- 1Institute of Military Cognitive and Brain Sciences, Academy of Military Medical Sciences, Beijing, China
- 2School of Basic Medicine, Henan University, Kaifeng, China
- 3School of Pharmacy, Jiamusi University, Jiamusi, China
Disturbed sleep is closely associated with an increased risk of metabolic diseases. However, the underlying mechanisms of circadian clock genes linking sleep and lipid profile abnormalities have not been fully elucidated. This study aimed to explore the important role of the circadian clock in regulating impaired cholesterol metabolism at an early stage of sleep deprivation (SD). Sleep disturbance was conducted using an SD instrument. Our results showed that SD increased the serum cholesterol levels. Concentrations of serum leptin and resistin were much lower after SD, but other metabolic hormone concentrations (adiponectin, glucagon, insulin, thyroxine, norepinephrine, and epinephrine) were unchanged before and after SD. Warning signs of cardiovascular diseases [decreased high density lipoprotein (HDL)-cholesterol and increased corticosterone and 8-hydroxyguanosine levels] and hepatic cholestasis (elevated total bile acids and bilirubin levels) were observed after SD. Cholesterol accumulation was also observed in the liver after SD. The expression levels of HMGCR, the critical enzyme for cholesterol synthesis, remained unchanged in the liver. However, the expression levels of liver CYP7A1, the enzyme responsible for the conversion of cholesterol into bile acids, significantly reduced after SD. Furthermore, expression of NR1D1, a circadian oscillator and transcriptional regulator of CYP7A1, strikingly decreased after SD. Moreover, NR1D1 deficiency decreased liver CYP7A1 levels, and SD could exacerbate the reduction of CYP7A1 expression in NR1D1−/− mouse livers. Additionally, NR1D1 deficiency could further increase serum cholesterol levels under SD. These results suggest that sleep disturbance can induce increased serum cholesterol levels and liver cholesterol accumulation by NR1D1 mediated CYP7A1 inhibition.
Introduction
Maintenance of sleep homoeostasis regulated by the homoeostatic and circadian process plays important roles in the balance of psychological and physical health (Saper et al., 2005; Kon et al., 2017). However, short sleep duration or disturbed sleep due to shift work, jet lag, and insomnia is becoming a widely social and medical problem in modern society (Kecklund and Axelsson, 2016; Bertisch et al., 2018). Numerous epidemiological studies have suggested that adverse effects of sleep loss are closely associated with increased risk of obesity, type 2 diabetes, and cardiovascular diseases (CVDs; Reutrakul and Van Cauter, 2018; Liu and Chen, 2019). The association of metabolic disorders with sleep loss caused by total sleep deprivation (SD), chronic sleep restriction, or sleep fragmentation has gained attention (Killick et al., 2012; Nedeltcheva and Scheer, 2014). It has been suggested that the development of metabolic disorders or CVDs may be associated with long term or immediate effects of sleep loss or disruption. Thus, it is important to explore the potential mechanism underlying this association.
The circadian clock is regarded as the endogenous timekeeper of many day to day biological and physiological processes, including the sleep-wake cycle and cyclic metabolic changes (Panda, 2016; Kon et al., 2017). The circadian clock is located in the suprachiasmatic nucleus and peripheral tissues, and it synchronizes physiology and behavior throughout the 24-h day-night cycle (Takahashi, 2017). It has been suggested by some experimental and epidemiological reports that misalignment or disruption of the circadian clock can increase the risk of metabolic diseases. For example, the genetic model of clock mutant mice is susceptible to obesity, hyperlipidaemia, hyperglycaemia, and hepatic steatosis (Turek et al., 2005). Additionally, knockout of clock genes, such as aryl hydrocarbon receptor nuclear translocator-like (ARNTL), PERIOD (PER2), and cryptochrome (CRY1/CRY2), increased their susceptibility to obesity and metabolic disorders (Yang et al., 2009; Marcheva et al., 2010; Hemmeryckx et al., 2011; Shimba et al., 2011; Barclay et al., 2013). Polymorphisms of human clock genes, such as ARNTL, circadian locomotor output cycles kaput (CLOCK), CRY2, and nuclear receptor subfamily 1, group D, member 1 (NR1D1), have been linked to obesity or some other features of metabolic syndromes (Woon et al., 2007; Sookoian et al., 2008; Ruano et al., 2014; Machicao et al., 2016). Clinical studies on chronic circadian misalignment due to shift work reported an increased incidence of diabetes, obesity, and CVDs in the study participants (Khan et al., 2018; Torquati et al., 2018; Chellappa et al., 2019). Thus, disruption of the circadian clock may contribute to the development of obesity and metabolic diseases.
Emerging evidence has revealed that total sleep deprivation or sleep restriction influences concentrations of lipid profiles in plasma (e.g., triglyceride, cholesterol, and lipoproteins), suggesting the role of sleep in regulating lipid metabolism (Chua et al., 2015; Brianza-Padilla et al., 2016). Additionally, it has been reported that insufficient or disturbed sleep can lead to an impairment of rhythmic transcriptomes of central and peripheral tissues, including the genes associated with circadian rhythm and metabolism (Archer and Oster, 2015). However, the underlying mechanisms of circadian clock genes linked to sleep duration and lipid profile abnormalities have not been fully understood.
Therefore, this study aimed to explore the role of the circadian clock in regulating impaired lipid metabolism under disturbed sleep conditions. Sleep disturbance measurement was conducted by a sleep deprivation instrument. Total cholesterol, triglycerides, free fatty acid, some risk factors for hepatic cholestasis, and CVDs were detected first. Pathological staining of the liver, cholesterol synthesis, and conversion of cholesterol to bile acids were also assessed during the study. Finally, the NR1D1 function in liver cholesterol accumulation induced by sleep disturbance was analyzed.
Materials and Methods
Animals and Sleep Deprivation
The male Sprague-Dawley rats and C57BL/6J mice aged at 6–8 weeks were purchased from Charles River Laboratory Animal Technology Co. (Beijing, China). The NR1D1−/− mice were purchased from Cyagen Technology Co. (Guangzhou, China). The rats and mice were bred in animal facilities under specific pathogen-free conditions with a 12:12 h light-dark cycle (light on at 06:00 AM and light off at 18:00 PM). Morning 06:00 (zeitgeber time) was considered as the ZT 0. Food and water were available ad libitum. The rats and mice were randomly divided into experimental and control groups. Animals in the experimental group were subjected to the sleep deprivation instrument (DB036, BEIJING ZHISHU DUOBAO BIOLOGICAL TECHNOLOGY CO., LTD), which used a randomly moving platform to keep animals awake. The platform provides separate cages to allow animals free access to food and water. Parameters for the moving platform can be set by the touch screen of the controller. Additionally, the parameters can be changed by programming to reduce the adaptation of sleep deprived animals to the environment. After the parameters are set, sleep deprivation can be carried out in rats or mice without human intervention. Sleep deprivation was carried out at ZT 0, and animals were sacrificed after SD for 72 h. Then fresh blood samples and liver tissues were obtained for further analysis. The instrument does not need to train animals and can limit sleep in a mild way, which could greatly reduce the damage caused by the traditional water environment sleep deprivation method. Care, use, and treatment of rats and mice in this study were in strict agreement with international guidelines for the care and use of laboratory animals. This study was approved by the Animal Ethics Committee of the Beijing Institute of Basic Medical Sciences.
Serum Lipids and Metabolic Related Hormones Assay
Fresh blood samples were obtained and then centrifuged at 2000 g at 4°C for 20 min. The serum was transferred into a new tube and stored at −80°C until use. The serum levels of cholesterol (CEB701Ge), triglycerides (CEB687Ge), leptin (SEA084Ra), resistin (RETN) (SEA847Ra), adiponectin (ADP) (SEA605Ra), thyroxine (CEA452Ge), epinephrine (CEA858Ge), norepinephrine (CEA907Ge), insulin (CEA448Ra), glucagon (CEB266Ra), corticosterone (CEA540Ge), 8-hydroxyguanosine (8-OHdG) (CEA660Ge), alkaline phosphatase (ALP) (SEB472Ra), and bilirubin (CEK522Ge) were measured using the corresponding ELISA kits (CLOUD-CLONE CORP, China). Concentration of high density lipoprotein (HDL)-cholesterol (HDL-C; ab65390, Abcam), free fatty acid (ab65341, Abcam), and total bile acids (ab239702, Abcam) were analyzed with the corresponding assay kits. All the assay procedures were performed according to the protocols provided by the manufacturer.
Hepatic Lipids Assay
Rat hepatic lipids were determined by the method as described previously (Becares et al., 2019). Briefly, liver tissues were homogenized in 10 mM Tris buffer with 250 mM sucrose and 2 mM EDTA. Hepatic lipids in the homogenates were extracted with chloroform:methanol solution (1:1) by shaking for 3–4 h at room temperature. The homogenates were vortexed and centrifuged at 15000 g to obtain the organic phase. The organic phase could be dried with nitrogen and dissolved in 0.01 M phosphate buffer saline (PBS, with 1% Triton X-100). Then, further analyses of cholesterol (CEB701Ge), HDL (SEB006Ra), triglycerides (CEB687Ge), and very low density lipoprotein (VLDL; E-EL-R1203c, Elabscience) were performed according to the protocols provided by the manufacturer.
Oil Red O Staining
The rats were anesthetized and then transcardially perfused with normal saline, 4% paraformaldehyde in 0.01 M PBS. Liver tissues were dissected out, fixed in 4% PFA in 0.01 M PBS at 4°C overnight, transferred into 30% (w/v) sucrose in PBS for 24 h, embedded in tissue freezing medium (JUNG, Leica Biosystems), and stored at −80°C. Livers samples were cryosectioned into 10 μm thick sections and mounted on glass slides. The sections were stained with 0.3% Oil red O solution, counterstained with hematoxylin, rinsed under running tap water, and coverslipped with aqueous mountant.
Hematoxylin-Eosin Staining
Hematoxylin-eosin (HE) staining was conducted according to routine protocols. Liver tissues of six rats from each group were fixed in 10% neutral buffered formalin for 48 h, and then embedded in paraffin and sectioned into 3–5 μm thick sections. After deparaffinization and rehydration, the sections were stained with hematoxylin, dipped in 1% acid ethanol, and rinsed in distilled water. Then, the sections were stained with eosin solution and followed by with graded alcohol dehydration. The mounted slides were photographed by an Olympus BX53 fluorescence microscope (Tokyo, Japan).
Western Blot Analysis
Fresh liver samples were harvested, washed with cold PBS, and lysed with ice-cold lysis buffer supplemented with protease inhibitors. Cell lysate was run on 10% SDS-PAGE, transferred to a polyvinylidene fluoride membrane (PVDF; ISEQ00010, Millipore), blocked with 5% skim milk, and incubated with primary antibodies at 4°C overnight. The primary antibodies against 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR; sc-271595, Santa Cruz Biotechnology), cytochrome P450 family 7 subfamily A member 1 (CYP7A1; sc-518007, Santa Cruz Biotechnology), NR1D1 (sc-323,915, Santa Cruz Biotechnology), and β-actin (sc-8432, Santa Cruz Biotechnology) were used. Then, the membrane was incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. After extensive washing three times, the bands were detected using an ECL detection system (4600SF, Tanon Science & Technology Co., Ltd).
RT-PCR Analysis
Total RNA was isolated from fresh liver tissues using Trizol reagent (T9424, Sigma Aldrich) according to the manufacturer’s instructions. After concentration determination, the cDNA products were obtained using All-in-One cDNA Synthesis SuperMix (B24403, Biotool) for reverse transcription. The amplification of target genes was performed using the standard protocol by Bio-Rad T100TM Thermal Cycler (1861096, Bio-Rad). The rat primers used were as follows: HMGCR-F: 5'-gcctccattgagtccggagg-3', HMGCR-R: 5'-gcaccgggttatcgtgagga-3'; CYP7A1-F: 5'-gaccaagtctttccggcac-3', CYP7A1-R: 5'-cagagaatagcgaggtgcgt-3'; FXR-F: 5'-aggaagtgcagagagatggga-3', FXR-R: 5'-agcttggtcgtggaggtcac-3'; RXRa-F: 5'-cgctcctcaggcaaacacta-3', RXRa-R: 5'-ggaggatgccgtctttcaca-3'; SHP-F: 5'-aggctcactgggcattgtgt-3', SHP-R 5'-acatctccgatgacagggcg-3'; HNF4a-F: 5'-aaaaccctcgccgacatgga-3', HNF4a-R: 5'-ctgacacccaggctgttgga-3'; C/EBPa-R: 5'-tggagggttcttagcccctt-3', C/EBPa-F: 5'ggctggcgacatacagtaca-3'; NR1D1-F: 5'-tacaagtggccatggaagaca-3', NR1D1-R: 5'-ttggccaagttcatggcgttc-3'; E4BP4-F: 5'-ctgatgcagctgagaaaaatgc-3', E4BP4-R: 5'-gggagagcagctcagctttta-3'; R-GAPDH-R: 5'-cagttccagcctcgtctcat-3', GAPDH-F: 5'-actgtgccgttgaacttgcc-3'. The mouse primers used were as follows: NR1D1-F: 5'-acgaccctggactccaataa-3', NR1D1-R: 5'-ccattggagctgtcactgtaga-3'; β-actin-F: 5'-ttgccctagacttcgagcaa-3', β-actin-R: 5'-caggaaggaaggctggaaga-3'. Gene relative expression was normalized to the mRNA levels of β-actin.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, United States). Data are presented as the mean ± SE and analyzed using the two-tailed, nonpaired Student’s t-test for comparison of two groups. The significant difference was set at p < 0.05.
Results
SD Induced Increased Cholesterol Concentration in Serum Lipid Profiles
Rat serum samples were analyzed to evaluate the metabolic effects induced by SD. SD for 72 h induced significant increasing of cholesterol level, while triglycerides and free fatty acids levels kept unchanged compared with the control group (Figures 1A–C). Hypercholesterolemia is a metabolic disorder characterized by increased cholesterol levels in circulation. The above results indicated that the tendency of hypercholesterolemia occurrence was much earlier than hypertriglyceridemia induced by SD. Some hormones secreted during the sleep-wake cycle contribute to the metabolism and energy balance of the body. Therefore, concentrations of some hormones involved in metabolism regulation and appetite control were determined. The results showed that concentration of serum leptin and resistin in the SD group were much lower than that in the control group (Figures 1D,E). However, serum levels of some other hormones, including adiponectin, glucagon, insulin, thyroxine, norepinephrine, and epinephrine, slightly differed in both groups (Figures 1F–H). The above results suggested that high cholesterol level in serum lipid profiles was induced by disturbed sleep stress.
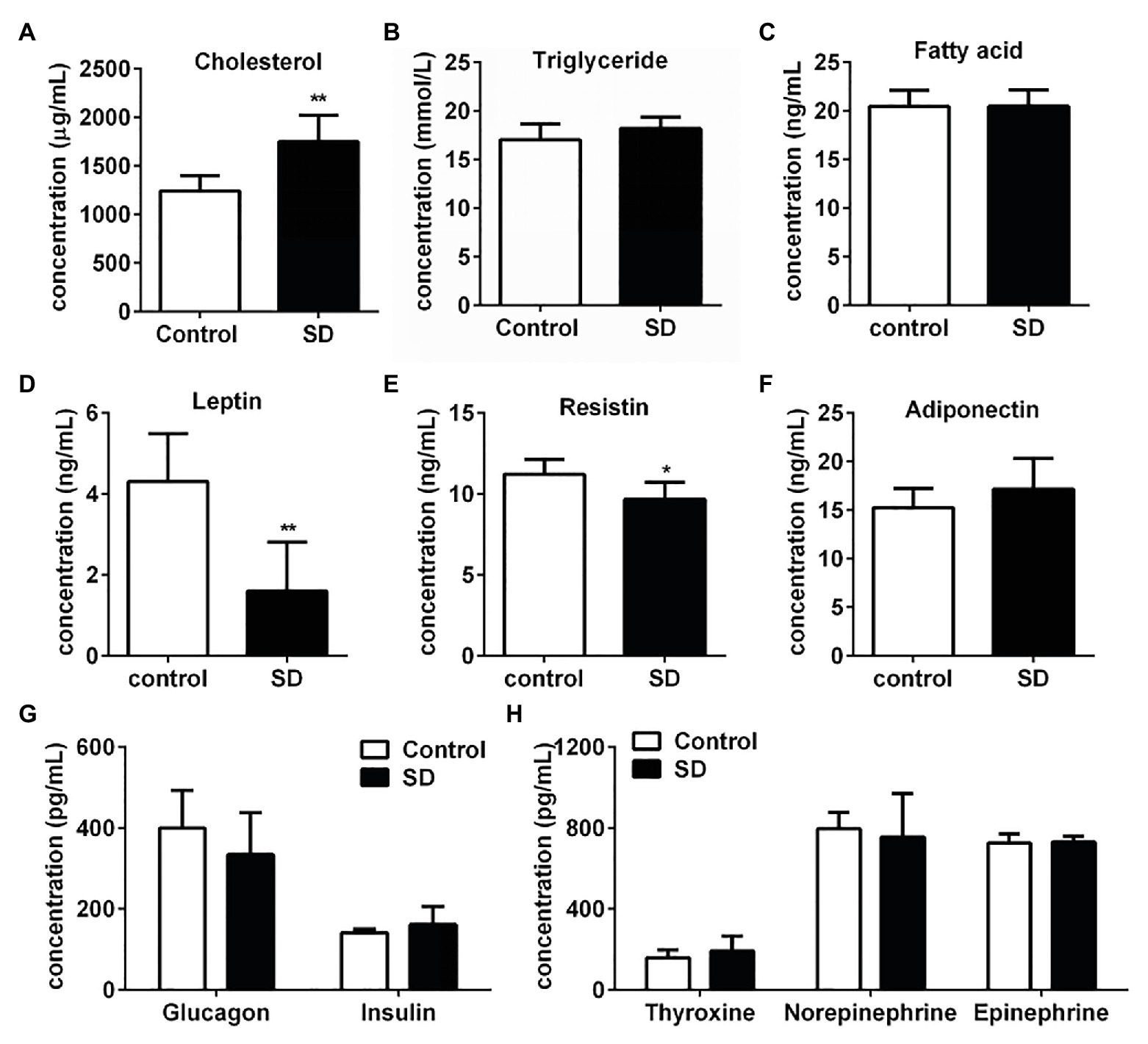
Figure 1. Sleep deprivation (SD) induced increased cholesterol concentration in serum lipid profiles. (A) Cholesterol, (B) triglyceride, and (C) free fatty acids concentrations in rat serums were analyzed under disturbed sleep condition for 72 h. Metabolism related hormones including (D) leptin, (E) resistin (RETN), (F) adiponectin (ADP), (G) glucagon and insulin, (H) thyroxine, norepinephrine, and epinephrine concentration in rat serums were detected. **p < 0.01; *p < 0.05.
SD Induced the Warning Signs of an Increased Risk of Cardiovascular and Hepatic Diseases
A high level of serum cholesterol or hypercholesterolemia is closely associated with increased risk of CVDs and hepatic diseases due to the induction of various stress pressures. In the experimental group, the total serum bile acid and bilirubin levels significantly increased after SD, but alkaline phosphatase (ALP) concentration remained unchanged in serum (Figures 2A–C). The results suggest the risk of hepatic cholestasis caused by SD. Simultaneously, the level of some risk factors for CVDs such as corticosterone and 8-OHdG in serum were found to be notably increased, while the concentration of protective component predicting cardiovascular risk HDL-C was markedly decreased in the SD group (Figures 2D–F). The above results together suggested the warning signs of future hepatic diseases and CVDs.
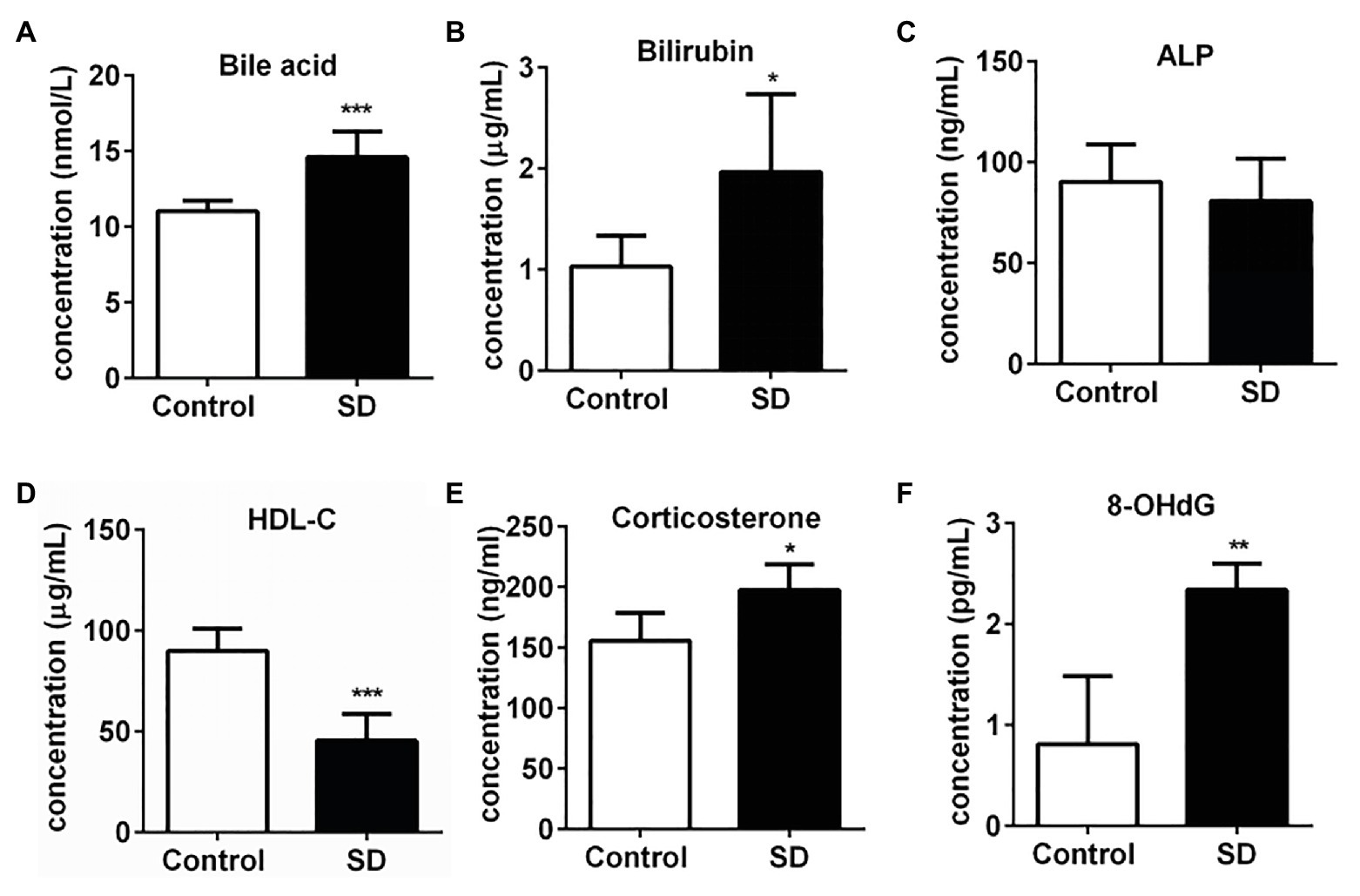
Figure 2. Sleep deprivation induced the warning signs of an increased risk of cardiovascular and hepatic diseases. (A) Total bile acids, (B) bilirubin, and (C) alkaline phosphatase (ALP) concentrations in rat serums increased after sleep disturbance for 72 h. The decreased concentration of (D) high density lipoprotein (HDL)-cholesterol (HDL-C) and increased concentration of (E) corticosterone and (F) 8-hydroxyguanosine (8-OHdG) in rat serums were induced by SD. ***p < 0.001; **p < 0.01; *p < 0.05.
SD Induced Accumulation of Cholesterol in the Liver
Synthesis and degradation of cholesterol mainly occurs in the liver. It was found that cholesterol and HDL levels in rat liver were significantly increased after SD (Figure 3A). Meanwhile, triglycerides and VLDL levels in liver remained unchanged by SD (Figure 3A). Moreover, no obvious lipid droplets were observed in the pathological results of oil red O staining, but the coloring degree of staining was seemingly slightly enhanced after SD (Figure 3B), and there was no distinct increasing of inflammatory cells invasion by HE staining after SD (Figure 3B). The above results indicate that sleep disturbance induced an abnormal cholesterol metabolism in the liver before the occurrence of other pathological changes.
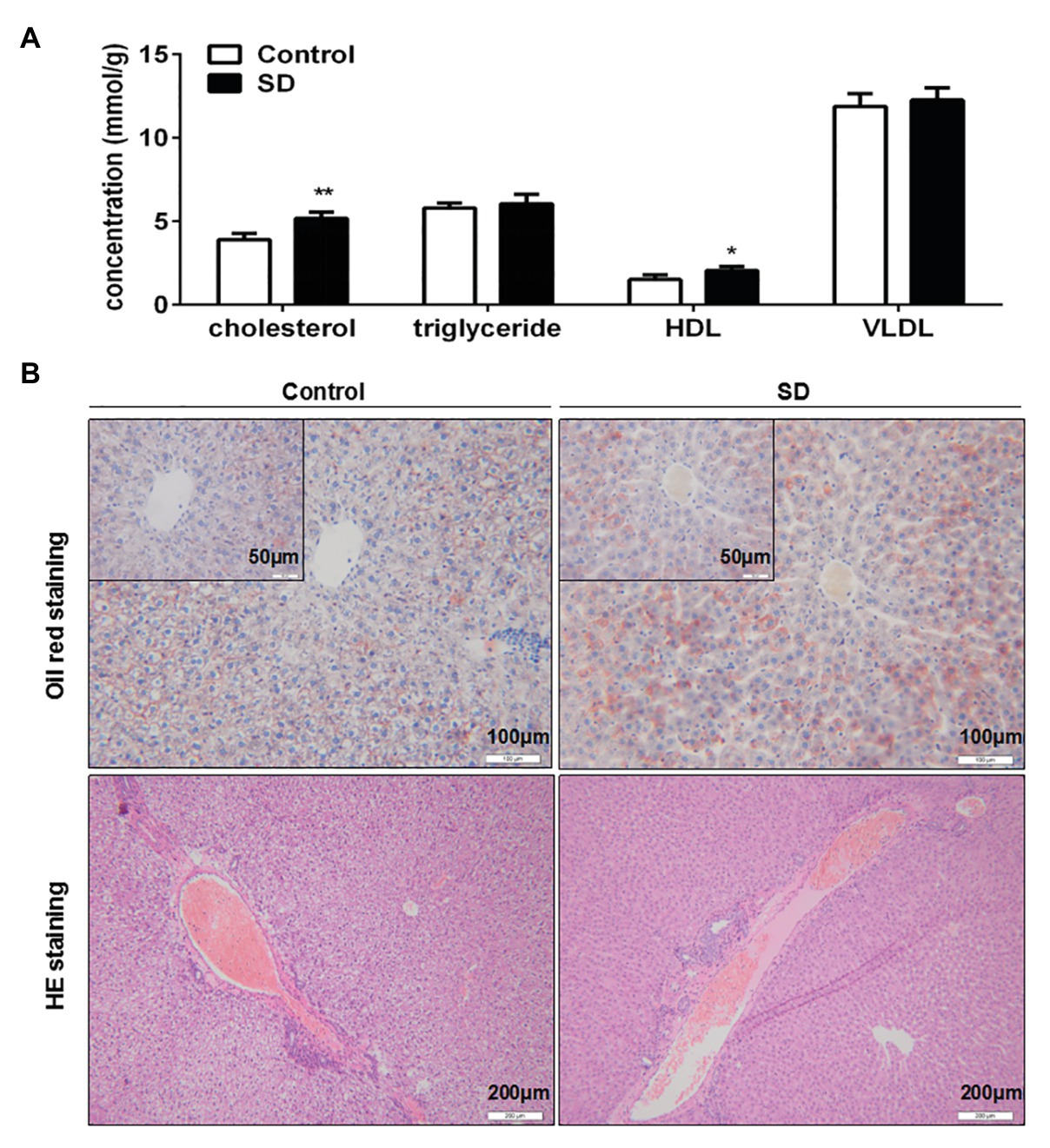
Figure 3. Sleep deprivation induced accumulation of cholesterol in liver. (A) The level of cholesterol, triglyceride, HDL, and very low density lipoprotein (VLDL) in rat liver lipid profiles were analyzed. (B) Pathological staining of rat livers was assessed by oil red O staining and hematoxylin-eosin (HE) staining. **p < 0.01; *p < 0.05.
Bile Acid Synthesis Was More Vulnerable to SD Than Cholesterol Synthesis
To determine the reason behind high serum cholesterol levels, the synthesis and degradation of cholesterol in the liver were analyzed. Cholesterol synthesis and its conversion to bile acids are necessary to ensure cholesterol homoeostasis. The two rate-controlling enzymes involved in cholesterol and bile acid synthesis are HMGCR and CYP7A1, respectively. The expression of CYP7A1 mRNA and protein in rat livers was downregulated but not HMGCR expression after SD (Figures 4A,B). In addition, liver CYP8B1 expression, a downstream enzyme of CYP7A1, was also weakened under the SD condition. However, expression of CYP27A1, a critical enzyme in the alternative pathway of bile acid synthesis, remained unchanged after SD (Supplementary Figure 1). The results indicate that bile acid synthesis was more vulnerable to SD than cholesterol synthesis. Additionally, bile acid synthesis regulators were analyzed. The results showed that expression levels of farnesoid-X-receptor (FXR), retinoid X receptor alpha (RXRa), liver receptor homolog 1 (LRH1), hepatocyte nuclear receptor 4 alpha (HNF4a), CCAAT enhancer binding protein α (C/EBPa), and forkhead box O1 (FOXO1) at transcription level were not strikingly affected by SD (Figure 4C). The above results suggest that bile acid synthesis was partially inhibited under disturbed sleep conditions, which indicates that the conversion of cholesterol to bile acids is more susceptible to SD than cholesterol synthesis.
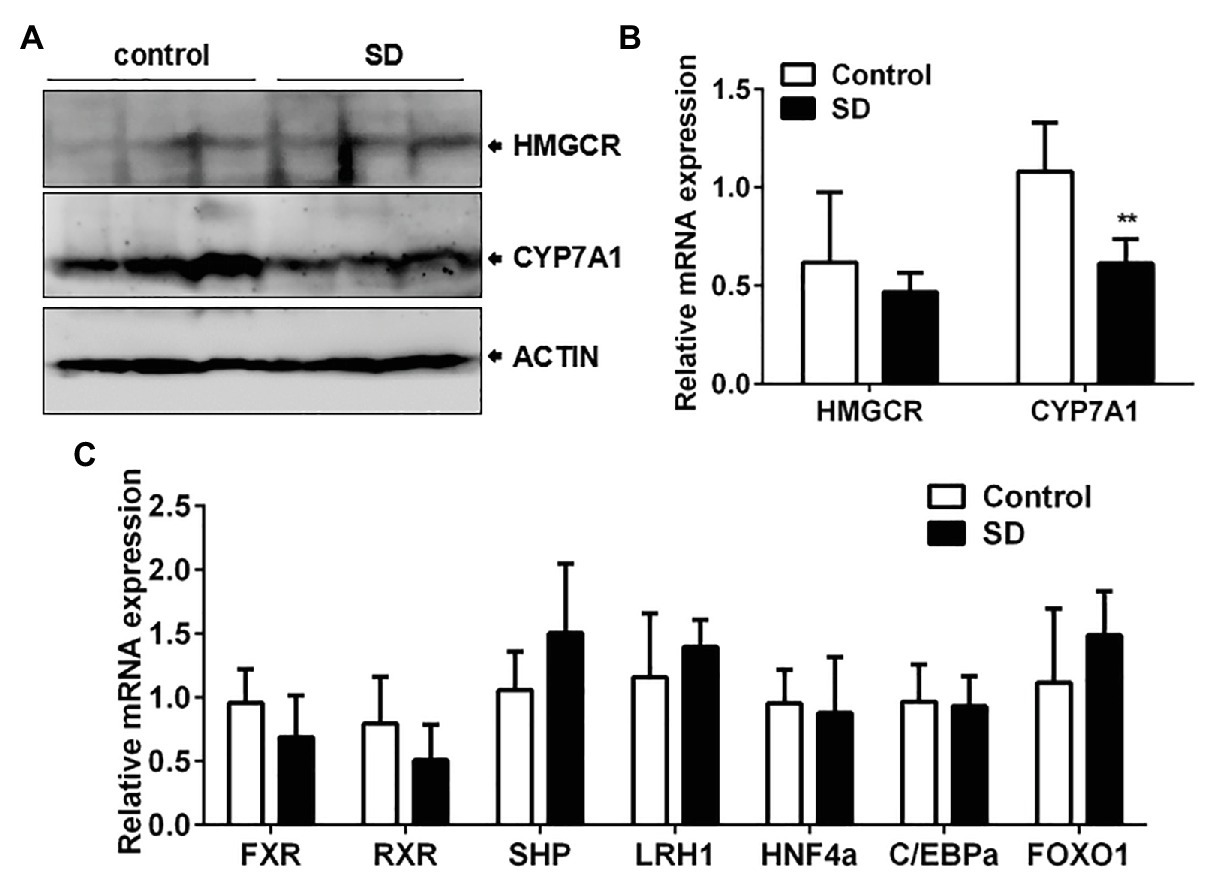
Figure 4. Conversion of cholesterol to bile acids was affected by SD. (A) The protein and (B) relative mRNA expression of cytochrome P450 family 7 subfamily A member 1 (CYP7A1) and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) in rat livers. (C) Expression of some reported bile acid synthesis regulators including farnesoid-X-receptor (FXR), retinoid X receptor alpha (RXRa), liver receptor homolog 1 (LRH1), hepatocyte nuclear receptor 4 alpha (HNF4a), CCAAT enhancer binding protein α (C/EBPa), and forkhead box O1 (FOXO1) were not strikingly affected by SD. **p < 0.01.
Correlation of NR1D1 With Bile Acid Synthesis Under Disturbed Sleep Conditions
Hepatic cholesterol and bile acid synthesis are controlled by the circadian clock. We found that SD for 24 h could induce obvious increasing of cholesterol level in rat serum (Figure 5A). The rapid increase in cholesterol levels induced by an acute SD of 24 h suggests the potential role of circadian clock in regulating cholesterol metabolism. The rapid increase in cholesterol level induced by an acute SD in 24 h suggests the potential role of the circadian clock in regulating cholesterol metabolism. NR1D1 and E4 promoter binding protein 4 (E4BP4) have been reported to participate in regulation of CYP7A1 expression. It was found that NR1D1 expression was downregulated after SD for 72 h but not E4BP4 expression, which remained unchanged (Figure 5B). The expression of CYP7A1 in liver displayed diurnal oscillation variation (data not shown). Further analyses demonstrated that amplitudes of NR1D1 expression were partially decreased by acute SD of 24 h. NR1D1 expression could exhibit a rapid response to SD at ZT4, but the pattern of E4BP4 expression remained unchanged by an acute SD of 24 h (Figures 5C,D). The above results suggested the role of NR1D1 in regulating CYP7A1 expression under disturbed sleep conditions.
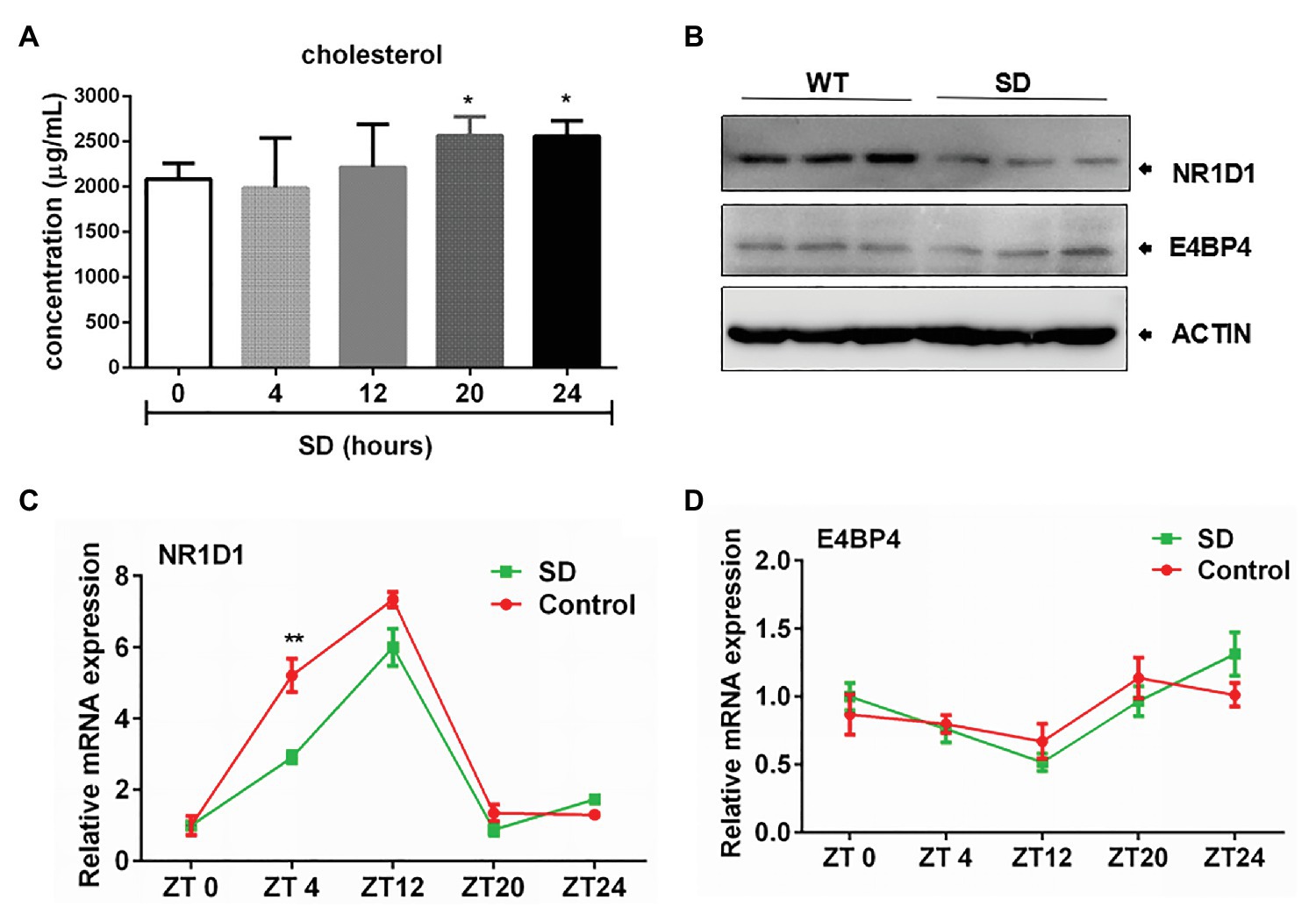
Figure 5. Correlation of nuclear receptor subfamily 1, group D, member 1 (NR1D1) with bile acid synthesis under sleep disturbance condition. (A) Increasing of cholesterol level in rat serum induced by acute SD in 24 h. (B) NR1D1 and E4, promoter binding protein 4 (E4BP4) expression in rat liver after SD for 72 h. Oscillation expressions of (C) NR1D1 and (D) E4BP4 in rat liver under acute SD stress. **p < 0.01; *p < 0.05.
High Cholesterol Level Induced by SD Correlated With Downregulation of CYP7A1 Expression by NR1D1
In order to figure out the potential role of NR1D1 in CYP7A1 expression, we compared the responses of wild type (WT) and NR1D1−/− mice after SD. Similarly, SD could induce an obvious increase in serum cholesterol levels along with downregulation of NR1D1 and CYP7A1 expression in the livers of WT mice (Figures 6A–C). Furthermore, NR1D1 deficiency downregulated the expression levels of CYP7A1, and sleep disturbance could exacerbate the decreased CYP7A1 expression (Figure 6D). Additionally, NR1D1 deficiency could also further increase serum cholesterol level and the tendency of cholesterol accumulation in the liver under sleep disturbance conditions (Figures 6E,F). The above data indicated that downregulation of NR1D1 expression by the SD-mediated transcriptional inhibition of CYP7A1, leading to the block of cholesterol degradation and high level cholesterol accumulation in serum.
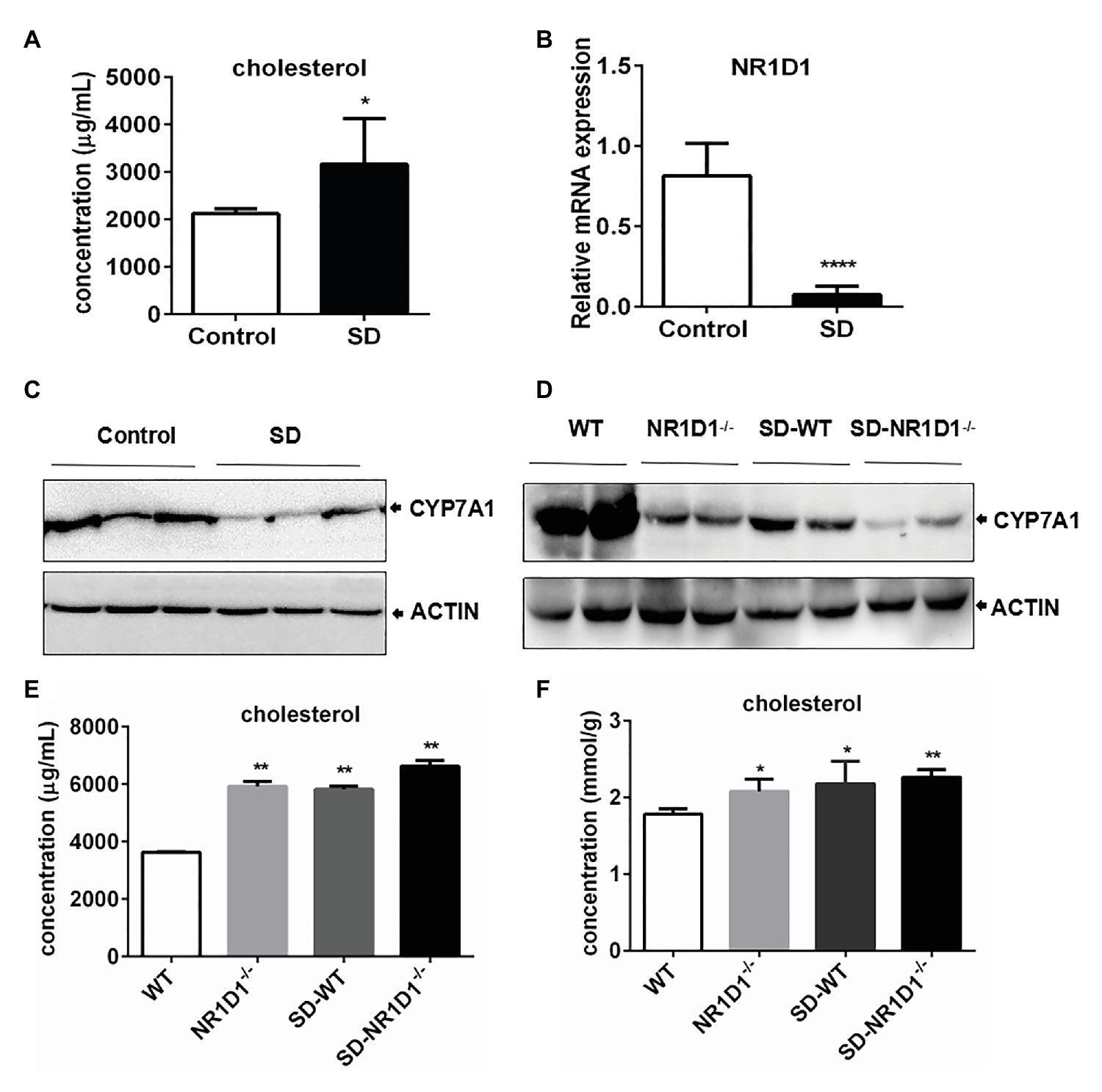
Figure 6. High level cholesterol induced by SD correlated with downregulation of CYP7A1 expression by NR1D1. (A) Increasing of cholesterol concentration in mice serums induced by SD. (B) Relative mRNA expression of NR1D1 in mice livers after SD. (C) Inhibition of CYP7A1 expression in mice livers after SD. (D) Reduction of CYP7A1 expression in sleep disturbed NR1D1−/− mice. (E) Serum cholesterol level in sleep disturbed NR1D1−/− mice. (F) liver cholesterol level in sleep disturbed NR1D1−/− mice. ****p < 0.0001; **p < 0.01; *p < 0.05.
Discussion
Accumulating evidence from the epidemiologic and laboratory studies have suggested the role of reduced or disturbed sleep in the metabolic dysfunction, leading to an increased risk of type 2 diabetes, obesity, or weight gain. Considering the early metabolic alterations caused by sleep disturbance, our results showed that a high cholesterol level in serum lipid profiles was rapidly induced by sleep deprivation exposure (24 and 72 h) in both rat and mice. Therefore, influences on cholesterol homeostasis regulation should be the early metabolic related events urged by sleep disturbance. Long term and uncontrolled exposure to hypercholesterolemia can lead to a slow and progressive occurrence of cardiovascular events (Jeong et al., 2018). In our study, sleep disturbance induced some early signs of future CVDs. Plasma HDL-C was decreased under disturbed sleep stress. The cholesterol contained in HDL is verified to be inversely associated with the risk of coronary heart disease and a predictor of cardiovascular risk for its central role in reverse cholesterol transport (Rader and Hovingh, 2014; Ouimet et al., 2019). Besides, HDL has also been proved to exert inhibitory roles on oxidative pathways in CVDs (Kontush and Chapman, 2010; Brites et al., 2017). High levels of 8-OHdG, a biomarker of oxidative stress, have been observed in patients with severe atherosclerotic lesions and hypertension (Martinet et al., 2002; Xiang et al., 2011; Rosello-Lleti et al., 2012). Therefore, the elevated 8-OHdG level observed in this study reminds the future rising oxidative damage induced by SD. Accordingly, the above risk factors together suggest that hypercholesterolemia induced by SD might predict the warning sign of CVDs by affecting multiple blood parameters related to the cardiovascular system.
High plasma cholesterol can contribute to the development of liver disorders, such as non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatosis hepatitis (NASH) (Kerr and Davidson, 2012; Ioannou, 2016). Our results found the elevated level of bilirubin, bile acids, and cholesterol in serum under disturbed sleep stress. Bilirubin, bile acids, and cholesterol are important components of bile, and impairment in their formation or excretion can lead to cholestasis (Paumgartner, 2010). Particularly, the accumulation of bile acids in the serum and hepatocytes suggests the increased risk of cholestasis occurrence at early stage of liver diseases under disturbed sleep stress. Cholestasis is usually caused by alterations of hepatobiliary transport systems in hereditary cholestasis or acquired cholestatic liver diseases (Lee and Boyer, 2000; Sticova et al., 2018). Apart from mutations in transporter genes, pro-inflammatory cytokines, hormones, and drugs are major pathogenic factors for acquired forms of cholestasis (Stieger et al., 2000; Zollner and Trauner, 2006). Thus, the results of our study indicate that short sleep duration or sleep disturbance also should be a risk factor for cholestatic liver diseases.
Various epidemiological studies have reported that short sleep duration is associated with an elevated weight and body mass index (BMI; Kobayashi et al., 2012; Bayon et al., 2014). Being overweight is associated with greater energy intake than expenditure. Though some reported hormones involved in regulation of lipid and glucose metabolism and triglycerides levels kept unchanged, leptin concentration was found to be decreased under disturbed sleep stress for 72 h in this study. Leptin plays a major role in energy homoeostasis through appetite control and food intake limitation. It has been reported that insufficient sleep promotes dietary intake and contributes to the development of obesity and metabolic alteration (Prinz, 2004; Taheri et al., 2004). Thus, the decline in serum leptin levels induced by SD is in agreement with earlier reports on the links between short sleep duration and metabolic alterations by regulation on dietary intake. Therefore, exogenous dietary intake by appetite regulation is likely to be an important source of high cholesterol level in serum induced by sleep disturbance.
Regulation of hepatic cholesterol metabolism is controlled by the circadian clock (Pan et al., 2013; Ma et al., 2015). Oscillatory transcription of genes encoding key enzymes regulating cholesterol homoeostasis is tightly controlled during day and night. The results in this study showed that CYP7A1 expression was inhibited but with HMGCR expression unchanged induced by sleep disturbance. Therefore, conversion of cholesterol to bile acids rather than cholesterol synthesis was more susceptible to disturbed sleep stress. CYP7A1 expression exhibits a strong circadian rhythm, which is negatively regulated by peroxisome proliferator-activated receptor a, DEC1/2, and E4BP4, and positively regulated by NR1D1 and DBP (Noshiro et al., 2007; Duez et al., 2008). In this study, expressions NR1D1 were downregulated, and exhibit rapid response to SD at ZT4. Moreover, sleep disturbance could exacerbate the reduction of NR1D1 mediated CYP7A1 expression. Accordingly, the results provided a way to understand how circadian clocks link sleep disturbance to cholesterol metabolism by NR1D1. Additionally, the results partially explained the endogenous regulatory mechanism of liver cholesterol accumulation. However, NR1D1 acts as a transcriptional repressor in the inhibition of BMAL1 transcription and regulation of biological processes, such as adipocyte differentiation and lipid metabolism (Wang and Lazar, 2008; Le Martelot et al., 2009). Since both NR1D1 and CYP7A1 expression were downregulated in this study, regulation of CYP7A1 expression should be an indirect mechanism mediated by NR1D1 through negative regulators of CYP7A1 rather than positive regulators. The involvements of negative regulators of CYP7A1 in this study remains to be explored in our future work.
In summary, our study has demonstrated that an increased serum and liver cholesterol level induced by sleep disturbance is an early pathological event, which closely correlates with a decreased conversion of cholesterol to bile acids by NR1D1 mediated CYP7A1 inhibition. Thus, our finding explained the important role of the circadian clock in linking sleep loss and lipid profile abnormality.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material and further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of the Beijing Institute of Basic Medical Sciences.
Author Contributions
XH and YZ were the co-first authors who performed most of the experiments and data analysis. WW, CZ, and LW performed the animal experiments. MD and MZ contributed to animal breeding. CX designed, performed, and supervised the study and wrote the paper. LS provided suggestions and guidance of this study and revised the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Nature Science Foundation of China (no. 81700759) and the National Military Sciences Foundation of China (no. AWS17J014 and no. BWS17J025).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.610496/full#supplementary-material
Abbreviations
CVDs, Cardiovascular diseases; SD, Sleep deprivation; ARNTL, Aryl hydrocarbon receptor nuclear translocator-like; PER2, PERIOD2; CRY1/CRY2, Cryptochrom1/2; NR1D1, Nuclear receptor subfamily 1, group D, member 1; HDL, High density lipoprotein; VLDL, Very low density lipoprotein; 8-OHdG, 8-Hydroxyguanosine; ALP, Alkaline phosphatase; HMGCR, 3-Hydroxy-3-methylglutaryl-CoA reductase; CYP7A1, Cytochrome P450 family 7 subfamily A member 1; FXR, Farnesoid-X-receptor; RXRa, Retinoid X receptor alpha; LRH1, Liver receptor homolog 1; HNF4a, Hepatocyte nuclear receptor 4 alpha; C/EBPa, CCAAT enhancer binding protein α; FOXO1, Forkhead box O1; E4BP4, E4, promoter binding protein 4.
References
Archer, S. N., and Oster, H. (2015). How sleep and wakefulness influence circadian rhythmicity: effects of insufficient and mistimed sleep on the animal and human transcriptome. J. Sleep Res. 24, 476–493. doi: 10.1111/jsr.12307
Barclay, J. L., Shostak, A., Leliavski, A., Tsang, A. H., Johren, O., Muller-Fielitz, H., et al. (2013). High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am. J. Physiol. Endocrinol. Metab. 304, E1053–E1063. doi: 10.1152/ajpendo.00512.2012
Bayon, V., Leger, D., Gomez-Merino, D., Vecchierini, M. F., and Chennaoui, M. (2014). Sleep debt and obesity. Ann. Med. 46, 264–272. doi: 10.3109/07853890.2014.931103
Becares, N., Gage, M. C., Voisin, M., Shrestha, E., Martin-Gutierrez, L., Liang, N., et al. (2019). Impaired LXRalpha phosphorylation attenuates progression of fatty liver disease. Cell Rep. 26, 984.e986–995.e986. doi: 10.1016/j.celrep.2018.12.094
Bertisch, S. M., Pollock, B. D., Mittleman, M. A., Buysse, D. J., Bazzano, L. A., Gottlieb, D. J., et al. (2018). Insomnia with objective short sleep duration and risk of incident cardiovascular disease and all-cause mortality: sleep heart health Sstudy. Sleep 41, 984.e986–995.e986. doi: 10.1093/sleep/zsy047
Brianza-Padilla, M., Bonilla-Jaime, H., Almanza-Perez, J. C., Lopez-Lopez, A. L., Sanchez-Munoz, F., and Vazquez-Palacios, G. (2016). Effects of different periods of paradoxical sleep deprivation and sleep recovery on lipid and glucose metabolism and appetite hormones in rats. Appl. Physiol. Nutr. Metab. 41, 235–243. doi: 10.1139/apnm-2015-0337
Brites, F., Martin, M., Guillas, I., and Kontush, A. (2017). Antioxidative activity of high-density lipoprotein (HDL): mechanistic insights into potential clinical benefit. BBA Clin. 8, 66–77. doi: 10.1016/j.bbacli.2017.07.002
Chellappa, S. L., Vujovic, N., Williams, J. S., and Scheer, F. (2019). Impact of circadian disruption on cardiovascular function and disease. Trends Endocrinol. Metab. 30, 767–779. doi: 10.1016/j.tem.2019.07.008
Chua, E. C., Shui, G., Cazenave-Gassiot, A., Wenk, M. R., and Gooley, J. J. (2015). Changes in plasma lipids during exposure to total sleep deprivation. Sleep 38, 1683–1691. doi: 10.5665/sleep.5142
Duez, H., van der Veen, J. N., Duhem, C., Pourcet, B., Touvier, T., Fontaine, C., et al. (2008). Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology 135, 689–698. doi: 10.1053/j.gastro.2008.05.035
Hemmeryckx, B., Himmelreich, U., Hoylaerts, M. F., and Lijnen, H. R. (2011). Impact of clock gene Bmal1 deficiency on nutritionally induced obesity in mice. Obesity 19, 659–661. doi: 10.1038/oby.2010.266
Ioannou, G. N. (2016). The role of cholesterol in the pathogenesis of NASH. Trends Endocrinol. Metab. 27, 84–95. doi: 10.1016/j.tem.2015.11.008
Jeong, S. M., Choi, S., Kim, K., Kim, S. M., Lee, G., Park, S. Y., et al. (2018). Effect of change in total cholesterol levels on cardiovascular disease among young adults. J. Am. Heart Assoc. 7:e008819. doi: 10.1161/JAHA.118.008819
Kecklund, G., and Axelsson, J. (2016). Health consequences of shift work and insufficient sleep. BMJ 355:i5210. doi: 10.1136/bmj.i5210
Kerr, T. A., and Davidson, N. O. (2012). Cholesterol and nonalcoholic fatty liver disease: renewed focus on an old villain. Hepatology 56, 1995–1998. doi: 10.1002/hep.26088
Khan, S., Duan, P., Yao, L., and Hou, H. (2018). Shiftwork-mediated disruptions of circadian rhythms and sleep homeostasis cause serious health problems. Int. J. Genomics 2018:8576890. doi: 10.1155/2018/8576890
Killick, R., Banks, S., and Liu, P. Y. (2012). Implications of sleep restriction and recovery on metabolic outcomes. J. Clin. Endocrinol. Metab. 97, 3876–3890. doi: 10.1210/jc.2012-1845
Kobayashi, D., Takahashi, O., Deshpande, G. A., Shimbo, T., and Fukui, T. (2012). Association between weight gain, obesity, and sleep duration: a large-scale 3-year cohort study. Sleep Breath. 16, 829–833. doi: 10.1007/s11325-011-0583-0
Kon, K., Ode, K. L., and Ueda, H. R. (2017). Molecular mechanisms of circadian rhythm and sleep homeostasis. Brain Nerve 69, 257–264. doi: 10.11477/mf.1416200735
Kontush, A., and Chapman, M. J. (2010). Antiatherogenic function of HDL particle subpopulations: focus on antioxidative activities. Curr. Opin. Lipidol. 21, 312–318. doi: 10.1097/MOL.0b013e32833bcdc1
Le Martelot, G., Claudel, T., Gatfield, D., Schaad, O., Kornmann, B., Lo Sasso, G., et al. (2009). REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 7:e1000181. doi: 10.1371/journal.pbio.1000181
Lee, J., and Boyer, J. L. (2000). Molecular alterations in hepatocyte transport mechanisms in acquired cholestatic liver disorders. Semin. Liver Dis. 20, 373–384. doi: 10.1055/s-2000-9390
Liu, H., and Chen, A. (2019). Roles of sleep deprivation in cardiovascular dysfunctions. Life Sci. 219, 231–237. doi: 10.1016/j.lfs.2019.01.006
Ma, D., Liu, T., Chang, L., Rui, C., Xiao, Y., Li, S., et al. (2015). The liver clock controls cholesterol homeostasis through Trib1 protein-mediated regulation of PCSK9/low density lipoprotein receptor (LDLR) axis. J. Biol. Chem. 290, 31003–31012. doi: 10.1074/jbc.M115.685982
Machicao, F., Peter, A., Machann, J., Konigsrainer, I., Bohm, A., Lutz, S. Z., et al. (2016). Glucose-raising polymorphisms in the human clock gene cryptochrome 2 (CRY2) affect hepatic lipid content. PLoS One 11:e0145563. doi: 10.1371/journal.pone.0145563
Marcheva, B., Ramsey, K. M., Buhr, E. D., Kobayashi, Y., Su, H., Ko, C. H., et al. (2010). Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631. doi: 10.1038/nature09253
Martinet, W., Knaapen, M. W., De Meyer, G. R., Herman, A. G., and Kockx, M. M. (2002). Elevated levels of oxidative DNA damage and DNA repair enzymes in human atherosclerotic plaques. Circulation 106, 927–932. doi: 10.1161/01.cir.0000026393.47805.21
Nedeltcheva, A. V., and Scheer, F. A. (2014). Metabolic effects of sleep disruption, links to obesity and diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 21, 293–298. doi: 10.1097/MED.0000000000000082
Noshiro, M., Usui, E., Kawamoto, T., Kubo, H., Fujimoto, K., Furukawa, M., et al. (2007). Multiple mechanisms regulate circadian expression of the gene for cholesterol 7alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J. Biol. Rhythm. 22, 299–311. doi: 10.1177/0748730407302461
Ouimet, M., Barrett, T. J., and Fisher, E. A. (2019). HDL and reverse cholesterol transport. Circ. Res. 124, 1505–1518. doi: 10.1161/CIRCRESAHA.119.312617
Pan, X., Jiang, X. C., and Hussain, M. M. (2013). Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation 128, 1758–1769. doi: 10.1161/CIRCULATIONAHA.113.002885
Panda, S. (2016). Circadian physiology of metabolism. Science 354, 1008–1015. doi: 10.1126/science.aah4967
Paumgartner, G. (2010). Biliary physiology and disease: reflections of a physician-scientist. Hepatology 51, 1095–1106. doi: 10.1002/hep.23472
Prinz, P. (2004). Sleep, appetite, and obesity—what is the link? PLoS Med. 1:e61. doi: 10.1371/journal.pmed.0010061
Rader, D. J., and Hovingh, G. K. (2014). HDL and cardiovascular disease. Lancet 384, 618–625. doi: 10.1016/S0140-6736(14)61217-4
Reutrakul, S., and Van Cauter, E. (2018). Sleep influences on obesity, insulin resistance, and risk of type 2 diabetes. Metabolism 84, 56–66. doi: 10.1016/j.metabol.2018.02.010
Rosello-Lleti, E., de Burgos, F. G., Morillas, P., Cortes, R., Martinez-Dolz, L., Almenar, L., et al. (2012). Impact of cardiovascular risk factors and inflammatory status on urinary 8-OHdG in essential hypertension. Am. J. Hypertens. 25, 236–242. doi: 10.1038/ajh.2011.202
Ruano, E. G., Canivell, S., and Vieira, E. (2014). REV-ERB ALPHA polymorphism is associated with obesity in the Spanish obese male population. PLoS One 9:e104065. doi: 10.1371/journal.pone.0104065
Saper, C. B., Scammell, T. E., and Lu, J. (2005). Hypothalamic regulation of sleep and circadian rhythms. Nature 437, 1257–1263. doi: 10.1038/nature04284
Shimba, S., Ogawa, T., Hitosugi, S., Ichihashi, Y., Nakadaira, Y., Kobayashi, M., et al. (2011). Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One 6:e25231. doi: 10.1371/journal.pone.0025231
Sookoian, S., Gemma, C., Gianotti, T. F., Burgueno, A., Castano, G., and Pirola, C. J. (2008). Genetic variants of clock transcription factor are associated with individual susceptibility to obesity. Am. J. Clin. Nutr. 87, 1606–1615. doi: 10.1093/ajcn/87.6.1606
Sticova, E., Jirsa, M., and Pawlowska, J. (2018). New insights in genetic cholestasis: from molecular mechanisms to clinical implications. Can. J. Gastroenterol. Hepatol. 2018:2313675. doi: 10.1155/2018/2313675
Stieger, B., Fattinger, K., Madon, J., Kullak-Ublick, G. A., and Meier, P. J. (2000). Drug‐ and estrogen-induced cholestasis through inhibition of the hepatocellular bile salt export pump (Bsep) of rat liver. Gastroenterology 118, 422–430. doi: 10.1016/s0016-5085(00)70224-1
Taheri, S., Lin, L., Austin, D., Young, T., and Mignot, E. (2004). Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 1:e62. doi: 10.1371/journal.pmed.0010062
Takahashi, J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. doi: 10.1038/nrg.2016.150
Torquati, L., Mielke, G. I., Brown, W. J., and Kolbe-Alexander, T. (2018). Shift work and the risk of cardiovascular disease. A systematic review and meta-analysis including dose-response relationship. Scand. J. Work Environ. Health 44, 229–238. doi: 10.5271/sjweh.3700
Turek, F. W., Joshu, C., Kohsaka, A., Lin, E., Ivanova, G., McDearmon, E., et al. (2005). Obesity and metabolic syndrome in circadian clock mutant mice. Science 308, 1043–1045. doi: 10.1126/science.1108750
Wang, J., and Lazar, M. A. (2008). Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol. Cell. Biol. 28, 2213–2220. doi: 10.1128/MCB.01608-07
Woon, P. Y., Kaisaki, P. J., Braganca, J., Bihoreau, M. T., Levy, J. C., Farrall, M., et al. (2007). Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc. Natl. Acad. Sci. U. S. A. 104, 14412–14417. doi: 10.1073/pnas.0703247104
Xiang, F., Shuanglun, X., Jingfeng, W., Ruqiong, N., Yuan, Z., Yongqing, L., et al. (2011). Association of serum 8-hydroxy-2'-deoxyguanosine levels with the presence and severity of coronary artery disease. Coron. Artery Dis. 22, 223–227. doi: 10.1097/MCA.0b013e328344b615
Yang, S., Liu, A., Weidenhammer, A., Cooksey, R. C., McClain, D., Kim, M. K., et al. (2009). The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology 150, 2153–2160. doi: 10.1210/en.2008-0705
Keywords: CYP7A1, circadian clock, cholesterol, sleep disturbance, NR1D1
Citation: Xing C, Huang X, Zhang Y, Zhang C, Wang W, Wu L, Ding M, Zhang M and Song L (2020) Sleep Disturbance Induces Increased Cholesterol Level by NR1D1 Mediated CYP7A1 Inhibition. Front. Genet. 11:610496. doi: 10.3389/fgene.2020.610496
Edited by:
Luoying Zhang, Huazhong University of Science and Technology, ChinaReviewed by:
Mitsuyuki Nakao, Tohoku University, JapanXiaoyue Pan, New York University, United States
Copyright © 2020 Xing, Huang, Zhang, Zhang, Wang, Wu, Ding, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Xing, eGluZ2NoZW54YzAxMjFAMTYzLmNvbQ==; Lun Song, bHVuc29uZzA3NTJAMTYzLmNvbQ==
†These authors share first authorship
 Chen Xing
Chen Xing Xin Huang1
†
Xin Huang1
†