- 1Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
- 2Federal Research Center for Bast Fiber Crops, Torzhok, Russia
- 3Moscow Institute of Physics and Technology, Dolgoprudny, Russia
In the present work, a highly pathogenic isolate of Fusarium oxysporum f. sp. lini, which is the most harmful pathogen affecting flax (Linum usitatissimum L.), was sequenced for the first time. To achieve a high-quality genome assembly, we used the combination of two sequencing platforms – Oxford Nanopore Technologies (MinION system) with long noisy reads and Illumina (HiSeq 2500 instrument) with short accurate reads. Given the quality of DNA is crucial for Nanopore sequencing, we developed the protocol for extraction of pure high-molecular-weight DNA from fungi. Sequencing of DNA extracted using this protocol allowed us to obtain about 85x genome coverage with long (N50 = 29 kb) MinION reads and 30x coverage with 2 × 250 bp HiSeq reads. Several tools were developed for genome assembly; however, they provide different results depending on genome complexity, sequencing data volume, read length and quality. We benchmarked the most requested assemblers (Canu, Flye, Shasta, wtdbg2, and MaSuRCA), Nanopore polishers (Medaka and Racon), and Illumina polishers (Pilon and POLCA) on our sequencing data. The assembly performed with Canu and polished with Medaka and POLCA was considered the most full and accurate. After further elimination of redundant contigs using Purge Haplotigs, we achieved a high-quality genome of F. oxysporum f. sp. lini with a total length of 59 Mb, N50 of 3.3 Mb, and 99.5% completeness according to BUSCO. We also obtained a complete circular mitochondrial genome with a length of 38.7 kb. The achieved assembly expands studies on F. oxysporum and plant-pathogen interaction in flax.
Introduction
Flax (Linum usitatissimum L.) is widely used for the production of seeds and fiber. Flax seeds are rich in healthy alpha-linolenic acid (omega-3), lignans, and soluble dietary fibers. They are of great medicinal and nutraceutical value (Goyal et al., 2014; Kezimana et al., 2018; Cullis, 2019) and are potential functional animal feed for improving reproductive capacity and quality of meat, eggs, and milk (Yi et al., 2014; Dutra et al., 2019; Head et al., 2019; Isenberg et al., 2019; Marino et al., 2019; Mattioli et al., 2019). Linseed oil is used in the production of paints, enamels, and resins (Baroncini et al., 2016; Fombuena et al., 2019), while flax fiber is used in textile and composite industries (Muir and Westcott, 2003; Costa et al., 2018; Fombuena et al., 2019; Hu et al., 2019). Fungal pathogens cause diseases in flax that have the greatest negative impact on yield and quality of products worldwide. Fusarium oxysporum f. sp. lini is responsible for one of the most widespread diseases of flax – fusarium wilt, which leads to estimated yield losses of 20% and in some cases up to 100% (Nyvall, 1989). The pathogen is soil-borne, and infection occurs mainly through the roots. Then it spreads inside the vascular tissues, causing water and nutrient blocking, and eventually leading to wilting, yellowing and browning of top parts of the plants, and finally death (Nair and Kommedahl, 1957; Kroes et al., 1998; Michielse and Rep, 2009). Isolates of F. oxysporum f. sp. lini are varied and can differ in morphology, physiology, and pathogenicity (Kommedahl et al., 1970; Saharan et al., 2005; Loshakova et al., 2014). Molecular genetic studies are the basis for revealing the origins of the diversities in these traits. In the present study, using the combination of Oxford Nanopore Technologies (ONT) and Illumina platforms, a highly pathogenic isolate of F. oxysporum f. sp. lini, the most harmful pathogen of flax, was sequenced for the first time.
Materials and Methods
Materials
Pathogenic isolate #39 of F. oxysporum f. sp. lini was provided by the Institute for Flax (Torzhok, Russia). The fungal mycelium was grown on potato dextrose agar (Alpha Biosciences, United States) for 3 weeks.
DNA Extraction and Purification
Using a sterile scalpel, 1 g of the upper layer of agar with germinated mycelia of F. oxysporum was collected and triturated in a mortar with liquid nitrogen. Then, 10 ml of a Carlson lysis buffer [100 mM Tris–HCl pH 9.5 (VWR Life Science, United States); 2% CTAB (VWR Life Science); 1.4 M NaCl (Scharlab, Spain); 1% PEG 8000 (PanReac AppliChem, Germany); 20 mM EDTA (Promega, United States)] that was prewarmed to 65°C and supplemented with 20 μl of β-mercaptoethanol (Bio-Rad, United States) and 0.1 g of PVP K30 (PanReac AppliChem) were added. The homogenate was incubated at 65°C for 1 h 30 min, with stirring every 15 min. Next, an equal volume of chloroform (Acros Organics, United States) was added to the homogenate, stirred on a Thermolyne Maxi Mix III Type 65800 shaker (Thermo Fisher Scientific, United States) at 800 rpm for 10 min, followed by centrifugation at 12000 g for 10 min at 4°C. The aqueous phase was transferred to a clean tube with the addition of 0.2 volume of 5x CTAB buffer (5% CTAB, 350 mM EDTA) and incubated at 65°C for 10 min. After that, an equal volume of chloroform was introduced, stirred on a shaker for 10 min, and centrifuged at 12000 g for 10 min at 4°C. The aqueous phase was transferred to clean tubes with the addition of 0.7 volume of cold isopropanol (VWR Chemicals, United States), stirred 20 times, and incubated at 72°C for 20 min. It was then centrifuged at 13000 g for 10 min at 4°C. Next, the alcohol was collected gently without disturbing the precipitate. The DNA pellet was air-dried for 5 min and dissolved in 2 ml of prewarmed 60°C G-buffer from the Blood & Cell Culture DNA Mini Kit (Qiagen, United States) and incubated at 60°C for 10 min.
To the DNA sample in G-buffer, 4 μl of RNase A (100 mg/ml; 7000 units/ml; Qiagen) was added and incubated at 37°C for 30 min. To this, 25 μl of proteinase K (>600 mAU/ml; Qiagen) was introduced and incubated at 50°C for 40 min. Further, DNA was purified according to the Blood & Cell Culture DNA Mini Kit (Qiagen) protocol. To the DNA elution, 0.7 volume of isopropanol was added and stirred until DNA strands appeared, then, the strands were neatly wrapped around a glass rod. The DNA was transferred to a tube containing a DNA dilution buffer (Evrogen, Russia) and incubated at 50°C for 60 min. The DNA quality and concentration were evaluated on a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific) and a Qubit 2.0 fluorometer (Life Technologies, United States). The assessment of DNA length and the control of RNA absence were performed by electrophoresis in a 0.8% agarose gel (Lonza, Switzerland).
DNA Library Preparation and Sequencing on the Illumina Platform
DNA was fragmented on a S220 ultrasonic homogenizer (Covaris, United States), and 1 μg of fragmented DNA was used to prepare the library with the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, United Kingdom) according to the manufacturer’s protocol with size selection of adaptor-ligated DNA of about 600–800 bp. The quality and concentration of the DNA library were evaluated using a 2100 Bioanalyzer instrument (Agilent Technologies, United States) and a Qubit 2.0 fluorometer (Life Technologies), respectively. The resulting DNA library was sequenced on a HiSeq 2500 instrument (Illumina, United States) with a read length of 250 + 250 bp.
DNA Library Preparation and Sequencing on the ONT Platform
To remove short DNA fragments (up to 10 kb), a Short Read Eliminator Kit (Circulomics, United States) was used. Then, the DNA sample was purified with AMPure XP beads (Beckman Coulter, United States) in a ratio of 1:0.7 (sample:beads).
Preparation of the library was performed using the SQK-LSK109 Ligation Sequencing Kit (ONT, United Kingdom) for 1D genomic DNA sequencing. Minor modifications were introduced to the recommended protocol for library preparation by increasing the incubation time to 20 min at 20°C at the step of DNA recovery and to 60 min at the step of ligation. Sequencing was performed on MinION (ONT) with a FLO-MIN-106 R9.4 flow-cell (ONT).
Genome Assembly
First, we derived fastq read sequences from MinION raw electric signal fast5 files using guppy 3.2.2 with the high accuracy flip-flop algorithm (dna_r9.4.1_450bps_hac.cfg configuration file). Adapters were trimmed out with Porechop (https://github.com/rrwick/Porechop). Low quality reads (with average Q < 6) were filtered out using trimmomatic 0.38 (Bolger et al., 2014). Illumina reads were trimmed (trailing Q at least 28) and filtered (length < 50) with trimmomatic 0.38.
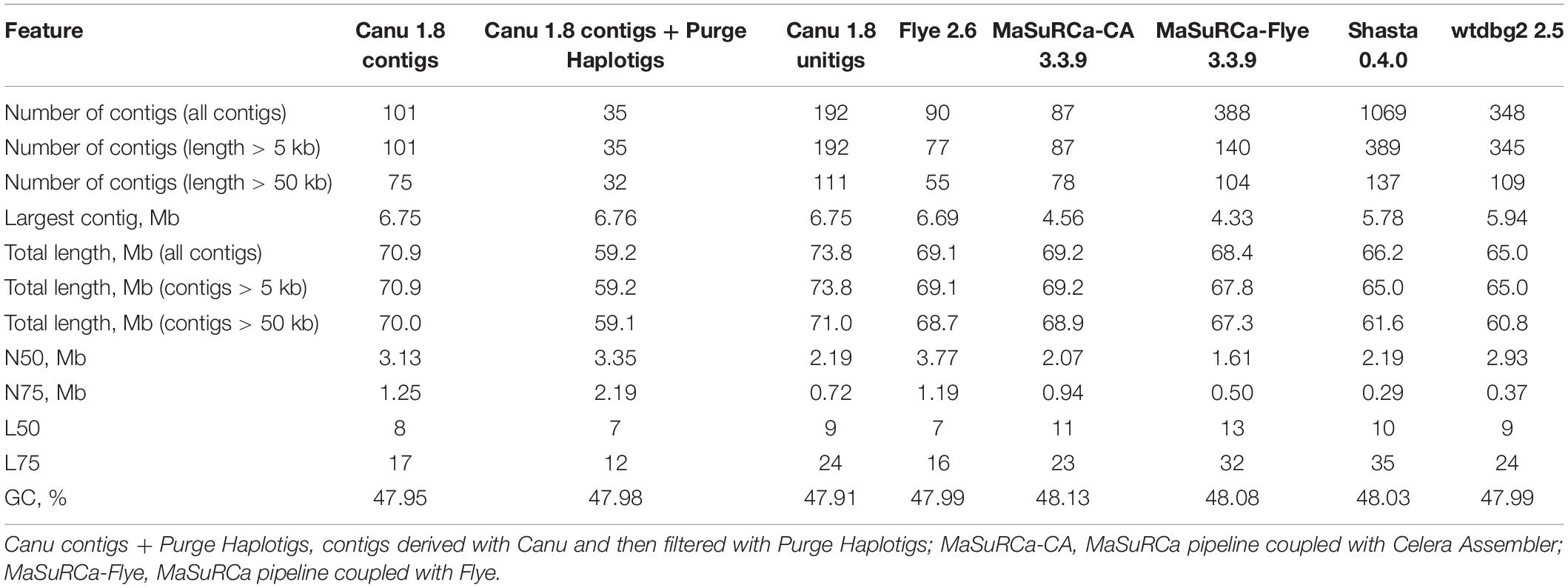
We used five different assemblers to perform the initial genome assemblies: Canu 1.8 (Koren et al., 2017), Flye 2.6 (Kolmogorov et al., 2019), Shasta 0.4.0 (Shafin et al., 2019), wtdbg2 2.5 (Ruan and Li, 2020), and MaSuRCA 3.3.9 (Zimin et al., 2017). The first four use Nanopore reads and the last one performs a hybrid assembly with Nanopore and Illumina reads. All the parameters were set by default except for the following: minimal read length was set as 3000 bp for Shasta, expected genome size was set as 60 Mb for Flye and wtdbg2. The obtained assemblies were polished using Nanopore reads by Medaka 0.12.11 or Racon 1.4.3 (1–4 iterations) (Vaser et al., 2017). Next, the polished sequences were additionally corrected using Illumina reads by Pilon 1.23 (Walker et al., 2014) or POLCA (built-in MaSuRCA 3.3.9 polisher) (Zimin et al., 2017). Nanopore read mapping to the genome assembly was performed with minimap2 2.17 (Li, 2018), Illumina read mapping – with BWA 0.7.17 (Li, 2013) or bowtie2 2.3.4.1 (Langmead and Salzberg, 2012).
Nx statistics for F. oxysporum f. sp. lini genome assemblies were calculated using QUAST 5.0.2 (Gurevich et al., 2013). Nx is a specific length for which the subset of contigs of that length or longer covers at least x percent of the assembly (i.e., 50% for N50, 75% for N75, etc.). Using QUAST, we also evaluated the misassembly rates relatively to F. oxysporum genomes assembled up to chromosome level and available in the NCBI Genome database.2 The completeness of assemblies was evaluated using BUSCO v3 (Pezizomycotina odb9 dataset, total 3156 BUSCOs) (Seppey et al., 2019). To visually compare the newly assembled genome of F. oxysporum f. sp. lini to NCBI chromosome-level assemblies, we created dotplots with the LAST v.1066 aligner (Kielbasa et al., 2011). The percentage of repetitive elements in genome assemblies was identified using RepeatMasker 4.0.9 (rmblast engine).
The best one of the obtained assemblies was chosen for further annotation using the funannotate 1.7.4 pipeline3 and previously obtained transcriptome sequencing data (BioProject accession – PRJNA412801). Before annotating, we eliminated redundant contigs using Purge Haplotigs 1.0.4 (Roach et al., 2018).
Results
The purity of DNA is crucial for Nanopore sequencing, so the protocol was developed to extract DNA of sufficient purity from Fusarium fungi. The CTAB-DNA precipitation and Blood & Cell Culture DNA Mini Kit (Qiagen) allowed us to obtain the DNA sample with A260/280 = 1.9 and A260/230 = 2.4. For this sample, DNA concentrations measured with a Qubit fluorometer (Life Technologies) and a Nanodrop spectrophotometer (Thermo Fisher Scientific) had close values that is an important criterion of DNA purity. When DNA was extracted using the CTAB method only, the concentration values differed up to 10-fold and A260/230 values were down to 0.3, indicating significant contamination. The length of Nanopore reads depends on the DNA length and is a key feature affecting the quality of genome assembly. The application of Short Read Eliminator Kit (Circulomics) enabled the removal of short DNA fragments and the average DNA length was about 50 kb. Thus, using the developed protocol of DNA extraction, we obtained long high-purity DNA of F. oxysporum f. sp. lini that allowed us to receive 312 thousand reads (5.1 Gb) with N50 = 29 kb on MinION (ONT) and 3.5 million 2 × 250 bp reads on HiSeq (Illumina). This corresponds to about 85x coverage with Nanopore reads and about 30x coverage with Illumina reads.
Using the obtained sequencing data, we benchmarked the five most requested genome assemblers: Canu, Flye, Shasta, wtdbg2, and MaSuRCA. Statistics across all assemblies are represented in Table 1 and Figure 1. Canu produced the longest assembly: 71 Mb (contigs) and 74 Mb (unitigs) with N50 values equal to 3.1 Mb and 2.2 Mb, respectively (according to Canu’s terminology, unitigs are high-confidence contigs). N50 value was the greatest for Flye (3.7 Mb, total length – 69 Mb). However, the first (largest) Flye contig seemed to be misassembled. It represented a combination of several contigs produced by other assemblers – four Canu contigs, three Shasta contigs, or two wtdbg2 contigs. Moreover, this contig did not completely align to any of five NCBI chromosome-level assemblies of F. oxysporum strains (Figure 2), and the breakpoint exactly matched to the boundaries of contigs assembled by Canu, Shasta, or wtdbg2. These facts suggested that the first Flye contig was not assembled correctly. One of the possible causes is chimeric Nanopore reads (Xu et al., 2018; Martin and Leggett, 2019). Remarkably, there are repeats upstream of the breakpoint region (Figure 2).
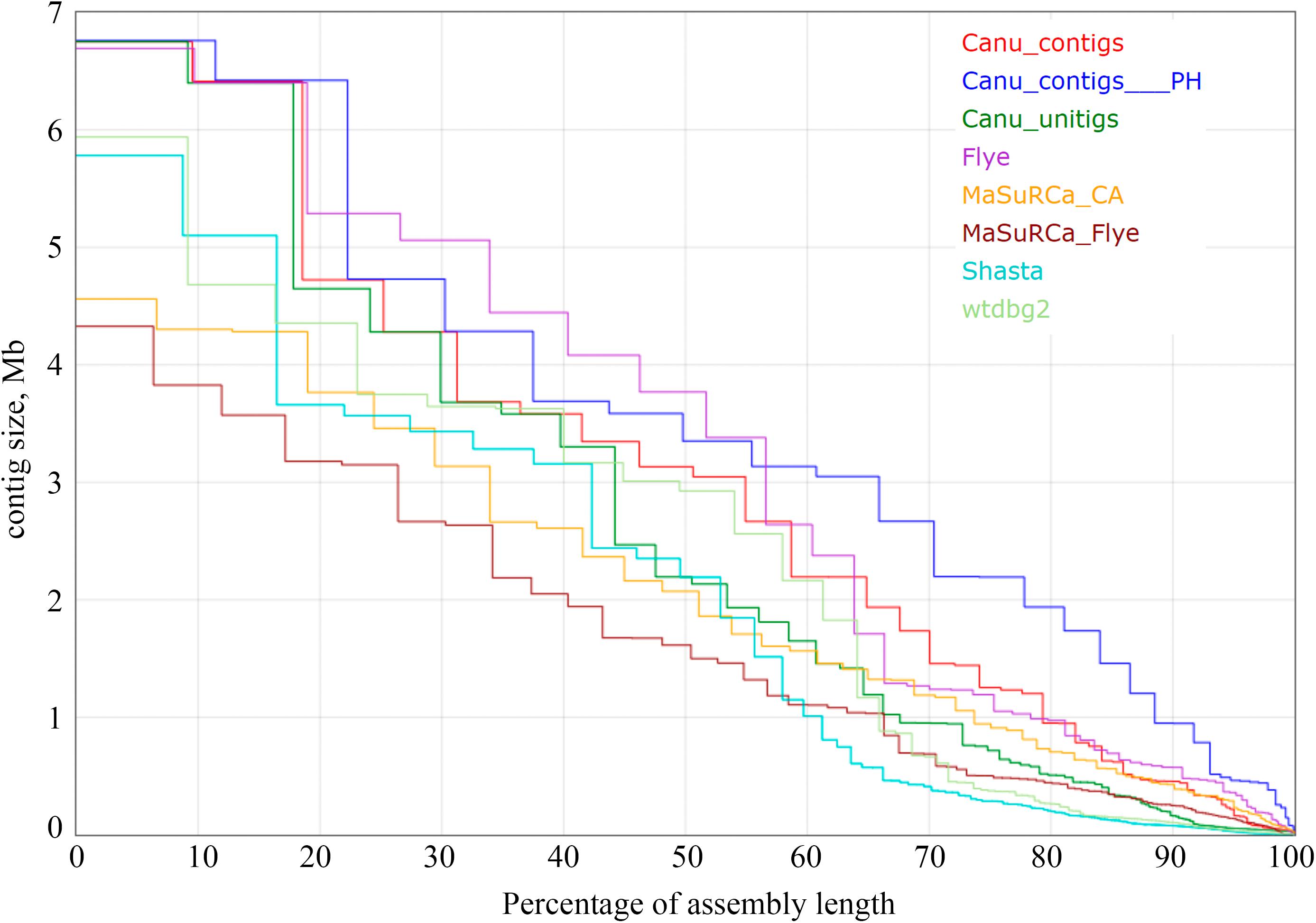
Figure 1. The Nx statistics for Fusarium oxysporum f. sp. lini assemblies. Results for the following assemblers are presented: Canu 1.8 contigs, Canu 1.8 contigs filtered with Purge Haplotigs (Canu_contigs__PH), Canu 1.8 unitigs, Flye 2.6, MaSuRCA 3.3.9 coupled with Celera Assembler (MaSuRCA_CA), MaSuRCA 3.3.9 coupled with Flye (MaSuRCA_Flye), Shasta 0.4.0, wtdbg2 2.5. Medaka, Racon, Pilon, POLCA polishing did not impact Nx statistics.
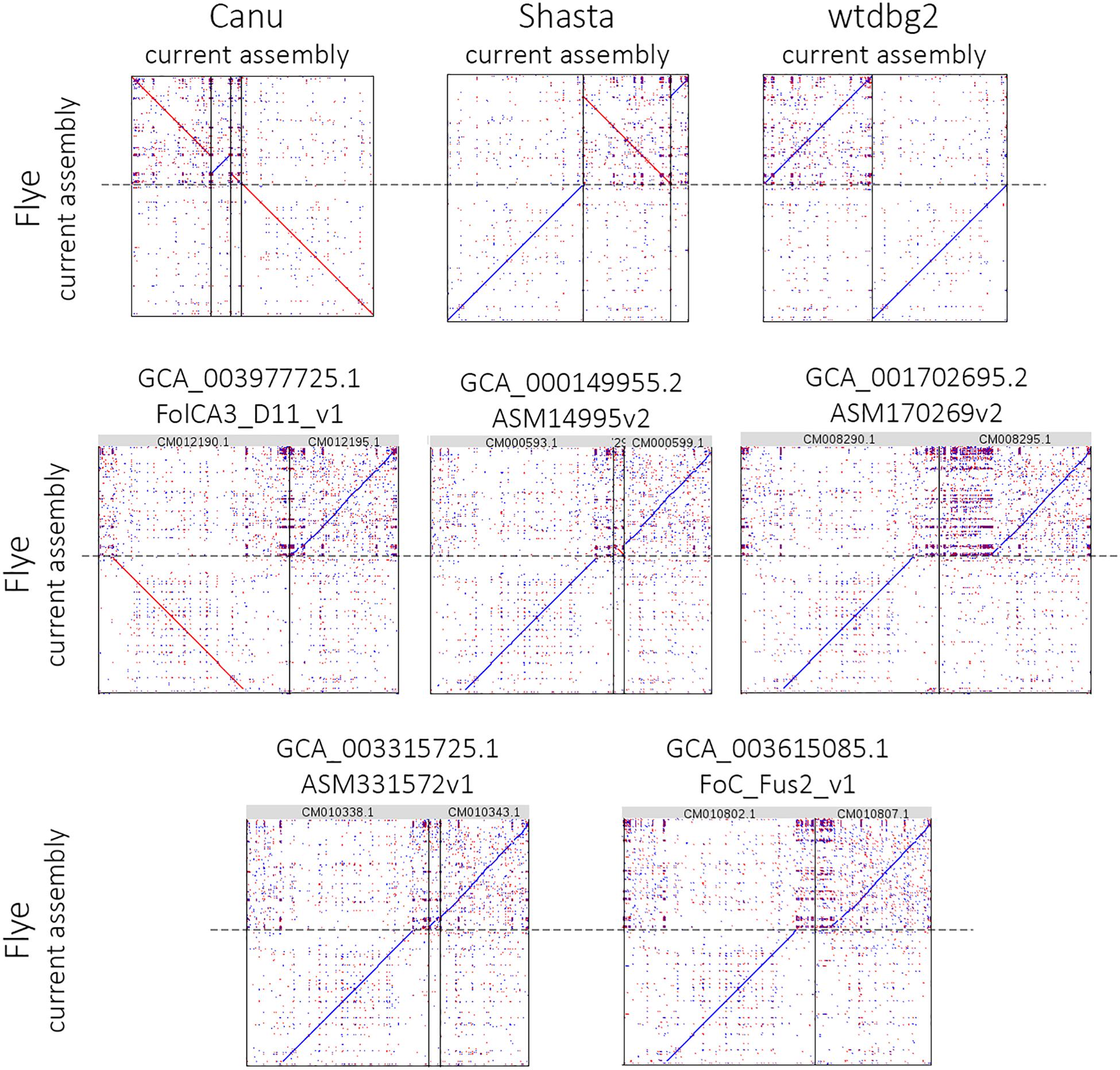
Figure 2. Alignment of the largest Flye 2.6 contig to Canu 1.8, Shasta 0.4.0, wtdbg2 2.5 assemblies and five NCBI chromosome-level assemblies of Fusarium oxysporum strains. The chromosome-level assemblies of F. oxysporum strains can be found in the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome/genomes/707).
Using QUAST, we evaluated the consistency between our assemblies and five NCBI chromosome-level assemblies of F. oxysporum strains (Supplementary Data S1). It should be noted that we can only compare the number of misassemblies between different assemblers within the same reference genome. Judging by the minimum number of misassemblies, the closest to our genome was the assembly of strain GCA_001702695.2/ASM170269v2 (F. oxysporum f. sp. radicis-cucumerinum). Comparing our assemblies with the reference ones, it was difficult to give preference to any assembler: different tools led in different parameters. According to one of the key parameters, NA50 (analog of N50 for fragments successfully mapped to reference), Shasta was the most accurate. However, Shasta gave an increased number of contigs and worse statistics without reference.
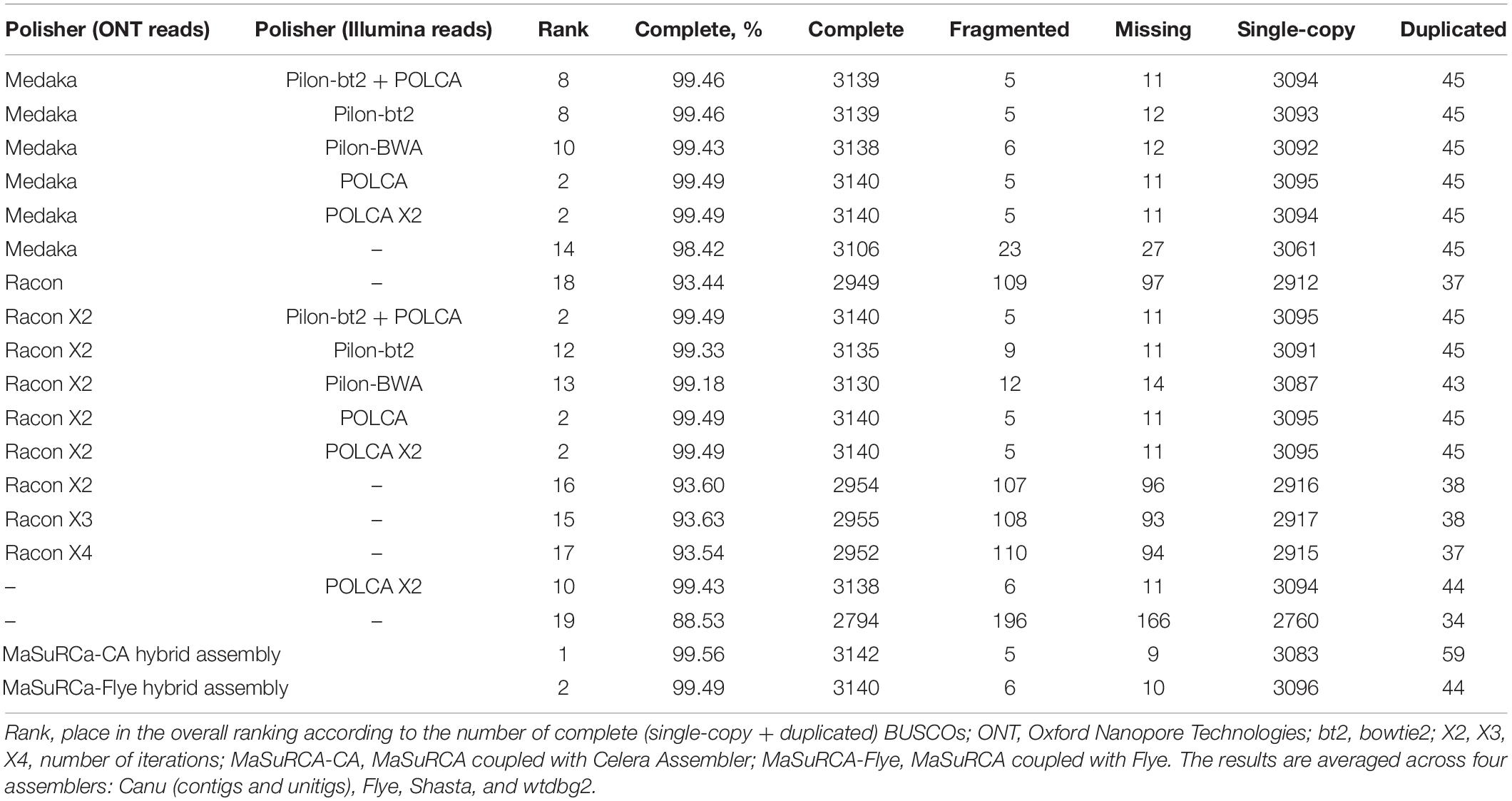
Next, to improve the contig accuracy, raw assemblies were polished with Nanopore reads, Illumina reads, or a combination of these approaches. To polish contigs with Nanopore reads, we applied Medaka or Racon (1–4 iterations). To evaluate the efficacy of polishing, we assessed contig accuracy by matching them to high-quality Illumina reads (statistics were calculated using FreeBayes/POLCA) or by the presence of highly conservative ORFs characteristic for all fungi (by BUSCO). Racon (2 iterations) slightly outperformed Medaka in the number of point substitutions (Supplementary Data S2). However, Medaka won several times by the number of indels – about 40 thousand after Racon and about 10–15 thousand after Medaka. We saw this reflected in the BUSCO metrics, where Medaka greatly outperformed Racon – 98.4% versus 93.4% (Table 2). Indeed, the presence of frameshift insertions/deletions inside ORF in most cases leads to the fact that the protein is no longer detected by BUSCO (or considered as “fragmented BUSCO”). Medaka handled homopolymers well, and the contigs polished with it contained much less false indels. We also noticed that the 2nd iteration of Racon made a much smaller contribution compared to the 1st one. The 3rd and 4th iterations did not make any contribution and even refuse a negative impact somewhere (Table 2 and Supplementary Data S3). Among four assemblers (Canu, Flye, Shasta, and wtdbg2), after polishing with Medaka or Racon, the smallest number of both point substitutions and indels was revealed for Canu assemblies (Supplementary Data S2).
After the initial polishing with Medaka or Racon, we performed the secondary polishing with Pilon and POLCA using Illumina reads. This allowed us to improve the result of BUSCO analysis from 98.4% (Medaka) and 93.4% (Racon) to 99.2–99.5%. POLCA performed better than Pilon both in terms of speed (a difference of about 10 times) and BUSCO completeness. Nevertheless, based on this, it cannot be said that POLCA is superior to Pilon. The question remains how correctly these polishers work with haplotypes, paralogs, and repetitive elements.
Pilon requires BAM files containing Illumina reads mapped to the genome to be polished. We compared two read mappers, BWA and bowtie2, on the efficacy of polishing. BWA may produce more mapping errors compared to bowtie2 (Thankaswamy-Kosalai et al., 2017). This difference may be greater or smaller depending on read length, genome complexity, and richness in paralogs or repeats. Indeed, bowtie2 performed slightly better than BWA in terms of BUSCO metrics (Table 2). However, these values were lower than those achieved by POLCA (99.49%), which implements only BWA. Thus, “Medaka + POLCA” (or only Medaka if you wish to preserve haplotypes) seems to be the best polishing scheme. However, MaSuRCa, a hybrid assembler utilizing both Nanopore and Illumina reads, provided the best results, surpassing all other assemblers and polishing schemes (99.56% BUSCO completeness; Table 2). But MaSuRCa did not demonstrate high N50 values (Table 1).
Based on Nx and BUSCO statistics and the presence of possible misassemblies, we decided that Canu assembly polished with Medaka and POLCA is the best for further analysis. The length of this assembly was 71 Mb that seemed to be excessive. For comparison, the length of 329 chromosome- and scaffold-level assemblies of F. oxysporum strains deposited in the NCBI Genome database (see text footnote 2) varied from 38 to 64 Mb (median length = 53 Mb) and the length of representative F. oxysporum genome GCA_000149955.2/ASM14995v2 is 61 Mb.
At the same time, BUSCO analysis revealed a very low number of duplicated BUSCOs (1.4%) in our assembly, and therefore the increased genome length (+15–30%) cannot be ascribed to the haplotype separation. To find out possible reasons, we tested our F. oxysporum sample for bacterial contamination. However, among 13400 bacterial genomes from the NCBI Genome database deposited before May 2017 (total 28 Gb), we did not find any significant matches. The abundance of repetitive elements could abrogate or inflate the F. oxysporum f. sp. lini assembly, but RepeatMasker identified only 0.7% of repeats, so that was not the case also.
Next, we mapped our assembly to itself and five NCBI chromosome-level assemblies of F. oxysporum strains (Supplementary Data S4–S8) and found that the vast part of contigs greatly mapped to the reference sequences, but many other genomic regions (whole contigs or fragments of contigs) had multiple short homologies to themselves and other genomic regions. The presence of such regions was the reason for the increased length of our assembly. To eliminate this redundancy, we used the Purge Haplotigs tool, which analyzes read coverage density distribution and filters out haplotypes. It is designed to work with bimodal distribution: one peak (minor, as a rule) corresponds to genomic regions with separated haplotypes, and one peak (major) corresponds to regions with merged haplotypes. Even though we obtained only unimodal coverage density distribution, we managed to exclude the redundant haplotigs by tweaking Purge Haplotigs options (setting “align_cov” as 55% and “m” to the median coverage value). The number of complete BUSCOs remained unchanged. The length of the assembly had successfully reduced from 71 Mb to 59 Mb, L50 from 8 to 7, the total number of contigs from 101 to 35; N50 was increased from 3.1 Mb to 3.3 Mb. It should be noted that several assembled contigs represented whole chromosomes. We also assembled a complete circular mitochondrial genome with a length of 38.7 kb.
The obtained assembly was annotated using the funannotate pipeline. As a result, a total of 19351 gene models containing 19607 transcripts were predicted. About 11 thousand models came from PASA based on our RNA-Seq reads and mapping of Trinity transcripts to the assembled genome (from PASA). These models were used for training Augustus, GlimmerHMM, and SNAP, which along with GeneMark and CodingQuarry predicted the rest gene models included in the annotation (Supplementary Data S9). Among 19351 gene models, 311 refer to tRNA. 11841 gene models were successfully annotated using the Pfam database, 12423 – eggNOG, 1334 – BUSCO (Supplementary Data S10). Only 1032 genes were assigned with gene names (it’s normal since such annotation is based only on curated data of UniProt/SwissProt).
The assembled genome was deposited at DDBJ/ENA/GenBank under the accession WHMS00000000 (BioProject accession – PRJNA578147).
Discussion
The F. oxysporum species includes a wide variety of strains and exhibits a high genetic and functional diversity. There are no morphological differences among pathogenic and non-pathogenic strains, while pathogenic strains have a narrow specificity to the host plant (Nelson et al., 1981; Steinberg et al., 2016). Understanding the molecular basis of pathogenicity in the genus Fusarium largely depends on the degree of knowledge on the source material that is achieved through molecular genetic studies.
Genome sequencing of fungal plant pathogens enables understanding of the mechanisms of pathogenesis and identification of the key effector genes that enhance infection (Moller and Stukenbrock, 2017; Plissonneau et al., 2017). Obtaining high-quality genomes is a crucial task. The first F. oxysporum genome assembly (f. sp. lycopersici strain) was done by Broad Institute in 2007 using Sanger sequencing with 6x coverage. The genome consists of 15 chromosomes and has a total length of about 62 Mb (Ma et al., 2013). In 2018, a new version of the assembly of the F. oxysporum f. sp. lycopersici genome was obtained using the Illumina platform with 76x coverage (Ayhan et al., 2018). This assembly is used as a representative (default) genome for F. oxysporum in the NCBI Genome database. It is known that genotypes of different strains, even within the same species, can significantly differ in the gene copy number, presence or absence of specific genes, structure of repeating non-coding regions, and single nucleotide polymorphisms (Schatz et al., 2014; Thudi et al., 2016; Bayer et al., 2017). Till now, there were no sequenced genomes of F. oxysporum f. sp. lini, which is a harmful pathogen of flax. To date, within specific for flax pathogens, the genome sequence was available only for Melampsora lini, which causes rust. Although the assembly of the Melampsora lini genome is far from chromosome level (21130 scaffolds were obtained with N50 equal to 31 kb), this is a valuable source to study the molecular mechanisms of pathogenesis (Nemri et al., 2014). Obtained by us high-quality genome assembly of F. oxysporum f. sp. lini consists of 35 contigs with a total length of 59 Mb and has 99.5% completeness according to BUSCO that became possible due to the development of sequencing technologies and approaches for genome assembly.
Revolution in sequencing methods began from Sanger sequencing that enabled obtaining complete genomes, then high-throughput next-generation sequencing (NGS) methods were developed and made genome sequencing faster and cheaper, and finally, third-generation technologies allowed single-molecule and long-read sequencing that is crucial for high-quality genome assemblies (van Dijk et al., 2018; Arboleda and Xian, 2019). NGS platforms, such as Illumina, BGI, and Ion Torrent, are cost-effective and produce short reads (up to 600 bp), while third-generation platforms, such as Pacific Biosciences (PacBio) and ONT, result in read length of tens and even hundreds of thousands of nucleotides. Currently, genome assembly using long reads is at its peak (Kono and Arakawa, 2019; Amarasinghe et al., 2020; Giani et al., 2020). Over the past 5 years, since the widespread of ONT and PacBio sequencing, several genome assembly tools based on long noisy reads were developed: miniasm, HGAP (only PacBio), Canu, Falcon, ABruijn, wtdbg, Unicycler (the last one is mainly applied for prokaryotic genomes). Among them, Canu seems to be the most accurate assembler, as it was previously shown (Jayakumar and Sakakibara, 2019). However, it requires lots of CPU time (for example, the present F. oxysporum assembly required about 3000 CPU hours; ∼2 days with 64 CPU cores) and can hardly be used for the large genomes. To solve this issue, several novel assemblers were developed during the last year, for example, Shasta assembly took only 11.3 min (5.6 CPU hours), Flye – 98 min (∼75 CPU hours). In some cases, Shasta and Flye can outperform Canu in terms of the number of misassemblies and speed but not Nx stats (Shafin et al., 2019). Strictly speaking, the choice of genomic assembler may depend on the genome (its size, ploidy and allele differences, complexity, presence of repetitive elements, etc.), and various assemblers will perform differently for different genomes.
Hence, in this study, we tested five assemblers to choose the most suitable for our purpose. In general, Canu assembly may be considered the best. Compared to other ONT assemblers (Flye, Shasta, and wtdbg2), it is known for a relatively long time (since 2016). Along with Nx statistics, the most important parameters for assessing the assembly quality are misassembly rate, resolution of repeats and tandem duplications, which are rather difficult to evaluate without close reference. At the raw assembly stage, it is these indicators that are important. The accuracy of the assembled contig sequences (i.e., presence of substitutions and indels) is entirely corrected at the polishing stage.
While the BUSCO completeness of raw contigs produced by four assemblers varied significantly (some assemblers have their polishing modules), after processing with Medaka or Racon (Nanopore reads), the results of BUSCO analysis were almost equal, and after subsequent polishing with POLCA or Pilon (Illumina reads), the differences completely smoothed out. Polishing with Illumina reads has both pros and cons. This procedure allows one to quickly decrease the number of short mismatches and indels, achieving sequence accuracy higher than that obtained by polishing with Nanopore reads only. Nevertheless, some authors write that short reads used for correction will homogenize repeats, mix up haplotypes and close paralogs. Even though Illumina reads are of very high quality, their length isn’t sufficient for the true alignment to be identified, and reads from other repeat instances (or paralogs) are used for correction, resulting in incorrect edits.4
Conclusion
In the present study, we for the first time sequenced the genome of a highly pathogenic isolate of F. oxysporum f. sp. lini using the Nanopore system generating long noisy reads and the Illumina one with short accurate reads and obtained a high-quality genome assembly by testing the most requested assemblers and polishing tools and choosing the best ones for received data. Besides, the protocol for extraction of pure high-molecular-weight DNA from fungi was developed. The obtained genome will significantly improve the studies on F. oxysporum and flax response to this pathogen.
Data Availability Statement
The datasets generated for this study can be found in the DDBJ/ENA/GenBank under the accession WHMS00000000 (BioProject accession – PRJNA578147).
Author Contributions
TR, AD, and NM conceived and designed the work. EP, RN, LK, ED, LP, and NM performed the experiments. GK, AK, AD, and NM analyzed the data. GK, EP, AD, and NM wrote the manuscript. All authors agreed with the final version of the manuscript and all aspects of the work.
Funding
Part of this work (F. oxysporum f. sp. lini genome sequencing, assembly, and annotation) was financially supported by the Russian Science Foundation, grant 16-16-00114. Part of this work (the development of the protocol for extraction of pure high-molecular-weight DNA from fungi) was funded by RFBR, project number 19-34-90055.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, EIMB RAS for providing the computing power and techniques for the data analysis. This work was performed using the equipment of EIMB RAS “Genome” center (http://www.eimb.ru/ru1/ckp/ccu_genome _c.php).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00959/full#supplementary-material
DATA S1 | The consistency between the obtained F. oxysporum f. sp. lini assemblies and five chromosome-level assemblies of F. oxysporum strains available in the NCBI Genome database.
DATA S2 | The accuracy of contigs polished by Medaka (1 iteration) or Racon (2 iterations).
DATA S3 | BUSCO results for various combinations of assemblers, Nanopore polishers, and/or Illumina polishers.
DATA S4 | Dotplot illustrating homology between the current F. oxysporum f. sp. lini assembly (before filtering redundant contigs with the Purge Haplotigs tool) and GCA_000149955.2/ASM14995v2 deposited in the NCBI Genome database (vertical axis).
DATA S5 | Dotplot illustrating homology between the current F. oxysporum f. sp. lini assembly (before filtering redundant contigs with the Purge Haplotigs tool) and GCA_001702695.2/ASM170269v2 deposited in the NCBI Genome database (vertical axis).
DATA S6 | Dotplot illustrating homology between the current F. oxysporum f. sp. lini assembly (before filtering redundant contigs with the Purge Haplotigs tool) and GCA_003315725.1/ASM331572v1 deposited in the NCBI Genome database (vertical axis).
DATA S7 | Dotplot illustrating homology between the current F. oxysporum f. sp. lini assembly (before filtering redundant contigs with the Purge Haplotigs tool) and GCA_003615085.1/FoC_Fus2_v1 deposited in the NCBI Genome database (vertical axis).
DATA S8 | Dotplot illustrating homology between the current F. oxysporum f. sp. lini assembly (horizontal axis; before filtering redundant contigs with the Purge Haplotigs tool) and GCA_003977725.1/FolCA3_D11_v1 deposited in the NCBI Genome database (vertical axis).
DATA S9 | Sources of gene models.
DATA S10 | Annotation statistics for gene models.
Footnotes
- ^ https://github.com/nanoporetech/medaka
- ^ https://www.ncbi.nlm.nih.gov/genome/genomes/707
- ^ https://funannotate.readthedocs.io/en/latest/
- ^ https://canu.readthedocs.io/en/latest/faq.html
References
Amarasinghe, S. L., Su, S., Dong, X., Zappia, L., Ritchie, M. E., and Gouil, Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21:30. doi: 10.1186/s13059-020-1935-5
Arboleda, V. A., and Xian, R. R. (2019). An overview of DNA analytical methods. Methods Mol. Biol. 1897, 385–402. doi: 10.1007/978-1-4939-8935-5_31
Ayhan, D. H., Lopez-Diaz, C., Di Pietro, A., and Ma, L. J. (2018). Improved assembly of reference genome Fusarium oxysporum F. sp. lycopersici strain Fol4287. Microbiol. Resour. Announc. 7:e0910-18. doi: 10.1128/MRA.00910-18
Baroncini, E. A., Yadav, S. K., Palmese, G. R., and Stanzione, J. F. I. I. I. (2016). Recent advances in bio-based epoxy resins and bio-based epoxy curing agents. J. Appl. Polym. Sci. 133:103. doi: 10.1002/app.44103
Bayer, P. E., Hurgobin, B., Golicz, A. A., Chan, C. K., Yuan, Y., Lee, H., et al. (2017). Assembly and comparison of two closely related Brassica napus genomes. Plant Biotechnol. J. 15, 1602–1610. doi: 10.1111/pbi.12742
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Costa, S. M., Ferreira, D. P., Ferreira, A., Vaz, F., and Fangueiro, R. (2018). Multifunctional Flax fibres based on the combined effect of Silver and Zinc Oxide (Ag/ZnO) Nanostructures. Nanomaterials 8:1069. doi: 10.3390/nano8121069
Dutra, P. A., Pinto, L. F. B., Cardoso Neto, B. M., Gobikrushanth, M., Barbosa, A. M., and Barbosa, L. P. (2019). Flaxseed improves embryo production in Boer goats. Theriogenology 127, 26–31. doi: 10.1016/j.theriogenology.2018.12.038
Fombuena, V., Petrucci, R., Dominici, F., Jorda-Vilaplana, A., Montanes, N., and Torre, L. (2019). Maleinized linseed oil as epoxy resin hardener for composites with high bio content obtained from linen byproducts. Polymers 11:301. doi: 10.3390/polym11020301
Giani, A. M., Gallo, G. R., Gianfranceschi, L., and Formenti, G. (2020). Long walk to genomics: history and current approaches to genome sequencing and assembly. Comput. Struct. Biotechnol. J. 18, 9–19. doi: 10.1016/j.csbj.2019.11.002
Goyal, A., Sharma, V., Upadhyay, N., Gill, S., and Sihag, M. (2014). Flax and flaxseed oil: an ancient medicine & modern functional food. J. Food Sci. Technol. 51, 1633–1653. doi: 10.1007/s13197-013-1247-9
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Head, B., Bionaz, M., and Cherian, G. (2019). Flaxseed and carbohydrase enzyme supplementation alters hepatic n-3 polyunsaturated fatty acid molecular species and expression of genes associated with lipid metabolism in broiler chickens. Vet. Sci. 6:25. doi: 10.3390/vetsci6010025
Hu, D., Dang, L., Zhang, C., and Zhang, Z. (2019). Mechanical behaviors of flax fiber-reinforced composites at different strain rates and rate-dependent constitutive model. Materials 12:854. doi: 10.3390/ma12060854
Isenberg, B. J., Soder, K. J., Pereira, A. B. D., Standish, R., and Brito, A. F. (2019). Production, milk fatty acid profile, and nutrient utilization in grazing dairy cows supplemented with ground flaxseed. J. Dairy Sci. 102, 1294–1311. doi: 10.3168/jds.2018-15376
Jayakumar, V., and Sakakibara, Y. (2019). Comprehensive evaluation of non-hybrid genome assembly tools for third-generation PacBio long-read sequence data. Brief. Bioinform. 20, 866–876. doi: 10.1093/bib/bbx147
Kezimana, P., Dmitriev, A. A., Kudryavtseva, A. V., Romanova, E. V., and Melnikova, N. V. (2018). Secoisolariciresinol diglucoside of flaxseed and its metabolites: biosynthesis and potential for nutraceuticals. Front. Genet. 9:641. doi: 10.3389/fgene.2018.00641
Kielbasa, S. M., Wan, R., Sato, K., Horton, P., and Frith, M. C. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493. doi: 10.1101/gr.113985.110
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Kommedahl, T., Christensen, J. J., and Frederiksen, R. A. (1970). A Half Century of Research in Minnesota on Flax Wilt Caused by Fusarium oxyporum. Minneapolis: University of Minnesota.
Kono, N., and Arakawa, K. (2019). Nanopore sequencing: review of potential applications in functional genomics. Dev. Growth Differ. 61, 316–326. doi: 10.1111/dgd.12608
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kroes, G. M. L. W., Baayen, R. P., and Lange, W. (1998). Histology of root rot of flax seedlings (Linum usitatissimum) infected by Fusarium oxysporum f.sp. lini. Eur. J. Plant Pathol. 104, 725–736. doi: 10.1023/A:1008604417614
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint], Available at: https://arxiv.org/abs/1303.3997 (accessed July 20, 2020).
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Loshakova, N. I., Kudryavtseva, L. P., Pavlova, L. N., and Rozhmina, T. A. (2014). The role of “Collection of the phytopathogenic microorganisms - agents of flax diseases” in flax breeding on group resistance to diseases. Oil Crops 2, 159–160.
Ma, L. J., Geiser, D. M., Proctor, R. H., Rooney, A. P., O’Donnell, K., Trail, F., et al. (2013). Fusarium pathogenomics. Annu. Rev. Microbiol. 67, 399–416. doi: 10.1146/annurev-micro-092412-155650
Marino, R., Della Malva, A., Caroprese, M., de Palo, P., Santillo, A., Sevi, A., et al. (2019). Effects of whole linseed supplementation and treatment duration on fatty acid profile and endogenous bioactive compounds of beef muscle. Animal 13, 444–452. doi: 10.1017/S1751731118001635
Martin, S., and Leggett, R. M. (2019). Alvis: a tool for contig and read ALignment VISualisation and chimera detection. bioRxiv [Preprint]. doi: 10.1101/663401
Mattioli, S., Dal Bosco, A., Maranesi, M., Petrucci, L., Rebollar, P. G., and Castellini, C. (2019). Dietary fish oil and flaxseed for rabbit does: fatty acids distribution and Delta6-desaturase enzyme expression of different tissues. Animal 13, 1934–1942. doi: 10.1017/S175173111900020X
Michielse, C. B., and Rep, M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10, 311–324. doi: 10.1111/j.1364-3703.2009.00538.x
Moller, M., and Stukenbrock, E. H. (2017). Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Microbiol. 15, 756–771. doi: 10.1038/nrmicro.2017.76
Nair, P. N., and Kommedahl, T. (1957). The establishment and growth of Fusarium lini in flax tissues. Phytopathology 47:25.
Nelson, P. E., Toussoun, T. A., and Cook, R. J. (1981). Fusarium: Diseases, Biology, and Taxonomy. University Park, PA: Pennsylvania State University Press.
Nemri, A., Saunders, D. G., Anderson, C., Upadhyaya, N. M., Win, J., Lawrence, G. J., et al. (2014). The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front. Plant Sci. 5:98. doi: 10.3389/fpls.2014.00098
Nyvall, R. F. (1989). “Diseases of flax,” in Field Crop Diseases Handbook, ed. R. F. Nyvall (Boston, MA: Springer), 251–264. doi: 10.1007/978-1-4757-5221-2_8
Plissonneau, C., Benevenuto, J., Mohd-Assaad, N., Fouche, S., Hartmann, F. E., and Croll, D. (2017). Using population and comparative genomics to understand the genetic basis of effector-driven fungal pathogen evolution. Front. Plant Sci. 8:119. doi: 10.3389/fpls.2017.00119
Roach, M. J., Schmidt, S. A., and Borneman, A. R. (2018). Purge haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 19:460. doi: 10.1186/s12859-018-2485-7
Ruan, J., and Li, H. (2020). Fast and accurate long-read assembly with wtdbg2. Nat. Methods 17, 155–158. doi: 10.1038/s41592-019-0669-3
Saharan, G. S., Naresh, M., and Sangwan, M. S. (2005). “Fungal diseases of linseed,” in Diseases of Oilseed Crops, eds G. S. Saharan, M. Naresh, and M. S. Sangwan (New Delhi: Indus Publishing Company), 176–201.
Schatz, M. C., Maron, L. G., Stein, J. C., Hernandez Wences, A., Gurtowski, J., Biggers, E., et al. (2014). Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 15:506. doi: 10.1186/PREACCEPT-2784872521277375
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Shafin, K., Pesout, T., Lorig-Roach, R., Haukness, M., Olsen, H. E., Bosworth, C., et al. (2019). Efficient de novo assembly of eleven human genomes using PromethION sequencing and a novel nanopore toolkit. bioRxiv [Preprint], doi: 10.1101/715722
Steinberg, C., Lecomte, C., Alabouvette, C., and Edel-Hermann, V. (2016). “Root interactions with nonpathogenic Fusarium oxysporum. Hey Fusarium oxysporum, what do you do in life when you do not infect a plant?,” in Belowground Defence Strategies in Plants, ed. C. M. F. Vos (Cham: Springer), 281–299.
Thankaswamy-Kosalai, S., Sen, P., and Nookaew, I. (2017). Evaluation and assessment of read-mapping by multiple next-generation sequencing aligners based on genome-wide characteristics. Genomics 109, 186–191. doi: 10.1016/j.ygeno.2017.03.001
Thudi, M., Khan, A. W., Kumar, V., Gaur, P. M., Katta, K., Garg, V., et al. (2016). Whole genome re-sequencing reveals genome-wide variations among parental lines of 16 mapping populations in chickpea (Cicer arietinum L.). BMC Plant Biol. 16(Suppl. 1):10. doi: 10.1186/s12870-015-0690-3
van Dijk, E. L., Jaszczyszyn, Y., Naquin, D., and Thermes, C. (2018). The third revolution in sequencing technology. Trends Genet. 34, 666–681. doi: 10.1016/j.tig.2018.05.008
Vaser, R., Sovic, I., Nagarajan, N., and Sikic, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Xu, Y., Lewandowski, K., Lumley, S., Pullan, S., Vipond, R., Carroll, M., et al. (2018). Detection of viral pathogens with multiplex nanopore MinION sequencing: be careful with cross-talk. Front. Microbiol. 9:2225. doi: 10.3389/fmicb.2018.02225
Yi, H., Hwang, K. T., Regenstein, J. M., and Shin, S. W. (2014). Fatty acid composition and sensory characteristics of eggs obtained from hens fed flaxseed oil, dried whitebait and/or fructo-oligosaccharide. Asian Austr. J. Anim. Sci. 27, 1026–1034. doi: 10.5713/ajas.2013.13775
Keywords: Fusarium oxysporum f. sp. lini, Linum usitatissimum L., de novo genome assembly, Nanopore, Illumina, genome assemblers, assembly polishers, pure high-molecular-weight DNA
Citation: Krasnov GS, Pushkova EN, Novakovskiy RO, Kudryavtseva LP, Rozhmina TA, Dvorianinova EM, Povkhova LV, Kudryavtseva AV, Dmitriev AA and Melnikova NV (2020) High-Quality Genome Assembly of Fusarium oxysporum f. sp. lini. Front. Genet. 11:959. doi: 10.3389/fgene.2020.00959
Received: 30 November 2019; Accepted: 30 July 2020;
Published: 27 August 2020.
Edited by:
Jianxin Wang, Central South University, ChinaReviewed by:
Milind B. Ratnaparkhe, ICAR-Indian Institute of Soybean Research, IndiaXingyu Liao, Central South University, China
Copyright © 2020 Krasnov, Pushkova, Novakovskiy, Kudryavtseva, Rozhmina, Dvorianinova, Povkhova, Kudryavtseva, Dmitriev and Melnikova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nataliya V. Melnikova, bW52LTQ1MjkyNjRAeWFuZGV4LnJ1
†These authors have contributed equally to this work
 George S. Krasnov
George S. Krasnov Elena N. Pushkova
Elena N. Pushkova Roman O. Novakovskiy
Roman O. Novakovskiy Ludmila P. Kudryavtseva2
Ludmila P. Kudryavtseva2 Ekaterina M. Dvorianinova
Ekaterina M. Dvorianinova Liubov V. Povkhova
Liubov V. Povkhova Anna V. Kudryavtseva
Anna V. Kudryavtseva Alexey A. Dmitriev
Alexey A. Dmitriev Nataliya V. Melnikova
Nataliya V. Melnikova