- 1School of Food and Biological Engineering, Jiangsu University, Zhenjiang, China
- 2National Engineering Laboratory for Crop Molecular Breeding, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 3Crop Research Institution, Shandong Academy of Agricultural Sciences, Jinan, China
- 4Yangzhou Academy of Agricultural Sciences, Yangzhou, China
- 5School of Environment, Jiangsu University, Zhenjiang, China
Wheat powdery mildew caused by Blumeria graminis f. sp. tritici (Bgt) is a devastating disease that threatens wheat production and yield worldwide. The powdery mildew resistance gene Pm21, originating from wheat wild relative Dasypyrum villosum, encodes a coiled-coil, nucleotide-binding site, leucine-rich repeat (CC-NBS-LRR) protein and confers broad-spectrum resistance to wheat powdery mildew. In the present study, we isolated 73 Pm21 alleles from different powdery mildew-resistant D. villosum accessions, among which, 38 alleles were non-redundant. Sequence analysis identified seven minor insertion-deletion (InDel) polymorphisms and 400 single nucleotide polymorphisms (SNPs) among the 38 non-redundant Pm21 alleles. The nucleotide diversity of the LRR domain was significantly higher than those of the CC and NB-ARC domains. Further evolutionary analysis indicated that the solvent-exposed LRR residues of Pm21 alleles had undergone diversifying selection (dN/dS = 3.19734). In addition, eight LRR motifs and four amino acid sites in the LRR domain were also experienced positive selection, indicating that these motifs and sites play critical roles in resistance specificity. The phylogenetic tree showed that 38 Pm21 alleles were divided into seven classes. Classes A (including original Pm21), B and C were the major classes, including 26 alleles (68.4%). We also identified three non-functional Pm21 alleles from four susceptible homozygous D. villosum lines (DvSus-1 to DvSus-4) and two susceptible wheat-D. villosum chromosome addition lines (DA6V#1 and DA6V#3). The genetic variations of non-functional Pm21 alleles involved point mutation, deletion and insertion, respectively. The results also showed that the non-functional Pm21 alleles in the two chromosome addition lines both came from the susceptible donors of D. villosum. This study gives a new insight into the evolutionary characteristics of Pm21 alleles and discusses how to sustainably utilize Pm21 in wheat production. This study also reveals the sequence variants and origins of non-functional Pm21 alleles in D. villosum populations.
Introduction
Dasypyrum villosum L. Candargy (2n = 2x = 14, VV), a diploid species native to the Mediterranean region, is an important wild resource for the improvement of common wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD). D. villosum possesses good resistance to multiple wheat diseases, such as wheat spindle streak mosaic disease, eyespot, take-all, stem rust, stripe rust, and powdery mildew (Li and Zhu, 1999; De Pace et al., 2011; Wang et al., 2017). Four powdery mildew resistance (Pm) genes, Pm21 (Chen et al., 1995), PmV (Li et al., 2005), Pm55 (Zhang et al., 2016), and Pm62 (Zhang et al., 2018), have been found in D. villosum. Among them, both Pm21 and PmV are located on the short arm of chromosome 6V (6VS) and confer immunity to powdery mildew at the whole growth stages of wheat. Pm55 and Pm62 are mapped to the short arm of chromosome 5V (5VS) and the long arm of chromosome 2V (2VL), respectively, which provide powdery mildew resistance at the adult-plant stage.
Pm21 was originally transferred from an accession of D. villosum, collected from Cambridge Botanical Garden, United Kingdom, to durum wheat (T. turgidum var. durum L.), and then a translocation line of wheat-D. villosum T6AL·6VS carrying Pm21 was further developed (Chen et al., 1995). Using this translocation line as the powdery mildew resistance source, more than 20 varieties have been developed and released in the middle and lower reaches of the Yangtze River Valley and the southwest wheat-producing area, the most rampant areas of powdery mildew in China, where some Pm genes, such as Pm2a and Pm4a, are gradually losing their resistance (Bie et al., 2015a).
Undoubtedly, Pm21 is a very valuable gene that confers highly effective resistance to tested isolates of Blumeria graminis f. sp. tritici (Bgt). However, no recombination occurs between the alien chromosome arm 6VS carrying Pm21 and the wheat homoeologous chromosome arms, which limits the genetic mapping and the cloning of Pm21 in the wheat backgrounds (Zhu et al., 2018). Recently, four seedling-susceptible D. villosum lines were identified from the natural populations. Based on the fine genetic map constructed, the gene Pm21 was cloned and confirmed to encode a single coiled-coil, nucleotide-binding site, leucine-rich repeat (CC-NBS-LRR) protein (He et al., 2017, 2018).
In the present study, we isolated the Pm21 alleles from different resistant D. villosum accessions and determined their genetic diversity, non-synonymous and synonymous substitution rates and positive selection sites. On the other hand, D. villosum germplasms susceptible to powdery mildew are rare, and only four susceptible D. villosum lines (DvSus-1 to DvSus-4) and two wheat-D. villosum chromosome 6V disomic addition lines (DA6V#1 and DA6V#3) were identified (Qi et al., 1998; Liu et al., 2011; He et al., 2017). Understanding the reason that these D. villosum germplasms keep or lose their resistance to powdery mildew will be useful to extend the effective duration of Pm21 in agriculture. We also detected the sequence variations of Pm21 alleles in the above germplasms for tracing their origins in natural population of D. villosum.
Materials and Methods
Plant Materials
Dasypyrum villosum accessions were gifted from Germplasm Resources Information Network (GRIN), GRIN Czech, Genebank Information System of the IPK Gatersleben (GBIS-IPK), and Nordic Genetic Resource Center (NordGen). The wheat-D. villosum chromosome 6V disomic addition lines DA6V#1 and DA6V#3 were provided by GRIN and Dr. Bernd Friebe (Kansas State University, Manhattan, KS, USA), respectively (Table S1). The D. villosum line DvRes-1 carries the original Pm21 gene. DvRes-2 and DvRes-3 were derived from the powdery mildew resistant individuals of the accessions GRA961 and GRA1114, respectively. Lines DvSus-1 to DvSus-4 were derived from the susceptible individuals of the accessions GRA2738, GRA962, GRA1105, and PI 598390, respectively. The wheat variety (cv.) Yangmai 18 was a wheat-D. villosum translocation line that carries Pm21. The wheat cv. Yangmai 9 was susceptible to powdery mildew. Both of them were developed in Yangzhou Academy of Agricultural Sciences, Yangzhou, China. Plants were grown under a daily cycle of 16 h of light and 8 h of darkness at 24°C in a greenhouse.
Evaluation of Powdery Mildew Resistance
Blumeria graminis f. sp. tritici (Bgt) isolate YZ01 is a virulent isolate collected from Yangzhou region (Jiangsu Province, China). All plants, D. villosum accessions or lines and wheat varieties, were inoculated with Bgt isolate YZ01 at one-leaf stage (He et al., 2016). The powdery mildew responses of plants were evaluated at 8 d after inoculation.
Allelic Test
The susceptible homozygous D. villosum line DvSus-1 was used as female parent to cross with other susceptible lines, DvSus-2, DvSus-3, and DvSus-4, to produce three F1 hybrids, DvSus-1/DvSus-2, DvSus-1/DvSus-3, and DvSus-1/DvSus-4, respectively. The wheat-D. villosum chromosome 6V disomic addition line DA6V#1 susceptible to powdery mildew was crossed with another susceptible chromosome 6V addition line DA6V#3 to result in F1 hybrid DA6V#1/DA6V#3. All F1 plants derived from different crosses were inoculated with Bgt isolate YZ01 at one-leaf stage for investigation of their responses.
DNA Isolation and Molecular Analysis of Pm21 Alleles
Genomic DNA was extracted from leaves of one-leaf-stage plants by the TE-boiling method (He et al., 2017). The marker MBH1, developed from the promoter region of Pm21 gene (Bie et al., 2015b), was used to detect genetic diversity of different D. villosum individuals. PCR amplification was carried out according to our previous description (He et al., 2017). PCR products with different sizes were T/A-cloned and sequenced.
Isolation of Pm21 Alleles
Total RNA of different D. villosum accessions/lines and wheat materials was extracted from seedlings leaves using the TRIzol solution (Life Technologies, Carlsbad, California, USA). About 2 μg of total RNA was used for synthesis of cDNA using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Shiga, Japan) according to the manufacturer's guidelines. Pm21 alleles were isolated from the cDNAs by PCR using the high fidelity PrimeSTAR Max Premix (TaKaRa, Shiga, Japan) and the primer pair (forward primer: 5′-TTACCCGGGCTCACCCGTTGGACTTGGACT-3′; reverse primer: 5′-CCCACTAGTCTCTCTTCGTTACATAATGTAGTGCCT-3′). PCR products were digested with SmaI and SpeI, inserted into pAHC25-MCS1 and sequenced. The genomic DNA of the alleles in the susceptible materials, DvSus-1 to DvSus-4, DA6V#1, and DA6V#3, were also isolated using PCR with LA Taq DNA polymerase (TaKaRa, Shiga, Japan) and the above primer pair. Each Pm21 allele was amplified from its donor material by three independent PCR, followed by cloning and Sanger sequencing.
Sequence Data Analysis
Multiple alignment analysis was carried out using the CLUSTAL W tool (Thompson et al., 1994). Nucleotide diversity of Pm21 alleles and their coding sequences of different domains or non-domain regions was analyzed using the MEGA7 software (Kumar et al., 2016) and assessed by Tajima's test of neutrality (Tajima, 1989). π meant the average number of nucleotide differences per site between two sequences. θ represented Watterson's nucleotide diversity estimator based on the value of π. Synonymous substitution rate (dS), non-synonymous substitution rate (dN), and natural selection for each codon were estimated by the HyPhy program in the MEGA7 software. Sequence logos of LRR motifs were created by the WebLogo tool (Crooks et al., 2004). For evolutionary analyses, all positions containing gaps were eliminated. So, there were a total of 2,718 positions in the final dataset. A phylogenetic tree based on the cDNA sequences of the Pm21 alleles was constructed using the Neighbor-Joining method in the MEGA7 software (Kumar et al., 2016).
Accession Numbers
The accession number of Pm21 gene in the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) is MF370199. The Pm21 alleles obtained have been deposited in the GenBank under the accession numbers MG831524–MG831526, MG831528–MG831561 and MH184801–MH184806.
Results
Powdery Mildew Responses of Different Germplasms
The D. villosum accessions provided by different germplasm resource institutions were collected from the Mediterranean region, mainly from Greece and Italy (Figure 1; Table S1). A total of 62 accessions were used to detect the responses to Bgt isolate YZ01. All plants of the 58 accessions were immune to Bgt isolate YZ01, whereas in each of the other four accessions (GRA2738, GRA962, GRA1105, and PI 598390), several individuals (2–5%) were susceptible despite that most plants were resistant. The four susceptible homozygous lines derived from the above accessions were then designated as DvSus-1 to DvSus-4, respectively. The results also showed that the wheat-D. villosum chromosome 6V disomic addition lines DA6V#1 and DA6V#3 were susceptible to powdery mildew (Figure 2).
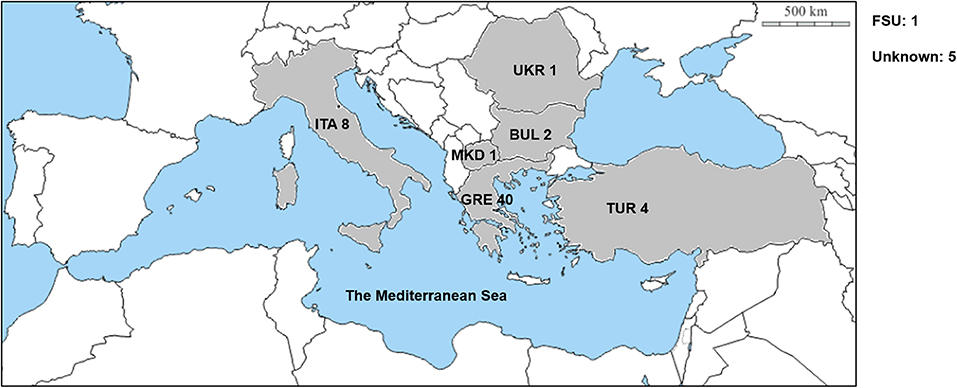
Figure 1. Geographic distribution of the D. villosum accessions used in this study. GRE, Greece. ITA, Italy. TUR, Turkey. BUL, Bulgaria. UKR, Ukraine. MKD, Macedonia. FSU, former Soviet Union. Unknown, the origins of the accessions are unclear.
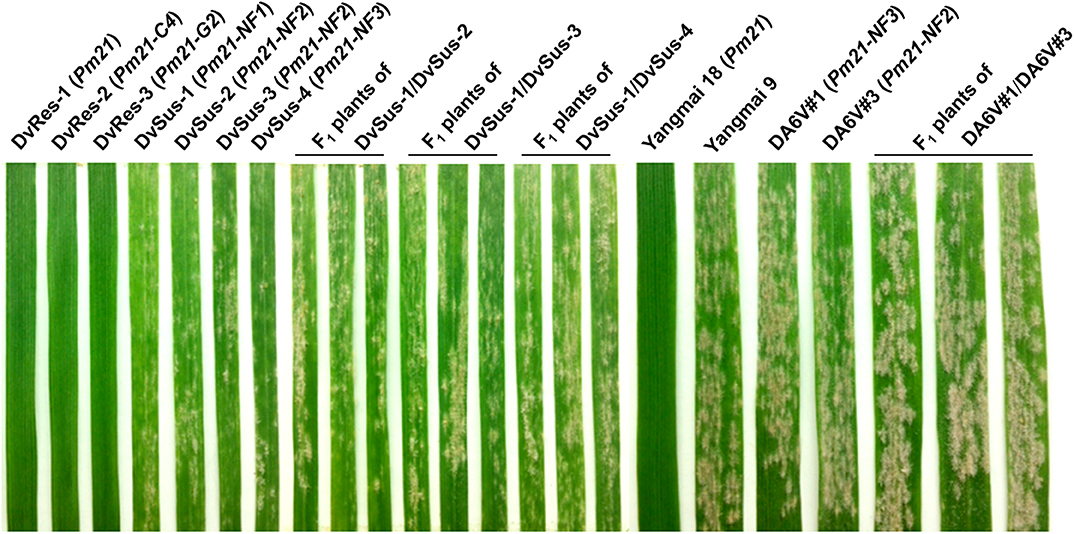
Figure 2. Powdery mildew responses of different D. villosum lines, wheat genetic stocks, and F1 plants of the crosses DvSus-1/DvSus-2, DvSus-1/DvSus-3, DvSus-1/DvSus-4, Dv6V#1/ Dv6V#3. The plants were inoculated with Bgt isolate YZ01 at one-leaf stage. The resistant wheat cv. Yangmai 18 and the susceptible cv. Yangmai 9 were used as the controls. Line DvRes-1 carries Pm21. Lines DvRes-2 and DvRes-3, carrying Pm21-C4 and Pm21-G2, were the resistant individuals of the accessions GRA961 and GRA1114, respectively. Lines DvSus-1 to DvSus-4, carrying non-functional Pm21 genes, were the susceptible individuals in the accessions GRA2738, GRA962, GRA1105, and PI598390, respectively.
The powdery mildew responses of the F1 plants derived from four different crosses, DvSus-1/DvSus-2, DvSus-1/DvSus-3, DvSus-1/DvSus-4, and DA6V#1/DA6V#3, were also assessed. The data showed that all the F1 hybrids displayed high susceptibility to Bgt isolate YZ01 (Figure 2). It was indicated that there was no obvious allelic complementation in any of the above crosses. Therefore, it was suggested that the potential mutation(s), which led to susceptibility of the four D. villosum lines (DvSus-1, DvSus-2, DvSus-3, and DvSus-4) and the two wheat-D. villosum chromosome 6V disomic addition lines (DA6V#1 and DA6V#3), may all occur in the alleles of Pm21.
Molecular and Nucleotide Diversity of the Pm21 Alleles
To understand the diversity at the Pm21 loci, MBH1, designed based on the promoter sequence of Pm21 (Bie et al., 2015b), was used to detect the resistant individuals from 62 different D. villosum accessions. The PCR products were sequenced and eight representative bands with different sizes, 271, 339, 340, 341, 342, 344, 396, and 467 bp, were found. This indicated that insertion-deletion (InDel) polymorphisms exist at the promoter regions of different Pm21 alleles. Given that all MBH1 sequences were isolated from resistant individuals, it was suggested that the variations in the promoter regions have no obviously adverse impact on the expression of Pm21 alleles. In some individuals, two specific DNA bands were observed (Figures S1, S2), suggesting that these individuals might be heterozygous at the Pm21 loci.
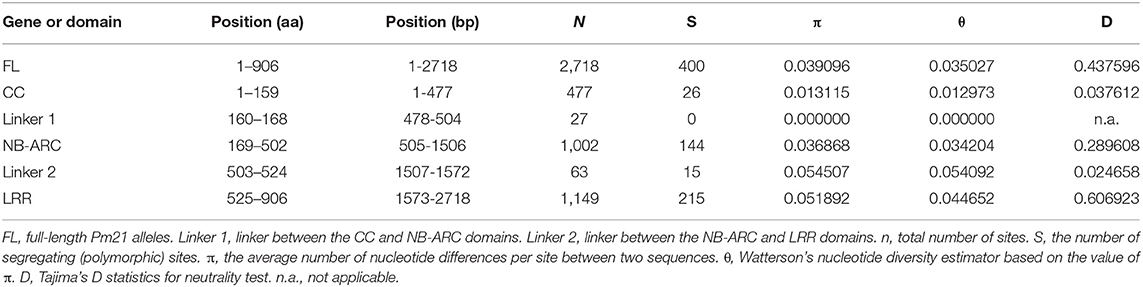
We then isolated Pm21 alleles from the resistant individuals of 62 D. villosum accessions. Each of the individuals of 52 accessions had one copy of Pm21 allele. However, due to open pollination of D. villosum species, each of the tested individuals of 9 accessions (PI 368886, W619414, W67270, GRA960, GRA1109, GRA1114, GRA2711, GRA2716, and 01C2300013) had two copies of Pm21 alleles. In addition, three different alleles were, respectively, isolated from three individuals of the accession PI 251478. As a result, a total of 73 Pm21 alleles were isolated in this study (Table S1). Among them, 38 alleles were non-redundant, sharing 91.7–100% identities with each other. In general, a total of seven InDels (Table S2), including three 3-bp insertions, one 30-bp insertion and three 3-bp deletions, and 400 single nucleotide polymorphism (SNP) sites were identified among these alleles. The 38 non-redundant Pm21 alleles and their coding sequences of different domains were further used to determine the nucleotide diversity. The average pairwise nucleotide diversity π and Watterson's nucleotide diversity estimator θ of the full-length Pm21 alleles were 0.039096 and 0.035027, respectively. Compared with the full-length alleles, the values of π and θ of the NB-ARC domain-encoding sequences were slightly lower (π = 0.036868 and θ = 0.034204), whereas those of the CC domain-encoding sequences were significantly lower (π = 0.013115 and θ = 0.012973) and those of the LRR domain-encoding sequences were obviously higher (π = 0.051892 and θ = 0.044652). These results indicated that the CC domain was more conserved than other domains whereas the LRR domain was more variable. We also analyzed the π and θ values of Linker 1 and Linker 2, the regions between the CC and NB-ARC domains, and between the NB-ARC and LRR domains, respectively. The data showed that Linker 1 had no nucleotide diversity. Contrarily, Linker 2 had the highest nucleotide diversity (π = 0.054507 and θ = 0.054092) in different domains or regions of Pm21 alleles (Figure 3; Table 1). Up to now, the function of Linker 2 is unclear yet. One reasonable explanation for its high variation is that Linker 2 may be an extension of the LRR domain.
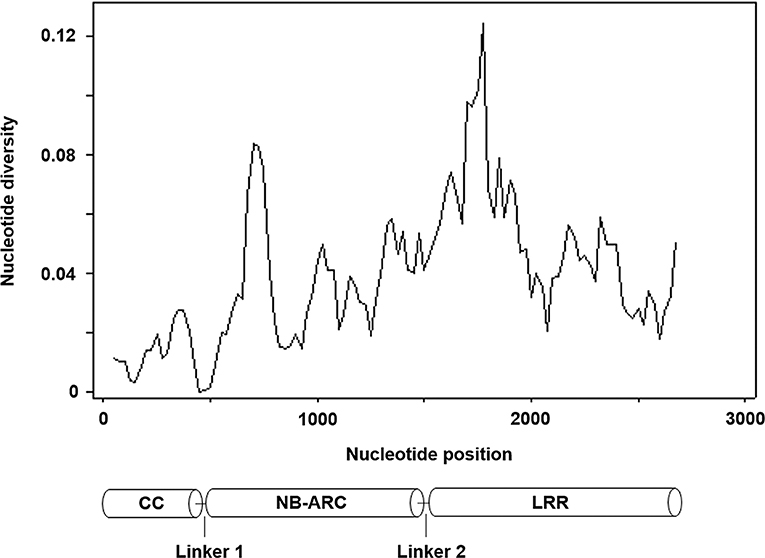
Figure 3. Nucleotide diversity of 38 non-redundant Pm21 alleles isolated from the resistant D. villosum accessions. All the positions containing gaps were eliminated. Therefore, there were a total of 2,718 positions in the final dataset. The predicted protein structure is shown at the bottom.
Selection Pressure Analysis
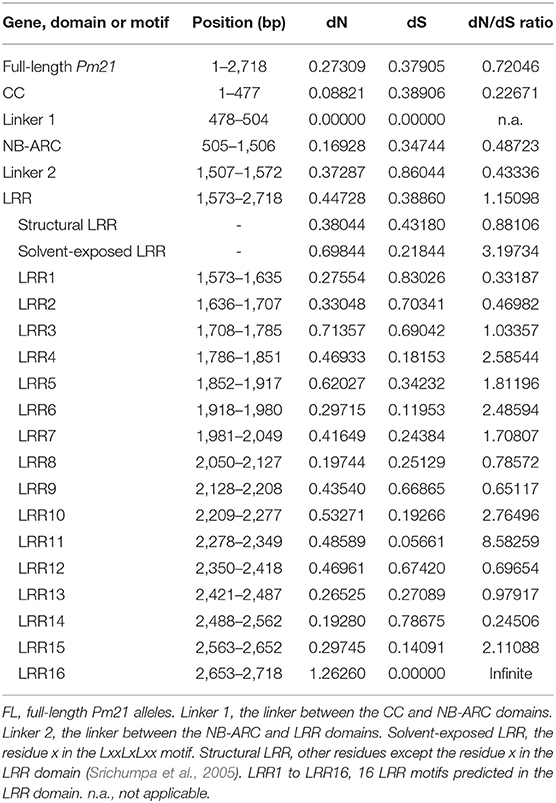
To determine the potential evolutionary selection occurred in Pm21 alleles, dN and dS rates were assessed using the HyPhy program. The dN/dS ratio of full-length Pm21, CC-, NB-ARC-, and LRR-encoding sequences were 0.72046, 0.22671, 0.48723, and 1.15098, respectively, which suggested that the LRR domain might be under positive selection. The dN/dS ratio of the structural LRR residues and the solvent-exposed LRR residues, the two parts of the LRR domain, were 0.88106 and 3.19734, respectively (Table 2). This indicated diversifying selection acting on the solvent-exposed residues in the LRR domain of Pm21 alleles.
The LRR domain of Pm21 consists of 16 LRR motifs. The dN/dS ratios of 8 LRR motifs (LRR4-LRR7, LRR10, LRR11, LRR15, and LRR16) were greater than 1. Among them, the dN/dS ratio of LRR11 was 8.58259 and that of LRR16 was infinite because its dS value was zero (Figure 4; Table 2). These results indicated that the above 8 LRR motifs have undergone positive selection. In the LRR domain, four sites at the positions 628, 885, 903, and 905 were subject to positive selection, detected by four different models (Felsensten 1981 model, Hasegawa-kishino-Yano model, Tamura-Nei model, and General Time Reversible model). Position 628 lied in the LRR5 motif and positions 885, 903, and 905 were all located in the LRR16 motif (Figure 4; Table S3).
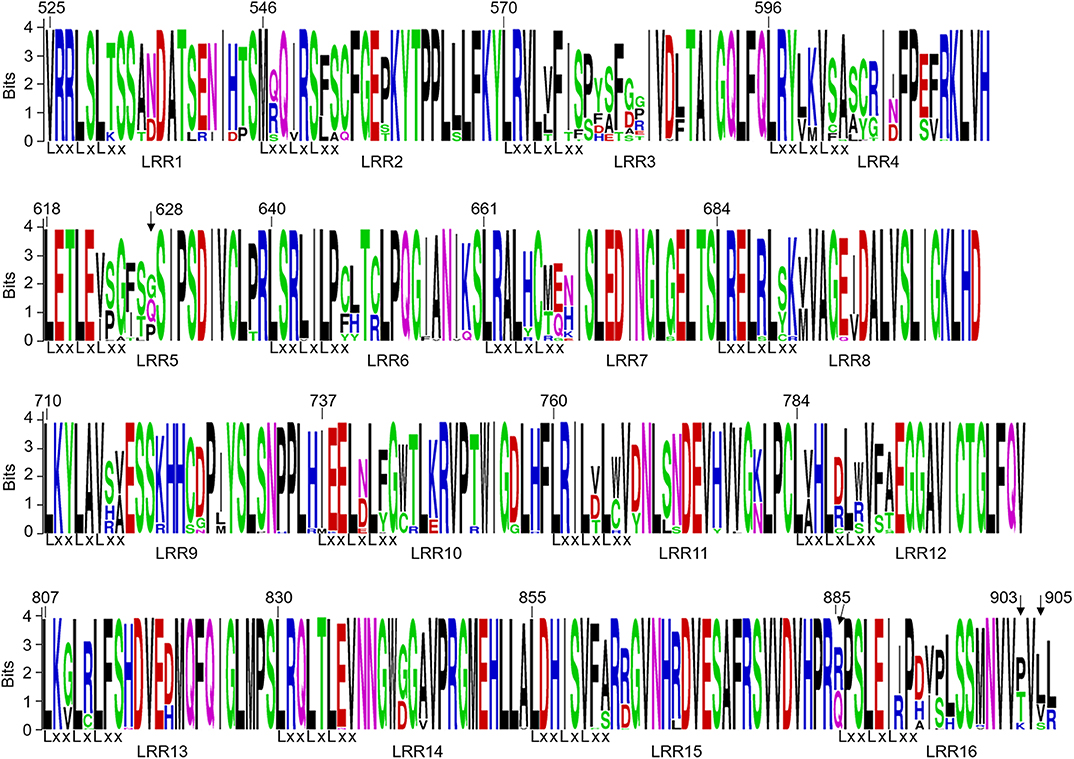
Figure 4. Sequence logos of 16 LRR motifs encoded by Pm21 alleles. In the LxxLxLxx motifs, x represents the predicted solvent-exposed LRR residues, and L represents a leucine or another aliphatic amino acid residue. The sites at positions 628, 885, 903, and 905 pointed by arrows are predicted to be under positive selection.
Phylogenetic Analysis and Classification of the Pm21 Alleles
The phylogenetic tree for Pm21 alleles showed that 38 non-redundant Pm21 alleles were clustered into seven clades (Clade A to G). Among these clades, Clades A, B, and C were the major types in the D. villosum populations, which included 26 members, accounting for 68.4% (Figure 5).
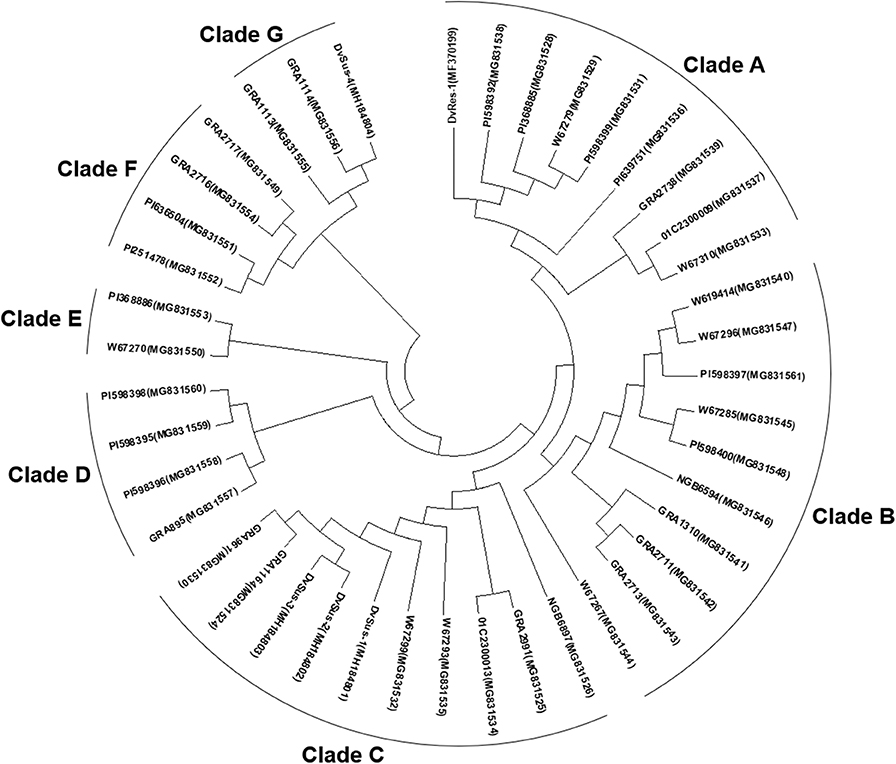
Figure 5. Phylogenetic tree of Pm21 alleles constructed by the Neighbor-Joining method. The GenBank accession numbers are shown in brackets.
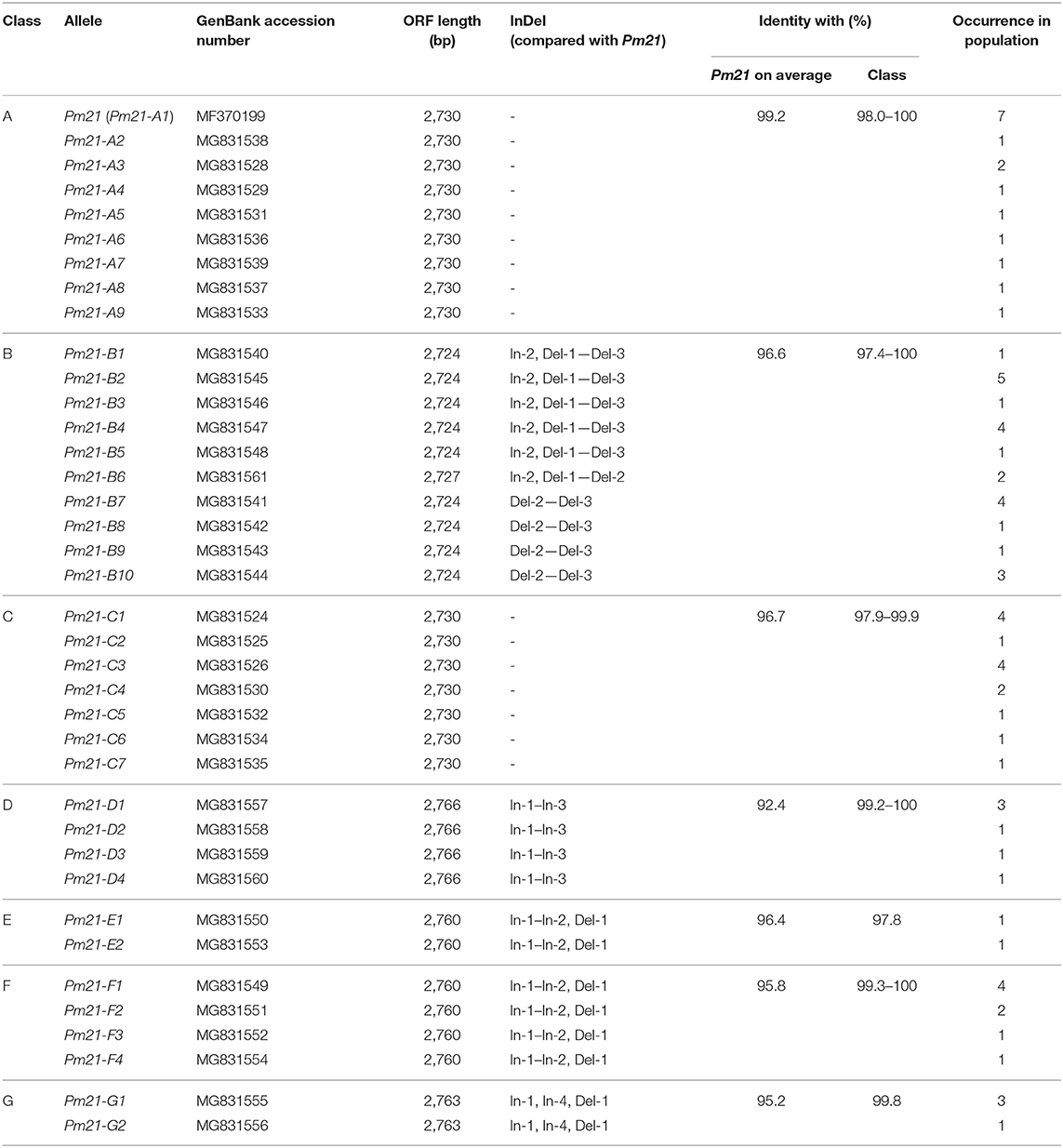
According to the clades categorized in the phylogenetic tree, the Pm21 alleles isolated from the resistant D. villosum accessions were correspondingly divided into seven classes (Class A to G). Class A consisted of 9 alleles, Pm21-A1 to Pm21-A9, whose open reading frames (ORFs) were 2,730 bp in length sharing the highest identities with Pm21 (99.2% on average). Class B contained 10 alleles, Pm21-B1 to Pm21-B10, most of which were 2,724 bp sharing 96.6% identity with Pm21 on average. Class C harbored 7 alleles, Pm21-C1 to Pm21-C7, with 2,730 bp in length and had 96.7% identity with Pm21 on average. The remaining 12 alleles, sharing 92.1–97.0% identities with Pm21, were divided into four classes, Class D to G, whose obvious sequence characteristics was a 30-bp insertion compared with Pm21 (Table 3).
Natural Variations of Pm21 Alleles in Susceptible Germplasms
To test the rare natural variations leading to lose of resistance to powdery mildew, we isolated Pm21 alleles from the susceptible D. villosum lines DvSus-1 to DvSus-4, derived from the accessions GRA2738, GRA962, GRA1105, and PI 598390, respectively. The non-functional allele Pm21-NF1 isolated from the genome of DvSus-1 was 3,699 bp in length, whose ORF was 2,730 bp. Compared with Pm21, Pm21-NF1 had 98 SNPs; however, compared with the 38 non-redundant alleles isolated from the resistant D. villosum accessions, Pm21-NF1 only had two specific variations. The first variation was a transversion G61T leading to the amino acid change A21S in the CC domain. The second variation was a transition A821G resulting in the change D274G (Figure S3A), corresponding to the latter aspartate (D) in kinase-2 motif (also called Walker B motif; consensus sequence: LLVLDDVW) in the NB-ARC domain. The latter D is considered to act as the catalytic site for ATP hydrolysis and activation of disease resistance protein (Meyers et al., 1999; Tameling et al., 2006). Here, bioinformatic analysis showed that the latter D was highly conserved in all the tested disease resistance proteins from Arabidopsis thaliana, barley (Hordeum vulgare L.) and wheat (Figure S4), suggesting that the amino acid change D274G might lead to loss-of-function of Pm21-NF1.
The genomic sequence of the non-functional allele Pm21-NF2 isolated from the susceptible DvSus-2 was 3,698 bp in length, whose ORF contained a 1-bp deletion after position 876, leading to frame shift and resulting in a truncated protein (296 aa). The variations of Pm21 alleles isolated from DvSus-3 and DA6V#3 were both identical to that of Pm21-NF2. In DvSus-4 and DA6V#1, the sequences of the alleles were identical (4,988 bp) and designated as Pm21-NF3. Pm21-NF3 harbored an insertion of 1281 bp that caused a premature stop codon (Figure S3B) and led to loss of the last four LRR motifs. These results suggested that the non-functional Pm21 alleles in DA6V#1 and DA6V#3 both directly originated from their D. villosum donors susceptible to powdery mildew.
Molecular Tracing of the Origins of Non-functional Pm21 Alleles
Phylogenetic analysis showed that DvSus-1, DvSus-2, DvSus-3, GRA961, and GRA1164 were clustered in Clade C (Figure 5). In contrast to the alleles, Pm21-C4 in GRA961 and Pm21-C1 in GRA1164, the non-functional allele Pm21-NF1 in DvSus-1 had 8 and 10 SNPs, and Pm21-NF2 in DvSus2/DvSus-3 had 1 and 3 SNPs, respectively (Figure S3B). This suggested that the non-functional allele Pm21-NF2 originated from the allele Pm21-C4 in the resistant accession GRA961 (Figures 5, 6; Table 3). In the tested accessions, the origin of Pm21-NF1 could not be well-traced yet.
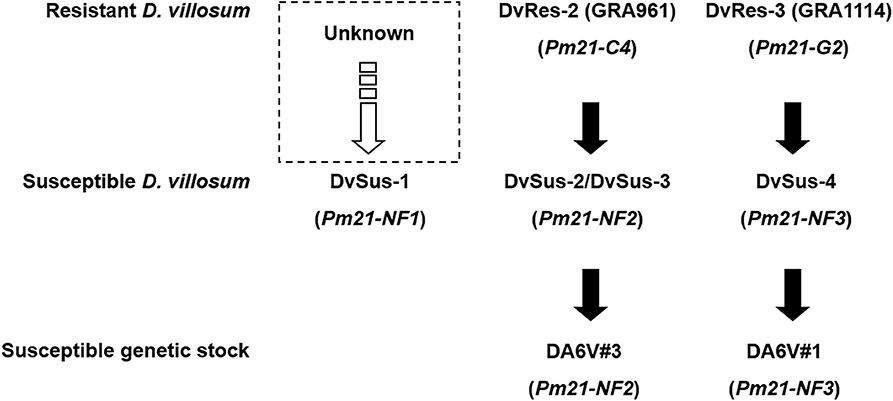
Figure 6. Origins of the non-functional alleles Pm21-NF1 to Pm21-NF3. Among them, the origin of Pm21-NF1 is unclear yet.
The data also indicated that lines DvSus-4, GRA1113, and GRA1114 were clustered in Clade G (Figure 5). Except the 1281-bp insertion, Pm21-NF3 in DvSus-4 had no difference from Pm21-G2 in GRA1114 (Figure S3C). This result revealed that the non-functional allele Pm21-NF3 came from the variation of the allele Pm21-G2 in the resistant accession GRA1114 (Figures 5, 6; Table 3).
Discussion
Diversity, Classification and Geographic Distribution of Pm21 Alleles
As a wild relative of wheat, D. villosum possesses several powdery mildew resistance genes that have important potential for controlling wheat powdery mildew disease (He et al., 2017). Among them, Pm21 and PmV, located on chromosome 6VS derived from different D. villosum accessions, confer powdery mildew resistance at whole-plant growth stages. It seems that Pm21 and PmV may be allelic (Bie et al., 2015b). Both Pm55 and Pm62 confer resistance at adult-plant stage but not at the seedling stage (Zhang et al., 2016, 2018). In this study, Bgt-responses of all D. villosum accessions were detected at one-leaf stage, which could exclude the resistance conferred by Pm55 and Pm62. Therefore, the seedling-resistance in these materials was considered to be provided by Pm21 alleles.
Recently, the broad-spectrum powdery mildew resistance gene Pm21 was isolated from D. villosum using the map-based cloning strategy (He et al., 2018). Based on the investigation of powdery mildew responses of different D. villosum accessions collected from the Mediterranean countries, we isolated 73 Pm21-like sequences from the resistant individuals. The previous work showed that Pm21 is adjacent to another CC-NBS-LRR-encoding gene DvRGA1 (He et al., 2018). Although DvRGA1 is the highest matched gene of Pm21 in Genbank database, they had only 72.7% nucleotide sequence identity. Here, the isolated Pm21-like genes shared 91.7–100% identities with each other, indicating that all the sequences are identical or allelic to Pm21. Of the 73 sequences, 38 were different from each other. Compared with Pm21, the other 37 non-redundant alleles have seven InDels involved in 3-bp, 6-bp, 30-bp, 33-bp, or 36-bp, which make the alleles maintain correct ORFs and encode full-length proteins. The alleles also had many SNPs and the average pairwise nucleotide diversity of the LRR-encoding region was significantly higher than those of the CC- or NB-ARC-encoding regions. Compared with other domains, the LRR domain were supposed to have undergone faster evolution. Because all of the individuals containing these alleles were still effective against the highly virulent Bgt isolate YZ01, it was proposed that the wide variations of Pm21 alleles have no obviously adverse effect on the disease resistance. However, whether they still keep broad-spectrum resistance remains to be disclosed.
Phylogenetic analysis identified seven independent clades that involved all the Pm21 alleles. Among them, Classes A to C represented the three major classes. The functional Pm21 gene was originally found in an accession provided by Cambridge Botanic Garden in the United Kingdom, but the exact collection site of this accession was unclear. Pm21, with the systemic name Pm21-A1 here, belongs to Class A whose members were only found in the accessions of Greece or Turkey. In particular, among the six isolated sequences identical to Pm21, five came from independent Greece accessions and one from a Turkey accession. Therefore, based on the present data, it was proposed that the original D. villosum donor of Pm21 might come from Greece or Turkey.
Geographic distributions of different Pm21 alleles were further investigated in this study. It is indicated that the Pm21 alleles isolated from Greece D. villosum accessions had more genetic diversity and covered the most members of all the seven classes (Class A to G). In addition, Pm21-A8, Pm21-E2, and Pm21-F3 were only detected in Turkey accessions, and Pm21-B7 and Pm21-G2 were only detected in Italy accessions (Table S1). The characteristics of geographic distributions of the Pm21 alleles may help to search the accessions carrying specific Pm21 alleles as donors for future breeding purpose.
Variations and Origins of Non-functional Pm21 Alleles in Susceptible D. villosum Lines and Wheat Genetic Stocks
It has been believed that D. villosum resources are all resistant to wheat powdery mildew (Qi et al., 1998). In our previous work, four D. villosum lines DvSus-1 to DvSus-4 susceptible to powdery mildew were identified from different accessions of D. villosum, which made it possible to clone Pm21 using the map-based cloning strategy (He et al., 2017, 2018). In this study, we demonstrated that the variations of Pm21 alleles, Pm21-NF1 to Pm21-NF3, isolated from the four susceptible D. villosum lines, involved point mutation, deletion and insertion, respectively. Among them, Pm21-NF1 had an important amino acid change (D274G) in the highly conserved kinase-2 motif of the NB-ARC domain that might hamper the function of ATP hydrolysis (Meyers et al., 1999; Tameling et al., 2006), while Pm21-NF2 and Pm21-NF3 both encoded truncated proteins caused by premature stop codons.
Previously, the wheat-D. villosum chromosome 6V disomic addition lines DA6V#1 and DA6V#3 were reported to be highly susceptible to powdery mildew (Qi et al., 1998; Liu et al., 2011). During the creation of the two addition lines, colchicine was used for chromosome doubling, which is proved to be an effective mutagen in fact (Gilbert and Patterson, 1965). So, researchers did not know if the susceptibilities of DA6V#1 and DA6V#3 came from colchicine treatment or the D. villosum donors. Since Pm21 has been cloned, through sequencing of allele genes here, we demonstrated that Pm21 alleles isolated from DA6V#1 and DA6V#3 had identical variations to Pm21-NF3 (DvSus-4) and Pm21-NF2 (DvSus-2 and DvSus-3), respectively. Therefore, it was suggested that the variations of the Pm21 alleles from DA6V#1 and DA6V#3 both originated from their D. villosum donors, rather than colchicine treatment.
The non-functional alleles, Pm21-NF1, Pm21-NF2, and Pm21-NF3, were found in the accessions GRA2738, GRA962, PI 598390, respectively. In theory, their wild-type genes could be isolated from the corresponding accessions above. We tried to do so but not succeeded. The major reason may be that D. villosum is highly outcrossing which causes that the pollen with a mutated gene is subject to separate from the one carrying a corresponding wild-type gene. Therefore, we attempted to trace the origins of the non-functional alleles through evolutionary analysis. The origins of the two non-functional alleles, Pm21-NF2 and Pm21-NF3, were both traceable in the natural populations of D. villosum. Except the identified mutations, the sequences of Pm21-NF2 and Pm21-NF3 were entirely identical to those of Pm21-C4 and Pm21-G2 that were cloned from the resistant individuals of the accessions GRA961 and GRA1114, respectively. Hence, we concluded that the non-functional alleles Pm21-NF2 and Pm21-NF3 originated from Pm21-C4 and Pm21-G2, respectively. However, the origin of Pm21-NF1 remains unclear yet.
Diversifying Selection Acting on the Solvent-Exposed LRR Residues of Pm21 Alleles
It was confirmed that the broad-spectrum resistance of Pm21 is conferred by a single CC-NBS-LRR-encoding gene (He et al., 2018). However, it is believed that the resistance provided by such kind of genes is most likely race-specific, which is prone to be overcome by fast-evolving pathogens. For instance, Pm8 from rye (Secale cereale L.), also encoding a CC-NBS-LRR protein, previously provided effective resistance to wheat powdery mildew (Hurni et al., 2013), has lost its resistance in most wheat producing regions with the worldwide utilization. In this study, the value of dN/dS (3.19734) significantly exceeded 1 in the solvent-exposed LRR residues, which is considered to take part in the specific recognition of pathogens (Meyers et al., 1999). This result suggested that the solvent-exposed LRR residues of Pm21 have been undergone diversifying selection and may play critical roles in resistance specificity. This situation is similar to those of race-specific powdery mildew resistance gene Pm3 from wheat (Srichumpa et al., 2005) and Mla from barley (Seeholzer et al., 2010). In several works, the researchers reported that the wheat varieties carrying Pm21 could be infected by Bgt pathogens in different regions (Shi et al., 2009; Yang et al., 2009). Therefore, combined the data given by evolutionary analysis, it is speculated that Pm21 may be a race-specific resistance gene although it still provides broad-spectrum resistance to the most Bgt isolates so far.
Since 1995 when the translocation line of wheat-D. villosum T6AL.6VS was released, many wheat varieties carrying Pm21 have been commercialized in China, mainly in the middle and lower reaches of the Yangtze River Valley and the southwest wheat-producing regions, where Bgt pathogen is prevailing (Jiang et al., 2014; Bie et al., 2015a; Cheng et al., 2020). The long-time and wide-range application of Pm21 in agriculture would accelerate the evolution of Bgt pathogens. Correspondingly, Pm21 would face to an increasing risk of losing its resistance to powdery mildew. Consequently, it will be a great challenge to sustainably utilize the Pm21 resistance in the future. In this study, a total of 38 non-redundant Pm21 alleles were obtained, which allows to comparatively analyze their fine functions against Bgt pathogens in further researches. Utilization of different Pm21 alleles with functional diversity would be a way to extend the lifespan of Pm21 resistance in wheat production. The marker MBH1, which can reveal genetic diversity of Pm21 alleles in some degree, will be a useful tool when transferring them from D. villosum into common wheat. Other reasonable means would be diversifying use of Pm genes in field, such as pyramiding other effective Pm gene(s) into Pm21-carrying varieties or exploring new Pm genes and developing wheat varieties carrying different Pm genes.
Data Availability Statement
The datasets generated for this study can be found in the GenBank, MG831524–MG831526, MG831528–MG831561, MH184801–MH184806.
Author Contributions
HH, CL, and SZ conceived and designed the experiments. HH, JJ, JT, YF, XW, RH, and TB performed the experiments. HH, JJ, and HL analyzed the data and wrote the paper.
Funding
This study was supported by grants from Shandong Agricultural Seed Improvement Project (2019LZGC016), National Natural Science Foundation of China (31971874), Jiangsu Agricultural Science and Technology Innovation Fund [CX(19)2042], Priority Academic Program Development of Jiangsu Higher Education Institutions, Jiangsu Education Department and Taishan Scholars Project (tsqn201812123).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been released as a pre-print at bioRxiv (Zhu and He, 2018). The authors are thankful to Germplasm Resources Information Network (GRIN), GRIN Czech, and Genebank Information System of the IPK Gatersleben (GBIS-IPK), Nordic Genetic Resource Center (NordGen) and Dr. Bernd Friebe (Kansas State University) for providing the Dasypyrum villosum accessions and wheat genetic stocks.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00489/full#supplementary-material
Figure S1. Molecular analysis of the diversity of D. villosum by the marker MBH1 that was developed from the promoter region of Pm21. M, DNA marker DL2000. Line 1 to 24, PCR products obtained from resistant individuals of different D. villosum accessions.
Figure S2. Multiple sequence alignment of different representative products PCR-amplified with the marker MBH1.
Figure S3. Detection of mutations in the non-functional Pm21 alleles. (A) Mutations of Pm21-NF1 in DvSus-1 contrasted to Pm21. (B) Mutations of Pm21-NF2 in DvSus-2, DvSus-3, and DA6V#3 contrasted to Pm21-C4 in DvRes-2 (derived from GRA961). (C) Mutations of Pm21-NF3 in DvSus-4 and DA6V#1 in contrast to Pm21-G2 in DvRes-3 (derived from GRA1114). SNPs, tandem premature stop codons and insertion sequences are shown by arrows, underlines and brackets, respectively.
Figure S4. Multiple sequence alignment of the surrounding sequences of kinase-2 motif (consensus sequence: LLVLDDVW) of plant disease resistance proteins. The conserved second aspartate (D) of kinase-2 motif is marked by an arrow.
Table S1. Pm21 alleles and the corresponding germplasms.
Table S2. InDel polymorphisms in the Pm21 alleles. All InDels, compared with Pm21, occur after the positions showed in the brackets.
Table S3. Amino acid sites under positive selection.
References
Bie, T., Zhao, R., Jiang, Z., Gao, D., Zhang, B., and He, H. (2015a). Efficient marker-assisted screening of structural changes involving Haynaldia villosa chromosome 6V using a double-distal-marker strategy. Mol. Breeding 35:34. doi: 10.1007/s11032-015-0211-y
Bie, T., Zhao, R., Zhu, S., Chen, S., Cen, B., Zhang, B., et al. (2015b). Development and characterization of an efficient breeding-practical marker MBH1 simultaneously tagging Pm21 and PmV genes conferring resistance to wheat powdery mildew. Mol. Breeding 35:189. doi: 10.1007/s11032-015-0385-3
Chen, P. D., Qi, L. L., Zhou, B., Zhang, S. Z., and Liu, D. J. (1995). Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 91, 1125–1128. doi: 10.1007/BF00223930
Cheng, B., Ding, Y., Gao, X., Cao, N., Xin, Z., and Zhang, L. (2020). The diversity of powdery mildew resistance gene loci among wheat germplasm in Southwest China. Cereal Res. Commun. 48, 65–70. doi: 10.1007/s42976-020-00015-2
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
De Pace, C., Vaccino, P., Cionini, P. G., Pasquini, M., Bizzarri, M., and Qualset, C. O. (2011). “Dasypyrum,” in Wild Crop Relatives: Genomic And Breeding Resources, Cereals ed C. Kole (Berlin: Springer), 185–292. doi: 10.1007/978-3-642-14228-4_4
Gilbert, S. K., and Patterson, F. L. (1965). Colchicine-induced mutants in Decatur barley. Crop Sci. 5, 44–47. doi: 10.2135/cropsci1965.0011183X000500010014x
He, H., Ji, Y., Zhu, S., Li, B., Zhao, R., Jiang, Z., et al. (2017). Genetic, physical and comparative mapping of the powdery mildew resistance gene Pm21 originating from Dasypyrum villosum. Front. Plant Sci. 8:1914. doi: 10.3389/fpls.2017.01914
He, H., Zhu, S., Jiang, Z., Ji, Y., Wang, F., Zhao, R., et al. (2016). Comparative mapping of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor. Appl. Genet. 129, 819–829. doi: 10.1007/s00122-016-2668-4
He, H., Zhu, S., Zhao, R., Jiang, Z., Ji, Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882. doi: 10.1016/j.molp.2018.03.004
Hurni, S., Brunner, S., Buchmann, G., Herren, G., Jordan, T., Krukowski, P., et al. (2013). Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–969. doi: 10.1111/tpj.12345
Jiang, Z., Wang, Q., Wu, J., Xue, W., Zeng, Q., Huang, L., et al. (2014). Distribution of powdery mildew resistance gene Pm21 in Chinese winter wheat cultivars and breeding lines based on gene-specific marker. Scientia Agric. Sin. 47, 2078–2087. (in Chinese with English abstract)
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, H., Chen, X., Xin, Z., Ma, Y., Xu, H., Chen, X., et al. (2005). Development and identification of wheat–Haynaldia villosa T6DL.6VS chromosome translocation lines conferring resistance to powdery mildew. Plant Breed. 124, 203–205. doi: 10.1111/j.1439-0523.2004.01062.x
Li, H., and Zhu, Z. (1999). The potential value of Dasypyrum villosum in breeding and its chromosome manipulation. Chin Bull Bot. 16, 504–510. (in Chinese with English abstract)
Liu, C., Qi, L., Liu, W., Zhao, W., Wilson, J., Friebe, B., et al. (2011). Development of a set of compensating Triticum aestivum – Dasypyrum villosum robertsonian translocation lines. Genome 54, 836–844. doi: 10.1139/g11-051
Meyers, B. C., Dickerman, A. W., Michelmore, R. W., Sivaramakrishnan, S., Sobral, B. W., and Yong, N. D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–332. doi: 10.1046/j.1365-313X.1999.t01-1-00606.x
Qi, L. L., Wang, S. L., Chen, P. D., Liu, D. J., and Gill, B. S. (1998). Identification and physical mapping of three Haynaldia villosa chromosome-6V deletion lines. Theor. Appl. Genet. 97, 1042–1046. doi: 10.1007/s001220050989
Seeholzer, S., Tsuchimatsu, T., Jordan, T., Bieri, S., Pajonk, S., Yang, W., et al. (2010). Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant Microbe Interact. 23, 497–509. doi: 10.1094/MPMI-23-4-0497
Shi, Y., Wang, B., Li, Q., Wu, X., Wang, F., Liu, H., et al. (2009). Analysis of the virulent genes of Erysiphe graminis f. sp. tritici and the resistance genes of wheat commercial cultivars in shaanxi province. J. Triticeae Crop 29, 706–711. (in Chinese with English abstract)
Srichumpa, P., Brunner, S., Keller, B., and Yahiaoui, N. (2005). Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 139, 885–895. doi: 10.1104/pp.105.062406
Tajima, F. (1989). Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tameling, W. I. L., Vossen, J. H., Albrecht, M., Lengauer, T., Berden, J. A., Michel, A., et al. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140, 1233–1245. doi: 10.1104/pp.105.073510
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Wang, H., Dai, K., Xiao, J., Yuan, C., Zhao, R., DoleŽel, J., et al. (2017). Development of intron targeting (IT) markers specific for chromosome arm 4VS of Haynaldia villosa by chromosome sorting and next-generation sequencing. BMC Genomics 18:167. doi: 10.1186/s12864-017-3567-z
Yang, L., Xiang, L., Zeng, F., Wang, H., Shi, W., and Yu, D. (2009). Virulence gene structure analysis of Blumeria graminis f. sp. tritici in Hubei. Plant Protect. 35, 76–79. (in Chinese with English abstract)
Zhang, R., Fan, Y., Kong, L., Wang, Z., Wu, J., Xing, L., et al. (2018). Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 131, 2613–2620. doi: 10.1007/s00122-018-3176-5
Zhang, R., Sun, B., Chen, J., Cao, A., Xing, L., Feng, Y., et al. (2016). Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 129, 1975–1984. doi: 10.1007/s00122-016-2753-8
Zhu, S., Ji, Y., Bie, T., Gao, A., and He, H. (2018). Fine physical bin mapping of the powdery mildew resistance gene Pm21 based on chromosomal structural variations in wheat. Int. J. Mol. Sci. 19:643. doi: 10.3390/ijms19020643
Keywords: Dasypyrum villosum, Pm21 allele, genetic diversity, evolutionary analysis, diversifying selection, wheat powdery mildew resistance
Citation: He H, Ji J, Li H, Tong J, Feng Y, Wang X, Han R, Bie T, Liu C and Zhu S (2020) Genetic Diversity and Evolutionary Analyses Reveal the Powdery Mildew Resistance Gene Pm21 Undergoing Diversifying Selection. Front. Genet. 11:489. doi: 10.3389/fgene.2020.00489
Received: 17 March 2020; Accepted: 20 April 2020;
Published: 12 May 2020.
Edited by:
Zhu-Qing Shao, Nanjing University, ChinaReviewed by:
Diaoguo An, Institute of Genetics and Developmental Biology(CAS), ChinaZujun Yang, University of Electronic Science and Technology of China, China
Dale Zhang, Henan University, China
Copyright © 2020 He, Ji, Li, Tong, Feng, Wang, Han, Bie, Liu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huagang He, aGdoZUBtYWlsLnVqcy5lZHUuY24=; Cheng Liu, bGNoNjY4ODQwN0AxNjMuY29t; Shanying Zhu, emh1c2hhbnlpbmdAbWFpbC51anMuZWR1LmNu
†These authors have contributed equally to this work
 Huagang He
Huagang He Jian Ji1†
Jian Ji1† Hongjie Li
Hongjie Li Xiaolu Wang
Xiaolu Wang Ran Han
Ran Han Cheng Liu
Cheng Liu