
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 28 July 2020
Sec. Toxicogenomics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00818
This article is part of the Research Topic Environmental Genomics and Epigenomics: Response, Development and Disease View all 10 articles
Nano silicon dioxide (Nano-SiO2) has been widely used in industries such as the field of biomedical engineering. Despite the existing evidence that Nano-SiO2 exposure could induce oxidative stress and inflammatory responses in multiple organ systems, the carcinogenicity of Nano-SiO2 exposure has rarely been investigated. Thus in this study, two types of human bronchial epithelial cell lines (16HBE and BEAS-2B) were selected as in vitro models to investigate the carcinogenicity of Nano-SiO2. Our results revealed that Nano-SiO2 induces a malignant cellular transformation in human bronchial epithelial cells according to the soft agar colony formation assay. The carcinogenesis induced by Nano-SiO2 was also confirmed in nude mice. By using immunofluorescence assay and high-performance capillary electrophoresis (HPCE), we observed a genome-wide DNA hypomethylation induced by Nano-SiO2. Besides the reduced enzyme activity of total DNMTs upon Nano-SiO2 treatment, altered expression of DNMTs and methyl-CpG binding proteins were observed. Besides, we found that the expression of NRF2 was activated by demethylation of CpG islands within the NRF2 promoter region and the overexpression of NRF2 could alleviate the carcinogenesis induced by Nano-SiO2. Taken together, our results suggested that Nano-SiO2 induces malignant cellular transformation with a global DNA hypomethylation, and the demethylation of NRF2 promoter activates the expression of NRF2, which plays an important role in protecting against the carcinogenesis induced by Nano-SiO2.
Nano silicon dioxide (Silica nanoparticles, Nano-SiO2), due to its special characteristics of large surface area and optical transparency, has been widely used in various fields including biomedical imaging, drug delivery, cosmetics, and electronics industry (Vivero-Escoto et al., 2012; Sweeney et al., 2016; Mohajerani et al., 2019). Meanwhile, its widespread applications raise potential health risks to humans through occupational and environmental exposure. Recent studies have demonstrated that Nano-SiO2 exposure in a short term could induce oxidative stress, mitochondria dysfunction, and inflammatory responses in multiple organ systems (Song et al., 2009; Napierska et al., 2010; Fruijtier-Polloth, 2012; Lu et al., 2013), among which respiratory system is the primary site that exposed to the airborne Nano-SiO2 particles (Song et al., 2009; Yu et al., 2015).
Long-lasting nanomaterials in certain tissues have been reported to cause chronic adverse effects such as carcinogenesis due to their special physicochemical properties (Hirose et al., 2011), and nano-sized particles might be more carcinogenic than micron-sized particles concerning long-term exposure (Gebel, 2012). Inhalation of TiO2 nanoparticles was shown to increase the risk of lung cancers in rats (Bermudez et al., 2004) and carbon nanotube exposure through inhalation is carcinogenic to the lungs of male and female rats (Kasai et al., 2016). Although SiO2 has been classified as carcinogenic to humans by the International Agency for Research on Cancer (IARC) in 1997 (Iarc., 1997), there is limited data on carcinogenicity following chronic exposure to Nano-SiO2. Nano-SiO2 can be localized to the nucleus thereby affecting nuclear integrity and causing DNA damage (Chen and von Mikecz, 2005; Wang et al., 2007).
Alterations in DNA methylation, a typical epigenetic modification, have been reported in cells exposed to Nano-SiO2. A global genomic hypomethylation induced by Nano-SiO2 was observed in HaCaT cells and the CpGs in the promoter of PARP1 gene were demethylated, resulting in DNA repair dysfunction (Gong et al., 2010, 2012b). Abnormal DNA methylation occurs frequently in the process of carcinogenesis (Miousse et al., 2018; Dreval et al., 2019), through devastating genome stability and activating aberrant transcription (Gama-Sosa et al., 1983). Decreased global DNA methylation and increased expression of DNA methyltransferases (DNMTs) were demonstrated in the early stage of Nano-SiO2-induced malignant cellular transformation (Seidel et al., 2017), indicating that DNA methylation is involved in the Nano-SiO2-induced carcinogenesis.
Nuclear factor erythroid-2-related factor 2 (NRF2) is a key transcription factor involved in cellular responses to stresses induced by electrophiles, oxidants, and chemicals (Motohashi and Yamamoto, 2004). In addition to stress responses, NRF2 plays a contradictory role in cancers (Wu et al., 2019). Loss of NRF2 ultimately leads to malignant cellular transformation in the prostate gland of murine models (Frohlich et al., 2008). NRF2 dysfunction in mice accelerates the acetylhydrolase-induced hepatocarcinogenesis (Marhenke et al., 2008). On the contrary, accumulating evidence indicated that increased activation of NRF2 frequently occurs in multiple types of tumors, which promotes cancer cell growth and metastasis formation (Liu et al., 2016; Zhang et al., 2016; Lu et al., 2017). The hypomethylation of CpG islands within the NRF2 promoter region was reported in colorectal cancer (Kang et al., 2014; Zhao et al., 2015). Decreased NRF2 expression due to hypermethylation of CpG islands within the NRF2 promoter region was found to be associated with prostate cancer (Yu et al., 2010; Khor et al., 2014).
In the present study, we demonstrated that Nano-SiO2 induces a malignant cellular transformation and a global DNA hypomethylation in human bronchial epithelial cells. Intriguingly, we found that demethylation of CpG islands within the NRF2 promoter region increases the NRF2 gene expression, which inhibits the carcinogenesis induced by Nano-SiO2.
The 15 nm Nano-SiO2 particles were purchased from Wan Jing New Material Co., Ltd. (Hangzhou, Zhejiang, China) and were characterized as previously described (Gong et al., 2012a). Nano-SiO2 samples were sonicated to distribute in the solution as evenly as possible. Dosing solutions were prepared by dissolving the calculated amount of Nano-SiO2 in the cell culture medium, following the approved standard operating procedures for handling toxic agents. All other reagents were obtained from commercial sources and were of the highest available grade.
Human bronchial epithelial cell lines (16HBE and BEAS-2B) and adenocarcinomic human alveolar basal epithelial cell line (A549) were purchased from the Cell Bank in Chinese Academy of Sciences Cell Bank in Shanghai. Frozen cells were thawed and expanded in MEM medium supplemented with 10% fetal bovine serum (FBS), 10 U/ml penicillin, and 10 U/ml streptomycin. Cells were incubated in a humidified atmosphere with 5% CO2 at 37°C and passaged at about 80% confluence.
Human bronchial epithelial cell lines were treated with Nano-SiO2 for several passages and harvested at certain time points as shown in Figure 2A. In detail, 16HBE and BEAS-2B cells were grown in complete MEM medium until they reached a confluence of about 70–80%. Cells were then treated with Nano-SiO2 (10.0 μg/ml for 16HBE cells, 40.0 μg/ml for BEAS-2B cells) for 24 h and changed into the complete MEM medium until they are ready for subculture, these cells treated with Nano-SiO2 for one passage are referred to P1. Accordingly, cells treated as above for n passages are named as P(n). Cells cultured in complete MEM medium for the same passages without any Nano-SiO2 treatment are taken as the negative control. A549, an epithelial cell line derived from human lung carcinoma, was used as a positive control.
Cell viability was determined by MTT assay. 16HBE and BEAS-2B cells growing at the exponential phase were seeded in 96-well plates with a density of 5 × 104cells/ml. After treated with various dosages of Nano-SiO2 (0, 1.0, 2.5, 5.0, 10.0, 25.0, and 50.0 μg/ml for 16HBE, 0, 1.0, 5.0, 10.0, 20.0, 40.0, 80.0, and 160.0 μg/ml for BEAS-2B) for 24 h, 50 μl MTT solution was added to each well and incubated for another 4 h. After adding 150 μl DMSO, the absorbance of each well was measured at 490 nm using a spectrophotometer. Each treatment group has at least three replicates. Cell viability was obtained as a percentage of the value of viable cells in the control groups.
Anchorage-independent growth of Nano-SiO2-treated human bronchial epithelial cells (16HBE and BEAS-2B) was measured by soft agar colony formation assay. Cells treated with Nano-SiO2 for various passages (P0, P8, P16, and P32 of 16HBE cells, P0, P15, P30, and P45 of BEAS-2B cells) were trypsinized. Cells were resuspended at a density of 1 × 104/ml in MEM medium with 0.3% agar and plated over 3 ml of a solidified complete MEM culture medium containing 0.5% agar. Cells were then incubated in a humidified atmosphere with 5% CO2 at 37°C for 3 weeks. The colonies were photographed and counted by using ImageJ software.
The tumorigenicity of the Nano-SiO2-treated human bronchial epithelial cells (16HBE and BEAS-2B) was determined in nude mice. Balb/c-nu male nude mice (4 weeks old) purchased from the Laboratory Animal Center in Shanghai, were housed in ventilated microisolators with sterile food and water. After one week of acclimation, mice were randomly divided into eight treatment groups, with three mice in each group. Specifically, four groups of mice were injected with different passages of 16HBE cells (P0, P8, P16, and P32), another four groups of mice were injected with different passages of BEAS-2B cells (P0, P15, P30, and P45). Nude mice injected with cells cultured in complete MEM medium for the same passages without any Nano-SiO2 treatment were taken as the negative control. Cells were harvested by trypsinization, washed, and resuspended in PBS. Cell suspensions were then injected (0.2 ml) subcutaneously to mice at a density of 1 × 107 cells/ml. Tumor growth was monitored for 90 consecutive days after injection by measuring the major diameter of the tumor externally with a slide caliper. Mice were sacrificed by cervical dislocation if the major diameter of its tumor reaches about 2 cm. Mice were kept for more than 90 days after injection. The tumor sizes were measured at the fifth week after injection and the incidence and latency period of tumorigenesis were recorded. The incidence was presented as the number of mice with a tumor at the site of injection versus the total number of mice. The latency period is defined as the time required for a tumor to be able to visually detect.
Total RNAs from three cell lines (16HBE, BEAS-2B, and A549) were isolated and reverse transcribed into cDNAs by using the PrimeScriptTM RT reagent kit with gDNA Eraser (Takara Bio Inc., Japan). Four genes related to the NRF2 signaling pathway were selected for qRT-PCR assay. Primers are listed in Supplementary Table S1. All qPCR reactions were performed on an Applied BiosystemsTM StepOneTM Real-time PCR system using iTaqTM Universal SYBR Green Supermix, with three technical replicates. The amplification procedure was as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 20 s. Relative quantification of target genes was performed using the ΔΔCt method with GAPDH as a reference gene.
Cells were washed with ice-cold PBS and lysed on ice with a protease inhibitor cocktail. Protein concentrations were measured by BCA method. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. The membranes were probed with NRF2 (1:500), HO1 (1:1000), SOD1 (1:1000), GST (1:1000), DNMT1 (1:500), DNMT3a (1:500), DNMT3b (1:500), MeCP2 (1:500), MBD2 (1:500), GAPDH (1:10000) antibodies (Santa Cruz Biotechnology, Inc., Texas, United States) at 4°C overnight. The bands were visualized after incubation with a chemiluminescent substrate. Quantification of the band density was determined by densitometric analysis.
Cells were fixed with 4% paraformaldehyde for 15 min and then fixed with cold formaldehyde for 5 min, washed with PBST, and then incubated with hydrochloric acid at 37°C for 30 min. After blocked with 3% PBST-BSA for 30 min, samples were incubated with anti-5-mC antibody (1:1000) at 37°C for 1 h and incubated with FITC-conjugated secondary antibody (1:400) for 30 min. Finally, coverslips were then incubated with DAPI for double staining and then mounted on glass slides by using Fluo-Antifading medium II. The fluorescence intensity was detected by using an inverted fluorescent microscope.
Cells treated with Nano-SiO2 for different passages (P0, P8, P16, and P32 of 16HBE cells, P0, P15, P30, and P45 of BEAS-2B cells) and A549 cells were harvested by trypsinization. Nuclear was extracted and total DNMT enzymes activity was determined by EpiQuikTM DNA Methyltransferase Activity/Inhibition Assay Kit (EpiGentek, United States) according to the procedures provided by the manufacturers.
Genomic DNA was extracted and treated with bisulfite as previously described (Gong et al., 2012a). After sodium bisulfite conversion, PCR assay was conducted to analyze the DNA methylation level in specific NRF2 gene loci quantitatively. Primer sets M1 and M2 were designed and synthesized to amplify the methylated DNA, primer sets U1 and U2 were designed and synthesized to amplify the unmethylated DNA. PCR products were separated by electrophoresis at 100 V for 40 min on 2.2% agarose gels. Primers were listed in Supplementary Table S1.
The cells (16HBE and BEAS-2B) were separately transfected with NRF2 shRNA(h) lentiviral particles (sc-37030-V) or plasmid pEGFP-NRF2 to construct NRF-2 knockdown (KD) or overexpression (OE) cell lines, according to the manufacturer’s instructions. For the knockdown of NRF-2, NRF2 shRNA(h) lentiviral particles (shRNA-NRF2) and control shRNA Lentiviral Particles (shRNA-Ctrl) were transfected into cells. After transfected for 24 h, the transfection culture medium was replaced with complete culture medium and incubated for 72 h. Then, 10 μg/ml puromycin dihydrochloride was used to select and achieve stable knockdown cell lines. For the overexpression of NRF-2, pEGFP containing NRF-2 gene sequence (pEGFP-NRF2) or empty pEGFP (pEGFP) were transfected into cells. Full length cDNAs of NRF-2 were amplified through RT-PCR using specific forward 5′-CACCATGGGAATGGACTTGGAGCTGCC-3′ and reverse 5′-CTAGTTTTTCTTAACATCTGGCTTCTTAC-3′ primers. After transfected for 24 h, cells were selected by G418. Selected cells were harvested for transfection efficiency confirmation and subsequent experiments (Supplementary Figure S1).
Data were represented by the means ± SD of at least three independent experiments. Statistical significances among experimental groups were evaluated by ANOVA followed by the Tukey post hoc test performed with the GraphPad Prism (version 8.0.2; San Diego, CA, United States).
Since the respiratory system is the primary site that exposed to the airborne Nano-SiO2 particles (Song et al., 2009; Yu et al., 2015), two types of human bronchial epithelial cell lines (16HBE and BEAS-2B) were selected as our in vitro models. To investigate the cytotoxicity of Nano-SiO2, we determined the effects of Nano-SiO2 on the viability of human bronchial epithelial cells by MTT assay. The survival rate of cells treated with Nano-SiO2 was expressed as the percentage of that of cells in the control group without nano-SiO2 treatment. For 16HBE cells, no significant change in cell viability was observed when treated with 1.0 or 2.5 μg/ml Nano-SiO2 for 24 h. After exposure to 10.0 μg/ml Nano-SiO2 for 24 h, cell viability was reduced to 77.04% of the control group (p < 0.05) (Figure 1A). For BEAS-2B cells, no significant change in cell viability was observed when treated with 1.0, 5.0, or 10.0 μg/ml Nano-SiO2 for 24 h (Figure 1B). Cell viability was significantly decreased when treated with Nano-SiO2 no less than 20.0 μg/ml. At a concentration of 40.0 μg/ml Nano-SiO2, cell viability was reduced to 75.18% of the control group after 24 h (p < 0.05) (Figure 1B).
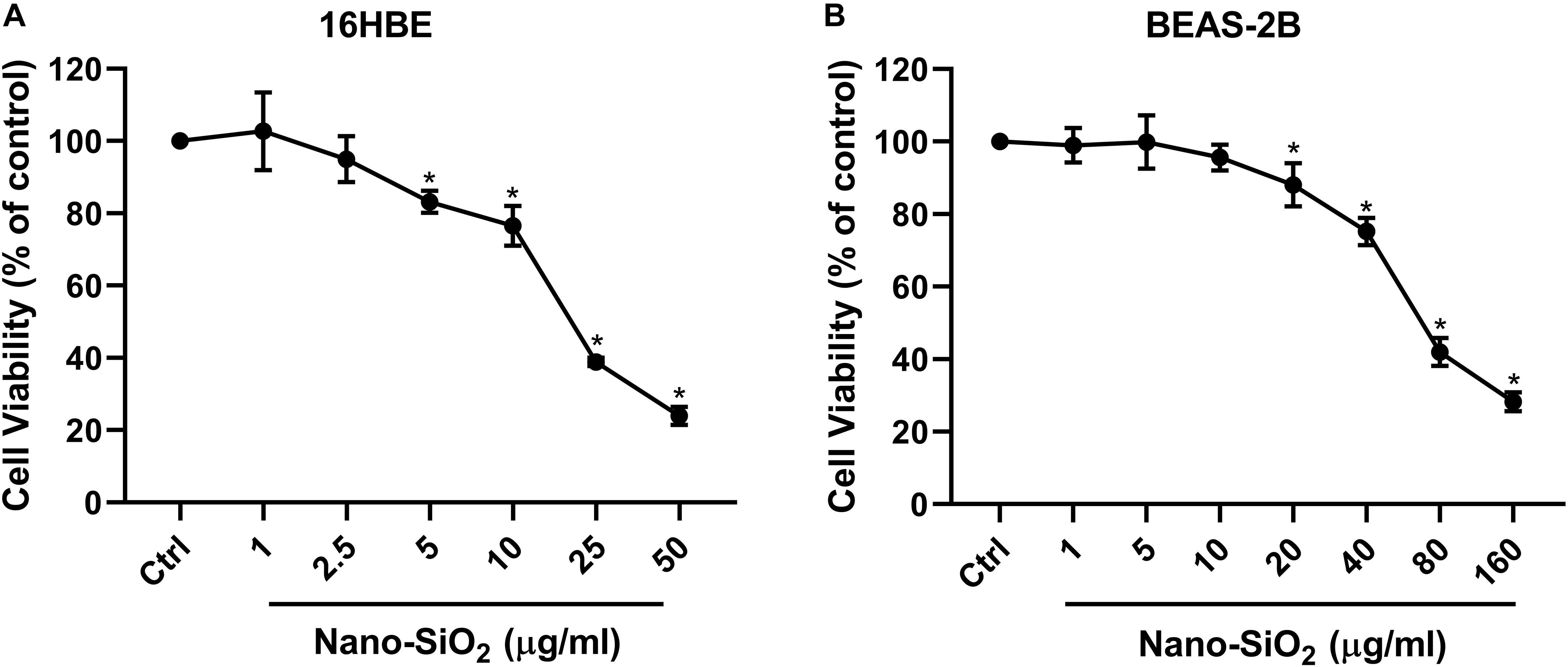
Figure 1. Cell viability of two types of human bronchial epithelial cells (A: 16HBE cell line, B: BEAS-2B cell line) upon treatment with Nano-SiO2 for 24 h. Values were mean ± SD (n = 3); *p < 0.05, control versus Nano-SiO2 treatment.
With the increase of Nano-SiO2 concentration, the reduction of cell viability in both cell lines became more severe, showing a dose-dependent manner. Based on the cell viability results, 10 μg/ml and 40 μg/ml Nano-SiO2 were used as the treatment dosage in subsequent experiments for 16HBE and BEAS-2B cells, respectively.
To confirm the malignant cellular transformation induced by Nano-SiO2, a soft agar colony formation assay was performed to evaluate the ability of Nano-SiO2-treated cells to grow independently on a solid surface (anchorage-independent growth). The number of colonies grown from 16HBE cells cultured in complete MEM medium (control) for 8, 16, and 32 passages without Nano-SiO2 exposure was first compared, and no obvious difference was observed (data not shown). Similarly, no obvious difference in the numbers of the colony was observed among the 15th, 30th, and 45th passage of BEAS-2B cells in the control group (data not shown). Hereafter, when compared with treatment groups, the control group was named as 16HBE-P0 or BEAS-2B-P0.
We observed that with the prolonged exposure to Nano-SiO2, both 16HBE and BEAS-2B cells developed a significant amount of colonies in the soft agar (Figure 2B). Specifically, after treated with 10 μg/ml Nano-SiO2 for 8 passages, ten thousand 16HBE cells developed 555 colonies in the soft agar, which is approximately twice that from 16HBE-P0 cells which are not exposed to Nano-SiO2. As the Nano-SiO2 exposure continued for 32 passages, ten thousand 16HBE cells developed 2161 colonies, which is eight times as many as those growing from 16HBE-P0 cells (Figure 2C).
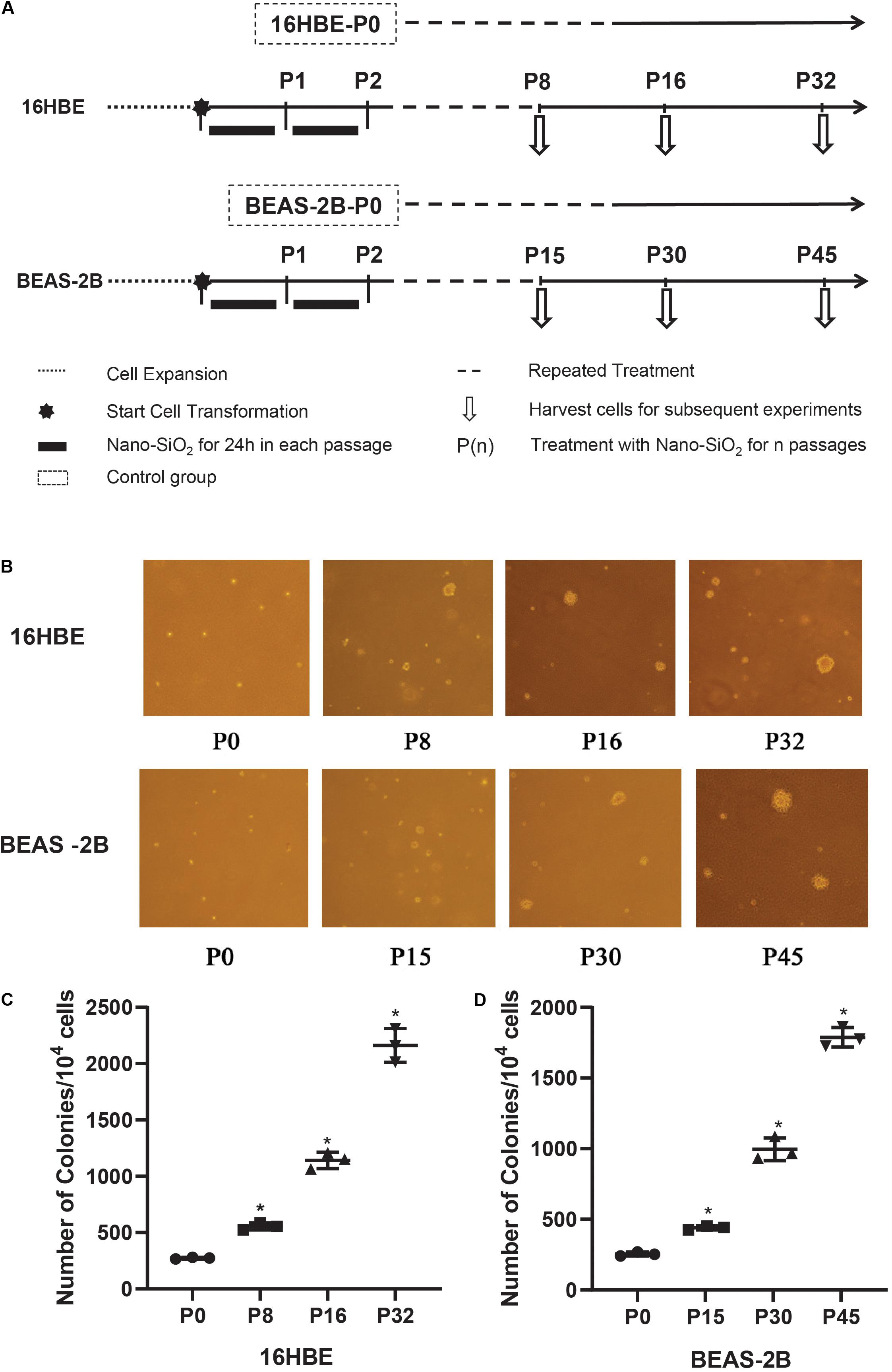
Figure 2. The effects of Nano-SiO2 on soft agar colony formation of 16HBE and BEAS-2B cells. (A) The experimental protocol indicates the Nano-SiO2 treatment timeline of 16HBE and BEAS-2B cells. (B) Representative colonies on soft agar plates. Cells treated with Nano-SiO2 (10 μg/ml for 16HBE cells, and 40 μg/ml for BEAS-2B cells) or without Nano-SiO2 (16HBE-P0, BEAS-2B-P0) were grown on soft agar plates after indicated passages. (C,D) Dot plots showing the number of colonies developed from 16HBE and BEAS-2B cells. The data are shown as mean ± SD (n = 3); *p < 0.05, control (P0) vs. Nano-SiO2 treatment.
Similarly, after treated with 40 μg/ml Nano-SiO2 for 15 passages, ten thousand BEAS-2B cells developed 440 colonies, which is approximately twice as many as the colonies growing from BEAS-2B-P0 cells. As the Nano-SiO2 exposure continued for 45 passages, ten thousand BEAS-2B cells developed 1788 colonies, which is seven times as many as the colonies growing from BEAS-2B-P0 cells (Figure 2D). The results indicated that Nano-SiO2 could induce malignant cell transformation in human bronchial epithelial cells and promote tumorigenesis in vitro.
To assess the in vivo carcinogenicity of Nano-SiO2-treated cells, twenty-four nude mice received subcutaneous injections of human bronchial epithelial cells with various treatments (three mice in each group). The features of the tumor formed at the site of cell injection are summarized in Table 1. We did not observe any tumor when 16HBE or BEAS-2B cells were not exposed to Nano-SiO2 (P0). In contrast, both 16HBE and BEAS-2B cells treated with Nano-SiO2 formed tumors in nude mice within five weeks. The latent period of tumorigenesis induced by cells treated with Nano-SiO2 for a longer period was generally shorter than cells treated for a shorter period. Also, the size of tumors growing in nude mice correlated with the duration of Nano-SiO2 treatment. Specifically, at the fifth week after injection, P8-, P16-, and P32- 16HBE cells formed tumors with a mean diameter of 2, 4, and 7 mm, respectively. Similarly, at the fifth week after injection, P15-, P30-, and P45- BEAS-2B cells formed tumors with a mean diameter of 3, 4, and 8 mm, respectively. Cells treated with Nano-SiO2 for a longer time tend to induce larger tumors in nude mice. These results indicated that human bronchial epithelial cells could be malignantly transformed by Nano-SiO2 exposure and acquire tumorigenicity in vivo.
Since global changes in DNA methylation are a hallmark of carcinogenesis, the levels of genome-wide DNA methylation in Nano-SiO2 treated cells were determined. Immunofluorescence assay was first performed to estimate the relative intensity of 5-mC in human bronchial epithelial cells (16HBE and BEAS-2B cells) and A549 cells (human lung carcinoma cell line, as a positive control).
A significant decrease in the fluorescence intensity of 5-mC was observed in both 16HBE and BEAS-2B cells treated with Nano-SiO2. For 16HBE cells, the mean fluorescence intensity of 5-mC was reduced significantly by 39.85%, 60.14%, and 72.18% in P8, P16, and P32 cells, respectively, when compared with the control group (16HBE-P0) (Figure 3A). For BEAS-2B cells, the mean fluorescence intensity of 5-mC was reduced significantly by 41.40%, 52.18%, and 59.23% in P15, P30, and P45 cells, respectively, when compared with the control group (BEAS-2B-P0) (Figure 3B).
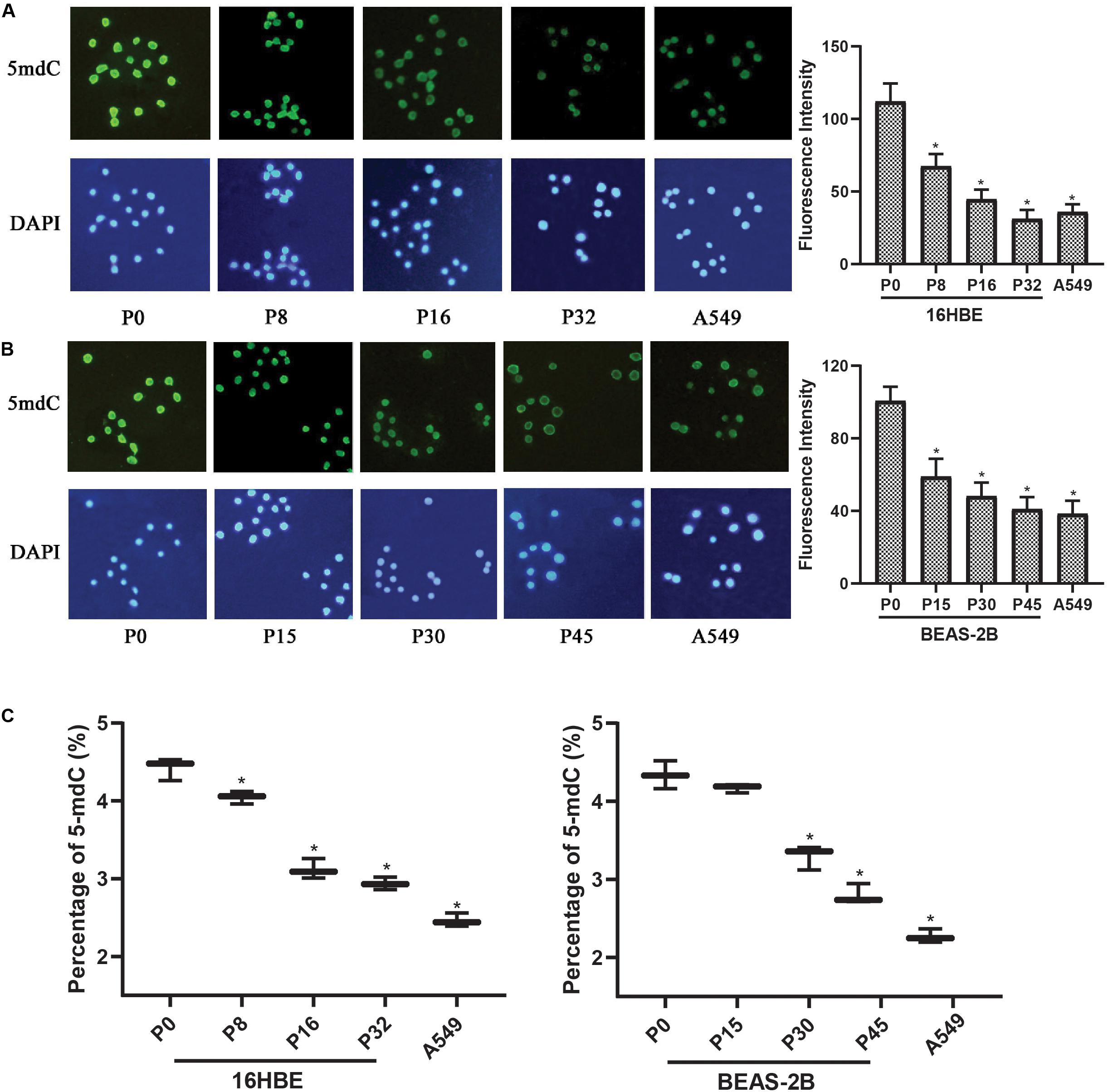
Figure 3. Nano-SiO2 exposure induces global DNA hypomethylation in human bronchial epithelial cells. 16HBE cells were treated with 10 μg/ml Nano-SiO2 for 8, 16, or 32 passages and BEAS-2B cells were treated with 40 μg/ml Nano-SiO2 for 15, 30, or 45 passages. (A,B) Immunofluorescence assay was performed to detect 5-mC levels in 16HBE and BEAS-2B cells. 5-mC was visualized by immunostaining with the antibodies against 5-mC (green) and nuclei were visualized by staining with DAPI (blue). Quantification of relative fluorescence intensity was shown on the right. (C) The levels of 5-mdC normalized to that of cytosine were shown in percentage. The data are presented as mean ± SD (n = 3). *p < 0.05, compared with the control group (P0). A549, an epithelial cell line derived from human lung carcinoma, was used as a positive control.
High-performance capillary electrophoresis (HPCE) assays were performed to further investigate the levels of genome DNA methylation in cells treated with Nano-SiO2. We found that the levels of 5-mdC were reduced in cells exposed to Nano-SiO2 in both 16HBE and BEAS-2B cells when compared with the non-exposed cells in P0, the control group (Figure 3C). Besides, the extent of 5-mdC reduction correlated with the duration of Nano-SiO2 exposure (Figure 3C). These results suggested that Nano-SiO2 exposure induces genome-wide DNA hypomethylation of human bronchial epithelial cells, and the decrease of DNA methylation is aggravated with prolonged Nano-SiO2 exposure.
To characterize the global DNA hypomethylation in cells treated with Nano-SiO2, the total activity of DNA methyltransferases (DNMTs) catalyzing DNA methylation was investigated. We found that the total activity of DNMTs in both cell lines (16HBE and BEAS-2B) was gradually inhibited with the prolonged treatment with Nano-SiO2 (Figures 4A,B). We further determined the levels of DNMTs in cells treated with Nano-SiO2. According to the results of western blot analysis, we found that the levels of DNMT1 are increased while DNMT3a protein expression is decreased in cells treated with Nano-SiO2 (Figures 4C,D). Further investigation on levels of methyl CpG binding proteins (MeCP-2 and MBD2) revealed that Nano-SiO2 exposure significantly increases MBD2 protein expression in both 16HBE and BEAS-2B cells. No significant change was observed in levels of MeCP-2 in both cell lines after Nano-SiO2 treatment (Figures 4C,D).
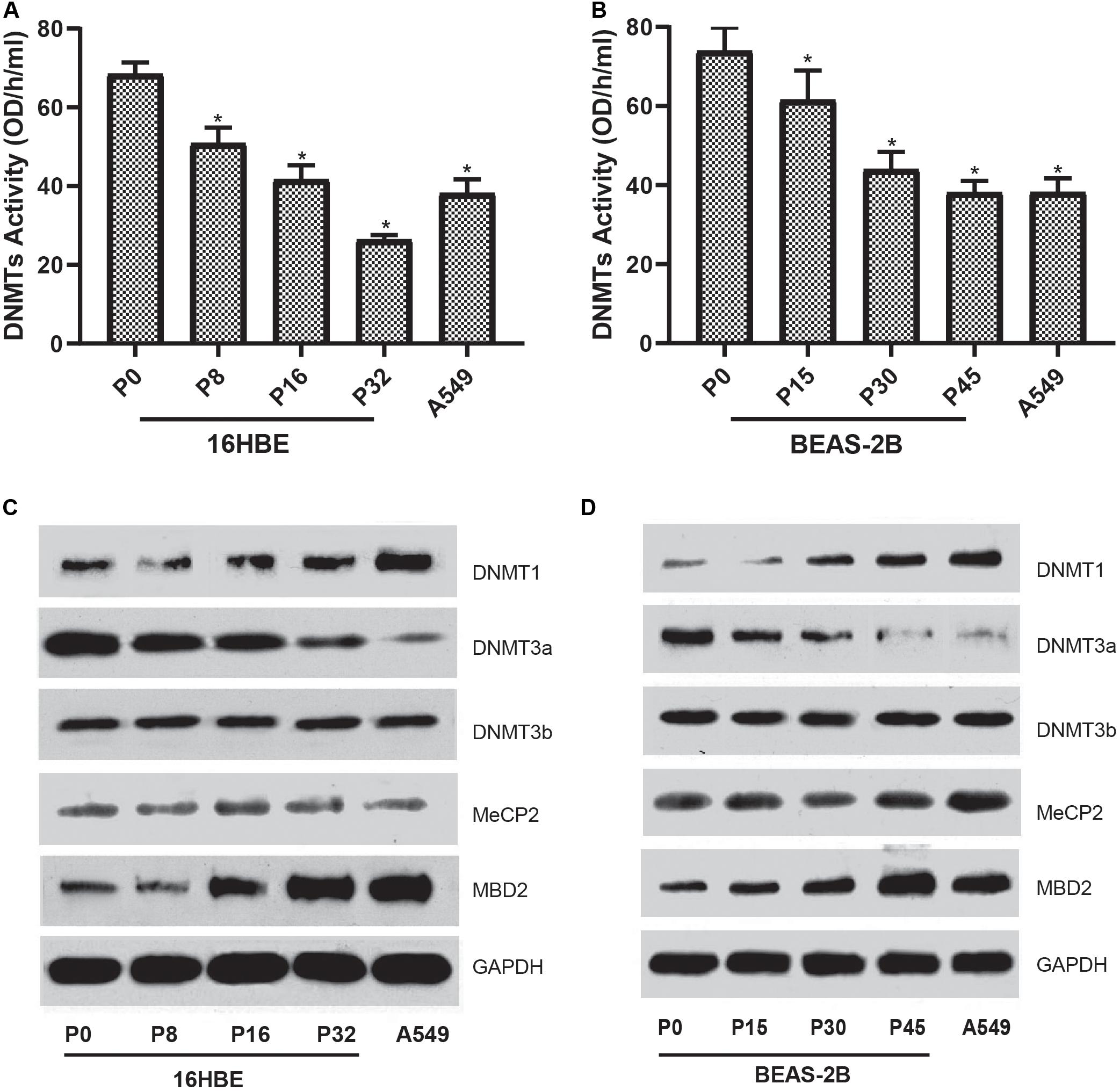
Figure 4. Effects of Nano-SiO2 on the expression of DNA methylation-associated proteins in human bronchial epithelial cells. DNMTs activity of 16HBE cells (A) and BEAS-2B cells (B) treated with Nano-SiO2 for various durations were determined. The data are shown as mean ± SD (n = 3). *p < 0.05, compared with the control group (P0). A549, an epithelial cell line derived from human lung carcinoma, was used as a positive control. Levels of DNMT-1, DNMT-3a, DNMT-3b, MeCP-2, and MBD-2 in 16HBE cells (C) and BEAS-2B cells (D) treated with Nano-SiO2 for different passages were evaluated using western blot. GAPDH was used as the loading control.
The transcription factor NRF2 involved in stress response plays a role in cancer development. Expression of the NRF2 gene is known to be influenced by the status of methylation of CpG islands in the promoter region in cancer cells (Kang et al., 2014; Zhao et al., 2015). Therefore, we asked whether DNA methylation of the promoter region of the NRF2 gene is influenced by Nano-SiO2 exposure. We conducted a methylation-specific PCR (MSP) to assess the methylation status of CpG sites at the NRF2 promoter region. As illustrated in Figure 5A, two sets of primers (Supplementary Table S2) were designed to detect the methylation status in different CpG islands (region A and region B). The methylation level at region A of 16HBE cells treated with Nano-SiO2 for 16 (P16) and 32 (P32) passages were decreased when compared with the untreated group (16HBE-P0). Similarly, the methylation level at region A of BEAS-2B cells treated with Nano-SiO2 for 15, 30, and 45 passages was also reduced when compared with the untreated group (BEAS-2B-P0). However, no obvious changes were found at region B in 16HBE or BEAS-2B cells after Nano-SiO2 treatment (Figures 5B,C).
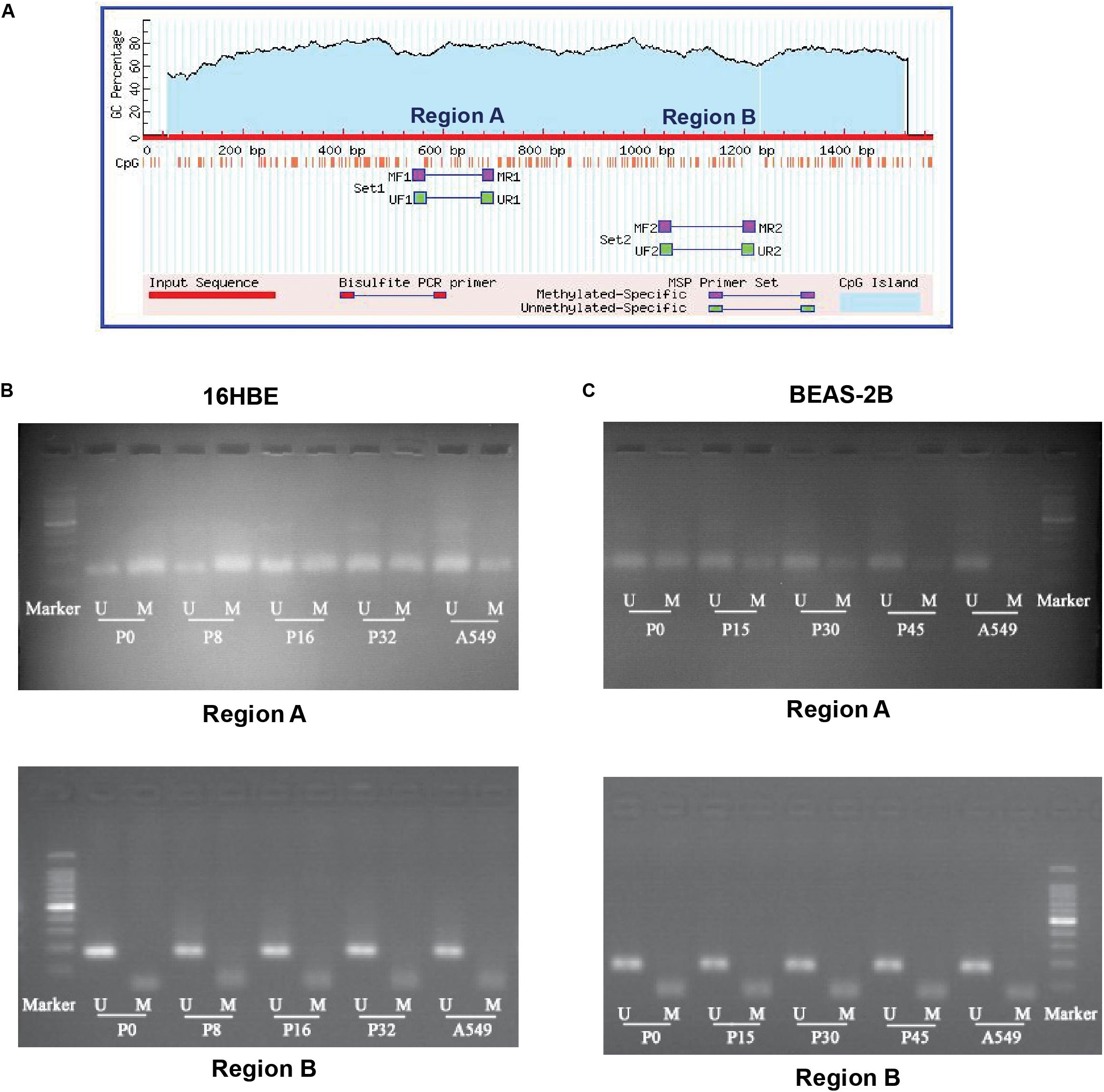
Figure 5. MSP analysis of DNA methylation of the promoter of the NRF2 gene. (A) Schematic graph showing CpG islands in the NRF2 promoter region and binding sites for MSP PCR primers. (B,C) Representative electrophoresis results of PCR products of CpG islands in the NRF2 promoter region using primers illustrated in (A). M: methylated alleles. U: unmethylated alleles. A549 was used as a positive control.
We further investigated the expression of NRF2 gene using qRT-PCR for mRNA and western blot analysis for protein in cells treated with Nano-SiO2. Our results showed that Nano-SiO2 exposure significantly increases the mRNA and protein expression of the NRF2 gene in both cell lines (Figures 6A–D), which is consistent with the hypomethylation of global DNA induced by Nano-SiO2 (Figures 3A,C). Furthermore, we also determined the expression levels of several gene targets (HO-1, SOD1, and GST) of the transcription factor NRF2 upon Nano-SiO2 exposure. Analysis using qPCR and western blot showed that the mRNA and protein levels of HO-1, SOD1, and GST were also significantly increased by Nano-SiO2 treatment. The mRNA and protein expression of these anti-oxidative genes regulated by NRF2 largely correlated with the duration of Nano-SiO2 exposure (Figure 5). These results revealed that the NRF2 gene and its target genes were up-regulated by Nano-SiO2 exposure.
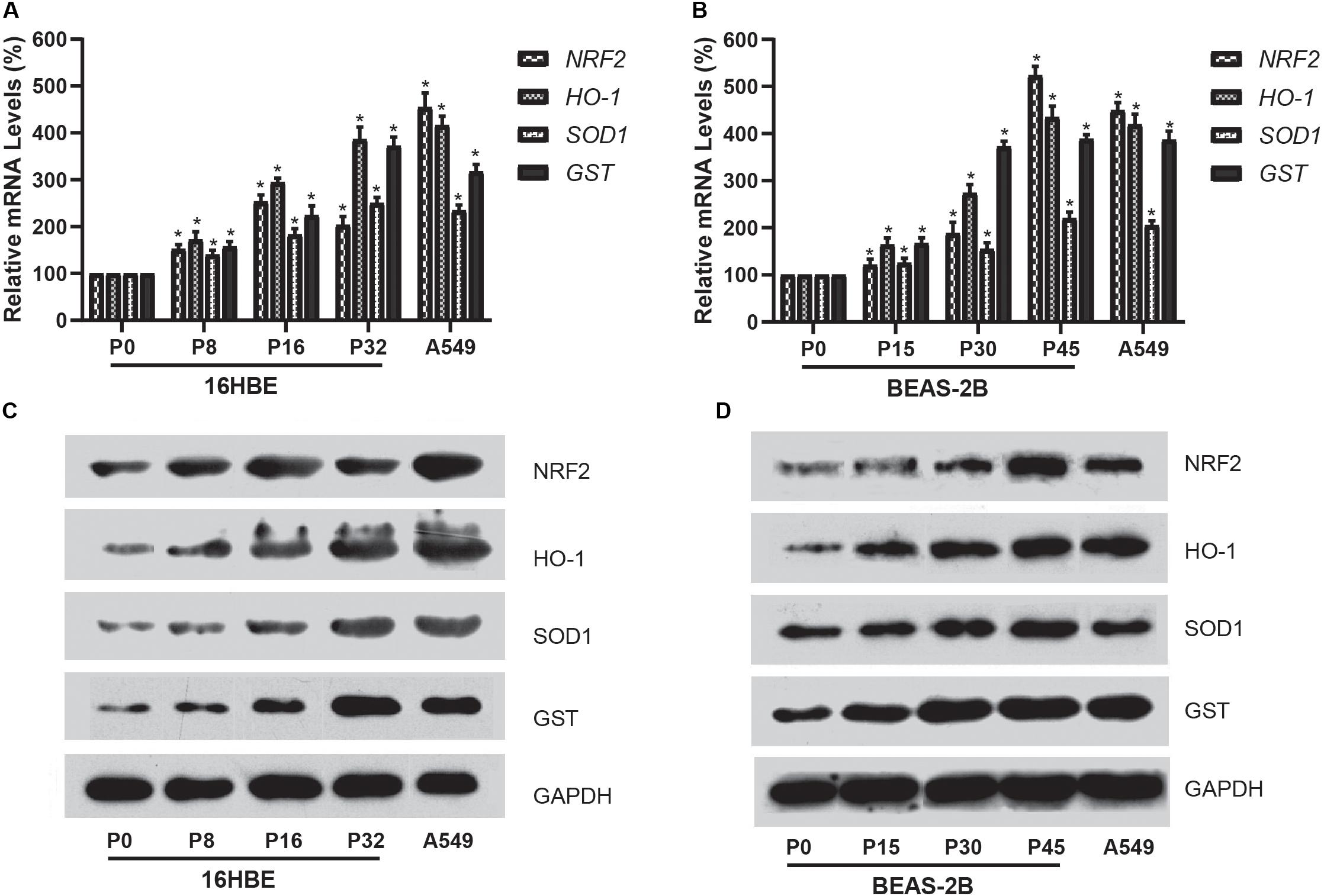
Figure 6. Effects of Nano-SiO2 on the expression of NRF2 and its target genes in human bronchial epithelial cells. (A,B) mRNA expression of NRF2, HO-1, SOD1, and GST genes in 16HBE or BEAS-2B cells were determined by qRT-PCR. (C,D) Protein expression levels of NRF2, HO-1, SOD1, and GST in 16HBE or BEAS-2B cells were evaluated by western blotting analysis. GAPDH was used as the internal control for normalization. The data are presented as mean ± SD (n = 3). *p < 0.05, compared with the untreated control group (P0). A549 was used as a positive control.
Since the expression of the NRF2 gene is up-regulated by Nano-SiO2, we further investigated the role of NRF2 in carcinogenesis induced by Nano-SiO2 by knockdown or overexpression of the NRF2 gene. Cells were transfected with vector harboring the NRF2 gene or the NRF2 gene shRNA to achieve overexpression (NRF2-OE) or knockdown (NRF2-KD) of the NRF2 gene and levels of NRF2 protein were determined using western blot (Supplementary Figures S1A–D). Cell viability assay showed that neither knockdown nor overexpression of the NRF2 gene introduces cytotoxicity to 16HBE and BEAS-2B cells (Supplementary Figures S1E,F). Thereafter, the effects of NRF2-OE and NRF2-KD on the viability of cells treated with Nano-SiO2 were determined using the MTT method. We observed that NRF2-OE increases the viability of both 16HBE and BEAS-2B cells after treated with Nano-SiO2, whereas NRF2-KD tends to reduce the cell viability upon Nano-SiO2 treatment (Figure 7). These results suggest that the NRF2 attenuates the cytotoxicity induced by Nano-SiO2 exposure. To investigate the role of NRF2 protein in the tumorigenicity induced by Nano-SiO2, the tumorigenicity of cells with NRF2-OE or NRF2-KD exposed to Nano-SiO2 were determined both in vitro and in vivo. The 16HBE and BEAS-2B cells with NRF2-OE or NRF2-KD were treated with Nano-SiO2 (10.0 μg/ml for 16HBE cells, 40.0 μg/ml for BEAS-2B cells) for various passages and followed with soft agar colony formation assay and nude mice injection. With the prolonged Nano-SiO2 exposure, both NRF2-OE and NRF2-KD cells (16HBE and BEAS-2B) developed a significant amount of colonies in the soft agar (Tables 2, 3). Interestingly, we observed that cells with NRF2-OE formed a lower number of colonies than cells of the control group, while NRF2-KD was able to increase the formation of colonies on soft agar. Consistent with the in vitro assay, both 16HBE (P8, P16, P32) and BEAS-2B (P15, P30, P45) cells with NRF2-OE formed tumors of a smaller size than the control group, while cells with NRF2-KD tend to increase the average tumor size (Figure 8). Taken together, results from both the in vitro and in vivo assay suggest that the NRF2 protein plays an inhibitory role in the tumorigenesis induced by Nano-SiO2 exposure.
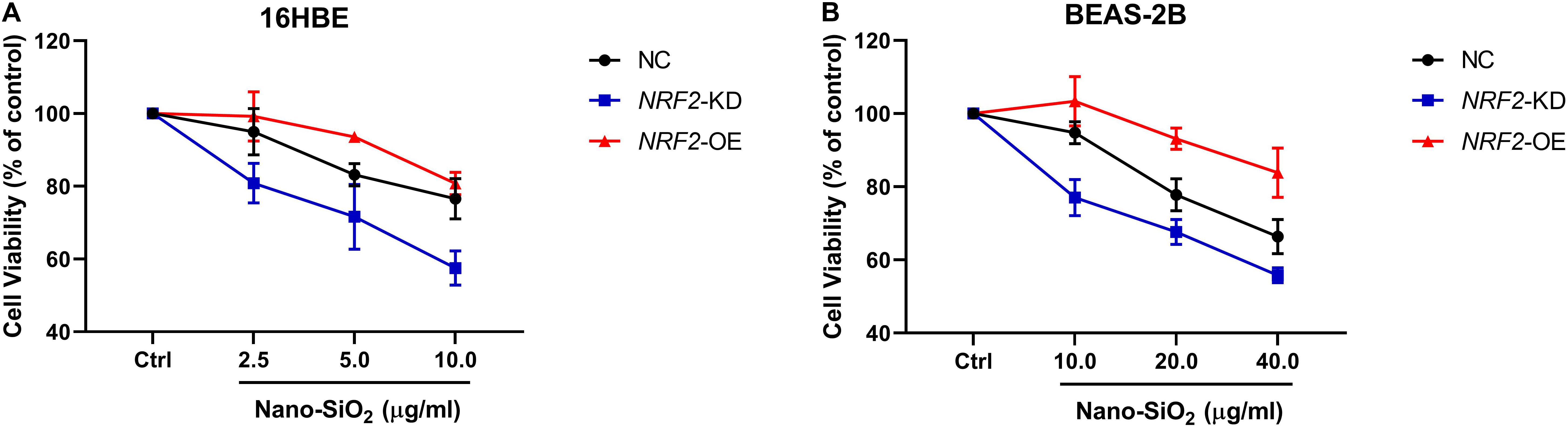
Figure 7. The NRF2 protein protects against the cytotoxicity of Nano-SiO2 on human bronchial epithelial cells. Cells (A) 16HBE, (B) BEAS-2B) were treated with Nano-SiO2 particles for 24 h. Values were mean ± SD (n = 3). NC: cells (16HBE or BEAS-2B) transfected with negative control; NRF2-KD: knockdown of the NRF2 gene; NRF2-OE: NRF2 overexpression.
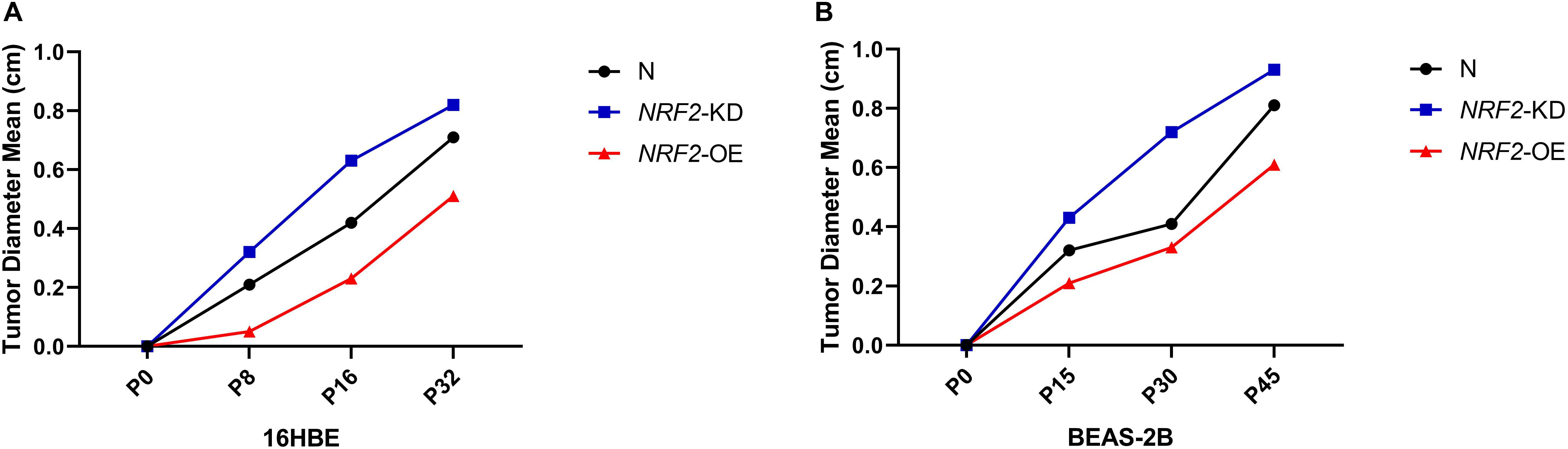
Figure 8. NRF2 expression affects the tumorigenicity of Nano-SiO2-transformed cells in nude mice. (A,B) The major diameter of the tumor formed at the subcutaneous injection site in nude mice. Tumor sizes were measured precisely 5 weeks after injection. The data are presented as a mean of tumor diameter. N: 16HBE or BEAS-2B cells without transfection; NRF2-KD: knockdown of NRF2; NRF2-OE: overexpression of NRF2.
In this study, two types of human bronchial epithelial cell lines (16HBE and BEAS-2B) were selected as the in vitro models to investigate the carcinogenicity of Nano-SiO2. We observed the dose-dependent cytotoxic effects of Nano-SiO2 on the human bronchial epithelial cells. Also, we showed the tumorigenicity of Nano-SiO2 in human bronchial epithelial cells.
As a hallmark of carcinogenesis, alterations in DNA methylation occur frequently in a wide range of cancers (Miousse et al., 2018; Dreval et al., 2019). Our results demonstrated a global genomic hypomethylation in cells exposed to the Nano-SiO2, which is consistent with the reduction of the total enzyme activity of DNA methyltransferases. Although the total activity of DNMTs is reduced, we found that the expression of DNMT1 protein is elevated and the expression of DNMT3a protein is downregulated. The DNMT1 is responsible for the methylation of tumor suppressor genes (Jair et al., 2006; Ting et al., 2006), the increased expression of which might induce hypermethylation of the tumor suppressor genes, thereby inhibiting the expression of tumor suppressors and promoting carcinogenesis induced by Nano-SiO2. The DNMT3a protein is known for the de novo CpG methylation independent of replication, which is consistent with the global hypomethylation induced by Nano-SiO2. Loss of DNMT3a has been reported to be associated with leukemia pathogenesis and poor prognosis (Shivarov et al., 2013). Moreover, MBD2 and MeCP2 that can bind to and mediate the repression of methylated tumor suppressor genes (Klose and Bird, 2006; Lopez-Serra et al., 2008; Mian et al., 2011), were also induced by the Nano-SiO2 exposure.
In addition to the global DNA hypomethylation, we observed a decreased methylation of CpG islands within the NRF2 promoter region in human bronchial epithelial cells exposed to Nano-SiO2. The NRF2 protein is a key transcription factor involved in cellular defensive mechanisms against stresses induced by electrophiles, oxidants, and chemicals (Motohashi and Yamamoto, 2004). Under the basal condition, NRF2 is localized in the cytoplasm by binding to its cytosolic repressor Kelch-like ECH-associated protein 1 (KEAP1) (Motohashi and Yamamoto, 2004). In response to stress, NRF2 dissociates from KEAP1 and translocates to the nucleus, where it activates the transcription of its target genes to maintain cellular homeostasis (Panieri and Saso, 2019). Short-term exposure to Nano-SiO2 can increase the expression of the NRF2 gene (Liu et al., 2017), which is consistent with our results that the NRF2 gene expression is up-regulated in both mRNA and protein levels in cells exposed to Nano-SiO2 exposure. Moreover, our findings revealed that levels of HO1, SOD1, and GST, which are encoded by antioxidative gene targets of the NRF2 protein (Itoh et al., 1997), are increased upon Nano-SiO2 exposure. These results suggest that NRF2 promotes the stress responses of cells treated with Nano-SiO2.
Activation of the NRF2 protein is suggested to suppress carcinogenesis, especially in its early stage (Wu et al., 2019). Loss of NRF2 disrupts the antioxidant axis resulting in increased oxidative stress, ultimately leading to DNA damage and the initiation of malignant cellular transformation (Frohlich et al., 2008). In the tumor microenvironment, the tumor suppressor gene BRCA1 activates NRF2 (Gorrini et al., 2013), whereas the oncogene Fyn mediates nuclear export and induces degradation of NRF2 (Niture et al., 2014). NRF2-deficient mice are more susceptible to carcinogens and develop severer tumors in the urinary bladder and liver (Iida et al., 2007; Kitamura et al., 2007).
Collectively, our results showed that NRF2 plays an important role in protecting against the carcinogenesis induced by Nano-SiO2 exposure. Moreover, hypomethylation at the promoter region of the NRF2 gene contributed to the alterations of NRF2 upon Nano-SiO2 exposure. It should, however, be noted that histone modifications are also important for NRF2 gene transcription and it cannot be excluded that the alterations of NRF2 at both transcription and protein levels are caused by a more intrinsic mechanism.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Shanghai Municipal Center for Disease Prevention and Control.
DL, GT, and YS jointly conceived this project and supervised the experiments. DL and XW designed the research. DL, XW, PX, QH, JS, and XH performed the experiments and analyzed the experimental results. DL, XW, YS, and GT prepared the manuscript. All authors contributed to the article and approved the submitted version.
The present study was supported by the National Natural Science Foundation of China (81302468), the Project of Shanghai Municipal Health Commission (20124080), and the Three-year action plan of Shanghai Public Health System Construction (GWTD2015S03).
YS is employed by the Syngenta (China) Investment Company Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00818/full#supplementary-material
Bermudez, E., Mangum, J. B., Wong, B. A., Asgharian, B., Hext, P. M., Warheit, D. B., et al. (2004). Pulmonary responses of mice, rats, and hamsters to subchronic inhalation of ultrafine titanium dioxide particles. Toxicol. Sci. 77, 347–357. doi: 10.1093/toxsci/kfh019
Chen, M., and von Mikecz, A. (2005). Formation of nucleoplasmic protein aggregates impairs nuclear function in response to SiO2 nanoparticles. Exp. Cell Res. 305, 51–62. doi: 10.1016/j.yexcr.2004.12.021
Dreval, K., Tryndyak, V., De Conti, A., Beland, F. A., and Pogribny, I. P. (2019). Gene expression and DNA methylation alterations during non-alcoholic steatohepatitis-associated liver carcinogenesis. Front. Genet. 10:486. doi: 10.3389/fgene.2019.00486
Frohlich, D. A., Mccabe, M. T., Arnold, R. S., and Day, M. L. (2008). The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene 27, 4353–4362. doi: 10.1038/onc.2008.79
Fruijtier-Polloth, C. (2012). The toxicological mode of action and the safety of synthetic amorphous silica-a nanostructured material. Toxicology 294, 61–79. doi: 10.1016/j.tox.2012.02.001
Gama-Sosa, M. A., Slagel, V. A., Trewyn, R. W., Oxenhandler, R., Kuo, K. C., Gehrke, C. W., et al. (1983). The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 11, 6883–6894. doi: 10.1093/nar/11.19.6883
Gebel, T. (2012). Small difference in carcinogenic potency between GBP nanomaterials and GBP micromaterials. Arch. Toxicol. 86, 995–1007. doi: 10.1007/s00204-012-0835-1
Gong, C., Tao, G., Yang, L., Liu, J., He, H., and Zhuang, Z. (2012a). The role of reactive oxygen species in silicon dioxide nanoparticle-induced cytotoxicity and DNA damage in HaCaT cells. Mol. Biol. Rep. 39, 4915–4925. doi: 10.1007/s11033-011-1287-z
Gong, C., Tao, G., Yang, L., Liu, J., Liu, Q., Li, W., et al. (2012b). Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicol. Lett. 209, 264–269. doi: 10.1016/j.toxlet.2012.01.007
Gong, C., Tao, G., Yang, L., Liu, J., Liu, Q., and Zhuang, Z. (2010). SiO(2) nanoparticles induce global genomic hypomethylation in HaCaT cells. Biochem. Biophys. Res. Commun. 397, 397–400. doi: 10.1016/j.bbrc.2010.05.076
Gorrini, C., Baniasadi, P. S., Harris, I. S., Silvester, J., Inoue, S., Snow, B., et al. (2013). BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J. Exp. Med. 210, 1529–1544. doi: 10.1084/jem.20121337
Hirose, A., Takagi, A., Nishimura, T., Tsuda, H., Sakamoto, Y., Ogata, A., et al. (2011). [Importance of researches on chronic effects by manufactured nanomaterials]. Yakugaku Zasshi 131, 195–201. doi: 10.1248/yakushi.131.195
Iarc. (1997). Silica, some silicates, coal dust and para-aramid fibrils. IARC Monogr. Eval. Carcinog. Risks Hum. 68, 1–475.
Iida, K., Itoh, K., Maher, J. M., Kumagai, Y., Oyasu, R., Mori, Y., et al. (2007). Nrf2 and p53 cooperatively protect against BBN-induced urinary bladder carcinogenesis. Carcinogenesis 28, 2398–2403. doi: 10.1093/carcin/bgm146
Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., et al. (1997). An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236, 313–322. doi: 10.1006/bbrc.1997.6943
Jair, K. W., Bachman, K. E., Suzuki, H., Ting, A. H., Rhee, I., Yen, R. W., et al. (2006). De novo CpG island methylation in human cancer cells. Cancer Res. 66, 682–692. doi: 10.1158/0008-5472.can-05-1980
Kang, K. A., Piao, M. J., Kim, K. C., Kang, H. K., Chang, W. Y., Park, I. C., et al. (2014). Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis. 5:e1183. doi: 10.1038/cddis.2014.149
Kasai, T., Umeda, Y., Ohnishi, M., Mine, T., Kondo, H., Takeuchi, T., et al. (2016). Lung carcinogenicity of inhaled multi-walled carbon nanotube in rats. Part Fibre Toxicol. 13:53.
Khor, T. O., Fuentes, F., Shu, L., Paredes-Gonzalez, X., Yang, A. Y., Liu, Y., et al. (2014). Epigenetic DNA methylation of antioxidative stress regulator NRF2 in human prostate cancer. Cancer Prev. Res. (Phila) 7, 1186–1197. doi: 10.1158/1940-6207.capr-14-0127
Kitamura, Y., Umemura, T., Kanki, K., Kodama, Y., Kitamoto, S., Saito, K., et al. (2007). Increased susceptibility to hepatocarcinogenicity of Nrf2-deficient mice exposed to 2-amino-3-methylimidazo[4,5-f]quinoline. Cancer Sci. 98, 19–24. doi: 10.1111/j.1349-7006.2006.00352.x
Klose, R. J., and Bird, A. P. (2006). Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97. doi: 10.1016/j.tibs.2005.12.008
Liu, D., Zhang, Y., Wei, Y., Liu, G., Liu, Y., Gao, Q., et al. (2016). Activation of AKT pathway by Nrf2/PDGFA feedback loop contributes to HCC progression. Oncotarget 7, 65389–65402. doi: 10.18632/oncotarget.11700
Liu, W., Hu, T., Zhou, L., Wu, D., Huang, X., Ren, X., et al. (2017). Nrf2 protects against oxidative stress induced by SiO2 nanoparticles. Nanomedicine (Lond) 12, 2303–2318. doi: 10.2217/nnm-2017-0046
Lopez-Serra, L., Ballestar, E., Ropero, S., Setien, F., Billard, L. M., Fraga, M. F., et al. (2008). Unmasking of epigenetically silenced candidate tumor suppressor genes by removal of methyl-CpG-binding domain proteins. Oncogene 27, 3556–3566. doi: 10.1038/sj.onc.1211022
Lu, K., Alcivar, A. L., Ma, J., Foo, T. K., Zywea, S., Mahdi, A., et al. (2017). NRF2 induction supporting breast cancer cell survival is enabled by oxidative stress-induced DPP3-KEAP1 interaction. Cancer Res. 77, 2881–2892. doi: 10.1158/0008-5472.can-16-2204
Lu, X., Jin, T., Jin, Y., Wu, L., Hu, B., Tian, Y., et al. (2013). Toxicogenomic analysis of the particle dose- and size-response relationship of silica particles-induced toxicity in mice. Nanotechnology 24, 015106. doi: 10.1088/0957-4484/24/1/015106
Marhenke, S., Lamle, J., Buitrago-Molina, L. E., Canon, J. M., Geffers, R., Finegold, M., et al. (2008). Activation of nuclear factor E2-related factor 2 in hereditary tyrosinemia type 1 and its role in survival and tumor development. Hepatology 48, 487–496. doi: 10.1002/hep.22391
Mian, O. Y., Wang, S. Z., Zhu, S. Z., Gnanapragasam, M. N., Graham, L., Bear, H. D., et al. (2011). Methyl-binding domain protein 2-dependent proliferation and survival of breast cancer cells. Mol. Cancer Res. 9, 1152–1162. doi: 10.1158/1541-7786.mcr-11-0252
Miousse, I. R., Ewing, L. E., Kutanzi, K. R., Griffin, R. J., and Koturbash, I. (2018). DNA methylation in radiation-induced carcinogenesis: experimental evidence and clinical perspectives. Crit. Rev. Oncogen. 23, 1–11. doi: 10.1615/critrevoncog.2018025687
Mohajerani, A., Burnett, L., Smith, J. V., Kurmus, H., Milas, J., Arulrajah, A., et al. (2019). Nanoparticles in construction materials and other applications, and implications of nanoparticle use. Materials (Basel) 12, 3052. doi: 10.3390/ma12193052
Motohashi, H., and Yamamoto, M. (2004). Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10, 549–557. doi: 10.1016/j.molmed.2004.09.003
Napierska, D., Thomassen, L. C., Lison, D., Martens, J. A., and Hoet, P. H. (2010). The nanosilica hazard: another variable entity. Part Fibre Toxicol. 7:39. doi: 10.1186/1743-8977-7-39
Niture, S. K., Khatri, R., and Jaiswal, A. K. (2014). Regulation of Nrf2-an update. Free Radic. Biol. Med. 66, 36–44. doi: 10.1016/j.freeradbiomed.2013.02.008
Panieri, E., and Saso, L. (2019). Potential applications of NRF2 inhibitors in cancer therapy. Oxid. Med. Cell Longev. 2019:8592348.
Seidel, C., Kirsch, A., Fontana, C., Visvikis, A., Remy, A., Gate, L., et al. (2017). Epigenetic changes in the early stage of silica-induced cell transformation. Nanotoxicology 11, 923–935. doi: 10.1080/17435390.2017.1382599
Shivarov, V., Gueorguieva, R., Stoimenov, A., and Tiu, R. (2013). DNMT3A mutation is a poor prognosis biomarker in AML: results of a meta-analysis of 4500 AML patients. Leuk Res. 37, 1445–1450. doi: 10.1016/j.leukres.2013.07.032
Song, Y., Li, X., and Du, X. (2009). Exposure to nanoparticles is related to pleural effusion, pulmonary fibrosis and granuloma. Eur. Respir. J. 34, 559–567. doi: 10.1183/09031936.00178308
Sweeney, S. K., Luo, Y., O’donnell, M. A., and Assouline, J. (2016). Nanotechnology and cancer: improving real-time monitoring and staging of bladder cancer with multimodal mesoporous silica nanoparticles. Cancer Nanotechnol. 7:3.
Ting, A. H., Jair, K. W., Schuebel, K. E., and Baylin, S. B. (2006). Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res. 66, 729–735. doi: 10.1158/0008-5472.can-05-1537
Vivero-Escoto, J. L., Huxford-Phillips, R. C., and Lin, W. (2012). Silica-based nanoprobes for biomedical imaging and theranostic applications. Chem. Soc. Rev 41, 2673–2685.
Wang, J. J., Sanderson, B. J., and Wang, H. (2007). Cytotoxicity and genotoxicity of ultrafine crystalline SiO2 particulate in cultured human lymphoblastoid cells. Environ. Mol. Mutagen. 48, 151–157. doi: 10.1002/em.20287
Wu, S., Lu, H., and Bai, Y. (2019). Nrf2 in cancers: a double-edged sword. Cancer Med. 8, 2252–2267. doi: 10.1002/cam4.2101
Yu, S., Khor, T. O., Cheung, K. L., Li, W., Wu, T. Y., Huang, Y., et al. (2010). Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS ONE 5:e8579. doi: 10.1371/journal.pone.0008579
Yu, Y., Duan, J., Li, Y., Yu, Y., Jin, M., Li, C., et al. (2015). Combined toxicity of amorphous silica nanoparticles and methylmercury to human lung epithelial cells. Ecotoxicol. Environ. Saf. 112, 144–152. doi: 10.1016/j.ecoenv.2014.10.026
Zhang, C., Wang, H. J., Bao, Q. C., Wang, L., Guo, T. K., Chen, W. L., et al. (2016). NRF2 promotes breast cancer cell proliferation and metastasis by increasing RhoA/ROCK pathway signal transduction. Oncotarget 7, 73593–73606. doi: 10.18632/oncotarget.12435
Keywords: Nano-SiO2, malignant transformation, carcinogenesis, DNA methylation, NRF2
Citation: Lou D, Wei X, Xiao P, Huo Q, Hong X, Sun J, Shuai Y and Tao G (2020) Demethylation of the NRF2 Promoter Protects Against Carcinogenesis Induced by Nano-SiO2. Front. Genet. 11:818. doi: 10.3389/fgene.2020.00818
Received: 07 May 2020; Accepted: 08 July 2020;
Published: 28 July 2020.
Edited by:
Qiang Zhang, Tianjin Medical University, ChinaReviewed by:
Yongmei Xiao, Sun Yat-sen University, ChinaCopyright © 2020 Lou, Wei, Xiao, Huo, Hong, Sun, Shuai and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Lou. ZGxvdTFAamh1LmVkdQ==; Yi Shuai, eWkuc2h1YWlAc3luZ2VudGEuY29t; Gonghua Tao, dGFvZ29uZ2h1YUBzY2RjLnNoLmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.