
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 17 February 2021
Sec. Computational Genomics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.609405
This article is part of the Research Topic Computational tools in inferring cancer tissue-of-origin and molecular classification towards personalized cancer therapy, Volume II View all 13 articles
 Yong Cheng1†
Yong Cheng1† Yanxiang Zhang2†
Yanxiang Zhang2† Yuwei Yuan2
Yuwei Yuan2 Jiao Wang1
Jiao Wang1 Ke Liu2
Ke Liu2 Bin Yu1
Bin Yu1 Li Xie1
Li Xie1 Chao Ou-Yang1
Chao Ou-Yang1 Lin Wu2*
Lin Wu2* Xiaoqun Ye1*
Xiaoqun Ye1*The poor prognosis and fewer treatment option is a current clinical challenge for patients with lung adenosquamous carcinoma (ASC). The previous studies reported that tumor mutational burden (TMB, numbers of mutation per Megabase) is a predictor of clinical response in trials of multiple cancer types, while fewer studies assessed the relationship between TMB level and clinical features and outcomes of lung ASC. Herein, the present study enrolled Chinese patients with lung ASC. DNA was extracted from formalin-fixed paraffin-embedded tumor samples and subjected to next generation sequencing (NGS), and the 457 cancer related genes were evaluated. The results demonstrated that 95 unique genes with somatic variations were identified in the enrolled patients. The top three of high frequency gene mutations were TP53, EGFR, PIK3CA with rates of 62% (13 cases), 48% (10 cases), and 14% (3 cases), respectively. We identified TMB value was significantly correlated with pathological stages (p < 0.05) and invasion of lymph node (p < 0.05). However, TMB value was not significantly correlated to other clinicopathologic indexes, for examples, age, sex, smoking history, tumor size, as well as TP53 and EGFR mutations in lung ASC. Moreover, TMB value was associated with the overall survival (p < 0.01), but not with the relapse-free survival (p = 0.23). In conclusion, this study indicated that lung ASC with high TMB might be associated with the invasion of lymph node and short overall survival. Immunotherapy might be a promising treatment option for lung ASC patients with high TMB.
Worldwide, lung cancer is the most prevalent cause of cancer related death (Siegel et al., 2018). Adenosquamous carcinoma (ASC) is a small subtype of non-small-cell lung cancer (NSCLC), accounting for <4% of all patients with NSCLC (Uramoto et al., 2010; Li and Lu, 2018). It is defined that lung ASC is a mixed-type tumor, comprised of adenocarcinoma and squamous cell carcinoma. Each component accounts for at least 10% of the total tumor cells, according to the tumor classification by the fifth edition of world health organization (WHO) (Travis et al., 2015). It is reported that lung ASC displays the worse prognosis than other types of NSCLC. Lung ASC is resistant to the treatment of adjuvant chemotherapy, and more probably to occur local recurrence or distant metastasis in comparison with other histologic types of NSCLC (Hsia et al., 1999; Nakagawa et al., 2003; Maeda et al., 2012).
In recent years, the important advancements have been achieved in NSCLC treatments (Herbst et al., 2018; Testa et al., 2018). For example, the small molecule tyrosine kinase inhibitors (TKIs) were effective for patients with advanced lung adenocarcinoma with the somatic mutation of epidermal growth factor receptor (EGFR) and the rearrangement of echinoderm microtubule-associated protein-like 4 (EML4) with anaplastic lymphoma kinase (ALK) (Paez et al., 2004; Soda et al., 2007; Robichaux et al., 2018; Ramalingam et al., 2020). Interestingly, a few case reports and retrospective studies have demonstrated that EGFR-TKIs therapies were effective for the selected patients with advanced ASC of the lung (Song et al., 2013; Kurishima et al., 2014; Fan et al., 2017; Zhang et al., 2018; Lin et al., 2020). Therefore, besides of EGFR mutation, the continued research is required to identify more cancer-related gene mutations and the corresponding targeted agents or combined therapies to improve outcomes for lung ASC.
Immune checkpoint inhibitor (ICI) therapies have shown significant benefit in treatment of patients with NSCLC (Herbst et al., 2018), for example, pembrolizumab treatment achieved better clinical outcomes compared to platinum-based chemotherapy in advanced NSCLC patients with high expression of programmed death ligand 1 (PD-L1) in tumor cells (Herbst et al., 2016; Reck et al., 2016). Besides of programmed death 1 (PD-1) and its ligand PD-L1, the resent studies indicated that tumor mutational burden (TMB) could predict clinical outcomes in multiple cancer types, including lung cancer patients receiving immunotherapy (Rizvi et al., 2015, 2018; Samstein et al., 2019). However, there is still lack of prospective data and retrospective study to comprehensively depict the genomic landscape and immune biomarkers, as well as their association with the clinicopathologic features in patients with lung ASC.
To address the limited knowledge, we performed this study in patients with surgically resected lung ASC to evaluate (1) the genomic variations and its correlation with TMB and PD-L1 expression and (2) the clinical relevance of TMB and PD-L1 expression, including clinicopathologic features, relapse-free survival (RFS), and overall survival (OS). Meanwhile, we compared the data of lung ASC to other ethnicities as well as other subtypes such as adenocarcinoma and squamous cell carcinoma.
All the enrolled patients with lung adenosquamous carcinoma (ASC) underwent surgical resection from the Second Affiliated Hospital of Nanchang University between April, 2014 and May, 2019. The criteria of the enrolled patients were as follows: (1) pathological diagnosis of lung ASC according to the tumor classification in the fifth edition of WHO, each component of adenocarcinoma and squamous cell carcinoma at least 10% of the tumor cells; (2) patients without anticancer treatment before surgery; (3) availability of complete medical records, including patient's age, gender, smoking history, immunohistochemistry results, pathological reports, operation time and surgical approach, medication records, tumor response assessment. All the enrolled patients accepted and signed the informed consent, the protocol was approved by the Ethics Committee of medical research, the Second Affiliated Hospital of Nanchang University.
After surgical resection, tumor tissues and normal tissues (incision margin 5 cm away from the tumor) were fixed with formalin, subsequently embedded in paraffin (FFPE). Genomic DNA was extracted from each FFPE sample using the GeneRead DNA FFPE Kit (Qiagen, USA) according to the manufacturer's protocol, respectively.
To construct the pre-library, genomic DNA was digested into ~200 bp fragments by enzymatic method, then subjected to end repairing, A-tailing, adapter ligation and universal amplification. Purified pre-library was hybridized with a customized biotin probe pool (the 457 genes panel, Berry Oncology, Peking, China) to capture target fragments (Supplementary Table 1). Captured fragments were amplified with universal primers and purified to acquire the final library. The library of paired-end multiplex samples were sequenced with the NovaSeq 6000 System. Sequencing depth was ~2,000 x per sample.
The generated sequences were trimmed, low-quality-filtered, and subjected for variant calling. Variants was filtered for nonsynonymous SNPs, indels and spliced variants. Somatic variations were identified with variant allele frequency (cutoff ≥ 3%) and cancer hotspots were screened with variant allele frequency (cutoff ≥ 1%) and at least 20 high-quality reads.
The tumor mutation burden (TMB) was determined by the number of all the nonsynonymous mutation and indel variants per magabase of coding regions. The 457 gene panels cover the coding region of 1,141,951 bp. Hence, TMB was calculated with the number of all the nonsynonymous mutations and indel variants/1.14 Mb. The threshold of high TMB was set to 10 according to the previous studies (Hellmann et al., 2018; Barroso-Sousa et al., 2020).
Immunohistochemistry assays were performed on FFPE sections using a primary anti-PD-L1 (SP263) rabbit monoclonal antibody (Roche) and a secondary anti-rabbit-IgG antibody (ZSGB-BIO, Beijing, China), then detected with DBA detection kit (ZSGB-BIO, Beijing, China). PD-L1-positive was determined if membrane staining exhibited in ≥ 25% of tumor cells in the tumor sample, as described in the previous study (Shi et al., 2017).
R Foundation for Statistics Computing, R script (v3.6.0) was used to perform the statistics analysis. Fisher's exact test was used to analyze the relationship between TMB and clinical indexes. Kaplane-Meier method was used to estimate progress-free survival and overall survival. p < 0.05 was defined as statistically significant.
In total, 21 patients with lung adenosquamous carcinoma (ASC) were finally enrolled in this study. The median age at diagnosis was 63 years (range: 49–75), eleven were male and ten were female, seven were smokers and 14 were non-smokers. The tumors were stage I, II, and III in nine (42.9%), three (14.3%), and nine (42.9%) cases, respectively. Ten patients were with adeno cells accounted for 10%-50% and eleven patients were with adeno cells accounted for 51–90%. The clinical information of patients was overviewed in Table 1. The characteristics of each patient were listed in Supplementary Table 2.
To discover somatic variation in ASC, DNA was extracted from formalin-fixed paraffin-embedded samples and subjected to NGS based large-gene panel test. The somatic mutations of each tumor sample were analyzed and summarized in Figure 1 and Supplemental Table 3. Our study identified 95 unique genes with somatic variations. Among those, the top three of high frequency gene mutations were TP53, EGFR, PIK3CA with rates of 61.9% (13 cases), 47.6% (10 cases), and 14.3% (3 cases), respectively. Coexisting mutations were detected in TP53 and EGFR with rate of 33.3% (7 cases), EGFR, and PIK3CA with rate of 4.8% (1 case).
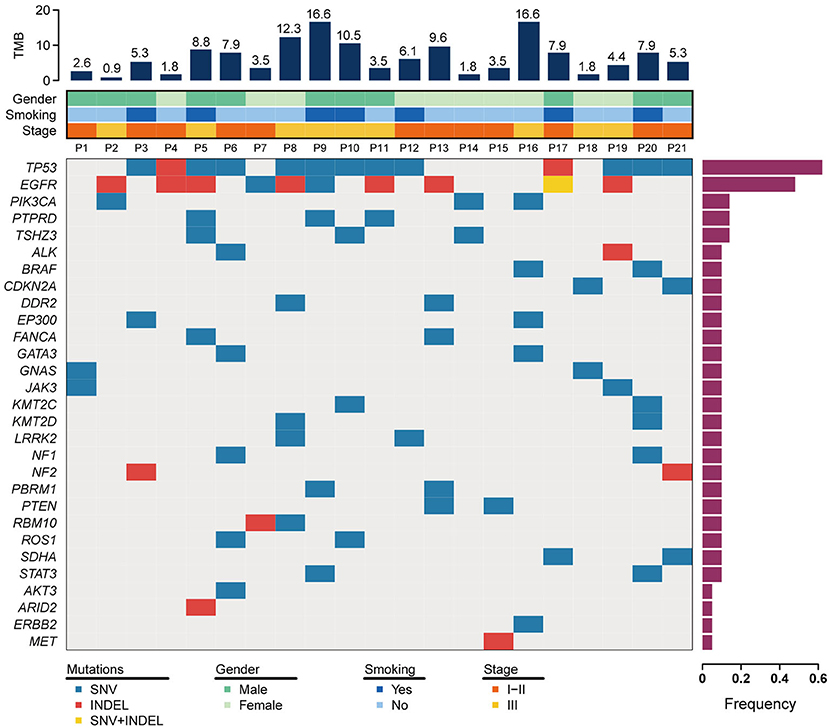
Figure 1. Somatic mutation landscape for 21 patients with lung ASC. The top 30 mutant genes are displayed. The frequency of mutated genes are shown in a histogram on the right. Tumor mutation burden (TMB) and clinical features are illustrated in the upper two panels. SNV+INDEL represents the case carrying both of mutation types of SNV and INDEL.
Mutations of the TP53 gene are universal in lung cancer, with mutation rate of about 50% in NSCLC (Mogi and Kuwano, 2011). In lung ASC, our study indicated TP53 was a highest frequency mutation gene, with a mutation rate of 62%. The common mutation was detected in exon 8 (5 cases), exon 7 (4 cases), exon 6 (2 cases), and exon 4 (1 case). The hotspot mutations of TP53 p.R248Q and TP53 p.R248W were detected in p11 and p19, respectively, which are the target of APR-246 drug. At present, FDA has approved APR-246 in combination therapies to treat myelodysplastic syndromes with TP53 mutation, while more clinical trials are required to prove the efficacy of the drug in treating patients with lung cancer.
For EGFR gene, deletion in exon 19 was the most common mutation (7 cases), and the single nucleotide variation in exon 21 resulting in EGFR p.L858R variant was observed only in one case. These mutations are related to the increase of sensitivity to tyrosine kinase inhibitors. The drug resistant mutation was detected in one case harboring insertion mutation in exon 20, while none of T790M variant was observed in this study. Two variants of in-frame deletion in exon 19 and single nucleotide variation resulting in EGFR p.E758D were coexisted in one case, as shown in Figure 1 and Supplemental Table 3.
In addition to EGFR mutations, a set of genes involved in the PI3K signaling pathway were observed in seven lung ACS cases in our study. Two cases (p2 and p16) harbored a single nucleotide variation in exon 9 of PIK3CA, resulting in a p.E545K variant in the helical region, and one case (p14) harbored a mutation in exon 21 of PIK3CA which generated a p.H1047L variant in p110α catalytic subunit. The variant of PTEN p.H123Y was present in one case (p13). These variants were sensitive to class I PI3K inhibitor. We also found PIK3C2B p.E545K variant in one case (p9), and PIK3C2G p.M1047I variant in another case (p3), both variants belong to class II PI3K. In addition, gene mutations were observed in CDKN2A in two cases (p18 and p21), NF1 in two cases (p6 and p20), DDR2 in two cases (p8 and p13), PBRM1 in two cases (p9 and p13), WHSC1L1 in one case (p10), IRF4 in one case (p16), and HRAS in one case (p21).
The rearrangement of anaplastic lymphoma kinase (ALK), c-ros oncogene 1, receptor tyrosine kinase (ROS1), and ret proto-oncogene (RET) play a role on driving the occurrence and development of NSCLC (Takeuchi et al., 2012; Cancer Genome Atlas Research, 2014). These gene translocations were detected by NGS based large-gene panel test and RNA amplification assays in the present study. However, none of ALK, ROS1, and RET rearrangements were detected in the enrolled patients with lung ASC (data not shown).
The somatic copy number alterations (CNA) play an important role in the development of lung cancer. In this study, CNA was detected in the enrolled patients with lung ASC. The results indicated that the amplification of six genes in tumor tissues were at least twice than in the normal tissues (Figure 2 and Table 2). Among those, the amplification of cyclin dependent kinase 4 (CDK4) was observed in two cases (p7 and p12). Meanwhile, the coexisting amplification of cyclin D1 (CCND1) and CDK4 were detected in one case (p12). CDK4 and CCND1 are very important components involved in regulating the progress of G1/S phase in cell cycle. The complex of CKD4 and CCND1 has been studied as a therapeutic target for cancer (Malumbres and Barbacid, 2009; Musgrove et al., 2011). In addition, the tyrosine kinase receptors EGFR and FGFR1 were amplified for eleven times in p8 and p10 cases, respectively. The present study also indicated p1 and p7 cases harboring MDM2 and MYCN with copy number gains, respectively. Overexpression or amplification of MDM2 occurs in a variety of cancer types.
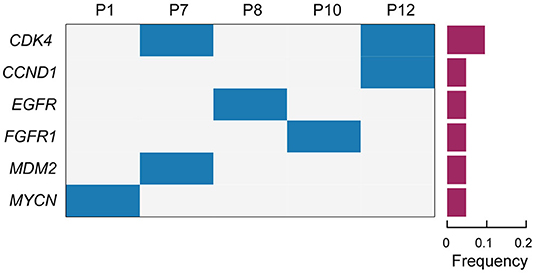
Figure 2. The profiles of somatic copy number alterations for the enrolled patients. Cases with somatic copy number alterations (CNA) are shown. Patient numbers are displayed in the upper lane. Gene names are listed on the left. The frequency of CNA are shown in a histogram on the right.
Immune checkpoint inhibitor (ICI) therapies have earned its spurs in the treatment of malignant tumors in recent years (Gandhi et al., 2018). Tumor mutation burden (TMB) is a promising marker to predict survival after immunotherapy across multiple cancer types (Samstein et al., 2019). In our study, TMB value was determined in the enrolled patients. The results indicated that the median TMB was 5.25 mutations per megabases, with a range from 0.88 to 16.64 (Figure 1). We analyzed the association between TMB value and the proportion of adeno and squamous cells carcinoma of ASC (Figure 3). The results indicated that the high level of TMB was not significantly related to the high proportion of squamous cells in ASC of the lung. Meanwhile, the relationships between TMB level and the clinicopathologic features of adenosquamous cell carcinoma of the lung were analyzed. The results demonstrated that TMB value correlated significantly with pathological stages (p = 0.03) and lymph node (p = 0.03). The higher TMB value (cutoff ≥ 10 mut/Mb) was related to invasion of lymph node, while the lower TMB value (cutoff < 10 mut/Mb) was related to none of invasion of lymph node. However, there was no significant relationship between TMB and other clinicopathologic indexes, for instances, age, sex, smoking history, as well as TP53 and EGFR mutations in lung ASC (Table 3). It was no distant metastasis in the enrolled patients. Therefore, we did not analyze the relationship between TMB level and the index of distant metastasis.
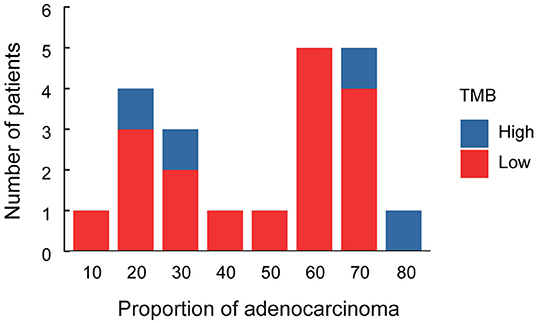
Figure 3. Histogram of proportion of adenocarcinoma cells in lung ASC. Red column represents patients with the low level of TMB, blue column represents patients with the high level of TMB.
In this study, the enrolled patients underwent operation to completely resect the primary tumor tissues and accomplish the lymphadenectomy. In total, five (23.8 %) of 21 patients did not occur disease recurrence, 14 (66.7%) of 21 patients had local recurrence or distant metastasis, two (9.5) of 21 patients were out of contact. The Kaplan–Meier survival curves of relapse-free survival, and overall survival were displayed in Figure 4, with the median of 6 and 10 months, respectively. Among the enrolled patients, one case (p8) harboring EGFR exon 19del and amplification of EGFR, received the EGFR-TKI therapy. The patient developed brain metastases after 14 months of treatment with gefitinib. None of the enrolled patients treated with immune checkpoint inhibitor therapies.
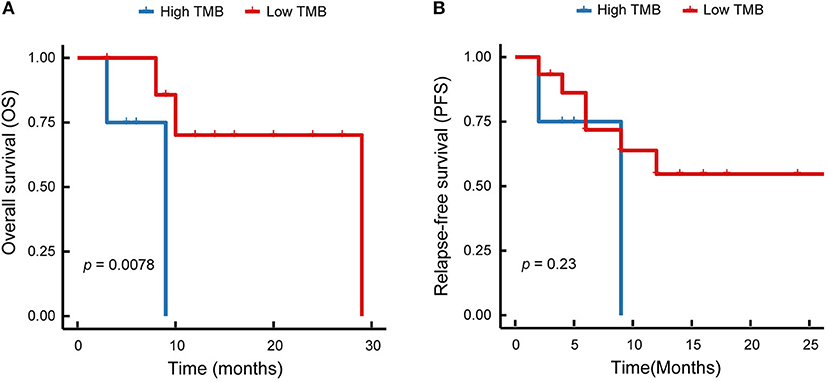
Figure 4. Kaplan–Meier survival curves of patients with lung ASC. (A) Overall survival of lung ASCs with high TMB and low TMB are shown. (B) Relapse-free survival of lung ASCs with high TMB and low TMB are displayed. Red solid line represents patients with low TMB, blue solid line represents patients with high TMB.
We analyzed the relationship between TMB and survival time. The results demonstrated that TMB value was significantly correlated with the overall survival (p = 0.0078, Figure 4A), but not with the relapse-free survival (p = 0.23, Figure 4B). As shown in Figure 4, the high level of TMB was related to the short survival time. Therefore, immunotherapy might be a promising treatment option to improve the outcomes in lung ASC patients with high TMB.
The poor prognosis and fewer treatment option is still a clinical challenge for lung ASC. So far, a few studies introduced the mutational profile of lung ASC (Sasaki et al., 2007; Tochigi et al., 2011; Morodomi et al., 2015; Vassella et al., 2015; Shi et al., 2016; Lin et al., 2020), while most analyses were restricted to small gene panels. The continued studies are required to investigate genetic alterations and explore the potential therapies for lung ASC. The present study displayed the comprehensive analyses of somatic variations in lung ASC. In addition, it is the first study to reveal the clinical relevance of TMB level and PD-L1 expression in lung ASC.
Our study showed a high frequency of EGFR mutations in lung ASC, the mutation rate was 48%. However, in contrast to our observation, a lower prevalence of EGFR mutations was reported in lung ASC of Caucasian ethnic group, with a mutation rate of 13% (Tochigi et al., 2011). That might be due to the ethnicity differences between Asians and Caucasians. Moreover, consistent with the incidence of EGFR mutation in lung adenocarcinoma, the mutation rate was 46.7% in the Asian population and 15% in the white population (Liu et al., 2017). Furthermore, the current study revealed that the landscape of somatic variations of ASC was similar to that of lung adenocarcinomas, and supported the hypothesis that adenocarcinoma components and squamous cell carcinoma components of ASC shared a monoclonal origin (Lin et al., 2020). Therefore, considering the similar profile of somatic variations in ASC and lung adenocarcinoma, TKI might be an effective targeted agents for lung ASC with EGFR mutations. In our study, one resectable patient (pT2aN2M0) received EGFR-TKI therapy after four cycles of adjuvant chemotherapy, and had a clinical benefit from the treatment of gefitinib, with progress-free survival (PFS) of 14 months. In line with the observation, a current multicenter retrospective study also indicated that EGFR-TKIs were effective for patients with advanced ASC of lung, with the median PSF being 10.1 months (Lin et al., 2020). None of ALK, ROS1, and RET rearrangements were detected in our study. In line with our results, the previous study also did not find gene rearrangements in Caucasian patients with lung ASC (Vassella et al., 2015). That might be due to the lower prevalence of gene translocations in lung cancers and the small size of enrolled patients with lung ASC.
In addition, ICI therapies have been applied in treatment of malignant tumors in recent years. We valuated PD-L1 expression in tumor cells of the enrolled patients, while there were no significant associations between PD-L1 expression and the clinicopathologic features of lung ASC (Supplemental Table 4). Besides of PD-L1, TMB is a promising marker to predict clinical outcomes of patients with NSCLC to immunotherapy (Carbone et al., 2017; Hellmann et al., 2018; Samstein et al., 2019). The previous studies indicated lung squamous cell carcinoma harboring higher TMB than other solid cancer types (Vogelstein et al., 2013; Zhang et al., 2019). However, our results indicated that the high level of TMB was not significantly related to the high proportion of squamous cells in lung ASC. The current study displayed that TMB was lower in adenocarcinoma component than in squamous cell carcinoma component, with the median of 6.5 and 7.2 mutations/Mb, respectively (Lin et al., 2020). However, it is difficult to distinguish such small differences of TMB value in adenocarcinoma component and squamous cell carcinoma component.
In the present study, we also evaluated the relationships between TMB level and the clinicopathologic features and outcomes of lung ASC, though none of enrolled patients received ICI therapy. Our results indicated that the high level of TMB was related to the invasion of lymph node and the short survival time. Patients with the short survival time might be due to the invasion of lymph node. In line with the results, the previous study indicated that high TMB is a poor prognostic factor for the advanced NSCLC, as well as patient in early stage (Owada-Ozaki et al., 2018). However, as the limited numbers of enrolled patients, we did not obtain lung ASC with high TMB in early stage.
In conclusion, the lung ASC with high TMB might be associated with invasion of lymph node and short overall survival. Therefore, immunotherapy might be a potential treatment option for lung ASC patients with the high level of TMB.
The datasets presented in this study can be found in online repositories. The accession number is CNP0001586, the link is followed as: https://db.cngb.org/search/project/CNP0001586/.
The studies involving human participants were reviewed and approved by Ethics Committee of Medical Research, the Second Affiliated Hospital of Nanchang University. The patients/participants provided their written informed consent to participate in this study.
XY and LW designed the project. YZ, YY, KL and YC performed the work. YZ wrote the paper. JW, BY, LX and CO-Y reviewed the paper. All authors accepted the final version of this manuscript for publication.
XY is supported by National Nature Science Foundation of China (No. 81660493) and Natural Science Foundation of Jiang Xi Province (No. 20171BAB205053).
YY, LW, and YZ are currently employed by Berry Oncology Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.609405/full#supplementary-material
Barroso-Sousa, R., Jain, E., Cohen, O., Kim, D., Buendia-Buendia, J., Winer, E., et al. (2020). Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann. Oncol 31, 387–394. doi: 10.1016/j.annonc.2019.11.010
Cancer Genome Atlas Research, N. (2014). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. doi: 10.1038/nature13385
Carbone, D. P., Reck, M., Paz-Ares, L., Creelan, B., Horn, L., Steins, M., et al. (2017). First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med 376, 2415–2426. doi: 10.1056/NEJMoa1613493
Fan, L., Yang, H., Yao, F., Zhao, Y., Gu, H., Han, K., et al. (2017). Clinical outcomes of epidermal growth factor receptor tyrosine kinase inhibitors in recurrent adenosquamous carcinoma of the lung after resection. Onco. Targets. Ther. 10, 239–245. doi: 10.2147/OTT.S114451
Gandhi, L., Rodriguez-Abreu, D., Gadgeel, S., Esteban, E., Felip, E., De Angelis, F., et al. (2018) Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med 378, 2078–2092. doi: 10.1056/NEJMoa1801005.
Hellmann, M. D., Ciuleanu, T. E., Pluzanski, A., Lee, J. S., Otterson, G. A., Audigier-Valette, C., et al. (2018). Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 378, 2093–2104. doi: 10.1056/NEJMoa1801946
Herbst, R. S., Baas, P., Kim, D. W., Felip, E., Perez-Gracia, J. L., Han, J. Y., et al. (2016). Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550. doi: 10.1016/S0140-6736(15)01281-7
Herbst, R. S., Morgensztern, D., and Boshoff, C. (2018). The biology and management of non-small cell lung cancer. Nature 553, 446–454. doi: 10.1038/nature25183
Hsia, J. Y., Chen, C. Y., Hsu, C. P., Shai, S. E., and Wang, P. Y. (1999). Adenosquamous carcinoma of the lung. Surgical results compared with squamous cell and adenocarcinoma. Scand. Cardiovasc. J. 33, 29–32. doi: 10.1080/14017439950142000
Kurishima, K., Ohara, G., Kagohashi, K., Watanabe, H., Takayashiki, N., Ishibashi, A., et al. (2014). Adenosquamous cell lung cancer successfully treated with gefitinib: a case report. Mol. Clin. Oncol. 2, 282–284. doi: 10.3892/mco.2013.221
Li, C., and Lu, H. (2018). Adenosquamous carcinoma of the lung. Onco. Targets. Ther. 11, 4829–4835. doi: 10.2147/OTT.S164574
Lin, G., Li, C., Li, P. S., Fang, W. Z., Xu, H. P., Gong, Y. H., et al. (2020). Genomic origin and EGFR-TKI treatments of pulmonary adenosquamous carcinoma. Ann. Oncol. 31, 517–524. doi: 10.1016/j.annonc.2020.01.014
Liu, L., Liu, J., Shao, D., Deng, Q., Tang, H., Liu, Z., et al. (2017). Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in East Asian patients. Cancer Sci. 108, 2487–2494. doi: 10.1111/cas.13410
Maeda, H., Matsumura, A., Kawabata, T., Suito, T., Kawashima, O., Watanabe, T., et al. (2012). Adenosquamous carcinoma of the lung: surgical results as compared with squamous cell and adenocarcinoma cases. Eur. J. Cardiothorac. Surg. 41, 357–361. doi: 10.1016/j.ejcts.2011.05.050
Malumbres, M., and Barbacid, M. (2009). Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166. doi: 10.1038/nrc2602
Mogi, A., and Kuwano, H. (2011). TP53 mutations in nonsmall cell lung cancer. J. Biomed. Biotechnol. 2011:583929. doi: 10.1155/2011/583929
Morodomi, Y., Okamoto, T., Takenoyama, M., Takada, K., Katsura, M., Suzuki, Y., et al. (2015). Clinical significance of detecting somatic gene mutations in surgically resected adenosquamous cell carcinoma of the lung in Japanese patients. Ann. Surg. Oncol. 22, 2593–2598. doi: 10.1245/s10434-014-4218-0
Musgrove, E. A., Caldon, C. E., Barraclough, J., Stone, A., and Sutherland, R. L. (2011). Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558–572. doi: 10.1038/nrc3090
Nakagawa, K., Yasumitu, T., Fukuhara, K., Shiono, H., and Kikui, M. (2003). Poor prognosis after lung resection for patients with adenosquamous carcinoma of the lung. Ann. Thorac. Surg. 75, 1740–1744. doi: 10.1016/s0003-4975(03)00022-5
Owada-Ozaki, Y., Muto, S., Takagi, H., Inoue, T., Watanabe, Y., Fukuhara, M., et al. (2018). Prognostic impact of tumor mutation burden in patients with completely resected non-small cell lung cancer: brief report. J. Thorac. Oncol. 13, 1217–1221. doi: 10.1016/j.jtho.2018.04.003
Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., et al. (2004). EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500. doi: 10.1126/science.1099314
Ramalingam, S. S., Vansteenkiste, J., Planchard, D., Cho, B. C., Gray, J. E., Ohe, Y., et al. (2020). Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med 382, 41–50. doi: 10.1056/NEJMoa1913662
Reck, M., Rodriguez-Abreu, D., Robinson, A. G., Hui, R., Csoszi, T., Fulop, A., et al. (2016). Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med 375, 1823–1833. doi: 10.1056/NEJMoa1606774
Rizvi, H., Sanchez-Vega, F., La, K., Chatila, W., Jonsson, P., Halpenny, D., et al. (2018). Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 36, 633–641. doi: 10.1200/JCO.2017.75.3384
Rizvi, N. A., Hellmann, M. D., Snyder, A., Kvistborg, P., Makarov, V., Havel, J. J., et al. (2015). Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128. doi: 10.1126/science.aaa1348
Robichaux, J. P., Elamin, Y. Y., Tan, Z., Carter, B. W., Zhang, S., Liu, S., et al. (2018). Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat. Med. 24, 638–646. doi: 10.1038/s41591-018-0007-9
Samstein, R. M., Lee, C. H., Shoushtari, A. N., Hellmann, M. D., Shen, R., Janjigian, Y. Y., et al. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206. doi: 10.1038/s41588-018-0312-8
Sasaki, H., Endo, K., Yukiue, H., Kobayashi, Y., Yano, M., and Fujii, Y. (2007). Mutation of epidermal growth factor receptor gene in adenosquamous carcinoma of the lung. Lung Cancer 55, 129–130. doi: 10.1016/j.lungcan.2006.09.003
Shi, X., Wu, H., Lu, J., Duan, H., Liu, X., and Liang, Z. (2016). Screening for major driver oncogene alterations in adenosquamous lung carcinoma using PCR coupled with next-generation and Sanger sequencing methods. Sci. Rep. 6:22297. doi: 10.1038/srep22297
Shi, X., Wu, S., Sun, J., Liu, Y., Zeng, X., and Liang, Z. (2017). PD-L1 expression in lung adenosquamous carcinomas compared with the more common variants of non-small cell lung cancer. Sci. Rep. 7:46209. doi: 10.1038/srep46209
Siegel, R. L., Miller, K. D., and Jemal, A. (2018). Cancer statistics, 2018. Cancer J. Clin. 68, 7–30. doi: 10.3322/caac.21442
Soda, M., Choi, Y. L., Enomoto, M., Takada, S., Yamashita, Y., Ishikawa, S., et al. (2007). Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566. doi: 10.1038/nature05945
Song, Z., Lin, B., Shao, L., and Zhang, Y. (2013). Therapeutic efficacy of gefitinib and erlotinib in patients with advanced lung adenosquamous carcinoma. J. Chin. Med. Assoc. 76, 481–485. doi: 10.1016/j.jcma.2013.05.007
Takeuchi, K., Soda, M., Togashi, Y., Suzuki, R., Sakata, S., Hatano, S., et al. (2012). RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–381. doi: 10.1038/nm.2658
Testa, U., Castelli, G., and Pelosi, E. (2018). Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers 10:248. doi: 10.3390/cancers10080248
Tochigi, N., Dacic, S., Nikiforova, M., Cieply, K. M., and Yousem, S. A. (2011). Adenosquamous carcinoma of the lung: a microdissection study of KRAS and EGFR mutational and amplification status in a western patient population. Am. J. Clin. Pathol. 135, 783–789. doi: 10.1309/AJCP08IQZAOGYLFL
Travis, W. D., Brambilla, E., Burke, A. P., Marx, A., and Nicholson, A. G. (2015). Introduction to the 2015 world health organization classification of tumors of the lung, pleura, thymus, and heart. J. Thorac. Oncol. 10, 1240–1242. doi: 10.1097/JTO.0000000000000663
Uramoto, H., Yamada, S., and Hanagiri, T. (2010). Clinicopathological characteristics of resected adenosquamous cell carcinoma of the lung: risk of coexistent double cancer. J. Cardiothorac. Surg. 5:92. doi: 10.1186/1749-8090-5-92
Vassella, E., Langsch, S., Dettmer, M. S., Schlup, C., Neuenschwander, M., Frattini, M., et al. (2015). Molecular profiling of lung adenosquamous carcinoma: hybrid or genuine type? Oncotarget 6, 23905–23916. doi: 10.18632/oncotarget.4163
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A. Jr., and Kinzler, K. W. (2013). Cancer genome landscapes. Science 339, 1546–1558. doi: 10.1126/science.1235122
Zhang, C., Yang, H., Lang, B., Yu, X., Xiao, P., Zhang, D., et al. (2018). Surgical significance and efficacy of epidermal growth factor receptor tyrosine kinase inhibitors in patients with primary lung adenosquamous carcinoma. Cancer Manag. Res. 10, 2401–2407. doi: 10.2147/CMAR.S165660
Keywords: adenosquamous carcinoma, EGFR, lung, next generation sequencing, PD-L1, somatic variations, TMB
Citation: Cheng Y, Zhang YX, Yuan YW, Wang J, Liu K, Yu B, Xie L, Ou-Yang C, Wu L and Ye XQ (2021) The Comprehensive Analyses of Genomic Variations and Assessment of TMB and PD-L1 Expression in Chinese Lung Adenosquamous Carcinoma. Front. Genet. 11:609405. doi: 10.3389/fgene.2020.609405
Received: 23 September 2020; Accepted: 18 December 2020;
Published: 17 February 2021.
Edited by:
Cheng Guo, Columbia University, United StatesCopyright © 2021 Cheng, Zhang, Yuan, Wang, Liu, Yu, Xie, Ou-Yang, Wu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqun Ye, NTExMjAxNjYzQHFxLmNvbQ==; Lin Wu, d3VsaW5AYmVycnlvbmNvbG9neS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.