- 1School of Agricultural Biotechnology, Punjab Agricultural University, Ludhiana, India
- 2Department of Plant Breeding and Genetics, Punjab Agricultural University, Ludhiana, India
Wheat (Triticum aestivum L.) is an important cereal crop globally as well as in India and yield improvement programs encounter a strong impediment from ever-evolving rust pathogens. Hence, durable rust resistance is always a priority trait for wheat breeders globally. Grain weight, represented as thousand grain weight (TGW), is the most important yield-contributing trait in wheat. In the present study high TGW has been transferred into two elite Indian wheat cultivars PBW343 and PBW550 from a high TGW genotype, Rye selection 111, selected from local germplasm. In the background of PBW343 and PBW550, an increase in TGW upto 27.34 and 18% was observed, respectively (with respect to recipient parents), through conventional backcross breeding with phenotypic selections in 3 years replicated RBD trials. Resistance to leaf rust and stripe rust has been incorporated in the high TGW version of PBW550 through marker assisted pyramiding of stripe rust resistance gene Yr15 using marker Xuhw302, and a pair of linked leaf rust and stripe rust resistance genes Lr57-Yr40 using marker Ta5DS-2754099_kasp23. Improved versions of PBW550 with increased TGW ranging from 45.0 to 46.2 g (up to a 9% increase) and stacked genes for stripe and leaf rust resistance have been developed. This study serves as proof of utilizing conventional breeding and phenotypic selection combined with modern marker assisted selection in improvement of important wheat cultivars as a symbiont of conventional and moderan techniques.
Introduction
Wheat (Triticum aestivum L.) is an important cereal crop in India, ranking second after rice for the area (29.31 million hectares) and production (103.6 million metrics) with the state of Punjab sharing 18% of production. With global per capita consumption of 67.4 kg/year, wheat is the most widely consumed food grain (Djanaguiraman et al., 2019). In India only, the population is projected to cross the 1.70 billion mark by 2050 with a domestic demand of wheat exceeding 140 million tons (Nagarajan, 2005)1. A consistent increase in the wheat yields is a primary goal for food security of the growing population (Singh et al., 2007; Ye and Smith, 2008). Crop yield is a complex quantitative trait determined by different parameters of tiller number, grain number, grain weight, etc. Grain or kernel weight (1,000 grain weight in g, TGW), consisting of grain length, width, and area has high heritability (> 0.68) (Giura and Saulescu, 1996; Kuchel et al., 2007b) which not only translates into higher yields but also has a favorable effect over flour yield (Gegas et al., 2010). Moreover, uniform and larger sized grains are visually appealing and fetch higher market prices.
As rusts are major diseases in India, keeping pace of increasing yields with rust resistance is one of the most challenging tasks. The three species of rust viz., stripe rust caused by Puccinia striiformis, leaf rust caused by Puccinia triticina, and stem rust caused by Puccinia graminis are severely affecting wheat yield. An estimated loss of 200 million rupees occur every year due to rusts (Mehta, 1950). Leaf rust is prevalent in all the wheat growing zones of India and its widespread occurrence was observed during periods of 1971–1973, 1993–1994 (Joshi et al., 1975; Nayar et al., 1997). The 70% of the total area under wheat cultivation in cooler parts Northern India is under constant threat of ever evolving epidemic stripe rust pathogenic races. It occurred almost every year from 1967 to 1974, with high incidence recorded in 2001 and 2011 (Nayar et al., 1997; Prashar et al., 2007; Pannu et al., 2010; Tomar et al., 2014; Pal et al., 2015). An approximate loss of rupees 236 crore have been recorded in Punjab state during the epidemic year of 2009–2010 (Jindal et al., 2012). Stem rust on other hand is important central and peninsular India since its first epidemic reported in 1786AD (Nagarajan and Joshi, 1975).
Despite the devastating nature and continuous occurrence of rusts, wheat production is growing linearly due to the continuous addition of new rust resistance genes in the wheat gene pool. Use of resistant wheat cultivars is not only effective and economical but also environment friendly (Peng et al., 2003; Singh et al., 2020). Due to pathogen evolution, combination of resistance genes is being pursued by the crop breeders world over for increasing durability of resistance. Resistance gene pyramiding has been found to increase the life of each gene though the synergistic effect of pyramided genes (Klymiuk et al., 2018; Mundt, 2018). One of the greatest successes in the history of resistance breeding is with the pyramiding of resistance genes in controlling stem rust (Singh et al., 2015). The Ug99 race group of pathotypes of stem rust defeated rust resistance genes Sr31 and Sr38 but their differential pyramiding combinations of different resistant genes Sr22, Sr25, Sr26, Sr33, Sr35, Sr45, and Sr50 were found to be effective. For leaf rust, pyramiding of different genes in combination with leaf rust resistance genes provided long lasting resistance (Kolmer, 1996; Bhawar et al., 2011; Aboukhaddour et al., 2020; Babu et al., 2020). Resistance to stripe rust has also been significantly improved by pyramiding of different resistance genes (Zheng et al., 2017; Randhawa et al., 2019).
The present study reports the introgression of high thousand grain weight (TGW) to wheat varieties PBW343 and PBW550 from a local selection named “Rye Selection 111.” Pyramiding of three rust resistance genes viz Yr15 and linked Lr57-Yr40 into improved TGW version of PBW550 was done using “transfer first and assemble later” strategy (Ishii and Yonezawa, 2007a,b). Wheat cultivar PBW343, released in India in 1995, was the most widely grown cultivar in the country and soon was recognized as the new miracle genotype with wider adaptability and yield potential (Gupta et al., 2018). PBW550 was released in 2008 and farmers’ receptivity to this short duration variety with bold grains has accelerated its spread in the first few years of its release (Gupta et al., 2018). However, both of these hexaploid elite cultivars succumbed to emerging races of YR thus it became imperative to restructure the genetic makeup of these cultivars for improved yield and rust resistance.
Materials and Methods
Plant Material
The plant material used in the present study included two cultivated hexaploid bread wheat varieties named PBW343 (ND/VG9144//KAL/BB/3/YACO’S’/4/VEE#5 “S,” notification number 1(E) dated 01.01.1996, Indian Council of Agricultural Research, Government of India) and PBW550 (WH 594/RAJ 3856//W 485 notification number 72(E) dated 10.01.2008, Indian Council of Agricultural Research, Government of India), two near isogenic lines (NILs) developed at Punjab Agricultural University (PAU) in PBW550 background viz PBW550 + Yr15 and PBW550 + Lr57-Yr40, a high thousand grain weight selection from local germplasm named Rye selection 111 (RyeSel 111) and the progenies from crosses of RyeSel 111 × PBW343, RyeSel 111 × PBW550, HGW343 × PBW550, HGW550 × PBW550 + Yr15 and HGW550 + Yr15 × PBW550 + Lr57-Yr40. (The “PBW” stands for Punjab Bread Wheat followed by numeric value of varietial release number, HGW = high grain weight).
Introgression of High Grain Weight in PBW343 and PBW550
For transfer of high TGW to wheat variety PBW343, cross was made with RyeSel 111 as male parent and the phenotypic selection was done in subsequent backcross generations (Figure 1a). The BC2F5 progenies of cross PBW343-RyeSel 111 with high TGW will be referred to as HGW343 hereon. Selected HGW343 plants were used as donor to transfer high TGW to wheat variety PBW550 and BC2F5 progenies with high TGW (Figure 1b) thus obtained will be termed as HGW550 hereon. All the crosses and selections were done at PAU, Ludhiana, India.
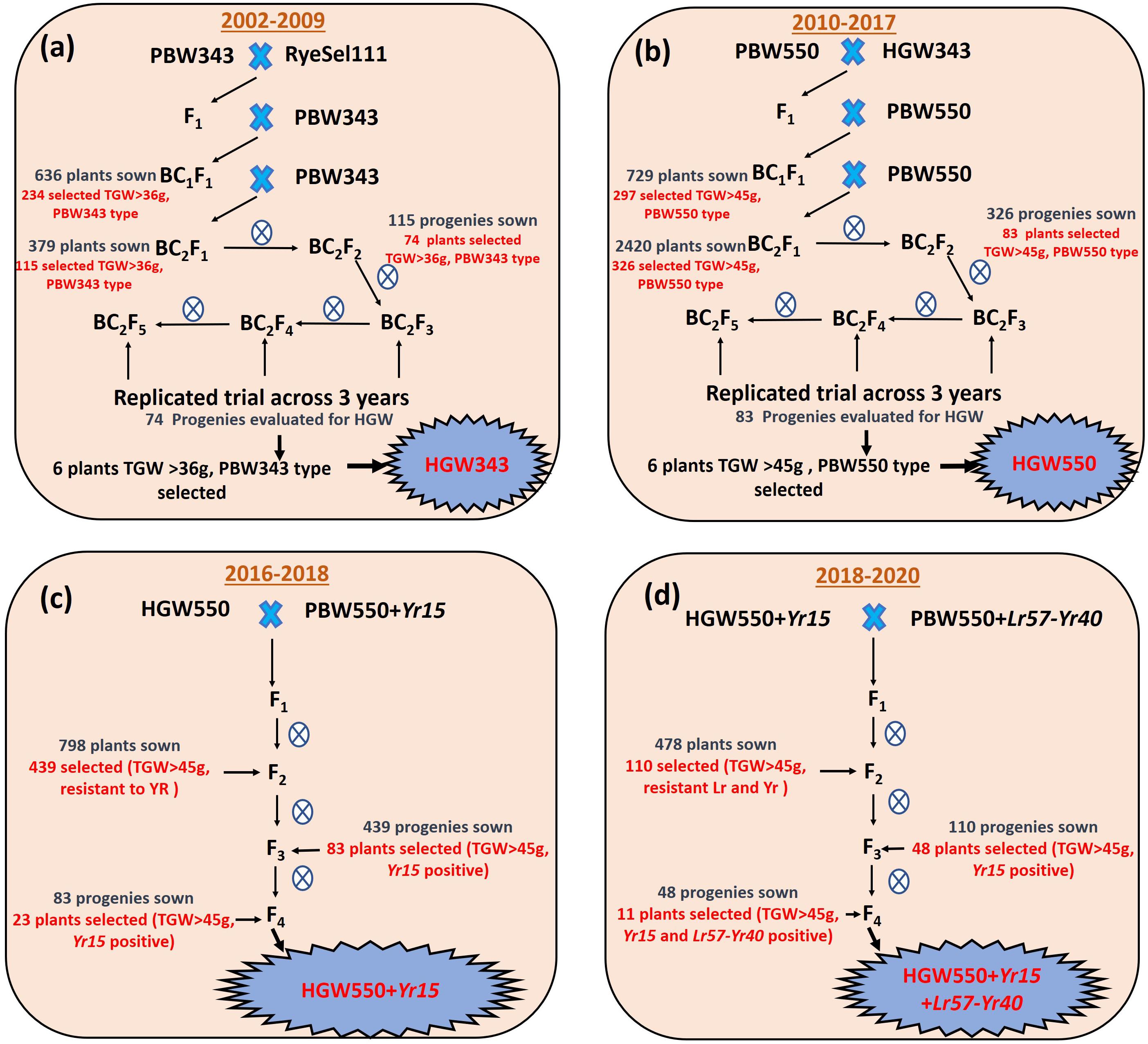
Figure 1. Schematic view of the transfer of high thousand grain weight to (a) wheat variety PBW343 to obtain HGW343. (b) Wheat variety PBW550 to obtain HGW550. (c) Combining HGW550 with stripe rust resistance gene Yr15. (d) Combining HGW550 + Yr15 with linked leaf rust and stripe rust resistance gene Lr57-Yr40.
Phenotyping and Experimental Design
In both the above crosses, traits were recorded on single plants in BC2-F1/F2 generations while in BC2-F3/F4/F5 progenies, traits were recorded on 10 single plants of each progeny as well as bulk of progeny. Single plants from uniform and promising progenies were selected and carried forward. Evaluation of each of the BC2-F3/F4/F5 progenies along with parental lines were done in three replications (1.5 m paired rows with the plant to plant distance of 10 cm and row to row distance of 20 cm) in randomized block design. Wherever single plant selections were done, the weight of 300–500 grains (as available per plant) was recorded and converted into TGW, while the weight of thousand grains was recorded for progeny bulks. Selections were done for plant/progenies having TGW higher than recurrent parent. Similarly other phenotypic traits of plant height, tiller number, spikelets per spike, spike length, and grains per spike were also recorded and plants/progenies having either similar value to or higher than recurrent parent were selected (data not given).
For each progeny the mean values were calculated for recorded traits in each replication and adjusted means of replications were finalized for each year. Three years data was again used to calculate adjusted means across the years for final selections.
Combining High Grain Weight and Rust Resistance in PBW550
For pyramiding HGW with stripe rust resistance gene Yr15, first the selected HGW550 progenies obtained from the above cross were crossed with NIL PBW550 + Yr15. The HGW550 + Yr15 thus obtained was again crossed with another NIL, PBW550 + Lr57-Yr40, to develop the HGW550 + Yr15 + Lr57-Yr40 line. Selections were done for high TGW and rust resistance as for single plant in F1/F2 generations, while in F3/F4/F5 progenies 10 single plants along with progeny bulks recorded to select single plants from best performing progenies. Since both the parental lines were from the same background, no backcrossing was done.
Marker Assisted Selection for Yr15 and Lr57-Yr40 Genes
Selection for rust resistance was done in a combined phenotypic–genotypic manner. For marker assisted selection, DNA from parental lines and segregating progenies was extracted using CTAB method (Saghai-Maroof et al., 1984) with the small modification of reducing incubation time to 30 min at 65°C with CTAB buffer and to 20 min at room temperature for solvent extraction using chloroform: isoamyl alcohol (24:1). Yr15 positive plants were selected by amplifying gene specific marker Xuhw302 (Klymiuk et al., 2018). For the selection of Lr57-Yr40 gene, linked Kasp marker Ta5DS-2754099_kasp23 (Bansal et al., 2020) and caps marker Lr57-Yr40_caps16 (Toor et al., 2016) were used. The PCR reaction for gel based marker Xuhw302 and Lr57-Yr40_caps16 was done in 10 μl reaction volume (60 ng DNA, 5 μl of 2× EmeraldAmp® GT PCR Master Mix, 0.75 μl of 5 μM each primer) in 384 well microtiter plate in an Applied Biosciences 384 thermal cyclers. The PCR products were resolved using 2.5% agarose gel electrophoresis and visualized and photographed using gel documentation system. The scoring was done identifying the presence of gene specific amplicon as per the positive control. The Kasp marker Ta5DS-2754099_kasp23 was amplified in 4 μl reaction (20 ng DNA, 1.944 μl of 2X KASP V3.0 master mix from LGC, Biosearch Technologies, 0.056 μl of primer mix in ratio of 12:12:30:46 Allele specific Primer I:Allele specific II primer: Common primer: Water) in a 384 well microtiter plate. The amplicons were identified by measuring allele specific flourescence in a high throughput TECAN infinite F200 PRO plate reader. Kluster Caller software was used to view the allele specific calls in an x–y plot to identify the positive and negative allels against the positive control.
Screening Against Stripe Rust and Leaf Rust
Screening for stripe rust and leaf rust was also done in the field along with marker selection to validate the effectiveness of gene pyramiding. F3 and F4 progenies of HGW550 X PBW550 + Yr15 were screened against stripe rust at the adult plant stage in the field during season 2016–2017 and 2017–2018. Similarly, F3 and F4 progenies of HGW550 + Yr15 X PBW550 + Lr57-Yr40 were screened both against stripe rust and leaf rust at the adult plant stage in the field during season 2018–2019 and 2019–2020. For screening, artificial epiphytotic conditions for rust were created by spraying the urediniospores mixed and diluted in water, containing Tween-20, of Pst pathotypes (100S119, 78S84), and Pt pathotypes (77-1, 77-2, 77-5, 104-2) and a mixture of leaf rust and stripe rust inoculum collected from farmers’ fields. For the uniform spread of disease, highly susceptible cultivar WL711 was planted as spreader rows all around the field and after every 20 rows. Rust data was recorded when WL711 showed complete susceptibility. Rust was recorded using Cobbs scale, as illustrated in McIntosh et al. (1995) where types of spores were recorded as zero (immune); TR (traces of severity); MR (moderately resistant), MS (moderately susceptible); and S (susceptible), and numeric numbers associated with these scores signified the percentage of leaf area covered by rust. The TR score is given to a plant when there are no lesions, MR when there are no visual postules of spores on the leaf but apoptotic lesions are visible, MS when there are minute visual postules of spores on the leaf with apoptotic lesions while S score is given to a plant when advanced postules of spores are visible to the leaf.
Results
Development and Selection of HGW343
Different number of plants phenotypically similar to PBW343 with TGW > 36g were selected from the cross of PBW343-Rye Sel 111 across different generations. In the crop season 2003–2004, 636 BC1F1 plants were sown and BC2 progenies of only 234 BC1F1 plants (Figure 1a) were carried forward. In BC2F1, 115 plants were selected and planted as single plant-to-row BC2F2 progenies. During the years 2005–2006, from 943 well-established BC2F2 single plants, only 74 superior plants were selected.
For three consecutive seasons (2006–2009), 74 BC2F3–5 progenies, along with RyeSel 111 and PBW343, were evaluated in replicated trials. Adjusted means for 3 years showed that TGW ranged from 36.66 to 46.81 g (0.30–28.10% increase), where TGW for PBW343 was 36.54 g and for RyeSel 111 was 54.17 g (Table 1 and Figure 2). Six BC2F5 plants of progenies with significantly high TGW, namely HGW343-3 (TGW-45.36 g, 24.13% increment), HGW343-6 (TGW-45.54 g, 24.63% increment), HGW343-8 (TGW-46.08 g, 26.11% increment), HGW343-60 (TGW-46.48 g, 27.20% increment), HGW343-61 (TGW-46.71 g, 27.83% increment), and HGW343-11 (TGW-46.81 g, 28.10% increment) were selected (Figure 3). The popularity of PBW343 in the late 1990s and early 2000s led to its widespread sowing which facilitated the selection of virulence for a super aggressive stripe rust race 78S84 (Prashar et al., 2007), breaking down resistance of its predominant gene Yr27, crumbling the wheat production in India. For restructuring the resistant version of PBW343, the high TGW progenies in PBW343 background were selected and disease resistance gene pyramiding carried out under a separate breeding program (not included in the present manuscript).
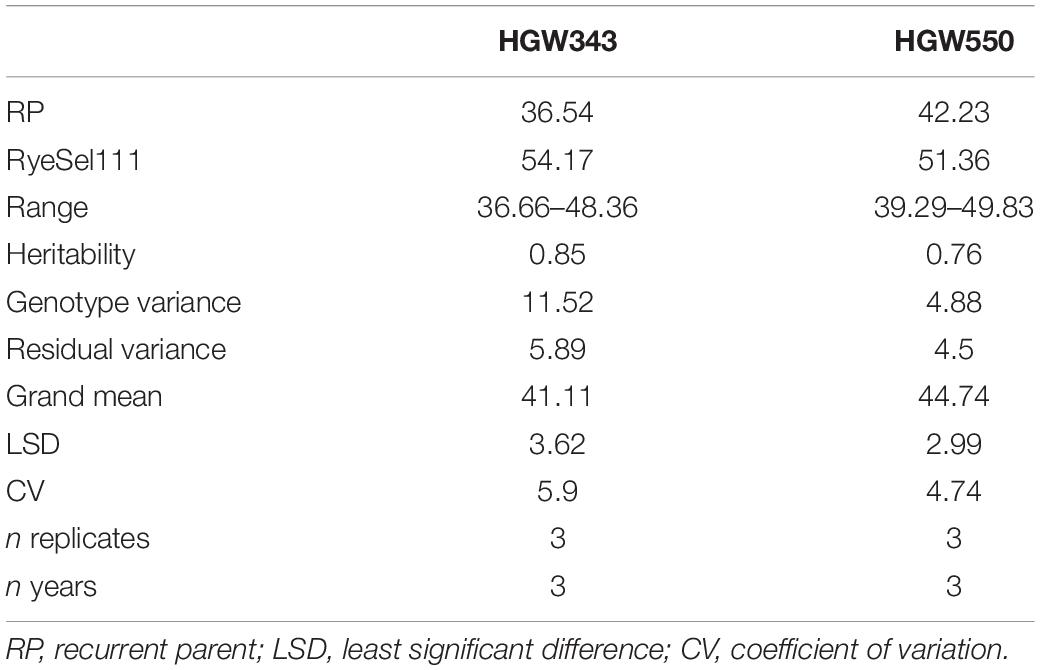
Table 1. Statistical analysis of HGW343 and HGW550 progenies across F3, F4, and F5 generations planted in RBD design.
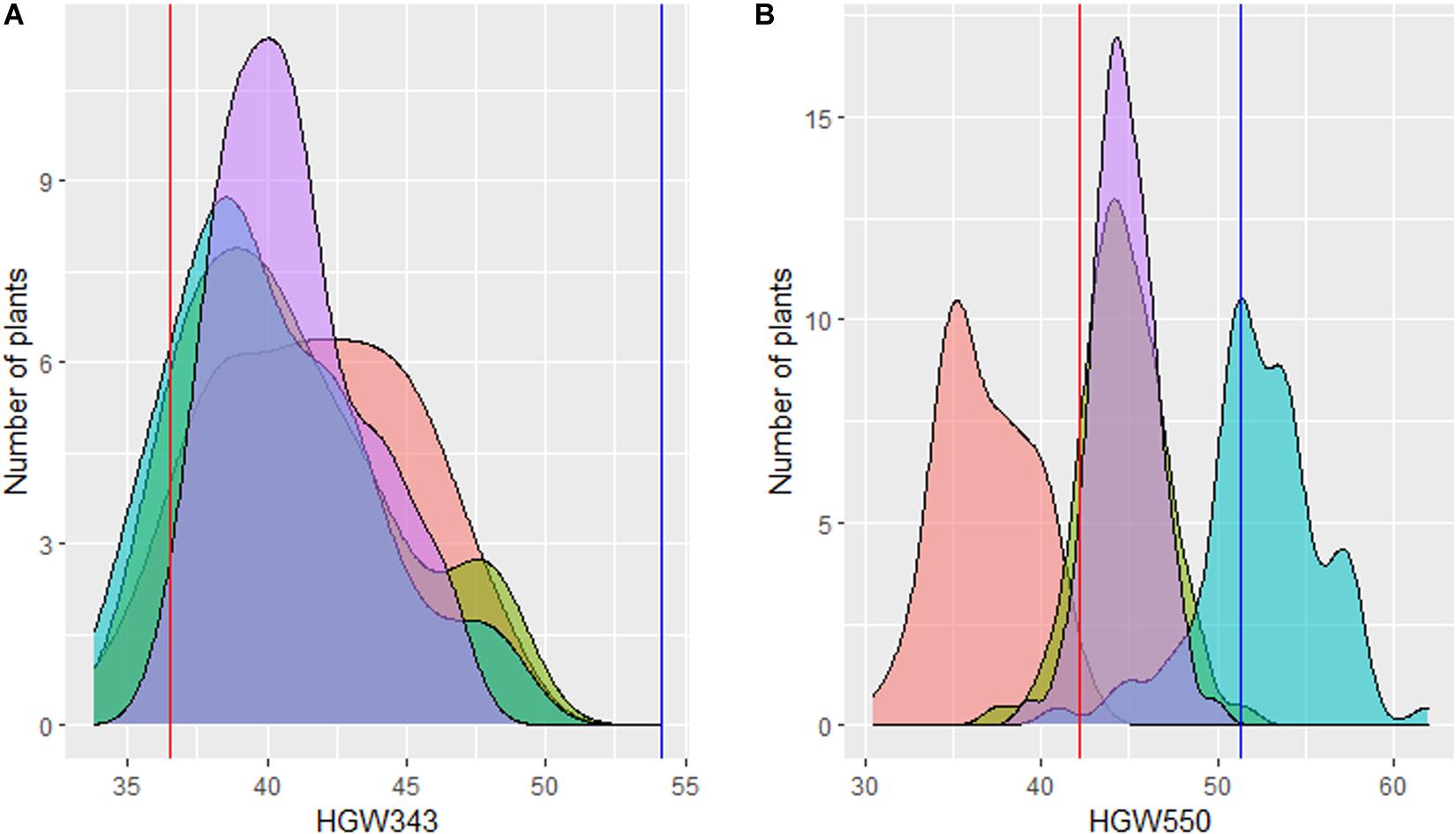
Figure 2. Graphical representation of distribution of thousand grain weight of derived BC2F3–5. (A) HGW343 progenies and (B) HGW550 progenies with the BLUPs in each environment (purple). The BLUPs of Rye Selection111 and recurrent parents (PBW343 and PBW550) are given as vertical red lines and blue lines, respectively.

Figure 3. Representative grains from the field grown BC2F5 progenies along with parental genotypes. (a) PBW343. (b) Rye selection 111. (c–i) BC2F5 progenies obtained from cross of PBW343/Rye selection 111.
Development and Selection of HGW550
Four HGW343 plants named HGW343-8, HGW343-60, HGW343-61, and HGW343-11 were crossed and backcrossed with PBW550 (Figure 1b). Phenotypic selections for high TGW were done in BC1F1, BC2F1, and BC2F2. From 729 BC1F1 plants, backcrosses from 297 single plants were selected to obtain 2,420 BC2F1 plants. A total of 83 BC2F2 plants were selected and BC2F3, BC2F4, and BC2F5 progenies were evaluated across 3 years (2014–2017) in replicated trials, each having three replications (Table 1 and Figure 2). Overall adjusted means for 3 years showed that TGW for the progenies ranged from 39.29 to 49.83 g where TGW for PBW550 was 42.23 g and for RyeSel 111 was 51.36. Six plants from progenies with TGW higher than 47.0 g, namely HGW550-8 (TGW-49.83 g, 17.99% increment), HGW550-6 (TGW-48.1 g, 13.90% increment), HGW550-63 (TGW-47.93 g, 13.50% increment), HGW550-3 (TGW-47.64 g, 12.81% increment), HGW550-7 (TGW-47.54 g, 12.57% increment), and HGW550-21 (TGW-47.5 g, 12.48% increment) were selected (Figure 4) for introducing rust resistance.
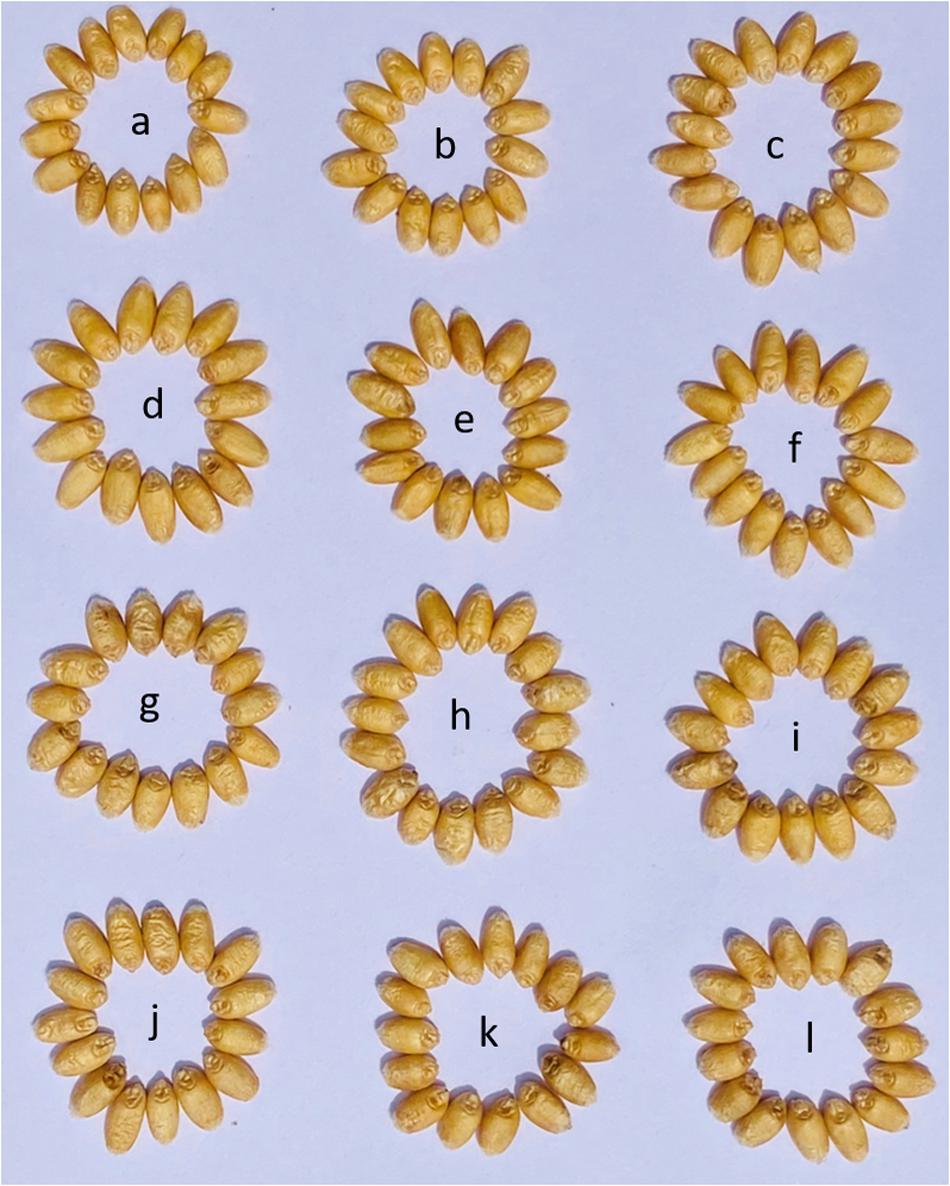
Figure 4. Representative grains from the field grown pyramided lines along with parental genotypes. (a) PBW550. (b) PBW550 + Yr15. (c) PBW550 + Yr15 + Lr57-Yr40. (d–f) BC2F5 progenies obtained from cross of HGW343 × PBW550. (g–i) F4 progenies obtained from cross of HGW550 × Yr15; (j–l) F4 progenies obtained from cross of HGW550 + Yr15 X Lr57-Yr40.
Marker Assisted Pyramiding of HGW550 and Yr15
Three selected HGW550 plants; HGW550-8, HGW550-6, and HGW550-63 were crossed with NIL PBW550 + Yr15 (Figure 1c). F1s thus obtained were selfed to develop 798 F2 plants and 439 stripe rust resistant plants with TGW of more than 42 g were selected. In F3, plants having a TGW equal to or more than 45.0 g were selected from promising uniform stripe rust resistant progenies. The selected plants were screened with molecular marker Xuhw302, and 83 plants, homozygous for Yr15 were selected (Supplementary Figure S1A). These 83 plants were further sown in plant to progeny rows in F4 and 23 single F4 plants with TGW ranging between 45.0 and 48.0 g and positive for Yr15 were selected (Figure 4). These plants were also completely resistant to stripe rust in the field.
Marker Assisted Pyramiding of HGW550 + Yr15 and Lr57-Yr40
F1s generated by crossing four selected single F4 plants HGW550 + Yr15-34 (TGW-46.3), HGW550 + Yr15-29 (TGW-47.1), HGW550 + Yr15-3 (TGW-47.3), HGW550 + Yr15-34 (TGW-47.4) with PBW550 + Lr57-Yr40 were selfed to generate 478 F2 plants, of which 110 individual F2 plants were selected with TGW > 45 g (Figure 1d). These 110 plants were sown as plants to progeny rows and 48 single F3 plants with the positive allele for Yr15 and Lr57-Yr40 markers were selected (Supplementary Figure S1B). These plants were also screened against stripe rust and leaf rust at the adult plant stage and found to be completely resistant to both diseases. These 48 plants were again evaluated in F4 generation in plant to row progenies and 11 F4 plants from superior progenies with TGW between 45.0 and 46.2 g and complete resistance to stripe rust and leaf rust have been selected (Figure 4).
Discussion
The stacking of genes governing multiple traits enhances the value of breeding material besides improving its durability. Yield is a trait of foremost priority for the commercial success of a variety, but combining improved grain yield with disease resistance, and high grain quality is a need of the changing time and environment. The introgression and pyramiding of major genes/QTL for different traits through marker-assisted selection (MAS) has been reported in wheat (Gupta et al., 2010; Kumar et al., 2010; Tyagi et al., 2014; Gautam et al., 2020).
We used RyeSel 111 as the donor to transfer the high TGW trait to elite cultivar PBW343 and PBW550 through conventional phenotypic selection. Though QTLs for high grain weight have been reported in the RyeSel 111-Chinese Spring RIL population (Kumar et al., 2006), the reported linked markers were found to be non-polymorphic between the receipient and donor combinations used in the present study. Thus a conventional route is followed through phenotypic selection for high TGW. Mapping and transfer of QTLs for yield related traits often sound more logical theoretically, but these complex traits are controlled by many QTLs of large and small effects and pyramiding all these QTLs in one background through MAS to retrieve the donor effect is not always achieved. This is more often pronounced in cases where the minor QTLs additively contribute toward the trait along with major ones. Thus phenotypic selection especially for an easily quantified trait like grain weight, gives more realistic results. Wheat yield is controlled by traits of yield per area (includes grains per spike, grain weight, and spikes per area) and yield per spike (spikelet number per spike, grain number, and grain size) (Slafer et al., 2014). Several QTLs related to grain yield related traits have been mapped on different chromosomes but none have been cloned or effectively utilized for marker assisted selection. From different factors influencing wheat yield, grain weight was found to be one of the most stably inherited (Sidwell et al., 1976; Kuchel et al., 2007a,b; Aisawi et al., 2015) suggesting selection for heavier grains lead to effective yield improvement. Raising plant yield by conventional breeding methods (Reynolds et al., 2009) has remained very successful in the past, though new emerging technologies are creating a different niche. High TGW was introgressed effectively into two popular wheat cultivars cv. PBW343 and PBW550 through the phenotypic selection, and improved progenies with an increase of TGW by 27.34 and 18% were obtained, respectively. Initially, direct crosses of RyeSel 111 were done with PBW343 then using improved HGW343 progenies as the donor, HGW introgressed into PBW 550. Similarly improvement in grain yield by improving TGW has been reported by several studies (Giunta et al., 2007; Beche et al., 2014; Qin et al., 2015; Zhang et al., 2016; Gao et al., 2017).
PBW343 has been one of the most popular wheat varieties and was grown in about 25% of the 27 million hectares under the wheat cultivation in the country and contribute roughly 55% of the total wheat output in the country (Pavithra et al., 2017). Due to its wider adaptability to a range of environments, it has been a variety of choice for many improvement programs and was selected in this program for improving grain weight. After the PBW343 succumbed to 78S84 pathotype of stripe rust in 2004 and so did the whole breeding pipeline at PAU, it became mandatory to quickly mobilize some known stripe rust resistance genes to PBW343 and other advance germplasms to have a rust resistant variety for the farmers of the region. It was then that the development of rust resistant version of PBW343 became the main mandate of the wheat breeders, and reintroduction of its rust resistant version led to the shift of this work to a separate program.
PBW550, on the other hand, has been recognized as a good quality cultivar with bold grains and was spread to whole wheat growing regions of the country in the few years since its release in the year 2008. This variety was tested and also recommended for cultivation in Eastern and Central India for processing the commercial wheat flour under the PAU-ITC (Indian Tobacco Company) agreement. However, the evolution of the 78S84 pathogen was so targeted and led to the development of a very aggressive strain which not only rendered PBW343 susceptible to stripe rust but also slowly wrapped up PBW550 including all the newly released varieties during that time. As a result, PBW550 too became susceptible to yellow rust after 6–7 years of release despite having multi-pathogen resistance gene loci Lr34/Yr18/Sr57. The short-lived resistance makes it of utmost importance to introgress rust resistance in addition to yield improvement in these cultivars.
Though HGW has been introduced with backcross breeding only while the rust resistance was facilitated through MAS. Increased adoption of combining conventional breeding with the MAS approach (Gupta et al., 2010) in recent years has given multifold advantages, the major one being accelerated mobilization of the desirable gene(s) and their efficient stacking in elite backgrounds. Two different NILs carrying stripe rust resistance gene Yr15 and linked leaf rust-stripe rust resistance gene Lr57-Yr40 have been developed by PAU. NIL PBW550 + Yr15 has also been released as a variety for cultivation in Punjab under timely sown irrigated conditions as Unnat PBW550. Yr15 mapped near distal Nor-B1 on the short arm of chromosome 1B was first discovered in the 1980s in wild emmer wheat T. turgidum var. dicoccoides. Avocet + Yr15 was initially used as a donor to transfer this gene in PBW550 background. As per the Global Rust Reference Centre2, Yr15 has been providing a complete and broad spectrum resistance (Klymiuk et al., 2018). Linked Lr57-Yr40 (on chromosome 5DS) has been transferred from an introgression line INGR15046, developed in the wheat wide hybridization program of PAU by transfer of leaf rust and stripe rust resistance from Aegilopes geniculata to wheat cultivar WL711 (Kuraparthy et al., 2007). This linked gene provides complete resistance to leaf rust and partial resistance (20MR) to stripe rust in field.
For pyramiding, the “transfer first and assemble later” approach (Ishii and Yonezawa, 2007a,b) already given above was followed where PBW550 + Yr15 and PBW550 + Lr57-Yr40, NILs were used as donors of respective genes. Since both the parental genotypes were in the PBW550 background, backcrossing was not required. Similarly, HGW550 + Yr15 and PBW550 + Lr57-Yr40 were crossed, and selfing generations F2, F3, and F4 were evaluated for HGW and two resistances to leaf and stripe rust leading to the selection of 11 HGW550 + Yr15 + Lr57-Yr40 progenies with three rust resistance genes and high TGW. Gene pyramiding of disease resistance genes reported improving the durability of resistance, though the combined effect depends upon the nature of the individual genes and their synergism. Resistance breeding by marker led pyramid in the last decade is being used in wheat programs globally. Moreover, the combination of major and minor genes was also found to have more significant effects. Although Yr15 has already succeeded in conferring stripe rust resistance for many years in different introgression lines around the world, marker assisted gene pyramiding of Yr15, and Lr57-Yr40 genes provides durable resistance to stripe rust in combination with leaf rust resistance. Advanced breeding lines of PBW550 with three rust resistance genes and high TGW can lead to the development of new varieties and also serve as valuable germplasm for breeders to be used in the varietal development program to aid future breeding programs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s. The raw data for trials has been added as Supplementary Table S1.
Author Contributions
SK conceived and designed the research, planned, and conducted the experiments, did rust screening, compiled the work, and produced the final draft of the manuscript. JK and RS conducted field experiments and did marker assisted selection. GM and AS conducted the field experiments. GD did statistical analysis and compilation of the manuscript. UD helped in field experiments. PC conceived and designed the research, and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Department of Biotechnology (DBT) under the grant number BT/PR21024/AGIII/103/925/2016.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.593426/full#supplementary-material
Supplementary Figure 1 | (A) PCR amplification profile of marker Xuhw302 amplifying Yr15 gene in F3 plants derived from cross HGW550 × PBW550 + Yr15 as on agarose gel. (B) KlusterCaller output view of segregation of Co-dominant Ta5DS-2754099_kasp23 markers in F3 plants derived from cross HGW550 + Yr15 × PBW550 + Lr57-Yr40. FAM tailed Lr57-Yr40 (blue color) allele on X-axis, HEX tailed PBW550 alleles (red color) on Y-axis, heterozygous individuals on mid-axis (green color). Black color represents non-template control and pink color represents unamplified or unclustered samples.
Supplementary Table 1 | Raw data of thousand grain weight in three-year replicated trials as recorded on BC2F3–5 progenies of cross PBW343 X Rye Sel 111 and BC2F3–5 progenies of cross PBW550 × HGW343 as Env1, Env2 and Env3. The Env 4 Represents the adjusted means of the three years.
Footnotes
- ^ https://www.thehindubusinessline.com; https://www.eastasiaforum.org
- ^ https://agro.au.dk/forskning/internationale-platforme/wheatrust/
References
Aboukhaddour, R., Fetch, T., McCallum, B. D., Harding, M. W., Beres, B. L., and Graf, R. J. (2020). Wheat diseases on the prairies: a Canadian story. Plant Pathol. 69, 418–432. doi: 10.1111/ppa.13147
Aisawi, K. A. B., Reynolds, M. P., Singh, R. P., and Foulkes, M. J. (2015). The physiological basis of the genetic progress in yield potential of CIMMYT spring wheat cultivars from 1966 to 2009. Crop Sci. 55, 1749–1764. doi: 10.2135/cropsci2014.09.0601
Babu, P., Baranwal, D. K., Harikrishna Pal, D., Bharti, H., Joshi, P., et al. (2020). Application of genomics tools in wheat breeding to attain durable rust resistance. Front. Plant Sci. 11:567147. doi: 10.3389/fpls.2020.567147
Bansal, M., Adamski, N. M., Toor, P. I., Kaur, S., Molnár, I., Holušová, K., et al. (2020). Aegilops umbellulata introgression carrying leaf rust and stripe rust resistance genes Lr76 and Yr70 located to 9.47-Mb region on 5DS telomeric end through a combination of chromosome sorting and sequencing. Theor. Appl. Genet. 133, 903–915. doi: 10.1007/s00122-019-03514-x
Beche, E., Benin, D., Da Silva, C. L., Munaro, L. B., and Marchese, J. A. (2014). Genetic gain in yield and changes associated with physiological traits in Brazilian wheat during the 20th century. Eur. J. Agron. 61, 49–59. doi: 10.1016/j.eja.2014.08.005
Bhawar, K., Vinod Sharma, J., Singh, A., Sivasamy, M., Singh, M., Prabhu, K., et al. (2011). Molecular marker assisted pyramiding of leaf rust resistance genes Lr19 and Lr28 in wheat variety HD2687. Ind. J. Genet. Plant Breed. 71, 304–311.
Djanaguiraman, M., Prasad, P.V. V., Kumari, J., Sehgal, S. K., Friebe, B., Djalovic, I., et al. (2019). Alien chromosome segment from Aegilops speltoides and Dasypyrum villosum increases drought tolerance in wheat via profuse and deep root system. BMC Plant Biol. 19, 242–257. doi: 10.1186/s12870-019-1833-8
Gao, F., Ma, D., Yin, G., Rasheed, A., Dong, Y., Xiao, Y., et al. (2017). Genetic progress in grain yield and physiological traits in Chinese wheat cultivars of southern Yellow and Huai Valley since 1950. Crop Sci. 57, 760–773. doi: 10.2135/cropsci2016.05.0362
Gautam, T., Dhillon, G. S., Saripalli, G., Rakhi, Singh, V. P., Prasad, P., et al. (2020). Marker-assisted pyramiding of genes/QTL for grain quality and rust resistance in wheat (Triticum aestivum L.). Mol. Breed. 40:49. doi: 10.1007/s11032-020-01125-9
Gegas, V. C., Nazari, A., Griffiths, S., Simmonds, J., Fish, L., Orford, S., et al. (2010). A genetic framework for grain size and shape variation in wheat. Plant Cell 22, 1046–1056. doi: 10.1105/tpc.110.074153
Giunta, F., Motzo, R., and Pruneddu, G. (2007). Trends since 1900 in the yield potential of Italian-bred durum wheat cultivars. Eur. J. Agron. 27, 12–24. doi: 10.1016/j.eja.2007.01.009
Giura, A., and Saulescu, N. N. (1996). Chromosomal location of genes controlling grain size in a large grained selection of wheat (Triticum aestivum L.). Euphytica 89, 77–80. doi: 10.1007/bf00015722
Gupta, A., Singh, C., Kumar, V., Tyagi, B. S., Tiwari, V., Chatrathand, R., et al. (2018). Wheat Varieties Notified in India Since 1965. Karnal: ICAR- Indian Institute of Wheat & Barley Research, 101.
Gupta, P. K., Kumar, J., Mir, R. R., and Kumar, A. (2010). Marker-assisted selection as a component of conventional plant breeding. Plant Breed. Rev. 33, 145–217. doi: 10.1002/9780470535486.ch4
Ishii, T., and Yonezawa, K. (2007a). Optimization of the marker-based procedures for pyramiding genes from multiple donor lines: I. Schedule of crossing between the donor lines. Crop Sci. 47, 537–546. doi: 10.2135/cropsci2006.06.0435
Ishii, T., and Yonezawa, K. (2007b). Optimization of the marker-based procedures for pyramiding genes from multiple donor lines: II. Strategies for selecting the objective homozygous plant. Crop Sci. 47, 1878–1886. doi: 10.2135/cropsci2006.11.0750
Joshi, L. M., Srivastava, K. D., and Ramanujam, K. (1975). An analysis of brown rust epidemics of 1971- 72 and 1972-73. Indian Phytopathol. 28:137.
Jindal, M. M., Sharma, I., and Bains, N. S. (2012). Losses due to stripe rust caused by Puccinia striiformis in different varieties of wheat. J. Wheat Res. 4, 33–36.
Klymiuk, V., Yaniv, E., Huang, L., Raats, D., Fatiukha, A., Chen, S., et al. (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 9:3735. doi: 10.1038/s41467-018-06138-9
Kolmer, J. A. (1996). Genetics of resistance to wheat leaf rust. Annu. Rev. Phytopathol. 34, 435–455. doi: 10.1146/annurev.phyto.34.1.435
Kuchel, H., Fox, R., Reinheimer, J., Mosionek, L., Willey, N., Bariana, H., et al. (2007a). The successful application of a marker-assisted wheat breeding strategy. Mol. Breed. 20, 295–308. doi: 10.1007/s11032-007-9092-z
Kuchel, H., Williams, K. J., Langridge, P., Eagles, H. A., and Jefferies, S. P. (2007b). Genetic dissection of grain yield in bread wheat. I. QTL analysis. Theor. Appl. Genet. 115, 1029–1041.
Kumar, J., Mir, R. R., Kumar, N., Kumar, A., Mohan, A., Prabhu, K. V., et al. (2010). Marker-assisted selection for pre-harvest sprouting tolerance and leaf rust resistance in bread wheat. Plant Breed. 129, 617–621. doi: 10.1111/j.1439-0523.2009.01758.x
Kumar, N., Kulwal, P. L., Gaur, A., Tyagi, A. K., Khurana, J. P., Khurana, P., et al. (2006). QTL analysis for grain weight in common wheat. Euphytica 151, 135–144. doi: 10.1007/s10681-006-9133-4
Kuraparthy, V., Chhuneja, P., Dhaliwal, H. S., Kaur, S., Bowden, R. L., and Gill, B. S. (2007). Characterization and mapping of cryptic alien introgression from Aegilops geniculata with new leaf rust and stripe rust resistance genes Lr57 and Yr40 in wheat. Theor. Appl. Genet. 114, 1379–1389. doi: 10.1007/s00122-007-0524-2
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts: An Atlas of Resistance Genes CSIRO Publications, Vol. 1. Melbourne: CSIRO Publications, doi: 10.1007/bf03214019
Mehta, K. C. (1950). Control of rust epidemics of wheat in India- a national emergency. Sci. Cult. 15, 263–270.
Mundt, C. C. (2018). Pyramiding for resistance durability: theory and practice. Phytopathology 108, 792–802. doi: 10.1094/phyto-12-17-0426-rvw
Nagarajan, S., and Joshi, L. M. (1975). A historical account of wheat rust epidemics in India and their significance. Cereal Rusts Bull. 3, 29–33.
Nayar, S. K., Prashar, M., and Bhardwaj, S. C. (1997). Mannual of current techniques in wheat rusts. Res. Bull. 2, 1–32. doi: 10.1007/978-94-011-0083-0_1
Pal, D., Bhardwaj, S. C., Sharma, D., Kumari, S., Patial, M., Vinod, P., et al. (2015). Assessment of genetic diversity and validating rust resistance gene sources using molecular markers in wheat (Triticum aestivum L.). SABRAOJ. Breed. Genet. 47, 89–98.
Pannu, P. P. S., Mohan, C., Singh, G., Kaur, J., Mann, S. K., Bala, G. K., et al. (2010). Occurrence of yellow rust of wheat, its impact on yield viz-a-viz its management. Plant Dis. Res. 25, 144–150.
Pavithra, S., Mittal, S., Bhat, S. A., Birthal, P. S., Shah, S. A., and Hariharan, V. K. (2017). Spatial and temporal diversity in adoption of modern wheat varieties in India. Agric. Econ. Res. Rev. 30, 57–72. doi: 10.5958/0974-0279.2017.00005.2
Peng, J., Ronin, Y., Fahima, T., Röder, M. S., Li, Y., Nevo, E., et al. (2003). Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. U.S.A. 100, 2489–2494. doi: 10.1073/pnas.252763199
Prashar, M., Bhardwaj, S. C., Jain, S. K., and Datta, D. (2007). Pathotypic evolution in Puccinia striiformis in India during 1995-2004. Aust. J. Agric. Res. 58, 603–604.
Qin, X., Zhang, F., Liu, C., Yu, H., Cao, B., Tian, S., et al. (2015). Wheat yield improvements in China: past trends and future directions. Field Crops Res. 177, 117–124. doi: 10.1016/j.fcr.2015.03.013
Randhawa, M. S., Bains, N. S., Sohu, V. S., Chhuneja, P., Trethowan, R. M., Bariana, H. S., et al. (2019). Marker assisted transfer of stripe rust and stem rust resistance genes into four wheat cultivars. Agronomy 9:497. doi: 10.3390/agronomy9090497
Reynolds, M., Foulkes, M. J., Slafer, G. A., Berry, P., Parry, M. A. J., Snape, J. W., et al. (2009). Raising yield potential in wheat. J. Exp. Bot. 60, 1899–1918. doi: 10.1093/jxb/erp016
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8028. doi: 10.1073/pnas.81.24.8014
Sidwell, R. J., Smith, E. L., and McNew, R. W. (1976). Inheritance and interrelationships of grain yield and selected yield-related traits in a hard red winter wheat cross. Crop Sci. 16, 650–654. doi: 10.2135/cropsci1976.0011183x001600050013x
Singh, H., Kaur, J., Bala, R., Srivastava, P., and Bains, N. S. (2020). Virulence and genetic diversity of Puccinia striiformis f. sp. tritici isolates in sub-mountainous area of Punjab, India. Phytoparasitica. 48, 383–395. doi: 10.1007/s12600-020-00809-4
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884. doi: 10.1094/phyto-01-15-0030-fi
Singh, R. P., Huerta-Espino, J., Sharma, R., Joshi, A. K., and Trethowan, R. (2007). High yielding spring bread wheat germplasm for global irrigated and rainfed production systems. Euphytica 157, 351–363. doi: 10.1007/s10681-006-9346-6
Slafer, G. A., Savin, R., and Sadras, V. O. (2014). Coarse and fine regulation of wheat yield components in response to genotype and environment. Field Crops Res. 157, 71–83. doi: 10.1016/j.fcr.2013.12.004
Tomar, S. M. S., Singh, S. K., Sivasamy, M., and Vinod (2014). Wheat rusts in India: resistance breeding and gene deployment - A review. Indian J. Genet. 74, 129–156. doi: 10.5958/0975-6906.2014.00150.3
Toor, P. I., Kaur, S., Bansal, M., Yadav, B., and Chhuneja, P. (2016). Mapping of stripe rust resistance gene in an Aegilops caudata introgression line in wheat and its genetic association with leaf rust resistance. J. Genet. 95, 933–938. doi: 10.1007/s12041-016-0718-y
Tyagi, S., Mir, R. R., Kaur, H., Chhuneja, P., Ramesh, B., Balyan, H. S., et al. (2014). Marker-assisted pyramiding of eight QTLs/genes for seven different traits in common wheat (Triticum aestivum L.). Mol. Breed. 34, 167–175. doi: 10.1007/s11032-014-0027-1
Ye, G. B., and Smith, K. (2008). Marker-assisted gene pyramiding for inbred line development: basic principles and practical guidelines. Int. J. Plant Breed. 2, 1–10. doi: 10.17221/51/2016-cjgpb
Zhang, Y., Xu, W., Wang, W., Dong, H., Qi, X., Zhao, M., et al. (2016). Progress in genetic improvement of grain yield and related physiological traits of Chinese wheat in henan province. Field Crops Res. 199, 117–128. doi: 10.1016/j.fcr.2016.09.022
Keywords: wheat, PBW550, PBW343, pyramiding, grain weight, rust resistance
Citation: Kaur S, Kaur J, Mavi GS, Dhillon GS, Sharma A, Singh R, Devi U and Chhuneja P (2020) Pyramiding of High Grain Weight With Stripe Rust and Leaf Rust Resistance in Elite Indian Wheat Cultivar Using a Combination of Marker Assisted and Phenotypic Selection. Front. Genet. 11:593426. doi: 10.3389/fgene.2020.593426
Received: 10 August 2020; Accepted: 30 November 2020;
Published: 22 December 2020.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaCopyright © 2020 Kaur, Kaur, Mavi, Dhillon, Sharma, Singh, Devi and Chhuneja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satinder Kaur, c2F0aW5kZXIuYmlvdGVjaEBwYXUuZWR1
†ORCID: Satinder Kaur, orcid.org/0000-0003-3704-3074; Guriqbal Singh Dhillon, orcid.org/0000-0001-6766-810X; Parveen Chhuneja, orcid.org/0000-0002-8599-9479
 Satinder Kaur
Satinder Kaur Jaspreet Kaur
Jaspreet Kaur G. S. Mavi2
G. S. Mavi2 Guriqbal Singh Dhillon
Guriqbal Singh Dhillon Achla Sharma
Achla Sharma Rohtas Singh
Rohtas Singh Urmila Devi
Urmila Devi Parveen Chhuneja
Parveen Chhuneja