
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 23 November 2020
Sec. Evolutionary and Population Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.569314
This article is part of the Research Topic The Evolution of Biomineralization in Metazoans View all 16 articles
 Jian Ming Khor
Jian Ming Khor Charles A. Ettensohn*
Charles A. Ettensohn*Members of the alx gene family encode transcription factors that contain a highly conserved Paired-class, DNA-binding homeodomain, and a C-terminal OAR/Aristaless domain. Phylogenetic and comparative genomic studies have revealed complex patterns of alx gene duplications during deuterostome evolution. Remarkably, alx genes have been implicated in skeletogenesis in both echinoderms and vertebrates. In this review, we provide an overview of current knowledge concerning alx genes in deuterostomes. We highlight their evolutionarily conserved role in skeletogenesis and draw parallels and distinctions between the skeletogenic gene regulatory circuitries of diverse groups within the superphylum.
Biomineralization, the formation of mineral by living organisms, is an exceptionally widespread phenomenon and is thought to have evolved independently and rapidly in many different metazoan phyla through the deployment of a wide range of biomineralization mechanisms and chemistries. Depending on the type and extent of the mineral components, biomineralized tissues are used for structural support, resource acquisition, and protection. There are three predominant classes of biogenic mineral in metazoans: calcium carbonates, calcium phosphates, and silica. The carbonate and phosphate salts of calcium are widely used as skeletal material by vertebrates and invertebrates, while silica biomineralization is prevalent in sponges (Wang et al., 2010b). The emergence of biomineralization during the Cambrian Explosion, followed by evolutionary modifications of these biomineralization programs, gave rise to the diverse biomineralized structures found in modern metazoans (Knoll, 2003; Zhuravlev and Wood, 2018).
Within the deuterostome superphylum, only vertebrates and echinoderms produce extensive biomineralized skeletal structures. The vertebrate endoskeleton consists primarily of the skull, vertebrae, ribs, and limb bones all of which are composed of matrix proteins (e.g., collagens) and calcium phosphate crystals. Vertebrate biomineralization is predominantly orchestrated by chondrogenic cells (chondrocytes) and osteogenic cells (osteoblasts and osteoclasts). The vertebrate skeleton is formed during early development by cartilage and/or connective tissue membranes, which are subsequently replaced by bony tissues through the process of ossification. There are two forms of ossification, endochondral and intramembranous ossification. Endochondral ossification is associated with the formation of long bones and requires the presence of a hyaline cartilage template formed by chondrocytes (Mackie et al., 2008). During vertebrate embryonic development, chondrocytes are derived from neural crest cells, somitic mesodermal cells, and lateral plate mesodermal cells (see review by Hirasawa and Kuratani, 2015). Developmental cues signal the cartilage matrix to calcify. This prevents the diffusion of nutrients into the matrix and results in chondrocyte apoptosis, allowing blood vessels to invade the cartilage cavities. Osteoblasts, derived from common osteochondroprogenitor or directly from chondrocytes (Yang et al., 2014), and osteoclasts, derived from erythron-myeloid progenitors (Jacome-Galarza et al., 2019), then transform the calcified cartilage into biomineralized bone (Mackie et al., 2008). During intramembranous ossification, spongy bones are formed when osteoblasts directly deposit biomineral on extracellular sheets of mesenchymal connective tissues (Percival and Richtsmeier, 2013). This process is commonly involved in the formation of flat bones found in the skull, mandible, and clavicles. Whether intramembranous or endochondral ossification arose first during vertebrate evolution remains unclear (Cervantes-Diaz et al., 2017; Wood and Nakamura, 2018; Brazeau et al., 2020).
All adult echinoderms produce calcite-based endoskeletons that consist of the test, teeth, and spines. In most species, the adult form arises from a swimming, feeding larva via metamorphosis, and these two life history stages bear little morphological resemblance to one another. In some echinoderm clades, specifically echinoids (sea urchins) and ophiuroids (brittle stars), the feeding larva also possesses an intricate and extensive calcitic endoskeleton, which is first laid down during embryonic development and further elaborated after feeding begins. The founder cells of the embryonic skeletogenic lineage, the large micromeres, arise early in development and are specified by a combination of localized maternal factors and unequal cell division. At the mesenchyme blastula stage, the large micromere descendants undergo an epithelial-to-mesenchyme transition (EMT) and ingress into the blastocoel as primary mesenchyme cells, or PMCs (see reviews by Ettensohn, 2020; McClay et al., 2020). After ingression, PMCs extend filopodia and migrate along the blastocoel wall, gradually adopting a ring-like configuration near the equator of the embryo. As the PMCs migrate, their filopodia fuse, forming a cable-like cytoplasmic strand that connects the cells in a syncytial network. Amorphous calcium carbonate and associated proteins are then secreted into an intercellular space within the cytoplasmic cable, where the biomineral matures and grows, eventually producing the elaborate, branched skeletal elements (spicules) of the larva (Wilt, 2002; McIntyre et al., 2014; Shashikant et al., 2018).
Due to differences in mechanisms underlying axial patterning, developmental timing, and embryological structures, it is often difficult to deduce morphological homology. Although the biomineralized tissues found in different metazoan phyla are not considered homologous in the strictest sense, recent comparative studies have revealed common elements across different biomineralization systems. This has led to the recognition of a possible “biomineralization toolkit;” an ancestral gene regulatory network (GRN) consisting of signaling and gene regulatory pathways that was independently co-opted and fine-tuned for biomineralization in diverse animal taxa. One common regulator of deuterostome skeletogenesis is the Alx transcription factor family, which has been shown to have an ancient, conserved role in this process in both vertebrates and echinoderms. In this review, we examine the current state of knowledge concerning deuterostome alx genes, with a focus on their role in skeletogenesis.
The alx gene family encodes Paired-class homeodomain transcription factors that contain a highly conserved DNA-binding homeodomain and a C-terminal Otp, Aristaless, and Rax (OAR) domain, features that are shared by many Paired-class homeodomain proteins. Phylogenetic and comparative genomic studies have revealed considerable variability in the number of alx genes in different deuterostomes, pointing to a complex evolutionary pattern of lineage-specific gene duplication and loss (Figure 1; adapted from McGonnell et al., 2011; Koga et al., 2016). Hemichordates possess a single alx gene (Koga et al., 2016) while echinoderms have two (alx1 and alx4; Ettensohn et al., 2003; Koga et al., 2016). In contrast, humans and mammals possess three alx genes (alx1/cart1, alx3, and alx4) that arose through two duplication events. Through the course of evolution, one of the paralogues, alx3, was lost from amphibian and reptile lineages (McGonnell et al., 2011). Additionally, ray-finned fishes such as zebrafish acquired two paralogues of alx4, designated alx4a and alx4b, as a result of a separate, whole genome duplication event (McGonnell et al., 2011). The lancelets have two alx genes. In Branchiostoma floridae, these two genes (Bf-alx1 and Bf-alx2) are located close to each other in the genome and have very similar intron-exon organizations. Molecular phylogenetic analysis of Alx proteins indicate that Bf-Alx1 and Bf-Alx2 form a monophyletic group, providing further support for the view that they arose from a lineage-specific gene duplication event (Figure 1; Koga et al., 2016).
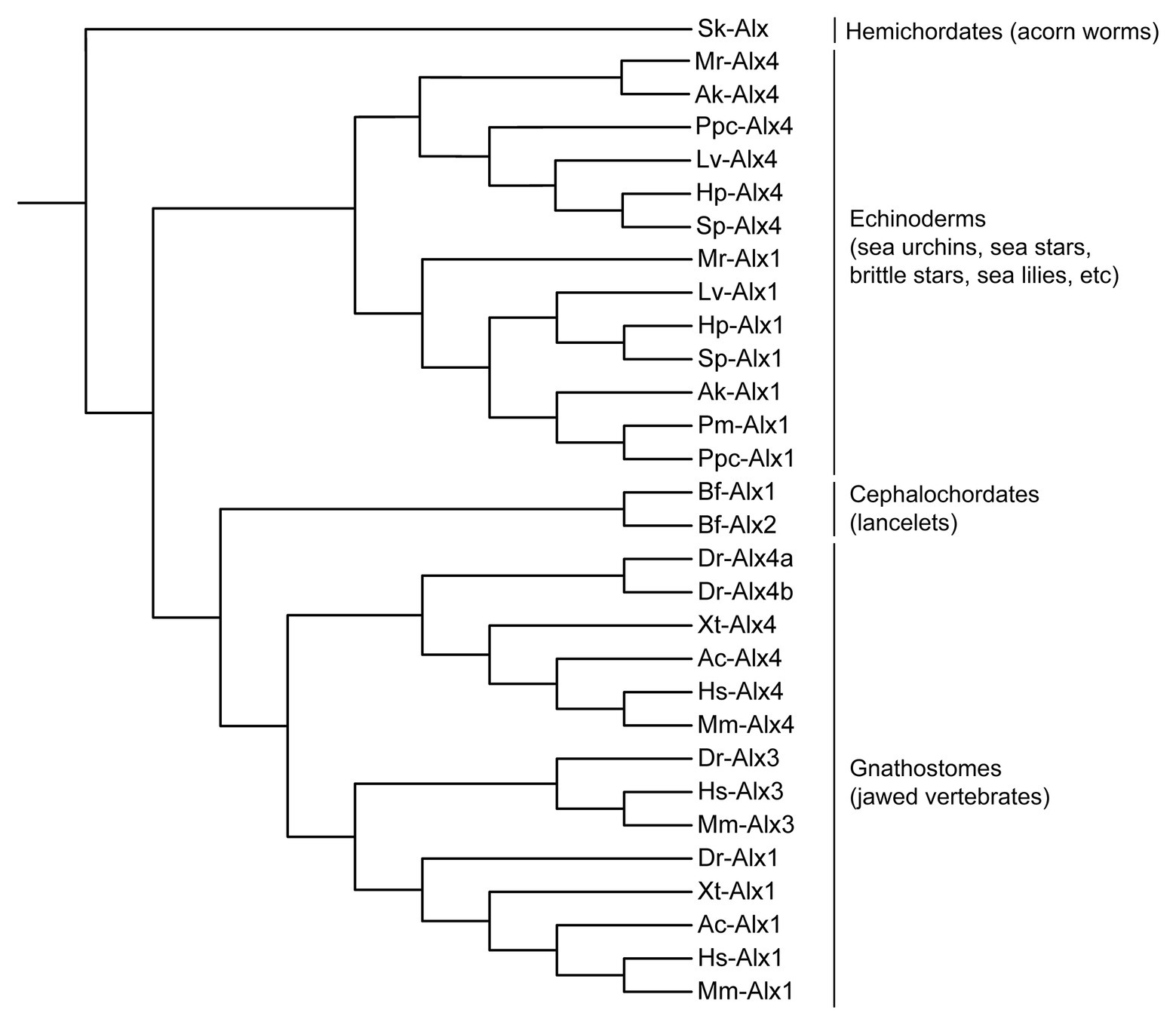
Figure 1. Molecular phylogeny of Alx proteins (adapted from McGonnell et al., 2011; Koga et al., 2016). Branch lengths are arbitrary. Sk, Saccoglossus kowalevskii (acorn worm); Lv, Lytechinus variegatus (euechinoid sea urchin); Hp, Hemicentrotus pulcherrimus (euechinoid sea urchin); Sp, Strongylocentrotus purpuratus (euechinoid sea urchin); Mr, Metacrinus rotundus (sea lily); Ak, Amphipholis kochii (brittle star); Pm, Patiria miniata (sea star); Ppc, Patiria pectinifera (sea star); Bf, Branchiostoma floridae (lancelet); Dr, Danio rerio (zebrafish); Xt, Xenopus tropicalis (frog); Ac, Anolis carolinensis (lizard); Hs, Homo sapiens (human); Mm, Mus musculus (mouse).
Members of the alx gene family are expressed in several mesenchymal tissues during the embryogenesis of jawed vertebrates (gnathostomes), a group that includes most of the vertebrate species used for developmental studies. These genes are expressed most prominently in distinct but partially overlapping patterns in neural crest-derived craniofacial mesenchyme and in mesenchyme of the limb bud, both of which are sources of cartilage and bone (Zhao et al., 1994; Qu et al., 1997a; ten Berge et al., 1998; Beverdam and Meijlink, 2001). Other sites of embryonic expression have also been reported, including the head mesoderm, sclerotome of the somite (another tissue that produces cartilage and bone), hair follicles, dental papillae of teeth, and parts of the developing urogenital system (Zhao et al., 1994; Hudson et al., 1998; ten Berge et al., 1998; Bothe et al., 2011; Wang et al., 2019).
In the developing head, genes of the alx family are expressed by neural crest cells, which give rise to cartilages and bones of the skull, jaw, and middle ear, as well as other derivatives (see reviews by Santagati and Rijli, 2003; Noden and Trainor, 2005). Consistent with this pattern of expression, perturbations of alx genes commonly result in severe craniofacial malformations, including frontonasal dysplasia and the reduction or malformation of many neural crest-derived skeletal elements (Table 1). In mice, loss-of-function mutations of alx1/cart1 or alx4 also lead to other cranial abnormalities such as anencephaly and lacrimal gland aplasia (Zhao et al., 1996; Garg et al., 2017), although these effects are likely to be secondary consequences of defects in neural crest cells, which provide essential signals that regulate the development of the brain and eye (Zhao et al., 1996; Bhattacherjee et al., 2009; Le Douarin, 2012; Garg et al., 2017). While alx3-null mice appear normal, alx3/alx4 double mutant mice exhibit severe frontonasal dysplasia and cranial skeletal defects that are more extreme than those observed in alx4 mutant mice, revealing non-equivalent but overlapping functions of these highly similar proteins (Beverdam et al., 2001).
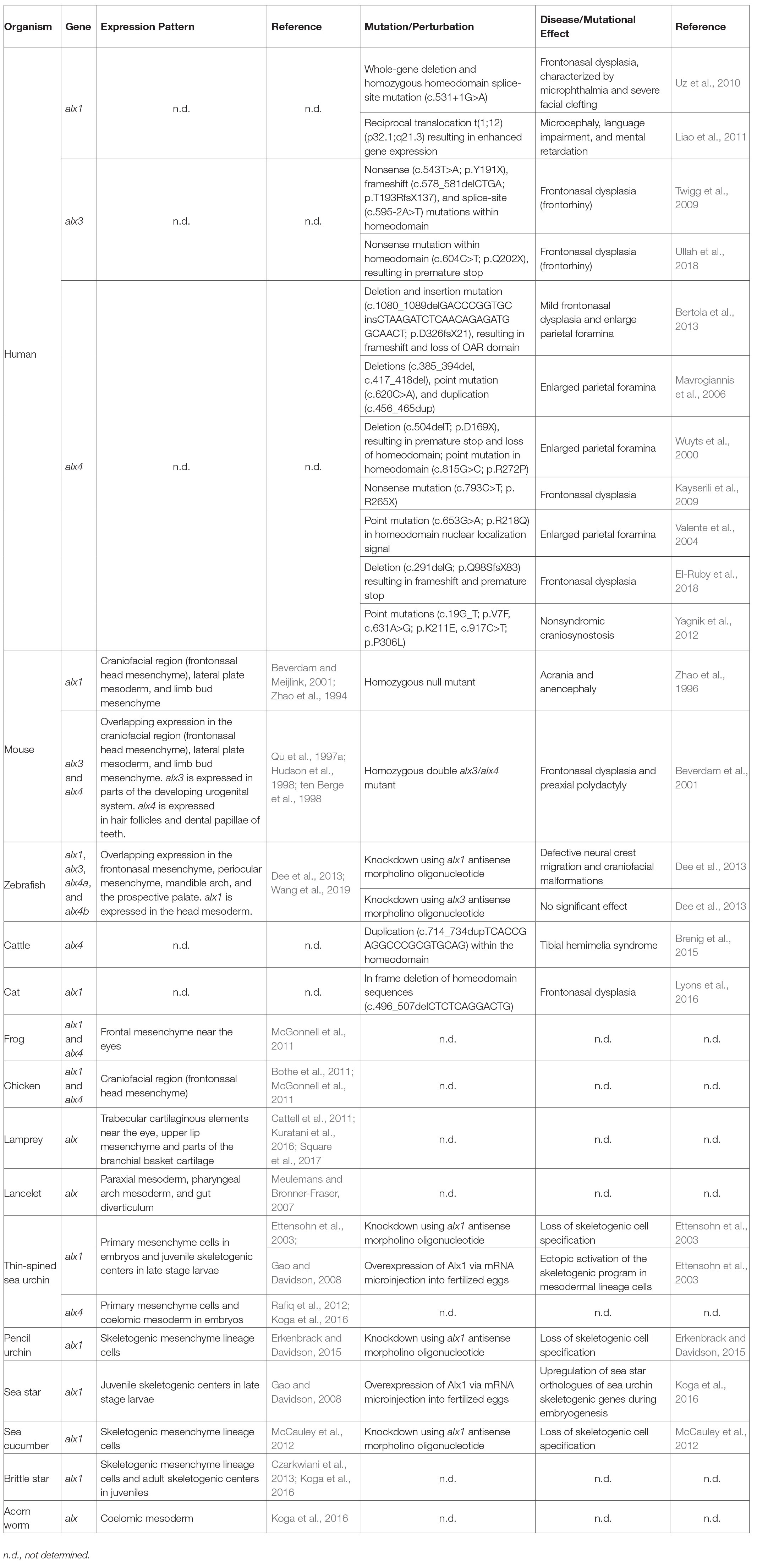
Table 1. Summary of expression patterns, mutations, perturbations, and diseases associated with alx genes across different deuterostome phyla.
During early zebrafish development, the expression of alx1 alone is detected in migrating neural crest cells, while at later stages, alx1, alx3, alx4a, and alx4b exhibit overlapping patterns of expression in the craniofacial mesenchyme (Dee et al., 2013; Wang et al., 2019). Alx1 is also transiently expressed in the cranial paraxial mesoderm at early developmental stages (Wang et al., 2019). Perturbation of Alx1 expression using antisense morpholino oligonucleotides (MOs) produces severe craniofacial defects in zebrafish, similar to results seen in the mouse, inhibition of alx3 alone results in no significant craniofacial abnormalities (Dee et al., 2013). In developing frog and chick embryos, both alx1 and alx4 are expressed robustly in the craniofacial mesenchyme (Bothe and Dietrich, 2006; McGonnell et al., 2011; Square et al., 2015).
Genes of the alx family are also expressed in the mesodermal compartment of the limb buds. At early embryonic stages, these genes are expressed specifically in an anterior, proximal zone while later in development they are also expressed at the distal margin (Qu et al., 1997b). The anterior, proximal zone of expression may include sites where skeletal elements of the shoulder and pelvic girdles (the scapula and pelvis, respectively) form, although this has not been shown directly. The skeletal elements of the limb girdles have complex embryological origins that are only partially understood. The scapula may arise from three sources: somatic mesoderm of the lateral plate, somite-derived dermamyotome, and neural crest, while the pelvis likely arises from somatic mesoderm and sclerotome (Young et al., 2019). Genetic knockouts in mice have revealed essential and partially redundant roles for alx1, alx3, and alx4 in the formation of the superior/anterior portion of the scapula blade (and in the development of the clavicle) and have shown that alx1 expression in this region is under the direct control of the transcription factors Emx2 and Pbx1 (Kuijper et al., 2005a,b; Capellini et al., 2010). Similarly, compound alx1:alx4 and alx3:alx4 double mutants reveal overlapping roles for these genes in the formation of the pelvic skeleton (Kuijper et al., 2005b; Young et al., 2019). Unlike the neural crest-derived skeleton of the head, the scapula and pelvis both form by endochondral ossification, and defects are observed in both the cartilaginous and bony compartments of these skeletal elements when the function of alx family genes is compromised.
A striking developmental consequence of alx4 null mutations is preaxial polydactyly – the formation of one or more supernumerary anterior digits (Forsthoefel, 1963; Qu et al., 1997b). This effect is associated with the formation of an ectopic, anterior zone of polarizing activity (ZPA) in the limb bud and concomitant, anterior expression of sonic hedgehog (shh; Chan et al., 1995; Qu et al., 1997a,b; Takahashi et al., 1998). At relatively late developmental stages, Shh signaling is required for polydactyly to develop in alx4-null mutants, but it has been proposed that alx4 also plays an earlier, Shh-independent role in anterior-posterior patterning (Kuijper et al., 2005b). The expression domains of alx4 and shh during limp outgrowth are established, in part, by mutual repression (Kuijper et al., 2005b; Matsubara et al., 2017).
Consistent with the results of experimental gene perturbations, genetic association studies in several vertebrate species have shown that polymorphisms in alx genes are associated with phenotypic variations in skeletal development. A genome-wide scan of genetic diversity between two closely related species of Darwin’s finches has revealed that polymorphism within the alx1 gene is strongly associated with beak morphology (Lamichhaney et al., 2015). Linkage analysis and genome-wide association studies have also identified a small 12 bp deletion in the alx1 gene that is associated with frontonasal dysplasia in Burmese cats (Lyons et al., 2016). Furthermore, variations in the number of repeats in the coding region of alx4 are quantitatively associated with polydactyly in the Great Pyrenees dog breed (Fondon and Garner, 2004), and a 20 bp duplication in the alx4 gene is linked to congenital tibial hemimelia (loss or shortening of the tibia) in Gallow cattle (Brenig et al., 2015). Taken together, these findings suggest that an ancient alx gene may have constituted a conserved, core element of the ancestral vertebrate skeletogenic GRN and that gene duplication followed by divergence of the paralogs with respect to their developmental expression and/or biochemical properties has produced multiple alx family members with overlapping functions.
Considered as a whole, these studies show that members of the vertebrate alx gene family play a conserved, prominent role in the development of the cranial and appendicular skeletons. In contrast, they do not appear to mediate the development of the sclerotome-derived, axial skeleton of the trunk (the vertebrae and ribs). Members of the alx gene family may also have other, less well-characterized, developmental functions, although some of the effects of mutations in these genes on non-skeletal tissues are likely to be indirect. In the cranial region, it is well-established that alx-family genes are expressed robustly and selectively by neural crest cells (Rice et al., 2003; Dee et al., 2013; Garg et al., 2017), a cell population that gives rise to both cartilage and membranous bone. Expression of alx family genes is not uniform in all regions of the developing head, however, and it has been hypothesized that this contributes to a regulatory code that controls the region-specific identity of the cranial neural crest (Square et al., 2017). With respect to appendage development, the expression of alx-related genes is associated with skeleton-forming potential of mesenchymal cell that will form proximal elements of the limb girdles (clavicle, scapula, and pelvis; Young et al., 2019). The embryological origins and the precise developmental fates of these cells, as well as that of other cells of the developing limb that express alx-related genes, are not well-characterized.
In basally-derived (jawless) vertebrates and cephalochordates (amphioxus), animals that possess only cartilaginous skeletons, alx-family genes are expressed in patterns consistent with a role in skeletogenesis. The single lamprey alx gene is expressed at high levels in the trabecular cartilaginous elements near the eye, in a region that may be derived from mesoderm or from the cranial neural crest (Kuratani et al., 2016; Square et al., 2017). Cephalochordates have stiff, acellular pharyngeal endoskeletons that contain fibrillar collagen, and the adult form has a cartilaginous oral skeleton that supports the cirri (Jandzik et al., 2015). Amphioxus lacks a neural crest, and the embryonic cell lineage that produces the oral skeleton has not been identified. One study has examined the expression of alx-related genes in cephalochordates and reported expression in the somites and right gut diverticulum at neurula/early larval stages (Meulemans and Bronner-Fraser, 2007). At present, the function of alx-related genes in jawless vertebrates and amphioxus has not been explored through gene perturbation studies.
In echinoderm clades that form larval skeletons, alx1 is one of the earliest regulatory genes expressed during development, and it plays a pivotal role in specifying the fate of PMCs, the embryonic skeletogenic cells (Ettensohn et al., 2003; Erkenbrack and Davidson, 2015; Dylus et al., 2016; Shashikant et al., 2018). Transcription of alx1 can be detected as early as the 56-cell stage specifically in the large micromeres (the progenitors of PMCs), and expression remains restricted to this cell lineage throughout embryogenesis (Ettensohn et al., 2003). Perturbation of Alx1 expression using MOs inhibits PMC specification while overexpression of Alx1 results in ectopic activation of the skeletogenic program in other mesodermal lineages. Furthermore, experimental ablation of PMCs leads to the activation of alx1 and downstream components of the skeletogenic GRN by non-skeletogenic mesoderm (NSM) cells, which ultimately reform a larval skeleton (Ettensohn et al., 2007). The ectopic activation of alx1 is essential for NSM cells to acquire skeletogenic properties, although this activation occurs by a mechanism distinct from that which normally operates in the large micromeres (Oliveri et al., 2008; Sharma and Ettensohn, 2011; Ettensohn and Adomako-Ankomah, 2019). Remarkably, the removal of NSM cells via microsurgical removal of the archenteron as well as PMCs results in the activation of alx1 and formation of a skeleton by presumptive endoderm cells (Sharma and Ettensohn, 2011).
The role of alx1 in the skeletogenic GRN in euechinoid sea urchins has been especially well-characterized (Figure 2). Alx1 provides positive inputs into almost half of the ~420 genes that are differentially expressed by PMCs, highlighting the pivotal role of Alx1 in establishing skeletogenic cell identity (Rafiq et al., 2014). A recent chromatin immunoprecipitation sequencing (ChIP-seq) study determined that many of these genes, including both regulatory (i.e., transcription factor-encoding) and effector (i.e., differentiation) genes, are direct targets of alx1 (Khor et al., 2019). A second transcription factor, Ets1, collaborates with Alx1 in the co-regulation of a large fraction of genes differentially expressed by PMCs (Rafiq et al., 2014), in many cases through a feed-forward mechanism (i.e., Ets1 > Alx1, Ets + Alx1 > effector gene; Yamasu and Wilt, 1999; Amore and Davidson, 2006; Oliveri et al., 2008; Yajima et al., 2010; Shashikant et al., 2018; Khor et al., 2019). Downstream effector genes that are regulated by Alx1 include those that directly mediate biomineralization (e.g., those that encode secreted spicule matrix proteins that are incorporated into the biomineral) and those that mediate skeletogenesis through signaling pathways and morphogenetic cell behaviors (Figure 2).
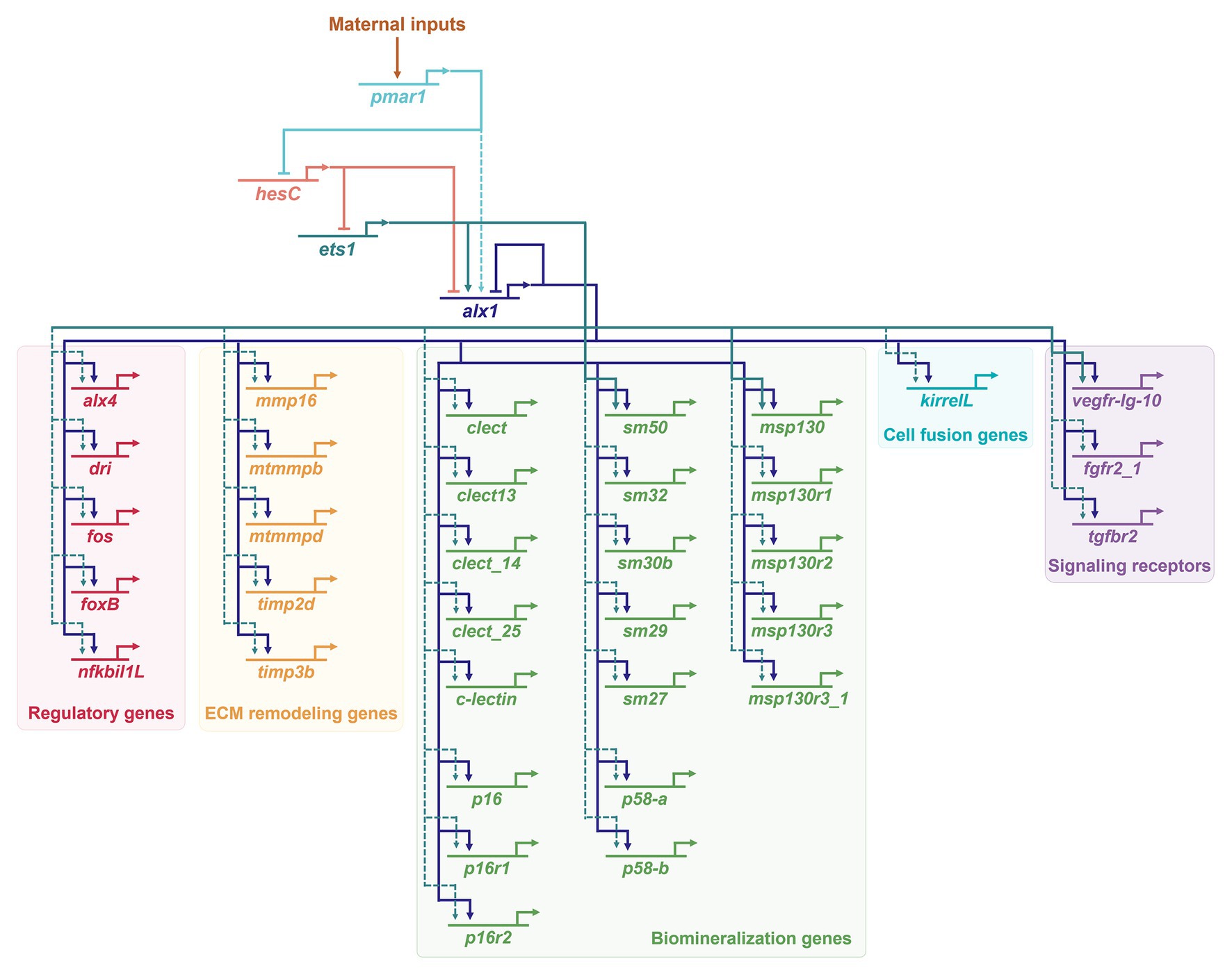
Figure 2. Activation of Alx1 in euechinoids (S. purpuratus) and regulatory inputs into primary mesenchyme cell (PMC) effector genes. Only a small number of more than 420 effector genes differentially expressed in PMCs (Rafiq et al., 2014) is shown here. A large subset of effector genes receives regulatory inputs from both Ets1 and Alx1 (Rafiq et al., 2014). Positive regulatory inputs by Ets1 and Alx1 into msp130, sm50, and vegf-Ig-10 are described in (Oliveri et al., 2008). Direct targets of the sea urchin Alx1 (Khor et al., 2019) define a genetic subcircuit that impinges on almost all aspect of PMC morphogenesis, including directional cell migration, extracellular matrix (ECM) remodeling, cell-cell fusion, and biomineralization. Dashed arrows indicate interactions that may be indirect. For additional information regarding the developmental functions of the specific effector genes shown here, see Shashikant et al. (2018) and references therein.
The alx1 gene is also expressed specifically in skeletogenic cells of cidaroids (pencil urchins) and holothuroids (sea cucumbers), and is required and for skeletogenesis in these species (McCauley et al., 2012; Erkenbrack and Davidson, 2015). The alx1 gene is robustly expressed in adult skeletogenic centers, even in sea stars, which lack a larval skeleton (Gao and Davidson, 2008; Czarkwiani et al., 2013; Gao et al., 2015). Comparative studies have revealed many similarities in the gene regulatory programs of skeletogenic cells in the larva and adult (Richardson et al., 1989; Gao and Davidson, 2008; Killian et al., 2010; Czarkwiani et al., 2013; Gao et al., 2015). Hence, it is widely thought that the larval skeleton arose within the echinoderms by co-option of an adult skeletogenic program. Moreover, ectopic expression of sea urchin or sea star alx1 in sea star embryos is sufficient to activate several sea star orthologs of sea urchin skeletogenic genes (Koga et al., 2016). These findings confirm the critical role that Alx1 plays in establishing skeletogenic identity across all echinoderms at all life history stages, supporting the view that this function was present in the last common ancestor of echinoderms.
Echinoderms also possess a paralog of alx1, known as alx4. The two genes are directly adjacent to one another in the sea urchin genome, suggesting that they arose through gene duplication. The sister group to echinoderms, the hemichordates, possess a single alx gene, suggesting that the gene duplication occurred after the divergence of echinoderms from hemichordates (Koga et al., 2016). The alx4 gene, like alx1, is expressed by skeletogenic PMCs, but alx4 is also expressed by presumptive coelomic pouch cells at the tip of the archenteron (Rafiq et al., 2012; Koga et al., 2016). The function of alx4 has not been experimentally determined but it has been proposed to be involved in coelom development as the single alx gene in hemichordates is expressed in the coelomic mesoderm. As adult hemichordates possess only small biomineralized elements (Cameron and Bishop, 2012), these observations suggest that alx1 gained enhanced skeletogenic function in echinoderms secondarily. Structure-function analysis of Alx1 and Alx4 in euechinoid sea urchins has revealed that the gene duplication event permitted the functional specialization of Alx1 through changes in intron-exon organization and the acquisition of a novel protein motif known as the D2 domain (Khor and Ettensohn, 2017). As noted above, a recent genome-wide ChIP-seq study showed that a large part of the embryonic skeletogenic GRN of sea urchins is directly regulated by Alx1, including many morphoeffector genes that are also expressed in adult skeletogenic centers. Hence, a heterochronic shift in alx1 expression from adult skeletogenic centers to the embryonic skeletogenic cells may have been sufficient to co-opt a substantial subcircuit of biomineralization genes and ultimately transfer skeletogenesis into the embryo (Khor et al., 2019).
Studies on vertebrates and echinoderms have identified many examples of closely related genes that mediate biomineralization in both taxa, such as collagens and carbonic anhydrases (see reviews by Veis, 2011; Le Roy et al., 2014). Here, we focus on effector genes that have been identified as direct targets of Alx1 in echinoderms (sea urchins) and that have vertebrate counterparts implicated in chondrogenesis or osteogenesis. Though much is known about the interactions between regulatory genes and signaling pathways in vertebrate neural crest and chondrogenic GRNs (Cole, 2011; Simoes-Costa and Bronner, 2015), direct transcriptional inputs into biomineralization genes that are the downstream effectors of these networks have not been elucidated. Such information will be crucial to definitively assess homology between echinoderm and vertebrate skeletogenic GRNs.
One of the direct targets of sea urchin alx1 in biomineralizing cells is the vascular endothelial growth factor (VEGF) receptor, vegfr-Ig-10, one of the two vegfr genes in sea urchins (Duloquin et al., 2007; Rafiq et al., 2014; Khor et al., 2019). During embryonic development, vegfr-Ig-10 expression is restricted to PMCs, while its ligand, vegf3 is expressed in the ectoderm specifically in the regions that lie adjacent to two ventro-lateral clusters of PMCs that initiate biomineral formation. MO-based knockdown of Vegf3 or Vegfr-Ig-10 results in the downregulation of skeletogenic genes and lack of embryonic skeleton formation, while ectopic expression of Vegf3 results in supernumerary skeletal elements and irregular branching (Duloquin et al., 2007; Adomako-Ankomah and Ettensohn, 2013). The vegfr-Ig-10 gene is also expressed in adult skeletogenic centers, even in clades that lack a larval skeleton (Gao and Davidson, 2008; Morino et al., 2012). Other comparative studies in echinoderms have found a strict correlation between the expression of vegf3/vegfr-Ig-10 and the formation of an embryonic skeleton (Duloquin et al., 2007; Morino et al., 2012; Adomako-Ankomah and Ettensohn, 2013; Erkenbrack and Petsios, 2017; Erkenbrack and Thompson, 2019). Remarkably, human VEGFA is able to rescue skeleton formation in sea urchin embryos that lack endogenous Vegf3 expression (Morgulis et al., 2019).
During vertebrate endochondral ossification, the cartilage intermediate is replaced by bone in a process that is partly regulated by the formation of a vascular network (see review by Green et al., 2015). Chondrocytes stimulate vasculogenesis through the secretion of VEGF ligands (Carlevaro et al., 2000). In vitro studies show that VEGF ligands (VEGFA, VEGFB, and VEGFC) and VEGF receptors (VEGFR2 and VEGFR3) are expressed by chondrocytes and chondrogenic cells, and autocrine signaling through this pathway regulates morphogenesis and differentiation (Carlevaro et al., 2000; Bluteau et al., 2007). Inhibition of Vegf signaling perturbs ossification and bone elongation by promoting chondrocyte proliferation rather than osteoblast differentiation (Gerber et al., 1999; Jacobsen et al., 2008). Mice with conditional deletion of vegfa in skeletal lineage cells exhibit thinner bones and decreased skeletal mineralization (Duan et al., 2015). Moreover, conditional deletion of vegfr2 results in reduced osteogenic differentiation (Duan et al., 2015).
Another class of effector protein common to echinoderm and vertebrate biomineralization consists of matrix remodeling proteins such as matrix metalloproteases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs). MMPs constitute a class of enzymes that function in the degradation of extracellular matrix (ECM) proteins (see review by Rose and Kooyman, 2016). In sea urchins, chemical inhibition of MMPs reversibly blocks spiculogenesis by PMCs in vivo and in vitro (Roe et al., 1989; Ingersoll and Wilt, 1998). In vertebrates, mmp-13 (collagenase-3) is expressed specifically in chondrocytes (Tuckermann et al., 2000). Additionally, in vitro studies have shown that silencing of mmp-2 by siRNA disrupts chondrogenic differentiation of mesenchymal stem cells while treatment with a MMP-2 activator stimulates chondrogenesis (Jin et al., 2007). TIMPs have been reported to be the primary endogenous inhibitors of MMPs and are involved in regulating the function of MMPs in many systems (Brew and Nagase, 2010). Overexpression of timp-3 in mice induces defects in skeletal development and growth (Poulet et al., 2016). In contrast, knockdown of timp-1 results in upregulated proliferation of mesenchymal stem cells while delaying osteogenic differentiation (Liang et al., 2019).
Many members of the solute carrier (SLC) family of membrane transport proteins are differentially expressed in the PMCs (Rafiq et al., 2014; Barsi et al., 2015). In addition, Alx1 directly regulates the expression of several members of the SLC5 and SLC26 sub-families, including Slc26a5/1 and Slc5a11/2 (Rafiq et al., 2014; Khor et al., 2019). While there are data pointing to SLCs that are essential for echinoderm skeletogenesis, mainly Slc4a10 (Hu et al., 2018) and Slc26a2/7 (Piacentino et al., 2016), the functions of the proteins that are directly regulated by Alx1 have not been tested. In vertebrates, Slc26a2, a sulfate transporter, has been shown to be highly expressed in developing and mature cartilage (Haila et al., 2001). Mice homozygous for mutations in Slc26a2 exhibit chondrodysplasia, a condition characterized by growth defects and skeletal dysplasia due to reduced chondrocyte proliferation (Forlino et al., 2005). Similarly, mutations in human Slc26a2 also results in chondrodysplasia (Superti-Furga et al., 1996; Jackson et al., 2012).
One of the direct targets of sea urchin Alx1 is fam20C, which encodes a kinase of the FAM20 (family with sequence similarity 20) family (Rafiq et al., 2014; Khor et al., 2019). In vertebrates, members of this family are highly expressed in mineralized tissues, such as teeth and bone (Hao et al., 2007; Wang et al., 2010a). Fam20C is a secreted kinase responsible for the phosphorylation of secreted proteins, many of which are known to be involved in biomineralization (Tagliabracci et al., 2012). Mutations in the human fam20C gene cause Raine syndrome, an autosomal recessive disorder characterized by defects in bone development, including microcephaly, cleft palate, and osteosclerosis (Simpson et al., 2007; Rafaelsen et al., 2013; Takeyari et al., 2014; Seidahmed et al., 2015). In vitro mutational analyses suggest that Fam20C is involved in the differentiation and mineralization of mouse mesenchymal cells (Hao et al., 2007; Liu et al., 2017), and fam20C-null mice exhibit severe biomineralization defects, such as lesions in bones and teeth (Vogel et al., 2012; Wang et al., 2012; Du et al., 2015).
Sea urchin Alx1 also provides positive inputs directly into otop2L, the single sea urchin ortholog of the vertebrate otopetrin genes (Rafiq et al., 2014; Khor et al., 2019). Otopetrins are multi-pass transmembrane proteins that function as proton channels (Saotome et al., 2019). In vertebrates, these proteins play an essential role in regulating the timing, size, and shape of the developing otoconia, extracellular calcium carbonate biominerals that are required for vestibular functions (Hughes et al., 2004; Sollner et al., 2004; Kim et al., 2010). During mouse and zebrafish embryogenesis, otop1 is highly expressed in the developing sensory epithelium of the ear (Hurle et al., 2003; Hughes et al., 2004). In zebrafish, MO-based knockdown of Otop1 results in otolith malformations (Hughes et al., 2004; Sollner et al., 2004). Moreover, otop1 knockout mice also lack otoconia, a phenotype that has been attributed to mis-regulation of intracellular calcium levels (Hughes et al., 2007; Kim et al., 2010). The function of the echinoderm Otop2L protein has not been examined.
Among present-day deuterostomes, extensive biomineralized skeletons are found only in echinoderms and vertebrates. It is inherently difficult to reconstruct the underlying evolutionary relationships between the skeletogenic programs of these two groups, which diverged >600 million years ago (Peterson and Eernisse, 2016). It is widely accepted that the ancestral chordate possessed only a cartilaginous skeleton (Rychel et al., 2006; Murdock and Donoghue, 2011; Jandzik et al., 2015; Keating et al., 2018), strongly supporting the view that biomineralized skeletons appeared independently in vertebrates and echinoderms, and therefore, are not homologous in the strictest sense. This does not, of course, resolve the question of whether common embryological and/or genetic mechanisms were deployed to create a biomineralized skeleton in these two groups; i.e., whether skeletogenesis in the two clades is an example of “deep homology” (Shubin et al., 2009). The presence of collagenous pharyngeal cartilage in both cephalochordates and hemichordates supports the view that this was an ancestral feature of deuterostomes that was later lost in echinoderms (Rychel and Swalla, 2007; Jandzik et al., 2015). Moreover, a recent analysis of chondrogenesis in protostomes (horseshoe crabs and cuttlefish) suggests that a more ancient, SoxE and collagen-based chondrogenic gene network was present in the last common ancestor of all Bilateria (Tarazona et al., 2016), providing further support for the view that echinoderm ancestors at one time also possessed cartilage-forming cells. It should be noted that although there is no evidence for definitive cartilage in modern echinoderms, there are mesoderm-derived populations of mesenchymal cells that produce connective tissue containing fibrillar collagen (Suzuki et al., 1997; Whittaker et al., 2006; Goh and Holmes, 2017).
The evolutionary relationships among the skeletogenic cell lineages of vertebrates that express alx-related genes and the alx1-expressing cells of echinoderms are uncertain. With respect to echinoderms, considerable evidence supports the view that alx1 arose very early in echinoderm evolution through gene duplication, relatively quickly acquired a robust, biomineralization-related function, and was subsequently co-opted into the early embryo in echinoderm taxa that possess larval skeletons (echinoids and ophiuroids; Khor and Ettensohn, 2017; Shashikant et al., 2018). The biomineralizing cells of the ancestral echinoderm, which were likely of mesodermal origins, expressed alx1, ets1, erg, vegfr, and other components of a core skeletogenic program, as well as an assortment of more rapidly evolving biomineralization effector proteins (Gao and Davidson, 2008; Dylus et al., 2018; Erkenbrack and Thompson, 2019; Li et al., 2020). To draw inferences concerning the evolution of alx gene expression and function more deeply within Ambulacraria (echinoderms and hemichordates), it will be important to learn more about the single alx gene of hemichordates, including its pattern of expression, gene targets, and role in the formation of the small, calcareous skeletal elements of adult hemichordates (Cameron and Bishop, 2012) and to more precisely determine the embryological origins of the alx1-expressing cells of adult echinoderms, which are more relevant to the ancestral echinoderm condition than the more commonly studied larval forms.
In vertebrates, as noted above, the embryonic lineages of cells in the limbs and limb girdles that express alx1-related genes have not been mapped precisely, although many of these cells are presumably derived from the somatic layer of the lateral plate mesoderm, a major source of limb skeletal tissue. There is evidence that chondrocytes and osteoblasts of the limb are derived from a common, mesenchymal precursor cell and that the specialization of these two cell types depends upon regulatory functions of sox9 (a member of a small number of paralogous, soxE-family genes in vertebrates) and other sox genes in the chondrogenic lineage, and runx2 and osterix in the osteoblast lineage (Akiyama et al., 2005; Cervantes-Diaz et al., 2017; Lefebvre, 2019; Marín-Llera et al., 2019). Because alx-related genes have not been linked directly to the regulatory network that underlies limb skeletogenesis, and because Sox and Runx proteins are not currently known to be associated with skeleton formation in echinoderms, there is presently no obvious similarity between the GRN circuitry that controls skeletal development in the vertebrate limb and the echinoderm skeleton. As noted above, during limb girdle (scapula) development, alx1 is co-regulated by Emx2 and Pbx1, but the orthologous echinoderm genes have not been studied in detail.
Perhaps the best-characterized cell population in vertebrates that employs alx-related genes in biomineralization is the cranial neural crest. There is agreement that a definitive neural crest is found only in vertebrates, but the evolutionary history of this cell population, particularly the origins of the skeletogenic (cranial) compartment, remains a subject of much debate (Jandzik et al., 2015; Rothstein et al., 2018; Cheung et al., 2019; York and McCauley, 2020). Like the program of skeletogenesis in the limb, the formation of cranial neural crest-derived cartilage and bone is believed to progress through the specification of a common osteochondral progenitor, with important contributions by Sox9 and Runx2 in chondrocyte and osteoblast differentiation, respectively (Martik and Bronner, 2017; Dash and Trainor, 2020). The regulatory inputs into alx-family genes in the cranial neural crest are unknown, however, and only one direct target (fgf10) has been identified (Garg et al., 2017). Thus, the precise role of alx-related genes in the dynamic differentiation program of skeletogenic cranial neural crest cells and their connections to the underlying gene regulatory circuitry remain to be elucidated.
As noted above, in jawless vertebrates and cephalochordates, the expression patterns of alx-family genes are consistent with a possible function in the formation of the cartilaginous, pharyngeal skeletons of these animals. A detailed comparison of the expression patterns of alx-family genes in lampreys and jawed vertebrates has led to the hypothesis that an expansion of the domain of alx-expressing cells may have supported the expansion of the cranial skeleton during vertebrate evolution (Square et al., 2017). With the important caveat that expression data are sparse in these taxa and function studies are lacking, these observations are consistent with the hypothesis that alx-related genes were expressed (at least) in the anterior, pharyngeal mesoderm of ancient chordates, in cells that produced pharyngeal cartilage (Kaucka and Adameyko, 2019).
A hypothesis that emerges from these comparative studies is that a rudimentary, ancestral program of chondrogenesis, perhaps deployed in mesenchyme cells derived from embryonic mesoderm, was present in the ancestral deuterostome and provided a suitable gene regulatory system onto which biomineralization-promoting circuitry could be layered. We propose that in echinoderms, gene duplication was followed by the neo-functionalization of alx1; i.e., the acquisition of a new role in robustly mediating biomineralization, as reflected by the direct transcriptional inputs this transcription factor provides into a large fraction of biomineralization effector genes (Rafiq et al., 2014; Khor et al., 2019). A similar (and presumably independent) neo-functionalization may have occurred in vertebrates, but the transcriptional targets of vertebrate alx-family genes have not been characterized, and therefore, it is not known whether they include effectors of biomineralization. It should be noted that possible signals of evolutionary conservation between echinoderms and vertebrates in this context would likely be obscured by the well-documented, rapid evolution of many biomineralization-related proteins (Kawasaki et al., 2004; Livingston et al., 2006; Marin et al., 2016; McDougall and Degnan, 2018). Presumably, the independent duplication of alx-family genes in echinoderms and vertebrates initially involved the sharing and/or duplication of cis-regulatory elements among paralogs, as indicated by the overlapping patterns of expression of paralogous alx-family genes in both taxa. The recruitment of duplicated, alx-related genes to a biomineralization-related function would likely have been facilitated if the ancestral gene was already expressed in an embryonic tissue that produced an extensive extracellular matrix, a prerequisite for the assembly and growth of biomineral (Bolean et al., 2017; Murshed, 2018). In this regard, it will be valuable to characterize more completely in representative deuterostomes the cell lineages that express alx-family genes and to better reconstruct the evolutionary relationships among those cell lineages.
JK and CE contributed to the conception of the review and co-wrote the paper. Both the authors contributed to the article and approved the submitted version.
This work was supported by National Science Foundation (IOS-1354973 to C.A.E.).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adomako-Ankomah, A., and Ettensohn, C. A. (2013). Growth factor-mediated mesodermal cell guidance and skeletogenesis during sea urchin gastrulation. Development 140, 4214–4225. doi: 10.1242/dev.100479
Akiyama, H., Kim, J. -E., Nakashima, K., Balmes, G., Iwai, N., Deng, J. M., et al. (2005). Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U. S. A. 102, 14665–14670. doi: 10.1073/pnas.0504750102
Amore, G., and Davidson, E. H. (2006). cis-regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev. Biol. 293, 555–564. doi: 10.1016/j.ydbio.2006.02.024
Barsi, J. C., Tu, Q., Calestani, C., and Davidson, E. H. (2015). Genome-wide assessment of differential effector gene use in embryogenesis. Development 142, 3892–3901. doi: 10.1242/dev.127746
Bertola, D. R., Rodrigues, M. G., Quaio, C. R. D. C., Kim, C. A., and Passos-Bueno, M. R. (2013). Vertical transmission of a frontonasal phenotype caused by a novel ALX4 mutation. Am. J. Med. Genet. 161, 600–604. doi: 10.1002/ajmg.a.35762
Beverdam, A., Brouwer, A., Reijnen, M., Korving, J., and Meijlink, F. (2001). Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 128, 3975–3986.
Beverdam, A., and Meijlink, F. (2001). Expression patterns of group-I aristaless-related genes during craniofacial and limb development. Mech. Dev. 107, 163–167. doi: 10.1016/S0925-4773(01)00450-6
Bhattacherjee, V., Horn, K. H., Singh, S., Webb, C. L., Pisano, M. M., and Greene, R. M. (2009). CBP/p300 and associated transcriptional co-activators exhibit distinct expression patterns during murine craniofacial and neural tube development. Int. J. Dev. Biol. 53, 1097–1104. doi: 10.1387/ijdb.072489vb
Bluteau, G., Julien, M., Magne, D., Mallein-Gerin, F., Weiss, P., Daculsi, G., et al. (2007). VEGF and VEGF receptors are differentially expressed in chondrocytes. Bone 40, 568–576. doi: 10.1016/j.bone.2006.09.024
Bolean, M., Simão, A. M. S., Barioni, M. B., Favarin, B. Z., Sebinelli, H. G., Veschi, E. A., et al. (2017). Biophysical aspects of biomineralization. Biophys. Rev. 9, 747–760. doi: 10.1007/s12551-017-0315-1
Bothe, I., and Dietrich, S. (2006). The molecular setup of the avian head mesoderm and its implication for craniofacial myogenesis. Dev. Dyn. 235, 2845–2860. doi: 10.1002/dvdy.20903
Bothe, I., Tenin, G., Oseni, A., and Dietrich, S. (2011). Dynamic control of head mesoderm patterning. Development 138, 2807–2821. doi: 10.1242/dev.062737
Brazeau, M. D., Giles, S., Dearden, R. P., Jerve, A., Ariunchimeg, Y., Zorig, E., et al. (2020). Endochondral bone in an early Devonian ‘placoderm’ from Mongolia. Nat. Ecol. Evol. doi: 10.1038/s41559-020-01290-2 [Epub ahead of print]
Brenig, B., Schütz, E., Hardt, M., Scheuermann, P., and Freick, M. (2015). A 20 bp duplication in exon 2 of the aristaless-like homeobox 4 gene (ALX4) is the candidate causative mutation for tibial hemimelia syndrome in galloway cattle. PLoS One 10:e0129208. doi: 10.1371/journal.pone.0129208
Brew, K., and Nagase, H. (2010). The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim. Biophys. Acta 1803, 55–71. doi: 10.1016/j.bbamcr.2010.01.003
Cameron, C. B., and Bishop, C. D. (2012). Biomineral ultrastructure, elemental constitution and genomic analysis of biomineralization-related proteins in hemichordates. Proc. Biol. Sci. 279, 3041–3048. doi: 10.1098/rspb.2012.0335
Capellini, T. D., Vaccari, G., Ferretti, E., Fantini, S., He, M., Pellegrini, M., et al. (2010). Scapula development is governed by genetic interactions of Pbx1 with its family members and with Emx2 via their cooperative control of Alx1. Development 137, 2559–2569. doi: 10.1242/dev.048819
Carlevaro, M. F., Cermelli, S., Cancedda, R., and Cancedda, F. D. (2000). Vascular endothelial growth factor (VEGF) in cartilage neovascularization andchondrocyte differentiation: auto-paracrine role during endochondral boneformation. J. Cell Sci. 113, 59–69.
Cattell, M., Lai, S., Cerny, R., and Medeiros, D. M. (2011). A New Mechanistic Scenario for the origin and evolution of vertebrate cartilage. PLoS One 6:e22474. doi: 10.1371/journal.pone.0022474
Cervantes-Diaz, F., Contreras, P., and Marcellini, S. (2017). Evolutionary origin of endochondral ossification: the transdifferentiation hypothesis. Dev. Genes Evol. 227, 121–127. doi: 10.1007/s00427-016-0567-y
Chan, D. C., Laufer, E., Tabin, C., and Leder, P. (1995). Polydactylous limbs in strong’s Luxoid mice result from ectopic polarizing activity. Development 121, 1971–1978.
Cheung, M., Tai, A., Lu, P. J., and Cheah, K. S. (2019). Acquisition of multipotent and migratory neural crest cells in vertebrate evolution. Curr. Opin. Genet. Dev. 57, 84–90. doi: 10.1016/j.gde.2019.07.018
Cole, A. G. (2011). A review of diversity in the evolution and development of cartilage: the search for the origin of the chondrocyte. Eur. Cells Mater. 21, 122–129. doi: 10.22203/eCM.v021a10
Czarkwiani, A., Dylus, D. V., and Oliveri, P. (2013). Expression of skeletogenic genes during arm regeneration in the brittle star Amphiura filiformis. Gene Expr. Patterns 13, 464–472. doi: 10.1016/j.gep.2013.09.002
Dash, S., and Trainor, P. A. (2020). The development, patterning and evolution of neural crest cell differentiation into cartilage and bone. Bone 137:115409. doi: 10.1016/j.bone.2020.115409
Dee, C. T., Szymoniuk, C. R., Mills, P. E. D., and Takahashi, T. (2013). Defective neural crest migration revealed by a zebrafish model of Alx1-related frontonasal dysplasia. Hum. Mol. Genet. 22, 239–251. doi: 10.1093/hmg/dds423
Du, E. -X., Wang, X. -F., Yang, W. -C., Kaback, D., Yee, S. -P., Qin, C. -L., et al. (2015). Characterization of Fam20C expression in odontogenesis and osteogenesis using transgenic mice. Int. J. Oral Sci. 7, 89–94. doi: 10.1038/ijos.2014.67
Duan, X., Murata, Y., Liu, Y., Nicolae, C., Olsen, B. R., and Berendsen, A. D. (2015). Vegfa regulates perichondrial vascularity and osteoblast differentiation in bone development. Development 142, 1984–1991. doi: 10.1242/dev.117952
Duloquin, L., Lhomond, G., and Gache, C. (2007). Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 134, 2293–2302. doi: 10.1242/dev.005108
Dylus, D. V., Czarkwiani, A., Blowes, L. M., Elphick, M. R., and Oliveri, P. (2018). Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. Genome Biol. 19:26. doi: 10.1186/s13059-018-1402-8
Dylus, D. V., Czarkwiani, A., Stångberg, J., Ortega-Martinez, O., Dupont, S., and Oliveri, P. (2016). Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. EvoDevo 7:2. doi: 10.1186/s13227-015-0039-x
El-Ruby, M., El-Din Fayez, A., El-Dessouky, S. H., Aglan, M. S., Mazen, I., Ismail, N., et al. (2018). Identification of a novel homozygous ALX4 mutation in two unrelated patients with frontonasal dysplasia type-2. Am. J. Med. Genet. 176, 1190–1194. doi: 10.1002/ajmg.a.38655
Erkenbrack, E. M., and Davidson, E. H. (2015). Evolutionary rewiring of gene regulatory network linkages at divergence of the echinoid subclasses. Proc. Natl. Acad. Sci. U. S. A. 112, E4075–E4084. doi: 10.1073/pnas.1509845112
Erkenbrack, E. M., and Petsios, E. (2017). A conserved role for VEGF signaling in specification of homologous mesenchymal cell types positioned at spatially distinct developmental addresses in early development of sea urchins. J. Exp. Zool. B Mol. Dev. Evol. 328, 423–432. doi: 10.1002/jez.b.22743
Erkenbrack, E. M., and Thompson, J. R. (2019). Cell type phylogenetics informs the evolutionary origin of echinoderm larval skeletogenic cell identity. Commun. Biol. 2:160. doi: 10.1038/s42003-019-0417-3
Ettensohn, C. A. (2020). The gene regulatory control of sea urchin gastrulation. Mech. Dev. 162:103599. doi: 10.1016/j.mod.2020.103599
Ettensohn, C. A., and Adomako-Ankomah, A. (2019). The evolution of a new cell type was associated with competition for a signaling ligand. PLoS Biol. 17:e3000460. doi: 10.1371/journal.pbio.3000460
Ettensohn, C. A., Illies, M. R., Oliveri, P., and De Jong, D. L. (2003). Alx1, a member of the Cart1/Alx3/Alx4 subfamily of paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development 130, 2917–2928. doi: 10.1242/dev.00511
Ettensohn, C. A., Kitazawa, C., Cheers, M. S., Leonard, J. D., and Sharma, T. (2007). Gene regulatory networks and developmental plasticity in the early sea urchin embryo: alternative deployment of the skeletogenic gene regulatory network. Development 134, 3077–3087. doi: 10.1242/dev.009092
Fondon, J. W., and Garner, H. R. (2004). Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. U. S. A. 101, 18058–18063. doi: 10.1073/pnas.0408118101
Forlino, A., Piazza, R., Tiveron, C., Torre, S. D., Tatangelo, L., Bonafè, L., et al. (2005). A diastrophic dysplasia sulfate transporter (SLC26A2) mutant mouse: morphological and biochemical characterization of the resulting chondrodysplasia phenotype. Hum. Mol. Genet. 14, 859–871. doi: 10.1093/hmg/ddi079
Forsthoefel, P. F. (1963). The embryological development of the effects of strong’s luxoid gene in the mouse. J. Morphol. 113, 427–451. doi: 10.1002/jmor.1051130307
Gao, F., and Davidson, E. H. (2008). Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc. Natl. Acad. Sci. U. S. A. 105, 6091–6096. doi: 10.1073/pnas.0801201105
Gao, F., Thompson, J. R., Petsios, E., Erkenbrack, E., Moats, R. A., Bottjer, D. J., et al. (2015). Juvenile skeletogenesis in anciently diverged sea urchin clades. Dev. Biol. 400, 148–158. doi: 10.1016/j.ydbio.2015.01.017
Garg, A., Bansal, M., Gotoh, N., Feng, G. -S., Zhong, J., Wang, F., et al. (2017). Alx4 relays sequential FGF signaling to induce lacrimal gland morphogenesis. PLoS Genet. 13:e1007047. doi: 10.1371/journal.pgen.1007047
Gerber, H. -P., Vu, T. H., Ryan, A. M., Kowalski, J., Werb, Z., and Ferrara, N. (1999). VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med. 5, 623–628. doi: 10.1038/9467
Goh, K., and Holmes, D. (2017). Collagenous extracellular matrix biomaterials for tissue engineering: lessons from the common sea urchin tissue. Int. J. Mol. Sci. 18:901. doi: 10.3390/ijms18050901
Green, J. D., Tollemar, V., Dougherty, M., Yan, Z., Yin, L., Ye, J., et al. (2015). Multifaceted signaling regulators of chondrogenesis: implications in cartilage regeneration and tissue engineering. Genes Dis. 2, 307–327. doi: 10.1016/j.gendis.2015.09.003
Haila, S., Hästbacka, J., Böhling, T., Karjalainen–Lindsberg, M. -L., Kere, J., and Saarialho–Kere, U. (2001). SLC26A2 (diastrophic dysplasia sulfate transporter) is expressed in developing and mature cartilage but also in other tissues and cell types. J. Histochem. Cytochem. 49, 973–982. doi: 10.1177/002215540104900805
Hao, J., Narayanan, K., Muni, T., Ramachandran, A., and George, A. (2007). Dentin matrix protein 4, a novel secretory calcium-binding protein that modulates odontoblast differentiation. J. Biol. Chem. 282, 15357–15365. doi: 10.1074/jbc.M701547200
Hirasawa, T., and Kuratani, S. (2015). Evolution of the vertebrate skeleton: morphology, embryology, and development. Zool. Lett. 1:2. doi: 10.1186/s40851-014-0007-7
Hu, M. Y., Yan, J. -J., Petersen, I., Himmerkus, N., Bleich, M., and Stumpp, M. (2018). A SLC4 family bicarbonate transporter is critical for intracellular pH regulation and biomineralization in sea urchin embryos. eLife 7:e36600. doi: 10.7554/eLife.36600
Hudson, R., Taniguchi-Sidle, A., Boras, K., Wiggan, O., and Hamel, P. A. (1998). Alx-4, a transcriptional activator whose expression is restricted to sites of epithelial-mesenchymal interactions. Dev. Dyn. 213, 159–169. doi: 10.1002/(SICI)1097-0177(199810)213:2<159::AID-AJA1>3.0.CO;2-F
Hughes, I., Blasiole, B., Huss, D., Warchol, M. E., Rath, N. P., Hurle, B., et al. (2004). Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev. Biol. 276, 391–402. doi: 10.1016/j.ydbio.2004.09.001
Hughes, I., Saito, M., Schlesinger, P. H., and Ornitz, D. M. (2007). Otopetrin 1 activation by purinergic nucleotides regulates intracellular calcium. Proc. Natl. Acad. Sci. U. S. A. 104, 12023–12028. doi: 10.1073/pnas.0705182104
Hurle, B., Ignatova, E., Massironi, S. M., Mashimo, T., Rios, X., Thalmann, I., et al. (2003). Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum. Mol. Genet. 12, 777–789. doi: 10.1093/hmg/ddg087
Ingersoll, E. P., and Wilt, F. H. (1998). Matrix metalloproteinase inhibitors disrupt spicule formation by primary mesenchyme cells in the sea urchin embryo. Dev. Biol. 196, 95–106. doi: 10.1006/dbio.1998.8857
Jackson, G. C., Mittaz-Crettol, L., Taylor, J. A., Mortier, G. R., Spranger, J., Zabel, B., et al. (2012). Pseudoachondroplasia and multiple epiphyseal dysplasia: a 7-year comprehensive analysis of the known disease genes identify novel and recurrent mutations and provides an accurate assessment of their relative contribution. Hum. Mutat. 33, 144–157. doi: 10.1002/humu.21611
Jacobsen, K. A., Al-Aql, Z. S., Wan, C., Fitch, J. L., Stapleton, S. N., Mason, Z. D., et al. (2008). Bone formation during distraction osteogenesis is dependent on both VEGFR1 and VEGFR2 signaling. J. Bone Miner. Res. 23, 596–609. doi: 10.1359/jbmr.080103
Jacome-Galarza, C. E., Percin, G. I., Muller, J. T., Mass, E., Lazarov, T., Eitler, J., et al. (2019). Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature 568, 541–545. doi: 10.1038/s41586-019-1105-7
Jandzik, D., Garnett, A. T., Square, T. A., Cattell, M. V., Yu, J. -K., and Medeiros, D. M. (2015). Evolution of the new vertebrate head by co-option of an ancient chordate skeletal tissue. Nature 518, 534–537. doi: 10.1038/nature14000
Jin, E. -J., Choi, Y. -A., Kyun Park, E., Bang, O. -S., and Kang, S. -S. (2007). MMP-2 functions as a negative regulator of chondrogenic cell condensation via down-regulation of the FAK-integrin beta1 interaction. Dev. Biol. 308, 474–484. doi: 10.1016/j.ydbio.2007.06.003
Kaucka, M., and Adameyko, I. (2019). Evolution and development of the cartilaginous skull: from a lancelet towards a human face. Semin. Cell Dev. Biol. 91, 2–12. doi: 10.1016/j.semcdb.2017.12.007
Kawasaki, K., Suzuki, T., and Weiss, K. M. (2004). Genetic basis for the evolution of vertebrate mineralized tissue. Proc. Natl. Acad. Sci. U. S. A. 101, 11356–11361. doi: 10.1073/pnas.0404279101
Kayserili, H., Uz, E., Niessen, C., Vargel, I., Alanay, Y., Tuncbilek, G., et al. (2009). ALX4 dysfunction disrupts craniofacial and epidermal development. Hum. Mol. Genet. 18, 4357–4366. doi: 10.1093/hmg/ddp391
Keating, J. N., Marquart, C. L., Marone, F., and Donoghue, P. C. J. (2018). The nature of aspidin and the evolutionary origin of bone. Nat. Ecol. Evol. 2, 1501–1506. doi: 10.1038/s41559-018-0624-1
Khor, J. M., and Ettensohn, C. A. (2017). Functional divergence of paralogous transcription factors supported the evolution of biomineralization in echinoderms. eLife 6:e32728. doi: 10.7554/eLife.32728
Khor, J. M., Guerrero-Santoro, J., and Ettensohn, C. A. (2019). Genome-wide identification of binding sites and gene targets of Alx1, a pivotal regulator of echinoderm skeletogenesis. Development 146:dev180653. doi: 10.1242/dev.180653
Killian, C. E., Croker, L., and Wilt, F. H. (2010). SpSM30 gene family expression patterns in embryonic and adult biomineralized tissues of the sea urchin, Strongylocentrotus purpuratus. Gene Expr. Patterns 10, 135–139. doi: 10.1016/j.gep.2010.01.002
Kim, E., Hyrc, K. L., Speck, J., Lundberg, Y. W., Salles, F. T., Kachar, B., et al. (2010). Regulation of cellular calcium in vestibular supporting cells by otopetrin 1. J. Neurophysiol. 104, 3439–3450. doi: 10.1152/jn.00525.2010
Knoll, A. H. (2003). Biomineralization and evolutionary history. Rev. Mineral. Geochem. 54, 329–356. doi: 10.2113/0540329
Koga, H., Fujitani, H., Morino, Y., Miyamoto, N., Tsuchimoto, J., Shibata, T. F., et al. (2016). Experimental approach reveals the role of alx1 in the evolution of the echinoderm larval skeleton. PLoS One 11:e0149067. doi: 10.1371/journal.pone.0149067
Kuijper, S., Beverdam, A., Kroor, C., Brouwer, A., Candille, S., Barsh, G., et al. (2005a). Genetics of shoulder girdle formation: roles of Tbx15 and aristaless-like genes. Development 132, 1601–1610. doi: 10.1242/dev.01735
Kuijper, S., Feitsma, H., Sheth, R., Korving, J., Reijnen, M., and Meijlink, F. (2005b). Function and regulation of Alx4 in limb development: complex genetic interactions with Gli3 and Shh. Dev. Biol. 285, 533–544. doi: 10.1016/j.ydbio.2005.06.017
Kuratani, S., Oisi, Y., and Ota, K. G. (2016). Evolution of the vertebrate cranium: viewed from hagfish developmental studies. Zool. Sci. 33, 229–238. doi: 10.2108/zs150187
Lamichhaney, S., Berglund, J., Almén, M. S., Maqbool, K., Grabherr, M., Martinez-Barrio, A., et al. (2015). Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518, 371–375. doi: 10.1038/nature14181
Le Douarin, N. M. (2012). Piecing together the vertebrate skull. Development 139, 4293–4296. doi: 10.1242/dev.085191
Le Roy, N., Jackson, D. J., Marie, B., Ramos-Silva, P., and Marin, F. (2014). The evolution of metazoan α-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front. Zool. 11:75. doi: 10.1186/s12983-014-0075-8
Lefebvre, V. (2019). Roles and regulation of SOX transcription factors in skeletogenesis. Curr. Top. Dev. Biol. 133, 171–193. doi: 10.1016/bs.ctdb.2019.01.007
Li, Y., Omori, A., Flores, R. L., Satterfield, S., Nguyen, C., Ota, T., et al. (2020). Genomic insights of body plan transitions from bilateral to pentameral symmetry in echinoderms. Commun. Biol. 3:371. doi: 10.1038/s42003-020-1091-1
Liang, T., Gao, W., Zhu, L., Ren, J., Yao, H., Wang, K., et al. (2019). TIMP-1 inhibits proliferation and osteogenic differentiation of hBMSCs through Wnt/β-catenin signaling. Biosci. Rep. 39:BSR20181290. doi: 10.1042/BSR20181290
Liao, H. -M., Fang, J. -S., Chen, Y. -J., Wu, K. -L., Lee, K. -F., and Chen, C. -H. (2011). Clinical and molecular characterization of a transmitted reciprocal translocation t(1;12)(p32.1;q21.3) in a family co-segregating with mental retardation, language delay, and microcephaly. BMC Med. Genet. 12:70. doi: 10.1186/1471-2350-12-70
Liu, P., Ma, S., Zhang, H., Liu, C., Lu, Y., Chen, L., et al. (2017). Specific ablation of mouse Fam20C in cells expressing type I collagen leads to skeletal defects and hypophosphatemia. Sci. Rep. 7:3590. doi: 10.1038/s41598-017-03960-x
Livingston, B. T., Killian, C. E., Wilt, F., Cameron, A., Landrum, M. J., Ermolaeva, O., et al. (2006). A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 300, 335–348. doi: 10.1016/j.ydbio.2006.07.047
Lyons, L. A., Erdman, C. A., Grahn, R. A., Hamilton, M. J., Carter, M. J., Helps, C. R., et al. (2016). Aristaless-like homeobox protein 1 (ALX1) variant associated with craniofacial structure and frontonasal dysplasia in Burmese cats. Dev. Biol. 409, 451–458. doi: 10.1016/j.ydbio.2015.11.015
Mackie, E. J., Ahmed, Y. A., Tatarczuch, L., Chen, K. -S., and Mirams, M. (2008). Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int. J. Biochem. Cell Biol. 40, 46–62. doi: 10.1016/j.biocel.2007.06.009
Marin, F., Bundeleva, I., Takeuchi, T., Immel, F., and Medakovic, D. (2016). Organic matrices in metazoan calcium carbonate skeletons: composition, functions, evolution. J. Struct. Biol. 196, 98–106. doi: 10.1016/j.jsb.2016.04.006
Marín-Llera, J. C., Garciadiego-Cázares, D., and Chimal-Monroy, J. (2019). Understanding the cellular and molecular mechanisms that control early cell fate decisions during appendicular skeletogenesis. Front. Genet. 10:977. doi: 10.3389/fgene.2019.00977
Martik, M. L., and Bronner, M. E. (2017). Regulatory logic underlying diversification of the neural crest. Trends Genet. 33, 715–727. doi: 10.1016/j.tig.2017.07.015
Matsubara, H., Saito, D., Abe, G., Yokoyama, H., Suzuki, T., and Tamura, K. (2017). Upstream regulation for initiation of restricted Shh expression in the chick limb bud. Dev. Dyn. 246, 417–430. doi: 10.1002/dvdy.24493
Mavrogiannis, L. A., Taylor, I. B., Davies, S. J., Ramos, F. J., Olivares, J. L., and Wilkie, A. O. M. (2006). Enlarged parietal foramina caused by mutations in the homeobox genes ALX4 and MSX2: from genotype to phenotype. Eur. J. Hum. Genet. 14, 151–158. doi: 10.1038/sj.ejhg.5201526
McCauley, B. S., Wright, E. P., Exner, C., Kitazawa, C., and Hinman, V. F. (2012). Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. EvoDevo 3:17. doi: 10.1186/2041-9139-3-17
McClay, D. R., Warner, J., Martik, M., Miranda, E., and Slota, L. (2020). Gastrulation in the sea urchin. Curr. Top. Dev. Biol. 136, 195–218. doi: 10.1016/bs.ctdb.2019.08.004
McDougall, C., and Degnan, B. M. (2018). The evolution of mollusc shells. Wiley Interdiscip. Rev. Dev. Biol. 7:e313. doi: 10.1002/wdev.313
McGonnell, I. M., Graham, A., Richardson, J., Fish, J. L., Depew, M. J., Dee, C. T., et al. (2011). Evolution of the Alx homeobox gene family: parallel retention and independent loss of the vertebrate Alx3 gene. Evol. Dev. 13, 343–351. doi: 10.1111/j.1525-142X.2011.00489.x
McIntyre, D. C., Lyons, D. C., Martik, M., and McClay, D. R. (2014). Branching out: origins of the sea urchin larval skeleton in development and evolution. Genesis 52, 173–185. doi: 10.1002/dvg.22756
Meulemans, D., and Bronner-Fraser, M. (2007). Insights from amphioxus into the evolution of vertebrate cartilage. PLoS One 2:e787. doi: 10.1371/journal.pone.0000787
Morgulis, M., Gildor, T., Roopin, M., Sher, N., Malik, A., Lalzar, M., et al. (2019). Possible cooption of a VEGF-driven tubulogenesis program for biomineralization in echinoderms. Proc. Natl. Acad. Sci. U. S. A. 116, 12353–12362. doi: 10.1073/pnas.1902126116
Morino, Y., Koga, H., Tachibana, K., Shoguchi, E., Kiyomoto, M., and Wada, H. (2012). Heterochronic activation of VEGF signaling and the evolution of the skeleton in echinoderm pluteus larvae. Evol. Dev. 14, 428–436. doi: 10.1111/j.1525-142X.2012.00563.x
Murdock, D. J. E., and Donoghue, P. C. J. (2011). Evolutionary origins of animal skeletal biomineralization. Cells Tissues Organs 194, 98–102. doi: 10.1159/000324245
Murshed, M. (2018). Mechanism of bone mineralization. Cold Spring Harb. Perspect. Med. 8:a031229. doi: 10.1101/cshperspect.a031229
Noden, D. M., and Trainor, P. A. (2005). Relations and interactions between cranial mesoderm and neural crest populations: neural crest populations. J. Anat. 207, 575–601. doi: 10.1111/j.1469-7580.2005.00473.x
Oliveri, P., Tu, Q., and Davidson, E. H. (2008). Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. U. S. A. 105, 5955–5962. doi: 10.1073/pnas.0711220105
Percival, C. J., and Richtsmeier, J. T. (2013). Angiogenesis and intramembranous osteogenesis. Dev. Dyn. 242, 909–922. doi: 10.1002/dvdy.23992
Peterson, K. J., and Eernisse, D. J. (2016). The phylogeny, evolutionary developmental biology, and paleobiology of the deuterostomia: 25 years of new techniques, new discoveries, and new ideas. Org. Divers. Evol. 16, 401–418. doi: 10.1007/s13127-016-0270-x
Piacentino, M. L., Zuch, D. T., Fishman, J., Rose, S., Speranza, E. E., Li, C., et al. (2016). RNA-Seq identifies SPGs as a ventral skeletal patterning cue in sea urchins. Development 143, 703–714. doi: 10.1242/dev.129312
Poulet, B., Liu, K., Plumb, D., Vo, P., Shah, M., Staines, K., et al. (2016). Overexpression of TIMP-3 in chondrocytes produces transient reduction in growth plate length but permanently reduces adult bone quality and quantity. PLoS One 11:e0167971. doi: 10.1371/journal.pone.0167971
Qu, S., Li, L., and Wisdom, R. (1997a). Alx-4: cDNA cloning and characterization of a novel paired-type homeodomain protein. Gene 203, 217–223. doi: 10.1016/S0378-1119(97)00497-6
Qu, S., Niswender, K. D., Ji, Q., Meer, R.van der, Keeney, D., and Magnuson, M. A. (1997b). Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development 124, 3999–4008.
Rafaelsen, S. H., Raeder, H., Fagerheim, A. K., Knappskog, P., Carpenter, T. O., Johansson, S., et al. (2013). Exome sequencing reveals FAM20c mutations associated with fibroblast growth factor 23-related hypophosphatemia, dental anomalies, and ectopic calcification. J. Bone Miner. Res. 28, 1378–1385. doi: 10.1002/jbmr.1850
Rafiq, K., Cheers, M. S., and Ettensohn, C. A. (2012). The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development 139, 579–590. doi: 10.1242/dev.073049
Rafiq, K., Shashikant, T., McManus, C. J., and Ettensohn, C. A. (2014). Genome-wide analysis of the skeletogenic gene regulatory network of sea urchins. Development 141, 950–961. doi: 10.1242/dev.112763
Rice, R., Rice, D. P. C., Olsen, B. R., and Thesleff, I. (2003). Progression of calvarial bone development requires Foxc1 regulation of Msx2 and Alx4. Dev. Biol. 262, 75–87. doi: 10.1016/S0012-1606(03)00355-5
Richardson, W., Kitajima, T., Wilt, F., and Benson, S. (1989). Expression of an embryonic spicule matrix gene in calcified tissues of adult sea urchins. Dev. Biol. 132, 266–269. doi: 10.1016/0012-1606(89)90222-4
Roe, J. L., Park, H. R., Strittmatter, W. J., and Lennarz, W. J. (1989). Inhibitors of metalloendoproteases block spiculogenesis in sea urchin primary mesenchyme cells. Exp. Cell Res. 181, 542–550. doi: 10.1016/0014-4827(89)90110-9
Rose, B. J., and Kooyman, D. L. (2016). A tale of two joints: the role of matrix metalloproteases in cartilage biology. Dis. Markers 2016:4895050. doi: 10.1155/2016/4895050
Rothstein, M., Bhattacharya, D., and Simoes-Costa, M. (2018). The molecular basis of neural crest axial identity. Dev. Biol. 444, S170–S180. doi: 10.1016/j.ydbio.2018.07.026
Rychel, A. L., Smith, S. E., Shimamoto, H. T., and Swalla, B. J. (2006). Evolution and development of the chordates: collagen and pharyngeal cartilage. Mol. Biol. Evol. 23, 541–549. doi: 10.1093/molbev/msj055
Rychel, A. L., and Swalla, B. J. (2007). Development and evolution of chordate cartilage. J. Exp. Zool. B Mol. Dev. Evol. 308, 325–335. doi: 10.1002/jez.b.21157
Santagati, F., and Rijli, F. M. (2003). Cranial neural crest and the building of the vertebrate head. Nat. Rev. Neurosci. 4, 806–818. doi: 10.1038/nrn1221
Saotome, K., Teng, B., Tsui, C. C., Lee, W. -H., Tu, Y. -H., Kaplan, J. P., et al. (2019). Structures of the otopetrin proton channels Otop1 and Otop3. Nat. Struct. Mol. Biol. 26, 518–525. doi: 10.1038/s41594-019-0235-9
Seidahmed, M. Z., Alazami, A. M., Abdelbasit, O. B., Al Hussein, K., Miqdad, A. M., Abu-Sa’da, O., et al. (2015). Report of a case of Raine syndrome and literature review. Am. J. Med. Genet. A 167, 2394–2398. doi: 10.1002/ajmg.a.37159
Sharma, T., and Ettensohn, C. A. (2011). Regulative deployment of the skeletogenic gene regulatory network during sea urchin development. Development 138, 2581–2590. doi: 10.1242/dev.065193
Shashikant, T., Khor, J. M., and Ettensohn, C. A. (2018). From genome to anatomy: the architecture and evolution of the skeletogenic gene regulatory network of sea urchins and other echinoderms. Genesis 56:e23253. doi: 10.1002/dvg.23253
Shubin, N., Tabin, C., and Carroll, S. (2009). Deep homology and the origins of evolutionary novelty. Nature 457, 818–823. doi: 10.1038/nature07891
Simoes-Costa, M., and Bronner, M. E. (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257. doi: 10.1242/dev.105445
Simpson, M. A., Hsu, R., Keir, L. S., Hao, J., Sivapalan, G., Ernst, L. M., et al. (2007). Mutations in FAM20C are associated with lethal osteosclerotic bone dysplasia (Raine syndrome), highlighting a crucial molecule in bone development. Am. J. Hum. Genet. 81, 906–912. doi: 10.1086/522240
Sollner, C., Schwarz, H., Geisler, R., and Nicolson, T. (2004). Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev. Genes Evol. 214, 582–590. doi: 10.1007/s00427-004-0440-2
Square, T., Jandzik, D., Cattell, M., Coe, A., Doherty, J., and Medeiros, D. M. (2015). A gene expression map of the larval Xenopus laevis head reveals developmental changes underlying the evolution of new skeletal elements. Dev. Biol. 397, 293–304. doi: 10.1016/j.ydbio.2014.10.016
Square, T., Jandzik, D., Romášek, M., Cerny, R., and Medeiros, D. M. (2017). The origin and diversification of the developmental mechanisms that pattern the vertebrate head skeleton. Dev. Biol. 427, 219–229. doi: 10.1016/j.ydbio.2016.11.014
Superti-Furga, A., Rossi, A., Steinmann, B., and Gitzelmann, R. (1996). A chondrodysplasia family produced by mutations in the diastrophic dysplasia sulfate transporter gene: genotype/phenotype correlations. Am. J. Med. Genet. 63, 144–147. doi: 10.1002/(SICI)1096-8628(19960503)63:1<144::AID-AJMG25>3.0.CO;2-N
Suzuki, H. R., Reiter, R. S., D’Alessio, M., Di Liberto, M., Ramirez, F., Exposito, J. -Y., et al. (1997). Comparative analysis of fibrillar and basement membrane collagen expression in embryos of the sea urchin, Strongylocentrotus purpuratus. Zool. Sci. 14, 449–454. doi: 10.2108/zsj.14.449
Tagliabracci, V. S., Engel, J. L., Wen, J., Wiley, S. E., Worby, C. A., Kinch, L. N., et al. (2012). Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 336, 1150–1153. doi: 10.1126/science.1217817
Takahashi, M., Tamura, K., Buscher, D., Masuya, H., Yonei-Tamura, S., Matsumoto, K., et al. (1998). The role of Alx-4 in the establishment of anteroposterior polarity during vertebrate limb development. Development 125, 4417–4425.
Takeyari, S., Yamamoto, T., Kinoshita, Y., Fukumoto, S., Glorieux, F. H., Michigami, T., et al. (2014). Hypophosphatemic osteomalacia and bone sclerosis caused by a novel homozygous mutation of the FAM20C gene in an elderly man with a mild variant of Raine syndrome. Bone 67, 56–62. doi: 10.1016/j.bone.2014.06.026
Tarazona, O. A., Slota, L. A., Lopez, D. H., Zhang, G., and Cohn, M. J. (2016). The genetic program for cartilage development has deep homology within Bilateria. Nature 533, 86–89. doi: 10.1038/nature17398
ten Berge, D., Brouwer, A., El Bahi, S., Guénet, J. -L., Robert, B., and Meijlink, F. (1998). Mouse Alx3: an aristaless-like homeobox gene expressed during embryogenesis in ectomesenchyme and lateral plate mesoderm. Dev. Biol. 199, 11–25. doi: 10.1006/dbio.1998.8921
Tuckermann, J. P., Pittois, K., Partridge, N. C., Merregaert, J., and Angel, P. (2000). Collagenase-3 (MMP-13) and integral membrane protein 2a (Itm2a) are marker genes of chondrogenic/osteoblastic cells in bone formation: sequential temporal, and spatial expression of Itm2a, alkaline phosphatase, MMP-13, and osteocalcin in the mouse. J. Bone Miner. Res. 15, 1257–1265. doi: 10.1359/jbmr.2000.15.7.1257
Twigg, S. R. F., Versnel, S. L., Nürnberg, G., Lees, M. M., Bhat, M., Hammond, P., et al. (2009). Frontorhiny, a distinctive presentation of frontonasal dysplasia caused by recessive mutations in the ALX3 homeobox gene. Am. J. Hum. Genet. 84, 698–705. doi: 10.1016/j.ajhg.2009.04.009
Ullah, A., Umair, M., E-Kalsoom, U., Shahzad, S., Basit, S., and Ahmad, W. (2018). Exome sequencing revealed a novel nonsense variant in ALX3 gene underlying frontorhiny. J. Hum. Genet. 63, 97–100. doi: 10.1038/s10038-017-0358-y
Uz, E., Alanay, Y., Aktas, D., Vargel, I., Gucer, S., Tuncbilek, G., et al. (2010). Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am. J. Hum. Genet. 86, 789–796. doi: 10.1016/j.ajhg.2010.04.002
Valente, M., Valente, K. D., Sugayama, S. S. M., and Kim, C. A. (2004). Malformation of cortical and vascular development in one family with parietal foramina determined by an ALX4 homeobox gene mutation. AJNR Am. J. Neuroradiol. 25, 1836–1839.
Veis, A. (2011). Organic matrix-related mineralization of sea urchin spicules, spines, test and teeth. Front. Biosci. 16, 2540–2560. doi: 10.2741/3871
Vogel, P., Hansen, G. M., Read, R. W., Vance, R. B., Thiel, M., Liu, J., et al. (2012). Amelogenesis imperfecta and other biomineralization defects in Fam20a and Fam20c null mice. Vet. Pathol. 49, 998–1017. doi: 10.1177/0300985812453177
Wang, X., Hao, J., Xie, Y., Sun, Y., Hernandez, B., Yamoah, A. K., et al. (2010a). Expression of FAM20C in the osteogenesis and odontogenesis of mouse. J. Histochem. Cytochem. 58, 957–967. doi: 10.1369/jhc.2010.956565
Wang, H., Holland, P. W. H., and Takahashi, T. (2019). Gene profiling of head mesoderm in early zebrafish development: insights into the evolution of cranial mesoderm. EvoDevo 10:14. doi: 10.1186/s13227-019-0128-3
Wang, X., Wang, S., Li, C., Gao, T., Liu, Y., Rangiani, A., et al. (2012). Inactivation of a novel FGF23 regulator, FAM20C, leads to hypophosphatemic rickets in mice. PLoS Genet. 8:e1002708. doi: 10.1371/journal.pgen.1002708
Wang, X., Wiens, M., Schröder, H. C., Hu, S., Mugnaioli, E., Kolb, U., et al. (2010b). Morphology of sponge spicules: silicatein a structural protein for bio-silica formation. Adv. Eng. Mater. 12, B422–B437. doi: 10.1002/adem.200980042
Whittaker, C. A., Bergeron, K. -F., Whittle, J., Brandhorst, B. P., Burke, R. D., and Hynes, R. O. (2006). The echinoderm adhesome. Dev. Biol. 300, 252–266. doi: 10.1016/j.ydbio.2006.07.044
Wilt, F. H. (2002). Biomineralization of the spicules of sea urchin embryos. Zool. Sci. 19, 253–261. doi: 10.2108/zsj.19.253
Wood, T. W. P., and Nakamura, T. (2018). Problems in fish-to-tetrapod transition: genetic expeditions into old specimens. Front. Cell Dev. Biol. 6:70. doi: 10.3389/fcell.2018.00070
Wuyts, W., Cleiren, E., Homfray, T., Rasore-Quartino, A., Vanhoenacker, F., and Van Hul, W. (2000). The ALX4 homeobox gene is mutated in patients with ossification defects of the skull (foramina parietalia permagna, OMIM 168500). J. Med. Genet. 37, 916–920. doi: 10.1136/jmg.37.12.916
Yagnik, G., Ghuman, A., Kim, S., Stevens, C. G., Kimonis, V., Stoler, J., et al. (2012). ALX4 gain-of-function mutations in nonsyndromic craniosynostosis. Hum. Mutat. 33, 1626–1629. doi: 10.1002/humu.22166
Yajima, M., Umeda, R., Fuchikami, T., Kataoka, M., Sakamoto, N., Yamamoto, T., et al. (2010). Implication of HpEts in gene regulatory networks responsible for specification of sea urchin skeletogenic primary mesenchyme cells. Zool. Sci. 27, 638–646. doi: 10.2108/zsj.27.638
Yamasu, K., and Wilt, F. H. (1999). Functional organization of DNA elements regulating SM30alpha, a spicule matrix gene of sea urchin embryos. Dev. Growth Differ. 41, 81–91. doi: 10.1046/j.1440-169x.1999.00407.x
Yang, L., Tsang, K. Y., Tang, H. C., Chan, D., and Cheah, K. S. E. (2014). Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. U. S. A. 111, 12097–12102. doi: 10.1073/pnas.1302703111
York, J. R., and McCauley, D. W. (2020). The origin and evolution of vertebrate neural crest cells. Open Biol. 10:190285. doi: 10.1098/rsob.190285
Young, M., Selleri, L., and Capellini, T. D. (2019). Genetics of scapula and pelvis development: an evolutionary perspective. Curr. Top. Dev. Biol. 132, 311–349. doi: 10.1016/bs.ctdb.2018.12.007
Zhao, Q., Behringer, R. R., and de Crombrugghe, B. (1996). Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat. Genet. 13, 275–283. doi: 10.1038/ng0796-275
Zhao, G. -Q., Eberspaecher, H., Seldin, M. F., and de Crombrugghe, B. (1994). The gene for the homeodomain-containing protein Cart-1 is expressed in cells that have a chondrogenic potential during embryonic development. Mech. Dev. 8, 245–254. doi: 10.1016/0925-4773(94)90063-9
Keywords: Alx transcription factors, skeletogenesis, chondrogenesis, osteogenesis, deuterostome evolution, neural crest cell, biomineralization, calcification
Citation: Khor JM and Ettensohn CA (2020) Transcription Factors of the Alx Family: Evolutionarily Conserved Regulators of Deuterostome Skeletogenesis. Front. Genet. 11:569314. doi: 10.3389/fgene.2020.569314
Received: 03 June 2020; Accepted: 19 October 2020;
Published: 23 November 2020.
Edited by:
Sylvain Marcellini, University of Concepcion, ChileReviewed by:
Robert A. Haney, Ball State University, United StatesCopyright © 2020 Khor and Ettensohn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles A. Ettensohn, ZXR0ZW5zb2huQGNtdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.