- Laboratory of Cellular and Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, United States
Cellular identity in multicellular organisms is maintained by characteristic transcriptional networks, nutrient consumption, energy production and metabolite utilization. Integrating these cell-specific programs are epigenetic modifiers, whose activity is often dependent on nutrients and their metabolites to function as substrates and co-factors. Emerging data has highlighted the role of the nutrient-sensing enzyme O-GlcNAc transferase (OGT) as an epigenetic modifier essential in coordinating cellular transcriptional programs and metabolic homeostasis. OGT utilizes the end-product of the hexosamine biosynthetic pathway to modify proteins with O-linked β-D-N-acetylglucosamine (O-GlcNAc). The levels of the modification are held in check by the O-GlcNAcase (OGA). Studies from model organisms and human disease underscore the conserved function these two enzymes of O-GlcNAc cycling play in transcriptional regulation, cellular plasticity and mitochondrial reprogramming. Here, we review these findings and present an integrated view of how O-GlcNAc cycling may contribute to cellular memory and transgenerational inheritance of responses to parental stress. We focus on a rare human genetic disorder where mutant forms of OGT are inherited or acquired de novo. Ongoing analysis of this disorder, OGT- X-linked intellectual disability (OGT-XLID), provides a window into how epigenetic factors linked to O-GlcNAc cycling may influence neurodevelopment.
Introduction
Throughout the lifetime of an organism there is a requirement to be able to adapt to environmental changes, whether that be development, stress, or nutritional state. This adaptation requires changes in transcriptional programs that allow an appropriate gene regulatory network response. One way this is achieved is through epigenetic regulation: heritable modifications of DNA and histones that influence complex networks impacting transcription, DNA replication, and DNA repair (Janke et al., 2015). Importantly, the activity of all epigenetic modifying enzymes relies on the availability of specific metabolites. This sets up signaling pathways in which cells have evolved the ability to detect nutrient alterations and respond with epigenetic changes to coordinate appropriate transcriptional programs. Some of the most well defined metabolites and their influence on epigenetic modifications include: acetyl-CoA and histone acetylation; sirtuins, NAD+, and histone deacetylation; S-adenosylmethionine and DNA/histone methylation; FAD, α-ketoglutarate, and DNA/histone demethylation (Janke et al., 2015; Etchegaray and Mostoslavsky, 2016; Wong et al., 2017; Schvartzman et al., 2018). Here, we focus on the conserved epigenetic role that the nutrient-sensitive post-translational modification (PTM) O-GlcNAc, and the enzymes involved in O-GlcNAc cycling, the O-GlcNAc transferase (OGT) and the O-GlcNAcase (MGEA5 or OGA) have in coordinating transcriptional responses to environmental changes.
Neurodevelopment is one such process that is heavily coordinated by the crosstalk between the genome, environment and metabolic flux. Alterations during this process could lead to neurodevelopmental disorders, which constitute a broad range of disorders that originate during development of the central nervous system. As more and more sequences become available from neurodevelopmental disorder patients, it is becoming increasingly clear the essential role chromatin modifiers and epigenetic regulators play in these diseases. Of the hundreds of genes with mutations or copy number variations that are causative of neurodevelopmental disorders, chromatin regulation is the second most common association behind synaptic function (Gabriele et al., 2018). Interestingly, recent data has identified placental OGT levels as a biomarker for neurodevelopmental disorders (Howerton et al., 2013; Howerton and Bale, 2014) and mutations in the OGT gene as being causative to a subset of X-linked intellectual disability (XLID) patients (Pravata et al., 2020b).
O-GlcNAc transferase uses the end-product of the hexosamine biosynthetic pathway (HBP), UDP-GlcNAc, to add a single GlcNAc monosaccharide onto serines and threonines of target intracellular proteins. To form UDP-GlcNAc, the HBP incorporates intermediate metabolites derived from carbohydrates, amino acids, fat, and nucleotides (Figure 1). Thus, OGT is uniquely positioned to sense environmental changes and respond through modifying key targets. A diverse array of more than 4,000 proteins have been identified to be O-GlcNAc modified (Ma and Hart, 2014), including transcription factors, epigenetic modulators, enzymes, kinases, mitochondrial proteins, structural proteins, nuclear porins, and components of vesicular trafficking pathways. How OGT recognizes specific target proteins remains relatively unclear, although it has preference for intrinsically disordered domains (Nishikawa et al., 2010). O-GlcNAcylation can influence modified proteins in various ways such as through crosstalk or competition with other PTMs like phosphorylation, altering enzyme activity, impacting protein stability, influencing subcellular localization, or altering binding partners (Bond and Hanover, 2013, 2015). Through this broad range of targets, O-GlcNAc influences many basic molecular processes including transcription, translation, proteostasis, and signaling [for more extensive reviews on the broad roles of O-GlcNAc and its targets see Bond and Hanover (2015) and Yang and Qian (2017)]. Regulation of O-GlcNAcylation has proven to be essential as too little or too much O-GlcNAc is associated with a number of diseases such as cancer, metabolic syndromes, and neurodegenerative diseases. Recently, OGT has been linked to a rare neurodevelopmental disorder known as OGT-XLID, which we hypothesize could be caused by dysfunction of the essential role of O-GlcNAc as an epigenetic regulator, the focus of this review.
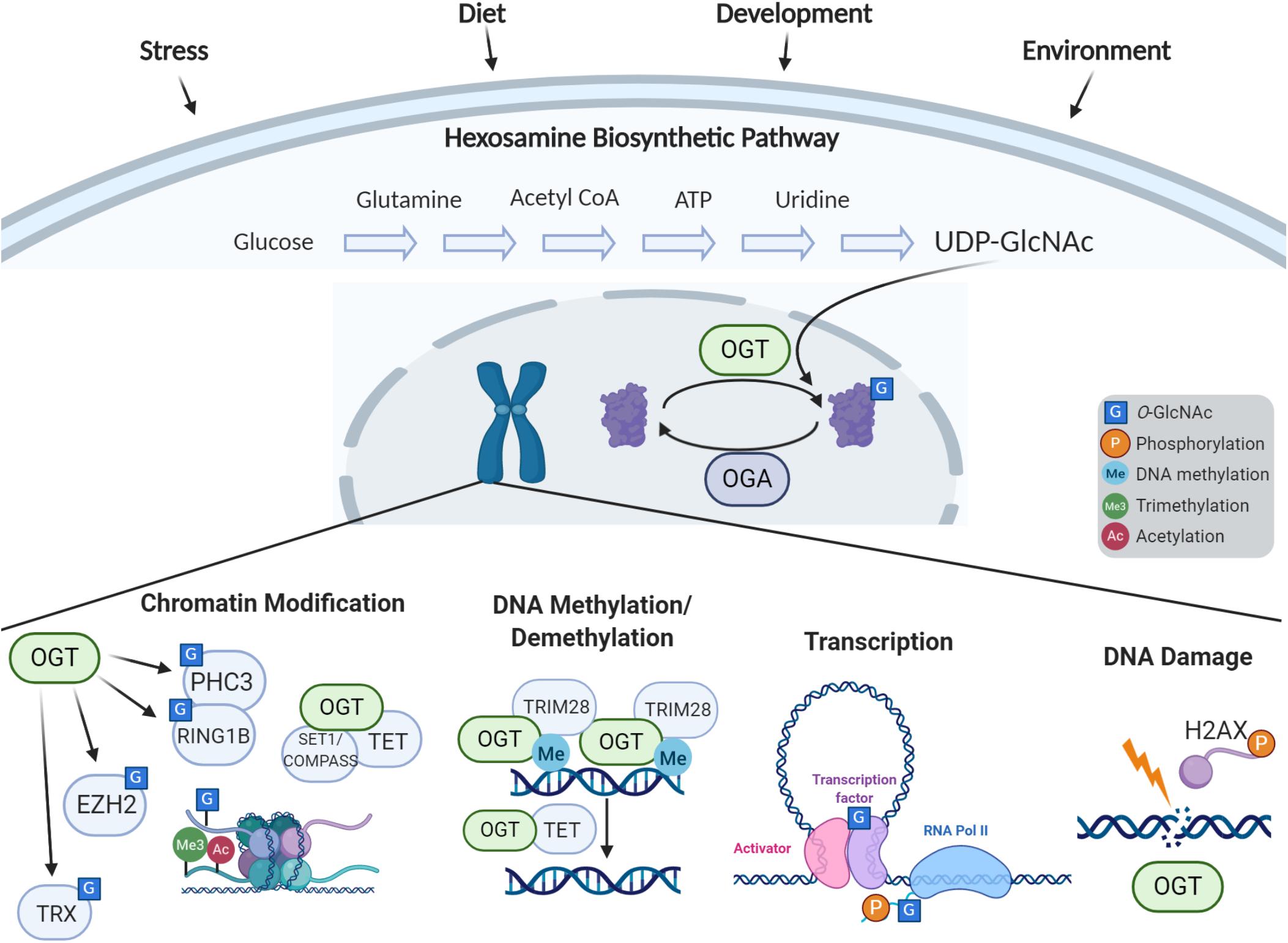
Figure 1. Cellular response to stress via O-GlcNAc-dependent epigenetic regulation. The hexosamine biosynthetic pathway incorporates nutrients and intermediate metabolites to form the nucleotide sugar UDP-GlcNAc. The O-GlcNAc Transferase (OGT) utilizes this nucleotide sugar as the sugar donor to modify diverse nucleocytoplasmic proteins including transcription factors, chromatin modifiers, and histones. This modification can be removed by O-GlcNAcase (OGA), whose levels are also implicated in regulation of gene expression. The process of O-GlcNAc cycling enables the cell to sense and integrate nutrient and signaling information and respond to extracellular stressors. O-GlcNAc influences histone modifications through the interactions of OGT with the Polycomb complex and SET1/COMPASS, OGA’s interactions with the trithorax complex, and direct O-GlcNAcylation of histones. OGT associates with TRIM28 to induce DNA methylation-dependent repression, and OGT is also involved in demethylation of DNA through the TET proteins. O-GlcNAc influences transcription by modification of transcription factors and the C-terminal domain of RNA Polymerase II. O-GlcNAc is also involved in the DNA damage response.
OGT, OGA, and O-GlcNAc itself have all emerged as critical epigenetic regulators (Figure 1) essential for stem cell maintenance, development, and in the nervous system. Studies in model organisms have highlighted the conserved role OGT plays in gene regulation and cellular identity. Since the initial findings in Drosophila which defined the gene encoding OGT as a Polycomb group member critical for Hox gene silencing (Gambetta et al., 2009), O-GlcNAcylation has been recognized to play multifaceted roles in epigenetic regulation. Recent studies in C. elegans have demonstrated that OGT plays a critical role in preventing transitions between terminal cell fates. Advances in mass spectrometry technology have uncovered O-GlcNAcylation of histones, and is now widely believed to be part of the “histone code.” Studies have also indicated that O-GlcNAc plays a role in histone exchange and is essential for the DNA damage response (Na et al., 2020). Further, OGT has been found in complex with TET proteins, signifying a potential role in DNA demethylation (Chen et al., 2013; Deplus et al., 2013; Vella et al., 2013). O-GlcNac has also been defined to regulate transcription through modification of key transcription factors as well as the RNA polymerase II C-terminal repeat domain (Lewis and Hanover, 2014).
In this review we dissect the conserved and wide-ranging roles O-GlcNAc cycling has in regulating transcriptional networks which contribute toward maintaining cellular identity. We discuss how placental OGT could be a marker for placental stress contributing toward neurodevelopmental disorders. Lastly, we examine how OGT’s role in epigenetic regulation contributes toward mutations manifesting as an XLID in human patients.
O-GlcNAc Cycling, Epigenetics and Cell Fate Determination: Lessons From Model Organisms
Super Sex Combs: OGT and Hox Gene Repression
Across species, O-GlcNAc has a key role to play in development. Though its functions are numerous, OGT is particularly important in the spatiotemporal control of gene expression through its regulation of Hox genes. Conserved across bilaterians, the Hox genes encode critical transcription factors which specify the body plan along the anterior-posterior axis. Inappropriate regulation of these genes results in homeotic transformations, developmental errors in which one region of an animal inappropriately adopts the characteristics of another region. Two major groups of genes control the spatial expression of Hox genes: transcriptional repression by the Polycomb group (PcG) and activation by the trithorax group (trxG). In Drosophila, the gene encoding OGT was first described genetically by mutations that caused homeotic phenotypes including antenna-to-leg and wing-to-haltere transformations (Butler et al., 2019). Without knowing the enzymatic function of the protein it encodes, the gene was named super sex combs (sxc) for its loss of function phenotype. Like PcG genes Polycomb (Pc) and extra sex combs (esc), sxc mutations affect the development of the sex combs, a structure on the forelimbs of Drosophila males. Similar to other components of the PcG, sxc was required for the repression of Hox genes in tissues where they should not be expressed. Derepression of Hox genes could explain the homeotic transformations associated with sxc mutations. Epistasis experiments confirmed this, as sxc flies lacking the Hox gene Ubx had more normal wing development (Ingham, 1984). Later, immunostaining of embryos showed aberrant expression of at least six PcG target genes including Ubx and Abd-B (Gambetta et al., 2009; Gambetta and Müller, 2014).
Experiments on the PcG gene polyhomeotic (ph) demonstrated genetic interactions with sxc. Flies carrying sxc and ph mutations had enhanced sxc phenotypes, with greater numbers of sex combs, stronger developmental defects, and lethality even earlier in development (Cheng et al., 1994). These findings uncovered further genetic interactions and demonstrated that sxc functions with the PcG to prevent ectopic expression of Hox genes.
In 2009, the protein product of sxc was determined to be the O-GlcNAc transferase, with expression of an Ogt transgene rescuing the pupal lethality of sxc mutant animals (Sinclair et al., 2009). Further studies better defined the interaction between sxc/Ogt and ph. In fact, co-IP experiments indicated that PH was O-GlcNAc modified (Gambetta et al., 2009). In extracts of larvae with homozygous sxc/Ogt mutations and no maternal Ogt contribution, PH was shown to aggregate into high molecular weight assemblies, which likely impaired its function (Gambetta and Müller, 2014). Loss of O-GlcNAcylation of PH resulted in sxc phenotypes, demonstrating modification was required to prevent aggregation, allowing PH to function with the PcG (Gambetta and Müller, 2014). Later, mass spectrometry of mouse embryonic stem cell (ESC) lysates demonstrated PHC3, the mammalian homolog of PH, is O-GlcNAc modified, suggesting the regulation of the PcG by O-GlcNAc is conserved (Myers et al., 2011). In human ESCs, the core subunit of PRC1, RING1B, is also modified and plays a role in targeting the complex (Maury et al., 2015). These findings clearly demonstrate the necessity of O-GlcNAc for the proper repression of Hox genes through the PcG component PH.
In mammals, the role of O-GlcNAc in PcG repression has diverged in certain ways. While PRC2 protein E(z) is not O-GlcNAc modified in Drosophila (Gambetta et al., 2009), several studies have identified its mammalian homolog EZH2 as modified, resulting in increased protein stability (Chu et al., 2014; Jiang et al., 2019). The effect of O-GlcNAc modification on PRC2-mediated H3K27me3 is controversial, with several publications describing a decrease associated with OGT depletion (Chu et al., 2014; Butler et al., 2019) and others noting no change (Myers et al., 2011; Forma et al., 2018; Jiang et al., 2019). These discrepancies likely arise from different cell lines being used, though one study found OGT knockdown prevented H3K27me3 changes associated with learning at specific gene promoters in mouse hippocampal tissue (Butler et al., 2019). This suggests the effects of O-GlcNAc may play roles in targeting PcG repression which are only apparent in certain developmental and genomic contexts.
O-GlcNAc transferase additionally interacts directly with the mammalian HOXA1 protein, as was found in a yeast-two-hybrid experiment (Lambert et al., 2012) and confirmed by co-IP (Draime et al., 2018). Though the authors did not find evidence for O-GlcNAc affecting localization, stability, or transcription factor activity (Draime et al., 2018), they only tested OGT and HOXA1 overexpression, so further experimentation may reveal additional layers of Hox gene regulation by O-GlcNAc. Although the phenotypes of sxc/Ogt flies are mostly consistent with those of other PcG genes, several findings suggest sxc/Ogt is also involved in Hox gene activation through trxG. Alone, both sxc/Ogt and Asx mutations produce anterior-to-posterior transformations associated with PcG mutations, but sxc/Ogt;Asx double heterozygotes display posterior-to-anterior transformations, which are typically associated with trxG mutations (Milne et al., 1999). This suggests sxc/Ogt acts not only as a Hox gene repressor through PcG but also as an activator through trxG. In fact, TRX and related transcriptional activators SET1/COMPASS and ASH1 have been shown to be O-GlcNAc modifiable, and staining for these proteins overlaps with O-GlcNAc on polytene chromosomes (Akan et al., 2016). The interaction between O-GlcNAc and trxG also appears in mammals, with O-GlcNAc modification of the H3K4 methyltransferase MLL5 working to stabilize this trxG enzyme (Ding et al., 2015). Thus, in addition to the necessary role of O-GlcNAc in the Polycomb repressive complex, this modification is also involved in transcriptional activation, highlighting the diverse routes in which O-GlcNAc regulates gene expression.
The appropriate spatiotemporal expression of Hox genes is crucial to the proper development of an organism. These regulatory systems which repress and activate Hox genes can cause severe developmental disorders when out of balance (Quinonez and Innis, 2014). Common clinical presentation for PcG mutations include intellectual disability and growth defects (Deevy and Bracken, 2019), which mirror those of OGT-XLID. Thus, the role of OGT in developmental regulation may contribute to symptoms of this disorder.
O-GlcNAc Transferase in Cell Fate Plasticity
Developmental plasticity is necessary for the development of multicellular organisms, but this plasticity must be restricted when cells reach their terminal fates. Several labs have worked to define which factors are involved in the epigenetic processes that impose barriers between cell fates using the nematode C. elegans. The worm is a model system perfectly suited to study the genetics of development, as C. elegans are relatively simple multicellular animals, with a rigidly regulated series of cell divisions and fate decisions. Several studies have employed genetic screens to determine which genes are required to maintain proper cell fates. Two independent studies have found the sole nemotode OGT ortholog ogt-1 is involved in the restriction of cellular plasticity.
Both experiments are based around genetic screens using strains sensitized to cell fate transformation by ectopic overexpression of the CHE-1 transcription factor. CHE-1 is a master regulator which is necessary to establish the fate of a specific pair of neurons called the ASE neurons, which can be monitored with a cell-specific reporter: gcy-5::gfp (Tursun et al., 2011). Due to mechanisms that impose barriers between cell fates, ectopic expression of che-1 later in development does not have any effects on gene expression or cell fate. Even when overexpressed in tissues that would not normally transcribe che-1, neither activation of CHE-1 target gene expression nor induction of ASE fate are observed in otherwise wild-type animals (Tursun et al., 2011). However, they found that mutation of the histone chaperone lin-53 allowed che-1 overexpression to implement neural fate in germ cells (Tursun et al., 2011), suggesting a role for histones and potentially histone modifications in maintaining cellular identity. Using similar genetic systems, more recent studies have uncovered additional factors, including ogt-1 (Hajduskova et al., 2019; Rahe and Hobert, 2019), that are required for maintenance of cellular identity.
Hajduskova et al. (2019) performed an RNAi screen of 730 candidate genes with known or predicted roles in chromatin modifications and remodeling. For this screen, expression of che-1 was induced with a heat-shock promoter and reporter GFP signal was monitored. Ectopic GFP expression was detected with knockdown of 10 of the 730 tested genes. Different gene knockdowns allowed GFP expression in different tissues, suggesting tissue-specific regulatory mechanisms. This study focused on one hit in particular: mrg-1, an ortholog of the mammalian gene MRG15. Knockdown of mrg-1 enabled che-1 overexpression to reprogram germline cells to neuron-like cells. To further study the process by which MRG-1 works as a barrier between cell fates, the authors performed IP-MS to identify MRG-1 binding partners, and uncovered OGT-1. The IP-MS experiments also identified other known chromatin interactors such as set-26 (H3K9 methyltransferase) and sin-3 (histone deacetylase), which is itself a predicted interactor with OGT-1 (Yang et al., 2002).
A similar screen was performed in an independent lab which looked for genes involved in protecting epidermal cells from transdifferentiation (Rahe and Hobert, 2019). In this study, they overexpressed che-1, but restricted its expression to the epidermis starting at the end of the final larval stage (L4) and continuing into adulthood. Without mutagenesis, gcy-5::gfp expression was limited to ASE neurons, but mutations in seven genes caused ectopic expression of the reporter. Three independent missense mutations were isolated in the ogt-1 gene. These three mutations were at highly conserved residues within the catalytic domains of OGT-1, in close proximity to sites previously reported to be necessary for enzymatic activity in human OGT (Lazarus et al., 2011). Other genes this screen identified included dot-1.1 (an H3K79 methyltransferase) and pmk-1/p38-alpha MAPK. In mammalian neuroblastoma cells, OGT and p38 MAPK have been shown to physically interact to drive O-GlcNAcylation of specific targets (Cheung and Hart, 2008).
Though these studies were performed in nematodes in the context of ectopic gene expression, these results point toward an important role for O-GlcNAcylation in the control of cell fate plasticity. ogt-1 came out of these two independent unbiased methods in the context of two different tissue types, suggesting it plays this role broadly. Though it has not been studied in as much depth in mammalian systems, some studies implicate O-GlcNAc in related processes. Much evidence points toward O-GlcNAc homeostasis being critical in stem and progenitor cells, which we will discuss further through its role in DNA damage and transcription factor regulation. In addition to its role restricting plasticity late in development, O-GlcNAc is critical in the networks of transcription factors which enable pluripotency in early development and adult stem cells which we will discuss in detail in its own section below. The developmental phenotypes of OGT-XLID patients and related transcriptomic data demonstrate the importance of O-GlcNAc in regulating sensitive cell fate specification events (Selvan et al., 2018). The importance of O-GlcNAc in cell fate may be intimately related to its roles in histone dynamics.
O-GlcNAc Cycling and Histones
Direct Modification of Histones by O-GlcNAc
O-GlcNAc affects chromatin and histone dynamics in a number of different ways. As described above, major complexes that modify histones are regulated by O-GlcNAc cycling. O-GlcNAc is also involved in the histone exchange required for DNA repair, and can modify histones directly.
Sakabe et al. (2010) first demonstrated direct evidence linking O-GlcNAcylation to the histone code, the holy grail of modern epigenetics (Jenuwein and Allis, 2001). Using an arduous combination of techniques they found O-GlcNAc on histones H2A, H2B, and H4. Acetylated histones were modified by O-GlcNAc and O-GlcNAcylation increased upon heat stress. Heat shock and OGT overexpression also modestly increased chromatin condensation. Using a chemical enrichment procedure, they mapped three O-GlcNAc sites on histones: H2AT101, H2BS36, and H4S47 (Sakabe et al., 2010). Additionally, they also provided evidence that other sites, including probable sites for modification of the remaining histone H3 must exist in the histone preparations. The identification of O-GlcNAc sites near known DNA interaction sites lead the authors to speculate that O-GlcNAcylation could induce major changes in chromatin structure not only by regulating peptide backbone conformation but also due to it being considerably larger than other common PTMs. One particularly intriguing O-GlcNAcylated site discussed was H4S47 (Sakabe et al., 2010). In yeast, mutation of this site to a cysteine induced activation of SWI/SNF targets independent of the SWI/SNF chromatin remodeling complex.
Since then, a total 17 different histone O-GlcNAcylation sites on H2A, H2AX, H2B, H3, H3.3, and H4 have been reported (see Table 1 for the full list with references). Evidence for these sites primarily comes from identification techniques such as chemoenzymatic detection, immunoblotting, selective enzymatic labeling, and lectin staining, in combinations with various mutation experiments. Due to the indirect nature of these methods, some skepticism has been raised about the existence of histone O-GlcNAcylation (Fujiki et al., 2011; Schouppe et al., 2011; Zhang et al., 2011; Hahne et al., 2012; Deplus et al., 2013; Lercher et al., 2015; Ronningen et al., 2015; Chen and Yu, 2016; Hirosawa et al., 2016; Hayakawa et al., 2017). However, the presence of O-GlcNAc at the two sites, H2AS40 and H3.3T32, have been confirmed based on the recognition of endogenous O-GlcNAc by tandem mass spectrometry (MS) analysis of histones isolated from mammalian cells (Sakabe et al., 2010; Fong et al., 2012), providing the most robust evidence for the O-GlcNAcylation of histones. Direct identification of peptidyl O-GlcNAcylated Ser/Thr by MS is burdensome due to its unstable nature, limiting attempts to confirm additional modification sites. Some studies have disputed the existence of histone O-GlcNAcylation, including one which reported an inability to detect modified histones in cultured mammalian cells (Gagnon et al., 2015; Gambetta and Muller, 2015). Still, indirect evidence suggests that O-GlcNAcylation is an important form of histone PTM.
The 17 identified histone O-GlcNAcylation sites play various important roles in different biological functions. O-GlcNAc is identified at higher levels on H3 during interphase than mitosis, inversely related with phosphorylation, suggesting PTM crosstalk. Also, an increase in O-GlcNAcylation was observed to reduce mitosis specific phosphorylation at Ser10, Ser28, and Thr32. Inhibition of OGA hindered the transition from G2 to M phase of the cell cycle, showing a phenotype similar to hindering mitosis-specific phosphorylation on H3 delivering a mechanistic switch that orchestrates the G2-M transition of the cell cycle (Fong et al., 2012).
The combination of the extensive diversity of histone modifications allows for the complexity and flexibility of epigenetic regulation (Chen and Yu, 2016). It is now possible to map the genome- wide distribution and colocalization of histone modifications at high resolutions using ChIP-seq, revealing the many amalgamations of histone modification crosstalk, such as precondition, mutual exclusion, or coexistence (Pick et al., 2014). H2BS112 O-GlcNAcylation functions as a precondition for H2BK120 monoubiquitination, with GlcNAc acting as an anchor for ubiquitin ligase, ultimately resulting in transcriptional activation via H3K4me3 (Fujiki et al., 2011). An increase in the intracellular level of UDP-GlcNAc induces an increase in histone O-GlcNAcylation and a partial suppression in H3S10ph, suggesting these modifications are mutually exclusive. Further examples of competition between O-GlcNAcylation and phosphorylation have been reported for the H3T32, H3S10, and H2AXS139 sites (Zhang et al., 2011; Fong et al., 2012; Chen and Yu, 2016). Future studies should, therefore, investigate phosphorylation of other residues for which O-GlcNAc modification has been reported. The crosstalk between O-GlcNAcylation and acetylation should also be validated considering the presence of a HAT-like domain in OGA, although the enzymatic activity of this domain is a point of controversy (Torres and Hart, 1984; Toleman et al., 2006; Kim et al., 2007; Butkinaree et al., 2008; Hayakawa et al., 2013; Rao et al., 2013).
O-GlcNAcylation of site-specific adapter proteins directly regulate the stability of H2A/H2B dimers in the nucleosome in synthetic O-GlcNAcylated histones (Lercher et al., 2015). To generate homogenous O-GlcNAc modified nucleosomes, one study generated H2AT101 O-GlcNAc mimics by replacing the threonine with cysteine and using a series of chemical reactions in vitro to stably link GlcNAc to the thiol. The authors showed that H2AT101 GlcNAcylation destabilized the H3/H4 tetramer-H2A/B dimer interface reducing nucleosome stability. Thus, regulation of nucleosome stability by OGT-dependent GlcNAc transfer may contribute to transcriptional regulation. O-GlcNAc’s role in histone dynamics and in modifying variant histones make up an additional layer of histone regulation.
H2AX and the DNA Damage Response
Histone modifications play a crucial role in chromatin organization through processes including DNA metabolism, replication, transcription, and repair. Modification and exchange of histones can also reorganize chromatin to allow DNA repair machinery to access damaged chromosomal DNA (Downs et al., 2004). H2AX is a histone variant that differs from H2A at various amino acid residues along the entire protein and in its C-terminal extensions (Bonisch and Hake, 2012). The importance of this histone variant is highlighted by the phenotypes of knockout mice, which show radiation sensitivity, developmental delay, and male infertility (Celeste et al., 2002). H2AX is a central player in the DNA damage response (DDR) when phosphorylated at serine 139 (γH2AX) (Wahl and Carr, 2001), and as mentioned previously, can also be O-GlcNAc modified at this site (Liu and Li, 2018). γH2AX is incorporated into nucleosomes at double strand break sites, where it promotes accumulation of DNA repair proteins (Wahl and Carr, 2001). γ-phosphorylation is an early event in the DSB damage response induced by the ATM and ATR kinases, which additionally activate kinases Chk1 and Chk2 (Wahl and Carr, 2001).
In the growth and development of an organism, DNA damage poses a serious risk, as mutations will propogate from progenitors to their daughter cells. To maintain genome integrity, the cell cycle is regulated by the DDR pathway following DNA damage stress, with O-GlcNAc involved by modifying the arrangement of histones and kinases (Hanover et al., 2018; Liu and Li, 2018). Blocking O-GlcNAc transferase activity leads to delayed DSB repair, reduced cell proliferation, and increased cell senescence in vivo, while increased O-GlcNAc promotes DSB repair and hyper-proliferation in vivo and in vitro (Efimova et al., 2019). These findings suggest O-GlcNAc is necessary to protect the genome and for proper cell cycle progression. These effects are likely related to OGT’s recruitment to sites of DNA damage, where it modifies H2A and mediator of DNA damage checkpoint 1 (MDC1) (Chen and Yu, 2016). In addition, one report has suggested OGT transfers GlcNAc onto H2AXS139, the same site as γ-phosphorylation (Chen and Yu, 2016). The authors suggest O-GlcNAc inhibits the DDR, though other studies suggest O-GlcNAc activates the DDR pathway (Efimova et al., 2019; Na et al., 2020).
The development of a multicellular organism is an energy-intensive and error-prone process. Stem and progenitor cells require high levels of glucose to grow and proliferate. A consequence of this energy use is the generation of reactive oxygen species (ROS), which cause damage, cellular stress, and DNA breaks. As such, DDR-related factors are prominent among proteins that accumulate O-GlcNAc when cells are stressed by ROS (Katai et al., 2016). Reciprocally, the DDR pathway has been shown to increase ROS levels (Rowe et al., 2008), pointing to a complex interplay between these processes. For example, high glucose has been demonstrated to elevate levels of O-GlcNAc, ROS, and DNA damage (Hu et al., 2019). The elevation of ROS in high glucose conditions may be linked to O-GlcNAc, as inhibition of OGT decreases ROS levels in a dose-dependent manner, and ultimately reduces neural tube defects in embryos of diabetic mice (Kim et al., 2017). Recently, we have reported that stress induces hyper-proliferation, O-GlcNAcylation, and DDR in Drosophila intestinal stem cells (Na et al., 2020). Likewise, genetic elevation of O-GlcNAc by deletion of Oga induced proliferation and DDR in fly intestinal stem cells, mouse embryonic fibroblasts, and mouse ESCs (Na et al., 2020). Previous work had shown that Chk1 phosphorylates OGT, which stabilizes the protein (Li et al., 2017). Through this interaction, we demonstrate that O-GlcNAc participates in an autoregulatory feedback loop where CHK1/CHK2 stabilizes OGT, allowing further O-GlcNAcylation that continues to activate the DDR pathway (Na et al., 2020) (Figure 2).
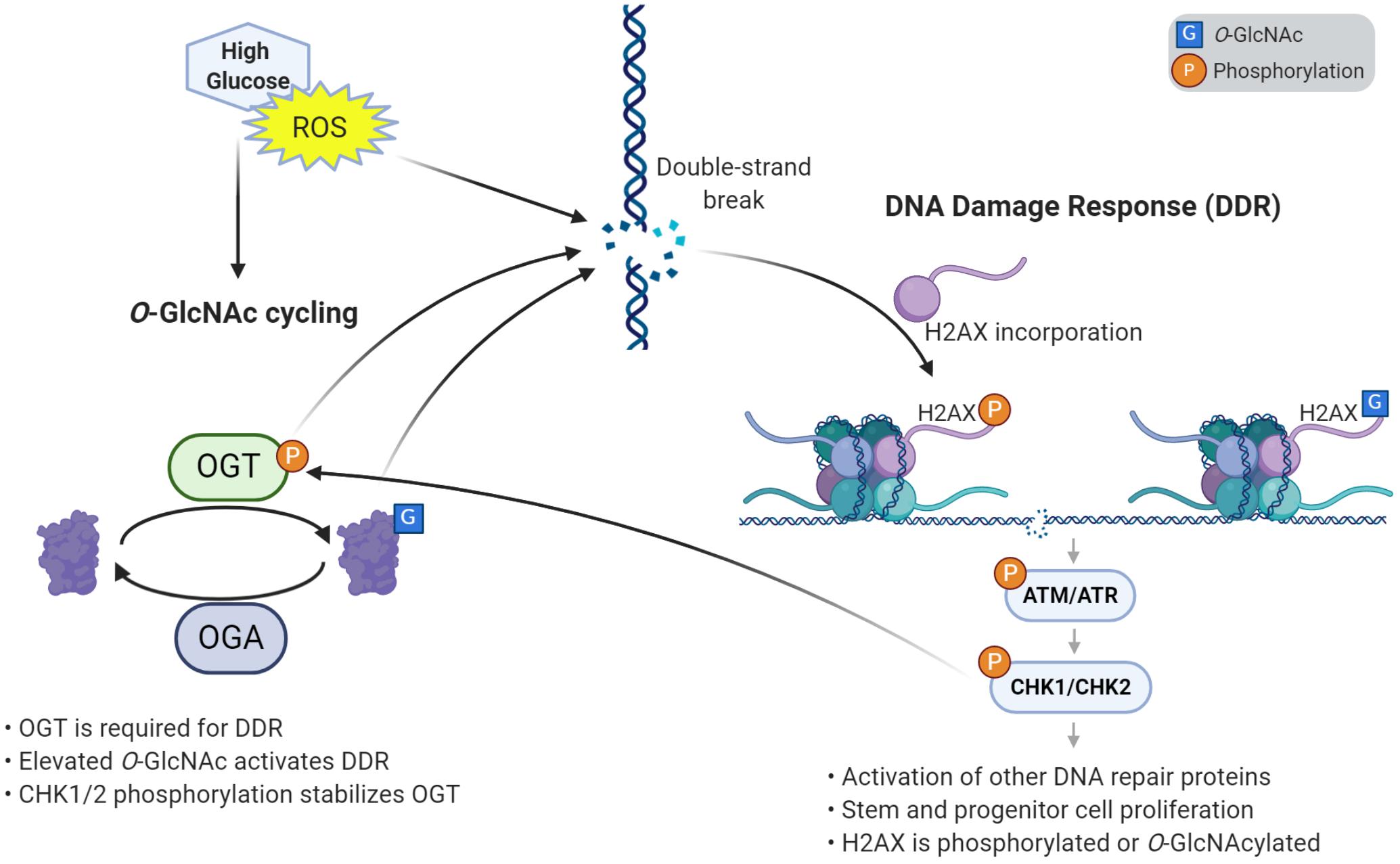
Figure 2. O-GlcNAcylation induces the DNA damage response and proliferation in stem cells. High glucose and reactive oxygen species (ROS) cause damage to DNA such as double strand breaks. O-GlcNAcylation is required to respond to DNA damage, through its accumulation at damage sites and activation of the DNA damage response (DDR) pathway. Response to DNA damage also requires the incorporation of variant histone H2AX, which is phosphorylated and can also be O-GlcNAc modified. Elevated O-GlcNAc induces stem and progenitor cell proliferation and the DNA damage response through γH2AX and a phosphorylation cascade of effector kinases. One of the downstream kinases, CHK1/2, phosphorylates OGT, which stabilizes the enzyme. This autoregulatory loop helps further promote the DNA damage response and proliferation.
Thus, O-GlcNAcylation is a key regulator of the DDR pathway, which is crucial in supporting the development of a healthy organism. This further establishes O-GlcNAc’s role in cell identity discussed above, which will be explored below in our discussion of transcription factor modification. Observations of abnormal neural proliferation and developmental delay in mice harboring an Oga deletion in the brain (Olivier-Van Stichelen et al., 2017) suggest O-GlcNAc homeostasis is needed for the proper development of sensitive tissues such as the brain. Thus, the pathways discussed above may contribute to the developmental defects associated with OGT-XLID. Beyond the effects O-GlcNAc has on chromatin dynamics, O-GlcNAc interacts with DNA methylation pathways to regulate gene expression.
OGT and DNA Methylation
DNA methylation is a critical epigenetic modification in mammals which occurs predominantly at the 5-position carbon on cytosine residues (5mC) followed by guanines. This epigenetic mark is involved in a variety of functions in the mammalian genome, including X-chromosome inactivation, gene silencing, genomic stability, cellular identity, and genomic imprinting (SanMiguel and Bartolomei, 2018). Two models for DNA methylation-dependent repression have been described. The first is a direct mechanism in which the presence of 5mC inhibits binding of transcription factors to DNA, thereby silencing gene expression. The second model is an indirect mechanism that involves recruitment of proteins that bind methylated DNA and associate with chromatin modifiers. These models are not mutually exclusive and can work in concert (Klose and Bird, 2006). Despite being stable and heritable, DNA methylation is also highly dynamic, particularly during development. Active DNA demethylation is mediated by the TET family proteins TET1, TET2, and TET3. These proteins iteratively oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009; Ito et al., 2010, 2011; He et al., 2011). These modifications can be transient intermediates in the demethylation process which are ultimately removed by base excision repair (Cortellino et al., 2011; He et al., 2011), or can act on their own as a stable modifications (Bachman et al., 2014, 2015). Importantly, OGT is a well established binding partner of the TET proteins and has recently been described to play a critical role in DNA methylation-dependent repression.
Recent studies have revealed that OGT is found in complex with the three TET proteins. OGT was found to be associated with TET1 at gene promoters of transcriptionally active genes in mouse ESCs. This interaction was required for OGT binding to chromatin and enhanced TET1 activity (Vella et al., 2013). Further, O-GlcNAcylation of TET1 was shown to regulate TET1 stability (Shi et al., 2013a), and OGT enhanced TET1 activity in vitro (Hrit et al., 2018). Disrupting the OGT-TET1 interaction in mouse ESCs resulted in increased 5mC and compensatory increases in TET2, accompanied by transcriptional changes (Hrit et al., 2018). These data indicate that the TET1-OGT complex is critical for proper pluripotency gene regulatory networks which maintain stem cell identity.
TET2 and TET3 also form complexes with OGT. Interaction of TET2 with OGT associates at transcriptional start sites and facilitates histone O-GlcNAcylation. HCF-1 as part of the SET1/COMPASS complexes is a specific target of the TET2/3-OGT complex and promotes H3K4me3. A closer look at the TET3-OGT interaction indicated that this interaction stabilized OGT and enhanced chromatin association (Ito et al., 2014). However, another study found that O-GlcNAcylation of TET3 promoted its export from the nucleus, thereby inhibiting TET3 function (Zhang et al., 2014). Further complicating the regulation of TET proteins by OGT is the fact that they are also highly phosphorylated (Bauer et al., 2015) setting up the possibility of PTM crosstalk. While the functional relationship between OGT and TET proteins still remains controversial, there are some common themes in these papers. TET, OGT, and H2BS112 O-GlcNAc are colocalized in the genome, largely at CpG islands containing promoters of actively transcribed genes (Chen et al., 2013; Deplus et al., 2013; Vella et al., 2013). Thus this interaction likely reinforces active transcription to maintain cell intrinsic transcriptional program, possibly impacting development and gene expression in OGT-XLID.
In addition to its effects on the process of DNA demethylation, OGT has been found to be involved in gene silencing mediated by DNA methylation. OGT selectively associates with the scaffolding protein TRIM28 only in the presence of methylated DNA (Boulard et al., 2020). It has been proposed that O-GlcNAcylation of chromatin modifiers that interact with TRIM28 is required at the sites of retrotransposon promoters to repress their transcription (Boulard et al., 2020). This suggests disruption of O-GlcNAc cycling may lead to increased genome instability in addition to the contribution of impaired DNA damage response described above.
Transcription Factor O-GlcNAcylation
Gene expression is largely regulated by transcription factors, which themselves are heavily regulated by PTMs. O-GlcNAcylation is one of the major modifications that affects transcription factor functions, modulating their localization, stability, interacting partners, resulting in gene activation or silencing. While many studies have defined the role of O-GlcNAc modification of key transcription factors regulating processes like immune activation (Chang et al., 2020), here we focus on the role O-GlcNAc has in pluripotency and differentiation as well as in the nervous system and glucose and lipid metabolism in the liver as it relates more directly to neurodevelopmental disorders that will be described (Figure 3).
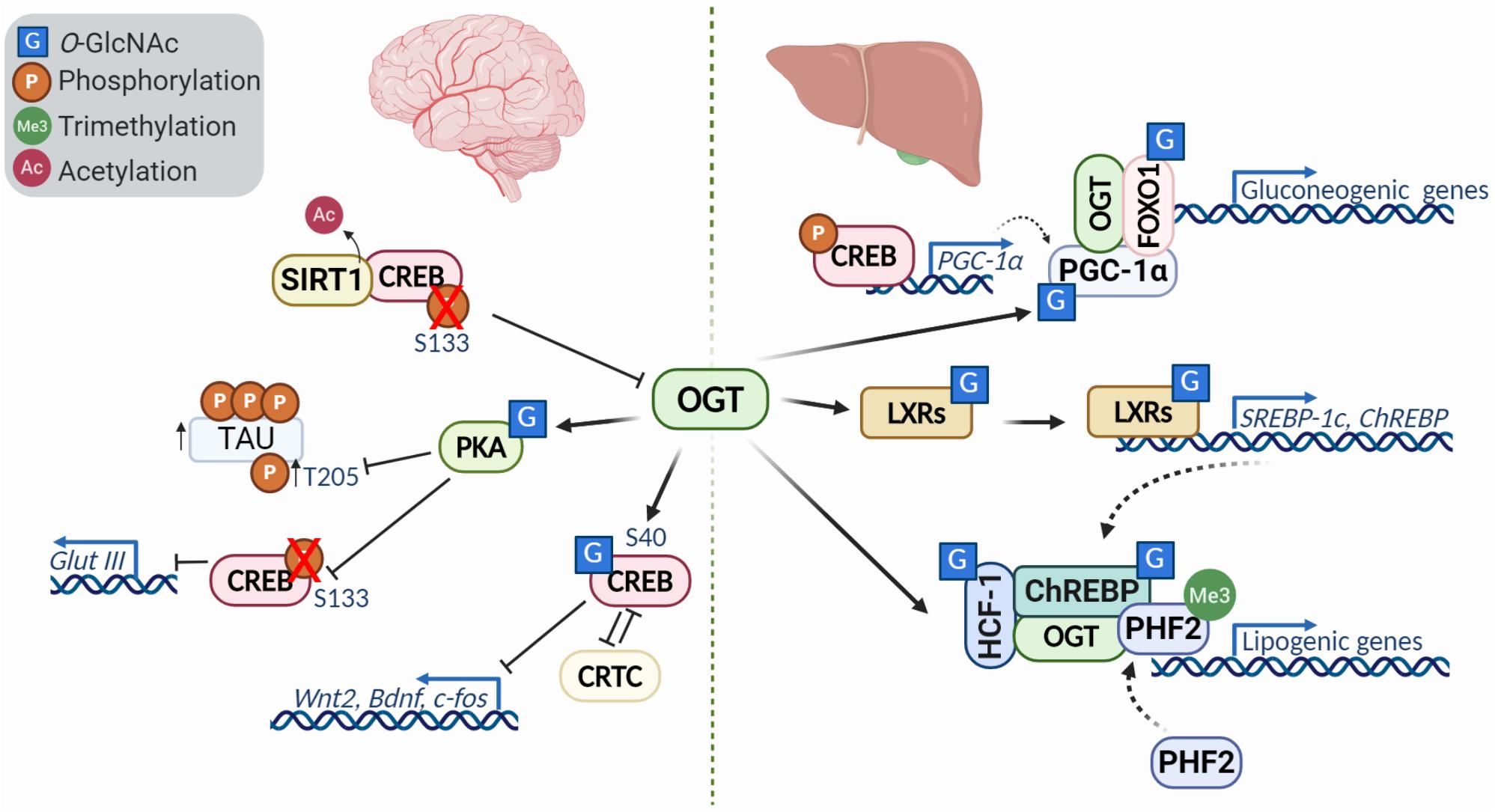
Figure 3. Diversity of transcription factor O-GlcNAcylation in the brain and liver. O-GlcNacylation of transcription factors can act to modulate their activity, localization, stability, binding partners and other post translational modifications. In the brain (left), O-GlcNAcylation of CREB represses its activity and downregulates transcriptional targets like Wnt2 and Bdnf in a CRTC (cAMP-regulated transcriptional co-activators)-dependent manner. Reduced O-GlcNAc of protein kinase A (PKA) is characteristic of Alzheimer’s disease, and leads to decreased phosphorylation of CREB at S133 and increased phosphorylation of tau at T205. Further, deacetylation of CREB by SIRT1, causes downregulation of OGT. In the liver (right), modification of PGC-1a and association with OGT promotes FOXO1 O-GlcNAcylation, enhancing activity and promoting transcription gluconeogenic genes. The liver X receptors (LXRs) are nutrient sensors with critical roles in lipid metabolism, glucose homeostasis, and metabolism. O-GlcNAc modification of LXRs induces expression of SREBP-1 and ChREBP, key factors involved in hepatic lipogenesis and energy metabolism. HCF-1 O-GlcNAcylation stimulates OGT recruitment and modification of ChREBP, enhancing ChREBP activity and promoting recruitment of the activator PHF2 which binds the activating histone mark H3K4me3.
O-GlcNAcylation in Development and Cell Differentiation
Throughout development, the process of differentiation allows a multicellular organism to form a complex system of multiple tissues and cell types. In adulthood, stem cells retain the capacity to differentiate allowing for tissue repair. O-GlcNAcylation during development is essential, as deletion of Ogt in mouse ESCs is lethal (Shafi et al., 2000). Further, reduced expression of Ogt disrupts stem cell self-renewal through deregulation of pluripotency transcriptional networks (Jang et al., 2012). This is likely due to OGT’s role in chromatin remodeling as described above, as well as through direct modification of transcription factors involved in pluripotency. In fact, the Yamanaka factors OCT4, SOX2, and C-MYC are all O-GlcNAc modified (Chou et al., 1995; Jang et al., 2012), and increasing O-GlcNAc in ESCs hampers normal differentiation (Jang et al., 2012). In fact, deleting Oga in the mouse is largely perinatal lethal (Keembiyehetty et al., 2015). Mice in which Oga was conditionally deleted in the brain exhibited microcephaly, high body fat percentage, hypotonia, and delayed development of the brain associated with abnormal cell proliferation and migration. This phenotype is related to transcriptional changes of pluripotency markers, including Sox2, Nanog, and Otx2 (Olivier-Van Stichelen et al., 2017). The transcription factor Sox2 and other transcription factors involved in the maintenance of mouse ESC pluripotency and stem cell self-renewal all require O-GlcNAcylation (Myers et al., 2011; Jang et al., 2012).
Cellular differentiation is regulated by O-GlcNAcylation, as lowered UDP-GlcNAc levels by HBP inhibition blocks the differentiation of adipocytes in vitro (Ishihara et al., 2010). Normal adipocyte differentiation in cell culture requires the transcription factors C/EBPα and C/EBPβ. It has been reported that C/EBPβ is modified by O-GlcNAc, and diminished O-GlcNAcylation reduces C/EBPα protein levels, suggesting the modification stabilizes the protein (Ishihara et al., 2010). Mass spectrometry analysis indicated two O-GlcNAc sites on Ser180 and Ser181, which are very close to C/EBPβ phosphorylation sites at Thr188, Ser184, and Thr179 (Li et al., 2009). The sequential phosphorylation of C/EBPβ at Thr188 then Ser184 by MAPK or CDK2, and Thr179 by GSK3β is required for DNA binding and transcriptional activity (Kim et al., 2007). Increased O-GlcNAcylation during adipocyte differentiation prevents C/EBPβ phosphorylation and subsequently delays adipocyte differentiation (Li et al., 2009). In addition, mutations of Ser180/181 rescued the phenotype induced by O-GlcNAcylation which suggests that the transcriptional activity of C/EBPβ is regulated by phosphorylation and O-GlcNAcylation in a competitive manner by alternative occupancy at adjacent sites (Li et al., 2009).
Hematopoietic stem cell (HSC) maintenance also requires balanced O-GlcNAc cycling. In fact, when Oga has been deleted in HSCs in mice, these mice exhibit diminished HSC pools as well as reduced intermediate progenitor populations. The elevated O-GlcNAcylation in progenitor cells of these mice were correlated with transcriptional changes in factors involved in adult stem cell maintenance, lineage specification and nutrient uptake (Abramowitz et al., 2019).
Transcription Factor O-GlcNAcylation in the Nervous System
O-GlcNAcylation has been extensively studied in the brain due to its critical role during development described above, and in neurodegenerative diseases like Alzheimer’s (Griffith and Schmitz, 1995; Olivier-Van Stichelen et al., 2017). Emerging data has highlighted the role of transcription factor O-GlcNAcylation during neuronal development, synaptic plasticity and memory. CREB, a key transcription factor involved in learning and memory, is O-GlcNAc modified in the TAFII130 binding domain, a component of the TFIID transcriptional complex (Lamarre-Vincent and Hsieh-Wilson, 2003). O-GlcNAcylation of CREB impairs its interaction with TAFII130 and represses CREB transcriptional activity in vitro (Lamarre-Vincent and Hsieh-Wilson, 2003). Further studies demonstrated that O-GlcNAcylation of CREB at Ser40 modulates dendrite and axonal elongation with downregulation of Wnt2 and BDNF signaling (Rexach et al., 2012). It has also been suggested that glycosylation of CREB has a significant impact on long-term memory consolidation (Rexach et al., 2012). In accordance with this hypothesis, an independent research group has provided a link between O-GlcNAcylation, Protein Kinase A (PKA)-CREB signaling, and memory loss in Alzheimer’s disease. Alzheimer’s disease is associated with a decrease in O-GlcNAcylation, which has been shown to influence aggregating proteins such as tau (Akan et al., 2018). PKAs can be O-GlcNAc modified, which influences their localization, activity, and phosphorylation (Xie et al., 2016). The inhibition of PKA-CREB signaling by O-GlcNAcylation was associated with impaired learning and memory in mice.
While CREB itself is regulated by O-GlcNAc, it can also regulate OGT expression. CREB can be deactivated by the deacetylase SIRT1, thereby reducing OGT expression and promoting tau phosphorylation, one of the major events in the course of Alzheimer’s disease (Lu et al., 2020).
With important roles in regulating the transcriptional networks required for proper development, differentiation and within the nervous system, it is unsurprising that deregulation in O-GlcNAc homeostasis is associated with neurodevelopmental diseases.
Transcription Factor O-GlcNAcylation in Carbohydrate and Lipid Metabolism
Beyond the brain, CREB also plays an important role in the liver where it regulates hepatic glucose and lipid metabolism (Herzig et al., 2001; Dentin et al., 2008). During prolonged fasting, CREB stimulates the gluconeogenic program with the coactivator PGC-1 (Herzig et al., 2001). PGC-1α (peroxisome proliferator-activated receptor gamma, co-activator 1 alpha) is a transcriptional co-activator that controls energy and nutrient homeostasis by coordinating gene expression. PGC-1α has been shown to form a complex with OGT and be O-GlcNAcylated at Ser333. Moreover, increased glucose levels lead to FOXO1 (Forkhead box other 1) O-GlcNAcylation via the PGC-1α/OGT complex, enhancing transcriptional activity (Housley et al., 2008). In addition, increased O-GlcNAc, either by addition of glucosamine or an OGA inhibitor, enhanced FOXO1 target gene expression in HepG2 cells (Kuo et al., 2008).
Insulin has two main functions within the liver: (1) downregulation of gluconeogenesis genes by initiating inhibitory phosphorylation of FOXO1, and (2) promotion of lipogenic pathways through activation of SHREBP-1c (Brown and Goldstein, 2008). O-GlcNAcylation regulates both pathways by attenuating insulin signaling and activating lipogenic pathways. The liver X receptors (LXRs) are described as nutritional sensors for lipid metabolism, glucose homeostasis and inflammation, and are posttranslationally modified by phosphorylation, acetylation, and O-GlcNAcylation (Anthonisen et al., 2010). Increased glucose levels leads to LXR O-GlcNAcylation, inducing SREBP-1c (sterol regulatory element binding protein 1c) expression. SREBP-1c is a major player of gene expression in hepatic lipogenesis (Anthonisen et al., 2010). LXR has been shown to interact and co-localize with OGT in vitro and in vivo. Additionally, LXR enhanced the expression of SREBP-1c and ChREBPα/β under hyperglycemic conditions (Bindesbøll et al., 2015). The effects of O-GlcNAc on the LXR pathway may extend beyond the liver, as this was the most dysregulated pathway found by transcriptomics of OGT-XLID mutant cells (Selvan et al., 2018). The LXR pathway has been shown to be critical in the development of dopaminergic neurons from stem cells (Ma et al., 2009), providing a possible link between this pathway and the developmental disorder.
ChREBP (carbohydrate responsive element binding protein) is a key factor of energy metabolism in the liver and is regulated by O-GlcNAcylation. High glucose or OGT expression increased ChREBP O-GlcNAcylation, stabilizing the protein and increasing expression of its target genes. Oga overexpression in mouse livers markedly reduced ChREBP O-GlcNAcylation and decreased abundance of ChREBP targets (Guinez et al., 2011). These results clearly demonstrated that O-GlcNAcylation of ChREBP increases its stability and activity. In addition, a recent study identified HCF-1 as a modulator of ChREBP activity and the lipogenic program, which is glucose dependent. Elevated glucose induced HCF-1 O-GlcNAcylation and HCF-1/ChREBP complex formation, where HCF-1 recruits OGT to further promote O-GlcNAcylation of ChREBP (Lane et al., 2019). Moreover, HCF-1/ChREBP complex formation was associated with the recruitment of epigenetic activator PHF2, which binds to H3K4me3 and enhances transcription (Lane et al., 2019). These data demonstrated that lipogenic gene expression is under the control of epigenetic modulations, ChREBP O-GlcNAcylation, and activation by HCF-1.
Through regulation of transcription factors, O-GlcNAc is able to alter patterns of gene expression in response to nutrient status and stress. Mutations in OGT may impair its targeting of specific pathways and disrupt their normal function in responding to the environment and coordinating development.
Placental OGT as a Biomarker for Neurodevelopmental Disease: a Model of Transgenerational Inheritance
Prenatal development is a particularly vulnerable time, when tight regulation of transcriptional networks is required to transform a fertilized egg into a multicellular organism requiring complex tissue development. In mammals, energy flow from the mother to the fetus is mediated by the placenta, which transfers macronutrients, gases, and metabolites into fetal circulation. Thus, the placenta transmits nutritional and stress information from the mother to the developing fetus. Glucose is the primary fuel for the fetus, which is provided by maternal transfer through the placenta. The fetal brain is a particularly nutritionally demanding tissue during fetal development. Brain regions are more sensitive to nutrient availability at particular gestational stages. Interestingly, a recent hypothesis to explain the male-biased presentation of neurodevelopmental diseases focuses on sex differences in the placenta in relaying signals to the developing brain regarding maternal perturbations (Charil et al., 2010; Howerton and Bale, 2012, 2014; Gabory et al., 2013; Howerton et al., 2013; Davis and Pfaff, 2014; Nugent and Bale, 2015; Bale, 2016).
As an X-linked gene and having the ability to transmit nutritional information to the nucleus, OGT is a unique candidate to signal maternal stress through the placenta to the developing fetus in a sex-dependent manner. Interestingly, O-GlcNAcylation of placental proteins correlates with maternal glycemic index (Dela Justina et al., 2018). In fact, a genome-wide screen looking for sex-specific changes in placental transcription after exposure to early prenatal stress identified Ogt as a top candidate for exhibiting sexually dimorphic expression in the placenta and changes in expression as a response to maternal stress (Howerton et al., 2013). This study found that male mouse placentas had about half the amount of OGT protein, corresponding to decreased total O-GlcNAc levels and even lower levels of OGT and O-GlcNAc upon prenatal stress, as compared to their female counterparts (Howerton et al., 2013). Hemizygous and homozygous mice that had Ogt deleted specifically in the placenta recapitulated models of early prenatal stress presenting with hypothylamic mitochondrial dysfunction characterized by transcriptional changes and altered cytochrome c oxidase activity (Howerton and Bale, 2014).
As discussed previously, OGT plays a critical role in regulating gene expression, particularly as a key regulator of Polycomb repression. Analysis into how placental expression of Ogt could causally contribute to neurodevelopmental disorders in the offspring have highlighted OGT’s role in regulating the H3K27me3 repressive histone mark. Interestingly, Nugent et al. (2018), found that OGT establishes sex differences in placental H3K27me3, with an enrichment found in females. This sex difference was Ogt dependent, as genetic reduction of Ogt in mice masculinized female placental H3K27me3 levels (Nugent et al., 2018). It was hypothesized that the higher levels of H3K27me3 allow for resiliency and less transcriptional response to maternal stress in the female than the male placenta. One deregulated gene of particular interest was Hsd17b3 (17-β-hydroxysteroid dehydrogenase-3). Through a ChIP-seq experiment on placental tissue, the researchers identified a correlation between O-GlcNAc occupancy and Hsd17b3 expression and that O-GlcNAc is significantly reduced by exposure to early prenatal stress (Howerton and Bale, 2014). This gene codes a key enzyme in testosterone biosynthesis. Examination of testosterone in a model of early prenatal stress male placentas showed a significant reduction in testosterone levels, potentially contributing to hypothalamic changes in offspring (Howerton and Bale, 2014). Taken together, these observations supports the possibility that acting as an epigenetic modifier in the placenta, OGT is able to pass transgenerational stress signals from the mother to the offspring in a sex-specific manner.
OGT and X-Linked Intellectual Disability (OGT-XLID): a Disease of O-GlcNAc Imbalance
X-linked intellectual disabilities are a group of a neurodevelopmental disorders representing about 5-10% of all cases of intellectual disability (Pravata et al., 2020b). Over 200 genes have been linked to XLID, although some of the candidates remain controversial. Over the past few years, a syndromic form of XLID affecting multiple families has been described which co-segregates with variants in the human OGT gene (Vaidyanathan et al., 2017; Willems et al., 2017; Selvan et al., 2018; Pravata et al., 2019, 2020a, 2020b). At present, some 14 patients from 8 families have been analyzed which show non-synonymous variants in the OGT gene (Pravata et al., 2020a). All the patients carrying OGT variants were found to have decreased intellectual ability with IQ scores well below 70. In addition, the patients exhibit both mental and physical developmental delay, intrauterine growth retardation, low birth weight, short stature, restricted language skills, and drooling. A summary of XLID-causative variants and their position in the proposed OGT structure is shown in Figure 4A. It is notable that all of the inherited variants are in the TPR repeats (Bouazzi et al., 2015; Vaidyanathan et al., 2017; Willems et al., 2017; Selvan et al., 2018), which are implicated in both substrate binding and complex formation with other epigenetic regulators. Two mutations appeared de novo and are present in the catalytic domain, such as the N567L variant found in two female twins (Pravata et al., 2019), and the N648Y variant found in a male patient (Pravata et al., 2020a). On the basis of the common features of these patients and the fact that the clinical features co-segregate with the OGT gene it has been proposed that this syndrome be classified as a congenital disorder of glycosylation (CDG) termed OGT-CDG (Pravata et al., 2020b). Identification of this disorder strongly reinforces a growing body of evidence that O-GlcNAcylation plays a key role in development, particularly neurodevelopment. The involvement of OGT in numerous epigenetic pathways, with the propensity of epigenetic disorders to manifest in neurodevelopmental disorders (Gabriele et al., 2018) suggests possible mechanisms causing this form of XLID. Thus, the features of this disorder allows a dissection and discussion of the role of OGT and O-GlcNAcylation in many aspects of human physiology (Figure 4B).
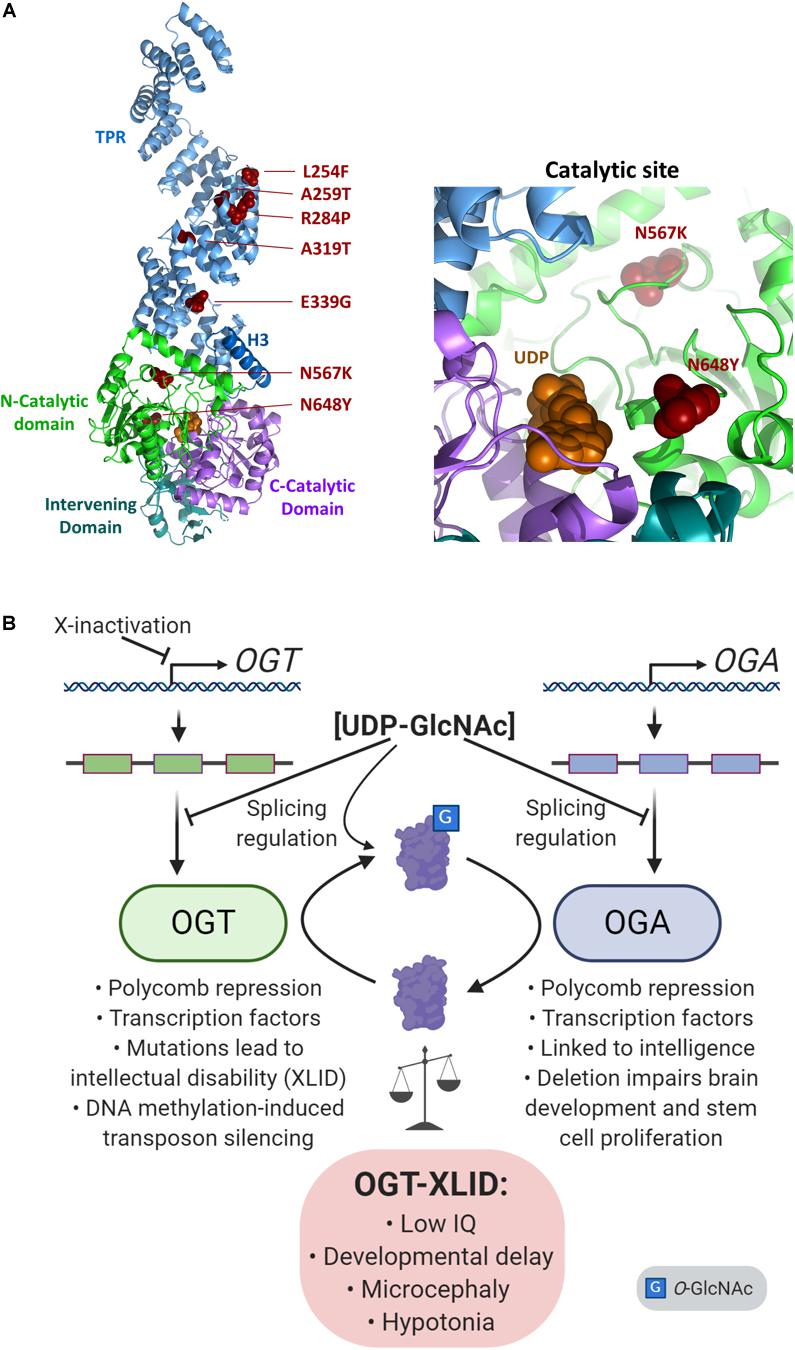
Figure 4. Imbalance of O-GlcNAc cycling leads to epigenetic changes and disease. (A) The presumed structure of OGT (Lazarus et al., 2011) shown as a cartoon in complex with UDP (orange spheres), with sites of OGT-XLID causative variants highlighted (red spheres). Each domain is colored-coded and labeled with its name. To better show the variants in the catalytic domain, a zoomed panel rotated 180° about the y-axis from the full structure is shown on the right. (B) Expression of both OGT and OGA are regulated by cellular concentrations of UDP-GlcNAc through splicing mechanisms. As an X-chromosome gene, OGT is also regulated by X-inactivation. OGT and OGA proteins dynamically cycle O-GlcNAc to regulate many epigenetic mechanisms such as the Polycomb repressive complexes and regulation of transcription factors. In addition, both genes are linked to intelligence: OGT mutations cause intellectual disability, and a GWAS meta-analysis found OGA is associated with intelligence (Savage et al., 2018). Deletion of Oga disrupts proper neural development and the proliferation of mouse embryonic stem cells (Olivier-Van Stichelen et al., 2017).
OGT Activity: Enzyme Stability, Target Recognition and Responsiveness to Hexosamine Flux
The previous examination of XLID patients have revealed changes in OGT activity and stability (Vaidyanathan et al., 2017; Willems et al., 2017; Selvan et al., 2018). Interestingly, several reports have suggested little to no change in O-GlcNAc levels (Vaidyanathan et al., 2017; Willems et al., 2017). One exception is the N648Y mutation which shows reduced enzymatic activity (Pravata et al., 2019). However, the tools we have available may be too insensitive to detect small changes or changes in subsets of substrates. In particular, the mutations associated with TPR repeats have the potential to affect substrate or complex recognition without direct effect on the catalytic domain. Our previous structural work on the TPRs allowed us to propose a role for an asparagine ladder motif in substrate recognition (Jinek et al., 2004). Recently this hypothesis was confirmed by additional structural studies (Levine et al., 2018). Thus substrate recognition may be affected by the XLID mutations.
The mutations in OGT may also destabilize the OGT protein. This was observed with the OGT L254F mutation in particular (Pravata et al., 2020a, b). However, current data reveal that while OGT-XLID variants may be destabilized, OGT protein levels in the majority of cell lines carrying the XLID variants were minimally altered (Pravata et al., 2020b). It is yet to be determined what effect these variants might have on levels of UDP-GlcNAc derived from hexosamine flux. Since OGT is a critical regulator of glucose utilization and metabolic reprogramming, changes in both glucose utilization and synthesis of UDP-GlcNAc may be altered in OGT-XLID (Saeed et al., 2016; Hanover et al., 2018).
X-inactivation and XLID
The X chromosome has a unique pattern of inheritance compared to autosomal chromosomes. The X chromosome in males is inherited from the mother making recessive X-linked mutations predominant in males. The significance of the X-chromosomal location OGT has been previously discussed (Shafi et al., 2000; Hanover et al., 2003, 2012; Love et al., 2003, 2010; O’Donnell et al., 2004; Abramowitz et al., 2014; Olivier-Van Stichelen and Hanover, 2014, 2015; Olivier-Van Stichelen et al., 2014). In females, X-inactivation is primarily random, with individual cells inactivating one of the X chromosome employing a long non-coding RNA (Xist) and chromatin modifiers such as the Polycomb complexes (Love et al., 2010; Brockdorff, 2013; Froberg et al., 2013; Shi et al., 2013b; Monfort and Wutz, 2020). Interestingly, female XLID patients with de novo mutations at OGT N567K have been shown to exhibit extreme skewing in X-inactivation (98%) although it is unclear which of the two X chromosome are inactivated (Pravata et al., 2019). In these patients, the levels of O-GlcNAc are affected and this reduction can be recapitulated in vitro and in other model systems (Pravata et al., 2019). Given these findings, it is unclear why such skewing occurs in the context of this de novo mutation arising in female probands. OGT could contribute to DNA methylation/demethylation associated with the X-inactivation process. In addition, the role of OGT in Polycomb repression previously discussed raises the possibility that alterations in O-GlcNAc cycling could contribute to the skewing observed.
The OGA Gene and Compensation for OGT Perturbations
In many instances, variants in OGT associated with XLID resulted in a reduction in levels of OGA (Vaidyanathan et al., 2017; Willems et al., 2017; Selvan et al., 2018; Pravata et al., 2020a, b). OGA can upregulate gene expression of OGT through activation of the transcription factor C/EBP-β (Vaidyanathan et al., 2017), in addition to this protein’s role in differentiation discussed above. Similarly, loss of OGA leads to an elevation in OGT protein levels (Keembiyehetty et al., 2015). The genes encoding OGA and OGT exhibit complex splicing patterns and recent findings suggest that mechanisms exist which serve to limit the translation of alternatively spliced species of the enzymes of O-GlcNAc cycling (Figure 4B) (Hanover et al., 2003; Park et al., 2017; Parra et al., 2020; Willems et al., 2017; Tan et al., 2020). O-GlcNAc regulates this process, suggesting a distinct feedback mechanism limiting the production of certain isoforms in response to O-GlcNAc elevation. In one study, an internal silencing site has been identified which appears to alter splicing of OGT mRNA, limiting its translation (Park et al., 2017). A similar mechanism is at play with OGA, where abundance of the enzyme is regulated by alternative splicing in response to O-GlcNAc levels (Tan et al., 2020). Thus, homeostatic systems are in place to limit the deregulation of total O-GlcNAc. However, lowering OGA levels can have a dramatic effect on neurodevelopment. We recently reported that mice in which Oga was deleted in the brain showed numerous phenotypes including microcephaly, enlarged ventricles, hypotonia, and developmental delay, strongly suggesting a possible link between OGT-XLID variants and perturbations of OGA levels (Olivier-Van Stichelen et al., 2017). In addition, a genome-wide association meta-analysis in 269,867 individuals identified MGEA5/OGA as one of the genes closely associated with intelligence (Savage et al., 2018). Studies on several OGT-XLID variants report a decrease in OGA levels (Vaidyanathan et al., 2017; Willems et al., 2017), although others have shown no evidence for a change in OGA levels (Selvan et al., 2018; Pravata et al., 2020a). So clear is this linkage between OGT variants and OGA that lowered OGA levels have been suggested as a diagnostic for XLID (Pravata et al., 2020b).
XLID and Other OGT Functions: HCF-1 Cleavage
O-GlcNAc transferase strongly associates with HCF-1 and heavily modifies it. In combination, OGT-HCF-1 forms interactions with numerous epigenetic complexes (Lazarus et al., 2013; Janetzko et al., 2016; Kapuria et al., 2016, 2018; Levine and Walker, 2016; Leturcq et al., 2017; Vaidyanathan et al., 2017; Gao et al., 2018). In addition, OGT cleaves HCF-1 using a catalytic mechanism which has been recently studied (Lazarus et al., 2013; Bhuiyan et al., 2015; Janetzko et al., 2016; Kapuria et al., 2016, 2018). Some of the OGT variants show changes in HCF-1 cleavage, but this does not seem to be universal to the disorder (Vaidyanathan et al., 2017; Willems et al., 2017; Selvan et al., 2018; Pravata et al., 2020a, b). Mutations in the gene encoding HCF-1 have been found to cause another form of XLID (Koufaris et al., 2016), suggesting a possible overlap in the mechanism of these disorders. Unlike OGT-XLID, this disorder does not include obvious developmental and morphological abnormalities, but HCF-1 dysregulation may still be an important contributor to key features of OGT-XLID.
OGT Interactions With Epigenetic Complexes
O-GlcNAc transferase interacts with numerous protein complexes associated with epigenetic regulation including the Polycomb repressive complexes, the pluripotency network associated with Oct4, Sin3A-HDAC complexes, and many others (Love et al., 2010; Hanover et al., 2012; Lewis and Hanover, 2014; Levine and Walker, 2016; Leturcq et al., 2017; Gao et al., 2018). The mutations seen in OGT-XLID could disrupt subsets of these interactions with accompanying changes in O-GlcNAc modification leading to a more pleiotropic deregulation of development. Improper regulation of these epigenetic complexes is known to impact development. For example, mutations in PRC2 proteins EZH2, SUZ12, and EED can cause Weaver Syndrome, an autism spectrum disorder that presents with developmental delay (Imagawa et al., 2017). These overlapping phenotypes may suggest common molecular pathways are at play with OGT-XLID.
At the present, it is difficult to examine these interactions quantitatively, but sensitive methods to examine both protein-protein interactions and O-GlcNAc turnover in those complexes are under continuous development.
OGT and Neurodevelopmental Disease: Summary and Implications
The identification of OGT-XLID as a rare human disorder suggests that non-synonymous variants of OGT can be tolerated to a limited degree. These OGT variants are hypomorphic, leading to only modest changes in O-GlcNAc levels due to compensatory changes in the O-GlcNAcase expression. Patients with OGT-XLID show numerous developmental and neurodevelopmental deficits resulting in a form of intellectual disability. This intellectual disability phenotype results from changes in neurodevelopment which strongly suggests that O-GlcNAc addition and removal may play a particularly important role in the functioning and development of the brain. This is perhaps not surprising given the importance of glucose and its metabolites in the physiology of the central nervous system. In addition, the complexity of the human brain originates from a finely tuned developmental process influenced by both genome and environment (Gabriele et al., 2018). A particularly sensitive time of brain development occurs in utero when glucose is provided primarily by maternal transfer through the placenta. This underscores the importance of maternal nutritional and environmental state and proper placental glucose metabolism for fetal brain development.
Development of the human cortex is incredibly sensitive, in part due to the requirement for two waves of rapid proliferation of progenitor cells (Florio and Huttner, 2014). During these developmental periods, cells are particularly prone to the accumulation of genetic lesions. Errors in DNA replication and repair can induce single nucleotide variants and insertions-deletions (Ernst, 2016). Transposable elements are another major source of genome instability which have been linked to neurological disorders (Doyle et al., 2017). Mutations in OGT may derepress transposable elements considering the recent findings of OGT working with TRIM28 to silence retrotransposons in a DNA methylation-dependant manner (Boulard et al., 2020). TRIM28 silencing is active in neural progenitor cells, and heterozygous mice present behavioral phenotypes which suggest an important role in brain development (Whitelaw et al., 2010; Grassi et al., 2019). The roles OGT plays both in activating the DNA damage response and in the silencing of retrotransposons help maintain genome integrity in the developing brain. The contribution of DNA damage and transposable element activity to OGT-XLID phenotypes should be investigated further.
Thus, the functional and morphological features of the human brain render it highly vulnerable to both genetically and environmentally induced alterations. In addition, there is an increasing awareness that chromatin regulation may be central to understanding neurodevelopmental disorders (Gabriele et al., 2018). The insights gained from analysis of both OGT-XLID patients and deregulated placental OGT, are likely to provide a platform for understanding how the O-GlcNAc pathway is integrated in human physiology. We have highlighted the role of O-GlcNAc cycling in numerous epigenetic complexes regulating development and differentiation. We have also examined the role of O-GlcNAc cycling in stem cell differentiation and the regulation of DNA damage response signaling. Finally, we have argued that compensatory mechanisms may be in place to limit the impact of the OGT mutations including X-inactivation of OGT, intron retention of OGT transcripts, OGA down regulation by Polycomb repression, intron retention, and transcription. These highly varied modes of regulation serve to buffer the effects of the OGT mutations, but may themselves have phenotypic consequences.
Author Contributions
DK, LA, and JH edited the manuscript. All authors contributed to the literature search, wrote specific sections of the manuscript, and agreed on the final version.
Funding
This review was supported by the Intramural NIDDK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Figures 1–3 and 4B were created with BioRender.com. Figure 4A was created using PyMol.
References
Abramowitz, L., Olivier-Van Stichelen, S., and Hanover, J. (2014). Chromosome imbalance as a driver of sex disparity in disease. J. Genomics 2, 77–88. doi: 10.7150/jgen.8123
Abramowitz, L. K., Harly, C., Das, A., Bhandoola, A., and Hanover, J. A. (2019). Blocked O-GlcNAc cycling disrupts mouse hematopoeitic stem cell maintenance and early T cell development. Sci. Rep. 9:12569. doi: 10.1038/s41598-019-48991-8
Akan, I., Love, D. C., Harwood, K. R., Bond, M. R., and Hanover, J. A. (2016). Drosophila O-GlcNAcase Deletion Globally Perturbs Chromatin O-GlcNAcylation. J. Biol. Chem. 291, 9906–9919. doi: 10.1074/jbc.M115.704783
Akan, I., Olivier-Van Stichelen, S., Bond, M. R., and Hanover, J. A. (2018). Nutrient-driven O-GlcNAc in proteostasis and neurodegeneration. J. Neurochem. 144, 7–34. doi: 10.1111/jnc.14242
Anthonisen, E. H., Berven, L., Holm, S., Nygard, M., Nebb, H. I., and Gronning-Wang, L. M. (2010). Nuclear receptor liver X receptor is O-GlcNAc-modified in response to glucose. J. Biol. Chem. 285, 1607–1615. doi: 10.1074/jbc.M109.082685
Bachman, M., Uribe-Lewis, S., Yang, X., Burgess, H. E., Iurlaro, M., Reik, W., et al. (2015). 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 11, 555–557. doi: 10.1038/nchembio.1848
Bachman, M., Uribe-Lewis, S., Yang, X., Williams, M., Murrell, A., and Balasubramanian, S. (2014). 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6, 1049–1055. doi: 10.1038/nchem.2064
Bale, T. L. (2016). The placenta and neurodevelopment: sex differences in prenatal vulnerability. Dialogues Clin. Neurosci. 18, 459–464. doi: 10.31887/dcns.2016.18.4/tbale
Bauer, C., Göbel, K., Nagaraj, N., Colantuoni, C., Wang, M., Müller, U., et al. (2015). Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). J. Biol. Chem. 290, 4801–4812. doi: 10.1074/jbc.M114.605881
Bhuiyan, T., Waridel, P., Kapuria, V., Zoete, V., and Herr, W. (2015). Distinct OGT-Binding Sites Promote HCF-1 Cleavage. PLoS One 10:e0136636. doi: 10.1371/journal.pone.0136636
Bindesbøll, C., Fan, Q., Nørgaard, R. C., MacPherson, L., Ruan, H. B., Wu, J., et al. (2015). Liver X receptor regulates hepatic nuclear O-GlcNAc signaling and carbohydrate responsive element-binding protein activity. J. Lipid Res. 56, 771–785. doi: 10.1194/jlr.M049130
Bond, M. R., and Hanover, J. A. (2013). O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33, 205–229. doi: 10.1146/annurev-nutr-071812-161240
Bond, M. R., and Hanover, J. A. (2015). A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880. doi: 10.1083/jcb.201501101
Bonisch, C., and Hake, S. B. (2012). Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 40, 10719–10741. doi: 10.1093/nar/gks865
Bouazzi, H., Lesca, G., Trujillo, C., Alwasiyah, M. K., and Munnich, A. (2015). Nonsyndromic X-linked intellectual deficiency in three brothers with a novel MED12 missense mutation [c.5922G>T (p.Glu1974His)]. Clin. Case Rep. 3, 604–609. doi: 10.1002/ccr3.301
Boulard, M., Rucli, S., Edwards, J. R., and Bestor, T. H. (2020). Methylation-directed glycosylation of chromatin factors represses retrotransposon promoters. Proc. Natl. Acad. Sci. U.S.A. 117, 14292–14298. doi: 10.1073/pnas.1912074117
Brockdorff, N. (2013). Noncoding RNA and Polycomb recruitment. RNA 19, 429–442. doi: 10.1261/rna.037598.112
Brown, M. S., and Goldstein, J. L. (2008). Selective versus total insulin resistance: a pathogenic paradox. Cell Metab. 7, 95–96. doi: 10.1016/j.cmet.2007.12.009
Butkinaree, C., Cheung, W. D., Park, S., Park, K., Barber, M., and Hart, G. W. (2008). Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J. Biol. Chem. 283, 23557–23566. doi: 10.1074/jbc.M804116200
Butler, A. A., Sanchez, R. G., Jarome, T. J., Webb, W. M., and Lubin, F. D. (2019). O-GlcNAc and EZH2-mediated epigenetic regulation of gene expression during consolidation of fear memories. Learn. Mem. 26, 373–379. doi: 10.1101/lm.049023.118
Celeste, A., Petersen, S., Romanienko, P. J., Fernandez-Capetillo, O., Chen, H. T., Sedelnikova, O. A., et al. (2002). Genomic instability in mice lacking histone H2AX. Science 296, 922–927. doi: 10.1126/science.1069398
Chang, Y. H., Weng, C. L., and Lin, K. I. (2020). O-GlcNAcylation and its role in the immune system. J. Biomed. Sci. 27:57. doi: 10.1186/s12929-020-00648-9
Charil, A., Laplante, D. P., Vaillancourt, C., and King, S. (2010). Prenatal stress and brain development. Brain Res. Rev. 65, 56–79. doi: 10.1016/j.brainresrev.2010.06.002
Chen, Q., Chen, Y., Bian, C., Fujiki, R., and Yu, X. (2013). TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564. doi: 10.1038/nature11742
Chen, Q., and Yu, X. (2016). OGT restrains the expansion of DNA damage signaling. Nucleic Acids Res. 44, 9266–9278. doi: 10.1093/nar/gkw663
Cheng, N. N., Sinclair, D. A., Campbell, R. B., and Brock, H. W. (1994). Interactions of polyhomeotic with Polycomb group genes of Drosophila melanogaster. Genetics 138, 1151–1162.
Cheung, W. D., and Hart, G. W. (2008). AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J. Biol. Chem. 283, 13009–13020. doi: 10.1074/jbc.M801222200
Chou, T. Y., Dang, C. V., and Hart, G. W. (1995). Glycosylation of the c-Myc transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 92, 4417–4421. doi: 10.1073/pnas.92.10.4417
Chu, C. S., Lo, P. W., Yeh, Y. H., Hsu, P. H., Peng, S. H., Teng, Y. C., et al. (2014). O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. U.S.A. 111, 1355–1360. doi: 10.1073/pnas.1323226111
Cortellino, S., Xu, J., Sannai, M., Moore, R., Caretti, E., Cigliano, A., et al. (2011). Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79. doi: 10.1016/j.cell.2011.06.020
Davis, E. P., and Pfaff, D. (2014). Sexually dimorphic responses to early adversity: implications for affective problems and autism spectrum disorder. Psychoneuroendocrinology 49, 11–25. doi: 10.1016/j.psyneuen.2014.06.014
Deevy, O., and Bracken, A. P. (2019). PRC2 functions in development and congenital disorders. Development 146:dev181354. doi: 10.1242/dev.181354
Dela Justina, V., Dos Passos Junior, R. R., Bressan, A. F., Tostes, R. C., Carneiro, F. S., Soares, T. S., et al. (2018). O-linked N-acetyl-glucosamine deposition in placental proteins varies according to maternal glycemic levels. Life Sci. 205, 18–25. doi: 10.1016/j.lfs.2018.05.013
Dentin, R., Hedrick, S., Xie, J., Yates, J. III, and Montminy, M. (2008). Hepatic glucose sensing via the CREB coactivator CRTC2. Science 319, 1402–1405. doi: 10.1126/science.1151363
Deplus, R., Delatte, B., Schwinn, M. K., Defrance, M., Mendez, J., Murphy, N., et al. (2013). TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655. doi: 10.1038/emboj.2012.357
Ding, X., Jiang, W., Zhou, P., Liu, L., Wan, X., Yuan, X., et al. (2015). Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PLoS One 10:e0145023. doi: 10.1371/journal.pone.0145023
Downs, J. A., Allard, S., Jobin-Robitaille, O., Javaheri, A., Auger, A., Bouchard, N., et al. (2004). Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990. doi: 10.1016/j.molcel.2004.12.003
Doyle, G. A., Crist, R. C., Karatas, E. T., Hammond, M. J., Ewing, A. D., Ferraro, T. N., et al. (2017). Analysis of LINE-1 Elements in DNA from Postmortem Brains of Individuals with Schizophrenia. Neuropsychopharmacology 42, 2602–2611. doi: 10.1038/npp.2017.115
Draime, A., Bridoux, L., Belpaire, M., Pringels, T., Degand, H., Morsomme, P., et al. (2018). The O-GlcNAc transferase OGT interacts with and post-translationally modifies the transcription factor HOXA1. FEBS Lett. 592, 1185–1201. doi: 10.1002/1873-3468.13015
Efimova, E. V., Appelbe, O. K., Ricco, N., Lee, S. S., Liu, Y., Wolfgeher, D. J., et al. (2019). O-GlcNAcylation enhances double-strand break repair, promotes cancer cell proliferation, and prevents therapy-induced senescence in irradiated tumors. Mol. Cancer Res. 17, 1338–1350. doi: 10.1158/1541-7786.mcr-18-1025
Ernst, C. (2016). Proliferation and differentiation deficits are a major convergence point for neurodevelopmental disorders. Trends Neurosci. 39, 290–299. doi: 10.1016/j.tins.2016.03.001
Etchegaray, J. P., and Mostoslavsky, R. (2016). Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol. Cell 62, 695–711. doi: 10.1016/j.molcel.2016.05.029
Florio, M., and Huttner, W. B. (2014). Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194. doi: 10.1242/dev.090571
Fong, J. J., Nguyen, B. L., Bridger, R., Medrano, E. E., Wells, L., Pan, S., et al. (2012). beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J. Biol. Chem. 287, 12195–12203. doi: 10.1074/jbc.M111.315804
Forma, E., Jóźwiak, P., Ciesielski, P., Zaczek, A., Starska, K., Bryś, M., et al. (2018). Impact of OGT deregulation on EZH2 target genes FOXA1 and FOXC1 expression in breast cancer cells. PLoS One 13:e0198351. doi: 10.1371/journal.pone.0198351
Froberg, J., Yang, L., and Lee, J. (2013). Guided by RNAs: X-inactivation as a model for lncRNA function. J. Mol. Biol. 425, 3698–3706. doi: 10.1016/j.jmb.2013.06.031
Fujiki, R., Hashiba, W., Sekine, H., Yokoyama, A., Chikanishi, T., Ito, S., et al. (2011). GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, 557–560. doi: 10.1038/nature10656
Gabory, A., Roseboom, T. J., Moore, T., Moore, L. G., and Junien, C. (2013). Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics. Biol. Sex Differ. 4:5. doi: 10.1186/2042-6410-4-5
Gabriele, M., Lopez Tobon, A., D’Agostino, G., and Testa, G. (2018). The chromatin basis of neurodevelopmental disorders: rethinking dysfunction along the molecular and temporal axes. Prog. Neuropsychopharmacol. Biol. Psychiatry 84(Pt B), 306–327. doi: 10.1016/j.pnpbp.2017.12.013
Gagnon, J., Daou, S., Zamorano, N., Iannantuono, N. V., Hammond-Martel, I., Mashtalir, N., et al. (2015). Undetectable histone O-GlcNAcylation in mammalian cells. Epigenetics 10, 677–691. doi: 10.1080/15592294.2015.1060387
Gambetta, M. C., and Müller, J. (2014). O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev. Cell 31, 629–639. doi: 10.1016/j.devcel.2014.10.020
Gambetta, M. C., and Muller, J. (2015). A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma 124, 429–442. doi: 10.1007/s00412-015-0513-1
Gambetta, M. C., Oktaba, K., and Muller, J. (2009). Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96. doi: 10.1126/science.1169727
Gao, J., Yang, Y., Qiu, R., Zhang, K., Teng, X., Liu, R., et al. (2018). Proteomic analysis of the OGT interactome: novel links to epithelial-mesenchymal transition and metastasis of cervical cancer. Carcinogenesis 39, 1222–1234. doi: 10.1093/carcin/bgy097
Grassi, D. A., Jönsson, M. E., Brattås, P. L., and Jakobsson, J. (2019). TRIM28 and the control of transposable elements in the brain. Brain Res. 1705, 43–47. doi: 10.1016/j.brainres.2018.02.043
Griffith, L. S., and Schmitz, B. (1995). O-linked N-acetylglucosamine is upregulated in Alzheimer brains. Biochem. Biophys. Res. Commun. 213, 424–431. doi: 10.1006/bbrc.1995.2149
Guinez, C., Filhoulaud, G., Rayah-Benhamed, F., Marmier, S., Dubuquoy, C., Dentin, R., et al. (2011). O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 60, 1399–1413. doi: 10.2337/db10-0452
Hahne, H., Gholami, A. M., and Kuster, B. (2012). Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol. Cell. Proteomics 11, 843–850. doi: 10.1074/mcp.M112.019463
Hajduskova, M., Baytek, G., Kolundzic, E., Gosdschan, A., Kazmierczak, M., Ofenbauer, A., et al. (2019). MRG-1/MRG15 Is a Barrier for Germ Cell to Neuron Reprogramming in Caenorhabditis elegans. Genetics 211, 121–139. doi: 10.1534/genetics.118.301674
Hanover, J., Chen, W., and Bond, M. (2018). O-GlcNAc in cancer: an Oncometabolism-fueled vicious cycle. J. Bioenerg. Biomembr. 50, 155–173. doi: 10.1007/s10863-018-9751-2
Hanover, J., Krause, M., and Love, D. (2012). Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321. doi: 10.1038/nrm3334
Hanover, J., Yu, S., Lubas, W., Shin, S., Ragano-Caracciola, M., Kochran, J., et al. (2003). Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–297. doi: 10.1016/s0003-9861(02)00578-7
Hayakawa, K., Hirosawa, M., Tabei, Y., Arai, D., Tanaka, S., Murakami, N., et al. (2013). Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J. Biol. Chem. 288, 17099–17110. doi: 10.1074/jbc.M113.455899
Hayakawa, K., Hirosawa, M., Tani, R., Yoneda, C., Tanaka, S., and Shiota, K. (2017). H2A O-GlcNAcylation at serine 40 functions genomic protection in association with acetylated H2AZ or gammaH2AX. Epigenetics Chromatin 10:51. doi: 10.1186/s13072-017-0157-x
He, Y. F., Li, B. Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. doi: 10.1126/science.1210944
Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., et al. (2001). CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183. doi: 10.1038/35093131
Hirosawa, M., Hayakawa, K., Yoneda, C., Arai, D., Shiota, H., Suzuki, T., et al. (2016). Novel O-GlcNAcylation on Ser(40) of canonical H2A isoforms specific to viviparity. Sci. Rep. 6:31785. doi: 10.1038/srep31785
Housley, M. P., Rodgers, J. T., Udeshi, N. D., Kelly, T. J., Shabanowitz, J., Hunt, D. F., et al. (2008). O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283, 16283–16292. doi: 10.1074/jbc.M802240200
Howerton, C. L., and Bale, T. L. (2012). Prenatal programing: at the intersection of maternal stress and immune activation. Horm. Behav. 62, 237–242. doi: 10.1016/j.yhbeh.2012.03.007
Howerton, C. L., and Bale, T. L. (2014). Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc. Natl. Acad. Sci. U.S.A. 111, 9639–9644. doi: 10.1073/pnas.1401203111
Howerton, C. L., Morgan, C. P., Fischer, D. B., and Bale, T. L. (2013). O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc. Natl. Acad. Sci. U.S.A. 110, 5169–5174. doi: 10.1073/pnas.1300065110
Hrit, J., Goodrich, L., Li, C., Wang, B. A., Nie, J., Cui, X., et al. (2018). OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development. eLife 7:e34870. doi: 10.7554/eLife.34870
Hu, C. M., Tien, S. C., Hsieh, P. K., Jeng, Y. M., Chang, M. C., Chang, Y. T., et al. (2019). High Glucose Triggers Nucleotide Imbalance through O-GlcNAcylation of Key Enzymes and Induces KRAS Mutation in Pancreatic Cells. Cell Metab. 29, 1334–1349.e10. doi: 10.1016/j.cmet.2019.02.005
Imagawa, E., Higashimoto, K., Sakai, Y., Numakura, C., Okamoto, N., Matsunaga, S., et al. (2017). Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum. Mutat. 38, 637–648. doi: 10.1002/humu.23200
Ingham, P. W. (1984). A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell 37, 815–823. doi: 10.1016/0092-8674(84)90416-1
Ishihara, K., Takahashi, I., Tsuchiya, Y., Hasegawa, M., and Kamemura, K. (2010). Characteristic increase in nucleocytoplasmic protein glycosylation by O-GlcNAc in 3T3-L1 adipocyte differentiation. Biochem. Biophys. Res. Commun. 398, 489–494. doi: 10.1016/j.bbrc.2010.06.105
Ito, R., Katsura, S., Shimada, H., Tsuchiya, H., Hada, M., Okumura, T., et al. (2014). TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells 19, 52–65. doi: 10.1111/gtc.12107
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi: 10.1038/nature09303
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi: 10.1126/science.1210597
Janetzko, J., Trauger, S., Lazarus, M., and Walker, S. (2016). How the glycosyltransferase OGT catalyzes amide bond cleavage. Nat. Chem. Biol. 12, 899–901. doi: 10.1038/nchembio.2173
Jang, H., Kim, T. W., Yoon, S., Choi, S. Y., Kang, T. W., Kim, S. Y., et al. (2012). O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 11, 62–74. doi: 10.1016/j.stem.2012.03.001
Janke, R., Dodson, A. E., and Rine, J. (2015). Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 31, 473–496. doi: 10.1146/annurev-cellbio-100814-125544
Jenuwein, T., and Allis, C. D. (2001). Translating the histone code. Science 293, 1074–1080. doi: 10.1126/science.1063127
Jiang, M., Xu, B., Li, X., Shang, Y., Chu, Y., Wang, W., et al. (2019). O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene 38, 301–316. doi: 10.1038/s41388-018-0435-5
Jinek, M., Rehwinkel, J., Lazarus, B. D., Izaurralde, E., Hanover, J. A., and Conti, E. (2004). The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 11, 1001–1007. doi: 10.1038/nsmb833
Kapuria, V., Röhrig, U., Bhuiyan, T., Borodkin, V., van Aalten, D., Zoete, V., et al. (2016). Proteolysis of HCF-1 by Ser/Thr glycosylation-incompetent O-GlcNAc transferase:UDP-GlcNAc complexes. Genes Dev. 30, 960–972. doi: 10.1101/gad.275925.115
Kapuria, V., Röhrig, U., Waridel, P., Lammers, F., Borodkin, V., van Aalten, D., et al. (2018). The conserved threonine-rich region of the HCF-1PRO repeat activates promiscuous OGT:UDP-GlcNAc glycosylation and proteolysis activities. J. Biol. Chem. 293, 17754–17768. doi: 10.1074/jbc.RA118.004185
Katai, E., Pal, J., Poor, V. S., Purewal, R., Miseta, A., and Nagy, T. (2016). Oxidative stress induces transient O-GlcNAc elevation and tau dephosphorylation in SH-SY5Y cells. J. Cell. Mol. Med. 20, 2269–2277. doi: 10.1111/jcmm.12910
Keembiyehetty, C., Love, D., Harwood, K., Gavrilova, O., Comly, M., and Hanover, J. (2015). Conditional knock-out reveals a requirement for O-linked N-Acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J. Biol. Chem. 290, 7097–7113. doi: 10.1074/jbc.M114.617779
Kim, E. J., Amorelli, B., Abdo, M., Thomas, C. J., Love, D. C., Knapp, S., et al. (2007). Distinctive Inhibition of O-GlcNAcase Isoforms by an α-GlcNAc Thiolsulfonate. J. Am. Chem. Soc. 129, 14854–14855. doi: 10.1021/ja076038u
Kim, G., Cao, L., Reece, E. A., and Zhao, Z. (2017). Impact of protein O-GlcNAcylation on neural tube malformation in diabetic embryopathy. Sci. Rep. 7:11107. doi: 10.1038/s41598-017-11655-6
Klose, R. J., and Bird, A. P. (2006). Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97. doi: 10.1016/j.tibs.2005.12.008
Koufaris, C., Alexandrou, A., Tanteles, G. A., Anastasiadou, V., and Sismani, C. (2016). A novel HCFC1 variant in male siblings with intellectual disability and microcephaly in the absence of cobalamin disorder. Biomed. Rep. 4, 215–218. doi: 10.3892/br.2015.559
Kriaucionis, S., and Heintz, N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. doi: 10.1126/science.1169786
Kuo, M., Zilberfarb, V., Gangneux, N., Christeff, N., and Issad, T. (2008). O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 582, 829–834. doi: 10.1016/j.febslet.2008.02.010
Lamarre-Vincent, N., and Hsieh-Wilson, L. C. (2003). Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J. Am. Chem. Soc. 125, 6612–6613. doi: 10.1021/ja028200t
Lambert, B., Vandeputte, J., Remacle, S., Bergiers, I., Simonis, N., Twizere, J. C., et al. (2012). Protein interactions of the transcription factor Hoxa1. BMC Dev. Biol. 12:29. doi: 10.1186/1471-213x-12-29
Lane, E. A., Choi, D. W., Garcia-Haro, L., Levine, Z. G., Tedoldi, M., Walker, S., et al. (2019). HCF-1 Regulates De Novo Lipogenesis through a Nutrient-Sensitive Complex with ChREBP. Mol. Cell 75, 357–371.e7. doi: 10.1016/j.molcel.2019.05.019
Lazarus, M., Jiang, J., Kapuria, V., Bhuiyan, T., Janetzko, J., Zandberg, W., et al. (2013). HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342, 1235–1239. doi: 10.1126/science.1243990
Lazarus, M. B., Nam, Y., Jiang, J., Sliz, P., and Walker, S. (2011). Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567. doi: 10.1038/nature09638
Lercher, L., Raj, R., Patel, N. A., Price, J., Mohammed, S., Robinson, C. V., et al. (2015). Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun. 6:7978. doi: 10.1038/ncomms8978
Leturcq, M., Lefebvre, T., and Vercoutter-Edouart, A. (2017). O-GlcNAcylation and chromatin remodeling in mammals: an up-to-date overview. Biochem. Soc. Trans. 45, 323–338. doi: 10.1042/BST20160388
Levine, Z., Fan, C., Melicher, M., Orman, M., Benjamin, T., and Walker, S. (2018). O-GlcNAc Transferase Recognizes Protein Substrates Using an Asparagine Ladder in the Tetratricopeptide Repeat (TPR) Superhelix. J. Am. Chem. Soc. 140, 3510–3513. doi: 10.1021/jacs.7b13546
Levine, Z., and Walker, S. (2016). The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells. Annu. Rev. Biochem. 85, 631–657. doi: 10.1146/annurev-biochem-060713-035344
Lewis, B., and Hanover, J. (2014). O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 289, 34440–34448. doi: 10.1074/jbc.R114.595439
Li, X., Molina, H., Huang, H., Zhang, Y. Y., Liu, M., Qian, S. W., et al. (2009). O-linked N-acetylglucosamine modification on CCAAT enhancer-binding protein beta: role during adipocyte differentiation. J. Biol. Chem. 284, 19248–19254. doi: 10.1074/jbc.M109.005678
Li, Z., Li, X., Nai, S., Geng, Q., Liao, J., Xu, X., et al. (2017). Checkpoint kinase 1-induced phosphorylation of O-linked beta-N-acetylglucosamine transferase regulates the intermediate filament network during cytokinesis. J. Biol. Chem. 292, 19548–19555. doi: 10.1074/jbc.M117.811646
Liu, C., and Li, J. (2018). O-GlcNAc: a sweetheart of the cell cycle and DNA Damage Response. Front. Endocrinol. 9:415. doi: 10.3389/fendo.2018.00415
Love, D., Kochan, J., Cathey, R., Shin, S., Hanover, J., and Kochran, J. (2003). Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 116(Pt 4), 647–654. doi: 10.1242/jcs.00246
Love, D., Krause, M., and Hanover, J. (2010). O-GlcNAc cycling: emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 21, 646–654. doi: 10.1016/j.semcdb.2010.05.001
Lu, S., Yin, X., Wang, J., Gu, Q., Huang, Q., Jin, N., et al. (2020). SIRT1 regulates O-GlcNAcylation of tau through OGT. Aging 12, 7042–7055. doi: 10.18632/aging.103062
Ma, D. K., Ming, G. L., and Song, H. (2009). Oxysterols drive dopaminergic neurogenesis from stem cells. Cell Stem Cell 5, 343–344. doi: 10.1016/j.stem.2009.09.001
Ma, J., and Hart, G. W. (2014). O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics 11:8. doi: 10.1186/1559-0275-11-8
Maury, J. J., El Farran, C. A., Ng, D., Loh, Y. H., Bi, X., Bardor, M., et al. (2015). RING1B O-GlcNAcylation regulates gene targeting of polycomb repressive complex 1 in human embryonic stem cells. Stem Cell Res. 15, 182–189. doi: 10.1016/j.scr.2015.06.007
Milne, T. A., Sinclair, D. A., and Brock, H. W. (1999). The Additional sex combs gene of Drosophila is required for activation and repression of homeotic loci, and interacts specifically with Polycomb and super sex combs. Mol. Gen. Genet. 261, 753–761. doi: 10.1007/s004380050018
Monfort, A., and Wutz, A. (2020). The B-side of Xist. F1000Res 9:F1000 Faculty Rev-55. doi: 10.12688/f1000research.21362.1
Myers, S. A., Panning, B., and Burlingame, A. L. (2011). Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9490–9495. doi: 10.1073/pnas.1019289108
Na, H.-J., Akan, I., Abramowitz, L. K., and Hanover, J. A. (2020). Nutrient-Driven O-GlcNAcylation Controls DNA Damage Repair Signaling and Stem/Progenitor Cell Homeostasis. Cell Rep. 31:107632. doi: 10.1016/j.celrep.2020.107632
Nishikawa, I., Nakajima, Y., Ito, M., Fukuchi, S., Homma, K., and Nishikawa, K. (2010). Computational prediction of O-linked glycosylation sites that preferentially map on intrinsically disordered regions of extracellular proteins. Int. J. Mol. Sci. 11, 4991–5008. doi: 10.3390/ijms11124991
Nugent, B. M., and Bale, T. L. (2015). The omniscient placenta: metabolic and epigenetic regulation of fetal programming. Front. Neuroendocrinol. 39:28–37. doi: 10.1016/j.yfrne.2015.09.001
Nugent, B. M., O’Donnell, C. M., Epperson, C. N., and Bale, T. L. (2018). Placental H3K27me3 establishes female resilience to prenatal insults. Nat. Commun. 9:2555. doi: 10.1038/s41467-018-04992-1
O’Donnell, N., Zachara, N., Hart, G., and Marth, J. (2004). Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690. doi: 10.1128/mcb.24.4.1680-1690.2004
Olivier-Van Stichelen, S., Abramowitz, L., and Hanover, J. (2014). X marks the spot: does it matter that O-GlcNAc transferase is an X-linked gene. Biochem. Biophys. Res. Commun. 453, 201–207. doi: 10.1016/j.bbrc.2014.06.068
Olivier-Van Stichelen, S., and Hanover, J. (2014). X-inactivation normalizes O-GlcNAc transferase levels and generates an O-GlcNAc-depleted Barr body. Front. Genet. 5:256. doi: 10.3389/fgene.2014.00256
Olivier-Van Stichelen, S., and Hanover, J. (2015). You are what you eat: O-linked N-acetylglucosamine in disease, development and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care 18, 339–345. doi: 10.1097/MCO.0000000000000188
Olivier-Van Stichelen, S., Wang, P., Comly, M., Love, D., and Hanover, J. (2017). Nutrient-driven O-linked N-acetylglucosamine (O-GlcNAc) cycling impacts neurodevelopmental timing and metabolism. J. Biol. Chem. 292, 6076–6085. doi: 10.1074/jbc.M116.774042
Park, S., Zhou, X., Pendleton, K., Hunter, O., Kohler, J., O’Donnell, K., et al. (2017). A Conserved Splicing Silencer Dynamically Regulates O-GlcNAc Transferase Intron Retention and O-GlcNAc Homeostasis. Cell Rep. 20, 1088–1099. doi: 10.1016/j.celrep.2017.07.017
Parra, M., Zhang, W., Vu, J., DeWitt, M., and Conboy, J. (2020). Antisense targeting of decoy exons can reduce intron retention and increase protein expression in human erythroblasts. RNA 26, 996–1005. doi: 10.1261/rna.075028.120
Pick, H., Kilic, S., and Fierz, B. (2014). Engineering chromatin states: chemical and synthetic biology approaches to investigate histone modification function. Biochim. Biophys. Acta 1839, 644–656. doi: 10.1016/j.bbagrm.2014.04.016
Pravata, V., Gundogdu, M., Bartual, S., Ferenbach, A., Stavridis, M., Õunap, K., et al. (2020a). A missense mutation in the catalytic domain of O-GlcNAc transferase links perturbations in protein O-GlcNAcylation to X-linked intellectual disability. FEBS Lett. 594, 717–727. doi: 10.1002/1873-3468.13640
Pravata, V., Muha, V., Gundogdu, M., Ferenbach, A., Kakade, P., Vandadi, V., et al. (2019). Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability. Proc. Natl. Acad. Sci. U.S.A. 116, 14961–14970. doi: 10.1073/pnas.1900065116
Pravata, V., Omelková, M., Stavridis, M., Desbiens, C., Stephen, H., Lefeber, D., et al. (2020b). An intellectual disability syndrome with single-nucleotide variants in O-GlcNAc transferase. Eur. J. Hum. Genet. 28, 706–714. doi: 10.1038/s41431-020-0589-9
Quinonez, S. C., and Innis, J. W. (2014). Human HOX gene disorders. Mol. Genet. Metab. 111, 4–15. doi: 10.1016/j.ymgme.2013.10.012
Rahe, D. P., and Hobert, O. (2019). Restriction of Cellular Plasticity of Differentiated Cells Mediated by Chromatin Modifiers, Transcription Factors and Protein Kinases. G3 9, 2287–2302. doi: 10.1534/g3.119.400328
Rao, F. V., Schuttelkopf, A. W., Dorfmueller, H. C., Ferenbach, A. T., Navratilova, I., and van Aalten, D. M. (2013). Structure of a bacterial putative acetyltransferase defines the fold of the human O-GlcNAcase C-terminal domain. Open Biol. 3:130021. doi: 10.1098/rsob.130021
Rexach, J. E., Clark, P. M., Mason, D. E., Neve, R. L., Peters, E. C., and Hsieh-Wilson, L. C. (2012). Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 8, 253–261. doi: 10.1038/nchembio.770
Ronningen, T., Shah, A., Oldenburg, A. R., Vekterud, K., Delbarre, E., Moskaug, J. O., et al. (2015). Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 25, 1825–1835. doi: 10.1101/gr.193748.115
Rowe, L. A., Degtyareva, N., and Doetsch, P. W. (2008). DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free Radic. Biol. Med. 45, 1167–1177. doi: 10.1016/j.freeradbiomed.2008.07.018
Saeed, M., Ahmad, J., Kanwal, S., Holowatyj, A., Sheikh, I., Zafar Paracha, R., et al. (2016). Formal modeling and analysis of the hexosamine biosynthetic pathway: role of O-linked N-acetylglucosamine transferase in oncogenesis and cancer progression. PeerJ 4:e2348. doi: 10.7717/peerj.2348
Sakabe, K., Wang, Z., and Hart, G. W. (2010). Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920. doi: 10.1073/pnas.1009023107
SanMiguel, J. M., and Bartolomei, M. S. (2018). DNA methylation dynamics of genomic imprinting in mouse development. Biol. Reprod. 99, 252–262. doi: 10.1093/biolre/ioy036
Savage, J. E., Jansen, P. R., Stringer, S., Watanabe, K., Bryois, J., de Leeuw, C. A., et al. (2018). Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919. doi: 10.1038/s41588-018-0152-6
Schouppe, D., Ghesquiere, B., Menschaert, G., De Vos, W. H., Bourque, S., Trooskens, G., et al. (2011). Interaction of the tobacco lectin with histone proteins. Plant Physiol. 155, 1091–1102. doi: 10.1104/pp.110.170134
Schvartzman, J. M., Thompson, C. B., and Finley, L. W. S. (2018). Metabolic regulation of chromatin modifications and gene expression. J. Cell Biol. 217, 2247–2259. doi: 10.1083/jcb.201803061
Selvan, N., George, S., Serajee, F., Shaw, M., Hobson, L., Kalscheuer, V., et al. (2018). O-GlcNAc transferase missense mutations linked to X-linked intellectual disability deregulate genes involved in cell fate determination and signaling. J. Biol. Chem. 293, 10810–10824. doi: 10.1074/jbc.RA118.002583
Shafi, R., Iyer, S., Ellies, L., O’Donnell, N., Marek, K., Chui, D., et al. (2000). The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739. doi: 10.1073/pnas.100471497
Shi, F. T., Kim, H., Lu, W., He, Q., Liu, D., Goodell, M. A., et al. (2013a). Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J. Biol. Chem. 288, 20776–20784. doi: 10.1074/jbc.M113.460386
Shi, X., Sun, M., Liu, H., Yao, Y., and Song, Y. (2013b). Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 339, 159–166. doi: 10.1016/j.canlet.2013.06.013
Sinclair, D. A., Syrzycka, M., Macauley, M. S., Rastgardani, T., Komljenovic, I., Vocadlo, D. J., et al. (2009). Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432. doi: 10.1073/pnas.0904638106
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Tan, Z., Fei, G., Paulo, J., Bellaousov, S., Martin, S., Duveau, D., et al. (2020). O-GlcNAc regulates gene expression by controlling detained intron splicing. Nucleic Acids Res. 48, 5656–5669. doi: 10.1093/nar/gkaa263
Toleman, C. A., Paterson, A. J., and Kudlow, J. E. (2006). The Histone Acetyltransferase NCOAT Contains a Zinc Finger-like Motif Involved in Substrate Recognition. J. Biol. Chem. 281, 3918–3925. doi: 10.1074/jbc.m510485200
Torres, C. R., and Hart, G. W. (1984). Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317.
Tursun, B., Patel, T., Kratsios, P., and Hobert, O. (2011). Direct conversion of C. elegans germ cells into specific neuron types. Science 331, 304–308. doi: 10.1126/science.1199082
Vaidyanathan, K., Niranjan, T., Selvan, N., Teo, C., May, M., Patel, S., et al. (2017). Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 292, 8948–8963. doi: 10.1074/jbc.M116.771030
Vella, P., Scelfo, A., Jammula, S., Chiacchiera, F., Williams, K., Cuomo, A., et al. (2013). Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656. doi: 10.1016/j.molcel.2012.12.019
Wahl, G. M., and Carr, A. M. (2001). The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat. Cell Biol. 3, E277–E286. doi: 10.1038/ncb1201-e277
Whitelaw, N. C., Chong, S., Morgan, D. K., Nestor, C., Bruxner, T. J., Ashe, A., et al. (2010). Reduced levels of two modifiers of epigenetic gene silencing, Dnmt3a and Trim28, cause increased phenotypic noise. Genome Biol. 11:R111. doi: 10.1186/gb-2010-11-11-r111
Willems, A., Gundogdu, M., Kempers, M., Giltay, J., Pfundt, R., Elferink, M., et al. (2017). Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J. Biol. Chem. 292, 12621–12631. doi: 10.1074/jbc.M117.790097
Wong, C. C., Qian, Y., and Yu, J. (2017). Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene 36, 3359–3374. doi: 10.1038/onc.2016.485
Xie, S., Jin, N., Gu, J., Shi, J., Sun, J., Chu, D., et al. (2016). O-GlcNAcylation of protein kinase A catalytic subunits enhances its activity: a mechanism linked to learning and memory deficits in Alzheimer’s disease. Aging Cell 15, 455–464. doi: 10.1111/acel.12449
Yang, X., and Qian, K. (2017). Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465. doi: 10.1038/nrm.2017.22
Yang, X., Zhang, F., and Kudlow, J. E. (2002). Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80. doi: 10.1016/s0092-8674(02)00810-3
Zhang, Q., Liu, X., Gao, W., Li, P., Hou, J., Li, J., et al. (2014). Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked beta-N-acetylglucosamine transferase (OGT). J. Biol. Chem. 289, 5986–5996. doi: 10.1074/jbc.M113.524140
Keywords: O-linked β-D-N-acetylglucosamine (O-GlcNAc), X-linked intellectual disability (XLID), epigenetics, histone modification, DNA methylation, nutrient-sensing
Citation: Konzman D, Abramowitz LK, Steenackers A, Mukherjee MM, Na H-J and Hanover JA (2020) O-GlcNAc: Regulator of Signaling and Epigenetics Linked to X-linked Intellectual Disability. Front. Genet. 11:605263. doi: 10.3389/fgene.2020.605263
Received: 11 September 2020; Accepted: 20 October 2020;
Published: 23 November 2020.
Edited by:
Dag H. Yasui, University of California, Davis, United StatesReviewed by:
Richard Alan Katz, Fox Chase Cancer Center, United StatesBeisi Xu, St. Jude Children’s Research Hospital, United States
Copyright © 2020 Konzman, Abramowitz, Steenackers, Mukherjee, Na and Hanover. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John A. Hanover, am9obi5oYW5vdmVyQG5paC5nb3Y=; amFoQGhlbGl4Lm5paC5nb3Y=
 Daniel Konzman
Daniel Konzman Lara K. Abramowitz
Lara K. Abramowitz Agata Steenackers
Agata Steenackers Mana Mohan Mukherjee
Mana Mohan Mukherjee Hyun-Jin Na
Hyun-Jin Na John A. Hanover
John A. Hanover