock, C. H., El Jarroudi, M., Delfosse, P., Lee, I. H., and Liu, H. I. (2016). Effects of rater bias and assessment method on disease severity estimation with regard to hypothesis testing. Plant Pathol. 65, 523–535. doi: 10.1111/ppa.12435
Chisenga, S. M., Workneh, T. S., Bultosa, G., and Laing, M. (2019). Characterization of physicochemical properties of starches from improved cassava varieties grown in Zambia. AIMS Agric. Food 4, 939–966. doi: 10.3934/agrfood.2019.4.939
Choperena, E. D. P. M., Ospina, C., Fregene, M., Montoya-Lerma, J., and Bellotti, A. C. (2012). Identificación de microsatélites en yuca asociados con la resistencia al ácaro Mononychellus tanajoa (acari: Tetranychidae). Rev. Colomb. Entomol. 38, 70–75.
Chueamchaitrakun, P., Chompreeda, P., Haruthaithanasan, V., Suwonsichon, T., and Kasemsamran, S. (2011). Prediction of pasting and thermal properties of mixed Hom-Mali and glutinous rice flours using near infrared spectroscopy. Kasetsart J. Nat. Sci. 45, 481–489.
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Oladosu, Y., Okporie, E., et al. (2019). Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 33, 440–455. doi: 10.1080/13102818.2019.1584054
Cobb, J. N., Biswas, P. S., and Platten, J. D. (2019). Back to the future: revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 132, 647–667. doi: 10.1007/s00122-018-32663264
Collard, B. C. Y., Jahufer, M. Z. Z., Brouwer, J. B., and Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
Collard, B. C., and Mackill, D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 363, 557–572. doi: 10.1098/rstb.2007.2170
Cozzolino, D., Roumeliotis, S., and Eglinton, J. (2013). Relationships between swelling power, water solubility and near-infrared spectra in whole grain barley: A feasibility study. Food Bioprocess Technol. 6, 2732–2738. doi: 10.1007/s11947-012-0948-9
de Andrade, L. R. B., Bandeirae Sousa, M., Oliveira, E. J., de Resende, M. D. V., and Azevedo, C. F. (2019). Cassava yield traits predicted by genomic selection methods. PLoS One 14:e0224920. doi: 10.1371/journal.pone.0224920
de Oliveira, E. J., de Resende, M. D. V., da Silva, Santos, V., Ferreira, C. F., Oliveira, G. A. F., et al. (2012). Genome-wide selection in cassava. Euphytica 187, 263–276. doi: 10.1007/s10681-012-0722-0
Delgado, A., Hays, D. B., Bruton, R. K., Ceballos, H., Novo, A., Boi, E., et al. (2017). Ground penetrating radar: A case study for estimating root bulking rate in cassava (Manihot esculenta Crantz). Plant Methods 13:65. doi: 10.1186/s13007-017-0216-0
Delgado, A., Novo, A., and Hays, D. B. (2019). Data acquisition methodologies utilizing ground penetrating radar for cassava (Manihot esculenta Crantz) root architecture. Geosci 9:171. doi: 10.3390/geosciences9040171
do Carmo, C. D., da Silva, M. S., Oliveira, G. A. F., and de Oliveira, E. J. (2015). Molecular-assisted selection for resistance to cassava mosaic disease in Manihot esculenta Crantz. Sci. Agric. 72, 520–527. doi: 10.1590/0103-9016-2014-0348
do Carmo, C. D., e Sousa, M. B., Brito, A. C., and de Oliveira, E. J. (2020). Genome-wide association studies for waxy starch in cassava. Euphytica 216:82. doi: 10.1007/s10681-020-02615-9
Donkor, E., Onakuse, S., Bogue, J., de Los Rios, and Carmenado, I. (2017). The impact of the presidential cassava initiative on cassava productivity in Nigeria: Implication for sustainable food supply and food security. Cogent Food Agric. 3:1368857. doi: 10.1080/23311932.2017.1368857
Echefu, S. U., Rabbi, I. Y., Nwachukwu, E. C., and Adamu, C. L. Y. (2016). Mapping of Quantitative Trait Loci Associated With Cassava Mosaic Disease (Cmd) Resistance. Int. J. Sci. Environ. 5, 2637–2645
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi: 10.1371/journal.pone.0019379
Esuma, W., Kawuki, R. S., Herselman, L., and Labuschagne, M. T. (2016). Diallel analysis of provitamin A carotenoid and dry matter content in cassava (Manihot esculenta Crantz). Breed. Sci. 66, 627–635. doi: 10.1270/jsbbs.15159
Ezenwaka, L., Carpio, D. P., Del, Jannink, J. L., Rabbi, I., Danquah, E., et al. (2018). Genome-wide association study of resistance to cassava green mite pest and related traits in cassava. Crop Sci. 58, 1–12. doi: 10.2135/cropsci2018.01.0024
Ezenwaka, L., Rabbi, I., Onyeka, J., Kulakow, P., and Egesi, C. (2020). Identification of additional/novel QTL associated with resistance to cassava green mite in a biparental mapping population. PLoS One 15:e0231008. doi: 10.1371/journal.pone.0231008
Fanou, A. A., Zinsou, V. A., and Wydra, K. (2018). Cassava bacterial blight: a devastating disease of cassava. London: Intechopen, 14–36.
FAOSTAT. (2020). http://www.fao.org/faostat/en/#data/QC (Accessed January 24, 2020).
Feleke, S., Manyong, V., Abdoulaye, T., and Alene, A. D. (2016). Assessing the impacts of cassava technology on poverty reduction in Africa. Stud. Agric. Econ. 118, 101–111. doi: 10.7896/j.1612
Ferguson, M. E., Hearne, S. J., Close, T. J., Wanamaker, S., Moskal, W. A., Town, C. D., et al. (2012). Identification, validation and high-throughput genotyping of transcribed gene SNPs in cassava. Theor. Appl. Genet. 124, 685–695. doi: 10.1007/s00122-011-1739-9
Ferguson, M. E., Shah, T., Kulakow, P., and Ceballos, H. (2019). A global overview of cassava genetic diversity. PLoS One 14:e0224763. doi: 10.1371/journal.pone.0224763
Ferguson, M., Rabbi, I., Kim, D., Gedil, M., Lopez-Lavalle, L. A. B., and Okogbenin, E. (2011). Molecular Markers and their Application to Cassava Breeding: Past, Present and Future. Trop. Plant Biol. 5, 95–109.
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The Sol Genomics Network (SGN)-from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041. doi: 10.1093/nar/gku1195
Fregene, M., Angel, F., Gomez, R., Rodriguez, F., Chavarriaga, P., Roca, W., et al. (1997). A molecular genetic map of cassava (Manihot esculenta crantz). Theor. Appl. Genet. 95, 431–441.
FSIN (2019). Global report on food crises. Available online at: https://www.wfp.org/publications/2020-global-report-food-crises (Accessed on October 23, 2020)
Fukuda, W. M. G., Guevara, C. L., Kawuki, R., and Ferguson, M. E. (2010). Selected morphological and agronomic descriptors for the characterization of cassava. Ibadan:, NigeriaIITA, 19.
Fukushima, A. R., Nicoletti, M. A., Rodrigues, A. J., Pressutti, C., Almeida, J., Brandão, T., et al. (2016). Cassava flour: Quantification of cyanide content. Food Nutr. Sci. 7, 592–599. doi: 10.4236/fns.2016.77060
Garcia-Oliveira, A. L., Kimata, B., Kasele, S., Kapinga, F., Masumba, E., Mkamilo, G., et al. (2020). Genetic analysis and QTL mapping for multiple biotic stress resistance in cassava. PLoS One 15:e0236674. doi: 10.1371/journal.pone.0236674
Gegios, A., Amthor, R., Maziya-Dixon, B., Egesi, C., Mallowa, S., Nungo, R., et al. (2010). Children consuming cassava as a staple food are at risk for inadequate zinc, iron, and vitamin A intake. Plant Foods Hum. Nutr. 65, 64–70. doi: 10.1007/s11130-010-0157155
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
ICGMC. (2015). High-resolution linkage map and chromosome-scale genome assembly for cassava (Manihot esculenta crantz) from 10 populations. G3 Genes Genomes Genet 5, 133–144. doi: 10.1534/g3.114.015008
Iglesias, C., Mayer, J., Chavez, L., and Calle, F. (1997). Genetic potential and stability of carotene content in cassava roots. Euphytica 94, 367–373. doi: 10.1023/A:1002962108315
Ikeogu, U. N., Akdemir, D., Wolfe, M. D., Okeke, U. G., Chinedozi, A., Jannink, J. L., et al. (2019). Genetic correlation, genome-wide association and genomic prediction of portable nirs predicted carotenoids in cassava roots. Front. Plant Sci. 10:1570. doi: 10.3389/fpls.2019.01570
Ikeogu, U. N., Davrieux, F., Dufour, D., Ceballos, H., Egesi, C. N., and Jannink, J. L. (2017). Rapid analyses of dry matter content and carotenoids in fresh cassava roots using a portable visible and near infrared spectrometer (Vis/NIRS). PLoS One 12:e0188918.
Iliya, J. M., and Madumelu, M. (2019). Determination of cyanide conetent in cassava tubers (Manihot esculenta) and apple seed (Pyrus malus). Int. J. Curr. Res. Chem. Pharm. Sci. 6, 14–20. doi: 10.22192/ijcr
Jaramillo, A. M., Londoño, L. F., Orozco, J. C., Patiño, G., Belalcazar, J., Davrieux, F., et al. (2018). A comparison study of five different methods to measure carotenoids in biofortified yellow cassava (Manihot esculenta). PLoS One. 13:e0209702. doi: 10.1371/journal.pone.0209702
Jonas, E., and de Koning, D. J. (2013). Does genomic selection have a future in plant breeding? Trends Biotechnol. 31, 497–504. doi: 10.1016/j.tibtech.2013.06.003
Jorge, V., Fregene, M. A., Duque, M. C., Bonierbale, M. W., Tohme, J., and Verdier, V. (2000). Genetic mapping of resistance to bacterial blight disease in cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 101, 865–872. doi: 10.1007/s001220051554
Jorge, V., Fregene, M., Vélez, C. M., Duque, M. C., Tohme, J., and Verdier, V. (2001). QTL analysis of field resistance to Xanthomonas axonopodis pv. manihotis in cassava. Theor. Appl. Genet. 102, 564–571. doi: 10.1007/s001220051683
Kainuma, K., Odat, T., and Cuzuki, S. (1967). Study of starch phosphates monoesters. J. Tech. Soc. Starch 14, 24–28
Kalyebi, A., MacFadyen, S., Parry, H., Tay, W. T., De Barro, P., and Colvin, J. (2018). African cassava whitefly, Bemisia tabaci, cassava colonization preferences and control implications. PLoS One 13:e0204862. doi: 10.1371/journal.pone.0204862
Kamanda, I., Blay, E. T., Asante, I. K., Danquah, A., Ifie, B. E., Parkes, E., et al. (2020). Genetic diversity of provitamin-A cassava (Manihot esculenta Crantz) in Sierra Leone. Genet. Resour. Crop Evol. 67, 1193–1208. doi: 10.1007/s10722-020-00905-8
Kaur, N., Alok, A. S., Kaur, N., Pandey, P., Awasthi, P., et al. (2018). CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 18, 89–99. doi: 10.1007/s10142-017-0577-5
Kawuki, R. S., Esuma, W., Ozimati, A., Kayondo, I. S., Nandudu, L., and Wolfe, M. (2019). Alternative approaches for assessing cassava brown streak root necrosis to guide resistance breeding and selection. Front. Plant Sci. 10:1461. doi: 10.3389/fpls.2019.01461
Kawuki, R. S., Ferguson, M., Labuschagne, M., Herselman, L., and Kim, D. J. (2009). Identification, characterisation and application of single nucleotide polymorphisms for diversity assessment in cassava (Manihot esculenta Crantz). Mol. Breed. 23, 669–684. doi: 10.1007/s11032-009-9264-0
Kayondo, S. I., Pino Del, Carpio, D., Lozano, R., Ozimati, A., Wolfe, M., et al. (2018). Genome-wide association mapping and genomic prediction for CBSD resistance in Manihot esculenta. Sci. Rep. 8:1549. doi: 10.1038/s41598-018-19696-1
Kengkanna, J., Jakaew, P., Amawan, S., Busener, N., Bucksch, A., and Saengwilai, P. (2019). Phenotypic variation of cassava root traits and their responses to drought. Appl. Plant Sci. 7:e1238. doi: 10.1002/aps3.1238
Khandare, V., and Choomsook, P. (2019). Cassava export of Thailand: Growth performance and composition. Int. J. Res. Anal. Rev. 6, 847–857.
Kizito, E. B., Rönnberg-Wästljung, A.-C., Egwang, T., Gullberg, U., Fregene, M., and Westerbergh, A. (2007). Quantitative trait loci controlling cyanogenic glucoside and dry matter content in cassava (Manihot esculenta Crantz) roots. Hereditas 144, 129–136. doi: 10.1111/j.2007.0018-0661.01975.x
Koros, J. C., Runo, S. M., Yusuf, M., and Orek, C. O. (2018). Screening selected cassava cultivars for resistance against cassava viruses and cassava green mites under advanced yield trials in Kenya. IOSR J. Biotechnol. Biochem. 4, 37–52. doi: 10.9790/264X-0405023752
Kretzschmar, T., Mbanjo, E. G. N., Magalit, G. A., Dwiyanti, M. S., Habib, M. A., Diaz, M. G., et al. (2018). DNA fingerprinting at farm level maps rice biodiversity across Bangladesh and reveals regional varietal preferences. Sci. Rep. 8:14920. doi: 10.1038/s41598-018-33080-z
Kuon, J. E., Qi, W., Schläpfer, P., Hirsch-Hoffmann, M., Von Bieberstein, P. R., Patrignani, A., et al. (2019). Haplotype-resolved genomes of geminivirus-resistant and geminivirus-susceptible African cassava cultivars. BMC Biol. 17:75. doi: 10.1186/s12915-019-0697-6
Lan, S., Zheng, C., Hauck, K., McCausland, M., Duguid, S. D., Booker, H. M., et al. (2020). Genomic prediction accuracy of seven breeding selection traits improved by QTL identification in flax. Int. J. Mol. Sci. 21:1577. doi: 10.3390/ijms21051577
Leach, H. W. (1959). Structure of the starch granule I swelling and solubility patterns of various starches. Cereal Chem. 36, 534–544.
Lu, G., Huang, H., and Zhang, D. (2006a). Application of near-infrared spectroscopy to predict sweetpotato starch thermal properties and noodle quality. J. Zhejiang Univ. Sci. B. 7, 475–481. doi: 10.1631/jzus.2006.B0475
Lu, G., Huang, H., and Zhang, D. (2006b). Prediction of sweetpotato starch physiochemical quality and pasting properties using near-infrared reflectance spectroscopy. Food Chem. 94, 632–639. doi: 10.1016/j.foodchem.2005.02.006
Luo, X., Tomlins, K. I., Carvalho, L. J. C. B., Li, K., and Chen, S. (2018). The analysis of candidate genes and loci involved with carotenoid metabolism in cassava (Manihot esculenta Crantz) using SLAF-seq. Acta Physiol. Plant. 40:66. doi: 10.1007/s11738-018-2634-7
Ly, D., Hamblin, M., Rabbi, I., Melaku, G., Bakare, M., Gauch, H. G., et al. (2013). Relatedness and genotype × environment interaction affect prediction accuracies in genomic selection: A study in cassava. Crop Sci. 53, 1312–1325. doi: 10.2135/cropsci2012.11.0653
Ma’Aruf, A. G., and Abdul, H. R. (2020). Efficient processing of cassava starch: Physicochemical characterization at different processing parameters. Food Res. 4, 143–151. doi: 10.26656/fr.2017.4(1).235
Maieves, H. A., Oliveira, D. C., De, Bernardo, C., Müller, C. M. D. O., and Amante, E. R. (2012). Microscopy and texture of raw and cooked cassava (Manihot esculenta Crantz) roots. J. Texture Stud. 43, 164–173. doi: 10.1111/j.1745-4603.2011.00327.x
Malik, A. I., Kongsil, P., Nguyễn, V. A., Ou, W., and Srean, P. (2020). Cassava breeding and agronomy in Asia: 50 years of history and future directions. Breed. Sci. 70, 145–166. doi: 10.1270/jsbbs.18180
Masumba, E. A., Kapinga, F., Mkamilo, G., Salum, K., Kulembeka, H., Rounsley, S., et al. (2017). QTL associated with resistance to cassava brown streak and cassava mosaic diseases in a bi-parental cross of two Tanzanian farmer varieties, Namikonga and Albert. Theor. Appl. Genet. 130, 2069–2090. doi: 10.1007/s00122-017-2943-z
Mehta, D., Stürchler, A., Anjanappa, R. B., Zaidi, S. S. E. A., Hirsch-Hoffmann, M., Gruissem, W., et al. (2019). Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 20:80. doi: 10.1186/s13059-019-1678-3
Meullenet, J. F., Mauromoustakos, A., Horner, T. B., and Marks, B. P. (2002). Prediction of texture of cooked white rice by near-infrared reflectance analysis of whole-grain milled samples. Cereal Chem. 79, 52–57. doi: 10.1094/CCHEM.2002.79.1.52
Meuwissen, T. H. E., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
Moriasi, G. A., Olela, B. O., Waiganjo, B. W., Wakori, E. W. T., and Onyancha, J. M. (2017). Evaluation of cyanide levels in two cassava varieties (Mariwa and Nyakatanegi) grown in Bar-agulu, Siaya County, Kenya. J. Food Nutr. Res. 5, 817–823. doi: 10.12691/jfnr-5-1114
Morillo, A. C. C., Morillo, Yacenia, C., and Ceballos, H. L. (2013). Identification of QTLs for carotene content in the genome of cassava (Manihot esculenta Crantz) and S1 population validation. Acta Agron. 62, 196–206.
Mtunguja, M. K., Beckles, D. M., Laswai, H. S., Ndunguru, J. C., and Sinha, N. J. (2019). Opportunities to commercialize cassava production for poverty alleviation and improved food security in Tanzania. African J. Food, Agric. Nutr. Dev. 19, 13928–13946. doi: 10.18697/AJFAND.84.BLFB1037
Naim, F., Dugdale, B., Kleidon, J., Brinin, A., Shand, K., Waterhouse, P., et al. (2018). Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 27, 451–460. doi: 10.1007/s11248-018-0083-0
Nakatumba-Nabende, J., Akera, B., Tusubira, J. F., Nsumba, S., and Mwebaze, E. (2020). A dataset of necrotized cassava root cross-section images. Data Br. 32:106170. doi: 10.1016/j.dib.2020.106170
Nassar, N., and Ortiz, R. (2010). Breeding Cassava to Feed the Poor. Sci. Am. 302, 78–85. doi: 10.1002/9781118060995.ch3
Ni, J., Pujar, A., Youens-Clark, K., Yap, I., Jaiswal, P., Tecle, I., et al. (2009). Gramene QTL database: Development, content and applications. Database 2009:ba005. doi: 10.1093/database/bap005
Noranizan, M. A., Dzulkifly, M. H., and Russly, A. R. (2010). Effect of heat treatment on the physico-chemical properties of starch from different botanical sources. Int. Food Res. J. 17, 127–135.
Ntui, V. O., Tripathi, J. N., and Tripathi, L. (2020). Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 2020:100128. doi: 10.1016/j.cpb.2019.100128
Nzuki, I., Katari, M. S., Bredeson, J. V., Masumba, E., Kapinga, F., Salum, K., et al. (2017). QTL mapping for pest and disease resistance in cassava and coincidence of some QTL with introgression regions derived from Manihot Glaziovii. Front. Plant Sci. 8:1168. doi: 10.3389/fpls.2017.01168
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., and Taylor, N. J. (2017). Efficient CRISPR/cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 8:1780. doi: 10.3389/fpls.2017.01780
Ogbonna, A. C., de Andrade, L. R., Rabbi, I. Y., Mueller, L. A., de Oliveira, E. J., and Bauchet, G. J. (2020). Genetic architecture and gene mapping of cyanide in cassava (Manihot esculenta Crantz). bioRxiv preprint doi: 10.1101/2020.06.19.159160
Ojijo, N., Franzel, S., Simtowe, F., Madakadze, R., Nkwake, A., and Moleko, L. (2016). “The role for agricultural Research Systems, Advisory services and capacity developement and knowledge transfer,” in Africa agriculture status report 2016: progress towards agricultural transformation (AGRA). (Uttar Pradesh: AGRA)200–230.
Okogbenin, E., Egesi, C. N., Olasanmi, B., Ogundapo, O., Kahya, S., Hurtado, P., et al. (2012). Molecular marker analysis and validation of resistance to cassava mosaic disease in elite cassava genotypes in Nigeria. Crop Sci. 52, 2576–2586. doi: 10.2135/cropsci2011.11.0586
Okogbenin, E., Marin, J., and Fregene, M. (2006). An SSR-based molecular genetic map of cassava. Euphytica 147, 433–440
Okogbenin, E., Marin, J., and Fregene, M. (2008). QTL analysis for early yield in a pseudo F2 population of cassava. Afr. J. Biotechnol. 7, 131–138. doi: 10.5897/AJB2008.000-5017
Otekunrin, O. A., and Sawicka, B. (2019). Cassava, a 21st century staple crop: How can Nigeria harness its enormous trade potentials? Acta Sci. Agric. 3, 194–202. doi: 10.31080/asag.2019.03.0586
Owomugisha, G., and Mwebaze, E. (2016). Machine learning for plant disease incidence and severity measurements from leaf images. in Proceedings - 2016 15th IEEE International Conference on Machine Learning and Applications. ICMLA 2016, 158–163. doi: 10.1109/ICMLA.2016.126
Ozimati, A., Kawuki, R., Esuma, W., Kayondo, I. S., Wolfe, M., Lozano, R., et al. (2018). Training population optimization for prediction of cassava brown streak disease resistance in West African Clones. G3 Genes Genomes Genet. 8, 3903–3913. doi: 10.1534/g3.118.200710
Patil, B. L., Legg, J. P., Kanju, E., and Fauquet, C. M. (2015). Cassava brown streak disease: a threat to food security in Africa. J. Gen. Virol. 96, 956–968. doi: 10.1099/jgv.0.000014
Ping, H., Wang, J., and Ren, G. (2013). Prediction of the total starch and amylose content in barley using near-infrared reflectance spectroscopy. Intell. Autom. Soft. Comput. 19, 231–237. doi: 10.1080/10798587.2013.823719
Pootakham, W., Shearman, J. R., Ruang-Areerate, P., Sonthirod, C., Sangsrakru, D., Jomchai, N., et al. (2014). Large-scale SNP discovery through RNA sequencing and SNP genotyping by targeted enrichment sequencing in cassava (Manihot esculenta Crantz). PLoS One 9:e116028. doi: 10.1371/journal.pone.0116028
Prohens, J. (2011). Plant breeding: a success story to be continued thanks to the advances in genomics. Front. Plant Sci. 2:51. doi: 10.3389/fpls.2011.00051
Rabbi, I. Y., Hamblin, M. T., Kumar, P. L., Gedil, M. A., Ikpan, A. S., Jannink, J. L., et al. (2014). High-resolution mapping of resistance to cassava mosaic geminiviruses in cassava using genotyping-by-sequencing and its implications for breeding. Virus Res. 186, 87–96. doi: 10.1016/j.virusres.2013.12.028
Rabbi, I. Y., Kayondo, S. I., Bauchet, G., Yusuf, M., Aghogho, C. I., Ogunpaimo, K., et al. (2020). Genome-wide association analysis reveals new insights into the genetic architecture of defensive, agro-morphological and quality-related traits in cassava. Plant Mol. Biol. 30:20. doi: 10.1007/s11103-020-01038-3
Rabbi, I. Y., Kulakow, P. A., Manu-Aduening, J. A., Dankyi, A. A., Asibuo, J. Y., Parkes, E. Y., et al. (2015). Tracking crop varieties using genotyping-by-sequencing markers: A case study using cassava (Manihot esculenta Crantz). BMC Genet. 16:115. doi: 10.1186/s12863-015-0273-1
Rabbi, I. Y., Kulembeka, H. P., Masumba, E., Marri, P. R., and Ferguson, M. (2012). An EST-derived SNP and SSR genetic linkage map of cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 125, 329–342. doi: 10.1007/s00122-012-18361834
Rabbi, I. Y., Udoh, L. I., Wolfe, M., Parkes, E. Y., Gedil, M. A., Dixon, A., et al. (2017). Genome-wide association mapping of correlated traits in cassava: Dry matter and total carotenoid content. Plant Genome 3:10. doi: 10.3835/plantgenome2016.09.0094
Ragot, M., Bonierbale, M., Weltzein, E., and Genetics, N. F. (2018). From market demand to breeding decisions: A framework. Lima: CGIAR Gender and Breedinfg Initiative.
Ramcharan, A., Baranowski, K., McCloskey, P., Ahmed, B., Legg, J., and Hughes, D. P. (2017). Deep learning for image-based cassava disease detection. Front. Plant Sci. 8:1852. doi: 10.3389/fpls.2017.01852
Ramcharan, A., McCloskey, P., Baranowski, K., Mbilinyi, N., Mrisho, L., Ndalahwa, M., et al. (2019). A mobile-based deep learning model for cassava disease diagnosis. Front. Plant Sci. 10:272. doi: 10.3389/fpls.2019.00272
Ramu, P., Esuma, W., Kawuki, R., Rabbi, I. Y., Egesi, C., Bredeson, J. V., et al. (2017). Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat. Genet. 49, 959–963. doi: 10.1038/ng.3845
Resende, M. D., Resende, M. F. Jr., Sansaloni, C. P., Petroli, C. D., Missiaggia, A. A., Aguiar, A. M., et al. (2012). Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol. 194, 116–128.
Rodríguez-Sandoval, E., Fernández-Quintero, A., Sandoval-Aldana, A., and Cuvelier, G. (2008). Effect of processing conditions on the texture of reconstituted cassava dough. Brazilian J. Chem. Eng. 25, 713–722. doi: 10.1590/S0104-66322008000400008
Rosales-soto, M. U., Ross, C. F., Younce, F., John, K., Mattinson, D. S., Huber, K., et al. (2016). Advances in food technology and nutritional sciences physico-chemical and sensory evaluation cassava (Manihot Esculenta Crantz) Flour. Adv. Food Technol. Nutr. Sci. Open J. 2, 9–18. doi: 10.17140/AFTnSOJ-2-126
Said, J. I., Knapka, J. A., Song, M., and Zhang, J. (2015). Cotton QTLdb: a cotton QTL database for QTL analysis, visualization, and comparison between Gossypium hirsutum and G. hirsutum × G. barbadense populations. Mol. Genet. Genom. 290, 1615–1625. doi: 10.1007/s00438-015-1021-y
Sakurai, T., Mochida, K., Yoshida, T., Akiyama, K., Ishitani, M., Seki, M., et al. (2013). Genome-wide discovery and information resource development of DNA polymorphisms in cassava. PLoS One 8:e74056. doi: 10.1371/journal.pone.0074056
Sambasivam, G., and Opiyo, G. D. (2020). A predictive machine learning application in agriculture: Cassava disease detection and classification with imbalanced dataset using convolutional neural networks. Egypt. Inform. J. 3:119. doi: 10.1016/j.eij.2020.02.007
Sánchez, T., Ceballos, H., Dufour, D., Ortiz, D., Morante, N., Calle, F., et al. (2014). Prediction of carotenoids, cyanide and dry matter contents in fresh cassava root using NIRS and Hunter color techniques. Food Chem. 151, 444–451. doi: 10.1016/j.foodchem.2013.11.081
Sandoval-Aldana, A., and Fernandez, Q. A. (2013). Physicochemical characterization of two cassava (Manihot esculenta Crantz) starches and flours. Sci. Agroaliment. 1, 19–25.
Santantonio, N., Atanda, S. A., Beyene, Y., Varshney, R. K., Olsen, M., Jones, E., et al. (2020). Strategies for effective use of genomic information in crop breeding programs serving Africa and South Asia. Front. Plant Sci. 11:353. doi: 10.3389/fpls.2020.00353
SASHA (2019). The sweetpotato breeding community of practice: Breeding in Africa for Africa (2009-2019). Available online at: https://www.sweetpotatoknowledge.org/wp-content/uploads/2019/12/01_SASHA_BREEDING_The-Sweetpotato-Breeding-Community-Practice-Breeding-Africa-2009-2019-4p_web.pdf (accessed on October 23, 2020).
Sedano, J. C. S., Moreno, R. E. M., Mathew, B., Léon, J., Cano, F. A. G., and Ballvora, A., et al. (2017). Major novel QTL for resistance to Cassava bacterial blight identified through a multi-environmental analysis. Front. Plant Sci. 8:1169. doi: 10.3389/fpls.2017.01169
Sedano, J. S., Eduardo, R., Moreno, M., Calle, F., Ernesto, C., and Carrascal, L. (2017). QTL identification for cassava bacterial blight under natural infection conditions. Acta Biol. Colomb. 22, 19–26
Singh, A. K., Krishnan, S. G., Vinod, K. K., Ellur, R. K., Bollinedi, H., Bhowmick, P. K., et al. (2019). Precision breeding with genomic tools: A decade long journey of molecular breeding in rice. Indian J. Genet. 79, 181–191. doi: 10.31742/ijgpb.79s.1.7
Somo, M., Kulembeka, H., Mtunda, K., Mrema, E., Salum, K., Wolfe, M. D., et al. (2020). Genomic prediction and quantitative trait locus discovery in a cassava training population constructed from multiple breeding stages. Crop Sci. 60, 896–913. doi: 10.1002/csc2.20003
Sonnewald, U., Fernie, A. R., Gruissem, W., Schläpfer, P., Anjanappa, R. B., Chang, S. H., et al. (2020). The Cassava Source-Sink project: opportunities and challenges for crop improvement by metabolic engineering. Plant J. 103, 1655–1665. doi: 10.1111/tpj.14865
Soto, J. C., Ortiz, J. F., Perlaza-Jiménez, L., Vásquez, A. X., Lopez-Lavalle, L. A. B., Mathew, B., et al. (2015). A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes. BMC Genom. 16:190. doi: 10.1186/s12864-015-13971394
Sraphet, S., Boonchanawiwat, A., Thanyasiriwat, T., Boonseng, O., Tabata, S., Sasamoto, S., et al. (2011). SSR and EST-SSR-based genetic linkage map of cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 122, 1161–1170. doi: 10.1007/s00122-010-1520-5
Sraphet, S., Boonchanawiwat, A., Thanyasiriwat, T., Thaikert, R., Whankaew, S., Smith, D. R., et al. (2017). Quantitative trait loci underlying root yield and starch content in an F1 derived cassava population (Manihot esculenta Crantz). J. Agric. Sci. 155, 569–581. doi: 10.1017/S0021859616000678
Stephenson, K., Amthor, R., Mallowa, S., Nungo, R., Maziya-Dixon, B., Gichuki, S., et al. (2010). Consuming cassava as a staple food places children 2-5 years old at risk for inadequate protein intake, an observational study in Kenya and Nigeria. Nutr. J. 9:9. doi: 10.1186/1475-2891-9-9
Tappiban, P., Ying, Y., Pang, Y., Sraphet, S., Srisawad, N., Smith, D. R., et al. (2020). Gelatinization, pasting and retrogradation properties and molecular fine structure of starches from seven cassava cultivars. Int. J. Biol. Macromol. 150, 831–838. doi: 10.1016/j.ijbiomac.2020.02.119
Teeken, B., Agbona, A., Abolore, B., Olaosebikan, O., Alamu, E., Adesokan, M., et al. (2020). Understanding cassava varietal preferences through pairwise ranking of gari−eba and fufu prepared by local farmer−processors. New York: Wiley.
Tester, M., and Langridge, P. (2010). Breeding technologies to increase crop production in a changing world. Science 327, 818–822. doi: 10.1126/science.1183700
Teye, E., Asare, A. P., Amoah, R. S., and Tetteh, J. P. (2011). Determination of the dry matter content of cassava (Manihot esculenta Crantz) tubers using specific gravity method. APRN J. Agric. Biol. Sci. 6, 23–28.
Thanyasiriwat, T., Sraphet, S., Whankaew, S., Boonseng, O., Bao, J., Lightfoot, D. A., et al. (2013). Quantitative trait loci and candidate genes associated with starch pasting viscosity characteristics in cassava (Manihot esculenta Crantz). Plant Biol. 16, 197–207. doi: 10.1111/plb.12022
Thirathumthavorn, D., and Trisuth, T. (2008). Gelatinization and retrogradation properties of native and hydroxypropylated crosslinked tapioca starches with added sucrose and sodium chloride. Int. J. Food Prop. 11, 858–864. doi: 10.1080/10942910701659567
Tivana, L. D., Da Cruz, Francisco, J., Zelder, F., Bergenståhl, B., and Dejmek, P. (2014). Straightforward rapid spectrophotometric quantification of total cyanogenic glycosides in fresh and processed cassava products. Food Chem. 158, 20–27. doi: 10.1016/j.foodchem.2014.02.066
Tonukari, N. J., Tonukari, N. J., Ezedom, T., Enuma, C. C., Sakpa, S. O., Avwioroko, O. J., et al. (2015). White gold: Cassava as an industrial base. Am. J. Plant Sci. 6, 972–979. doi: 10.4236/ajps.2015.67103
Torres, L. G., De Resende, M. D. V., Azevedo, C. F., Fonseca, E., Silva, F., and De Oliveira, E. J. (2019). Genomic selection for productive traits in biparental cassava breeding populations. PLoS One 14:e0220245. doi: 10.1371/journal.pone.0220245
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., and Tripathi, L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2:46. doi: 10.1038/s42003-019-0288-7
Tripathi, L., Ntui, V. O., and Tripathi, J. N. (2020). CRISPR/Cas9-based genome editing of banana for disease resistance. Curr. Opin. Plant Biol. 56, 118–126.
Turyagyenda, L. F., Kizito, E. B., Ferguson, M. E., Baguma, Y., Harvey, J. W., Gibson, P., et al. (2012). Genetic diversity among farmer-preferred cassava landraces in Uganda. Afr. Crop Sci. J. 20, 15–30.
Udoh, L. I., Gedil, M., Parkes, E. Y., Kulakow, P., Adesoye, A., Nwuba, C., et al. (2017). Candidate gene sequencing and validation of SNP markers linked to carotenoid content in cassava (Manihot esculenta Crantz). Mol. Breed 37:123. doi: 10.1007/s11032-017-0718-5
Waisundara, V. Y. (2018). Introductory Chapter: Cassava as a Staple Food. doi: 10.5772/intechopen.70324
Wang, W., Feng, B., Xiao, J., Xia, Z., Zhou, X., Li, P., et al. (2014). Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 5:5110. doi: 10.1038/ncomms6110
Whankaew, S., Poopear, S., Kanjanawattanawong, S., Tangphatsornruang, S., Boonseng, O., Lightfoot, D. A., et al. (2011). A genome scan for quantitative trait loci affecting cyanogenic potential of cassava root in an outbred population. BMC Genomics 12:266. doi: 10.1186/1471-2164-12-266
Wolfe, M. D., Del Carpio, D. P., Alabi, O., Ezenwaka, L. C., Ikeogu, U. N., Kayondo, I. S., et al. (2017). Prospects for genomic selection in cassava breeding. Plant Genome 10:3. doi: 10.3835/plantgenome2017.03.0015
Wolfe, M. D., Rabbi, I. Y., Egesi, C., Hamblin, M., Kawuki, R., Kulakow, P., et al. (2016). Genome-wide association and prediction reveals genetic architecture of cassava mosaic disease resistance and prospects for rapid genetic improvement. Plant Genome 9:2. doi: 10.3835/plantgenome2015.11.0118
Wossen, T., Girma, G., Abdoulaye, T., Rabbi, I., Olanrewaju, A., Alene, A., et al. (2017). The Cassava Monitoring Survey in Nigeria.IITA. Ibadan: Nigeria, 66.
Xia, Z., Zou, M., Zhang, S., Feng, B., and Wang, W. (2014). AFSM sequencing approach: A simple and rapid method for genome-wide SNP and methylation site discovery and genetic mapping. Sci. Rep. 4:7300. doi: 10.1038/srep07300
Xu, J., Hua, K., and Lang, Z. (2019). Genome editing for horticultural crop improvement. Hortic. Res. 6:113. doi: 10.1038/s41438-019-0196-5
Yonemaru, J. i., Yamamoto, T., Fukuoka, S., Uga, Y., Hori, K., and Yano, M. (2010). Q-TARO: QTL annotation rice online database. Rice 3, 194 203. doi: 10.1007/s12284-010-9041-z
Yonis, B. O., Pino, del Carpio, D., Wolfe, M., Jannink, J. L., Kulakow, P., et al. (2020). Improving root characterisation for genomic prediction in cassava. Sci. Rep. 10:80003. doi: 10.1038/s41598-020-64963-9
Keywords: cassava, innovations, QTL mapping, association mapping, marker-assisted selection, genomic selection, genome-editing
Citation: Mbanjo EGN, Rabbi IY, Ferguson ME, Kayondo SI, Eng NH, Tripathi L, Kulakow P and Egesi C (2021) Technological Innovations for Improving Cassava Production in Sub-Saharan Africa. Front. Genet. 11:623736. doi: 10.3389/fgene.2020.623736
Received: 30 October 2020; Accepted: 23 December 2020;
Published: 21 January 2021.
Edited by:
Deepmala Sehgal, International Maize and Wheat Improvement Center, MexicoReviewed by:
Zhi Zou, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, ChinaJose Sebastian, Indian Institute of Science Education and Research Berhampur (IISER), India
Copyright © 2021 Mbanjo, Rabbi, Ferguson, Kayondo, Eng, Tripathi, Kulakow and Egesi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiedozie Egesi, Qy5FZ2VzaUBjZ2lhci5vcmc=
 Edwige Gaby Nkouaya Mbanjo
Edwige Gaby Nkouaya Mbanjo Ismail Yusuf Rabbi
Ismail Yusuf Rabbi Morag Elizabeth Ferguson
Morag Elizabeth Ferguson Siraj Ismail Kayondo
Siraj Ismail Kayondo Ng Hwa Eng3
Ng Hwa Eng3 Leena Tripathi
Leena Tripathi Peter Kulakow
Peter Kulakow Chiedozie Egesi
Chiedozie Egesi