- 1State Key Laboratory of Animal Nutrition, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 2State Key Laboratory of Stem cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 3College of Life Science, University of Science and Technology of China, Hefei, China
- 4Savaid Medical School, University of Chinese Academy of Sciences, Beijing, China
- 5Sun Yat-sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, China
- 6College of Life Sciences, Qingdao Agricultural University, Qingdao, China
Pig is an important agricultural economic animal, providing large amount of meat products. With the development of functional genomics and bioinformatics, lots of genes and functional single nucleotide polymorphisms (SNPs) related to disease resistance and (or) economic traits in pigs have been identified, which provides the targets for genetic improvement by genome editing. Base editors (BEs), combining Cas9 nickase and cytidine or adenine deaminase, achieve all four possible transition mutations (C-to-T, A-to-G, T-to-C, and G-to-A) efficiently and accurately without double strand breaks (DSBs) under the protospacer adjacent motif (PAM) sequence of NGG. However, the NGG PAM in canonical CRISPR-Cas9 can only cover approximately 8.27% in the whole genome which limits its broad application. In the current study, hA3A-BE3-NG system was constructed with the fusion of SpCas9-NG variant and hA3A-BE3 to create C-to-T conversion at NGN PAM sites efficiently. The editing efficiency and scope of hA3A-BE3-NG were confirmed in HEK293T cells and porcine fetal fibroblast (PFF) cells. Results showed that the efficiency of hA3A-BE3-NG was much higher than that of hA3A-BE3 on NGH (H = A, C, or T) PAM sites (21.27 vs. 2.81% at average). Further, nonsense and missense mutations were introduced efficiently and precisely via hA3A-BE3-NG in multiple pig economic trait-related genes (CD163, APN, MSTN, and MC4R) in PFF cells by one transfection. The current work indicates the potential applications of hA3A-BE3-NG for pyramid breeding studies in livestock.
Introduction
As an agricultural animal, pig is an important meat resource with great economic value. The conventional pig breeding is to pyramid desirable traits by cross breeding with cost and long breeding cycle. The genome-editing technology is an effective approach for pig improvement in growth, meat quality, reproductive capacity, and disease resistance (Song et al., 2020). It is the desired goal to exploit efficient and precise genome-editing tools to achieve rapid pyramid breeding through modifying multiple agriculture-related functional genes simultaneously.
Base editors (BEs), combining Cas9 nickase and cytidine or adenine deaminase, perform efficient and accurate base substitutions (C-to-T, A-to-G, T-to-C, and G-to-A) without double strand breaks (DSBs) at target sites, which provides an alternative strategy for precise genome editing (Komor et al., 2016; Gaudelli et al., 2017). Recently, various versions of BEs were exploited to optimize the specificity, sensitivity, and safety of base conversions (Rees and Liu, 2018). One of the many versions, the hA3A-BE3 system, replaces the rat cytidine deaminase (APOBEC1) with human cytidine deaminase (APOBEC3A), which performs C-to-T conversion more efficiently with expanded activity windows at target sites in human cells, plants, rabbits, and pigs than the original BE3 (Wang et al., 2018; Zong et al., 2018; Liu et al., 2019; Xie et al., 2019). However, the targetable scope of hA3A-BE3 is restricted for use with conventional SpCas9, which recognizes target loci through NGG as its protospacer adjacent motif (PAM) sequence.
This limitation can be overcome by using Cas9 variants with targeting preferences other than NGG PAM, which have been validated using conventional BEs. For example, BE variants, such as VQR-BE3, EQR-BE3, VRER-BE3, SaBE3, and SaKKH-BE3, have been developed to target NGAN, NGA, NGCG, NNGRRT, and NNNRRT PAM sites, circumventing the need for NGG PAM sequences in human cells (Kim et al., 2017). CRISPR-Cpf1-based BEs have even been developed that recognize and target T-rich PAM sequences (TTTV; Li et al., 2018; Kleinstiver et al., 2019). Recently, three newly engineered SpCas9 variants, xCas9, SpCas9-NG, and SpG, were reported to expand the targetable scope of NGN PAM sites in cultured cells, plants, and animals (Hu et al., 2018; Nishimasu et al., 2018; Endo et al., 2019; Fujii et al., 2019; Walton et al., 2020). Further, the SpCas9-NG system has been applied only in bacteria, human cells, plants, and rabbits (Huang et al., 2019; Thuronyi et al., 2019; Wang Y, et al., 2019; Zhong et al., 2019; Li et al., 2020; Liu et al., 2020b).
CRISPR/Cas9 mediated clusters of differentiation 163 (CD163)-deletion conferred the ability of effective resistance to porcine reproduction and respiratory syndrome virus (PRRSVs) infection on pigs (Whitworth et al., 2016; Wang H, et al., 2019). Aminopeptidase N (APN) gene deletion gave the ability of neonatal piglets to resist infection with the highly virulent transmissible gastroenteritis virus (TGEVs; Luo et al., 2019; Whitworth et al., 2019). For meat production, deletion of the porcine myostatin (MSTN) gene has been shown to improve muscle growth, resulting in a double-muscled phenotype (Qian et al., 2015). Many of these targeted gene deletions could potentially be achieved by generating a premature terminal codon (iStop-codon) through precise C-to-T mutations via cytosine base editors (CBEs; Billon et al., 2017; Kuscu et al., 2017). Precision single-base editing provides a strategy to manipulate functional single nucleotide polymorphisms (SNPs) for accurate genetic improvement in pig production. For example, porcine melanocortin-4 receptor (MC4R) c.893G>A was reported to be associated with fatness, growth, and feed intake traits (Kim et al., 2000). In the current study, in order to increase the efficiency of base editing at expanded target sites in pigs, hA3A-BE3-NG system was constructed and used to produce C-to-T mutation with high efficiency and expanded editable scope in human cells and porcine cells. Economic related genes including CD163, MSTN, APN, and MC4R, were simultaneously targeted via hA3A-BE3-NG. To our knowledge, this is the first study to precisely edit multiple genes responsible for economic traits in the porcine genome using BEs, and suggest the incredible potential of using BEs to accelerate molecular pyramid breeding in livestock.
Materials and Methods
Plasmid Construction
The hA3A-BE3-NG vector was constructed in this study through in-fusion cloning to transfer the DNA fragment containing VRVRFRR variants of SpCas9-NG from Target-AID-NG (119861#; Addgene, Watertown, MA, United States) to hA3A-BE3 (113410#; Addgene, Watertown, MA, United States). For construction of sgRNAs, oligos were synthesized, annealed, and cloned into the BsaI site of the sgRNA-expressing vector, pGL3-U6-sgRNA-PGK-puromycin (51133#; Addgene, Watertown, MA, United States). The fragment pCAG-tdTomato was cloned into the BspQI linearized pGL3-U6-sgRNA-PGK-puromycin to construct the sgRNA-tdTomato-expressing vector, pGL3-U6-sgRNA-tdTomato. The sgRNAs used in this study are summarized in Supplementary Table S1. The primers used in the construction of hA3A-BE3-NG are listed in Supplementary Table S2.
Cell Culture and Transfection
HEK293T cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, United States), supplemented with 10% fetal bovine serum (FBS; v/v; HyClone, Logan, UT, United States) and 1% Penicillin Streptomycin (v/v; Gibco, Grand Island, NY, United States). HEK293T cells were seeded 1 day prior to transfection in 24-well plates (Corning, Corning, NY, United States), at a density of 1 × 105 cells per well. Cells were transfected with 1 μg base editor plasmid (hA3A-BE3, Target-AID-NG or hA3A-BE3-NG), 500 ng pGL3-U6-sgRNA-PGK-puromycin, and 50 ng pCMV-GFP (11153#; Addgene, Watertown, MA, United States) per well, using Lipofectamine LTX (Life Technologies, Gaithersburg, MD, United States) according to the manufacturer’s recommended protocol. HEK293T cells were cultured at 37°C with 5% of CO2.
Porcine fetal fibroblast (PFF) cells were isolated from 35-day-old fetuses of Bama pigs. A day before transfection, PFF cells were thawed and cultured in the Minimum Essential Medium (MEM Alpha; Gibco, Grand Island, NY, United States), supplemented with 15% FBS (v/v; HyClone, Logan, Utah, United States), 1% nonessential amino acids (NEAA; v/v; Gibco, Grand Island, NY, United States), 2 mM GlutaMAX (Gibco, Grand Island, NY, United States), and bFGF (Life Technologies, Gaithersburg, MD, United States). PFF cells were seeded 1 day prior to transfection in 6-well plates (Corning, Corning, NY, United States). Four microgram base editor vector (hA3A-BE3 or hA3A-BE3-NG) and 2.73 μg pGL3-U6-sgRNA-tdTomato were co-transfected into 5 × 105 PFF cells by nucleofection with Lonza/Amaxa Nucleofector 2B (Lonza, Basel, Switzerland) according to the manufacturer’s recommended protocol. Cells were harvested approximately 48 h post-transfection. PFF cells were cultured at 38.5°C with 5% of CO2.
Fluorescence-Activated Cell Sorting
HEK293T and PFF cells were harvested and subjected to flow cytometry 48 h after transfection. A total of 10,000 cell events were collected and analyzed using FlowJo software. Single PFF cell with positive signal was seeded into 96-cell plates and cultured for 8 days to form colonies.
Base Editing Analysis and Single Cell Line Genotyping
Genomic DNA of HEK293T and PFF cells was extracted using One Step Mouse GenoTyping Kit (Vazyme, Nanjing, China). The cell lysate was then used as the PCR template. PCR fragments for Sanger sequencing were generated in one step PCR reaction. The editing efficiency was analyzed by an online tool, EditR 1.0.9.1 The primers are listed in Supplementary Table S3.
Reverse Transcription-PCR
Total RNA was extracted from cultured cells by using TRIzol reagent (Invitrogen, Carlsbad, CA, United States), according to manufacturer’s protocol. Complementary DNA (cDNA) was generated by using Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, United States). The PCR reaction with 25 ng cDNA template was performed for 30 cycles. The housekeeping gene, GAPDH, was used as an internal control. Relative expression of Cas9 was detected by gel electrophoresis. All the primer sequences were shown in Supplementary Table S4.
Statistical Analysis
The statistical data are expressed as mean ± SEM, and at least three individual replicates were conducted in all experiments. Statistical significance was analyzed with unpaired Student’s t-tests using GraphPad prism software 6.0 (GraphPad Prism, La Jolla, CA, United States). A value of p < 0.05 was considered statistically significant.
Results
The Successful Construction of hA3A-BE3-NG Targeting Plasmid
The targetable scope of traditional BEs was restricted for the conventional SpCas9 preferred to recognize the target loci with NGG PAM. Approximately 205,013,891 NGG and CCN sites exist in the pig genome, which accounts for only 8.27% of the total genome sites (Figure 1A) within the approximately 2,478,444,698 base pairs estimated by Sscrofa11.1 assembly (Li et al., 2017). Overall, the percentage of NGN and NCN sites in the porcine genome was about 33.04%, which is four times higher than that of NGG and CCN sites (Figure 1A). To expand the targeting scope of hA3A-BE3, we fused SpCas9-NG with hA3A-BE3 to generate a new BE named hA3A-BE3-NG by in-fusion strategy (Figure 1B and Supplementary Figure S1). Our construct incorporated three fragments: a restriction fragment of 5,570 bp digested by BsrGI and PmeI from hA3A-BE3, and two PCR fragments amplified from hA3A-BE3 and Target-AID-NG, respectively (Figures 1B,C). The successful construction of the vector was confirmed by PCR, gel electrophoresis (Figure 1C), and Sanger sequencing (Figure 1D).
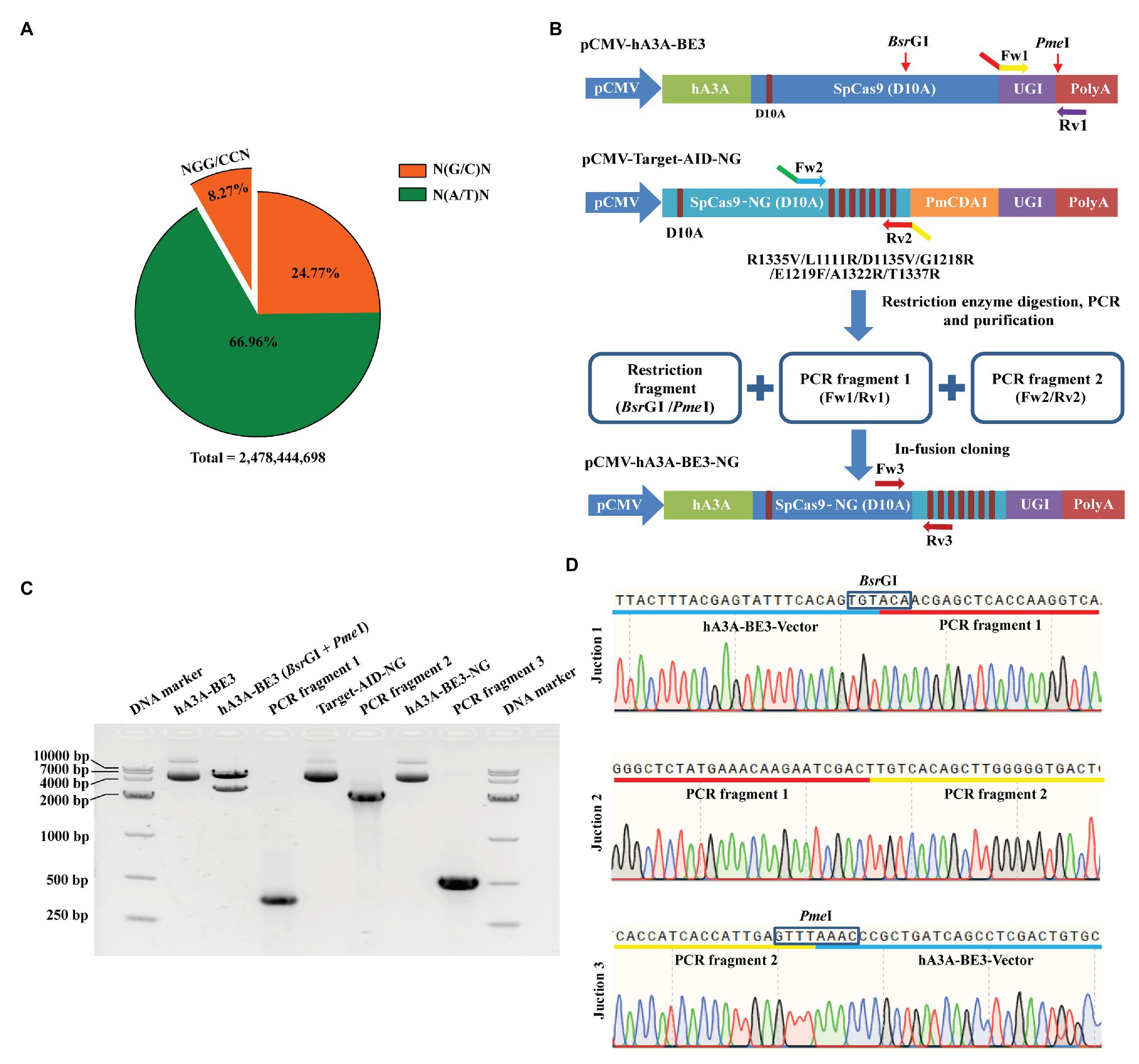
Figure 1. Construction of hA3A-BE3-NG vector for expanded targeting scope. (A) Pie chart shows the proportion of porcine genomic sites that can be targeted by SpCas9 or SpCas9-NG with distinct protospacer adjacent motif (PAM) specificities (NGG or NG). Pig reference genome (Sscrofa11.1) was used for analysis. (B) Schematic of the pCMV-hA3A-BE3-NG vector. Compared to SpCas9 (D10A) in hA3A-BE3, SpCas9-NG (D10A) in Target-AID-NG contained seven amino acids variants: R1335V, L1111R, D1135V, G1218R, E1219F, A1322R, and T1337R. hA3A-BE3-NG was constructed by in-fusion cloning of a restriction fragment digested via BsrGI and PmeI from hA3A-BE3, PCR fragment 1 amplified from hA3A-BE3 via Fw1/Rv1 primers, and PCR fragment 2 amplified from Target-AID-NG via Fw2/Rv2 primers. Overlapping sequences exist in the junction of the three different fragments. (C) The gel image indicates that hA3A-BE3 was digested into two fragments by BsrGI and PmeI. The PCR fragment 1 (387 bp) was amplified from hA3A-BE3 via Fw1/Rv1 primers, and the PCR fragment 2 (2,556 bp) was amplified from Target-AID-NG via Fw2/Rv2 primers. The large fragment (5,570 bp) from hA3A-BE3, PCR fragment 1 and PCR fragment 2 were fused into a recombinant vector, hA3A-BE3-NG, which was confirmed by a PCR product (540 bp) amplified via Fw3/Rv3 primers. (D) The chromatograms of Sanger sequencing show the junctional sequence was accurate among the above three fragments in recombinant hA3A-BE3-NG.
hA3A-BE3-NG-Mediated Gene Editing at NGN PAM Sites in Human Cells
One study revealed that Target-AID-NG was another superior base editor for introducing C-to-T conversion at NGN PAM sites efficiently in human cells (Nishimasu et al., 2018). To further validate the editing capacity of hA3A-BE3-NG, fused with different cytosine deaminase, four sgRNAs that targeted AGA, GGT, GGG, and AGC PAMs sites in human empty spiracles homeobox 1 (EMX1) loci were designed. The hA3A-BE3, hA3A-BE3-NG, or Target-AID-NG plasmid were co-transfected with sgRNAs‐ and GFP-expressing plasmids into HEK293T cells, respectively. All GFP-positive cells (no less than 25% of total cells) were isolated via flow cytometry for further characterization (Supplementary Figures S2A,B). The expression of hA3A-BE3-NG was confirmed by reverse transcription PCR (RT-PCR) in 48 h post-transfected HEK293T cells (Supplementary Figure S2C). Mutation frequencies by different BEs at NGN PAM sites were quantified using Sanger sequencing and EditR software (Figures 2A–C and Supplementary Figure S2D). Results showed that hA3A-BE3-NG achieved a C-to-T editing frequency of at least 15% at AGA and GGT PAM sites when compared with the mutation frequency of hA3A-BE3 showed less than 5% (Figures 2A,B and Supplementary Figure S2D). The hA3A-BE3-NG induced slightly lower C-to-T conversion, compared to those of hA3A-BE3 (14.27 vs. 19.00%) in the activity window (C3, C4, C5, C6, and C12) at GGG PAM (Figure 2A and Supplementary Figure S2D). This is in line with a recently observed phenomenon, SpCas9-NG shows slightly reduced activity at NGG PAM sites in human cells (Nishimasu et al., 2018). In addition, hA3A-BE3-NG also showed relatively low conversion efficiency at AGC PAM sites (Figure 2A and Supplementary Figure S2D), which is consistent with a previous report (Nishimasu et al., 2018). Overall, hA3A-BE3-NG-mediated C-to-T conversion was more efficient than hA3A-BE3 at NGH PAM sites (21.27 vs. 2.81% at average; Figure 2C). In addition, Target-AID-NG showed efficient editing of C3 and C4 at GGG PAM site (Figure 2A and Supplementary Figure S2D), highlighting differences in editing windows, base preference, and efficiencies between hA3A‐ and PmCDA1-derived BEs. However, hA3A-BE3-NG achieved a higher mutation frequency than that of Target-AID-NG at AGA and GGT PAM sites (Figures 2A,B and Supplementary Figure S2D), indicating that hA3A-BE3-NG could be considered a more efficient BE with an expanded targetable scope for gene editing in the mammalian genome.
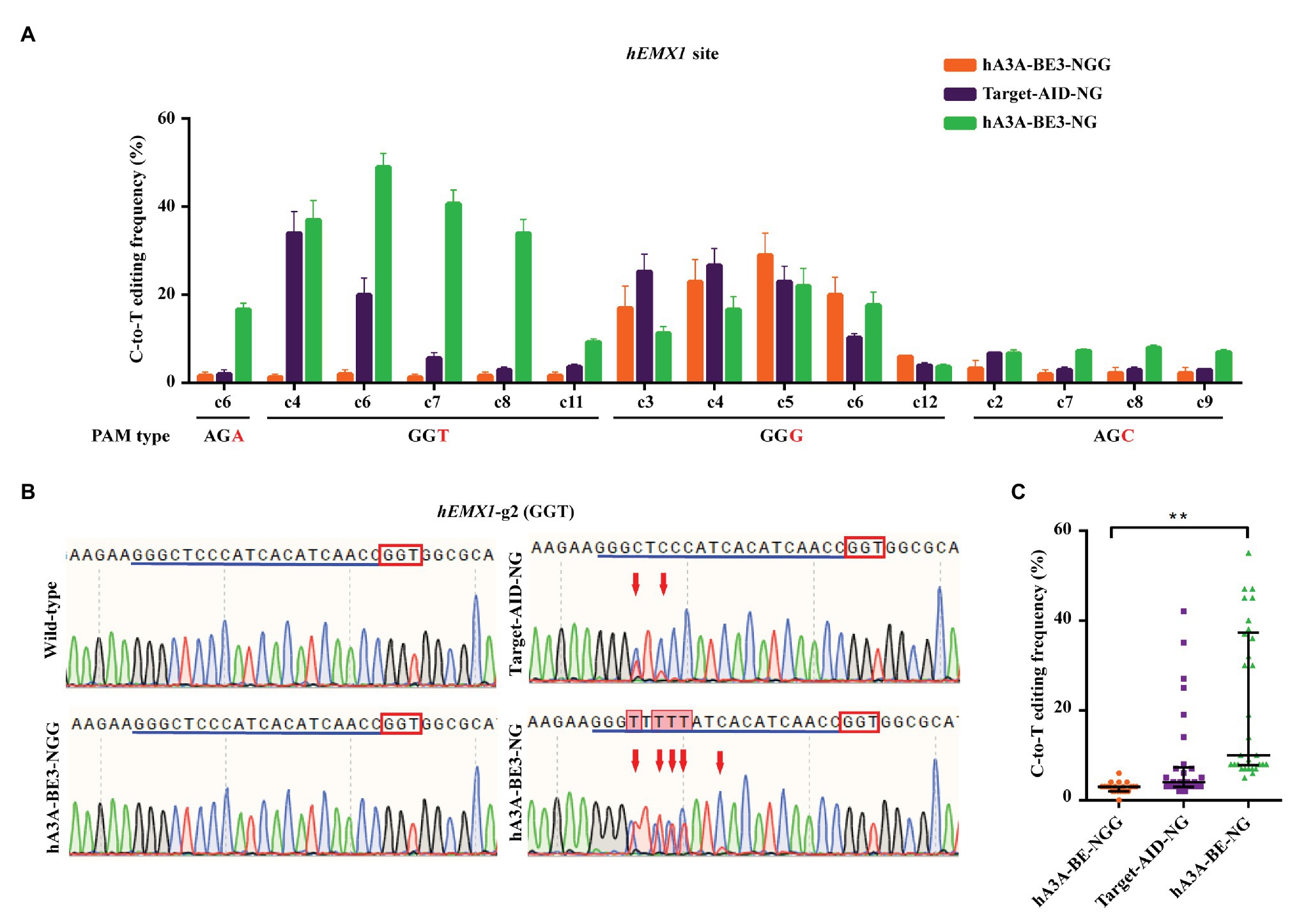
Figure 2. Efficient C-to-T conversion in human HEK293T cells by hA3A-BE3-NG. (A) C-to-T editing by hA3A-BE3, hA3A-BE3-NG, and Target-AID-NG at four endogenous EMX1 gene sites in human HEK293T cells. The target base in the editing window is shown, counting the end distal to the PAM as position 1. Data are represented as the mean ± SEM (n = 3). (B) Sanger sequencing results of HEK293T cells transfected with hA3A-BE3, hA3A-BE3-NG, or Target-AID-NG. Red boxes indicate PAMs and blue lines indicate sgRNA sequences. Red arrows indicate substituted nucleotides. (C) Statistical analysis of the C-to-T editing frequency induced by hA3A-BE3, hA3A-BE3-NG, or Target-AID-NG at NGH PAM sites in (A). The median and interquartile range (IQR) are shown; ** p < 0.01.
Expanded Editable Scope Through hA3A-BE3-NG to Generate Premature Terminal Codon in PFF Cells
Stop codons (TAG, TGA, or TAA) could be produced by a C-to-T conversion of the CAG, CGA, or CAA codons on the sense strand and the G-to-A conversion of the TGG codon caused by C-to-T mutation on the anti-sense strand (Figure 3A). The loss of function mutation in various genes was reported to confer the elite traits in pigs, such as CD163 gene for PRRSVs resistant (Whitworth et al., 2016; Burkard et al., 2017; Wells et al., 2017), APN gene for TGEVs resistant (Luo et al., 2019; Whitworth et al., 2019; Zhang et al., 2019), and MSTN for increased lean meat production (Qian et al., 2015; Wang K, et al., 2015). Thus, we explore the possibility and editing efficiency of inducing stop codons over these loci at expanded targetable sites by hA3A-BE3-NG in pigs. We designed a total of 32 sgRNAs (A1–19, C1–7, and M1–6) with NGN PAM in porcine CD163, APN, and MSTN genomic loci. Of the 32 sgRNAs, 28 (A2–18, C2–7, and M1–5) could produce premature terminal codons in the targeted activity windows if C-to-T conversion occurs (positions 2–13, counting the PAM as positions 21–23; Figure 3B). We firstly evaluated hA3A-BE3-NG-mediated editing efficiency on 32 NGN PAM sites (Supplementary Figure S3A). hA3A-BE3-NG showed comparable activity to hA3A-BE3 at 6 NGG PAM sites (A2, A6, A11, A13, A15, and C6) and reduced activity at 2 NGG PAM sites (A3 and M3), suggesting that hA3A-BE3-NG was also a useful BE at NGG PAM sites in pigs (Figure 3C and Supplementary Figure S3B). With NGH PAM sites, hA3A-BE3-NG showed at least a 3% mutation frequency at 21 of the 24 sites and at least a 10% mutation frequency at half of the 21 sites (Figure 3C and Supplementary Figure S3C). By contrast, hA3A-BE3 only edited the AGA PAM site (M4) with a low mutation frequency of 4% and had no efficiency at other 23 NGH PAM sites (Figure 3C). Interestingly, as shown in Figure 3C, hA3A-BE3-NG was editing ineffective at modifying TGA PAM sites (M1 and M2), which might be resulted from sequence signatures and nucleotide preferences (Xue et al., 2019). In brief, compared with hA3A-BE3 that induced C-to-T conversion efficiently at NGG PAM sites, hA3A-BE3-NG showed efficient editing at a variety of PAM sites (Figures 3C,D and Supplementary Figures S3B,C). Particularly, at 25 of 28 target sites that sgRNAs could generate premature stop codons to knockout target genes, hA3A-BE3-NG achieved detectable C-to-T mutation frequency if 3% was used as the cutoff threshold. By contrast, only 9 of these 28 sgRNAs were functional with hA3A-BE3 (Figure 3C). Besides the above three genes that was designed to induce loss-of-function mutations, we further exploited to introduce a beneficial SNP (c.893 G>A) into MC4R gene that was reported to be a marker for decreased fat deposition trait (Kim et al., 2000; Schroyen et al., 2015). The sgRNA was designed on the reverse strand of TGA PAM site, positioning the targeted cytosine in the activity window of hA3A-BE3-NG to produce MC4R c.893G>A on the sense strand (Figure 3E). hA3A-BE3-NG mediated higher mutation frequency than hA3A-BE3 (21.67 vs. 10.33%; Figure 3F and Supplementary Figure S3D).
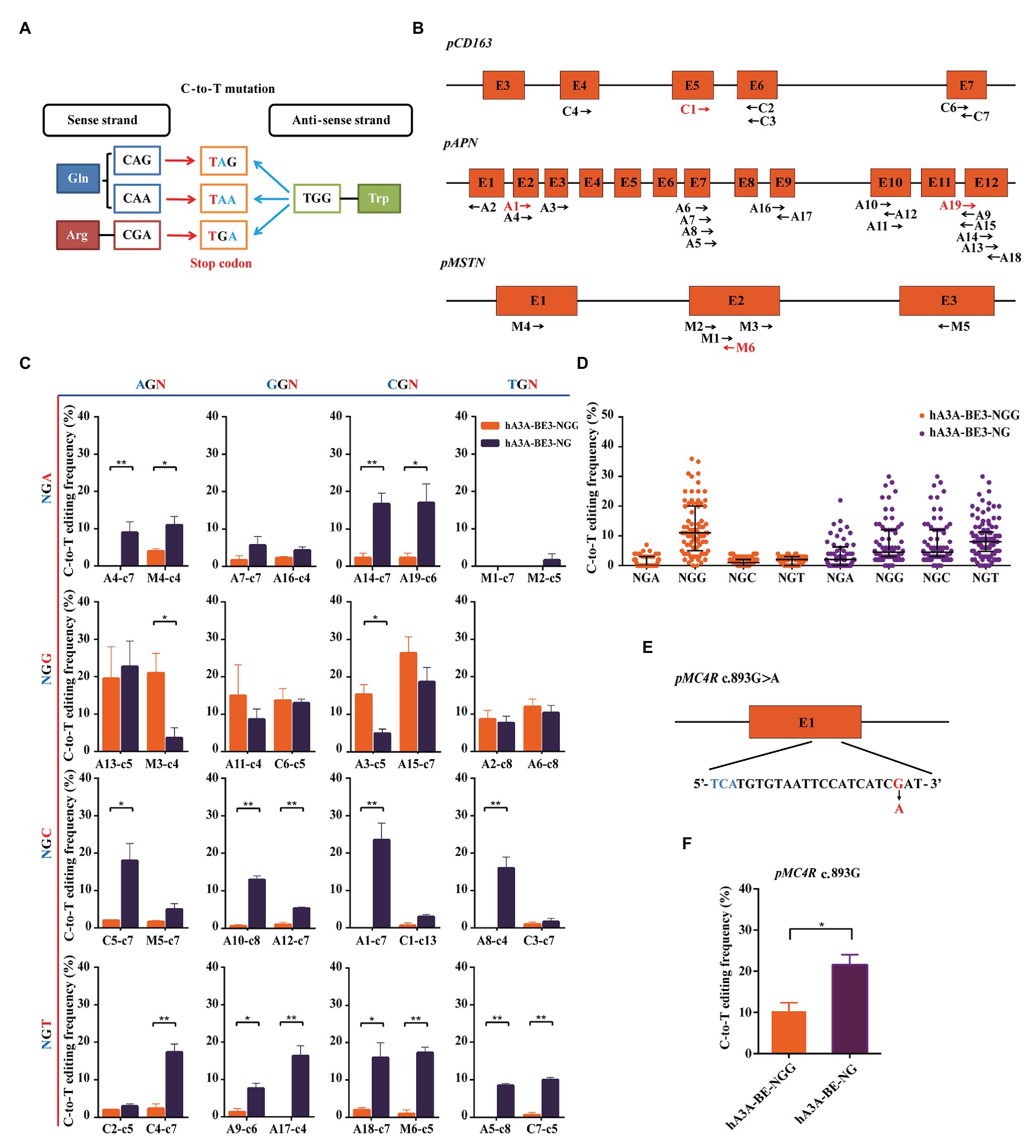
Figure 3. Precision missense mutation using hA3A-BE3-NG to expand the editing scope in porcine fetal fibroblast (PFF) cells. (A) Representation of the C-to-T conversion induced by base editors to generate stop codons. The base editors convert CAA, CAG, and CGA codons to stop codons (red) in the sense strand. The TGG codon is converted to stop codons (blue) through G-to-A conversion. (B) Schematic of the target sites at the porcine CD163, APN, and MSTN loci. The target sites indicated by the black arrows can generate stop codons using base editors (BEs). The forward direction of arrow indicates sgRNA-matched anti-sense strand, and vice versa. Total of 32 sgRNAs were designed (A1–19, C1–7, and M1–6). (C) Base editing at 32 NGN PAM sites by hA3A-BE3 and hA3A-BE3-NG. The target sites covered all 16 possible NGN PAM combinations, counting the end distal to the PAM as position 1. (D) Statistical analysis of the C-to-T editing frequency induced by hA3A-BE3 or hA3A-BE3-NG at a total of 32 endogenous target sites. The median and IQR are shown. (E) Schematic of the target site at MC4R locus. MC4R c.893G>A could be produced by hA3A-BE3-NG. The PAM sequence and substituted base are shown in blue and red, respectively. (F) Base editing at the MC4R locus by hA3A-BE3 and hA3A-BE3-NG. In (C,F) values were shown as mean ± SEM (n = 3); ** p < 0.01 and * p < 0.05.
hA3A-BE3-NG-Mediated Base Editing in Multiple Loci
In livestock, most of the economic traits were considered to be regulated by a massive number of SNPs in various genes (Song et al., 2020). Thus, the ability to create precise and multiple genetic modification in various loci across the pig genome simultaneously is necessary for successful pyramid breeding. To investigate the feasibility of hA3A-BE3-NG for base editing in multiple loci, we simultaneously co-transfected hA3A-BE3-NG and sgRNAs-tdTomato-expressing plasmid that targeted APN, CD163, MC4R, and MSTN into PFF cells. After 48 h of transfection, tdTomato-positive single PFF cell was isolated and seeded into 96-cell plates via FACS, and then cultured for another 8 days to form single-cell colonies. A total of 54 colonies were obtained and genotyped by Sanger sequencing (Figures 4A,B). Results showed that 21 out of 54 (38.89%), 23 out of 54 (42.59%), 3 out of 54 (5.56%), and 25 out of 54 (46.30%) colonies had mutations in the APN, CD163, MC4R, and MSTN genes, respectively, and most of them had effective C-to-T conversion at the target sites (Figure 4A). Due to the wide activity window of hA3A-BE3-NG, we also found that a number of colonies had bystander mutations with C-to-T substitution existing in the vicinity of the targeted cytosine (35.19, 12.96, and 18.52% colonies in APN, CD163, and MSTN genes, respectively; Figure 4A). Moreover, 12 out of 54 (22.22%) colonies showed a proximal off-target mutation at position −4 (with the base distal from the PAM set as position 1) in CD163 (Figure 4A). Importantly, 35 out of 54 colonies had mutations, and therein, two single-cell colonies (3.70%, 2/54) showed targeted mutations of all four genes (APN, CD163, MSTN, and MC4R; Figures 4A,B). In addition, we identified 14 colonies (25.93%, 14/54) with triple-gene mutations, 8 (14.81%, 8/54) with double-gene mutations, and 11 (20.37%, 11/54) with single-gene mutation (Figures 4A,B).
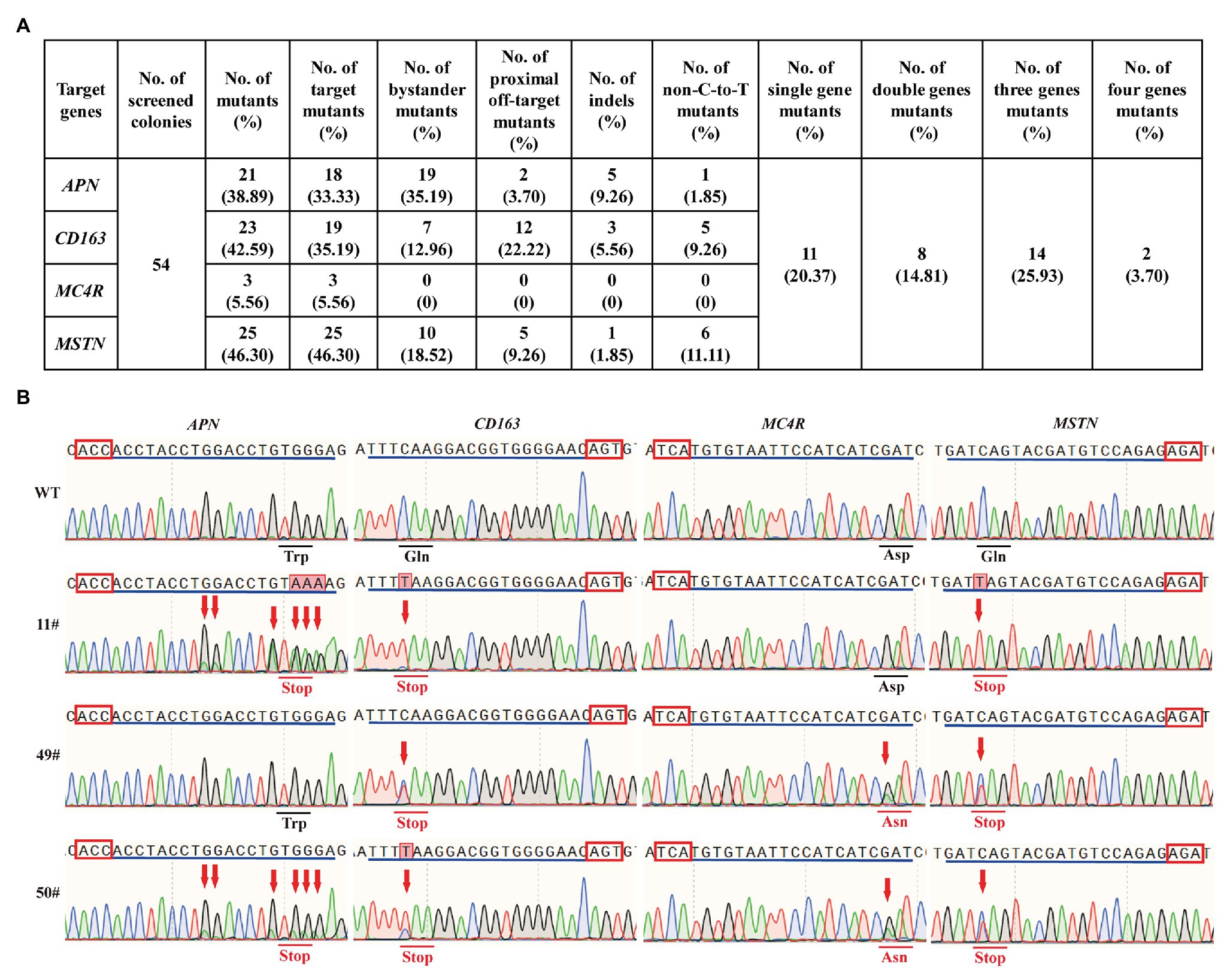
Figure 4. hA3A-BE3-NG-mediated base editing at multiple genes in PFF cells. (A) Summary of multiple sites base editing by hA3A-BE3-NG in PFF cells. (B) Sanger sequencing results of selected single-cell colonies. 11# and 49# colonies have mutations on three genes, and 50# colony has mutations on all four genes. The red box indicates the PAMs and the blue line indicates the sgRNA sequence. The red arrow indicates the substituted nucleotide. The amino acid in the red line indicates expected substitutions at target sites.
Discussion
Genome editing technologies have provided a revolutionary strategy for making genetic improvements in pig breeding. Compared to conventional cross breeding in livestock, the molecular breeding to accurately modify the agriculture-related functional genes will save a lot of time, money, and manpower. The focus of recent genome editing research is to modify the genome efficiently, accurately, and safely. In the current study, hA3A-BE3-NG was constructed and proved to be a powerful base editor to improve the editing efficiency and expand the targeting scope in pigs. It has been reported that, 20 endogenous target sites (including EMX1, VEGFA, GRIN2B, etc.) with different PAM have been used to compare C-to-T conversion efficiency between Target-AID and Target-AID-NG in HEK293T cells (Nishimasu et al., 2018). Therefore, we also selected the EMX1 targets from the study above to analyze the targeting efficiency of hA3A-BE3, Target-AID-NG, and hA3A-BE3-NG in the current study. Here, hA3A-BE3-NG was confirmed to show editing activity comparable with or even higher than Target-AID-NG at the four target sites in human cells. And it could induce C-to-T mutation in a broader activity window in human and porcine cells efficiently, which is consistent with a previous study that hA3A-BE3 had an approximately 12 nucleotides activity window (Wang et al., 2018).
To avoid potential chimeric issues and long-time frame of breeding, the generation of genetically modified large animals was mostly created by genome editing technology combined with somatic cell nuclear transfer (SCNT) instead of embryo injection (Zhao et al., 2019). So how to obtain the cell colonies with desired modification efficiently is one of the key steps. Here, we found that hA3A-BE3-NG could induce C-to-T conversion efficiently not only at NGG PAM sites as hA3A-BE3 but also exhibited expand targeting scope at NGN PAM sites. For the MC4R c.893G>A mutation, hA3A-BE3-NG showed more efficient than hA3A-BE3 at the TGA PAM site (21.67 vs. 10.33% at average). Thus, applications of hA3A-BE3-NG could expand the editing scope at NGN PAM sites, possibly facilitating breeding improvements in pigs.
With the development of functional genomics and bioinformatics, more and more SNPs responsible for economic traits have been identified in livestock (Song et al., 2020). And many economic traits are majorly controlled or orchestrated by combinations of SNPs. Therefore, it is of importance to create precise and multiple genome-editing livestock for exploring the function of SNPs and evaluate their potential breeding value. In addition, the potential of chromosomal structural abnormalities would increase when multiplex target loci were cut simultaneously by conventional CRISPR-Cas systems, causing genomic instability, chromosome elimination, and even cell death (Wang T, et al., 2015; Aguirre et al., 2016; Zuo et al., 2017). BEs provided a safe strategy to edit multiple gene sites efficiently and accurately without DSBs. Recently, the multiplex base editing was accomplished by BE3 at NGG PAM sites in pigs (Xie et al., 2019; Yuan et al., 2019). Using BE3 and hA3A-BE3, Xie et al. (2019) simultaneously mutated the porcine RAG1, RAG2, and IL2RG or DMD, TYR, and LMNA triple gene in PFF cells with high efficiency, and subsequently generated a triple gene knockout pig model with immunodeficiency for applications in regenerative medicine. Yuan et al. (2019) prepared GGTA1/B4GAlNT2/CMAH triple gene knockout pigs which could be used as organ donors for xenotransplantation by BE4-Gam. hA3A-BE3-NG could simultaneously introduce targeted mutations at multiple sites of four genes, APN, CD163, MSTN, and MC4R in PFF cells, suggesting the great potential of hA3A-BE3-NG in animal pyramid breeding.
Previous studies have suggested that CBEs could cause DNA off-target effects in mouse embryos and plants (Jin et al., 2019; Zuo et al., 2019); however, BE variants are continuously being improved and exploited to improve targeted specificity (Doman et al., 2020). In this current study, bystander and proximal off-target mutations were also found at APN, CD163, and MSTN gene sites, resulting from the wide editing window of hA3A-BE3-NG. Some engineered precise hA3A variants have been developed to reduce bystander mutations such as hA3A-Y130F via narrowing the width of the editing window and eA3A (hA3A-N57G) according to the preferential target base motif (Gehrke et al., 2018; Wang et al., 2018; Liu et al., 2020a). These off-target effects are less crucial when using base editing to introduce premature terminal codons, generating loss-of-function mutations and inactivating protein function. In agricultural breeding, the unpredicted editing byproducts through BEs might be more tolerated and could provide a new source of mutations with favorable economic characteristics. Recently, it has been reported that some new engineering variant of the Cas9, SpRY, which is free of PAM restriction (Walton et al., 2020). In the future, the combine of BEs and the new Cas9 variant will further expand the editing scope to improve base editing tools for pyramid breeding and genetic improvement in livestock.
In summary, we generated hA3A-BE3-NG, a versatile CBEs, that substantially expands the scope and capability of base editing at NGN PAM sites. To our knowledge, this is the first study to precisely edit multiple genes responsible for economic traits in the porcine genome using BEs, suggesting the incredible potential of using BEs to accelerate molecular pyramid breeding in livestock.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material and further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by Laboratory Animal Welfare and Ethics Committee Institute of Zoology, Chinese Academy of Sciences.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This research was funded by the National Natural Science Foundation of China (31672387, 31925036, and 31601008), the China National Key R&D Program (2020YFC1316600), the National Transgenic Project of China (2016ZX08009003-006-007), and the Agricultural Science and Technology Innovation Program of CAAS (ASTIP-IAS05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Xingxu Huang (Shanghai Tech University, China) for providing the hA3A-BE3 plasmid and Hao Yin (Wuhan University, China) for providing the Target-AID-NG plasmid.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.592623/full#supplementary-material
Footnotes
References
Aguirre, A. J., Meyers, R. M., Weir, B. A., Vazquez, F., Zhang, C. Z., Ben-David, U., et al. (2016). Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 6, 914–929. doi: 10.1158/2159-8290.CD-16-0154
Billon, P., Bryant, E. E., Joseph, S. A., Nambiar, T. S., Hayward, S. B., Rothstein, R., et al. (2017). CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell 67:1068.e1064–1079.e1064. doi: 10.1016/j.molcel.2017.08.008
Burkard, C., Lillico, S. G., Reid, E., Jackson, B., Mileham, A. J., Ait-Ali, T., et al. (2017). Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 13:e1006206. doi: 10.1371/journal.ppat.1006206
Doman, J. L., Raguram, A., Newby, G. A., and Liu, D. R. (2020). Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628. doi: 10.1038/s41587-020-0414-6
Endo, M., Mikami, M., Endo, A., Kaya, H., Itoh, T., Nishimasu, H., et al. (2019). Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat. Plants 5, 14–17. doi: 10.1038/s41477-018-0321-8
Fujii, W., Ito, H., Kanke, T., Ikeda, A., Sugiura, K., and Naito, K. (2019). Generation of genetically modified mice using SpCas9-NG engineered nuclease. Sci. Rep. 9:12878. doi: 10.1038/s41598-019-49394-5
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi: 10.1038/nature24644
Gehrke, J. M., Cervantes, O., Clement, M. K., Wu, Y., Zeng, J., Bauer, D. E., et al. (2018). An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 36, 977–982. doi: 10.1038/nbt.4199
Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., et al. (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. doi: 10.1038/nature26155
Huang, S., Liao, Z., Li, X., Liu, Z., Li, G., Li, J., et al. (2019). Developing ABEmax-NG with precise targeting and expanded editing scope to model pathogenic splice site mutations in vivo. Science 15, 640–648. doi: 10.1016/j.isci.2019.05.008
Jin, S., Zong, Y., Gao, Q., Zhu, Z., Wang, Y., Qin, P., et al. (2019). Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295. doi: 10.1126/science.aaw7166
Kim, Y. B., Komor, A. C., Levy, J. M., Packer, M. S., Zhao, K. T., and Liu, D. R. (2017). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35, 371–376. doi: 10.1038/nbt.3803
Kim, K. S., Larsen, N., Short, T., Plastow, G., and Rothschild, M. F. (2000). A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm. Genome 11, 131–135. doi: 10.1007/s003350010025
Kleinstiver, B. P., Sousa, A. A., Walton, R. T., Tak, Y. E., Hsu, J. Y., Clement, K., et al. (2019). Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282. doi: 10.1038/s41587-018-0011-0
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Kuscu, C., Parlak, M., Tufan, T., Yang, J., Szlachta, K., Wei, X., et al. (2017). CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods 14, 710–712. doi: 10.1038/nmeth.4327
Li, M., Chen, L., Tian, S., Lin, Y., Tang, Q., Zhou, X., et al. (2017). Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies. Genome Res. 27, 865–874. doi: 10.1101/gr.207456.116
Li, X., Wang, Y., Liu, Y., Yang, B., Wang, X., Wei, J., et al. (2018). Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 36, 324–327. doi: 10.1038/nbt.4102
Li, C., Zhang, R., Meng, X., Chen, S., Zong, Y., Lu, C., et al. (2020). Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38, 875–882. doi: 10.1038/s41587-019-0393-7
Liu, Z., Chen, S., Shan, H., Jia, Y., Chen, M., Song, Y., et al. (2020a). Precise base editing with CC context-specificity using engineered human APOBEC3G-nCas9 fusions. BMC Biol. 18:111. doi: 10.1186/s12915-020-00849-6
Liu, Z., Chen, S., Shan, H., Zhang, Q., Chen, M., Lai, L., et al. (2019). Efficient and precise base editing in rabbits using human APOBEC3A-nCas9 fusions. Cell Discov. 5:31. doi: 10.1038/s41421-019-0099-5
Liu, Z., Shan, H., Chen, S., Chen, M., Song, Y., Lai, L., et al. (2020b). Highly efficient base editing with expanded targeting scope using SpCas9-NG in rabbits. FASEB J. 34, 588–596. doi: 10.1096/fj.201901587R
Luo, L., Wang, S., Zhu, L., Fan, B., Liu, T., Wang, L., et al. (2019). Aminopeptidase N-null neonatal piglets are protected from transmissible gastroenteritis virus but not porcine epidemic diarrhea virus. Sci. Rep. 9:13186. doi: 10.1038/s41598-019-49838-y
Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., et al. (2018). Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262. doi: 10.1126/science.aas9129
Qian, L., Tang, M., Yang, J., Wang, Q., Cai, C., Jiang, S., et al. (2015). Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci. Rep. 5:14435. doi: 10.1038/srep14435
Rees, H. A., and Liu, D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. doi: 10.1038/s41576-018-0059-1
Schroyen, M., Janssens, S., Stinckens, A., Brebels, M., Bertolini, F., Lamberigts, C., et al. (2015). The MC4R c.893G>A mutation: a marker for growth and leanness associated with boar taint odour in Belgian pig breeds. Meat Sci. 101, 1–4. doi: 10.1016/j.meatsci.2014.08.009
Song, R., Wang, Y., Wang, Y., and Zhao, J. (2020). Base editing in pigs for precision breeding. Front. Agric. Sci. Eng. 7, 161–170. doi: 10.15302/j-fase-2019308
Thuronyi, B. W., Koblan, L. W., Levy, J. M., Yeh, W. H., Zheng, C., Newby, G. A., et al. (2019). Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 37, 1070–1079. doi: 10.1038/s41587-019-0193-0
Walton, R. T., Christie, K. A., Whittaker, M. N., and Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296. doi: 10.1126/science.aba8853
Wang, T., Birsoy, K., Hughes, N. W., Krupczak, K. M., Post, Y., Wei, J. J., et al. (2015). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101. doi: 10.1126/science.aac7041
Wang, X., Li, J., Wang, Y., Yang, B., Wei, J., Wu, J., et al. (2018). Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 36, 946–949. doi: 10.1038/nbt.4198
Wang, Y., Liu, Y., Li, J., Yang, Y., Ni, X., Cheng, H., et al. (2019). Expanding targeting scope, editing window, and base transition capability of base editing in Corynebacterium glutamicum. Biotechnol. Bioeng. 116, 3016–3029. doi: 10.1002/bit.27121
Wang, K., Ouyang, H., Xie, Z., Yao, C., Guo, N., Li, M., et al. (2015). Efficient generation of myostatin mutations in pigs using the CRISPR/Cas9 system. Sci. Rep. 5:16623. doi: 10.1038/srep16623
Wang, H., Shen, L., Chen, J., Liu, X., Tan, T., Hu, Y., et al. (2019). Deletion of CD163 exon 7 confers resistance to highly pathogenic porcine reproductive and respiratory viruses on pigs. Int. J. Biol. Sci. 15, 1993–2005. doi: 10.7150/ijbs.34269
Wells, K. D., Bardot, R., Whitworth, K. M., Trible, B. R., Fang, Y., Mileham, A., et al. (2017). Replacement of porcine CD163 scavenger receptor cysteine-rich domain 5 with a CD163-like homolog confers resistance of pigs to genotype 1 but not genotype 2 porcine reproductive and respiratory syndrome virus. J. Virol. 91, e01521–e01516. doi: 10.1128/JVI.01521-16
Whitworth, K. M., Rowland, R. R., Ewen, C. L., Trible, B. R., Kerrigan, M. A., Cino-Ozuna, A. G., et al. (2016). Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22. doi: 10.1038/nbt.3434
Whitworth, K. M., Rowland, R. R. R., Petrovan, V., Sheahan, M., Cino-Ozuna, A. G., Fang, Y., et al. (2019). Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic Res. 28, 21–32. doi: 10.1007/s11248-018-0100-3
Xie, J., Ge, W., Li, N., Liu, Q., Chen, F., Yang, X., et al. (2019). Efficient base editing for multiple genes and loci in pigs using base editors. Nat. Commun. 10:2852. doi: 10.1038/s41467-019-10421-8
Xue, L., Tang, B., Chen, W., and Luo, J. (2019). Prediction of CRISPR sgRNA activity using a deep convolutional neural network. J. Chem. Inf. Model. 59, 615–624. doi: 10.1021/acs.jcim.8b00368
Yuan, H., Yu, T., Wang, L., Yang, L., Zhang, Y., Liu, H., et al. (2019). Efficient base editing by RNA-guided cytidine base editors (CBEs) in pigs. Cell. Mol. Life Sci. 77, 719–733. doi: 10.1007/s00018-019-03205-2
Zhang, J., Wu, Z., and Yang, H. (2019). Aminopeptidase N knockout pigs are not resistant to porcine epidemic diarrhea virus infection. Virol. Sin. 34, 592–595. doi: 10.1007/s12250-019-00127-y
Zhao, J., Lai, L., Ji, W., and Zhou, Q. (2019). Genome editing in large animals: current status and future prospects. Natl. Sci. Rev. 6, 402–420. doi: 10.1093/nsr/nwz013
Zhong, Z., Sretenovic, S., Ren, Q., Yang, L., Bao, Y., Qi, C., et al. (2019). Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant 12, 1027–1036. doi: 10.1016/j.molp.2019.03.011
Zong, Y., Song, Q., Li, C., Jin, S., Zhang, D., Wang, Y., et al. (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. doi: 10.1038/nbt.4261 [Epub ahead of print]
Zuo, E., Huo, X., Yao, X., Hu, X., Sun, Y., Yin, J., et al. (2017). CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 18:224. doi: 10.1186/s13059-017-1354-4
Keywords: base editing, NGN PAM, hA3A-BE3-NG, multiple gene editing, pyramid breeding
Citation: Wang Y, Bi D, Qin G, Song R, Yao J, Cao C, Zheng Q, Hou N, Wang Y and Zhao J (2020) Cytosine Base Editor (hA3A-BE3-NG)-Mediated Multiple Gene Editing for Pyramid Breeding in Pigs. Front. Genet. 11:592623. doi: 10.3389/fgene.2020.592623
Edited by:
Zhanjun Li, Jilin University, ChinaCopyright © 2020 Wang, Bi, Qin, Song, Yao, Cao, Zheng, Hou, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfang Wang, d2FuZ3lhbmZhbmdAY2Fhcy5jbg==; Jianguo Zhao, emhhb2pnQGlvei5hYy5jbg==
 Yu Wang1
,2
Yu Wang1
,2
 Qiantao Zheng
Qiantao Zheng Jianguo Zhao
Jianguo Zhao