Corrigendum: Genetic Variation in an Experimental Goldfish Derived From Hybridization
- 1State Key Laboratory of Developmental Biology of Freshwater Fish, College of Life Sciences, Hunan Normal University, Changsha, China
- 2Department of Histology and Embryology, Clinical Anatomy and Reproductive Medicine Application Institute, Hengyang Medical School, University of South China, Hengyang, China
- 3Hunan Province Cooperative Innovation Center for Molecular Target New Drug Study Institute of Pharmacy and Pharmacology, University of South China, Hengyang, China
Owning to the extreme difficulty in identifying the primary generation (G0), the common ancestor of various twin-tail goldfish strains remains unclear. However, several authors have hypothesized that this ancestor may have been the crucian carp (Carassius auratus). Previously, we generated an experimental hybrid goldfish (EG) from the interspecific hybridization of red crucian carp (Carassius auratus ♀, RCC) × common carp (Cyprinus carpio ♂, CC). Unlike either parent, EG possessed twin caudal fins similar to those of natural goldfish (Carassius auratus, NG). The genetic characteristics of EG, as well as the mechanisms underlying its formation, are largely unknown. Here, we identified the genetic variation in the chordin gene that was associated with the formation of the twin-tail phenotype in EG: a stop codon mutation at the 127th amino acid. Furthermore, simple sequence repeat (SSR) genotyping indicated that, among the six alleles, all of the EG alleles were also present in female parent (RCC), but alleles specific to the male parent (CC) were completely lost. At some loci, EG and NG alleles differed, showing that these morphologically similar goldfish were genetically dissimilar. Collectively, our results demonstrated that genetic variations and differentiation contributed to the changes of morphological characteristics in hybrid offspring. This analysis of genetic variation in EG sheds new light on the common ancestor of NG, as well as on the role of hybridization and artificial breeding in NG speciation.
Introduction
The basic architecture of the vertebrate tail is highly conserved across all vertebrates (Janvier, 1996; Kardong, 2011), including fish, even though fish caudal fins possess divergent and complex skeletal elements (Abe et al., 2014). Interestingly, in certain natural goldfish (Carassius auratus, NG) lineages, this conserved architecture has undergone extreme modifications due to artificial selection (Watase, 1887; Bateson, 1894; Matsui, 1934; Chen, 1956; Smartt, 2001). Indeed, some NG strains have twin-tail, which are of considerable ornamental interest (Matsui, 1934; Smartt, 2001). Historical accounts indicate that NG domestication in China began at around 1000 C.E., but the earliest record of twin-tail NG dates from 1596 C.E., suggesting that twin-tail NG emerged approximately 600 years after initial domestication (Chen, 1956; Smartt, 2001). It has been shown that a mutation at the 127th amino-acid position of the chordin gene, wherein the glutamic codon is mutated to a stop codon, gives rise to the bifurcated caudal fin (the twin-tail) in NG during ventralization in early embryonic development (Abe et al., 2014). This mutation of the chordin gene may have appeared during the domestication period (Abe et al., 2014).
Hybridization, defined as reproduction between members of genetically distinct populations that produces offspring with mixed ancestry, is common in nature and has wide-ranging effects on speciation and population evolution (Barton and Hewitt, 1989; Barton, 2001; Mallet, 2007, 2008; Abbott et al., 2013). Hybridization typically produces heterozygous offspring, which inherit genetic material from both parent genetic material and express biological characteristics intermediate between parents. Numerous species have hybrid ancestry, presumably due to hybridization between closely related species (Cui et al., 2013; Schumer et al., 2016; Meier et al., 2017). In fish, the common carp (CC) and the crucian carp (Carassius carassius) belong to the same subfamily, Cyprininae. Previous phylogenic studies have shown that NG are closely related to crucian carp (Komiyama et al., 2009; Gao et al., 2012; Wang et al., 2013). Thus, it has been hypothesized that NG are derived from crucian carp, and may represent a variant of this species (Wheeler, 2000; Smith and McVeagh, 2005). However, as NG is not recently derived, it is extremely difficult or even impossible to reconstruct its exact origin and evolutionary history (James and Abbott, 2005). Thus, we still relatively know little about the precise processes of NG evolution and domestication following hybridization; the exact point at which twin-tail arose, as well as the underlying causes of this phenotype, also remain elusive.
Previously, we obtained an experimental hybrid goldfish (EG) from the hybridization of female red crucian carp (Carassius carassius, RCC) × male common carp (Cyprinus carpio, CC) after sequential selective breeding. A stable population of EG was successfully established by self-mating (Wang et al., 2014). EG provides an ideal model to study not only the origin and evolution of NG, but also the effects of hybridization on the generation of new species. Consequently, it is important to carefully investigate the genetic characteristics and formation mechanisms of EG.
Here, we identified a stop codon mutation at the 127th amino acid of chordin gene, which is associated with the formation of twin-tail, in EG. We also found that the SSR alleles of EG were biased toward the female parent (RCC), and alleles differed between EG and NG at some loci. Collectively, our results showed that certain genetic characteristics of EG, including the chordin gene mutation and the RCC-aligned SSR alleles, originated from the distant hybridization, and contributed to the observed difference in morphology. The process of EG generation, as well as the genetic characters of this hybrid, shed new light on the common ancestor of NG, as well as on the role of hybridization and artificial breeding in NG speciation.
Materials and Methods
Experimental Fish
All RCC, CC, and EG were raised in ponds at the State Key Laboratory of Developmental Biology of Freshwater Fish, Hunan Normal University, Changsha, China. NG were purchased at a local market. All experiments were approved by the Animal Care Committee of Hunan Normal University and followed the guidelines of the Administration of Affairs Concerning Animal Experimentation of China.
Cloning and Sequence Analysis of the Chordin Gene
To compare the sequences of the chordin gene between single-tail and twin-tail fish, we isolated and sequenced the 1st to 6th exons of chordin homologues from the embryonic cDNA pools of four fish: single-tail RCC and CC, and twin-tail EG and NG. After self-mating, total RNA was extracted from the gastrula-stage embryos of all four fish using Trizol (Invitrogen, Camarillo, CA, United States). The first-strand cDNA of the chordin gene was synthesized using ReverTra Ace (Toyobo, Osaka, Japan). The forward primer 5′-GCGTTACCCATCCAACC-3′ and the reverse primer 5′-TCTGTRTCCGCTTGTGGT-3′ were designed based on CDS of the chordin genes from Carassius auratus (AB874473.1) and Cyprinus carpio (LC092194.1), which were downloaded from GenBank. Each PCR (25 μL) contained 20 ng of cDNA template, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.4 μM of each primer, 1 × PCR buffer, and 1.25 U Taq polymerase (Takara, Dalian, China). The cycling conditions were as follows: an initial denaturation at 94°C for 4 min; followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 10 min. PCR products were separated and purified using 1.2% agarose gels and Gel Extraction Kits (Sangon, Shanghai, China), respectively. Purified products were ligated into pMD18-T vectors and transfected into Escherichia coli DH5α. Positive clones were sequenced and further analyzed using BLAST1 and CLUSTAL W2.
SSR Amplification and Sequencing
We randomly selected eight fish of each type (RCC, CC, NG, and EG) for SSR testing. Total genomic DNA was extracted from the fin tissue of each fish with a DNA extraction kit (Sangon, Shanghai, China). The microsatellite regions were PCR amplified using six florescent-labeled microsatellite primers (synthesized by Sangon, Shanghai, China): three from the common carp (MWF 4, MWF 5, and MWF 16) (Crooijmans et al., 1997); two from the bighead carp (HLJY 3940 and HLJY 2526) (Geng et al., 2006); and one from the crucian carp (MFW1, developed for this study). Primer sequences and annealing temperatures are listed in Supplementary Table 1. PCR amplifications were performed in a total volume of 20 μL, containing 1 μL of genomic DNA (5 ng/μL), 2 μL of 10 × Taq Buffer (Mg2+ Plus), 1 μL of 2.5 mM dNTPs, 0.4 μL of each primer (5 μM), 0.4 μL of Taq DNA Polymerase (5 U/μL), and 14.8 μL of ddH2O. The PCR cycling conditions were as follows: an initial denaturation at 94°C for 3 min; followed by 35 cycles of 94°C for 30 s, a primer-specific annealing temperature for 30 s, and 72°C for 45 s; a final extension step of 72°C for 7 min. PCR products were sequenced using capillary electrophoresis on an ABI 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, United States) using BigDye Terminator Cycle Sequencing kits (Applied Biosystems, Foster City, CA, United States).
Genetic Analysis
Genetic distance and genetic polymorphism indexes, including major allele frequency, numbers of genotypes, numbers of alleles, heterozygosity, and gene diversity, were calculated using Popgene32 (Yeh et al., 2000). SSR genotypes, polymorphic information content (PIC) values and the cluster analysis of the 4 populations, which was performed by UPGMA method basing on the genetic distance, were determined with PowerMarker 3.25 (Liu and Muse, 2005).
Results
Genetic Variation in the Chordin Gene
Both RCC and CC have single-tail (Figures 1A,B), while EG and NG have twin-tail (Figures 1C,D). To investigate the formation of the twin-tail phenotype in EG, the 1st to 6th exons of chordin homologues were cloned from RCC, CC, EG, and NG. All cloned sequences were identified as chordin and were submitted to GenBank (RCC, MN918649; CC, MN918650; EG, MN918651; NG, MN918652). There was high nucleotide sequence similarity (> 99%) among the chordin genes from all four fish (Supplementary Figure 1). Further analysis showed that chordin gene sequences from RCC and CC, the single-tail fish, were identical, as were the chordin sequences of EG and NG, the twin-tail fish. However, the chordin sequences between the twin-tail and single-tail fish differed: the twin-tail fish had a stop codon (TAG) at the 127th amino acid position of the chordin protein, while the single-tail fish had a glutamic acid codon (GAG) (Figure 2).

Figure 1. Lateral and dorsal views of phenotypically-representative examples of the four fish taxa included in this study. (A) Red crucian carp (RCC, Carassius auratus). (B) Common carp (CC, Cyprinus carpio). (C) Natural/wild-type goldfish (NG, Carassius auratus). (D) Experimental hybrid goldfish (EG). Scale bars, 2 cm (A,C,D), 5 cm (B).
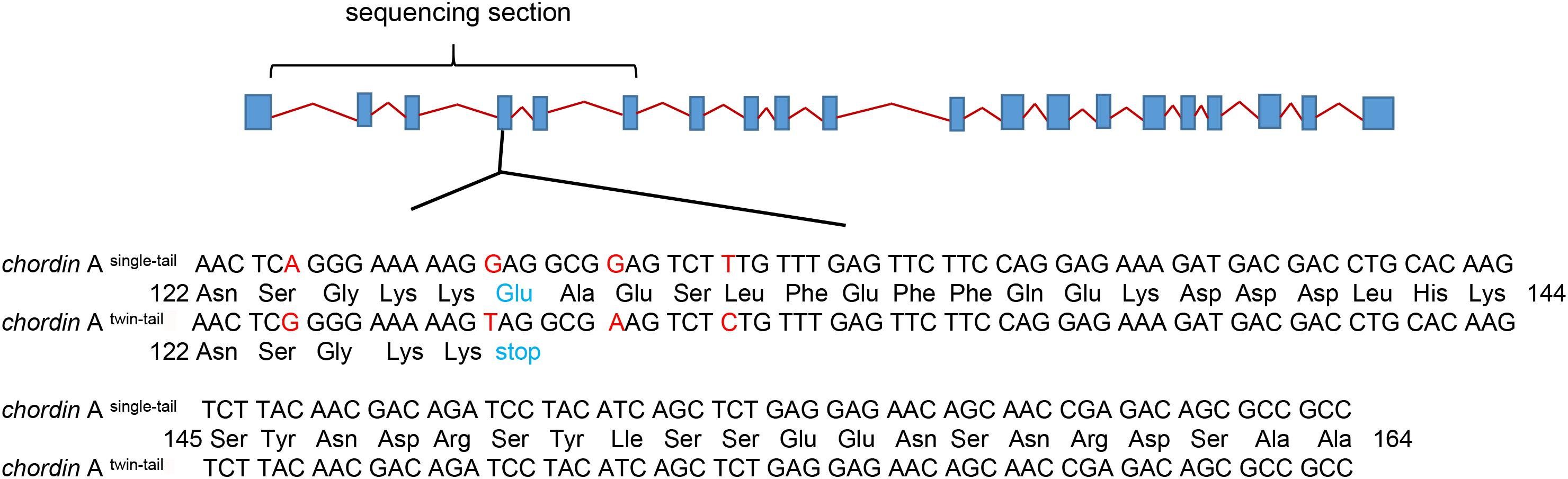
Figure 2. The chordin Awt and chordin AE127X amino acid sequences in single-tail and twin-tail fish. Diagram shows the composition of chordin A; exons are shown as light green boxes. The twin-tail-specific mutation changes a glutamic acid codon (GAG) to a stop codon (TAG) at amino acid position 127 of the 4th exon; the mutated amino acid is indicated by red letters. The sequenced exons (1st to 6th) are located in the horizontal bracket. The lower panel shows the amino acid composition of the 4th exon, which includes the mutation.
SSR Sequencing and Genotyping
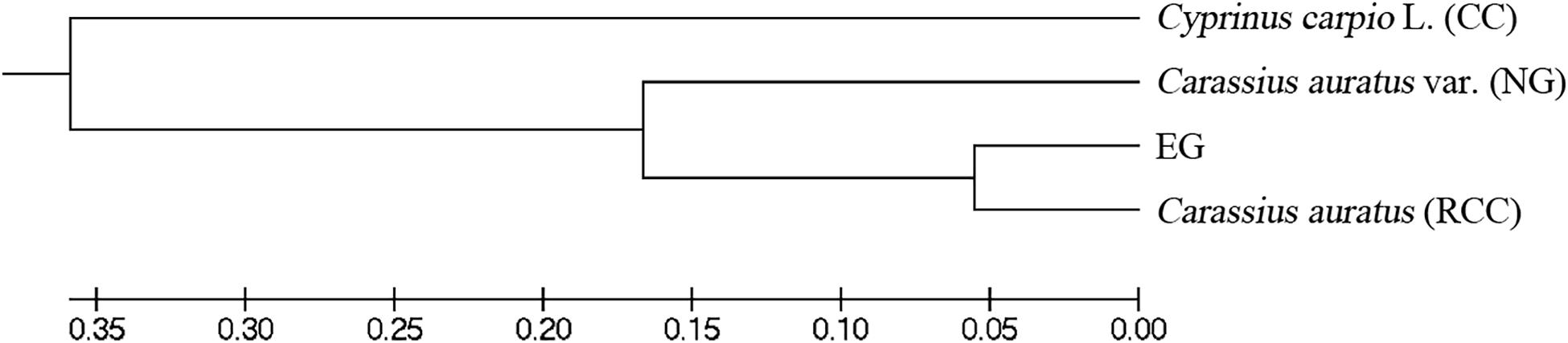
Across the four fish (EG, NG, RCC, and CC), each of the six amplified SSR loci (121–302 bp) had 1–8 alleles (Table 1). Almost all the alleles identified in EG, as well as most in NG, were also found in RCC (Table 1). However, nearly all the alleles (except one in HLJY 2526) identified in CC were absent in EG and RCC. The peaks at the HLJY 2526 loci were identical across all four fish (Figure 3A). However, at the MWF 16 locus, RCC, EG, and NG had a peak at 275 bp, while CC had a peak at 133 bp instead (Figure 3B). The peak patterns at the MFW 1 loci presented similar situation as MWF 16 where CC was different from RCC, NG and EG (Figure 3F), and higher similarity to HLJY3940 (Figure 3E). Like RCC and NG, EG had a peak at 206 bp at allele HLJY 3940; however, unlike RCC and NG, EG lacked a peak at 241 bp (Figure 3E). Indeed, all EG alleles also appeared in RCC. In contrast, NG exhibited a specific allele of MWF 5 (at 185 bp) (Figure 3D). In addition, NG respectively presented one allele similar to CC at MWF 4 (at 164 bp) (Figure 3C) and MWF 5 (at 156 bp) (Figure 3D). These results indicated that, although EG and NG appeared morphologically similar, these fish differed genetically. Genetic polymorphism analyses indicated that, of the four populations investigated (RCC, CC, NG, and EG), EG had the lowest polymorphism indexes, corresponding to the highest homogeneity (Table 2). In addition, across all pairs of taxa, genetic distance was lowest between EG and RCC (0.1103; Table 3). Consistent with this, the UPGMA phylogenetic tree recovered EG and RCC as a sister taxa. NG, EG, and RCC formed a single cluster, distinct from CC (Figure 4).
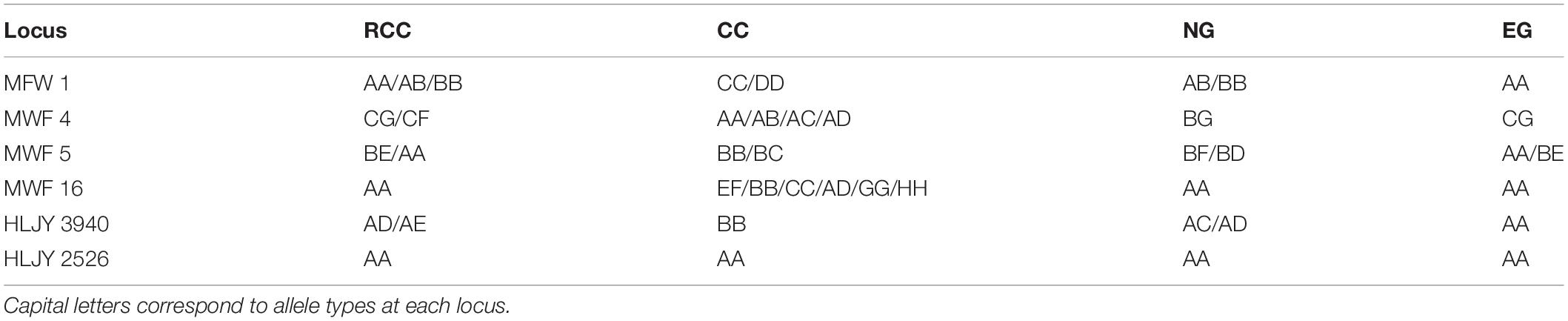
Table 1. Simple sequence repeat (SSR) genotypes of the four fish lines included in this study: red crucian carp (RCC, Carassius auratus); common carp (CC, Cyprinus carpio); natural goldfish (NG, Carassius auratus); experimental hybrid goldfish (EG).
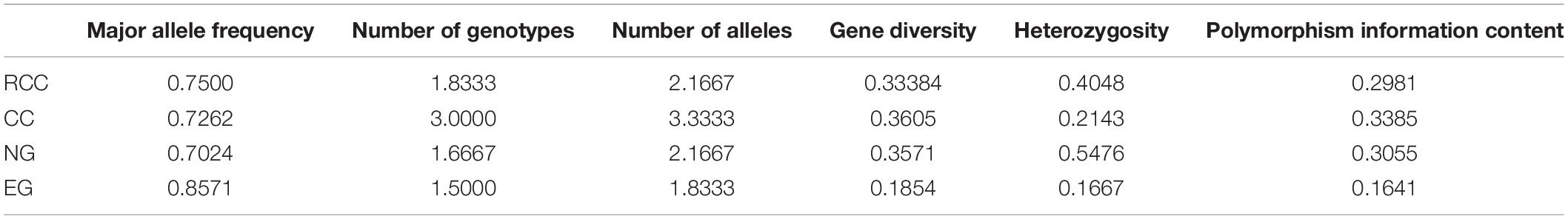
Table 2. Genetic polymorphism indexes for the four fish lines included in this study: RCC, CC, EG, NG.
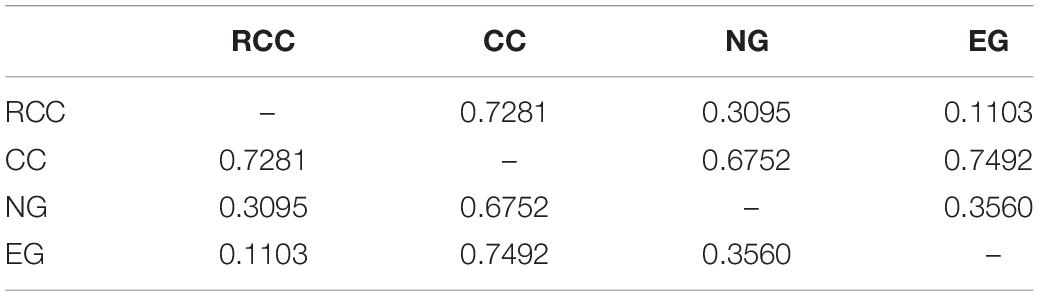
Table 3. The average genetic distances between each pair of fish taxa included in this study: RCC, CC, EG, NG.
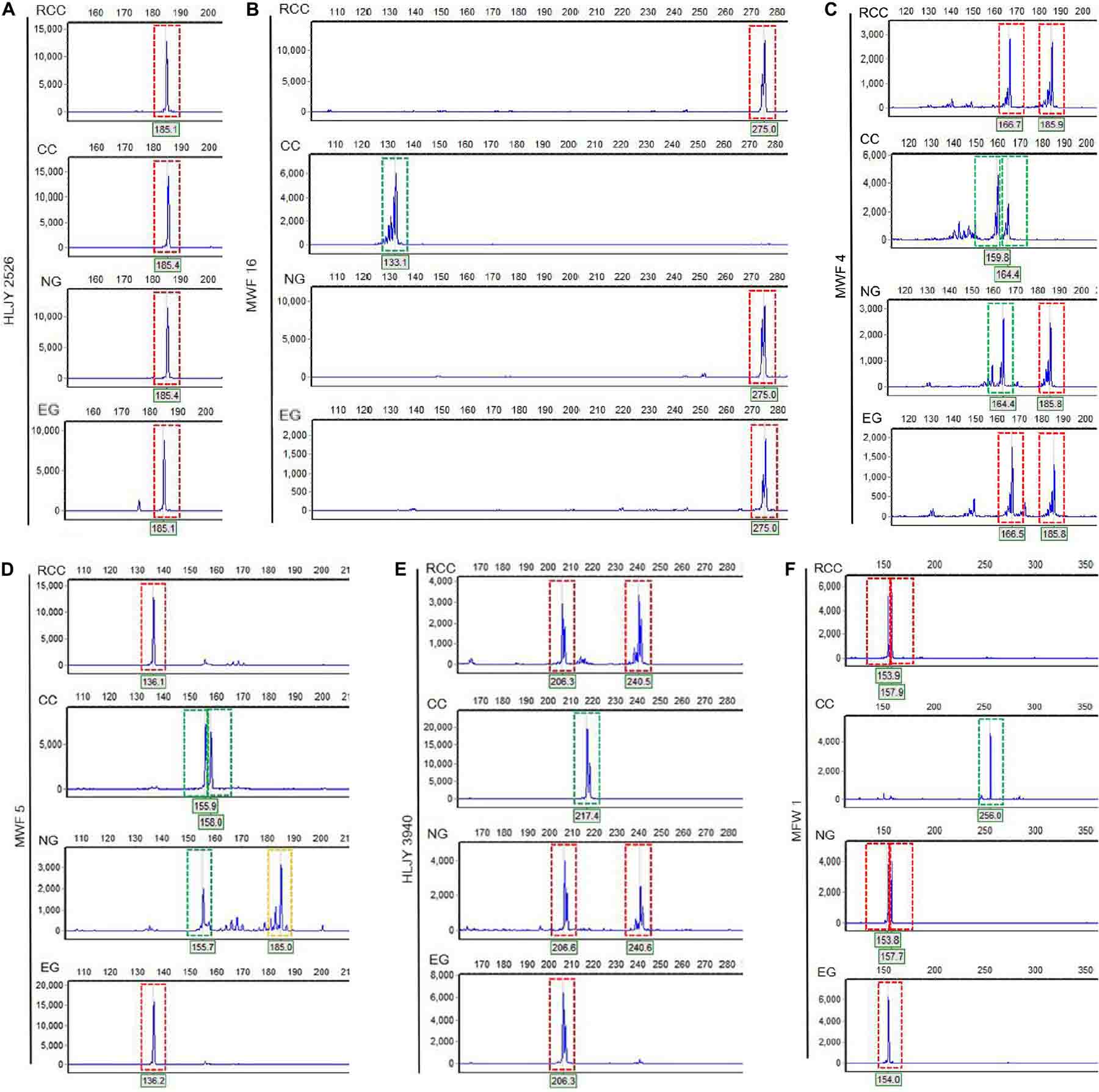
Figure 3. Electropherograms showing allelic peaks at six loci across the four fish lines. (A) HLJY 2526 locus. (B) MWF 16 locus. (C) MWF 4 locus. (D) MWF 5 locus. (E) HLJY 3940 locus. (F) MFW 1 locus. Peaks boxed in red are unique to the crucian carp; peaks boxed in green are unique to the common carp; peaks boxed in yellow are unique to the natural goldfish. The x-axes show the size of each segment, and the y-axes indicate the strength of the corresponding signal.
Discussion
Given the uncertainty surrounding the common ancestor of NG strains, as well as the evolutionary processes underlying their emergence (James and Abbott, 2005), the formation and subsequentially stable inheritance of transgressive phenotypes in EG provides a unique opportunity to study the origin and evolution of NG (Wang et al., 2014). That is, studies of the genetic characteristics and formation mechanisms of EG will improve our understanding of the genetic characteristics and formation mechanisms of NG.
Hybridization and the Formation of Twin-Tail
Hybridization is an important method of animal breeding because this process increases genetic variation (Bullini, 1994), and because hybrids frequently present transgressive segregation exceeding the range between the parental means (Slatkin and Lande, 1994; Rieseberg et al., 1999). The morphological characteristics of EG were highly similar to those of NG, but differed from the original parents RCC and CC: unlike RCC and CC, EG had twin-tail, a spherical body, a short caudal peduncle, and a range of body coloration patterns (Wang et al., 2014). Of these morphological characters, the most recognizable is the twin-tail, as most other fish species have a single tail.
In NG, the twin-tail trait is associated with mutations in the 4th exon of the chordin gene (Takashima et al., 2007; Abe et al., 2014). In NG, two alleles of the chordin A gene have been identified: the wild-type allele (chordin Awt) includes a glutamic codon at the 127th amino-acid position, while the other allele (chordinAE127X) includes a stop codon at the same position. The allele chordinAE127X is predicted to encode a truncated protein that contributes to the formation of the twin-tail (Abe et al., 2014). In zebrafish, dysfunction in the chordin gene gave rise to a twin-tail phenotype (Schulte-Merker et al., 1997; Fisher and Halpern, 1999). In EG, the potential primary generation (G0) of NG, we identified the twin-tail-associated mutation at the 127th amino-acid position in the chordin gene: a stop codon (TAG) instead of a glutamic acid codon (GAG). This mutation was identical to that found in NG. In contrast, the chordin genes of RCC and CC had a wild-type glutamic acid codon at the same position (Figure 2). These results in the G0 generation of EG are consistent with previous studies of twin-tail formation (Abe et al., 2014), and provide direct evidence that this mutation, as well as the associated phenotype (i.e., twin-tail), might have arisen due to hybridization.
Genetic Bias in EG
SSRs (or microsatellite markers) have been broadly used in studies of molecular evolution (Chistiakov et al., 2006). These simple tandem-repeats are abundantly and randomly distributed throughout genomes, and due to their high polymorphism and rapid evolutionary rate, microsatellite loci are useful for analyses of genetic diversity, population structure, gene flow, and hybridization (Pritchard et al., 2000; Wang et al., 2014). SSR genotyping, which is based on the numbers of repeats, may improve our understanding of genetic relationships within populations or among close relatives (Roy et al., 1994). SSR allele inheritance proceeds in accordance with Mendelian laws of segregation and independent assortment. That is, in diploids, each offspring inherits one copy of each microsatellite segment from each parent (Tesson et al., 2013). Thus, EG, derived from the hybridization of RCC and CC, should have inherited both sets of parental microsatellites. However, among the six alleles, which showed good reproducibility and stability between RCC and CC, we only identified RCC alleles in EG; all alleles unique to CC were completely lost (Table 1 and Figure 3). It may be that the inheritance of SSR alleles by EG did not conform to Mendelian laws because the EG lineage was subject to intense artificial selection. As a result of this selective pressure, the EG population did not contain all the alleles from RCC and CC.
Large-scale morphological changes, which require extensive modifications of developmental mechanisms, are often presumed to require relatively long periods of evolutionary time (Janvier, 2002; Kardong, 2011). However, previous studies have shown that gene duplication and subsequent artificial selection generated dramatic morphological and developmental changes in NG strains within 600 years (Abe et al., 2014). Previously, using a combination of hybridization and artificial selection, we established a stable EG lineage in even less time (F1–F6, within 6 years) (Wang et al., 2014). Here, our results suggested that the hybridization of female RCC and male CC led to a mutation in the chordin gene, which contributed to the expression of the twin-tail phenotype. During hybridization, the separation, recombination, and mutation of genetic materials leads to the formation of new varieties (Schumer et al., 2016). Thus, our results reveal the effects of hybridization on the formation of the twin-tail phenotype in EG, and SSR inheritance patterns, shedding new light on the origin and evolution of NG.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Animal Care Committee of Hunan Normal University and followed the guidelines statement of the Administration of Affairs Concerning Animal Experimentation of China. All samples were raised in natural ponds, all dissections were performed with 100 mg/L MS-222 (Sigma-Aldrich, St Louis, MO, United States), and all efforts were made to minimize suffering.
Author Contributions
JW and WH conceived the research, analyzed the data, and wrote the manuscript. JZ, LL, GZ, TL, CX, and MC performed the research and writing-reviewed the manuscript. SL provided substantial contributions to conception and coordination. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Key R&D Program of China (2018YFD0900200), the National Natural Science Foundation of China (Grant Nos. 31872549, 31730098, and U19A2040), Natural Science Foundation of Hunan Province (Grant No. 2018JJ3431), Outstanding Youth Fund of The Education Department of Hunan Province (Grant No. 2017SJQ13), Open fund of State Key Laboratory of Developmental Biology of Freshwater Fish (Grant No. 2017KF008), the earmarked fund for China Agriculture Research System (Grant No. CARS-45), 111 Project (D20007), and Hunan Provincial Natural Science and Technology Major Project (Grant No. 2017NK1031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance and scientific consultation during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.595959/full#supplementary-material
Supplementary Figure 1 | Nucleotide alignment of the sequenced exons (1st to 6th) of the chordin gene from the four fish lines (RCC, CC, NG, EG). Asterisks (∗) indicate bases that differ among lines. The site of termination codon mutation is highlighted in blue.
Supplementary Table 1 | Primer pairs and corresponding annealing temperatures for the six SSR loci used in this study.
Footnotes
References
Abbott, R., Albach, D., Ansell, S., Arntzen, J. W., Baird, S. J., Bierne, N., et al. (2013). Hybridization and speciation. J. Evol. Biol. 26, 229–246.
Abe, G., Lee, S. H., Chang, M., Liu, S. C., Tsai, H. Y., and Ota, K. G. (2014). The origin of the bifurcated axial skeletal system in the twin-tail goldfish. Nat. Commun. 5:3360.
Barton, N. H. (2001). The role of hybridization in evolution. Mol. Ecol. 10, 551–568. doi: 10.1046/j.1365-294x.2001.01216.x
Barton, N. H., and Hewitt, G. M. (1989). Adaptation, speciation and hybrid zones. Nature 341, 497–503. doi: 10.1038/341497a0
Bateson, W. (1894). Materials for the Study of Variation: Treated with Special Regard to Discontinuity in the Origin of Species. London: Macmillan.
Bullini, L. (1994). Origin and evolution of animal hybrid species. Trends Ecol. Evol. 9, 422–426. doi: 10.1016/0169-5347(94)90124-4
Chen, S. C. (1956). A history of the domestication and the factors of the varietal formation of the common goldfish, Carassius auratus. Sci. Sin. 5, 287–321.
Chistiakov, D. A., Hellemans, B., and Volckaert, F. A. M. (2006). Microsatellites and their genomic distribution, evolution, function and applications: a review with special reference to fish genetics. Aquaculture 255, 1–29. doi: 10.1016/j.aquaculture.2005.11.031
Crooijmans, R. P. M. A., Poel, J. J. V. D., Groenen, M. A. M., Bierbooms, V. A. F., and Komen, J. (1997). Microsatellite markers in common carp (Cyprinus carpio L.). Anim. Genet. 28, 129–134.
Cui, R., Schumer, M., Kruesi, K., Walter, R., Andolfatto, P., and Rosenthal, G. G. (2013). Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evol. Int. J. Org. Evol. 67, 2166–2179. doi: 10.1111/evo.12099
Fisher, S., and Halpern, M. E. (1999). Patterning the zebrafish axial skeleton requires early chordin function. Nat. Genet. 23, 442–446. doi: 10.1038/70557
Gao, Y., Wang, S., Luo, J., Murphy, R. W., Du, R., Wu, S., et al. (2012). Quaternary palaeoenvironmental oscillations drove the evolution of the Eurasian Carassius auratus complex (Cypriniformes, Cyprinidae). J. Biogeogr. 39, 2264–2278. doi: 10.1111/j.1365-2699.2012.02755.x
Geng, B., Sun, X. W., Liang, L. Q., Ouyang, H. S., and Tong, J. G. (2006). Microsatellite analysis of genetic diversity of Aristichthys nobilis in China. Yi Chuan Hereditas 28, 683–688.
James, J. K., and Abbott, R. J. (2005). Recent, allopatric, homoploid hybrid speciation: the origin of Senecio squalidus (Asteraceae) in the British Isles from a hybrid zone on Mount Etna. Sicily. Evol. Int. J. Org. Evol. 59, 2533–2547. doi: 10.1554/05-306.1
Kardong, K. V. (2011). Vertebrates: Comparative Anatomy, Function, Evolution, 6th Edn. New York: McGraw-Hill, 816.
Komiyama, T., Kobayashi, H., Tateno, Y., Inoko, H., Gojobori, T., and Ikeo, K. (2009). An evolutionary origin and selection process of goldfish. Gene 430, 5–11. doi: 10.1016/j.gene.2008.10.019
Liu, K., and Muse, S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis [J]. Bioinformatics 21, 2128–2129. doi: 10.1093/bioinformatics/bti282
Mallet, J. (2008). Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 363, 2971–2986. doi: 10.1098/rstb.2008.0081
Meier, J. I., Marques, D. A., Mwaiko, S., Wagner, C. E., Excoffier, L., and Seehausen, O. (2017). Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8:14363.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Rieseberg, L. H., Archer, M. A., and Wayne, R. K. (1999). Transgressive segregation, adaptation and speciation. Heredity 83(Pt 4), 363–372. doi: 10.1038/sj.hdy.6886170
Roy, M. S., Geffen, E., Smith, D., Ostrander, E. A., and Wayne, R. K. (1994). Patterns of differentiation and hybridization in North American wolflike canids, revealed by analysis of microsatellite loci. Mol. Biol. Evol. 11, 553–570. doi: 10.3398/064.071.0413
Schulte-Merker, S., Lee, K. J., McMahon, A. P., and Hammerschmidt, M. (1997). The zebrafish organizer requires chordino. Nature 387, 862–863. doi: 10.1038/43092
Schumer, M., Cui, R., Powell, D. L., Rosenthal, G. G., and Andolfatto, P. (2016). Ancient hybridization and genomic stabilization in a swordtail fish. Mol. Ecol. 25, 2661–2679. doi: 10.1111/mec.13602
Slatkin, M., and Lande, R. (1994). Segregation variance after hybridization of isolated populations. Genet. Res. 64, 51–56. doi: 10.1017/s0016672300032547
Smartt, J. (2001). Goldfish Varieties and Genetics: Handbook for Breeders. Oxford: Blackwell Science, 216.
Smith, P. J., and McVeagh, S. M. (2005). Genetic Analyses of Carp, Goldfish, and Carp-Goldfish Hybrids in New Zealand. Wellington: Science & technical Publishing Department of Conservation, 5–20.
Takashima, S., Shimada, A., Kobayashi, D., Yokoi, H., Narita, T., Jindo, T., et al. (2007). Phenotypic analysis of a novel chordin mutant in medaka. Dev. Dyn. 236, 2298–2310.
Tesson, S. V., Legrand, C., van Oosterhout, C., Montresor, M., Kooistra, W. H., and Procaccini, G. (2013). Mendelian inheritance pattern and high mutation rates of microsatellite alleles in the diatom Pseudo-nitzschia multistriata. Protist 164, 89–100. doi: 10.1016/j.protis.2012.07.001
Wang, J., Liu, S., Xiao, J., Tao, M., Zhang, C., Luo, K., et al. (2014). Evidence for the evolutionary origin of goldfish derived from the distant crossing of red crucian carp x common carp. BMC Genet. 15:33. doi: 10.1186/1471-2156-15-33
Wang, S. Y., Luo, J., Murphy, R. W., Wu, S. F., Zhu, C. L., Gao, Y., et al. (2013). Origin of Chinese goldfish and sequential loss of genetic diversity accompanies new breeds. PLoS One 8:e59571. doi: 10.1371/journal.pone.0059571
Wheeler, A. (2000). Status of the crucian carp, Carassius carassius (L.), in the UK. Fish. Manag. Ecol. 7, 315–322. doi: 10.1046/j.1365-2400.2000.00205.x
Keywords: hybridization, goldfish, genetic variation, speciation, evolution
Citation: Wang J, He W, Zeng J, Li L, Zhang G, Li T, Xiang C, Chai M and Liu S (2020) Genetic Variation in an Experimental Goldfish Derived From Hybridization. Front. Genet. 11:595959. doi: 10.3389/fgene.2020.595959
Received: 18 August 2020; Accepted: 25 November 2020;
Published: 15 December 2020.
Edited by:
Guo-Bo Chen, Zhejiang Provincial People’s Hospital, ChinaReviewed by:
Jisen Zhang, Fujian Agriculture and Forestry University, ChinaYuan Lu, Texas State University, United States
Copyright © 2020 Wang, He, Zeng, Li, Zhang, Li, Xiang, Chai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaojun Liu, bHNqQGh1bm51LmVkdS5jbg==
†These authors have contributed equally to this work
 Jing Wang
Jing Wang Weiguo He
Weiguo He Jinfeng Zeng1
Jinfeng Zeng1 Shaojun Liu
Shaojun Liu