
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 02 November 2020
Sec. Evolutionary and Population Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.536640
 Yue-Tian Li1,2,3†
Yue-Tian Li1,2,3† Zhao-Zhe Xin1,4,5†
Zhao-Zhe Xin1,4,5† Ying-Yu Tang1,5†
Ying-Yu Tang1,5† Ting-Ting Yang1,5†
Ting-Ting Yang1,5† Bo-Ping Tang1*†
Bo-Ping Tang1*† Yue Sun1
Yue Sun1 Dai-Zhen Zhang1
Dai-Zhen Zhang1 Chun-Lin Zhou1
Chun-Lin Zhou1 Qiu-Ning Liu1*
Qiu-Ning Liu1* Xiao-Min Yu2*
Xiao-Min Yu2*Mitochondrial genomes (mitogenomes) are important for understanding molecular evolution and phylogenetic relationships. The complete mitogenome of Perisesarma bidens was determined, which is 15,641 bp in length. The A + T content of P. bidens mitogenome was 74.81%. The AT skew was slightly negative (−0.021). The 22 tRNAs ranged from 65 to 73 bp and were highly A + T biased. All tRNA genes had typical cloverleaf structures, except for the trnS1 gene, which lacked a dihydrouridine (DHU) arm. The gene order within the P. bidens mitogenome was identical to the pancrustacean ground pattern, except for the translocation of the trnH. Additionally, the gene order of trnI-trnQ-trnM in pancrustacean ground pattern became trnQ-trnI-trnM in P. bidens. Phylogenetic analyses supported the inclusion of P. bidens in Sesarmidae and the promotion of Sesarminae to Sesarmidae. The results will help us to better understand the status and evolutionary history of Grapsoidea crabs.
Decapoda is the most diverse, species-rich group of crustaceans, containing many well-known animals, such as crayfish, lobsters, shrimps, hermit crabs, and “true” crabs (Shen et al., 2013; Basso et al., 2017). The true crabs belong to Brachyura, which is a diverse, economically important group, with about 7200 described species (De Grave et al., 2009; Ahyong et al., 2011). Brachyura is highly adaptable and can live on land and in both marine and fresh water. Therefore, crabs have become important groups for the study of evolution (Castro et al., 2015). Some Brachyura are edible and medicinal and have economic importance (Carpenter and Niem, 1998).
Most Brachyura are grouped into the Podotremata, Heterotremata, and Thoracotremata, with the latter two referred to as the Eubrachyura. However, the phylogenetic relationships within Eubrachyura remain controversial, particularly the relationships of the Sesarmidae and Varunidae, and between these two and Grapsoidea (Schubart et al., 2000, 2002; Kitaura et al., 2002). The traditional classification of Grapsidae contains four subfamilies: Grapsinae, Plagusiinae, Sesarminae, and Varuninae (Schubart et al., 2000). Traditional methods place the following in the Sesarminae: Perisesarma bidens, Sesarmops sinensis, Clistocoeloma sinensis, Helice tientsinensis, Helice latimera, Helice wuana, and Metaplax longipes. Of these, P. bidens and S. sinensis should be Sesarmops crabs; C. sinensis should be a Clistocoeloma crab; H. tientsinensis, H. latimera, and H. wuana should be Helice crabs and M. longipes should be a Metaplax crab (Schubart et al., 2000). However, some scholars have suggested that Grapsidae should be promoted to Grapsoidea, promoting the four subfamilies to families, i.e., Grapsidae, Varunidae, Sesarmidae and Plagusiidae. Other studies have advised that Sesarmops and Clistocoeloma crabs, which originally belonged to Sesarminae, should belong to Sesarmidae, and that Helice and Metaplax crabs that originally belonged to Sesarminae should be transferred from the Sesarminae to the Varunidae (Kitaura et al., 2002; Schubart et al., 2002). The classification of these taxa remains unresolved. Sesarmid crabs are common in mangroves and can tolerate great variation in salinity along the environment (Theuerkauff et al., 2018). They are very good experimental research objects.
Many studies have investigated these relationships using nuclear DNA, mitochondrial DNA (mtDNA), and morphological character analyses. Some studies combined mtDNA and nuclear genes to reconstruct more reliable phylogenetic trees. However, the combination of these genes made alignment and model selection quite difficult (Foster, 2004; Cox et al., 2008). This has led to the conclusion that the taxon sampling is insufficient and unbalanced (Bergsten, 2005; Wägele and Mayer, 2007). It is evident that more species are necessary to improve the quality of the analyses and stability of phylogenetic trees (Brinkmann and Philippe, 2008).
The mitochondrial genome (mitogenome) has been widely used in phylogenetic analyses, due to its rich signals from sequence information and gene arrangement (Xin et al., 2017a, b). The mitogenome has a simple structure, haploid nature, maternal inheritance, and rapid evolutionary rate (Liu et al., 2015). The mitogenomes are closed circular double-stranded molecules in the range of 14–18 kb in most bilaterian animals, including 13 protein-coding genes (PCGs; cox1–3, cob, nad1–6 and nad4L, atp6, and atp8), 2 rRNA genes, 22 tRNA genes, and an AT-rich region (control region) (Tang et al., 2003, 2017, 2018; Xin et al., 2017a, b). The taxonomy of Sesarmid crabs has been studied extensively and benefited from recent refinements in species of Perisesarma and Sesarmops (Li et al., 2019; Shih et al., 2019; Ng et al., 2020). However, the mitogenome of P. bidens has not been analyzed. Here, we determined the mitogenome of P. bidens and used the mitogenomes of 65 species to construct phylogenetic trees to discuss the systemic status and genetic relationships of the controversial taxa, Sesarmidae and Grapsoidea.
We have taken a close look at the website1. We found that the species Perisesarma bidens is not considered endangered or protected species, the IUCN status for this species is “Not evaluated.” Similarly, the species Perisesarma bidens is also Not endangered or protected species in China. No special permit Is required to collect crabs at selected sites in China. the sampling locations are Not privately-owned or natural protected areas, the collection of this species is legal in China. So we can use this species for experiments and subsequent analysis.
Specimens of P. bidens were collected from the seaside of Zhangzhou City, Fujian Province, China, identified using the morphological methods of Dai (1999) and molecular identification with COI marker, and preserved in 95% ethanol at –20°C until DNA extraction. Voucher specimens of P. bidens were deposited in the Jiangsu Provincial Key Laboratory of Coastal Wetland Bioresources and Environmental Protection, School of Ocean and Biological Engineering, Yancheng Teachers University, Yancheng, China.
Total genomic DNA was extracted from muscle using a genomic DNA extraction kit (Sangon, China), following the manufacturer’s instructions, and was visualized on 1.0% agarose gels. The complete mitogenome was obtained using a combination of conventional PCR and long PCR to amplify overlapping fragments spanning the entire mitogenome. Initially, conserved sequences, such as cox1, cox3, nad5, nad4, and rrnS, were amplified by conventional PCR using universal primers synthesized by Beijing Sunbiotech (Tang et al., 2003, 2017, 2018; Liu et al., 2015; Xin et al., 2017a, b).
We designed species-specific primers to amplify large overlapping regions of the mitogenome based on conserved sequences using Primer Premier 5 (Supplementary Table S1). All amplifications were performed on a Mastercycler (Eppendorf) and Mastercycler gradient. The reactions were 50 μL and contained 34.65 μL ddH2O, 5 μL 10 × LA PCR buffer II (Mg2+ Plus, Aidlab), 4 μL dNTPs (10 mM), 2 μL each primer (10 μM), 0.35 μL red Taq DNA Polymerase (5 U/μL, Aidlab), and 2 μL DNA template (∼30 ng).
The PCR conditions for conserved sequences followed a standard three-step protocol, with an initial denaturing at 96°C for 3 min, then 40 cycles of 94°C for 30 s, annealing at the recommended temperature for each primer for 30 s, and elongation at 72°C for 45 s, with a final 5 min extension at 72°C. The PCR conditions for large overlapping regions followed a standard two-step protocol with 3 min at 94°C, followed by 35 cycles of 35 s at 94°C, 3–6 min at 50–56°C, and 10 min at 72°C. All PCR products were sent to General Biosystems, Anhui for Sanger sequencing.
The sequence was annotated using DNASTAR (DNASTAR, Madison, WI, United States). The locations of the PCGs, rRNA genes, tRNA genes, and CR were initially identified using the MITOS Web Server2. The PCG coding regions were further identified using the NCBI ORF Finder3. Two rRNA genes were identified by alignment with published brachyuran sequences. Codon usage and the nucleotide composition of the mitogenomes were determined using MEGA6 (Tamura et al., 2013). The nucleotide sequence of the complete P. bidens mitogenome was deposited in the NCBI database under accession no. KY808394. Gene orders in the complete mitogenome were also inferred through the MITOS Web Server.
We used nucleotide (NT) sequences for phylogenetic analyses. The sequences were aligned with MAFFT using the default settings (Katoh et al., 2002). Gaps in the sequences were removed using Gblocks (Castresana, 2000), and the saturation of the sequences was examined using DAMBE (Xia and Xie, 2001), which indicated that the sequences were not saturated and were suitable for phylogenetic analyses. Complete mitogenomes of 65 decapods (60 crabs plus 5 outgroups) were downloaded from NCBI (Table 1). The five outgroups were Cherax destructor, Cambaroides similis, Neopetrolisthes maculatus, Paralithodes camtschaticus, and Pagurus longicarpus.
Phylogenetic analyses were performed using Bayesian inference (BI) and maximum likelihood (ML) methods using MrBayes v 3.2.2 (Ronquist et al., 2012) and IQ-Tree (Nguyen et al., 2014; Kalyaanamoorthy et al., 2017; Hoang et al., 2018), respectively. The GTRmodel was selected by MrModeltest 2.3 (Nylander, 2004). The BI analyses ran four independent chains for 10,000,000 generations, sampled every 100 generations, with a burn-in of 25,000 generations. The average standard deviation of split frequencies was < 0.01. Convergence was assessed using Tracer v1.6 and the effective sampling size for all parameters was > 200. The ML analyses were performed on 1000 bootstrap replications. The resulting phylogenetic trees were visualized using FigTree v1.4.2.
The complete mitogenome of P. bidens was a circular of 15,641 bp (GenBank accession no. KY808394). Its size was within the range observed in completely sequenced brachyuran species. The mitogenome composition (A: 36.61%, T: 38.20%, C: 15.13%, G: 10.06%) was strongly A + T biased which accounts for 74.81%, and exhibited with negative AT-skew (–0.021). The AT-skew of the mitogenomes of most crabs were negative, for example, H. wuana (Tang et al., 2018), S. sinensis (Tang et al., 2017), H. tientsinensis (Xin et al., 2017b), C. sinensis (Xin et al., 2017a), the AT-skew value of mitogenomes in other crabs had also been calculated and counted in related studies (Xin et al., 2017a, b). The genes were typical of animal mitogenomes, with 22 tRNA genes, 13 PCGs, 2 rRNA genes, and a CR (Table 2). Overall, 4 of the 13 PCGs (nad5, nad4, nad4L, and nad1), 8 tRNAs [trnQ, trnC, trnY, trnF, trnH, trnP, trnL (CUN), and trnV], and 2 rRNAs (rrnL and rrnS) were encoded by the minority strand, while the other 23 genes were encoded by the majority strand (Table 2 and Figure 1). The 13 PCGs ranged from 159 to 1731 bp. Of 22 tRNA genes, 8 were encoded by the L-strand and the remaining 14 by the H-strand. All tRNAs had the typical clover-leaf secondary structures observed in mitochondrial tRNA genes, except for trnS1 (AGN), which lacked a stable dihydrouridine (DHU) arm; this has been observed in several animals, including insect and brachyuran mitogenomes (Liu et al., 2015; Xin et al., 2017a, b). Figure 2 shows the relative synonymous codon usage (RSCU) of P. bidens. The codon usage was biased with a high frequency of AT compared to GC in the third codon position. The codon usage analysis revealed that the leucine 2 (Leu2), isoleucine (Ile), phenylalanine (Phe) codon families were most frequently utilized, while cysteine (Cys) family was the least used (Figure 3).
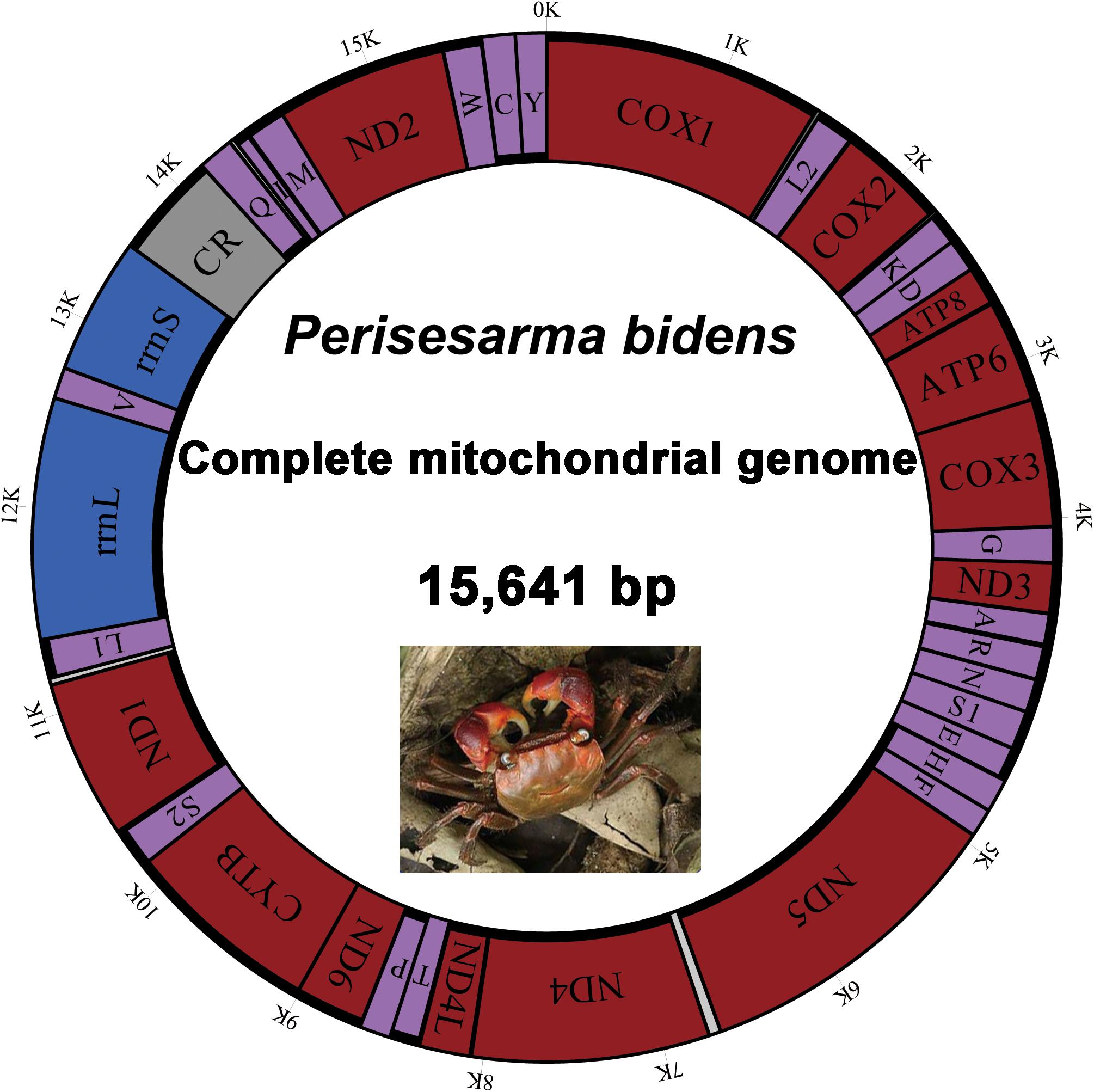
Figure 1. Map of the P. bidens mitogenome. Protein-coding and ribosomal genes are shown with standard abbreviations. Genes for tRNAs are abbreviated by single letters, with S1 = AGN, S2 = UCN, L1 = CUN, and L2 = UUR. CR, control region.
The gene order of P. bidens was identical to other Sesarmidae species in our study. In contrast to the inferred ancestral gene sequences of Pancrustaceans, where trnH was located between nad5 and nad4, here it was found between trnE and trnF. In Pancrustaceans, the tRNA gene sequences between CR and trnM was trnI-trnQ, but here was trnQ-trnI (Figure 4A).
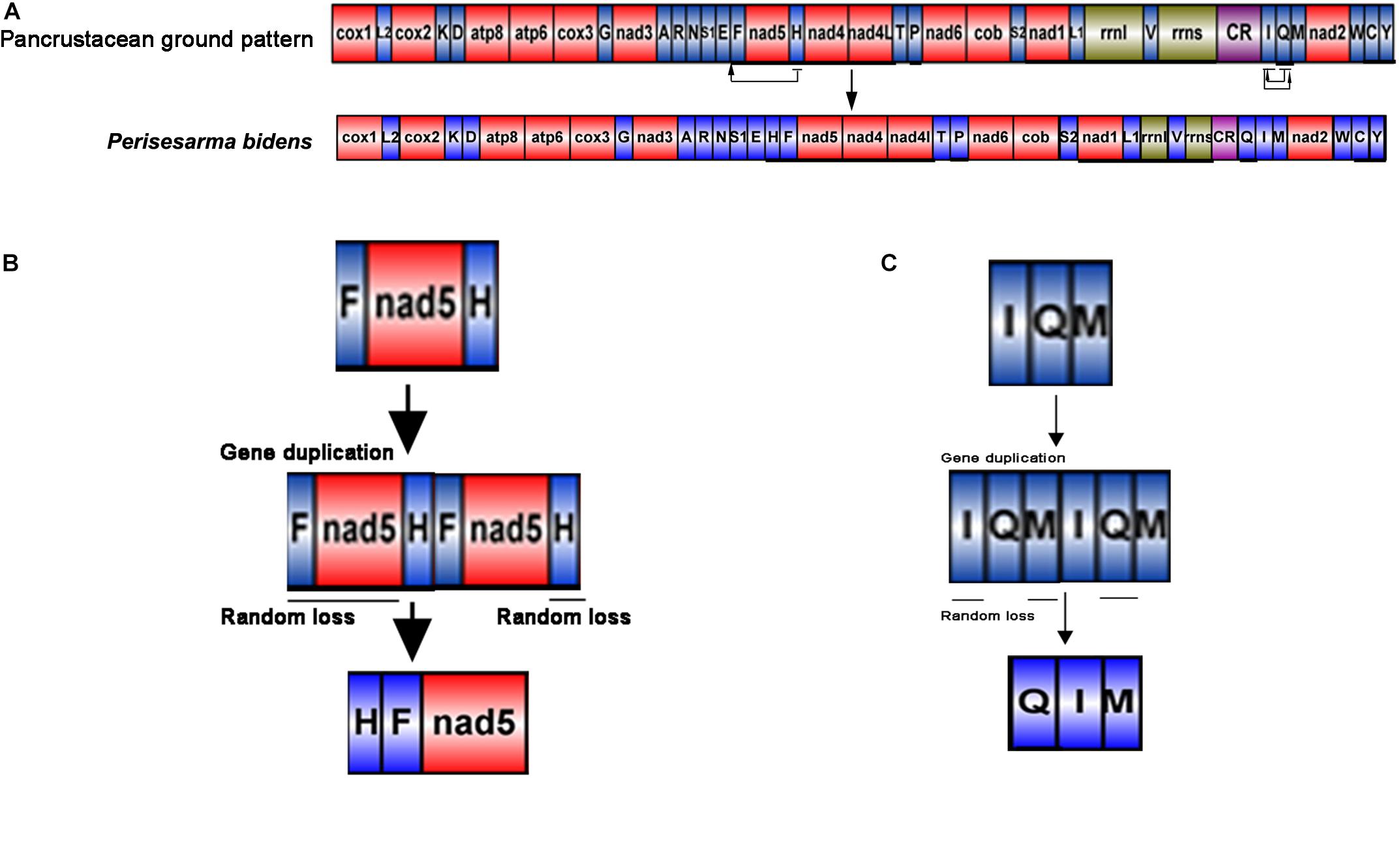
Figure 4. Generation of the P. bidens mitochondrial gene arrangement. The duplication/random loss, recombination, and duplication/non-random loss models were used to explain the principle of gene rearrangement. (A) Comparison of gene order in mitogenome of Perisesarma bidens and Pancrustacean ground pattern. tRNA genes are indicated by the singer letter IUPAC-IUB abbreviation with S1 = AGN, S2 = UCN, L1 = CUN, and L2 = UUR, where as protein and rRNA genes are labled with three letter codes. (B) Gene duplication occurred in trnF, nad5, and trnH, changing the arrangement of trnF-nad5-trnH to trnF-nad5-trnH -trnF-nad5-trnH. Then, the redundant trnF, nad5, and trnH genes were lost at random. Finally, the new gene order of trnH-trnF-nad5 was formed. (C) Gene duplication occurred in trnI, trnQ, and trnM, changing the arrangement of trnI-trnQ-trnM to trnI-trnQ-trnM-trnI-trnQ-trnM. Then, the redundant trnI, trnM, and trnQ genes were lost at random. Finally, the new gene order of trnQ-trnI-trnM was formed.
The duplication/random loss model was used to explain the rearrangements seen in Sesarmidae (Moritz and Brown, 1987; Macey et al., 1997; Boore and Brown, 1998). The movement of trnH can be explained as follows. First, gene duplication occurred in trnF, nad5, and trnH, changing the arrangement of trnF-nad5-trnH to trnF-nad5-trnH-trnF-nad5-trnH. Then, the redundant trnF, nad5, and trnH genes were lost at random. Finally, the new gene order of trnH-trnF-nad5 was formed (Figure 4B). The order principles of trnQ moving from the junction between trnI and trnM to between the CR and trnI could also be explained similarly (Figure 4C).
The gene orders of all species are shown in Figure 5. The gene sequences within 13 families were the same. The gene order pattern of Macrophthalmus japonicus (Ocypodoidea, Macrophthalmidae) was identical to that of other Varunidae. The gene orders of Damithrax spinosissimus (Majoidea, Mithracidae) and Dynomene pilumnoides (Dromioidea, Dynomenidae) were different, as were those of two Xenograpsidae crabs (Xenograpsus testudinatus and X. ngatama). However, two Majidae crabs (Maja squinado and M. crispata) had the same gene order. Interestingly, although there were only four species of Potamoide, each showed a different gene order.
The phylogenetic trees were constructed based on 13 PCGs under ML and BI methods, which resulted in congruent tree topologies, except for minor differences within “Grapsoidea + Ocypodoidea” (Figure 6). As shown in Figure 6, P. bidens formed a well-supported clade with Parasesarmops tripectinis (BP = 100; BPP = 1). (P. bidens + P. tripectinis) clade, (S. sinensis + S. neglectum) clade, (C. sinensis + M. depressus) clade were well supported with each other; these results were in accordance with the information provided by the same genes orders of P. bidens, P. tripectinis, S. sinensis, S. neglectum, C. sinensis, and M. depressus. Moreover, S. neglectum, M. depressus, and P. tripectinis all belonged to Sesarmidae (Park et al., 2018). Therefore, P. bidens, S. sinensis, and C. sinensis should belong to Sesarmidae rather than to Sesarminae. The species originally belonging to the Sesarminae should belong to the Sesarmidae. These results agree with previous analyses using the mitogenome of one species (Tang et al., 2017; Xin et al., 2017a, b).
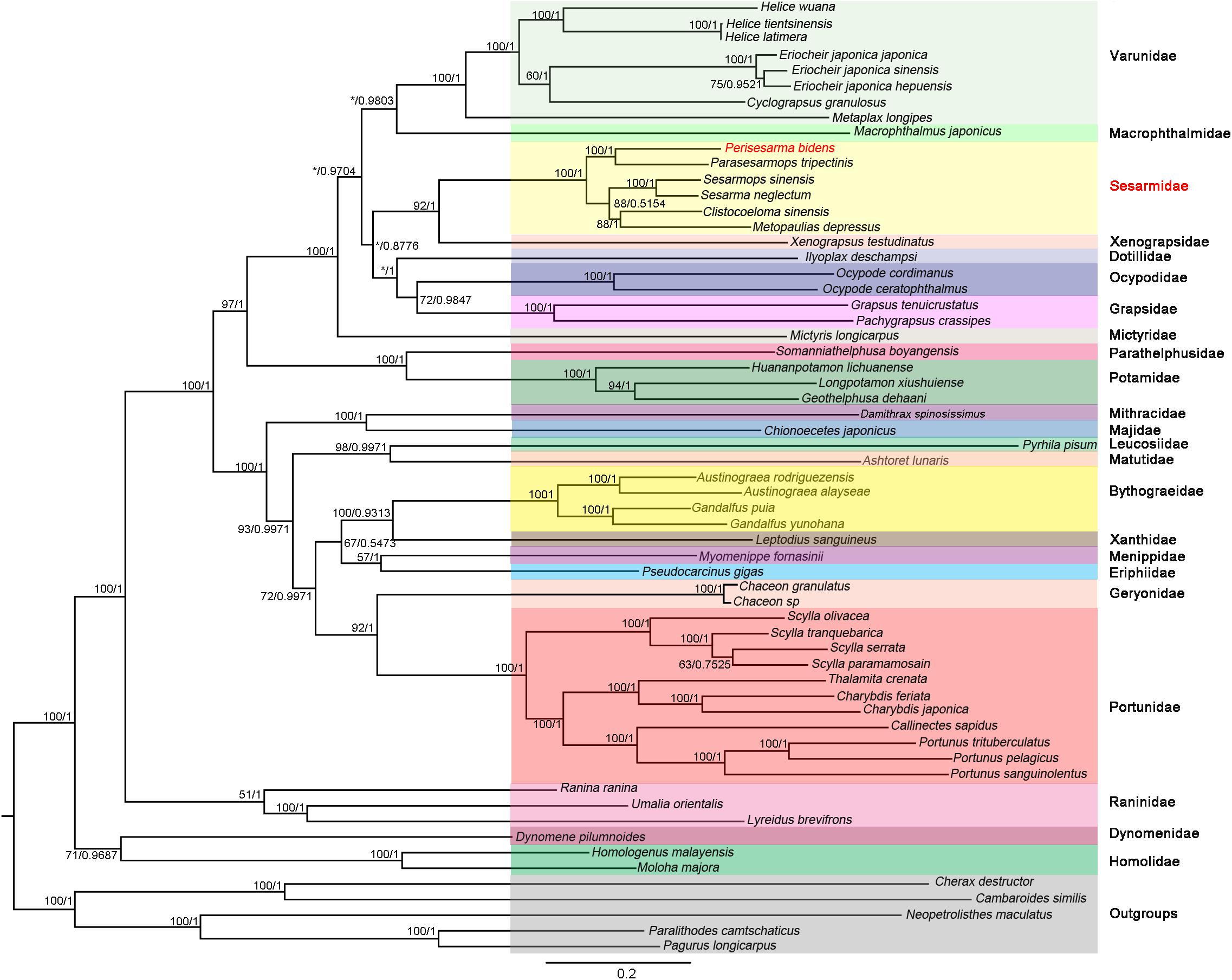
Figure 6. Phylogenetic trees were constructed using BI and ML methods based on NT dataset. Bootstrap values (BP) (IQ-Tree) and Bayesian posterior probability (BPP) of each node are shown as BP based on NT dataset/BPP based on NT dataset. C. destructor, C. similis, N. maculatus, P. camtschaticus, and P. longicarpus were used as outgroups. The supermatrix underlying this figure is as a Supplementary File.
In our study, two families (Potamidae and Parathelphusidae) were primarily freshwater crabs and were recognized as true heterotremes (Guinot et al., 2013). The systemic status of primary freshwater crabs had stimulated interest because of their high value and diversity (Cumberlidge et al., 2009; Klaus et al., 2010). The monophyly of Potamidae and Parathelphusidae was confirmed based on morphological and molecular analyses. However, there still were uncertainties regarding the phylogenetic placement of Potamidae and Parathelphusidae (Xing et al., 2017). Von Sternberg and Cumberlidge (2001) suggested that these two families Potamidae and Parathelphusidae should be placed in Thoracotremata. Here, the Thoracotremata contained Grapsoidea and Ocypodoidea crabs. Our results showed that four heterotreme crabs (Geothelphusa dehaani, Longpotamon xiushuiense, Huananpotamon lichuanense, and Somanniathelphusa boyangensis) were actually more closely associated with thoracotreme crabs, showing that Heterotremata was not monophyletic; this result was in accordance with that inferred from 23 brachyuran crabs, in which the author use the two mitogenomes (Ji et al., 2014). Within Podotremata, the clade was monophyletic. The six crabs formed a robust clade [(Homolidae + Dynomenidae) + Raninidae]. Within Heterotremata, the phylogenetic relationships were clear, with the exception of the four potamid crabs, which were outside of the heterotreme crabs.
The datasets generated for this study can be found in the GenBank accession no. KY808394.
Q-NL, B-PT, and X-MY conceived and designed the study. Z-ZX, Q-NL, Y-YT, and T-TY conducted the molecular work and data analysis. Z-ZX and Y-TL drafted the manuscript. Z-ZX, Q-NL, Y-YT, YS, D-ZZ, C-LZ, and T-TY prepared all figures and tables. Z-ZX and Q-NL performed the phylogenetic analyses. Z-ZX, Q-NL, B-PT, and X-MY contributed to drafting the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31672267), the Jiangsu Agriculture Science and Technology Innovation Fund (CX(18)3027), the China Postdoctoral Science Foundation (2018M642105), the Research Start-up Fund of Wenzhou Medical University (QTJ16017), the Opening Project of Zhejiang Provincial Top Key Discipline of Pharmaceutical Sciences (201713), the 16th Six Talents Peak Project of Jiangsu Province (NY-126), and the “Outstanding Young Talents” of YCTU for Q-NL, and sponsored by Qing Lan Project of Jiangsu Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.536640/full#supplementary-material
A, adenine; Atp6 and Atp8, genes for the ATPase subunits 6 and 8; BI, Bayesian inference; BP, Base pair; C, cytosine; Cox1-cox3, genes for cytochrome C oxidase subunits I–III; G, guanine; l-rRNA (large), rRNA subunit; Mitogenomes, Mitochondrial genomes; ML, maximum likelihood; mtDNA, mitochondrial DNA; Nad1–nad6 and nad4L, genes for NADH dehydrogenase subunits 1–6 and 4L; PCGs, protein-coding genes; rRNA, ribosomal RNA genes subunit; s-rRNA, (small); T, thymine; tRNAx, transfer RNA, where X is replaced by three letters amino acid code of the corresponding amino acid.
Ahyong, S. T., Lowry, J. K., Alonso, M., Bamber, R. N., Boxshall, G. A., Castro, P., et al. (2011). “Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness,” in Zootaxa, Vol. 3148, ed. Z.-Q. Zhang (Auckland: Magnolia Press), 165–191. doi: 10.11646/zootaxa.3148.1.33
Basso, A., Massimiliano, B., Marianna, P., Emilio, R., Tomaso, P., Enrico, N., et al. (2017). The highly rearranged mitochondrial genomes of the crabs Maja crispata and Maja squinado (Majidae) and gene order evolution in Brachyura. Sci. Rep. 7:4096.
Bergsten, J. (2005). A review of long-branch attraction. Cladistics 21, 163–193. doi: 10.1111/j.1096-0031.2005.00059.x
Boore, J. L., and Brown, W. M. (1998). Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 8, 668–674. doi: 10.1016/s0959-437x(98)80035-x
Brinkmann, H., and Philippe, H. (2008). Animal phylogeny and large-scale sequencing: progress and pitfalls. J. Syst. Evol. 46, 274–286. doi: 10.1017/9781316882313.013
Carpenter, K. E., and Niem, V. H. (1998). The living marine resources of the Western Central Pacific. Cephalop. Crustac. Holothur. Sharks 2, 687–1396.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Castro, P., Davie, P. J. F., Guinot, D., Schram, F. R., von Vaupel, and Klein, J. C. (2015). The Crustacea; Complementary to the Volumes Translated from the French of the Traite de Zoologie. Leiden: Brill, 163.
Cox, C. J., Foster, P. G., Hirt, R. P., Harris, S. R., and Embley, T. M. (2008). The archaebacterial origin of eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 105, 20356–20361. doi: 10.1073/pnas.0810647105
Cumberlidge, N., Ng, P. K. L., Yeo, D. C. J., Magalhães, C., Campos, M. R., Fernando, A., et al. (2009). Freshwater crabs and the biodiversity crisis: importance, threats, status, and conservation challenges. Biol. Conserv. 142, 1665–1673.
Dai, A. Y. (1999). Fauna Sinica. Arthropoda: Crustacea: Malacostraca: Decapoda: Parathelphusidae, Potamidae. Editorial Committee of Fauna Sinica, Academia Sinica. Beijing: Science Press, 501.
De Grave, S., Dean, P. N., Shane, A., Tin-Yam, C., Keith, C. A., Peter, D. C., et al. (2009). A classification of living and fossil genera of decapod crustaceans. Raffles Bull. Zool. 21, 1–109. doi: 10.5479/si.00963801.78-2851.1
Foster, P. G. (2004). Modeling compositional heterogeneity. Syst. Biol. 53, 485–495. doi: 10.1080/10635150490445779
Guinot, D., Tavares, M., and Castro, P. (2013). Significance of the sexual openings and supplementary structures on the phylogeny of brachyuran crabs (Crustacea, Decapoda, Brachyura), with new nomina for higher-ranked podotreme taxa. Zootaxa 3665, 1–414. doi: 10.11646/zootaxa.3665.1.1
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Ji, Y. K., An, W., Xiu-Ling, L., Dan-Hong, S., Yu-Hong, J., Jian-Jian, L., et al. (2014). Mitochondrial genomes of two brachyuran crabs (Crustacea: Decapoda) and phylogenetic analysis. J. Crustacean Biol. 34, 494–503. doi: 10.1163/1937240x-00002252
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: 2002. a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kitaura, J., Wada, K., and Nishida, M. (2002). Molecular phylogeny of grapsoid and ocypodoid crabs with special reference to the genera Metaplax and Macrophthalmus. J. Crustacean Biol. 22, 682–693. doi: 10.1651/0278-0372(2002)022[0682:mpogao]2.0.co;2
Klaus, S., Schubart, C. D., Streit, B., and Pfenninger, M. (2010). When Indian crabs were not yet Asian-biogeographic evidence for Eocene proximity of India and Southeast Asia. BMC Evol. Biol. 10:287. doi: 10.1186/1471-2148-10-287
Li, J. J., Shih, H. T., and Ng, P. K. L. (2019). Three new species and two new records of Parasesarma De Man, 1895 (Crustacea: Brachyura: Sesarmidae) from Taiwan and the Philippines from morphological and molecular evidence. Zool. Stud. 58:40.
Liu, Q. N., Zhang, H. B., Jiang, S., Fujun, X., Li, C.-F., Zhang, D., et al. (2015). The complete mitochondrial genome of Eriocheir japonica sinensis, (Decapoda: Varunidae) and its phylogenetic analysis. Biochem. Syst. Ecol. 62, 241–248. doi: 10.1016/j.bse.2015.09.008
Macey, J. R., Larson, A., Ananjeva, N. B., Fang, Z., and Papenfuss, T. J. (1997). Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 14, 91–104. doi: 10.1093/oxfordjournals.molbev.a025706
Moritz, C., and Brown, W. M. (1987). Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc. Natl. Acad. Sci. U.S.A. 84, 7183–7187. doi: 10.1073/pnas.84.20.7183
Ng, P. K. L., Li, J. J., and Shih, H. T. (2020). What is Sesarmops impressus (H. Milne Edwards, 1837) (Crustacea: Brachyura: Sesarmidae)? Zool. Stud. 59:27.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nylander, J. (2004). MrModeltest v2. Program Distributed by the Author, Vol. 2. Uppsala: Evolutionary Biology Centre, Uppsala University.
Park, Y. J., Park, C., Lee, S. H., Ko, H. S., Ullah, I., Hwang, U. W., et al. (2018). The complete mitochondrial genome sequence of the intertidal crab Parasesarma Tripectinis (Arthropoda, Decapoda, Sesarmidae). Mitochond. DNA Part B 3, 193–194. doi: 10.1080/23802359.2018.1437804
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schubart, C. D., Cuesta, J. A., Diesel, R., and Felder, D. L. (2000). Molecular phylogeny, taxonomy, and evolution of nonmarine lineages within the American grapsoid crabs (Crustacea: Brachyura). Mol. Phylogenet Evol. 15, 179–190. doi: 10.1006/mpev.1999.0754
Schubart, C. D., Cuesta, J. A., and Felder, D. L. (2002). Glyptograpsidae, a new brachyuran family from Central America: larval and adult morphology, and a molecular phylogeny of the Grapsoidea. J. Crustacean Biol. 22, 28–44. doi: 10.1651/0278-0372(2002)022[0028:ganbff]2.0.co;2
Shen, H., Braband, A., and Scholtz, G. (2013). Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol. Phylogenet. Evol. 66, 776–789. doi: 10.1016/j.ympev.2012.11.002
Shih, H. T., Hsu, P. Y., Shahdadi, A., Schubart, C. D., and Li, J. J. (2019). The synonymy of the supratidal crab species Parasesarma cognatum Rahayu & Li, 2013 with P. liho Koller, Liu & Schubart, 2010 (Decapoda: Brachyura: Sesarmidae) based on morphological and molecular evidence, with a note on P. paucitorum Rahayu & Ng, 2009. Zool. Stud. 58:21.
Tamura, K., Stecher, G., and Peterson, D. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, B. P., Liu, Y., Xin, Z.-Z., Zhang, D.-Z., Wang, Z.-F., Zhu, X. Y., et al. (2018). Characterisation of the complete mitochondrial genome of Helice wuana (Grapsoidea: Varunidae) and comparison with other Brachyuran crabs. Genomics 110, 221–230. doi: 10.1016/j.ygeno.2017.10.001
Tang, B. P., Xin, Z.-Z., Liu, Y., Zhang, D.-Z., Wang, Z.-F., Zhang, H.-B., et al. (2017). The complete mitochondrial genome of Sesarmops sinensis reveals gene rearrangements and phylogenetic relationships in brachyura. PLoS One 12:e0179800. doi: 10.1371/journal.pone.0179800
Tang, B. P., Zhou, K. Y., Song, D. Y., Yang, G., and Dai, A. Y. (2003). Molecular systematics of the Asian mitten crabs, genus Eriocheir (Crustacea: Brachyura). Mol. Phylogenet. Evol. 29, 309–316. doi: 10.1016/s1055-7903(03)00112-x
Theuerkauff, D., Rivera-Ingraham, G. A., Roques, J. A. C., Azzopardi, L., Bertini, M., Lejeune, M., et al. (2018). Salinity variation in a mangrove ecosystem: a physiological investigation to assess potential consequences of salinity disturbances on mangrove crabs. Zool. Stud. 57:36.
Von Sternberg, R., and Cumberlidge, N. (2001). On the Heterotreme-Thoracotreme distinction in the Eubrachyura de Saint Laurent, 1980 (Decapoda. Brachyura). Crustaceana 74, 321–338. doi: 10.1163/156854001300104417
Wägele, J. W., and Mayer, C. (2007). Visualizing differences in phylogenetic information content of alignments and distinction of three classes of long-branch effects. BMC Evol. Biol. 7:147. doi: 10.1186/1471-2148-7-147
Xia, X., and Xie, Z. (2001). DAMBE: 2001. software package for data analysis in molecular biology and evolution. J. Hered. 92, 371–373. doi: 10.1093/jhered/92.4.371
Xin, Z. Z., Liu, Y., Zhang, D.-Z., Chai, X.-Y., Wang, Z.-F., Zhang, H.-B., et al. (2017a). Complete mitochondrial genome of Clistocoeloma sinensis (Brachyura: Grapsoidea): gene rearrangements and higher-level phylogeny of the Brachyura. Sci. Rep. 7:4128.
Xin, Z. Z., Liu, Y., Zhang, D.-Z., Wang, Z. F., Zhang, H.-B., Tang, B.-P., et al. (2017b). Mitochondrial genome of Helice tientsinensis (Brachyura: Grapsoidea: Varunidae): gene rearrangements and higher-level phylogeny of the Brachyura. Gene 627, 307–314. doi: 10.1016/j.gene.2017.06.036
Xing, Y. H., Lijun, Z., Yue, H., Xiaoqi, W., Chen, Z., Huilun, Z., et al. (2017). Complete mitochondrial genomes from two species of Chinese freshwater crabs of the genus Sinopotamon recovered using next-generation sequencing reveal a novel gene order (Brachyura, Potamidae). ZooKeys 705, 41–60. doi: 10.3897/zookeys.705.11852
Keywords: mitochondrial genomes, phylogeny, gene order, crustacean, Perisesarma bidens
Citation: Li Y-T, Xin Z-Z, Tang Y-Y, Yang T-T, Tang B-P, Sun Y, Zhang D-Z, Zhou C-L, Liu Q-N and Yu X-M (2020) Comparative Mitochondrial Genome Analyses of Sesarmid and Other Brachyuran Crabs Reveal Gene Rearrangements and Phylogeny. Front. Genet. 11:536640. doi: 10.3389/fgene.2020.536640
Received: 20 February 2020; Accepted: 28 August 2020;
Published: 02 November 2020.
Edited by:
Denis Baurain, University of Liège, BelgiumReviewed by:
Beng Kah Song, Monash University Malaysia, MalaysiaCopyright © 2020 Li, Xin, Tang, Yang, Tang, Sun, Zhang, Zhou, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo-Ping Tang, Ym9wdGFuZ0AxNjMuY29t; Qiu-Ning Liu, bGl1cW5AeWN0dS5lZHUuY24=; eWN0dTE5ODlAMTYzLmNvbQ==; Xiao-Min Yu, eHhpYW8zMzZAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.