- 1Department of Geriatrics and Neurology, The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Department of Preventive Medicine, Wenzhou Medical University, Wenzhou, China
Objective: A novel functional cis-regulatory element (CRE) located at SNCA intron 4 has recently been identified in association with Parkinson’s disease (PD) risk in European descendants. We aimed to investigate whether this CRE is associated with PD in Han Chinese ethnicity.
Methods: A Chinese cohort comprising 513 sporadic PD patients and 517 controls was recruited. CRE variants were identified by sequencing and then analyzed.
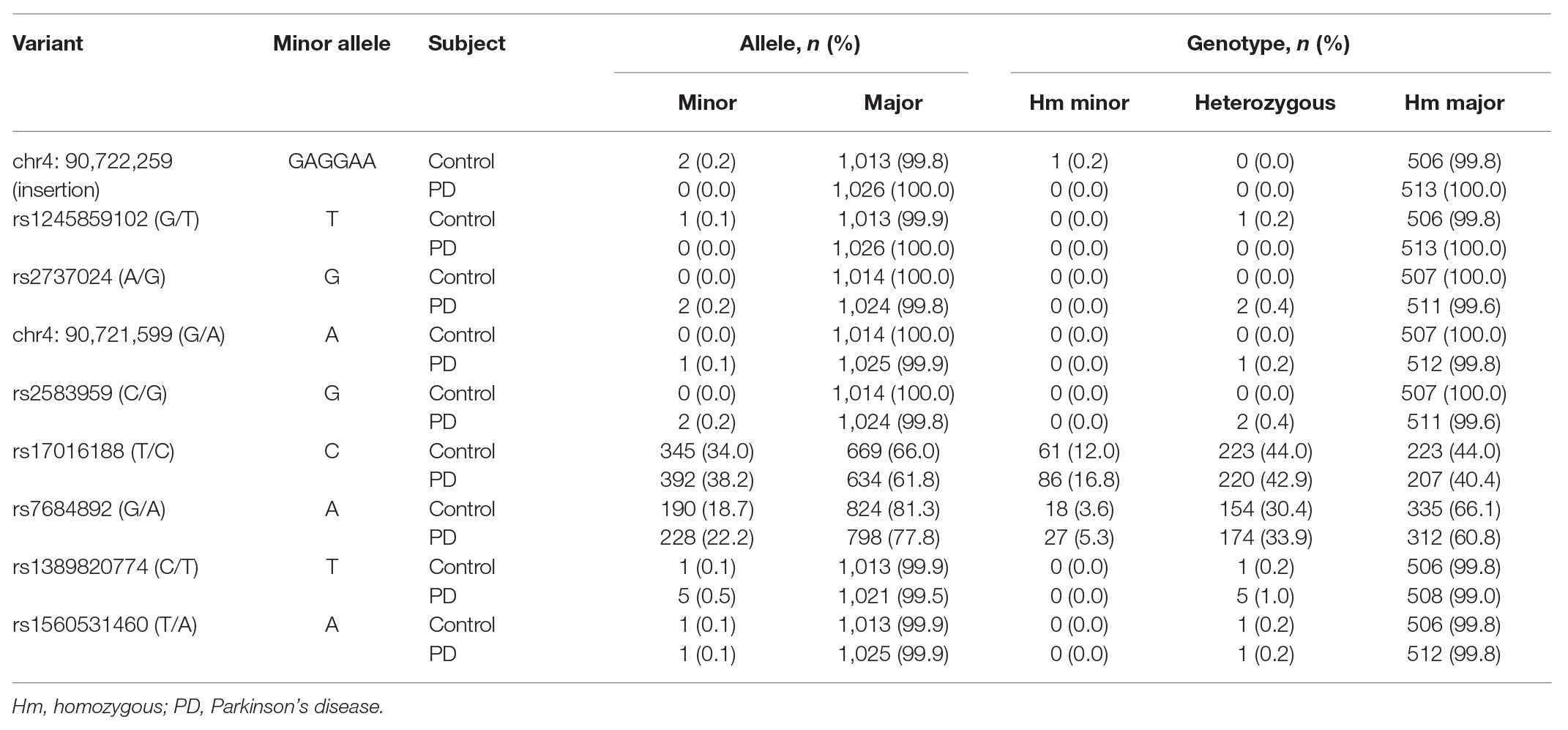
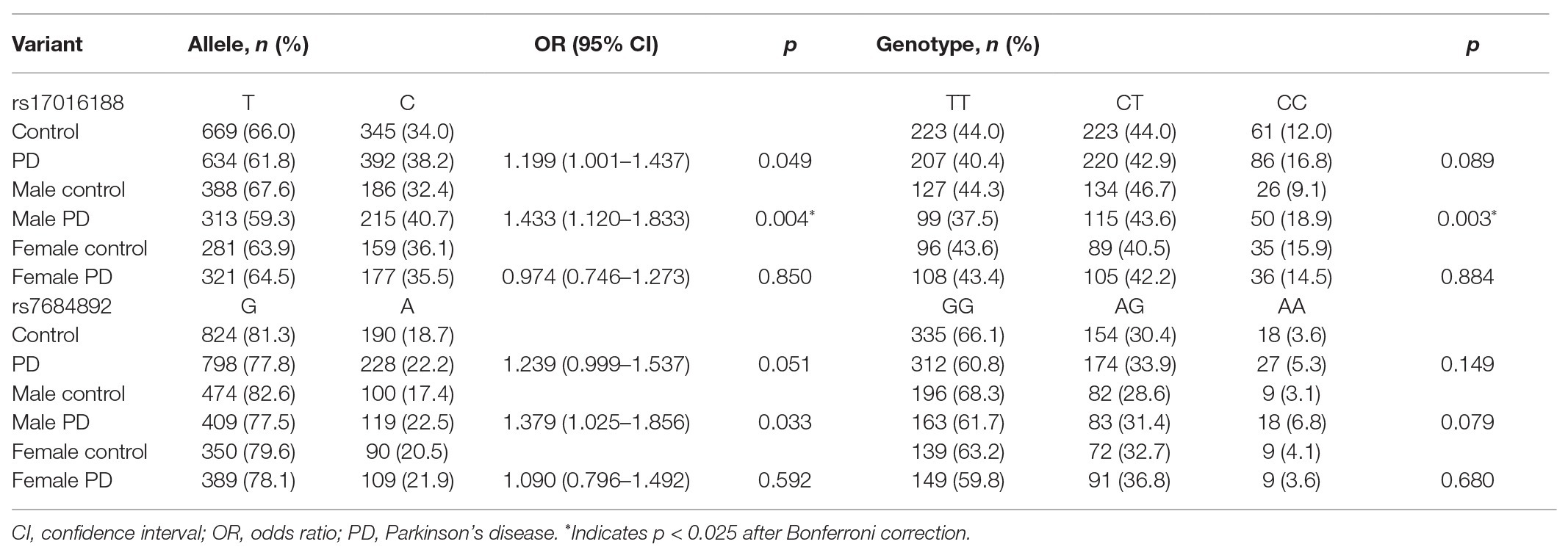
Results: A total of nine variants were detected, namely eight single nucleotide variants and one new insertion variant. Two variants, rs17016188 and rs7684892, had minor allele frequency greater than 5%. A difference of rs17016188 was observed in males with the C allele serving as a recessive risk factor (p = 0.001, OR = 2.349, 95% CI = 1.414–3.901) following Bonferroni correction. Haplotypes of rs17016188 and rs7684892 showed distribution differences in the total and the male populations (p = 0.002 and 4.08 × 10−5, respectively). Among the haplotypes, rs17016188/T-rs7684892/G was associated with a reduced risk for PD (p = 4.8 × 10−4, OR = 0.731, 95% CI = 0.614–0.872).
Conclusions: Our results provide insight into how the SNCA intron 4 CRE harbors variants and its contribution to PD risk in Chinese ethnicity.
Introduction
Parkinson’s disease (PD) is a common neurodegenerative disease characterized by the progressive loss of dopaminergic neurons in the substantia nigra and the formation of Lewy bodies. Considerable progress has been made to understand the genetic underpinning of PD. Monogenic mutations such as in genes of SNCA, LRRK2, and Parkin have been well defined to cause hereditary PD (Obeso et al., 2017). A number of independent risk variants including SNCA/rs356182, RIT2/rs12456492, and SIPA1L2/rs10797576 have been consistently identified to associate with sporadic PD (Nalls et al., 2014; Wang et al., 2015; Zou et al., 2018).
SNCA and its product α-synuclein are at the convergence of PD genetically and pathologically. It has been intensively investigated in regard with its causative genetic mutations and risk variants. Patients with SNCA duplication or triplication exhibit a dosage effect in disease onset, severity, and progression (Obeso et al., 2017). The encoded α-synuclein is the main component of Lewy body inclusion, and dopaminergic neurons are particularly vulnerable to the toxic α-synuclein aggregates (Alegre-Abarrategui et al., 2019). In recent years, misfolded α-synuclein is considered possessing prion-like properties, propagating between neurons, neurons and glia cells, and even between animals (Makin, 2016).
By analyzing open chromatins of midbrain dopaminergic neurons and their correlations with putative enhancers, McClymont et al. (2018) recently revealed a novel functional cis-regulatory element (CRE; chr4: 90,721,063–90,722,122; hg19) located at SNCA intron 4. Variants of this CRE were shown to be associated with elevated PD risk in non-Hispanic Caucasians of European descent and be able to impact DNA-binding capacity of a set of proteins. Also, DNase hypersensitivity site analysis showed an interaction between the CRE and the SNCA promoter (Thurman et al., 2012; Wang et al., 2018). However, whether this SNCA CRE is associated with PD risk in other ethnicities remains unknown. In this study, we recruited a total of 1,020 individuals of Han Chinese descent and aimed to investigate those bearing CRE variants and their associations with PD risk.
Materials and Methods
Subjects
Enrolled in this study were 513 sporadic PD patients (264 males and 249 females) and 507 controls (287 males and 220 females) of Han Chinese ethnicity from southeast China. The median ages of the patients and controls were 64 (interquartile range, 55–71) and 65 (interquartile range, 60–73) years, respectively, following a normality test. PD patients were diagnosed by two movement disorder specialists according to the UK Parkinson’s disease Society Brain Bank Criteria. Patients with a family history of PD or with secondary and atypical Parkinsonism were excluded. Control subjects were free of neurological disorders according to medical history and physical and laboratory examinations. All subjects participating in the study signed written informed consent. The study was approved by the Ethics Committee of the Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University (No. KYKT2018-15, 03/02/2018).
Genotyping
Genomic DNA was extracted from human peripheral blood using an extraction kit from Tiangen (Beijing, China). The CRE fragment was amplified by polymerase chain reaction (PCR) in 20 μl of reaction mixture that contained 2 μl DNA template, 10 μl 2 × PCR Master Mix (Vazyme, Nanjing, China), and 1 μl of each primer. The primer pair was 5'-GGG AGG GAA CTA TGG AGG CAT C-3' and 5'-GGC CTA GCA GGG CGA GAA TAG-3'. The PCR condition was initial denaturation at 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 60 s, and a final extension at 72°C for 5 min. Variants were identified by bidirectional Sanger sequencing (BGI, Shanghai, China).
Data Analysis
Sequence data were analyzed using Lasergene (DNASTAR). Statistical analyses were performed using the statistical package of Predictive Analytics Software 18.0 (PASW, version 18.0) for Windows. Hardy-Weinberg equilibrium in genotype distribution, as well as differences in gender, genotype, allele, and haplotype frequencies, was assessed using the χ2 test. Mann-Whitney U test was used to evaluate age difference. Variants with significant difference were further analyzed using binary logistic regression model with gender and age as covariates. Haplotype association analysis was evaluated using SHEsis (Shi and He, 2005; Li et al., 2009). Linkage disequilibrium structure and r2 values of the SNCA loci were extracted from LDlink with the LDmatrix tool in the 1,000 Genomes Chinese population (Machiela and Chanock, 2015) and were plotted with R.1 Statistical significance was considered when p < 0.05.
Results
Identification of Variants in the CRE Region of SNCA Intron 4
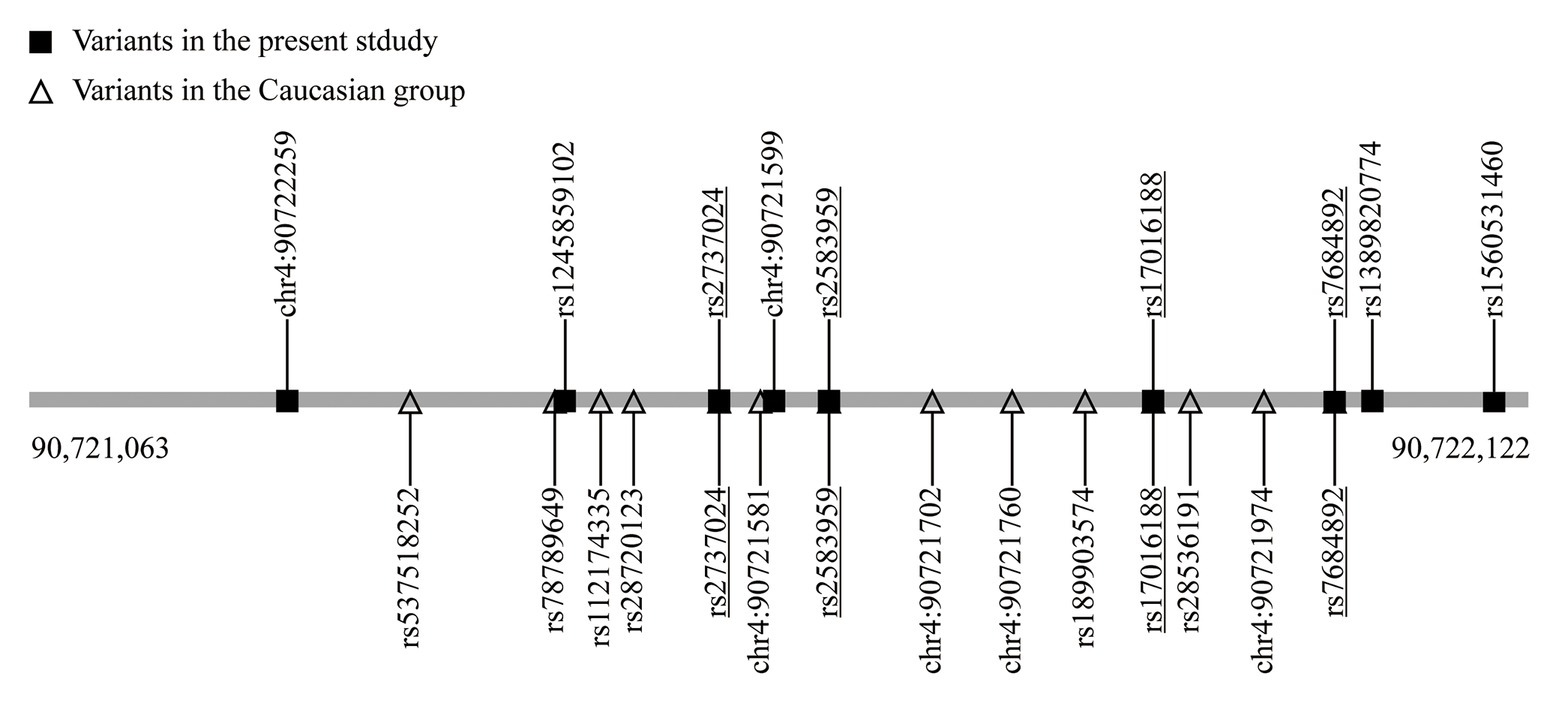
A total of nine variants were detected in the CRE region out of the 1,020 subjects (Table 1). Among these variants, eight were single nucleotide variation and one was insertion variation. Two single nucleotide variants, rs17016188 and rs7684892, had minor allele frequencies greater than 5%. The other six single nucleotide variants were found in heterozygous carriers. Based on the TOPMed project,2 the chr4: 90,721,599 (G > A) discovered in a PD patient was a new variant, and the rs1245859102 discovered in a control was a new G > T transversion instead of the usual G > A transition. The homozygous insertion variant, chr4: 90,722,259, was discovered in a control subject (Table 1).
Association Analysis of the Variants rs1701688 and rs7684892 With PD Risk
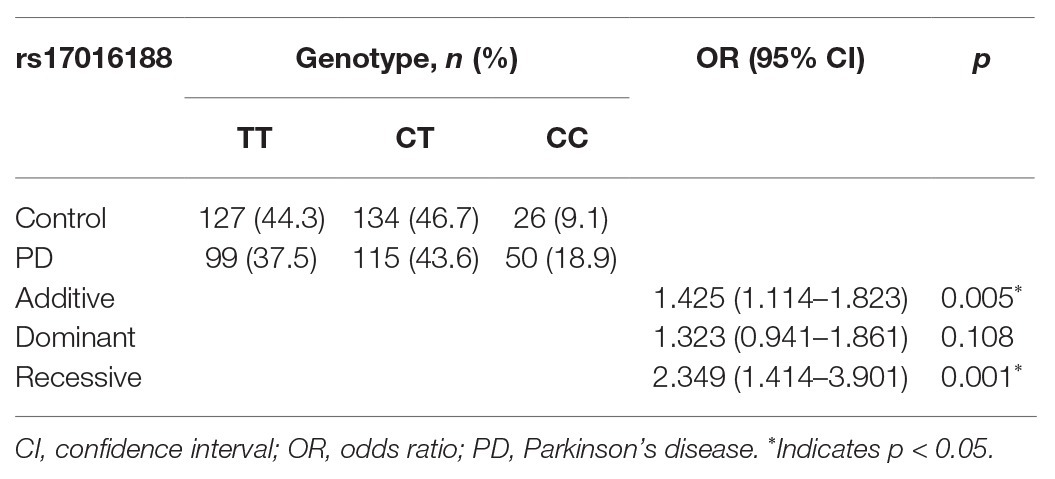
Genotype distribution of both rs17016188 and rs7684892 met with Hardy-Weinberg equilibrium (p > 0.05). The gender frequencies in the PD cases and controls were comparable (p > 0.05). There was a difference in age between the two groups (p < 0.05). Allele frequencies of both rs17016188 and rs7684892 showed marginal associations with PD risk in the total population (p = 0.049 and 0.051, respectively; Table 2). After Bonferroni correction, the difference, that is p < 0.025, was found in the male group in rs17016188 (allele: p = 0.004, OR = 1.433, 95% CI = 1.120–1.833; genotype: p = 0.003). Further multivariate analysis of rs17016188 with age adjusted confirmed the difference in the males with the C allele serving as a recessive risk factor (p = 0.001, OR = 2.349, 95% CI = 1.414–3.901; Table 3).
Haplotype Analysis of rs1701688 and rs7684892 in Association With PD
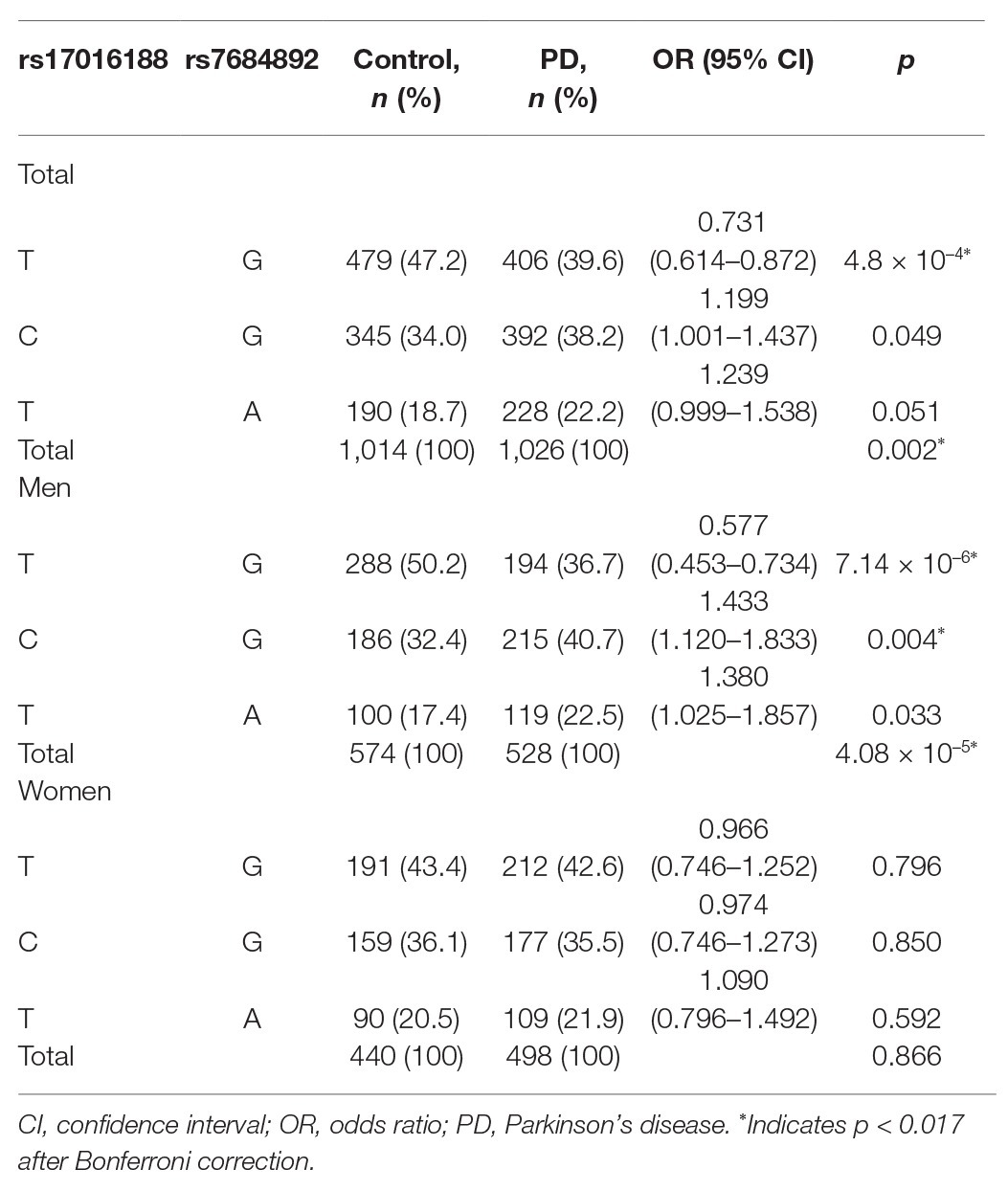
Haplotypes were constructed to understand the combined effect of rs17016188 and rs7684892 on PD risk (Table 4). The two variants displayed a linkage disequilibrium with r2 = 0.146 and D’ = 0.9993 in the cohort. Out of four potential combinations, three haplotypes were detected except for the rs17016188/C-rs7684892/A. An overall significance in haplotype distribution was found in the total and male populations (p = 0.002 and 4.08 × 10−5, respectively), but not in the females. The haplotype, rs17016188/T-rs7684892/G, exhibited a protective effect against PD in the total population (p = 4.8 × 10−4, OR = 0.731, 95% CI = 0.614–0.872) and in the males (p = 7.14 × 10−6, OR = 0.577, 95% CI = 0.453–0.734).
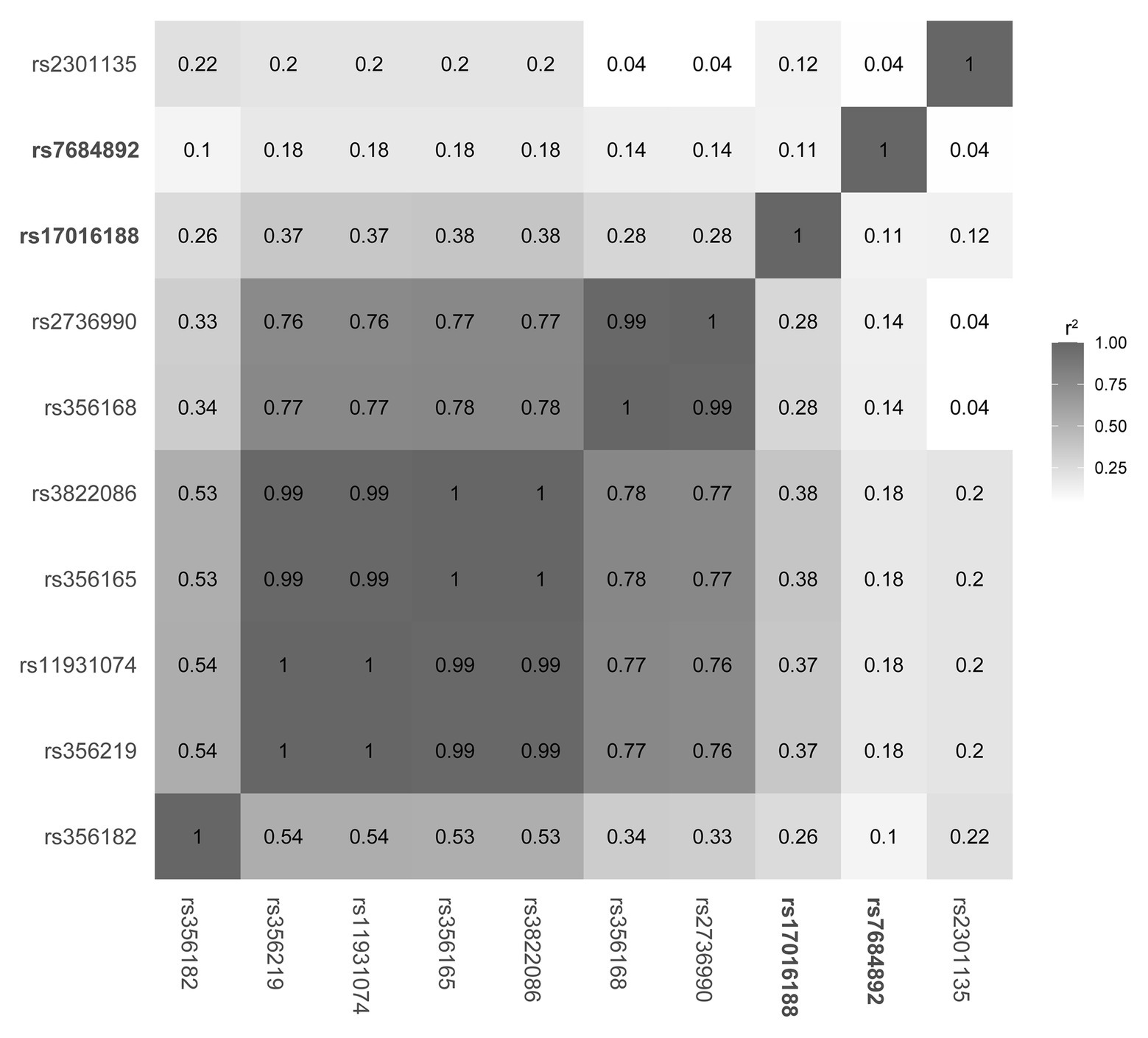
Linkage disequilibrium structure was subsequently constructed in the 1,000 Genomes data to understand how the two CRE variants might be interacting with other SNCA variants (Figure 1). Of the variants, rs356168 is a previous functionally validated variant (Soldner et al., 2016) and rs356182 is the lead variant of genome-wide associated studies (GWAS; Nalls et al., 2014; Chang et al., 2017). The rest are a panel of PD-associated variants identified across SNCA in Chinese ethnicity (Li et al., 2019). The linkage disequilibrium analysis showed a low linkage disequilibrium structure between the CRE variants (rs1701688 and rs7684892) and the previously reported SNCA sites.
Discussion
SNCA is a central piece of the genetic puzzle leading to PD pathogenesis. The recent advance of the SNCA intron 4 CRE in association with PD risk warrants further identification in additional ethnicities. The current study involving a Chinese population reveals nine variants, including one new rare variant (chr4: 90,721,599; G > A) and one variant with different nucleotide replacement (rs124589102; G > T). Four variants, namely rs17016188, rs7684892, rs2737024, and rs2583959, are common between ours and the 14 variants discovered in the European descendants (McClymont et al., 2018; Figure 2).
Current knowledge of biologically related noncoding variation is limited. It is considered that CRE candidates may serve functional roles in regulating gene expression (Snyder et al., 2020). Recent studies have highlighted the importance of CREs harboring functional variation as well as their regulated genes (Maurano et al., 2012; Thurman et al., 2012; Soldner et al., 2016; McClymont et al., 2018). For instance, the CRE in SNCA intron 4 may affect the midbrain-specific expression of SNCA, and the variants within the CRE can disturb binding affinity of a set of proteins, including NOVA1, APOBEC3C, PEG10, SNRPA, and CHMP5. Three of them, PEG10, SNRPA, and CHMP5, show increased affinities of binding to the minor risk allele, that is, G of rs2583959 or G of rs2737024. To understand whether the CRE variants are independently connected to PD, eight known PD-associated SNCA risk variants, including those functionally validated and leading in GWAS studies (Nalls et al., 2014; Soldner et al., 2016; Chang et al., 2017), and those identified in a Chinese population (Li et al., 2019), are used for linkage disequilibrium analysis. Results suggest that the CRE variants rs1701688 and rs7684892 and the resulting haplotypes are independent in association with PD. Though detailed mechanisms and consequences remain to be further explored, it is possible that the variants within the CRE, independently or in concert with additional variation, may influence SNCA expression and/or susceptibility of catecholaminergic neurons.
In line with the previous results (McClymont et al., 2018), both rs17016188 and rs7684892 show minor allele frequency over 5% and are not associated with PD risk in the overall population. While the gender-stratified analysis was not presented in the earlier study, our results suggest an impact of rs17016188 on PD risk in males. Similar results were also found in the PD-associated protective haplotype, rs17016188/T-rs7684892/G. It is not clear how gender bias occurs in the SNCA CRE variant and haplotype, which may need further verification in other populations. Nonetheless, the gender bias per se is not surprising for genetic association with PD risk (Palacios et al., 2010; Klebe et al., 2013; Wang et al., 2015). The male-to-female ratio of PD prevalence is approximately at 3:2 (de Lau and Breteler, 2006), probably due to genes in the sex chromosomes and sex hormone-related mechanisms (Jurado-Coronel et al., 2018).
The other two variants, rs2737024 and rs2583959, whose minor allele frequencies are above 5% in the Caucasian population, are significantly associated with PD risk (Heckman et al., 2012; McClymont et al., 2018). In contrast, it is reported that rs2583959 is not associated with multiple system atrophy and essential tremor (Al-Chalabi et al., 2009; Ross et al., 2011). Interestingly, both variants are rare single nucleotide polymorphisms in the Chinese population. Each variant is only found in two PD patients in heterozygous form, and coincidentally, their carriers are identical individuals. Although their frequencies are distinct between the Chinese and Caucasian populations, our results support the previous findings that the minor alleles of rs2737024 and rs2583959 aggravated PD risk and the two variants were in strong linkage disequilibrium (McClymont et al., 2018).
In conclusion, the current study reveals nine variants in the SNCA intron 4 CRE and demonstrates their associations with PD risk in a Chinese population. Our results provide further insight into how this specific gene regulatory region of SNCA harbors variants contributing to PD risk in different ethnicities.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
J-HZ and XZ designed the research. XZ, S-GZ, and BO collected and managed the samples. S-GZ, HL, and MM conducted the experiments. S-GZ, Z-FL, and LC analyzed the data. J-HZ and S-GZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funding from Zhejiang Provincial Natural Science Foundation (LD19H090001 and LZ19H090002), National Natural Science Foundation of China (81771380 and 81771510), Zhejiang Provincial Medical Technology Program (2019KY445), Wenzhou Municipal Science and Technology Bureau (Y20190074 and C20170003), and Wenzhou Medical University (QTJ10024 and 89218014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all of the subjects participating in this study and the colleagues for the management of the patients.
Footnotes
References
Al-Chalabi, A., Dürr, A., Wood, N. W., Parkinson, M. H., Camuzat, A., Hulot, J. S., et al. (2009). Genetic variants of the alpha-synuclein gene SNCA are associated with multiple system atrophy. PLoS One 4:e7114. doi: 10.1371/journal.pone.0007114
Alegre-Abarrategui, J., Brimblecombe, K. R., Roberts, R. F., Velentza-Almpani, E., Tilley, B. S., Bengoa-Vergniory, N., et al. (2019). Selective vulnerability in alpha-synucleinopathies. Acta Neuropathol. 138, 681–704. doi: 10.1007/s00401-019-02010-2
Chang, D., Nalls, M. A., Hallgrímsdóttir, I. B., Hunkapiller, J., van der Brug, M., Cai, F., et al. (2017). A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 49, 1511–1516. doi: 10.1038/ng.3955
de Lau, L. M., and Breteler, M. M. (2006). Epidemiology of Parkinson’s disease. Lancet Neurol. 5, 525–535. doi: 10.1016/s1474-4422(06)70471-9
Heckman, M. G., Soto-Ortolaza, A. I., Diehl, N. N., Carrasquillo, M. M., Uitti, R. J., Wszolek, Z. K., et al. (2012). Evaluation of the role of SNCA variants in survival without neurological disease. PLoS One 7:e42877. doi: 10.1371/journal.pone.0042877
Jurado-Coronel, J. C., Cabezas, R., Avila Rodriguez, M. F., Echeverria, V., Garcia-Segura, L. M., and Barreto, G. E. (2018). Sex differences in Parkinson’s disease: features on clinical symptoms, treatment outcome, sexual hormones and genetics. Front. Neuroendocrinol. 50, 18–30. doi: 10.1016/j.yfrne.2017.09.002
Klebe, S., Golmard, J. L., Nalls, M. A., Saad, M., Singleton, A. B., Bras, J. M., et al. (2013). The Val158Met COMT polymorphism is a modifier of the age at onset in Parkinson’s disease with a sexual dimorphism. J. Neurol. Neurosurg. Psychiatry 84, 666–673. doi: 10.1136/jnnp-2012-304475
Li, G., Ma, J., Cui, S., He, Y., Xiao, Q., Liu, J., et al. (2019). Parkinson’s disease in China: a forty-year growing track of bedside work. Transl. Neurodegener. 8:22. doi: 10.1186/s40035-019-0162-z
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., et al. (2009). A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 19, 519–523. doi: 10.1038/cr.2009.33
Machiela, M. J., and Chanock, S. J. (2015). LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557. doi: 10.1093/bioinformatics/btv402
Maurano, M. T., Humbert, R., Rynes, E., Thurman, R. E., Haugen, E., Wang, H., et al. (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. doi: 10.1126/science.1222794
McClymont, S. A., Hook, P. W., Soto, A. I., Reed, X., Law, W. D., Kerans, S. J., et al. (2018). Parkinson-associated SNCA enhancer variants revealed by open chromatin in mouse dopamine neurons. Am. J. Hum. Genet. 103, 874–892. doi: 10.1016/j.ajhg.2018.10.018
Nalls, M. A., Pankratz, N., Lill, C. M., Do, C. B., Hernandez, D. G., Saad, M., et al. (2014). Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993. doi: 10.1038/ng.3043
Obeso, J. A., Stamelou, M., Goetz, C. G., Poewe, W., Lang, A. E., Weintraub, D., et al. (2017). Past, present, and future of Parkinson’s disease: a special essay on the 200th Anniversary of the Shaking Palsy. Mov. Disord. 32, 1264–1310. doi: 10.1002/mds.27115
Palacios, N., Weisskopf, M., Simon, K., Gao, X., Schwarzschild, M., and Ascherio, A. (2010). Polymorphisms of caffeine metabolism and estrogen receptor genes and risk of Parkinson’s disease in men and women. Parkinsonism Relat. Disord. 16, 370–375. doi: 10.1016/j.parkreldis.2010.02.012
Ross, O. A., Conneely, K. N., Wang, T., Vilarino-Guell, C., Soto-Ortolaza, A. I., Rajput, A., et al. (2011). Genetic variants of α-synuclein are not associated with essential tremor. Mov. Disord. 26, 2552–2556. doi: 10.1002/mds.23909
Shi, Y. Y., and He, L. (2005). SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15, 97–98. doi: 10.1038/sj.cr.7290272
Snyder, M. P., Gingeras, T. R., Moore, J. E., Weng, Z., Gerstein, M. B., Ren, B., et al. (2020). Perspectives on ENCODE. Nature 583, 693–698. doi: 10.1038/s41586-020-2449-8
Soldner, F., Stelzer, Y., Shivalila, C. S., Abraham, B. J., Latourelle, J. C., Barrasa, M. I., et al. (2016). Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99. doi: 10.1038/nature17939
Thurman, R. E., Rynes, E., Humbert, R., Vierstra, J., Maurano, M. T., Haugen, E., et al. (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. doi: 10.1038/nature11232
Wang, J. Y., Gong, M. Y., Ye, Y. L., Ye, J. M., Lin, G. L., Zhuang, Q. Q., et al. (2015). The RIT2 and STX1B polymorphisms are associated with Parkinson’s disease. Parkinsonism Relat. Disord. 21, 300–302. doi: 10.1016/j.parkreldis.2014.12.006
Wang, Y., Song, F., Zhang, B., Zhang, L., Xu, J., Kuang, D., et al. (2018). The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19:151. doi: 10.1186/s13059-018-1519-9
Keywords: Parkinson’s disease, SNCA, cis-regulatory element, association, variant
Citation: Zhu S-G, Lu H, Mao M, Li Z-F, Cui L, Ovlyakulov B, Zhang X and Zhu J-H (2020) The cis-Regulatory Element of SNCA Intron 4 Modulates Susceptibility to Parkinson’s Disease in Han Chinese. Front. Genet. 11:590365. doi: 10.3389/fgene.2020.590365
Edited by:
Hao Deng, Central South University, ChinaReviewed by:
Qiying Sun, Central South University, ChinaAtif Adnan, China Medical University, China
Copyright © 2020 Zhu, Lu, Mao, Li, Cui, Ovlyakulov, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Hong Zhu, amh6aHVAd211LmVkdS5jbg==; Xiong Zhang, emhhbmd4aW9uZzk4QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Shi-Guo Zhu1,2†
Shi-Guo Zhu1,2† Hui Lu
Hui Lu Xiong Zhang
Xiong Zhang Jian-Hong Zhu
Jian-Hong Zhu