
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 07 August 2020
Sec. Computational Genomics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00896
 Zhaopeng Cai1†
Zhaopeng Cai1† Wenjie Liu1,2†
Wenjie Liu1,2† Keng Chen1
Keng Chen1 Peng Wang1
Peng Wang1 Zhongyu Xie1
Zhongyu Xie1 Jinteng Li1
Jinteng Li1 Ming Li2
Ming Li2 Shuizhong Cen2
Shuizhong Cen2 Guiwen Ye2
Guiwen Ye2 Zhaofeng Li2
Zhaofeng Li2 Zepeng Su1
Zepeng Su1 Mengjun Ma1
Mengjun Ma1 Yanfeng Wu3*
Yanfeng Wu3* Huiyong Shen1,2*
Huiyong Shen1,2*Ectopic bone formation is the chief characteristic of ossification of the posterior longitudinal ligament (OPLL). Emerging evidence has revealed that long non-coding RNAs (lncRNAs) can regulate the osteogenic differentiation of mesenchymal stem cells (MSCs), which are the main cells responsible for bone formation. However, the role of lncRNAs in the pathogenesis of OPLL remains unclear. In this study, 725 aberrantly expressed lncRNAs and 664 mRNAs in osteogenically differentiated MSCs from OPLL patients (OPLL MSCs) were identified by microarrays and confirmed by qRT-PCR assays. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses showed that the most enriched pathways included the p53, JAK-STAT, and PI3K-Akt signaling pathways. The co-expression network showed the interactions between the aberrantly expressed lncRNAs and mRNAs in OPLL MSCs, and the potential targets and transcription factors of the lncRNAs were predicted. Our research demonstrated the aberrantly expressed lncRNA and mRNA and the potential regulatory networks involved in the ectopic bone formation of OPLL. These findings imply that lncRNAs may play a vital role in OPLL, which provides a new perspective on the pathogenesis of OPLL.
Ossification of the posterior longitudinal ligament (OPLL) is a common disease characterized by the ectopic bone formation of spinal ligaments (Xu et al., 2016; Yan et al., 2017). In the past several decades, studies have revealed many factors contributing to the development of OPLL, including diabetes and genetic, environmental, hormonal and lifestyle factors (Nakajima et al., 2014; Nakajima et al., 2016; Chen et al., 2018). However, the pathogenesis of OPLL remains unclear. The ossified ligaments enlarge over time and eventually compress the spinal cord, causing serious neurological problems (Tanaka et al., 2011; Xu et al., 2018). There are no effective therapies for preventing the formation and progression of ossified ligaments other than surgery (Shi et al., 2019). Therefore, it is of great significance to elucidate the pathogenesis of OPLL and look for new therapeutic targets for OPLL.
Mesenchymal stem cells (MSCs) can be isolated from a variety of other tissues, including umbilical cord blood, dental pulp, adipose tissue, tendon, skin and muscle, but they are mainly derived from bone marrow (Ding et al., 2011; Bernardo and Fibbe, 2013). MSCs can differentiate into osteoblasts, which are responsible for bone formation (Crane and Cao, 2014). In recent years, MSCs have been demonstrated to be involved in the pathogenesis of many diseases, including fibrotic diseases, systemic lupus erythematosus, ankylosing spondylitis and osteoporosis (Tan et al., 2015; Xie et al., 2016b; El Agha et al., 2017; Wu et al., 2018; Geng et al., 2019; Liu w. et al., 2019). Recent studies suggested an increase in the osteogenic potential of MSCs from OPLL patients, which may be a pathogenic factor of OPLL (Harada et al., 2014; Liu X. et al., 2017). However, the concrete mechanism underlying the role of MSCs in the abnormal osteogenic differentiation of OPLL is less known.
Long non-coding RNAs (lncRNAs) are defined as transcripts that are more than 200 nucleotides in length that do not encode proteins (Chen et al., 2017). The non-coding genome was previously considered junk DNA (Kopp and Mendell, 2018). However, with the development and application of high-throughput technologies, it is surprising that more than 98% of the human genome does not encode proteins (Kopp and Mendell, 2018). Many studies have demonstrated that lncRNAs play an important role in cell proliferation, differentiation and apoptosis (Li M. et al., 2018; Tang S et al., 2018). Recent studies have shown that lncRNAs are involved in the occurrence and progression of many diseases, such as non-small cell lung cancer, hepatocellular carcinoma, and ankylosing spondylitis (Xie et al., 2016a, 2018; Huang et al., 2019). However, little is known about the functions of lncRNAs in the pathogenesis of OPLL.
In this study, aberrantly expressed lncRNAs and mRNAs in osteogenically differentiated MSCs from OPLL patients (OPLL MSCs) were detected by microarrays and confirmed by qRT-PCR assays. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to identify the cellular events and biological pathways involved in the abnormal osteogenic differentiation of OPLL MSCs. The co-expression network showed the interactions between the aberrantly expressed lncRNAs and mRNAs in OPLL MSCs, and the potential targets and transcription factors of the lncRNAs were predicted. Our findings provide a new perspective on the mechanisms of the ectopic bone formation of spinal ligaments in OPLL.
In this study, 30 healthy donors (HDs) and 30 OPLL patients were recruited. The OPLL was diagnosed based on clinical examinations including cervical radiographs, computed tomography (CT), and magnetic resonance imaging (MRI). Details of the study subjects were presented in Supplementary Table S1. The study was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital (Sun Yat-sen University, Guangzhou, China), and informed consent was obtained from all participants. MSCs were isolated from bone marrow and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Sijiqing Biological Engineering Material Company). All cells were cultured in 5% CO2 at 37°C, and the culture medium was changed every 3 days.
MSCs were digested with 0.25% trypsin containing 0.53 mM EDTA and washed thoroughly with phosphate-buffered saline (PBS). After incubation with specific antibodies for 30 min, the MSCs were washed again in preparation for flow cytometry with a BD Biosciences Influx cell sorter (BD Biosciences). Antibodies against human phycoerythrin (PE)-conjugated CD105, PE-conjugated CD73, PE-conjugated CD 90, fluorescein isothiocyanate (FITC)-conjugated CD45, FITC-conjugated CD34, FITC-conjugated CD14, FITC-conjugated 79-a, and FITC-conjugated HLA-DR were used in the study (all from BD PharMingen).
To induce osteogenic differentiation, MSCs (1.5 × 104 cells/cm2) were seeded in 12-well plates and cultured in osteogenic differentiation medium containing DMEM with 10% FBS, 100 IU/ml penicillin, 100 IU/ml streptomycin, 0.1 μM dexamethasone, 10 mM β-glycerol phosphate and 50 μM ascorbic acid (Sigma-Aldrich, St. Louis, MO, United States). The medium was changed every 3 days.
To detect ALP activity, the cells were cultured in osteogenic differentiation medium for 10 days. Protein was extracted from the cells using RIPA buffer (Sigma), and the protein concentration was determined with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Then, ALP activity was detected using an ALP activity kit (Nanjing Jiancheng Biotech, Nanjing, China) according the manufacturer’s protocol. For ALP staining, the cells were fixed and stained using the BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime) according the manufacturer’s protocols.
To perform ARS staining, the cells were cultured in osteogenic differentiation medium for 14 days. The cells were fixed with 4% paraformaldehyde and stained with 1% ARS for 15 min. After washing thoroughly with PBS, the cells were observed under a microscope. For ARS quantification, the above cells were destained using 10% cetylpyridinium chloride (CPC) monohydrate (Sigma-Aldrich). Then, 200 μl of supernatant was taken from the above mixture and transferred to a 96-well plate to measure the absorbance at 562 nm.
After adding TRIzol (Invitrogen, Massachusetts, United States), total RNA was extracted from the cells. Then, cDNA was transcribed from 1 μg of RNA with the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. qRT-PCR was performed with the LightCycler®480 PCR system (Roche, Basel, Switzerland) using SYBR Premix Ex Taq (TaKaRa) according to the manufacturer’s protocol. GAPDH was used as the internal reference, and the relative expression levels of all genes were calculated using the 2−ΔΔCt method. The primer sequences are shown in Supplementary Table S2.
The cells used for microarray analysis were cultured in osteogenic differentiation medium for 10 days. After adding TRIzol (Invitrogen), total RNA was extracted from three HDMSC samples (control group; sample H1–H3) and three OPLLMSC samples (experimental group; sample O1–O3). RNA integrity was detected by 1% formaldehyde denaturing gel electrophoresis. Using the CbcScript reverse transcriptase with cDNA synthesis system, double-stranded cDNAs (dsDNAs) were synthesized from 1 μg RNA according to the manufacturer’s instruction (CapitalBio). Then the dsDNA products were purified with a PCR NucleoSpin Extract II Kit (MN) after completion of dsDNA synthesis using DNA polymerase and RNase H. The dsDNA were amplified by PCR and purified using the RNA Clean-up Kit (MN). Finally, microarrays (CapitalBio Co.) were performed according to the manufacturer’s protocol.
The microarray data were analyzed using GeneSpring software V13.0 (Agilent). To identify the differentially expressed genes, genes with fold changes ≥ 2 and p < 0.05 were considered significant. The data were log2 transformed and median centered (by genes) using the Adjust Data function of CLUSTER 3.0 software and then further analyzed with hierarchical clustering with average linkage.
GO analysis was performed to provide a label classification for gene function and gene product attributes in three domains (cellular component, molecular function and biological process) using KOBAS 3.0 software. KEGG pathway analysis was performed to identify the significant enrichment of different pathways using KOBAS 3.0 software.
Based on the correlations between the differentially expressed lncRNAs and mRNAs, a co-expression network was constructed. The Pearson correlation for each pair of genes was calculated, and the significant correlation pairs were selected to construct the network using Cytoscape software. Pearson correlation coefficient ≥ 0.99 was considered significant.
The target prediction included cis-acting lncRNA prediction and trans-acting lncRNA prediction. The cis-acting lncRNA prediction was performed by assessing the close correlation of the lncRNA (Pearson correlation coefficient ≥ 0.99) to coding genes. The lncRNA must reside at genomic loci where a coding gene and the lncRNA gene were within 10 kb of each other along the genome. The trans-prediction was carried out using BLAT tools (Standalone BLAT v. 35 × 1) to compare the full sequence of the lncRNA with the 3′ untranslated region (UTR) of its co-expression mRNAs.
Transcription factor prediction was performed using the Match-1.0 Public transcription factor prediction tool. Based on the results of the co-expression network, the binding of the upstream 2000 bp region and the downstream 500 bp region of the starting site of lncRNAs with transcription factors was predicted.
Statistical analysis was performed with SPSS 22.0 software (Chicago, IL, United States). Student’s t-test was used to compare the differences between two groups. A value of p < 0.05 was considered significant.
Both HD MSCs and OPLL MSCs expressed typical MSC surface markers, with positivity for CD105, CD73, and CD90 and negativity for CD45, CD34, CD14, CD79a, and HLA-DR (Figure 1A). ALP staining assays showed that the ALP activity of OPLL MSCs was stronger than that of HD MSCs after osteogenic differentiation (Figure 1B). In addition, OPLL MSCs showed greater ARS staining than HD MSCs after osteogenic differentiation (Figure 1C). These results indicate that OPLL MSCs have a greater capacity for osteogenic differentiation than HD MSCs.
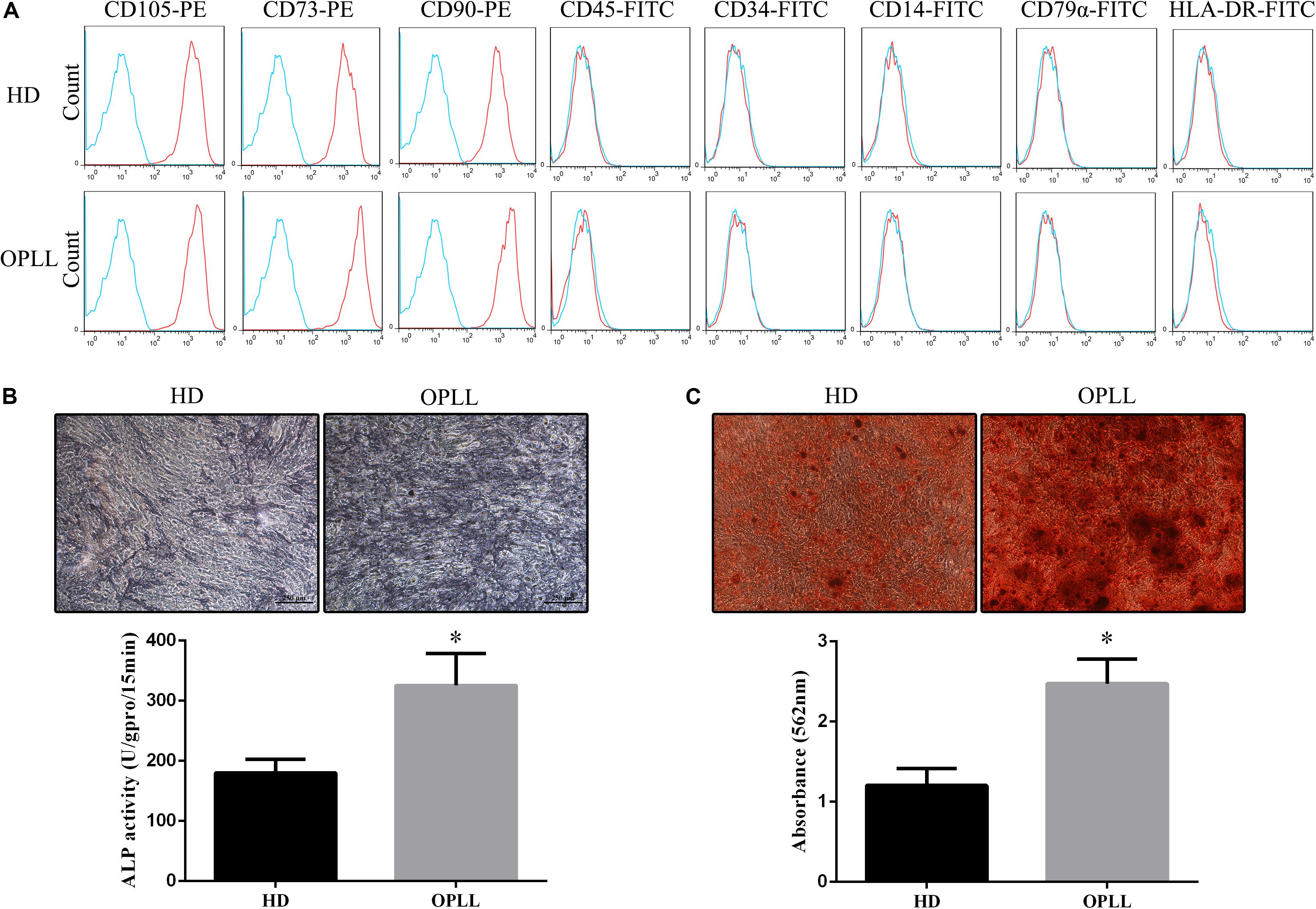
Figure 1. Phenotypes and osteogenic differentiation capacities of HD MSCs and OPLL MSCs. (A) Both HD MSCs and OPLL MSCs expressed typical MSC surface markers, with positivity for CD105, CD73, and CD90 and negativity for CD45, CD34, CD14, CD79a, and HLA-DR. (B) Alkaline phosphatase (ALP) assays were performed to assess the osteogenic differentiation capacity of MSCs. OPLL MSCs showed greater ALP staining than HD MSCs after 10 days of induction. The absorbance of ALP quantification of OPLL MSCs was also greater than that of HD MSCs. (C) Alizarin red S (ARS) staining assays were performed to assess the osteogenic differentiation capacity of MSCs. OPLL MSCs showed greater ARS staining than HD MSCs after 14 days of induction. The absorbance of ARS quantification of OPLL MSCs was also greater than that of HD MSCs. n = 15. ∗p < 0.05. HD MSC: mesenchymal stem cells from healthy donors; OPLL MSC: MSCs from patients with ossification of the posterior longitudinal ligament.
A microarray was used to identify differentially expressed lncRNAs and mRNAs between osteogenically differentiated OPLL MSCs and HD MSCs, and a total of 28,156 lncRNAs and 28,339 mRNAs expressed in MSCs were detected. Hierarchical clustering analysis showed homogeneous lncRNA and mRNA expression profiles within the same group and distinct expression profiles between the two groups (Figures 2A,C). Compared to that in osteogenically differentiated HD MSCs, in OPLL MSCs, 725 lncRNAs were differentially expressed, including 651 upregulated lncRNAs and 74 downregulated lncRNAs (fold change > 2.0, P < 0.05) (Figure 2B). In addition, 664 mRNAs were differentially expressed in osteoclasts, including 453 upregulated mRNAs and 211 downregulated mRNAs (fold change > 2.0, P < 0.05) (Figure 2D). The top 10 differentially expressed lncRNAs and mRNAs are shown in Tables 1, 2, respectively.
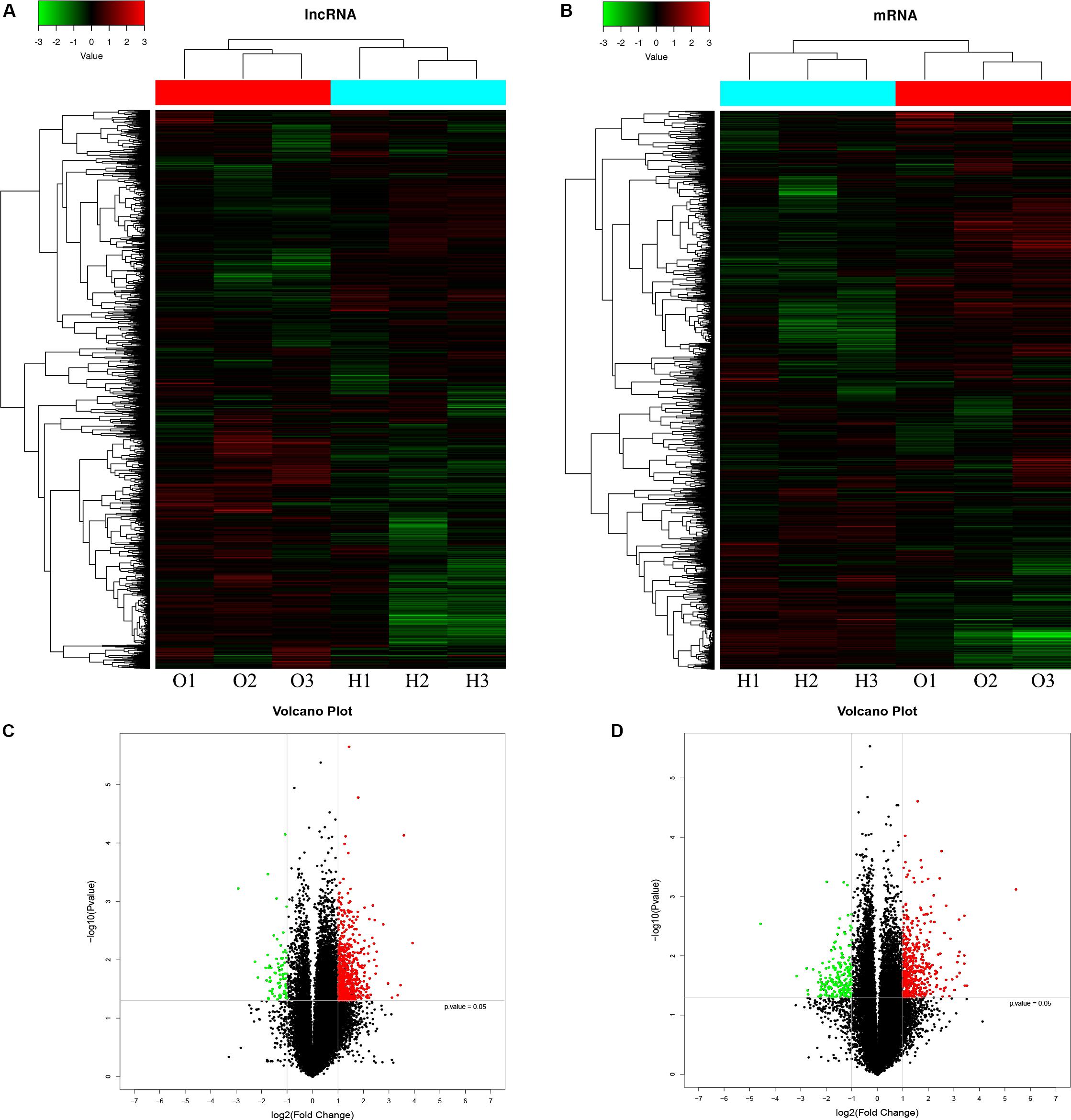
Figure 2. LncRNA and mRNA expression profiles in osteogenically differentiated OPLL MSCs compared to HD MSCs. Cluster heat map shows differentially expressed lncRNAs (A) and mRNAs (C) in OPLL MSCs. The names of the sample groups are on the x-axis, and the different probes are on the y-axis. The red strip indicates high relative expression, and the green strip indicates low relative expression. O1–O3: group with OPLL MSCs; H1–H3: group with HD MSCs. Volcano plots of lncRNAs (B) and mRNAs (D) in OPLL MSCs vs. HD MSCs. The red squares in the plots represent the upregulated transcripts with statistical significance, while the green squares represent the downregulated transcripts (fold change ≥ 2.0, p < 0.05).
To confirm the microarray data, the top 10 differentially expressed lncRNAs and mRNAs were selected for the qRT-PCR assays. Their expression levels in HD MSCs and OPLL MSCs are shown in Figures 3A,B. For the lncRNAs, p11878, p23569, p22941, p20197, p13631, p13632, p1957, p8129 and p4982 were upregulated in OPLL MSCs compared to HD MSCs, while p26842 was downregulated. For the mRNAs, CST2, CLSTN2, NDUFA4L2, CST1, OLFM2, CYP26B1, ADH1C, and ADH1B were upregulated in OPLL MSCs compared to HD MSCs, while HAPLN1 and MMP13 were downregulated. These results of the qRT-PCR assays were almost consistent with the microarray data (Supplementary Table S3).
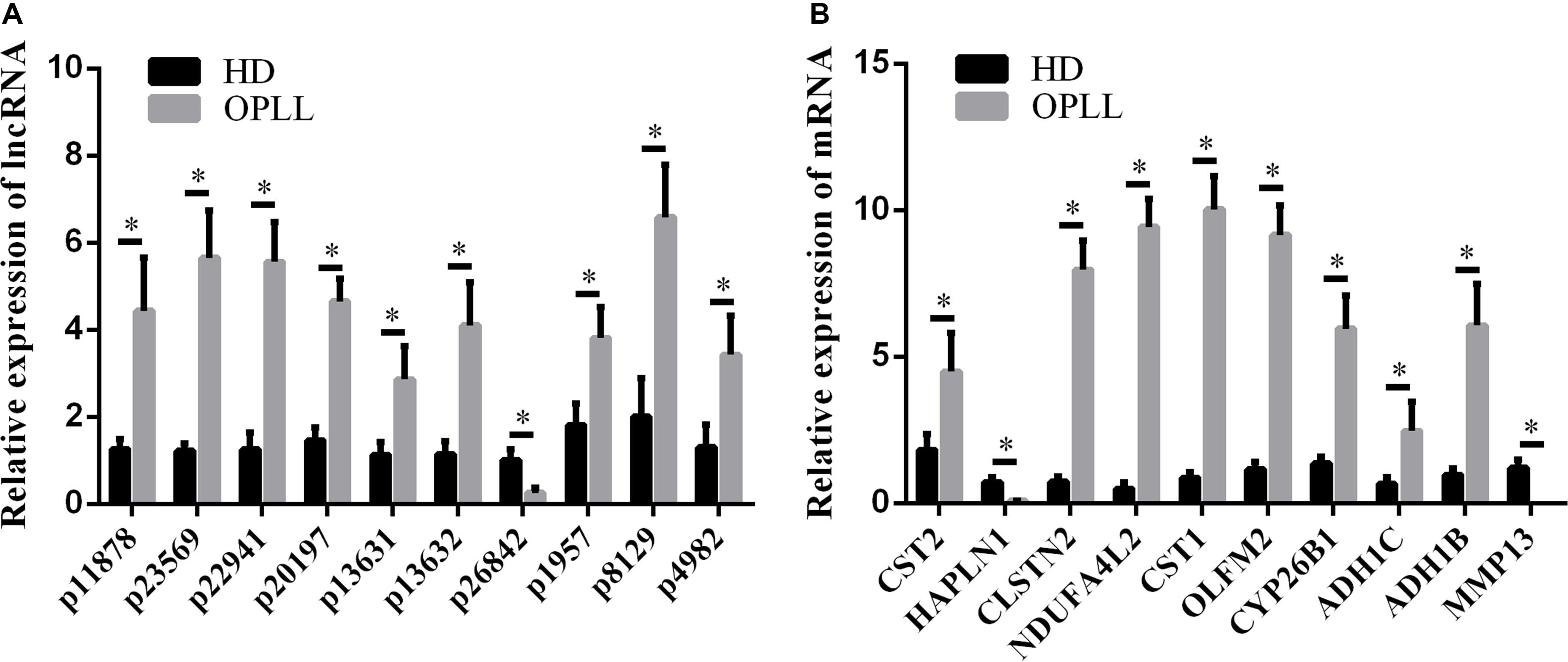
Figure 3. Validation of the expression profiles by qRT-PCR. Several genes were selected for the qRT-PCR assays. (A) Several lncRNAs were selected for the qRT-PCR assays. (B) Several mRNAs were selected for the qRT-PCR assays. The data are presented as the mean ± SD (n = 30). The results represent three independent experiments. ∗p < 0.05.
The differentially expressed mRNAs were used in GO analysis. A total of 2,156 GO terms in the biological process category were differentially expressed between HD MSCs and OPLL MSCs (P < 0.05), including cellular component organization or biogenesis (GO: 0071840). There were 249 GO terms in the cellular component category that were differentially expressed between HD MSCs and OPLL MSCs (P < 0.05), including organelles (GO: 0043226). A total of 372 GO terms in the molecular function category were differentially expressed between HD MSCs and OPLL MSCs (P < 0.05), including receptor regulator activity (GO: 0030545). The significantly enriched GO terms are presented in Figure 4A.
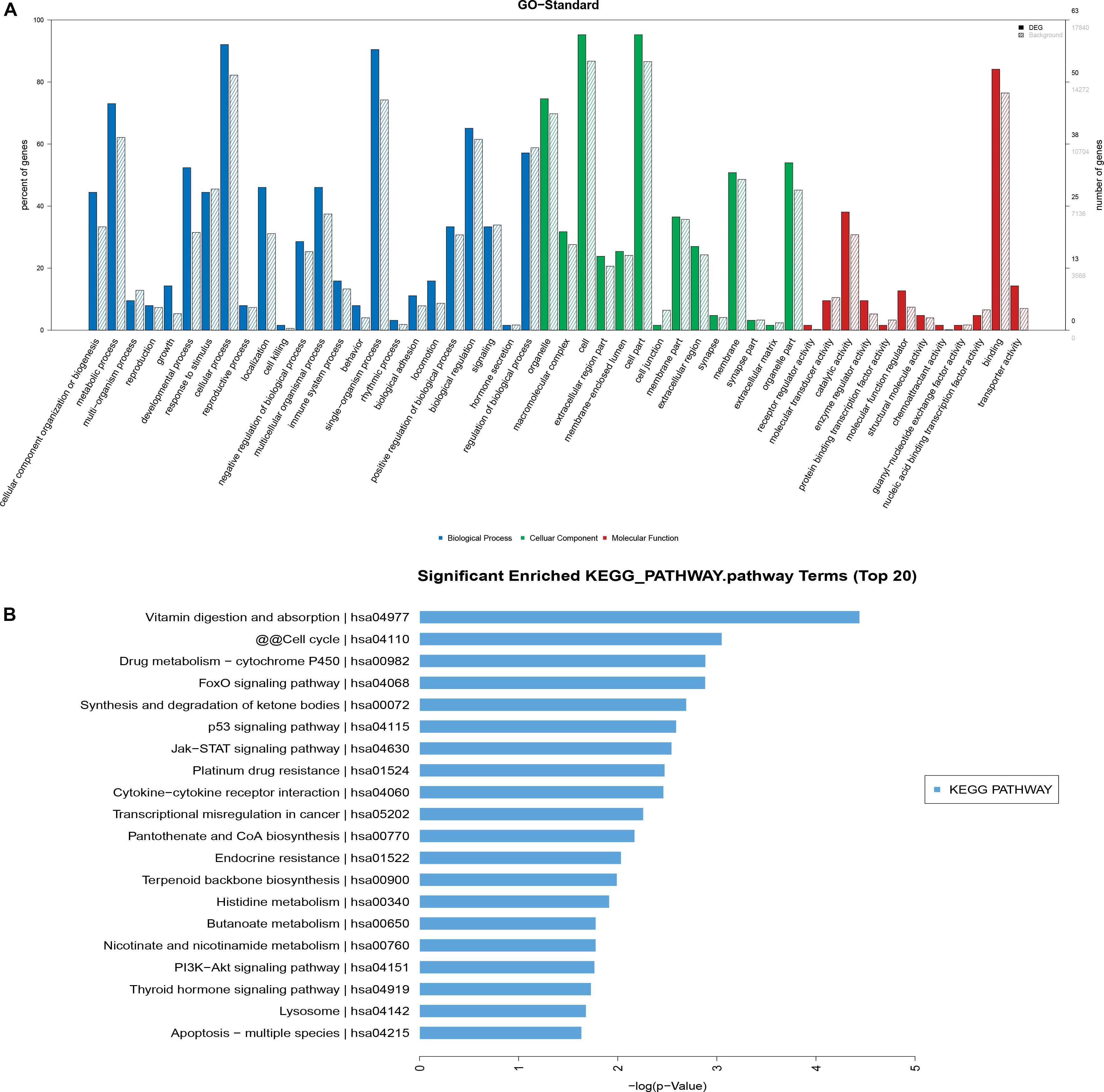
Figure 4. GO and KEGG pathway analyses. (A) The GO terms with significantly differential expression from GO analysis of the biological process, cellular component and molecular function categories are shown. (B) The top 20 KEGG pathways with significantly differential expression are shown. Higher -log P (LgP) values indicate higher significance, and lower -LgP values indicate lower significance.
KEGG pathway analysis showed that 119 pathways were statistically significantly enriched. The most enriched pathways included the p53, JAK-STAT and PI3K-Akt signaling pathways (Figure 4B), suggesting that these signaling pathways may play important roles in the osteogenic differentiation of OPLL MSCs. The top 20 significantly enriched pathways are presented in Figure 4B.
To evaluate the interactions among the key differentially expressed mRNAs and lncRNAs, a co-expression network was constructed. The results showed that lnc-KRT18-1, TSPEAR, CLIC2, ANKRD33B, TWIST2, C19orf73, TBX19, TWIST2, LOC101926975, and ABLIM1 had the highest number of interactions, which revealed the vital roles of these genes in the pathogenesis of OPLL (Figure 5).
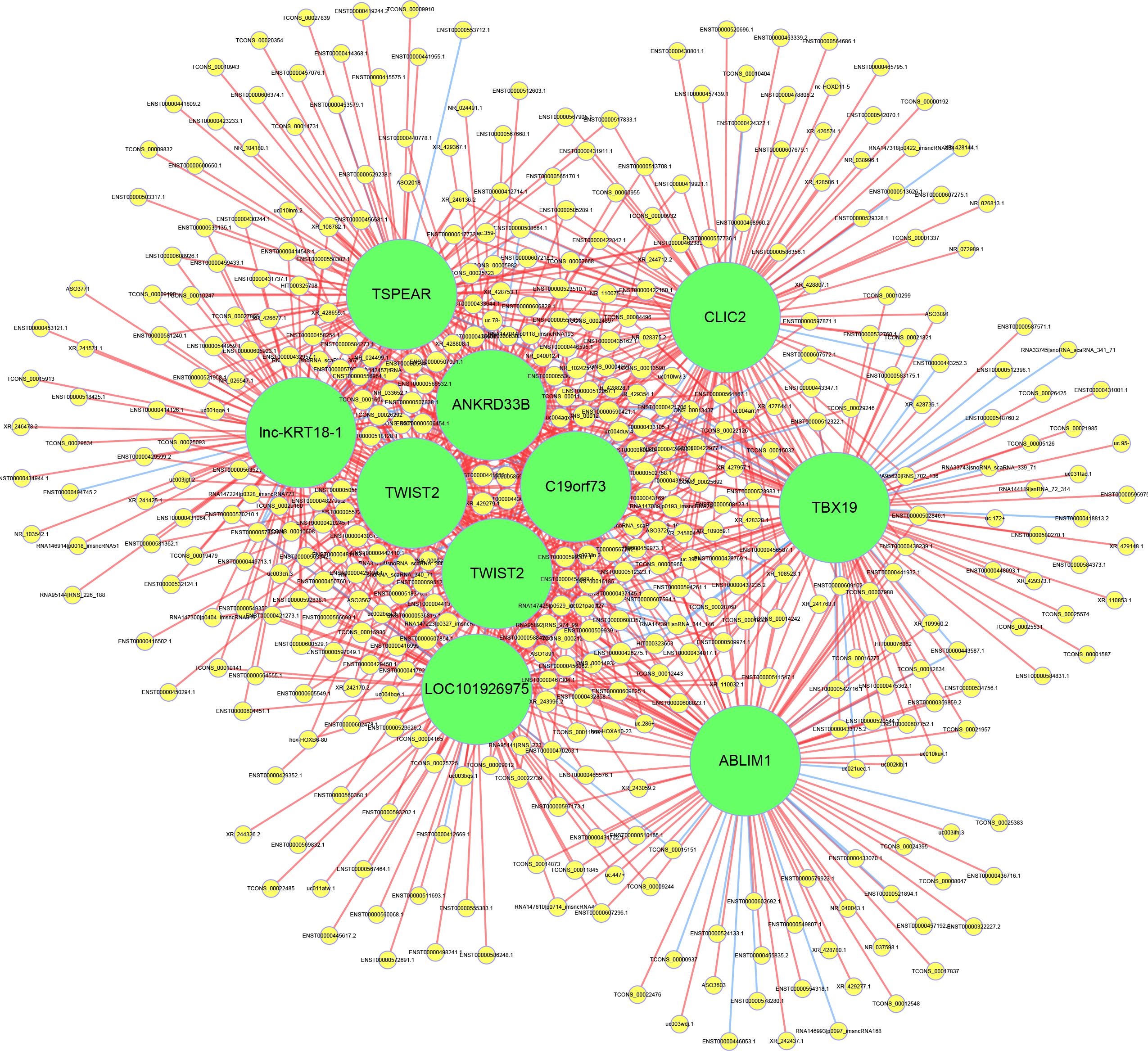
Figure 5. Construction of the lncRNA-mRNA coexpression network. The yellow circles represent lncRNAs, and the green circles represent mRNAs. The size of the circle is determined by the number of other genes that interact with this gene. The red line represents a positive correlation, and the blue line represents a negative correlation.
To predict the potential target mRNAs of the differentially expressed lncRNAs, cis-acting lncRNA prediction and trans-acting lncRNA prediction were used, and gene target networks were created. In the potential target network, MDM2 was the most important target for the lncRNAs since MDM2 had the most interactions with lncRNAs. The gene target network is shown in Figure 6.
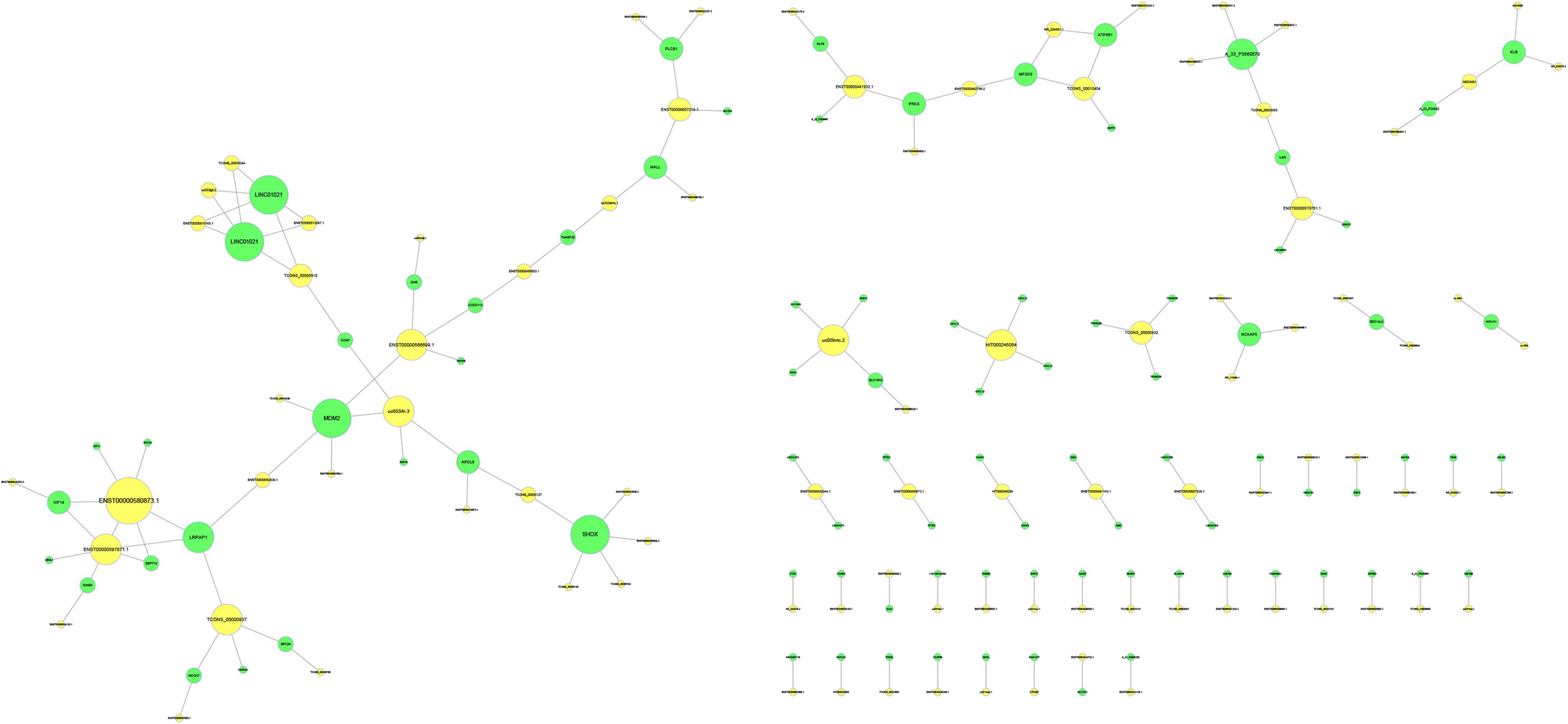
Figure 6. Target prediction of lncRNAs. The yellow circles represent lncRNAs, and the green circles represent the target genes. The size of the circle is determined by the number of other genes that interact with this gene.
To predict potential transcription factors of the differentially expressed lncRNAs, the Match-1.0 Public transcription factor prediction tool was used. As shown in Figure 7, Oct-1 was the most important transcription factor in the pathogenesis of OPLL since it had the highest number of interactions.
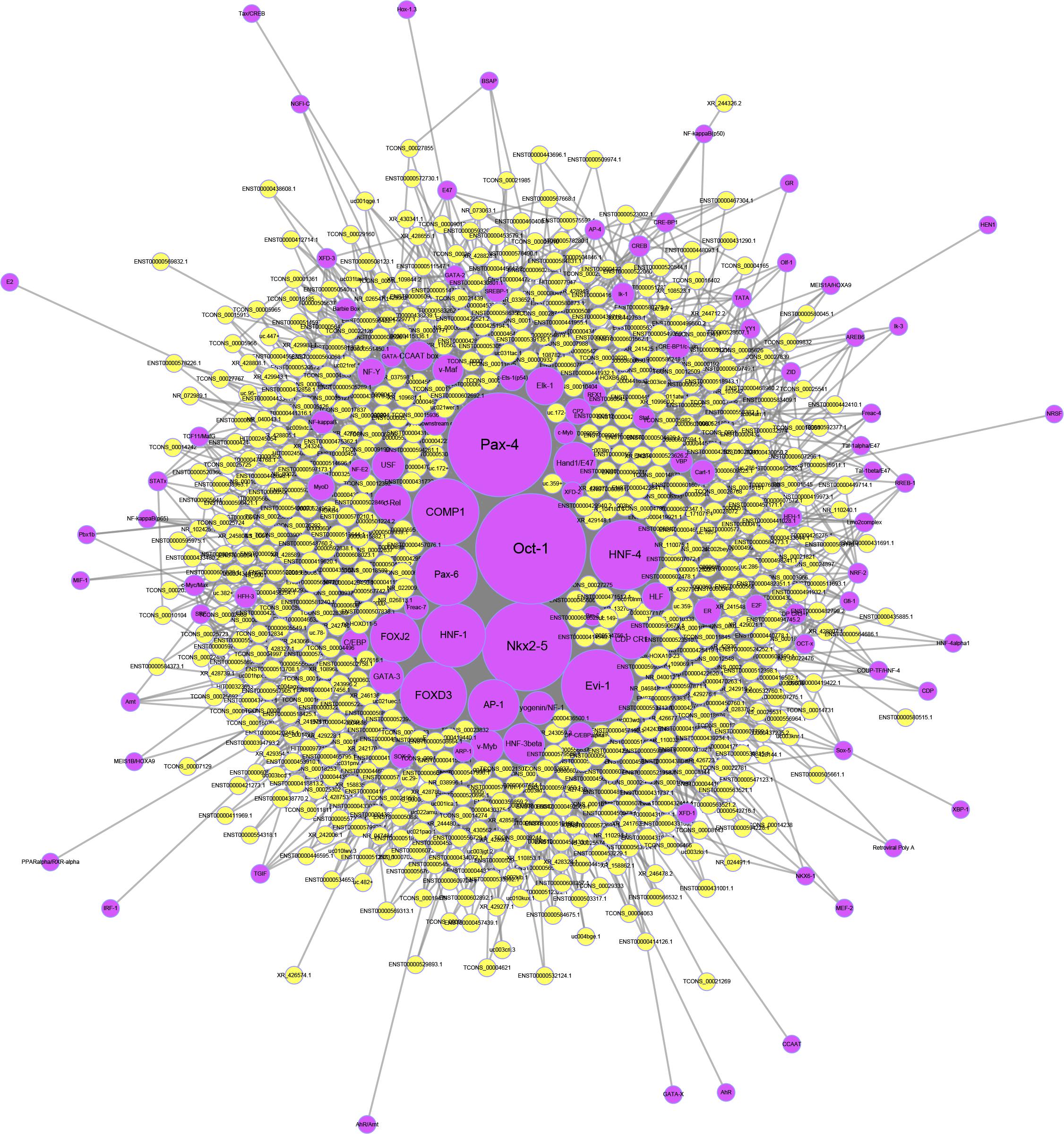
Figure 7. Transcription factor prediction of lncRNAs. The yellow circles represent lncRNAs, and the purple circles represent transcription factors. The size of each circle is determined by the number of other genes that interact with this gene.
In this study, we performed microarrays to identify a number of lncRNAs and mRNAs aberrantly expressed in OPLL MSCs when they differentiated into osteoblasts. The cellular events and biological pathways involved in the abnormal osteogenic differentiation of OPLL MSCs were identified by GO and KEGG analyses. We further investigated the interactions between the aberrantly expressed lncRNAs and mRNAs in OPLL MSCs and identified core regulatory factors by constructing a co-expression network. Finally, we predicted the targets and transcription factors of the lncRNAs. Our results provide a basis for further understanding the role and mechanism of lncRNAs in the pathogenesis of OPLL.
Ectopic ossification in spinal ligament tissues is the chief characteristic of OPLL, and its progression leads to serious neurologic impairment (Xu et al., 2016). In recent years, the etiology of OPLL has been extensively investigated (Inamasu et al., 2006; Furukawa, 2008). Yoshifumi Harada suggested an increase in the osteogenic potential of MSCs from OPLL patients, which may be a pathogenic factor in OPLL (Harada et al., 2014; Imagama et al., 2017). Gentaro Kumagai showed that the MSCs from a spinal ossification mouse model had a higher osteogenic potential than those from normal mice (Liu X. et al., 2017). Our results confirmed that MSCs from OPLL patients had an increase in the osteogenic potential. However, the underlying mechanism of MSCs participating in the ectopic ossification of OPLL remains unclear.
Emerging evidence has revealed that lncRNAs can regulate osteogenic differentiation (He et al., 2018; Li J. et al., 2018; Tang Z et al., 2018). With the development of high-throughput technologies, an in-depth examination of the non-coding genome can be achieved (Kopp and Mendell, 2018). In our study, aberrantly expressed lncRNAs and mRNAs in the osteogenic differentiation of OPLL MSCs were identified by microarrays and confirmed by qRT-PCR assays. The results of the qRT-PCR assays were almost in complete agreement with the microarray data, further confirming the high credibility of the microarray data analysis. Among these aberrantly expressed lncRNAs and mRNAs, we found that ENPP1 was down-regulated in OPLL. A previous study has showed that Enpp1 knockout mouse is a model of OPLL (Saito et al., 2011), which supports our finding. Further studies are needed to explore the functions of other aberrantly expressed lncRNAs and mRNAs in OPLL. LncRNA expression profiles are important symbols for many diseases, including laryngeal cancer, systematic lupus erythematosus, rheumatoid arthritis, lung adenocarcinoma and intrahepatic cholangiocarcinoma (Zhang et al., 2016; Yang et al., 2017; Wang et al., 2018; Liu H. et al., 2019). Together with previous studies, our study highlights the importance of lncRNAs in the development of diseases.
To investigate the molecular function and pathways involved in the osteogenic differentiation of OPLL MSCs, GO and KEGG analyses were performed based on the differentially expressed mRNAs. KEGG analysis demonstrated that the p53, JAK-STAT and PI3K/Akt signaling pathways may be involved in the abnormal osteogenic differentiation of OPLL MSCs. A recent study showed that the activation of the JAK2-STAT3 pathway and the PI3K/Akt signaling pathway promoted osteogenic differentiation in OPLL (Chen et al., 2018). In addition, many researchers have found that the p53 signaling pathway mediates the osteogenic differentiation of MSCs (Yoon et al., 2016; Haffner-Luntzer et al., 2018). However, there are no studies on the role of the p53 signaling pathway in OPLL. Our results revealed the importance of the p53 signaling pathway in OPLL, and further studies need to explore its function in regulating the osteogenic differentiation of OPLL.
To identify the core regulatory factors involved in the osteogenic differentiation of OPLL, we constructed a lncRNA-mRNA co-expression network. Our results indicated that lnc-KRT18-1, TSPEAR, CLIC2, ANKRD33B, TWIST2, C19orf73, TBX19, TWIST2, LOC101926975, and ABLIM1 may play a central role in regulating the osteogenic differentiation of OPLL MSCs. There are no studies reporting their significant roles in the osteogenic differentiation of OPLL, and further experiments are needed.
To further investigate the regulatory mechanism of the aberrantly expressed lncRNAs, we predicted the potential targets and transcription factors of the lncRNAs. In the potential target network, MDM2 was the most important target for the lncRNAs since MDM2 had the most interactions with lncRNAs. Previous studies showed that osteoblast differentiation was regulated by the MDM2-p53 signaling pathway (Jin et al., 2017). Together with KEGG analysis, we proposed that the MDM2-p53 signaling pathway may play an important role in the osteogenic differentiation of OPLL. In addition to downstream target gene prediction, we also predicted the upstream transcription factors of the lncRNAs. We found that Oct-1 was the most important transcription factors since it had the highest number of interactions. Recently, Anirudha Karvande demonstrated that Oct-1 promoted osteoblast differentiation (Karvande et al., 2018). Therefore, we regard Oct-1 as an important transcription factor for lncRNAs in OPLL, and further studies are necessary to confirm its role in OPLL.
In conclusion, we detected the aberrantly expressed lncRNAs and mRNAs in the osteogenic differentiation of OPLL MSCs compared to HD MSCs and identified potential regulatory mechanisms with bioinformatic analyses. We aimed to reveal the role of lncRNAs in the abnormal osteogenic differentiation of OPLL. These results may help illuminate the pathogenesis of OPLL and provide new ideas for the diagnosis and treatment of OPLL. However, there are some limitations in our research. For example, we did not confirm the concrete functions of the aberrantly expressed lncRNAs in OPLL. Further studies are needed to explore the role of these aberrantly expressed lncRNAs in the osteogenic differentiation of OPLL MSCs.
Details of the microarray data can be found on the GEO website (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153829).
This study was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. The MSCs used in this study were obtained from the Center for Biotherapy, Sun Yat-sen Memorial Hospital, Sun Yat-sen University.
ZC, WL, KC, PW, ZX, JL, ML, SC, GY, ZL, ZS, and MM performed the trials. ZC and WL analyzed the data. YW and HS contributed the reagents. ZC wrote the manuscript. YW and HS revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was financially supported by the Key-Area Research and Development Program of Guangdong Province (2019B020236001), the National Natural Science Foundation of China (81971518), the National Natural Science Foundation of China (81871750), the Natural Science Foundation of Guangdong Province (2018A030313232), and the Fundamental Research Funds for the Central Universities (19ykpy01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00896/full#supplementary-material
Bernardo, M. E., and Fibbe, W. E. (2013). Mesenchymal stromal cells: sensors and switchers of inflammation. Cell Stem Cell 13, 392–402. doi: 10.1016/j.stem.2013.09.006
Chen, S., Zhu, H., Wang, G., Xie, Z., Wang, J., and Chen, J. (2018). Combined use of leptin and mechanical stress has osteogenic effects on ossification of the posterior longitudinal ligament. Eur. Spine J. 27, 1757–1766. doi: 10.1007/s00586-018-5663-4
Chen, Y. G., Satpathy, A. T., and Chang, H. Y. (2017). Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 18, 962–972. doi: 10.1038/ni.3771
Crane, J. L., and Cao, X. (2014). Bone marrow mesenchymal stem cells and TGF-beta signaling in bone remodeling. J. Clin. Invest. 124, 466–472. doi: 10.1172/jci70050
El Agha, E., Kramann, R., Schneider, R. K., Li, X., Seeger, W., Humphreys, B. D., et al. (2017). Mesenchymal stem cells in fibrotic Disease. Cell Stem Cell 21, 166–177. doi: 10.1016/j.stem.2017.07.011
Furukawa, K. (2008). Pharmacological aspect of ectopic ossification in spinal ligament tissues. Pharmacol. Ther. 118, 352–358. doi: 10.1016/j.pharmthera.2008.03.007
Geng, L., Tang, X., Zhou, K., Wang, D., Wang, S., Yao, G., et al. (2019). MicroRNA-663 induces immune dysregulation by inhibiting TGF-beta1 production in bone marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Cell Mol Immunol 16, 260–274. doi: 10.1038/cmi.2018.1
Haffner-Luntzer, M., Kovtun, A., Fischer, V., Prystaz, K., Hainzl, A., Kroeger, C. M., et al. (2018). Loss of p53 compensates osteopenia in murine Mysm1 deficiency. FASEB J. 32, 1957–1968. doi: 10.1096/fj.201700871r
Harada, Y., Furukawa, K., Asari, T., Chin, S., Ono, A., Tanaka, T., et al. (2014). Osteogenic lineage commitment of mesenchymal stem cells from patients with ossification of the posterior longitudinal ligament. Biochem. Biophys. Res. Commun. 443, 1014–1020. doi: 10.1016/j.bbrc.2013.12.080
He, Q., Yang, S., Gu, X., Li, M., Wang, C., and Wei, F. (2018). Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis. 9:455.
Huang, J. F., Jiang, H. Y., Cai, H., Liu, Y., Zhu, Y. Q., Lin, S. S., et al. (2019). Genome-wide screening identifies oncofetal lncRNA Ptn-dt promoting the proliferation of hepatocellular carcinoma cells by regulating the Ptn receptor. Oncogene 38, 3428–3445. doi: 10.1038/s41388-018-0643-z
Imagama, S., Ando, K., Ito, Z., Kobayashi, K., Hida, T., Ito, K., et al. (2017). Risk factors for ineffectiveness of posterior decompression and dekyphotic corrective fusion with instrumentation for beak-type thoracic ossification of the posterior longitudinal ligament: a single institute study. Neurosurgery 80, 800–808. doi: 10.1093/neuros/nyw130
Inamasu, J., Guiot, B. H., and Sachs, D. C. (2006). Ossification of the posterior longitudinal ligament: an update on its biology, epidemiology, and natural history. Neurosurgery 58, 1027–1039. doi: 10.1227/01.neu.0000215867.87770.73
Jin, F., Wang, Y., Wang, X., Wu, Y., Wang, X., Liu, Q., et al. (2017). Bre enhances osteoblastic differentiation by promoting the Mdm2-mediated degradation of p53. Stem Cells 35, 1760–1772. doi: 10.1002/stem.2620
Karvande, A., Kushwaha, P., Ahmad, N., Adhikary, S., Kothari, P., Tripathi, A. K., et al. (2018). Glucose dependent miR-451a expression contributes to parathyroid hormone mediated osteoblast differentiation. Bone 117, 98–115. doi: 10.1016/j.bone.2018.09.007
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Li, C. J., Xiao, Y., Yang, M., Su, T., Sun, X., Guo, Q., et al. (2018). Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. J. Clin. Invest. 128, 5251–5266. doi: 10.1172/jci99044
Li, M., Xie, Z., Wang, P., Li, J., Liu, W., Tang, S., et al. (2018). The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis. 9:554.
Liu, H., Sun, Y., Tian, H., Xiao, X., Zhang, J., Wang, Y., et al. (2019). Characterization of long non-coding RNA and messenger RNA profiles in laryngeal cancer by weighted gene co-expression network analysis. Aging 11, 10074–10099. doi: 10.18632/aging.102419
Liu, W., Wang, P., Xie, Z., Wang, S., Ma, M., Li, J., et al. (2019). Abnormal inhibition of osteoclastogenesis by mesenchymal stem cells through the miR-4284/CXCL5 axis in ankylosing spondylitis. Cell Death Dis. 10:188.
Liu, X., Kumagai, G., Wada, K., Tanaka, T., Asari, T., Oishi, K., et al. (2017). High osteogenic potential of adipose- and muscle-derived mesenchymal stem cells in spinal-ossification model mice. Spine 42, E1342–E1349.
Nakajima, M., Kou, I., and Ohashi, H. Genetic Study Group of the Investigation Committee on the Ossification of Spinal Ligaments, and Ikegawa, S. (2016). Identification and functional characterization of RSPO2 as a susceptibility gene for ossification of the posterior longitudinal ligament of the spine. Am. J. Hum. Genet. 99, 202–207. doi: 10.1016/j.ajhg.2016.05.018
Nakajima, M., Takahashi, A., Tsuji, T., Karasugi, T., Baba, H., Uchida, K., et al. (2014). A genome-wide association study identifies susceptibility loci for ossification of the posterior longitudinal ligament of the spine. Nat. Genet. 46, 1012–1016. doi: 10.1038/ng.3045
Saito, T., Shimizu, Y., Hori, M., Taguchi, M., Igarashi, T., Fukumoto, S., et al. (2011). A patient with hypophosphatemic rickets and ossification of posterior longitudinal ligament caused by a novel homozygous mutation in ENPP1 gene. Bone 49, 913–916. doi: 10.1016/j.bone.2011.06.029
Shi, L., Shi, G., Li, T., Luo, Y., Chen, D., Miao, J., et al. (2019). The endoplasmic reticulum stress response participates in connexin 43-mediated ossification of the posterior longitudinal ligament. Am. J. Transl. Res. 11, 4113–4125.
Tan, J., Xu, X., Tong, Z., Lin, J., Yu, Q., Lin, Y., et al. (2015). Decreased osteogenesis of adult mesenchymal stem cells by reactive oxygen species under cyclic stretch: a possible mechanism of age related osteoporosis. Bone Res. 3:15003.
Tanaka, S., Kudo, H., Asari, T., Ono, A., Motomura, S., Toh, S., et al. (2011). P2Y1 transient overexpression induced mineralization in spinal ligament cells derived from patients with ossification of the posterior longitudinal ligament of the cervical spine. Calcif. Tissue Int. 88, 263–271. doi: 10.1007/s00223-010-9456-y
Tang, S., Xie, Z., Wang, P., Li, J., Wang, S., Liu, W., et al. (2018). LncRNA-OG promotes the osteogenic differentiation of bone marrow-derived mesenchymal stem cells under the regulation of hnRNPK. Stem Cells 37, 270–283. doi: 10.1002/stem.2937
Tang, Z., Gong, Z., and Sun, X. (2018). LncRNA DANCR involved osteolysis after total hip arthroplasty by regulating FOXO1 expression to inhibit osteoblast differentiation. J. Biomed. Sci. 25:4.
Wang, Y., Chen, S., Chen, S., Du, J., Lin, J., Qin, H., et al. (2018). Long noncoding RNA expression profile and association with SLEDAI score in monocyte-derived dendritic cells from patients with systematic lupus erythematosus. Arthrit. Res. Ther. 20:138.
Wu, Y., Xie, L., Wang, M., Xiong, Q., Guo, Y., Liang, Y., et al. (2018). Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 9:4772.
Xie, Y., Zhang, Y., Du, L., Jiang, X., Yan, S., Duan, W., et al. (2018). Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 12, 648–658. doi: 10.1002/1878-0261.12188
Xie, Z., Li, J., Wang, P., Li, Y., Wu, X., Wang, S., et al. (2016a). Differential expression profiles of long noncoding RNA and mRNA of osteogenically differentiated mesenchymal stem cells in ankylosing spondylitis. J. Rheumatol. 43, 1523–1531. doi: 10.3899/jrheum.151181
Xie, Z., Wang, P., Li, Y., Deng, W., Zhang, X., Su, H., et al. (2016b). Imbalance between bone morphogenetic protein 2 and noggin induces abnormal osteogenic differentiation of mesenchymal stem cells in ankylosing spondylitis. Arthrit. Rheumatol. 68, 430–440. doi: 10.1002/art.39433
Xu, C., Chen, Y., Zhang, H., Chen, Y., Shen, X., Shi, C., et al. (2016). Integrated microRNA-mRNA analyses reveal OPLL specific microRNA regulatory network using high-throughput sequencing. Sci. Rep. 6:21580.
Xu, C., Zhang, H., Gu, W., Wu, H., Chen, Y., Zhou, W., et al. (2018). The microRNA-10a/ID3/RUNX2 axis modulates the development of ossification of posterior longitudinal ligament. Sci. Rep. 8:9225.
Yan, L., Gao, R., Liu, Y., He, B., Lv, S., and Hao, D. (2017). The pathogenesis of ossification of the posterior longitudinal ligament. Aging Dis. 8, 570–582.
Yang, W., Li, Y., Song, X., Xu, J., and Xie, J. (2017). Genome-wide analysis of long noncoding RNA and mRNA co-expression profile in intrahepatic cholangiocarcinoma tissue by RNA sequencing. Oncotarget 8, 26591–26599. doi: 10.18632/oncotarget.15721
Yoon, D. S., Choi, Y., and Lee, J. W. (2016). Cellular localization of NRF2 determines the self-renewal and osteogenic differentiation potential of human MSCs via the P53-SIRT1 axis. Cell Death Dis. 7:e2093. doi: 10.1038/cddis.2016.3
Keywords: ossification of the posterior longitudinal ligament, long non-coding RNA, mesenchymal stem cells, osteogenic differentiation, mRNA
Citation: Cai Z, Liu W, Chen K, Wang P, Xie Z, Li J, Li M, Cen S, Ye G, Li Z, Su Z, Ma M, Wu Y and Shen H (2020) Aberrantly Expressed lncRNAs and mRNAs of Osteogenically Differentiated Mesenchymal Stem Cells in Ossification of the Posterior Longitudinal Ligament. Front. Genet. 11:896. doi: 10.3389/fgene.2020.00896
Received: 02 April 2020; Accepted: 20 July 2020;
Published: 07 August 2020.
Edited by:
Jie Sun, Wenzhou Medical University, ChinaReviewed by:
Buling Wu, Southern Medical University, ChinaCopyright © 2020 Cai, Liu, Chen, Wang, Xie, Li, Li, Cen, Ye, Li, Su, Ma, Wu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfeng Wu, d3V5ZkBtYWlsLnN5c3UuZWR1LmNu; Huiyong Shen, c2hlbmh1aXlAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.