- 1Key Laboratory of Birth Defects and Related Diseases of Women and Children, Department of Pediatric Hematology/Oncology, West China Second Hospital, Sichuan University, Ministry of Education, Chengdu, China
- 2State Key Laboratory of Biotherapy, Department of Laboratory Medicine, Precision Medicine Center, West China Hospital, Sichuan University, Chengdu, China
- 3Department of Radiology, West China Hospital, Sichuan University, Chengdu, China
- 4Department of Hematology and Hematology Research Laboratory, West China Hospital, Sichuan University, Chengdu, China
- 5State Key Laboratory of Biotherapy, Department of Thoracic Oncology, Cancer Center, West China Hospital, Sichuan University, Chengdu, China
Through genome-wide association studies (GWAS), multiple inherited predispositions to acute lymphoblastic leukemia (ALL) have been identified in children. Most recently, a novel susceptibility locus at ERG was localized, exhibiting Hispanic-specific manner. In this study, we conducted a replication study to in all-age Chinese patients (N = 451), not only validating the novel ERG locus, but also systematically determining the impact of age on association status of the top GWAS signals. We found that single nucleotide polymorphisms at ARID5B, IKZF1, CEBPE, PIP4K2A were only significantly associated with ALL susceptibility in childhood patients with no BCR-ABL fusion, while GATA3 signal exhibited its significance in adults no matter carrying BCR-ABL fusion or not. Moreover, the novel ERG SNP can be validated in pediatric patients without both BCR-ABL and ETV6-RUNX1 fusion. Our finding suggests the modifying effects of age on genetic predisposition to ALL, and highlights the impact of ERG SNP in Chinese patients.
Introduction
Acute lymphoblastic leukemia (ALL) is a leading cause of disease-induced death, especially in children aged between 2 and 5 years old, and age is negatively related to prognosis and survival rates of ALL patients according to the clinical statistics. The inferior prognosis of adult ALL patients is likely to be multifactorial, including the age-related differences in leukemia blasts and host genomic alterations. Leukemogenesis mechanisms were considered to be different between children and adults. For example, more frequent somatic BCR-ABL fusion (also known as Philadelphia chromosome positive, Ph+) were observed in adults’ (Inaba et al., 2013). Additionally, racial disparities were also noticed in terms of incidence and treatment outcomes, highlighting the importance of age- and ethnicity- related study on leukemogenesis and clinical practice.
By conducting genome-wide association studies (GWASs), we and other independent groups identified multiple top inherited predispositions to acute lymphoblastic leukemia (ALL) susceptibility, including single nucleotide polymorphisms (SNPs) at ARID5B, IKZF1, GATA3 and etc. (Papaemmanuil et al., 2009; Trevino et al., 2009; Perez-Andreu et al., 2013, 2015; Xu et al., 2013, 2015; Vijayakrishnan and Studd, 2018; Wiemels et al., 2018). Association of these SNPs are greatly impacted by clinical characteristics (e.g., age, ethnicity, and subtypes) (Xu et al., 2012; Perez-Andreu et al., 2015; Liao et al., 2016). For instance, we and another independent group noticed that association of ARID5B and GATA3 SNPs with ALL risk were oppositely influenced by age in Caucasians and Hispanics (Migliorini et al., 2013; Xu et al., 2013). Also, we recently identified a novel locus at rs2836365 in ERG gene, which tends to exhibit Hispanic specific manner and varied in different subtypes (Qian et al., 2018). This novel site has been recently validated in independent studies (de Smith et al., 2019; Vijayakrishnan et al., 2019), but predominately performed in childhood without BCR-ABL fusion, with limited systematical investigation in Ph+ adolescents/adults and Chinese patients.
In this study, we sought to investigate association of the reported top GWAS signals (i.e., ARID5B, IKZF1, GATA3, PIP4K2A, CEBPE, and ERG) with ALL susceptibility in all-age Chinese patients, and also estimate the impact of BCR-ABL fusion and age at diagnosis.
Materials and Methods
Subjects and Genotyping
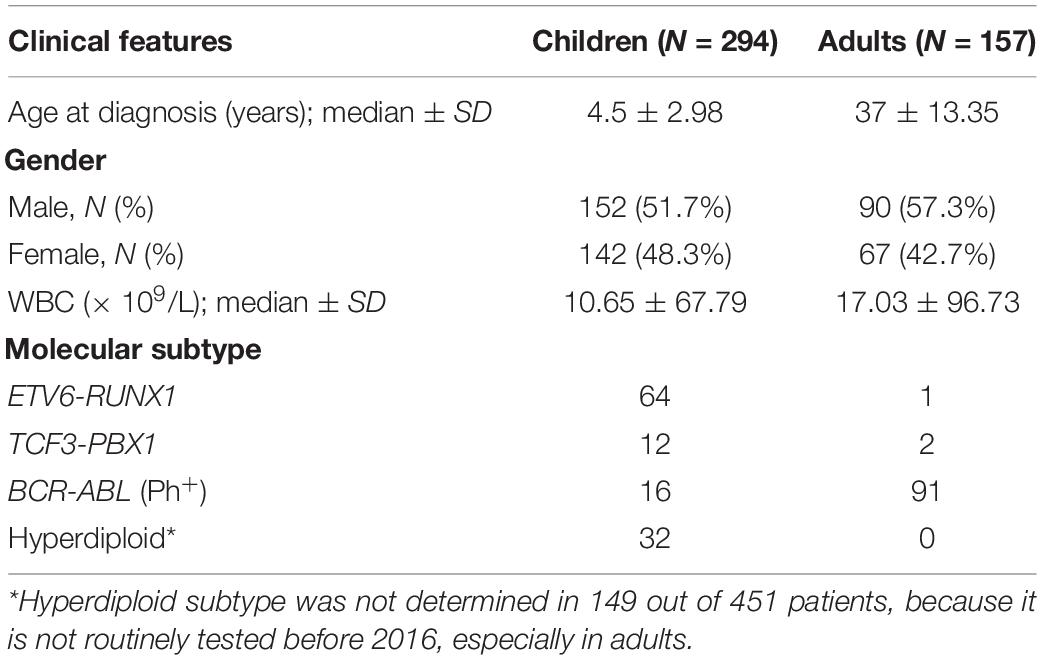
Peripheral blood was obtained from 456 non-ALL individuals, as well as 451 all-age B-lineage ALL patients, who were treated with standard protocol in Department of Hematology/Oncology, West China Hospital and West China Second Hospital (e.g., CCGC-ALL2015, registered in http://www.chictr.org.cn/with ID: ChiCTR-IPR-14005706) (Zhu et al., 2018; Cao et al., 2020). Clinical information of each patient was obtained from the electronic records system at our hospitals, including gender, age and white blood cell (WBC) at diagnosis, molecular subtypes. Fusion-based molecular subtypes were determined by Fluorescence in situ hybridization, while hyperdiploid was determined by flow cytometry-based DNA index. Patients (14 years is considered as children, while the rest were considered as adolescents/adults (following referred as “Adults”) in this study.
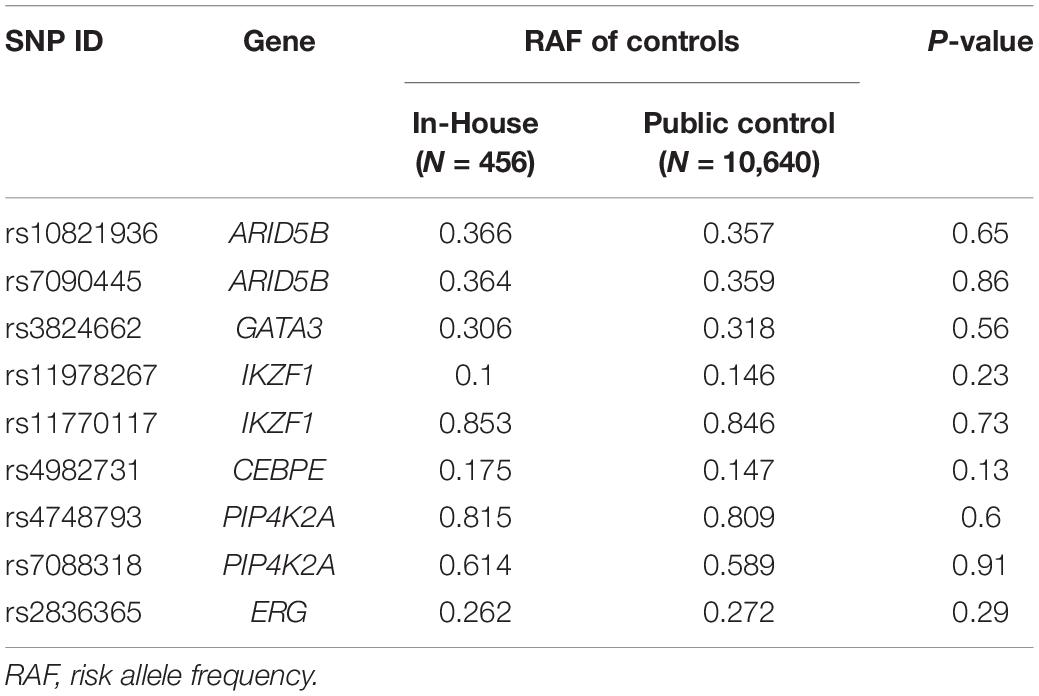
Nine SNPs at 6 loci (ARID5B, IKZF1, GATA3, PIP4K2A, CEBPE, and ERG) were directly genotyped through Sanger sequencing. Additionally, we also retrieved genotype information of a large Chinese Han population from the public dataset to increase the statistical power (Cai et al., 2017). After filtered out the individuals with missing information of either SNPs analyzed in this study, genotype information of 10,640 out of 11,670 individuals from this public database were used for further association analyses. Comparison were conducted between these two control cohorts in terms of risk allele frequencies (RAF). Because the prevalence of patients with ALL is less than 1 in 10,000 in China, these two control cohorts were combined and considered as non-ALL controls (totally 11,096) for SNPs with no significant difference.
This study was approved by Ethnics Committee of West China Hospital and West China Second Hospital, and informed consent was obtained from patients or their guardians, as appropriate.
Statistical Analysis
All SNPs have passed the quality control based on call rate of the SNPs (at least 95% patients have been successfully genotyped for each SNP), and Hardy-Weinberg equilibrium (P > 0.05 for χ2-test). The association of each SNP with ALL susceptibility was estimated by comparing the genotype frequency between ALL cases and non-ALL controls by using logistic regression model. P-value, odds ratio (OR) and 95% confidence interval (95% CI) was estimated by using R (version 3.5), and a two-sided P < 0.05 was considered as statistically significant.
Results
Totally 451 B lineage all-age ALL patients were included in this study, and were subsequently divided into two age groups, including 294 childhood (median age = 4.5; ≤ 14 years), and 157 adolescents/adults (median age = 37; range 14–68). Baseline characteristics of ALL patients are summarized in Table 1. BCR-ABL fusion has been taken it into account in the association test, because it is enriched in adults and barely considered in previous association studies. Totally nine SNPs were directly genotyped in all cases and 456 non-ALL controls, with overall call rate >95%. To increase the statistical power, genotypes of these nine SNPs were also retrieved from public dataset with 10,640 Han Chinese, and compared with our non-ALL controls in terms of risk allele frequencies (RAF). No significantly difference were observed in any of these SNPs, we therefore combined the two control cohorts (N = 11,096) to conduct association analyses (Table 2).
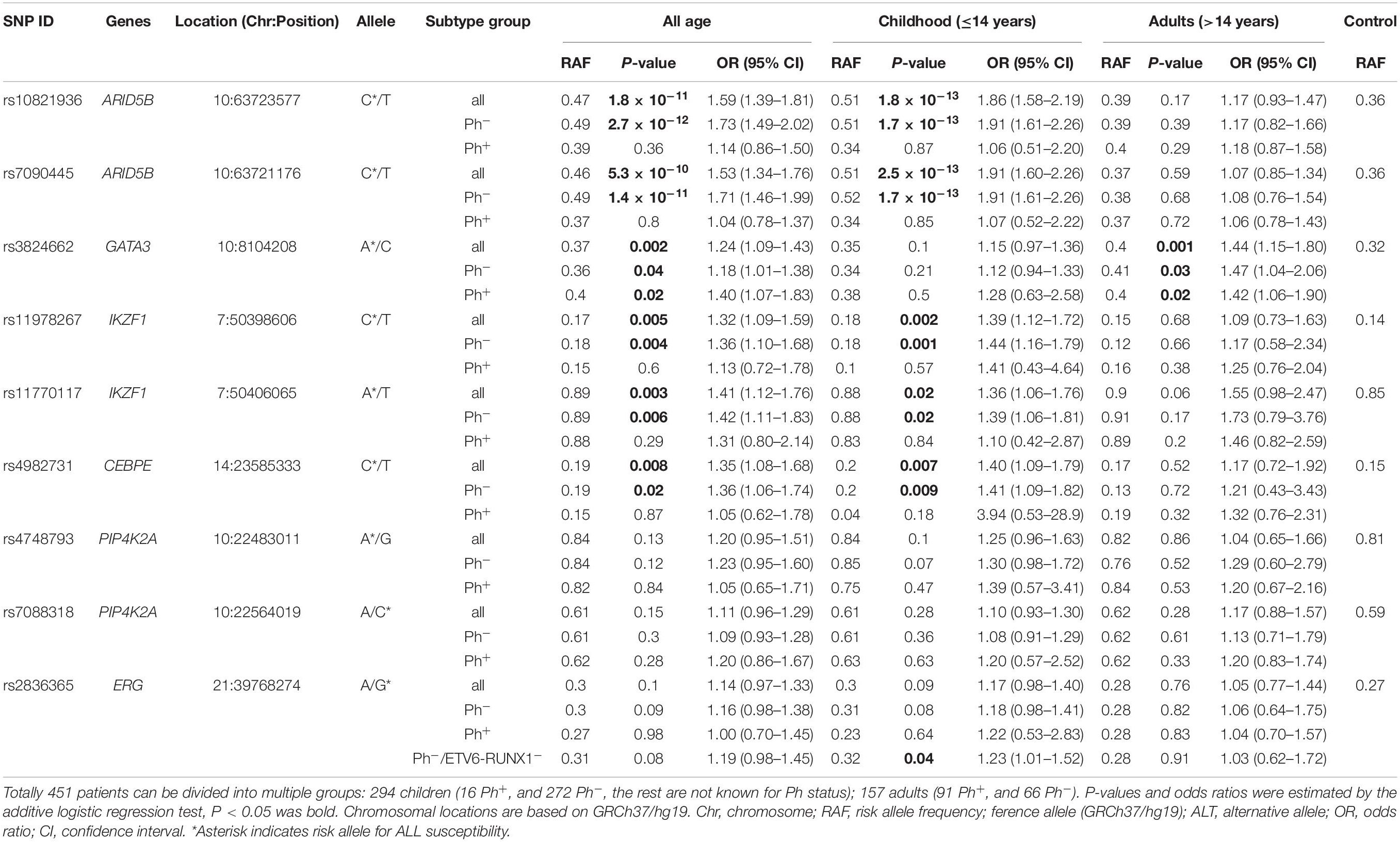
After comparing the genotype frequency between ALL cases and non-ALL controls, six SNPs at 4 loci (i.e., GATA3, ARID5B, IKZF1, and CEBPE) were significantly associate with ALL susceptibility in all-age patients (Table 3). rs10821936 at ARID5B locus [P = 1.8 × 10–11, OR = 1.59 (1.39–1.81)] was in high lineage disequilibrium (r2 = 0.98) with rs7090445 [P = 5.3 × 10–10, OR = 1.53 (1.34–1.76)], which was considered to be the potential causal variant for ARID5B locus (Studd et al., 2017), whereas rs11770117 [P = 0.003, OR = 1.41 (1.12–1.76)] at IKZF1 exhibit as an independent signal even after adjust for rs11978267 [P = 0.005, OR = 1.32 (1.09–1.59)] with Padjust = 0.02, which is consistent with our previous observation in Hispanics (Xu et al., 2013). when BCR-ABL fusion was taken into accounts, SNPs at ARID5B, IKZF1 and CEBPE loci lost their significance in Ph+ patients, while risk allele of rs3824662 at GATA3 locus was overrepresented in both Ph+ patients [P = 0.02, OR = 1.40 (1.07–1.83)] and Ph– patients [P = 0.04, OR = 1.18 (1.01–1.38)] (Table 3).
We next performed association analyses in two age groups separately. Interestingly, ARID5B and GATA3 SNPs were only significantly associated with ALL susceptibility in childhood and adolescent/adult patients, respectively (Table 3). Additionally, the independent IKZF1 signal exhibit its association in both age group, with even higher odds ratio in adult than childhood Ph– patients (1.55 vs. 1.36), while the top IKZF1 SNP (i.e., rs11978267) was only significant in childhood Ph– patients (Table 3). SNPs at PIP4K2A locus also tend to be associated with ALL susceptibility in childhood patients, especially with hyperdiploid subtype [e.g., rs4748793, P = 0.02, OR = 1.93 (1.09–3.40)].
Particularly, we focused on the novel Hispanic-specific ALL risk signal at ERG locus, observing only marginally significant association of this SNP with ALL susceptibility in childhood patients [P = 0.09, OR = 1.17 (0.98–1.40)], but not adults (P = 0.76). Because RAF of rs2836365 was observed to be under-represented in patients with ETV6-RUNX1 fusion (Qian et al., 2018), the association reached significance after excluding ETV6-RUNX1 and Ph+ subtypes of patients [P = 0.04, OR = 1.23 (1.01–1.52)], Next, RAF of rs2836365 was determined in each ALL subtype and age group. Consistent with observation in Hispanics (Qian et al., 2018), enriched risk allele was observed in TCF3-PBX1 subtype at rs2836365 (RAF = 0.36, Figure 1). However, the difference was not significant probably because of the small sample size.
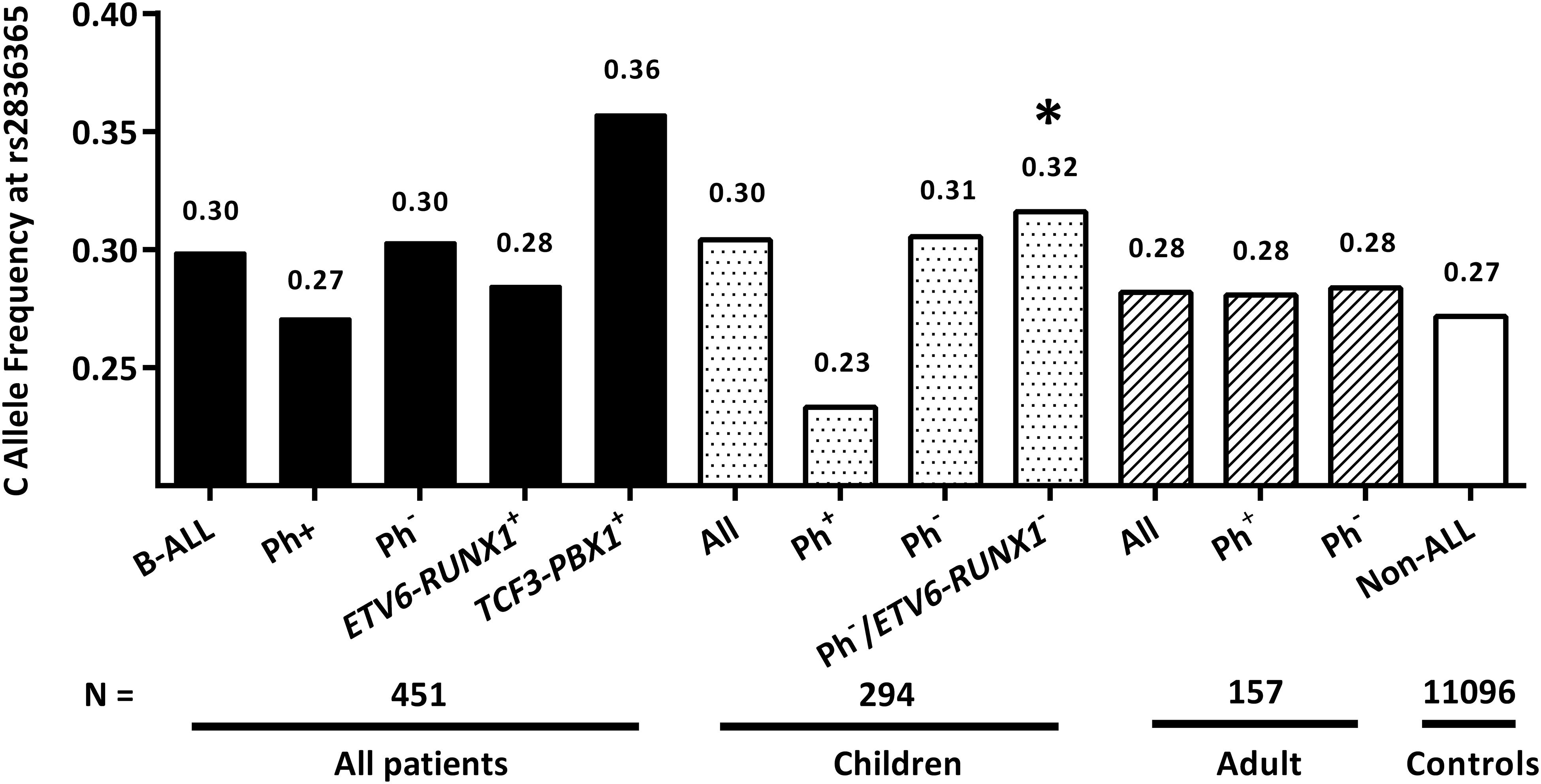
Figure 1. The frequency of ERG SNP in age-aged ALL patients. Risk allele frequencies of rs2836365 were illustrated in all patients/non-ALL controls, and separately in each group divided in terms of age and subtypes. Logistic regression tests were conducted by comparing risk allele frequencies of rs2836365 in each patient group with non-ALL controls. ∗P < 0.05.
Discussion
Racial differences in cancer risk and survivals are well documented, but with poor biological basis explored (Schafer and Hunger, 2011). Although increased genomic screenings were conducted in non-Caucasians, the most comprehensive records and novel findings in cancer genomics are still predominated based on Caucasian but limited in Chinese, including ALL.
In this study, we systematically analyzed the association status of the reported top and novel GWAS signals in all-age Chinese ALL patients, demonstrating the large effect of age on genetic predispositions to ALL susceptibility. The top signal at ARID5B locus exhibited the strongest association only in childhood patients, which further support its importance in Chinese patients (Wang et al., 2013). However, no significant association was found in adolescents/adults, which is consistent with the findings in Caucasians (Peyrouze et al., 2012; Burmeister et al., 2014), as well as the fact of negative correlation of ARID5B SNP effect with age at diagnosis in pediatric patients (Xu et al., 2013). In contrast, only GATA3 SNP exhibited its significance in adolescents/adults, validating previous Caucasian population based GWAS performed by us and another independent group. On the other hand, we validated the association of IKZF1-rs11978267 and CEBPE SNP with ALL risk in childhood patients, standing by the side of replication study with positive validation result (Urayama et al., 2018), rather than the race-specific assumption (Wang et al., 2013). Moreover, at least two independent association signals were located at IKZF1 locus, and higher impact of rs11770117 than rs11978267 was noticed in Hispanics but not Caucasians. In this case, IKZF1-rs11770117 was also evaluated in our cohort, and exhibits not only independent association with childhood ALL susceptibility after adjusting for rs11978267, but also marginally significance in adolescents/adults, indicating different regulation patterns of IKZF1 may be involved in leukemogenesis between children and adults.
Effects of the reported inherited predispositions to ALL varied among different subtypes. For instance, ARID5B and PIP4K2A SNPs were more enriched in hyperdiploid subtype, which was also validated in our study. However, Ph+ subtype has barely mentioned because of its low frequency in childhood ALL. In this study, 70 Ph+ patients were enrolled for association analyses, and mostly in adolescents/adults. Although no clear conclusion can be drawn in childhood ALL due to the limited statistical power, we observed significant association of GATA3 SNP with ALL risk in adults with Ph+ subtype, indicating its broad effect in both Ph+ and Ph-like ALL in elder patients (Perez-Andreu et al., 2013). To our knowledge, this is the first report that established the correlation of GATA3 SNP with ALL risk in adult Ph+ patients, which therefore requiring more evidences in independent validation cohorts.
Importantly, we recently identified a novel Hispanic-specific signal at ERG locus, which is highly related to Native Americans ancestry (Qian et al., 2018). Since ancestors of Native Americans is considered to descend from a single founding population initially split from East Asians (Moreno-Mayar et al., 2018), we sought to evaluate its effect in Chinses ALL patients. Actually, the trend of ERG-rs2836365 RAF distribution in each subtype is similar to that in childhood Hispanic patients, which is high in TCF3-PBX1+ subtype and low in ETV6-RUNX1– subtype, while no difference was observed in adults with any subtypes. Association of ERG-rs2836365 can only reach statistically significance in childhood patients with Ph–/ETV6-RUNX1–, probably because of the small sample size.
Collectively, our results not only described association of the top GWAS hits with ALL susceptibility in all-age Chinese patient (including Ph+ patients), but also provided independent confirmation that the novel signal at ERG locus conferred risk of childhood ALL in an age and subtype specific manner. Additionally, the fact of consistent significant race-specific association of ERG and IKZF1-rs11770117 with ALL susceptibility, suggests the similarity of East Asians and Hispanics for leukemogenesis. However, since the relatively small sample size in our study, additional independent cohorts with larger sample size are needed to confirm the effect of ERG SNP, especially in different molecular subtypes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of West China Hospital and West China Second Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
YS, YiZ, and XL designed and supervised this study. FL, DY, JZ, ZD, and YW conducted the experiments and data analyses, and interpreted the data. YY, YQ, WZ, YaZ, BY, LW, and JG collected the clinical information. XL contributed to the conception of the study and drafted the manuscript. All authors contributed to writing of the manuscript and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81903735, 81902872, and 81902437), and the National Key Research and Development Program of China [No. 2016YFC0905000 (2016YFC0905001)].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Burmeister, T., Bartels, G., Groger, D., Trautmann, H., Schwartz, S., and Lenz, K. (2014). Germline variants in IKZF1, ARID5B, and CEBPE as risk factors for adult-onset acute lymphoblastic leukemia: an analysis from the GMALL study group. Haematologica 99, e23–e25. doi: 10.3324/haematol.2013.090720
Cai, N., Bigdeli, T. B., Kretzschmar, W. W., Li, Y., Liang, J., and Hu, J. (2017). 11,670 whole-genome sequences representative of the Han Chinese population from the CONVERGE project. Sci. Data 4:170011. doi: 10.1038/sdata.2017.11
Cao, M., Yin, D., Qin, Y., Liao, F., Su, Y., and Xia, X. (2020). Screening of novel pharmacogenetic candidates for Mercaptopurine-induced toxicity in patients with acute lymphoblastic leukemia. Front. Pharmacol. 11:267. doi: 10.3389/fphar.2020.00267
de Smith, A. J., Walsh, K. M., Morimoto, L. M., Francis, S. S., Hansen, H. M., and Jeon, S. (2019). Heritable variation at the chromosome 21 gene ERG is associated with acute lymphoblastic leukemia risk in children with and without Down syndrome. Leukemia 33, 2746–2751. doi: 10.1038/s41375-019-0514-9
Inaba, H., Greaves, M., and Mullighan, C. G. (2013). Acute lymphoblastic leukaemia. Lancet 381, 1943–1955. doi: 10.1016/S0140-6736(12)62187-4
Liao, F., Yin, D., Zhang, Y., Hou, Q., Zheng, Z., Yang, L., et al. (2016). Association between PIP4K2A polymorphisms and acute lymphoblastic leukemia susceptibility. Medicine 95:e3542. doi: 10.1097/md.0000000000003542
Migliorini, G., Fiege, B., Hosking, F. J., Ma, Y., Kumar, R., and Sherborne, A. L. (2013). Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood 122, 3298–3307. doi: 10.1182/blood-2013-03-491316
Moreno-Mayar, J. V., Potter, B. A., Vinner, L., Steinrucken, M., Rasmussen, S., and Terhorst, J. (2018). Terminal pleistocene alaskan genome reveals first founding population of Native Americans. Nature 553, 203–207. doi: 10.1038/nature25173
Papaemmanuil, E., Hosking, F. J., Vijayakrishnan, J., Price, A., Olver, B., and Sheridan, E. (2009). Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 41, 1006–1010. doi: 10.1038/ng.430
Perez-Andreu, V., Roberts, K. G., Harvey, R. C., Yang, W., Cheng, C., and Pei, D. (2013). Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat. Genet. 45, 1494–1498. doi: 10.1038/ng.2803
Perez-Andreu, V., Roberts, K. G., Xu, H., Smith, C., Zhang, H., and Yang, W. (2015). A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood 125, 680–686. doi: 10.1182/blood-2014-09-595744
Peyrouze, P., Guihard, S., Grardel, N., Berthon, C., Pottier, N., and Pigneux, A. (2012). Genetic polymorphisms in ARID5B, CEBPE, IKZF1 and CDKN2A in relation with risk of acute lymphoblastic leukaemia in adults: a group for research on adult acute Lymphoblastic leukaemia (GRAALL) study. Br. J. Haematol. 159, 599–602. doi: 10.1111/bjh.12063
Qian, M., Xu, H., Perez-Andreu, V., Roberts, K. G., Zhang, H., and Yang, W. (2018). Novel susceptibility variants at the ERG locus for childhood acute lymphoblastic leukemia in hispanics. Blood 133, 724–729. doi: 10.1182/blood-2018-07-862946
Schafer, E. S., and Hunger, S. P. (2011). Optimal therapy for acute lymphoblastic leukemia in adolescents and young adults. Nat. Rev. Clin. Oncol. 8:417. doi: 10.1038/nrclinonc.2011.77
Studd, J. B., Vijayakrishnan, J., Yang, M., Migliorini, G., and Paulsson, K. (2017). Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2. Nat. Commun. 8:14616. doi: 10.1038/ncomms14616.
Trevino, L. R., Yang, W., French, D., Hunger, S. P., Carroll, W. L., and Devidas, M. (2009). Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 41, 1001–1005. doi: 10.1038/ng.432
Urayama, K. Y., Takagi, M., Kawaguchi, T., Matsuo, K., Tanaka, Y., and Ayukawa, Y. (2018). Regional evaluation of childhood acute lymphoblastic leukemia genetic susceptibility loci among Japanese. Sci. Rep. 8:789. doi: 10.1038/s41598-017-19127-7
Vijayakrishnan, J., Qian, M., Studd, J. B., Yang, W., Kinnersley, B., and Law, P. J. (2019). Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat. Commun. 10:5348. doi: 10.1038/s41467-019-13069-6
Vijayakrishnan, J., and Studd, J. (2018). Genome-wide association study identifies susceptibility loci for B-cell childhood acute lymphoblastic leukemia. Nat. Commun. 9:1340. doi: 10.1038/s41467-018-03178-z
Wang, Y., Chen, J., Li, J., Deng, J., Rui, Y., and Lu, Q. (2013). Association of three polymorphisms in ARID5B, IKZF1 and CEBPE with the risk of childhood acute lymphoblastic leukemia in a Chinese population. Gene 524, 203–207. doi: 10.1016/j.gene.2013.04.028
Wiemels, J. L., Walsh, K. M., De Smith, A. J., Metayer, C., and Gonseth, S. (2018). GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nat. Commun. 9:286. doi: 10.1038/s41467-017-02596-9
Xu, H., Cheng, C., Devidas, M., Pei, D., Fan, Y., and Yang, W. (2012). ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 30, 751–757. doi: 10.1200/jco.2011.38.0345
Xu, H., Yang, W., Perez-Andreu, V., Devidas, M., Fan, Y., and Cheng, C. (2013). Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J. Natl. Cancer Inst. 105, 733–742. doi: 10.1093/jnci/djt042
Xu, H., Zhang, H., Yang, W., Yadav, R., Morrison, A. C., and Qian, M. (2015). Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nat. Commun. 6:7553. doi: 10.1038/ncomms8553
Zhu, Y., Yin, D., Su, Y., Xia, X., Moriyama, T., and Nishii, R. (2018). Combination of common and novel rare NUDT15 variants improves predictive sensitivity of thiopurine-induced leukopenia in children with acute lymphoblastic leukemia. Haematologica 103, e293–e295. doi: 10.3324/haematol.2018.187658
Keywords: acute lymphoblastic leukemia, genetic susceptibility, ERG, all-age patients, subtype specific
Citation: Liao F, Ye Y, Yin D, Qin Y, Zhao J, Zhang W, Zhang Y, Deng Z, Wang Y, Ying B, Wang L, Gao J, Shu Y, Zhu Y and Lu X (2020) Validations of Top and Novel Susceptibility Variants in All-Age Chinese Patients With Acute Lymphoblastic Leukemia. Front. Genet. 11:1004. doi: 10.3389/fgene.2020.01004
Received: 07 August 2019; Accepted: 06 August 2020;
Published: 25 August 2020.
Edited by:
Annalisa Lonetti, University of Bologna, ItalyReviewed by:
Maoxiang Qian, Fudan University, ChinaWentao Yang, St. Jude Children’s Research Hospital, United States
Copyright © 2020 Liao, Ye, Yin, Qin, Zhao, Zhang, Zhang, Deng, Wang, Ying, Wang, Gao, Shu, Zhu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Shu, shuyang1986@gmail.com; Yiping Zhu, yipingzhu428@yahoo.com; Xiaoxi Lu, lu_helena@sina.com
†These authors have contributed equally to this work
 Fei Liao
Fei Liao Yuanxin Ye2†
Yuanxin Ye2† Dandan Yin
Dandan Yin Yun Qin
Yun Qin Yuelan Wang
Yuelan Wang Binwu Ying
Binwu Ying Yang Shu
Yang Shu