
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 14 October 2020
Sec. Livestock Genomics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.557846
This article is part of the Research TopicEpigenetic Variation Influences on Livestock Production and Disease TraitsView all 24 articles
The bovine represents an important agriculture species and dairy breeds have experienced intense genetic selection over the last decades. The selection of breeders focused initially on milk production, but now includes feed efficiency, health, and fertility, although these traits show lower heritability. The non-genetic paternal and maternal effects on the next generation represent a new research topic that is part of epigenetics. The evidence for embryo programming from both parents is increasing. Both oocytes and spermatozoa carry methylation marks, histones modifications, small RNAs, and chromatin state variations. These epigenetic modifications may remain active in the early zygote and influence the embryonic period and beyond. In this paper, we review parental non-genetic effects retained in gametes on early embryo development of dairy cows, with emphasis on parental age (around puberty), the metabolism of the mother at the time of conception and in vitro culture (IVC) conditions. In our recent findings, transcriptomic signatures and DNA methylation patterns of blastocysts and gametes originating from various parental and IVC conditions revealed surprisingly similar results. Embryos from all these experiments displayed a metabolic signature that could be described as an “economy” mode where protein synthesis is reduced, mitochondria are considered less functional. In the absence of any significant phenotype, these results indicated a possible similar adaptation of the embryo to younger parental age, post-partum metabolic status and IVC conditions mediated by epigenetic factors.
Phenotype is the term that refers to the observable characteristics or traits of an organism. It is determined by the genotype of an organism, and by the growing environment, but precisely predicting the actual outcome for a particular individual is still a challenge (Burga and Lehner, 2012). The idea that phenotype changes caused by environmental alterations can be passed to next generations has existed for centuries (Roberts and Hay, 2019) but only in recent decades the advances in epigenetics knowledge gave us insights to the underlying mechanism of how environment affects the phenotype of an organism. In this review, we will discuss the parental epigenetic effects on programming in gametes and embryos, mainly focusing on dairy cattle.
The definition of epigenetics is evolving to accommodate our increasing knowledge of mechanisms that regulate gene expression. Presently, the widely accepted definition of epigenetics is “heritable changes in gene expression without altering the DNA sequence” with two main characteristics: inheritable and reprogrammable (Allis and Jenuwein, 2016). Inheritability of epigenetics mainly refers to the maintenance of epigenetic modifications in a short term, such as during mitosis of differentiated somatic cells, while reprogramming of epigenetics describes the ability to erase and rebuild modifications in a long term, such as across generations (Ji and Khurana Hershey, 2012). However, reprogrammable epigenetic information does not mean it will be completely removed at each generation, on the contrary, a small portion of epigenetic modifications can be transferred to next generation (intergenerational inheritance) or even beyond them (transgenerational inheritance) (Perez and Lehner, 2019).
The understanding of epigenetics is constantly modulated by the advance in our knowledge of evolution and development (Felsenfeld, 2014). There are four main epigenetic components: DNA methylation, histone modifications, non-coding RNAs, and chromatin state. In contrast to genetic information, epigenetic factors are vulnerable to be influenced by environmental changes. These alterations have been reported to be transgenerationally and/or intergenerationally inherited and affect the phenotype of the subsequent generations (Xavier et al., 2019).
DNA methylation is an epigenetic modification catalyzed by DNA methyltransferases (DNMTs) which transfer the methyl from the donor s-adenosyl methionine (SAM) to specific bases (Lyko, 2018). In mammals, DNA methylation predominantly occurs in the dinucleotide sequence 5′CpG3′, generating 5-methylcytosine (5mC) (Lyko, 2018). Most bulk genomic methylation status are steady over lifetime, with minor changes during specific cellular activities (Smith and Meissner, 2013). The two exceptions occur in embryo development, i.e., pre-implantation stage and primordial germ cell (PGC) stage, when most of the CpGs undergo global demethylation and remethylation (Wang et al., 2014). However, in mammals, a small portion of DNA methylation marks are protected from two waves of genome-wide DNA methylation reprogramming, which is not only indispensable for embryogenesis but can also inherit parental acquired traits to next generations (Smith et al., 2014; Wang et al., 2014; Jiang et al., 2018). The dynamic DNA methylome landscape carries important regulatory roles in gene expression, genomic imprinting, embryo development, and chromosome structure (Smith and Meissner, 2013).
Histones are basic proteins in eukaryotic cell nuclei that package DNA into nucleosomes. Different covalent post-translational modifications of histones bind to different genomic elements and have diverse functions in modulating the chromatin structure or recruiting other proteins to regulate gene expression. Modifications such as histone H3 lysine 4 di-/tri-methylation (H3K4me2/me3), H3K36me3, H3K79me3, and histone acetylation are linked to active expression, while H3K9me2/me3, H3K27me3 and histone deacetylation are associated with transcriptional repression (Zhang et al., 2015). During spermatogenesis and maturation, histones are gradually replaced with protamine to reduce the size of the sperm head. However, 1–15% of the histones are retained and are associated with DNA regions accessible soon after fertilization (Siklenka et al., 2015). In contrast, this replacement does not occur in oocytes, but the histones go through spatial and temporal post-translational modifications, including acetylation, methylation, phosphorylation, etc., which is critical for oocyte maturation (Gu et al., 2010) and maternal-to-zygotic transition after fertilization (Dahl et al., 2016; Zheng et al., 2016; Xia et al., 2019). A novel imprinting mechanism was reported to be dependent on maternal H3K27me3 rather than DNA methylation (Inoue et al., 2017).
Non-coding RNAs (ncRNAs) are transcripts that are not translated into proteins: such as ribosomal RNA (rRNA), ribozyme, transfer RNA (tRNA), and small nuclear RNA (snRNA). They are involved in the regulation of numerous bioactivities: such as cell proliferation, cell differentiation, cell apoptosis, cell metabolism, and chromosome remodeling. Moreover, ncRNA from both parental gametes are required for normal embryonic development (Yuan et al., 2016; Conine et al., 2018; McJunkin, 2018). Before the embryonic genome activation (EGA), early embryos mainly rely on transcript reservoir from maternal origin, thus the importance of maternal ncRNAs during early embryogenesis is taken for granted. Plenty of maternal ncRNAs were discovered to be involved in EGA, hence transfer maternal environmental influences to offspring (Perez and Lehner, 2019).
Sperm of germline-specific Dicer and Drosha conditional knockout mice have deficient miRNAs and/or endo-siRNAs profiles (Yuan et al., 2016). Although these sperms could still fertilize wild-type oocytes by intracytoplasmic sperm injection (ICSI), developmental potential of embryos produced were significantly impaired. Injecting wild-type sperm-derived total or small RNAs was able to rescue the developmental deficiency of these embryos (Yuan et al., 2016). Further studies identified that these transcripts especially small non-coding RNAs were gained during later maturation in epididymis (Conine et al., 2018). Embryos generated by ICSI using sperm from caput failed after implantation, while this deficiency could be rescued by microinjection of cauda-specific small RNAs (Conine et al., 2018).
Chromatin state is used to describe various conformations of chromatin structure which reflects its dynamic spatial changes during embryo reprogramming and other cellular activities (Zhou and Dean, 2015). Multiple epigenetic factors are involved in determining chromatin state, including histone modification, DNA methylation and non-coding RNAs (Gupta et al., 2010; Kundaje et al., 2015). The complex relationship between epigenetic modifications, chromatin state and transcriptional activities are still largely unknown, but our understanding of these associations is improving, based on the increasing experimental evidence (Ernst et al., 2011; Ernst and Kellis, 2012, 2017). Basically, genes with higher expression are most likely located at open chromatin regions which are generally modified by active epigenetic marks: such as H3K4me3 on proximal promoters, H3K27-ac on enhancers, and low DNA methylated promoters (Gaspar-Maia et al., 2011; Tian et al., 2011). Repressed expression genes are found in closed chromatin regions bound with inactive epigenetic marks including H3K37me3, hypermethylation at transcription start sites, and long non-coding RNA bound regions (Saxena and Carninci, 2011; Ando et al., 2019; Igolkina et al., 2019). During gametogenesis and embryogenesis, chromatin states undergo extensive remodeling to reach totipotency. However, the erasure is not complete, a complex pattern of 3D interactions between chromatin and transcription factors in both oocytes and sperm can still be transmitted to zygote according to ATAC-seq and ChIP-seq results (Jung et al., 2017, 2019). In bovine, retained histones were also identified by Mnase-seq in spermatozoa and seem required for spermatogenesis and fertilization (Sillaste et al., 2017). Overall, these retained accessible chromatin are indispensable for early embryogenesis and may also be potential candidates to transmit parental effects to next generations.
Throughout mammalian life cycle, individual experiences two waves of global epigenetic reprogramming, during preimplantation stages and germ cell development (Zeng and Chen, 2019). After fertilization, murine parental genomes were both observed to be actively (TETs-dependent) and passively (replication-dependent) demethylated during early embryogenesis, with exception of certain genomic loci, including imprinted genes (Guo F. et al., 2014; Wang et al., 2014). Recent single-cell chromatin overall omic-scale landscape sequencing (scCOOL-seq) of mouse preimplantation embryos further comprehensively described a heterogeneous but highly coordinated features of epigenetic reprogramming (Guo et al., 2017). Global DNA demethylation occurs within 12 h of fertilization, while distinct gene regions were resistant to reprogramming in parental genomes (Guo et al., 2017). Comparable chromatin accessibility was observed between paternal and maternal genomes from the late zygote to the blastocyst stage (Wu et al., 2016; Guo et al., 2017). Genomic regions that are resistant to global epigenetic reprogramming are potential factors to inherit parental environmental influences on offspring, but this requires to be further distinguished from de novo modifications after fertilization.
Epigenetic reprogramming during early embryo development is conserved between human and mice, while the kinetics of human embryos is relatively slower (Eckersley-Maslin et al., 2018). In human, major wave of global DNA demethylation was completed at 2-cell stage, and further reduced to 29% as the bottom at blastocyst stage in ICM (Guo H. et al., 2014). Besides imprinted genes, evolutionarily young transposable elements with more active transcription retain their DNA methylation status in early embryos (Guo H. et al., 2014). scCOOL-seq of human preimplantation embryos identified a complex, yet highly ordered epigenetic reprogramming process (Li et al., 2018). In contrary to mice, the paternal genomes in human early embryos are already more open than maternal genomes from the mid-zygote to the 4-cell stage (Li et al., 2018).
In bovine, the genome-wide demethylation is closely related to EGA since the major wave reduction of DNA methylation is completed by the 8-cell stage (Jiang et al., 2018). During that period, promoter methylation is negatively correlated with the expression levels of genes at preimplantation stages; gametes-specific differentially methylated regions (DMRs) are enriched in different regions and demethylated in different manners (Jiang et al., 2018). However, a small portion of DNA methylation will be maintained during the global reprogramming after fertilization, including imprinted genes, which could become a legacy between parental environment effects with offspring phenotype (Jiang et al., 2018). Moreover, dynamic landscape of accessible chromatin in bovine preimplantation embryos were revealed recently by ATAC-seq. Chromatin accessibility is dramatically increased in coordination with EGA and reached to peak in elongating embryos at day 14 (Ming et al., 2020). Combined with bovine transcriptomic and DNA methylation data, accessible promoters were related to genes with high expression and the accessibility is closely related to DNA methylation level and CpG density (Ming et al., 2020). Although the expression of histone modification enzymes was profiled during early embryo development (McGraw et al., 2003; Glanzner et al., 2018), global reprogramming of modified histones throughout bovine preimplantation stages is not known.
During the two waves, somatic epigenetic modifications in PGCs are erased and established as sex-specific patterns, including the genome-wide DNA methylation reprogramming and chromatin reorganization (Hackett et al., 2013). Less than 10% CpG sites were protected from demethylation in mouse PGC at E13.5, which is predominantly located in long terminal repeats (LTRs) at intergenic regions (Hackett et al., 2013; Wang et al., 2014). However, conflicting results were reported as to whether IAP elements (Intracisternal A-type particle) were resistant to DNA demethylation (Seisenberger et al., 2012; Hackett et al., 2013) or not (Wang et al., 2014). Along with the global DNA demethylation, chromatin structure undergoes actively remodeling by both extensive erasure of various histone modifications and exchange of histone variants (Hajkova et al., 2008). Although genome-wide distribution of retained histone modification is still not clear in mice, it has been identified that several silencing marks, such as H3K9me3 and H4K20me3, were retained on pericentric heterochromatin during PGC development (Magaraki et al., 2017). Diverse piRNAs were expressed in mice PGCs and involved in not only silencing of transposable elements but also translation regulations (Barreñada et al., 2020), which can be regarded as another transgenerational inheritance factors as reported in Caenorhabditis elegans (Ashe et al., 2012).
In human, the lowest DNA methylation was observed in the female PGCs of 10-week embryos, with an average of 6% methylation remaining (Guo et al., 2015). Similar to the observations in mice, the retained loci are particularly enriched in evolutionarily younger and more active repeat and transposable elements (Gkountela et al., 2015; Guo et al., 2015). Moreover, H3K9me3 can escape from global reprogramming to repress the constitutive heterochromatin (Gkountela et al., 2015; Guo et al., 2015), while the involvement of non-coding RNAs in human PGC development is still not clear.
Studies on PGC reprogramming were mainly focusing on model animals and human, although bovine PGCs were already identified and isolated by AP staining in embryos at E18-E39 in 1990s (Lavoir et al., 1994; Wrobel and Süß, 1998). Thus, it is urgently needed to characterize the landscapes of DNA methylation, histone modifications/chromatin states, ncRNAs, etc. during PGC development in bovine. This will enlighten our knowledge of the most extensively reprogramming process in large animals and point out the potential transgenerational inheritance factors which are able to escape from this global erasure.
Due to the vast amount of information conveyed from female gametes to zygotes and the exposure to the uterus environment during pregnancy, epigenetic influences of maternal origin were studied extensively. The effects of maternal nutrition status, age, stress, lifestyle, disease, and others were reported to be transmitted to next generations. However, research on paternal non-genetic effects has been long neglected compared to the numerous studies undertaken on the maternal side. Benefitting from studies to identify the molecules that sperm transfer during fertilization, paternal epigenetic influences are now gaining more and more attention. Here, we will firstly illustrate parental non-genetic effects focusing on mouse and human studies.
Due to the short lifespan, transgenerational studies of maternal effects in mice are quite common and informative. Pre-conceptional and gestational maternal obesity induced cardiac dysfunction and hypertension in offspring (Loche et al., 2018). Also, exposure to bisphenol A (BPA, an endocrine disruptor) induced metabolic defects transgenerationally up to the F3 offspring (Bansal et al., 2019). However, the phenotype was less severe with increasing generations, probably due to the diluting effects of epigenetic reprogramming during early embryo development (Bansal et al., 2019). Pre-conceptional maternal exposure to cyclophosphamide (an agent for breast cancer therapy) altered DNA methylation levels in F1 and F2 mouse oocytes, resulting in delayed growth in these two generations (Di Emidio et al., 2019).
In humans, the mother condition has a profound impact on offspring across generations through epigenetic modifications. Epidemiological studies demonstrated that F2 generations of mothers who experienced famine periods had a higher tendency toward metabolic disorders (Aiken et al., 2016), partially due to the alteration of methylation levels in genomic DNA and histones (Uchiyama et al., 2018; Zimmet et al., 2018). Maternal age is another factor that can influence the developmental outcomes of progeny. Offspring of younger mothers tended to take more time to get pregnant (Reynolds et al., 2020); while offspring of advanced-age mothers were more likely to have metabolism or neurodevelopmental disorders with reduced methylation levels of several specific CpG sites (Markunas et al., 2016). Hence, epigenetic modifications are involved in the massive non-genetic maternal influence on offspring.
Several comprehensive studies on paternal epigenetic effects on offspring were generated with the mouse model. The effects of metabolic disorders in male mice (Wei et al., 2014) and toxic exposure (Guerrero-Bosagna et al., 2012) can be transgenerationally inherited by disturbing DNA methylation levels in sperm. Disruption of histone methylation during spermatogenesis resulted in modified chromatin states in sperm and impaired development and survivability over two generations (Siklenka et al., 2015). Transfer RNA-derived small RNAs (tsRNAs), the major component of ncRNAs in mature spermatozoa apparently acquired during the transit in the epididymis (Chen et al., 2016b; Sharma et al., 2018), is believed to mediate paternal diet-induced metabolic disorders in offspring (Chen et al., 2016a; Sharma et al., 2016). These results clearly demonstrated the transgenerational inheritability of paternal epigenetic information in mice.
In human, the paternal contribution to an embryo is required for the embryo development and non-genetic components are involved in the transmission of paternal acquired traits to their children. Alteration of DNA methylation level, histone modifications, non-coding RNA expression and chromatin status were observed in the sperm of men who smokes, are obese, or experience mental stresses indicating an intergenerational inheritance through them (Soubry et al., 2016; Jenkins et al., 2017; Rowold et al., 2017; Dupont et al., 2019). Moreover, some of these aberrant changes were found in the sperm of their offspring and transgenerationally influenced the health of next generation (Soubry et al., 2013; Craig et al., 2017).
In bovine, prenatal maternal conditions were significantly correlated with the daughters’ fertility and milk production. For example, daughters of dam that calved early for the first time produced more first-lactation daily milk, had higher body condition score (BCS), but experienced difficulties conceiving (Banos et al., 2007). Meanwhile, gestating dams with higher BCS tended to give birth to calves with higher BCS, had lower return rates, but slightly lower daily milk yields (Banos et al., 2007). The prenatal environment of grand-dam can also somewhat influence the milk production in subsequent daughters potentially in a transgenerationally manner by epigenetic mechanisms (Singh et al., 2012; Gudex et al., 2014). Additionally, the separation of heifers from their mother shortly after birth, which is a widely applied procedure in dairy cow management, will result in stress analogous to the maternal separation and unpredictable maternal stress (MSUS) model in rodents. Thus, this practice makes dairy cows a model to study the non-genetic inheritance of MSUS (Engmann, 2018).
As mentioned above, the maternal metabolic status can influence production and reproductive traits of subsequent generations through non-genetic pathways. However, there is a paucity of studies on the specific mechanisms underlying the effects of the maternal metabolism pre-conception or during gestation on offspring. Recently, embryos from dairy cows experiencing different levels of negative energy balance (NEB) were collected to study the influence of the maternal metabolic environment on the transcriptome and epigenome of early embryos by microarrays (Chaput and Sirard, 2019). Transcriptomic data highlighted that the most significantly affected pathways were metabolism-related: such as protein synthesis (EIF2 Signaling and eIF4, translation factors), mitochondrial function (oxidative phosphorylation and mitochondrial dysfunction), and metabolism (Sirtuin signaling and mTOR Signaling) (Chaput and Sirard, 2019). Gene expression levels do not provide any information about the amount of protein generated, or the extent of further modifications: such as phosphorylation, acylation, and/or methylation. Transcriptomic studies can be used to evaluate the immediate impact of environmental stresses and the embryonic responses (Cagnone and Sirard, 2016). Meanwhile, to study the long-term effects of stressors, studies on epigenetic modifications are required, especially for those regions that are affected according to the transcriptomic results. Using a DNA methylation microarray, 462 DMRs were identified between embryos from cows in high and low NEB, with many of them being located in gene regions, including introns, exons, proximal promoters, promoters, and distal promoters (Chaput and Sirard, 2019). Most of these genic regions, except exons, were more hypermethylated in embryos from cows experiencing severe NEB (Chaput and Sirard, 2019). Functional analysis of DMRs located in gene regions was consistent with transcriptomic results and pointed toward metabolic related pathways (AMPK signaling and mTOR signaling) which are significantly affected by maternal energy deficits (Chaput and Sirard, 2019). As a key switch for keep the energy balance, AMPK were also regulated by small non-coding RNAs to control cellular anabolic and catabolic processes in dairy cows (Mahmoudi et al., 2015). These changes demonstrated the metabolic adaptations of embryos to the maternal gestational environment by the regulation of mitochondrial functions, cell growth, and other protective pathways. These NEB-associated DMRs were retained at the time of the global reprogramming after fertilization, thus it is highly possible that they could affect the phenotype of offspring intergenerationally. The DNA methylation analysis of blood from eight calves produced from high and low BHB mothers resulted in 1675 DMRs (p < 0.05) indicating a post-natal legacy. This study highlighted how embryos interact with the maternal environment and the potential for intergenerationally inherited phenotype transmission through epigenetic modifications.
Multiple epigenetic factors conveyed by sperm are affected by the environmental or physical conditions of bulls. Not only maternal and fetal effects influence gestation length, a total of 66,318 DMRs in sperm were correlated with gestation length as well as days to first breeding after calving, somatic cell score, body type, milk production, and other traits (Fang et al., 2019). Reactome pathways analysis further validated that DMRs were mainly related to pregnancy, embryonic development, and lipid metabolism pathways (Fang et al., 2019). Moreover, some of these DMR were mapped to genes that are transcriptionally active during preimplantation stages, suggesting their potential role in early embryo development (Fang et al., 2019). Similarly, age-related DMRs were observed in bovine spermatozoa (Takeda et al., 2017, 2019; Lambert et al., 2018), and 57 of 2223 DMRs (2.56%) were retained in blastocysts (Lambert et al., 2018; Wu et al., 2020). Among the genes that are mapped with these DMRs, some of them were involved in spermatogenesis (FKBP6) and embryonic preimplantation development (AKT2). Chromatin condensation status can also be altered under heat-stress and further influence DNA methylation reprogramming of paternal pronuclei, which may be responsible for the reduced fertilization rates after IVF (Rahman et al., 2014). In vitro exposure to Chlorpyrifos, a pesticide, significantly affected bovine sperm DNA methylation patterns, resulted in reduced sperm motility and IVF rates, and increased chromatin structure abnormalities (Pallotta et al., 2019). Although large epidemiologic studies are missing, these changes in the epigenetic marks could potentially alter the phenotype of offspring.
Similar to cows, paternal metabolic status significantly influences semen quality. Enhanced pre-pubertal nutrition elevates the percentage of progressively motile and upregulates mitochondrial function in sperm of post-pubertal dairy bulls (Johnson et al., 2020). In contrast, low planes of nutrition of young Holstein-Friesian bulls resulted in the retarded onset of puberty (Byrne et al., 2018). Although epigenetic studies related to bull metabolic status is still missing, DNA methylation and histone modifications patterns were both reported to be associated with bull fertility (Verma et al., 2015; Kropp et al., 2017; Kutchy et al., 2018; Capra et al., 2019; Ugur et al., 2019). Functional annotation of these alterations indicate that they might be involved in spermatogenesis and embryo development (Verma et al., 2015; Ugur et al., 2019).
Another tool to study embryo programming is in vitro culture (IVC) in different types of environment. Assisted reproductive technologies (ARTs) have been widely used to either overcome reproductive difficulties (for human) or increase the genetic gain of elite sires (for cattle). In vitro culture, an indispensable ART procedure, allows zygotes to divide to a transferrable stage at around day 7 for bovine embryos. Each aspect of the IVC medium, including physicochemical, oxidative, and energetic conditions (Summers and Biggers, 2003), has profound effects on embryo development, and these effects could be maintained to adulthood or even subsequent generations. The physiochemical parameters increased osmolality (Etienne and Martin, 1979), decreased local pH (Dagilgan et al., 2015), and heat shock (Sakatani et al., 2008) were all shown to compromise embryo development. Additionally, specific levels of reactive oxygen species (ROS) are required for normal embryo development; however, it is inevitable that embryos will be exposed to higher concentrations of oxygen in vitro, which is detrimental as a result of increased H2O2 production, DNA fragmentation, and mitochondrial dysfunction-induced apoptosis (Van Soom et al., 2002; Harvey et al., 2004; Kitagawa et al., 2004). The presence of nutrients in concentrations that mimic in vivo conditions is fundamental to early embryogenesis as demonstrated by the deleterious effects on embryo development of high concentrations of glucose and lipids in the culture medium (Díaz-Ruiz et al., 2008). An excessive inflammatory response was observed in IVC blastocysts, and this could interfere with the embryo-maternal recognition process following transfer (Cagnone and Sirard, 2014). Based on the metabolic pathways affected by culture conditions, embryos can either enhance a Warburg-like effect to adapt to minor stresses or induce apoptosis under severe stress. As a result, even though blastocyst rates may not be significantly impacted in modified media, embryo loss rates may be higher compared to control conditions. Moreover, mitochondrial dysfunction is involved in the embryonic response to IVC stresses which may impact its main role as energy factory, as well as the production of acetyl-CoA and methyl groups associated with one-carbon metabolism (Steegers-Theunissen et al., 2013), which controls histone acetylation and DNA methylation. Thus, suboptimal IVC conditions do disturb the embryonic epigenome which could contribute to the sometime altered phenotype of offspring (Cagnone and Sirard, 2016).
Potential genetically elite sires can be identified a few days postnatally by genomic selection. In combination with ARTs, these approaches greatly reduce generation interval to increase the rate of genetic gain (Kasinathan et al., 2015). Additionally, there is increased demand for the use of juvenile calves or heifers for embryo production in combination with genomic selection and ARTs, which can further shorten generation interval (Landry et al., 2016). However, gametes from young donors are suboptimum compared to those from adults. Recent studies also indicated the potential parental age effects on early embryo development.
On the male side, semen quality and quantity are significantly correlated with the age of the bull (Takeda et al., 2017). The ontogeny of the male reproductive system is initiated during the fetal period and the onset of bull puberty is governed by complex neuroendocrine networks (Plant, 2015) which are responsible for the tight coupling between metabolic status and reproductive system development (Roa et al., 2010). The early transient rise in LH pulsatility marks the initiation of puberty (Evans et al., 1995), which is observed in bull calves between 10 and 20 weeks of age (Rawlings and Evans, 1995). Leydig cells proliferate rapidly with an elevated responsiveness to LH, resulting in increased testosterone concentration to initiate the differentiation of unmatured Sertoli cells and further spermatogenesis (Amann and Walker, 1983). The first unmatured spermatids are generated between 25 and 35 weeks of age, while mature spermatozoa are obvious in seminiferous tubules at 32–40 weeks of age (Abdel-Raouf, 1960; Macmillan and Hafs, 1968; Evans et al., 1996; Bagu et al., 2006). A bull is then considered to have reached puberty when the first ejaculation containing over 50 million spermatozoa with at least 10% progressively motile spermatozoa is observed at around 45 weeks of age (Wolf et al., 1965). Hence, it is expected that semen collected at prepubertal stages is often sub-optimal in terms of sperm concentration, motility, and IVF performance compared to semen collected from adult bulls (Takeda et al., 2017). Moreover, this sub-standard sperm quality is associated with lower body weight in sexually immature bulls (Devkota et al., 2008), indicating that the paternal age effects on reproductive outcome may resemble the restricted diet effects (Brito et al., 2007).
Several studies demonstrated that enhanced early-life nutrition of bull calves positively affects several key metabolic and reproductive hormones related to the hypothalamic-pituitary-gonadal (HPG) axis, as reviewed by Kenny and Byrne (2018), and successfully hastens puberty without interfering with post-pubertal semen quality (Dance et al., 2015, 2016). In the commercial environment (optimized for breeders) setting used for the bulls in our study, semen ejaculated at the age of 10 months performed similarly to semen from post-pubertal animals regarding sperm concentration, motility, and blastocyst rates; although the semen from younger animals contained fewer spermatozoa (Wu et al., 2020).
To study the epigenetic programming of sperm from young bulls, a new bovine specific platform, EmbryoGENE1, which can simultaneously evaluate the genome-wide epigenome and transcriptome of small samples, such as sperm, oocytes, and early embryos (Shojaei Saadi et al., 2014), was used to evaluate the influence of environmental and parental effects on embryo development (Salilew-Wondim et al., 2015; Desmet et al., 2016; Pagé-Larivière et al., 2016; Morin-Doré et al., 2017; Tremblay et al., 2018; Chaput and Sirard, 2019; Wu et al., 2020).
Different DNA methylation patterns were observed in sperm collected either from the same bulls at different pubertal stages (Lambert et al., 2018) or from different bulls of different ages (Takeda et al., 2017, 2019). Approximately 69% of the DMRs were located in genic regions, including one associated with the paternally imprinted gene MEST (mesoderm specific transcript) (Lambert et al., 2018). Methylation levels of most DMRs were higher with increasing age (Lambert et al., 2018; Takeda et al., 2019); interestingly, these levels changed rapidly especially at younger ages (Takeda et al., 2019). Network analysis of these DMRs revealed that sperm function related pathways, such as PKA signaling, sperm motility, calcium signaling, and protein G signaling pathways were significantly affected by paternal age (Lambert et al., 2018). Hence, although young bulls can produce functional semen, epigenetic factors transmitted by spermatozoa could potentially impact embryo or offspring development.
Epigenetic modifications are known to occur as paternal inter or transgenerational inheritance factors (Rando, 2016; Spadafora, 2017; Bošković and Rando, 2018). To further study paternal age effects on embryos, blastocysts were produced by IVF with spermatozoa from the same bulls at different pubertal periods (10, 12, and 16 months) and oocytes from several matched (the same cow for each individual bull) adult cows (Wu et al., 2020). Transcriptomic and epigenetic analysis were performed on four pairs, where the only difference between embryos was the age of the bull. The results revealed elevated mitochondrial dysfunction, suppressed oxidative phosphorylation, and reduced protein synthesis in blastocysts generated from younger bulls suggesting a low energy status in these blastocysts (Wu et al., 2020). Moreover, the affected metabolic and sperm function pathways observed in blastocysts were consistent with the sperm studies mentioned above (Lambert et al., 2018), suggesting paternal age effects on embryos mediated by epigenetic factors in sperm.
On the female side, the same selection pressure pushes the breeding industry to collect oocytes from dairy heifers to reduce generational interval and increase genetic gain. It has been reported that in vitro produced embryos can be obtained using oocytes from heifers as young as 2–4 months; however, few cumulus oocyte complexes (COCs) could be recovered from non-stimulated heifer ovaries (Majerus et al., 1999; Palma et al., 2001; Kauffold et al., 2005). Even though these COCs had similar performance in maturation, fertilization, and early cleavage rates after IVF compared to COCs from adult cows, reduced blastocyst yields and greater embryo loss following embryo transfer were observed, which may be a consequence of increased apoptosis in embryos from young donors (Zaraza et al., 2010). Ovarian stimulation is widely accepted for generating oocytes of high quality and for increasing blastocyst yields from adult cows (Nivet et al., 2012; Labrecque et al., 2013). More follicles were aspirated from young calves following stimulation, and higher numbers of mature oocytes and cleaved embryos were obtained (Landry et al., 2016). However, there was no difference in the total number of morula or viable embryos from heifers 5–18 months old, due to the significant lower morula and blastocyst rates in heifer groups which neutralized the larger number of oocytes recovered (Landry et al., 2016). Further analysis of granulosa cells collected from heifers demonstrated that cell differentiation, inflammation, and apoptosis pathways were inhibited indicating a suboptimal environment for oocytes in young donors (Landry et al., 2018).
To further investigate the maternal age effects on embryos, oocytes from the same heifers at different pubertal stages were collected and in vitro fertilized with spermatozoa from matched adult bulls (Morin-Doré et al., 2017). Transcriptomic analysis revealed that mitochondrial function was impacted in blastocysts from younger heifers, with inhibited mTOR, NRF2, and PPAR signaling (Morin-Doré et al., 2017). Thus, we can obtain more oocytes and comparable numbers of transferable embryos from young donors following ovarian stimulation; however, young maternal age impairs metabolic functions during early embryo development and may cause embryo loss at later stages or induce offspring health disorders. The identification of genes affected in blastocysts from younger females revealed several gene pathways similarly affected in embryos originating from younger males making the results even more convincing and suggesting that there might be an evolutionary conserved mechanism involved.
Genotype (G) × Environment (E) interaction is defined as genotype-specific phenotypic responses to different environments (Falconer, 1952). Taking G × E into consideration in dairy cow breeding reduces the error variance and hence increases the accuracy of genetic evaluation of sires and cows compared to classical approach, which considers the effects of genotype and environment only (Figure 1B) (Ron and Hillel, 1983). Two formations of G × E interactions can be taken to induce either unchanged ranking, i.e., a scaling effects, or reranking of sires across environments (Cromie et al., 1998). Plenty of across countries and within a country dairy cattle studies concluded that scale effects, which is due to the unequal scale of differences in sire proofs in the two environments, account majorly for the G × E interaction (McDaniel and Corley, 1967; Stanton et al., 1991; Boettcher et al., 2003). However, emerging evidence using genetic correlation proved the existence of re-ranking effects of G × E interactions, especially between the traits expressed in environments with large differences as reviewed by Hammami et al. (2009b) and Wakchaure et al. (2016). Genetic correlations < 0.80 were observed between milk yield across countries (Ojango and Pollott, 2002; Cerón-Muñoz et al., 2004b; Hammami et al., 2009a), between age at first calving across countries (Cerón-Muñoz et al., 2004a), between two feeding systems within country (Ramírez-Valverde et al., 2010), indicating the presence of genotype by environment interactions and at least some re-ranking of the animals. In this case, the breeders are required to optimize the breeding programs to accommodate the various environments.
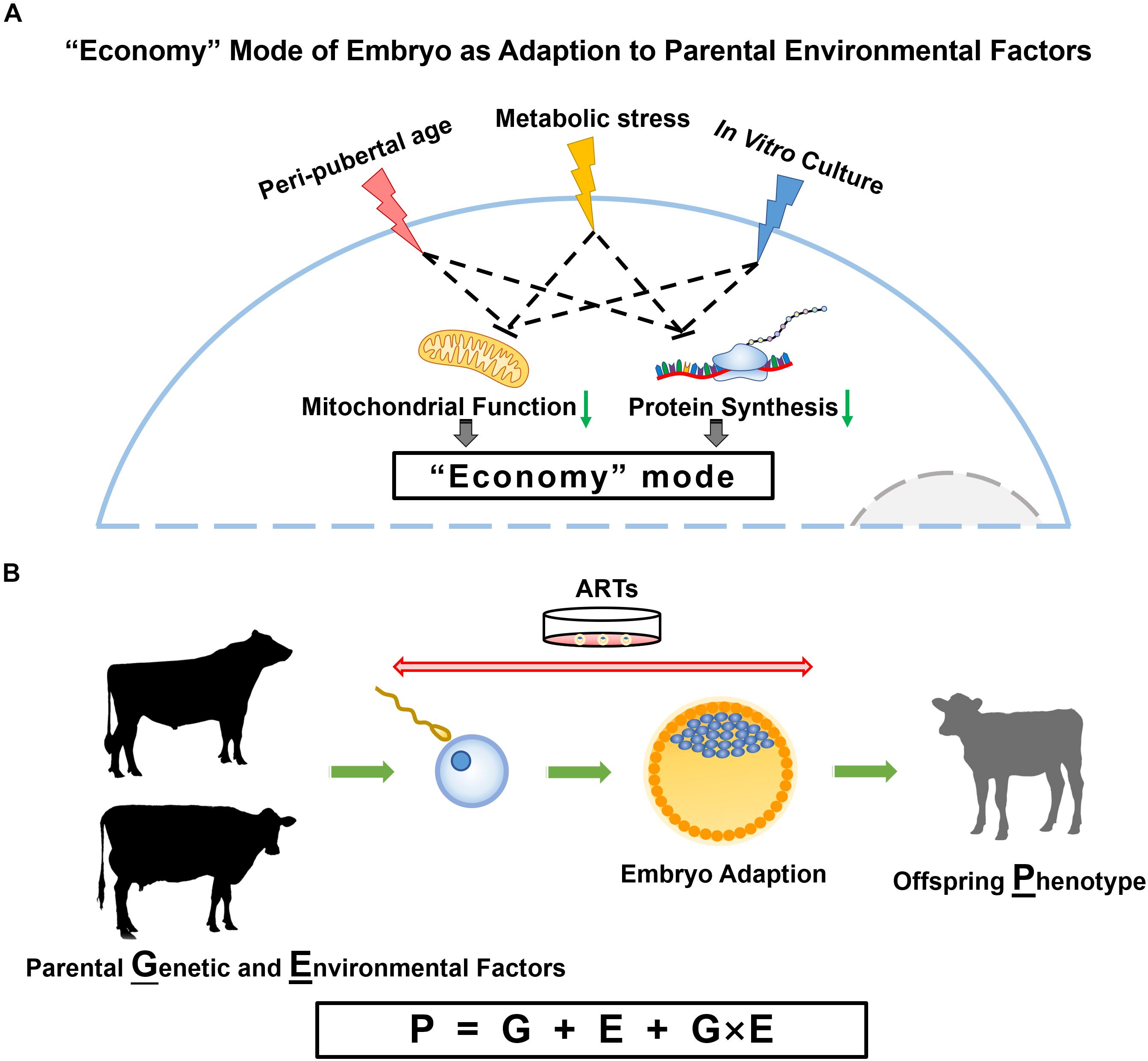
Figure 1. Parental influences on embryo and offspring. (A) Proposed model for embryo adaption of parental environmental factors. In this model, peri-pubertal age, metabolic status and IVC conditions result in embryos turning to an “economy” mode with slowed down cellular activities, especially the reduced mitochondrial function and protein synthesis. (B) The phenotype of offspring is determined by genotype, environment, and the interplay of genetic and environmental factors.
Genes may be expressed in different patterns under different environment, and this could be one of the molecular mechanisms of G × E interactions (Hammami et al., 2009b). Epigenetic factors are involved in changing gene expression under varied environments. For example, DNA methylation at promoter regions can affect the binding of transcription factors, small non-coding RNAs can also regulate the transcription and translation in different ways, histone modification and chromatin states can determine the accessibility of chromatin for gene expression. Epigenetic regulation of multiple traits of dairy cow under different environments have been studied broadly and deeply (Singh et al., 2010; Ibeagha-Awemu and Zhao, 2015; Thompson et al., 2020). However, direct study of involvement of epigenetic factors in G × E interactions is still missing. Nevertheless, genomic bias were presented in relation to gene with G × E interactions, as reviewed by Grishkevich and Yanai (2013) based on whole-genome approaches in model organism and concluded that gene having long promoters with high concentration of regulatory motifs showed high correlation with distant-acting loci. Thus, epigenetic variations in different environments are highly possible to be responsible for G × E interactions effects, while substantial and systematic studies are required.
Epigenetic information conveyed by gametes represent non-genetic factors that may explain why gametes of donors at different ages have similar reproductive performance, but result in different gene expression and DNA methylation patterns in embryos they produced. Counterintuitively, parental nutritional status, age, and IVC environment have similar consequences on embryo programming, i.e., alterations in metabolic pathways, especially mitochondrial signaling, which proves that cellular energy production is central in the response to environmental changes (Figure 1A). The most important task now becomes the analysis of post-natal phenotypes to identify the phenotypical consequences of all these epigenetic modifications in offspring. Fortunately, in sub-species like the dairy cow, data are accumulating on the genetic side (G) as more and more animals get genotyped, and on the phenotype (P) side using manual or electronic data generation at the farm. The combination of information from the farm environment (E) with genotype (G) and phenotype (P) information will allow the modeling of optimal environmental conditions for each animal or the optimal genotype for a defined environment (Figure 1B).
M-AS designed the manuscript. CW and M-AS wrote the manuscript. All authors read and approved the final manuscript.
This work was funded by the Natural Sciences and Engineering Research Council (NSERC grant no. 445230-12), Semex and DairyGen of Canada. CW received studentship support from China Scholarship Council-Université Laval (CSC-UL) joint scholarship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Sylvie Bilodeau-Goeseels for proofreading this article.
Abdel-Raouf, M. (1960). The postnatal development of the reproductive organs in bulls with special reference to puberty. Acta Endocrinol. 34(2 Suppl.), 1–109. doi: 10.1530/acta.0.XXXIVS009
Aiken, C. E., Tarry-Adkins, J. L., and Ozanne, S. E. (2016). Transgenerational effects of maternal diet on metabolic and reproductive ageing. Mamm. Genome 27, 430–439. doi: 10.1007/s00335-016-9631-1
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. doi: 10.1038/nrg.2016.59
Amann, R. P., and Walker, O. A. (1983). Changes in the pituitary-gonadal axis associated with puberty in Holstein bulls. J. Anim. Sci. 57, 433–442. doi: 10.2527/jas1983.572433x
Ando, M., Saito, Y., Xu, G., Bui, N. Q., Medetgul-Ernar, K., Pu, M., et al. (2019). Chromatin dysregulation and DNA methylation at transcription start sites associated with transcriptional repression in cancers. Nat. Commun. 10:2188. doi: 10.1038/s41467-019-09937-w
Ashe, A., Sapetschnig, A., Weick, E.-M., Mitchell, J., Bagijn, Marloes, P., et al. (2012). piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99. doi: 10.1016/j.cell.2012.06.018
Bagu, E., Cook, S., Gratton, C., and Rawlings, N. (2006). Postnatal changes in testicular gonadotropin receptors, serum gonadotropin, and testosterone concentrations and functional development of the testes in bulls. Reproduction 132, 403–411. doi: 10.1530/rep.1.00768
Banos, G., Brotherstone, S., and Coffey, M. P. (2007). Prenatal maternal effects on body condition score, female fertility, and milk yield of dairy cows. J. Dairy Sci. 90, 3490–3499. doi: 10.3168/jds.2006-809
Bansal, A., Li, C., Xin, F., Duemler, A., Li, W., Rashid, C., et al. (2019). Transgenerational effects of maternal bisphenol: a exposure on offspring metabolic health. J. Dev. Orig. Health Dis. 10, 164–175. doi: 10.1017/S2040174418000764
Barreñada, O., Fernández-Pérez, D., Larriba, E., Brieño-Enriquez, M., and del Mazo, J. (2020). Diversification of piRNAs expressed in PGCs and somatic cells during embryonic gonadal development. RNA Biol. 17, 1309–1323. doi: 10.1080/15476286.2020.1757908
Boettcher, P. J., Fatehi, J., and Schutz, M. M. (2003). Genotype×environment interactions in conventional versus pasture-based dairies in Canada. J. Dairy Sci. 86, 383–389. doi: 10.3168/jds.S0022-0302(03)73617-0
Bošković, A., and Rando, O. J. (2018). Transgenerational epigenetic inheritance. Annu. Rev. Genet. 52, 21–41. doi: 10.1146/annurev-genet-120417-031404
Brito, L. F. C., Barth, A. D., Rawlings, N. C., Wilde, R. E., Crews, D. H., Boisclair, Y. R., et al. (2007). Effect of feed restriction during calfhood on serum concentrations of metabolic hormones, gonadotropins, testosterone, and on sexual development in bulls. Reproduction 134, 171–181. doi: 10.1530/rep-06-0353
Burga, A., and Lehner, B. (2012). Beyond genotype to phenotype: why the phenotype of an individual cannot always be predicted from their genome sequence and the environment that they experience. FEBS J. 279, 3765–3775. doi: 10.1111/j.1742-4658.2012.08810.x
Byrne, C. J., Fair, S., English, A. M., Cirot, M., Staub, C., Lonergan, P., et al. (2018). Plane of nutrition before and after 6 months of age in Holstein-Friesian bulls: I. Effects on performance, body composition, age at puberty, and postpubertal semen production. J. Dairy Sci. 101, 3447–3459. doi: 10.3168/jds.2017-13719
Cagnone, G., and Sirard, M.-A. (2014). The impact of exposure to serum lipids during in vitro culture on the transcriptome of bovine blastocysts. Theriogenology 81, 712.e3–722.e3. doi: 10.1016/j.theriogenology.2013.12.005
Cagnone, G., and Sirard, M. A. (2016). The embryonic stress response to in vitro culture: insight from genomic analysis. Reproduction 152, R247–R261. doi: 10.1530/REP-16-0391
Capra, E., Lazzari, B., Turri, F., Cremonesi, P., Portela, A. M. R., Ajmone-Marsan, P., et al. (2019). Epigenetic analysis of high and low motile sperm populations reveals methylation variation in satellite regions within the pericentromeric position and in genes functionally related to sperm DNA organization and maintenance in Bos taurus. BMC Genomics 20:940. doi: 10.1186/s12864-019-6317-6
Cerón-Muñoz, M. F., Tonhati, H., Costa, C. N., Maldonado-Estrada, J., and Rojas-Sarmiento, D. (2004a). Genotype×environment interaction for age at first calving in brazilian and colombian holsteins. J. Dairy Sci. 87, 2455–2458. doi: 10.3168/jds.S0022-0302(04)73369-X
Cerón-Muñoz, M. F., Tonhati, H., Costa, C. N., Rojas-Sarmiento, D., and Echeverri Echeverri, D. M. (2004b). Factors that cause genotype by environment interaction and use of a multiple-trait herd-cluster model for milk yield of holstein cattle from Brazil and Colombia. J. Dairy Sci. 87, 2687–2692. doi: 10.3168/jds.S0022-0302(04)73395-0
Chaput, C., and Sirard, M. A. (2019). Embryonic response to high beta-hydroxybutyrate (BHB) levels in postpartum dairy cows. Domest. Anim. Endocrinol. 72:106431. doi: 10.1016/j.domaniend.2019.106431
Chen, Q., Yan, M., Cao, Z., Li, X., Zhang, Y., Shi, J., et al. (2016a). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400. doi: 10.1126/science.aad7977
Chen, Q., Yan, W., and Duan, E. (2016b). Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 17, 733–743. doi: 10.1038/nrg.2016.106
Conine, C. C., Sun, F., Song, L., Rivera-Pérez, J. A., and Rando, O. J. (2018). Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell 46, 470.e3–480.e3. doi: 10.1016/j.devcel.2018.06.024
Craig, J. R., Jenkins, T. G., Carrell, D. T., and Hotaling, J. M. (2017). Obesity, male infertility, and the sperm epigenome. Fertil. Steril. 107, 848–859. doi: 10.1016/j.fertnstert.2017.02.115
Cromie, A., Kelleher, D., Gordon, F., and Rath, M. (1998). Genotype by environment interaction for milk production traits in Holstein Friesian dairy cattle in Ireland. Interbull. Bull. 15, 100–104.
Dagilgan, S., Dundar-Yenilmez, E., Tuli, A., Urunsak, I. F., and Erdogan, S. (2015). Evaluation of intracellular pH regulation and alkalosis defense mechanisms in preimplantation embryos. Theriogenology 83, 1075–1084. doi: 10.1016/j.theriogenology.2014.12.011
Dahl, J. A., Jung, I., Aanes, H., Greggains, G. D., Manaf, A., Lerdrup, M., et al. (2016). Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552. doi: 10.1038/nature19360
Dance, A., Thundathil, J., Blondin, P., and Kastelic, J. (2016). Enhanced early-life nutrition of Holstein bulls increases sperm production potential without decreasing postpubertal semen quality. Theriogenology 86, 687.e2–694.e2.
Dance, A., Thundathil, J., Wilde, R., Blondin, P., and Kastelic, J. (2015). Enhanced early-life nutrition promotes hormone production and reproductive development in Holstein bulls. J. Dairy Sci. 98, 987–998. doi: 10.3168/jds.2014-8564
Desmet, K. L. J., Van Hoeck, V., Gagné, D., Fournier, E., Thakur, A., O’Doherty, A. M., et al. (2016). Exposure of bovine oocytes and embryos to elevated non-esterified fatty acid concentrations: integration of epigenetic and transcriptomic signatures in resultant blastocysts. BMC Genomics 17:1004. doi: 10.1186/s12864-016-3366-y
Devkota, B., Koseki, T., Matsui, M., Sasaki, M., Kaneko, E., Miyamoto, A., et al. (2008). Relationships among age, body weight, scrotal circumference, semen quality and peripheral testosterone and estradiol concentrations in pubertal and postpubertal Holstein bulls. J. Vet. Med. Sci. 70, 119–121. doi: 10.1292/jvms.70.119
Di Emidio, G., D’Aurora, M., Placidi, M., Franchi, S., Rossi, G., Stuppia, L., et al. (2019). Pre-conceptional maternal exposure to cyclophosphamide results in modifications of DNA methylation in F1 and F2 mouse oocytes: evidence for transgenerational effects. Epigenetics 14, 1057–1064. doi: 10.1080/15592294.2019.1631111
Díaz-Ruiz, R., Avéret, N., Araiza, D., Pinson, B., Uribe-Carvajal, S., Devin, A., et al. (2008). Mitochondrial oxidative phosphorylation is regulated by fructose 1,6-bisphosphate. A possible role in Crabtree effect induction? J. Biol. Chem. 283, 26948–26955. doi: 10.1074/jbc.M800408200
Dupont, C., Kappeler, L., Saget, S., Grandjean, V., and Lévy, R. (2019). Role of miRNA in the transmission of metabolic diseases associated with paternal diet-induced obesity. Front. Genet. 10:337. doi: 10.3389/fgene.2019.00337
Eckersley-Maslin, M. A., Alda-Catalinas, C., and Reik, W. (2018). Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450. doi: 10.1038/s41580-018-0008-z
Engmann, O. (2018). Dairy cows – an opportunity in the research field of non-genetic inheritance? Environ. Epigenet. 4:dvy014. doi: 10.1093/eep/dvy014
Ernst, J., and Kellis, M. (2012). ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216. doi: 10.1038/nmeth.1906
Ernst, J., and Kellis, M. (2017). Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 12, 2478–2492. doi: 10.1038/nprot.2017.124
Ernst, J., Kheradpour, P., Mikkelsen, T. S., Shoresh, N., Ward, L. D., Epstein, C. B., et al. (2011). Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49. doi: 10.1038/nature09906
Etienne, S. E., and Martin, T. G. (1979). Secondary selection differentials for cow productivity traits associated with seven criteria of selecting replacement beef heifers. J. Anim. Sci. 49, 26–38. doi: 10.2527/jas1979.49126x
Evans, A., Pierson, R., Garcia, A., McDougall, L., Hrudka, F., and Rawlings, N. (1996). Changes in circulating hormone concentrations, testes histology and testes ultrasonography during sexual maturation in beef bulls. Theriogenology 46, 345–357. doi: 10.1016/0093-691x(96)00190-2
Evans, A. C. O., Davies, F. J., Nasser, L. F., Bowman, P., and Rawlings, N. C. (1995). Differences in early patterns of gonadotrophin secretion between early and late maturing bulls, and changes in semen characteristics at puberty. Theriogenology 43, 569–578. doi: 10.1016/0093-691x(94)00062-y
Falconer, D. S. (1952). The problem of environment and selection. Am. Nat. 86, 293–298. doi: 10.1086/281736
Fang, L., Jiang, J., Li, B., Zhou, Y., Freebern, E., Vanraden, P. M., et al. (2019). Genetic and epigenetic architecture of paternal origin contribute to gestation length in cattle. Commun. Biol. 2:100. doi: 10.1038/s42003-019-0341-6
Felsenfeld, G. (2014). A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 6:a018200. doi: 10.1101/cshperspect.a018200
Gaspar-Maia, A., Alajem, A., Meshorer, E., and Ramalho-Santos, M. (2011). Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47. doi: 10.1038/nrm3036
Gkountela, S., Zhang, K. X., Shafiq, T. A., Liao, W.-W., Hargan-Calvopiña, J., Chen, P.-Y., et al. (2015). DNA demethylation dynamics in the human prenatal germline. Cell 161, 1425–1436. doi: 10.1016/j.cell.2015.05.012
Glanzner, W. G., Rissi, V. B., de Macedo, M. P., Mujica, L. K. S., Gutierrez, K., Bridi, A., et al. (2018). Histone 3 lysine 4, 9, and 27 demethylases expression profile in fertilized and cloned bovine and porcine embryos†. Biol. Reprod. 98, 742–751. doi: 10.1093/biolre/ioy054
Grishkevich, V., and Yanai, I. (2013). The genomic determinants of genotype x environment interactions in gene expression. Trends Genet. 29, 479–487. doi: 10.1016/j.tig.2013.05.006
Gu, L., Wang, Q., and Sun, Q.-Y. (2010). Histone modifications during mammalian oocyte maturation: dynamics, regulation and functions. Cell Cycle 9, 1942–1950. doi: 10.4161/cc.9.10.11599
Gudex, B., Johnson, D., and Singh, K. (2014). Prenatal maternal and possible transgenerational epigenetic effects on milk production. PLoS One 9:e98928. doi: 10.1371/journal.pone.0098928
Guerrero-Bosagna, C., Covert, T. R., Haque, M. M., Settles, M., Nilsson, E. E., Anway, M. D., et al. (2012). Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod. Toxicol. 34, 694–707. doi: 10.1016/j.reprotox.2012.09.005
Guo, F., Li, L., Li, J., Wu, X., Hu, B., Zhu, P., et al. (2017). Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988. doi: 10.1038/cr.2017.82
Guo, F., Li, X., Liang, D., Li, T., Zhu, P., Guo, H., et al. (2014). Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459. doi: 10.1016/j.stem.2014.08.003
Guo, F., Yan, L., Guo, H., Li, L., Hu, B., Zhao, Y., et al. (2015). The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161, 1437–1452. doi: 10.1016/j.cell.2015.05.015
Guo, H., Zhu, P., Yan, L., Li, R., Hu, B., Lian, Y., et al. (2014). The DNA methylation landscape of human early embryos. Nature 511, 606–610. doi: 10.1038/nature13544
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. doi: 10.1038/nature08975
Hackett, J. A., Sengupta, R., Zylicz, J. J., Murakami, K., Lee, C., Down, T. A., et al. (2013). Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine. Science 339, 448–452.
Hajkova, P., Ancelin, K., Waldmann, T., Lacoste, N., Lange, U. C., Cesari, F., et al. (2008). Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452, 877–881. doi: 10.1038/nature06714
Hammami, H., Rekik, B., Bastin, C., Soyeurt, H., Bormann, J., Stoll, J., et al. (2009a). Environmental sensitivity for milk yield in Luxembourg and Tunisian Holsteins by herd management level. J. Dairy Sci. 92, 4604–4612. doi: 10.3168/jds.2008-1513
Hammami, H., Rekik, B., and Gengler, N. (2009b). Genotype by environment interaction in dairy cattle. Biotechnol. Agron. Soc. Environ. 13, 155–164.
Harvey, A. J., Kind, K. L., Pantaleon, M., Armstrong, D. T., and Thompson, J. G. (2004). Oxygen-regulated gene expression in bovine blastocysts. Biol. Reprod. 71, 1108–1119. doi: 10.1095/biolreprod.104.028639
Ibeagha-Awemu, E. M., and Zhao, X. (2015). Epigenetic marks: regulators of livestock phenotypes and conceivable sources of missing variation in livestock improvement programs. Front. Genet. 6:302. doi: 10.3389/fgene.2015.00302
Igolkina, A. A., Zinkevich, A., Karandasheva, K. O., Popov, A. A., Selifanova, M. V., Nikolaeva, D., et al. (2019). H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 histone tags suggest distinct regulatory evolution of open and condensed chromatin landmarks. Cells 8:1034.
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424. doi: 10.1038/nature23262
Jenkins, T. G., James, E. R., Alonso, D. F., Hoidal, J. R., Murphy, P. J., Hotaling, J. M., et al. (2017). Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 5, 1089–1099. doi: 10.1111/andr.12416
Ji, H., and Khurana Hershey, G. K. (2012). Genetic and epigenetic influence on the response to environmental particulate matter. J. Allergy Clin. Immunol. 129, 33–41. doi: 10.1016/j.jaci.2011.11.008
Jiang, Z., Lin, J., Dong, H., Zheng, X., Marjani, S. L., Duan, J., et al. (2018). DNA methylomes of bovine gametes and in vivo produced preimplantation embryos. Biol. Reprod. 99, 949–959. doi: 10.1093/biolre/ioy138
Johnson, C., Dance, A., Kovalchuk, I., Kastelic, J., and Thundathil, J. (2020). Enhanced pre-pubertal nutrition upregulates mitochondrial function in testes and sperm of post-pubertal Holstein bulls. Sci. Rep. 10:2235. doi: 10.1038/s41598-020-59067-3
Jung, Y. H., Kremsky, I., Gold, H. B., Rowley, M. J., Punyawai, K., Buonanotte, A., et al. (2019). Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol. Cell. 75, 154.e5–171.e5. doi: 10.1016/j.molcel.2019.04.014
Jung, Y. H., Sauria, M. E. G., Lyu, X., Cheema, M. S., Ausio, J., Taylor, J., et al. (2017). Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep. 18, 1366–1382. doi: 10.1016/j.celrep.2017.01.034
Kasinathan, P., Wei, H., Xiang, T., Molina, J. A., Metzger, J., Broek, D., et al. (2015). Acceleration of genetic gain in cattle by reduction of generation interval. Sci. Rep. 5:8674. doi: 10.1038/srep08674
Kauffold, J., Amer, H. A. H., Bergfeld, U., Weber, W., and Sobiraj, A. (2005). The in vitro developmental competence of oocytes from juvenile calves is related to follicular diameter. J. Reprod. Dev. 51, 325–332. doi: 10.1262/jrd.17002
Kenny, D. A., and Byrne, C. J. (2018). Review: the effect of nutrition on timing of pubertal onset and subsequent fertility in the bull. Animal 12, s36–s44. doi: 10.1017/S1751731118000514
Kitagawa, Y., Suzuki, K., Yoneda, A., and Watanabe, T. (2004). Effects of oxygen concentration and antioxidants on the in vitro developmental ability, production of reactive oxygen species (ROS), and DNA fragmentation in porcine embryos. Theriogenology 62, 1186–1197. doi: 10.1016/j.theriogenology.2004.01.011
Kropp, J., Carrillo, J. A., Namous, H., Daniels, A., Salih, S. M., Song, J., et al. (2017). Male fertility status is associated with DNA methylation signatures in sperm and transcriptomic profiles of bovine preimplantation embryos. BMC Genomics 18:280. doi: 10.1186/s12864-017-3673-y
Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., Heravi-Moussavi, A., et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. doi: 10.1038/nature14248
Kutchy, N. A., Menezes, E. S. B., Chiappetta, A., Tan, W., Wills, R. W., Kaya, A., et al. (2018). Acetylation and methylation of sperm histone 3 lysine 27 (H3K27ac and H3K27me3) are associated with bull fertility. Andrologia 50:e12915. doi: 10.1111/and.12915
Labrecque, R., Vigneault, C., Blondin, P., and Sirard, M.-A. (2013). Gene expression analysis of bovine oocytes with high developmental competence obtained from FSH-stimulated animals. Mol. Reprod. Dev. 80, 428–440. doi: 10.1002/mrd.2217
Lambert, S., Blondin, P., Vigneault, C., Labrecque, R., Dufort, I., and Sirard, M.-A. (2018). Spermatozoa DNA methylation patterns differ due to peripubertal age in bulls. Theriogenology 106(Suppl. C), 21–29. doi: 10.1016/j.theriogenology.2017.10.006
Landry, D. A., Bellefleur, A.-M., Labrecque, R., Grand, F.-X., Vigneault, C., Blondin, P., et al. (2016). Effect of cow age on the in vitro developmental competence of oocytes obtained after FSH stimulation and coasting treatments. Theriogenology 86, 1240–1246. doi: 10.1016/j.theriogenology.2016.04.064
Landry, D. A., Labrecque, R., Grand, F.-X., Vigneault, C., Blondin, P., and Sirard, M.-A. (2018). Effect of heifer age on the granulosa cell transcriptome after ovarian stimulation. Reprod. Fertil. Dev. 30, 980–990. doi: 10.1071/RD17225
Lavoir, M.-C., Basrur, P. K., and Betteridge, K. J. (1994). Isolation and identification of germ cells from fetal bovine ovaries. Mol. Reprod. Dev. 37, 413–424. doi: 10.1002/mrd.1080370408
Li, L., Guo, F., Gao, Y., Ren, Y., Yuan, P., Yan, L., et al. (2018). Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 20, 847–858. doi: 10.1038/s41556-018-0123-2
Loche, E., Blackmore, H. L., Carpenter, A. A., Beeson, J. H., Pinnock, A., Ashmore, T. J., et al. (2018). Maternal diet-induced obesity programmes cardiac dysfunction in male mice independently of post-weaning diet. Cardiovasc. Res. 114, 1372–1384. doi: 10.1093/cvr/cvy082
Lyko, F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. doi: 10.1038/nrg.2017.80
Macmillan, K., and Hafs, H. (1968). Gonadal and extra gonadal sperm numbers during reproductive development of holstein bulls. J. Anim. Sci. 27, 697–700. doi: 10.2527/jas1968.273697x
Magaraki, A., van der Heijden, G., Sleddens-Linkels, E., Magarakis, L., van Cappellen, W. A., Peters, A. H. F. M., et al. (2017). Silencing markers are retained on pericentric heterochromatin during murine primordial germ cell development. Epigenet. Chromatin 10:11. doi: 10.1186/s13072-017-0119-3
Mahmoudi, A., Zargaran, A., Amini, H.-R., Assadi, A., Vajdi Hokmabad, R., and Eghbalsaied, S. (2015). A SNP in the 3′-untranslated region of AMPKγ1 may associate with serum ketone body and milk production of Holstein dairy cows. Gene 574, 48–52. doi: 10.1016/j.gene.2015.07.077
Majerus, V., De Roover, R., Etienne, D., Kaidi, S., Massip, A., Dessy, F., et al. (1999). Embryo production by ovum pick up in unstimulated calves before and after puberty. Theriogenology 52, 1169–1179. doi: 10.1016/s0093-691x(99)00209-5
Markunas, C. A., Wilcox, A. J., Xu, Z., Joubert, B. R., Harlid, S., Panduri, V., et al. (2016). Maternal age at delivery is associated with an epigenetic signature in both newborns and adults. PLoS One 11:e0156361. doi: 10.1371/journal.pone.0156361
McDaniel, B., and Corley, E. (1967). Relationships between sire evaluations at different herdmate levels. J. Dairy Sci. 50, 735–741.
McGraw, S., Robert, C., Massicotte, L., and Sirard, M.-A. (2003). Quantification of histone acetyltransferase and histone deacetylase transcripts during early bovine embryo development1. Biol. Reprod. 68, 383–389. doi: 10.1095/biolreprod.102.005991
McJunkin, K. (2018). Maternal effects of microRNAs in early embryogenesis. RNA Biol. 15, 165–169. doi: 10.1080/15476286.2017.1402999
Ming, H., Sun, J., Pasquariello, R., Gatenby, L., Herrick, J. R., Yuan, Y., et al. (2020). The landscape of accessible chromatin in bovine oocytes and early embryos. Epigenetics doi: 10.1080/15592294.2020.1795602 [Epub ahead of print].
Morin-Doré, L., Blondin, P., Vigneault, C., Grand, F.-X., Labrecque, R., and Sirard, M.-A. (2017). Transcriptomic evaluation of bovine blastocysts obtained from peri-pubertal oocyte donors. Theriogenology 93, 111–123. doi: 10.1016/j.theriogenology.2017.01.005
Nivet, A.-L., Bunel, A., Labrecque, R., Belanger, J., Vigneault, C., Blondin, P., et al. (2012). FSH withdrawal improves developmental competence of oocytes in the bovine model. Reproduction 143:165. doi: 10.1530/rep-11-0391
Ojango, J. M. K., and Pollott, G. E. (2002). The relationship between Holstein bull breeding values for milk yield derived in both the UK and Kenya. Livestock Prod. Sci. 74, 1–12. doi: 10.1016/S0301-6226(01)00282-2
Pagé-Larivière, F., Tremblay, A., Campagna, C., Rodriguez, M. J., and Sirard, M.-A. (2016). Low concentrations of bromodichloromethane induce a toxicogenomic response in porcine embryos in vitro. Reprod. Toxicol. 66, 44–55. doi: 10.1016/j.reprotox.2016.09.010
Pallotta, M. M., Barbato, V., Pinton, A., Acloque, H., Gualtieri, R., Talevi, R., et al. (2019). In vitro exposure to CPF affects bovine sperm epigenetic gene methylation pattern and the ability of sperm to support fertilization and embryo development. Environ. Mol. Mutagen. 60, 85–95. doi: 10.1002/em.22242
Palma, G. A., Tortonese, D. J., and Sinowatz, F. (2001). Developmental capacity in vitro of prepubertal oocytes. Anat. Histol. Embryol. 30, 295–300. doi: 10.1046/j.1439-0264.2001.00324.x
Perez, M. F., and Lehner, B. (2019). Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21, 143–151. doi: 10.1038/s41556-018-0242-9
Plant, T. M. (2015). Neuroendocrine control of the onset of puberty. Front. Neuroendocrinol. 38, 73–88. doi: 10.1016/j.yfrne.2015.04.002
Rahman, M. B., Kamal, M. M., Rijsselaere, T., Vandaele, L., Shamsuddin, M., and Van Soom, A. (2014). Altered chromatin condensation of heat-stressed spermatozoa perturbs the dynamics of DNA methylation reprogramming in the paternal genome after in vitro fertilisation in cattle. Reprod. Fertil. Dev. 26, 1107–1116. doi: 10.1071/RD13218
Ramírez-Valverde, R., Peralta-Aban, J. A., Núñez-Domínguez, R., Ruíz-Flores, A., García-Muñiz, J. G., and García-Peniche, T. B. (2010). Genotype by feeding system interaction in the genetic evaluation of Jersey cattle for milk yield. Animal 4, 1971–1975. doi: 10.1017/S175173111000128X
Rando, O. J. (2016). Intergenerational transfer of epigenetic information in sperm. Cold Spring Harb. Perspect. Med. 6:a022988. doi: 10.1101/cshperspect.a022988
Rawlings, N. C., and Evans, A. C. (1995). Androgen negative feedback during the early rise in LH secretion in bull calves. J. Endocrinol. 145, 243–249. doi: 10.1677/joe.0.1450243
Reynolds, T. S., Lynch, C. D., Hade, E. M., Allain, D. C., Westman, J. A., and Toland, A. E. (2020). Maternal age at delivery and fertility of the next generation. Paediatr. Perinatal Epidemiol. doi: 10.1111/ppe.12666 [Epub ahead of print].
Roa, J., García-Galiano, D., Castellano, J. M., Gaytan, F., Pinilla, L., and Tena-Sempere, M. (2010). Metabolic control of puberty onset: new players, new mechanisms. Mol. Cell. Endocrinol. 324, 87–94. doi: 10.1016/j.mce.2009.12.018
Roberts, A. J., and Hay, E. H. (2019). Multigenerational effects. Vet. Clin. Food Anim. Pract. 35, 355–364. doi: 10.1016/j.cvfa.2019.02.009
Ron, M., and Hillel, J. (1983). Genotype x environment interaction in dairy cattle and its role in breeding programmes. TAG. Theoretical and applied genetics. Theor. Angew. Genet. 66, 93–99. doi: 10.1007/bf00265180
Rowold, E. D. H., Schulze, L., Van der Auwera, S., and Grabe, H. J. (2017). Paternal transmission of early life traumatization through epigenetics: do fathers play a role? Med. Hypothes. 109, 59–64. doi: 10.1016/j.mehy.2017.09.011
Sakatani, M., Yamanaka, K., Kobayashi, S., and Takahashi, M. (2008). Heat shock-derived reactive oxygen species induce embryonic mortality in in vitro early stage bovine embryos. J. Reprod. Dev. 54, 496–501. doi: 10.1262/jrd.20017
Salilew-Wondim, D., Fournier, E., Hoelker, M., Saeed-Zidane, M., Tholen, E., Looft, C., et al. (2015). Genome-wide DNA methylation patterns of bovine blastocysts developed in vivo from embryos completed different stages of development in vitro. PLoS One 10:e0140467. doi: 10.1371/journal.pone.0140467
Saxena, A., and Carninci, P. (2011). Long non-coding RNA modifies chromatin. BioEssays 33, 830–839. doi: 10.1002/bies.201100084
Seisenberger, S., Andrews, S., Krueger, F., Arand, J., Walter, J., Santos, F., et al. (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 48, 849–862. doi: 10.1016/j.molcel.2012.11.001
Sharma, U., Conine, C. C., Shea, J. M., Boskovic, A., Derr, A. G., Bing, X. Y., et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. doi: 10.1126/science.aad6780
Sharma, U., Sun, F., Conine, C. C., Reichholf, B., Kukreja, S., Herzog, V. A., et al. (2018). Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 46, 481.e6–494.e6. doi: 10.1016/j.devcel.2018.06.023
Shojaei Saadi, H. A., O’Doherty, A. M., Gagne, D., Fournier, E., Grant, J. R., Sirard, M. A., et al. (2014). An integrated platform for bovine DNA methylome analysis suitable for small samples. BMC Genomics 15:451. doi: 10.1186/1471-2164-15-451
Siklenka, K., Erkek, S., Godmann, M., Lambrot, R., McGraw, S., Lafleur, C., et al. (2015). Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 350:aab2006. doi: 10.1126/science.aab2006
Sillaste, G., Kaplinski, L., Meier, R., Jaakma, Ü, Eriste, E., and Salumets, A. (2017). A novel hypothesis for histone-to-protamine transition in Bos taurus spermatozoa. Reproduction 153, 241–251.
Singh, K., Erdman, R. A., Swanson, K. M., Molenaar, A. J., Maqbool, N. J., Wheeler, T. T., et al. (2010). Epigenetic regulation of milk production in dairy cows. J. Mamm. Gland Biol. Neoplasia 15, 101–112. doi: 10.1007/s10911-010-9164-2
Singh, K., Molenaar, A. J., Swanson, K. M., Gudex, B., Arias, J. A., Erdman, R. A., et al. (2012). Epigenetics: a possible role in acute and transgenerational regulation of dairy cow milk production. Animal 6, 375–381. doi: 10.1017/S1751731111002564
Smith, Z. D., Chan, M. M., Humm, K. C., Karnik, R., Mekhoubad, S., Regev, A., et al. (2014). DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615. doi: 10.1038/nature13581
Smith, Z. D., and Meissner, A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220. doi: 10.1038/nrg3354
Soubry, A., Guo, L., Huang, Z., Hoyo, C., Romanus, S., Price, T., et al. (2016). Obesity-related DNA methylation at imprinted genes in human sperm: results from the TIEGER study. Clin. Epigenet. 8:51. doi: 10.1186/s13148-016-0217-2
Soubry, A., Schildkraut, J. M., Murtha, A., Wang, F., Huang, Z., Bernal, A., et al. (2013). Paternal obesity is associated with IGF2 hypomethylation in newborns: results from a newborn epigenetics Study (NEST) cohort. BMC Med. 11:29. doi: 10.1186/1741-7015-11-29
Spadafora, C. (2017). Sperm-mediated transgenerational inheritance. Front. Microbiol. 8:2401. doi: 10.3389/fmicb.2017.02401
Stanton, T., Blake, R., Quaas, R., Van Vleck, L. D., and Carabano, M. (1991). Genotype by environment interaction for Holstein milk yield in Colombia, Mexico, and Puerto Rico. J. Dairy Sci. 74, 1700–1714.
Steegers-Theunissen, R. P. M., Twigt, J., Pestinger, V., and Sinclair, K. D. (2013). The periconceptional period, reproduction and long-term health of offspring: the importance of one-carbon metabolism. Hum. Reprod. Update 19, 640–655. doi: 10.1093/humupd/dmt041
Summers, M. C., and Biggers, J. D. (2003). Chemically defined media and the culture of mammalian preimplantation embryos: historical perspective and current issues. Hum. Reprod. Update 9, 557–582. doi: 10.1093/humupd/dmg039
Takeda, K., Kobayashi, E., Akagi, S., Nishino, K., Kaneda, M., and Watanabe, S. (2017). Differentially methylated CpG sites in bull spermatozoa revealed by human DNA methylation arrays and bisulfite analysis. J. Reprod. Dev. 63, 279–287. doi: 10.1262/jrd.2016-160
Takeda, K., Kobayashi, E., Nishino, K., Imai, A., Adachi, H., Hoshino, Y., et al. (2019). Age-related changes in DNA methylation levels at CpG sites in bull spermatozoa and in vitro fertilization-derived blastocyst-stage embryos revealed by combined bisulfite restriction analysis. J. Reprod. Dev. 65, 305–312. doi: 10.1262/jrd.2018-146
Thompson, R. P., Nilsson, E., and Skinner, M. K. (2020). Environmental epigenetics and epigenetic inheritance in domestic farm animals. Anim. Reprod. Sci. 18:106316. doi: 10.1016/j.anireprosci.2020.106316
Tian, Y., Jia, Z., Wang, J., Huang, Z., Tang, J., Zheng, Y., et al. (2011). Global Mapping of H3K4me1 and H3K4me3 reveals the chromatin state-based cell type-specific gene regulation in human treg cells. PLoS One 6:e27770. doi: 10.1371/journal.pone.0027770
Tremblay, R., Dufort, I., and Sirard, M.-A. (2018). Metabolic stress induces modifications in the epigenetic program of preimplantation bovine embryos. Mol. Reprod. Dev. 85, 117–127. doi: 10.1002/mrd.22941
Uchiyama, R., Kupkova, K., Shetty, S. J., Linford, A. S., Pray-Grant, M. G., Wagar, L. E., et al. (2018). Histone H3 lysine 4 methylation signature associated with human undernutrition. Proc. Natl. Acad. Sci. U.S.A. 115, E11264–E11273. doi: 10.1073/pnas.1722125115
Ugur, M. R., Kutchy, N. A., de Menezes, E. B., Ul-Husna, A., Haynes, B. P., Uzun, A., et al. (2019). Retained acetylated histone four in bull sperm associated with fertility. Front. Vet. Sci. 6:223. doi: 10.3389/fvets.2019.00223
Van Soom, A., Yuan, Y. Q., Peelman, L. J., de Matos, D. G., Dewulf, J., Laevens, H., et al. (2002). Prevalence of apoptosis and inner cell allocation in bovine embryos cultured under different oxygen tensions with or without cysteine addition. Theriogenology 57, 1453–1465. doi: 10.1016/s0093-691x(01)00726-9
Verma, A., Rajput, S., Kumar, S., De, S., Chakravarty, A. K., Kumar, R., et al. (2015). Differential histone modification status of spermatozoa in relation to fertility of buffalo bulls. J. Cell. Biochem. 116, 743–753. doi: 10.1002/jcb.25029
Wakchaure, R., Ganguly, S., and Praveen, K. (2016). Genotype x environment interaction in animal breeding: a review. Biodiversity conservation in changing climate. Cap 3, 60–73.
Wang, L., Zhang, J., Duan, J., Gao, X., Zhu, W., Lu, X., et al. (2014). Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991.
Wei, Y., Yang, C.-R., Wei, Y.-P., Zhao, Z.-A., Yi, H., Schatten, H., et al. (2014). Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc. Natl. Acad. Sci. U.S.A. 111, 1873–1878. doi: 10.1073/pnas.1321195111
Wolf, F., Almquist, J., and Hale, E. (1965). Prepuberal behavior and puberal characteristics of beef bulls on high nutrient allowance. J. Anim. Sci. 24, 761–765. doi: 10.2527/jas1965.243761x
Wrobel, K. H., and Süß, F. (1998). Identification and temporospatial distribution of bovine primordial germ cells prior to gonadal sexual differentiation. Anat. Embryol. 197, 451–467. doi: 10.1007/s004290050156
Wu, C., Blondin, P., Vigneault, C., Labrecque, R., and Sirard, M.-A. (2020). The age of the bull influences the transcriptome and epigenome of blastocysts produced by IVF. Theriogenology 144, 122–131. doi: 10.1016/j.theriogenology.2019.12.020
Wu, J., Huang, B., Chen, H., Yin, Q., Liu, Y., Xiang, Y., et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657. doi: 10.1038/nature18606
Xavier, M. J., Roman, S. D., Aitken, R. J., and Nixon, B. (2019). Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health. Hum. Reprod. Update 25, 519–541. doi: 10.1093/humupd/dmz017
Xia, W., Xu, J., Yu, G., Yao, G., Xu, K., Ma, X., et al. (2019). Resetting histone modifications during human parental-to-zygotic transition. Science 365, 353–360. doi: 10.1126/science.aaw5118
Yuan, S., Schuster, A., Tang, C., Yu, T., Ortogero, N., Bao, J., et al. (2016). Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 143, 635–647. doi: 10.1242/dev.131755
Zaraza, J., Oropeza, A., Velazquez, M. A., Korsawe, K., Herrmann, D., Carnwath, J. W., et al. (2010). Developmental competence and mRNA expression of preimplantation in vitro–produced embryos from prepubertal and postpubertal cattle and their relationship with apoptosis after intraovarian administration of IGF-1. Theriogenology 74, 75–89. doi: 10.1016/j.theriogenology.2009.11.033
Zeng, Y., and Chen, T. (2019). DNA methylation reprogramming during mammalian development. Genes 10:257.
Zhang, T., Cooper, S., and Brockdorff, N. (2015). The interplay of histone modifications – writers that read. EMBO Rep. 16, 1467–1481. doi: 10.15252/embr.201540945
Zheng, H., Huang, B., Zhang, B., Xiang, Y., Du, Z., Xu, Q., et al. (2016). Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell. 63, 1066–1079. doi: 10.1016/j.molcel.2016.08.032
Zhou, L.-Q., and Dean, J. (2015). Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol. 25, 82–91. doi: 10.1016/j.tcb.2014.09.006
Keywords: parental age, embryonic development, dairy cows, epigenetics, metabolism, transgenerational inheritance, intergenerational inheritance
Citation: Wu C and Sirard M-A (2020) Parental Effects on Epigenetic Programming in Gametes and Embryos of Dairy Cows. Front. Genet. 11:557846. doi: 10.3389/fgene.2020.557846
Received: 30 April 2020; Accepted: 18 September 2020;
Published: 14 October 2020.
Edited by:
Stephanie McKay, University of Vermont, United StatesReviewed by:
George E. Liu, United States Department of Agriculture, United StatesCopyright © 2020 Wu and Sirard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc-André Sirard, bWFyYy1hbmRyZS5zaXJhcmRAZnNhYS51bGF2YWwuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.