
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 01 February 2021
Sec. Genomic Medicine
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.600227
 Wei Liu1
Wei Liu1 Jin-Mou Yi1
Jin-Mou Yi1 Yi Liu1
Yi Liu1 Cong Chen1
Cong Chen1 Kai-Xuan Zhang1
Kai-Xuan Zhang1 Cheng Zhou1
Cheng Zhou1 Hui-En Zhan2
Hui-En Zhan2 Liang Zhao1
Liang Zhao1 Stephanie Morales3
Stephanie Morales3 Xie-Lan Zhao1
Xie-Lan Zhao1 Hui Zeng2*
Hui Zeng2*Acute myeloid leukemia (AML) is a threatening hematological malignant disease in which new successful approaches in therapy are needed. Cyclin-dependent kinase 6 (CDK6), a regulatory enzyme of the cell cycle that plays an important role in leukemogenesis and the maintenance of leukemia stem cells (LSC), has the potential to predict the prognosis of AML. By analyzing public databases, we observed that the messenger RNA (mRNA) levels of CDK6 were significantly overexpressed in AML cell lines and non-acute promyelocytic leukemia (non-APL) AML patients when compared to healthy donors. Furthermore, CDK6 expression was significantly reduced in AML patients who achieved complete remission (CR) compared to that at the time of diagnosis in our validated cohort. The expression of CDK6 was tightly correlated with peripheral blood blasts, French–American–British (FAB) subtypes, CCAAT-enhancer-binding protein α (CEBPA) mutation, and chromosomal abnormalities of t(8;21). However, the clinical significance and effects of CDK6 expression on the prognosis of non-APL AML patients remain uncertain. We found that CDK6 expression was inversely correlated with overall survival (OS) among non-APL AML patients using the Kaplan–Meier analysis. CDK6 was also found to be positively associated with genes identified to contribute to the development of leukemia, including CCND2, DNMT3B, SOX4, and IKZF2, as well as being negatively associated with anticancer microRNAs, including miR-187, miR-9, miR-582, miR708, and miR-362. In summary, our study revealed that CDK6 might be a potential diagnostic and prognostic biomarker in non-APL AML patients.
Acute myeloid leukemia (AML) is a malignant hematological disease with poor prognosis characterized by cytogenetic and molecular heterogeneity. Most APL patients could be cured under all-trans retinoic acid (ATRA) and arsenic trioxide (ATO) chemo-combination regimen. Due to the heterogeneity of leukemia, the general therapeutic strategy of non-APL AML is still a challenge (Walter et al., 2015). Even with transplantations or intensive chemotherapy, some patients still face the risk of relapse. Emerging new therapeutic compounds for AML hold promising prospects, such as targeted inhibition of mutated molecules, tyrosine kinases, and key components of signaling pathways (Abdel-Wahab and Levine, 2013; Gasiorowski et al., 2014).
Compelling research on the biology of AML has provided us with a more comprehensive understanding of cancer-targeted therapies to improve the clinical outcome of AML. Researchers have identified 23 genetic mutations, including FLT3, IDH1/2, TET2, TP53, and DNMT3A, which have been closely linked to prognosis. Clinically, FLT3 and IDH1/2 mutations occur in 20–30% AML patients (Leung et al., 2013; Dohner et al., 2015). With further studies emerging, the presence of FLT3-ITD or IDH1/2 mutation is linked to reduced overall survival rates (Reindl et al., 2006; Eisfeld et al., 2020). In an advanced breakthrough in hematological cancer therapy, the Food and Drug Administration (FDA) has approved the following drugs for AML patients with specific mutations: multitargeted kinase inhibitor midostaurin (Stone et al., 2017), FLT3-ITD, FLT3-TKD, c-Kit activity inhibitor gilteritinib (Perl et al., 2019), IDH1 mutant enzymes inhibitor ivosidenib (DiNardo et al., 2018), and IDH2 mutant enzymes inhibitor enasidenib (Stein et al., 2017).
Aberrant cell cycle control is also a significant hallmark of cancer cells (Deshpande et al., 2005). During mitotic cell division, cyclin-dependent kinases (CDKs) are critical regulators of G1-S transition. Researchers found that CDK6 and CDK4 have approximately 70% homology in their sequences and closely related biochemical properties (Chan et al., 1995). CDK4 and CDK6 are important regulators for initiation of the cell cycle. Lacking CDK4 and CDK6 resulted in late embryonic lethality due to defects in hematopoiesis (Kozar and Sicinski, 2005). However, evidence shows that the two kinases have different functions beyond cell cycle regulation. CDK6, not CDK4, is involved in both cell cycle and tumor-promoting progression (Kollmann et al., 2013). During cell cycle progression, changes in oncogene expression are regulated by CDK6 activity (Meyerson and Harlow, 1994). Previous studies have found that CDK6 is often overexpressed in both leukemia and lymphoma (Chilosi et al., 1998).
Up until now, the complete role of CDK6 expression in non-APL AML has not been elucidated. In this study, we sought to investigate the pattern of CKD6 expression and to infer the clinical implications of CDK6 in AML patients. To accomplish this, we conducted an analysis on data obtained from Gene Expression Omnibus (GEO) (Barrett et al., 2013), Oncomine (Rhodes et al., 2004), and The Cancer Genome Atlas (TCGA) database (Weinstein et al., 2013) and found that CDK6 is overexpressed in AML patients. We also validated the expression of CDK6 in bone marrow (BM) samples of AML patients and healthy donors via quantitative real-time PCR (qRT-PCR). The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses were utilized to investigate the underlying molecular mechanisms of CDK6. Exploring the clinical features of CDK6 can help us target AML synergistically with other potential therapies and reduce the frequency of resistance.
The gene expression databases ONCOMINE1, Gene Expression Profiling Interactive Analysis (GEPIA)2, GEO3, Xena4, Cancer Cell line Encyclopedia (CCLE)5, Human Protein Atlas (HPA)6, and TCGA7 are publicly accessible. The analyses of messenger RNA (mRNA) expressions of CDK6 in human cancer cells were assessed using online tools associated with HPA and CCLE databases. Comparisons between mRNA expressions of CDK6 in subjects with AML cancer and healthy donors were analyzed using ONCOMINE, GEO, and GEPIA. The TCGA database was screened for 173 adult AML patients with CDK6 expression data, complete corresponding clinical features, and non-zero overall survival time. Among them, 135 non-APL AML patients who met the criteria were included in the study; 72 received chemotherapy treatment only, and 63 patients received auto-/allo-HSCT. The major clinical features used in the diagnosis of AML patients are presented in Table 2. In addition, the AML gene expression datasets GSE13159, GSE15061, and GSE34577 from the GEO database were also included. The online web tool GenomicScape8 was used to explore the prognostic value of CDK6 expression in CN-AML patients(GSE12417).
A cohort of 127 newly diagnosed non-APL AML patients, 48 AML-RR patients, 146 AML-CR patients, and 54 healthy donors were enrolled between March 2016 and December 2019 in this study. The French–American–British (FAB) classification of AML patients was according to the 2016 World Health Organization (WHO) criteria. This study was approved by the Xiangya Hospital, Central South University and The First Affiliated Hospital of Jinan University.
Bone marrow mononuclear cells (BMMNCs) were separated using Ficoll-Hypaque (GE Healthcare, United States). Total RNA was extracted from BMMNCs with Trizol reagent (Life Technologies, United States) and reverse transcription to complementary DNA (cDNA) was performed using PrimeScript Kit (TaKaRa, Japan) as described in our previous reports (Liang et al., 2017b). Real-time PCR using ChamQ Universal SYBR Green Master Mix (Vazyme, China) was completed on the ViiATM7 RT-PCR system (Applied Biosystems, United States). The primers used for CDK6 expression were the following: forward, 5′–3′ CTGAATGCTCTTGCTCCTTT; reverse, 5′–3′ AAAGTTTTGGTGGTCCTTGA. Relative CDK6 expression mRNA levels were calculated by 2–ΔΔCT and were normalized to internal control (β-actin).
The differentially expressed genes (DEGs) of RNA and microRNA expression data were analyzed by Rstudio (“edgeR package”). Biological process, molecular function, and cellular component analysis of DEGs were performed using STRING9; genomes (KEGG) pathways of CDK6 showed enrichment when analyzed using Gene Set Enrichment Analysis (GSEA)10. The microRNAs that could target CDK6 were predicted by online tools miRDB11, miRwalk12, TargetScan13, mirDIP14, and DIANA15.
All statistical analyses were completed using SPSS 22.0 and GraphPad Prism 8.0. Either Pearson chi-square analysis or Fisher’s exact test was used for the comparison of categorical variables, whereas Mann–Whitney’s U-test was used for the comparison of continuous variables. The prognostic effect of CDK6 expression was analyzed through Kaplan–Meier analysis using the log-rank test. Univariate and multivariate proportional hazard regression analysis was performed using Cox regression. The p < 0.05 (two-tailed) in all statistical analyses was defined as statistically significant.
By analyzing 40 different types of the human cancer cell lines in the CCLE database, the expression of CDK6 was found to be highly expressed in both acute lymphoblastic leukemia (ALL) and AML cell lines (Figure 1A). The HPA public database also presented CDK6 overexpression in AML cell lines (Figure 1B). We further screened for CDK6 expression by analyzing the Gene Expression Profiling Interactive Analysis (GEPIA) database and found that the aberrant expression of CDK6 was observed in AML patients among 33 types of human cancer (Figure 1C). The mRNA expression of CDK6 in 173 newly diagnosed AML patients was significantly increased compared to the 70 GTEx normal samples (p < 0.01) (Figure 1D). In Haferlach leukemia 2, Valk leukemia, and Stegmaier leukemia statistics, the CDK6 expression was significantly higher in AML patients than in normal samples, using the ONCOMINE database (Figures 1H–J). To further validate the expression of CDK6 in non-APL AML patients, we analyzed the three independent validation cohorts of the GEO database. Among the GSE13159, GSE34577, and GSE15061-DS GEO sets, the CDK6 levels were significantly upregulated in non-APL AML patients when compared with normal ones (Figures 1E–G).
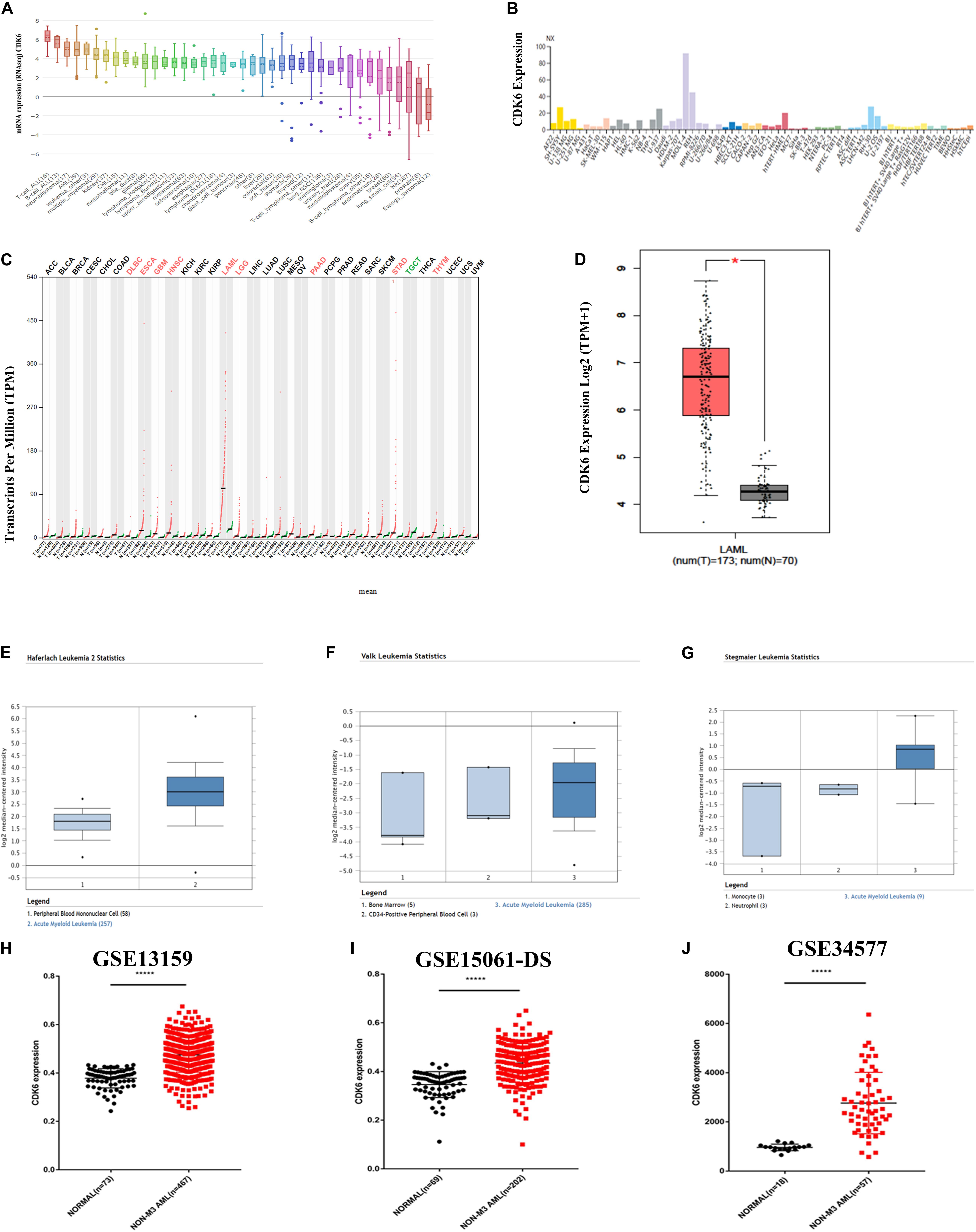
Figure 1. CDK6 overexpression in non-APL AML in the public database. (A) The expression of CDK6 in 40 types of human cancer cell line in the Cancer Cell Line Encyclopedia (CCLE) database. (B) The expression of CDK6 in human cancer cell lines in Human Protein Atlas (HPA) database. (C) The expression of CDK6 in 33 types compared with normal subjects in Gene Expression Profiling Interactive Analysis (GEPIA) database. (D) The expression of CDK6 in AML patients (n = 173) from TCGA and normal (n = 70) from GTX database by using (GEPIA) (p < 0.01). (E–G) In Haferlach leukemia2, Valk Leukemia, and Stegmaier leukemia statistics, CDK6 was overexpressed in AML patients than normal with a fold change = 2.388, p = 2.88E-30, fold change = 1.786, p = 0.025 and fold change = 3.179, p = 0.011, respectively. (H) The expression levels of CDK6 mRNA in non-APL AML patients(n = 467) compared with the normal samples (n = 73) (mean ± SEM level: 0.4748 ± 0.003325 vs. 0.3783 ± 0.004646, p < 0.0001) in GSE13159. (I) The expression level of CDK6 mRNA in non-APL AML patients (n = 202) compared with the normal samples (n = 69) (mean ± SEM level: 613.6 ± 30.5 vs. 253.2 ± 12.21, n = 69, p < 0.0001) in GSE15061-DS. (J) The expression level of CDK6 mRNA in non-APL AML patients (n = 57) compared with normal (n = 18) (mean ± SEM level: 961.7 ± 31.09 vs. 961.7 ± 31.09, p < 0.0001) in GSE34577.
Multiple datasets were utilized as validate sets, and the expression of CDK6 was elevated in patients with non-APL AML. In order to confirm the public database results, we analyzed another cohort consisting of 54 healthy donors, 127 de novo non-APL patients, 146 AML patients who achieved complete remission (CR), and 46 AML refractory remission (RR) patients from Xiangya Hospital and The First Affiliated Hospital of Jinan University. The disease state of AML patients were classified according to the 2016 World Health Organization (WHO) criteria. The relative CDK6 expression was higher in newly diagnosed AML patients than healthy donors (p = 0.0007). The relative CDK6 expression was noticeably downregulated in AML-CR patients compared to AML-RR patients and AML-de novo patients (p < 0.0001 and p = 0.0002) (Figure 2A). Moreover, we observed that the CDK6 expression in AML CR patients was significantly reduced compared to the original measurement when first diagnosed (p = 0.0001) (Figure 2B). To monitor the dynamic change of CDK6 expression in non-APL AML patients in different clinical stages, we tested CDK6 expression in 11 paired patients with available follow-up data at first diagnosis, complete remission (CR), and refractory remission (RR) time (de novo vs. CR: p = 0.002; RR vs. CR: p = 0.0086, respectively) (Figure 2C).
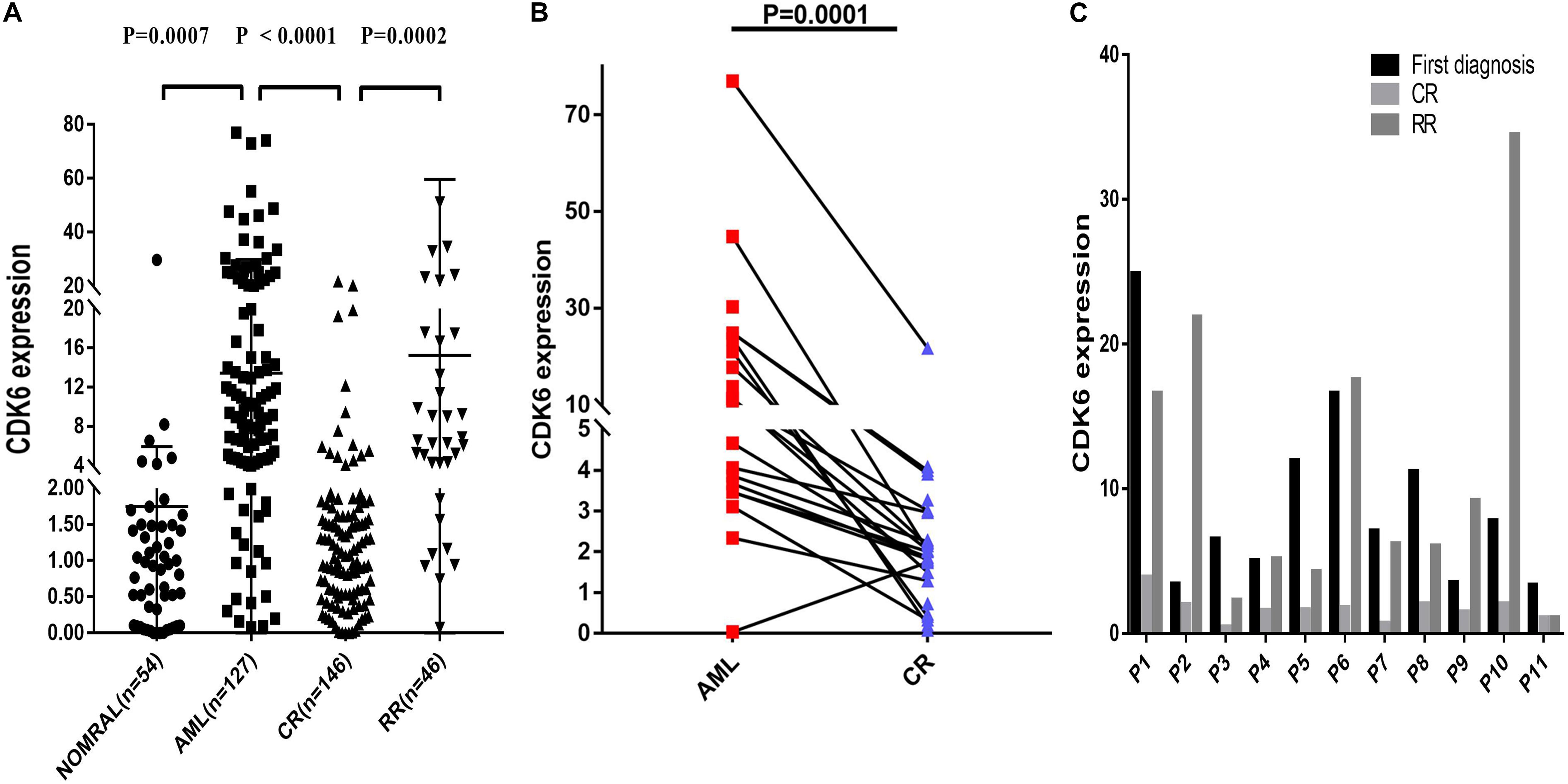
Figure 2. Relative levels of CDK6 expression in the validated non-APL AML patients. (A) The CDK6 expression in healthy donors (n = 54), de novo non-APL patients (n = 127), AML-CR patients (n = 146), and AML-RR (n = 48) patients from Xiangya Hospital and The First Affiliated Hospital of Jinan University Hospital. The distributions of CDK6 expression were presented with scatter plots. The expression level of CDK6 mRNA in de novo non-APL AML patients (n = 127) compared with the normal samples (n = 54) (mean ± SEM level: 13.42 ± 1.46 vs. 1.75 ± 0.5703, p = 0.007). The expression level of CDK6 mRNA in de novo non-APL AML patients (n = 127) compared with AML-CR patients (n = 146) (mean ± SEM level: 13.42 ± 1.46 vs. 2.257 ± 0.2894, p < 0.0001). The expression level of CDK6 mRNA in AML-RR patients (n = 48) compared with AML-CR patients (n = 146) (mean ± SEM level: 15.11 ± 6.256 vs. 2.257 ± 0.2894, p = 0.0002). (B) The CDK6 expression at the time of diagnosis higher than at the time of evaluation for response following standard induction (3 + 7) chemotherapy in patients with AML who achieved CR (p = 0.0001) (n = paired groups of 29). (C) P1–P11 represented 11 paired AML non-APL patients with available follow-up data in first diagnosed time, complete remission (CR) and refractory remission (RR) time. (First diagnosed time vs. CR time: p = 0.002; RR vs. CR time: p = 0.0086, respectively) (p-Values were adjusted by the Holm–Sidak method. HD, healthy donor).
Based on the above observations of increased CDK6 in AML patients, we hypothesized that there was a relationship between abnormal CDK6 expression and the therapeutic outcomes of AML patients. We analyzed the relative expression of CDK6 to AML patient survival in previously published datasets. Among the TCGA AML patients, 72 non-APL patients received only standard induction chemotherapy, whereas the 63 non-APL patients received auto-/allo-HSCT after induction chemotherapy. In both groups, CDK6 expression did not affect overall prognosis based on Kaplan–Meier analysis. The OS of CDK6high patients showed no significant difference compared with the OS of CDK6low patients in auto-/allo-HSCT and chemotherapy only groups (OS median: 822 vs. 854 days, p = 0.79; 245 vs. 304 days, p = 0.76, respectively). In cytogenetically normal AML (CN-AML) patients, there was also no difference between CDK6high and CDK6low group, no matter with auto-/allo-HSCT or chemotherapy only (chemotherapy only group: OS median, 245 vs. 485 days, p = 0.3; auto-/allo-HSCT group: OS median, 731 vs. 761 days, p = 0.31) (Figures 3A–D). Cox regression analysis also identified that CDK6 expression could not be an independent factor of OS in whole-TCGA-AML patients (Table 1). To further explore the impact of high CDK6 in CN-AML, we analyzed the overall survival of three CN-AML patient cohorts from the GEO database (GSE12417) by using an online tool GenomicScape. Each set’s patients were divided into two groups: those with above-cutoff CDK6 expression and those with below-cutoff CDK6 expression. Above-cutoff CDK6 expression was associated with negative OS in CN-AML patients (Figures 3E–H; Supplementary Figure 1).
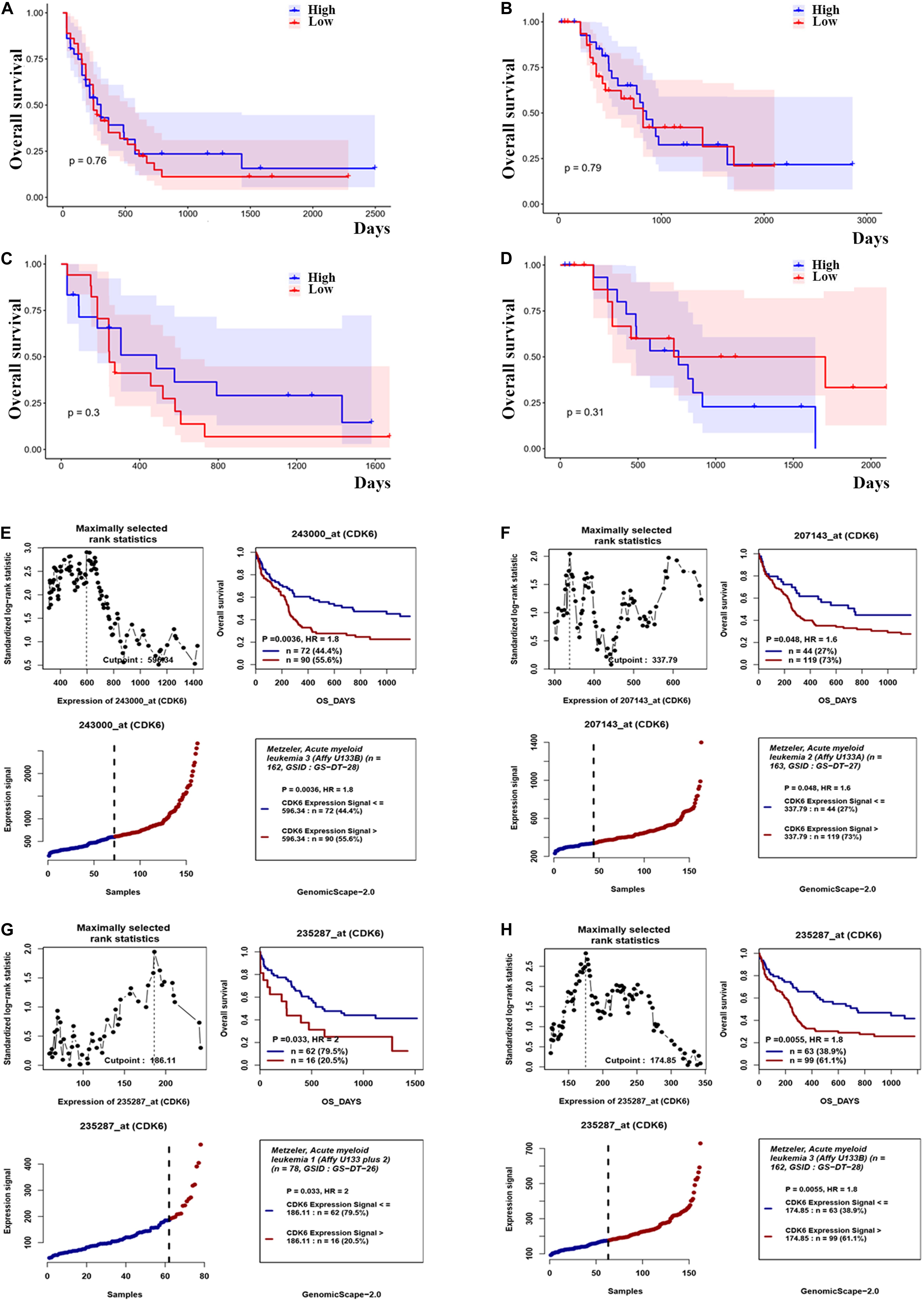
Figure 3. The effect of CDK6 expression on overall survival in non-APL AML patients. (A) Kaplan–Meier analysis of OS in chemotherapy only group in non-APL AML patients [CDK6high (n = 36) vs. CDK6low (n = 36): p = 0.76] (cut point: median CDK6 expression level). (B) Kaplan–Meier analysis of OS in auto-/allo-HSCT group in non-APL AML patients [CDK6high (n = 31) vs. CDK6low (n = 32): p = 0.79] (cut point: median CDK6 expression level). (C) Kaplan–Meier analysis of OS in chemotherapy only group in cytogenetically normal AML patients [CDK6high (n = 17) vs. CDK6low (n = 18): p = 0.3] (cut point: median CDK6 expression level). (D) Kaplan–Meier analysis of OS in auto-/allo-HSCT group in cytogenetically normal AML patients [CDK6high (n = 17) vs. CDK6low (n = 18): p = 0.3] (cut point: median CDK6 expression level). (E) The survival analysis of probe 243000_at of 162 cytogenetically normal AML (CN-AML) patients using the online web tool GenomicScape (cut point: 596.34, p = 0.0036, HR = 1.8); (F) the survival analysis of probe 207143_at of 162 cytogenetically normal AML (CN-AML) patient (cut point: 337.79, p = 0.048, HR = 1.6). (G) The survival analysis of probe 235287_at of 78 cytogenetically normal AML (CN-AML) patients using the online web tool Genomicscape (cut off: 596.34, p = 0.033, HR = 2). (H) The survival analysis of probe 235287_at of 162 cytogenetically normal AML (CN-AML) patients using the online web tool Genomicscape (cut point: 596.34, p = 0.0055, HR = 1.8).
To explore the clinical feature of CDK6 expression in non-APL AML patients, we compared the clinical characteristics of non-APL AML patients between CDK6high and CDK6low groups (divided by the median level of CDK6 expression) (Table 2). CDK6high patients had higher peripheral blood blasts compared with CDK6low group (p < 0.0001). However, no significant differences were found in gender, white blood cell (WBC), percentage of BM blasts, treatment, and risk cytogenetics between the two groups. In addition, significant differences were observed in the distributions of FAB subtypes. The CDK6high group frequently occurred in FAB-M0/M1/M2 (p < 0.0001, p = 0.005, and p = 0.001, respectively) and less frequently occurred in FAB M4/M5 (p < 0.0001 and p < 0.0001). Among cytogenetics and gene mutations, high CDK6 expression was associated with CEBPA mutations and chromosomal abnormalities of t(8;21) (p = 0.01 and p = 0.039), and low CDK6expression was associated with inv(16) (p = 0.021).
To investigate the biological function of CDK6, we compared the transcriptome between CDK6high and CDK6low group divided by median CDK6 expression. The comparison resulted in 1,280 DEGs [false discovery rate (FDR) < 0.05, Log2FC > 1, Figures 4A,B and Supplementary Table 1], in which 331 positively correlated with CDK6 expression and 949 negatively correlated with CDK6 expression. CCND2, DNMT3B, SOX4, and IKZF2, all previously reported to associate with leukemia development, were found within the positively correlated genes (Hanamura et al., 2006; Zhang et al., 2013; Niederwieser et al., 2015; Park et al., 2019). To further explore the role of CDK6 in AML, Gene Ontology analysis showed that DEGs were involved in signaling receptor activity, cell activation, immune response, and molecular transducer activity (Figure 4C). GSEA between CDK6high and CDK6low group identified more noticeable changes in the aminoacyl transfer RNA (tRNA) biosynthesis pathway, cell cycle pathway, and ubiquitin-mediated proteolysis pathway (Figure 4D). MicroRNAs are known as regulators of the oncogenesis of AML. The comparison between two groups also yielded 33 correlated microRNAs (Supplementary Table 2); among them, negatively correlated microRNAs, including miR-187, miR-9, miR-582, miR708, and miR-362, have been reported to have anticancer effects in previous studies (Liang et al., 2017a; Shi and Zhang, 2017; Li et al., 2019; Schneider et al., 2019; Xu et al., 2019) and were predicted to be able to directly target CDK6 (Figures 4E,F).
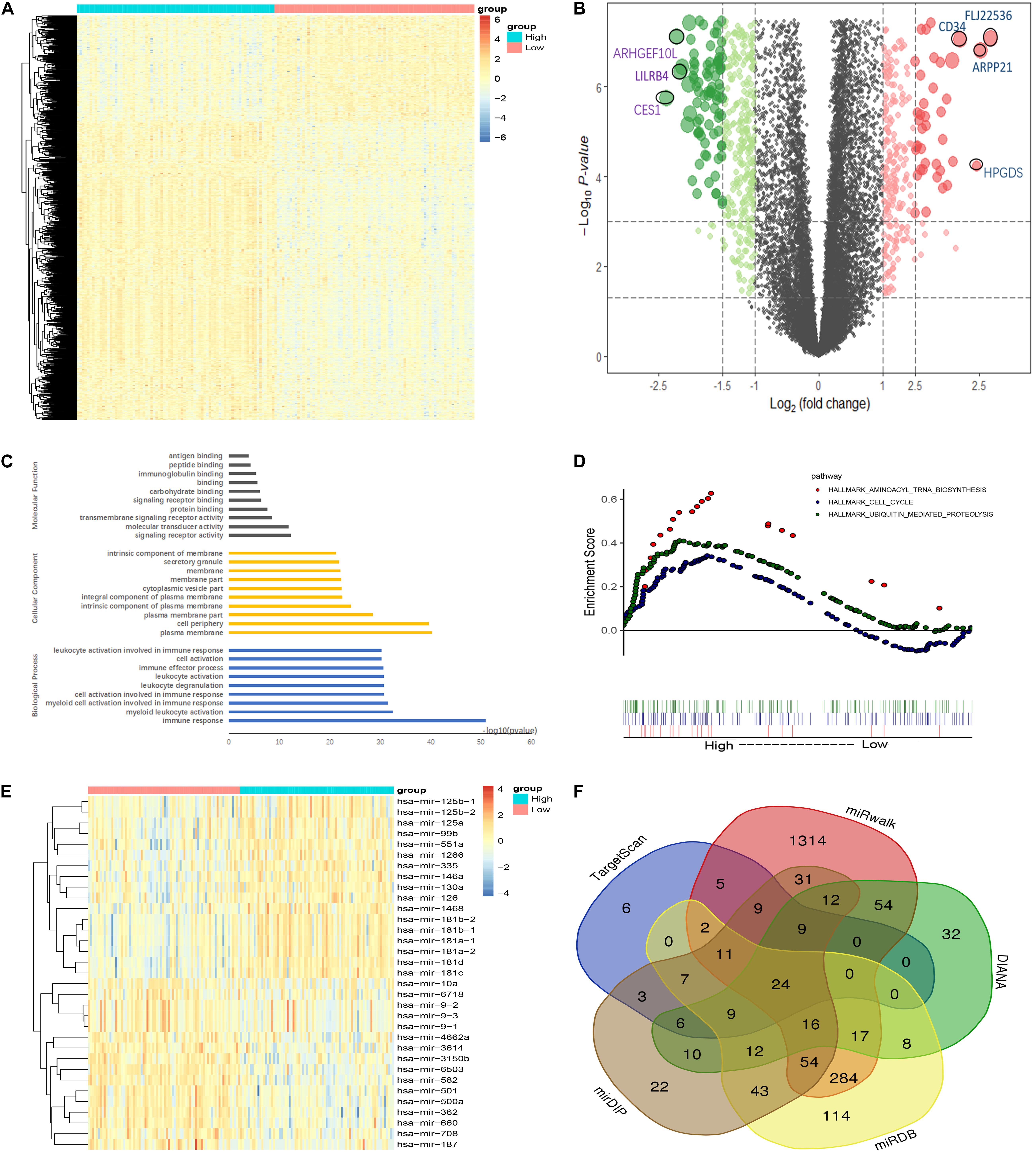
Figure 4. Molecular signatures associated with CDK6 in non-APL AML. (A) Heatmap of differentially expressed genes (DEGs) between CDK6high and CDK6low non-APL AML patients. (B) Volcano plot of DEGs between CDK6high and CDK6low non-APL AML patients. (C) Biological process, molecular functions, and cellular component analysis of DEGs using the online website of Search Tool for the Retrieval of Interacting Genes/Proteins. (D) KEGG pathway analysis between CDK6high and CDK6low group by using the online tool Gene Set Enrichment Analysis (GSEA); the red dot represents aminoacyl tRNA biosynthesis pathway (NES = 1.78, p = 0.003), the blue dot represents the cell cycle pathway (NES = 1.35, p = 0.021), the green dot represents ubiquitin-mediated proteolysis pathway (NES = 1.64, p < 0.0001). (E) Heatmap of DEGs miRNAs CDK6high and CDK6low AML patients. (F) Venn results of microRNAs that could target CDK6 precited by online tools miRDB, miRwalk, TargetScan, mirDIP, and DIANA.
Abnormal regulation of CDK6 is a hallmark of cancer. Increased CDK6 expression has been found in several human cancer. In our study, the significance of CDK6 expression in prognosis and the clinical relevance were investigated in non-APL AML. We determined that the expression of CDK6 was positively upregulated in de novo AML patients compared to healthy donors found in the public databases. The results of our study cohort remained consistent with those of previous studies, as well.
CDK6 has critical roles in multiple aspects of cell biology. It has been shown that CDK6 is involved in the biological function of leukemogenesis such as cell proliferation, cell apoptosis, and cell cycle regulation. CDK6 was not only known to be a cell-cycle kinase but also has a unique role of directly regulating transcription factors, which include AP-1 and vascular endothelial growth factor (VEGF-A), promoting tumor angiogenesis (Otto and Sicinski, 2013). CDK6 also plays a crucial role in malignant hematopoietic tumors. It is not only required for maintenance of NUP98-fusion AML but also could act an important role in MLL (KMT2A) fusions-mediated myeloid leukemogenesis (Schmoellerl et al., 2020). Some AML patients with FLT3-ITD mutation failed to induce a persistent response via FLT3 inhibitor. However, CDK6 showed a regulatory role in cell survival and cell apoptosis of FLT3 + AML cells (Uras et al., 2016). Reduced CDK6 kinase activity represents an encouraging target for anticancer drugs. Palbociclib, a CDK4/CDK6 inhibitor, demonstrated a potent effect in some cancer drugs. In patients with estrogen-receptor (ER)-positive breast cancer, treatment using palbociclib with fulvestrant resulted in prolonged progression-free survival (Turner et al., 2018). Based on the result of a phase Ib clinical trial, palbociclib has a positive effect in refractory/relapsed MLL (KMT2A)-rearranged leukemia patients with no incidence of limiting toxicities (NCT02310243) (Fröhling et al., 2016). A clinical trial is ongoing to estimate the efficacy and safety of palbociclib alone or in combination with sorafenib, decitabine, and dexamethasone in AML patients with relapsed and refractory (RR) leukemias (NCT03132454) (Kadia et al., 2018). Targeting CDK6 can act as a novel approach to increase chemotherapy response in AML patients.
MicroRNAs were recognized to act as oncomiRs and disease biomarkers in hematological malignancy (Wallace and O’Connell, 2017). CDK6 is targeted by many miRNAs, such as miR-29b, miR-218, miR-582, and miR-187. A recent report found miR-29b, which upregulates CDK6, to be a tumor suppressor via targeting multiple important oncogenic pathways (Huang et al., 2013). The expression of CDK6 was found to be negatively correlated with miR-187and miR-582.
The correlation between the expression of CDK6 and clinical features of AML patients has not yet been reported. The expression of CDK6 and correlation with AML clinical feature was also not examined in non-APL AML patients. In our study, we aimed to identify the clinical significance of CDK6 in non-APL AML. A significant adverse effect of high CDK6 expression on OS was observed among CN-AML patients, which indicated that CDK6 could be a potential prognostic and therapeutic value in AML.
Publicly available datasets were analyzed in this study. This data can be found here: ONCOMINE (https://www.oncomine.org/), GEPIA(http://gepia.cancer-pku.cn/), GEO (https://www.ncbi.nlm.nih.gov/geo/), Xena (https://xenabrowser.net/), CCLE (https://portals.broadinstitute. org/ccle), HPA (https://www.proteinatlas.org/), and TCGA (https://www.cancer.gov/tcga).
The studies involving human participants were reviewed and approved by the Xiangya Hospital, Central South University and Huaqiao Hospital, The First Affiliated Hospital of Jinan University. The patients/participants provided their written informed consent to participate in this study.
HZ designed the study and approved the final manuscript. WL performed the experiments and analyzed the data. J-MY, YL, CC, K-XZ, and CZ collected the clinical sample and data. H-EZ, LZ, and X-LZ performed the rest of necessary experiments. WL, SM, and HZ wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
The project was supported by the National Natural Science Foundation of China (Grant Nos. 81770184 and 81970143).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Jiang Xie, Dr. Li-Ge Xiao, and Dr. Guan-Teng Yang for collecting the clinical data. We also thank Dr. Juan Du for providing helpful comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.600227/full#supplementary-material
AML, acute myeloid leukemia; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GEPIA, Gene Expression Profiling Interactive Analysis; TCGA, The Cancer Genome Atlas; HSCT, hematopoietic stem cell transplantation; OS, overall survival; CDKs, cyclin-dependent kinases; CR, complete remission; RR, refractory remission; WHO, World Health Organization; CCLE, Cancer Cell line Encyclopedia; HPA, Human Protein Atlas; APL, acute promyelocytic leukemia; ATRA, all-trans retinoic acid; ATO, arsenic trioxide; CEBPA, CCAAT-enhancer-binding protein α.
Abdel-Wahab, O., and Levine, R. L. (2013). Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood 121, 3563–3572. doi: 10.1182/blood-2013-01-451781
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995. doi: 10.1093/nar/gks1193
Chan, F. K., Zhang, J., Cheng, L., Shapiro, D. N., and Winoto, A. (1995). Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol. Cell. Biol. 15, 2682–2688. doi: 10.1128/mcb.15.5.2682
Chilosi, M., Doglioni, C., Yan, Z., Lestani, M., Menestrina, F., Sorio, C., et al. (1998). Differential expression of cyclin-dependent kinase 6 in cortical thymocytes and T-cell lymphoblastic lymphoma/leukemia. Am. J. Pathol. 152, 209–217.
Deshpande, A., Sicinski, P., and Hinds, P. W. (2005). Cyclins and cdks in development and cancer: a perspective. Oncogene 24, 2909–2915. doi: 10.1038/sj.onc.1208618
DiNardo, C. D., Stein, E. M., de Botton, S., Roboz, G. J., Altman, J. K., Mims, A. S., et al. (2018). Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 378, 2386–2398. doi: 10.1056/NEJMoa1716984
Dohner, H., Weisdorf, D. J., and Bloomfield, C. D. (2015). Acute myeloid leukemia. N. Engl. J. Med. 373, 1136–1152. doi: 10.1056/NEJMra1406184
Eisfeld, A. K., Kohlschmidt, J., Mims, A., Nicolet, D., Walker, C. J., Blachly, J. S., et al. (2020). Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia 34, 3215–3227. doi: 10.1038/s41375-020-0872-873
Fröhling, S., Agrawal, M., Jahn, N., Fransecky, L. R., Baldus, C. D., Wäsch, R., et al. (2016). CDK4/6 inhibitor palbociclib for treatment of KMT2A-rearranged acute myeloid leukemia: interim analysis of the AMLSG 23-14 trial. Blood 128, 1608–1608. doi: 10.1182/blood.V128.22.1608.1608
Gasiorowski, R. E., Clark, G. J., Bradstock, K., and Hart, D. N. (2014). Antibody therapy for acute myeloid leukaemia. Br. J. Haematol. 164, 481–495. doi: 10.1111/bjh.12691
Hanamura, I., Huang, Y., Zhan, F., Barlogie, B., and Shaughnessy, J. (2006). Prognostic value of cyclin D2 mRNA expression in newly diagnosed multiple myeloma treated with high-dose chemotherapy and tandem autologous stem cell transplantations. Leukemia 20, 1288–1290. doi: 10.1038/sj.leu.2404253
Huang, X., Schwind, S., Yu, B., Santhanam, R., Wang, H., Hoellerbauer, P., et al. (2013). Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 19, 2355–2367. doi: 10.1158/1078-0432.CCR-12-3191
Kadia, T. M., Konopleva, M. Y., Garcia-Manero, G., Benton, C. B., Wierda, W. G., Bose, P., et al. (2018). Phase I study of palbociclib alone and in combination in patients with relapsed and refractory (R/R) leukemias. Blood 132(Suppl. 1), 4057–4057.
Kollmann, K., Heller, G., Schneckenleithner, C., Warsch, W., Scheicher, R., Ott, R. G., et al. (2013). A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 24, 167–181. doi: 10.1016/j.ccr.2013.07.012
Kozar, K., and Sicinski, P. (2005). Cell cycle progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell Cycle 4, 388–391. doi: 10.4161/cc.4.3.1551
Leung, A. Y., Man, C. H., and Kwong, Y. L. (2013). FLT3 inhibition: a moving and evolving target in acute myeloid leukaemia. Leukemia 27, 260–268. doi: 10.1038/leu.2012.195
Li, H., Tian, X., Wang, P., Huang, M., Xu, R., and Nie, T. (2019). MicroRNA-582-3p negatively regulates cell proliferation and cell cycle progression in acute myeloid leukemia by targeting cyclin B2. Cell Mol. Biol. Lett. 24:66. doi: 10.1186/s11658-019-0184-7
Liang, H., Luo, R., Chen, X., Zhao, Y., and Tan, A. (2017a). miR-187 inhibits the growth of cervical cancer cells by targeting FGF9. Oncol. Rep. 38, 1977–1984. doi: 10.3892/or.2017.5916
Liang, H., Zheng, Q. L., Fang, P., Zhang, J., Zhang, T., Liu, W., et al. (2017b). Targeting the PI3K/AKT pathway via GLI1 inhibition enhanced the drug sensitivity of acute myeloid leukemia cells. Sci. Rep. 7:40361. doi: 10.1038/srep40361
Meyerson, M., and Harlow, E. (1994). Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Biol. 14, 2077–2086. doi: 10.1128/mcb.14.3.2077
Niederwieser, C., Kohlschmidt, J., Volinia, S., Whitman, S. P., Metzeler, K. H., Eisfeld, A. K., et al. (2015). Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia. Leukemia 29, 567–575. doi: 10.1038/leu.2014.267
Otto, T., and Sicinski, P. (2013). The kinase-independent, second life of CDK6 in transcription. Cancer Cell 24, 141–143. doi: 10.1016/j.ccr.2013.07.019
Park, S. M., Cho, H., Thornton, A. M., Barlowe, T. S., Chou, T., Chhangawala, S., et al. (2019). IKZF2 drives leukemia stem cell self-renewal and inhibits myeloid differentiation. Cell Stem Cell 24, 153.e7–165.e7. doi: 10.1016/j.stem.2018.10.016
Perl, A. E., Martinelli, G., Cortes, J. E., Neubauer, A., Berman, E., Paolini, S., et al. (2019). Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N. Engl. J. Med. 381, 1728–1740. doi: 10.1056/NEJMoa1902688
Reindl, C., Bagrintseva, K., Vempati, S., Schnittger, S., Ellwart, J. W., Wenig, K., et al. (2006). Point mutations in the juxtamembrane domain of FLT3 define a new class of activating mutations in AML. Blood 107, 3700–3707. doi: 10.1182/blood-2005-06-2596
Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6. doi: 10.1016/s1476-5586(04)80047-2
Schmoellerl, J., Barbosa, I. A. M., Eder, T., Brandstoetter, T., Schmidt, L., Maurer, B., et al. (2020). CDK6 is an essential direct target of NUP98 fusion proteins in acute myeloid leukemia. Blood 136, 387–400. doi: 10.1182/blood.2019003267
Schneider, E., Pochert, N., Ruess, C., MacPhee, L., Escano, L., Miller, C., et al. (2019). MicroRNA-708 is a novel regulator of the Hoxa9 program in myeloid cells. Leukemia 34, 1253–1265. doi: 10.1038/s41375-019-0651-1
Shi, C., and Zhang, Z. (2017). MicroRNA-362 is downregulated in cervical cancer and inhibits cell proliferation, migration and invasion by directly targeting SIX1. Oncol. Rep. 37, 501–509. doi: 10.3892/or.2016.5242
Stein, E. M., DiNardo, C. D., Pollyea, D. A., Fathi, A. T., Roboz, G. J., Altman, J. K., et al. (2017). Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731. doi: 10.1182/blood-2017-04-779405
Stone, R. M., Mandrekar, S. J., Sanford, B. L., Laumann, K., Geyer, S., Bloomfield, C. D., et al. (2017). Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N. Engl. J. Med. 377, 454–464. doi: 10.1056/NEJMoa1614359
Turner, N. C., Slamon, D. J., Ro, J., Bondarenko, I., Im, S. A., Masuda, N., et al. (2018). Overall survival with palbociclib and fulvestrant in advanced breast cancer. N. Engl. J. Med. 379, 1926–1936. doi: 10.1056/NEJMoa1810527
Uras, I. Z., Walter, G. J., Scheicher, R., Bellutti, F., Prchal-Murphy, M., Tigan, A. S., et al. (2016). Palbociclib treatment of FLT3-ITD+ AML cells uncovers a kinase-dependent transcriptional regulation of FLT3 and PIM1 by CDK6. Blood 127, 2890–2902. doi: 10.1182/blood-2015-11-683581
Wallace, J. A., and O’Connell, R. M. (2017). MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood 130, 1290–1301. doi: 10.1182/blood-2016-10-697698
Walter, R. B., Othus, M., Paietta, E. M., Racevskis, J., Fernandez, H. F., Lee, J. W., et al. (2015). Effect of genetic profiling on prediction of therapeutic resistance and survival in adult acute myeloid leukemia. Leukemia 29, 2104–2107. doi: 10.1038/leu.2015.76
Weinstein, J. N., Collisson, E. A., Mills, G. B., Shaw, K. R., Ozenberger, B. A., Ellrott, K., et al. (2013). The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120. doi: 10.1038/ng.2764
Xu, X., Zou, H., Luo, L., Wang, X., and Wang, G. (2019). MicroRNA-9 exerts antitumor effects on hepatocellular carcinoma progression by targeting HMGA2. FEBS Open Biol. 9, 1784–1797. doi: 10.1002/2211-5463.12716
Keywords: AML, prognosis, CDK6, expression, target therapy
Citation: Liu W, Yi J-M, Liu Y, Chen C, Zhang K-X, Zhou C, Zhan H-E, Zhao L, Morales S, Zhao X-L and Zeng H (2021) CDK6 Is a Potential Prognostic Biomarker in Acute Myeloid Leukemia. Front. Genet. 11:600227. doi: 10.3389/fgene.2020.600227
Received: 29 August 2020; Accepted: 15 December 2020;
Published: 01 February 2021.
Edited by:
Liang-In Lin, National Taiwan University, TaiwanReviewed by:
Ming-Hsien Chien, Taipei Medical University, TaiwanCopyright © 2021 Liu, Yi, Liu, Chen, Zhang, Zhou, Zhan, Zhao, Morales, Zhao and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Zeng, YW5kcm9wczIwMTFAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.