
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet., 27 August 2020
Sec. Evolutionary and Genomic Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00916
This article is part of the Research TopicNew Advances in Genetic Studies to Understand Yeast Adaptation to Extreme and Fermentative EnvironmentsView all 13 articles
The budding yeast has been extensively studied for its physiological performance in fermentative environments and, due to its remarkable plasticity, is used in numerous industrial applications like in brewing, baking and wine fermentations. Furthermore, thanks to its small and relatively simple eukaryotic genome, the molecular mechanisms behind its evolution and domestication are more easily explored. Considerable work has been directed into examining the industrial adaptation processes that shaped the genotypes of species and hybrids belonging to the Saccharomyces group, specifically in relation to beverage fermentation performances. A variety of genetic mechanisms are responsible for the yeast response to stress conditions, such as genome duplication, chromosomal re-arrangements, hybridization and horizontal gene transfer, and these genetic alterations are also contributing to the diversity in the Saccharomyces industrial strains. Here, we review the recent genetic and evolutionary studies exploring domestication and biodiversity of yeast strains.
Beer brewing and winemaking have been rapidly changing over the years involving the development of several fermentation protocols and the use of starter cultures. In the past, these processes were mainly occurring naturally. For example, grape juice and fresh hoppy wort were exposed to open air microorganisms to spontaneously ferment wine and beer.
While this natural process still find application in some beers (traditional lambic style, Spitaels et al., 2014) and specific type of wines (Walker, 2014; Chen et al., 2020), commercial products now largely employ the usage of starter cultures. The first pure yeast starter was created and used in beer production back in 1880s by E. C. Hansen from the Carlsberg laboratory in Denmark. In 1890s, also the first inoculation of a grape must with a yeast starter was performed. These practices became more common and Saccharomyces cerevisiae starters are primarily being used in wine and beer fermentations, to facilitate the consistency of fermented beverages resulting in products with stable characteristics, aromas and flavors as well as ensuring rapid fermentation times (Valero et al., 2007).
Yeast species belonging to the Saccharomyces genus have been extensively used in fermentation, and throughout the years the ability to ferment has evolved from the exposure to stressful conditions (Gallone et al., 2016). Saccharomyces “make-accumulate-tolerate-consume” strategy enables fast growth during anaerobic conditions, maximizes ethanol and flavor metabolites production together with preventing growth of antagonistic microbes by creating a hostile environment for them to survive (Piškur et al., 2006; Goold et al., 2017). During wine and beer fermentation yeasts are exposed to numerous stresses such as high osmotic pressure, oxidative stress, temperature shifts, low oxygen availability, CO2 accumulation, nutrient restraint and high ethanol concentration (Legras et al., 2018). Typically, Saccharomyces spp. expresses its fermentative ability either in mixture of high sugar environments such as brewing wort and grape juice or even in hydrolyzed lactose in fermented milk. As a result of this environmental variation, yeast strains diversified extensively in industrial settings and became adapted to the production of specific beverages (Table 1).
The extent of these adaptations can be easily detected in S. cerevisiae and S. pastorianus, the workhorses of beer fermentation, that have shown wide phenotypic variation to different beer fermentation conditions. These species have evolved so that their unique complexity and diversity can generate different beer related products. Some characteristics of industrial isolates are high consumption rate of complex sugars like maltose and maltotriose, enhanced tolerance to hyperosmotic stress, high ethanol production and simultaneous repression of undesired metabolites such as beer off-flavors (Gibson et al., 2013; Steensels et al., 2014; Hill, 2015; Gallone et al., 2018). Normally, brewing strains are able to ferment adequately a 12–14 P wort and produce 5–6% ethanol (v/v). However, some yeasts are suitable for “high gravity” brewing (i.e., high amount of total sugars diluted in water) as they are able to utilize the elevated sugar content and tolerate the higher ethanol concentrations. In high gravity brewing, highly concentrated wort is fermented to beer and then diluted to the desired ethanol concentration. It is a sustainable approach for increasing brewery yields, reduce production costs, and produce a variety of different products with higher or lower alcohol levels (Pátková et al., 2000; Piddocke et al., 2009; Caspeta et al., 2019). Wort gravity also affects the final beer flavor and the formation of volatiles and thus not all yeast strains are suitable for this fermentation (Piddocke et al., 2009; Lei et al., 2012). Industrial isolates from different sources have phenotypes associated with adaptation to that specific source. For example, rapid maltose utilization and fermentation is found in baker’s yeast (Bell et al., 2001) and higher ethanol production rates in sake and wine yeast (Uebayashi et al., 2018), however, the same ability varies significantly in non-industrial yeast strains.
The environmental discontinuity has facilitated genetic diversification and phenotypic plasticity of yeast strains. Molecular patterns of domestication have now been explored in industrial yeasts, and significant variation have been shown among Saccharomyces beer, wine, sake and cider strains. S. cerevisiae brewing strains have shown remarkable population differentiation and are polyphyletic deriving from different geographical beer clades such as German, Belgian, and British ale. On the contrary wine, sake, and bread yeast have not shown much phenotypic diversity and are monophyletic (Almeida et al., 2015; Gonçalves et al., 2016). Population genomic studies of hundreds of Saccharomyces yeast strains reveal a remarkable level of variation in recombination rates and patterns even across very closely related lineages which can potentially translate in different phenotypic characteristics (Liti et al., 2009; Schacherer et al., 2009; Almeida et al., 2015). Advances in sequencing technologies have helped identification of yeast’s genetic traits underpinning different phenotypes from a plethora of environments (Sniegowski, 1999; Cromie et al., 2013; David et al., 2014). For instance, High- Throughput Sequencing approaches facilitated the understanding of the grape microbiome in different environmental conditions. In grapes and berries the identified microbiome included species belonging to the family of Dothioraceae, Pleosporaceae, Saccharomycodaceae, Enterobacteriales, Pseudomonadales, Bacillales, and Rhodospirillales and differences in field origin were examined for its relevance to wine fermentation and production of flavor metabolites in Cannonau wine from Sardinia (Mezzasalma et al., 2017). Sequencing of polyploidy beer strains revealed a common genetic ancestry with wine strains from European and Asian lineages. Polyploidization facilitated the gain or loss of genetic variation related with brewing characteristics and also indicates the usage of co-cultures has been employed in fermented beverages (Fay et al., 2019).
Several mechanisms that accelerate evolution in environmental challenges and stressful conditions have been studied. Yeast adapt to new environments via smaller and/or larger genetic changes (Peter et al., 2018). Small variations can be caused from single nucleotide and frame shift mutations, insertions or deletions, which will end up creating alterations in the structure/function of the encoding protein or alterations in the gene expression. Larger changes include structural variation such as chromosomal rearrangements (duplications, translocations, and inversions), segmental duplication and gene copy number variation. Genetic variations can also be driven from interspecific hybridization with introgression and horizontal gene transfer events. This enables the generation of novel characteristics to the genome that it could not occur with other nucleotide arrangements.
Recent whole genome sequencing and large-scale phenotyping data for 157 S. cerevisiae industrial brewing strains revealed that these yeasts are genetically and phenotypically separated from their wild ancestors through complex domestication events in the man-made environments (Gallone et al., 2016). The analysis shows that industrial strains can tolerance various stresses, have better performance and lack sexual reproduction. Insertions and deletions of small or large fragments were detected in most of the strains analyzed with the size of the fragment varying from a few base pairs to complete chromosomes. Deletion patterns and copy number variations commonly found in beer strains are connected with the high aneuploidy or polyploidy characteristic of brewing strains. It was also observed that all the industrial strains even from different beer clades show clear marks of domestication and adaptation to industrial niches. This is consistent with the fact that the brewing industry is following common practices distinct from the wineries. In fact, wine yeast differs from the brewing ones, because of the seasonality of the wine production, the nutritional fluctuations and their sexual cycles. This can explain the population diversity (both in population size and in genome variety) of beer yeasts compared to wine yeasts. Chromosomal re-arrangements play an important role in the phenotypic variation of yeast strains both in the laboratory environment (Colson et al., 2004; Naseeb et al., 2016, 2017a) and in nature, where karyotypic instability is found in wild strains and can affect their performance when transferred to industrial settings (Carro et al., 2003; Dujon, 2010; Peter et al., 2018).
Genome re-arrangements and copy number variations results also in extensive alterations of the gene expression network (Naseeb et al., 2016) and it is not only limited to the function of a specific duplicated gene. Any fitness improvement is resulting from both environmental and genomic conditions and multiple changes in the transcriptome (Guan et al., 2007; Hakes et al., 2007; Naseeb et al., 2017a). Furthermore, differences have been identified in the copy number of genes involved in the metabolism of fermentable sugars such as maltose and maltotriose. Copy number variations are often reported as an adaptation mechanism to environmental changes (Figure 1A; Magadum et al., 2013). The maltose metabolism consists of 3 gene families: MALT, MALS, and MALR which comprise maltose transporters, maltases and regulator proteins, respectively (Brown et al., 2010). Fluctuations in chromosomal location and copy number of the involved MAL genes are present in many industrial strains. Typically, beer strains contain six or more copies of the MAL3 locus (Gonçalves et al., 2016). Some German yeast strains were found to contain 15 copies of the MAL31 gene while wine Saccharomyces strains contains only three copies (Gonçalves et al., 2016). Uptake and breakdown of maltose, the main carbon source in beer fermentation but not in grape/must, has been of great importance for the survival and performance of brewing yeast strains. This remarkable genetic alteration in maltose uptake is giving a great selection of brewing candidates that are able of fast utilization of this sugar. van der Broek and co-workers examined chromosomal number variation in S. pastorianus and its link with the phenotype. They analyzed three S. pastorianus W34/70 isolates that produced different diacetyl concentrations during beer fermentation. Diacetyl, a vicinal diketone, with butter flavor is considered an undesirable metabolite occurring through yeast valine metabolism during beer fermentation (Kusunoki and Ogata, 2012). Analysis of the DNA sequence of the valine biosynthetic genes (ILV2, ILV6, ILV5, and ILV3) in the three isolates did not reveal any single nucleotide polymorphism, however, copy number variation in the chromosomes carrying those genes was identified. This resulted in one isolate containing extra copies of the ILV5 and ILV3 genes, that responsible for the downstream catalysis of a-acetolactate the precursor of diacetyl. Thus, this strain was a low-diacetyl producing isolate of W34/70 (van den Broek et al., 2015).
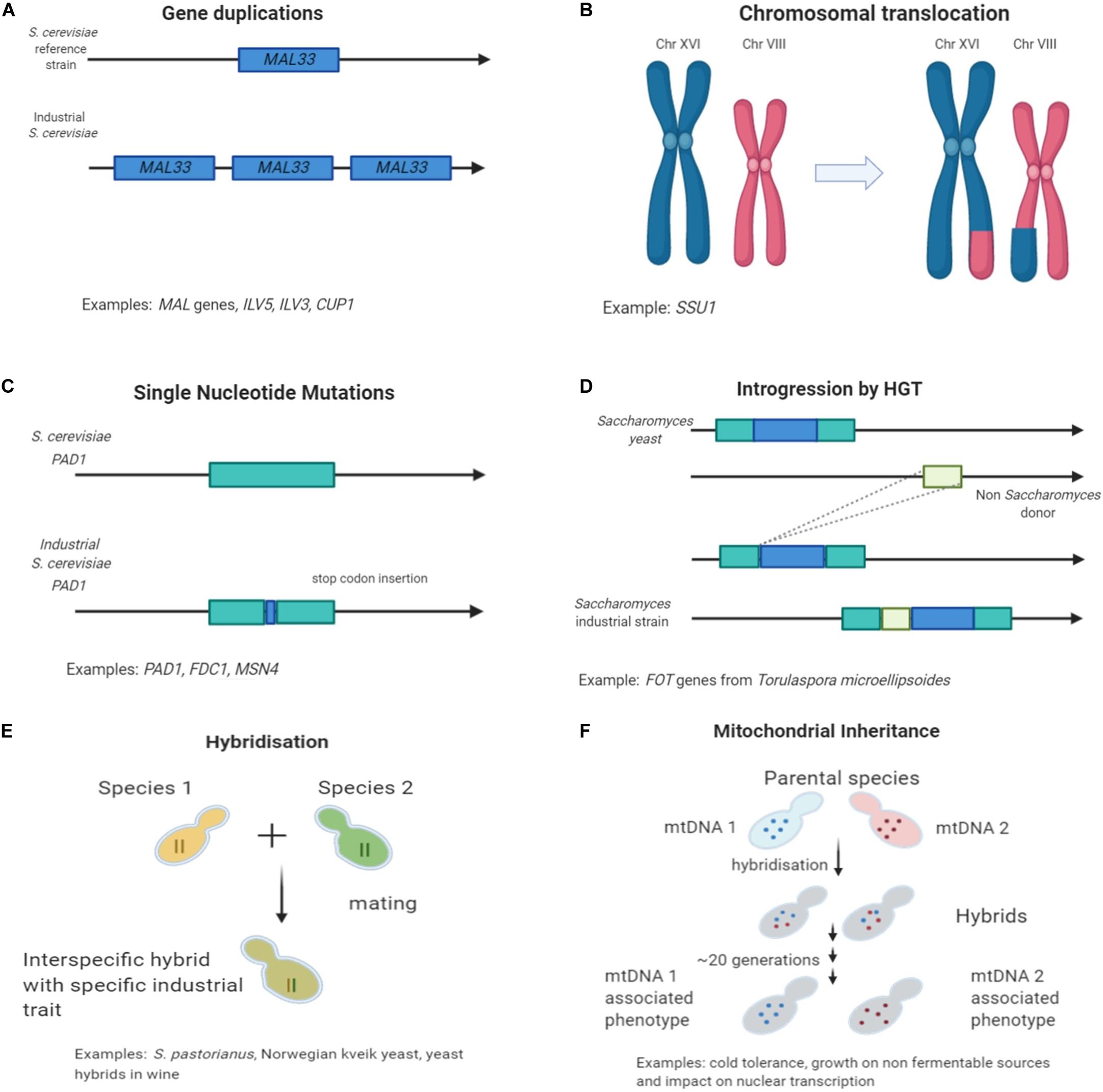
Figure 1. Mechanisms of genetic variation found in industrial Saccharomyces strains in fermentative environments. (A) Gene duplication and copy number variation; (B) Chromosomal re-arrangements; (C) Single nucleotide polymorphism causing loss of function; (D) Horizontal gene transfer events; (E) Interspecific hybridization; and (F) Mitochondrial inheritance.
In wine yeast, studies have shown significant adaptation motifs to sulfite compounds. Sulfite contained chemicals are used extensively as preservatives in wineries. A way that yeast can tolerate the excess levels of sulfite is by increasing the regulation of the sulfite uptake and efflux through the SSU1 plasma membrane pump encoded gene (Pérez-Ortín et al., 2002). An increase in expression of SSU1 was observed in wine yeast strains compared to laboratory strains. Studies about the mechanism of sulfite resistance, identified a chromosomal translocation (Figure 1B) and non-homologous recombination of the SSU1 gene promoter (Pérez-Ortín et al., 2002). Moreover, a chromosomal inversion between XVI and VIII connected with the SSU1 regulatory region also result in overexpression of SSU1 and thus in sulfite resistance to commercial wine strains (García-Ríos et al., 2019b).
Another adaptation in wine yeast has been triggered by the copper-based pesticides used in wineries. The CUP1 gene, responsible for copper binding and mediating resistance to high concentrations, was found in a higher copy number in wine strains associated with higher resistance to CuSO4 compared with natural isolates (Almeida et al., 2015). Liu et al. (2015) identified a promoter variant of CUP1 gene with increased expression suggesting that this benefit is involved in an adapting mechanism of the strains into a stressful condition. Interestingly, in organic vineyards where the usage of pesticides is strictly limited, a lower number of different yeast strains has been detected, as other wild micro-organisms naturally resistant to copper such as Aureobasidium pullulans and Starmerella bacillaris dominate (Grangeteau et al., 2017).
Yeast domestication studies for beer and wine using sequencing technologies have unravel traits and performance improvements in different populations. In beer, an example illustrating well the process of trait improvement through selection and domestication, is the loss of function of genes related with ferulic acid decarboxylation. 4-vinylguaiacol is a phenolic compound with a distinct clove-like aroma. The decarboxylation of ferulic acid to 4-vinylguaiacol is occurring through yeast metabolism under the regulation of genes PAD1 (phenylacrylic acid decarboxylase) and FDC1 (ferulic acid decarboxylase) (Gallone et al., 2016; Gonçalves et al., 2016). The production of 4-vinylguaiacol during beer fermentation is considered a phenolic off-flavor (POF), and the strains are described as POF+. The clove-like aroma is considered characteristic only in some specific style of beers, such as Belgian and German wheat beers, but even for those a low threshold of POF would be desirable (Scholtes et al., 2014). Yeasts used for the production of alcoholic beverages produce an amount of undesired metabolites characterized as off-flavors and ideally that accumulation should be limited. The biological role of PAD1 and FDC1 is to help detoxifying phenylacrylic acids from the cell walls of plants (Mukai et al., 2014), which explains why wild yeast express those genes in order to survive and proliferate in natural habitats. Genomic studies revealed that in many industrial brewing strains the genes appear to be inactive and have acquired a frameshift mutation or a premature stop codon in the PAD1 gene sequence (Figure 1C; Mukai et al., 2014; Chen et al., 2015). Interestingly, these type of mutations are not present in strains used in German like wheat beers. Such data show that different strains acquired different disruptive mutations, related to the presence of varied adaptive strategies in response to human selection against production of the POF+ character (Gallone et al., 2016).
Adaptation mechanisms due to different stressful conditions are also found yeast used in sake fermentation. These strains belong to the Saccharomyces cerevisiae Kyokai no. 7 group (K7). In sake brewing, the final ethanol content reaches almost 20%. Therefore, sake yeast genome has evolved to produce and accumulate high ethanol concentrations. The Kyokai strains express high fermentation rates. Both rapid fermentation and high ethanol production has been linked with environmental stress responses (Zhao and Bai, 2009). Studies in K7 yeast revealed a loss of function mutation in the genes MSN4, and MSN2 that are responsible for transcription factors regulation during different types of stresses. Interestingly, the K7 group acquired a dysfunctional MSN4 genes and results in high initial fermentation rate despite its lower stress tolerance compared to reference laboratory strains (Urbanczyk et al., 2011; Watanabe et al., 2011). The variant Km67 strain, belonging to the K7 sake group, has also been recently studied for its distinct characteristic of stress tolerance among the group. This strain has been used extensively and repeatedly as a starter culture for sake fermentation and surprisingly it doesn’t acquire the same loss-of-function mutation in stress response related genes and also confers unique sensory characteristics and high production of ethanol as the rest of the Kyokai group. This suggests that other underlying genomic adaptations have contributed to the phenotype and performance of Km67 and the strain represents a genetically distinguished isolate within the group (Takao et al., 2018). The same strain was also recently reported for high folate production compared to other strains in the K7 group but the mechanisms underlying this accumulation are yet to be determined (Shibata et al., 2019).
Horizontal gene transfer (HGT), including introgression of DNA fragments from one species to another, is a known mechanism to generate variation in prokaryotes and eukaryotes (Keeling and Palmer, 2008). In yeast, HGT has been proposed as a mechanism of genetic adaptation to a particular niche (Hall et al., 2005). For example, a wine yeast S. cerevisiae strain gained 2 FOT genes –responsible for encoding oligopeptide transporters- from Torulaspora microellipsoides by several HGT and re-arrangement events (Figure 1D). Gaining these genes conferred Saccharomyces a competitive advantage during wine fermentation as the strain could utilize more nitrogen sources and oligopeptides, enabling its cell viability and proliferation. As mentioned before, grape juice is often a nitrogen-limited environment thus the wine yeast are challenged from the availability (Marsit et al., 2015). In another study, in the wine strain S. cerevisiae EC1118, a unique gene has been identified contributing to glucose and fructose metabolism and adaptation to low nitrogen conditions. The genes were acquired from non S. cerevisiae donors mostly closely related to Zygosaccharomyces rouxii a wild species commonly found in wineries (Novo et al., 2009). HGT is a mechanism of acquisition of new genetic material between different species, not yet fully explored in eukaryotes, and further future studies will help understanding the causes, the likelihood and the environmental background of this genetic exchange.
Another mechanism, that has facilitated the evolution of industrial species, is the generation of interspecific hybrids (Figure 1E). The novel resulting combinations of genes (Nakao et al., 2009; Hewitt et al., 2014) and proteins (Piatkowska et al., 2013)is contributing to unique characteristics and advantages for the progenies compared to the parental strains. These unique phenotypes enable survival and proliferation in a new environment with a better performance over the parental species.
Interspecific hybridization has been extensively studied in the Saccharomyces. The lager yeast S. pastorianus, is a hybrid between S. cerevisiae and S. eubayanus and it is the most widely known and used in the beer industry. The history of S. pastorianus can be tied to lager beer brewing during the winter months requiring a cooler fermentation temperature. A hybridization between S. cerevisiae, a great fermenter strain, and S. eubayanus, a cryotolerant isolate, created a new yeast suitable for adapting and performing in the new demanding of the beer making (Martini and Martini, 1987; Nakao et al., 2009; Hewitt et al., 2014; Monerawela and Bond, 2018). Several other hybrids have been isolated from industrial applications such as, hybrids of S. cerevisiae and S. kudriavzevii in beer, cider and wine (Masneuf et al., 1998; González et al., 2008) and hybrids of S. cerevisiae and S. uvarum (Le Jeune et al., 2007). The molecular drivers and biochemical pathways important for the cryotolerance trait in S. kudriavzevii has been recently unveiled, showing that temperature-induced redox imbalances could be compensated by either increased glycerol accumulation or production of cytosolic acetaldehyde (Paget et al., 2014).
Recently, a S. cerevisiae × S. uvarum unique hybrid was isolated from the Norwegian kveik farmhouse yeast (Krogerus et al., 2018b). Sequencing and phenotypic investigation on the strain showed that, this hybrid has been generated in brewing conditions and results in desirable characteristics such as tolerance to a wide temperature range, tolerance to high ethanol and good production of ester-flavor compounds.
Hybridization events have also been reported in wine strains. Garcia-Rios and co-workers constructed a non-GMO S. cerevisiae x S. uvarum hybrid to improve the wine fermentation properties of the parental S. cerevisiae strain. They performed growth and fermentation tests in a variety of different temperatures and media to evaluate the cryotolerance character of the hybrid. It is know that wine fermentations in colder temperatures improve the character, quality and fruit-flavor of wine (Molina et al., 2007). The hybrids generated were evaluated and compared to their parental strains in competition experiments to generate phenotypic maps and identify the different recombination events from the expressed phenotypes (García-Ríos et al., 2019a). Wine hybrids were constructed in order to generate strains able to survive and proliferate in low nitrogen levels commonly found in grapes (Su et al., 2019). Nitrogen is essential for yeast metabolism and fermentation ability as it is also responsible for the accumulation of aroma-related compounds (Rollero et al., 2018). Occurrence of natural interspecific triploid hybrids is also found in fermentative environments: S. cerevisiae × S. cerevisiae × S. kudriavzevii hybrids have been isolated from wineries providing a growth advantage in cold temperatures and high production of volatile thiols (Borneman et al., 2012). Triploid and tetraploid hybrids are also common in beer, with S. pastorianus forming two groups based on its DNA content. Saaz (Group 1) lager yeast are allotriploid strains with fermentation phenotypic characteristic close to the S. eubayanus parent while the Frohberg strains are allotetraploid with a similar fermentation performance to the S. cerevisiae parent (Walther et al., 2014). Hybrids generation among the Saccharomyces species have therefore resulted in a variety of phenotypes with great industrial fermentative potential (Bellon et al., 2011). Stress responses associated with wine and beer fermentation seem to have influenced the spontaneous generation of natural hybrids with different physiological traits.
Another important factor related to the occurrence of specific characteristics in industrial strains is the inheritance of mitochondrial DNA (mtDNA). During hybridization, different hybrids can inherit the mitochondria from either one or the other parental species (Figure 1F).
Compared to the nuclear genomes, the mtDNA in Saccharomyces cerevisiae is more diverted and highly assorted. Structural rearrangements are rare in mtDNA resulting in few cases of DNA loss (De Chiara et al., 2020), however, mitochondrial recombination is common and can lead to phenotypic differentiation if enough divergence is present in the parental species (Leducq et al., 2017). Recent studies showed a strong influence on the different parental mtDNA in S. pastorianus strains related with adaptation to cold temperatures (Baker et al., 2019; Hewitt et al., 2020). Yeast mitochondria contribute to evolutionary divergence of cold tolerant strains (Li et al., 2019) and the type of mtDNA inherited in the hybrids affects both the cellular fitness in different nutritional conditions (Albertin et al., 2013; Hewitt et al., 2020), and the nuclear transcription in the hybrid (Hewitt et al., 2020).
The strong impact of molecular techniques and sequencing technologies have shed a light into evolutionary insights of industrial Saccharomyces species. Yeast domestication in man-made environments have been driven due to different stimuli that can include temperature and nutrient stresses, microbial competition, and ethanol and CO2 toxicity. Evidence of yeast adaptation in the fermentative environments show that an incredible variety of mechanisms, such as gene duplication events, chromosomal rearrangement, hybridization, HGT, and type of mitochondria inherited, contribute to re-shape the yeast genome for better survival traits.
Such acquired knowledge on yeast biology and evolution can now enable researchers to work on strain improvement and generate candidates that will facilitate the food and beverages industry. This include approaches such as selection of isolates with desirable characteristics, usage of non-conventional yeast and generation of hybrids.
Adaptive laboratory evolution experiments have introduced specific mutations in relevant traits or aneuploidy in domesticated yeast (Gorter De Vries et al., 2019), improved fermentation performance in polyploid strains and hybrids (Voordeckers et al., 2015; Krogerus et al., 2018a) and osmotic stress response resulted in shorter fermentation times (Ekberg et al., 2013). Laboratory adaptive evolution can lead to identification and observation of important fermentation characteristics as well as the ability to design and perform future experiments that will lead to yeast strain with desired industrial properties (Iattici et al., 2020).
Evolutionary selection is still the methodology of choice for yeast strain improvement and it’s usually preferred to classical breeding techniques. Breeding can generate progeny with desired phenotypic traits but is much more challenging because of yeast aneuploidy and poor sporulation efficiencies (Codon et al., 1995). The phenotypic characterization and evaluation of new strains improved via breeding can help the selection superior segregants that could be implemented in food and beverages industry (Sanchez et al., 2012; Figueiredo et al., 2017).
Interspecific hybrid sterility is a drawback in the generation of offspring with different combination of desirable traits. Several works have tried to overcome sterility through allotetraploidization. Through this method we are able to obtain fertile diploid spores, from an allotetraploid, allowing a recombination of traits between the different species (Greig et al., 2002; Sebastiani et al., 2002). The construction of complex de novo interspecific hybrids strains can result in phenotypic traits that weaken or strengthen under meiotic recombination and increase the diversity of the existing industrial candidates (Krogerus et al., 2017; Peris et al., 2020).
The fermentative potential of Saccharomyces species has not yet fully explored. S. kudriavzevii and S. uvarum are cold tolerant strains that have been isolated from fermentative environments. S. mikatae, S. paradoxus, S. jurei, yeast that possess traits such as cold tolerance and maltose utilization could be further exploited through interspecific hybridization (Fleet, 2006; Salvadó et al., 2011; Naseeb et al., 2017b, 2018). Hybrid strains can also address the need of aromatic novelty in fermented beverages (Nikulin et al., 2018).
Furthermore, applying population genomic studies will facilitate exploring the biodiversity of non-conventional yeast such as Brettanomyces bruxellensis, Torulaspora delbrueckii. These species, commonly found in wine and beer fermentations and characterized as spoilage yeast, have increasingly gained scientific and biotechnological interest (Avramova et al., 2018; Zhang et al., 2018). The occurrence and industrial potential of non-Saccharomyces yeast should further be considered as it will diversify the industrial strains suitable for generation of novel food products (Basso et al., 2016). In addition, the usage of mixed starter cultures of Saccharomyces and non-Saccharomyces strains can guide new product development with distinct flavors and performance characteristics. The exo-metabolites resulting from co-culturing strains during fermentation can influence yeast-yeast interactions and ultimately alter the population structure due to different competitive pressure (Ye et al., 2014).
Although there is now clear evidence for patterns of evolution and adaptation of Saccharomyces strains in the fermentative environments, not much is yet known on the survival of wild strains in natural habitats. Adaptation trajectories and mutations arising from extreme environmental conditions are still yet to be explored in wild yeast isolates (Aouizerat et al., 2019). Further exploration on the effect of harsh conditions and climate fluctuations on different yeast strains and species will broaden the understanding on how to maintain their biodiversity and on the importance of yeast-environment interactions.
KG performed the literature search with the inputs of DD and MC. KG and DD wrote the manuscript with the input of MC. All authors contributed to the article and approved the submitted version.
This work was supported by H2020-MSCA-ITN-2017 (764364). KG was supported by Innovate-UK grant between the University of Manchester and Cloudwater Brew.
MC was employed by the company Cloudwater Brew Co.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Federico Visinoni and Javier Pinto Aguirre for helpful comments and suggestions on the manuscript.
Albertin, W., da Silva, T., Rigoulet, M., Salin, B., Masneuf-Pomarede, I., de Vienne, D., et al. (2013). The mitochondrial genome impacts respiration but not fermentation in interspecific Saccharomyces hybrids. PLoS One 8:e75121. doi: 10.1371/journal.pone.0075121
Aldrete-Tapia, J. A., Miranda-Castilleja, D. E., Arvizu-Medrano, S. M., and Hernández-Iturriaga, M. (2018). Selection of yeast strains for tequila fermentation based on growth dynamics in combined fructose and ethanol media. J. Food Sci. 83, 419–423. doi: 10.1111/1750-3841.14031
Almeida, P., Barbosa, R., Zalar, P., Imanishi, Y., Shimizu, K., Turchetti, B., et al. (2015). A population genomics insight into the Mediterranean origins of wine yeast domestication. Mol. Ecol. 24, 5412–5427. doi: 10.1111/mec.13341
Aouizerat, T., Gelman, D., Szitenberg, A., Gutman, I., Glazer, S., Reich, E., et al. (2019). Eukaryotic adaptation to years-long starvation resembles that of bacteria. iScience 19, 545–558. doi: 10.1016/j.isci.2019.08.002
Avramova, M., Cibrario, A., Peltier, E., Coton, M., Coton, E., Schacherer, J., et al. (2018). Brettanomyces bruxellensis population survey reveals a diploid-triploid complex structured according to substrate of isolation and geographical distribution. Sci. Rep. 8:4136. doi: 10.1038/s41598-018-22580-7
Baker, E. C. P., Peris, D., Moriarty, R. V., Li, X. C., Fay, J. C., and Hittinger, C. T. (2019). Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 5:eaav1869. doi: 10.1126/sciadv.aav1869
Basso, R. F., Alcarde, A. R., and Portugal, C. B. (2016). Could non-Saccharomyces yeasts contribute on innovative brewing fermentations? Food Res. Int. 86, 112–120. doi: 10.1016/j.foodres.2016.06.002
Bell, P. J. L., Higgins, V. J., and Attfield, P. V. (2001). Comparison of fermentative capacities of industrial baking and wild-type yeasts of the species Saccharomyces cerevisiae in different sugar media. Lett. Appl. Microbiol. 32, 224–229. doi: 10.1046/j.1472-765X.2001.00894.x
Bellon, J. R., Eglinton, J. M., Siebert, T. E., Pollnitz, A. P., Rose, L., De Barros Lopes, M., et al. (2011). Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl. Microbiol. Biotechnol. 91, 603–612. doi: 10.1007/s00253-011-3294-3
Borneman, A. R., Desany, B. A., Riches, D., Affourtit, J. P., Forgan, A. H., Pretorius, I. S., et al. (2012). The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 12, 88–96. doi: 10.1111/j.1567-1364.2011.00773.x
Brown, C. A., Murray, A. W., and Verstrepen, K. J. (2010). Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 20, 895–903. doi: 10.1016/j.cub.2010.04.027
Carro, D., Bartra, E., and Piña, B. (2003). Karyotype rearrangements in a wine yeast strain by rad52-dependent and rad52-independent mechanisms. Appl. Environ. Microbiol. 69, 2161–2165. doi: 10.1128/AEM.69.4.2161-2165.2003
Caspeta, L., Coronel, J., Montes de Oca, A., Abarca, E., González, L., and Martínez, A. (2019). Engineering high−gravity fermentations for ethanol production at elevated temperature with Saccharomyces cerevisiae. Biotechnol. Bioeng. 116, 2587–2597. doi: 10.1002/bit.27103
Chen, P., Dong, J., Yin, H., Bao, X., Chen, L., He, Y., et al. (2015). Single nucleotide polymorphisms and transcription analysis of genes involved in ferulic acid decarboxylation among different beer yeasts. J. Inst. Brew. 121, 481–489. doi: 10.1002/jib.249
Chen, Y., Zhang, W., Yi, H., Wang, B., Xiao, J., Zhou, X., et al. (2020). Microbial community composition and its role in volatile compound formation during the spontaneous fermentation of ice wine made from Vidal grapes. Proc. Biochem. 92, 365–377. doi: 10.1016/j.procbio.2020.01.027
Codon, A. C., Gasent-Ramirez, J. M., and Benitez, T. (1995). Factors which affect the frequency of sporulation and tetrad formation in Saccharomyces cerevisiae baker’s yeasts. Appl. Environ. Microbiol. 61, 630–638. doi: 10.1128/aem.61.2.630-638.1995
Colson, I., Delneri, D., and Oliver, S. G. (2004). Effects of reciprocal chromosomal translocations on the fitness of Saccharomyces cerevisiae. EMBO Rep. 5, 392–398. doi: 10.1038/sj.embor.7400123
Cromie, G. A., Hyma, K. E., Ludlow, C. L., Garmendia-Torres, C., Gilbert, T. L., May, P., et al. (2013). Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 3, 2163–2171. doi: 10.1534/g3.113.007492
David, V., Terrat, S., Herzine, K., Claisse, O., Rousseaux, S., Tourdot-Maréchal, R., et al. (2014). High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biotechnol. 41, 811–821. doi: 10.1007/s10295-014-1427-2
De Chiara, M., Friedrich, A., Barré, B., Breitenbach, M., Schacherer, J., and Liti, G. (2020). Discordant evolution of mitochondrial and nuclear yeast genomes at population level. BMC Biol. 18:49. doi: 10.1186/s12915-020-00786-4
Dujon, B. (2010). Yeast Evolutionary Genomics. Berlin: Nature Publishing Group, doi: 10.1038/nrg2811
Eglinton, J. M., McWilliam, S. J., Fogarty, M. W., Francis, I. L., Kwiatkowski, M. J., Hoj, P. B., et al. (2000). The effect of Saccharomyces bayanus-mediated fermentation on the chemical composition and aroma profile of Chardonnay wine. Aust. J. Grape Wine Res. 6, 190–196. doi: 10.1111/j.1755-0238.2000.tb00178.x
Ekberg, J., Rautio, J., Mattinen, L., Vidgren, V., Londesborough, J., and Gibson, B. R. (2013). Adaptive evolution of the lager brewing yeast Saccharomyces pastorianus for improved growth under hyperosmotic conditions and its influence on fermentation performance. FEMS Yeast Res. 13, 335–349. doi: 10.1111/1567-1364.12038
Fay, J. C., Liu, P., Ong, G. T., Dunham, M. J., Cromie, G. A., Jeffery, E. W., et al. (2019). A polyploid admixed origin of beer yeasts derived from European and Asian wine populations. PLoS Biol. 17:e3000147. doi: 10.1371/journal.pbio.3000147
Figueiredo, B. I. C., Saraiva, M. A. F., de Souza Pimenta, P. P., de Souza Testasicca, M. C., Sampaio, G. M. S., da Cunha, A. C., et al. (2017). New lager brewery strains obtained by crossing techniques using cachaça (Brazilian spirit) yeasts. Appl. Environ. Microbiol. 83:e01582-17. doi: 10.1128/AEM.01582-17
Fleet, G. H. (2006). “Saccharomyces and related genera,” in Food Spoilage Microorganisms, ed. C. W. de Blackburn (Amsterdam: Elsevier), 306–335. doi: 10.1533/9781845691417.3.306
Gallone, B., Mertens, S., Gordon, J. L., Maere, S., Verstrepen, K. J., and Steensels, J. (2018). Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr. Opin. Biotechnol. 49, 148–155. doi: 10.1016/j.copbio.2017.08.005
Gallone, B., Steensels, J., Prahl, T., Soriaga, L., Saels, V., Herrera-Malaver, B., et al. (2016). Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397–1410.e16. doi: 10.1016/j.cell.2016.08.020
García-Ríos, E., Guillén, A., De La Cerda, R., Pérez-Través, L., Querol, A., and Guillamón, J. M. (2019a). Improving the cryotolerance of wine yeast by interspecific hybridization in the genus Saccharomyces. Front. Microbiol. 10:3232. doi: 10.3389/fmicb.2018.03232
García-Ríos, E., Nuévalos, M., Barrio, E., Puig, S., and Guillamón, J. M. (2019b). A new chromosomal rearrangement improves the adaptation of wine yeasts to sulfite. Environ. Microbiol. 21, 1771–1781. doi: 10.1111/1462-2920.14586
Gibson, B. R., Storgårds, E., Krogerus, K., and Vidgren, V. (2013). Comparative physiology and fermentation performance of Saaz and Frohberg lager yeast strains and the parental species Saccharomyces eubayanus. Yeast 30, 255–266. doi: 10.1002/yea.2960
Gonçalves, M., Pontes, A., Almeida, P., Barbosa, R., Serra, M., Libkind, D., et al. (2016). Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 26, 2750–2761. doi: 10.1016/j.cub.2016.08.040
González, S. S., Barrio, E., and Querol, A. (2008). Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 74, 2314–2320. doi: 10.1128/AEM.01867-07
Goold, H. D., Kroukamp, H., Williams, T. C., Paulsen, I. T., Varela, C., and Pretorius, I. S. (2017). Yeast’s balancing act between ethanol and glycerol production in low-alcohol wines. Microb. Biotechnol. 10, 264–278. doi: 10.1111/1751-7915.12488
Gorter De Vries, A. R., Voskamp, M. A., Van Aalst, A. C. A., Kristensen, L. H., Jansen, L., Van Den Broek, M., et al. (2019). Laboratory evolution of a Saccharomyces cerevisiae × Saccharomyces eubayanus hybrid under simulated lager-brewing conditions. Front. Genet. 10:242. doi: 10.3389/fgene.2019.00242
Grangeteau, C., David, V., Hervé, A., Guilloux-Benatier, M., and Rousseaux, S. (2017). The sensitivity of yeasts and yeasts-like fungi to copper and sulfur could explain lower yeast biodiversity in organic vineyards. FEMS Yeast Res. 17:fox092. doi: 10.1093/femsyr/fox092
Greig, D., Borts, R. H., Louis, E. J., and Travisano, M. (2002). Epistasis and hybrid sterility in Saccharomyces. Proc. R. Soc. Lond. Ser. B Biol. Sci. 269, 1167–1171. doi: 10.1098/rspb.2002.1989
Guan, Y., Dunham, M. J., and Troyanskaya, O. G. (2007). Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics 175, 933–943. doi: 10.1534/genetics.106.064329
Hakes, L., Pinney, J. W., Lovell, S. C., Oliver, S. G., and Robertson, D. L. (2007). All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 8:R209. doi: 10.1186/gb-2007-8-10-r209
Hall, C., Brachat, S., and Dietrich, F. S. (2005). Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryotic Cell 4, 1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005
Hewitt, S. K., Donaldson, I. J., Lovell, S. C., and Delneri, D. (2014). Sequencing and characterisation of rearrangements in three S. pastorianus strains reveals the presence of chimeric genes and gives evidence of breakpoint reuse. PLoS One 9:e0092203. doi: 10.1371/journal.pone.0092203
Hewitt, S. K., Duangrattanalert, K., Burgis, T., Zeef, L. A. H., Naseeb, S., and Delneri, D. (2020). Plasticity of mitochondrial DNA inheritance and its impact on nuclear gene transcription in yeast hybrids. Microorganisms 8:494. doi: 10.3390/microorganisms8040494
Hill, A. E. (2015). “Introduction to brewing microbiology,” in Brewing Microbiology: Managing Microbes, Ensuring Quality and Valorising Waste, ed. A. Hill (Cambridge: Woodhead Publishing), xxvii–xxix. doi: 10.1016/B978-1-78242-331-7.02001-3
Iattici, F., Catallo, M., and Solieri, L. (2020). Designing new yeasts for craft brewing: when natural biodiversity meets biotechnology. Beverages 6:3. doi: 10.3390/beverages6010003
Jackson, R. S. (2014). “Specific and distinctive wine styles,” in Wine Science, ed. R. Jackson (Amsterdam: Elsevier), 677–759. doi: 10.1016/b978-0-12-381468-5.00009-9
Katou, T., Namise, M., Kitagaki, H., Akao, T., and Shimoi, H. (2009). QTL mapping of sake brewing characteristics of yeast. J. Biosci. Bioeng. 107, 383–393. doi: 10.1016/j.jbiosc.2008.12.014
Keeling, P. J., and Palmer, J. D. (2008). Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618. doi: 10.1038/nrg2386
Kelly, J., Yang, F., Dowling, L., Nurgel, C., Beh, A., Di Profio, F., et al. (2018). Characterization of Saccharomyces bayanus CN1 for fermenting partially dehydrated grapes grown in cool climate winemaking regions. Fermentation 4, 1–13. doi: 10.3390/fermentation4030077
Krogerus, K., Holmström, S., and Gibson, B. (2018a). Enhanced wort fermentation with de novo lager hybrids adapted to high-ethanol environments. Appl. Environ. Microbiol. 84:e02302-17. doi: 10.1128/AEM.02302-17
Krogerus, K., Preiss, R., and Gibson, B. (2018b). A unique Saccharomyces cerevisiae × Saccharomyces uvarum hybrid isolated from norwegian farmhouse beer: characterization and reconstruction. Front. Microbiol. 9:2253. doi: 10.3389/fmicb.2018.02253
Krogerus, K., Seppänen-Laakso, T., Castillo, S., and Gibson, B. (2017). Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae×Saccharomyces eubayanus hybrids. Microb. Cell Fact. 16, 1–22. doi: 10.1186/s12934-017-0679-8
Kusunoki, K., and Ogata, T. (2012). Construction of self-cloning bottom-fermenting yeast with low vicinal diketone production by the homo-integration of ILV5. Yeast 29, 435–442. doi: 10.1002/yea.2922
Le Jeune, C., Lollier, M., Demuyter, C., Erny, C., Legras, J.-L., Aigle, M., et al. (2007). Characterization of natural hybrids of Saccharomyces cerevisiae and Saccharomyces bayanus var. uvarum. FEMS Yeast Res. 7, 540–549. doi: 10.1111/j.1567-1364.2007.00207.x
Leducq, J. B., Henault, M., Charron, G., Nielly-Thibault, L., Terrat, Y., Fiumera, H. L., et al. (2017). Mitochondrial recombination and introgression during speciation by hybridization. Mol. Biol. Evol. 34, 1947–1959. doi: 10.1093/molbev/msx139
Legras, J. L., Galeote, V., Bigey, F., Camarasa, C., Marsit, S., Nidelet, T., et al. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35, 1712–1727. doi: 10.1093/molbev/msy066
Lei, H., Zhao, H., Yu, Z., and Zhao, M. (2012). Effects of wort gravity and nitrogen level on fermentation performance of brewer’s yeast and the formation of flavor volatiles. Appl. Biochem. Biotechnol. 166, 1562–1574. doi: 10.1007/s12010-012-9560-8
Li, X. C., Peris, D., Hittinger, C. T., Sia, E. A., and Fay, J. C. (2019). Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci. Adv. 5:eaav1848. doi: 10.1126/sciadv.aav1848
Liti, G., Carter, D. M., Moses, A. M., Warringer, J., Parts, L., James, S. A., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337–341. doi: 10.1038/nature07743
Liu, J., Martin-Yken, H., Bigey, F., Dequin, S., François, J. M., and Capp, J. P. (2015). Natural yeast promoter variants reveal epistasis in the generation of transcriptional-mediated noise and its potential benefit in stressful conditions. Genome Biol. Evol. 7, 969–984. doi: 10.1093/gbe/evv047
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., and Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet. 92, 155–161. doi: 10.1007/s12041-013-0212-8
Marsit, S., Mena, A., Bigey, F., Sauvage, F.-X., Couloux, A., Guy, J., et al. (2015). Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol. Biol. Evol. 32, 1695–1707. doi: 10.1093/molbev/msv057
Martínez-Rodríguez, A., Carrascosa, A. V., Barcenilla, J. M., Angeles Pozo-Bayón, M., and Carmen Polo, M. (2001). Autolytic capacity and foam analysis as additional criteria for the selection of yeast strains for sparkling wine production. Food Microbiol. 18, 183–191. doi: 10.1006/fmic.2000.0390
Martini, A. V., and Martini, A. (1987). Three newly delimited species of Saccharomyces sensu stricto. Antonie Leeuwenhoek 53, 77–84. doi: 10.1007/bf00419503
Masneuf, I., Hansen, J., Groth, C., Piskur, J., and Dubourdieu, D. (1998). New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64, 3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998
Meier-Dörnberg, T., Kory, O. I, Jacob, F., Michel, M., and Hutzler, M. (2018). Saccharomyces cerevisiae variety diastaticus friend or foe?—spoilage potential and brewing ability of different Saccharomyces cerevisiae variety diastaticus yeast isolates by genetic, phenotypic and physiological characterizati. FEMS Yeast Res. 18:foy023. doi: 10.1093/femsyr/foy023
Mezzasalma, V., Sandionigi, A., Bruni, I., Bruno, A., Lovicu, G., Casiraghi, M., et al. (2017). Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS One 12:e0184615. doi: 10.1371/journal.pone.0184615
Molina, A. M., Swiegers, J. H., Varela, C., Pretorius, I. S., and Agosin, E. (2007). Influence of wine fermentation temperature on the synthesis of yeast-derived volatile aroma compounds. Appl. Microbiol. Biotechnol. 77, 675–687. doi: 10.1007/s00253-007-1194-3
Monerawela, C., and Bond, U. (2018). The hybrid genomes of Saccharomyces pastorianus?: a current perspective. Yeast 35, 39–50. doi: 10.1002/yea.3250
Mukai, N., Masaki, K., Fujii, T., and Iefuji, H. (2014). Single nucleotide polymorphisms of PAD1 and FDC1 show a positive relationship with ferulic acid decarboxylation ability among industrial yeasts used in alcoholic beverage production. J. Biosci. Bioeng. 118, 50–55. doi: 10.1016/j.jbiosc.2013.12.017
Nakao, Y., Kanamori, T., Itoh, T., Kodama, Y., Rainieri, S., Nakamura, N., et al. (2009). Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 16, 115–129. doi: 10.1093/dnares/dsp003
Naseeb, S., Alsammar, H., Burgis, T., Donaldson, I., Knyazev, N., Knight, C., et al. (2018). Whole genome sequencing, de novo assembly and phenotypic profiling for the new budding yeast species Saccharomyces jurei. G3 8, 2967–2977. doi: 10.1534/g3.118.200476
Naseeb, S., Ames, R. M., Delneri, D., and Lovell, S. C. (2017a). Rapid functional and evolutionary changes follow gene duplication in yeast. Proc. R. Soc. B Biol. Sci. 284:20171393. doi: 10.1098/rspb.2017.1393
Naseeb, S., Carter, Z., Minnis, D., Donaldson, I., Zeef, L., and Delneri, D. (2016). Widespread impact of chromosomal inversions on gene expression uncovers robustness via phenotypic buffering. Mol. Biol. Evol. 33, 1679–1696. doi: 10.1093/molbev/msw045
Naseeb, S., James, S. A., Alsammar, H., Michaels, C. J., Gini, B., Nueno-Palop, C., et al. (2017b). Saccharomyces jurei sp. Nov., isolation and genetic identification of a novel yeast species from Quercus robur. Int. J. Syst. Evol. Microbiol. 67, 2046–2052. doi: 10.1099/ijsem.0.002013
Nikulin, J., Krogerus, K., and Gibson, B. (2018). Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 35, 113–127. doi: 10.1002/yea.3246
Novo, M., Bigey, F., Beyne, E., Galeote, V., Gavory, F., Mallet, S., et al. (2009). Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U.S.A. 106, 16333–16338. doi: 10.1073/pnas.0904673106
Paget, C. M., Schwartz, J. M., and Delneri, D. (2014). Environmental systems biology of cold-tolerant phenotype in Saccharomyces species adapted to grow at different temperatures. Mol. Ecol. 23, 5241–5257. doi: 10.1111/mec.12930
Pando Bedriñana, R., Mangas Alonso, J. J., and Suárez Valles, B. (2017). Evaluation of autochthonous Saccharomyces bayanus strains under stress conditions for making ice ciders. Food Sci. Technol. 81, 217–225. doi: 10.1016/j.lwt.2017.03.055
Pátková, J., Šmogrovičová, D., Dömény, Z., and Bafrncová, P. (2000). Very high gravity wort fermentation by immobilised yeast. Biotechnol. Lett. 22, 1173–1177. doi: 10.1023/A:1005689313775
Pérez-Ortín, J. E., Querol, A., Puig, S., and Barrio, E. (2002). Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 12, 1533–1539. doi: 10.1101/gr.436602
Peris, D., Alexander, W. G., Fisher, K. J., Moriarty, R. V., Basuino, M. G., Ubbelohde, E. J., et al. (2020). Synthetic hybrids of six yeast species. Nat. Commun. 11:2085. doi: 10.1038/s41467-020-15559-4
Peter, J., De Chiara, M., Friedrich, A., Yue, J. X., Pflieger, D., Bergström, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Piatkowska, E. M., Naseeb, S., Knight, D., and Delneri, D. (2013). Chimeric protein complexes in hybrid species generate novel phenotypes. PLoS Genet. 9:e1003836. doi: 10.1371/journal.pgen.1003836
Piddocke, M. P., Kreisz, S., Heldt-Hansen, H. P., Nielsen, K. F., and Olsson, L. (2009). Physiological characterization of brewer’s yeast in high-gravity beer fermentations with glucose or maltose syrups as adjuncts. Appl. Microbiol. Biotechnol. 84, 453–464. doi: 10.1007/s00253-009-1930-y
Piškur, J., Rozpedowska, E., Polakova, S., Merico, A., and Compagno, C. (2006). How did Saccharomyces evolve to become a good brewer? Trends Genet. 22, 183–186. doi: 10.1016/j.tig.2006.02.002
Rollero, S., Bloem, A., Ortiz-Julien, A., Camarasa, C., and Divol, B. (2018). Fermentation performances and aroma production of non-conventional wine yeasts are influenced by nitrogen preferences. FEMS Yeast Res. 18:foy055. doi: 10.1093/femsyr/foy055
Salvadó, Z., Arroyo-López, F. N., Guillamón, J. M., Salazar, G., Querol, A., and Barrio, E. (2011). Temperature adaptation Markedly determines evolution within the genus Saccharomyces. Appl. Environ. Microbiol. 77, 2292–2302. doi: 10.1128/AEM.01861-10
Sanchez, R. G., Solodovnikova, N., and Wendland, J. (2012). Breeding of lager yeast with Saccharomyces cerevisiae improves stress resistance and fermentation performance. Yeast 29, 343–355. doi: 10.1002/yea.2914
Schacherer, J., Shapiro, J. A., Ruderfer, D. M., and Kruglyak, L. (2009). Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458, 342–345. doi: 10.1038/nature07670
Scholtes, C., Nizet, S., and Collin, S. (2014). Guaiacol and 4-Methylphenol as specific markers of torrefied malts. fate of volatile phenols in special beers through aging. J. Agric. Food Chem. 62, 9522–9528. doi: 10.1021/jf5015654
Sebastiani, F., Barberio, C., Casalone, E., Cavalieri, D., and Polsinelli, M. (2002). Crosses between Saccharomyces cerevisiae and Saccharomyces bayanus generate fertile hybrids. Res. Microbiol. 153, 53–58. doi: 10.1016/S0923-2508(01)01286-4
Shibata, Y., Yamada, T., Morimoto, T., Fujii, T., Akao, T., Goshima, T., et al. (2019). Mechanism of high folate accumulation in a sake yeast other than Kyokai yeasts. J. Biosci. Bioeng. 129, 1–5. doi: 10.1016/j.jbiosc.2019.07.008
Sniegowski, P. (1999). Evolution: the genomics of adaptation in yeast. Curr. Biol. 9, 897–898. doi: 10.1016/s0960-9822(00)80078-0
Spitaels, F., Wieme, A. D., Janssens, M., Aerts, M., Daniel, H.-M., Van Landschoot, A., et al. (2014). The microbial diversity of traditional spontaneously fermented lambic beer. PLoS One 9:e95384. doi: 10.1371/journal.pone.0095384
Steensels, J., Meersman, E., Snoek, T., Saels, V., and Verstrepen, K. J. (2014). Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 80, 6965–6975. doi: 10.1128/AEM.02235-14
Su, Y., Gamero, A., Rodríguez, M. E., Lopes, C. A., Querol, A., and Guillamón, J. M. (2019). Interspecific hybridisation among diverse Saccharomyces species: a combined biotechnological solution for low-temperature and nitrogen-limited wine fermentations. Int. J. Food Microbiol. 310:108331. doi: 10.1016/j.ijfoodmicro.2019.108331
Takao, Y., Takahashi, T., Yamada, T., Goshima, T., Isogai, A., Sueno, K., et al. (2018). Characteristic features of the unique house sake yeast strain Saccharomyces cerevisiae Km67 used for industrial sake brewing. J. Biosci. Bioeng. 126, 617–623. doi: 10.1016/j.jbiosc.2018.05.008
Uebayashi, K., Shimizu, H., and Matsuda, F. (2018). Comparative analysis of fermentation and enzyme expression profiles among industrial Saccharomyces cerevisiae strains. Appl. Microbiol. Biotechnol. 102, 7071–7081. doi: 10.1007/s00253-018-9128-9
Urbanczyk, H., Noguchi, C., Wu, H., Watanabe, D., Akao, T., Takagi, H., et al. (2011). Sake yeast strains have difficulty in entering a quiescent state after cell growth cessation. J. Biosci. Bioeng. 112, 44–48. doi: 10.1016/j.jbiosc.2011.03.001
Valero, E., Cambon, B., Schuller, D., Casal, M., and Dequin, S. (2007). Biodiversity of Saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Yeast Res. 7, 317–329. doi: 10.1111/j.1567-1364.2006.00161.x
van den Broek, M., Bolat, I., Nijkamp, J. F., Ramos, E., Luttik, M. A. H., Koopman, F., et al. (2015). Chromosomal copy number variation in Saccharomyces pastorianus is evidence for extensive genome dynamics in industrial lager brewing strains. Appl. Environ. Microbiol. 81, 6253–6267. doi: 10.1128/AEM.01263-15
Voordeckers, K., Kominek, J., Das, A., Espinosa-Cantú, A., De Maeyer, D., Arslan, A., et al. (2015). Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet. 11:e1005635. doi: 10.1371/journal.pgen.1005635
Walker, G., Brosnan, J., Bringhurst, T., and Jack, F. (2012). Chapter 16 Selecting new distilling yeasts for improved fermentation and for sustainability. Yeast 1–11.
Walker, G. M. (2014). “Wines: microbiology of winemaking,” in Encyclopedia of Food Microbiology: Second Edition, Vol. 3, eds C. Batt and C. A. Batt (New York, NY: Academic Press), 787–792. doi: 10.1016/B978-0-12-384730-0.00356-6
Walther, A., Hesselbart, A., and Wendland, J. (2014). Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. G3 4, 783–793. doi: 10.1534/g3.113.010090
Watanabe, D., Wu, H., Noguchi, C., Zhou, Y., Akao, T., and Shimoi, H. (2011). Enhancement of the initial rate of ethanol fermentation due to dysfunction of yeast stress response components Msn2p and/or Msn4p. Appl. Environ. Microbiol. 77, 934–941. doi: 10.1128/AEM.01869-10
Ye, M., Yue, T., and Yuan, Y. (2014). Effects of sequential mixed cultures of Wickerhamomyces anomalus and Saccharomyces cerevisiae on apple cider fermentation. FEMS Yeast Res. 14, 873–882. doi: 10.1111/1567-1364.12175
Zhang, B.-Q., Luan, Y., Duan, C.-Q., and Yan, G.-L. (2018). Use of Torulaspora delbrueckii Co-fermentation with two Saccharomyces cerevisiae strains with different aromatic characteristic to improve the diversity of red wine aroma profile. Front. Microbiol. 9:606. doi: 10.3389/fmicb.2018.00606
Keywords: fermentation, Saccharomyces, adaptation, diversity, evolution
Citation: Giannakou K, Cotterrell M and Delneri D (2020) Genomic Adaptation of Saccharomyces Species to Industrial Environments. Front. Genet. 11:916. doi: 10.3389/fgene.2020.00916
Received: 30 April 2020; Accepted: 23 July 2020;
Published: 27 August 2020.
Edited by:
Francisco A. Cubillos, University of Santiago, ChileReviewed by:
Alexander DeLuna, Instituto Politécnico Nacional de México (CINVESTAV), MexicoCopyright © 2020 Giannakou, Cotterrell and Delneri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Delneri, ZC5kZWxuZXJpQG1hbmNoZXN0ZXIuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.