- 1Institute of Insect Sciences, Zhejiang University, Hangzhou, China
- 2Ministry of Agriculture Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Zhejiang University, Hangzhou, China
- 3State Key Laboratory of Rice Biology, Zhejiang University, Hangzhou, China
Nuclear factor Y (NF-Y) is a heterotrimeric transcription factor with the ability to bind to a CCAAT box in nearly all eukaryotes. However, the function of NF-Y in the life-history traits of insects is unclear. Here, we identified three NF-Y subunits, NlNF-YA, NlNF-YB, and NlNF-YC, in the wing-dimorphic brown planthopper (BPH), Nilaparvata lugens. Spatio-temporal analysis indicated that NlNF-YA, NlNF-YB, and NlNF-YC distributed extensively in various body parts of fourth-instar nymphs, and were highly expressed at the egg stage. RNA interference (RNAi)-mediated silencing showed that knockdown of NlNF-YA, NlNF-YB, or NlNF-YC in third-instar nymphs significantly extended the fifth-instar duration, and decreased nymph-adult molting rate. The addition of 20-hydroxyecdysone could specifically rescue the defect in adult molting caused by NlNF-YARNAi, indicating that NlNF-Y might modulate the ecdysone signaling pathway in the BPH. In addition, NlNF-YARNAi, NlNF-YBRNAi, or NlNF-YCRNAi led to small and moderately malformed forewings and hindwings, and impaired the normal assembly of indirect flight muscles. Adult BPHs treated with NlNF-YARNAi, NlNF-YBRNAi, or NlNF-YCRNAi produced fewer eggs, and eggs laid by these BPHs had arrested embryogenesis. These findings deepen our understanding of NF-Y function in hemipteran insects.
Introduction
Nuclear transcription factor Y (NF-Y) is an evolutionarily conserved transcription factor that exists in almost all organisms, from prokaryotes to eukaryotes (Dorn et al., 1987; Laloum et al., 2013). NF-Y exerts differential regulation on a wide variety of genes through binding to a CCAAT box, one of the most ubiquitous elements in eukaryotic promoters (Li et al., 1992; Mantovani, 1998; Suzuki et al., 2001). NF-Y consists of three subunits, NF-YA, NF-YB, and NF-YC, all required for DNA-binding (McNabb et al., 1995; Sinha et al., 1995; Bi et al., 1997; Mantovani, 1999). Accumulated evidence indicates that animal NF-Y is essential for numerous biological processes involved in proliferation and apoptosis, cancer and tumorigenesis, stress responses, growth, and development (Li et al., 2018). For instance, the deletion of the mouse NF-YA homolog causes early embryo lethality (Bhattacharya et al., 2003). Down-regulation of mouse NF-YA was found to reduce the expression of several cell cycle control genes in differentiated muscle cells, suggesting NF-YA is involved in specifying myogenic fate (Gurtner et al., 2003). Knockdown or overexpression of the Drosophila melanogaster NF-YA homolog in all tissues results in lethality at the larval stage, indicating that a certain level of NF-YA is necessary for viability in flies (Yoshioka et al., 2007). However, he contribution of NF-Y homologs to the life-history traits of other insect species has yet to be reported.
The wing-dimorphic brown planthopper (BPH), Nilaparvata lugens (Hemiptera: Delphacidae), is the most destructive pest of rice in Asia (Xue et al., 2014), causing a loss in rice production of more than $300 million annually in Asia. It feeds exclusively on the phloem sap of the rice plant and transmits plant viruses such as rice ragged stunt virus and rice grassy stunt virus, resulting in loss of plant vigor and reduced yield (Backus et al., 2005; Otuka, 2013). The BPH is a hemimetabolous insect, the newly hatched first-instar nymphs look like miniature adults except for the absence of wings and sexual immaturity, and grow gradually within increasing stages. The BPH has five nymphal stages, and wing buds grow with each of these stages, but short- and long-winged morphs are externally indistinguishable until the adults emerge (Xu and Zhang, 2017). Long-winged BPH adults have well-developed forewings, hindwings, and indirect flight muscles (IFM), whereas, short-winged adults have undeveloped forewings and rudimentary hindwings (Xu and Zhang, 2017; Zhang et al., 2019). Wing dimorph and high fecundity are the two most important biological features contributing to the ecological success of the BPH (Zhang et al., 2019). Recently, facilitated by the development of genetic tools (Xu et al., 2013; Xue et al., 2018) and genomic information (Xue et al., 2014), the molecular basis underlying wing dimorphism (Xu et al., 2015) and population resurgence (Wu et al., 2020) are emerging, which offers a basis for the design of new control agents to combat BPH infestation.
In the present study, we identified the BPH NF-YA (NlNF-YA), NF-YB (NlNF-YB), and NF-YC (NlNF-YC) homologs by searching the BPH genomic and transcriptomic databases. RNA interference (RNAi)-mediated gene silencing showed that NlNF-YA, NlNF-YB, and NlNF-YC all played pivotal roles in nymph growth, wing formation, IFM development, and reproduction. In addition, we found that NF-Y regulated adult eclosion most likely through linking to the ecdysone signaling pathway. Our findings deepen our understanding of the function of NF-Y in hemipteran insects.
Materials and Methods
Insects
The BPH strain was originally collected from a rice field in Hangzhou, China. Insects were reared on rice seedlings (rice variety: Xiushui 134) in a walk-in chamber at 26 ± 0.5°C, with a light: dark photoperiod of 16: 8 h and relative humidity of 50 ± 5%.
Gene Identification and Sequence Analysis
The amino acid sequences of Drosophila NF-Y homologs were used to screen BPH genomic and transcriptomic databases for homologs. Total RNA was isolated from BPH nymphs using RNAiso plus (Takara #9109) according to the manufacturer’s protocol. For cDNA syntheses, 450 ng of total RNA was reversely transcribed in a 10 μL reaction with HiScript QRT SuperMix (Vazyme #R223-01). The NlNF-YA, NlNF-YB, and NlNF-YC sequences were amplified from cDNA using fidelity DNA polymerase (Toyobo #930700) with NlNF-YA-F/R, NlNF-YB-F/R, and NlNF-YC-F/R primer pairs, respectively (Supplementary Table 1). The PCR product was cloned and the sequence was determined by Sanger sequencing.
To identify the exon-intron construction of NlNF-Y, open reading frames (ORFs) of NlNF-YA, NlNF-YB, and NlNF-YC were used to search a BPH genomic database (Xue et al., 2014). For phylogenetic analysis, 39 NF-Y sequences from 13 species including D. melanogaster, N. lugens, Acyrthosiphon pisum, Apis cerana, Bemisia tabaci, Bombyx mori, Cryptotermes secundus, Halyomorpha halys, Monomorium pharaonis, Nasonia vitripennis, Nicrophorus vespilloides, Tribolium castaneum, and Zootermopsis nevadensis were used for sequence alignment. A phylogenetic tree was constructed with maximum-likelihood method and 1000 bootstraps using the MEGA 7 program (Kumar et al., 2016).
Spatio-Temporal Expression Pattern of NlNF-Y
Total RNA was isolated from eggs (n = 100), first-instar (n = 100), second-instar (n = 50), third-instar (n = 50), fourth-instar (n = 30), fifth-instar nymphs (n = 15), and adult females (n = 15), which were laid or ecdysed within 24 h. To examine tissue distribution, we dissected the head (n = 50), antenna (n = 200), tergum (n = 50), leg (n = 100), fat body (n = 30), cuticle (n = 50), and gut (n = 100) from four-instar nymphs for RNA extraction. First-strand cDNA was synthesized from total RNA (450 ng) using HiScript QRT SuperMix (Vazyme #R223-01). The synthesized cDNAs were 10-fold diluted and used as templates for quantitative real-time PCR (qPCR). The qPCR primers for NlNF-Y (Supplementary Table 1) were designed using Primer-Blast.1 The qPCR was conducted on a CFX96TM real-time PCR detection system (Bio-Rad) using the following conditions: denaturation for 3 min at 95°C, followed by 40 cycles at 95°C for 10 s, and 60°C for 30 s. The ribosomal protein S11 gene (rps11) was used as the internal reference gene (Yuan et al., 2014). The 2–ΔΔCt method (Ct represents the cycle threshold) was used to measure the relative expression level (Livak and Schmittgen, 2001). Three independent biological replicates with three technical replicates were conducted for each experiment.
RNAi and Microinjection
The dsRNAs were synthesized using a T7 High Yield Transcription Kit (Vazyme #TR101-02) according to the manufacturer’s instructions with primers containing the T7 RNA polymerase promoter at both ends (Supplementary Table 1). A dsRNA injection was carried out as in our previous study (Xu et al., 2015). Briefly, fourth-instar nymphs were anesthetized with carbon dioxide for 10–15 s. Approximately 150 ng dsRNA was microinjected into the mesothorax using a FemtoJet microinjection system (Eppendorf). To investigate whether disruption of NlNF-YA affected ecdysone signaling activity, fifth-instar nymphs (n = 10 for each of three replicates) were collected to examine the expression levels of NlE74A and NlE75B by qPCR using corresponding primers (Supplementary Table 1). To investigate RNAi efficiency, BPHs (n = 5 for each of three replicates) at 24 h after adult eclosion were collected for qPCR assay.
Forewing Size and Hind Tibiae Length
Fourth-instar nymphs (36–48 h after ecdysis, hAE) were microinjected with dsRNAs targeting NlNF-YA (dsNlNF-YA), NlNF-YB (dsNlNF-YB), NlNF-YC (dsNlNF-YC), or GFP (dsGFP). After adult eclosion (24 h), images of the forewings and hind tibiae were captured with a DFC320 digital camera attached to a Leica S8AP0 stereomicroscope using a LAS digital imaging system (v. 3.8). Digital images of forewings (n = 20) and hind tibias (n = 20) were collected for the measurement of forewing size and hind tibia length using ImageJ (v. 1.47).
Microinjection With 20-hydroxyecdysone (20E)
Fourth-instar nymphs (36–48 hAE) were microinjected with corresponding dsRNAs targeting each gene, and then nymphs were maintained on fresh rice seedings until the fifth-instar stage (within 48 h). Fifth-instar nymphs were microinjected with 20E (1 mg/ml) or distilled water, and the adult eclosion rate was calculated.
Transmission Electron Microscopy
Fourth-instar nymphs (36–48 hAE) were microinjected with dsRNAs targeting NlNF-YA, NlNF-YB, NlNF-YC, or GFP. The thoraxes were dissected from adult females (24 h after emergence) for Transmission Electron Microscopy (TEM) as in our previous study (Xue et al., 2013). Briefly, samples were fixed in 2.5% glutaraldehyde overnight at 4°C. After fixation, samples were post-fixed in 1% osmium tetroxide for 1.5 h. Then, samples were dehydrated in a standard ethanol/acetone series, infiltrated and embedded in Spurr medium, and then superthin sections were cut. The sections were stained with 5% uranyl acetate followed by Reynolds’ lead citrate solution and observed under a JEM-1230 transmission electron microscope (JEOL).
Fecundity Assay
Newly emerged females and males (within 12 h after eclosion) were microinjected with dsNF-YA, ds-NF-YB, dsNF-YC, or dsGfp. Then each female was allowed to match with two males in a glass tube. Insects were removed at 5 days after matching, and eggs were counted under a Leica S8AP0 stereomicroscope.
Data Analysis
Statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software). Means were compared using a two-tailed Student’s t-test and log-rank (Mantel-Cox) test at a significance level set at ∗P < 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001.
Results
NlNF-YA, NlNF-YB, and NlNF-YC Sequence Analysis
We identified three genes encoding BPH NF-Y homologs, NlNF-YA, NlNF-YB, and NlNF-YC, in a BLAST search against BPH genomic (Xue et al., 2014) and transcriptome databases using the D. melanogaster NF-Y proteins as query sequences. The ORFs of NlNF-YA, NlNF-YB, and NlNF-YC were 1002-, 609-, and 1011-bp in length, encoding 333, 202, and 336 amino acid residues, respectively (Supplementary Figures 1–3). Exon-intron construction analysis showed that the NlNF-YA, NlNF-YB, and NlNF-YC ORFs consist of eight, five, and eight exons, located in scaffolds 6450/1657, 3414, and 1809, respectively. A phylogenetic analysis based on NF-Y homologs from 13 species showed that NlNF-YA, NlNF-YB, and NlNF-YC together with their counterparts formed three distinct clusters (Figure 1), indicating that the BPH NF-Y genes we identified were authentic NF-Y homologs.
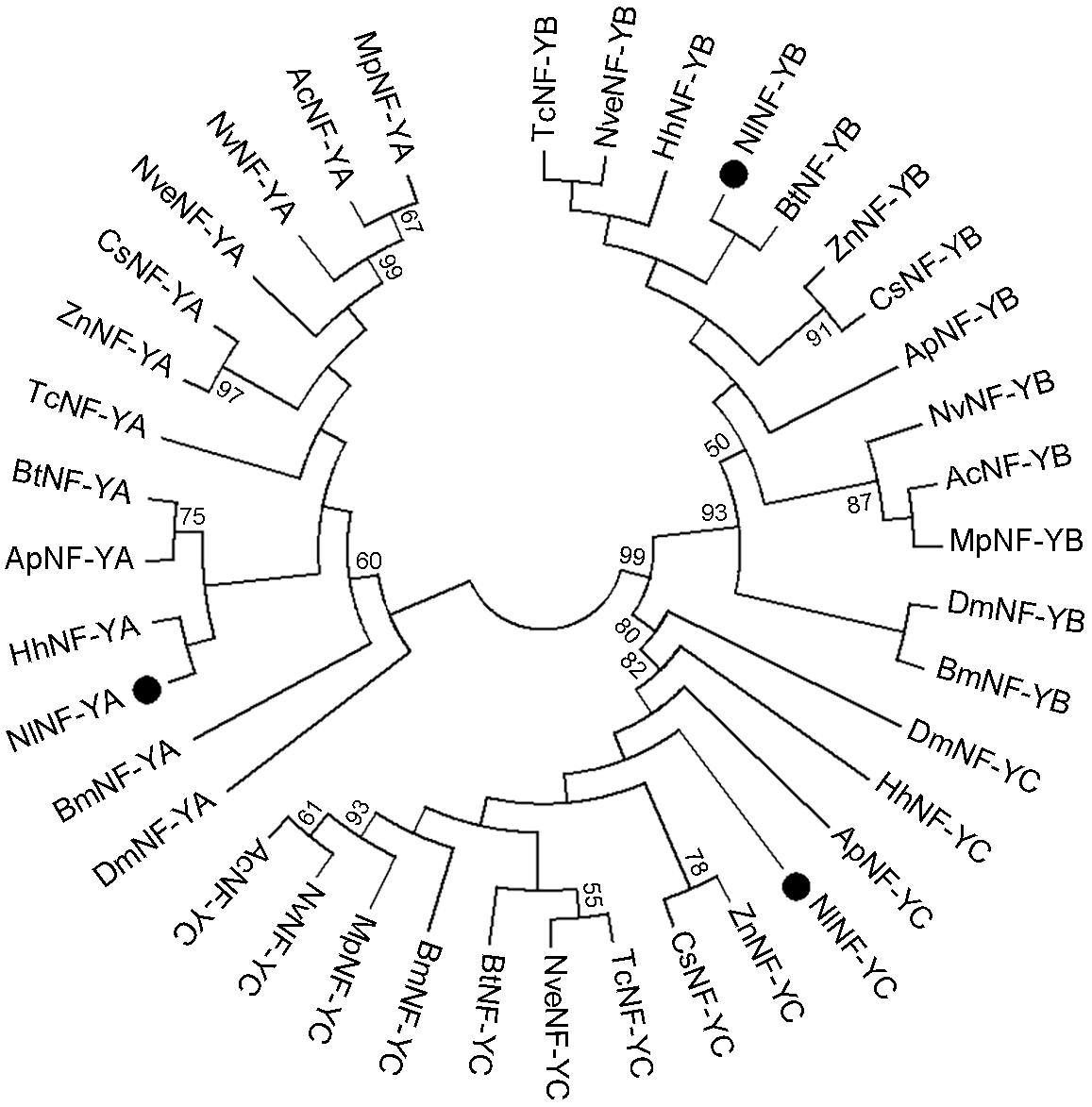
Figure 1. Phylogenetic analysis of NlNF-Y members. The phylogenetic tree was constructed with 39 full-length NlNF-Y members from 13 species using the maximum-likelihood method and bootstrapping was set to 1,000 replications. NlNF-YA, NlNF-YB, and NlNF-YC are indicated by dots. Nl, Nilaparvata lugens; Dm, Drosophila melanogaster; Ap, Acyrthosiphon pisum; Ac, Apis cerana; Bt, Bemisia tabaci; Bm, Bombyx mori; Cs, Cryptotermes secundus; Hh, Halyomorpha halys; Mp, Monomorium pharaonis; Nv, Nasonia vitripennis; Nve, Nicrophorus vespilloides; Tc, Tribolium castaneum, and Zn, Zootermopsis nevadensis.
Spatio-Temporal Analysis of NlNF-YA, NlNF-YB, and NlNF-YC
Spatio-temporal expression of NlNF-YA, NlNF-YB, and NlNF-YC was examined by qPCR. NlNF-YA, NlNF-YB, and NlNF-YC transcripts were readily detected from egg to adult stages, and relatively high levels were detected in eggs laid within 24 h (Egg-24 h) (Figure 2A), indicating that they might play important functions in egg development. Tissue distribution analysis showed that NlNF-YA, NlNF-YB, and NlNF-YC were evenly expressed in head, antenna, tergum, leg, fat body, cuticle, and gut of fourth-instar nymphs (Figure 2B), indicating NlNF-Y genes might be important for BPH growth and development.
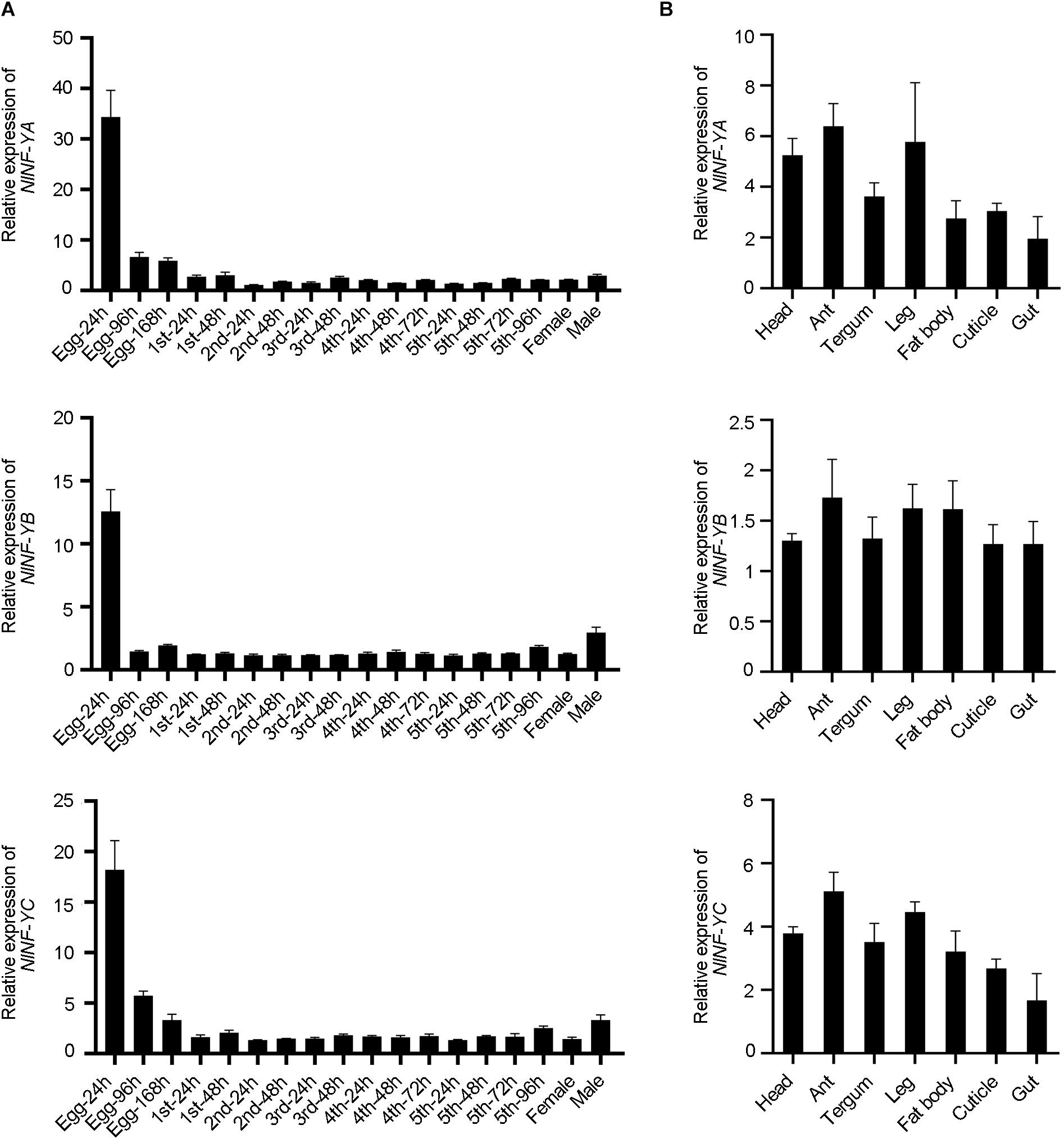
Figure 2. Spatio-temporal expression of NlNF-Y genes. (A) Developmental profile of NlNF-YA, NlNF-YB, and NlNF-YC. Total RNAs were isolated from eggs (n = 100), first-instar (n = 100), second-instar (n = 50), third-instar (n = 50), fourth-instar (n = 30), fifth-instar nymphs (n = 15), and adult females (n = 15). (B) Tissue distribution of NlNF-Y. Total RNAs were extracted from the head (n = 30), antenna (ant, n = 200), tergum (n = 50), leg (n = 100), fat body (n = 30), cuticle (n = 50), and gut (n = 100) from fourth-instar nymphs. First-strand cDNA was synthesized using random primers, and qPCR was conducted using specific primers corresponding to each gene. The relative expression level was normalized by the ribosomal S11 gene (rps11). Bars represent s.d. derived from three independent biological replicates.
Knockdown of NlNF-Y Genes Leads to Small and Malformed Wings
To investigate whether NlNF-YA, NlNF-YB, and NlNF-YC played any roles in BPH development, fourth-instar short-wing-destined nymphs were microinjected with corresponding dsRNAs targeting each gene. At 48 h after microinjection, RNAi efficiency was examined by qPCR, which showed that dsRNA treatments significantly down-regulated the expression levels of NlNF-YA, NlNF-YB, and NlNF-YC by 63.7, 99.9, and 82.7%, respectively, relative to the GFPRNAi treatment (Figure 3A). Notably, NlNF-YARNAi, NlNF-YBRNAi, and NlNF-YCRNAi caused high mortality, leading to approximately 60, 60, and 40% of nymphs died before adult eclosion (Figure 3B), respectively. The remaining nymphs could successfully molt into adults. However, these adults had moderately smaller and slightly malformed forewings relative to GFPRNAi BPHs (Figures 4A,B). NlNF-YARNAi, NlNF-YBRNAi, and NlNF-YCRNAi specifically reduced forewing size but had marginal roles on hind tibiae length (Figure 4C). These data indicated that NlNF-YA, NlNF-YB, and NlNF-YC were essential for normal wing growth in BPHs.
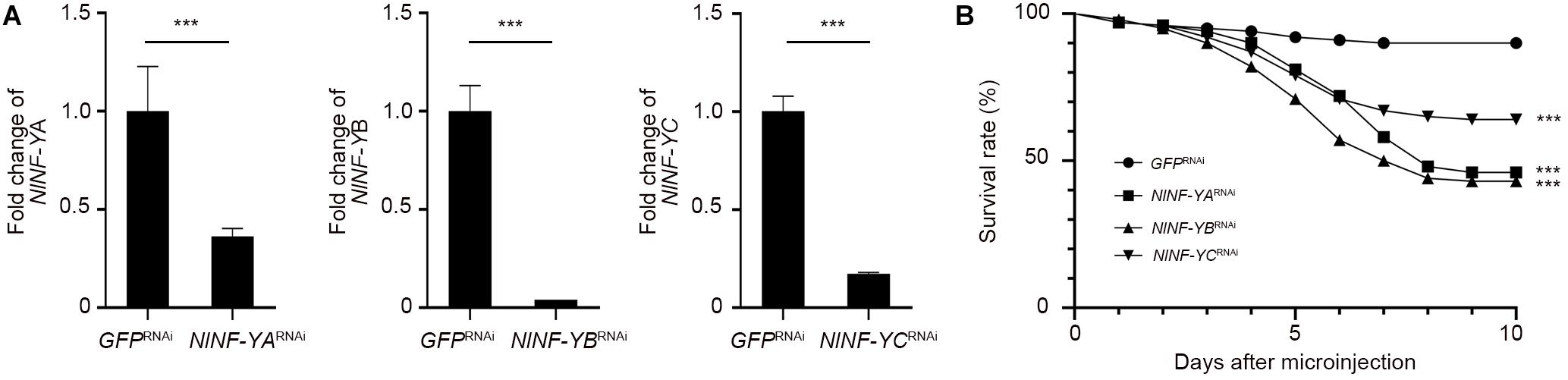
Figure 3. The survival rate of BPHs after dsRNA treatment. (A) Examination of RNAi efficiency. Fifth-instar nymphs (0–24 hAE) were microinjected with dsRNAs, and BPHs (n = 5 for each of three replicates) at 24 h after adult eclosion were collected for qPCR assay. The relative expression of NlNF-YA, NlNF-YB, and NlNF-YC was normalized to the expression of rps11. (B) The survival rate of BPHs after dsRNA treatment. Fourth-instar nymphs (n = 100) were microinjected with dsRNAs targeting NlNF-YA, NlNF-YB, NlNF-YC, or Gfp. Statistical analysis was performed by log-rank (Mantel-Cox) test. NlNF-YARNAi, dsNlNF-YBRNAi, or dsNlNF-YCRNAi significantly decreased the survivability of nymphs compared to GFPRNAi (***P < 0.001).
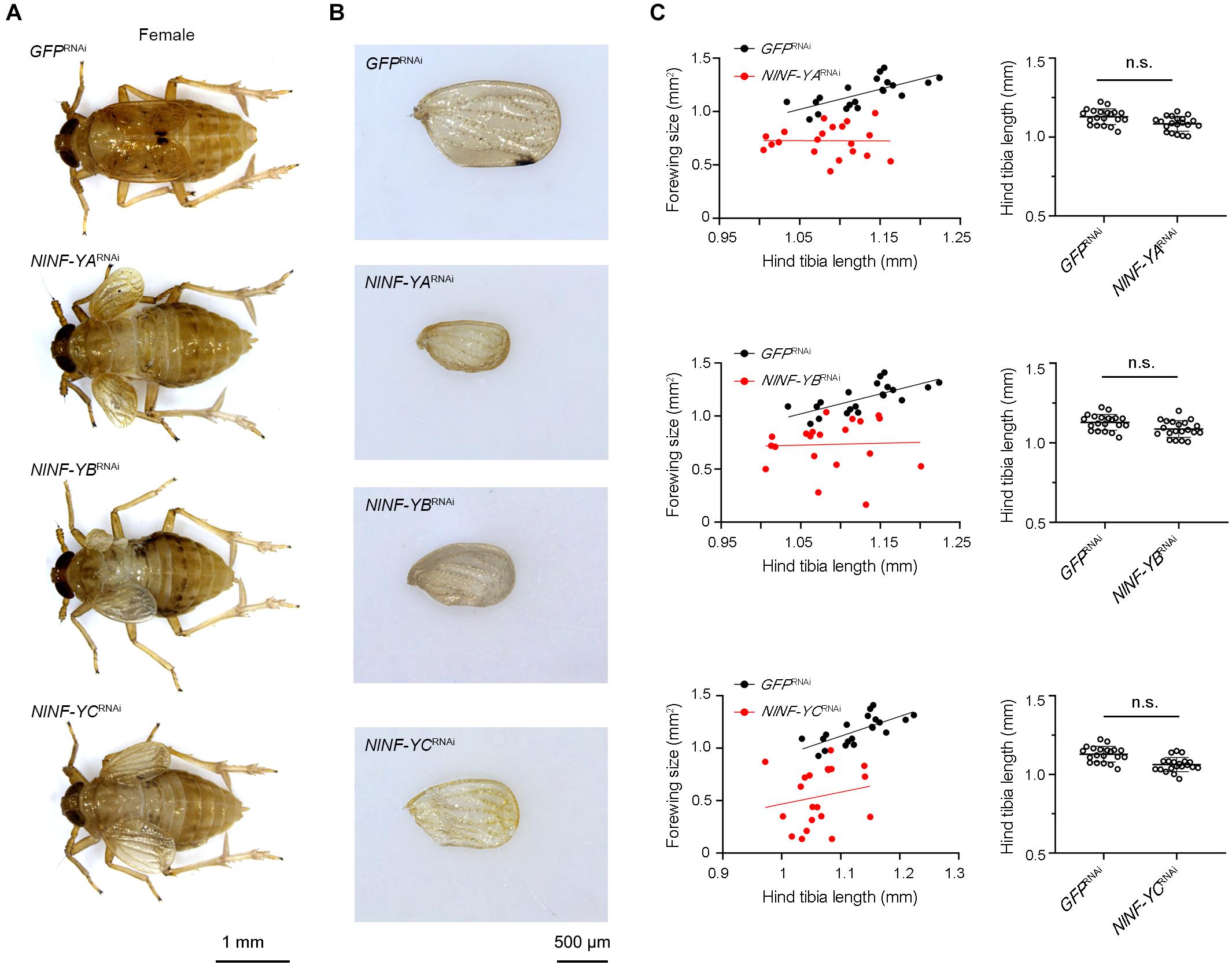
Figure 4. Knockdown of NlNF-Y reduces forewing size in short-winged BPHs. (A) Morphology of dsRNA-treated short-winged BPHs. (B) The size of forewings after dsRNA treatment. (C) Relative wing size and tibia length in BPHs with NlNF-Y or Gfp knockdown. Each dot represents the wing size and tibia length derived from an individual female (n = 20). Bars represent the mean ± s.d. derived from independent biological replicates (n = 20). Statistical comparisons between two groups were performed using a two-tailed Student’s t-test (n.s., non-significant).
Knockdown of NlNF-Y Genes Leads to Defective IFM
Because short-wing BPHs lack hindwings and IFM, we used long-wing BPHs to investigate whether NlNF-YA, NlNF-YB, and NlNF-YC had any effect on IFM development. For this, fourth-instar long-wing-destined nymphs were microinjected with corresponding dsRNAs targeting each gene. NlNF-YARNAi, NlNF-YBRNAi, and NlNF-YCRNAi gave rise to adults with curved forewings and hindwings (Figure 5A). In line with the phenotype in short-winged BPHs, smaller forewings and hindwings were observed in adults previously treated with either NlNF-YARNAi, NlNF-YBRNAi, or NlNF-YCRNAi (Figure 4B), although there was no discernable effect on hind tibia length (Figure 4C). Next, these adults were collected for IFM ultrastructure examination using TEM. In GFPRNAi adults, sarcomeres were clearly divided by Z discs and well-organized (Figure 6). In contrast, NlNF-YARNAi, NlNF-YBRNAi, or NlNF-YCRNAi led to defective sarcomeres, with deformed Z discs and weakly organized myofibrillar. These events indicated that the NlNF-Y complex might be essential for IFM assembly in BPH.
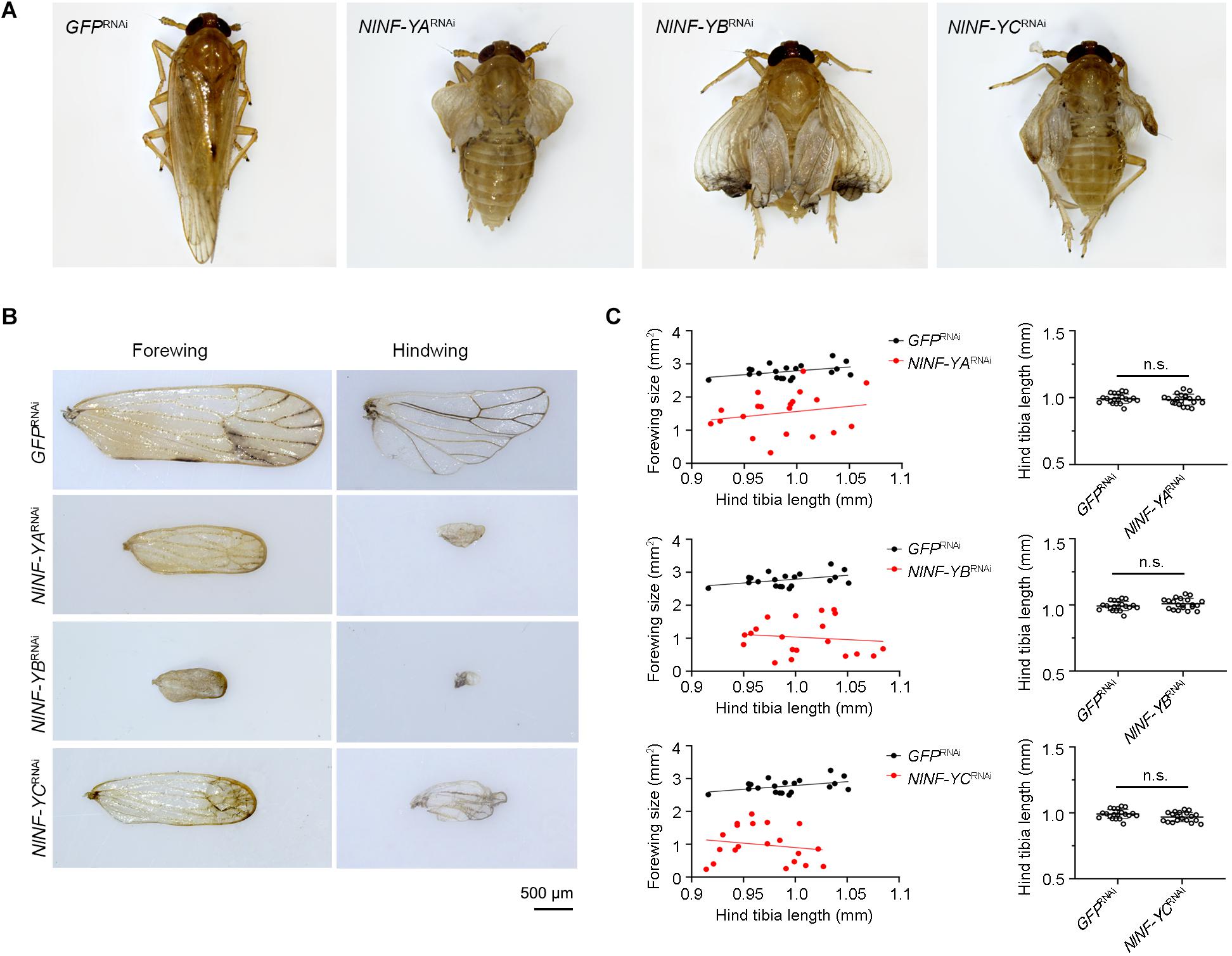
Figure 5. Knockdown of NlNF-Y reduces forewing size in long-winged BPHs. (A) Morphology of dsRNA-treated long-winged BPHs. (B) The size of forewings and hindwings after dsRNA treatment. (C) Relative wing size and tibia length in BPHs with NlNF-Y or Gfp knockdown. Each dot represents the wing size and tibia length derived from an individual female (n = 20). Bar represents mean ± s.d. derived from independent biological replicates (n = 20). Statistical comparisons between two groups were performed using a two-tailed Student’s t-test (n.s., non-significant).
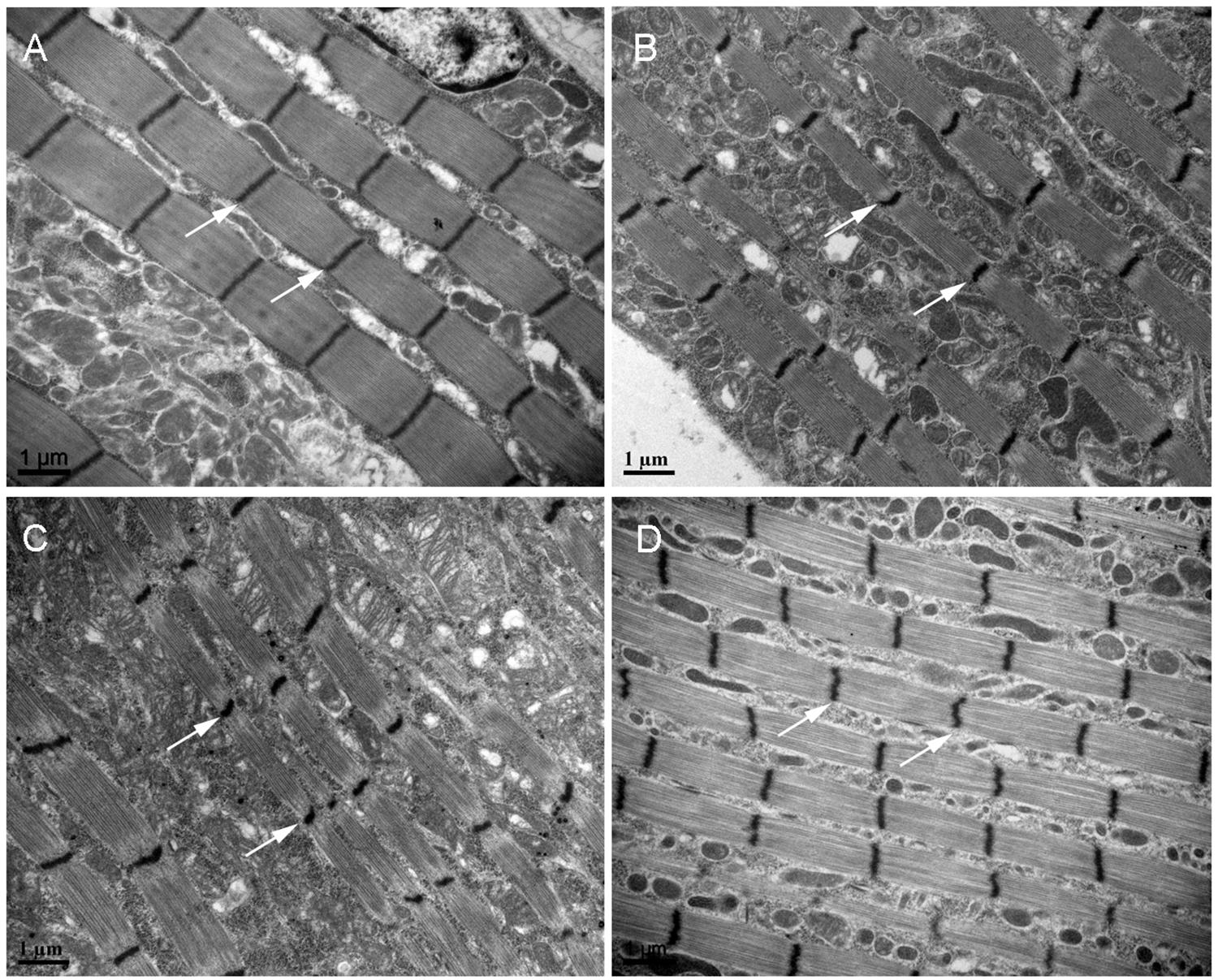
Figure 6. TEM of the IFM of dsRNA-treated long-winged BPHs. Fourth-instar nymphs were treated with dsRNAs targeting Gfp (A), NlNF-YA (B), NlNF-YB (C), or NlNF-YC (D). Thorax was dissected from female adults (24 h after eclosion) for TEM. The Z discs are indicated by arrows.
Knockdown of NlNF-Y Genes Impairs Nymphal Growth and Adult Molting
Given that NlNF-Y genes were evenly expressed across nymphal stages (Figure 2A), we investigated whether knockdown of NlNF-Y genes would affect nymph growth. For this purpose, third-instar nymphs were microinjected with corresponding dsRNAs targeting each gene. Nymphs treated with NlNF-YARNAi, NlNF-YBRNAi, or NlNF-YCRNAi had an extended fifth-instar duration compared with those treated with GFPRNAi (Figure 7A), with NlNF-YARNAi showing the most profound effect (Figure 7A). In addition, the majority of NlNF-YARNAi-treated nymphs died before adult emergence (Figure 7B), indicating that the disruption of the NF-Y complex impaired nymph-to-adult ecdysis.
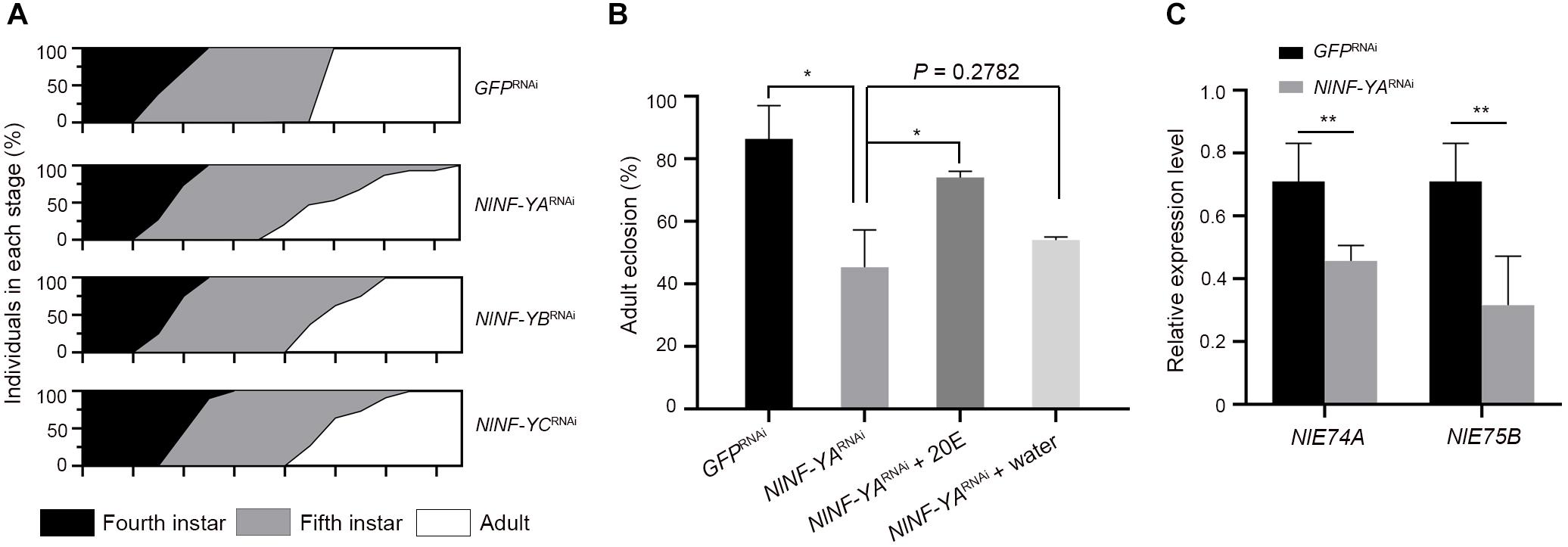
Figure 7. Nymphal duration and adult eclosion of dsRNA-treated BPHs. (A) Third-instar nymphs (0–24 hAE) were microinjected with dsRNAs targeting NlNF-YA (n = 660), NlNF-YB (n = 435), NlNF-YC (n = 480), or Gfp (n = 442). The curve in different developmental stages indicates the molting ratio. (B) The nymph-adult eclosion rate of BPHs. Fourth-instar nymphs (36–48 hAE) were microinjected with dsRNAs targeting NlNF-YA or Gfp, and then 20E or distilled water was microinjected when nymphs proceeded the fifth-instar stage. (C) The expression of NlE74A and NlE75B in context of NlNF-YA knockdown. The relative expressions of NlE74A and NlE75B were normalized to the expression of rps11. Statistical comparisons between two groups were performed using two-tailed Student’s t-test (*P < 0.05, and **P < 0.01).
Given that ecdysone is a hormone that initiates all major developmental transitions from the egg, to larva, to pupa, to adult in insects (Gilbert et al., 2002; Dubrovsky, 2005), we asked whether it mediated the effects of NlNF-YA on adult molting. For this purpose, we first assessed the expression levels of two early ecdysone response genes, NlE74A and NlE75E (Dubrovsky, 2005), in the context of NlNF-YA knockdown. NlNF-YARNAi significantly reduced both NlE74A and NlE75E transcripts compared with GFPRNAi (Figure 7C), indicating that knockdown of NlNF-YA might impair ecdyson signaling activity. Following this observation, we asked whether the addition of ecdysone could rescue the NlNF-YARNAi defect. To this end, fourth-instar nymphs were microinjected with dsNlNF-YA, followed by microinjection with 20E when nymphs proceeded to the fifth-instar at 48 h after eclosion. The addition of 20E could partially restore the molting defect caused by NlNF-YARNAi (Figure 7B) compared with the addition of water, indicating that NlNF-YARNAi decreased adult eclosion at least partially through the ecdysone signaling pathway.
Knockdown of NlNF-Y Genes Impairs Reproduction
Given that relatively high amounts of NlNF-YA, NlNF-YB, and NlNF-YC transcripts were detected at the early egg stage (Figure 2A), we asked whether NlNF-YA, NlNF-YB, and NlNF-YC contributed to BPH reproduction. Female and male adults at 12 h after eclosion were microinjected with corresponding dsRNAs targeting each gene, and then allowed to mate and to lay eggs for 5 days. BPHs treated with NlNF-YARNAi, NlNF-YBRNAi, and NlNF-YCRNAi laid substantially fewer eggs than those treated with GFPRNAi (Figure 8A). In addition, the eggs laid by BPHs previously treated with NlNF-YARNAi, NlNF-YBRNAi, or NlNF-YCRNAi failed to develop eye pigmentation, a characteristic hallmark of egg development, indicating NlNF-Y genes might be essential for embryogenesis (Figure 8B).
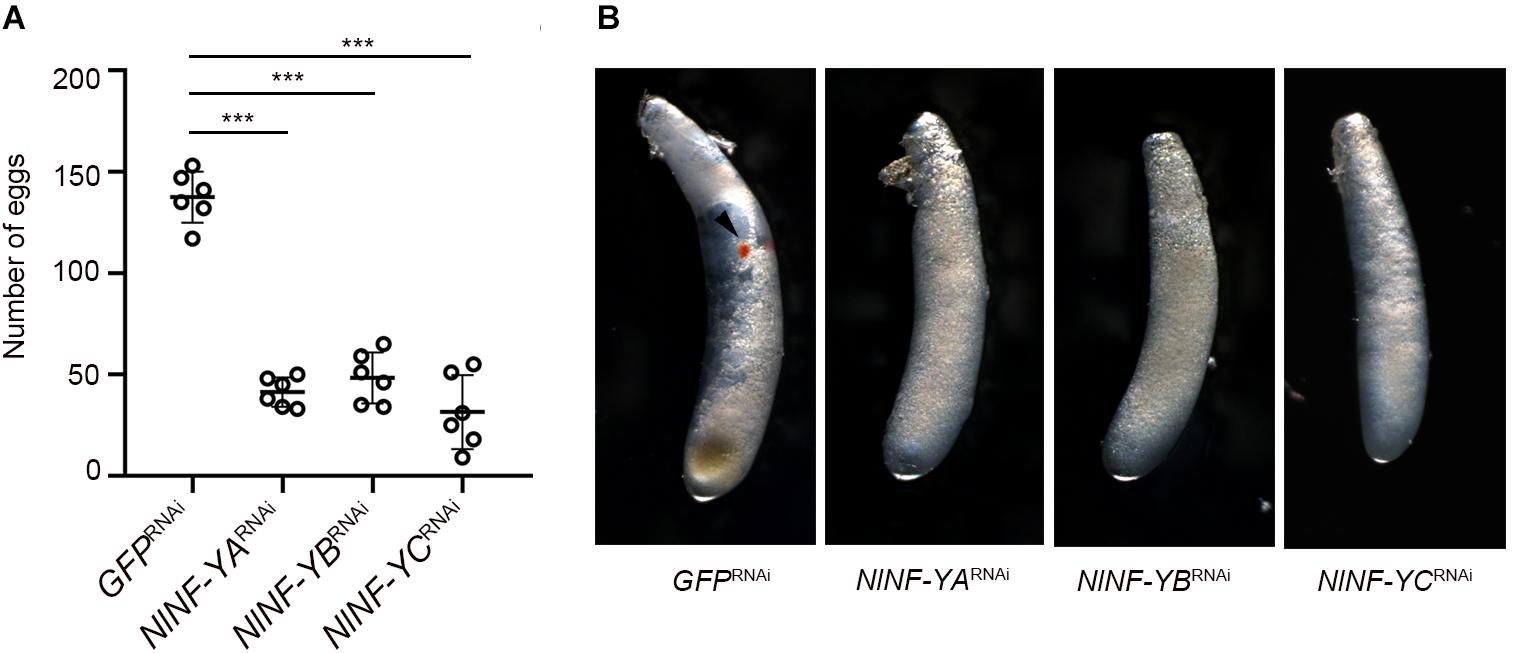
Figure 8. Fecundity of adults with gene knockdown and phenotype of eggs produced. (A) Newly emerged adults (0–12 hAE) were treated with dsRNAs targeting each gene. The dsRNA-treated females and males were allowed to mate and laid eggs for 5 days. Each circle represents eggs produced by an individual female (n = 6). Bars represent the mean ± s.d. Statistical comparisons between two groups were performed using a two-tailed Student’s t-test (***P < 0.001). (B) Morphology of eggs deposited after 5 days by dsRNA-treated BPHs. The eye pigmentation is indicated by an arrow head.
Discussion
In this study, we investigated the functions of individual NlNF-Y members in the BPH during nymphal development, adult molting, wing growth, IFM development, and reproduction. The results demonstrated that NlNF-Y played pivotal roles in these processes. Depletion of NlNF-YA, NlNF-YB, and NlNF-YC resulted in extended nymphal duration, affected nymph-adult molting progress, reduced wing size, disrupted IFM assembly, reduced egg production, and impaired egg development. These findings provide an impetus to understand the function of NF-Y in life-history traits of insects.
The CCAAT box is one of the most common cis-acting elements found in the promoter and enhancer regions of various genes in eukaryotes (Li et al., 1992). An analysis of a large database of 1,031 human promoters indicated that the CCAAT box is present in 63% of them (Suzuki et al., 2001). NF-Y is the major CCAAT box recognizing protein that binds to DNA with high specificity and affinity (Bi et al., 1997; Mantovani, 1998). Knockdown or overexpression of the Drosophila NF-YA homolog with different GAL4 also drives lethality in the Drosophila pharate adult stage, possibly by influencing disc specification (Yoshioka et al., 2007; Ly et al., 2013). A similar phenomenon was observed in BPH, depleting NlNF-YA, NlNF-YB, and NlNF-YC delayed fifth-instar development and led to high lethality, indicating that the CCAAT box might also be a common cis-acting element in the BPH. In addition, we noticed that depletion either NlNF-YA, NlNF-YB, or NlNF-YC led to small and malformed forewings and hindwings. As a wing-dimorphic insect, fifth-instar BPH nymphs have the ability to develop into either short-winged or long-winged adults. A recent finding showed that cells of short wing pads are largely in the G2/M phase of the cell cycle, whereas those of long wings are largely in G1, indicating that cell cycle progression is necessary for wing morph determination (Lin et al., 2020). These observations are in line with previous reports that cell cycle-related genes are the main target genes of NF-Y (Zwicker et al., 1995; Bolognese et al., 1999; Farina et al., 1999; Kao et al., 1999; Korner et al., 2001; Manni et al., 2001). In addition, the depletion of NlNF-Y not only affected wing size, but also led to an IFM defect, the latter tissue is only present in long-winged adults. Based on these events, we speculate that NF-Y might be tightly involved in regulating alternative wing morphs in the BPH although this needs to be further confirmed experimentally.
It is of interest that the depletion of NlNF-YA significantly decreased the adult molting rate, and this defect could be partially rescued by the addition of 20E. In addition, the depletion of NlNF-YA significantly reduced the expression levels of NlE74A and NlE75B, the downstream genes of the ecdysone pathway. These findings indicated that NlNF-Y might affect the ecdysone pathway although the underlying mechanism remains unknown. Although there remains much to be done, our findings provided a first glimpse of the function of NF-Y in hemipteran insects.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
H-HC and H-JX designed the experiment and wrote the manuscript. H-HC, Y-LL, X-YL, and J-LZ conducted the experiments. H-JX managed and directed the project. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31772158 and 31972261), and the National Natural Science Foundation of China for Excellent Young Scholars (31522047). The authors declare that they have no competing interest. We thank the International Science Editing company for polishing this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.585320/full#supplementary-material
Supplementary Table 1 | Primers used in this study.
Footnotes
References
Backus, E. A., Serrano, M. S., and Ranger, C. M. (2005). Mechanisms of hopperburn: an overview of insect taxonomy, behavior, and physiology. Annu. Rev. Entomol. 50, 125–151. doi: 10.1146/annurev.ento.49.061802.123310
Bhattacharya, A., Deng, J. M., Zhang, Z., Behringer, R., de Crombrugghe, B., and Maity, S. N. (2003). The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 63, 8167–8172.
Bi, W., Wu, L., Coustry, F., de Crombrugghe, B., and Maity, S. N. (1997). DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J. Biol. Chem. 272, 26562–26572. doi: 10.1074/jbc.272.42.26562
Bolognese, F., Wasner, M., Dohna, C. L., Gurtner, A., Muller, H., Manni, I., et al. (1999). The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT-binding activity is cell-cycle regulated. Oncogene 18, 1845–1853. doi: 10.1038/sj.onc.1202494
Dorn, A., Bollekens, J., Staub, A., Benoist, C., and Mathis, D. (1987). A multiplicity of CCAAT box-binding proteins. Cell 50, 863–872. doi: 10.1016/0092-8674(87)90513-7
Dubrovsky, E. B. (2005). Hormonal cross talk in insect development. Trands Endocrino. Metab. 16, 6–11. doi: 10.1016/j.tem.2004.11.003
Farina, A., Manni, I., Fontemaggi, G., Tlainen, M., Cenciarelli, C., Bellorini, M., et al. (1999). Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene 18, 2818–2827. doi: 10.1038/sj.onc.1202472
Gilbert, L. I., Rybczynski, R., and Warren, J. T. (2002). Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 47, 883–916. doi: 10.1146/annurev.ento.47.091201.145302
Gurtner, A., Manni, I., Fuschi, P., Mantovani, R., Guadagni, F., Sacchi, A., et al. (2003). Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol. Biol. Cell. 14, 2706–2715. doi: 10.1091/mbc.e02-09-0600
Kao, C. Y., Tanimoto, A., Arima, N., Sasaguri, Y., and Padmanabhan, R. (1999). Transactivation of the human cdc2 promoter by adenovirus E1A. E1A induces the expression and assembly of a heteromeric complex consisting of the CCAAT box binding factor, CBF/NF-Y, and a 110-kDa DNA-binding protein. J. Biol. Chem. 274, 23043–23051. doi: 10.1074/jbc.274.33.23043
Korner, K., Jerome, V., Schmidt, T., and Muller, R. (2001). Cell cycle regulation of the murine cdc25B promoter: essential role for nuclear factor-Y and a proximal repressor element. J. Biol. Chem. 276, 9662–9669. doi: 10.1074/jbc.M008696200
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laloum, T., De Mita, S., Gamas, P., Baudin, M., and Niebel, A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18, 157–166. doi: 10.1016/j.tplants.2012.07.004
Li, G., Zhao, H., Wang, L., Wang, Y., Guo, X., and Xu, B. (2018). The animal nuclear factor Y: an enigmatic and important heterotrimeric transcription factor. Am. J. Cancer Res. 8, 1106–1125.
Li, X. Y., Mantovani, R., Hooft van Huijsduijnen, R., Andre, I., Benoist, C., and Mathis, D. (1992). Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res. 20, 1087–1091. doi: 10.1093/nar/20.5.1087
Lin, X., Gao, H., Xu, Y., Zhang, Y., Li, Y., Lavine, M. D., et al. (2020). Cell cycle progression determine wing morph in the polyphenic insect Nilaparvata lugens. iScience 23:101040. doi: 10.1016/j.isci.2020.101040
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ly, L. L., Suyari, O., Yoshioka, Y., Tue, N. T., Yoshida, H., and Yamaguchi, M. (2013). dNF-YB plays dual roles in cell death and cell differentiation during Drosophila eye development. Gene 520, 106–118. doi: 10.1016/j.gene.2013.02.036
Manni, I., Mazzaro, G., Gurtner, A., Mantovani, R., Haugwitz, U., Krause, K., et al. (2001). NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 276, 5570–5576. doi: 10.1074/jbc.M006052200
Mantovani, R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic. Acids Res. 26, 1135–1143. doi: 10.1093/nar/26.5.1135
Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. doi: 10.1016/s0378-1119(99)00368-6
McNabb, D. S., Xing, Y., and Guarente, L. (1995). Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 9, 47–58. doi: 10.1101/gad.9.1.47
Otuka, A. (2013). Migration of rice planthoppers and their vectored re-emerging and novel rice viruses in East Asia. Front. Microbiol. 4:309. doi: 10.3389/fmicb.2013.00309
Sinha, S., Lu, J., Maity, S. N., and de Crombrugghe, B. (1995). Recombinant rat CBF-C, the third subunit of CBF-NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl. Acad. Sci. U.S.A. 92, 1624–1628. doi: 10.1073/pnas.92.5.1624
Suzuki, Y., Tsunoda, T., Sese, J., Taira, H., Mizushima-Sugano, J., Hata, H., et al. (2001). Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 11, 677–684. doi: 10.1101/gr.gr-1640r
Wu, J., Ge, L., Liu, F., Song, Q., and Stanley, D. (2020). Pesticide-induced planthopper population resurgence in rice cropping system. Annu. Rev. Entomol. 65, 409–429. doi: 10.1146/annurev-ento-011019-025215
Xu, H. J., Chen, T., Ma, X. F., Xue, J., Pan, P. L., Zhang, X. C., et al. (2013). Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 22, 635–647. doi: 10.1111/imb.12051
Xu, H. J., Xue, J., Lu, B., Zhang, X. C., Zhuo, J. C., He, S. F., et al. (2015). Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467. doi: 10.1038/nature14286
Xu, H. J., and Zhang, C. X. (2017). Insulin receptors and wing dimorphism in rice planthoppers. Philos. Trans. R. Soc. Lond B Biol. Sci. 372:20150489. doi: 10.1098/rstb.2015.0489
Xue, J., Zhang, X. Q., Xu, H. J., Fan, H. W., Huang, H. J., Ma, X. F., et al. (2013). Molecular characterization of the flightin gene in the wing-dimorphic planthopper, Nilaparvata lugens, and its evolution in Pancrustacea. Insect Biochem. Mol. Biol. 43, 433–443. doi: 10.1016/j.ibmb.2013.02.006
Xue, J., Zhou, X., Zhang, C. X., Yu, L. L., Fan, H. W., Wang, Z., et al. (2014). Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15:521. doi: 10.1186/s13059-014-0521-0
Xue, W. H., Liu, Y. L., Jiang, Y. Q., He, S. F., Wang, Q. Q., Yang, Z. N., et al. (2018). CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 93, 19–26. doi: 10.1016/j.ibmb.2017.12.003
Yoshioka, Y., Suyari, O., Yamada, M., Ohno, K., Hayashi, Y., and Yamaguchi, M. (2007). Complex interference in the eye developmental pathway by Drosophila NF-YA. Genesis 45, 21–31. doi: 10.1002/dvg.20260
Yuan, M., Lu, Y., Zhu, X., Wan, H., Shakeel, M., Zhan, S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One 9:e86503. doi: 10.1371/journal.pone.0086503
Zhang, C. X., Brisson, J. A., and Xu, H. J. (2019). Molecular mechanisms of wing polymorphism in insects. Annu. Rev. Entomol. 64, 297–314. doi: 10.1146/annurev-ento-011118-112448
Keywords: Nilaparvata lugens, nuclear factor Y, wing development, ecdysone, reproduction
Citation: Chen H-H, Liu Y-L, Liu X-Y, Zhang J-L and Xu H-J (2020) Functional Analysis of Nuclear Factor Y in the Wing-Dimorphic Brown Planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Front. Genet. 11:585320. doi: 10.3389/fgene.2020.585320
Received: 20 July 2020; Accepted: 12 October 2020;
Published: 03 November 2020.
Edited by:
Wei Guo, Chinese Academy of Sciences (CAS), ChinaReviewed by:
LinQuan Ge, Yangzhou University, ChinaJunzheng Zhang, China Agricultural University, China
Copyright © 2020 Chen, Liu, Liu, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai-Jun Xu, aGFpanVueHVAemp1LmVkdS5jbg==
 Hao-Hao Chen1
Hao-Hao Chen1 Xin-Yang Liu
Xin-Yang Liu Jin-Li Zhang
Jin-Li Zhang Hai-Jun Xu
Hai-Jun Xu