
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genet. , 12 May 2020
Sec. Evolutionary and Population Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00465
This article is part of the Research Topic Evolution of Mitochondrial Genomes View all 5 articles
Despite both originating from endosymbiotic bacteria, one does not typically expect mitochondrial DNA (mtDNA) to show strong sequence identity to plastid DNA (ptDNA). Nevertheless, a recent analysis of Haematococcus lacustris revealed exactly that. A common repeat element has proliferated throughout the mtDNA and ptDNA of this chlamydomonadalean green alga, resulting in the unprecedented situation whereby these two distinct organelle genomes are largely made up of nearly identical sequences. In this short update to the work on H. lacustris, I highlight another chlamydomonadalean species (Stephanosphaera pluvialis) for which matching repeats have spread throughout its organelle genomes (but to a lesser degree than in H. lacustris). What’s more, the organelle repeats from S. pluvialis are similar to those from H. lacustris, suggesting that they have a shared origin, and perhaps existed in the mtDNA and ptDNA of the most recent common ancestor of these two species. However, my examination of organelle genomes from other close relatives of H. lacustris and S. pluvialis did not uncover further compelling examples of common organelle repeat elements, meaning that the evolutionary history of these repeats might be more complicated than initially thought.
Mitochondrial and plastid DNAs (mtDNAs and ptDNAs) are no strangers to repeats. In fact, the organelle genomes of many diverse species are distended with non-coding repetitive DNA, which can come in a wide range of forms, from short direct repeats to long complex ones (Figueroa-Martinez et al., 2017; Brázda et al., 2018; Čechová et al., 2018; Wynn and Christensen, 2019). The repeats within mtDNA and ptDNA often share certain similarities with each other, such as a propensity for AT or GC nucleotides, but their sequences are typically unique. This is why the recent discovery of nearly identical repeats in the mitochondrial and plastid genomes of the chlamydomonadalean green alga Haematococcus lacustris, strain UTEX 2505, was so remarkable (Zhang et al., 2019).
A common family of GC-rich repeat elements, many of which are palindromic (i.e., a sequences that can be folded into hairpin structures), have spread throughout the H. lacustris mtDNA and ptDNA, resulting in the unprecedented situation whereby these two different genomes are largely made up of matching sequences (Zhang et al., 2019). The proliferation of these elements has resulted in extremely high organelle GC compositions (∼50%) (Smith, 2012) as well as severe genome expansion—the plastome of H. lacustris, at 1.35 Mb, is the largest on record (Bauman et al., 2018; Smith, 2018) and its mtDNA (124.6 kb) is among the biggest from green algae (Zhang et al., 2019; Repetti et al., 2020).
Repeat elements, particularly palindromic ones, are rampant in the organelle genomes of other chlamydomonadalean algae (Smith and Lee, 2009; Del Vasto et al., 2015; Figueroa-Martinez et al., 2017). To the best of my knowledge, however, H. lacustris is the only known species, of all eukaryotes, for which the same repeat has proliferated in both the mtDNA and ptDNA, notwithstanding examples of small regions of similarity between mitochondrial and chloroplast genomes (Pombert et al., 2005; Wang et al., 2007; Turmel et al., 2016). Here, in this brief update to the earlier work on the H. lacustris organelle DNAs, I highlight another chlamydomonadalean species that harbors nearly identical repeats in its mitochondrial and plastid genomes and then use these data to further explore the origins of such a strange phenomenon.
During the initial characterization of the H. lacustris organelle genomes, it was noted that the mitochondrial and plastid repeats show similarity not only to each other but also to the GC-rich repeats in the ptDNA of another chlamydomonadalean: Stephanosphaera pluvialis, strain SAG 78-1a (Zhang et al., 2019). The plastid genome of this colonial fresh-water alga, which is a close relative of H. lacustris (Buchheim et al., 2013), was sequenced as part of a large-scale phylogenetic analysis (Lemieux et al., 2015), but it remains in a highly fragmented state (111 contigs; accumulative length 220.8 kb; overall GC content 46%), likely because the wealth of repeats prevented its accurate assembly. The sequence identity between the H. lacustris repeats and those in the S. pluvialis ptDNA raise the obvious question: are these same repeats also found in the S. pluvialis mtDNA?
Currently, there are no publicly available mtDNA sequence data for S. pluvialis. However, this alga (specifically, the strain used for plastome sequencing, SAG 78-1a) was one of many species to have its transcriptome sequenced as part of the One Thousand Plant Transcriptomes Initiative (2019). RNA-sequencing (RNA-seq) data have proven to be an excellent resource for mining organelle transcripts from green algae (Sanitá Lima and Smith, 2017) and have even been used to reconstruct complete chlamydomonadalean organelle genomes (Tian and Smith, 2016). By downloading the S. pluvialis transcriptome and searching it via BLAST using chlamydomonadalean mitochondrial genes as queries, I was able to identify 18 contigs corresponding to putative mtDNA-derived transcripts (Table 1). These mitochondrial contigs range from 109 to 1,946 nt (avgerage length = 627 nt), have an accumulative length of 11,293 nt, and together contain the standard cohort of genes typically found in chlamydomonadalean mitochondrial genomes, including fragmented and scrambled rRNAs (Table 1). Half of the contigs appear to have mitochondrial introns and most include sections of transcribed intergenic DNA, providing 5,705 nt of non-coding sequence data to investigate the presence/absence of repeat elements.
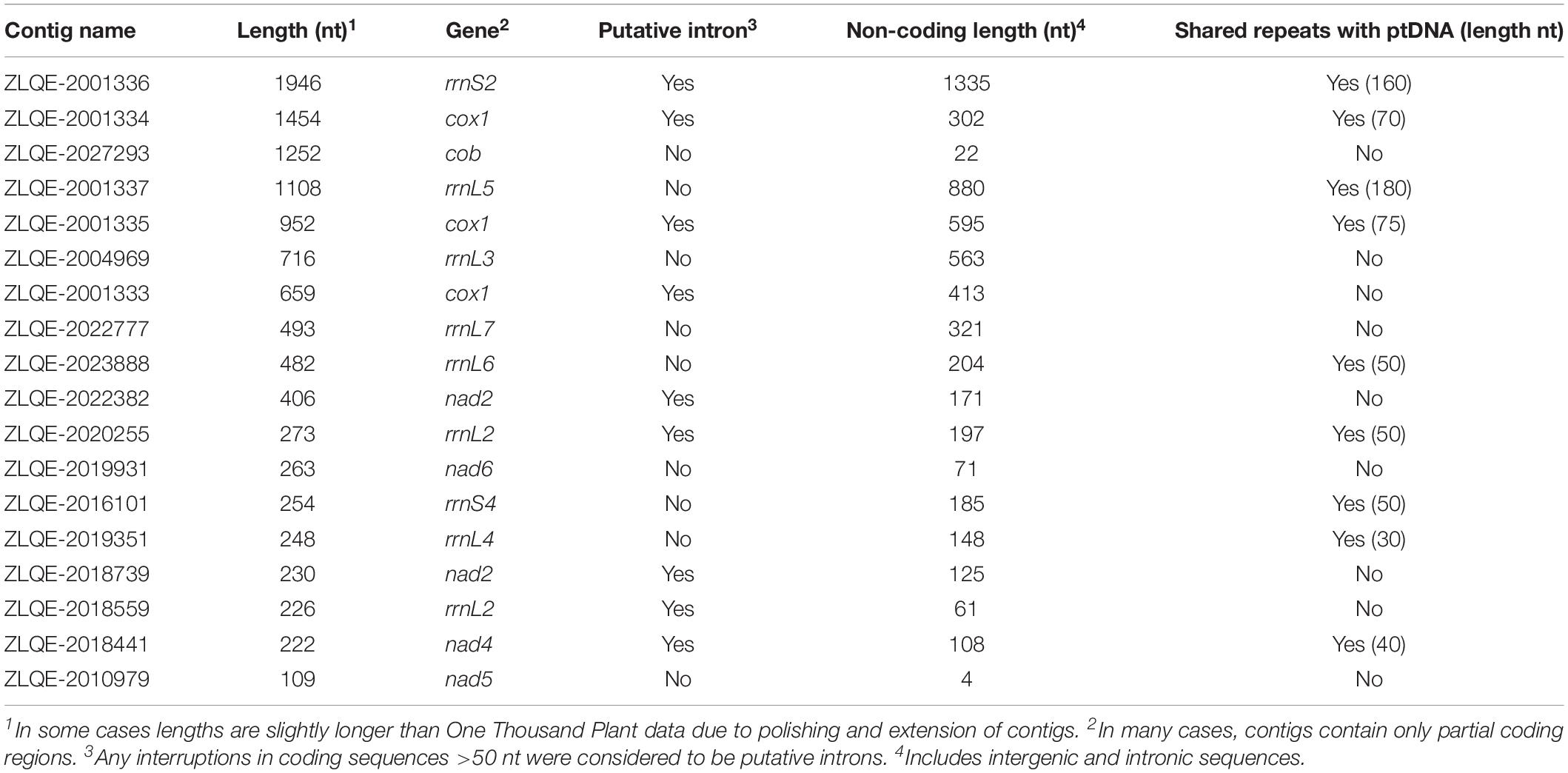
Table 1. Mitochondrial RNA-derived contigs identified from the Stephanosphaera pluvialis SAG 78-1a One Thousand Plant transcriptome data.
Sure enough, nine of the S. pluvialis mitochondrial transcripts contain repeats, including palindromes, that match to those from the neighboring ptDNA with ≥80% sequence identity (Table 1 and Figure 1A). In total, ∼700 nt of the mitochondrial contigs can be aligned to ptDNA repeats, meaning that the S. pluvialis organelle genomes harbor nearly identical repeat elements. These elements were identified by blasting the 18 mitochondrial contigs against a database made up of the S. pluvialis ptDNA (GenBank accessions KT625299-KT625409). As with H. lacustris, the S. pluvialis organelle repeats are GC-rich (>50%); moreover, a single repeat from the mitochondrial genome can match to hundreds of locations in the plastid genome, and palindromes were found in both intronic and intergenic regions. However, unlike H. lacustris, the segments of the S. pluvialis mitochondrial contigs that show similarity to ptDNA are relatively short (approximately 30–180 nt) and encompass only a small proportion (∼12%) of the analyzed regions (Table 1). The same cannot be said for the repeats in the S. pluvialis plastome, which appear to be widespread throughout much of the sequenced non-coding ptDNA (Lemieux et al., 2015). But keep in mind that these observations (which are intended to help direct future research) are based on partial mitogenome and plastome data and will need to be revised upon complete organelle genome sequencing of S. pluvialis.
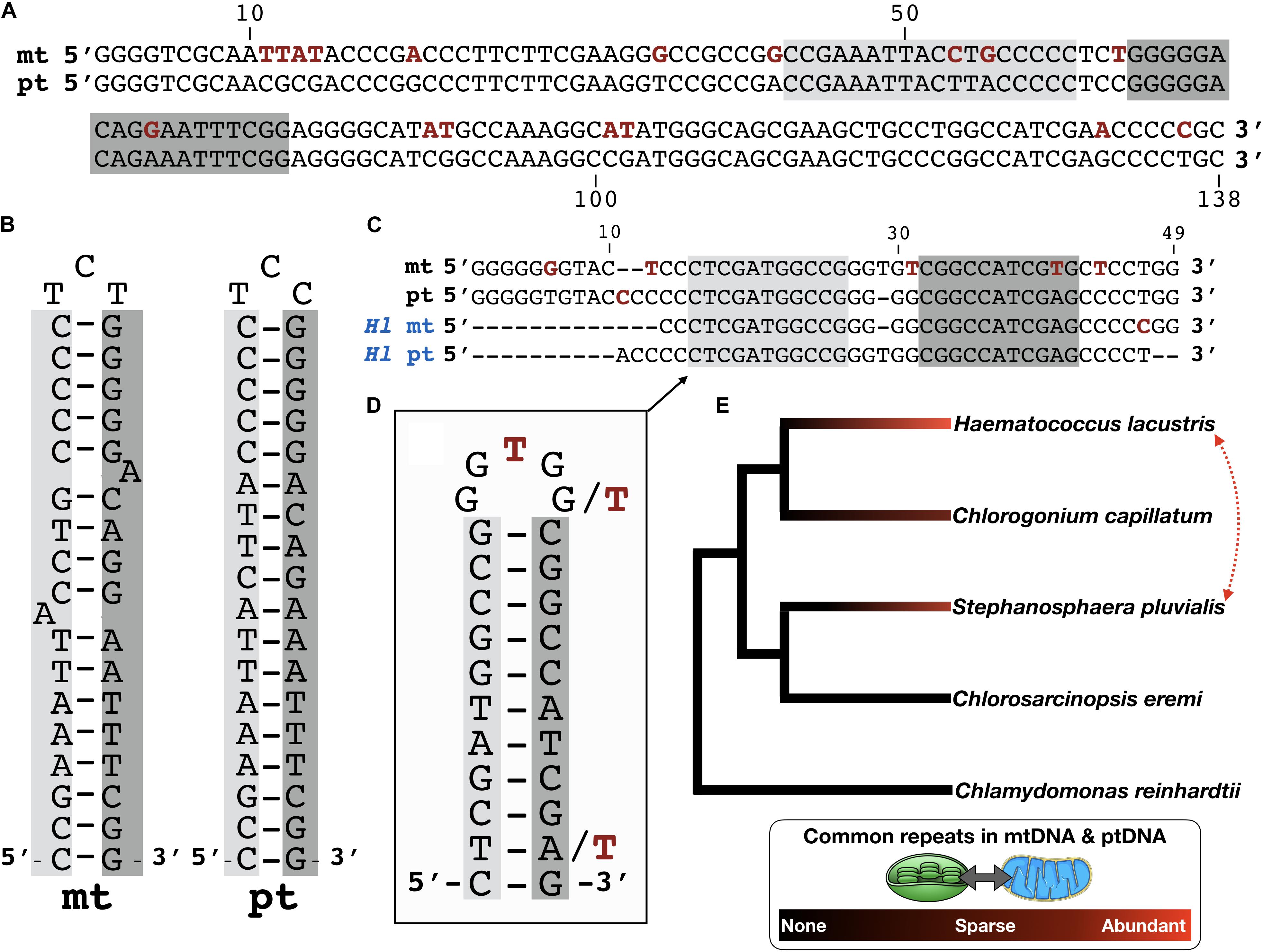
Figure 1. Common repeats in the organelle genomes of Stephanosphaera pluvialis. (A) Pairwise nucleotide alignment of a shared repeat element in the S. pluvialis mitochondrial (mt) and chloroplast (pt) genomes. Polymorphisms are highlighted in red. Palindromic repeat highlighted in light and dark gray. Region corresponds to nucleotides 95–232 of accession KT625314 and nucleotides 558–695 from contig ZLQE-2001337. Note, other smaller palindromes are also found in these sequences but are not shown. (B) Folded hairpin structure of the palindromic repeat shown in panel (A). (C) Pairwise alignment of a shared repeat element in the mitochondrial (mt) and chloroplast (pt) genomes of S. pluvialis and those of Haematococcus lacustris (Hl mt and Hl pt). Palindromic repeat highlighted in light and dark gray. (D) Folded hairpin structure of palindrome shown in panel (C). (E) Common repeats in the mitochondrial and chloroplast genomes of green algae. Branching order based on Fučíková et al. (2019). Dotted red line denotes similar repeats in different species.
Close inspection of the matching sequences between the S. pluvialis organelle genomes reveals an interesting trend: the palindromes from the mitochondrial genome typically contain more imperfections than their ptDNA counterparts (Figures 1A–D). This point can be easily interpreted by comparing the folded hairpin structures of mitochondrial palindromes to those from the plastome (Figures 1B,D). For example, the palindromes from the mitochondrial genome often contain mismatches and/or insertion-deletion mutations in the stem portion of the hairpin, which is not necessarily true for the corresponding ptDNA palindromes (Figures 1B,D). These imperfections could be an indication that the mitochondrial palindromic repeats are in a state of deterioration, or at least are not being as well maintained as those in the ptDNA. If this hypothesis is correct, it could explain why the palindromes are more widespread within the S. pluvialis ptDNA and might also indicate that they appeared first in the plastome and spread via intracellular DNA transfer to the mtDNA (Smith, 2011)—but see discussion below. Or it could just signal that the mtDNA has a higher rate of silent-site nucleotide substitution than the ptDNA, which is a common theme among eukaryotic algae (Smith, 2015). Keep in mind as well that GC-rich palindromic repeats are thought be transposable elements in certain organelle genomes (Wu and Hao, 2015).
Even more intriguing is the resemblance of the S. pluvialis mitochondrial and plastid repeats with those from H. lacustris (Figures 1C,D). Indeed, the organelle repeats from these two distinct species can share moderately strong sequence identity (>80%) with each other over regions that can exceed 100 nt. For S. pluvialis, however, the palindromes in the ptDNA show stronger similarity to those in the H. lacustris organelle genomes than the mitochondrial ones do (Figures 1C,D), further supporting the idea that the S. pluvialis mitochondrial palindromes are in disrepair. Nevertheless, these observations imply that the organelle DNA palindromes in S. pluvialis and H. lacustris have a shared origin, and perhaps existed in the mtDNA and ptDNA of the most recent common ancestor of these two species. To investigate this idea further, I explored the organelle genomes of close relatives of S. pluvialis and H. lacustris for palindromic elements.
The phylogenetic relationships among chlamydomonadalean algae are reasonably well resolved (Nakada et al., 2008a; Lemieux et al., 2015; Fučíková et al., 2019), including for species from the Chlorogonia and Stephanosphaerinia, the respective clades to which H. lacustris and S. pluvialis belong (Buchheim et al., 2013; Pegg et al., 2015). I cross-referenced members of these two clades against available organelle genome sequences, which, in turn, allowed me to investigate the mtDNA and ptDNA of an additional two species closely affiliated to H. lacustris and S. pluvialis for common organelle repeat elements (Figure 1E).
One of the closest known relatives of H. lacustris is the unicellular freshwater alga Chlorogonium capillatum (Nakada et al., 2010; Buchheim et al., 2013). This species has had its mitochondrial genome completely sequenced (Kroymann and Zetsche, 1998) and there is a nearly complete assembly of its ptDNA (Lemieux et al., 2015); these data come from two distinct but very closely related strains of C. capillatum: SAG 12-2e (mtDNA) and UTEX 11 (ptDNA) (Nozaki et al., 1998; Nakada et al., 2008b). [Note: SAG 12-2e was previously referred to as Chlorogonium elongatum.] The C. capillatum mitogenome is not particularly big or bloated (22.7 kb; ∼47% non-coding DNA), but it does contain repeats, including palindromic ones (Kroymann and Zetsche, 1998). The plastome, on the other hand, is large and expanded (>271 kb; >64% non-coding) and it, too, contains palindromic repeats, significantly more than the mtDNA. But are the mitochondrial and plastid repeats similar to one another?
Comparison of the C. capillatum organelle genomes using BLAST did uncover some regions of microhomology within non-coding regions. For example, two ∼60 nt segments from the mitogenome each match to a distinct location in the plastome with ∼75% sequence identity. Both of these segments correspond to GC-rich repeats (Kroymann and Zetsche, 1998), part of which can be folded into hairpin-like secondary structures. There were also dozens of short (20–30 nt) mtDNA regions showing high pairwise identity (90–95%) to the ptDNA, some of which represent palindromic elements. Thus, these two genomes do have some common repeats, but not to the same high degree as found in the organelle DNAs of H. lacustris and S. pluvialis. Moreover, the C. capillatum repeats show no obvious sequence similarity to those of the latter two species, which is surprising given that C. capillatum is believed to share a common ancestor with H. lacustris more recently than it does with S. pluvialis (Nakada et al., 2010; Buchheim et al., 2013).
I was also able to explore a close relative of S. pluvialis for common organelle repeat elements, namely Chlorosarcinopsis eremi strain MKA.28 (Figure 1E). This unicellular freshwater alga, which is normally found in desert environments (Juy-abad et al., 2018), has recently had its mitogenome and plastome completely sequenced (Fučíková et al., 2019; Juy-abad et al., 2019). The ptDNA is expanded (∼298 kb; ∼67% non-coding) and populated with hundreds of short palindromic repeats, which have been described in detail and do not show high sequence identity with those from other chlamydomonadalean species (Smith, 2020). Conversely, the mtDNA is small (24.9 kb) and essentially devoid of palindromes (Juy-abad et al., 2019; Smith, 2020). Nevertheless, I compared these two genomes using BLAST to see if any of the ptDNA palindromes matched to the mitogenome. Apart from short similarities among coding regions (e.g., a plastid rRNA gene matching to a mitochondrial one), the C. eremi organelle genomes are almost entirely made up of distinct non-coding sequences. This, again, is surprising given that C. eremi and S. pluvialis are more closely related to each other than to H. lacustris.
After all this, I feel like I am no further ahead in understanding how a common repeat element has proliferated throughout the mitochondrial and plastid genomes of H. lacustris and S. pluvialis. These data still leave open—but do not completely support—the scenario that the common ancestor of the Chlorogonia and Stephanosphaerinia clades had matching palindromes in its mtDNA and ptDNA and that these repeats have been preserved for millions of years. Perhaps more plausible is that the shared palindromes in these two species owe their origin to horizontal DNA transfer, both between species and between organelles within a cell. Precisely how this occurs is debated, but the lateral movement of DNA between distinct organelle genomes is well documented (Bergthorsson et al., 2003; Stegemann et al., 2012), particularly intracellular plastid-to-mitochondrion DNA transfer, which is especially prevalent in species with multiple plastids per cell (Smith, 2011). H. lacustris and S. pluvialis, however, have a single plastid per cell (Ettl, 1983; Nakada and Ota, 2016), which should greatly reduce the potential for ptDNA-to-mtDNA transmission. Their mitochondria, on the other hand, can apparently exist in multiple numbers per cell (Wayama et al., 2013), which should increase the probably of successful mtDNA-to-ptDNA transfers (Smith et al., 2011), contradicting my earlier suggestion above that the transfer might have occurred via ptDNA to mtDNA. Organelle introns are known to move between species and between mitochondria and plastids (Pombert et al., 2005), so it is possible that the repeats piggybacked on a mobile intron. In this context, it is noteworthy that all of the organelle genomes discussed here contain introns, and the mtDNA and ptDNA introns from H. lacustris and S. pluvialis do harbor palindromes. Complete organelle DNA sequences (including the intronic regions) from S. pluvialis might provide better insights into this hypothesis.
If anything, these data reinforce the notion that mitochondrial and plastid genomes can have similar repeat sequences. However, H. lacustris still stands out as an exceptionally extreme case of a common repeat expansion in two distinct compartments. It will be interesting to see if the organelle genomes of even closer relative of H. lacustris, such as Ettlia carotinosa (Buchheim et al., 2013), and other species of Haematococcus comprise shared palindromic repeats. Finally, the presence/absence of palindromes within an organelle genome may seem trivial from a broad biological perspective, but recent analyses have shown that these types of sequences can have significant impacts on the evolution of organelle DNAs (Smith, 2020) and, thus, should not be overlooked.
The One Thousand Plant Transcriptomes assembly for S. pluvialis can be found under accession number ZLQE; see Carpenter et al. (2019) for detailed instructions on accessing the data. Mitochondrial RNA-derived contigs were identified by blasting the C. eremi and H. lacustris mtDNAs (GenBank accessions NC_041430.1 and MK878592.1) against the S. pluvialis transcriptome with BlastN implemented through Geneious v10.2.6. (Biomatters Ltd., Auckland, New Zealand) using default settings. (Note: all other blast analyses described in the article were carried out using these same settings). Hits containing bona fide mitochondrial genes (Table 1) were polished by mapping the raw S. pluvialis RNA-seq data (GenBank accession ERX2100118) to the contigs using the Geneious read mapper (medium-low sensitivity; default settings); in a few instances, this resulted in minor (10–45 nt) extensions to the contigs.
The datasets used for this study can be found in The One Thousand Plant Transcriptomes assembly for S. pluvialis under accession number ZLQE.
DS wrote the manuscript and analyzed the data.
DS was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I thank all the authors of the primary sequencing data from which this study is based, including the One Thousand Plant Transcriptomes Initiative.
Bauman, N., Akella, S., Hann, E., Morey, R., and Schwartz, A. S. (2018). Next-generation sequencing of Haematococcus lacustris reveals an extremely large 1.35-megabase chloroplast genome. Genome Announc. 6:e00181-18. doi: 10.1128/genomeA.00181-18
Bergthorsson, U., Adams, K. L., Thomason, B., and Palmer, J. D. (2003). Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424, 197–201. doi: 10.1038/nature01743
Brázda, V., Lýsek, J., Bartas, M., and Fojta, M. (2018). Complex analyses of short inverted repeats in all sequenced chloroplast DNAs. BioMed. Res. Int. 2018:1097018. doi: 10.1155/2018/1097018
Buchheim, M. A., Sutherland, D. M., Buchheim, J. A., and Wolf, M. (2013). The blood alga: phylogeny of Haematococcus (Chlorophyceae) inferred from ribosomal RNA gene sequence data. Eur. J. Phycol. 48, 318–329.
Carpenter, E. J., Matasci, N., Ayyampalayam, S., Wu, S., Sun, J., Yu, J., et al. (2019). Access to RNA-sequencing data from 1,173 plant species: The 1000 Plant transcriptomes initiative (1KP). GigaScience 8:giz126. doi: 10.1093/gigascience/giz126
Čechová, J., Lýsek, J., Bartas, M., and Brázda, V. (2018). Complex analyses of inverted repeats in mitochondrial genomes revealed their importance and variability. Bioinformatics 34, 1081–1085. doi: 10.1093/bioinformatics/btx729
Del Vasto, M., Figueroa-Martinez, F., Featherston, J., Gonzalez, M. A., Reyes-Prieto, A., Durand, P. M., et al. (2015). Massive and widespread organelle genomic expansion in the green algal genus Dunaliella. Genome Biol. Evol. 7, 656–663. doi: 10.1093/gbe/evv027
Ettl, H. (1983). “Chlorophyta I. Phytomonadia,” in Süsswasserflora von Mitteleuropa, Vol. 9, eds H. Ettl, J. Gerloff, H. Heynig, and D. Mollenhauer (Stuttgart: Gustav Fischer Verlag), 807.
Figueroa-Martinez, F., Nedelcu, A. M., Smith, D. R., and Reyes-Prieto, A. (2017). The plastid genome of Polytoma uvella is the largest known among colorless algae and plants and reflects contrasting evolutionary paths to nonphotosynthetic lifestyles. Plant Phys. 173, 932–943. doi: 10.1104/pp.16.01628
Fučíková, K., Lewis, P. O., Neupane, S., Karol, K. G., and Lewis, L. A. (2019). Order, please! Uncertainty in the ordinal-level classification of Chlorophyceae. PeerJ 7:e6899. doi: 10.7717/peerj.6899
Juy-abad, F. K., Mohammadi, P., and Zarrabi, M. (2018). The identification of some phototrophic microorganisms from a semi-arid ecosystem in Iran. C. R. Acad. Bulg. Sci. 71, 1629–1637.
Juy-abad, F. K., Mohammadi, P., and Zarrabi, M. (2019). Comparative analysis of Chlorosarcinopsis eremi mitochondrial genome with some Chlamydomonadales algae. Physiol. Mol. Biol. Plants 25, 1301–1310. doi: 10.1007/s12298-019-00696-y
Kroymann, J., and Zetsche, K. (1998). The mitochondrial genome of Chlorogonium elongatum inferred from the complete sequence. J. Mol. Evol. 47, 431–440. doi: 10.1007/pl00006400
Lemieux, C., Vincent, A. T., Labarre, A., Otis, C., and Turmel, M. (2015). Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol. Biol. 15:264. doi: 10.1186/s12862-015-0544-5
Nakada, T., Misawa, K., and Nozaki, H. (2008a). Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol. Phylogenet. Evol. 48, 281–291. doi: 10.1016/j.ympev.2008.03.016
Nakada, T., Nozaki, H., and Pröschold, T. (2008b). Molecular phylogeny, ultrastructure, and taxonomic revision of Chlorogonium (Chlorophyta): emendation of Chlorogonium and description of Gungnir gen. nov. and Rusalka gen. nov. J. Phycol. 44, 751–760. doi: 10.1111/j.1529-8817.2008.00525.x
Nakada, T., and Ota, S. (2016). What is the correct name for the type of Haematococcus Flot. (Volvocales, Chlorophyceae)? Taxon 65, 343–348.
Nakada, T., Soga, T., Tomita, M., and Nozaki, H. (2010). Chlorogonium complexum sp. nov. (Volvocales, Chlorophyceae), and morphological evolution of Chlorogonium. Eur. J. Phycol. 45, 97–106.
Nozaki, H., Ohta, N., Morita, E., and Watanabe, M. M. (1998). Toward a natural system of species in Chlorogonium (Volvocales, Chlorophyta): a combined analysis of morphological and rbcL gene sequence data. J. Phycol. 34, 1024–1037.
One Thousand Plant Transcriptomes Initiative. (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679. doi: 10.1038/s41586-019-1693-2
Pegg, C., Wolf, M., Alanagreh, L. A., Portman, R., and Buchheim, M. A. (2015). Morphological diversity masks phylogenetic similarity of Ettlia and Haematococcus (Chlorophyceae). Phycologia 54, 385–397.
Pombert, J. F., Otis, C., Lemieux, C., and Turmel, M. (2005). The chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol. Biol. Evol. 22, 1903–1918. doi: 10.1093/molbev/msi182
Repetti, S. I, Jackson, C. J., Judd, L. M., Wick, R. R., and Holt, K. E. (2020). The inflated mitochondrial genomes of siphonous green algae reflect processes driving expansion of noncoding DNA and proliferation of introns. PeerJ 8:e8273. doi: 10.7717/peerj.8273
Sanitá Lima, M., and Smith (2017). Pervasive transcription of mitochondrial, plastid, and nucleomorph genomes across diverse plastid-bearing species. Genome Biol. Evol. 9, 2650–2657. doi: 10.1093/gbe/evx207
Smith, D. R. (2011). Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol. Evol. 3, 743–748. doi: 10.1093/gbe/evr068
Smith, D. R. (2012). Updating our view of organelle genome nucleotide landscape. Front. Genet. 3:175. doi: 10.3389/fgene.2012.00175
Smith, D. R. (2015). Mutation rates in plastid genomes: they are lower than you might think. Genome Biol. Evol. 7, 1227–1234. doi: 10.1093/gbe/evv069
Smith, D. R. (2018). Haematococcus lacustris: the makings of a giant-sized chloroplast genome. AoB Plants 10:ly058. doi: 10.1093/aobpla/ply058
Smith, D. R. (2020). Can green algal plastid genome size be explained by DNA repair mechanisms? Genome Biol. Evol. 12, 3797–3802. doi: 10.1093/gbe/evaa012
Smith, D. R., Crosby, K., and Lee, R. W. (2011). Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol. Evol. 3, 365–371. doi: 10.1093/gbe/evr001
Smith, D. R., and Lee, R. W. (2009). The mitochondrial and plastid genomes of Volvox carteri: bloated molecules rich in repetitive DNA. BMC Genomics 10:132. doi: 10.1186/1471-2164-10-132
Stegemann, S., Keuthe, M., Greiner, S., and Bock, R. (2012). Horizontal transfer of chloroplast genomes between plant species. Proc Natl. Acad. Sci. U.S.A. 109, 2434–2438. doi: 10.1073/pnas.1114076109
Tian, Y., and Smith, D. R. (2016). Recovering complete mitochondrial genome sequences from RNA-Seq: a case study of Polytomella non-photosynthetic green algae. Mol. Phylogenet. Evol. 98, 57–62. doi: 10.1016/j.ympev.2016.01.017
Turmel, M., Otis, C., and Lemieux, C. (2016). Mitochondrion-to-chloroplast DNA transfers and intragenomic proliferation of chloroplast group II introns in Gloeotilopsis green algae (Ulotrichales, Ulvophyceae). Genome Biol. Evol. 8, 2789–2805. doi: 10.1093/gbe/evw190
Wang, D., Wu, Y. W., Shih, A. C. C., Wu, C. S., Wang, Y. N., and Chaw, S. M. (2007). Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol. Biol. Evol. 24, 2040–2048. doi: 10.1093/molbev/msm133
Wayama, M., Ota, S., Matsuura, H., Nango, N., Hirata, A., and Kawano, S. (2013). Three-dimensional ultrastructural study of oil and astaxanthin accumulation during encystment in the green alga Haematococcus pluvialis. PLoS One 8:e53618. doi: 10.1371/journal.pone.0053618
Wu, B., and Hao, W. (2015). A dynamic mobile DNA family in the yeast mitochondrial genome. G3 5, 1273–1282. doi: 10.1534/g3.115.017822
Wynn, E. L., and Christensen, A. C. (2019). Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3 9, 549–559. doi: 10.1534/g3.118.200948
Keywords: Chlamydomonas, chloroplast DNA, genome size, Haematococcus, inverted repeat, mitochondrial DNA, palindrome, Stephanosphaera
Citation: Smith DR (2020) Common Repeat Elements in the Mitochondrial and Plastid Genomes of Green Algae. Front. Genet. 11:465. doi: 10.3389/fgene.2020.00465
Received: 22 January 2020; Accepted: 15 April 2020;
Published: 12 May 2020.
Edited by:
Eric Pante, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Ugo Cenci, Lille 1 University of Science and Technology, FranceCopyright © 2020 Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Roy Smith, ZHNtaXQyNDJAdXdvLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.