- College of Horticulture, Henan Agricultural University, Zhengzhou, China
Apple (Malus domestica Borkh.), an economically important tree fruit worldwide, frequently suffers from temperature stress during growth and development, which strongly affects the yield and quality. Heat shock protein 20 (HSP20) genes play crucial roles in protecting plants against abiotic stresses. However, they have not been systematically investigated in apple. In this study, we identified 41 HSP20 genes in the apple ‘Golden Delicious’ genome. These genes were unequally distributed on 15 different chromosomes and were classified into 10 subfamilies based on phylogenetic analysis and predicted subcellular localization. Chromosome mapping and synteny analysis indicated that three pairs of apple HSP20 genes were tandemly duplicated. Sequence analysis revealed that all apple HSP20 proteins reflected high structure conservation and most apple HSP20 genes (92.6%) possessed no introns, or only one intron. Numerous apple HSP20 gene promoter sequences contained stress and hormone response cis-elements. Transcriptome analysis revealed that 35 of 41 apple HSP20 genes were nearly unchanged or downregulated under normal temperature and cold stress, whereas these genes exhibited high-expression levels under heat stress. Subsequent qRT-PCR results showed that 12 of 29 selected apple HSP20 genes were extremely up-regulated (more than 1,000-fold) after 4 h of heat stress. However, the heat-upregulated genes were barely expressed or downregulated in response to cold stress, which indicated their potential function in mediating the response of apple to heat stress. Taken together, these findings lay the foundation to functionally characterize HSP20 genes to unravel their exact role in heat defense response in apple.
Introduction
Temperature is an important factor affecting plant growth and geographical distribution (Wang et al., 2016). Most plants undergo optimal growth and development within a narrow temperature range and can only tolerate minor fluctuations. Fluctuations beyond optimal range result in temperature stress, which is one of the most severe environmental stresses affecting plant growth, development and survival worldwide (Peleg and Blumwald, 2011). High and low temperature stresses have rapid and severe effects on plant cell physiology, altering gene expression, protein levels, and energy consumption (Wang et al., 2017; Shen et al., 2019; Suzuki, 2019). Plants have developed a series of physiological and molecular strategies to overcome temperature stress over evolutionary time (Asea et al., 2016; Huo et al., 2020). Heat shock proteins (HSPs) are one of the strategies, and HSPs are essential in regulating growth, development and stress response in plants (Waters et al., 1996; Wang et al., 2004; Asea et al., 2016; He et al., 2019).
HSPs can be divided into five categories according to their molecular weight: HSP100s, HSP90s, HSP70s, HSP60s, and HSP20s (Waters, 2013; Zhao et al., 2018). Of these groups, HSP20 is commonly associated with temperature stress in plants (Waters, 2013). As genomes for more species are sequenced, the HSP20 gene family has been identified in various plants. Nineteen HSP20 genes have been identified in Arabidopsis (Scharf et al., 2001), 39 in rice (Ouyang et al., 2009), 42 in tomato (Yu et al., 2016), 44 in watermelon (He et al., 2019), and 48 in grape (Ji et al., 2019). Previous studies have suggested that HSP20 genes are involved in regulating a diverse array of developmental processes and responses to abiotic stresses, especially in heat stress (Guo et al., 2015; He et al., 2019; Ji et al., 2019). Yu et al. (2016) identified tomato HSP20 family genes and analyzed their functions in abiotic-stress responses. Most pepper HSP20 genes were highly induced by heat stress (Guo et al., 2015). Among the GmHSP20 genes, five were shown to be involved in the soybean response to cold stress (Lopes-Caitar et al., 2013). Interestingly, the same HSP20 genes exhibited a different expression pattern in the heat tolerant and sensitive plants. These differences in expression pattern indicate the roles of HSP20 in heat tolerance. In addition, some studies have further verified the role of HSP20s in stress tolerance using transgenic methods. For example, overexpressing with WsHSP26 in Arabidopsis showed improved heat tolerance (Mu et al., 2013). Similarly, transgenic rice over-expressing OsHSP17.7 conferred enhanced tolerance to heat stress (Murakami et al., 2004). Together, these studies reveal the crucial role of HSP20 genes in mediating temperature stress tolerance.
Apple (Malus domestica Borkh.), an economically important fruit crop, is widely planted in temperate zones (Dobránszki and Teixeira da Silva, 2010). However, apple trees frequently suffer from both high and low temperature stresses during their life cycle, which strongly affect apple quality and yield. After suffering continuous heat stress in summer, the leaf and fruit of apple can be severely damaged; resulting in tissue discoloration and sunburn of the fruit surface (Torres et al., 2017). It is reported that fruit sunburn causes 10–40% yield losses in all major apple growing regions around the world (Wang et al., 2020). After suffering cold stress in early spring, the pollination, new leaves and shoots of apple can be severely damaged, thereby greatly reducing the yield and quality of apple. The entire genome of apple has been sequenced, providing powerful resource for the mining and identification of HSP20 gene family members at the whole genome level.
In the present study, we identified HSP20 genes from the apple genome using bioinformatics methods, and determined their chromosomal locations, gene duplication, phylogenetic relationships, gene structures, and conserved domains, as well as cis-elements. Furthermore, we analyzed the expression patterns of the apple HSP20 genes using qRT-PCR in order to determine their roles in response to heat and cold stresses. Our findings provide valuable information for subsequent research on the functions and regulatory mechanisms of potentially important HSP20 genes that are crucial in modulating heat stress tolerance in apple.
Materials and Methods
Plant Materials and Stress Treatments
Seeds of M. hupehensis were stratified using sand at 4°C for 40 days, as previously described (Bai et al., 2013). Then, one germinated seed was planted in one plastic pot (6 cm × 6 cm) filled with soil/organic substrate (1:5, v: v) in a greenhouse under natural light, 22–28°C (day) and 5–10°C (night), and a relative humidity of 60–70%. After growing for 70 days, uniform seedlings were randomly divided into three groups and transferred to different chambers for temperature stress: (1) control (CK), growth chamber were maintained at 25 ± 1°C, 16-h photoperiod (160 μmol m–2 s–1) and relative humidity of 60–70%; (2) heat stress (HS), growth chamber maintained at 40 ± 1°C; (3) cold stress (CS), growth chamber maintained at 4 ± 1°C, 16-h photoperiod (160 μmol m–2 s–1) and relative humidity of 60–70%. Each treatment contained three biological replicates and 30 plants for each replicate. At 0, 4, 8, 12, and 24 h after treatments, the samples were rapidly frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Genome-Wide Identification of the HSP20 Genes in Apple
The reference apple genome and protein sequences were downloaded from the Genome Database for Rosaceae (GDR1). The apple HSP20 candidates with an e-value ≤ 0.001 were identified based on the Hidden Markov Model (HMM) profile (PF00011) downloaded from Pfam protein family database2. The SMART database3 was used to further confirm the conserved HSP20 gene domain. ProtParam4 was used to predict the potential chemical characteristics of the HSP20 genes. ProtComp5 was used to predict the subcellular localization.
Phylogenetic Analysis and Classification of Apple HSP20 Genes
The full-length amino acid sequences of HSP20 genes derived from Arabidopsis and rice downloaded from the Ensembl Plants Database6 were combined with newly identified HSP20 genes in apple and used for phylogenetic analysis. The phylogenetic tree was constructed using MEGA 6.07.
Structure and Domain Analysis of Apple HSP20 Genes
The structures of HSP20 genes were identified using TBtools software (Chen et al., 2018). The conserved motifs of HSP20 genes were identified using MEME Suite 5.1.18, and the parameters were as follows: optimum motif width ranges from 6 to 200 amino acid residues and maximum of 10 misfits. The upstream 2.0 kb promoter sequence of the apple HSP20 genes was downloaded from the GDR and submitted to PlantCARE9 to identify the cis-elements (Lescot et al., 2002).
Chromosomal Location and Synteny Analysis
All identified HSP20 genes were mapped to apple chromosomes using TBtools software based on the information available at GDR. Synteny analysis of HSP20 genes was conducted using Circos v. 0.6310.
RNA Extraction, cDNA Library Construction, and Sequencing
Total RNA was extracted from the leaf tissues according to the CTAB method (Chang et al., 1993). Each sample was 0.5 g and three biological replicates were performed. RNA concentrations were determined using a NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, United States) and quality was assayed on a 1% agarose gel. The sample libraries were prepared according to the RNA-Seq library constructed flow path and sequenced on an Illumina HiSeq 4000 system. The raw sequence data from the sequence was used for analysis. After filtering the low quality reads and contaminant sequences, the clean reads were aligned to the apple genome (GDDH13 Version 1.111) (Daccord et al., 2017) using HISAT2 (Kim et al., 2015). Stringtie software was used to assemble the transcript (Pertea et al., 2016). Gene expression was calculated using the fragments per kilobase of transcript per million (FPKM) fragments mapped Reads method (Mortazavi et al., 2008). TBtools was used to generate the heatmap. The RNA-seq data were available at NCBI12.
qRT-PCR Analysis
Quantitative real-time PCR (qRT-PCR) was used to analyze the gene expression. Primers (Supplementary Table S1) were designed to amplify products of 150–250 bp using Primer 5.0 software. qRT-PCR was performed using ABI-7500 Connect Real-Time PCR Detection System. cDNAs were diluted to 200 ng and run in three technical replicates, with 1 μL template in a reaction volume of 20 μL. PCR amplification conditions were as follows: 95°C for 5 min for initial denaturation, then 45 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 10 s. Fluorescence was measured at the end of each cycle. A melting curve analysis was performed to determine whether a single product was amplified. The apple Actin gene was used as an internal standard in the analysis. The relative expression level of each gene was calculated according to the 2–ΔΔCT method (Livak and Schmittgen, 2001). Values for mean expression and standard error (SE) were calculated from the results of three independent biological replicates.
Results
Genome-Wide Identification of HSP20 Genes in Apple
A total of 45 HSP20 protein sequences were found in the apple cultivar “Golden Delicious” genome. Among 45 HSP20 sequences, four sequences lacked the conserved domain. Ultimately, 41 sequences were identified as genes in the apple HSP20 family and named HSP20-1 to HSP20-41 based on the position of the genes on the chromosomes (Supplementary Table S2). Gene name, gene ID, chromosomal location, open reading frame (ORF), amino acid (AA), molecular weight (MW) and isoelectric point (pI) for each gene are in Table 1. Sequence analysis showed that these HSP20 proteins vary widely in length, from 88 (HSP20-32) to 363 (HSP20-4) AAs, and predicted MWs from 9.98 kDa (HSP20-32) to 39.47 kDa (HSP20-4). The ORF lengths of the HSP20 genes ranged from 267 bp (HSP20-32) to 1,092 bp (HSP20-4), and the predicted pI-values of HSP20 proteins ranged from 4.60 (HSP20-5) to 9.85 (HSP20-10).
Phylogenetic Relationships of Plant HSP20 Genes
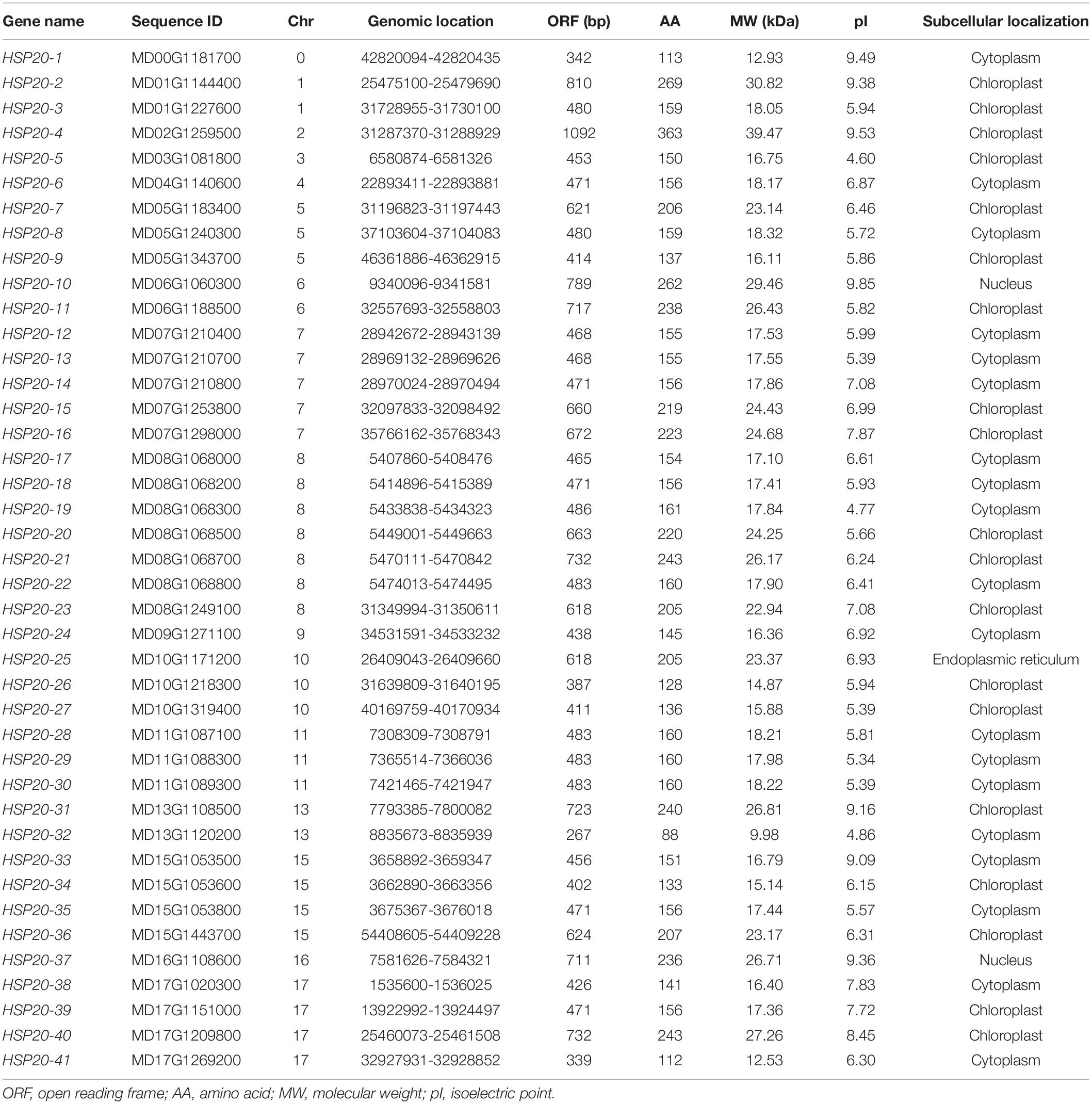
A phylogenetic tree was constructed based on the amino acid sequences of HSP20 genes (Figure 1), 19 from Arabidopsis, 22 from rice (Oryza sativa) and 41 from apple (M. domestica) (Supplementary Table S3). The 82 HSP20s were divided into 12 subfamilies, 25 cytosol Is (CIs), 14 CIIs, 4 CIIIs, 2 CIVs, 4 CVs, 1 CVI, 1 CVII, five mitochondria Is (MIs), three MIIs, four peroxisomes (Pos), 9 from the endoplasmic reticulum (ER), and 8 plastids (Ps) based on the phylogeny and the subcellular localization. However, two apple HSP20 genes (HSP20-4 and HSP20-11) failed to cluster into any subfamily and were thus unclassified. Of the 12 subfamilies, 10 (CIs, CIIs, CIIIs, CIVs, CVs, MIs, MIIs, Pos, ER, and Ps) contained apple HSP20 proteins. Except for the two unclassified HSP20s, 23 (59%) HSP20s were classified into CI-CVI, indicating that cytoplasm could be the primary functional area of the HSP20 family in apple.
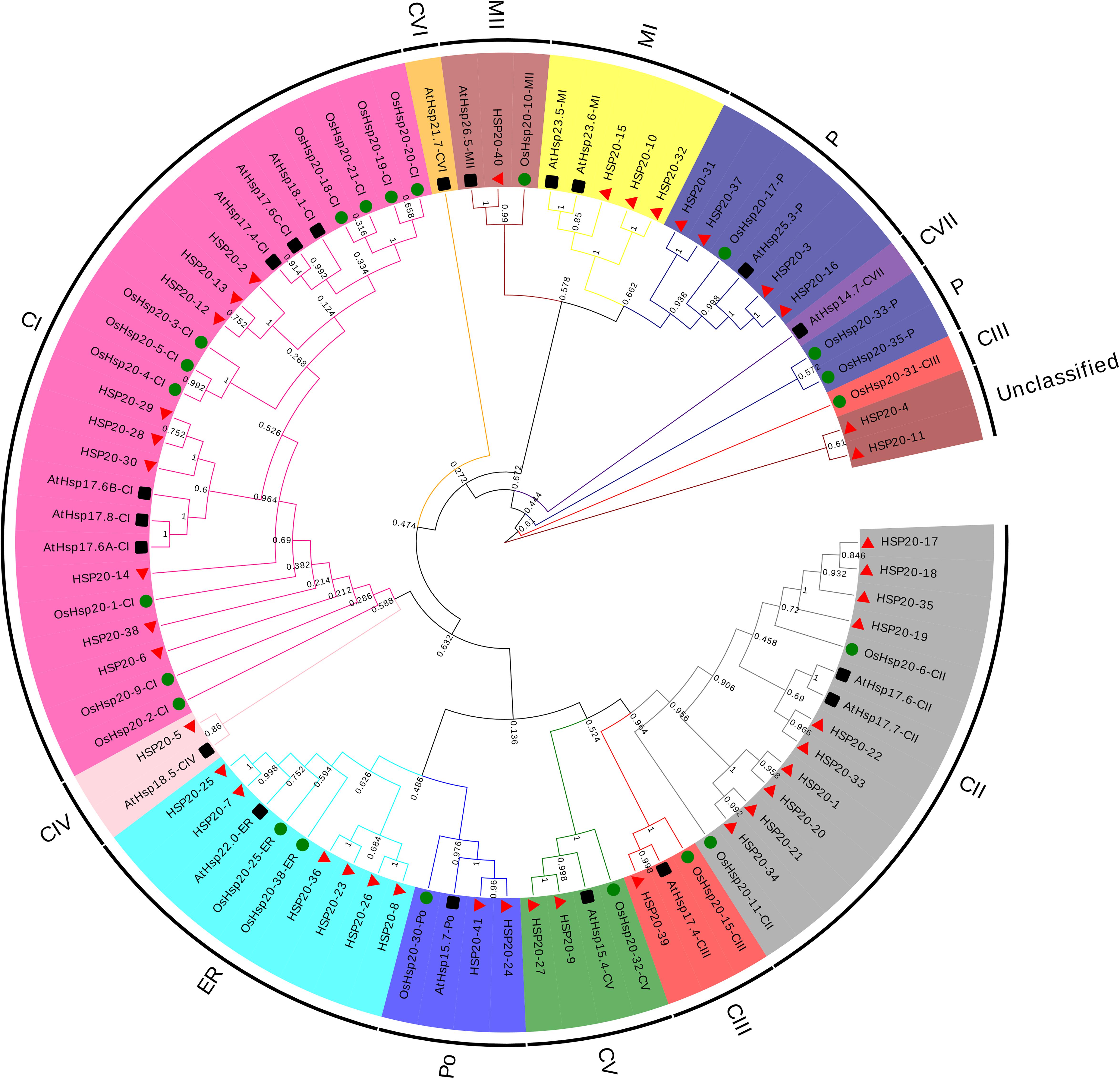
Figure 1. Phylogenetic analysis of HSP20 proteins from Arabidopsis, rice and apple. The phylogenetic tree was constructed using MEGA 6.0 based on the neighbor joining method with 1000 bootstrap replicates. The 12 subfamilies were distinguished with different colored arcs.
Conserved Motifs and Gene Structure of HSP20 Genes
The conserved motifs of apple HSP20 gene family were identified and divided into 10 motifs (Figure 2b). The lengths of the 10 motifs ranged from 6 to 50 AAs, with the longest motif (9) containing 50 AAs and the shortest motif (8) containing six AAs; motifs 4, motifs 5, and motifs 6 have 15 AAs (Supplementary Table S4). The number of the conserved motifs for each HSP20 gene ranged from 2 to 7. Most apple HSP20 genes had two to seven conserved motifs, however, HSP20-32 only contained two conserved motifs. The results suggested that the HSP20 genes exhibited extreme divergence during the evolutionary process.
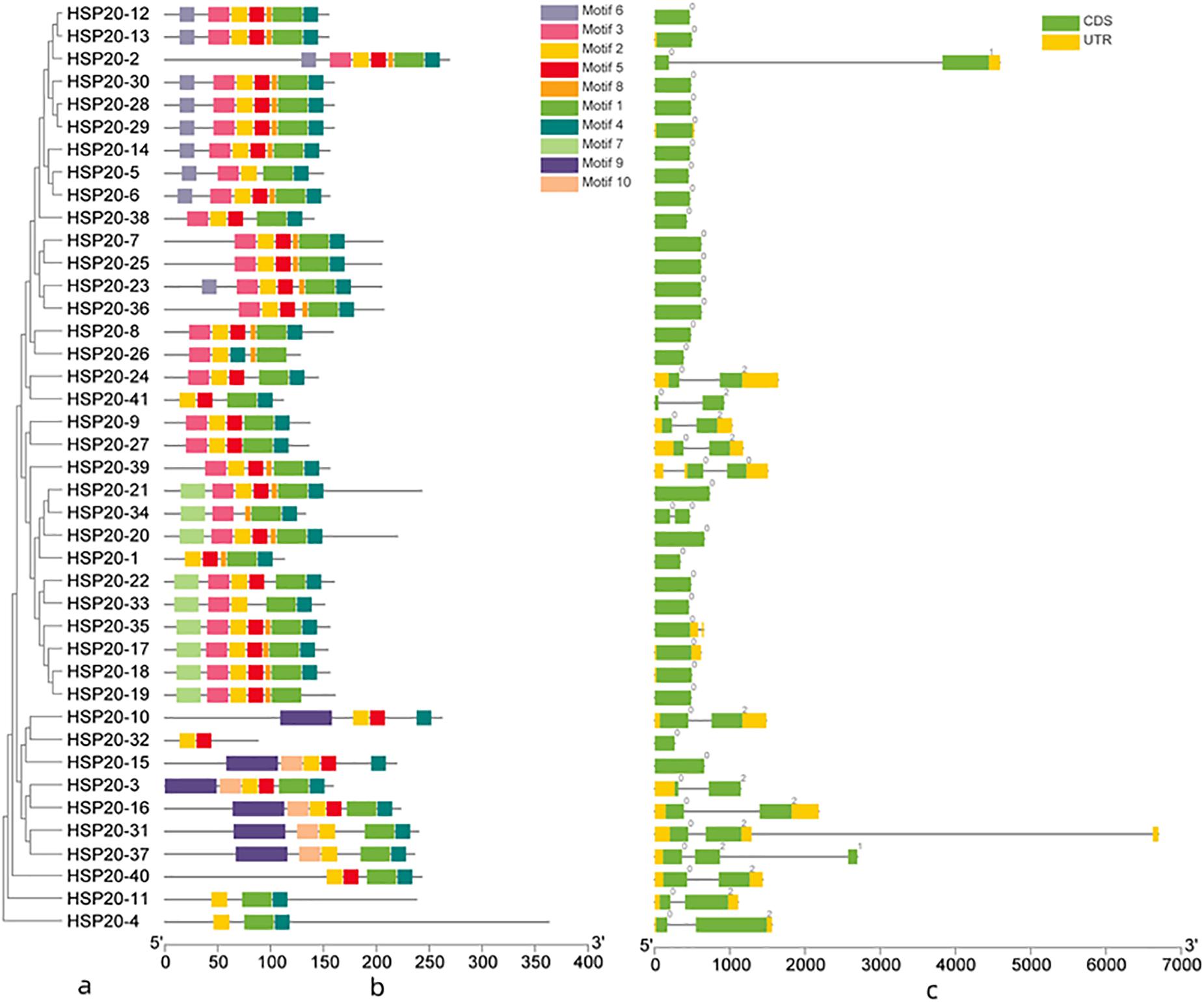
Figure 2. Phylogenetic tree, gene structure and domain analyses of HSP20s. (a) Phylogenetic tree of HSP20s. (b) Domain analyses of HSP20 proteins. (c) Gene structure of HSP20 genes. CDS sequences are represented with yellow round-corner rectangles and introns with gray lines, UTRs are shown with green boxes. Different colored boxes represent the different types of motifs.
To gain insight into the evolutionary relationships of apple HSP20 genes, the exon–intron structure of the HSP20 genes was analyzed (Figure 2c). Among the HSP20s, 25 (60.9%) were intronless, 13 (31.7%) possessed one intron, and three genes (7.3%) – HSP20-31, HSP20-37, and HSP20-38 – possessed two introns. Most HSP20 genes thus have no introns or only one intron, suggesting relatively simple gene structures. Gene structure analysis showed that the genes with similar exon–intron patterns were grouped in the same cluster (Figure 2a).
Chromosomal Location, Gene Duplication, and Synteny Analysis
A total of 41 apple HSP20 genes were mapped on 15 chromosomes (Chr), except Chr 12 and 14, with an obviously non-uniform distribution (Figure 3). One HSP20 gene (HSP20-1) could not be mapped on any of the apple chromosomes, so we mapped it on a pseudo-chromosome, named Chr00. In addition, most of the apple HSP20 genes were located on the distal ends of the chromosomes. The biggest cluster was seven HSP20 genes together on Chr 8, whereas the fewest HSP20s were found on Chrs 0, 2, 3, 4, 9, and 12 (one per Chr). Moreover, we analyzed the duplication events of apple HSP20 genes (Figure 4). In total, 37 (90.2%) HSP20 genes in apple exhibited segmental or tandem duplication. Twenty were segmentally duplicated and three pairs of genes (HSP20-13 and HSP20-14, HSP20-18 and HSP20-19, HSP20-21, and HSP20-22) were regarded as tandemly-duplicated genes (Supplementary Table S5). Chr 7 had the most duplication events, which could partly explain the larger numbers of HSP20 genes on Chr 7, while Chr 0 and 12 did not contain any duplicated genes.
Figure 3. Distribution of HSP20 genes in apple chromosomes. Forty-one HSP20 genes was mapped to the 16 linkage groups (Chr 01 through Chr 17, except Chr 12 and Chr 14), whereas one HSP20 gene were mapped on apseudo-chromosome, designated as Chr00.
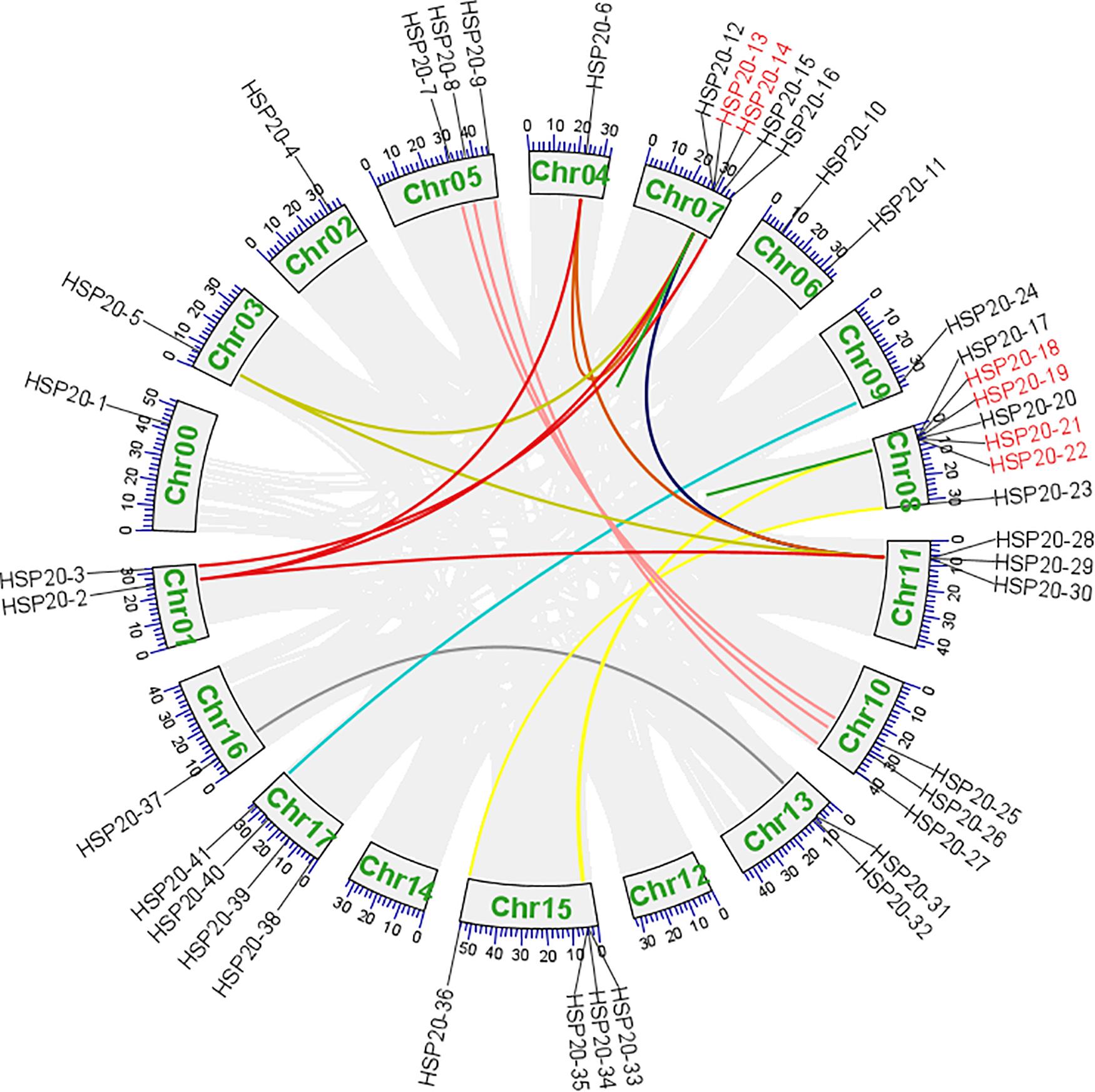
Figure 4. Syntenic relationships and gene duplications of the apple HSP20 genes. The segmental duplicated genes are indicated by differently colored lines and tandem duplicated genes are indicated by red gene names.
Analysis of Cis-Element in Apple HSP20 Gene Promoters
To further investigate the potential regulatory mechanisms of the apple HSP20 genes in response to temperature stress, the promoter in the upstream 2 kb region of 41 HSP20 genes was analyzed to detect the cis-regulatory element. The results showed that three categories of cis-elements, including stress-related (heat, defense and stress, low-temperature and light), hormone-related (abscisic acid, auxin, gibberellin, MeJa, and salicylic acid), and plant development-related cis-elements (meristem expression and circadian control), were identified (Figure 5a). Among the stress-related cis-elements, 13 apple HSP20 genes had the heat response elements (HRE) in their promoter regions, 26 apple HSP20 genes had the low temperature response elements (LTR), which suggests a potential stress response under temperature conditions (Figure 5b). Among the hormone-related cis-elements, abscisic acid responsive (ABRE), salicylic acid responsiveness (TCA-element), auxin responsive (AUXRR-core), and MeJA-responsiveness (CGTCA-motif) were identified in the promoters of apple HSP20 genes. All HSP20 genes contained light signal response elements, which indicate that HSP20s are essential in plant growth. The results indicate that the HSP20 gene family is not only involved in the stress response, but is also involved in other physiological response processes.
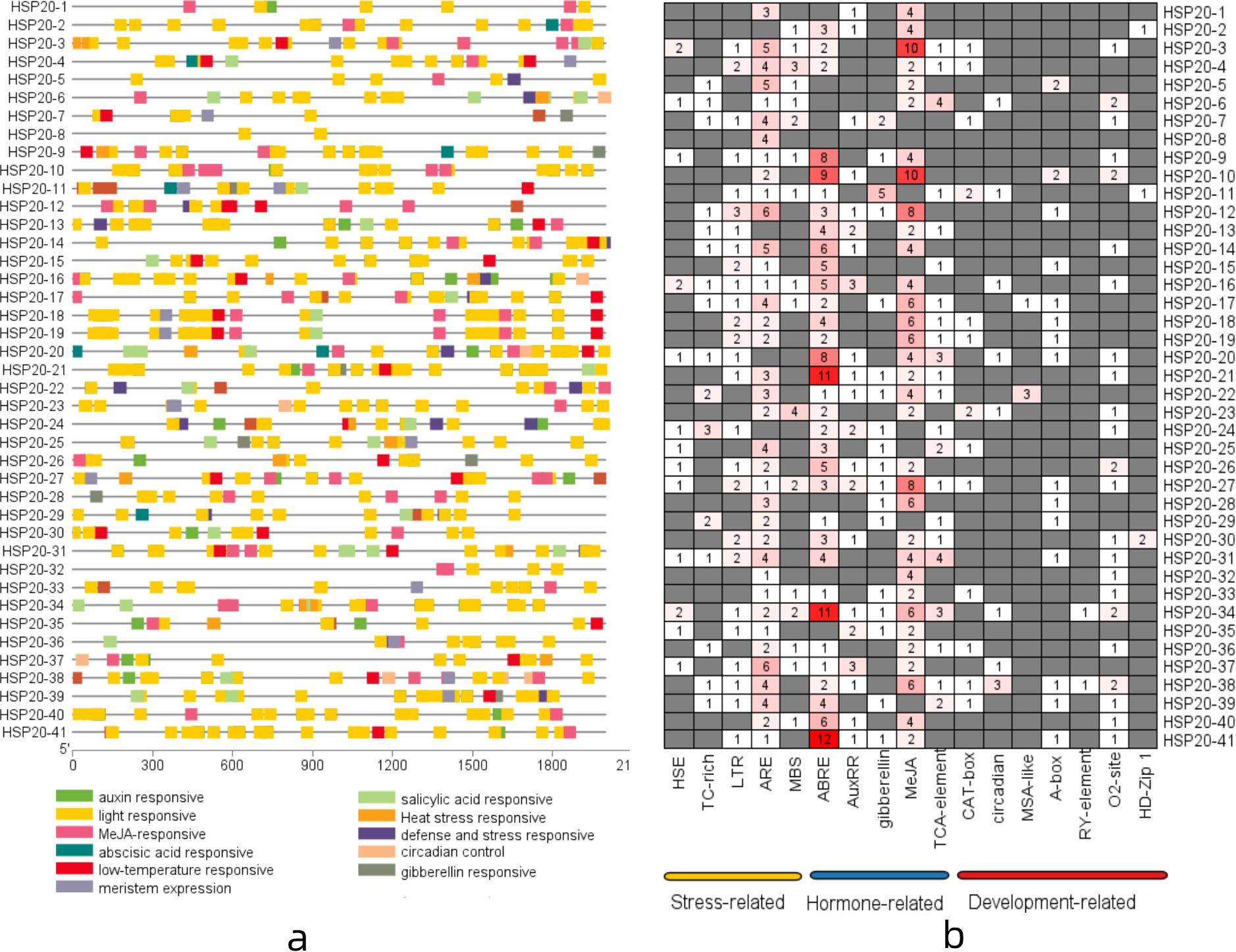
Figure 5. Cis-Element analysis of apple HSP20 gene promoters. (a) The different colored blocks represent the different types of cis-acting elements and their locations in each HSP20 gene. (b) The number of each cis-acting element in the promoter region of each apple HSP20 gene.
Expression Patterns of HSP20s in Response to Cold and Heat Stress
For a preliminary investigation of the functions of apple HSP20 genes in response to heat and cold stress, nine RNA-seq libraries, including three independent biological replicates for the control, cold-treated and heat-treated, were constructed and sequenced. A heatmap of 41 apple HSP20 genes was constructed using FPKM values from RNA-Seq data to estimate the expression levels of these genes (Figure 6). The heat map showed that the 41 HSP20 genes clustered in three groups. Cluster A contains one member (HSP20-33) of 41 detectable HSP20 genes, which was barely expressed after heat and cold stress treatment compared with the control. We found HSP20-33 has no HREs in promoter region. This may be why the gene does not respond to heat stress. All 35 members from cluster B were mainly upregulated after 4 h of heat stress. However, these genes were nearly unchanged or downregulated under cold treatment. Cluster C contains five members, which had similar expression with cluster A, which was barely expressed after heat and cold stress treatment compared with the control.
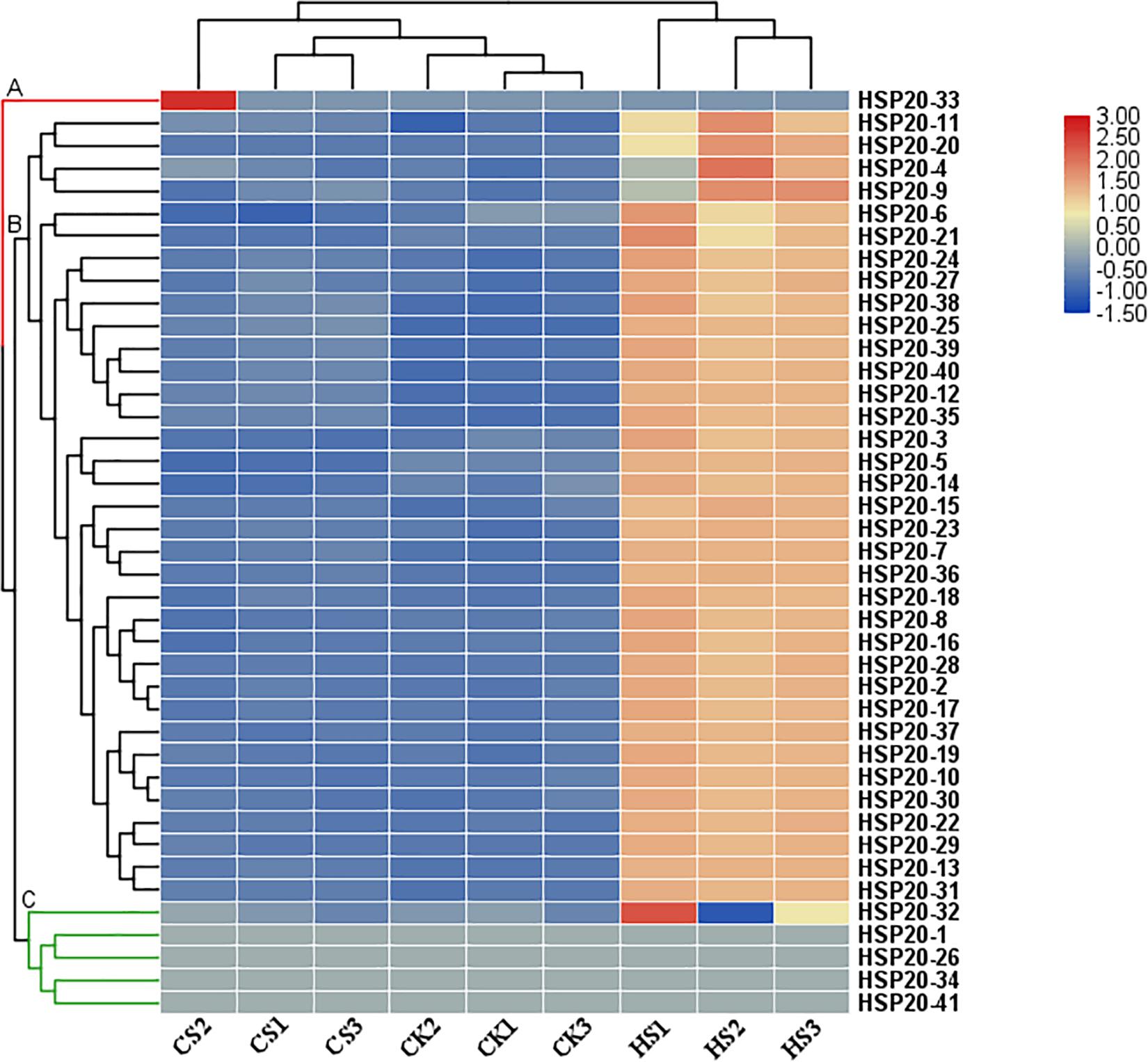
Figure 6. Heat map of the expression profiles of 41 apple HSP20 genes in response to heat and cold stresses based on RNA-Seq data. Log2 based FPKM value was used to create the heat map with clustering. The color scale representing the relative expression values is shown on the left. CK, control, plants were maintained at 25 ± 1°C; HS, heat stress, plants were maintained at 40 ± 1°C; CS, cold stress, plants were maintained at 4 ± 1°C.
To further identify which of these HSP20 genes are most important in mediating heat and cold stress tolerances, 29 differentially expressed HSP20 genes were selected to be further validated by qRT-PCR based RNA-Seq analysis (Figure 7 and Supplementary Table S6). Consistent with the RNA-Seq data, all 29 selected HSP20 genes were up-regulated under heat stress. The expression levels of 12 HSP20 genes (HSP20-8, 13, 16, 17, 18, 19, 23, 29, 35, 36, 37, and 38) were extremely up-regulated (more than 1,000-fold) after 4 h of heat stress. Under heat stress, 29 HSP20 genes were similarly expressed over time, with peak expression levels at 4 h, except for HSP20-40, with peak expression levels at 24 h, while most HSP20 (HSP20-2, 5, 7, 13, 17, 18, 19, 22, 23, 25, 28, 29, 30, 31, 35, 36, 37, and 38) genes were barely expressed in response to cold stress. However, eight apple HSP20 genes (HSP20-3, 10, 12, 14, 15, 16, 27, and 40) were up-regulated under cold stress.
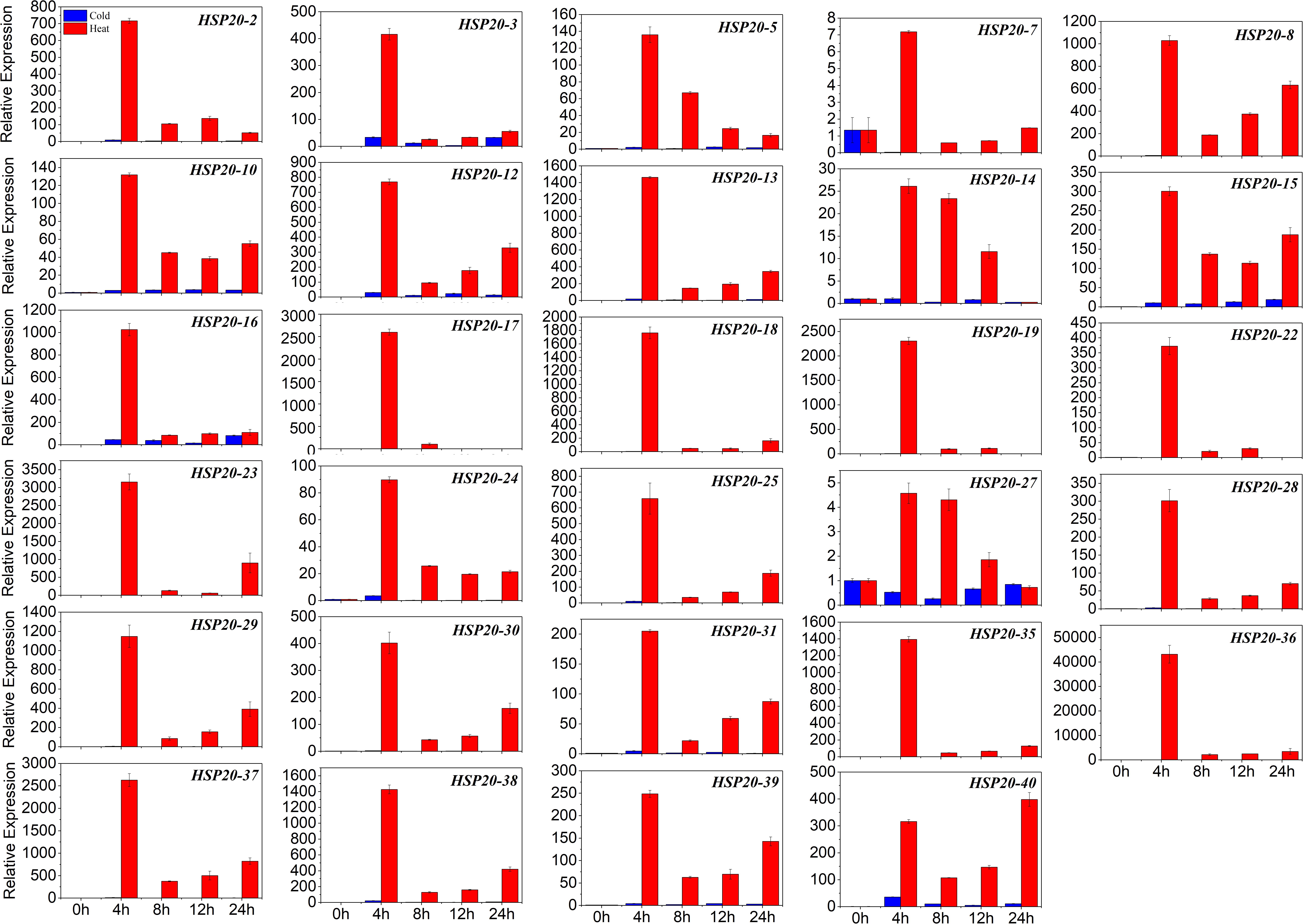
Figure 7. The relative expression levels of apple HSP20 genes under heat and cold stresses. Mean expression value was calculated from three independent replicates. Vertical bars indicate standard deviation from three independent technical replicates.
Discussion
HSP20s are considered to be the most abundant plant stress responsive class among HSPs (Waters, 2013). They have been identified in potato, pepper, tomato, and soybean in responding to temperature stress (Lopes-Caitar et al., 2013; Guo et al., 2015; Yu et al., 2016; Zhao et al., 2018). But not studies have conducted an overall identification and characterization of the apple HSP20 genes. Completion of high-quality apple genome sequencing has provided an opportunity to identify and characterize HSP20 genes at the whole-genome level.
In the present study, we identified 41 HSP20 genes and investigated their characteristics from the apple genome database. The number of apple HSP20 genes was higher than that of Arabidopsis (19) (Scharf et al., 2001), slightly higher than rice (39) (Ouyang et al., 2009) but lower than that of watermelon (44) (He et al., 2019), potato (48) (Zhao et al., 2018), and grape (48) (Ji et al., 2019). This difference is most likely due to the fact that apple had gene duplications during evolution (Velasco et al., 2010; Ma et al., 2018). Gene duplication was reported to play an important role in the expansion of the number of gene families in plants (Blanc and Wolfe, 2004; Han et al., 2011). In the current study, 41 apple HSP20 genes were unevenly mapped on 15 Chrs and most of the HSP20 genes were located on the distal ends of the Chrs, which might contribute to the occurrence of duplication events in the apple HSP20 gene family. We confirmed many tandem and segmental duplications in apple HSP20 genes: 37 of 41 HSP20 genes were affected by gene duplication, 20 of which were segmental duplication and 17 gene clusters were from tandem duplication. Ma et al. (2018) and Zuo et al. (2018) also found many tandem and segmental duplications in apple receptor-like kinase1-like kinase (CrRLK1L) genes and malate dehydrogenase (MDH) genes, respectively.
To determine the evolutionary relationships of HSP20 genes, a phylogenetic tree was constructed based on the amino acid sequences of apple, Arabidopsis and rice HSP20 genes. The phylogenetic analysis indicated that the apple HSP20 family could be divided into 10 subfamilies (CIs, CIIs, CIIIs, CIVs, CVs, MIs, MIIs, Pos, ER, and Ps), which is in line with previous evolutionary classification of HSP20 genes in Arabidopsis and rice (Scharf et al., 2001; Ouyang et al., 2009), indicating a close relationship among HSP20 genes from apple, Arabidopsis and rice. In addition, most apple HSP20 genes were classified into CI-CVI, indicating that cytoplasm could be the primary functional area of the HSP20 family in apple. Gene structure has been documented to function directly in the evolution of multiple gene families (Ji et al., 2019). Gene structure analysis indicated that most apple HSP20 genes have no introns (60.9%) or one intron (31.7%), suggesting relatively simple gene structures. Similarly, most (93.8%) grape HSP20 genes have no introns or one short intron (Ji et al., 2019).
Genes with few or no introns are considered to be rapidly activated in response to various stresses (Jeffares et al., 2008). In our study, most apple HSP20 genes were rapidly induced after 4 h of heat stress, which may support the rapid response. To more comprehensively investigate the evolution of HSP20 genes, the encoded conserved motifs were also studied. Our results showed that most of the apple HSP20 genes had five to seven conserved motifs and almost all the HSP20 genes contained motif 1. This indicates a slow evolutionary rate. Furthermore, we found that most HSP20 genes in the same subfamilies showed conserved motifs and similar exon/intron structures, supporting their close evolutionary relationship and the classification of subfamilies. Genes in the same subfamily tends to share similar motif and exon–intron organization, which was also reported in tomato (Yu et al., 2016).
Cis-elements in the promoters of genes have been documented as essential in plant physiological response and environmental stress (Yamaguchi-Shinozaki and Shinozaki, 2005). We identified cis-elements in the putative promoter regions of apple HSP20 genes. Numerous hormone responsive, stress-responsive and plant development-related cis-elements were found. Among these cis-elements, the hormone responsive elements accounted for the highest proportion. Most HSP20 genes contained stress-related response elements. The present results suggest that most apple HSP20 genes might be significantly related to stress response. Similar regulatory patterns of HSP20 genes were also found in pepper and grape (Guo et al., 2015; Ji et al., 2019). In addition, all apple HSP20 genes contain light signal response elements, which indicate that HSP20s were essential in plant growth and development.
Previous studies have revealed that HSP20s function directly in plant responses to various stresses (Waters et al., 1996; Guo et al., 2015; Zhao et al., 2018; He et al., 2019; Ji et al., 2019). In this study, the expression profiles of apple HSP20 genes under heat stress revealed that the apple HSP20 genes are involved in heat response. qRT-PCR analysis indicated that most apple HSP20 genes were up-regulated under heat stress. It is interesting to note that the relative expression levels of 12 HSP20 genes (HSP20-8, 13, 16, 17, 18, 19, 23, 29, 35, 36, 37, and 38) were extremely up-regulated after 4 h of heat stress. The results from this study suggest that these genes might be mainly involved in the heat stress biological pathway. Similarly, most of the pepper and potato HSP20s were also up-regulated in response to heat stress (Guo et al., 2015; Zhao et al., 2018).
Transgenic research has demonstrated the positive role of HSP20 genes in responding to heat, such as WsHSP26 in Arabidopsis (Mu et al., 2013), and OsHsp17.7 and OsHSP20 in rice (Murakami et al., 2004; Guo et al., 2020). In addition, some HSP20s showed the same expression profile in response to heat stress, being upregulated with peak expression levels at 4 h, suggesting that these HSP20s were co-expressed in response to heat stress. Furthermore, Guo et al. (2015) found that the inducibilities of HSP20 genes in response to heat stress were obviously different in pepper with different tolerance. Collectively, these results indicate that HSP20 genes may be positively involved in heat stress responses in plants. Induction of HSP20 genes by heat stress is well-known (Waters et al., 1996). However, some heat-regulated genes were barely expressed or downregulated in response to cold stress, which indicated that HSP20 genes were negatively or only slightly involved in the response to cold stress. These results imply that the signaling pathways in plant response to heat and cold stress might be different.
Conclusion
This study identified 41 HSP20 genes in apple. These genes are unequally distributed on 15 chromosomes and were classified into 10 subfamilies based on the phylogenetic tree and subcellular localization. The basic classification, genome distribution, gene structures, conserved motifs, and cis acting elements of these genes were analyzed, which will be helpful for a better understanding of the evolutionary relationships of the HSP20 gene family. Transcriptome analysis revealed that most apple HSP20 genes were highly induced by heat stress, whereas these genes were nearly unchanged or downregulated under cold stress, indicating that HSP20 genes were positively involved in heat stress responses in apple. Additionally, we identified several HSP20 genes that may be utilized as candidates for improving heat stress tolerance. The results presented here will lay a solid foundation for functional characterization of HSP20 genes through gene-transfer techniques to improve heat tolerance of apple.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TB designed the experiment, analyzed the data, and drafted the manuscript. FY, CS, and HW collected the public dataset and performed bioinformatics analysis. XZ and SS analyzed the data. JJ and MW assisted with revisions to the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD1000300 and 2019YFD1000100), the National Natural Science Foundation of China (31872058 and 31801844), Program of Young-Backbone Teacher of University in Henan Province (2018GGJS029), and the Special Fund for Henan Agriculture Research System (S2014-11-G02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Anita K. Snyder for critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.609184/full#supplementary-material
Supplementary Table 1 | qRT-PCR primers used in this study.
Supplementary Table 2 | The CDS sequences, deduced amino acid and promoter sequences of apple HSP20 genes.
Supplementary Table 3 | Information of HSP20 genes from apple, rice and Arabidopsis.
Supplementary Table 4 | List of the putative motifs of HSP20 proteins.
Supplementary Table 5 | Tandem and segmental duplicated HSP20 gene pairs within apple genome.
Supplementary Table 6 | Expression levels for selected apple HSP20 genes under heat and cold stress.
Footnotes
- ^ https://www.rosaceae.org/
- ^ http://pfam.xfam.org
- ^ http://smart.embl-heidelberg.de/
- ^ https://web.expasy.org/protparam/
- ^ http://linux1.softberry.com/
- ^ http://plants.ensembl.org/index.html
- ^ http://www.megasoftware.net/
- ^ http://meme-suite.org/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://circos.ca/
- ^ https://iris.angers.inra.fr/gddh13/the-apple-genome-downloads.html
- ^ https://www.ncbi.nlm.nih.gov/sra/PRJNA665791
References
Asea, A. A. A., Kaur, P., and Calderwood, S. K. (2016). Heat Shock Proteins and Plants. Berlin: Springer International Publishing.
Bai, T. H., Li, C. Y., Li, C., Liang, D., Ma, F. W., and Shu, H. R. (2013). Contrasting hypoxia tolerance and adaptation in Malus species linked to differences in the stomatal behavior and photosynthesis. Physiol. Planta 147, 514–523. doi: 10.1111/j.1399-3054.2012.01683.x
Blanc, G., and Wolfe, K. H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691. doi: 10.1105/tpc.021410
Chang, S. J., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1007/bf02670468
Chen, C. J., Chen, H., He, Y. H., and Xia, R. (2018). TBtools, a Toolkit for biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv [Preprint], doi: 10.1101/289660
Daccord, N., Celton, J. M., Linsmith, G., Becke, C., Choisne, N., Schijlen, E., et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 1099–1106. doi: 10.1038/ng.3886
Dobránszki, J., and Teixeira da Silva, J. A. (2010). Micropropagation of apple-A review. Biotechnol. Adv. 28, 462–488. doi: 10.1016/j.biotechadv.2010.02.008
Guo, L. M., Li, J., He, J., Liu, H., and Zhang, H. M. (2020). A class I cytosolic HSP20 of rice enhances heat and salt tolerance in different organisms. Sci. Rep. 10:1383.
Guo, M., Liu, J. H., Lu, J. P., Zhai, Y. F., Wang, H., Gong, Z. H., et al. (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 6:806. doi: 10.3389/fpls.2015.00806
Han, Y. P., Zheng, D. M., Vimolmangkang, S., Khan, M. A., Beever, J. E., and Korban, S. S. (2011). Integration of physical and genetic maps in apple confirms whole-genome and segmental duplications in the apple genome. J. Exp. Bot. 62, 5117–5130. doi: 10.1093/jxb/err215
He, Y. J., Fan, M., Sun, Y. Y., and Li, L. L. (2019). Genome-wide analysis of watermelon HSP20s and their expression profiles and subcellular locations under stresses. Int. J. Mol. Sci. 20:12. doi: 10.3390/ijms20010012
Huo, L. Q., Sun, X., Guo, Z. J., Jia, X., Che, R. M., Sun, Y. M., et al. (2020). MdATG18a overexpression improves basal thermotolerance in transgenic apple by decreasing damage to chloroplasts. Hortic. Res. 7:21.
Jeffares, D. C., Penkett, C. J., and Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Ji, X. R., Yu, Y. H., Ni, P. Y., Zhang, G. H., and Guo, D. L. (2019). Genome-wide identification of small heatshock protein (HSP20) gene family in grape and expression profile during berry development. BMC Plant Biol. 19:433. doi: 10.1186/s12870-019-2031-4
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopes-Caitar, V. S., De Carvalho, M. C., Darben, L. M., Kuwahara, M. K., Nepomuceno, A. L., Dias, W. P., et al. (2013). Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 14:577. doi: 10.1186/1471-2164-14-577
Ma, B. Q., Yuan, Y. Y., Gao, M., Qi, T. H., Li, M. J., and Ma, F. W. (2018). Genome-wide identification, molecular evolution, and expression divergence of aluminum-activated malate transporters in apples. Int. J. Mol. Sci. 19:2807. doi: 10.3390/ijms19092807
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mu, C. J., Zhang, S. J., Yu, G. Z., Chen, N., and Li, X. F. (2013). Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS One 8:e82264. doi: 10.1371/journal.pone.0082264
Murakami, T., Matsuba, S., Funatsuki, H., Kawaguchi, K., Saruyama, H., Tanida, M., et al. (2004). Overexpression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol. Breed. 13, 165–175. doi: 10.1023/b:molb.0000018764.30795.c1
Ouyang, Y., Chen, J. J., Xie, W. B., Wang, L., and Zhang, Q. F. (2009). Comprehensive sequence and expression profile analysis of hsp20 gene family in rice. Plant Mol. Biol. 70, 341–357. doi: 10.1007/s11103-009-9477-y
Peleg, Z., and Blumwald, E. (2011). Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 14, 290–295. doi: 10.1016/j.pbi.2011.02.001
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Scharf, K. D., Siddique, M., and Vierling, E. (2001). The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (acd proteins). Cell Stress Chaperon. 6:225. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2
Shen, J. Z., Zhang, D. Y., Zhou, L., Zhang, X. Z., Liao, J. R., Duan, Y., et al. (2019). Transcriptomic and metabolomic profiling of Camellia sinensis L.cv.‘Suchazao’ exposed to temperature stresses reveals modification in protein synthesis and photosynthetic and anthocyanin biosynthetic pathways. Tree Physiol. 39, 1583–1599. doi: 10.1093/treephys/tpz059
Suzuki, N. (2019). Temperature stress and responses in plants. Int. J. Mol. Sci. 20:2001. doi: 10.3390/ijms20082001
Torres, C. A., Sepúlveda, G., and Kahlaoui, B. (2017). Phytohormone interaction modulating fruit responses to photooxidative and heat stress on apple (Malus domestica Borkh.). Front. Plant Sci. 8:2129. doi: 10.3389/fpls.2017.02129
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus_domestica Borkh.). Nat. Genet. 42, 833–839.
Wang, B., Ranjan, R., Khot, L. R., and Peters, R. T. (2020). Smartphone application-enabled apple fruit surface temperature monitoring tool for in-field and real-time sunburn susceptibility prediction. Sensors 20:608. doi: 10.3390/s20030608
Wang, N., Guo, T. L., Sun, X., Jia, X., Wang, P., Shao, Y., et al. (2017). Functions of two Malus hupehensis (Pamp.) Rehd. YTPs (MhYTP1 and MhYTP2) in biotic-and abiotic-stress responses. Plant Sci. 261, 18–27. doi: 10.1016/j.plantsci.2017.05.002
Wang, Q. S., Liu, N. A., Yang, X. Y., Tu, L. L., and Zhang, X. L. (2016). Small RNA-mediated responses to low-and high-temperature stresses in cotton. Sci. Rep. 6:35558.
Wang, W. X., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Waters, E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. doi: 10.1093/jxb/ers355
Waters, E. R., Lee, G. J., and Vierling, E. (1996). Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 47, 325–338. doi: 10.1093/jxb/47.3.325
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yu, J. H., Cheng, Y., Feng, K., Ruan, M. Y., Ye, Q. J., Wang, R. Q., et al. (2016). Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Front. Plant Sci. 7:1215. doi: 10.3389/fpls.2016.01215
Zhao, P., Wang, D. D., Wang, R. Q., Kong, N. N., Zhang, C., Yang, C. H., et al. (2018). Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 19:61. doi: 10.1186/s12864-018-4443-1
Keywords: apple, HSP20 family, heat stress, genome-wide analysis, gene expression
Citation: Yao F, Song C, Wang H, Song S, Jiao J, Wang M, Zheng X and Bai T (2020) Genome-Wide Characterization of the HSP20 Gene Family Identifies Potential Members Involved in Temperature Stress Response in Apple. Front. Genet. 11:609184. doi: 10.3389/fgene.2020.609184
Received: 22 September 2020; Accepted: 20 October 2020;
Published: 06 November 2020.
Edited by:
Jian Ma, Sichuan Agricultural University, ChinaReviewed by:
Yanling Ma, Chinese Academy of Agricultural Sciences (CAAS), ChinaQuan Xie, Nanjing Agricultural University, China
Copyright © 2020 Yao, Song, Wang, Song, Jiao, Wang, Zheng and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tuanhui Bai, dHVhbmh1aWJhaTg4QDE2My5jb20=; Xianbo Zheng, WGlhbmJvekAxNjMuY29t
†These authors have contributed equally to this work
 Fuwen Yao†
Fuwen Yao† Chunhui Song
Chunhui Song Tuanhui Bai
Tuanhui Bai