
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 10 December 2020
Sec. Cancer Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.606274
This article is part of the Research Topic microRNAs and Muscle Cell Death in Cancer View all 5 articles
 Lucia Tombolan1*
Lucia Tombolan1* Caterina Millino2
Caterina Millino2 Beniamina Pacchioni2
Beniamina Pacchioni2 Manuela Cattelan3
Manuela Cattelan3 Angelica Zin1
Angelica Zin1 Paolo Bonvini1
Paolo Bonvini1 Gianni Bisogno1,4
Gianni Bisogno1,4Rhabdomyosarcoma (RMS) arises from myogenic precursors that fail to complete muscle differentiation and represents the most frequent soft tissue sarcoma in children. Two major histological subtypes are recognized: alveolar RMS, characterized by a more aggressive behavior and a greater proneness to metastasis, and embryonal RMS which accounts for the 80% of cases and carries a better prognosis. Despite the survival of patients with localized tumors has progressively improved, RMS remains a challenging disease especially for metastatic patients and in case of progressive or recurrent disease after front-line therapy. MicroRNAs, a class of small non-coding RNA, have emerged as crucial players in cancer development and progression, and their detection in plasma (circulating miRNAs) represents a promising minimally invasive approach that deserve to be exploited in clinical practice. We evaluated the utility of circulating miRNAs as diagnostic and prognostic biomarkers in children with RMS profiling miRNAs from plasma of a small cohort of RMS patients and healthy donors (HD) using a qPCR Cancer Panel. An assessment of hemolysis status of plasma using miR-451/miR-23a ratio was performed as pre-analytical analysis. Statistical analysis revealed that miRNAs expression pattern clearly distinguished RMS patients from HD (p < 0.05). Interestingly, plasma levels of muscle-specific miR-206 were found to be significantly increased in RMS patients compared to HD, whereas levels of three potential tumor-suppressor miRNAs, miR-26a and miR-30b/30c, were found lower. Reduced levels of circulating miR-26a and miR-30b/c were further measured in an independent larger cohort of patients (validation set) by digital droplet PCR. In particular, we evidenced that miR-26a absolute plasma levels were associated with fusion status and adverse outcome (p < 0.05). Taken together, these findings demonstrate the potential of circulating miRNA as diagnostic and prognostic biomarker in children affected by this malignancy and enforced the key role of miR-26a in pediatric rhabdomyosarcoma.
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma affecting children. It arises from myogenic precursor that are unable to complete muscle differentiation. Typical myogenic factors such as MyoD1 and myogenin are expressed by tumor cells and routinely used to define the diagnosis of RMS (Sebire and Malone, 2003). Two major histotypes are recognized: alveolar RMS (ARMS) and embryonal RMS (ERMS). About 80% of ARMS are characterized by a reciprocal translocation, the more frequent t (2;13) and the less common t (1;13) (Rudzinski et al., 2015). Both translocations involve PAX genes, PAX3 and PAX7 respectively, and the transcription factor FOXO1. Fusion-positive ARMS tumors are associated with a more aggressive phenotype and worse prognosis respect to the embryonal RMS(Sorensen et al., 2002). Despite the survival of RMS patients remarkably increased during the last years thanks to the adoption of a multidisciplinary treatment modalities, patients presenting metastatic disease at diagnosis or experiencing relapse after treatment still have a dismal prognosis, with chances to reach a long survival lower than 5% (Oberlin et al., 2012). Currently, detection of relapse in children with RMS includes bone marrow aspirate and biopsy and computed tomography (CT) scan (Huh and Skapek, 2010).
Herein, the detection of specific biomarkers in peripheral blood or other biological fluids may help to predict the probability of a relapse or a progressive disease in pediatric RMS.
The identification of biomarkers in biological fluids termed as liquid biopsy is a very promising field of investigation in solid tumors. Liquid biopsy is a minimally invasive approach with the potential of providing information on early diagnosis, prognosis and to monitor the response to treatment.
Circulating microRNAs (miRNAs) are endogenous non-coding RNA that have also been detected in many biological fluids, such as urine and plasma/serum.
MiRNAs can be released in the circulation as both cell-free miRNAs and packed into a secretory vesicles known as exosomes. Cell-free miRNAs are complex to proteins such as argonaute proteins (AGOs) or high density lipoproteins that confer to them an extremely stable form resistant to ribonuclease activity (Mitchell et al., 2008; Arroyo et al., 2011; Vickers et al., 2011).
MicroRNAs are able to finely modulate the expression of their target genes through a specific and well known mechanism which leads to target mRNA degradation or inhibition of protein translation (Bartel, 2004). MicroRNAs are involved in many physiological and pathological conditions that include tissue development and tumor-related processes. In RMS, several functional studies demonstrated that specific miRNAs can act both as tumor suppressor and oncomiRs thus regulating proliferation, invasion and apoptosis of cancer cells (Tombolan et al., 2015; Molist et al., 2020; Wang et al., 2020).
Numerous studies have demonstrated that miRNAs expression profiling varies between tumor and normal tissues as well as different tumors, like sarcomas, display a peculiar miRNAs expression pattern that contributes to define tumor phenotype (Varshney and Subramanian, 2015; Smolle et al., 2017).
MiRNAs play also a crucial role in skeletal muscle development in which tissue-specific miRNAs, called myomiRs (miR-1, miR133a, miR-133b, and miR-206) tightly regulate the differentiation and proliferation of myogenic precursor cells (McCarthy, 2008; Horak et al., 2016). Interestingly, in RMS myomiRs, that result significantly downregulated, act in concert with other non-muscle miRNAs (miR-29, miR-26a, and miR-183) impairing myogenesis and proliferation. MyomiRs in RMS interact directly with PAX3 and PAX7 genes as well as with cell cycle regulator Cyclin D2 and cMET oncogene (Rota et al., 2011; Li et al., 2012; Gasparini et al., 2019).
In this study, we investigated the expression of miRNAs in the plasma of two cohorts of RMS patients and healthy subjects to figure out whether miRNAs represent useful diagnostic and prognostic biomarkers in pediatric rhabdomyosarcoma. We used qPCR to test a panel of about 84 miRNAs in a small training group of patients and controls then we verified the expression of three miRNAs in a validation cohort of samples taking advantage of the droplets digital PCR (ddPCR) technique. Our findings unveiled that circulating miRNAs can distinguish RMS patients from healthy subjects and, as for miR-26a, can correlate with an increased risk of relapse and a poor prognosis.
A total of 30 blood samples from RMS patients enrolled in national and international pediatric sarcoma protocol (EpSSG RMS 2005) were analyzed in this study: 8 were processed with qPCR Cancer Panel (training group) and 22 with droplet digital PCR (validation group). All the clinical features are summarized in Table 1. In addition, 8 healthy donors were included in the study as controls. Peripheral blood of RMS patients was collected at the time of diagnosis, prior to any treatment, in sodium citrate tubes and processed within 24 hours. Plasma was obtained from peripheral blood after two steps of centrifugation: the first at 890 × g for 10 min and the second at high speed (16000 × g for 10 min) then the samples were stored at −80°C in the Pediatric-Oncology biobank (BBOP) (part of Italian Association of Pediatric Hematology and Oncology (AIEOP). This study was approved by Padua Hospital Ethics Committee (No. 988P, 31March 2005) and the patients have signed informed consent.
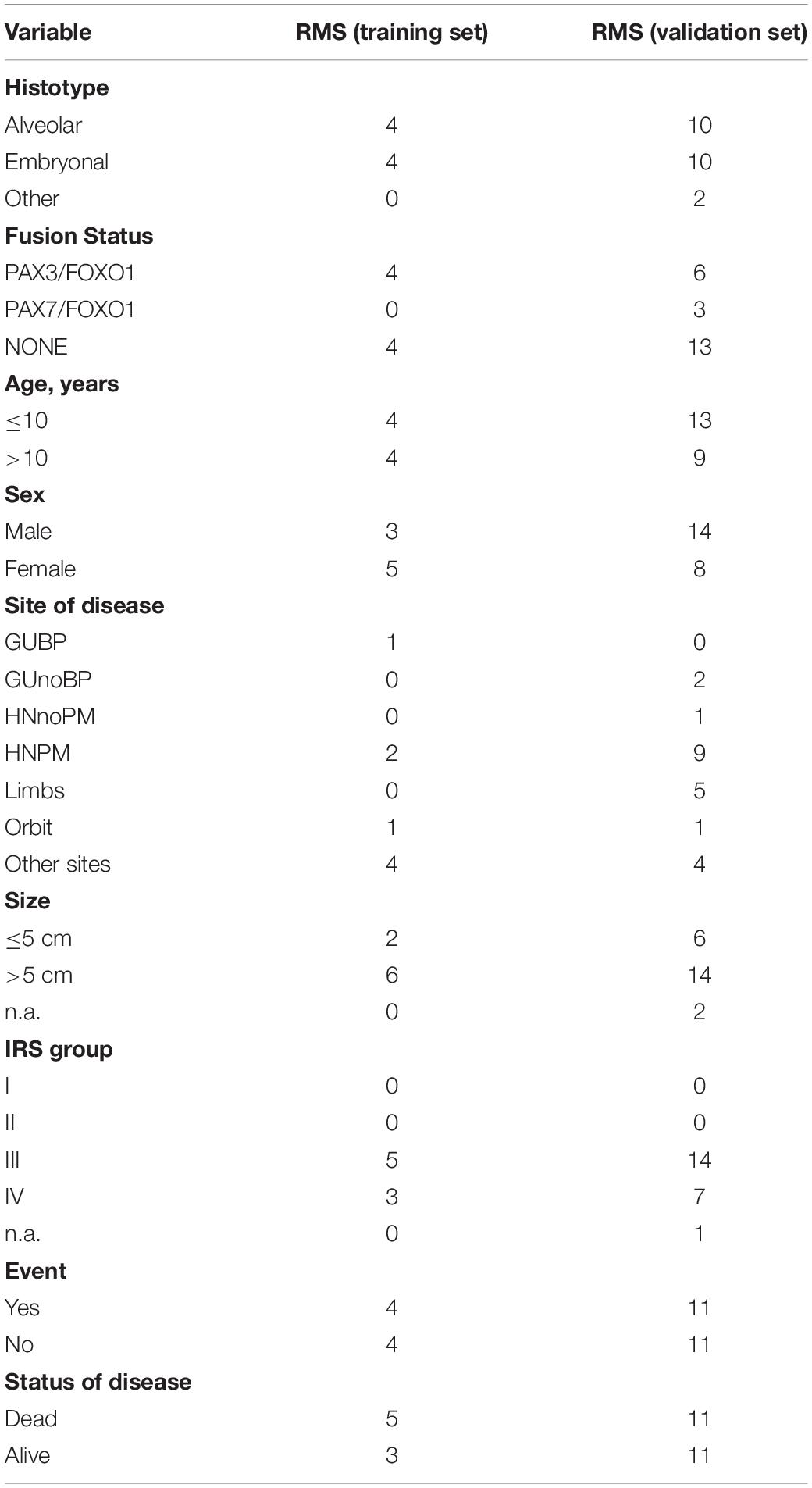
Table 1. Clinical features of RMS patients analyzed. GUBP = GenitoUrinary Bladder-Prostate, GUnoBP = GenitoUrinary non-Bladder-Prostate, HNnoPM = Haed Neck non-Parameningeal, HNPM = Haed Neck Parameningeal; IRS group, Intergroup Rhabdomyosarcoma Studies (Clinical Group), n.a. = data not available.
Before starting the miRNA extraction procedure, plasma samples were centrifuged at 3,000 × g for 5 min to remove all cellular debris. Then, 1 μl of Exiqon RNA spike-in mix (UniSp2, UniSp4, and UniSp5) and 2 μl 1 μM of ATH-miRNA159a were added to 200 μl of plasma (starting volume) to control the sample-to-sample variation in the RNA isolation procedure. Subsequently, isolation of plasma miRNAs was performed according to the manufacturer’s instructions of miRCURY RNA Isolation kit Exiqon.
Hemolysis in plasma samples was measured by two different methods. The first method uses a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) to measure the absorbance of hemoglobin at 414 nm (2 technical replicates per samples was performed). The second method is based on qPCR analysis of miRNA-451 and miRNA-23a-3p. A ratio between two miRNAs calculated as delta Cq (miRNA-23a-3p – miRNA-451) was used as hemolysis indicator. A ratio more than five indicates a high risk of hemolysis (Blondal et al., 2013). It was estimated that absorbance method has a hemolysis limit of detection of 0.004% while miRNA ratio has a limit of 0.001% (Shah et al., 2016).
cDNA was synthesized using a miRCURY LNA Universal cDNA Synthesis Kit II (Exiqon) starting from 6 μl of total RNA with the addition of 1 μl of UniSp6 and 1 μl of Cel-miR-39 as exogenous miRNA spiked-in control. The RT product was used as a template in the qPCR assays in Ready-to-use Human Cancer Focus microRNA PCR Panel V4 (Supplementary Table S1, Exiqon) and ExiLENT SYBR Green master mix (Exiqon). ROX (Invitrogen by Life Technologies, Carlsbad, CA, United States) was added to the master mix as the passive reference dye. The arrays were run in an ABI7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The qPCR cycling conditions were 95°C for 10 min and 40 cycles (95°C for 10 s and 60°C for 1 min).
For each ddPCR assay, 9 μl cDNA sample (diluted 1:20), 10 μl 2X ddPCR supermix for Eva Green (Bio-Rad, Hercules, CA, United States), 1 μl of LNA PCR primers set was mixed (miRCURY LNA miRNAs PCR assays, Qiagen, Hilden, Germany). The mixture and 70 μl of droplet generation oil for Eva Green were respectively loaded into the sample and oil wells of a disposable droplet generator cartridge (Bio-Rad, Hercules, CA, United States). Therefore, droplets were generated by QX200 droplet generator device (Bio-Rad) and transferred to a 96 plate (Bio-Rad, Hercules, CA, United States). The cycling condition were 95°C for 5 min and 40 cycles (95°C for 30 s 58°C for 1 min) 4°C for 5 min and 90°C for 5 min. At the end of the PCR reaction, droplets were loaded in the QX200 droplet reader and analyzed using Quantasoft version 1.7.4 software (Bio-Rad, Hercules, CA, United States). A no template control (NTC) was included in every assay. MiRCURY LNA miRNAs PCR assays used are: miRNA-26a, miRNA-26b, miRNA-30b, miRNA-30c (Qiagen, Hilden, Germany).
The Human Cancer Focus microRNA PCR Panel V4 (Exiqon) data analysis was carried out using GenEx Enterprise software (Exiqon). Both data exported form ABI7500 and the analysis were performed following the instruction reported in GenEx Enterprise software manual. In details, first we performed a pre-processing of the data that include an interpolate calibration using the spike UniSp3, a cut-off setting to exclude data with very high Ct (threshold cycle > 37), an estimation of frequency of missing data and a log conversion of the values. Then data were normalized using different methods: 1. Using 5 different reference miRNAs chosen based on the lowest standard deviation between samples; 2. Using the global mean normalization, a method often applied to gene expression data. Statistical analysis was performed on normalized data to compare RMS vs. healthy donors (HD) (t-test) or to compare 3 different groups: Alveolar (A), embryonal (E) and HD (ANOVA). The p-value (p < 0.05) was calculate. Cluster analysis was performed using morpheus software1, Broad Institute) while principal component analysis (PCA) plot was created with BioVinci software (BioTuring Inc., San Diego, CA, United States).
Droplet digital PCR data were analyzed using GraphPad Prism 6 and R statistical software. The association of miR-26a, miR-26b, miR30b, and miR-30c with clinical features were performed using non-parametric Mann-Whitney and Kruskal-Wallis tests. All p-values were considered statistically significant at the alpha level of 0.05. Kaplan–Meier survival curves were analyzed with log-rank test to estimate the prognostic value of miR-26a expression.
With the aim to explore the circulating miRNAs RMS profile in patients’ plasma samples we took advantage of a qPCR panel of 84 cancer-associated miRNAs (Cancer Panel). Starting from a small volume of plasma (200 μl) we initially tested the hemolysis status of each sample by using the miR-451/miR-23a ratio as described in the method section. It is known that hemolysis grade affects miRNAs plasma levels so this pre-analytical analysis is mandatory (Blondal et al., 2013; Shah et al., 2016). Only the samples that passed the hemolysis test were used in the following experiments (see Supplementary Table S2). We applied the qPCR Cancer Panel to a training group of plasma samples composed of 8 RMS patients (4 PAX3/FOXO1-positive ARMS and 4 fusion-negative embryonal RMS, ERMS) and 4 healthy donors (HD, controls), and quantified the differential expression of the 84-cancer associated miRNAs as reported in the Supplementary Table S3. Data normalization was performed applying two different methods: global mean (GB) and 5 reference genes (5RG) normalization. The Volcano plot displayed a good distribution of normalized data using either one of the two normalization approaches (see Supplementary Figures 1A,B), and when unsupervised hierarchical clustering analysis was performed using all miRNAs of the qPCR Cancer Panel a good separation between groups (ARMS, ERMS, and HD) was observed (Supplementary Figure 1C).
We identified a set of miRNAs differentially expressed between RMS patients and healthy donors with both normalization approaches, and observed a high percentage of common deregulated miRNAs (about 80%) between the two analysis, consistent with the high confidence of the data obtained (see Supplementary Figure 1D). The hierarchical clustering analysis performed using the differentially expressed miRNA demonstrated a clear separation between RMS cases and controls, as well as between alveolar and embryonal RMS patients (p-value < 0.05) (Figure 1A). The list of miRNAs differentially expressed was reported in Supplementary Table S4. Similarly, the Principal Component Analysis (PCA) confirmed the presence of distinct groups of samples with embryonal group more similar to controls with respect of alveolar ones (Figure 1B).
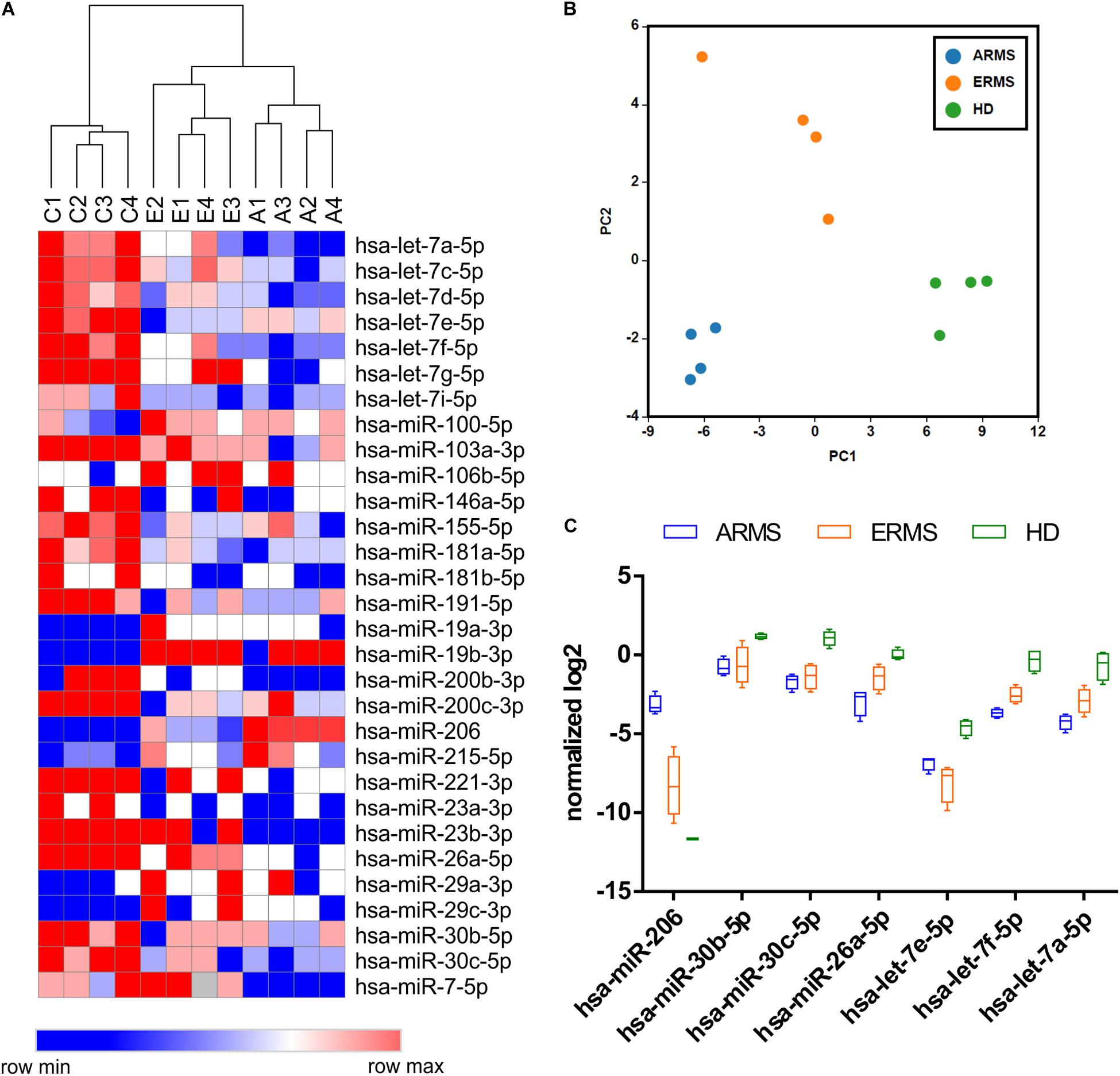
Figure 1. Circulating miRNAs expression profiles distinguish RMS patients from healthy donors. (A) The heatmap shows the circulating miRNAs expression profiles in 8 RMS samples (4 ERMS and 4 ARMS PAX3/FOXO1) and 4 healthy donors based on hierarchical clustering. The statistically different miRNAs were obtained using both global mean and 5 reference genes normalization. (B) The Principal component analysis (PCA) shows the clear separation of three groups (HD, ERMS, and ARMS) based on circulating miRNAs expression patterns. (C) Box plots of relevant miRNAs deregulated in RMS respect to HD are reported. C1–4 = healthy donors; A1–4 = alveolar RMS; E1–4 = embryonal RMS.
We, then, focused our attention on the miRNAs significantly different between RMS patients and HD. As expected, miR-206, a known muscle-related miRNA extensively studied in RMS, displayed higher levels in RMS plasma samples, especially in the alveolar subtype, respect to controls, whereas, like some members of the let-7 family, miR-26a and miR30b/30c, whose tumor suppressor role in other cancers has been described, were downregulated in RMS plasma samples compared to controls (Figure 1C). We also compared our results with a previously published dataset by Missiaglia et al. (2017) obtained analyzing the miRNAs profiling of 64 RMS samples by next-generation sequencing. Although the miRNAs profiling conducted by Missiaglia and colleagues was performed on tumor tissues and skeletal muscle cells as the normal counterpart, we observed that almost 50% (13 out of 30) of circulating miRNAs deregulated in plasma of RMS patients with respect to healthy donors was also deregulated in primary RMS tumors. Accordingly, miR-26a and miR30b/30c were downregulated in RMS samples of both datasets, whereas miR-206 and miR-29b/c expression levels were apparently in contrast. However, since it is known that both miR-206 and miR-29b/c are involved in myogenesis (Wang et al., 2008; Horak et al., 2016) their downregulation in differentiated skeletal muscles and overexpression in RMS plasma sample is consistent and reasonable.
Droplet digital PCR (ddPCR) is a relatively recent technique providing equal results to qPCR with the advantage to perform an absolute quantification without the need of normalization with internal/external references. Among its applications, the quantification of circulating miRNAs in blood is very promising.
Based on literature data we focused our study on three differentially expressed miRNAs, miR-26a, miR-30b and miR-30c that have a potential role as tumor suppressor. We also included in this analysis miR-26b which belongs to the same miRNA family, but was not found downregulated as miR-26a. We first optimized the ddPCR conditions such as primers annealing temperature and template dilution for each assay. We performed ddPCR on 26 plasma samples (22 RMS and 4 healthy donors) used as validation group of the data obtained with the qPCR miRNAs Cancer Panel described above. The clinical characteristics of the RMS patients studied are summarized in Table 1. Analyzing ddPCR data (expressed as copies of target/μl) of each miRNA we validated the different expression of miR-26a miR-30b/30c between RMS patients and controls, while miR-26b was not statistically different between the two groups (Figure 2, miR-26a p = 0.0329; miR-30b p = 0.0209; miR-30c p = 0.0209; miR-26b p = 0.0947, Mann-Whitney test). Therefore, we validated with an independent technique and in a novel cohort of patients the data obtained with qPCR Cancer Panel used as a discovery tool.
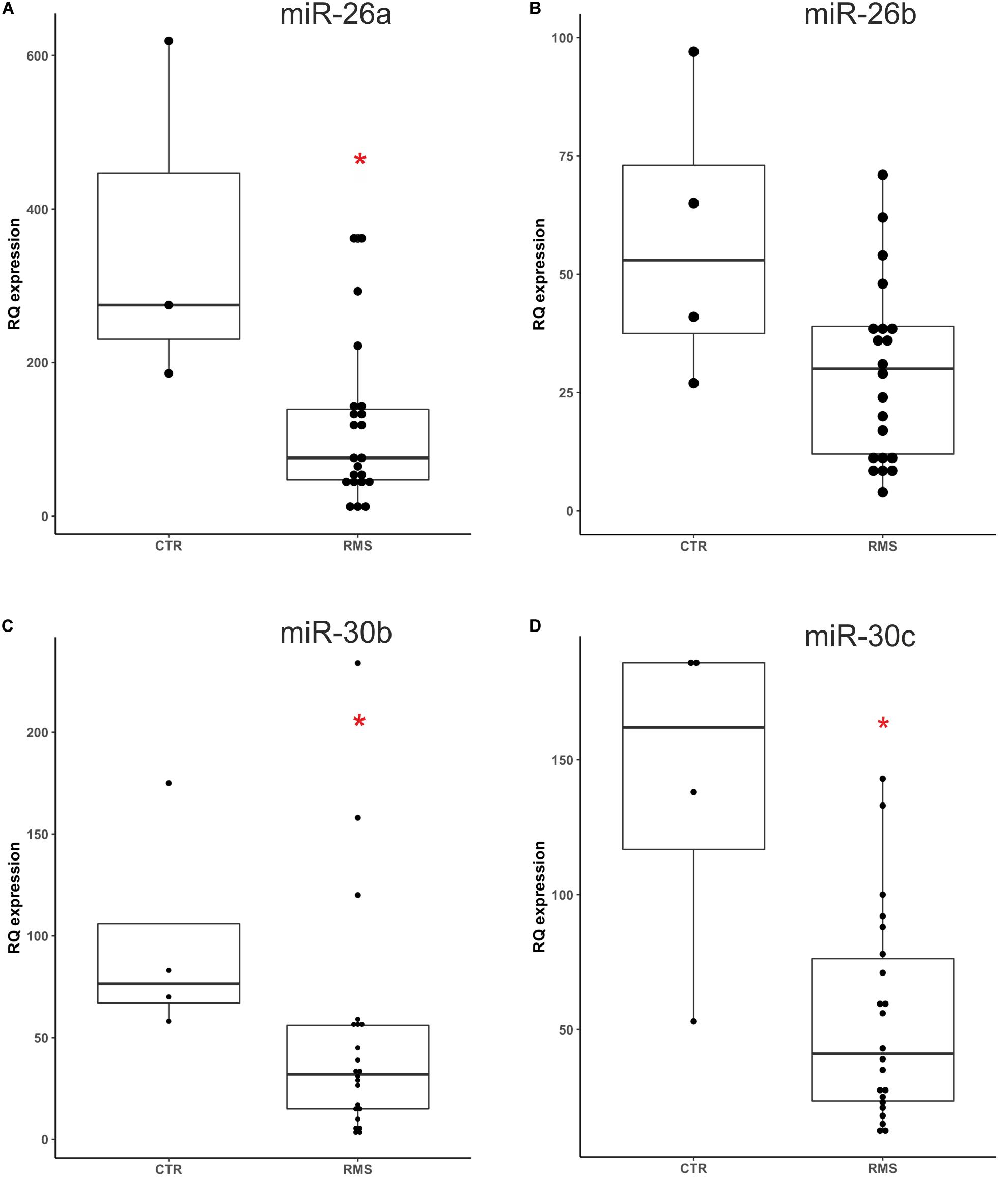
Figure 2. Absolute quantification of miR26a/b and miR-30b/c plasma levels in an independent cohort of RMS patients. The expression levels of four selected miRNAs were assessed by ddPCR in a novel group of RMS patients and healthy donors. Mann-Whitney test sustained miR-26a (A), miR30b (C), and miR-30c (D) different plasma levels in RMS patients compared to controls (CTR), whereas miR26b levels (B) resulted not statistically significant (miR-26a p = 0.0329*, miR-30b p = 0.0209*, miR-30c p = 0.0209*, and miR-26b p = 0.0947 n.s). *p < 0.05.
To investigate any correlation between miR26a and miR30b/30c plasma levels and known RMS prognostic factors we performed an association analysis including histology, fusion status, tumor size and clinical stage (according to IRS grouping) RMS clinical features.
We demonstrated that circulating miR-26a expression inversely correlated with RMS fusion status (p = 0.0177, Mann-Whitney test), since PAX3-7/FOXO1-positive RMS cases displayed lower levels of miR-26a compared to fusion-negative ones (Figure 3A), as it was in ARMS (mostly fusion-positive) than ERMS patients (all fusion-negative), although such a difference was not statistically significant (p = 0.0587 Mann-Whitney Figure 3C). When event occurrence (local relapse or distant metastasis at follow up) and circulating miR-26a levels were confronted, low levels of miR-26a characterized significantly RMS patients with progressive disease (p = 0.0417 Mann-Whitney Figure 3B), whereas clinical risk group (IRS group) and tumor size were not associated with circulating miR-26a expression (Figures 3D,E). Finally, no significant association was found between miR30b/30c plasma levels and RMS prognostic factors (Supplementary Figures 2, 3).
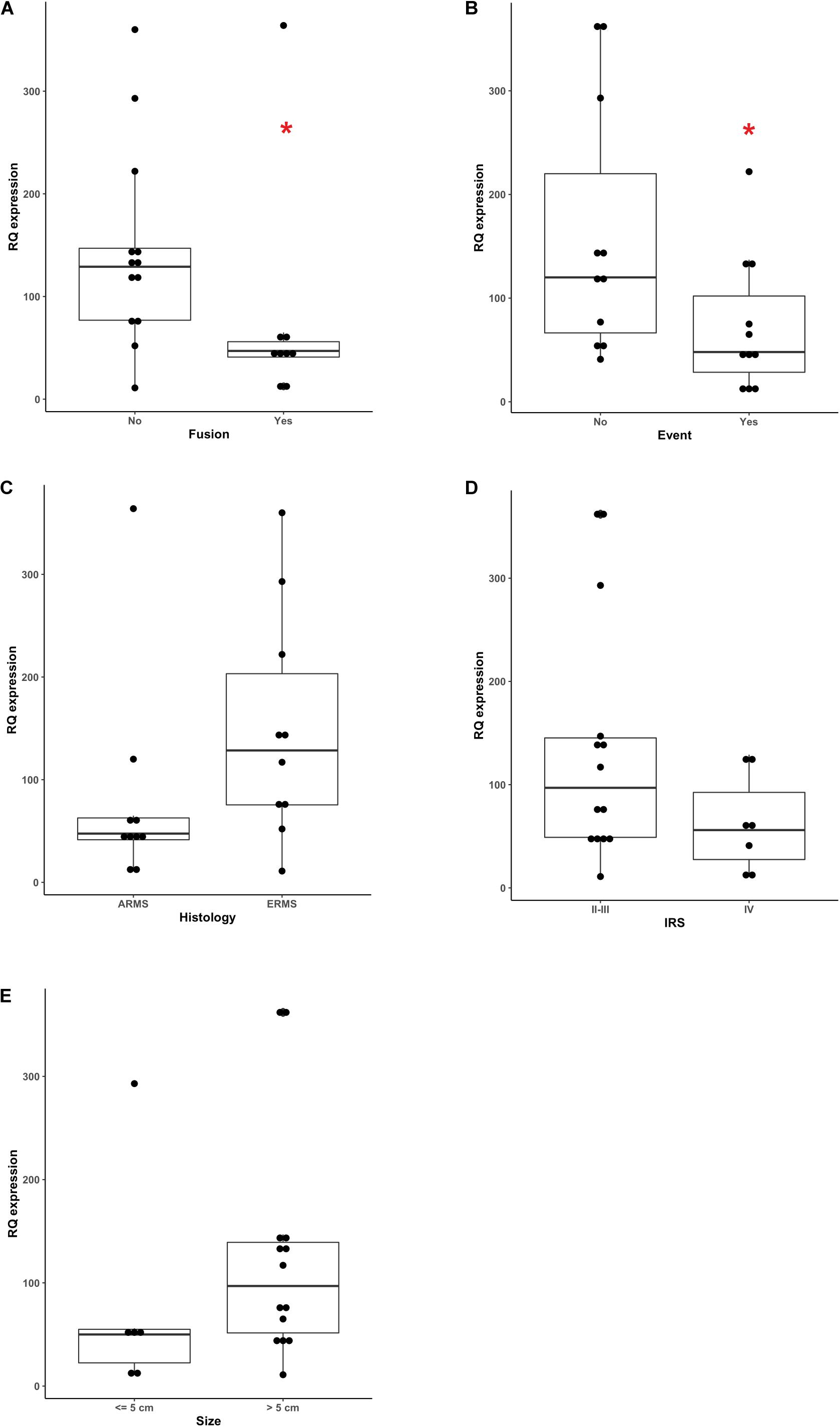
Figure 3. Correlation between miR-26a plasma levels and clinical prognostic factors. miR-26a levels assessed by ddPCR showed significant correlation with, (A) fusion status (p = 0.0177), (B) the occurrence of an event during the follow-up (p = 0.417) while no correlation was observed with (C) histology (p = 0.0587) (D) IRS group factors and (E) tumor size. Mann-Whitney test was used as non-parametric statistical test (*p < 0.05).
With the aim to test the prognostic value of miRNAs as well as the utility of measuring circulating miRNAs in plasma of RMS patients we performed survival analysis. Patients were divided according to median miRNA plasma levels. Remarkably, both overall (OS) and progression-free survival (PFS) analysis showed that patients low levels of miR-26a had poorer outcome compared to those with high miR-26a expression values, and this difference was statistically significant (OS p = 0.033; PFS p = 0.028, Figures 4A,B). Conversely, no survival correlation was found using miR-30b/30c median values (Supplementary Figures 2, 3). Univariate Cox regression analysis performed including clinical variables such as age, gender, fusion status, tumor size, IRS group confirmed these findings. According to it, circulating miR-26a was the only significant variable found to be associated with reduced OS and PFS in RMS patients (OS p = 0.047; PFS p = 0.041) (Supplementary Table 5).
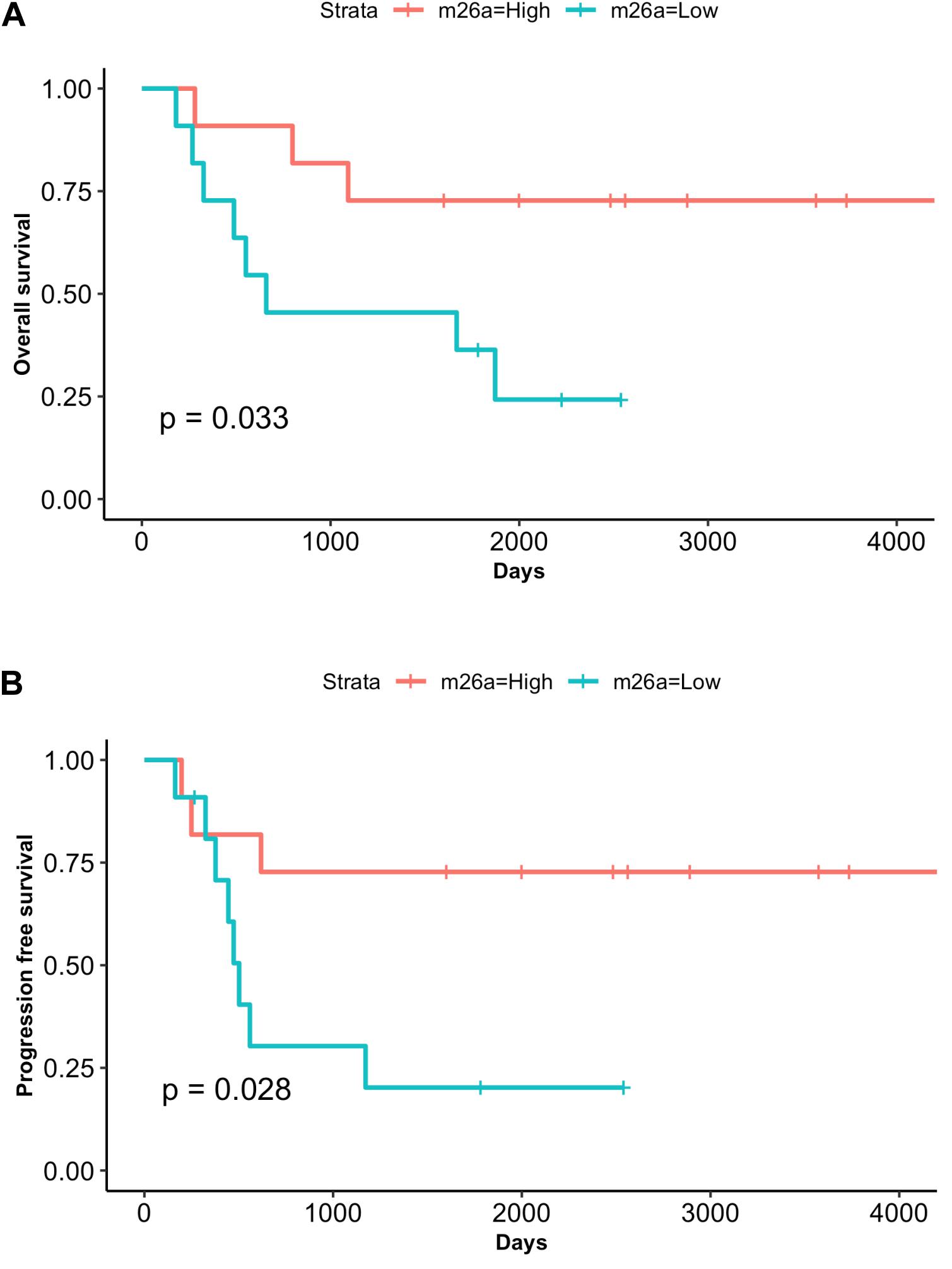
Figure 4. MiR-26a low plasma levels correlate with adverse outcome. Kaplan– Meier and log-rank analysis for overall survival (OS) (A) and progression-free survival (PFS) (B) were performed for the cohort of RMS patients (n = 22) based on low or high miR-26a plasma levels (median value). The group of patients with lower miR-26a plasma levels displays a significantly poor outcome (OS, P = 0.033; PFS, P = 0.028).
In this study, we explored the utility of circulating miRNAs as biomarkers in pediatric rhabdomyosarcoma. The possibility to use a non-invasive approach such as a liquid biopsy to track a tumor-related marker at diagnosis and during follow-up is very attractive in pediatric solid tumors.
Rhabdomyosarcoma, an aggressive pediatric tumor with a mesenchymal origin, remains a challenge for patients that show a progressive disease or a relapse after treatment.
MiRNAs are largely studied in RMS, in particular the muscle-specific miRNAs, termed myomiRs, involved in the regulation of both cell proliferation and muscle differentiation, two key processes that control muscle development (Chen et al., 2006; McCarthy, 2008; Horak et al., 2016). Herein we report that, among myomiRs, circulating miR-206 results differentially expressed between patients and healthy donors, consistent with the observation that miR-206 is overexpressed in RMS primary tumor specimens as well (Missiaglia et al., 2017) and correlates with the presence of metastasis at diagnosis and worse overall survival of RMS patients (Missiaglia et al., 2010). Besides, only miR-206 plasma levels were found able at distinguishing RMS patients from non-RMS patients and healthy donors (Miyachi et al., 2010) unlike other common circulating RMS miRNAs, such as miR-1, miR133a and miR-133b.
On the contrary, miR26a/26b and miR30b/30c, which are miRNAs ubiquitously expressed in many cancers that act as tumor suppressors, were found to be downregulated in RMS patients’ plasma samples and somehow correlated to known negative prognostic factors and poor patient’s survival.
These findings were further supported by previously reported miRNAs expression data in RMS primary tumors, when compared with differentiated skeletal muscles and normal tissues (Missiaglia et al., 2017).
Indeed, low levels of miR-30b and/or miR-30c have been found in gastric cancer, renal carcinoma (Liu et al., 2017) and colorectal cancer (Zhao et al., 2019; Fan et al., 2020) with respect to control tissues. MiR30b/30c exert their tumor suppressor function by targeting genes involved in key processes of tumorigenesis, such as growth, survival, invasion and migration (Cao et al., 2017; Liu et al., 2017; Fan et al., 2020).
Likewise, microRNA-26 family members, miR-26a and miR-26b, have been found to be downregulated in various cancers respect to normal tissues. In esophageal tumor cells miR-26a/b were shown to be involved in a feedback loop with c-MYC that causes a decrease of proliferation (Li J. et al., 2017), whereas in hepatocellular carcinoma they acted as tumor suppressor sensitizing chemoresistant cancer cells to drug-induced apoptosis. Drug-induced autophagy is also impaired by miR-26a/b: this increases cancer cells susceptibility to drug exposure and promotes apoptosis (Jin et al., 2017). A recent study on multiple myeloma (MM) confirmed the role of miR-26a as tumor suppressor. In this study miR-26a was found to act by targeting CD38, a cell-surface glycoprotein involved in MM cells invasion and metastasis, resulting both in the induction of apoptosis in vitro and drug sensitization of MM tumors in vivo (Hu et al., 2020). In c-MYC-induced lymphoma cell lines, downregulation of miR-26a has been reported to come along with cMYC-induced EZH2 expression, a histone methyltransferase member of the Polycomb-group (PcG) family of epigenetic gene silencers implicated in neoplastic development (Sander et al., 2008). Similarly, in C2C12 myoblasts, the increased expression of miR-26a occurs alongside the induction of cell differentiation, expression of muscle-specific genes myoD1 and myogenin, and Ezh2 downregulation of Chung and Tellam (2008), whereas in rhabdomyosarcoma tumor cells miR-26a expression is lower and EZH2 is higher (Ciarapica et al., 2009). Our findings provide further evidence the key role of miR-26a in rhabdomyosarcoma cells, proposing also a novel potential role as blood biomarker. The prospective to use miRNAs as novel biomarkers in liquid biopsies of cancer patients has been explored by many groups in the last years, proving that circulating miRNA levels are different in cancer patients and healthy subjects, and positively correlate with the presence of metastasis at diagnosis and poor overall survival of the former (Imaoka et al., 2016; Li H.Y. et al., 2017; Guo et al., 2018; Mazza et al., 2020). With respect to this, the usefulness of droplets digital PCR for the quantification of circulating miRNAs has been demonstrated, both when used to compare circulating miRNAs levels between cases and controls (Mangolini et al., 2015), as well as when used to monitor the absolute levels of tumor-related miRNAs in plasma of patients with solid tumors (Zhao et al., 2018; Mazza et al., 2020).
Taken together our study supports the importance to explore the use of liquid biopsies in rhabdomyosarcoma pediatric patients managing, in particular the clinical utility of circulating miRNAs as novel diagnostic and prognostic biomarkers for this disease.
By using a qPCR Cancer Panel to screen circulating miRNAs we confirmed three miRNAs significantly downregulated in RMS patients’ plasma sample, one of which (miR-26a) also associated with known negative prognostic factors (RMS histology and fusion status) and significantly correlated with worst outcome. Although we are aware that a wider analysis on a larger cohort of patients is warranted, these findings disclose for the first time the potential to detect the absolute levels of circulating miRNAs in RMS patients, for both diagnostic and prognostic purposes. In this context, our data enforced the key role of miR-26a as both tumor suppressor and regulator of myogenesis in pediatric rhabdomyosarcoma.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving human biological samples were reviewed and approved by the local Ethics Committee as part of a clinical trial currently carried out with the Associazione Italiana Ematologia Pediatrica (AIEOP = Italian Association of Pediatric Hematology and Oncology. The patients/participants provided their written informed consent to participate in this study.
LT conceptualized the study and wrote of the manuscript. CM and BP performed the experiments. AZ provided the biological samples. MC performed the statistical analysis. PB revised the manuscript. GB revised the manuscript, supervised the work, and funding acquisition. All authors reviewed and approved the final manuscript as submitted.
This work is supported by Fondazione AIRC per la Ricerca sul Cancro (AIRC) grant IG-15813 (GB) and Fondazione Cassa di Risparmio di Padova e Rovigo (CARIPARO), grant number ID 17/09. PB and AZ were supported by Fondazione Città della Speranza (the City of Hope Foundation).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Ilaria Zanetti, Angela Scagnellato, and Julia Daragjati for clinical data management. Thanks to Pediatric-Oncology Biobank (BBOP) for biological samples management.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.606274/full#supplementary-material
Arroyo, J. D., Chevillet, J. R., Kroh, E. M., Ruf, I. K., Pritchard, C. C., Gibson, D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008. doi: 10.1073/pnas.1019055108
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Blondal, T., Jensby Nielsen, S., Baker, A., Andreasen, D., Mouritzen, P., Wrang Teilum, M., et al. (2013). Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6. doi: 10.1016/j.ymeth.2012.09.015
Cao, J. M., Li, G. Z., Han, M., Xu, H. L., and Huang, K. M. (2017). MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed Pharmacother. 93, 554–560. doi: 10.1016/j.biopha.2017.06.084
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Chung, F. W., and Tellam, R. L. (2008). MicroRNA-26a targets the histone methyltransferase enhancer of zeste homolog 2 during myogenesis. J. Biol. Chem. 283, 9836–9843. doi: 10.1074/jbc.M709614200
Ciarapica, R., Russo, G., Verginelli, F., Raimondi, L., Donfrancesco, A., Rota, R., et al. (2009). Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle 8, 172–175. doi: 10.4161/cc.8.1.7292
Fan, M., Ma, X., Wang, F., Zhou, Z., Zhang, J., Zhou, D., et al. (2020). MicroRNA-30b-5p functions as a metastasis suppressor in colorectal cancer by targeting Rap1b. Cancer Lett. 477, 144–156. doi: 10.1016/j.canlet.2020.02.021
Gasparini, P., Ferrari, A., Casanova, M., Limido, F., Massimino, M., Sozzi, G., et al. (2019). MiRNAs as players in rhabdomyosarcoma development. Int. J. Mol. Sci. 20:5818. doi: 10.3390/ijms20225818
Guo, J., Liu, C., Wang, W., Liu, Y., He, H., Chen, C., et al. (2018). Identification of serum miR-1915-3p and miR-455-3p as biomarkers for breast cancer. PLoS One 13:e0200716. doi: 10.1371/journal.pone.0200716
Horak, M., Novak, J., and Bienertova-Vasku, J. (2016). Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 410, 1–13. doi: 10.1016/j.ydbio.2015.12.013
Hu, Y., Liu, H., Fang, C., Li, C., Xhyliu, F., Dysert, H., et al. (2020). Targeting of CD38 by the tumor suppressor miR-26a serves as a novel potential therapeutic agent in multiple myeloma. Cancer Res. 80, 2031–2044. doi: 10.1158/0008-5472.CAN-19-1077
Huh, W. W., and Skapek, S. X. (2010). Childhood rhabdomyosarcoma: new insight on biology and treatment. Curr. Oncol. Rep. 12, 402–410. doi: 10.1007/s11912-010-0130-3
Imaoka, H., Toiyama, Y., Fujikawa, H., Hiro, J., Saigusa, S., Tanaka, K., et al. (2016). Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. 27, 1879–1886. doi: 10.1093/annonc/mdw279
Jin, F., Wang, Y., Li, M., Zhu, Y., Liang, H., Wang, C., et al. (2017). MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 8:e2540. doi: 10.1038/cddis.2016.461
Li, H. Y., Liang, J. L., Kuo, Y. L., Lee, H. H., Calkins, M. J., Chang, H. T., et al. (2017). miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. 19:133. doi: 10.1186/s13058-017-0918-2
Li, J., Liang, Y., Lv, H., Meng, H., Xiong, G., Guan, X., et al. (2017). miR-26a and miR-26b inhibit esophageal squamous cancer cell proliferation through suppression of c-MYC pathway. Gene 625, 1–9. doi: 10.1016/j.gene.2017.05.001
Li, L., Sarver, A. L., Alamgir, S., and Subramanian, S. (2012). Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab Investig. 92, 571–583. doi: 10.1038/labinvest.2012.10
Liu, W., Li, H., Wang, Y., Zhao, X., Guo, Y., Jin, J., et al. (2017). MiR-30b-5p functions as a tumor suppressor in cell proliferation, metastasis and epithelial-to-mesenchymal transition by targeting G-protein subunit α-13 in renal cell carcinoma. Gene 626, 275–281. doi: 10.1016/j.gene.2017.05.040
Mangolini, A., Ferracin, M., Zanzi, M. V., Saccenti, E., Ebnaof, S. O., Poma, V. V., et al. (2015). Diagnostic and prognostic microRNAs in the serum of breast cancer patients measured by droplet digital PCR. Biomark Res. 3:12. doi: 10.1186/s40364-015-0037-0
Mazza, T., Gioffreda, D., Fontana, A., Biagini, T., Carella, M., Palumbo, O., et al. (2020). Clinical significance of circulating miR-1273g-3p and miR-122-5p in pancreatic cancer. Front. Oncol. 10:44. doi: 10.3389/fonc.2020.00044
McCarthy, J. J. (2008). MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta Gene Regul. Mech. 1779, 682–691. doi: 10.1016/j.bbagrm.2008.03.001
Missiaglia, E., Shepherd, C. J., Aladowicz, E., Olmos, D., Selfe, J., Pierron, G., et al. (2017). MicroRNA and gene co-expression networks characterize biological and clinical behavior of rhabdomyosarcomas. Cancer Lett. 385, 251–260. doi: 10.1016/j.canlet.2016.10.011
Missiaglia, E., Shepherd, C. J., Patel, S., Thway, K., Pierron, G., Pritchard-Jones, K., et al. (2010). MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br. J. Cancer 102, 1769–1777. doi: 10.1038/sj.bjc.6605684
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. doi: 10.1073/pnas.0804549105
Miyachi, M., Tsuchiya, K., Yoshida, H., Yagyu, S., Kikuchi, K., Misawa, A., et al. (2010). Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 400, 89–93. doi: 10.1016/j.bbrc.2010.08.015
Molist, C., Navarro, N., Giralt, I., Zarzosa, P., Gallo-Oller, G., Pons, G., et al. (2020). miRNA-7 and miRNA-324-5p regulate alpha9-Integrin expression and exert anti-oncogenic effects in rhabdomyosarcoma. Cancer Lett. 477, 49–59. doi: 10.1016/j.canlet.2020.02.035
Oberlin, O., Rey, A., Sanchez, De Toledo, J., Martelli, H., Jenney, M. E. M., et al. (2012). Randomized comparison of intensified six-drug versus standard three-drug chemotherapy for high-risk nonmetastatic rhabdomyosarcoma and other chemotherapy-sensitive childhood soft tissue sarcomas: long-term results from the international society of pediatric oncology MMT95 study. J. Clin. Oncol. 30, 2457–2465. doi: 10.1200/JCO.2011.40.3287
Rota, R., Ciarapica, R., Giordano, A., Miele, L., and Locatelli, F. (2011). MicroRNAs in rhabdomyosarcoma: pathogenetic implications and translational potentiality. Mol. Cancer 10:120. doi: 10.1186/1476-4598-10-120
Rudzinski, E. R., Anderson, J. R., Hawkins, D. S., Skapek, S. X., Parham, D. M., and Teot, L. A. (2015). The world health organization classification of skeletal muscle tumors in pediatric rhabdomyosarcoma a report from the children’s oncology group. Arch. Pathol. Lab. Med. 139, 1281–1287. doi: 10.5858/arpa.2014-0475-OA
Sander, S., Bullinger, L., Klapproth, K., Fiedler, K., Kestler, H. A., Barth, T. F. E., et al. (2008). MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 112, 4202–4212. doi: 10.1182/blood-2008-03-147645
Sebire, N. J., and Malone, M. (2003). Myogenin and MyoD1 expression in paediatric rhabdomyosarcomas. J. Clin. Pathol. 56, 412–416. doi: 10.1136/jcp.56.6.412
Shah, J. S., Soon, P. S., and Marsh, D. J. (2016). Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS One 11:e0153200. doi: 10.1371/journal.pone.0153200
Smolle, M. A., Leithner, A., Posch, F., Szkandera, J., Liegl-Atzwanger, B., and Pichler, M. (2017). MicroRNAs in different histologies of soft tissue sarcoma: a comprehensive review. Int. J. Mol. Sci. 18:1960. doi: 10.3390/ijms18091960
Sorensen, P. H. B., Lynch, J. C., Qualman, S. J., Tirabosco, R., Lim, J. F., Maurer, H. M., et al. (2002). PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J. Clin. Oncol. 20, 2672–2679. doi: 10.1200/jco.2002.03.137
Tombolan, L., Zampini, M., Casara, S., Boldrin, E., Zin, A., Bisogno, G., et al. (2015). MicroRNA-27a contributes to rhabdomyosarcoma cell proliferation by suppressing RARA and RXRA. PLoS One 10:e0125171. doi: 10.1371/journal.pone.0125171
Varshney, J., and Subramanian, S. (2015). MicroRNAs as potential target in human bone and soft tissue sarcoma therapeutics. Front. Mol. Biosci. 2:31. doi: 10.3389/fmolb.2015.00031
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–435. doi: 10.1038/ncb2210
Wang, H., Garzon, R., Sun, H., Ladner, K. J., Singh, R., Dahlman, J., et al. (2008). NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14, 369–381. doi: 10.1016/j.ccr.2008.10.006
Wang, Y., Zhang, L., Pang, Y., Song, L., Shang, H., Li, Z., et al. (2020). MicroRNA-29 family inhibits rhabdomyosarcoma formation and progression by regulating GEFT function. Am. J. Transl. Res. 12, 1136–1154.
Zhao, D. W., Li, M. M., Han, J. P., Wang, Y., Jiang, L. X., and Chang, H. L. (2019). MiR-30c exerts tumor suppressive functions in colorectal carcinoma by directly targeting BCL9. Eur. Rev. Med. Pharmacol. Sci. 23, 3335–3343. doi: 10.26355/eurrev_201904_17696
Keywords: miR-26a, circulating microRNAs, ddPCR, pediatric tumor, rhabdomyosarcoma
Citation: Tombolan L, Millino C, Pacchioni B, Cattelan M, Zin A, Bonvini P and Bisogno G (2020) Circulating miR-26a as Potential Prognostic Biomarkers in Pediatric Rhabdomyosarcoma. Front. Genet. 11:606274. doi: 10.3389/fgene.2020.606274
Received: 14 September 2020; Accepted: 17 November 2020;
Published: 10 December 2020.
Edited by:
Federica Calore, The Ohio State University, United StatesReviewed by:
Sandra Elie Ghayad, Lebanese University, LebanonCopyright © 2020 Tombolan, Millino, Pacchioni, Cattelan, Zin, Bonvini and Bisogno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucia Tombolan, bHVjaWEudG9tYm9sYW5AdW5pcGQuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.