
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 11 January 2021
Sec. Computational Genomics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.599834
This article is part of the Research Topic Multimodal and Integrative Analysis of Single-Cell or Bulk Sequencing Data View all 10 articles
 Jianhao Wang1†
Jianhao Wang1† Xiangyi Deng1†
Xiangyi Deng1† Yuan Xie2†
Yuan Xie2† Jiefu Tang3
Jiefu Tang3 Ziwei Zhou1
Ziwei Zhou1 Fan Yang1
Fan Yang1 Qiyuan He2
Qiyuan He2 Qingze Cao2
Qingze Cao2 Lei Zhang2,4*
Lei Zhang2,4* Liqun He1,5*
Liqun He1,5*Vasculature plays critical roles in the pathogenesis and neurological repair of traumatic brain injury (TBI). However, how vascular endothelial cells respond to TBI at the molecular level has not been systematically reviewed. Here, by integrating three transcriptome datasets including whole cortex of mouse brain, FACS-sorted mouse brain endothelial cells, and single cell sequencing of mouse brain hippocampus, we revealed the key molecular alteration of endothelial cells characterized by increased Myc targets and Epithelial-Mesenchymal Transition signatures. In addition, immunofluorescence staining of patients’ samples confirmed that IGFBP7 was up-regulated in vasculature in response to TBI. TGFβ1, mainly derived from microglia and endothelial cells, sufficiently induces IGFBP7 expression in cultured endothelial cells, and is significantly upregulated in response to TBI. Our results identified IGFBP7 as a potential biomarker of vasculature in response to TBI, and indicate that TGFβ signaling may contribute to the upregulation of IGFBP7 in the vasculature.
Traumatic brain injury (TBI), one of the leading cause of morbidity and disability, accounts for 30% of all injury-related deaths (Maas et al., 2008). It has been estimated that in the United States, an estimated 1.7 million people experience TBI annually, and 5.3 million people suffer TBI-related complications and sequela including long-term neurological and psychiatric disorders, chronic inflammation, and chronic traumatic encephalopathy (Nguyen et al., 2016; Jassam et al., 2017). TBI is a complicated pathophysiological process that can be divided into primary and secondary brain injuries (Sun et al., 2017). Primary injury is the direct damage of neural tissue caused by mechanical effect occurring at the moment of trauma (Silverberg et al., 2019). By contrast, secondary injury is the indirect injurious biochemical cascade initiated by primary insult, significantly contributing to the aggravation and high mortality of TBI (Bramlett and Dietrich, 2015; Silverberg et al., 2019). Secondary brain injury is mediated by several cellular and molecular pathways including excitotoxicity, inflammation, oxidative stress, and energy failure, and cerebral vasculature is a critical player to regulate these pathological processes (Jassam et al., 2017; Salehi et al., 2017; Simon et al., 2017). There is an increasing interest in the role that brain vasculature plays in the pathogenesis of TBI. Direct disruption of cerebral vasculature at the time of head impact leads to hemorrhage and blood flow abnormalities immediately after trauma, and dysfunction of vasculature leads to additional insults such as hypoxemia, hypoxia, hypo-perfusion, ischemia, and blood brain barrier (BBB) breakdown (Jullienne et al., 2016).
BBB is a specialized vascular structure in brain that maintains homeostasis and regulates the movement of molecular and cells across the brain vasculature (Yeoh et al., 2013). BBB disruption, as assessed by cerebrospinal fluid/serum albumin quotient, is found in 44% of non-penetrating TBI patients (Ho et al., 2014). Alteration of the vasculature in TBI causes vasogenic edema at both lesion and surrounding tissues resulting in tissue swelling and elevated intracranial pressure, contributing to about 50% of death in severe head injury (Marmarou, 2003). Moreover, increased vascular permeability observed in TBI patient allows harmful molecular and blood toxins into brain, which may lead to neuronal damage and long-lasting functional deficits (Badaut et al., 2011). In addition, up-regulation of cytokines/chemokines and leukocyte adhesion molecules in vascular endothelial cells (EC) augment inflammation and further increase the risk of edema formation and neuronal dysfunction (Ziebell and Morganti-Kossmann, 2010; Simon et al., 2017). Therefore, vasculature plays a critical role in the pathogenesis of TBI, and therapeutical manipulation of vasculature may represent a potential way for TBI treatment. However, how vascular EC respond to TBI at the molecular level has not been systematically reviewed.
Here, we integrated three transcriptome datasets in which the responses of whole cortex, FACS-sorted ECs, and isolated single ECs to brain injury were documented. Unbiased comparison by aligning three data sets to the same mouse genome revealed the key molecular alteration of ECs in response to brain injury. In addition, IGFBP7 was identified as a potential biomarker of vasculature in TBI pathogenesis.
The original sequencing data from three independent studies were obtained from the NCBI Sequence Read Archive (SRA) database1, including the mouse whole brain cortex RNAseq data (SRP072117, six samples), the FACS-sorted mouse brain endothelial RNAseq data (SRP100777, six samples), and the mouse brain hippocampus single cell RNAseq data (SRP113600, six samples). Each sample contains about 40–60 million RNA sequence reads.
The RNAseq sequencing data from the brain cortex and FACS-sorted EC were aligned to the mouse genome assembly (GRCm38) obtained from the Ensembl database using the TopHat2 software (version 2.1.1, with the following parameters: tophat2 -p 8 –keep-fasta-order –GTF < reference.gtf >< reference.genome >< read1.fastq >< read2.fastq >) (Kim et al., 2013). To quantify the differential expression between TBI and control groups, the default cuffdiff tests were performed using the Cufflinks tool (version 2.2.1, with the following parameters: cuffdiff -p 4 < reference.gtf >< control1.bam,control2.bam,control3.bam >< TBI1.bam,TBI2.bam,TBI3.bam >) (Trapnell et al., 2012). The genes with statistical multiple-test corrected (Benjamini-Hochberg) p-value smaller than 0.05 and more than 2-fold difference were selected as significant differentially expressed genes.
To compare the gene expression levels among different samples, heatmap visualization was performed using the pheatmap packages (version 1.0.12) with ward.D2 method. The genes were clustered using the Pearson correlation distance and the samples were clustered using the Euclidean distance.
The single cell RNAseq data were processed using the Drop-seq tools (version 2.3.0)2 with default parameters as described in the paper (Arneson et al., 2018). The fastq files were aligned to the mouse genome assembly (GRCm38) and the digital expression counts for each cell were quantified using the default parameters. The single cell expression counts data were then imported into the Seurat package (version: 3.1.1) for cell type clustering and cluster marker identifications (Butler et al., 2018).
The GSVA was performed using the R GSVA package (version 1.32.0). To identify the gene sets with significant changes, the 50 hallmark gene sets from the MSigDB collections were tested3 using the R limma package (version 3.40.6). The gene sets with statistical multiple-test (Benjamini-Hochberg) corrected p-value smaller than 0.05 were identified as significant.
The brain tissue was collected from the Department of Neurosurgery, Tianjin Medical University General Hospital, Tianjin 300052, PR China. The patient was an adult male. The preoperative Glasgow Coma Scale (GCS) was 11 points without underlying disease. The written informed consent from the patient was made before the surgery.
During the operation, the contusion tissue was extracted from the contusion area by bipolar electrocoagulation, and the brain contusion tissue was marked as TBI group. The control specimens were taken from the non-functional area about 1.5 cm from the tumor boundary of the glioblastoma patient. After removing the specimens, they were stored at -80°C. All human sample experiments were approved by the local hospital ethics committee.
After the specimens were taken out from -80°C, they were quickly embedded in optimal cutting temperature (O.C.T., Sakura, Oakland, CA, United States). Then, the specimens were cut into a thick coronal section of 6 μm, and immunofluorescence stained to determine the IGFBP7 and CD31 expression in the coronal brain section. The slices were first fixed in ice-cold methanol for 10 min and washed with phosphate buffered saline (PBS) for 10 min. Blocked with 3% BSA (Invitrogen) for 1 h at room temperature (RT). Then, the sections were incubated with the primary antibody against IGFBP7 (1:200, ab74169, Abcam) and CD31 (1:500, MA3105, Invitrogen) overnight at 4°C. The slides were washed three times with PBS plus 0.2% Tween (Sigma) and incubated with the secondary antibody Goat anti-Rabbit Alexa 555 (1:400, cat A-21428, Invitrogen) and Goat anti-Armenian hamster Alexa 488 (1:400, ab173003, Abcam) for 1 h at RT. After washing with PBS plus 0.2% Tween three times, the sections were mounted in Fluoromount (F4680, Sigma) containing 0.1% hoechst 33258 (14530, Sigma). Images were acquired by fluorescence microscope (Zeiss, Germany).
Murine brain EC (bEND.3) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (Biological Industries) and 1% 100 mM Sodium Pyruvate (Gibco). Human umbilical vein EC (HUVECs) were maintained up to passage 14 in the same culture medium as described above. The bEND.3 cells and HUVECs were starved in DMEM with 1% FBS or 0.5% FBS, respectively, overnight and stimulated with TGFβ1 (2 ng/mL; Peprotech) for 72 h, VEGFA (50 ng/mL; Peprotech) for 72 h, or cobalt chloride (400 μM CoCl2; Sigma Aldrich) for 24 h, respectively. The experiment was performed duplicates and repeated three times for both bEND.3 cells and HUVECs.
Total cell RNA was extracted with the RNeasy kit (Qiagen) and reverse-transcribed with the PrimeScript RT Master Mix (Takara). qPCR was performed on a Thermal Cycler iQ5 Multicolor Real-Time PCR Detection system (Bio-Rad) using TB Green Premix Ex taqTM II (Takara) and intron-spanning, gene-specific primers as listed below: mouse Hprt (forward: CAGTCCCAGCGTCGTGATTA, reverse: TGGCCTCCCATCTCCTTCAT); mouse Igfbp7 (forward: CTG GTGCCAAGGTGTTCTTGA, reverse: CTCCAGAGTGATCC CTTTTTACC); human HPRT (forward: CTTTGCTGACCT GCTGGATT, reverse: TCCCGTGTTGACTGGTCATT); human IGFBP7 (forward: GCGAGCAAGGTCCTTCCATA, reverse: TCTGAATGGCCAGGTTGTCC). Gene expression was normalized to the house keeping gene HPRT(Hprt).
In order to identify the genes that are differentially expressed in brain in TBI condition, we reanalyzed and integrated the data from three transcriptome sequencing profiling approaches (Table 1). These include one mouse whole cortex RNAseq study (Zhong et al., 2016), one mouse brain FACS-sorted endothelial RNAseq study (Munji et al., 2019), and one mouse brain single cell RNAseq study (Arneson et al., 2018). All three studies were performed with mouse TBI models and screened the gene expression changes at 24 h after TBI. The three datasets were processed differently in their corresponding studies. To achieve an unbiased comparison, we obtained the original sequence data for each sample and aligned them to the same mouse genome reference (Supplementary Table 1). Differentially expressed genes were identified by comparing the TBI and control in the respective studies. The complete gene analysis results from each of the three studies are listed in the supplements (whole cortex RNAseq study in Supplementary Table 2; FACS-sorted EC study in Supplementary Table 3; single cell RNAseq study in Supplementary Table 4).
In the whole cortex RNAseq study, 1,096 genes were tested as significantly differentially expressed (Supplementary Table 5). In the FACS-sorted EC study, 214 genes were identified (Supplementary Table 6).
For the single cell RNAseq study, all the qualified cells (N = 6,351, with 2,549 cells from the TBI samples and 3,370 cells from the control samples) were clustered and a pure endothelial cluster (n = 371) was identified (Supplementary Figure 1). Within the endothelial clusters, the comparison between the 185 cells in TBI samples and the 186 cells in the control samples yields 14 significantly differentially expressed genes (Supplementary Table 7).
We focused on the brain EC to evaluate and compare the results from three studies. First, we compared the two RNAseq studies (whole cortex and FACS-sorted EC). For a list of 10 well-known canonical EC markers, they showed a wild range of expression in RNAseq data, however, they all display significant enrichment in the FACS sorted EC samples (around 20–100-folds’ enrichment) (Figure 1A). In the results of differentially expressed genes, there were 184 genes differentially expressed in both studies (Figure 1B and Supplementary Table 8). Among these 184 genes, 170 genes were regulated in the same direction (both were up-regulated or down-regulated), and only 14 (8%) showed different regulation directions (Figure 1C).
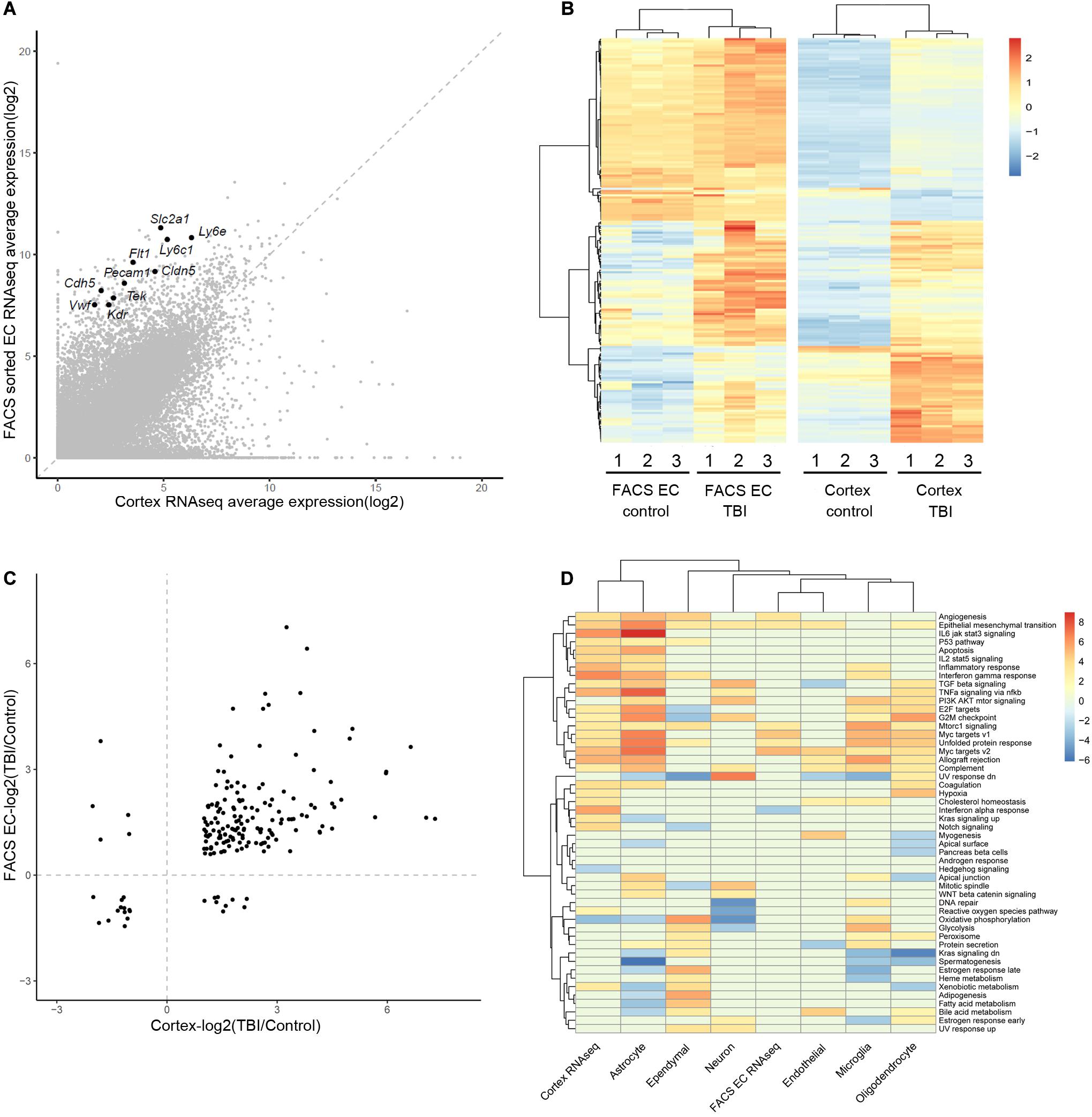
Figure 1. Comparison of the gene expression analysis results from the three studies. (A) Comparison of the average expression of whole cortex and FACS sorted EC RNAseq data. Ten well-known canonical EC markers are labeled. (B) Heatmap of the different expression of 184 DEGs in the two studies, using the pheatmap packages (version 1.0.12) with ward.D2 method. The color legend shows the relative expression. (C) Comparison of the log2 scaled fold change (TBI/Control) of the 184 DEGs in the two RNAseq studies. (D) Heatmap of the GSVA analysis of the three studies. The two RNAseq studies and the major cell types in the scRNAseq study are listed in the columns. The rows represent the 50 hallmark gene sets. The color legend shows the GSVA t statistics, and red color indicates up-regulation and blue color indicates down-regulation.
To identify the pathways that were affected during the TBI condition, Gene Set Variation Analysis (GSVA) was applied to each of the three datasets. The regulated hallmark gene sets in the two RNAseq studies as well as the major cell types in the single cell RNAseq studies were identified (Figure 1D). Myc targets and Epithelial-Mesenchymal Transition signature are enriched in samples from TBI in all three studies. Interestingly, both signatures are also enriched in tumor EC and associate with tumor angiogenesis (Lambrechts et al., 2018).
A comparison among the three different studies to identify differentially expressed genes (DEGs) in whole brain or brain EC is illustrated in Figure 2A. The DEGs of the three different studies are classified according to the up-regulated and down-regulated genes (Figure 2A). Totally, 1,135 different genes were identified by at least one study, and only three genes (Igfbp7, Fxyd5, and Itm2a) were consistently regulated in samples from TBI compared to control in all three studies. Among them, Igfbp7 and Fxyd5 are up-regulated, and Itm2a is down-regulated. Their detailed expression profile in the FACS-sorted EC samples is visualized in Figure 2B. All three genes showed EC specific expression in the single cell RNAseq study (Figure 2C). Expression of Igfbp7 and Itm2a in the vasculature can further be confirmed in the Allen Brain Atlas database (Figure 2D).
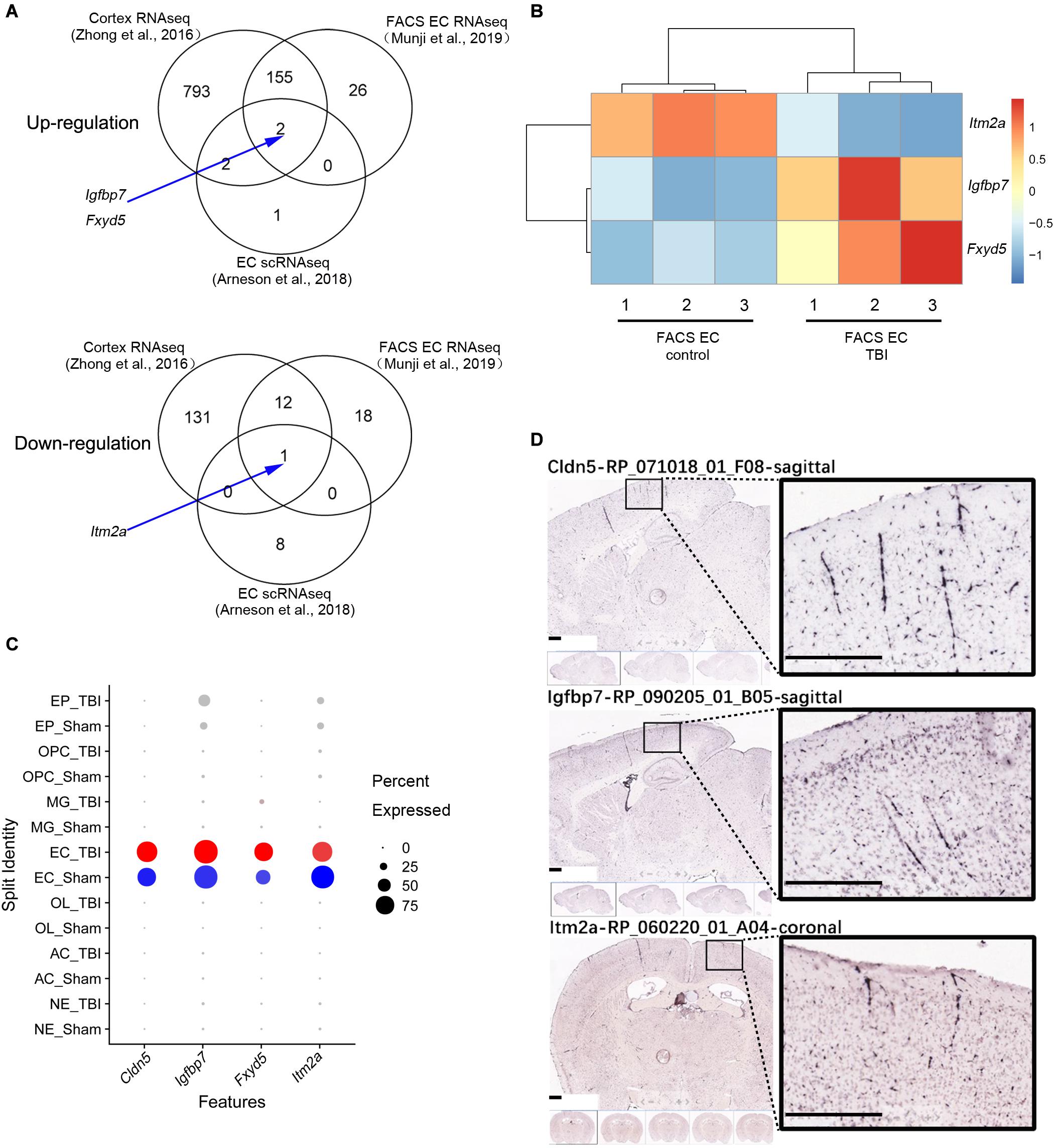
Figure 2. Analysis of the three shared DEGs. (A) Venn diagram illustrating the overlaps of the DEGs identified in the three studies, and the arrow indicate the three shared DEGs. (B) The details of the gene expression regulation of Itm2a, Igfbp7, and Fxyd5 in the TBI study from the FACS sorted EC samples. (C) The dot plot illustrating the expression level and the expression percentage of Cldn5, Igfbp7, Fxyd5, and Itm2a in the major cell types identified from the scRNAseq study. Cldn5 is a commonly used EC maker and used as a reference. The size of the dot represents the expression percentage in the cell type and the color intensity represents the expression level. The well-known EC marker Cldn5 is used as a reference. EP, Ependymocyte; OPC, Oligodendrocyte progenitor cell; MG, Microglia; EC, endothelial cell; OL, Oligodendrocyte; AC, Astrocyte; NE, Neuron. (D) The vascular expression of Igfbp7 and Itm2a genes expression using the Allen Brain Atlas database. Scale bar, 400 μm.
To uncover whether alteration of the three genes is TBI specific or reflect a broader response to brain injury, we analyzed the expression of Igfbp7/IGFBP7, Fxyd5/FXYD5, and Itm2a/ITM2A in EC in response to stoke, seizure, and EAE in Munji’s study (Munji et al., 2019), as well as in the human glioblastoma vasculature in our previous study (Dieterich et al., 2012). Upregulation of Fxyd5 in endothelial cells was observed in TBI and seizure. Down-regulation of Itm2a in endothelial cells was detected in TBI, seizure, and EAE (Supplementary Table 9). Igfbp7/IGFBP7 expression was up-regulated in all these disease models as well as glioblastoma vasculature (Supplementary Table 9), suggesting that its up-regulation is a universal response in EC to pathological alterations rather than TBI-specific.
Among the three commonly regulated genes in all three studies, Igfbp7 were up-regulated in all datasets (Figures 3A–C), and also showed endothelial enriched expression from single cell data (Figure 3D). IGFBP7 has been suggested as a critical regulator for angiogenesis, vessel integrity, and endothelial adhesion molecule (Hooper et al., 2009; Komiya et al., 2014; Rai et al., 2015), which all associate with the pathogenesis of TBI. Thus, we focused on IGFBP7, a matrix bound secreted protein, which belongs to insulin-like growth factor binding protein (IGFBP) family. Immunofluorescence co-staining of CD31 and IGFBP7 in human samples clearly confirms up-regulation of IGFBP7 in vasculature in response to TBI (Figure 3E).
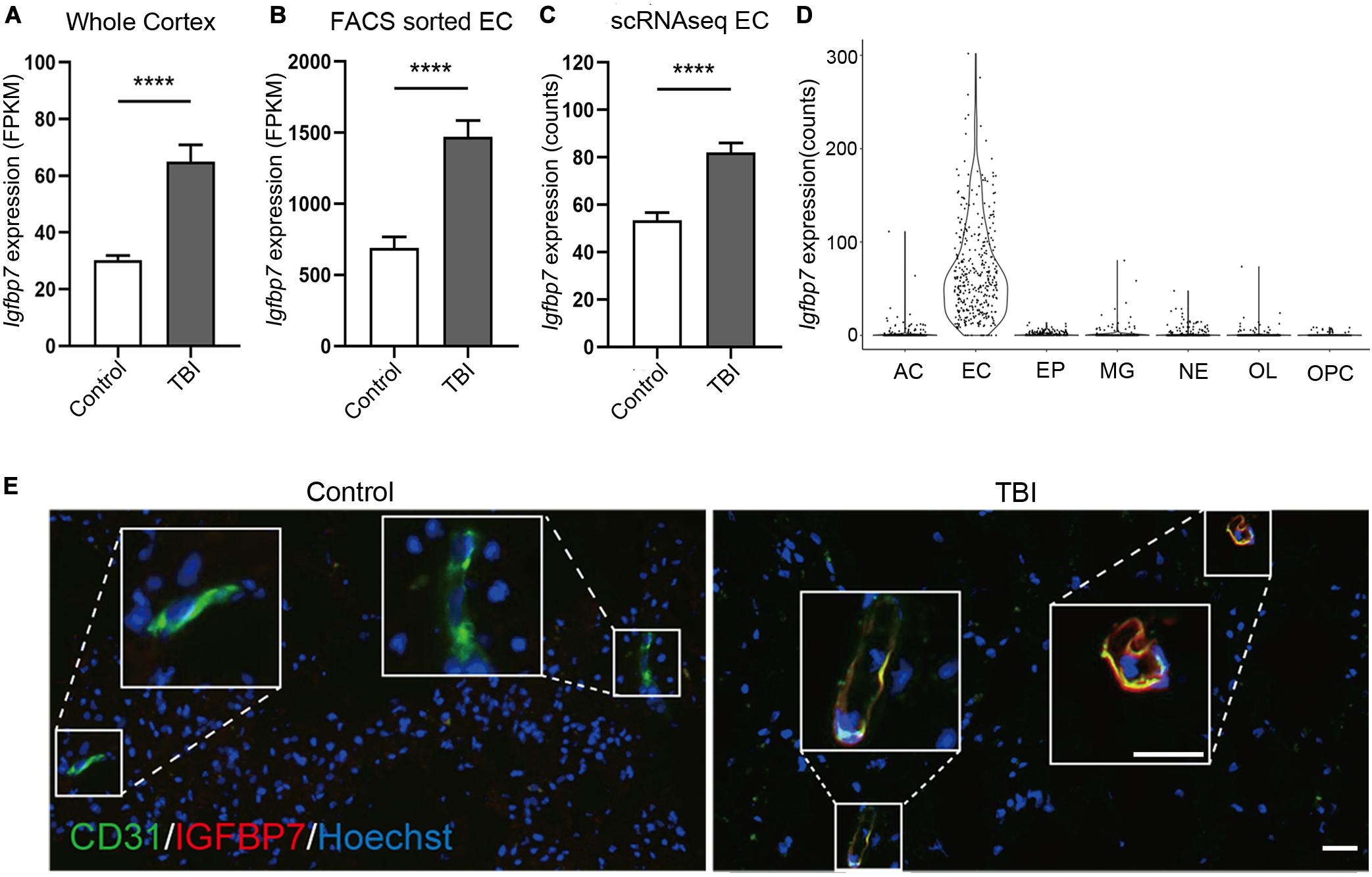
Figure 3. Igfbp7/IGFBP7 is upregulated in the vasculature in response TBI. (A–C) The expression level of Igfbp7 in the TBI and control groups from the three data sets. From left to right, whole cortex, FACS sorted EC RNAseq, and scRNAseq data sets. **** indicates p < 0.0001. (D) The expression level of Igfbp7 in the main cell types from the scRNAseq study. AC, Astrocyte; EC, endothelial cell; EP, Ependymocyte; MG, Microglia; NE, Neuron; OL, Oligodendrocyte; OPC, Oligodendrocyte progenitor cell. (E) Immunofluorescence co-staining of CD31 (green) and IGFBP7 (red) in human samples. The TBI and control group were both operated from the right frontal. Scale bar, 50 μm.
To uncover the signal pathway mediating IGFBP7 upregulation, we analyzed the expression of Igfbp7/IGFBP7 in EC (bEND.3 cells and HUVECs) upon the stimulation of VEGFA and TGFβ, which can increase IGFBP7 expression in breast cancer and glioblastoma vasculature respectively (Pen et al., 2008; Komiya et al., 2014). In addition, the effect of hypoxia, the most pronounced characteristic of brain injury causing pathogenesis, on Igfbp7/IGFBP7 expression in EC was determined by stimulating cells with CoCl2, which induces hypoxia stimulation through HIF1α stabilization (Xu et al., 2015). VEGFA stimulation failed to upregulate Igfbp7/IGFBP7 expression in either bEND.3 or HUVEC. Exposure of HUVECs to CoCl2 lead to a subtle upregulation of IGFBP7 expression, while this effect could not be observed in bEND.3 cells (Figures 4A,B). Notably, Igfbp7/IGFBP7 expression was significantly up-regulated in bEND.3 cells (7.2-folds) and HUVECs upon (1.6-folds) upon TGFβ stimulation (Figures 4A,B). In line with these findings, we observed a higher level of TGFβ1 in cortex from TBI compared to control in whole cortex RNAseq study (Zhong et al., 2016; Figure 4C), suggesting TGFβ signaling may potentiate vascular IGFBP7 upregulation in TBI. Interestingly, there is no significant difference in TGFβ2 level in cortex between TBI and control (Figure 4D). To identify the main source of TGFβ1, we have analyzed the TGFβ1 expression in single cell RNAseq dataset. TGFβ1 is mainly expressed in microglia and EC (Figure 4E). Based on these experiments in vitro, we proposed a potential mechanism that IGFBP7 up-regulation in endothelial cells in response to brain injury may be through TGFβ signaling. Experiments in vivo need to be further performed in the future to confirm the mechanism.
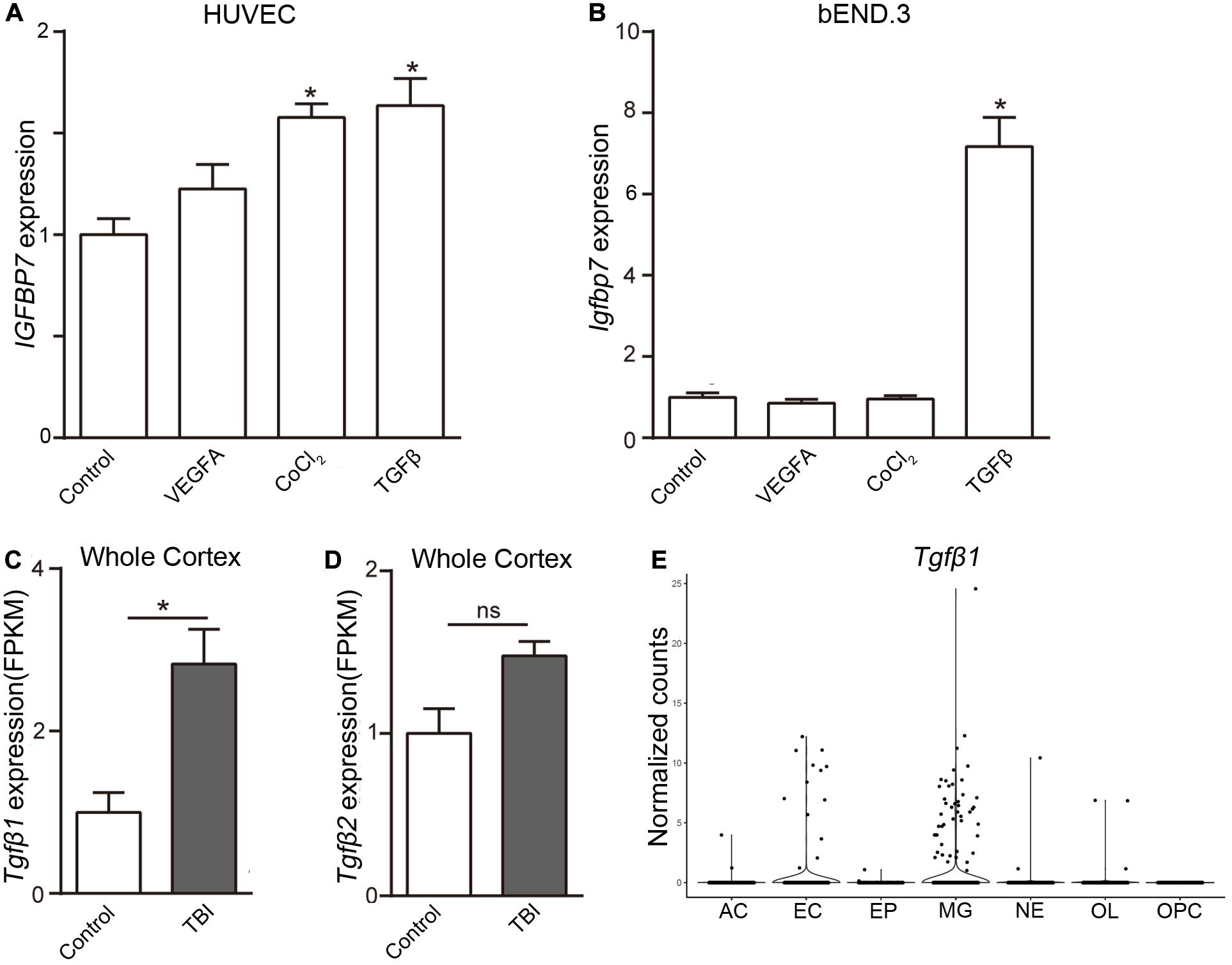
Figure 4. TGFβ induces the expression of Igfbp7/IGFBP7 in endothelial cells. The normalized Igfbp7/IGFBP7 expression levels in Control, VEGFA, CoCl2, and TGFβ stimulated HUVEC (A) and bEND.3 (B) cells are listed in the histogram, respectively. The expression levels of TGFβ1 (C) and TGFβ2 (D) from the whole cortex RNAseq data set. (E) The TGFβ1 expression in the main cell types from the scRNAseq study. AC, Astrocyte; EC, endothelial cell; EP, Ependymocyte; MG, Microglia; NE, Neuron; OL, Oligodendrocyte; OPC, Oligodendrocyte progenitor cell. * indicates p < 0.05.
Brain vasculature plays critical roles in brain physiology and pathology. In our previous studies, we have illustrated transcriptome profiles in the normal vasculature (He et al., 2016, 2018; Vanlandewijck et al., 2018). In this study, we focus on the gene expression changes in the brain vascular EC under TBI condition. To cover common EC alteration in TBI pathogenesis, we integrated the results from two bulk RANseq studies (Zhong et al., 2016; Munji et al., 2019) and one single cell RNAseq study (Arneson et al., 2018). The controlled cortical impact (CCI) model was employed in Zhang’s and Munji’s studies to generate focal injury, while the fluid percussion injury (FPI) model was used in Arneson’s study to induce mixed injury. All studies applied the mouse model and brain RNA expression analyses were performed at 24 h after injury. In the comparison of regulated genes from the three studies, despite the significant difference due to sample compositions and techniques, we identified three genes that were consistently regulated in all three analyses (Igfbp7, Fxyd5, and Itm2a).
IGFBP7 is highly expressed in the vasculature during development in the central nervous system. But, in adults, the IGFBP7 expression was reported to be only restricted to smooth muscle cells covering large vessels and choroid plexus vasculature, which is characterized by limited BBB property and high permeability (Hooper et al., 2009; Bar et al., 2020). Increased IGFBP7 expression in vasculature was observed in brain pathological conditions including glioblastoma and stroke, as well as other types of tumors (Hooper et al., 2009; Dieterich et al., 2012; Buga et al., 2014; Komiya et al., 2014). In this study, we showed that Igfbp7/IGFBP7 is up-regulated in the vasculature in both TBI mice models and surgical samples from patients with brain injury. Our results together with previous findings support the notion that Igfbp7/IGFBP7 may be a general marker of vasculature in response to pathological conditions in the brain. A higher level of TGFβ and hypoxia was observed in the brain tissue after TBI (Park et al., 2009), and they can sufficiently upregulate Igfbp7/IGFBP7 expression in EC, which may explain the molecular mechanism for Igfbp7/IGFBP7 up-regulation in vasculature in response to TBI.
The role of IGFBP7 on pathogenesis of brain injury remains largely unknown. Exposure of EC to IGFBP7 leads to stress fiber formation and disorganization of VE-cadherin mediated junctions, resulting in increased vascular permeability (Komiya et al., 2014), which indicates a role of IGFBP7 on BBB breakdown. In addition, stimulation of brain EC with IGFBP7 upregulated E-selectin, a crucial molecule in immune cell recruitment (Rai et al., 2015), indicating that IGFBP7 may regulate neuroinflammation in response to brain injury.
Emerging studies convincingly showed that the injury vasculature attempts to undergo repair by inducing angiogenesis (Park et al., 2009). IGFBP7 can act as both pro- and anti-angiogenic factors (Hooper et al., 2009; Pen et al., 2011; Tamura et al., 2009, 2014; Komiya et al., 2014). IGFBP7 could block VEGFA-induced tube formation, EC migration, proliferation, and vascular permeability (Tamura et al., 2009, 2014). In contrast, other studies suggested that IGFBP7 promotes angiogenesis by increasing EC adhesion and VEGFA bioavailability (Hooper et al., 2009; Komiya et al., 2014). Thus, the contradictory effect of IGFBP7 on angiogenesis in different systems may depend on cues in the microenvironment, such as distinct regional composition of ECM (Hooper et al., 2009). Whether IGFBP7 contributes to vascular repair by regulating angiogenesis after brain injury deserves further investigation.
Taken together, our study reveals the key molecular alteration of EC and identifies IGFBP7 as a potential biomarker of vasculature in response to brain injury.
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the ethics committee, Tianjin Medical University General Hospital. The patients/participants provided their written informed consent to participate in this study.
LH and LZ conceived the project. JW, YX, LZ, and ZZ performed the experiments. JW, XD, JT, FY, QH, QC, LZ, and LH analyzed the data. LZ and LH wrote the manuscript with significant input from JW and YX. All authors reviewed and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Nos. 81870978, 81702489, 81911530166, and 82002659), the National Key R&D Program of China (No. 2018YFC1313003), the Tianjin Natural Science Foundation (No. 18JCYBJC94000), the Natural Science Foundation of Shaanxi Province (No. 2020JQ-429), the Fundamental Research Funds for the Central University (Nos. GK202003050 and GK202003048), and the Natural Science Foundation of Huaihua City (Nos. 2020R3118 and 2020R3116).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.599834/full#supplementary-material
Supplementary Figure 1 | Clustering and annotation of the 6,351 cells from the single cell RNAseq study. (A) A Uniform Manifold Approximation and Projection (UMAP) overview of the whole dataset with main cell type annotated. OPC, Oligodendrocyte progenitor cell; EC, endothelial cell. (B) The detailed expression levels of typical maker genes to illustrate the main cell types. The size of the dot represents the expression percentage in the cell type and the color represents the expression level. Cluster 5 displays unique expression of EC markers (Pecam1, Cldn5, and Flt1) and is selected as EC cluster for downstream analysis. Cluster 8 shows lower expression of EC markers, but with specific expression of mural cells markers (Pdgfrb, Acta2, and Kcnj8), suggesting that it is likely an EC-contaminated mural cell cluster.
Arneson, D., Zhang, G., Ying, Z., Zhuang, Y., Byun, H. R., Ahn, I. S., et al. (2018). Single cell molecular alterations reveal target cells and pathways of concussive brain injury. Nat. Commun. 9:3894. doi: 10.1038/s41467-018-06222-0
Badaut, J., Ashwal, S., and Obenaus, A. (2011). Aquaporins in cerebrovascular disease: a target for treatment of brain edema? Cerebrovasc. Dis. 31, 521–531. doi: 10.1159/000324328
Bar, O., Gelb, S., Atamny, K., Anzi, S., and Ben-Zvi, A. (2020). Angiomodulin (IGFBP7) is a cerebral specific angiocrine factor, but is probably not a blood-brain barrier inducer. Fluids Barriers CNS 17:27. doi: 10.1186/s12987-020-00188-2
Bramlett, H. M., and Dietrich, W. D. (2015). Long-term consequences of traumatic brain injury: current status of potential mechanisms of injury and neurological outcomes. J. Neurotrauma 32, 1834–1848. doi: 10.1089/neu.2014.3352
Buga, A. M., Margaritescu, C., Scholz, C. J., Radu, E., Zelenak, C., and Popa-Wagner, A. (2014). Transcriptomics of post-stroke angiogenesis in the aged brain. Front. Aging Neurosci. 6:44. doi: 10.3389/fnagi.2014.00044
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. doi: 10.1038/nbt.4096
Dieterich, L. C., Mellberg, S., Langenkamp, E., Zhang, L., Zieba, A., Salomaki, H., et al. (2012). Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFbeta2 in vascular abnormalization. J. Pathol. 228, 378–390. doi: 10.1002/path.4072
He, L., Vanlandewijck, M., Mae, M. A., Andrae, J., Ando, K., Del Gaudio, F., et al. (2018). Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data 5:180160. doi: 10.1038/sdata.2018.160
He, L., Vanlandewijck, M., Raschperger, E., Andaloussi Mae, M., Jung, B., Lebouvier, T., et al. (2016). Analysis of the brain mural cell transcriptome. Sci. Rep. 6:35108. doi: 10.1038/srep35108
Ho, K. M., Honeybul, S., Yip, C. B., and Silbert, B. I. (2014). Prognostic significance of blood-brain barrier disruption in patients with severe nonpenetrating traumatic brain injury requiring decompressive craniectomy. J. Neurosurg. 121, 674–679. doi: 10.3171/2014.6.JNS132838
Hooper, A. T., Shmelkov, S. V., Gupta, S., Milde, T., Bambino, K., Gillen, K., et al. (2009). Angiomodulin is a specific marker of vasculature and regulates vascular endothelial growth factor-A-dependent neoangiogenesis. Circ. Res. 105, 201–208. doi: 10.1161/CIRCRESAHA.109.196790
Jassam, Y. N., Izzy, S., Whalen, M., McGavern, D. B., and El Khoury, J. (2017). Neuroimmunology of traumatic brain injury: time for a paradigm shift. Neuron 95, 1246–1265. doi: 10.1016/j.neuron.2017.07.010
Jullienne, A., Obenaus, A., Ichkova, A., Savona-Baron, C., Pearce, W. J., and Badaut, J. (2016). Chronic cerebrovascular dysfunction after traumatic brain injury. J. Neurosci. Res. 94, 609–622. doi: 10.1002/jnr.23732
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Komiya, E., Sato, H., Watanabe, N., Ise, M., Higashi, S., Miyagi, Y., et al. (2014). Angiomodulin, a marker of cancer vasculature, is upregulated by vascular endothelial growth factor and increases vascular permeability as a ligand of integrin alphavbeta3. Cancer Med. 3, 537–549. doi: 10.1002/cam4.216
Lambrechts, D., Wauters, E., Boeckx, B., Aibar, S., Nittner, D., Burton, O., et al. (2018). Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289. doi: 10.1038/s41591-018-0096-5
Maas, A. I., Stocchetti, N., and Bullock, R. (2008). Moderate and severe traumatic brain injury in adults. Lancet Neurol. 7, 728–741. doi: 10.1016/S1474-4422(08)70164-9
Marmarou, A. (2003). Pathophysiology of traumatic brain edema: current concepts. Acta Neurochir. Suppl. 86, 7–10. doi: 10.1007/978-3-7091-0651-8_2
Munji, R. N., Soung, A. L., Weiner, G. A., Sohet, F., Semple, B. D., Trivedi, A., et al. (2019). Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood-brain barrier dysfunction module. Nat. Neurosci. 22, 1892–1902. doi: 10.1038/s41593-019-0497-x
Nguyen, R., Fiest, K. M., McChesney, J., Kwon, C. S., Jette, N., Frolkis, A. D., et al. (2016). The international incidence of traumatic brain injury: a systematic review and meta-analysis. Can. J. Neurol. Sci. 43, 774–785. doi: 10.1017/cjn.2016.290
Park, E., Bell, J. D., Siddiq, I. P., and Baker, A. J. (2009). An analysis of regional microvascular loss and recovery following two grades of fluid percussion trauma: a role for hypoxia-inducible factors in traumatic brain injury. J. Cereb. Blood Flow Metab. 29, 575–584. doi: 10.1038/jcbfm.2008.151
Pen, A., Durocher, Y., Slinn, J., Rukhlova, M., Charlebois, C., Stanimirovic, D. B., et al. (2011). Insulin-like growth factor binding protein 7 exhibits tumor suppressive and vessel stabilization properties in U87MG and T98G glioblastoma cell lines. Cancer Biol. Ther. 12, 634–646. doi: 10.4161/cbt.12.7.17171
Pen, A., Moreno, M. J., Durocher, Y., Deb-Rinker, P., and Stanimirovic, D. B. (2008). Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-beta signaling. Oncogene 27, 6834–6844. doi: 10.1038/onc.2008.287
Rai, S., Nejadhamzeeigilani, Z., Gutowski, N. J., and Whatmore, J. L. (2015). Loss of the endothelial glycocalyx is associated with increased E-selectin mediated adhesion of lung tumour cells to the brain microvascular endothelium. J. Exp. Clin. Cancer Res. 34:105. doi: 10.1186/s13046-015-0223-9
Salehi, A., Zhang, J. H., and Obenaus, A. (2017). Response of the cerebral vasculature following traumatic brain injury. J. Cereb. Blood Flow Metab. 37, 2320–2339. doi: 10.1177/0271678X17701460
Silverberg, N. D., Duhaime, A. C., and Iaccarino, M. A. (2019). Mild traumatic brain injury in 2019-2020. JAMA 323, 177–178. doi: 10.1001/jama.2019.18134
Simon, D. W., McGeachy, M. J., Bayir, H., Clark, R. S., Loane, D. J., and Kochanek, P. M. (2017). The far-reaching scope of neuroinflammation after traumatic brain injury. Nat. Rev. Neurol. 13, 171–191. doi: 10.1038/nrneurol.2017.13
Sun, D., Gu, G., Wang, J., Chai, Y., Fan, Y., Yang, M., et al. (2017). Administration of tauroursodeoxycholic acid attenuates early brain injury via Akt pathway activation. Front. Cell. Neurosci. 11:193. doi: 10.3389/fncel.2017.00193
Tamura, K., Hashimoto, K., Suzuki, K., Yoshie, M., Kutsukake, M., and Sakurai, T. (2009). Insulin-like growth factor binding protein-7 (IGFBP7) blocks vascular endothelial cell growth factor (VEGF)-induced angiogenesis in human vascular endothelial cells. Eur. J. Pharmacol. 610, 61–67. doi: 10.1016/j.ejphar.2009.01.045
Tamura, K., Yoshie, M., Hashimoto, K., and Tachikawa, E. (2014). Inhibitory effect of insulin-like growth factor-binding protein-7 (IGFBP7) on in vitro angiogenesis of vascular endothelial cells in the rat corpus luteum. J. Reprod. Dev. 60, 447–453. doi: 10.1262/jrd.2014-069
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Vanlandewijck, M., He, L., Mae, M. A., Andrae, J., Ando, K., Del Gaudio, F., et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. doi: 10.1038/nature25739
Xu, X., Tan, X., Tampe, B., Sanchez, E., Zeisberg, M., and Zeisberg, E. M. (2015). Snail is a direct target of hypoxia-inducible factor 1alpha (HIF1alpha) in hypoxia-induced endothelial to mesenchymal transition of human coronary endothelial cells. J. Biol. Chem. 290, 16653–16664. doi: 10.1074/jbc.M115.636944
Yeoh, S., Bell, E. D., and Monson, K. L. (2013). Distribution of blood-brain barrier disruption in primary blast injury. Ann. Biomed. Eng. 41, 2206–2214. doi: 10.1007/s10439-013-0805-7
Zhong, J., Jiang, L., Cheng, C., Huang, Z., Zhang, H., Liu, H., et al. (2016). Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 1646, 589–600. doi: 10.1016/j.brainres.2016.07.002
Keywords: traumatic brain injury, vasculature, endothelial cell, IGFBP7, TGFβ
Citation: Wang J, Deng X, Xie Y, Tang J, Zhou Z, Yang F, He Q, Cao Q, Zhang L and He L (2021) An Integrated Transcriptome Analysis Reveals IGFBP7 Upregulation in Vasculature in Traumatic Brain Injury. Front. Genet. 11:599834. doi: 10.3389/fgene.2020.599834
Received: 29 August 2020; Accepted: 30 November 2020;
Published: 11 January 2021.
Edited by:
Cheng Peng, Yunnan University, ChinaReviewed by:
Andre Obenaus, University of California, Irvine, United StatesCopyright © 2021 Wang, Deng, Xie, Tang, Zhou, Yang, He, Cao, Zhang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Zhang, emxzbm51QGdtYWlsLmNvbQ==; Liqun He, bGlxdW4uaGVAaWdwLnV1LnNl
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.