
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 19 November 2020
Sec. Evolutionary and Population Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.592595
UDP-glucuronosyltransferases (UGTs), being multifunctional detoxification enzymes, play a major role in the process of resistance to various pesticides in insects. However, the mechanism underlying the molecular regulation of pesticide resistance remains unclear, especially in Apis cerana cerana. In this study, all of the UGTs in Apis cerana cerana (AccUGT) have been identified through the multiple alignment and phylogenetic analysis. Expression of AccUGT genes under different pesticides, and antioxidant genes after silencing of AccUGT2B20-like, were detected by qRT-PCR. The resistance of overexpressed AccUGT2B20-like to oxidative stress was investigated by an Escherichia coli overexpression system. Also, antioxidant-related enzyme activity was detected after silencing of the AccUGT2B20-like gene. Expression pattern analysis showed that almost all UGT genes were upregulated under different pesticide treatments. This result indicated that AccUGTs participate in the detoxification process of pesticides. AccUGT2B20-like was the major gene because it was more highly induced than the others. Overexpression of AccUGT2B20-like in E. coli could effectively improve oxidative stress resistance. Specifically, silencing the AccUGT2B20-like gene increased oxidative stress by repressing the expression of oxidation-related genes, decreasing antioxidant-related enzyme activity, and increasing malondialdehyde concentration. Taken together, our results indicate that AccUGTs are involved in pesticide resistance, among which, AccUGT2B20-like contributes to the detoxification of pesticides by eliminating oxidative stress in Apis cerana cerana. This study explains the molecular basis for the resistance of bees to pesticides and provides an important safeguard for maintaining ecological balance.
The Apis cerana cerana (A. cerana cerana) is an important pollinator that plays a critical role in maintaining the balance of regional ecologies in China. However, recently, the survival of A. cerana cerana has been seriously threatened because of pressure from multiple sources, including excessive use of pesticides, heavy metal pollution, and epidemics of honeybee diseases (Gegear et al., 2006; Li et al., 2012). It appears certain that exposure to these multiple stressors is the driving force behind A. cerana cerana colony losses (Goulson et al., 2015). Among these stressors, pesticides are the most controversial and debated. Extensive research has resulted in the detection of 161 different pesticides in honey bee colonies, and bees are often chronically exposed to pesticides throughout their development and adult life (Marie-Pierre et al., 2006; Krupke et al., 2012; Sanchez-Bayo et al., 2014). Several insecticides, such as neonicotinoids, are neurotoxins that target the central nervous system of insects, bind to postsynaptic nicotinic acetylcholine receptors, and can result in overstimulation, paralysis, and death (Tomizawa and Casida, 2004).
Considerable evidence has revealed a strong correlation between the levels of oxidative stress, reactive oxygen species (ROS) generation, antioxidase, and various types of toxicity associated with pesticides (Wang et al., 2018a). Oxidative stress occurs as a result of inadequate antioxidant defense or overproduction of free radicals and is initiated by ROS (Adams et al., 2015; Swomley and Butterfield, 2015). Once the honeybees are affected by the pesticides, the pesticides may induce oxidative stress leading to ROS or reactive nitrogen species generation and related toxic effects. Oxidative stress can increase the antioxidant defense system and lead to damage of cellular macromolecules, such as DNA, lipids, and proteins (Weidinger and Kozlov, 2015). Following oxidative stress, cell death can occur via apoptosis or necrotic mechanisms. Normally, the body maintains a steady balance between ROS generation and scavenging, which results in a relatively low level of intracellular ROS. However, stress caused by pesticides disrupts homeostasis and leads to excessive production or accumulation of ROS. To defend against oxidative damage, organisms have evolved complex mechanisms to maintain cellular homeostasis (Zhang et al., 2013). UDP-glucuronosyltransferases (UGTs) are a part of critical detoxification mechanisms (Hasegawa et al., 2010; Ahn et al., 2012). However, whether UGTs are involved in the regulation of oxidative stress caused by pesticides remains unknown.
UGTs (EC 2.4.1.17), being the major enzyme in the reaction of glycosylation, catalyze the conjugation of a diverse range of small lipophilic compounds with sugars to produce glycosides (Bock, 2003; Lairson et al., 2008; Ahn et al., 2012). UGTs represent major phase II drug-metabolizing enzymes that process and detoxify a large number of important endogenous compounds, drugs, and environmental agents (Mazerska et al., 2016). UGTs function by transforming their substrates into more polar metabolites, which are easier to remove from the cell by export transporters (Mazerska et al., 2016). The structure of a protein determines its biological function. The majority of UGT proteins are in the endoplasmic reticulum lumen (ER) and are composed of two functional domains, the N-terminal and C-terminal domains (Meech and Mackenzie, 1998). The N-terminal aglycone substrate-binding domain has a signal peptide responsible for targeting the membrane to the ER compartment. The C-terminal transmembrane domain anchors the protein to the ER membrane (Magdalou et al., 2010). The C-terminal is more conserved than the N-terminal. The UGT motif signature, (FVA)-(LIVMF)-(TS)-(HQ)-(SGAC)-G-X(2)-(STG)-X(2)-(DE)-X(6)-P-(LIVMFA)-(LIVMFA)-X(2)-P-(LMVFIQ)-X(2)-(DE)-Q, is at the C-terminal of different UGTs (Mackenzie et al., 1997; Ahn et al., 2012). UGTs, as a superfamily, have evolved in all kingdoms of life. Although the structure and functions of UGTs have been investigated in different species, UGTs still have specificity in different species.
UGTs are ubiquitous in all free-living organisms, including bacteria, fungi, plants, and animals (Bock, 2016). As major phase II enzymes in a detoxification system, UGTs catalyze the conjugation of a sugar, donated by a UDP-glycoside, typically to a lipophilic molecule, generating water-soluble products that can be easily excreted, as well as stably managed. Therefore, they can protect the cellular system from being damaged by toxic foreign compounds and regulate internal molecules more easily (Bock, 2003). In humans, the substrates for phase II metabolic enzymes are not only xenobiotics but also many endogenous compounds that play crucial functions in organisms. UGTs are involved in the conjugation of a variety of xenobiotics, including the phytoalexin quercetin and plant flavonoids, which are significant antioxidants in the human diet (Achour et al., 2014). In the process of catalyzing endogenous compounds, UGT8A1 is involved in the conjugation of ceramides and bile acids with galactose (Meech et al., 2015). UGT3A1 and UGT3A2 play crucial roles in the conjugation with N-acetylglucosamine and glucose, respectively (Meech et al., 2012). Previous studies have indicated that the glycosylation of small lipophilic compounds has long been considered an enzymatic detoxification mechanism in insects (Brattsten, 1988; Després et al., 2007). Li X. et al. (2018) provided evidence that the expression level of UGTs in Plutella xylostella was highly induced by more than five insecticides, whereas indoxacarb and metaflumizone significantly repressed the expression of most UGT genes. In Aphis gossypii, inhibitors of UGT enzymes significantly increased the toxicity of thiamethoxam against the resistant strain, and suppression of UGT transcripts by RNAi increased mortality (Pan et al., 2018). GST-4 and UGT-13 expression could be induced by low concentrations of allyl isothiocyanate in nematodes. This suggests that allyl isothiocyanate might confer tolerance to the nematode against oxidative stress, and their lifespan was considerably extended under the paraquat treatment (Hasegawa et al., 2010). Although many studies have been conducted on the function of UGTs in animals, there are still few published data on insects. To date, no research has been conducted on A. cerana cerana. Therefore, understanding the role of UGTs in resistance against pesticides could provide a basis for studying the detoxification system and a better understanding of the molecular regulation mechanism of UGTs in A. cerana cerana (AccUGTs).
In this study, we aim to investigate the molecular mechanism of AccUGTs under different pesticide treatments. We identified 10 AccUGT genes based on genome data. The phylogenetic relationships and tissue-specific expression profiles of these UGT genes were analyzed. Based on the expression profiling of AccUGT genes under different pesticide treatments, the AccUGT2B20-like gene was selected as the critical gene. Overexpression of AccUGT2B20-like in E. coli enhanced resistance to oxidative tolerance. Silencing of AccUGT2B20-like in adult bees reduced their ability to regulate oxidative stress. These results broadened our knowledge regarding the biological roles of UGTs in pesticide resistance and significantly improved our understanding of the relationship between UGTs and oxidation resistance in A. cerana cerana.
A. cerana cerana used in this study were obtained from the experimental base of Shandong Agricultural University (Tai’an, China). Fifteen-day-old worker bees were collected from the outdoor hives by marking newborn worker bees with paint 15 days before collection. Experimental honeybees were randomly placed into wooden cages, and maintenance was performed as previously described (Wang et al., 2014). The bees were divided into eight groups (n = 50/group). Each group was injected with 1 μl of different pesticides between the second and third abdominal segments of the bees. The pesticides included paraquat (0.6 mg/L), DDV (0.775 mg/L), acetamiprid (0.016 mg/L), lambda-cyhalothrin (0.012 mg/L), emamectin-benzoate (0.003 mg/L), mancozeb (0.002 mg/L), and abamectin (0.003 mg/L). The oxidative stress treatment consisted of H2O2 (0.5 μl/ml). Honeybees were collected at 0, 0.5, 1, 1.5, 2, or 2.5 h after treatment. Honeybees treated at 0 h were used as controls. The treated samples were immediately snap-frozen in liquid nitrogen and stored at −80°C.
For tissue-specific expression profile analysis, different kinds of tissues (head, antenna, muscle, feet, midgut, rectum, and cuticula) were dissected from 15-day-old worker bees on ice, frozen in liquid nitrogen, and stored at −80°C for total RNA isolation.
Total RNA for first-strand complementary DNA (cDNA) synthesis was extracted from experimental honeybees using the method described by Wang (Wang et al., 2018b). RNA was reverse transcribed into cDNA using 5 × All-In-One RT MasterMix with the AccuRT Genomic DNA Removal Kit (Applied Biological Materials Inc., Richmond, British Columbia, Canada), which facilitated the elimination of contaminating genomic DNA from RNA. qRT-PCR was performed using SYBR Premix Ex Taq (Takara) in 20-μl reaction volumes on a CFX96TM Real-time Detection System (Bio-Rad). The qRT-PCR program was set according to the manufacturer’s instructions (Wang et al., 2017). The 2–ΔΔCT method was used to determine the relative expression levels. The housekeeping gene, β-actin (No. HM640276), was used as an internal control to quantify relative transcript levels, which is stably expressed and has been chosen as the reference for many species, including A. cerana cerana (Lourenço et al., 2008). Each qRT-PCR experiment was repeated three times, with triplicates for each individual sample (n = 9). The sequences of the primers used for qRT-PCR are indicated in Supplementary Table 1.
The UGT mRNAs of A. cerana cerana were obtained from the NCBI database1 using BLAST searches according to the UGT gene motifs (Supplementary Table 2). Multiple alignments of the AccUGT proteins were generated using DNAMAN version 7.2 The conserved motifs were analyzed using MEME.3 The UGT amino acid sequences of Plutella xylostella (P. xylostella) (Li X. et al., 2018), Drosophila melanogaster (D. melanogaster) (Ahn et al., 2012), Helicoverpa armigera (H. armigera) (Ahn et al., 2012), and Bombyx mori (B. mori) (Huang et al., 2008) were recognized from previous studies and downloaded from the NCBI database. The amino acid sequences of UGT genes from different species are given in Supplementary Table 3. Amino acid sequences of UGTs from A. cerana cerana and five other insects were analyzed via ClustalW alignment using MEGA7 software.4 The alignment results were used to build a consensus phylogenetic tree using the neighbor-joining method. A total of 1,000 bootstrap replications were performed, and branches with bootstrap values above 50% are indicated.
To silence the AccUGT2B20-like gene, double-stranded RNAs (dsRNA) were synthesized using the T7 RiboMAXTM Express RNAi System (Promega, United States) following the manufacturer’s instructions (Wang et al., 2018b). The dsRNA of the green fluorescent protein (GFP; GenBank accession No. U87974) sequence, which does not exist in the genome of bees, was used as a control. The experimental group was injected with 1 μl (6 μg/individual) of dsAccUGT2B20-like, and the control groups were untreated but injected with an equal amount of dsRNA-GFP and water (1 μl). dsRNA or water was injected into the third and fourth abdominal segments as described by Li G. et al. (2018). Silenced efficiency was determined by qRT-PCR. The silenced bees were used to analyze oxidant- and antioxidant-related genes and enzymes.
To explore the changes in antioxidant genes after AccUGT2B20-like was silenced, the expression levels of 10 antioxidant genes were analyzed by qRT-PCR. These 10 genes included AccCAT (GenBank ID: KF765424), A. cerana cerana cytochrome P450 (Acccyp4g11, GenBank ID: KC243984), AccGrx1 (GenBank ID: JX844656), AccGSTO1 (GenBank ID: KF496073), AccGSTO2 (GenBank ID: JX434029), AccGSTD (GenBank ID: JF798572), AccGSTS4 (GenBank ID: JN008721), AccHsp22.6 (GenBank ID: KF150016), AccSOD1 (GenBank ID: JN700517), and AccTrx1 (GenBank ID: JX844651). These genes were selected based on our previous study (Zhang et al., 2016). The primers used in this experiment are shown in Supplementary Table 1.
Total protein was extracted from the silenced bees in a 1-ml isolation medium using the Tissue Protein Extraction Kit according to the manufacturer’s protocol. Total protein was quantified using the total protein assay kit, the BCA Protein Assay Kit (Nanjing Jiancheng Bioengineering Institute). Enzyme activities of superoxide dismutase (SOD), catalase (CAT), peroxidase (POD), and malondialdehyde (MDA) were detected using the SOD assay kit, CAT assay kit, POD assay kit, and MDA assay kit, respectively (all four kits were produced by Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
To construct an AccUGT2B20-like recombinant, the AccUGT2B20-like open reading frame was amplified using the primers eUF and eUR, and inserted into an expression vector, pET-30a(+). Then, the expression plasmid pET-30(+)-AccUGT2B20-like was transformed into E. coli strain BL21 (DE3) for protein expression. The recombinant was induced with 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown at 37°C for 8 h. The recombinant protein was analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie brilliant blue staining. To evaluate the activity of recombinant AccUGT2B20-like cells, disc diffusion assays were performed as described by Jia et al. (2017). E. coli cells overexpressing AccUGT2B20-like were overlaid onto LB-kanamycin agar and incubated at 37°C for 1 h. E. coli BL21 cells containing an empty pET-30a (+) vector were used as controls. Filter discs (6 mm diameter) soaked in different concentrations of cumene hydroperoxide (CHP) (0, 0.10, 0.14, 0.20, and 0.41 μmol) and tert-Butyl hydroperoxide (TBHP) (0, 0.16, 0.21, 0.32, and 0.64 μmol) were placed on the surface of agar. Following incubation at 37°C for 12 h, the killing zones around the paper discs were measured as described by Burmeister et al. (2008).
The results are presented as mean ± standard error of the mean (SEM) from three independent biological replicates. Statistical analysis was performed using SPSS (v. 25). Means were compared using Student’s t-test or one-way analysis of variance (ANOVA). Significance was set at ∗P < 0.05 and ∗∗P ≤ 0.01.
A signature sequence involved in the binding of the UDP moiety of the nucleotide sugar has been identified as a characteristic sequence in different organisms. To gain insight into the UGT family in A. cerana cerana, the amino acid sequence corresponding to this signature motif in insect UGTs was used to screen the predicted A. cerana cerana protein database. We ultimately identified 10 UGT genes from the A. cerana cerana genome, and the GenBank accession numbers are shown in Supplementary Table 2. Multiple alignments of 10 AccUGT amino acid sequences revealed the same signature sequence at the C-terminal (Figure 1A). The UGT motif signature sequence, (FVA)-(LIVMF)-(TS)-(HQ)-(SGAC)-G-X(2)-(STG)-X(2)-(DE)-X(6)-P-(LIVMFA)-(LIVMFA)-X(2)-P-(LMVFIQ)-X(2)-(DE)-Q, was also detected among the AccUGT genes. A phylogenetic tree constructed with amino acid sequences from A. cerana cerana (10), Apis mellifera (9), P. xylostella (22), B. mori (44), D. melanogaster (34), and H. armigera (40) revealed patterns of interspecific conservation and lineage-specific expansion of the gene families (Figure 1B). The 10 AccUGT genes were distributed into seven subfamilies, of which AccUGT1-6-like, AccUGT2C-1-like, and AccUGT1-1-like were clustered into one subfamily and AccUGT2B15-like and AccUGT2B17-like were clustered into another. The remaining five AccUGT genes were divided into five different subfamilies.
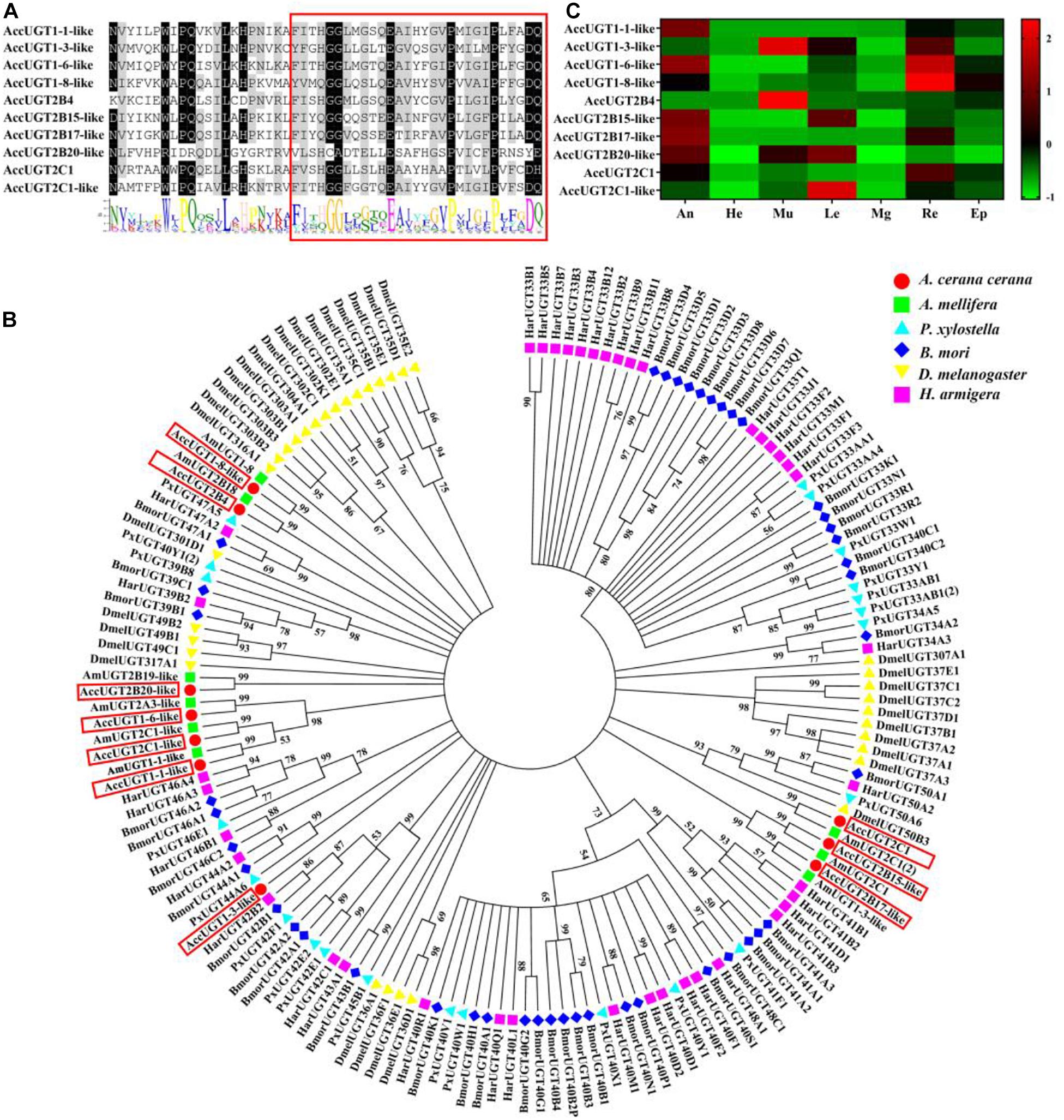
Figure 1. Phylogenetic analysis and tissue-specific expression patterns of Apis cerana cerana UDP-glucuronosyltransferases (UGT) genes. (A) Signature motif of AccUGT genes. Multiple alignment analysis of A. cerana cerana. The motif sequence discovered by MEME (http://meme-suite.org/) is shown at the bottom. The UGT signature motif is in the box. (B) The phylogenetic analysis involved 159 UGT amino acid sequences from Apis cerana cerana (10), Apis mellifera (9), Plutella xylostella (22), Bombyx mori (44), Drosophila melanogaster (34), and Helicoverpa armigera (40). Evolutionary analyses were conducted in MEGA7. (C) Tissue-specific expression patterns of AccUGTs from adult bees based on qRT-PCR. The mean expression value of each UGT gene was normalized using the Z-score method. He, head; An, antenna; Mu, muscle; Le, leg; Mg, midgut; Re, rectum; Ep, epidermis.
Tissue-specific expression patterns of 10 AccUGT genes were analyzed in adult bees by qRT-PCR. Among the different tissues, the antenna, muscle, and rectum had the highest level of expression. Five AccUGT genes exhibited high expression in the antenna and three did so in the muscle. Conversely, most UGTs exhibited lower expression in the head. Among the 10 UGT genes, AccUGT2B4, AccUGT1-3-like, AccUGT2C1-1-like, and AccUGT2B20-like exhibited high expression in different tissues, indicating they may have important regulatory functions. AccUGT2B4 showed the highest expression in all tested tissues, especially in the muscle and epidermis (Figure 1C).
To explore whether UGT genes in A. cerana cerana respond to pesticides, five different pesticides and H2O2 treatments were used to evaluate the expression of UGT genes in adult worker bees. After pesticide treatment, we chose the most highly upregulated gene based on expression-fold change relative to that of the control group. Thus, we could screen for genes in the UGT gene family that had a stronger response to pesticide treatment. The qRT-PCR results indicated that almost all the transcripts of the 10 UGT genes were induced by our treatments, of which AccUGT2B15-like, AccUGT2B17-like, and AccUGT2B20-like were significantly upregulated. Specifically, AccUGT2B20-like was upregulated in emamectin-benzoate, acetamiprid, and lambda-cyhalothrin treatments (Figure 2A). Therefore, based on the results of tissue-specific analysis and expression profiling analysis, the AccUGT2B20-like gene was identified as the major gene involved in resistance against the pesticides. We further chose AccUGT2B20-like to conduct the following experiment to investigate the function of AccUGTs.
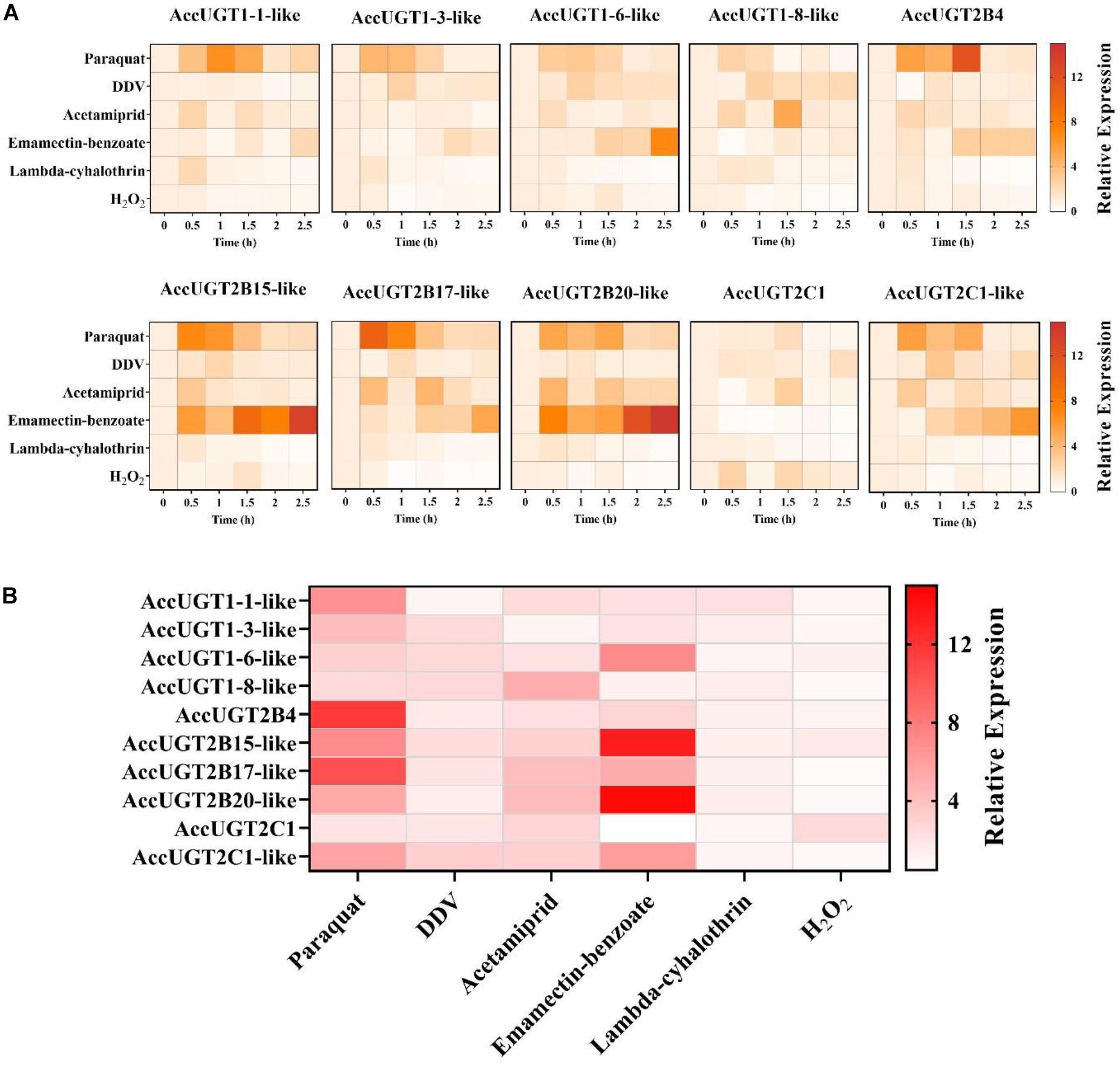
Figure 2. Expression pattern of AccUGT genes under different treatment conditions. (A) Expression profile of 10 AccUGT genes under pesticides and H2O2 stresses. (B) Most highly upregulated gene expression-fold change under different stresses. The most highly upregulated gene expression-fold change relative to that of the control (CK) group under different treatments. AccUGT mRNA expression was normalized using β-actin expression.
The full-length cDNA of AccUGT2B20-like (GenBank accession number XM_017063111) was 2,174 bp, which encodes a 494-amino acid protein with a predicted average mass of 56.804 kD. The full-length nucleotide and deduced amino acid sequences are shown in Figure 3A. The signal peptide was found at the N-terminal end of AccUGT2B20-like. The UGTs signature motif was found in the middle of the C-terminal domain, which exhibited higher conservation. A single short transmembrane domain, composed of 23 hydrophobic amino acid residues, occurred close to the end of the C-terminal domain. To further elucidate the protein structure of AccUGT2B20-like, we predicted its tertiary structure using the online database SWISS-MODEL (Figure 3B). We detected 14 α-helices and 12 β-sheets in AccUGT2B20-like, and the tertiary structure of proteins will provide strong evidence for our understanding of protein function.
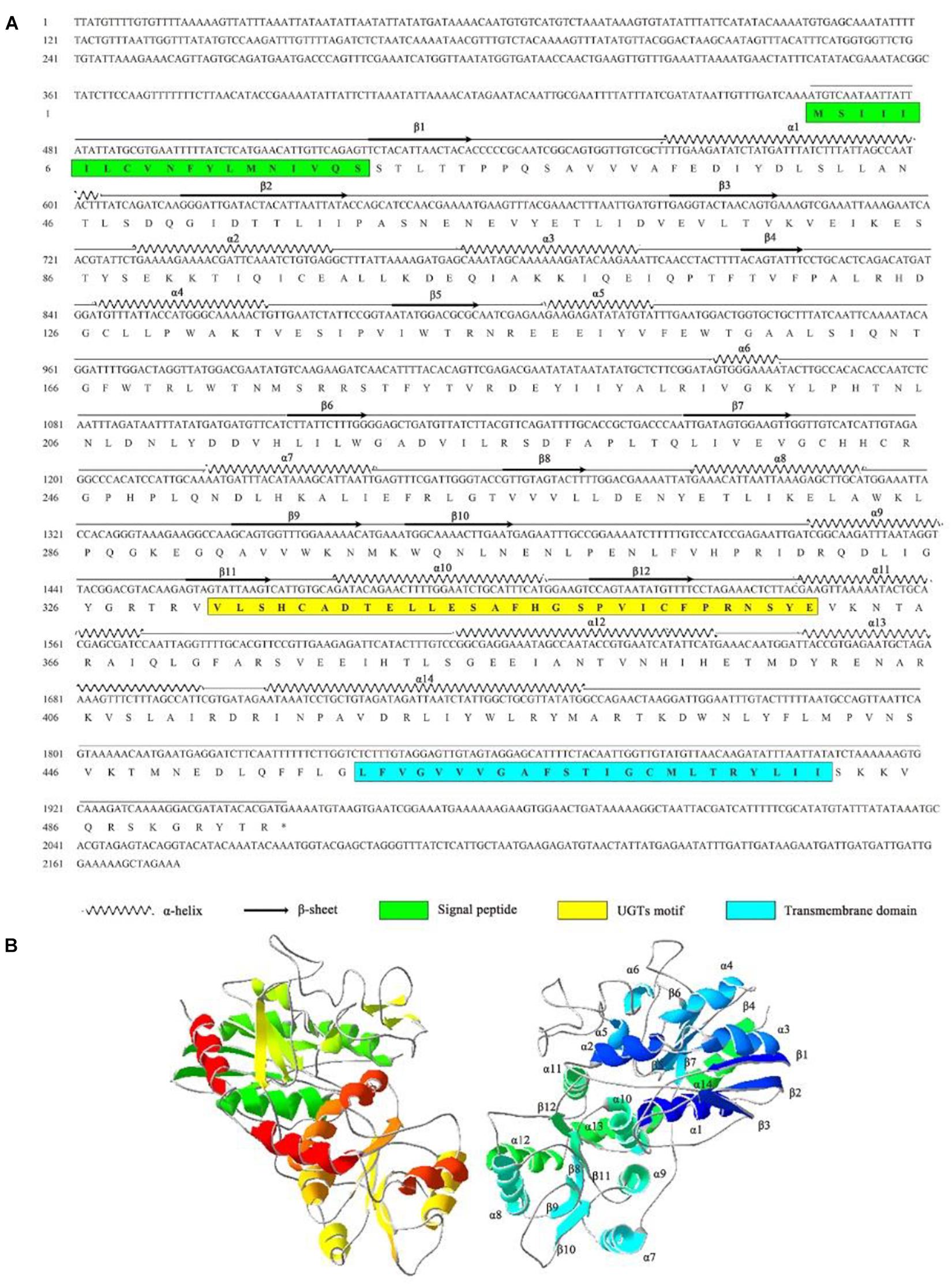
Figure 3. Characteristics of the AccUGT2B20-like gene. (A) cDNA and amino acid sequence of AccUGT2B20-like. The predicted protein secondary structure of AccUGT2B20-like is marked on the amino acid sequence. The predicted signal peptide in the N-terminus is in the green box, the UGT signature motif is in the yellow box, and the transmembrane domain in the C-terminal half and cytoplasmic tails are in the blue box. The sequence was deposited in GenBank (XM_017063111.1). (B) Putative tertiary structure characteristics of AccUGT2B20-like. The sequence was predicted using the SWISS-MODEL. The α-helices, β-sheets, coils were color-coded by secondary structure succession using SWISS-PDB VIEWER software, version 4.1.0 (Swiss Institute of Bioinformatics, Geneve, Suisse).
To investigate the specific role of AccUGT2B20-like under different pesticides, the expression profile of AccUGT2B20-like was examined by qRT-PCR. As shown in Figure 4, among six pesticide treatments, the application of paraquat, mancozeb, emamectin-benzoate, and lambda-cyhalothrin significantly induced the expression of AccUGT2B20-like. The response time was short, and the expression level of AccUGT2B20-like reached its peak within 2 h. AccUGT2B20-like was significantly upregulated under paraquat, acetamiprid, and lambda-cyhalothrin treatment, which was significantly different from that of the control group. The above results showed that AccUGT2B20-like is involved in protecting bees against damage from different pesticides.
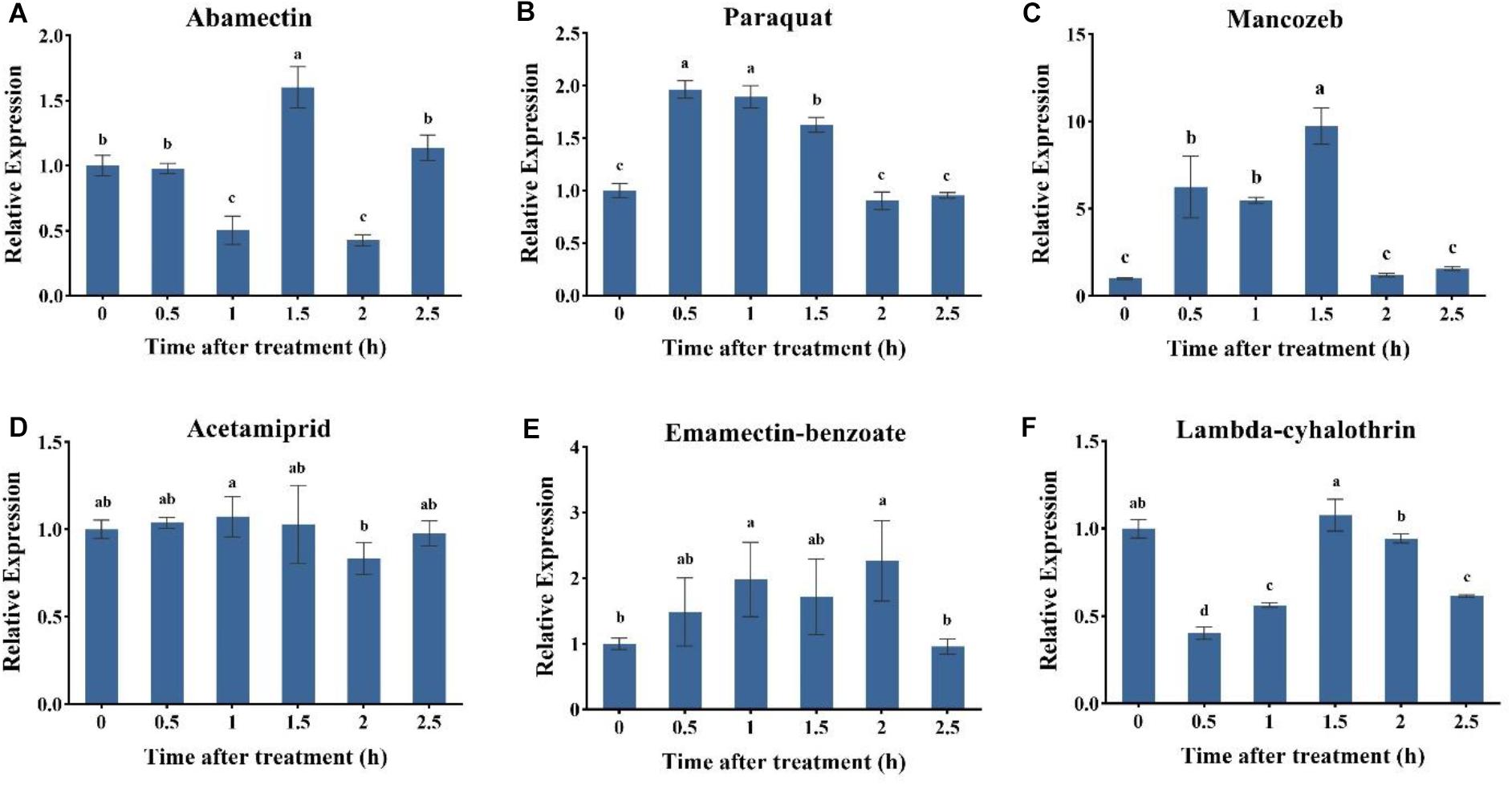
Figure 4. Expression profiles of the AccUGT2B20-like gene under different pesticides. qRT-PCR was performed on the total RNA extracted from adult bees at the indicated time points following the different stresses. The stressors include (A) abamectin, (B) paraquat, (C) acetamiprid, (D) acetamiprid treatment, (E) emamectin-benzoate, and (F) lambda-cyhalothrin treatment. The vertical bars represent the mean ± SEM (n = 9). β-actin (GenBank accession number: HM640276) was used as the control. The letters above the bars indicate significant differences (P < 0.05 in SPSS) based on Duncan’s multiple range test.
To explore the molecular regulation mechanism of AccUGTs in response to oxidative stress, we overexpressed AccUGT2B20-like in E. coli, and disk diffusion assays were conducted. After overnight exposure to cumene hydroperoxide (CHP) and tert-butyl hydroperoxide (TBHP), the killing zones around the drug-soaked filters were smaller on the plates containing E. coli overexpressing AccUGT2B20-like than on that of the control bacteria that were transfected with the vector only (Figure 5). These results provided further evidence that overexpression of AccUGT2B20-like can effectively improve resistance to oxidative stress.
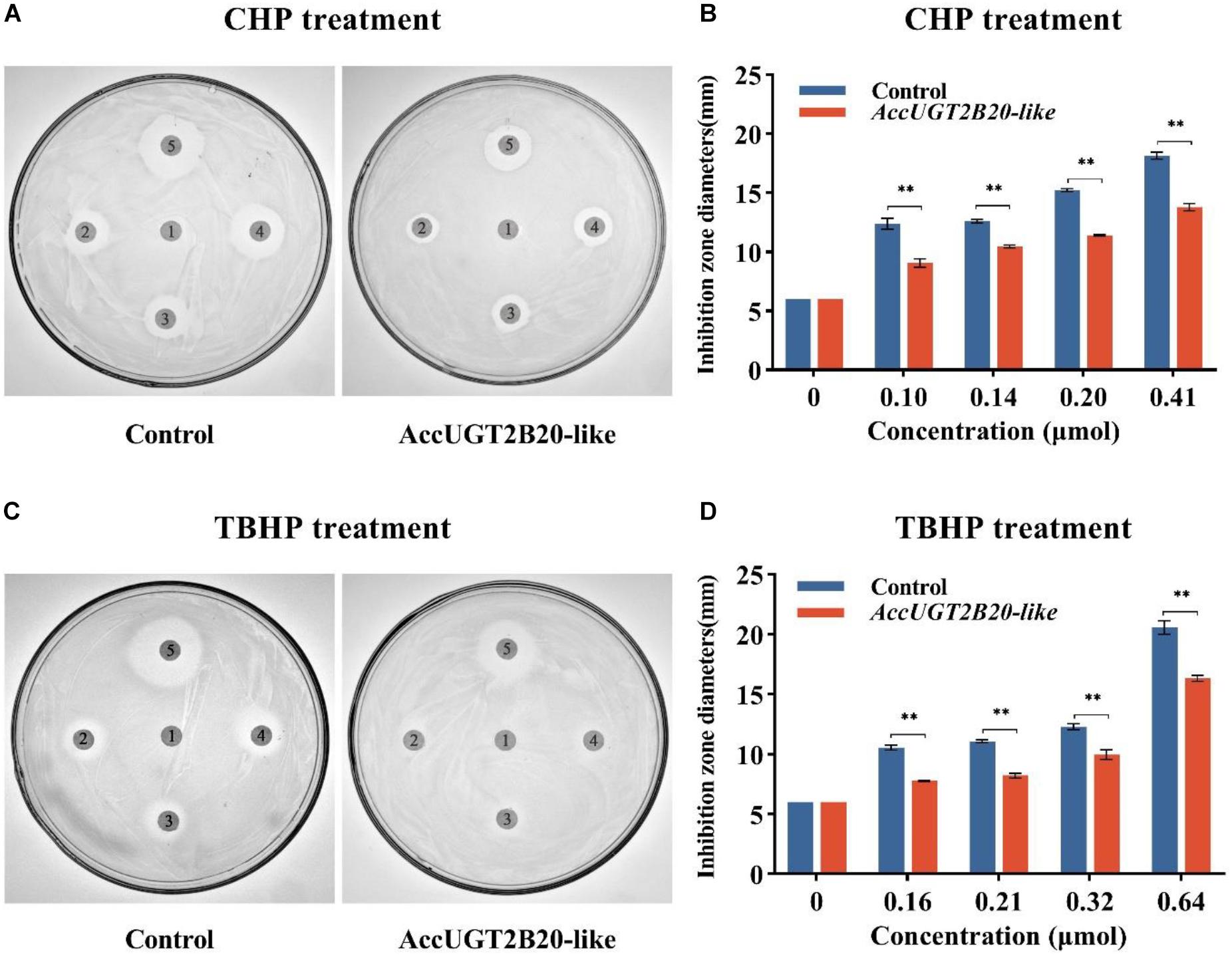
Figure 5. Disc diffusion assays using E. coli overexpressing AccUGT2B20-like. LB agar plates were inoculated with 5 × 108 cells. AccUGT2B20-like cells were overexpressed in E. coli, and bacteria transfected with pET-30a (+) were used as negative controls. Filter discs soaked with different concentrations of CHP (A,B) and TBHP (C,D) were placed on the agar plates. After overnight exposure, the killing zones around the drug-soaked filters were measured. *P < 0.05; **P < 0.01.
To further investigate the regulatory function of AccUGTs in response to oxidative stress induced by pesticides, we employed RNAi to knock down the expression of AccUGT2B20-like. dsRNA specific to AccUGT2B20-like was microinjected into adult workers to silence the target gene. For controls, adult workers were microinjected with dsGFP or water. qRT-PCR was used to detect the expression level of AccUGT2B20-like and determine the optimal silencing time. The transcription level of AccUGT2B20-like in the dsAccUGT2B20-injection group was significantly inhibited compared with that of the control groups, especially at 24 h (Figure 6A). The results indicated that dsRNA-mediated gene silencing was successful, and 24 h after dsRNA injection was selected for further study.
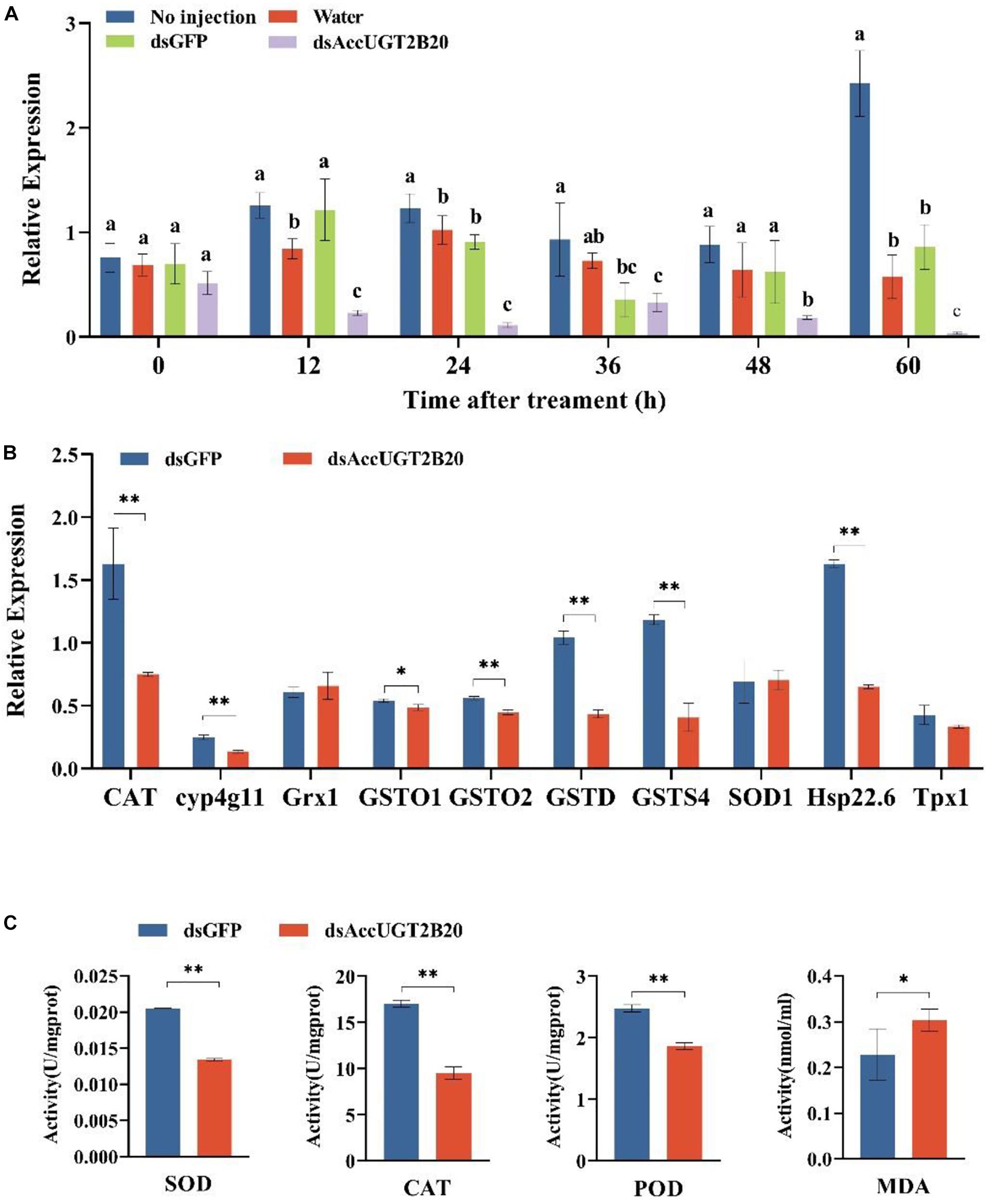
Figure 6. Effects of the RNAi-mediated silencing of AccUGT2B20-like. (A) qRT-PCR confirmation of RNAi knockdown of AccUGT2B20-like. (B) Expression patterns of antioxidant genes at 24 h after AccUGT2B20-like knocked down, as analyzed by qRT-PCR. (C) Determination of antioxidant enzyme activity. The error bars indicate 95% confidence intervals. ∗P < 0.05; ∗∗P < 0.01.
To elucidate the effects of AccUGT2B20-like knockdown, the transcriptional levels of other antioxidant genes were also investigated by qRT-PCR. We analyzed the expression levels of 10 different antioxidant genes 24 h after AccUGT2B20-like was silenced (Figure 6B). The results showed that AccCAT, Acccyp4g11, AccGSTO1, AccGSTO2, AccGSTD, AccGSTS4, and AccHsp22.6 were downregulated when AccUGT2B20-like was silenced. To confirm the effects of RNAi with AccUGT2B20-like on the antioxidant capacity of A. cerana cerana, we measured the enzymatic activities of SOD, CAT, POD, and MDA after RNAi treatment for 24 h. The activities of SOD, CAT, and POD declined compared with that of the control groups. Furthermore, MDA concentration increased after the AccUGT2B20-like expression was knocked down (Figure 6C). These results indicated that the knockdown of AccUGT2B20-like increased oxidative stress in A. cerana cerana.
In recent years, because of various environmental stresses, including pesticides, the number of A. cerana cerana has gradually decreased. However, the specific molecular mechanism of the resistance of Chinese honeybees to pesticide stress is still unclear. UGTs, as significant major phase II drug-metabolizing enzymes, play important roles in the detoxification process (Bock, 2003). Many UGT genes have been identified and exist in almost all living organisms. The UGT gene family is particularly interesting because it is involved in human disease, drug metabolism, and homeostasis maintenance (Rowland et al., 2013; Bock, 2016; Mazerska et al., 2016). However, information regarding the functions of AccUGT genes remains limited.
In this study, we identified the UGT genes in A. cerana cerana and investigated the molecular regulatory mechanism of UGTs in oxidative stress induced by pesticides. UGT genes have been identified in insects, such as D. melanogaster (Morello and Repetto, 1979), B. mori (Huang et al., 2008), Spodoptera littoralis (Bozzolan et al., 2014), H. armigera and Heliothis virescens (Krempl et al., 2016), and Aphis gossypii (Pan et al., 2018). Many studies have been conducted on the UGTs in insects; nevertheless, a few studies have been conducted using A. cerana cerana. In the present study, we identified 10 UGT sequences from the A. cerana cerana cDNA database using the UGT motif signature (Figure 1A). The C-terminal is highly conserved where the UGT motif signature is found. This result is consistent with that of Meech and Mackenzie (1998). Compared to that of other insects, A. cerana cerana has a lower number of UGT genes. This may be related to the highly specialized habitat, lack of exposure to food-derived toxins, and peculiar life history (Honeybee Genome Sequencing Consortium, 2006).
Understanding the tissue-specific expression patterns of AccUGTs may provide a better understanding of functional mechanisms. Therefore, we analyzed the relative expression of UGT genes in different tissues (Figure 1C). As mentioned in the literature, insect UGTs are preferentially expressed in the olfactory organ, the antenna, suggesting that UGTs may be involved in the deactivation of pheromones (Wang et al., 1999; Younus et al., 2014). Our results agree with those observed in earlier studies. Among 21 UGTs in P. xylostella, 8 exhibited high expressions in the midgut and 3 in the fat body (Li X. et al., 2018). The insect midgut is the largest part of the digestive tract and is the main target of pathogenic microorganisms, pesticides, and plant toxins (Li et al., 2008). In this study, the midgut did not exhibit high expression; however, higher gene expression was detected in the rectum. This result may be related to the method of injection. We injected the pesticide directly into the abdomen of the bees, but in their natural state, bees feed on pesticide-containing pollen or nectar, so the high expression of UGT in the midgut is reasonable. UGT genes with high relative expression in various tissues may play a critical role in honeybees. Highly expressed UGT genes, such as AccUGT2B4 and AccUGT2B20-like, may be candidates for the screening of major genes.
Previous studies have shown that the UGT genes are involved in the detoxification process of insects and respond to different pesticides or insecticides (Ahn et al., 2012; Bock, 2016). In a multiresistant population of P. xylostella, 10 of 21 UGT genes were upregulated, and the population was resistant to nine commonly used insecticides. The expression of UGT40V1, UGT45B1, and UGT33AA4 could be induced by more than five insecticides (Li X. et al., 2018). Comparison analysis of thiamethoxam-resistant cotton aphids and susceptible aphids showed that 13 UGT genes were increased by approximately 2.0-fold or greater in the thiamethoxam-resistant strain (Pan et al., 2018). According to previous research, we proposed that the UGT family may contribute to the protection of A. cerana cerana from pesticides. In this study, almost all the UGT genes were upregulated after exposure to different pesticides (Figure 2A). These results indicated that UGT genes are involved in the process of pesticide response in A. cerana cerana.
The UGT2B enzymes are important in the detoxification of a variety of endogenous and exogenous compounds. Previous studies have demonstrated that UGT2B15 exhibits high activity of BPA glucuronidation and is closely associated with the metabolism and toxicity of BPA, an endocrine-disrupting chemical that could result in reproductive and developmental toxicity at low doses (Richter et al., 2007; Hanioka et al., 2011). A study by Li et al. (2017) proved that overexpression of UGT2B17 decreased the toxicity of chlorantraniliprole and may play a critical role in chlorantraniliprole resistance in P. xylostella. However, the detoxification of UGT2B20 has not yet been reported. A. cerana cerana was subjected to 8 toxicity stressors in this study; all 10 UGT mRNAs were induced, among which the AccUGT2B family gene was significantly upregulated (Figure 2B). This result indicates that the UGT2B family may play an essential role in the resistance of the A. cerana cerana to external stresses. Interestingly, AccUGT2B20-like was induced in five of the eight treatments, and it was the most significantly upregulated gene under different treatments. We consequently chose AccUGT2B20-like as a representative of this gene family to explore their properties.
Many studies have demonstrated that pesticides, including insecticides and herbicides, can induce oxidative stress, leading to alteration in lipid peroxidation, free-radical scavengers, and a series of antioxidants (Banerjee et al., 1999; Etemadi-Aleagha et al., 2002; Abdollahi et al., 2004). Additionally, some studies have shown that UGTs are involved in the detoxification processes, including that of drugs, pesticides, and endogenous compounds (Mazerska et al., 2016). Therefore, to explore the relationship between UGTs and oxidative stress, we simulated several adverse life-threatening environmental conditions that could cause ROS damage in A. cerana cerana. Furthermore, we explored the molecular regulation mechanism of UGTs in response to oxidative stress. Previous studies have shown that overexpression of antioxidant-related genes, such as AccSCO2, AccGSTO2, and AccTpx4 in E. coli cells could protect them from oxidative stressors by performing disc diffusion assays (Huaxia et al., 2015; Zhang et al., 2016; Jia et al., 2017). In this study, we overexpressed AccUGT2B20-like in E. coli cells, and the results were consistent with those of earlier studies. Overexpression of AccUGT2B20-like could protect E. coli from oxidative stressors (Figure 5). This result provides preliminary evidence for the antioxidant function of UGTs.
High ROS concentrations in cells can cause oxidative stress and contribute to degenerative diseases and aging through the oxidation of nucleic acids, proteins, and lipid membranes (Jia et al., 2014). As byproducts of aerobic metabolism, ROS continuously exists during oxidative stress, whereas some antioxidant genes, such as those for SOD, ascorbate peroxidase (APX), glutathione peroxidase (GPX), and CAT, could serve as detoxication for ROS elimination (Apel and Hirt, 2004). Using antioxidant-related genes as marker genes to detect their expression can indirectly reflect their antioxidant properties. Previous reports have suggested that when AccGSTO2 is knocked down, most antioxidant-related gene expression is reduced (Zhang et al., 2016). Wang reported that AccSOD1, AccSOD2, AccGSTS4, AccCYP4G11, AccGSTO2, and AccGSTD was downregulated when AccMKK6 was knocked down (Wang et al., 2018b). To further investigate the gene function of AccUGT2B20-like and the relationship between AccUGT2B20-like and oxidation-related genes, AccUGT2B20-like gene was silenced. Then, we find that the expression levels of antioxidant genes were significantly reduced. This result suggested that AccUGT2B20-like may participate in the regulation of downstream antioxidant genes. The function of a gene is achieved through its expression product. Therefore, determining the activities of antioxidant enzymes will reflect the real function. After UGT enzymes were inhibited by two inhibitors (sulfinpyrazone and 5-nitrouracil), resistance to thiamethoxam by aphids was decreased (Pan et al., 2018). The activities of SOD, POD, and CAT were reduced, whereas MDA was increased in silenced bees (Figure 6C). This result indicated that the oxidative damage to the body was increased. Considering our results in the context of previous studies, we concluded that AccUGTs are involved in the detoxification process of pesticides. Furthermore, AccUGT2B20-like possesses potent antioxidant properties that contribute to the detoxification of pesticides by eliminating oxidative stress.
Our results indicate that UGTs are involved in pesticide resistance in A. cerana cerana, among which, AccUGT2B20-like contributes to the detoxification of pesticides by eliminating oxidative stress in A. cerana cerana. This study explains the molecular basis for the resistance of bees to pesticides and provides an essential safeguard for maintaining ecological balance.
All datasets generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Committee on Animals of Shandong Agricultural University Animal Care and Use Committee (SDAUA-2018-018).
XC was involved in the experimental design, data processing, and manuscript writing. CW, BX, and XG were involved in the experimental design and constructive amendments to the manuscript. XW participated in the qRT-PCR and gene silencing work. GL, ZL, and HW helped in the editing of the manuscript. All authors read and approved the final manuscript.
This work was financially supported by the special funds for the Taishan Industry Leading Talent Program, Shandong Agricultural Fine Varieties Breeding Projects (2017LZN006), and the earmarked fund for the China Agriculture Research System (No. CARS-44).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Xusheng Dong, Xiao Han, and Weixing Zhang from the College of Animal Science and Technology, Shandong Agricultural University, for the helpful and critical comments on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.592595/full#supplementary-material
Abdollahi, M., Ranjbar, A., Shadnia, S., Nikfar, S., and Rezaie, A. (2004). Pesticides and oxidative stress: a review. Med. Sci. Monit. 10, RA141–RA147. doi: 10.1051/medsci/2004206-7710
Achour, B., Rostami-Hodjegan, A., and Barber, J. (2014). Protein expression of various hepatic uridine 5′-diphosphate glucuronosyltransferase (UGT) enzymes and their inter-correlations: a meta-analysis. Biopharm. Drug Dispos. 35, 353–361. doi: 10.1002/bdd.1906
Adams, L., Franco, M. C., and Estevez, A. G. (2015). Reactive nitrogen species in cellular signaling. Exp. Biol. Med. 240, 711–717. doi: 10.1177/1535370215581314
Ahn, S. J., Vogel, H., and Heckel, D. G. (2012). Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect. Biochem. Mol. Biol. 42, 133–147. doi: 10.1016/j.ibmb.2011.11.006
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Banerjee, B. D., Seth, V., Bhattacharya, A., Pasha, S. T., and Chakraborty, A. K. (1999). Biochemical effects of some pesticides on lipid peroxidation and free-radical scavengers. Toxicol. Lett. 107, 33–47. doi: 10.1016/S0378-4274(99)00029-6
Bock, K. W. (2003). Vertebrate UDP-glucuronosyltransferases: functional and evolutionary aspects. Biochem. Pharmacol. 66, 691–696. doi: 10.1016/s0006-2952(03)00296-x
Bock, K. W. (2016). The UDP-glycosyltransferase (UGT) superfamily expressed in humans, insects and plants: animal-plant arms-race and co-evolution. Biochem. Pharmacol. 99, 11–17. doi: 10.1016/j.bcp.2015.10.001
Bozzolan, F., Siaussat, D., Maria, A., Durand, N., Pottier, M. A., Chertemps, T., et al. (2014). Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 23, 539–549. doi: 10.1111/imb.12100
Brattsten, L. B. (1988). Enzymic adaptations in leaf-feeding insects to host-plant allelochemicals. J. Chem. Ecol. 14, 1919–1939. doi: 10.1007/BF01013486
Burmeister, C., Luersen, K., Heinick, A., Hussein, A., Domagalski, M., Walter, R. D., et al. (2008). Oxidative stress in Caenorhabditis elegans: protective effects of the Omega class glutathione transferase (GSTO-1). FASEB J. 22, 343–354. doi: 10.1096/fj.06-7426com
Després, L., David, J. P., and Gallet, C. (2007). The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 22, 298–307. doi: 10.1016/j.tree.2007.02.010
Etemadi-Aleagha, A., Akhgari, M., and Abdollahi, M. (2002). A brief review on oxidative stress and cardiac diseases. Mid. East Pharmac. 10, 8–9.
Gegear, R. J., Otterstatter, M. C., and Thomson, J. D. (2006). Bumble-bee foragers infected by a gut parasite have an impaired ability to utilize floral information. Proc. R. Soc. B Biol. Sci. 273, 1073–1078. doi: 10.1098/rspb.2005.3423
Goulson, D., Nicholls, E., Botias, C., and Rotheray, E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347:1255957. doi: 10.1126/science.1255957
Hanioka, N., Oka, H., Nagaoka, K., Ikushiro, S., and Narimatsu, S. (2011). Effect of UDP-glucuronosyltransferase 2B15 polymorphism on bisphenol A glucuronidation. Arch. Toxicol. 85, 1373–1381. doi: 10.1007/s00204-011-0690-5
Hasegawa, K., Miwa, S., Tsutsumiuchi, K., and Miwa, J. (2010). Allyl isothiocyanate that induces GST and UGT expression confers oxidative stress resistance on C. elegans, as demonstrated by nematode biosensor. PLoS One 5:e9267. doi: 10.1371/journal.pone.0009267
Honeybee Genome Sequencing Consortium (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949. doi: 10.1038/nature05260
Huang, F. F., Chai, C. L., Zhang, Z., Liu, Z. H., Dai, F. Y., Lu, C., et al. (2008). The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics 9:563. doi: 10.1186/1471-2164-9-563
Huaxia, Y., Wang, F., Yan, Y., Liu, F., Wang, H., Guo, X., et al. (2015). A novel 1-Cys thioredoxin peroxidase gene in Apis cerana cerana: characterization of AccTpx4 and its role in oxidative stresses. Cell Stress Chaperones 20, 663–672. doi: 10.1007/s12192-015-0594-z
Jia, H., Ma, M., Zhai, N., Liu, Z., Wang, H., Guo, X., et al. (2017). Roles of a mitochondrial AccSCO2 gene from Apis cerana cerana in oxidative stress responses. J. Inorg. Biochem. 175, 9–19. doi: 10.1016/j.jinorgbio.2017.06.015
Jia, H., Sun, R., Shi, W., Yan, Y., Li, H., Guo, X., et al. (2014). Characterization of a mitochondrial manganese superoxide dismutase gene from Apis cerana cerana and its role in oxidative stress. J. Insect Physiol. 60, 68–79. doi: 10.1016/j.jinsphys.2013.11.004
Krempl, C., Sporer, T., Reichelt, M., Ahn, S.-J., Heidel-Fischer, H., Vogel, H., et al. (2016). Potential detoxification of gossypol by UDP-glycosyltransferases in the two Heliothine moth species Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. Biol. 71, 49–57. doi: 10.1016/j.ibmb.2016.02.005
Krupke, C. H., Hunt, G. J., Eitzer, B. D., Andino, G., and Given, K. (2012). Multiple routes of pesticide exposure for honey bees living near agricultural fields. PLoS One 7:e29268. doi: 10.1371/journal.pone.0029268
Lairson, L. L., Henrissat, B., Davies, G. J., and Withers, S. G. (2008). Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555. doi: 10.1146/annurev.biochem.76.061005.092322
Li, G., Zhang, Y., Ni, Y., Wang, Y., Xu, B., and Guo, X. (2018). Identification of a melatonin receptor type 1A gene (AccMTNR1A) in Apis cerana cerana and its possible involvement in the response to low temperature stress. Naturwissenschaften 105:24. doi: 10.1007/s00114-018-1546-0
Li, X., Shi, H., Gao, X., and Liang, P. (2018). Characterization of UDP-glucuronosyltransferase genes and their possible roles in multi-insecticide resistance in Plutella xylostella (L.). Pest Manag. Sci 74, 695–704. doi: 10.1002/ps.4765
Li, H. M., Buczkowski, G., Mittapalli, O., Xie, J., Wu, J., Westerman, R., et al. (2008). Transcriptomic profiles of Drosophila melanogaster third instar larval midgut and responses to oxidative stress. Insect Mol. Biol. 17, 325–339. doi: 10.1111/j.1365-2583.2008.00808.x
Li, J., Qin, H., Wu, J., Sadd, B. M., Wang, X., Evans, J. D., et al. (2012). The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PLoS One 7:e47955. doi: 10.1371/journal.pone.0047955
Li, X. X., Zhu, B., Gao, X. W., and Liang, P. (2017). Over-expression of UDP-glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 73, 1402–1409. doi: 10.1002/ps.4469
Lourenço, A. P., Mackert, A., Cristino, A. D. S., and Simões, Z. L. P. (2008). Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39, 372–385. doi: 10.1051/apido:2008015
Mackenzie, P. I., Owens, I. S., Burchell, B., Bock, K. W., Bairoch, A., Belanger, A., et al. (1997). The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7, 255–269. doi: 10.1097/00008571-199708000-00001
Magdalou, J., Fournel-Gigleux, S., and Ouzzine, M. (2010). Insights on membrane topology and structure/function of UDP-glucuronosyltransferases. Drug Metab. Rev. 42, 159–166. doi: 10.3109/03602530903209270
Marie-Pierre, C., Jean-Paul, F., Anne-Claire, M., Julie, L., Nicolas, C., and Michel, A. (2006). A survey of pesticide residues in pollen loads collected by honey bees in france. J. Econ. Entomol. 99, 253–262. doi: 10.1603/0022-0493-99.2.253
Mazerska, Z., Mroz, A., Pawlowska, M., and Augustin, E. (2016). The role of glucuronidation in drug resistance. Pharmacol. Ther. 159, 35–55. doi: 10.1016/j.pharmthera.2016.01.009
Meech, R., and Mackenzie, P. I. (1998). Determinants of UDP glucuronosyltransferase membrane association and residency in the endoplasmic reticulum. Arch. Biochem. Biophys. 356, 77–85. doi: 10.1006/abbi.1998.0750
Meech, R., Miners, J. O., Lewis, B. C., and Mackenzie, P. I. (2012). The glycosidation of xenobiotics and endogenous compounds: versatility and redundancy in the UDP glycosyltransferase superfamily. Pharmacol. Ther. 134, 200–218. doi: 10.1016/j.pharmthera.2012.01.009
Meech, R., Mubarokah, N., Shivasami, A., Rogers, A., Nair, P. C., Hu, D. G., et al. (2015). A novel function for UDP glycosyltransferase 8: galactosidation of bile acids. Mol. Pharmacol. 87, 442–450. doi: 10.1124/mol.114.093823
Morello, A., and Repetto, Y. (1979). UDP-glucosyltransferase activity of housefly microsomal fraction. Biochem. J. 177, 809–812. doi: 10.1042/bj1770809
Pan, Y., Tian, F., Wei, X., Wu, Y., Gao, X., Xi, J., et al. (2018). Thiamethoxam resistance in Aphis gossypii glover relies on multiple UDP-Glucuronosyltransferases. Front. Physiol. 9:322. doi: 10.3389/fphys.2018.00322
Richter, C. A., Birnbaum, L. S., Farabollini, F., Newbold, R. R., Rubin, B. S., Talsness, C. E., et al. (2007). In vivo effects of bisphenol A in laboratory rodent studies. Reprod Toxicol. 24, 199–224. doi: 10.1016/j.reprotox.2007.06.004
Rowland, A., Miners, J. O., and Mackenzie, P. I. (2013). The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 45, 1121–1132. doi: 10.1016/j.biocel.2013.02.019
Sanchez-Bayo, F., Goka, K., and Guedes, R. N. C. (2014). Pesticide residues and bees – A Risk assessment. PLoS One 9:e94482. doi: 10.1371/journal.pone.0094482
Swomley, A. M., and Butterfield, D. A. (2015). Oxidative stress in Alzheimer disease and mild cognitive impairment: evidence from human data provided by redox proteomics. Arch. Toxicol. 89, 1669–1680. doi: 10.1007/s00204-015-1556-z
Tomizawa, M., and Casida, J. E. (2004). Neonicotinoid insecticide toxicology: mechanisms of selective action. Annu. Rev. Pharmacol. Toxicol. 45, 247–268. doi: 10.1146/annurev.pharmtox.45.120403.095930
Wang, C., He, X., Wang, X., Zhang, S., and Guo, X. (2017). ghr-miR5272a-mediated regulation of GhMKK6 gene transcription contributes to the immune response in cotton. J. Exp. Bot. 68, 5895–5906. doi: 10.1093/jxb/erx373
Wang, Q., Hasan, G., and Pikielny, C. W. (1999). Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J. Biol. Chem. 274, 10309–10315. doi: 10.1074/jbc.274.15.10309
Wang, X., Anadon, A., Wu, Q., Qiao, F., Ares, I., Martinez-Larranaga, M. R., et al. (2018a). Mechanism of neonicotinoid toxicity: impact on oxidative stress and metabolism. Annu. Rev. Pharmacol. Toxicol. 58, 471–507. doi: 10.1146/annurev-pharmtox-010617-052429
Wang, X., Wang, C., Cui, X., Wang, L., Liu, Z., Xu, B., et al. (2018b). Molecular mechanism by which Apis cerana cerana MKK6 (AccMKK6)-Mediated MAPK cascades regulate the oxidative stress Response. Biosci Rep. 38:BSR20181301. doi: 10.1042/BSR20181301
Wang, Y., Ma, L.-T., Hang, X.-B., Yang, W.-R., Liu, F., and Xu, B.-H. (2014). Digestion of protein of two pollen types in China by the honeybee (Apis mellifera L). Apidologie 45, 590–600. doi: 10.1007/s13592-014-0278-1
Weidinger, A., and Kozlov, A. V. (2015). Biological activities of reactive oxygen and nitrogen species: oxidative stress versus signal transduction. Biomolecules 5, 472–484. doi: 10.3390/biom5020472
Younus, F., Chertemps, T., Pearce, S. L., Pandey, G., Bozzolan, F., Coppin, C. W., et al. (2014). Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 53, 30–43. doi: 10.1016/j.ibmb.2014.07.003
Zhang, Y., Yan, H., Lu, W., Li, Y., Guo, X., and Xu, B. (2013). A novel Omega-class glutathione S-transferase gene in Apis cerana cerana: molecular characterisation of GSTO2 and its protective effects in oxidative stress. Cell Stress Chaperones 18, 503–516. doi: 10.1007/s12192-013-0406-2
Keywords: UDP-glucuronosyltransferases, Apis cerana cerana, pesticide resistance, RNAi, oxidative stress
Citation: Cui X, Wang C, Wang X, Li G, Liu Z, Wang H, Guo X and Xu B (2020) Molecular Mechanism of the UDP-Glucuronosyltransferase 2B20-like Gene (AccUGT2B20-like) in Pesticide Resistance of Apis cerana cerana. Front. Genet. 11:592595. doi: 10.3389/fgene.2020.592595
Received: 07 August 2020; Accepted: 19 October 2020;
Published: 19 November 2020.
Edited by:
Jianke Li, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Bin Han, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2020 Cui, Wang, Wang, Li, Liu, Wang, Guo and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingqi Guo, eHFndW9Ac2RhdS5lZHUuY24=; Baohua Xu, Ymh4dUBzZGF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.