
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 03 December 2020
Sec. Evolutionary and Population Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.593577
This article is part of the Research Topic Comparative Genomics and Functional Genomics Analyses in Plants View all 37 articles
Class III peroxidases (PRXs) are plant-specific enzymes and play important roles in plant growth, development and stress response. In this study, a total of 102 non-redundant PRX gene members (StPRXs) were identified in potato (Solanum tuberosum L.). They were divided into 9 subfamilies based on phylogenetic analysis. The members of each subfamily were found to contain similar organizations of the exon/intron structures and protein motifs. The StPRX genes were not equally distributed among chromosomes. There were 57 gene pairs of segmental duplication and 26 gene pairs of tandem duplication. Expression pattern analysis based on the RNA-seq data of potato from public databases indicated that StPRX genes were expressed differently in various tissues and responded specifically to heat, salt and drought stresses. Most of the StPRX genes were expressed at significantly higher levels in root than in other tissues. In addition, real-time quantitative PCR (qRT-PCR) analysis for 7 selected StPRX genes indicated that these genes displayed various expression levels under abiotic stresses. Our results provide valuable information for better understanding the evolution of StPRX gene family in potato and lay the vital foundation for further exploration of PRX gene function in plants.
As a large family of isozymes, peroxidases (POD) play important roles in the growth, development and defense processes in plants (Hiraga et al., 2001). Peroxidases are divided into two major groups, hemoglobin peroxidases and non-hemoglobin peroxidases, according to their protein structures (Hiraga et al., 2000). Exception of animal peroxidases, the hemoglobin peroxidases have been further divided into three classes based on their sequences and catalytic properties, namely, I, II and III peroxidases. Class I peroxidases are widely distributed in most living organisms other than animals, and play an important role in removing excess H2O2 to prevent cell damage (Erman and Vitello, 2002; Shigeoka et al., 2002). Class II peroxidases only present in fungi and are mainly involved in lignin degradation (Piontek et al., 2001). Class III peroxidases (PRX, EC 1.11.1.7) exist in various plants as a multi-genic family (Tognolli et al., 2002; Duroux and Welinder, 2003; Passardi et al., 2004a). The PRX protein contains highly conserved amino acid residues, including a single peptide chain and the protoporphyrin IX domain (Welinder, 1993). Most plant PRXs fuse with carbohydrate side chains to form glycosylated proteins. This glycosylation prevents the protein being degraded by protease and maintains the enzyme stability (Zheng and Van Huystee, 1991). In addition, two histidine residues interact with a heme group and eight cysteine residues, forming disulfide bridges; the distal histidine is essential for catalytic activity (Passardi et al., 2004a). The functions of PRXs have been illustrated in several studies as important proteins for a wide range of physiological processes of plants, such as growth hormone metabolism (Gazaryan et al., 1996), formation of lignin and liposites, crosslinking of cell walls (Barcelo and Pomar, 2001; Passardi et al., 2004b), cell growth and elongation, and various defense processes against biotic and abiotic stresses (Schopfer et al., 2002; Liszkay et al., 2004; Bindschedler et al., 2006). For example, Arabidopsis peroxidases AtPrx33 and AtPrx34 are associated with root elongation (Passardi et al., 2006), and AtPrx72 has an important role in lignification (Herrero et al., 2013). Cotton GhPOX1 is involved in cotton fiber elongation by means of maintaining high levels of reactive oxygen species production (Mei et al., 2009). The expression of several ZmPRX is altered significantly in response to abiotic stress based on microarray analysis in maize, suggesting that these genes play important role in resistance to abiotic stress (Wang et al., 2015). It was also reported that either present or lack of specific peroxidase isoforms appears to be correlated to a particular cellular process or participating a particular protein localization (Loukili et al., 1999; Allison and Schultz, 2004). Genome-wide analysis of this large multigenic family will be greatly helpful to understand its physiological roles and characteristics. In recent years, the PRX family has been widely studied in many species, such as Arabidopsis thaliana (Tognolli et al., 2002), rice (Passardi et al., 2004a), and maize (Wang et al., 2015).
Potato (Solanum tuberosum) is one of the most important food crops in the world. Potato tubers are rich in nutrients and are valuable processing raw materials for food industry (Pang, 2019). However, potato contains rich phenolic substances, which are the substrates of peroxidase reaction and lead to an enzymatic browning reaction frequently caused by POD (peroxidase) in the storage and potato processing (Zhu and Hu, 2013), affecting the quality of products (Zhou et al., 2010; Zhu, 2017). In addition, potato plants are often subjected to various types of abiotic and biotic stresses during growth and development. Although PRX gene family members play important roles in the plant growth and development, their functions in potato are poorly deciphered. Thoroughly analyzing the PRX gene family in potato is a primary step to understand its physiological roles and characteristics.
In this study, a systematic investigation of the PRX gene family in potato, including PRX gene structure, chromosomal localization, gene duplication and phylogenetic relationship, was performed using the sequences from the genome database. Moreover, the tissue-specific expression profile and expression patterns under abiotic stresses such as drought, heat and salt treatments were also investigated. The objectives of this study were to identify and assess the sequence structures of the potato PRX gene family and analyze the evolutionary relationship of PRX gene family in plants. The results of this study provide a solid foundation for the next phase functional investigation of PRX genes in potato.
The protein sequences of the Arabidopsis thaliana PRX members (Tognolli et al., 2002) were downloaded from the Arabidopsis database1. These protein sequences of Arabidopsis were used as the queries to identify the PRX orthologs in potato using the BLASTP tool SpudDB2 and Phytozome v12.13. Proteins with more than 30% similarity to the query sequence and an E < E–10 were selected. The domains for PRX proteins were further confirmed using the Conserved Domain Database of NCBI4. The sequences possessing PRX conserved domain were selected as the final candidates of PRX genes (StPRXs) and were renamed according to their physical position in the potato genome. The information of these genes was obtained from Phytozome5, including gene IDs, physical position, gene sequence and protein sequence. The parameters for the predicted StPRX proteins were calculated using online ExPASy tools (Gasteiger et al., 2003)6.
The StPRX gene structures were identified using the Gene Structure Display Server (GSDS7; Guo et al., 2007). The conserved motifs were identified using the MEME software (version 5.0.38; Bailey et al., 2009). Parameters were set as 20 motifs with the optimum motif width of 50–300 residues. The conserved motifs were then further annotated with the CDD program9 (Marchler et al., 2017).
The protein sequences of StPRXs were aligned using the multiple sequence alignment tool ClustalX (Thompson et al., 1997). The phylogenetic tree of PRX family proteins was generated using the MEGA-X maximum-likelihood model (Kumar et al., 2016) with 1000 bootstrap replicates. Orthologs identification method is based on a report (Blanc, 2004). To identify putative orthologs between two different species, each sequence from potato was searched against all sequences from maize and Arabidopsis using the BLASTN tool (Altschul et al., 1997). For each query sequence, the best hit among those that met the criteria of alignment ≥ 300 bp and similarity ≥ 40% was considered as the ortholog.
Information of the chromosomal location image of StPRX genes was retrieved by the MapInspect tool10. To assess gene duplication, the parameters for the proportion of overlap and the similarity between the two sequences were set to be > 70% (Gu et al., 2002; Yang et al., 2008). Two nearby duplicated genes were defined as tandem duplicated genes when the physical space between them was less than 100 kb and contained less than three intervening genes (Wang et al., 2010), while any other two duplicated genes that did not meet the condition of tandem duplicated genes, including those located on the same chromosome or different chromosomes, were all defined as segmental duplicated genes.
The FPKM (fragments per kilobase per million) values of StRPX in various tissues and treatments (salt, drought and heat) and their control (CK) generated by RNA-seq (DM_v4.03) were extracted from the Potato Genome Database (see footnote). The expression profile of StRPX genes was generated using the R package11 of the heatmap function (Warnes et al., 2016).
T virus-free plantlets (S. tuberosum L. autotetraploid cultivar Zhongshu 3) were generated by in vitro nodal cutting method. Potato shots placed on full MS solid medium were cultured in a growth chamber under the condition of 22°C and 16 h light/8 h dark photoperiod for 1 month. The plantlets were transplanted into a tray with a half-strength modified Hoagland solution (Hammer et al., 1978) for 6 days, and then were exposed to the abiotic stress conditions, including heat (35°C), drought (260 mM mannitol) and salt (150 mM NaCl) treatments. Untreated plantlets were used as control (CK). The treated and control plantlets were collected 6 h after treatment and then stored at −80°C before RNA extraction.
The total RNA of the plantlets was extracted using TRIzol reagent (Invitrogen)12 according to the manufacturer’s instructions. The cDNA samples were then assessed by qRT-PCR using SYBR Premix Ex Taq (Takara). Actin was used as an internal control gene. Three biological replicates (each containing 6 plants) and three technical replicates were measured for each treatment. The relative expression level of a gene was calculated according to the 2–ΔΔCt method (Livak and Schmittgen, 2001). The primers used for qRT-PCR analysis are listed in Supplementary Table S1.
Using 73 Arabidopsis PRX sequences as queries (Tognolli et al., 2002) and validating the candidate sequences by conservative domain analysis based on CDD, a total of 102 PRX genes (StPRXs) were identified from potato genome and were renamed from StPRX1 to StPRX102 based on their physical position on chromosomes (Table 1). However, the location of StPRX1 on the chromosome could not yet be defined because it was located in the unmapped scaffold. The protein lengths of the 102 StPRX genes varied from 152 (StPRX29) to 592 (StPRX60) amino acids, with an average of 310.8 amino acids. The molecular weights ranged from 16866.8 Da (StPRX29) to 63735.06 Da (StPRX60). The theoretical isoelectric points (pI) of these StPRX genes varied from 4.43 (StPRX20) to 10.02 (StPRX81). The detailed information for StPRX genes was listed in Table 1.
To reveal the evolutionary relationship of the PRX gene family, an unrooted phylogenetic tree (Figure 1A) was obtained basing the MEGA-X maximum-likelihood model. The 102 StPRXs were classified into 9 subfamily (I-IX) with the bootstrap values (≥50%) on the phylogenetic tree. However, 2 StPRX genes (StPRX 8 and StPRX22) could not be assigned to any of the 9 subfamilies due to the low bootstrap values (<50%). Among these 9 groups, subfamily I possessed the largest clade, which contained 35 StPRX genes, followed by group VIII, which had 22 PRX members. These two subfamilies accounted for 55.88% of the total StPRXs. In contrast, groups III and IV only had two or three StPRX genes.
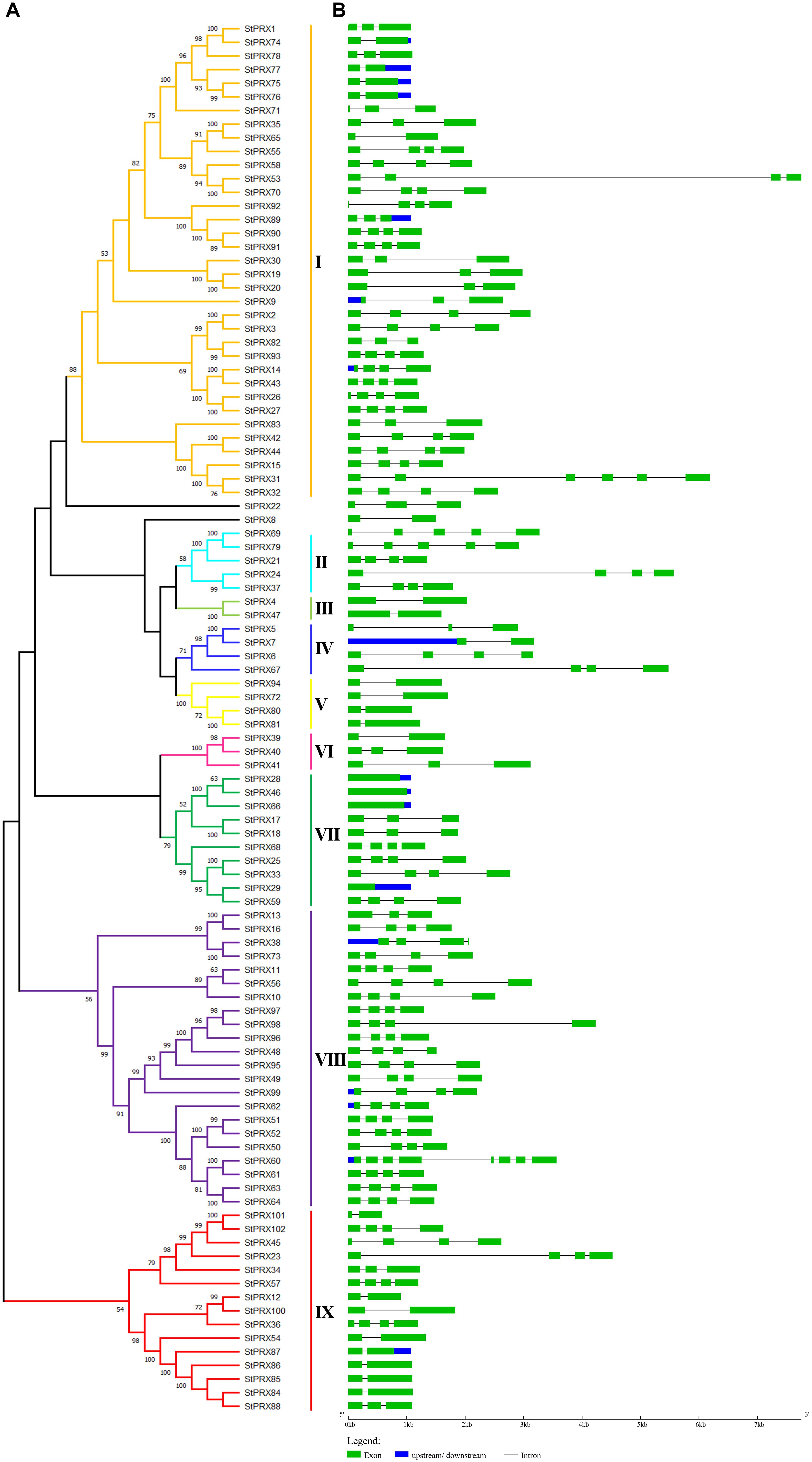
Figure 1. Phylogenetic tree and gene structure of StPRX genes. (A) The maximum-likelihood phylogenetic tree generated by MEGA X with bootstrapping analysis (1,000 replicates). Each subfamily is distinguished by different colors. (B) The exon-intron structure of StPRX genes generated by the online software GSDS. The horizontal black lines and the green boxes represent introns and exons, respectively. The blue boxes represent upstream or downstream sequences. The sizes of exons and introns can be estimated according to the scale at bottom.
To further explore the evolutionary process of the PRX family in potato, the 102 PRX protein sequences of potato were aligned with 73 PRX proteins of Arabidopsis thaliana and 119 PRX proteins of maize (Tognolli et al., 2002; Wang et al., 2015). The phylogenetic tree was divided into 12 different groups (groups A–L; Figure 2). Among these groups, group G was the largest, which contained 92 PRX members, including 35 of potato, 30 of maize and 27 of Arabidopsis. Groups A, D and K also had a large number of genes, containing 40, 35, and 40 members, respectively. In contrast, group F was the smallest, which contained only 5 members, including 1 of potato, 1 of maize and 3 of Arabidopsis. Interestingly, although group D was large, it had no members from Arabidopsis. Moreover, group I and L only contained 5 and 8 members from maize, respectively. A few groups were supported by low bootstrap values, which might be due to the relative less informative character positions beside the conserved PRX domains. This scenario has been also found in the analysis of other gene families (Li et al., 2006; Wang et al., 2018). In addition, a total of 82 ortholog pairs were identified between potato and Arabidopsis (St-At; Supplementary Table S4). However, only four ortholog pairs were retrieved between potato and maize (St-Zm; Supplementary Table S5). The orthologs between potato and Arabidopsis were much greater than that between potato and maize, probably because of the closer evolutionary distance between these eudicot species.
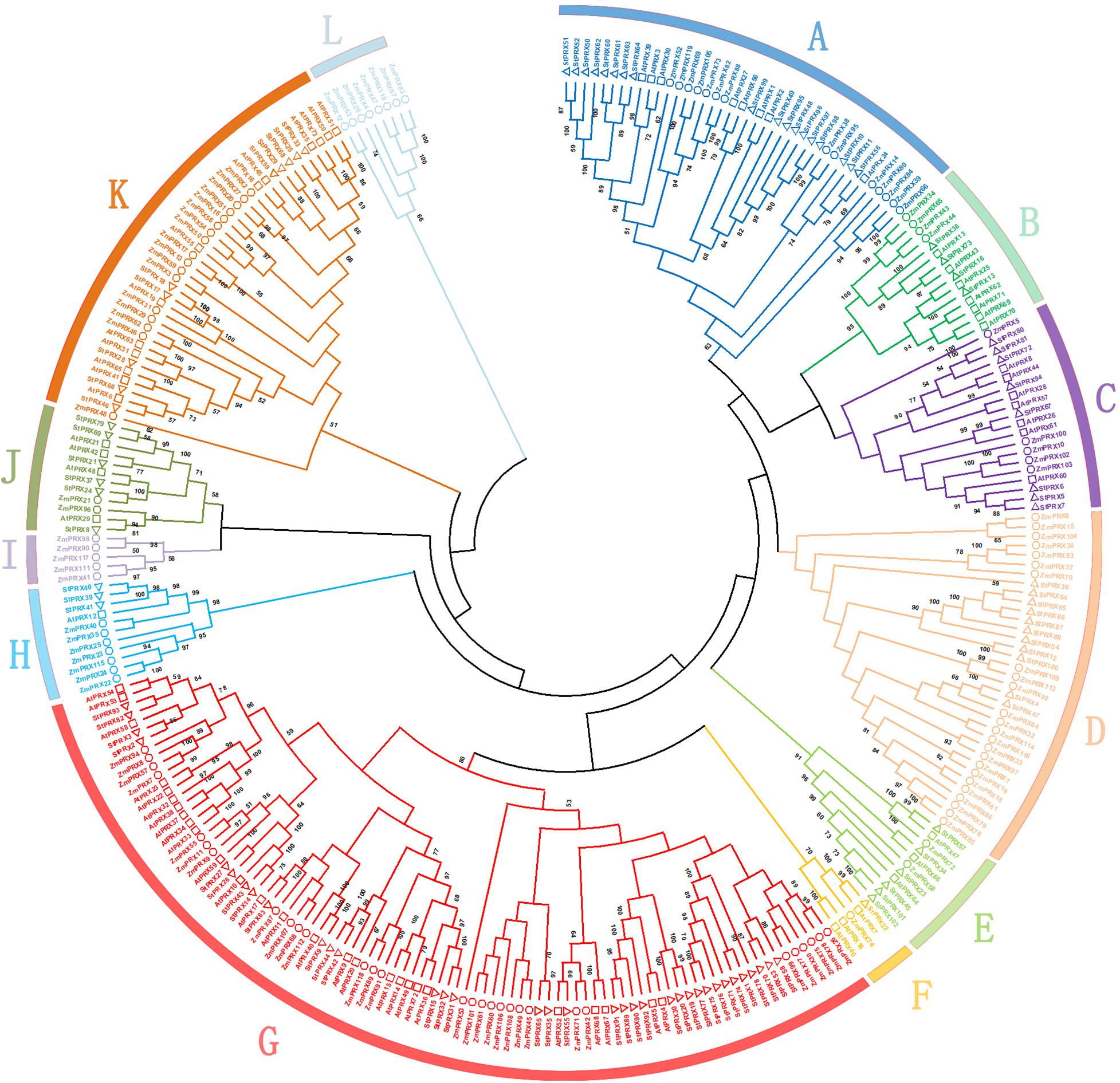
Figure 2. Phylogenetic tree of potato, maize and Arabidopsis PRXs. Each PRX subfamily is shown by a specific color. The phylogenetic tree was constructed by MEGA X with 1000 bootstrap replicates. The circles, squares, and triangles represent maize, Arabidopsis and potato PRX proteins, respectively.
Structure analysis of StPRX genes showed that the number of introns varied from 0 to 7 (Figure 1B). Most of them had 1–3 introns, with StPRX60 containing the maximum (7) while four genes lacked introns (StPRX28, StPRX29, StPRX46, and StPRX66) (Figure 1B). Genes with similar exon/intron structure were grouped together, but structural variation was also found among these StPRX genes (Figure 1).
To investigate the diversity of motif components among StPRXs, the motif distribution in 102 StPRX proteins was investigated using the online tool MEME program. A total of 18 conserved motifs were identified (Figure 3 and Supplementary Table S2). The majority of StPRX proteins contained two to three conserved motifs. The StPRX proteins on the same branch had similar conserved motif composition and sorting order, suggesting that StPRX proteins in the same branch might share similar function. Using the CDD tool, a total of 11 motifs (motif 1/2/3/4/5/6/7/9/10/12/15) were functionally annotated for the components of the conserved PRX domain (Figure 3 and Supplementary Table S2). All the members of the potato PRX family contained at least one motif belonging to the typical domains of PRX family. In addition, some motifs appeared to be unknown in function. For example, the functions of three motifs (motif 11/14/18) in subgroup III and of four motifs (motif 8/13/16/17) in subgroups I, III, V, and VIII were yet to be determined.
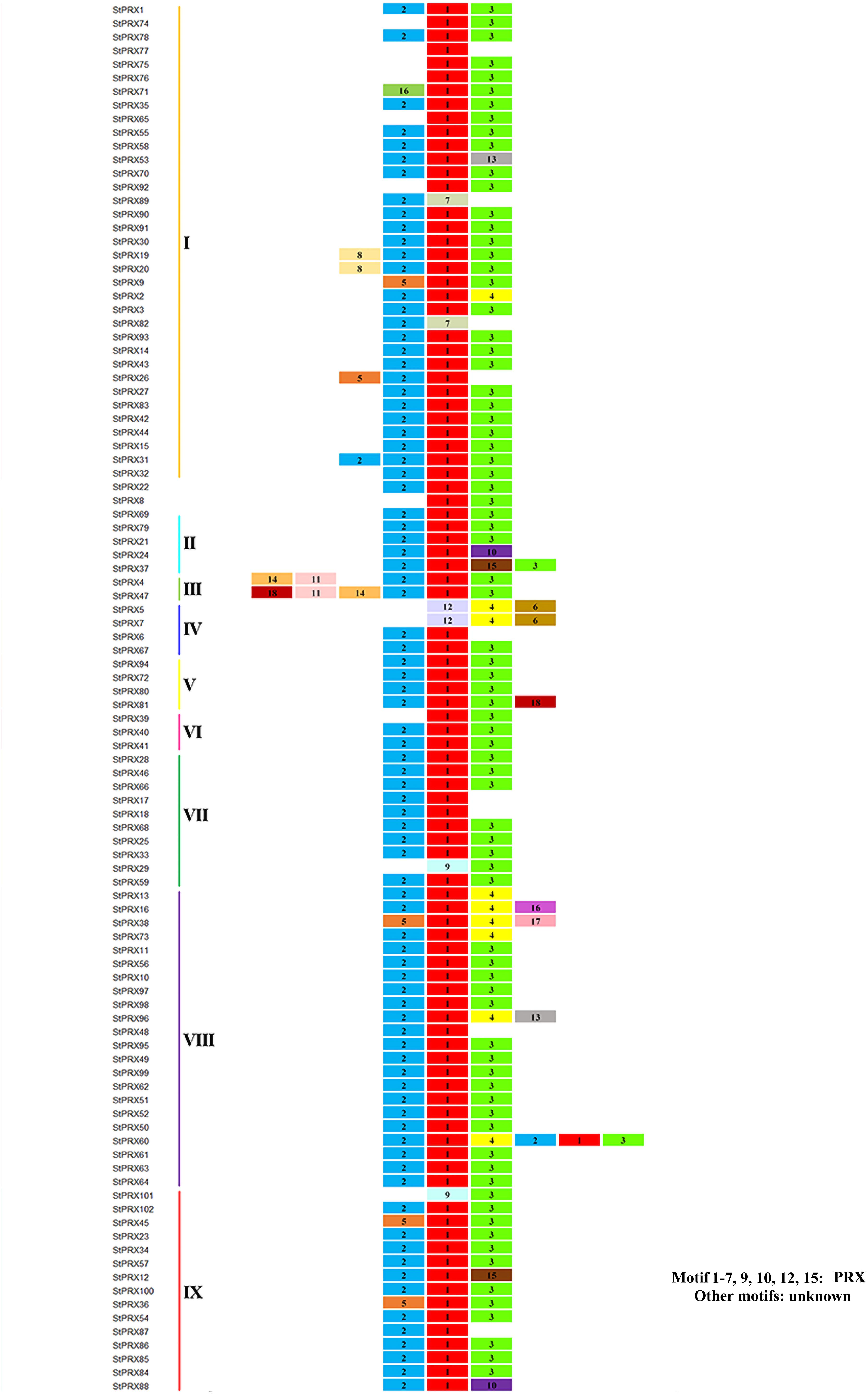
Figure 3. Conserved motifs of PRX proteins in potato. The protein list on the left is the same as that in Figure 1. The genes of different subfamilies are highlighted with different colors. Different motifs are exhibited with different colored boxes and numbers (1–18). The detailed sequences of motifs are listed in Supplementary Table S2.
To reveal the genome organization and distribution of StPRX on different chromosomes in potato, a graph of chromosomes was constructed using the MapInspect tool. A total of 101 out of the 102 StPRX genes were located on the 12 potato chromosomes (Figure 4). Among them, the largest number of StPRX genes (15) was located on chromosome 2, followed by chromosomes 1 (13), and chromosomes 11 (11) and 10 (10). In contrast, only a few StPRX genes were located on chromosomes 8 (3), 6 (4), and 9 (4). In addition, some chromosomes showed a dense cluster of StPRXs, such as near the telomeric region of chromosomes 2, 4, 5, 7, 10, 11, and 12. Gene duplication events, including segmental duplication and tandem duplication, are important for the expansion of the gene family during the process of the evolution (Cannon et al., 2004). In this study, a total of 83 StPRX gene pairs were identified from the phylogenetic and comparative analysis (Figure 4), among which 57 pairs were found to be involved in the segmental duplication events, and 26 pairs were confirmed to be tandem duplicated genes (Figure 4 and Supplementary Table S3). The number of segmental duplication gene pairs was twice as many as that of the tandem duplicated, and most of the tandem duplicated gene pairs were densely distributed at the end of chromosomes 5, 7, 10, 11, and 12.
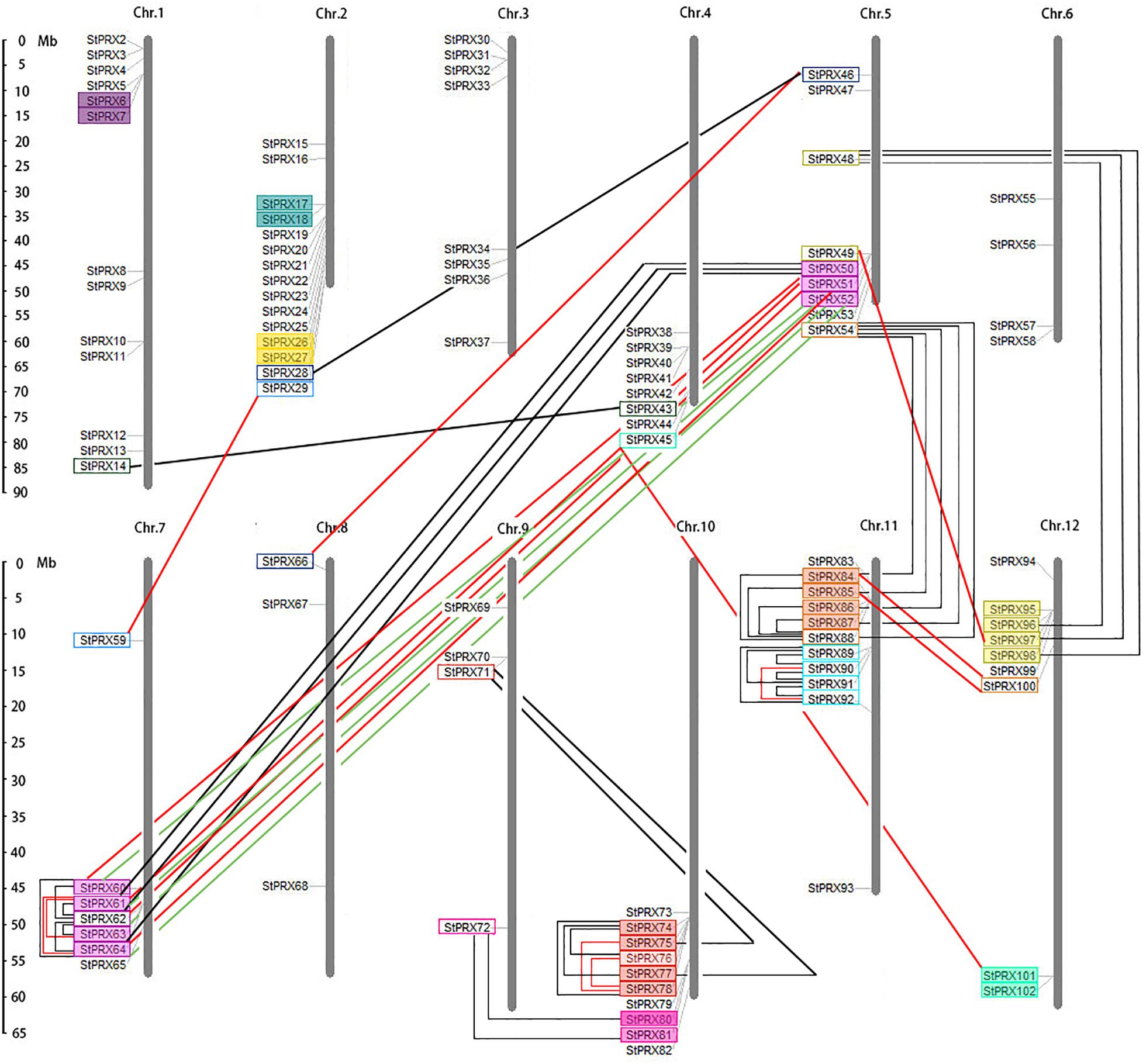
Figure 4. Chromosomal locations of potato PRX genes. The scale on the left presents the length of potato chromosomes (Mb). Tandem duplicated gene pairs are displayed with boxes of the same color. Segmented duplicated gene pairs are displayed with boxes connected by lines.
To further explore the expression patterns of the StPRX genes, the transcript data of major tissues was obtained from the public genome database, including root, shoot, petal, carpel, sepal, stamen, tuber, leaf, flower and petiole. A heatmap was generated based on the transcript data of 80 StPRX genes, and the other 22 genes were excluded from the heat map analysis due to the low expression level (FPKM < 0.5) or lack of expression in all tissues (Figure 5). As shown in Figure 5, some AtPRX genes exhibited distinct tissue-specific expression patterns, while others were active in the whole plant. The 80 StPRX genes were grouped into four groups (Figure 5). Six genes (StPRX41/19/28/25/40/21) were included in group IV, which had especially abundant expression level in all of the developmental stages, suggesting that these genes might play important basic roles in all development stages of plant. A total of 19 genes (StPRX51/13/20/30/29/55/39/62/23/2/3/34/9/69/83/68/35/57/33) were included in group III, which had high expression levels in most of the analyzed tissues. In contrast, 44 genes in group II exhibited low expression or no expression in the most of the tissues analyzed. However, most of them showed relatively high expression in root than in other tissues. This was also observed in the PRX family of maize (Wang et al., 2015) and Arabidopsis (Tognolli et al., 2002). Therefore, the expression patterns of StPRX genes may reflect the correlation with their functions.
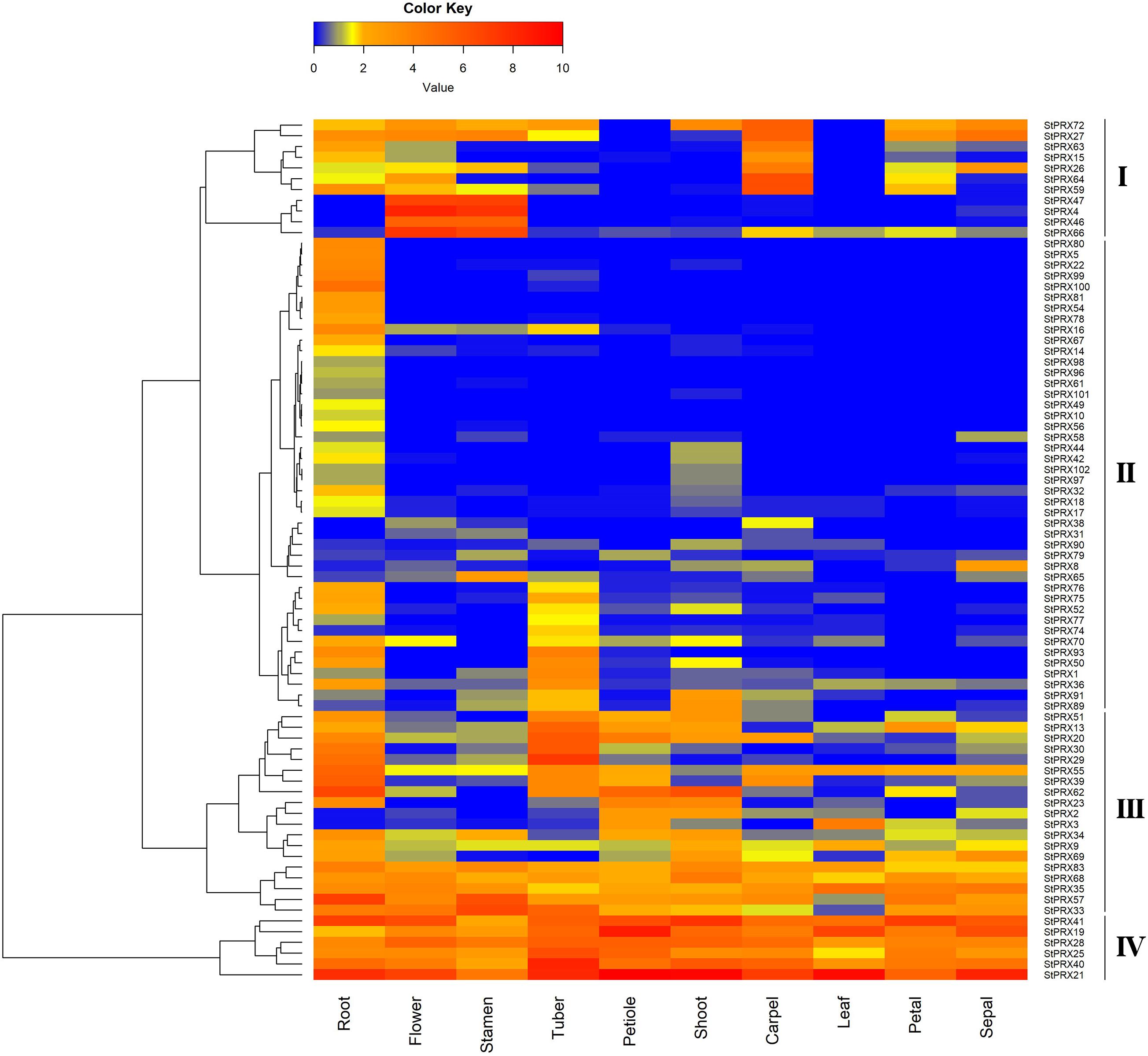
Figure 5. Expression profiles of StPRX genes in different tissues. FPKM values for StPRX genes were transformed by log2(FPKM+1). The heatmap was generated using the R package of the heatmap function.
To further investigate the response of the StPRX genes subjected to different stresses, including heat, salt and drought, the relative expression levels among the tissues were measured based on the expression FPKM values (stress/control) (Figure 6). The expression levels of different genes showed great variation under the various types of treatments. Under heat stress, most of the StPRX genes in groups III and IV were significant upregulated, such as StPRX93/33/39/74/31/100/36/22, whereas most of the StPRX genes in groups I and II exhibited significant downregulation, such as StPRX 28, -18 and -17. Most of the StPRX genes showed downregulation under the stress of drought, in contrast to the performance under heat stress, and some StPRX genes in groups II and III were extremely sensitive to the drought stress and exhibited significant downregulation, such as StPRX4/13/31/38/46/57/51/58/74/77/89/91. In terms of saline stress, the genes exhibited diverse responses, the genes in group IV were significantly upregulated, and the genes in group II showed significant downregulation, while the genes in groups I and III only showed slight changes, implying the functional dissimilation among the StPRX genes.
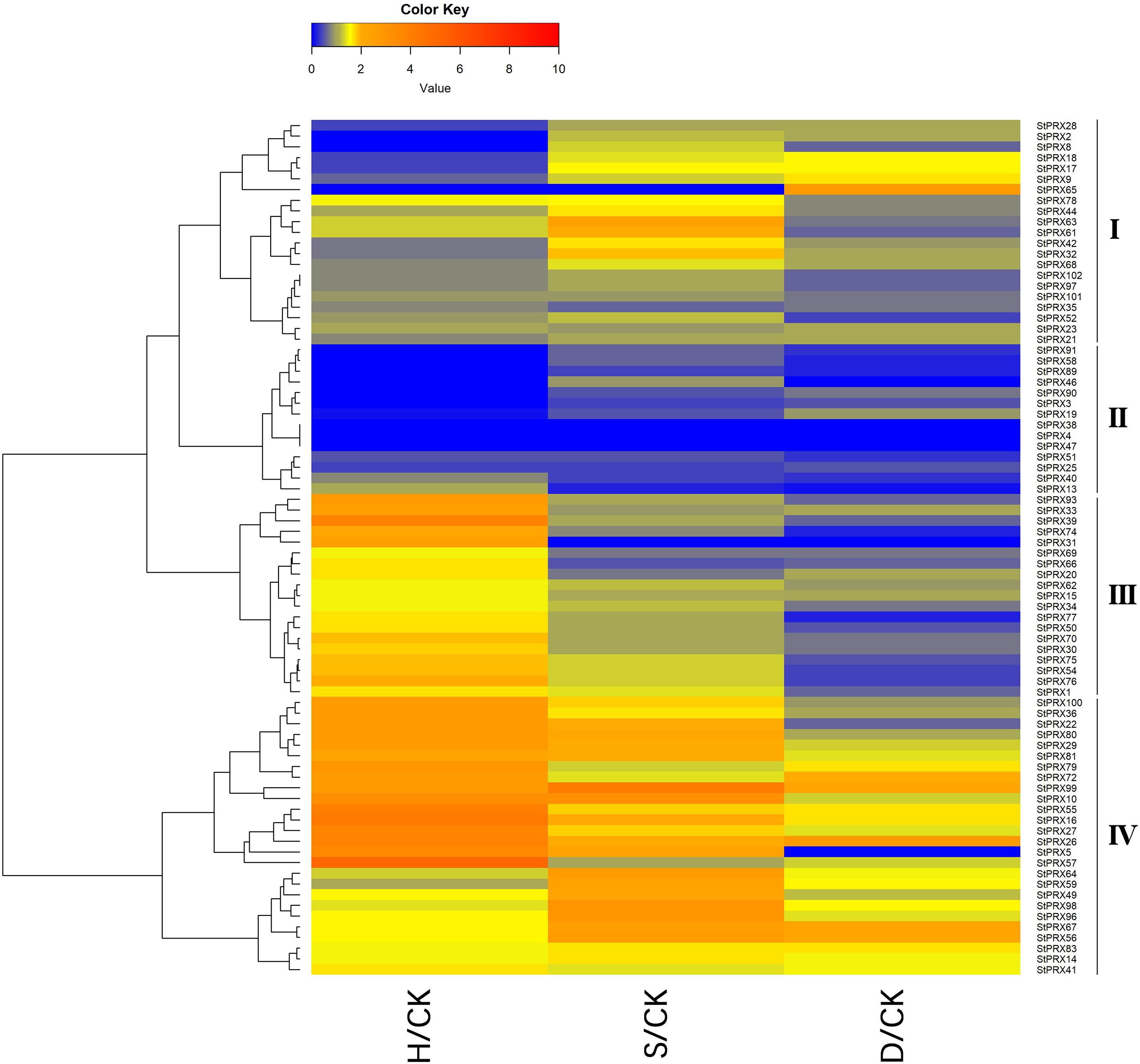
Figure 6. Expression changes of StPRX genes under heat (H), salt (S) and drought (D) stresses. The expression change is indicated by the ratio of FPKM value of the treatment to that of the control (CK). The heatmap was generated using the R package of the heatmap function.
Seven StPRX genes (StPRX19/28/33/35/40/41/57) with high expression levels in all organs were selected for further qRT-PCR analysis under different abiotic stresses. The 7 genes showed different levels of response to the three abiotic stress treatments (Figure 7). All of the selected genes were up-regulated under heat stress, and the expression change of StPRX57 was over twofold. For saline stress, most of the selected genes were up-regulated, and the upregulation of StPRX33 and StPRX57 was more than threefold. In response to drought stress, 5 genes (StPRX19/28/40/41/57) were upregulated, whereas StPRX33 and StPRX35 appeared to be slightly down-regulated. Notably, 2 genes (StPRX41 and StPRX57) were extremely sensitive to drought stress. Their expression levels increased 4 folds compared to that of control.
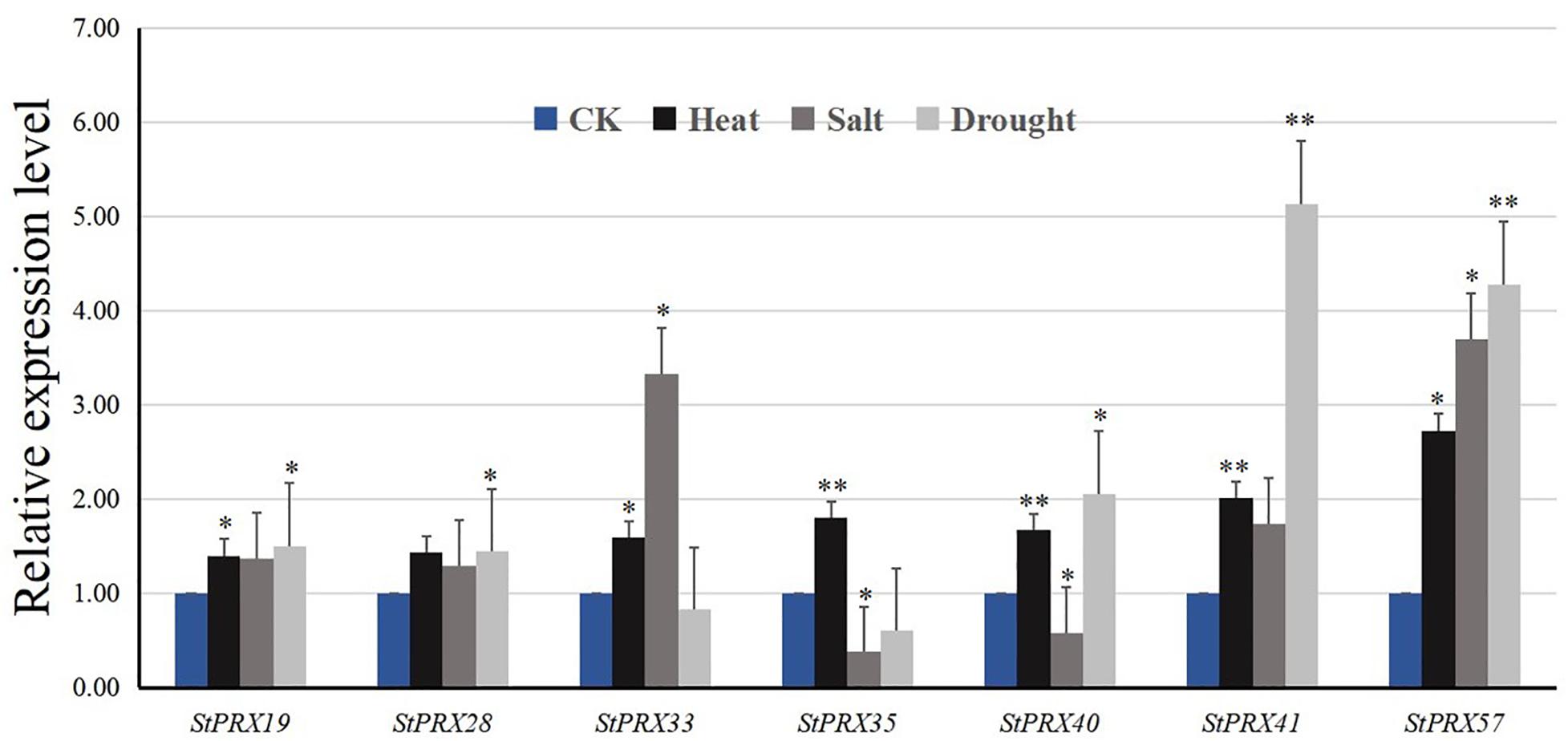
Figure 7. Relative expression levels of 7 StPRX genes in response to heat, salt and drought stresses after 6 h treatment compared with that of control (CK). Error bars are standard deviations of three biological replicates. *P < 0.05, **P < 0.01.
PRXs are plant-specific enzymes that have multiple functions in the growth and development of plants. It has been confirmed that PRX genes are widely involved in stress response in many species (Gray and Montgomery, 2003; Xue et al., 2008). To date, the comprehensive genome-wide analysis of the PRX gene family has been performed in many plant species, including Arabidopsis (Tognolli et al., 2002), rice (Passardi et al., 2004a), and maize (Wang et al., 2015). In this study, a total of 102 StPRX genes were identified. The number was more than that in Arabidopsis (containing 73 PRX genes), but slightly less than that of maize (119) and rice (138). According to the phylogenetic tree of the 288 PRX family members from Arabidopsis, maize and potato, we found that some groups only contained members from one or two species. In addition, orthologs between potato and Arabidopsis (82 pairs) were much more than those between potato and maize (4 pairs), suggesting that the PRX gene family underwent a specific expansion after the three species diverged each other in the evolutionary path of speciation, especially after the divergence of monocots and eudicots.
We have seen that the StPRX proteins showing similar domain architectures and motif constitutions were usually grouped in the same subfamily (Figure 3), and the structural constitutions of the StPRX genes in each group were basically in consistence with the result of phylogenetic analysis (Figure 1). Similar phenomenon have also been found in many other species, such as maize (Wang et al., 2015) and rice (Passardi et al., 2004a). As protein function is mostly determined by its domain structure, these results imply that the StPRX proteins with similar domain architectures and motif constitutions could probably perform similar functions.
It is known that the structural diversity of genes drives the evolution of multigene families. The intron/exon structure variation is a cause of gene diversity. Many studies have shown that introns are specifically inserted into and retained in the genome during evolution (Rogozin et al., 2003). The loss and insertion of new introns appear to be frequent events, which may result in diverse functional consequences in gene evolution. The duplication from an ancient gene formed by shuffling of small exons could be the reason that resulted in the genes with a relatively high number of introns (Gilbert et al., 1997). In maize, it is speculated that the intron variation among PRX genes might result from the depletion and duplication of single introns in the course of evolution (Wang et al., 2015). In this study, we found that the number of introns in StPRX genes was also quite variable (varying from 0 to 7), and the proteins in the same subfamily were not completely identical in terms of their intron/exon structure and motifs (Figures 1, 3), implying that exon shuffling might be a main pattern of StPRX evolution, which might be the main contributors to the functional diversity of the potato PRX family. Among the 102 StPRX genes, more than half of them consist of 3 introns and 4 exons. This 3-intron/4-exon model is also represented a significant proportion in Arabidopsis (Tognolli et al., 2002) and in rice (Passardi et al., 2004a), suggesting that it is an ancestral intronic model of PRX genes.
Gene duplication events, including segmental and tandem duplication are important for the expansion of the gene family during the process of the evolution (Cannon et al., 2004). Gene duplication events can theoretically produce two gene copies, and one or both copies can acquire the novel gene functions for adaptation under a smaller selective pressure of evolution (Van de Peer et al., 2009). Each paralog is specialized for a specific functional assignment (Zhang, 2003), which often leads to the expansion of gene family (Cannon et al., 2004). The segment duplication event refers to the duplication of large fragments of the genome, which may have derived from segmental, chromosomal or whole genome duplications with many losses and rearrangements (Zhang, 2003). Tandem duplication affects a limited number of genes (one or more neighboring genes); it often derives from unequal crossing-over (Achaz et al., 2000) and multiple episodes of unequal crossovers. In addition, the retrotransposition event of cDNA also contributes to the expansion of gene family, which is characterized by the loss of all introns and related regulatory sequences and by a random insertion within the genome. In our study, a total of 83 duplicated gene pairs were identified in potato PRX gene family, including 57 segmental duplication gene pairs and 26 tandem duplicated genes pairs (Supplementary Table S3), which were much more than those in maize (28 duplicated gene pairs; Wang et al., 2015) and rice (Passardi et al., 2004a). The segmental duplication gene pairs were twice as many as the tandem duplicated gene pairs, indicating that segmental duplication might play the dominant role in the expansion of the potato PRX family. Most of the tandem duplicated gene pairs were densely distributed in telomeric regions of chromosomes (such as on chromosomes 5 and 7), and many tandem duplicated genes shared high similarity with the same segmental duplication genes, implying that most of the tandem duplicated genes might appear after the segmental duplication events. Notably, two gene pairs (StPRX28/46, StPRX46/66) met the criteria of segmental duplication gene pairs but had no introns, suggesting that they might likely be generated by retrotransposition. Strictly speaking, therefore, they might not be segmental duplication gene pairs.
Gene expression pattern is an important aspect related to gene function. In this study, among the 102 StPRX genes, except for 22 with weak or without expression in all of the tissues examined, the rest all exhibited distinct patterns of tissue-specific expression (Figure 5) and response to stress (Figure 6), indicating the functional dissimilation of StPRX proteins. This is consistent with the results of phylogenetic and protein motif analyses. Several genes in group IV were expressed in all organs (Figure 5), suggesting that they might play basic roles for the plant. Notably, the largest number of StPRX genes with high expression levels was found in root (Figure 5). Similar observations were also reported in maize (Wang et al., 2015), Arabidopsis (Tognolli et al., 2002), and rice (Passardi et al., 2004a), suggesting that the PRX family might be critical for root function in plants. In maize, there are many cell wall or membrane-bound PRXs in root (Mika et al., 2008; Šukaloviæ et al., 2015); several ZmPRX genes from roots are regulated by methyl jasmonate, salicylic acid and pathogen elicitors (Mika et al., 2010); and some genes (ZmPRX26/42/71/75/78) highly expressed in root show significant responses to H2O2, SA, NaCl, and PEG treatments (Wang et al., 2015). In Arabidopsis, two AtPRX genes (AtPrx33/34) are associated with root elongation (Passardi et al., 2006). Interestingly, the five ZmPRX genes (ZmPRX26/42/71/75/78) and two AtPRX genes (AtPrx33/34) were all clustered in grouped G in this study (Figure 2). Most StPRX genes in group G (Figure 2) were included in the subfamily I (Figures 1, 3) with similar gene structure and motif components. The results imply that the StPRX genes clustered in group G might also function in root similar to their counterparts in maize and Arabidopsis. Therefore, the results of our study may provide a basis for the functional exploration of the potato PRX gene family members.
To analyze the trend of the gene expression derived from qRT-PCR (Figure 7) and the FPKM values, we compared the results from these two different platforms (Figure 6). Overall, similar propensity of gene expression was found between the two different approaches. However, the results of qRT-PCR did not totally agree with the pattern of the gene expression from RNA-seq data. There could be several reasons for the discrepancy. First, the genotypes of potato varieties used in the two experiments were different. A doubled monoploid potato variety (DM) was used in RNA-seq (Xu et al., 2011), whereas an autotetraploid cultivar Zhongshu 3 was used for qRT-PCR analysis. Second, the experimental treatments were different. The plantlets for qRT-PCR were grown under a photoperiod of 16 h light/8 h dark environment, while the materials for RNA-seq were grown in the dark (Xu et al., 2011).
In this study, a genome-wide investigation and comprehensive analysis of the PRX gene family in potato was conducted. The structural diversity of StPRXs may reflect their functional diversity. The analysis of expression patterns of StPRX genes showed that these genes were expressed distinctly in different tissues of potato, and some might be linked to stress responses. It is important to thoroughly investigate the biological functions of StPRX genes, especially the roles in the resistance to abiotic stresses. Our results provide the vital information for the exploration of the functional aspect of the gene family.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
XY and JYa performed the experiments and analyzed the data. WL collected plant materials and performed the experiments. MQ participated in handling figures and tables. XX designed this research and wrote the manuscript. WW and JYu helped to draft the manuscript. All authors read and approved the manuscript.
This study was supported by the National Natural Science Foundation of China (No. 31501085) and the Fujian Agriculture and Forestry University innovation foundation (No. CXZX2019052G).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.593577/full#supplementary-material
Achaz, G., Coissac, E., Viari, A., and Netter, P. (2000). Analysis of intrachromosomal repeats in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol. Biol. Evol. 17, 1268–1275. doi: 10.1093/oxfordjournals.molbev.a026410
Allison, S. D., and Schultz, J. C. (2004). Differential activity of peroxidase isozymes in response to wounding, gypsy moth, and plant hormones in northern red oak (Quercus rubra L.). J. Chem. Ecol. 30, 1363–1379. doi: 10.1023/b:joec.0000037745.66972.3e
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., and Miller, W. (1997). Gapped BLAST and PSI- BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Bailey, T. L., Boden, M., Buske, F. A., Frith, A., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208.
Barcelo, A. R., and Pomar, F. (2001). Oxidation of cinnamyl alcohols and aldehydes by a basic peroxidase from lignifying Zinnia elegans hypocotyls. Phytochemistry 57, 1105–1113. doi: 10.1016/s0031-9422(01)00050-4
Bindschedler, L. V., Julia, D., Julie, M., Blee, K. A., Julia, P., Dewi, R. D., et al. (2006). Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. doi: 10.1111/j.1365-313x.2006.02837.x
Blanc, K. H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. doi: 10.1105/tpc.021345
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Duroux, L., and Welinder, K. G. (2003). The peroxidase gene family in plants: a phylogenetic overview. J. Mol. Evol. 57, 397–407. doi: 10.1007/s00239-003-2489-3
Erman, J. E., and Vitello, L. B. (2002). Yeast cytochrome c peroxidase: mechanistic studies via protein engineering. Biochim. Biophys. Acta 1597, 193–220. doi: 10.1016/s0167-4838(02)00317-5
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gazaryan, I. G., Lagrimini, L. M., Ashby, G. A., and Thorneley, R. N. (1996). Mechanism of indole-3-acetic acid oxidation by plant peroxidases: anaerobic stopped-flow spectrophotometric studies on horseradish and tobacco peroxidases. Biochem. J. 313, 841–847. doi: 10.1042/bj3130841
Gilbert, W., de Souza, S. J., and Long, M. (1997). Origin of genes. Proc. Natl. Acad. Sci. U.S.A. 94, 7698–7703.
Gray, J. S., and Montgomery, R. (2003). Purification and characterization of a peroxidase from corn steep water. J. Agric. Food Chem. 51, 1592–1601. doi: 10.1021/jf025883n
Gu, Z. L., Cavalcanti, A., Chen, F. C., Bouman, P., and Li, W. H. (2002). Extent of gene duplication in the genomes of Dro-sophila. Nematode Yeast. Mol. Biol. Evol. 19, 256–262. doi: 10.1093/oxfordjournals.molbev.a004079
Guo, A. Y., Zhu, Q. H., and Chen, X. (2007). GSDS: a gene structure display server. Hereditas 29, 1023–1026. doi: 10.1360/yc-007-1023
Hammer, P. A., Tibbitts, T. W., Langhans, R. W., and Mcfarlane, J. C. (1978). Baseline growth studies of ‘Grand Rapids’ lettuce in controlled environments. J. Amer. Soc. Hor. Sci. 103, 649–655.
Herrero, J., Fernández-Pérez, F., Yebra, T., Novo-Uzal, E., Pomar, F., Pedreño, M. Á, et al. (2013). Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237, 1599–1612. doi: 10.1007/s00425-013-1865-5
Hiraga, S., Ichinose, C., Onogi, T., Niki, H., and Yamazoe, M. (2000). Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli. Genes Cells 5, 327–341. doi: 10.1046/j.1365-2443.2000.00334.x
Hiraga, S., Sasaki, K., Ito, H., Ohashi, Y., and Matsui, H. (2001). A large family of class III plant peroxidases. Plant Cell Physiol. 42, 462–468. doi: 10.1093/pcp/pce061
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, X. X., Duan, X., Jiang, H. X., Sun, Y. J., Tang, Y. P., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Liszkay, A., Vander, Z. E., and Schopfer, P. (2004). Production of reactive oxygen intermediates (O2-, H2O2, and (.)OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 136, 3114–3123. doi: 10.1104/pp.104.044784
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loukili, A., Limam, F., Ayadi, A., Boyer, N., and Ouelhazi, L. (1999). Purification and characterization of a neutral peroxidase induced by rubbing tomato internodes. Physiol. Plant. 105, 24–31. doi: 10.1034/j.1399-3054.1999.105105.x
Marchler, B. A., Bo, Y., Han, L., He, J., Lanczycki, C, J., Lu, S. N., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Mei, W., Qin, Y., and Song, W. (2009). Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. Genet. Genom. 36, 141–150. doi: 10.1016/s1673-8527(08)60101-0
Mika, A., Boenisch, M. J., Hopff, D., and Lüthje, S. (2010). Membrane-bound guaiacol peroxidases from maize (Zea mays L.) roots are regulated by methyl jasmonate, salicylic acid, and pathogen elicitors. J. Exp. Bot. 61, 831–841. doi: 10.1093/jxb/erp353
Mika, A., Buck, F., and Lüthje, S. (2008). Membrane-bound class III peroxidases: identification, biochemical properties and sequence analysis of isoenzymes purified from maize (Zea mays L.) roots. J. Proteome. 71, 412–424. doi: 10.1016/j.jprot.2008.06.006
Pang, W. Y. (2019). The significance and implementation of the potato main graining strategy. Grain Process. 44, 59–61.
Passardi, F., Longet, D., Penel, C., and Dunand, C. (2004a). The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65, 1879–1893. doi: 10.1016/j.phytochem.2004.06.023
Passardi, F., Penel, C., and Dunand, C. (2004b). Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9, 534–540. doi: 10.1016/j.tplants.2004.09.002
Passardi, F., Tognolli, M., and De Meyer, M. (2006). Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223, 965–974. doi: 10.1007/s00425-005-0153-4
Piontek, K., Smith, A. T., and Blodig, W. (2001). Lignin peroxidase structure and function. Biochem. Soc. Trans. 29, 111–116. doi: 10.1042/bst0290111
Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G., and Koonin, E. V. (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512–1517. doi: 10.1016/s0960-9822(03)00558-x
Schopfer, P., Liszkay, A., Bechtold, M., Frahry, G., and Wagner, A. (2002). Evidence that hydroxyl radicals mediate auxin-induced extension growth. Planta 214, 821–828. doi: 10.1007/s00425-001-0699-8
Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. doi: 10.1093/jexbot/53.372.1305
Šukaloviæ, V. H.-T., Vuletiæ, M., Markoviæ, K., Antiæ, T. C., and Vuèiniæ, Ž (2015). Comparative biochemical characterization of peroxidases (class III) tightly bound to the maize root cell walls and modulation of the enzyme properties as a result of covalent binding. Protoplasma 252, 335–343. doi: 10.1007/s00709-014-0684-2
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tognolli, M., Penel, C., Greppin, H., and Simon, P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288, 129–138. doi: 10.1016/s0378-1119(02)00465-1
Van de Peer, Y., Maere, S., and Meyer, A. (2009). The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732. doi: 10.1038/nrg2600
Wang, L. Q., Guo, K., Li, Y., Tu, Y. Y., Hu, H. Z., Wang, B. R., et al. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10:282. doi: 10.1186/1471-2229-10-282
Wang, R. Q., Zhao, P., Kong, N. N., Lu, R. Z., Pei, Y., Huang, C. X., et al. (2018). Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes 9:54. doi: 10.3390/genes9010054
Wang, Y., Wang, Q. Q., Zhao, Y., Han, G. M., and Zhu, S. W. (2015). Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 566, 95–108. doi: 10.1016/j.gene.2015.04.041
Warnes, G., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., et al. (2016). Various R Programming Tools for Plotting Data. Available online at: https://cran.r-project.org/
Welinder, K. G. (1993). “Plant peroxidases: structure–function relationships,” in Plant Peroxidases, eds C. Penel, T. Gaspar, and H. Greppin (Geneva: University of Geneva), 1–24. doi: 10.1385/1-59259-232-5:1
Xu, X., Pan, S., Cheng, S., Zhang, B., Mu, D., Ni, P., et al. (2011). Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Xue, Y. J., Tao, L., and Yang, Z. M. (2008). Aluminum-induced cell wall peroxidase activity and lignin synthesis are differentially regulated by jasmonate and nitric oxide. J. Agric. Food Chem. 56, 9676–9684. doi: 10.1021/jf802001v
Yang, S. H., Zhang, X. H., Yue, J. X., Tian, D. C., and Chen, J. Q. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics 280, 187–198. doi: 10.1007/s00438-008-0355-0
Zhang, J. Z. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/s0169-5347(03)00033-8
Zheng, X. H., and Van Huystee, R. B. (1991). Oxidation of tyrosine by peroxidase isozymes derived from peanut suspension culture medium and by isolated cell walls. Plant Cell Tissue Organ. Cult. 25, 35–43. doi: 10.1007/bf00033910
Zhou, X. J., Gao, Y. X., and Wang, Z. B. (2010). Study on the characteristics of “Black Beauty” potato peroxidase. Food Ferment. Ind. 36, 55–63.
Zhu, H. S. (2017). “Cloning and analysis of the family of common melon peroxidase (POD) gene,” in Proceedings of the 13th National Congress of the Chinese Horticultural Society and the 2017 Annual Conference of The Chinese Horticultural Society, Kunming.
Keywords: potato, class III peroxidase, phylogenetic analysis, expression pattern, abiotic stress
Citation: Yang X, Yuan J, Luo W, Qin M, Yang J, Wu W and Xie X (2020) Genome-Wide Identification and Expression Analysis of the Class III Peroxidase Gene Family in Potato (Solanum tuberosum L.). Front. Genet. 11:593577. doi: 10.3389/fgene.2020.593577
Received: 11 August 2020; Accepted: 09 November 2020;
Published: 03 December 2020.
Edited by:
Weike Duan, Huaiyin Institute of Technology, ChinaReviewed by:
Robert A. Haney, Ball State University, United StatesCopyright © 2020 Yang, Yuan, Luo, Qin, Yang, Wu and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofang Xie, eHhmMzE3QGZhZnUuZWR1LmNu; Weiren Wu, d3V3ckBmYWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.