
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 26 June 2020
Sec. Epigenomics and Epigenetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00636
This article is part of the Research Topic Genetic and Epigenetic Regulation of Insect Development, Reproduction, and Phenotypic Plasticity View all 24 articles
Steroid hormone signaling contributes to the development of multicellular organisms. In insects, ecdysteroids, like ecdysone and the more biologically-active derivative 20-hydroxyecdysone (20E), promote molting and metamorphosis. Ecdysone is biosynthesized in the prothoracic gland (PG), via several steps catalyzed by ecdysteroidogenic enzymes that are encoded by Halloween genes. The spatio-temporal expression pattern of ecdysteroidogenic genes is strictly controlled, resulting in a proper fluctuation of the 20E titer during insect development. However, their transcriptional regulatory mechanism is still elusive. A previous study has found that the polyadenylated tail [poly(A)] deadenylation complex, called Carbon catabolite repressor 4-Negative on TATA (CCR4-NOT) regulates the expression of spookier (spok), which encodes one of the ecdysteroidogenic enzymes in the fruit fly Drosophila melanogaster. Based on this finding, we speculated whether any other poly(A)-related protein also regulates spok expression. In this study, we reported that poly(A) binding protein (Pabp) is involved in spok expression by regulating nuclear localization of the transcription factor molting defective (Mld). When pabp was knocked down specifically in the PG by transgenic RNAi, both spok mRNA and Spok protein levels were significantly reduced. In addition, the spok promoter-driven green fluorescence protein (GFP) signal was also reduced in the pabp-RNAi PG, suggesting that Pabp is involved in the transcriptional regulation of spok. We next examined which transcription factors are responsible for Pabp-dependent transcriptional regulation. Among the transcription factors acting in the PG, we primarily focused on the zinc-finger transcription factor Mld, as Mld is essential for spok transcription. Mld was localized in the nucleus of the control PG cells, while Mld abnormally accumulated in the cytoplasm of pabp-RNAi PG cells. In contrast, pabp-RNAi did not affect the nuclear localization of other transcription factors, including ventral vein lacking (Vvl) and POU domain motif 3 (Pdm3), in PG cells. From these results, we propose that Pabp regulates subcellular localization in the PG, specifically of the transcription factor Mld, in the context of ecdysone biosynthesis.
Ecdysteroids, like ecdysone and the more biologically-active derivative 20-hydroxyecdysone (20E), regulate several biological events in insects (Niwa and Niwa, 2014; Uryu et al., 2015). The 20-hydroxyecdysone titers are temporally changed during insect development, where proper fluctuations of 20E titers are essential for insect development (Riddiford, 1993; Nijhout, 1994; Bellés, 2020). Ecdyone is biosynthesized in an endocrine organ named the prothoracic gland (PG) via several steps catalyzed by step-specific enzymes (Niwa and Niwa, 2014). The enzymes are encoded by a group of genes often called the Halloween gene (Rewitz et al., 2007). Therefore, the regulation of ecdysteroidogenic genes expression is essential to achieve proper fluctuations of 20E titers (Niwa and Niwa, 2016). However, the molecular mechanism by which the expression of ecdysteroidogenic genes is regulated is yet to be fully elucidated.
Previously, we have reported that polyadenylated tail [poly(A)] degradation complex, called Carbon catabolite repressor 4-Negative on TATA (CCR4-NOT) is involved in the regulation of ecdysteroidogenic gene expression in the fruit fly Drosophila melanogaster (Zeng et al., 2018). By knocking down the gene phosphoglycerate kinase promoter directed over production (Pop2), which encodes a vital component of the CCR4-NOT complex, specifically in the PG, D. melanogaster animals show a larval-arrested phenotype. Furthermore, the expression levels of some ecdysteroidogenic genes are strongly decreased in these animals. Based on this finding, we hypothesized that other poly(A) related protein(s) may also contribute to the expression of ecdysteroidogenic genes.
In this study, we revealed that poly(A) binding protein (Pabp) contributes to the expression of ecdysteroidogenic genes. The knockdown of the pabp gene in the PG caused first instar-arrest and a decrease in ecdysteroidogenic gene expression, especially spookier (spok). Interestingly, the nuclear localization of the transcription factor molting defective (Mld), a transcriptional activator of spok, was disrupted in PG cells of pabp-RNAi larvae. Our results suggest that Pabp positively regulates ecdysteroidogenic gene expression by regulating ecdysteroidogenic transcription factors.
D. melanogaster flies were reared on a standard agar-cornmeal medium at 25 or 17°C under a 12:12 h light/dark cycle. w 1118 served as a control strain. phm-GAL4#22 (a gift from Michael B. O’Connor, University of Minnesota, MN; McBrayer et al., 2007; Yamanaka et al., 2013) was used as the strain to drive forced gene expression in the PG. UAS-dicer2 (#24650) was obtained from the Bloomington Drosophila Stock Center. UAS-pabp-IR (#22007) and UAS-mld-IR (#17329) were obtained from the Vienna Drosophila Resource Center. Transgenic RNAi experiments were conducted by crossing these UAS RNAi lines with w, UAS-dicer2, and phm-GAL4#22/TM6 Ubi-GFP. A strain carrying the 1.45 kb spok promoter-fused GFP cassette (spok>GFP) was previously described (Komura-Kawa et al., 2015).
Rabbit polyclonal anti-Mld antibody was raised against the peptide: NH2-MSANRRRRSASAASSIAAET-COOH, which corresponds to 1–20 amino acid residues of the mature Mld protein (Neubueser et al., 2005).
Immunostaining of prothoracic glands was performed as described previously (Imura et al., 2017). Briefly, dissected larval tissues were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature. Samples were then washed with PBS + 0.3% Triton X-100 (Nacalai tesque, Kyoto Japan) and incubated overnight at 4°C with primary antibodies: rabbit anti-Mld antibody (1:200), guinea pig anti-Spok antibody (1:200; Gibbens et al., 2011), rabbit anti-Phantom (Phm) antibody (1:200; Parvy et al., 2005), rat anti-Ventral vein lacking (Vvl) antibody (1:3,000; Anderson et al., 1995), and guinea pig anti-POU domain motif 3 (Pdm3; 1:100; Chen et al., 2012). As fluorescent secondary antibodies, we used goat anti-guinea pig Alexa Fluor 488 (Life Technologies, Carlsbad, CA, USA) and goat anti-rabbit Alexa Fluor 555 (Life Technologies, Carlsbad, CA, USA). The secondary antibodies were diluted 1:200 and incubated for 2.5 h at room temperature. Nuclear stains used in this study were 4',6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO, USA) and TOPRO3 (Thermo Fisher Scientific, Waltham, MA, USA). For DAPI staining, after the incubation with the secondary antibodies, the samples were washed and then incubated with 1 μg/ml (final concentration) of DAPI for 1 h. For TOPRO3 staining, the samples were incubated with the secondary antibodies along with 10 μg/ml (final concentration) of RNase A (Takara Bio, Kusatsu, Japan). The samples were then washed, followed by the 1-h incubation with 10 μm TOPRO3 (Thermo Fisher Scientific, Waltham, MA, USA). Confocal images were captured using the LSM 700 laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany). Images were processed using the ImageJ software (Schneider et al., 2012).
RNA was isolated from the whole bodies of the second instar larvae using the RNAiso Plus reagent (TaKaRa, Shiga, Japan). Genomic DNA digestion and cDNA synthesis were performed using the ReverTra Ace qPCR RT Kit (TOYOBO, Tokyo, Japan). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using the THUNDERBIRD SYBR qPCR Mix (TOYOBO, Tokyo, Japan) with a Thermal Cycler Dic TP800 System (TaKaRa, Shiga, Japan). Serial dilutions of a plasmid containing the open reading frame (ORF) of each gene were used as a standard. The expression levels of the target genes were normalized to an endogenous control ribosomal protein 49 (rp49) in the same sample. Primers amplifying noppera-bo (nobo), neverland (nvd), shroud (sro), spok, phm, disembodied (dib), shadow (sad), and rp49 have been described previously (McBrayer et al., 2007; Niwa et al., 2010; Enya et al., 2014).
A spok promoter-driven GFP reporter assay was performed as described previously (Komura-Kawa et al., 2015). Briefly, spok>GFP/CyO Act-GFP; UAS-deicer2, phm22-GAL4/TM6 Ubi-GFP was established and crossed with UAS-pabp-IR. Eggs were laid on grape plates with yeast pastes at 25°C for 2 h. The GFP-negative first instar larvae were picked up and transferred into vials with standard cornmeal food. They were dissected 60 h after egg laying (AEL) and then immunostained.
To examine the importance of Pabp in the PG, we observed the developmental progress of pabp-RNAi larvae, for which we used a PG-specific driver (phm22-GAL4, hereafter phm>) to knock down pabp expression by transgenic RNAi. We found that PG-specific pabp-RNAi caused a larval-arrest phenotype. Eighty eight percent of phm>pabp-RNAi animals were arrested at the second larval instar and even 132 h AEL or later, while only few animals molted into the third larval instar or pupariated at the same time point (Figures 1A–C). The arrested second instar larvae failed to pupariate and died. This result suggests that the Pabp function in the PG is essential for larval development.
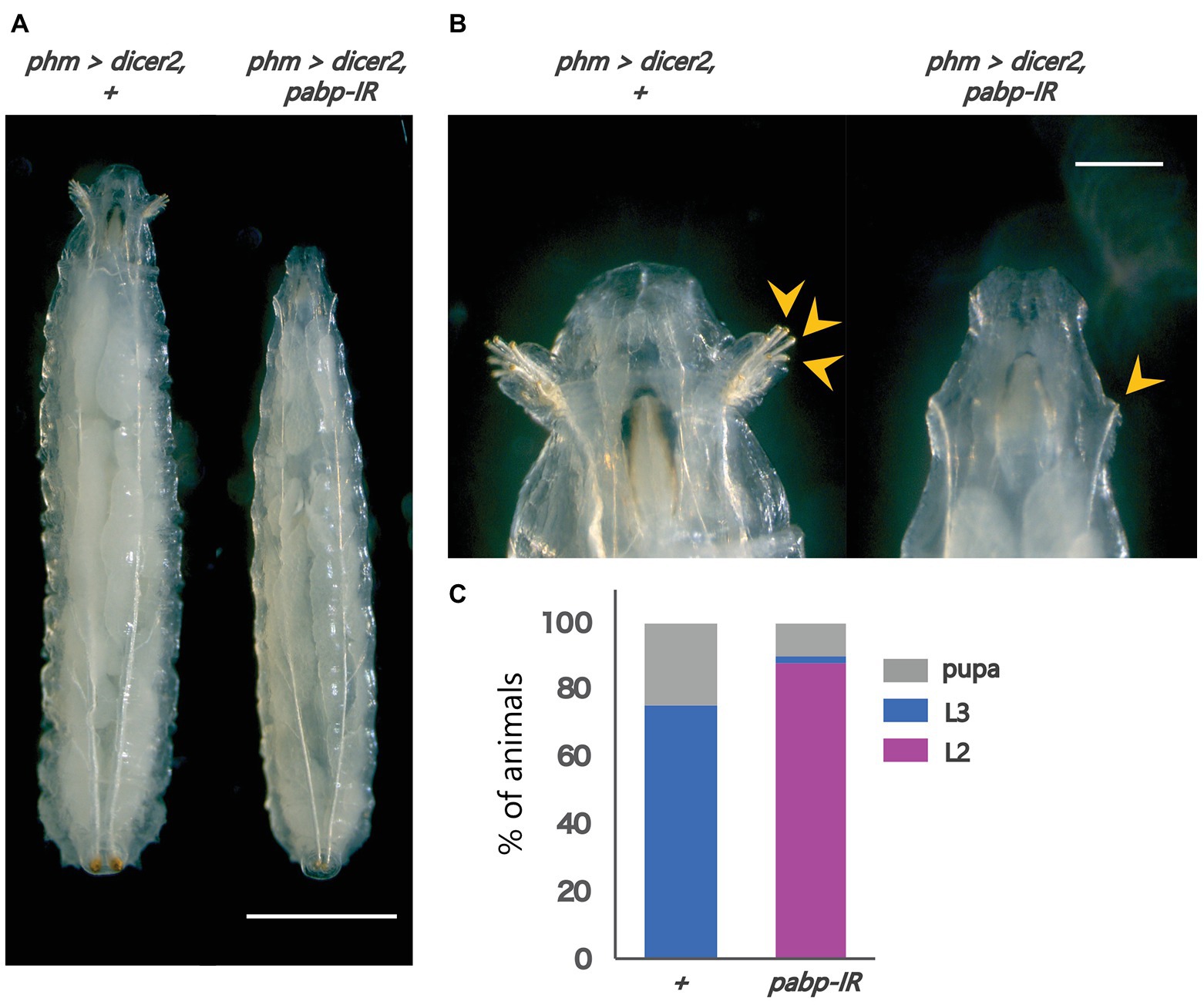
Figure 1. Pabp function is required in the PG for larval development. Phenotypes caused by PG-specific pabp-RNAi at 132 h AEL. (A) Whole bodies of the control and RNAi larvae. Scale bar: 5 mm (B) Magnified intensity of Figure 1A. Scale bar: 2 mm. (Left) phm>dicer2, + larvae molted in the third larval instar as judged by the branched morphology of the anterior tracheal pits (yellow arrowheads), typical features of third instar larvae. (Right) phm>dicer2, pabp-IR larvae raised at the second larval instar and exhibit singular insertions of anterior tracheal pits. (C) The developmental progression of phm>dicer2, + (N = 41) and phm>dicer2, and pabp-IR (N = 49) animals at 132 h AEL. L2: second instar larva, L3: third instar larva.
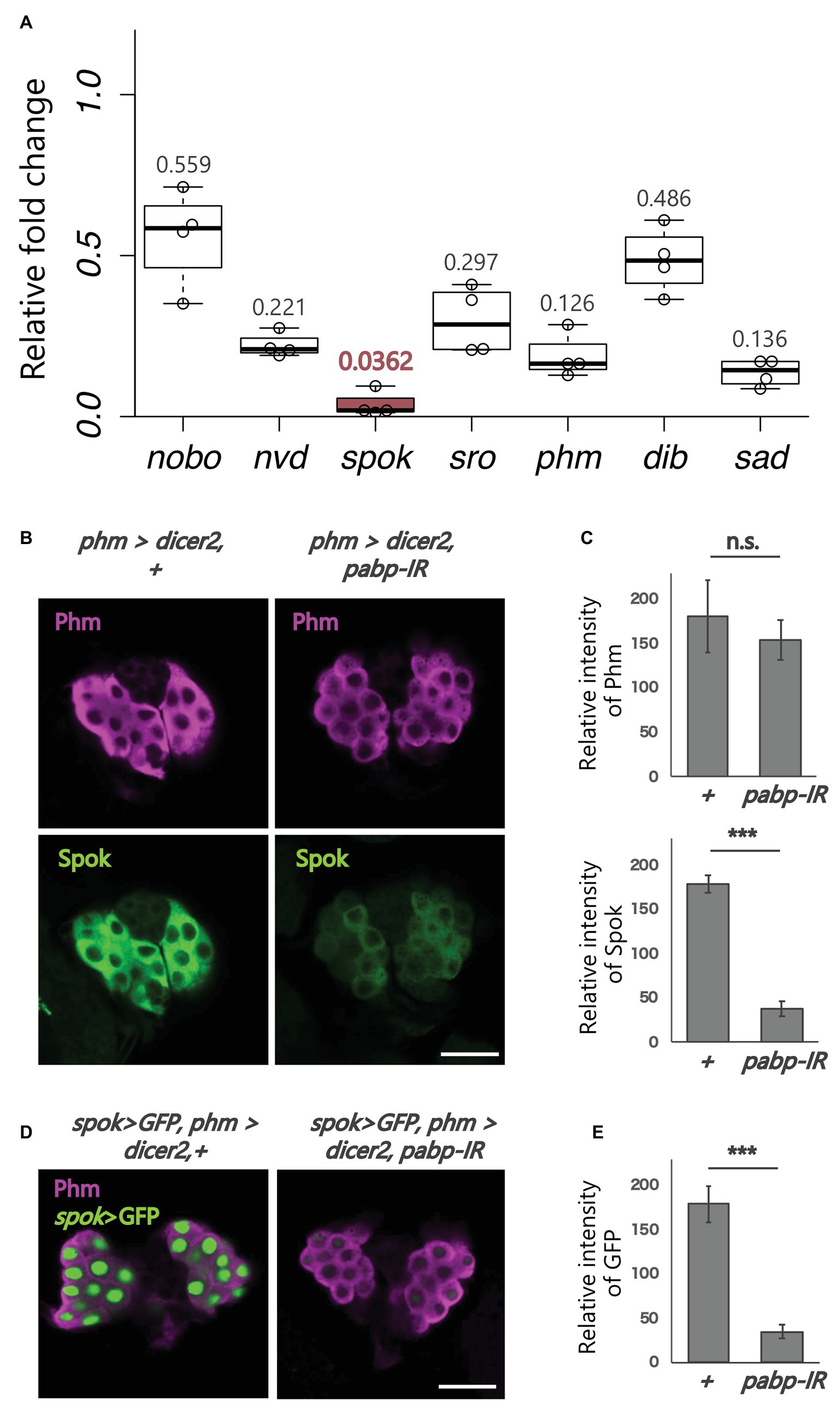
Figure 2. Expression analysis of ecdysteroidogenic genes. All photos are single-plane confocal images. (A) Relative expression levels of seven ecdysteroidogenic genes in phm>dicer2, pabp-IR at 60 h AEL compared to controls (phm>dicer2, +) based on the quantitative reverse transcription-polymerase chain reaction (qRT-PCR; N = 4). RNA was isolated from the whole bodies of the second instar larvae. (B) Immunostaining of the PG cells from phm>dicer2, + and phm>dicer2, and pabp-IR second instar larvae at 60 h AEL with antibodies against Phm (magenta) and Spok (green). (C) Quantifications of fluorescence intensities of Phm and Spok in the PG (each N = 3). Error bars represent standard deviations. *** and n.s. indicate p < 0.001 and non-significance (p > 0.05), respectively, by Student’s t-test. (D) Fluorescence images of the PG cells from phm>dicer2, + and phm>dicer2, pabp-IR larvae with spok enhancer/promoter-driven nuclear localized-GFP construct (spok>GFP) at 60 h AEL. (E) Quantification of GFP fluorescence intensity in the PG nuclei (N = 4). The bar plots are drawn in the same manner as (C). PG cells are immunostained with anti-Phm antibody (magenta). Scale bar: 20 μm.
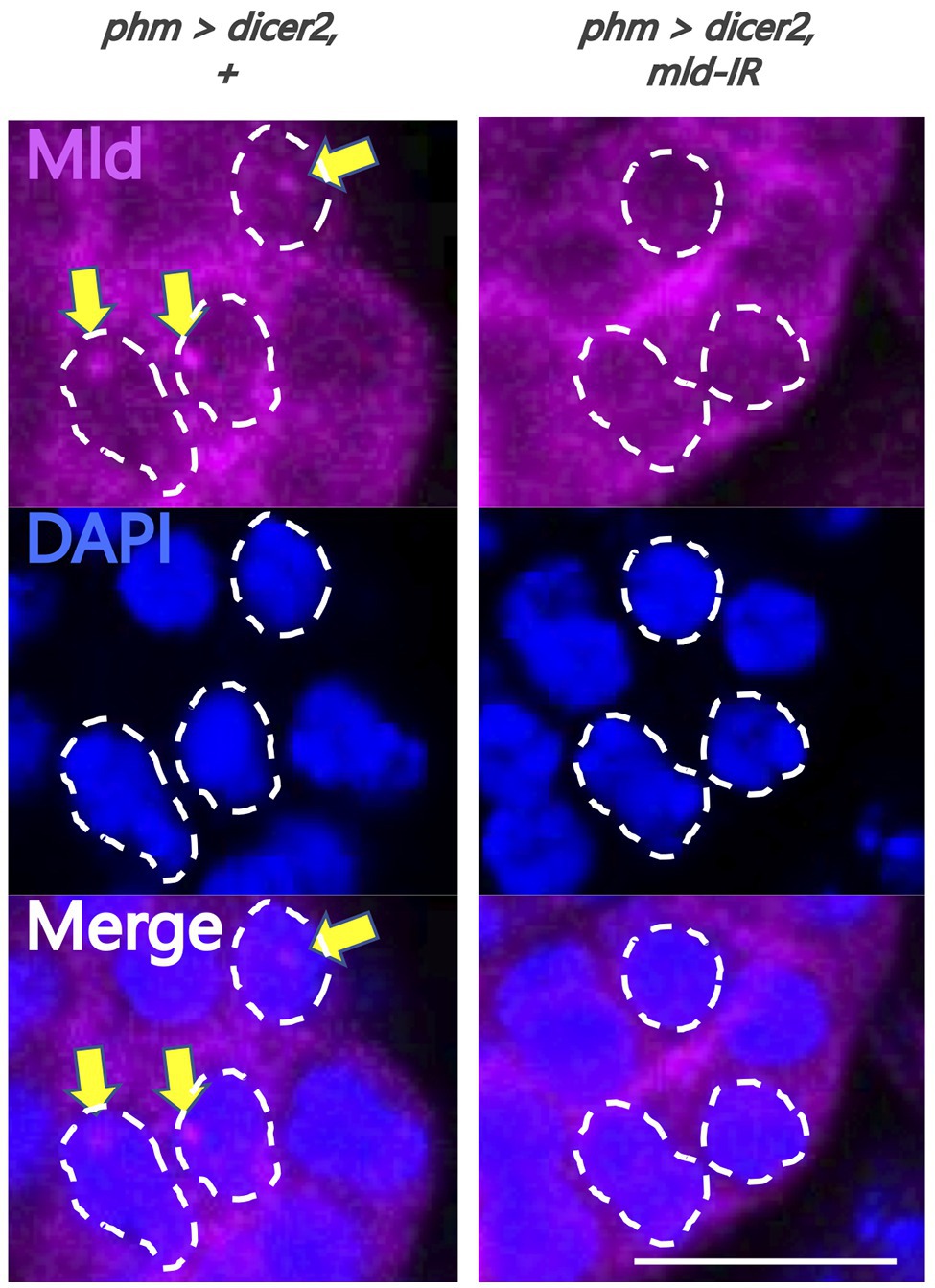
Figure 3. Evaluation of newly-generated anti-molting defective (Mld) antibody. Magnified image of prothoracic gland (PG) cells. All photos are single-plane confocal images. Tissues were stained with anti-Mld antibody (magenta) and 4',6-diamidino-2-phenylindole (DAPI; blue). Representative nuclei are outlined by dashed lines. Mld signals were detected in the nuclei of the PG cells of phm>dicer2, + (yellow arrow) at 36 h after egg laying (AEL). In contrast, the signal disappeared in phm>dicer2 and pabp-IR. Scale bar: 10 μm.
Next, we examined whether pabp knockdown in the PG changes the expression of ecdysteroidogenic genes. We conducted qRT-PCR to examine the expression levels of seven ecdysteroidogenic genes (Chávez et al., 2000; Warren et al., 2002, 2004; Niwa et al., 2004, 2010; Namiki et al., 2005; Ono et al., 2006; Yoshiyama et al., 2006; Chanut-delalande et al., 2014; Enya et al., 2014) in the second instar larvae of control and pabp-RNAi animals. The expression levels of all ecdysteroidogenic genes examined were suppressed in pabp-RNAi animals. However, specifically, the suppression levels were different among these seven genes. In particular, the suppression level of spok, encoding an ecdysteroidogenic cytochrome P450 enzyme, substantially decreased (Figure 2A). Consistent with this result, the Spok protein level also substantially decreased in the PG of pabp-RNAi animals as compared to control animals, while the protein level of another ecdysteroidogenic P450 enzyme Phm was only slightly affected (Figures 2B,C).
To determine whether spok expression is transcriptionally and/or translationally disrupted in the pabp-RNAi animals, we conducted a GFP reporter assay in vivo. The spok enhancer region, which is sufficient for spok transcription in the PG, has been identified in our previous study (Komura-Kawa et al., 2015). We found that the spok enhancer-driven GFP level was almost diminished in PG cells of pabp-RNAi animals, suggesting that pabp knockdown disrupts spok transcription (Figures 2D,E).
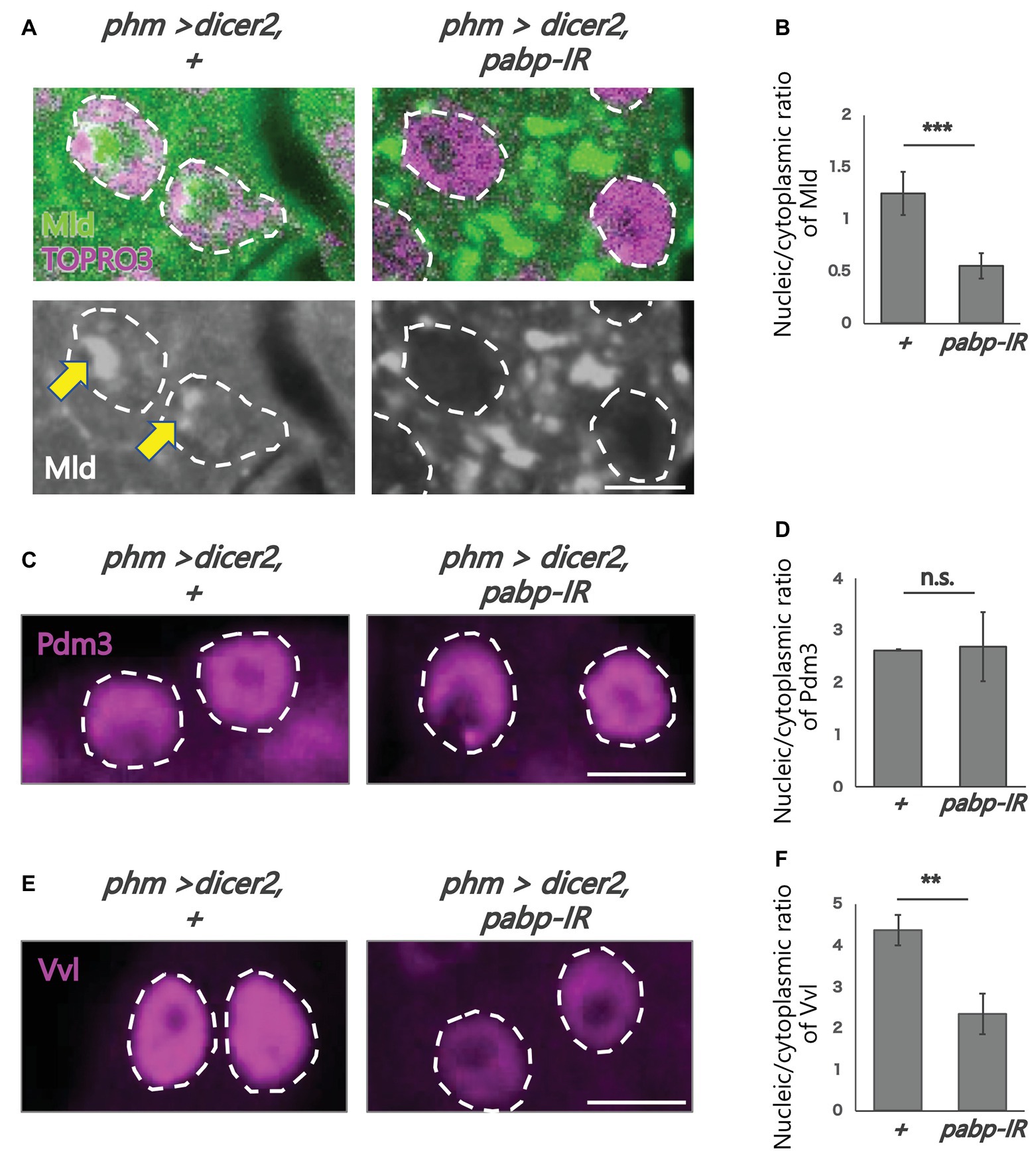
Figure 4. Subcellular localization of transcription factors in the PG cells. All photos are single-plane confocal images. (A,C,E) Immunostaining of the PG cells from phm>dicer2, + and phm>dicer2, pabp-IR second instar larvae at 60 h AEL with the antibodies against Mld (green), POU domain motif 3 (Pdm3; magenta), Ventral vein lacking (Vvl; magenta), respectively. Dashed lines indicate the shape of nuclei. (A) Nuclei are stained by TOPRO3 (magenta). Bottom panels are single channels of Mld (grayscale). Scale bar: 5 μm. (B,D,F) Quantifications of fluorescence intensities of Mld (N = 5), Pdm3 (N = 3), and Vvl (N = 3) in the PG nuclei. Error bars represent standard deviations. ***, ** and n.s. indicate p < 0.001, p < 0.005 and non-significance (p > 0.005), respectively, by Student’s t-test.
To analyze the molecular mechanism of suppression of spok transcription, we focused on the transcription factor Mld. Mld is a zinc-finger type DNA binding protein involved in ecdysone biosynthesis in the PG (Neubueser et al., 2005). Moreover, we have previously reported that Mld is crucial for transcription of spok, acting on the upstream region of the spok gene locus (Danielsen et al., 2014; Komura-Kawa et al., 2015). In this study, we newly-generated an anti-Mld antibody to visualize Mld protein in vivo by immunostaining. Before the immunostaining experiment, we examined the specificity of the newly-generated anti-Mld antibody. We observed that the Mld signal was observed in the nucleus of PG cells in control animals. In contrast, the Mld signal in the PG nucleus disappeared in mld-RNAi animals (Figure 3), confirming that the immunostaining signal in the PG nucleus corresponds to Mld protein localization. We then conducted immunostaining with an anti-Mld antibody against pabp-RNAi PG cells. Surprisingly, we found that nuclear localization of Mld was disrupted in the pabp-RNAi PG cells, although Mld is localized in the nucleus of control PG cells (Figures 4A,B). These results suggest that Pabp regulates the transcription of spok by mediating the nuclear localization of Mld.
Next, we examined whether such mislocalization is selective for Mld, but not for other transcription factors in PG cells. To address this issue, we examined the nuclear localization of two other transcription factors such as Vvl and Pdm3, in the pabp-RNAi PG cells. Vvl is a POU-domain transcription factor involved in the regulation of the transcription of all known ecdysteroidogenic genes in PG cells (Danielsen et al., 2016). Pdm3 is also a POU-domain transcription factor that is enriched in PG cells (Ou et al., 2016), whereas, its role in PG cells has not yet been elucidated. An immunohistological analysis using anti-Vvl and anti-Pdm3 antibodies revealed that nuclear localization of Vvl and Pdm3 was maintained in the nucleus, even in pabp-RNAi PG cells. Nevertheless, the Vvl signal did slightly decrease (Figures 4C–F). Taken together, these results suggest that Pabp regulates the nuclear localization specifically of Mld.
In this study, we revealed that Pabp is required for the nuclear localization of the ecdysteroidogenic transcription factor Mld. First, the PG-specific knockdown of pabp reduced ecdysteroidogenic gene expression, especially that of spok. Second, that reduction of spok expression correlated well with the mislocalization of its transcription factor Mld. Third, mislocalization did not occur for all transcription factors but did occur specifically for Mld. In conjunction with our previous data showing that Mld is crucial for inducing spok expression through the Mld-response element in spok promoter region (Uryu et al., 2018), we propose that Pabp positively regulates spok expression via mediating nuclear localization of Mld in PG cells (Figure 5).
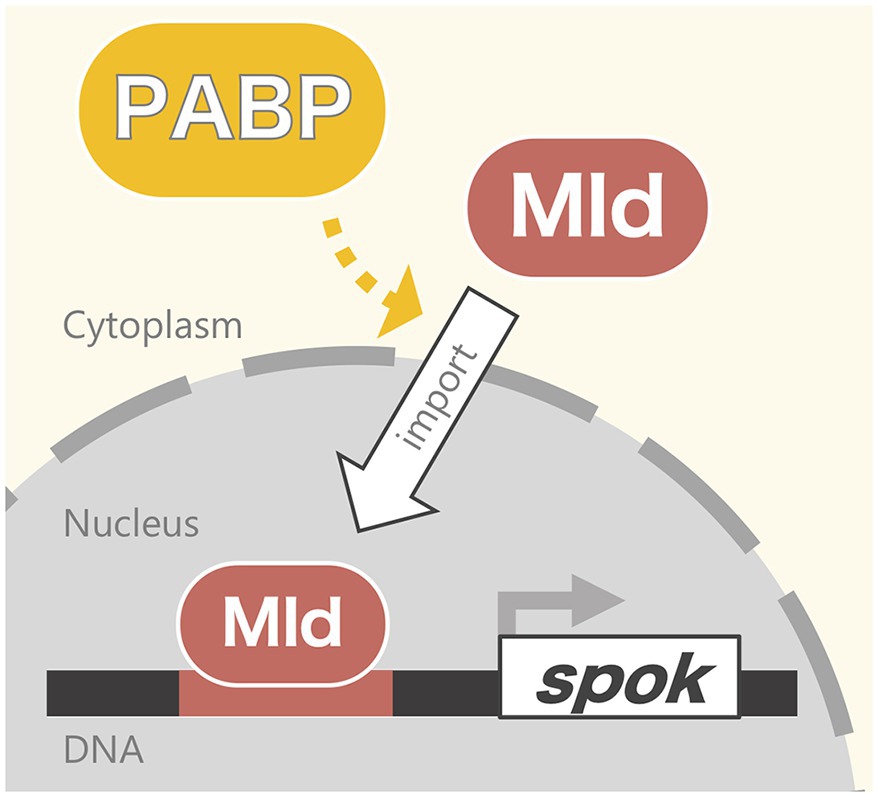
Figure 5. Model of the Pabp function contributing nuclear localization of Mld for spok transcription.
The predicted ORF of mld encodes a protein that belongs to the family of the zinc-finger associated domain (ZAD) containing C2H2 zinc-finger proteins (ZFPs; Neubueser et al., 2005). A previous study has shown that modification or depletion of ZAD disrupts nuclear localization of ZAD-ZFPs (Zolotarev et al., 2016). In conjunction with our observation, this fact raises the possibility that ZAD may be involved in the Pabp-dependent nuclear localization of ZAD-ZFPs, whereas, this hypothesis has yet been experimentally examined. It would also be intriguing to examine whether the subcellular localization of Ouija board (Ouib) and Séance (Séan), other ZAD-ZFP transcription factors of spok and nvd, respectively (Komura-Kawa et al., 2015; Uryu et al., 2018), are also affected by pabp-RNAi. Currently, we failed to generate anti-Ouib and anti-Séan specific antibodies.
This is the first report showing a novel function of Pabp in controlling the nuclear localization of a transcription factor for ecdysone biosynthesis. In the last decade, several transcription factors for ecdysone biosynthesis have been identified (Niwa and Niwa, 2016), while the regulation of subcellular localization of these transcription factors has not been rigorously studied; except DHR4 (Ou et al., 2011). Pabp and other poly(A)-related proteins might be critical research targets for understanding the nuclear and cytoplasmic translocation of ecdysteroidogenic transcription factors in the future.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Study conception: TK, WS, and RN. Investigation (performing of the experiments): TK, NT, and RN. Investigation (data analysis): TK, WS, NT, AN, and RN. Writing/manuscript preparation (writing the initial draft): TK and RN. Writing/manuscript preparation (critical review, commentary, or revision): TK, WS, NT, AN, and RN. Funding acquisition: AN and RN. All authors contributed to the article and approved the submitted version.
This work was supported by a grant from AMED-CREST, AMED to RN (19gm1110001h0003) and the program of the Joint Usage/Research Center for Developmental Medicine, Institute of Molecular Embryology and Genetics, Kumamoto University to RN. We received for an open access publication fee from our institution.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Cheng-Ting Chien, Michael B. O’Connor, Makoto Sato, Bloomington Stock Center, KYOTO Stock Center (DGRC), the National Institute of Genetics, the Vienna Drosophila RNAi Center, and the Drosophila Genomics Resource Center for stocks and reagents. We also thank Outa Uryu and Reiko Kise for their technical assistance.
Anderson, M. G., Perkins, G. L., Chittick, P., Shrigley, R. J., and Johnson, W. A. (1995). Drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline glia. Genes Dev. 9, 123–137. doi: 10.1101/GAD.9.1.123
Bellés, X. (2020). Insect metamorphosis: From natural history to regulation of development and evolution. 1st Edn. Cambridge, MA, USA: Academic Press.
Chanut-delalande, H., Hashimoto, Y., Pelissier-monier, A., Spokony, R., Dib, A., Kondo, T., et al. (2014). Pri peptides are mediators of ecdysone for the temporal control of development. Nat. Cell Biol. 16, 1035–1044. doi: 10.1038/ncb3052
Chávez, V. M., Marqués, G., Delbecque, J. P., Kobayashi, K., Hollingsworth, M., Burr, J., et al. (2000). The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 127, 4115–4126.
Chen, C. K., Chen, W. Y., and Chien, C. T. (2012). The POU-domain protein Pdm3 regulates axonal targeting of R neurons in the Drosophila ellipsoid body. Dev. Neurobiol. 72, 1422–1432. doi: 10.1002/dneu.22003
Danielsen, E. T., Moeller, M. E., Dorry, E., Komura-Kawa, T., Fujimoto, Y., Troelsen, J. T., et al. (2014). Transcriptional control of steroid biosynthesis genes in the Drosophila prothoracic gland by Ventral veins lacking and Knirps. PLoS Genet. 10:e1004343. doi: 10.1371/journal.pgen.1004343
Enya, S., Ameku, T., Igarashi, F., Iga, M., Kataoka, H., Shinoda, T., et al. (2014). A Halloween gene noppera-bo encodes a glutathione S-transferase essential for ecdysteroid biosynthesis via regulating the behaviour of cholesterol in Drosophila. Sci. Rep. 4:6586. doi: 10.1038/srep06586
Gibbens, Y. Y., Warren, J. T., Gilbert, L. I., and O’Connor, M. B. (2011). Neuroendocrine regulation of Drosophila metamorphosis requires TGFβ/activin signaling. Development 138, 2693–2703. doi: 10.1242/dev.063412
Imura, E., Yoshinari, Y., Shimada-Niwa, Y., and Niwa, R. (2017). Protocols for visualizing steroidogenic organs and their interactive organs with immunostaining in the fruit fly Drosophila melanogaster. J. Vis. Exp. 122:55519. doi: 10.3791/55519
Komura-Kawa, T., Hirota, K., Shimada-Niwa, Y., Yamauchi, R., Shimell, M. J., Shinoda, T., et al. (2015). The Drosophila zinc finger transcription factor Ouija board controls ecdysteroid biosynthesis through specific regulation of spookier. PLoS Genet. 11:e1005712. doi: 10.1371/journal.pgen.1005712
McBrayer, Z., Ono, H., Shimell, M., Parvy, J. P., Beckstead, R. B., Warren, J. T., et al. (2007). Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 13, 857–871. doi: 10.1016/j.devcel.2007.11.003
Namiki, T., Niwa, R., Sakudoh, T., Shirai, K. I., Takeuchi, H., and Kataoka, H. (2005). Cytochrome P450 CYP307A1/Spook: a regulator for ecdysone synthesis in insects. Biochem. Biophys. Res. Commun. 337, 367–374. doi: 10.1016/j.bbrc.2005.09.043
Neubueser, D., Warren, J. T., Gilbert, L. I., and Cohen, S. M. (2005). molting defective is required for ecdysone biosynthesis. Dev. Biol. 280, 362–372. doi: 10.1016/j.ydbio.2005.01.023
Niwa, R., Matsuda, T., Yoshiyama, T., Namiki, T., Mita, K., Fujimoto, Y., et al. (2004). CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 279, 35942–35949. doi: 10.1074/jbc.M404514200
Niwa, R., Namiki, T., Ito, K., Shimada-Niwa, Y., Kiuchi, M., Kawaoka, S., et al. (2010). Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the “Black Box” of the ecdysteroid biosynthesis pathway. Development 137, 1991–1999. doi: 10.1242/dev.045641
Niwa, R., and Niwa, Y. S. (2014). Enzymes for ecdysteroid biosynthesis: their biological functions in insects and beyond. Biosci. Biotechnol. Biochem. 78, 1283–1292. doi: 10.1080/09168451.2014.942250
Niwa, Y. S., and Niwa, R. (2016). Transcriptional regulation of insect steroid hormone biosynthesis and its role in controlling timing of molting and metamorphosis. Develop. Growth Differ. 58, 94–105. doi: 10.1111/dgd.12248
Ono, H., Rewitz, K. F., Shinoda, T., Itoyama, K., Petryk, A., Rybczynski, R., et al. (2006). Spook and spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 298, 555–570. doi: 10.1016/j.ydbio.2006.07.023
Ou, Q., Magico, A., and King-Jones, K. (2011). Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS Biol. 9:e1001160. doi: 10.1371/journal.pbio.1001160
Ou, Q., Zeng, J., Yamanaka, N., Brakken-Thal, C., O’Connor, M. B., and King-Jones, K. (2016). The insect prothoracic gland as a model for steroid hormone biosynthesis and regulation. Cell Rep. 16, 1–16. doi: 10.1016/j.celrep.2016.05.053
Parvy, J. P., Blais, C., Bernard, F., Warren, J. T., Petryk, A., Gilbert, L. I., et al. (2005). A role for βFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster. Dev. Biol. 282, 84–94. doi: 10.1016/j.ydbio.2005.02.028
Rewitz, K. F., O’Connor, M. B., and Gilbert, L. I. (2007). Molecular evolution of the insect Halloween family of cytochrome P450s: phylogeny, gene organization and functional conservation. Insect Biochem. Mol. Biol. 37, 741–753. doi: 10.1016/j.ibmb.2007.02.012
Riddiford, L. M. (1993). “Hormones and Drosophila development” in The development of Drosophila melanogaster. eds. M. Bate and A. Martinez-Arias (Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press), 899–939.
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to imageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Uryu, O., Ameku, T., and Niwa, R. (2015). Recent progress in understanding the role of ecdysteroids in adult insects: germline development and circadian clock in the fruit fly Drosophila melanogaster. Zoological Lett. 1:32. doi: 10.1186/s40851-015-0031-2
Uryu, O., Ou, Q., Komura-Kawa, T., Kamiyama, T., Iga, M., Syrzycka, M., et al. (2018). Cooperative control of ecdysone biosynthesis in Drosophila by transcription factors Séance, Ouija board, and Molting defective. Genetics 208, 605–622. doi: 10.1534/genetics.117.300268
Warren, J. T., Petryk, A., Marque, G., Jarcho, M., Parvy, J. P., Dauphin-villemant, C., et al. (2002). Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 99, 11043–11048. doi: 10.1073/pnas.162375799
Warren, J. T., Petryk, A., Marqués, G., Parvy, J. P., Shinoda, T., Itoyama, K., et al. (2004). Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol. 34, 991–1010. doi: 10.1016/j.ibmb.2004.06.009
Yamanaka, N., Romero, N. M., Martin, F. A., Rewitz, K. F., Sun, M., O’Connor, M. B., et al. (2013). Neuroendocrine control of Drosophila larval light preference. Science 341, 1113–1116. doi: 10.1126/science.1241210
Yoshiyama, T., Namiki, T., Mita, K., Kataoka, H., and Niwa, R. (2006). Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133, 2565–2574. doi: 10.1242/dev.02428
Zeng, J., Kamiyama, T., Niwa, R., and King-Jones, K. (2018). The Drosophila CCR4-NOT complex is required for cholesterol homeostasis and steroid hormone synthesis. Dev. Biol. 443, 10–18. doi: 10.1016/j.ydbio.2018.08.012
Keywords: poly(A) binding protein, nuclear localization, insect development, ecdysone biosynthesis, Halloween gene
Citation: Kamiyama T, Sun W, Tani N, Nakamura A and Niwa R (2020) Poly(A) Binding Protein Is Required for Nuclear Localization of the Ecdysteroidogenic Transcription Factor Molting Defective in the Prothoracic Gland of Drosophila melanogaster. Front. Genet. 11:636. doi: 10.3389/fgene.2020.00636
Received: 19 February 2020; Accepted: 26 May 2020;
Published: 26 June 2020.
Edited by:
Wei Guo, Institute of Zoology (CAS), ChinaReviewed by:
Xavier Belles, Instituto de Biología Evolutiva (IBE), SpainCopyright © 2020 Kamiyama, Sun, Tani, Nakamura and Niwa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryusuke Niwa, cnl1c3VrZS1uaXdhQHRhcmEudHN1a3ViYS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.