
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 30 September 2020
Sec. Evolutionary and Population Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.571188
This article is part of the Research Topic Domestication of Agronomic Traits in Legume Crops View all 22 articles
Agricultural production is greatly dependent on daylength, which is determined by latitude. Living organisms align their physiology to daylength through the circadian clock, which is made up of input sensors, core and peripheral clock components, and output. The light/dark cycle is the major input signal, moderated by temperature fluctuations and metabolic changes. The core clock in plants functions mainly through a number of transcription feedback loops. It is known that the circadian clock is not essential for survival. However, alterations in the clock components can lead to substantial changes in physiology. Thus, these clock components have become the de facto targets of artificial selection for crop improvement during domestication. Soybean was domesticated around 5,000 years ago. Although the circadian clock itself is not of particular interest to soybean breeders, specific alleles of the circadian clock components that affect agronomic traits, such as plant architecture, sensitivity to light/dark cycle, flowering time, maturation time, and yield, are. Consequently, compared to their wild relatives, cultivated soybeans have been bred to be more adaptive and productive at different latitudes and habitats for acreage expansion, even though the selection processes were made without any prior knowledge of the circadian clock. Now with the advances in comparative genomics, known modifications in the circadian clock component genes in cultivated soybean have been found, supporting the hypothesis that modifications of the clock are important for crop improvement. In this review, we will summarize the known modifications in soybean circadian clock components as a result of domestication and improvement. In addition to the well-studied effects on developmental timing, we will also discuss the potential of circadian clock modifications for improving other aspects of soybean productivity.
Studies have shown that plants with circadian clocks synchronized to their environmental conditions gain growth advantage over those not synchronized (Dodd et al., 2005). The circadian clock not only plays roles in growth and development, it is also involved in metabolism and stress responses (Fukushima et al., 2009; Dodd et al., 2015; Nakamichi et al., 2016; Nitschke et al., 2016; Kim et al., 2017; Gil and Park, 2019). Current knowledge of the plant circadian clock mostly came from studies on Arabidopsis, which is a relatively simple model plant. The Arabidopsis circadian clock consists of mainly three transcription feedback loops, namely the central loop, the morning loop, and the evening loop (Staiger et al., 2013; Foo et al., 2016; Gil and Park, 2019). The central loop is made up of a feedback loop made up of circadian clock associated 1/late elongated hypocotyl (CCA1/LHY) and timing of CAB (TOC1; also known as pseudo response regulator 1, PRR1), which are mutual repressors of each other (Alabadi et al., 2001). On the other hand, in the morning loop, CCA1/LHY activates PRR5, PRR7, and PRR9, which in turn inhibit CCA1/LHY in a negative feedback loop (Mizuno and Nakamichi, 2005). Early flowering 3 (ELF3), early flowering 4 (ELF4), and LUX ARRHYTHMO (LUX) make up the evening complex (EC) that represses PRRs, while they themselves are repressed by CCA1/LHY (Huang and Nusinow, 2016). This delicate configuration, together with the input sensors and other peripheral components, allows the clock to oscillate in a cycle of approximately 24 h. Detailed discussions on the Arabidopsis circadian clock could be found in some recent reviews (Staiger et al., 2013; Foo et al., 2016; Shim et al., 2017; Gil and Park, 2019).
Studies on some crop species suggested that domestication has imposed significant changes on the circadian clock, in terms of the phase, amplitude and period (Shor and Green, 2016). Shifting the circadian phase of important biological processes could have big impacts. For example, a 3-bp deletion in the Phytochrome A-associated F-box protein-encoding gene, EID1 (Empfindlicher im dunkelroten Licht protein 1), in the cultivated tomato has led to the lengthening of the clock period (Muller et al., 2016). This transition allowed the day-neutral tomatoes that originated from the equatorial Andean region of South America to adapt to longer daylengths in Mesoamerica and Europe (Muller et al., 2016). The deceleration of the clock reoriented the phase of some biological processes such as photosynthesis with the longer photoperiods in the higher latitudes to maximize the productivity of these processes. Other examples include the EAM8 from Barley (Faure et al., 2012), a homolog of Arabidopsis ELF3, and Eps3 from wheat (Gawronski et al., 2014), a homolog of Arabidopsis LUX. Alleles of these two genes have played important roles in affecting flowering time of the respective crops and altering the phase, period and amplitude of their circadian clocks (Faure et al., 2012; Gawronski et al., 2014). However, the physiological consequences of the altered clock in these two crops as a result of changes in these two genes are still largely unknown.
Up to now, specific studies on soybean circadian rhythm have been limited, but they can be dated back to the 1950s (Bunning, 1954; Brest, 1970). In those days, it has already been discovered that the circadian movement of trifoliate leaves of soybean, as measured with a kymograph, could last for two weeks after shifting to continuous light in a roughly 24-h fashion relying on the internal clock (Brest, 1970). A much more recent study examined the circadian fluctuations of chlorophyll contents using non-destructive multispectral imaging under drought conditions (Pan et al., 2015). Interestingly, circadian rhythm was not only observed in the aerial parts of the soybean plant. Expressing the luciferase reporter gene driven by the soybean LHY-CCA1-LIKE b2 (GmLCLb2) promoter or soybean PRR9b2 promoter in transgenic soybean hairy roots revealed that the circadian rhythm of the soybean root is out of sync with the clock in the leaf (Wang Y. et al., 2020), suggesting that the circadian clock may play different roles in different organs of the soybean plant. A recent circadian study on soybean observed that the free-running circadian period lengths of elite soybean cultivars are positively correlated to the latitudes associated with the maturity groups that these cultivars belong to (Greenham et al., 2017). Although the study did not involve any wild soybean, since maturity groups are largely the results of domestication and its subsequent diversification (Jiang et al., 2014; Zhou et al., 2015; Wang Y. et al., 2016; Miladinovic et al., 2018), the correlation between period length and latitude/maturity group suggests that alterations in the soybean circadian clock could be a by-product of these processes.
So far, there are a few transcriptomic studies regarding the soybean circadian clock. One study observed that only about 1.8% of the transcriptome from Glycine max cv. Williams 82 developing seeds cycled under constant light condition (Hudson, 2010), which was much lower than 6–40% in Arabidopsis seedlings (Covington et al., 2008). Another study identified 3,695 time-indicating genes in the unifoliate leaves of Williams 82, which amount to less than 10% of the total protein-coding genes in the soybean genome (Li M. et al., 2019). The discrepancy between the two species was probably due to organ-specific effects of the clock. However, we cannot rule out the possibility that the oscillation of the clock is normally less robust in cultivated varieties (Shor and Green, 2016), which makes rhythmic genes less likely to be detected.
In addition to the well-known input signals of light and temperature, stresses are also input signals that could greatly alter the clock. It has been found that mild drought, heat shock, iron deficiency, and alkaline stress could change the period and phase of the expression of core clock genes in soybean (Li M. et al., 2019). Under alkaline stress, the leaf movement of wild soybean (Glycine soja) was roughly aligned with the clock gene expressions which showed an advancement of phase (Li M. et al., 2019). However, the leaf movements under drought and heat stress, and in iron-deficient plants, went out of sync with the core clock gene expressions, suggesting that different stresses may have different physiological effects (Li M. et al., 2019). Treatment of soybean with arsenate (As[V]) and arsenite (As[III]) could lead to changes in the amplitude of diurnal expressions of GmLCL1, GmPRR9, GmELF4, and GmGI (Gigantea), depending on the tissue and the form of arsenic compounds (Vezza et al., 2020). Stomatal movements and expressions of antioxidative enzymes were also altered by the As(V) and As(III) treatments. Yet, the link between the changes in core clock gene expressions and physiological changes remain elusive (Vezza et al., 2020). All in all, the direct physiological consequences of the alteration of the soybean circadian clock as a result of stresses are mostly uncharacterized.
During the domestication and improvement processes, breeders have selected for new soybean varieties with the aim to improve yield by mainly modifying the plant architecture and ensuring the soybean plant will reach maturity within a desirable timeframe, especially when adapting to a new cultivation region or habitat. It is known that artificial selection has enriched some minor alleles compared to the gene pool of the progenitor populations (Lam et al., 2010). Using the latest technologies in genomic studies, it is discovered that breeders, without prior knowledge of the circadian clock, have inadvertently introduced a few important and dominant modifications to the core clock components. The soybean genome has recently undergone two rounds of whole-genome duplication. Some of the clock component genes have diversified in functions after duplication, which further complicates the studies on their functions. Genetic manipulation of the circadian clock components of soybean using the well-studied Arabidopsis circadian clock model as a guide could help further improve soybean productivity. With the recent population genomic data, three circadian clock components, GmPRR3, GmELF3, and GmGIa from soybean were well-supported to be under artificial selection. In the following sections, we have summarized the recent discoveries in the modifications of these three circadian clock components in soybean during domestication.
Earlier studies on the molecular controls of soybean maturation mainly focused on the E locus (Cober and Voldeng, 2001; Liu et al., 2008; Watanabe et al., 2009; Cober et al., 2010; Watanabe et al., 2011; Xia et al., 2012; Dissanayaka et al., 2016; Zhao et al., 2016; Lu et al., 2017; Samanfar et al., 2017; Yue et al., 2017). It was only recently that two major-effect quantitative trait loci (QTLs) on chromosomes 11 and 12 controlling maturation time and flowering time were found in wild soybeans or landraces (Qi et al., 2014; Lu et al., 2016, 2020; Fang et al., 2017; Pan et al., 2018; Li M. W. et al., 2019; Li C. et al., 2020; Ogiso-Tanaka et al., 2019). Dissection of these two QTLs identified two Pseudo Response Regulator 3 genes, GmPRR3a/Tof11 and GmPRR3b/Tof12 on chromosomes 11 and 12, respectively, as the causal genes for controlling these two functions (Li M. W. et al., 2019; Li Y. et al., 2019; Li C. et al., 2020; Li Y. H. et al., 2020; Lu et al., 2020).
There are five PRR genes in Arabidopsis, including PRR9, PRR7, PRR5, PRR3, and TOC1/PRR1, which are all expressed in a circadian fashion and their expressions peak sequentially (Staiger et al., 2013; Gil and Park, 2019). These PRR proteins are typified with a pseudo receiver (PR) domain, an ERF-associated amphiphilic repression (EAR) motif, and a CONSTANTS, CO-like and TOC1 (CCT) domain (Gendron et al., 2012). Compared with other Arabidopsis PRR proteins, two non-synonymous substitutions naturally occurred on the EAR motif and CCT domain, respectively, of AtPRR3. These two substitutions are basically fixed in all Arabidopsis thaliana ecotype (Alonso-Blanco et al., 2016). These substitutions made AtPRR3 somehow behaves differently from other PRRs (Para et al., 2007). Unlike other PRRs which play direct roles in transcript repression, AtPRR3 probably participates in the circadian clock through stabilizing TOC1/PRR1 but without directly interacting with DNA through the CCT domain (Para et al., 2007). However, the soybean genome preserved functional alleles of PRR3 with intact EAR motif and CCT domain (Li C. et al., 2020; Lu et al., 2020), making soybean a good model for the study of this clock component.
A recent investigation of the growth period QTLs on chromosomes 11 (Gp11) and 12 (Gp12) using a cultivated-wild soybean recombinant inbred lines (RIL) population identified major-effect mutations in GmPRR3a and GmPRR3b (Li M. W. et al., 2019). A frameshift mutation was found in GmPRR3a (GmPRR3aC08) and a nonsense mutation in GmPRR3b in the cultivated parent C08 (GmPRR3bC08) by comparing the reference genome of the soybean cultivar Williams 82 to that of the wild soybean parent W05 (GmPRR3aW05 and GmPRR3bW05) (Li M. W. et al., 2019). Both mutations bring about a pre-mature stop codon to the coding sequence and lead to the loss of the C-terminal CCT domain in the encoded proteins (Li M. W. et al., 2019). The end result was the shortening of the growth period of the soybean plant. Analysis of resequencing data suggested that the mutations are almost fixed in the improved cultivars, inferring that the mutations were strongly selected for and played important roles in domestication or during improvement (Li M. W. et al., 2019).
Genome-wide association mapping using 279 landraces discovered 16 flowering time and maturation time QTLs (Li C. et al., 2020). Linkage disequilibrium block analysis narrowed down the QTL on chromosome 12 to a single gene, GmPRR3b, confirming it to be the causal gene of the maturation and flowering time phenotypes (Li C. et al., 2020). In the same study, eight haplotypes of GmPRR3b were discovered (H1–H8). H1 has a stop-gain near the start codon (C43T), hence appearing to be a complete knockout of the GmPRR3b gene. H2, H3, and H5 have non-synonymous single-nucleotide polymorphisms (SNPs) outside of the conserved domains, which are assumed to be fully functional. H4 is the full-length GmPRR3b that is the same as GmPRR3bW05 and is the dominant allele in wild soybean. H6 is the sole haplotype in the elite cultivar which has an ochre mutation and the loss of the CCT domain (equivalent to GmPRR3bC08). Both H7 and H8 have a non-synonymous mutation in the PR domain (Li C. et al., 2020). Studies on the flowering time of landraces carrying the H4–H8 haplotypes showed that alleles with mutations in the PR domain (H7 and H8) or with a truncated CCT domain (H6) have an early flowering phenotype (Li C. et al., 2020).
A more recent genome-wide association mapping using a panel of 424 soybean accessions also identified GmPRR3a and GmPRR3b in the QTLs Tof11 and Tof12 as the major causal genes controlling flowering and maturity (Lu et al., 2020). Molecular dating suggested that both alleles of GmPRR3 in tof11 and tof12 lost their CCT domain ∼8,000 and ∼10,500 years ago, respectively (Lu et al., 2020). While only a tiny fraction of the tof11 allele occurred alone, it is proposed that the tof11 allele arose from the tof12 background (Lu et al., 2020).
Based on the above studies, it is agreed that the domestication has selected for the alleles of GmPRR3 without the CCT domain, leading to the early flowering phenotype and possibly altering the plant architecture related to yield (Figure 1) (Li M. W. et al., 2019; Li C. et al., 2020; Lu et al., 2020). GmPRR3s without the CCT domain probably promote flowering through derepressing the flower-promoting Florigen T gene, GmFT2a, under long-day conditions. It is interesting that a null allele with an 8.6% allele frequency in the wild soybean population was not selected (Li C. et al., 2020). This null allele was completely wiped out in the landrace (Li C. et al., 2020). It is highly possible that a complete loss of GmPRR3 functions may be detrimental. Knocking out the full-length GmPRR3b in the ZGDD background (equivalent to GmPRR3bH4) by CRISPR/cas9 led to a significant early flowering phenotype under long-day conditions (Wang L. et al., 2020). However, knocking out the GmPRR3bC08/GmPRR3bH6/tof12 allele in the Jack and Tianlong backgrounds did not result in any significant change in flowering time (Wang L. et al., 2020) and a late-flowering phenotype with diminished yield components (Li C. et al., 2020), respectively. Furthermore, the overexpression of full-length GmPRR3b in Williams 82 generated a late-flowering phenotype (Lu et al., 2020), and the overexpression of GmPRR3bC08/GmPRR3bH6/tof12 in Tianlong (Li C. et al., 2020) also produced a late-flowering phenotype and improved yield components. These observations suggest that the loss of the CCT domain in GmPRR3 in domesticated soybean might have gained new functions for the protein that are related to yield and plant architect which is not present in the null allele.
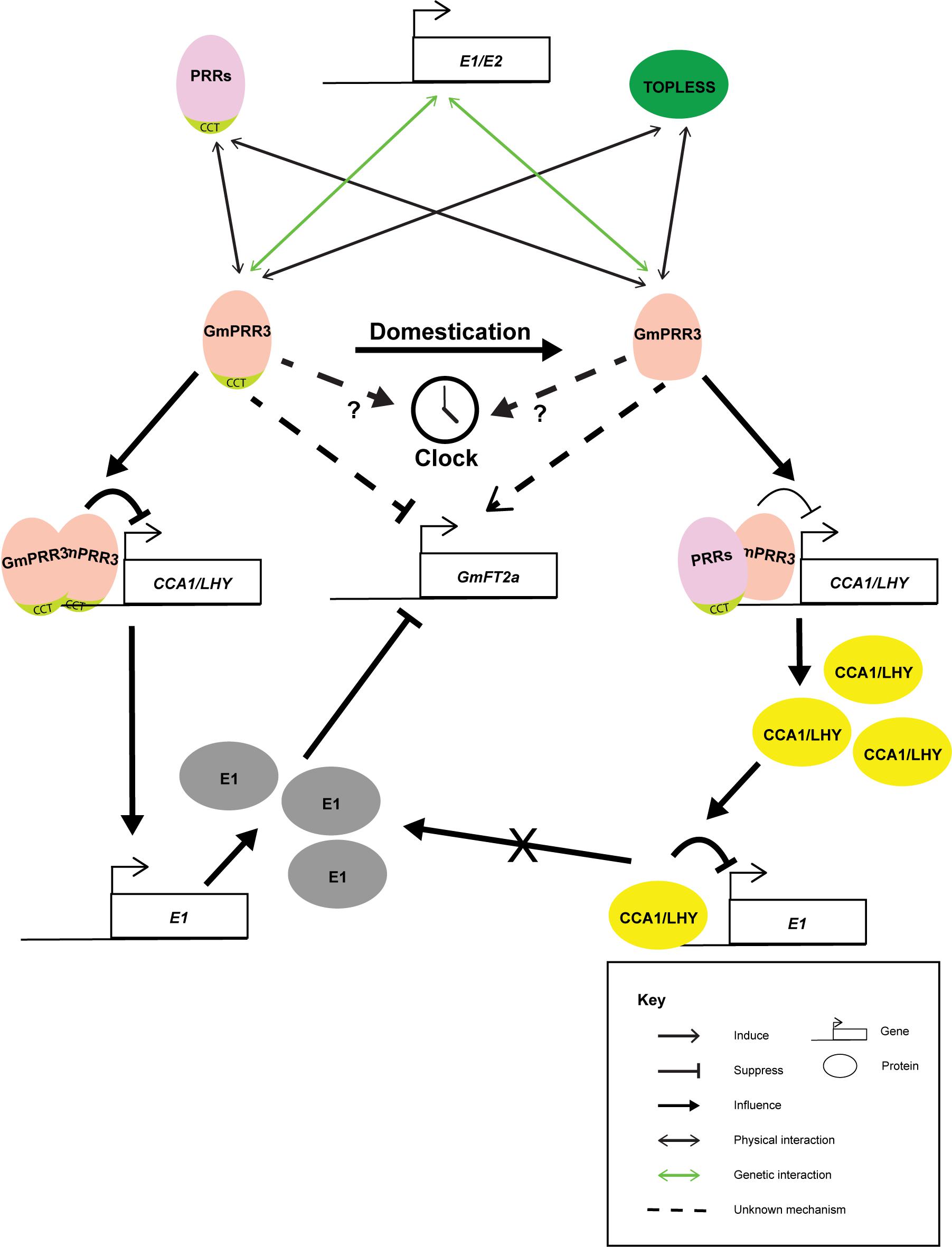
Figure 1. Cartoon summarizing the known interactions of GmPRR3a and GmPRR3b. GmPRR3s are genetically interacting with E1 or E2 loci in controlling flowering time and maturation of soybean probably through the regulation of GmFT2a expression. During domestication and soybean improvement, alleles of GmPRR3s without CCT domain were selected. GmPRR3s without CCT domain have weakened suppression of the expression of CCA1/LHY genes. The interaction of GmPRR3s without CCT domain may rely on other PRR proteins. Thus, the derepression of CCA1/LHY will in turn suppress the expression of E1. The suppression of E1 maybe one of the routes that GmPRR3s without CCT domain derepress GmFT2a.
The actual link between GmPRR3 and flowering time is largely unknown. There are also some controversies with the existing models. Expressions of GmPRR3a and GmPRR3b cycle robustly in both long-day and short-day conditions (Li M. W. et al., 2019; Li C. et al., 2020) similar to the other PRR genes. The expressions peak at the middle of the day no matter whether it was a long-day (the 8th hour in a 16/8 hour light/dark cycle) or a short-day regime (the 4th hour in a 8/16 hour light/dark cycle) (Li M. W. et al., 2019; Li C. et al., 2020). The amplitude of oscillation seems to be slightly affected by the E3 and E4 loci (Lu et al., 2020). It has been shown that GmPRR3 may be genetically interacting exclusively with either E1 (Lu et al., 2020) or E2 (Li M. W. et al., 2019), while transcriptomic network analyses suggested that GmPPR3 may interact with both (Pan et al., 2018). Overexpressed GmPRR3bH6, the allele without a CCT domain, was shown to bind to the GmCCA1a promoter and dampen the expression of GmCCA1a (Li C. et al., 2020), suggesting that GmPRR3b is playing a direct role in the soybean circadian clock. However, the CCT domain was known to be essential for PRR proteins to directly interact with DNA (Gendron et al., 2012). Thus, the interaction of GmPRR3bH6 with DNA may depend on its interactions with other PRR proteins or TOPLESS-like transcription factors (Li M. W. et al., 2019). On the other hand, the overexpression of GmPRR3bH4, the allele with a CCT domain, resulted in stronger suppression of GmCCA1a expression (Li C. et al., 2020), which could be the result of direct interactions with the GmCCA1a promoter. Tof11 was also shown to be potentially able to bind to the promoters of LHY/CCA1s and suppress their expressions (Lu et al., 2020). Thus, Lu et al. (2020) proposed that GmPRR3 with a functional CCT domain suppresses GmCCA1/LHY expression, which leads to the derepression of E1. The high E1 level in turn suppresses GmFT expressions, thus delaying flowering. Therefore, the mutated version of GmPRR3 without a CCT domain will lead to a higher expression of GmCCA1/LHY and the subsequent early flowering phenotype (Lu et al., 2020). However, this was soon questioned by the observation of Li et al. as the complete knockout of GmPRR3bH6 had led to higher expressions of GmCCA1a and GmFT2a and a lower expression of E1 (Li C. et al., 2020). Although this expression pattern is consistent with the assumption that GmPRR3b represses the expression of LHY/CCA1s, which act as repressors for E1 (Lu et al., 2020), but the lower E1 expression is not tally with the observed late-flowering phenotype (Li C. et al., 2020). Two possibilities on the delayed flowering were proposed. One is the involvement of pathway that is not mediated by CCA1/LHY-E1-FT2a module and the other is the consequence of alternated growth of PRR3b mutant (Li C. et al., 2020). The first possibility could be likely as E1 is a legume specific protein, while rice OsPRR37, a far homolog of GmPRR3, can influence the expression of Hd3a(FT) in the absent of E1 like protein (Matsubara et al., 2014). More delicate genetic studies will be needed to resolve these obstacles. Furthermore, studies suggested that GmPRR3 mainly regulated the expression of GmFT2a but not GmFT5a (Li M. W. et al., 2019; Li C. et al., 2020), which resembled the regulation of E2 (Lu et al., 2020). Nevertheless, GmFT5a may also be a target under certain circumstances. Therefore, further investigations into the interactions between GmPRR3 and GmFT5a is required to solve this puzzle. Actually, the rice OsPRR37 can either promote or suppress flowering depending on the genetic background (Zhang et al., 2019). Thus, controversies of GmPRR3 functions in previous studies were due to the inadequate study of these genes.
Soybean is a strict short-day species. Soybean plant normally requires staying vegetative for around 2 months to accumulate enough biomass to generate reasonable yield. Yet, a short-day photoperiod will trigger premature flowering of soybean plants and eventually result in low yield (Board and Hall, 1984). Thus, seasonal and latitudinal shifting of day-length have limited the growing season and acreage of soybean cultivation. However, according to model prediction, it is likely that more soybean will be grown in the tropics in 2100 (Fodor et al., 2017) where daylength is inductive for flowering in soybean.
Some late-flowering soybean cultivars in tropical regions were described as early as 1970s (Hartwig, 1970; Hartwig and Kiihl, 1979), especially in Brazil. These varieties could maintain a long juvenile (LJ) phenotype under short-day conditions, which enabled them to generate adequate seeds in the scale suitable for agricultural production (Destro et al., 2001; Carpentleri-Pipolo et al., 2002). The E6 (Bonato and Vello, 1999) and J (Ray et al., 1995) loci were independently identified to control the LJ phenotype and were later found to be possibly closely linked genes (Cober, 2011; Li et al., 2017). Although there are other potential loci controlling the LJ phenotype (Destro et al., 2001; Carpentleri-Pipolo et al., 2002), the J locus is the best characterized.
Study of the J locus using a RIL population from a conventional juvenile variety, Zhonghuang 24, and an LJ variety, Huaxia 3, narrowed the causal gene to an indel (AT > A) in Glyma.04G050200 (Yue et al., 2017). Glyma.04G050200 (GmELF3) encodes a homolog of the Arabidopsis Early Flowering 3 (AtELF3). The deletion of the nucleotide (AT > A) results in truncation of the protein corresponding to the recessive j in the LJ variety (Yue et al., 2017). Constitutive expression of the full-length GmELF3 promoted flowering in transgenic plants in the LJ parent, Huaxia 3, background (Yue et al., 2017). Another study identified a 10-bp deletion and a deletion of a cytosine in the ELF3 of BR121 and PI 159925, respectively, to be the causal mutations of the LJ phenotype in these varieties (Lu et al., 2017).
GmELF3 shows a circadian expression pattern and peaks at dusk (Lu et al., 2017), resembling the expression pattern of AtELF3. Interestingly, the loss-of-function allele of GmELF3 did not oscillate diurnally, suggesting that an intact GmELF3 per se is essential to sustain its own expression pattern (Lu et al., 2017). Nevertheless, the effect of this GmELF3 mutation on soybean circadian rhythm is not yet characterized. Like other E loci which are inter-dependent, the expression of GmELF3 is slightly suppressed by E3E4 (GmPHYA3 and GmPHYA2) compared to the e3e4 lines in the Harosoy background (Figure 2) (Lu et al., 2017). Yet, it is not known whether the higher expression of GmELF3 in the e3e4 background was due to the alteration in light input to the clock or due to a direct effect of the loss of GmPHYA functions. On the other hand, the functions of the J locus has been shown genetically to rely on E1 (Lu et al., 2017). Only an intact GmELF3 and not the truncated protein can physically interact with the promoter of E1, probably as part of the evening complex (Lu et al., 2017). The recessive j allele was thought to suppress FT expression through E1, as the expression of E1 was derepressed in the near-isogeneic line having a recessive j (NIL-j) with reference to NIL-J under inductive short-day conditions (Lu et al., 2017). Thus, although j allele was able to delay flowering in the e1as (a weak allele of E1) background, the effect was less prominent than in the E1 background (Lu et al., 2017). But still, it is possible that GmELF3 can alter flowering through alternative pathways. Taking rice Hd17(OsELF3) as an example. Hd17 mediates the expression of OsPRRs, OsLHY, OsGI, Ghd7, and Hd1(CO), which are acting in multiple pathways that affect Hd3a(FT) and flowering (Zhao et al., 2012; Yang et al., 2013; Matsubara et al., 2014).
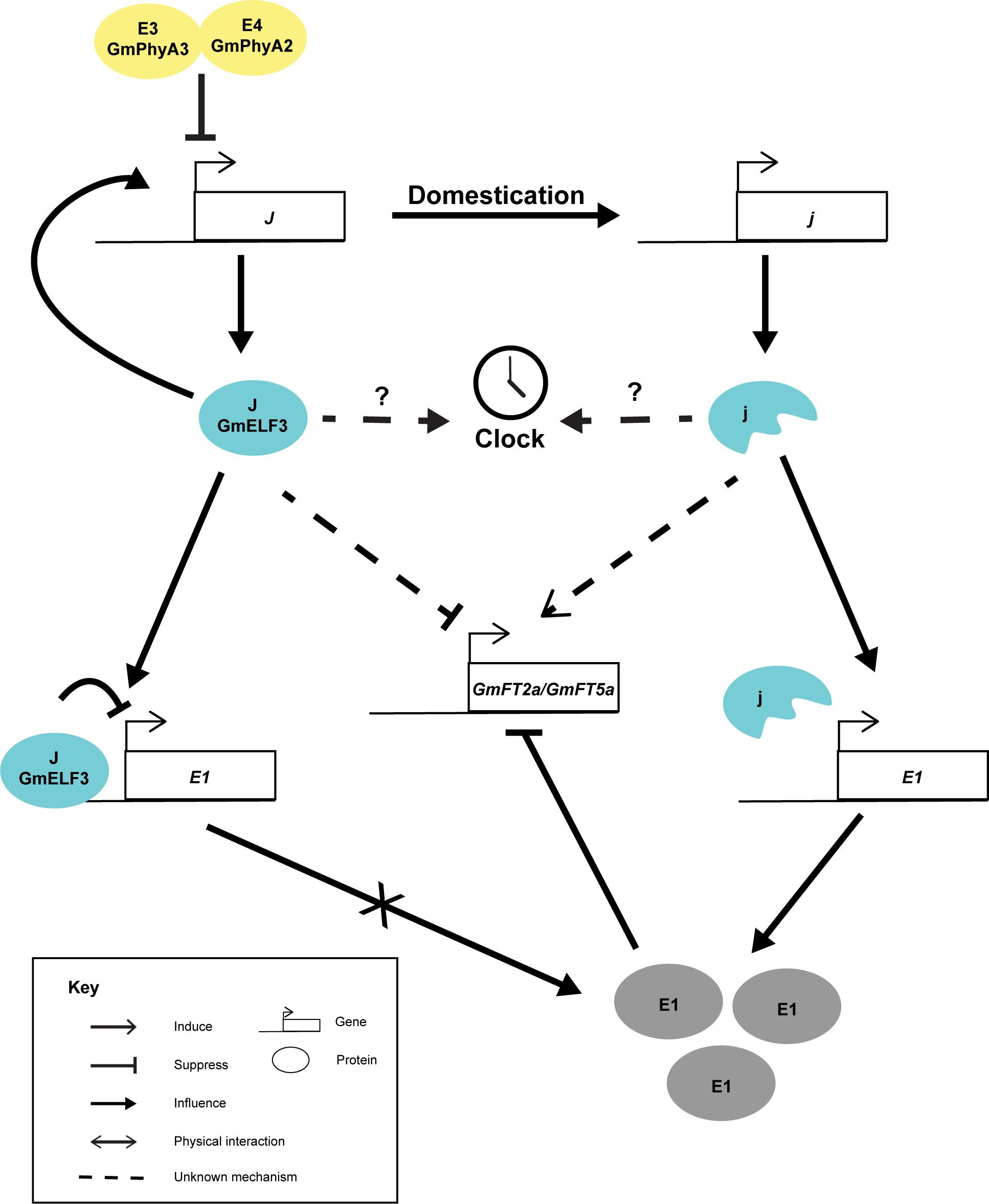
Figure 2. Cartoon summarizing the known interactions of J. The J locus possibly perceive light signal through E3 and E4 loci. The J locus encodes GmELF3 (J), which auto-regulates its own expression. GmELF3 can directly acts on E1 promoter and suppress its expression. Without E1 expression under short day conditions, GmFT2a and GmFT5a are highly induced and lead to unproductive flowering. In the opposite, domestication has selected j alleles in tropical regions. The j proteins are unable to auto-regulate its own expression and has no ability to bind to E1. Under this circumstance, E1 proteins are produced and suppresses the expression of GmFT2a and GmFT5a promoter under short day conditions which lead to the long juvenile phenotype.
Unlike other genes artificially selected, J is not widespread among soybean cultivars. Rather, J is tailored for soybean cultivation in tropical regions far away from the temperate origin of wild soybean (Lu et al., 2017). For instance, the Huaxia 3 allele (AT > A) of J can only be found in southern China (Yue et al., 2017) and eight other loss-of-function alleles (j-1 to j-8) were all found in low-latitude regions (Lu et al., 2017). One of these alleles was the result of induced mutation (Lu et al., 2017), which unintentionally mutated a core clock gene.
There is trade-off for having the recessive j alleles. A recent study has found that near isogenic lines with a recessive j were more sensitive to salt stress compared to those having the dominant J (Cheng et al., 2020). Transient overexpression of J in soybean hairy roots produced composite plants with higher salt tolerance, as expected (Cheng et al., 2020). A transcriptomic study comparing NIL-j and NIL-J showed that NIL-j had significantly lower expressions of ∼95 stress-related transcription factor genes (Cheng et al., 2020). Higher expressions of GmWRKY12, GmWRKY27, GmWRKY54, GmNAC11, and GmSIN1 were confirmed in J-overexpressing transgenic hairy roots (Cheng et al., 2020). It is not known whether J targeted the promoters of these genes directly, or if J altered the circadian clock, which led to different stress responses (Park et al., 2016; Coyne et al., 2019).
Gigantea (GI) is not directly involved in the circadian clock transcription feedback loop in the model plant, Arabidopsis. However, it has been found playing crucial roles in clock functions, seasonal flowering, and many other important biological processes (Mishra and Panigrahi, 2015). In brief, GI assisted the maturation and accumulation of ZEITLUPE (ZTL), an F-box E3 ligase that is responsible for the degradation of clock components including TOC1/PRR1, PRR5 and CCA1 HIKING EXPEDITION (CHE) (Cha et al., 2017; Lee et al., 2019), so as to derepress LHY/CCA1 and bring about the progression of the circadian rhythm. On the other hand, under inductive long-day conditions, the diurnal expression of Arabidopsis GI coincides with the circadian-controlled FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) (Mishra and Panigrahi, 2015). The GI-FKF1 thus is able to form a complex to degrade the flowering suppressors, cycling DOF factors (CDFs), allowing the transcription of CONSTANTS (CO). CO will then promote the expression of FT and result in flowering (Mishra and Panigrahi, 2015).
There are three copies of GI in the soybean genome, GmGIa, GmGIb, and GmGIc. GmGIa (Glyma.10g221500) was known as E2 (Watanabe et al., 2011), which played major roles in the flowering and maturation of soybean (Wang Y. et al., 2016). The nucleotide diversity of GIb and GIc were not different between wild and cultivated soybeans (Wang Y. et al., 2016), suggesting that they were not targets for human selection. Analyses of 104 wild soybeans and 203 Chinese landraces identified 47 haplotypes (H1–H47) of GmGIa in total. Due to artificial selection, there were only three haplotypes (H1–H3) of GmGIa in the 203 landraces. Furthermore, the GmGIa in cultivated soybeans retained only 4.7% nucleotide diversity and 2.9% nucleotide polymorphism compared to 66% nucleotide diversity and 49% nucleotide polymorphism of the entire genome, respectively (Hyten et al., 2006; Wang Y. et al., 2016), implying a strong bottleneck in this gene.
H2 and H3 encode the full-length, 1,177 (or 1,170; Watanabe et al., 2011) amino acids of the GmGIa protein and are different in a single amino acid substitution from V220 to I220 (Wang Y. et al., 2016). The substitution appeared to have no significant effect on the GI function on flowering. H1 (e2) has a 66.95% frequency in the Chinese landraces compared to 4.81% in wild soybeans. Due to a premature stop codon in the 10th exon, H1 encodes a protein with only 527 (or 521; Watanabe et al., 2011) amino acids and contributes to the early flowering and maturation phenotype (Wang Y. et al., 2016). So far, no Korean or Japanese wild soybean bearing the H1 haplotype has been found (Wang Y. et al., 2016). Therefore, it is proposed that H1 (e2) originated from China and was later introduced into the other East Asian regions (Kim et al., 2018). On the other hand, another haplotype with a premature stop codon in the 2nd exon has been specifically described in Korean early flowering varieties. This suggests the mutation arose independently and only spread locally (Kim et al., 2018).
Although GmGIa is well known as a gene controlling flowering and maturity, with the discrepancies between the short-day soybean and the long-day Arabidopsis, the actual molecular mechanism of how GmGIa regulates flowering and maturation is largely unknown. It is believed that the function of GmGIa has diverged from the GIs in other plant species. For instance, the full-length GmGIa serves as a flowering suppressor while its Arabidopsis counterpart acts as a flowering promoter (Watanabe et al., 2011). Interestingly, full-length OsGI from rice, a short-day monocot, serves as flowering suppressor (Hayama et al., 2003). Seemingly, GI functions in long-day and short-day plants are different. The expression of GmGIa was found to be correlated with the expression of GmFT2a but not that of GmFT5a under natural light conditions (Figure 3) (Watanabe et al., 2011). Yet, the mechanistic link between GmGIa and GmFT2a expressions is still missing. The expression of GmGIa is suppressed in the e3e4 background or by the overexpression of GmCOL1a (Cao et al., 2015). A combination of E2 and E3 alleles can synergistically improve seed yield in July sowing in Japan (Kawasaki et al., 2018). Under long-day conditions, the expression of GmGIa oscillates following a circadian rhythm but is slightly out of sync with the expressions of the two GmFKF1 genes (Li et al., 2013), while under short-day conditions, the expression of GmGIa is dampened and no longer oscillates (Li et al., 2013). This is something different from the expression of AtGI (Sawa et al., 2008) and OsGI (Hayama et al., 2003). Thus, the GI-FKF1 coincident model in Arabidopsis cannot be directly applied to soybean. Ectopic expressions of H2 and H3 in Arabidopsis delayed flowering while only the constitutive expression of H1 (a truncated GmGIa), but not those of H2 and H3, can rescue the early flowering phenotype of the gi-2 Arabidopsis mutant (Wang Y. et al., 2016). This may be explained by the fact that GmFKF1 could only interact with the N-terminus but not the full-length GmGIa in yeast two-hybrid experiments (Li et al., 2013). It is possible that the Arabidopsis FKF1 can only be stabilized by the truncated but not the full-length GmGIa. Thus, ectopic expressions of H2 and H3 could not rescue the late-flowering phenotype of gi-2 (Wang Y. et al., 2016). Furthermore, GmGIa has also been found to mediate the level of miR172a, which in turn regulates the stability of an AP2/TOE1 mRNA that affects flowering time (Wang T. et al., 2016).
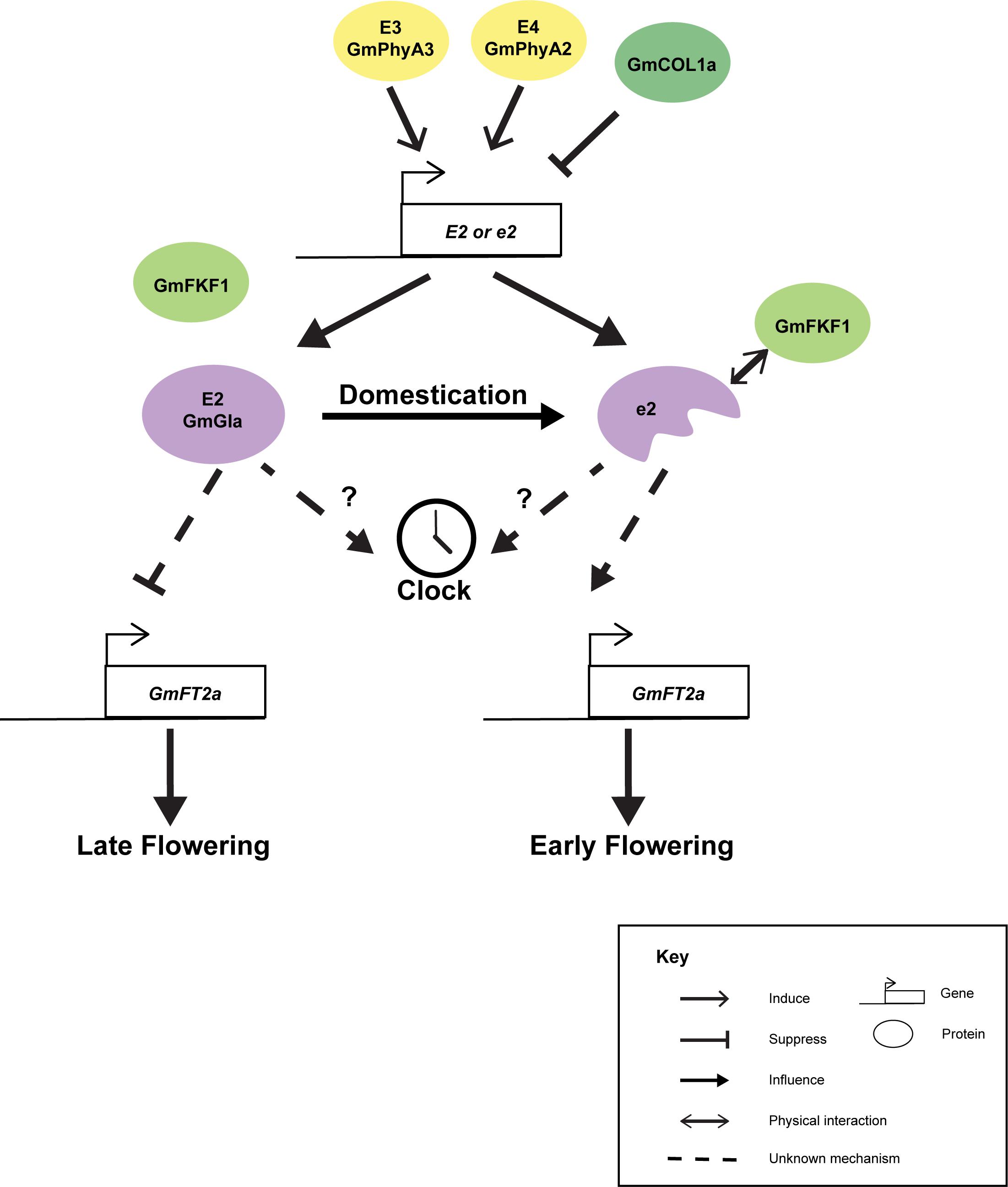
Figure 3. Cartoon summarizing the known interactions of GmGIa. Expression of GmGIa (E2) is controlled by E3, E4 loci and GmCOL1a. During domestication, a truncated GmGIa gene (e2) was selected. The truncated protein but not the full-length protein can interact with GmFKF1 in yeast 2 hybrid assay. The full-length GmGIa (E2) protein can suppress the expression of GmFT2a and leading late flowering phenotype. In the opposite, GmFT2a is derepressed in e2 background allowing early flowering phenotype.
Mutations in GI in other species have been shown to affect salt stress tolerance, oxidative stress tolerance, and water use efficiency (Kurepa et al., 1998; Kim et al., 2018; Simon et al., 2020), but it is still largely unknown whether the alteration of GmGIa during domestication has also contributed to stress-related phenotypes in addition to the well-known effects on flowering and maturation.
The modifications of GmPRR3, GmELF3, and GmGIa as a result of artificial selection seem to complement one another. The truncated alleles of GmPRR3a and GmPRR3b are widespread in landraces and are almost fixed in improved cultivars. This may be due to the possibility that the alteration of GmPRR3 confers an overall advantage in growth period as well as yield. On the other hand, the combination of different alleles of E2 with other E loci partially define the maturity groups of soybean cultivars. Maturity groups are useful for selecting the appropriate cultivars at different latitudes to maximize yield. However, the effective application of E2 is confined to the temperate and subtropical zones. In the tropics, GmELF3 serves a similar function to E2 for selecting the best adapted cultivars for that zone.
At the molecular level, the modifications of GmPPR3 and GmGIa are interesting processes. It involved more than simply altering the gene expressions or completely knocking out the gene functions because knockout alleles also exist in wild accessions. Instead, domestication has introduced new functions to these two proteins. Mutation by deletion of the CCT domain of both copies of GmPRR3 would not have happened purely by chance, since they showed a certain degree of redundancy, only modification of both could maximize the effect (Li M. W. et al., 2019). Similar mutations or deletions of the CCT domain in PRR37 alleles have also occurred during rice and sorghum domestication (Murphy et al., 2011; Koo et al., 2013). The plants having these CCT domain deletions in GmPRR3 behave differently from those complete GmPRR3-knockout lines (Li C. et al., 2020; Wang L. et al., 2020). The loss of the CCT domain abolished the DNA-binding ability of GmPPR3 but retained its protein–protein interaction capability. In this case, the truncated GmPRR3 may serve as an inhibitor in protein complexes and alter their functions. In the case of GmGIa, the truncation of the protein allows its interaction with GmFKF1. Interestingly, the morphologies of different Arabidopsis gi mutants differ drastically, implying that different parts of the GI protein are involved in diverse biological functions, and mutations in different parts of this protein could thus lead to quite different phenotypes (Mishra and Panigrahi, 2015). Based on this information, strategic modifications of GmGIa by genome editing may result in other possible changes to soybean as what researchers have done on the rice waxy gene (Chen and Mas, 2019; Huang et al., 2020). Although we have gained some knowledge of the soybean circadian clock components, in that they have crucial impacts on growth and development, the mechanisms of their actual involvement in the clock is still largely unknown. While the clock outputs are involved in diverse biological processes, ignorance of the mechanisms of how the clock components function while conducting genetic manipulation of the crop plant may result in undesired outcomes such as the alteration of metabolic profiles and stress responses. Besides, so far, there are many more circadian clock components identified in the soybean genome than the three discussed in this review (Liu et al., 2009; Liew et al., 2017; Marcolino-Gomes et al., 2017; Li M. et al., 2019). For example, in additional to GmPRR3a and GmPRR3b, there are 12 PRR homolog encoding genes in the soybean genome (Li M. W. et al., 2019). Nonetheless, their roles in soybean domestication and improvement are either less prominent or the genes have experienced less significant modification.
Modification of the circadian clock during domestication and improvement was entirely serendipitous. Selection was usually based on phenotypes at macroscopic levels. Thus, targets for modification are limited to those causing obvious phenotypic changes. However, with advanced knowledge in the circadian clock, soybean genomics, and genome editing technology, a second wave of modifications of the circadian clock components at the molecular level has become possible.
For example, isoflavones, a sub-group of flavonoids, are secondary metabolites unique to legumes and are beneficial to human either as food or nutraceuticals. Contrary to the model plant Arabidopsis and many other crop species, soybean and other legumes are able to fix nitrogen through a symbiotic relationship with rhizobia. The initiation of the host-symbiont interaction requires flavonoid signaling. Precursors of flavonoids are produced through the phenylpropanoid pathway, which is heavily regulated by the circadian clock at the transcriptional level (Harmer et al., 2000). Thus, well-designed genetic alterations of the circadian clock components could improve the production of these compounds, which raises the commercial values and also the nitrogen-fixing ability of soybean.
Although photosynthesis is dependent on daylength, light intensity, and temperature, the utilization of photosynthates at night is largely controlled by the clock (Graf and Smith, 2011). Since nitrogen fixation relies heavily on the availability of photosynthates supplied by the plant host, the ability of the microsymbiont to fix nitrogen could be impacted by the alteration in the circadian clock of the plant. At present, this is a completely unexplored area.
In addition to spreading soybean cultivation to lower latitudes, agriculture is also moving indoors. Advanced agricultural systems such as vertical farms or hydroponics are gaining popularity in more urban areas. Synchronization of the artificial light-dark cycle with the internal clock of the crop would be beneficial for maximizing crop production using these systems (Belbin et al., 2019). To achieve this goal, more in-depth knowledge of the circadian clock in soybean and other crop plants would be desirable.
In the past decade, since the official release of the first soybean genome assembly in 2010, identification of variants of soybean circadian clock components heavily relied on genome resequencing based on a single reference genome. Such strategy has limited power in the discovery of new genes and structural variations. With the advance of soybean genomics, more and more high-quality soybean genome assemblies are available (Shen et al., 2018; Valliyodan et al., 2019; Xie et al., 2019). A high-quality pan-genome of wild and cultivated soybeans are also reported recently (Liu et al., 2020). These genomes allow direct comparison of sequence of clock components from different soybean accessions which resulted in more accurate discovery of variants. Furthermore, better reference genomes also facilitate epigenetic study. It has been demonstrated that histone modifications have played crucial roles in regulating the core clock components in plant (reviewed in Chen and Mas, 2019; Du et al., 2019). Nevertheless, information of the roles of epigenetic on soybean domestication and soybean circadian clock function is scarce. Thus, exploration of the modification of circadian clock components and their regulation at epigenetic level could be the next chapter of soybean research.
M-WL drafted the manuscript. M-WL and H-ML edited and proofread the final manuscript. Both authors contributed to the article and approved the submitted version.
This work was supported by the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16), the Lo Kwee-Seong Biomedical Research Fund, and Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute (YFJGJS1.0). Any opinions, findings, conclusions, or recommendations expressed in this publication do not reflect the views of the Government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This manuscript was copy-edited by Jee-Yan Chu.
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Alonso-Blanco, C., Andrade, J., Becker, C., Bemm, F., Bergelson, J., Borgwardt, K. M., et al. (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491.
Belbin, F. E., Fraser, D. P., Comben, N. E., and Dodd, A. N. (2019). “Plant circadian rhythms and vertical agriculture,” in Plant Factory Using Artificial Light: Adapting to Environmental Disruption and Clues to Agricultural Innovation, eds M. Anpo, H. Fukuda, and T. Wada (Amsterdam: Elsevier), 79–88. doi: 10.1016/b978-0-12-813973-8.00007-5
Board, J. E., and Hall, W. (1984). Premature flowering in soybean yield reductions at nonoptimal planting dates as influenced by temperature and photoperiod. Agron. J. 76, 700–704. doi: 10.2134/agronj1984.00021962007600040043x
Bonato, E. R., and Vello, N. A. (1999). E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 22, 229–232. doi: 10.1590/s1415-47571999000200016
Brest, D. E. (1970). Circadian leaf movements in Biloxi soybeans. Plant Physiol. 45, 647–648. doi: 10.1104/pp.45.5.647
Bunning, E. (1954). Der verlauf der endogenen tagersrhythmik bei photoperiodischen storlicht-versuchen mit soja. Physiol. Plant 7, 538–547. doi: 10.1111/j.1399-3054.1954.tb07599.x
Cao, D., Li, Y., Lu, S. J., Wang, J. L., Nan, H. Y., Li, X. M., et al. (2015). GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. 56, 2409–2422. doi: 10.1093/pcp/pcv152
Carpentleri-Pipolo, V., De Almeida, L. A., and Kiihl, R. A. D. (2002). Inheritance of a long juvenile period under short-day conditions in soybean. Genet. Mol. Biol. 25, 463–469. doi: 10.1590/s1415-47572002000400016
Cha, J. Y., Kim, J., Kim, T. S., Zeng, Q., Wang, L., Lee, S. Y., et al. (2017). GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 8:3.
Chen, Z. J., and Mas, P. (2019). Interactive roles of chromatin regulation and circadian clock function in plants. Genome Biol. 20:62.
Cheng, Q., Gan, Z. R., Wang, Y. P., Lu, S. J., Hou, Z. H., Li, H. Y., et al. (2020). The soybean gene J contributes to salt stress tolerance by up-regulating salt-responsive genes. Front. Plant Sci. 11:272. doi: 10.3389/fpls.2020.00272
Cober, E. R. (2011). Long juvenile soybean flowering responses under very short photoperiods. Crop Sci. 51, 140–145. doi: 10.2135/cropsci2010.05.0262
Cober, E. R., Molnar, S. J., Charette, M., and Voldeng, H. D. (2010). A new locus for early maturity in soybean. Crop Sci. 50, 524–527. doi: 10.2135/cropsci2009.04.0174
Cober, E. R., and Voldeng, H. D. (2001). A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41, 698–701. doi: 10.2135/cropsci2001.413698x
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. doi: 10.1186/gb-2008-1189-1188-r1130
Coyne, K., Davis, M. M., Mizoguchi, T., and Hayama, R. (2019). Temporal restriction of salt inducibility in expression of salinity-stress related gene by the circadian clock in Solanum lycopersicum. Plant Biotechnol. 36, 195–200. doi: 10.5511/plantbiotechnology.19.0703a
Destro, D., Carpentieri-Pípolo, V., Afonso, R., Kiihl, S., and Almeida, L. (2001). Photoperiodism and genetic control of the long juvenile period in soybean: a review. Crop Breed. Appl. Biotechnol. 1, 72–92. doi: 10.13082/11984-17033.v13001n13001a13010
Dissanayaka, A., Rodriguez, T. O., Di, S., Yan, F., Githiri, S. M., Rodas, F. R., et al. (2016). Quantitative trait locus mapping of soybean maturity gene E5. Breed. Sci. 66, 407–415. doi: 10.1270/jsbbs.15160
Dodd, A. N., Bebin, F. E., Frank, A., and Webb, A. A. R. (2015). Interactions between circadian clocks and photosynthesis for the temporal and spatial coordination of metabolism. Front. Plant Sci. 6:245. doi: 10.3389/fpls.2015.00245
Dodd, A. N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Du, S. X., Chen, L., Ge, L. F., and Huang, W. (2019). A novel loop: mutual regulation between epigenetic modification and the circadian clock. Front. Plant Sci. 10:22. doi: 10.3389/fpls.2019.00022
Fang, C., Ma, Y. M., Wu, S. W., Liu, Z., Wang, Z., Yang, R., et al. (2017). Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 18:161.
Faure, S., Turner, A. S., Gruszka, D., Christodoulou, V., Davis, S. J., Von Korff, M., et al. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. U.S.A. 109, 8328–8333. doi: 10.1073/pnas.1120496109
Fodor, N., Challinor, A., Droutsas, I., Ramirez-Villegas, J., Zabel, F., Koehler, A. K., et al. (2017). Integrating plant science and crop modeling: assessment of the impact of climate change on soybean and maize production. Plant Cell Physiol. 58, 1833–1847. doi: 10.1093/pcp/pcx141
Foo, M., Somers, D. E., and Kim, P. J. (2016). Kernel architecture of the genetic circuitry of the Arabidopsis circadian system. PLoS Comput. Biol. 12:e1004748. doi: 10.1371/journal.pcbi.1004748
Fukushima, A., Kusano, M., Nakamichi, N., Kobayashi, M., Hayashi, N., Sakakibara, H., et al. (2009). Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. U.S.A. 106, 7251–7256. doi: 10.1073/pnas.0900952106
Gawronski, P., Ariyadasa, R., Himmelbach, A., Poursarebani, N., Kilian, B., Stein, N., et al. (2014). A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics 196, 1253–1261. doi: 10.1534/genetics.113.158444
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172.
Gil, K. E., and Park, C. M. (2019). Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 221, 1215–1229. doi: 10.1111/nph.15518
Graf, A., and Smith, A. M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16, 169–175. doi: 10.1016/j.tplants.2010.12.003
Greenham, K., Lou, P., Puzey, J. R., Kumar, G., Arnevik, C., Farid, H., et al. (2017). Geographic variation of plant circadian clock function in natural and agricultural settings. J. Biol. Rhythms 32, 26–34. doi: 10.1177/0748730416679307
Harmer, S. L., Hogenesch, L. B., Straume, M., Chang, H. S., Han, B., Zhu, T., et al. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. doi: 10.1126/science.290.5499.2110
Hartwig, E. E. (1970). Growth and reproductive characteristics of soybeans [Glycine max (L) Merr.] grown under short-day conditions. Trop. Sci. 12, 47–53.
Hartwig, E. E., and Kiihl, R. A. S. (1979). Identification and utilization of a delayed flowering character in soybeans for short-day conditions. Field Crops Res. 2, 145–151. doi: 10.1016/0378-4290(79)90017-0
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Huang, H., and Nusinow, D. A. (2016). Into the evening: complex interactions in the Arabidopsis circadian clock. Trends Genet. 32, 674–686. doi: 10.1016/j.tig.2016.08.002
Huang, L. C., Li, Q. F., Zhang, C. Q., Chu, R., Gu, Z. W., Tan, H. Y., et al. (2020). Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. doi: 10.1111/pbi.13391 [Epub ahead of print].
Hudson, K. A. (2010). The circadian clock-controlled transcriptome of developing soybean seeds. Plant Genome 3, 3–13. doi: 10.3835/plantgenome2009.08.0025
Hyten, D. L., Song, Q. J., Zhu, Y. L., Choi, I. Y., Nelson, R. L., Costa, J. M., et al. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. U.S.A. 103, 16666–16671. doi: 10.1073/pnas.0604379103
Jiang, B. J., Nan, H. Y., Gao, Y. F., Tang, L. L., Yue, Y. L., Lu, S. J., et al. (2014). Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS One 9:e106042. doi: 10.1371/journal.pone.0106042
Kawasaki, Y., Yamazaki, R., Katayama, K., Yamada, T., and Funatsuki, H. (2018). Effects of maturity genes E2 and E3 on yield formation in soybean cultivar Enrei in warm region, Fukuyama in Japan. Plant Prod. Sci. 21, 387–397. doi: 10.1080/1343943x.2018.1530063
Kim, J. A., Kim, H. S., Choi, S. H., Jang, J. Y., Jeong, M. J., and Lee, S. I. (2017). The importance of the circadian clock in regulating plant metabolism. Int. J. Mol. Sci. 18:2680. doi: 10.3390/ijms18122680
Kim, S. K., Kim, E. S., Kim, K. H., Jeong, N., Lee, J. S., and Kang, S. (2018). Genetic variance for flowering time conferring E2 gene in photoperiod-insensitive early-maturing soybean accessions and topological distribution in Korea peninsula. Mol. Breed. 38:148. doi: 10.1007/s11032-11018-10887-x
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Kurepa, J., Smalle, J., Van Montagu, M., and Inze, D. (1998). Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 14, 759–764. doi: 10.1046/j.1365-313x.1998.00168.x
Lam, H. M., Xu, X., Liu, X., Chen, W. B., Yang, G. H., Wong, F. L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. doi: 10.1038/ng.715
Lee, C. M., Li, M. W., Feke, A., Liu, W., Saffer, A. M., and Gendron, J. M. (2019). GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat. Commun. 10:3750.
Li, C., Li, Y. H., Li, Y., Lu, H., Hong, H., Tian, Y., et al. (2020). A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol. Plant 13, 745–759. doi: 10.1016/j.molp.2020.01.014
Li, Y. H., Li, D. L., Jiao, Y. Q., Schnable, J. C., Li, Y. F., Li, H. H., et al. (2020). Identification of loci controlling adaptation in Chinese soya bean landraces via a combination of conventional and bioclimatic GWAS. Plant Biotechnol. J. 18, 389–401. doi: 10.1111/pbi.13206
Li, F., Zhang, X. M., Hu, R. B., Wu, F. Q., Ma, J. H., Meng, Y., et al. (2013). Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS One 8:e79036. doi: 10.1371/journal.pone.0079036
Li, M., Cao, L. J., Mwimba, M., Zhou, Y., Li, L., Zhou, M., et al. (2019). Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Proc. Natl. Acad. Sci. U.S.A. 116, 23840–23849. doi: 10.1073/pnas.1708508116
Li, M. W., Liu, W., Lam, H. M., and Gendron, J. M. (2019). Characterization of two growth period QTLs reveals modification of PRR3 genes during soybean domestication. Plant Cell Physiol. 60, 407–420. doi: 10.1093/pcp/pcy215
Li, X. M., Fang, C., Xu, M. L., Zhang, F. G., Lu, S. J., Nan, H. Y., et al. (2017). Quantitative trait locus mapping of soybean maturity gene E6. Crop Sci. 57, 2547–2554. doi: 10.2135/cropsci2017.02.0106
Li, Y., Dong, Y. S., Wu, H. Y., Hu, B., Zhai, H., Yang, J. Y., et al. (2019). Positional cloning of the flowering time QTL qFT12-1 reveals the link between the clock related PRR homolog with photoperiodic response in soybeans. Front. Plant Sci. 10:1303. doi: 10.3389/fpls.2019.01303
Liew, L. C., Singh, M. B., and Bhalla, P. L. (2017). A novel role of the soybean clock gene LUX ARRHYTHMO in male reproductive development. Sci. Rep. 7:10605. doi: 10.1038/s41598-41017-10823-y
Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K., and Abe, J. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome a gene. Genetics 180, 995–1007. doi: 10.1534/genetics.108.092742
Liu, H., Wang, H. G., Gao, P. F., Xu, J. H., Xu, T. D., Wang, J. S., et al. (2009). Analysis of clock gene homologs using unifoliolates as target organs in soybean (Glycine max). J. Plant Physiol. 166, 278–289. doi: 10.1016/j.jplph.2008.06.003
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020). Pan-genome of wild and cultivated soybeans. Cell 182, 162–176.e13.
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Lu, S. J., Li, Y., Wang, J. L., Nan, H. Y., Cao, D., Li, X. M., et al. (2016). Identification of additional QTLs for flowering time by removing the effect of the maturity gene E1 in soybean. J. Integr. Agric. 15, 42–49. doi: 10.1016/s2095-3119(15)61046-2
Lu, S. J., Zhao, X. H., Hu, Y. L., Liu, S. L., Nan, H. Y., Li, X. M., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Marcolino-Gomes, J., Nakayama, T. J., Molinari, H. B. C., Basso, M. F., Henning, L. M. M., Fuganti-Pagliarini, R., et al. (2017). Functional characterization of a putative Glycine max ELF4 in transgenic Arabidopsis and its role during flowering control. Front. Plant Sci. 8:618. doi: 10.3389/fpls.2017.00618
Matsubara, K., Hori, K., Ogiso-Tanaka, E., and Yano, M. (2014). Cloning of quantitative trait genes from rice reveals conservation and divergence of photoperiod flowering pathways in Arabidopsis and rice. Front. Plant Sci. 5:193. doi: 10.3389/fpls.2014.00193
Miladinovic, J., Ceran, M., Dordevic, V., Balesevic-Tubic, S., Petrovic, K., Dukic, V., et al. (2018). Allelic variation and distribution of the major maturity genes in different soybean collections. Front. Plant Sci. 9:1286. doi: 10.3389/fpls.2018.01286
Mishra, P., and Panigrahi, K. C. (2015). GIGANTEA - an emerging story. Front. Plant Sci. 6:8. doi: 10.3389/fpls.2015.00008
Mizuno, T., and Nakamichi, N. (2005). Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 46, 677–685. doi: 10.1093/pcp/pci087
Muller, N. A., Wijnen, C. L., Srinivasan, A., Ryngajllo, M., Ofner, I., Lin, T., et al. (2016). Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 48, 89–93. doi: 10.1038/ng.3447
Murphy, R. L., Klein, R. R., Morishige, D. T., Brady, J. A., Rooney, W. L., Miller, F. R., et al. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. U.S.A. 108, 16469–16474. doi: 10.1073/pnas.1106212108
Nakamichi, N., Takao, S., Kudo, T., Kiba, T., Wang, Y., Kinoshita, T., et al. (2016). Improvement of Arabidopsis biomass and cold, drought and salinity stress tolerance by modified circadian clock-associated PSEUDO-RESPONSE REGULATORs. Plant Cell Physiol. 57, 1085–1097. doi: 10.1093/pcp/pcw057
Nitschke, S., Cortleven, A., Iven, T., Feussner, I., Havaux, M., Riefler, M., et al. (2016). Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28, 1616–1639.
Ogiso-Tanaka, E., Shimizu, T., Hajika, M., Kaga, A., and Ishimoto, M. (2019). Highly multiplexed AmpliSeq technology identifies novel variation of flowering time-related genes in soybean (Glycine max). DNA Res. 26, 243–260.
Pan, L. Y., He, J. B., Zhao, T. J., Xing, G. N., Wang, Y. F., Yu, D. Y., et al. (2018). Efficient QTL detection of flowering date in a soybean RIL population using the novel restricted two-stage multi-locus GWAS procedure. Theor. Appl. Genet. 131, 2581–2599. doi: 10.1007/s00122-018-3174-7
Pan, W. J., Wang, X., Deng, Y. R., Li, J. H., Chen, W., Chiang, J. Y., et al. (2015). Nondestructive and intuitive determination of circadian chlorophyll rhythms in soybean leaves using multispectral imaging. Sci. Rep. 5:11108.
Para, A., Farre, E. M., Imaizumi, T., Pruneda-Paz, J. L., Harmon, F. G., and Kay, S. A. (2007). PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19, 3462–3473. doi: 10.1105/tpc.107.054775
Park, H. J., Qiang, Z., Kim, W. Y., and Yun, D. J. (2016). Diurnal and circadian regulation of salt tolerance in Arabidopsis. J. Plant Biol. 59, 569–578. doi: 10.1007/s12374-016-0317-8
Qi, X. P., Li, M. W., Xie, M., Liu, X., Ni, M., Shao, G. H., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5:4340.
Ray, J. D., Hinson, K., Mankono, J. E. B., and Malo, M. F. (1995). Genetic-control of a long-juvenile trait in soybean. Crop Sci. 35, 1001–1006. doi: 10.2135/cropsci1995.0011183x003500040012x
Samanfar, B., Molnar, S. J., Charette, M., Schoenrock, A., Dehne, F., Golshani, A., et al. (2017). Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 130, 377–390. doi: 10.1007/s00122-016-2819-7
Sawa, M., Kay, S. A., and Imaizumi, T. (2008). Photoperiodic flowering occurs under internal and external coincidence. Plant Signal. Behav. 3, 269–271. doi: 10.4161/psb.3.4.5219
Shen, Y., Liu, J., Geng, H., Zhang, J., Liu, Y., Zhang, H., et al. (2018). De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 61, 871–884.
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Shor, E., and Green, R. M. (2016). The impact of domestication on the circadian clock. Trends Plant Sci. 21, 281–283. doi: 10.1016/j.tplants.2016.01.022
Simon, N. M. L., Graham, C. A., Comben, N. E., Hetherington, A. M., and Dodd, A. N. (2020). The circadian clock influences the long-term water use efficiency of Arabidopsis. Plant Physiol. 183, 317–330. doi: 10.1104/pp.20.00030
Staiger, D., Shin, J., Johansson, M., and Davis, S. J. (2013). The circadian clock goes genomic. Genome Biol 14:208.
Valliyodan, B., Cannon, S. B., Bayer, P. E., Shu, S. Q., Brown, A. V., Ren, L. H., et al. (2019). Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 100, 1066–1082. doi: 10.1111/tpj.14500
Vezza, M. E., Alderete, L. G. S., Agostini, E., and Talano, M. A. (2020). Expression of circadian clock genes and diurnal oscillations of key physiological events in response to AsV and AsIII in soybean plants. Environ. Exp. Bot. 174:104054. doi: 10.1016/j.envexpbot.2020.104054
Wang, L., Sun, S., Wu, T., Liu, L., Sun, X., Cai, Y., et al. (2020). Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 18, 1869–1881. doi: 10.1111/pbi.13346
Wang, T., Sun, M. Y., Wang, X. S., Li, W. B., and Li, Y. G. (2016). Over-expression of GmGIa-regulated soybean miR172a confers early flowering in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 17:645. doi: 10.3390/ijms17050645
Wang, Y., Gu, Y. Z., Gao, H. H., Qiu, L. J., Chang, R. Z., Chen, S. Y., et al. (2016). Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 16:79. doi: 10.1186/s12862-016-0653-9
Wang, Y., Yuan, L., Su, T., Wang, Q., Gao, Y., Zhang, S. Y., et al. (2020). Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 43, 637–648. doi: 10.1111/pce.13678
Watanabe, S., Hideshima, R., Xia, Z. J., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262. doi: 10.1534/genetics.108.098772
Watanabe, S., Xia, Z. J., Hideshima, R., Tsubokura, Y., Sato, S., Yamanaka, N., et al. (2011). A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407. doi: 10.1534/genetics.110.125062
Xia, Z. J., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109, E2155–E2164.
Xie, M., Chung, C. Y. L., Li, M. W., Wong, F. L., Wang, X., Liu, A. L., et al. (2019). A reference-grade wild soybean genome. Nat. Commun. 10:1216.
Yang, Y., Peng, Q., Chen, G. X., Li, X. H., and Wu, C. Y. (2013). OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol. Plant 6, 202–215. doi: 10.1093/mp/sss062
Yue, Y. L., Liu, N. X., Jiang, B. J., Li, M., Wang, H. J., Jiang, Z., et al. (2017). A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol. Plant 10, 656–658. doi: 10.1016/j.molp.2016.12.004
Zhang, B., Liu, H. Y., Qi, F. X., Zhang, Z. Y., Li, Q. P., Han, Z. M., et al. (2019). Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice. Rice 12:48. doi: 10.1186/s12284-12019-10314-x
Zhao, C., Takeshima, R., Zhu, J. H., Xu, M. L., Sato, M., Watanabe, S., et al. (2016). A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16:20. doi: 10.1186/s12870-016-0704-9
Zhao, J. M., Huang, X., Ouyang, X. H., Chen, W. L., Du, A. P., Zhu, L., et al. (2012). OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One 7:e43705. doi: 10.1371/journal.pone.0043705
Keywords: circadian clock, domestication, early flowering 3, Gigantea, pseudo response regulator 3, J locus, soybean
Citation: Li M-W and Lam H-M (2020) The Modification of Circadian Clock Components in Soybean During Domestication and Improvement. Front. Genet. 11:571188. doi: 10.3389/fgene.2020.571188
Received: 10 June 2020; Accepted: 19 August 2020;
Published: 30 September 2020.
Edited by:
Peerasak Srinives, Kasetsart University, ThailandReviewed by:
Jian Sun, Shenyang Agricultural University, ChinaCopyright © 2020 Li and Lam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hon-Ming Lam, aG9ubWluZ0BjdWhrLmVkdS5oaw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.