- Department of Integrative Oncology, BC Cancer Research Centre, Vancouver, BC, Canada
Background: Long non-coding RNAs (lncRNAs) have long been implicated in cancer-associated phenotypes. Recently, a class of lncRNAs, known as cis-acting, have been shown to regulate the expression of neighboring protein-coding genes and may represent undiscovered therapeutic action points. The chromatin architecture modification gene HMGA1 has recently been described to be aberrantly expressed in lung adenocarcinoma (LUAD). However, the mechanisms mediating the expression of HMGA1 in LUAD remain unknown. Here we investigate the deregulation of a putative cis-acting lncRNA in LUAD, and its effect on the oncogene HMGA1.
Methods: LncRNA expression was determined from RNA-sequencing data of tumor and matched non-malignant tissues from 36 LUAD patients. Transcripts with significantly deregulated expression were identified and validated in a secondary LUAD RNA-seq dataset (TCGA). SiRNA-mediated knockdown of a candidate cis-acting lncRNA was performed in BEAS-2B cells. Quantitative real-time PCR was used to observe the effects of lncRNA knockdown on the expression of HMGA1.
Results: We identified the lncRNA RP11.513I15.6, which we refer to as HMGA1-lnc, neighboring HMGA1 to be significantly downregulated in both LUAD cohorts. Conversely, we found HMGA1 significantly overexpressed in LUAD and anticorrelated with HMGA1-lnc. In vitro experiments demonstrated siRNA-mediated inhibition of HMGA1-lnc in immortalized non-malignant lung epithelial cells resulted in a significant increase in HMGA1 gene expression.
Conclusion: Our results suggest that HMGA1-lnc is a novel cis-acting lncRNA that negatively regulates HMGA1 gene expression in lung cells. Further characterization of this regulatory mechanism may advance our understanding of the maintenance of lung cancer phenotypes and uncover a novel therapeutic intervention point for tumors driven by HMGA1.
Introduction
Long non-coding RNAs (lncRNAs) are a previously-underappreciated class of transcripts with a wide variety of now-recognized functions in gene regulation. Since their functional roles have been uncovered, numerous lncRNAs have been implicated in the onset of many cancer-associated phenotypes, such as progression, tumorigenesis, and metastasis (Gibb et al., 2011; Schmitt and Chang, 2016). Further, as RNA represents the functional unit for lncRNAs, rather than an intermediate as is the case with mRNAs, these transcripts are promising targets for the development of future RNA-based therapies (Amodio et al., 2018). Despite these recognized roles in tumor development, a key challenge in the translational utility of lncRNA-based research is the effective identification of their downstream target genes. In order to harness their potential for disease-specific markers and potential therapeutic targets, a better understanding of lncRNA mechanisms of action in disease is required.
An emerging class of lncRNAs – cis-acting – has been shown to regulate the expression of neighboring protein-coding genes, frequently including protein-coding genes with oncogenic or tumor-suppressive functions. Through a variety of mechanisms primarily occurring in the nucleus, these cis-acting non-coding transcripts can activate or repress transcription of neighboring genes. For example, the lncRNA ANRIL (antisense non-coding RNA in the INK4 locus) interacts with polycomb repression complex 2 (PRC2), recruiting the complex to condense active chromatin and silence the genes around its transcriptional loci, including the well-known tumor suppressor INK4B (p15) (Yap et al., 2010).
Thus, cis-acting lncRNAs may represent novel mechanisms of cancer-gene regulation as well as potentially actionable intervention points in known cancer-driving pathways. Despite the prevalence of genetic and epigenetic deregulation events in lung adenocarcinoma (LUAD), the extent and consequences of aberrant cis-acting lncRNA expression on known cancer-driving genes is unknown.
High mobility group A1 (HMGA1) is part of a family of proteins involved in maintenance of chromatin architecture within the nucleus and as such, are implicated in tumorigenesis (Sgarra et al., 2018). Specifically, HMGA1 regulates a wide variety of genes through direct interactions with target sequences in promoter and enhancer regions. These downstream regulatory functions have been shown to lead to tumor development, particularly in breast cancer where the HMGA family has been shown to contribute, in part, to nearly every hallmark of cancer (Sgarra et al., 2018) (Hanahan and Weinberg, 2011). HMGA1 is enriched in several aggressive cancer types, including non-small cell lung cancer (NSCLC), where both mRNA and protein expression are substantially increased (Zhang et al., 2015). Further, overexpression of the HMGA1 gene has been shown to be a key factor driving lung metastasis (Lin and Peng, 2016; Fu et al., 2018). In LUAD, high HMGA1 gene expression has been associated with poor overall survival and chemotherapy resistance (Zhang et al., 2015). The oncogenic role of HMGA proteins stems from chromatin-mediated activation of cancer-driving genes such as E2F1, AP1, and CCNA1, as well as the repression of tumor suppressive genes such as TP53 (Fusco and Fedele, 2007). Interestingly, overexpression of HMGA proteins has also been shown to alter the expression of non-coding genes, such as miRNAs, leading to lung development through dysregulation of the cell cycle (Pallante et al., 2015). However, the role of lncRNAs in HMGA1 regulation has not been investigated. As such we hypothesized lncRNA expression changes may be a mechanism of HMGA1 deregulation in NSCLC. Our search for regulatory lncRNA has led to the identification of HMGA1-lnc, a cis-acting lncRNA neighboring the HMGA1 gene at 6p21.31 that acts as alternative mechanisms of HMGA1 deregulation.
Results
Cis-acting LncRNAs are deregulated in LUAD
We first sought to identify cis-acting lncRNAs with potential biological relevance in LUAD. We hypothesized that a biologically relevant cis-acting lncRNA would (i) be significantly deregulated in two LUAD datasets (Fold Change, FC: 1.5), (ii) would overlap or closely neighbor (within 1.5 Kb) protein coding genes, that (iii) were also deregulated in LUAD.
Putative cis-acting lncRNAs that fit these parameters were identified, and literature searches on PubMed were performed to identify neighboring protein coding genes with experimental evidence of roles in cancer biology (Supplementary Table 1). This gave us a list of deregulated putative cis-acting lncRNAs that may function to regulate known cancer driving genes. For example, we find a lncRNA RP11-122M14.1 neighboring protein-coding gene NEK2. NEK2 expression is associated with poor survival and inhibition results in anticancer effects in many cancer types, including lung cancer (Xia et al., 2015; Yao et al., 2019).
While many of these putative cis-acting lncRNAs are interesting, we decided to focus on cis-lncRNA-gene pairs that were deregulated in the opposite direction (discordant). We took this approach to avoid the false positives that can occur from regional “passenger” effects such as non-specific DNA copy number changes affecting multiple neighboring genes. Specifically, we decided to focus on a particular deregulated lncRNA, RP11.513I15.6, because of: (i) proximity to the known oncogene HMGA1, and (ii) the discordant expression relationship between the lncRNA and neighboring protein-coding gene. For simplicity we refer to this lncRNA as HMGA1-lnc (Figure 1).
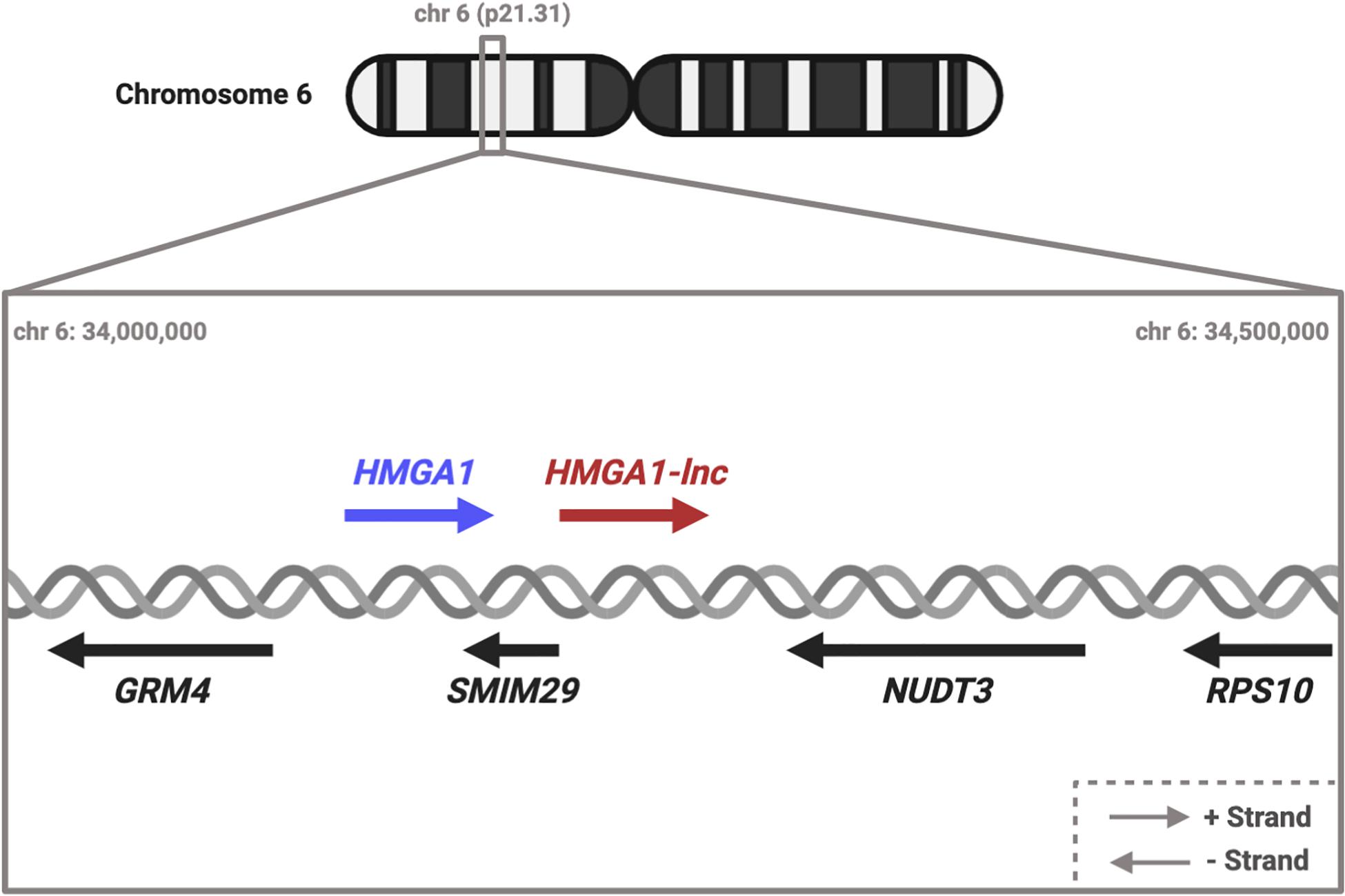
Figure 1. Diagram of HMGA1 transcriptional locus. HMGA1 and HMGA1-lnc are neighbors on the + strand. Located on the opposite strand are genes SMIM29, NUDT3, GRM4, and RPS10. #Dereg: Deregulation.
Expression of HMGA1-lnc and HMGA1 are deregulated in LUAD
We observed the lncRNA HMGA1-lnc to be significantly downregulated in tumors when compared to adjacent non-malignant tissues, which holds true in both datasets (Figure 2A and Supplementary Figure 1). In contrast, the neighboring protein-coding gene HMGA1 was found to be significantly overexpressed in both tumor datasets relative to matched non-malignant tissue (Figure 2B and Supplementary Figure 1). As mentioned above, the discordant expression in lung tumors between these genes lends further evidence to the existence of a possible regulatory relationship rather than a genomic-locus level alteration. To highlight this difference in expression, we compared the levels of HMGA1 between tertiles of tumors with the highest and lowest expression of HMGA1-lnc. Interestingly we found that levels of HMGA1 were significantly greater (p = 0.0326) in the low lncRNA expressing tumors (Figures 2C,D), and that HMGA1 and HMGA1-lnc were negatively correlated (p = 0.0153). To determine if HMGA1-lnc was affecting other genes in its transcriptional locus we performed correlation analysis between HMGA1-lnc expression and other neighboring genes. Interestingly we found SMIM29, NUDT3, and RPS10 to be significantly positively correlated with HMGA1-lnc expression in the BCCA dataset, however, we did not observe significant correlations with these genes in the TCGA dataset (Supplementary Table 2). We also tested whether genes known to regulated by HMGA1 were affected by HMGA1-lnc expression. Interestingly we found two genes known to be downregulated by HMGA1 (CAV1 and FOXP1) to be positively correlated with HMGA1-lnc expression (Supplementary Figure 2). As the expression of HMGA1 and HMGA1-lnc appears to be anti-correlated, this lncRNA may be involved in the inhibition of HMGA1 expression in normal lung contexts.
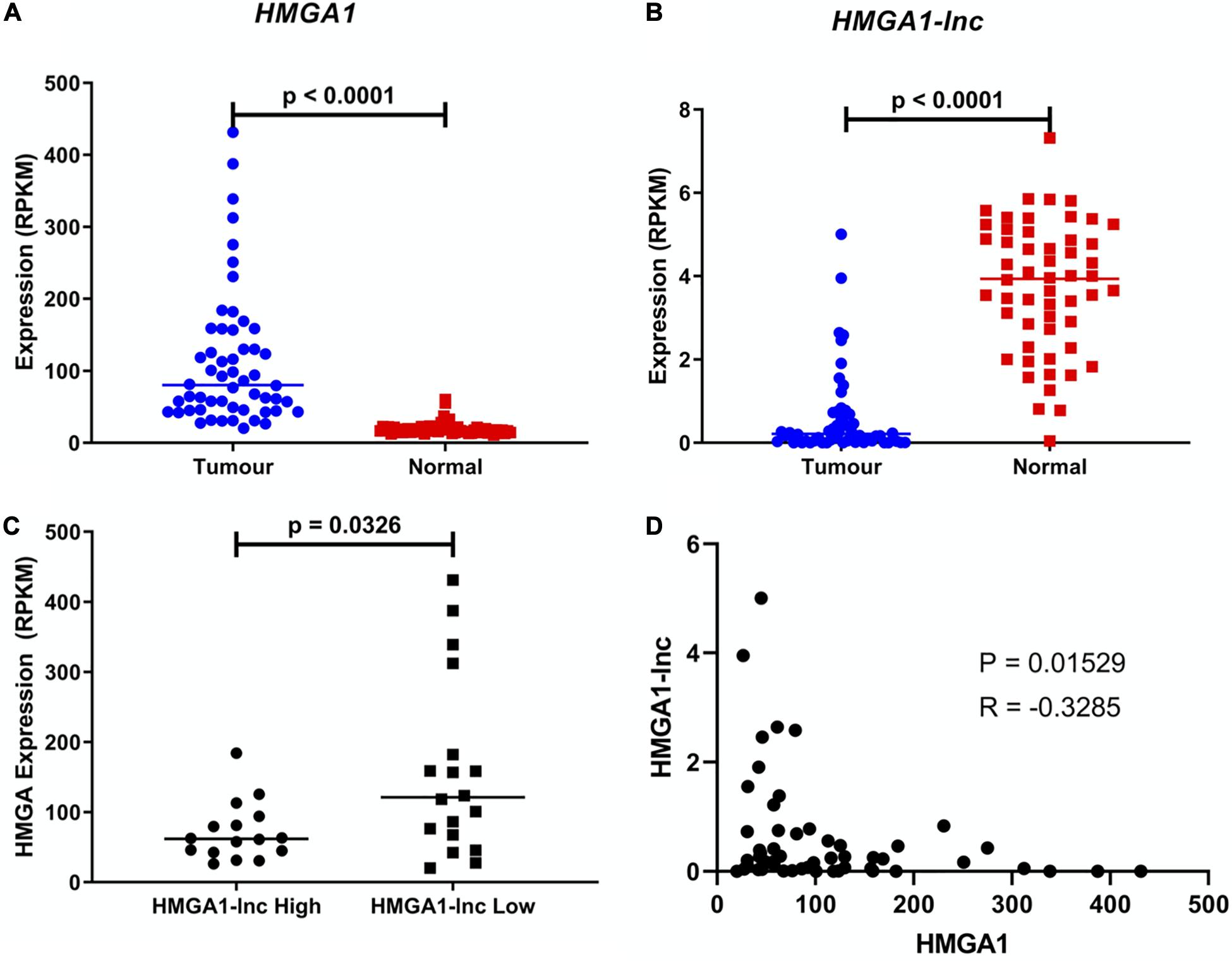
Figure 2. Expression of HMGA1 and HMGA1-lnc in LUAD. Expression of HMGA1 is upregulated in LUAD compared to adjacent non-malignant tissue (Students T-test) (A) while conversely, expression of HMGA1-lnc is downregulated in tumors (Students T-test) (B). Additionally, tumors with high levels of HMGA1-lnc, have significantly lower levels of HMGA1, when compared to tumors with low levels of the lncRNA (Mann-Whitney U-test) (C), and expression of HMGA1 and HMGA1-lnc are negatively correlated (Spearman’s Correlation) (D). All data shown above is from the TCGA cohort (n = 108, 54 paired samples).
As expression of HMGA1 has been previously described to increase with tumor stage and cancer aggressiveness, we examined whether expression of HMGA1-lnc was inversely associated with stage (Zhang et al., 2015). As the majority of our tumor samples were Stage | and || we performed a Mann-Whitney U-test between these two groups in our larger dataset (TCGA) to identify significant associations (Figures 3A,B). Interestingly, while HMGA1 was associated with increased tumor stage (p = 0.0011), the opposite was true for HMGA1-lnc, where expression of the lncRNA is significantly decreased in more advanced tumors (p = 0.0125). Further, as the TCGA dataset has paired DNA methylation data in the same tumors, we were able to investigate whether expression of HMGA1-lnc was associated with changes in DNA methylation to the HMGA1 locus. When we compared tumors with high and low expression of HMGA1-lnc (tertiles), we found that tumors with high HMGA1-lnc had significantly higher methylation of HMGA1 (Figure 3C). We also find that HMGA1 expression is significantly higher in tumors with low levels of HMGA1 methylation (Figure 3D). This may indicate that HMGA1-lnc plays a role in regulating the DNA methylation state of HMGA1.
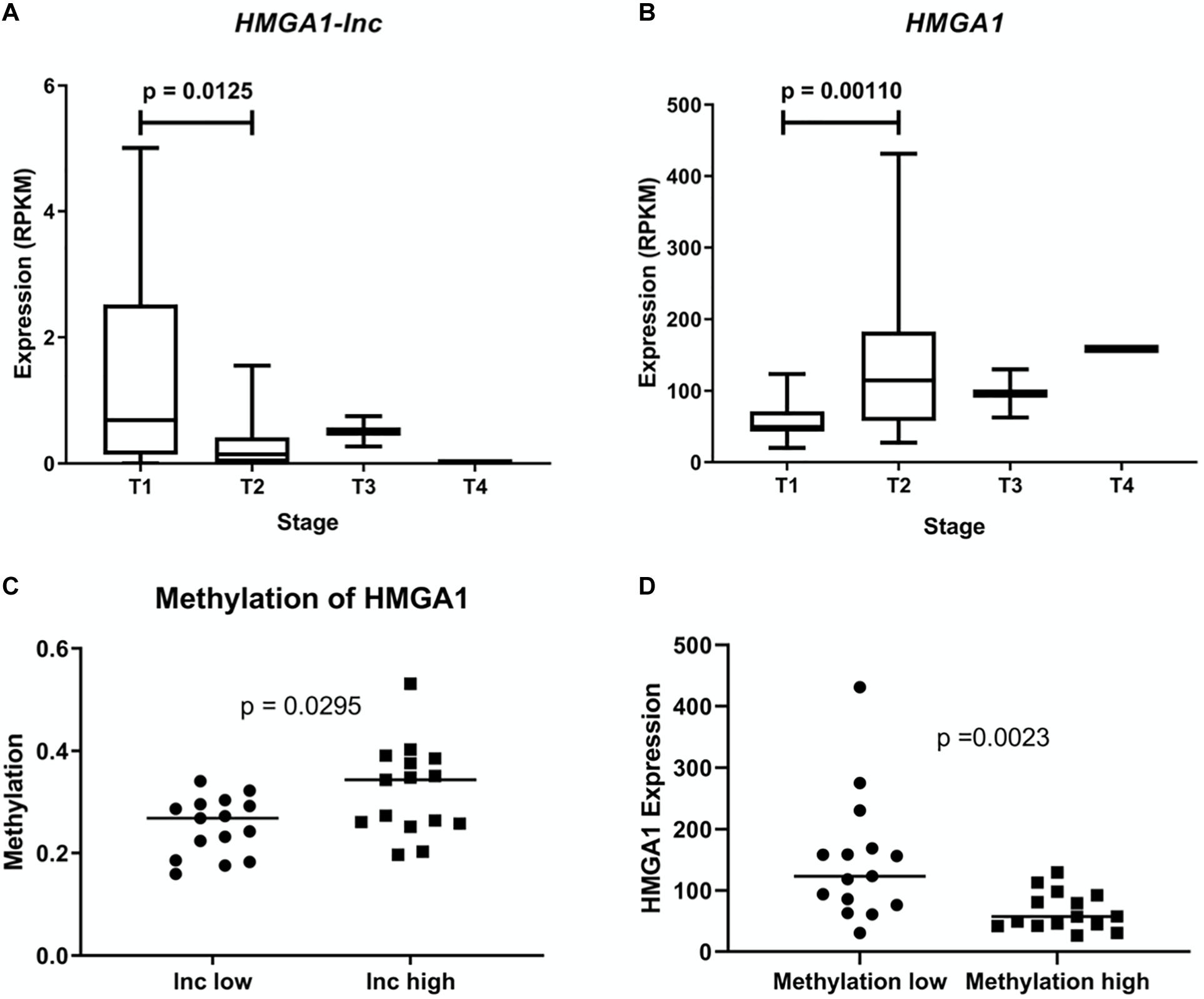
Figure 3. Expression of HMGA1 and HMGA1-lnc is associated with tumor stage. Expression of HMGA1-lnc decreases with increasing tumor stage (A), whereas HMGA1 expression increases with higher grade tumors (B). Tumors with high levels of HMGA1-lnc expression (Tertiles) had significantly higher levels of HMGA1 methylation than tumors with low levels of HMGA1-lnc (C). Expression of HMGA1 is higher in tumors with low levels of HMGA1 DNA methylation compared to tumors with high levels of DNA methylation (tertiles n = 30) (D).
HMGA1-lnc affects HMGA1 expression
To determine whether loss of HMGA1-lnc expression is a mechanism of HMGA1 overexpression in the lung, we performed a siRNA-mediated knockdown of HMGA1-lnc in the BEAS-2Bs using a pool of siRNAs specific to HMGA1-lnc. As HMGA1-lnc is downregulated in tumors, we used a non-malignant lung cell line (BEAS-2Bs) to demonstrate the effect of downregulation of HMGA1-lnc on HMGA1. We then quantified expression changes using qRT-PCR as described in Methods. From this, we observed a 3.42-fold reduction of the lncRNA after 48 h. Strikingly, in the cells with reduced HMGA1-lnc expression, the mRNA expression levels of HMGA1 were increased by 1.57-fold compared with cells transfected with non-targeting control siRNAs (Figure 4). This observed increase in HMGA1 levels in the lncRNA-inhibited cell lines, suggests that HMGA1-lnc may act to negatively regulate the expression of HMGA1, and that downregulation of this lncRNA in LUAD may be a mechanism for whereby this well-known cancer-driving gene becomes overexpressed in certain LUAD tumors.
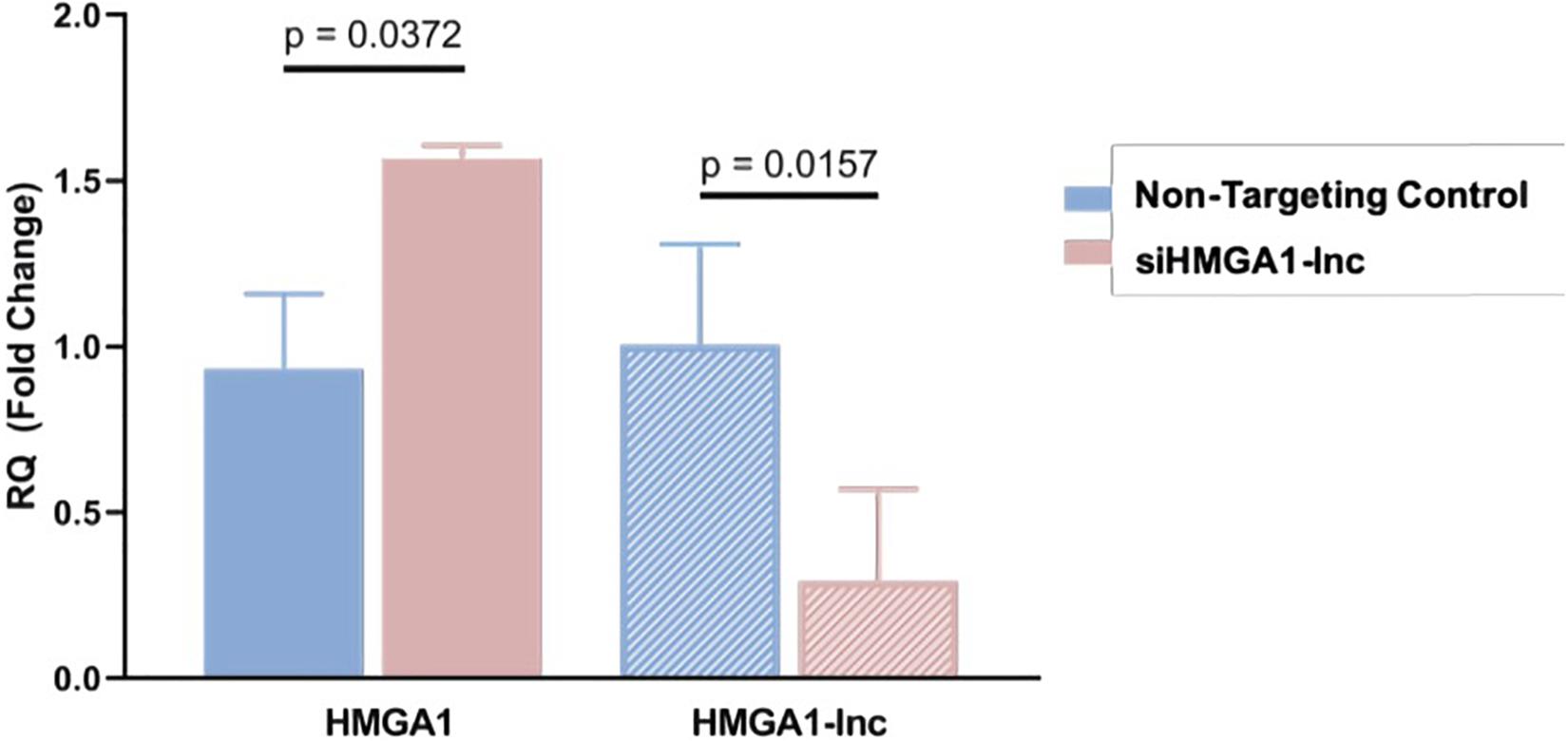
Figure 4. Inhibition of HMGA1-lnc results in increases of HMGA1 expression. SiRNA mediated inhibition of HMGA1-lnc was performed in normal bronchial epithelial cells and resulted in significant reduction of the lncRNA HMGA1-lnc. Conversely, cells where the lncRNA was inhibited showed significant increases in protein-coding HMGA1 expression.
Discussion
The role of protein-coding genes in the onset and progression of LUAD is well-established; however, there remains a lack of treatment options for patients who do not harbor one of the few clinically-actionable driver-gene alterations. LncRNAs have been shown to have important roles in the regulation of cancer-associated genes, but complex folding patterns and unknown binding motifs make lncRNAs particularly challenging to functionally characterize. Here, we used an approach that considered the genomic location, as well as the known function of neighboring oncogenic protein-coding genes, to find and characterize a novel cis-acting lncRNA deregulated in cancer, which we refer to as HMGA1-lnc.
Further, we found that this approach could be applied to other lncRNAs, finding several deregulated lncRNAs neighboring cancer associated protein-coding genes. This included well-known lung-cancer-associated genes such as NEK2, suggesting that there may be a selective pressure for the deregulation of these lncRNAs in order to release these cancer-promoting genes from negative regulation. Thus, alterations in lncRNA expression may consequently disrupt coding-gene expression as a means of promoting tumor development. This may be a useful methodology for researchers to use to identify other cis-acting lncRNAs that may be regulating genes of interest. However, it is worth noting that using a genomics-based approach to identify these cis-gene relationships is not without potential pitfalls. Previous studies have shown that many cis-acting lncRNAs have positive expression relationships with their neighbors in several tissue types (Balbin et al., 2015). However, genes in the same vicinity are often subject to regulation that can affect whole genomic regions, such as silencing through chromatin condensation. In particular, tumors often have significantly elevated levels of these broad genomic alterations to the DNA, which enables tumor suppressor gene silencing or oncogene activation. Genes neighboring these oncogenes and tumor suppressors are often caught in these regions of alteration, and display concordant expression with these genes, a phenomenon known as the passenger effect (Sheltzer and Amon, 2011; Pon and Marra, 2015). For example, frequent DNA amplification of the MET oncogene occurs in 5–20% of LUAD, leading the surrounding genes to display significantly increased DNA copy number (Zack et al., 2013; Presutti et al., 2015). While it is difficult to separate cis-acting concordant regulatory relationships from oncogene passengers without further verifying direct interactions via in vitro expression modulation, genes displaying discordant expression relationships with their neighbors are less susceptible to this effect.
We sought to avoid the passenger effect pitfall of concordant regulatory relationships by focusing the majority of our analysis on discordant relationships, particularly that between HMGA1-lnc and HMGA1. We found HMGA1-lnc to be significantly downregulated in LUAD, where its expression level is decreased 15-fold in tumors, compared to HMGA1 which has expression levels 5-fold greater in tumors (of the TCGA cohort). These observations in tandem with anti-correlated expression relationships within tumor samples led to our hypothesis that HMGA1-lnc acts to repress HMGA1 expression in non-malignant samples. HMGA1 is known to interact with large transcriptional networks in order to drive cancers (Sumter et al., 2016). Interestingly we found that FOXP1 and CAV1, genes that are commonly repressed by HMGA1 in cancer, were positively associated with HMGA1-lnc expression (Treff et al., 2004; Schuldenfrei et al., 2011). This may indicate that HMGA1-lnc expression is able to have the opposite effect on these cancer-associated transcriptional networks (Supplementary Figure 2). Consequently, the finding that HMGA1-lnc was downregulated with increasing stage, while HMGA1 expression increased with more advanced stages strengthened this putative regulatory relationship.
Methylation data for the TCGA samples allowed us to query whether expression of HMGA1-lnc affected the DNA methylation of HMGA1. Indeed, we found HMGA-lnc expression to be significantly correlated with DNA methylation of HMGA1. Further, other well-known cis-acting lncRNAs are known to function through regulation of the methylation state of their neighboring genes. For example, the lncRNA TARID which recruits GADD45A to actively demethylate the tumor suppressor TCF21 (Arab et al., 2014). This hints at one possible mechanism of HMGA1-lnc action, where expression may result in active methylation of HMGA1. We also tested the expression relationship between HMGA1-lnc and neighboring genes SMIM29, NUDT3, GRM4, and RPS10 to see if they were potentially affected by expression of this lncRNA. While we saw positive correlations for three of these genes in the BCCA dataset, these gene relationships were not consistent with the TCGA dataset (Supplementary Table 2). Further studies will be required to fully elucidate the effect of HMGA1-lnc on its neighboring genes, and the potential mechanism of its action.
To verify that HMGA1-lnc was able to affect HMGA1 expression levels, we performed siRNA-mediated knockdown of the lncRNA in cells derived from normal lung epithelium (BEAS-2B cells). We chose to use an inhibitory model to best recapitulate the phenotype we seen in our RNA sequencing datasets where HMGA1-lnc is downregulated in tumor samples. Additionally, some common methods of gene modulation such as exogenous over-expression may not work well to activate cis-acting lncRNAs. For example, the function of some cis-acting lncRNAs is tied to the act of transcription, rather than the produced transcript itself (Margaritis et al., 2012; Pelechano and Steinmetz, 2013). When the lncRNA was inhibited we noted significant increases in HMGA1 mRNA, confirming that this lncRNA directly regulates the expression of HMGA1 in vitro. Previous studies modulating expression levels of HMGA1 in these same normal lung epithelial cells (BEAS-2B) have shown that increased HMGA1 expression leads to transformed phenotypes and increases in anchorage-independent cell growth (Hillion et al., 2009). These results suggest that downregulation of this previously-uncharacterized lncRNA may lead to HMGA1 upregulation, potentially driving the onset of these same cancer phenotypes in normal human lung epithelial cells.
Our study shows that a novel cis-acting lncRNA, HMGA1-lnc, is deregulated in LUAD, which represents an alternative mechanism of activation of the oncogene HMGA1. The methodology used in this work was able to identify candidate cis-acting lncRNAs that may regulate cancer driving genes. This approach may be useful in the study of other cancer types, where with different genetic backgrounds, other cis-acting lncRNAs may be regulating other known oncogenes or tumor suppressors specific to other cancer types. Interestingly, this lncRNA has been described previously in hepatocellular carcinoma (HCC), where its expression was used in an RNA-based biomarker panel used to differentiate HCC patients from patients with chronic hepatitis C virus, and healthy controls (Abd El Gwad et al., 2018). As HCC is a malignancy known to be driven by HMGA1 it would be interesting to determine if HMGA1-lnc was similarly downregulated in these tumors in order to deregulate expression of HMGA1. Further, as HMGA1 is a known oncogene in other forms of malignancy, particularly breast cancer, it would be useful to determine if this lncRNA based mechanism is common across cancer types. If so, this interaction could represent a novel clinical intervention point in HMGA1-driven cancers.
Materials and Methods
Sample collection and processing
Collection and sequencing of both cohorts were performed in congruent manners as described in a previous publication (Stewart et al., 2019). Two separate cohorts of raw RNA sequencing reads from LUAD tumors with matched adjacent non-malignant tissues were used in this study: an in-house microdissected cohort collected at the BC Cancer Research Centre (BCCA, n = 72, or 36 pairs) (Supplementary Table 3), and a secondary set of LUAD tumors and matched non-malignant tissue (TCGA, n = 108, or 54 pairs) were downloaded from The Cancer Genome Atlas (TCGA) Data Portal1. The BCCA cohort was composed of fresh-frozen LUAD tumors and matched non-malignant lung parenchymal tissue collected from 36 patients at the Vancouver General Hospital. Consent obtained from the tissue donors of this study was both informed and written, and sample collection was approved from the University of British Columbia-BCCA Research Ethics Board. Matched non-malignant samples were collected from areas >2 cm away from the tumor. In order to reduce contaminating sequences derived from alternative cell types, tissue microdissection was guided by a pathologist. Samples used in this study contained >80% tumor cell or >80% non-malignant cell content. Total RNA was extracted using Trizol reagent and standard procedures.
RNA sequencing and processing
Libraries were constructed at Canada’s Michael Smith Genome Sciences Center using total RNA extracted from tumors and matched normal tissue (GSC, Vancouver, Canada). RNA quality was assessed using the Agilent Bioanalyzer RNA nanochip, and arrayed into a 96 well plate. RNA containing poly A sequences were then purified using the MultiMACS mRNA isolation kit 2 μg total RNA with on-column DNaseI-treatment (Miltenyi Biotec, Germany). The Superscript Double-Stranded cDNA Synthesis kit was used to synthesize DS cDNA (Life Technologies, United States). Library construction was done in a paired end, strand-specific manner following the GSC library preparation protocol, and the Illumina HiSeq 2,000 platform was used for RNA sequencing. Raw sequencing reads were subject to quality control based on length (<50 nt, under two thirds of maximum read length of 75 nt) and quality level (Phred < 20 were discarded). STAR aligned (version 2.4.1d) was then used to align reads (.fastq) to the human genome (NCBI GRCh37) (Dobin et al., 2013). Quantification of aligned reads (.bam) was performed using the Ensemble Transcripts reference (Release 75) (Flicek et al., 2014). As described in a previous manuscript, raw sequencing reads for the BCCA dataset were deposited at the Bioproject http://www.ncbi.nlm.nih.gov/bioproject/516232 (Stewart et al., 2019). RNA sequencing data was downloaded for TCGA dataset from The Cancer Genome Atlas (TCGA) Data Portal1. Sequencing data (.bam files) was then processed as described above. Quantification of both datasets was performed using the Partek Flow platform as reads per kilobase million (RPKM).
Gene expression analyses
To identify genes deregulated in LUAD, we performed a Wilcoxon signed-rank test on both lncRNA and protein-coding gene expression between tumor and matched non-malignant tissues. lncRNAs significantly deregulated in the same direction in both cohorts [Benjamini Hochberg-corrected p-value < 0.05; Fold Change (FC) > 1.5] were considered for further analyses (Supplementary Table 4). To assess potential cancer-relevant cis-acting lncRNAs, we identified deregulated lncRNAs neighboring protein-coding genes that were also deregulated. We then queried the literature (using PubMed) for experimental evidence of deregulated protein-coding genes with tumor biology. LncRNAs close enough to enact transcriptional or epigenetic changes (within 1.5 Kb) to protein-coding genes were considered as putative cis-acting lncRNAs (Supplementary Table 1; Gil and Ulitsky, 2020). To reduce the effect of passenger effects (such as DNA copy number alterations) on lncRNA:protein-coding-gene relationships, we focused on gene pairs with discordant (negatively correlated) expression patterns. Significant associations between HMGA1-lnc and neighboring protein-coding HMGA1 expression were determined using a Spearman’s correlation. These results were confirmed and visualized using a Mann-Whitney U-Test between the upper and lower tertiles of samples based on lncRNA expression. To determine if HMGA1-lnc had protein coding potential we used the coding potential assessment tool (CPAT2) which identified the gene as non-coding (Supplementary Figure 3; Wang et al., 2013).
Methylation analysis of HMGA1
DNA methylation data (HM450 beta-values) of HMGA1 for the TCGA LUAD dataset was downloaded from the cBioPortal for Cancer Genomics (www.cbioportal.org). Samples were then ranked by expression of HMGA1-lnc and separated into high and low tertiles. The methylation of HMGA1 was then compared between the high and low lncRNA expressing tertiles (Mann-Whitney U-Test). To determine if DNA methylation of HMGA1 had an effect on HMGA1 expression we compared tumors with high levels of methylation to those with low levels of methylation (tertiles). We found that tumors with low level of HMGA1 DNA methylation had significantly higher levels of HMGA1 expression (Mann-Whitney U-Test).
In vitro analyses
The immortalized non-malignant epithelial lung cell line BEAS-2Bs was used to assess the effect of inhibition of the candidate lncRNA identified in the above analysis (referred to as: HMGA1-lnc) on HMGA1 expression in vitro. Cells were cultured in serum-free medium: K-SFM supplemented with 30 μg/mL bovine pituitary extract (BPE) and 0.0002 ng/μL epidermal growth factor (EGF); maintained in an incubator at 37oC and 5% CO2. Once confluent, 2 mL of cell solution was seeded into each well of a 6 × 2 cm-well plate at a concentration of 50,000 cells/mL. DharmaFECT siRNAs were prepared for transfection as per manufacturer’s instructions in five conditions: (i) untreated control; (ii) a positive control siRNA targeting GAPDH (25 nM); (iii) a non-targeting control siRNA (25 nM); (iv) siRNA targeting HMGA1-lnc at a concentration of 12.5 nM; and (v) siRNA targeting HMGA1-lnc at a concentration of 25 nM (Supplementary Tables 4, 5). Non-targeting control was designed to target no known human genes and provide a baseline response for cellular exposure to siRNAs (Dharmacon, D-001210-01-D001210-05). RNA was harvested after both 48 and 72 h using the Quick-RNATM MiniPrep Kit (Zymo Research, Catalog number R1055). Total RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Catalogue number 4374967). Gene expression was assessed using real-time quantitative PCR with custom primers specific to HMGA1-lnc generated by Thermo Fisher, as well as established primers for the 18S ribosomal RNA (endogenous control), GAPDH (positive siRNA control), and HMGA1. RT-qPCR reactions were performed in triplicate as per Thermo Fisher recommended settings (denature 95°C for 15 s, anneal 60°C for 60 s, 40 PCR cycles). Relative expression was determined using the 2–ΔΔCt method.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by University of British Columbia-BCCA Research Ethics Board. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
GS and AS were responsible for manuscript conceptualization, study design, and performance of in silico and in vitro analyses. KE, EM, and DC contributed to data collection, interpretation, and manuscript preparation. WL was the principle investigator and was involved in manuscript conceptualization, study design, and manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Canadian Institutes for Health Research (CIHR FDN-143345). GS, AS, KE, EM, and DC are supported by Frederick Banting and Charles Best Canada Graduate Scholarships, and EAM is a Vanier Canada Graduate Scholar.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the support of Victor Martinez, Roland Hubaux, and Dave Rowbotham for helpful discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.615378/full#supplementary-material
Footnotes
References
Abd El Gwad, A., Matboli, M., El-Tawdi, A., Habib, E. K., Shehata, H., Ibrahim, D., et al. (2018). Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J. Cell. Biochem. 119, 8600–8610. doi: 10.1002/jcb.27109
Amodio, N., Stamato, M. A., Juli, G., Morelli, E., Fulciniti, M., Manzoni, M., et al. (2018). Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 32, 1948–1957. doi: 10.1038/s41375-018-0067-3
Arab, K., Park, Y. J., Lindroth, A. M., Schäfer, A., Oakes, C., Weichenhan, D., et al. (2014). Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 55, 604–614. doi: 10.1016/j.molcel.2014.06.031
Balbin, O. A., Malik, R., Dhanasekaran, S. M., Prensner, J. R., Cao, X., Wu, Y. M., et al. (2015). The landscape of antisense gene expression in human cancers. Genome Res. 25, 1068–1079. doi: 10.1101/gr.180596.114
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Flicek, P., Amode, M. R., Barrell, D., Beal, K., Billis, K., Brent, S., et al. (2014). Ensembl 2014. Nucleic Acids Res. 42, D749–D755. doi: 10.1093/nar/gkt1196
Fu, F., Wang, T., Wu, Z., Feng, Y., Wang, W., Zhou, S., et al. (2018). HMGA1 exacerbates tumor growth through regulating the cell cycle and accelerates migration/invasion via targeting miR-221/222 in cervical cancer. Cell Death Dis. 9:594. doi: 10.1038/s41419-018-0683-x
Fusco, A., and Fedele, M. (2007). Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7, 899–910. doi: 10.1038/nrc2271
Gibb, E. A., Vucic, E. A., Enfield, K. S., Stewart, G. L., Lonergan, K. M., Kennett, J. Y., et al. (2011). Human cancer long non-coding RNA transcriptomes. PLoS One 6:e25915. doi: 10.1371/journal.pone.0025915
Gil, N., and Ulitsky, I. (2020). Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117. doi: 10.1038/s41576-019-0184-5
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Hillion, J., Wood, L. J., Mukherjee, M., Bhattacharya, R., Di Cello, F., Kowalski, J., et al. (2009). Upregulation of MMP-2 by HMGA1 promotes transformation in undifferentiated, large-cell lung cancer. Mol. Cancer Res. 7, 1803–1812. doi: 10.1158/1541-7786.MCR-08-0336
Lin, S. Y., and Peng, F. (2016). Association of SIRT1 and HMGA1 expression in non-small cell lung cancer. Oncol. Lett. 11, 782–788. doi: 10.3892/ol.2015.3914
Margaritis, T., Oreal, V., Brabers, N., Maestroni, L., Vitaliano-Prunier, A., Benschop, J. J., et al. (2012). Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 8:e1002952. doi: 10.1371/journal.pgen.1002952
Pallante, P., Sepe, R., Puca, F., and Fusco, A. (2015). High mobility group a proteins as tumor markers. Front. Med. 2:15. doi: 10.3389/fmed.2015.00015
Pelechano, V., and Steinmetz, L. M. (2013). Gene regulation by antisense transcription. Nat. Rev. Genet. 14, 880–893. doi: 10.1038/nrg3594
Pon, J. R., and Marra, M. A. (2015). Driver and passenger mutations in cancer. Annu Rev. Pathol. 10, 25–50. doi: 10.1146/annurev-pathol-012414-040312
Presutti, D., Santini, S., Cardinali, B., Papoff, G., Lalli, C., Samperna, S., et al. (2015). MET Gene Amplification and MET Receptor Activation Are Not Sufficient to Predict Efficacy of Combined MET and EGFR Inhibitors in EGFR TKI-Resistant NSCLC Cells. PLoS One 10:e0143333. doi: 10.1371/journal.pone.0143333
Schmitt, A. M., and Chang, H. Y. (2016). Long Noncoding RNAs in Cancer Pathways. Cancer Cell 29, 452–463. doi: 10.1016/j.ccell.2016.03.010
Schuldenfrei, A., Belton, A., Kowalski, J., Talbot, C. C. Jr., Di Cello, F., Poh, W., et al. (2011). HMGA1 drives stem cell, inflammatory pathway, and cell cycle progression genes during lymphoid tumorigenesis. BMC Genom. 12:549. doi: 10.1186/1471-2164-12-549
Sgarra, R., Pegoraro, S., Ros, G., Penzo, C., Chiefari, E., Foti, D., et al. (2018). High Mobility Group A (HMGA) proteins: Molecular instigators of breast cancer onset and progression. Biochim. Biophys. Acta Rev. Cancer 1869, 216–229. doi: 10.1016/j.bbcan.2018.03.001
Sheltzer, J. M., and Amon, A. (2011). The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 27, 446–453. doi: 10.1016/j.tig.2011.07.003
Stewart, G. L., Enfield, K. S. S., Sage, A. P., Martinez, V. D., Minatel, B. C., Pewarchuk, M. E., et al. (2019). Aberrant Expression of Pseudogene-Derived lncRNAs as an Alternative Mechanism of Cancer Gene Regulation in Lung Adenocarcinoma. Front. Genet. 10:138. doi: 10.3389/fgene.2019.00138
Sumter, T. F., Xian, L., Huso, T., Koo, M., Chang, Y. T., Almasri, T. N., et al. (2016). The High Mobility Group A1 (HMGA1) Transcriptome in Cancer and Development. Curr. Molecular Med. 16, 353–393. doi: 10.2174/1566524016666160316152147
Treff, N. R., Pouchnik, D., Dement, G. A., Britt, R. L., and Reeves, R. (2004). High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human breast cancer cells. Oncogene 23, 777–785. doi: 10.1038/sj.onc.1207167
Wang, L., Park, H. J., Dasari, S., Wang, S., Kocher, J. P., and Li, W. (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41:e74. doi: 10.1093/nar/gkt006
Xia, J., Franqui Machin, R., Gu, Z., and Zhan, F. (2015). Role of NEK2A in human cancer and its therapeutic potentials. Biomed. Res. Int. 2015:862461. doi: 10.1155/2015/862461
Yao, Y., Su, J., Zhao, L., Luo, N., Long, L., and Zhu, X. (2019). NIMA-related kinase 2 overexpression is associated with poor survival in cancer patients: a systematic review and meta-analysis. Cancer Manag. Res. 11, 455–465. doi: 10.2147/CMAR.S188347
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674. doi: 10.1016/j.molcel.2010.03.021
Zack, T. I., Schumacher, S. E., Carter, S. L., Cherniack, A. D., Saksena, G., Tabak, B., et al. (2013). Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140. doi: 10.1038/ng.2760
Keywords: long non-coding RNA, HMGA1, gene regulation, cis-acting, non-coding RNA, lung cancer LncRNA-Mediated Control of HMGA1 Expression
Citation: Stewart GL, Sage AP, Enfield KSS, Marshall EA, Cohn DE and Lam WL (2021) Deregulation of a Cis-Acting lncRNA in Non-small Cell Lung Cancer May Control HMGA1 Expression. Front. Genet. 11:615378. doi: 10.3389/fgene.2020.615378
Received: 09 October 2020; Accepted: 11 December 2020;
Published: 11 January 2021.
Edited by:
Liang Chen, Wuhan University, ChinaReviewed by:
Matthias S. Leisegang, Goethe University Frankfurt, GermanyRui Xiao, Medical Research Institute, Wuhan University, China
Copyright © 2021 Stewart, Sage, Enfield, Marshall, Cohn and Lam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Greg L. Stewart, Z3N0ZXdhcnRAYmNjcmMuY2E=
†These authors have contributed equally to this work
‡Present address: Katey S. S. Enfield, The Francis Crick Institute, London, United Kingdom
 Greg L. Stewart
Greg L. Stewart Adam P. Sage
Adam P. Sage Katey S. S. Enfield‡
Katey S. S. Enfield‡ Erin A. Marshall
Erin A. Marshall David E. Cohn
David E. Cohn Wan L. Lam
Wan L. Lam