
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 09 April 2020
Sec. Epigenomics and Epigenetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00347
Recent discoveries have led to the development of novel ideas and techniques that have helped elucidate the correlation between epigenetics and tumor biology. Nowadays, the field of tumor genetics has evolved to include a new type of regulation by epigenetics. An increasing number of studies have demonstrated the importance of DNA methylation and hydroxymethylation in specific genes in the progression of cervical cancer. Determining the methylation and hydroxymethylation profiles of these genes will help in the early prevention and diagnosis, monitoring recurrence, prognosis, and treatment of patients with cervical cancer. In this review, we focus on the significance of aberrant DNA methylation and hydroxymethylation in cervical cancer and the use of these epigenetic signatures in clinical settings.
Cervical cancer closely correlates with high rates of infection with human papillomavirus (HPV), especially HPV16 and HPV18, and is the fourth most common gynecological malignancy and second leading cause of cancer-associated female mortality globally (da Costa et al., 2019). According to new statistics, that there are 528,000 new diagnoses and 266,000 deaths annually. Notably, the global incidence of cervical cancer is higher in the urban areas as compared to that in the rural area; however, the number of patients with cervical cancer is on the rise in both areas (Meng et al., 2018). Recent studies have shown the correlation between epigenetics and development and progression of cervical cancer. Epigenetic modifications, such as DNA methylation, hydroxymethylation, demethylation, chromatin remodeling, histone modification, regulation by non-coding RNAs, and gene imprinting, are inheritable and affect genetic information without interfering with mitosis or meiosis (Xie et al., 2018). Among the epigenetic modifications, the role of candidate gene DNA methylation in cervical cancer has been studied the most. Accumulated DNA methylation in specific genes is detected as early signatures of malignant cervical cancer (Li et al., 2019a). Like methylation, DNA hydroxymethylation has gained importance recently as an epigenetic regulator of gene expression. Hydroxymethylation is involved in DNA methylation homeostasis and is mediated by the synergism between DNA methyltransferases (DNMTs) and Ten-eleven translocation (TET).
DNA methylation is a complicated process that predominantly occurs in the CpG islands of genes. DNA methyltransferases (DNMTs) transfer a methyl group to alternate cytosines, thereby generating 5-methylcytosine (5-mC). Two classes of DNMTs have been identified: DNMT1 and DNMT3 (DNMT3a and DNMT3b). DNMT1 functions in maintaining the methylation status of genes; it recognizes hemi-methylated DNA. In contrast, DNMT3a and DNMT3b are responsible for methylating genes, i.e., de novo DNA methylation (Wu and Zhao, 2018). DNA methylation usually results in gene silencing, whereas DNA demethylation is associated with activating gene expression.
DNA hypermethylation occurs when multiple methyl groups are transferred to one cytosine that should not be methylated, thereby resulting in gene silencing. Studies on DNA hypermethylation have provided new insights into tumorigenesis (Su et al., 2018). DNMTs are important in DNA methylation as well as hypermethylation. DNMT3b activity is one of the main factors for DNA hypermethylation (Sandhu et al., 2015). DNMT3a has been reported to be downregulated in some cancers, but DNMT1 is not known to be involved in the deregulated expression of genes (Sen et al., 2017). Since the enzyme responsible for DNA hypermethylation has been elucidated, research has shifted to determine the target genes being methylated. Clarke et al. (2017) demonstrated that, compared with healthy controls, ADCYAP1, ASCL1, CADM1, DCC, ATP10, DBC1, HS3ST2, MOS, SOX1, MYOD1, SOX17, and TMEFF2 showed higher levels of methylation. ASCL1, LHX8, and ST6GALNAC5 exhibited increased DNA methylation in cervical cancer; DNA hypermethylation also increases with the severity of cervical cancer (Kremer et al., 2018). Moreover, Verlaat et al. showed that DNA methylation usually occurs at the pre-tumorigenic stage and reaches the highest level after tumorigenesis induced by hrHPV. Twelve genes (ANKRD18CP, C13orf18, EPB41L3, JAM3, SOX1, ZSCAN1, GHSR, SST, ZIC1, FAM19A4, PHACTR3, and PRDM14) are potential biomarkers for diagnosing cervical cancer (Verlaat et al., 2018b). Clarke et al. also found that DNA methylation occurs during the transition from HPV infection to the pre-cancer stage for all the 12 carcinogenic HPV types (Clarke et al., 2018). Promoter DNA methylation regulates miRNA expression that is an important mechanism employed during the development of cervical cancer. miR-424 has been shown to be hypermethylated in its promoter and is linked to the progression of cervical cancer (Varghese et al., 2018).
DNA methylation is performed by a complex system consisting of DNMT, DNA repair system, epigenetic regulatory factors, and environmental factors, etc. Abnormal DNA methylation (i.e., hypomethylation) affects replication and cell cycle in cells, thereby resulting in the development of tumors. During the progression of cervical cancer, rRNA levels increase. Moreover, epigenetic changes are also associated with rDNA promoter decondensation and hypomethylation. DNA methylation inhibits transcription by prohibiting the proteins from binding to methylated DNA. This can be observed in other cancers, such as lung or ovarian cancer. rRNA levels negatively correlate with the extent of methylation at the rDNA promoter; thus, rDNA hypomethylation influences cervical cancer development (Zhou et al., 2016). However, Poomipark et al. (2016) proposed a contrasting hypothesis: methyl donor status regulates DNA methylation and affects the incidence of cervical cancer. Human cervical cancer cells (C4-II) were incubated in the medium and after significant depletion of the methyl donor, DNMT3a and DNMT3b were reported to be downregulated, thereby leading to DNA hypomethylation. Thus, the status of methylation in the donor results in DNA hypomethylation; the effects of DNA hypomethylation on DNMT expression are reversible, suggesting a new idea that intake of methyl donor mediates gene expression and influences cervical cancer progression (Poomipark et al., 2016). Yin et al. (2016) have demonstrated that STK31 inhibits apoptosis by improving cell migration and invasiveness, thereby enabling cancer progression. STK31 is involved in hypermethylation and gene silencing. HPV16 E7 and E6/E7 oncoproteins epigenetically induce the expression of STK31 that causes DNA hypomethylation. Thus, STK31 is a potential candidate gene that may help the development of novel clinical approaches in the diagnosis and treatment of patients with cervical cancer (Yin et al., 2016). Varghese et al. (2018) have shown that miR-200b and miR-34c are hypomethylated during cervical cancer development.
There are target genes that undergo DNA hypermethylation and hypomethylation and can serve as cervical cancer biomarkers. However, the relevant pathways involved and other aspects of their biology remain to be understood more information still need to be studied in depth. These potential biomarkers show in Table 1.
The development of the HPV vaccine has greatly facilitated patients; however, the incidence of cervical cancer still remains high and primarily affects younger individuals. Therefore, early diagnosis and prevention of cervical cancer is imperative and can reduce the rate of mortality. Each HPV infection has a distinct gene signature, thus, different biomarkers may be of clinical significance for the different HPV types. It is important that the sensitivity, specificity, and rate of methylation of all the biomarkers are similar (Table 2).
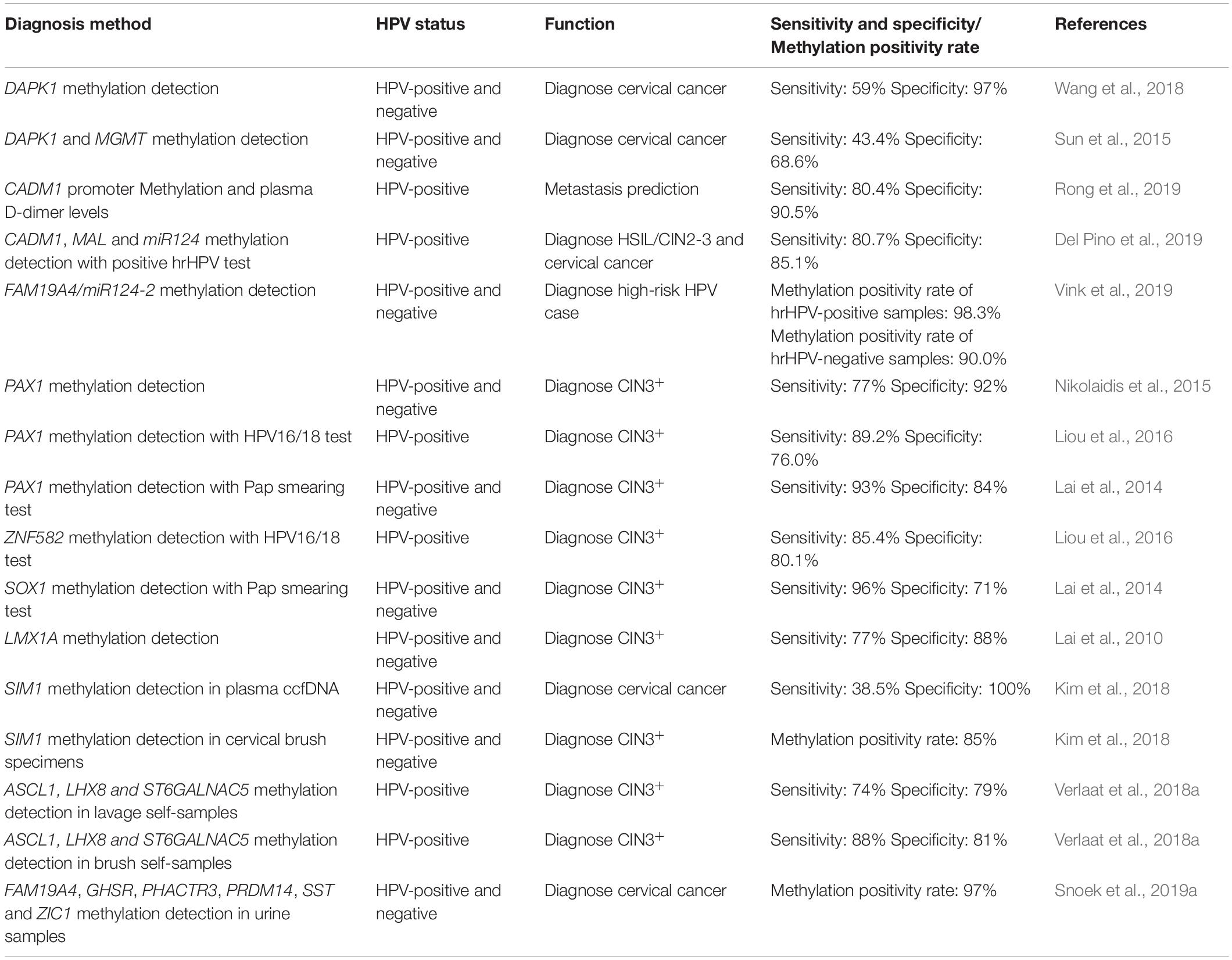
Table 2. Sensitivity and specificity or Methylation positivity rate of methylation diagnosis methods.
Mersakova et al. (2018) and Rong et al. (2019) have recently shown that CADM1 is a potential biomarker for cervical cancer. There was a significant difference in the promoter methylation of plasma CADM1 and its D-dimer between healthy individuals and those with cervical cancer. Combining these factors to predict metastasis revealed high specificity (90.5%) and sensitivity (80.4%) (Rong et al., 2019). Mersakova et al. (2018) speculated that CADM1 hypermethylation leads to suppressed Rb tumor suppressor signaling, but the exact mechanism remains to be understood. Combining the methylation of CADM1, MAL, and miR124 with a positive test for hrHPV increases the specificity and sensitivity for detecting HSIL/CIN2-3 and cervical cancer. Methylation of CADM1, MAL, and miR124 may be useful in estimating the risk of transformation. However, this requires further experiments to be proven conclusively (Del Pino et al., 2019).
Human papillomavirus infection, especially by HPV16 and HPV18, is a well-known cause for cervical cancer. However not all patients infected with HPV16 and/or HPV18 develop cervical cancer. Thus, screening for patients requiring therapy is problematic. High-risk HPV-infected specimens exhibit a high frequency of hypermethylation in the promoter of DcR1 (50.0%), whereas low-risk HPV-infected samples and non-HPV-infected samples contained 16.0 and 14.6% DcR1 hypermethylation, respectively. High-risk HPV infection silences DcR1 expression, thereby promoting the development of cervical cancer (Truong et al., 2018). Although the pathway or mechanism employed during cervical cancer are yet to be elucidated, the mechanism employed by DcR1 in other cancers has been reported. Vaitkiene et al. (2013) showed that DcR1 encodes a receptor incapable of inducing apoptosis; however, it protects glioblastoma multiforme (GBM) cells from TRAIL-induced apoptosis. DcR1 is hypermethylated and produces low levels of protein during the onset of cancer. This can explain specificity and sensitivity of cancer cells to the apoptosis-inducing activity of TRAIL (Vaitkiene et al., 2013). However, another study has identified the dependence of DcR1 on NF-κB/p50 and shown that DcR1 may be related to p53 and Bcl3 in GBM (Mansour et al., 2015). Taken together, DcR1 may be a promising biomarker for the early diagnosis of cervical cancer and help select patients requiring early treatment.
Kremer et al. (2018) have identified three related genes, including ASCL1, LHX8, and ST6GALNAC5. ASCL1 and LHX8 correlated well with CIN3+ (AUC 0.79 and 0.81, respectively) and ST6GALNAC5 correlated moderately with CIN3+ (AUC 0.71) of ASCL1, LHX8, and ST6GALNAC5 exhibited 72.1, 73.8, and 55.7% sensitivity with CIN3+, respectively. In conclusion, ASCL1 and LHX8 have better performance and higher sensitivity, thereby making them suitable biomarkers for early diagnosis (Kremer et al., 2018). Interestingly, ASCL1 functions as an oncogenic transcription factor in lung cancer (Lenhart et al., 2015). Other reports have not been successful in correlating LHX8 to cancer. However, Kremer et al. (2018) have reported increasing levels of hypermethylated ASCL1 and LHX8 with increasing severity of cervical cancer. This could be attributed to contamination by other factors like the human immunodeficiency virus (owing to the infected patient cohort). However, Kremer et al. (2018) also showed that using one of these three genes (ASCL1, LHX8, and ST6GALNAC5) was not sensitive. Verlaat et al. (2018a) have described a highly effective 3-gene methylation classifier (ASCL1, LHX8, and ST6GALNAC5) to detect the pre- and tumorigenic stage of cervical cancer. Combining these genes improves detection sensitivity and specificity. They detected methylation in hrHPV-positive lavage and brush self-samples; thus, the 3-gene methylation classifier (ASCL1, LHX8, and ST6GALNAC5) exhibited excellent clinical performance in the detection of CIN3. The sensitivity and specificity in hrHPV-positive lavage and brush self-samples were 74, 79, 88, and 81%, respectively (Verlaat et al., 2018a). Thus, this can be used as a prospective method for diagnosis.
In addition to putative biomarkers for the early diagnosis of cervical cancer lesions, there is an urgent need for the identification of biomarkers for CIN2/CIN3 hrHPV-positive patients to help determine the course of treatment. Some CIN2 and CIN3 lesions develop or regress spontaneously and do not need to be treated. Due to the lack of known biomarkers, these patients receive excessive treatment. Snoek et al. (2019b) showed that three genes (GHSR, SST, and ZIC1) are important for such patients. These genes were significantly methylated in tissue specimens and cervical scrapes with increasing severity of the disease. Especially in advanced CIN2/CIN3 and squamous cell carcinoma (SCC), these genes are highly methylated (Snoek et al., 2019b). Thus, methylated GHSR, SST, and ZIC1 may serve as prognostic markers for hrHPV-positive women.
Hypermethylated PAX1 is important in cancer progression (Huang et al., 2017) that functions in regulating cellular differentiation and proliferation (Fang et al., 2019). PAX1 can be used to detect cervical squamous cell carcinoma (CSCC) in 121 patients from eastern China with 80.9, 83.7, and 79.0% accuracy, specificity, and sensitivity, respectively (Xu et al., 2018). Combining methylated PAX1 and the presence of HPV16/18 results in an 89.2 and 76.0% sensitivity and specificity, respectively in detecting CIN3+ (Liou et al., 2016). Using the methylation status of PAX1 to determine the cancerous nature (and CIN1–3) of samples from Shanghai, China is associated with a sensitivity and specificity of 81 and 93%, respectively; however, the sensitivity and specificity for cancer versus CIN2/3 is 32% and 90%, respectively (Xu et al., 2015). Single mutations in PAX1 have been accurate in diagnosing cervical/HSIL in a group of 15 individuals with a specificity and sensitivity of 89% and 80%, respectively (Kong et al., 2015). Lai et al. (2014) have demonstrated that methylated PAX1 can be used for detecting CIN3+ lesions with a sensitivity and specificity of 64% and 91%, respectively in Taiwanese patients. Interestingly, combining results from Pap smears with parallel testing of PAX1 results in superior specificity (84%) and similar sensitivity (93%). Using a combination of Pap smear results with HPV testing leads to an increase in specificity and sensitivity of 66 and 97%, respectively. Thus, using Pap smears and PAX1 parallel testing results in better diagnosis (Lai et al., 2014). Kan et al. (2014) have also described the importance of combining PAX1 testing with Pap smears. Combining PAX1 with Pap testing, the sensitivity and specificity were 89 and 83%, respectively (Kan et al., 2014). A meta-analysis comprising Asian individuals revealed that the sensitivity and specificity of PAX1 methylation is 73 and 87%, respectively in HSIL/CIN3+/cervical cancer patients (Chen et al., 2016). Another meta-analysis used 1,385 individuals with different stages of CIN to show that the sensitivity and specificity of PAX1 methylation in CIN3+ samples are 77% and 92%, respectively (Nikolaidis et al., 2015). In summary, different geographical locations and various methods (especially using combinations) affect the sensitivity and specificity of detecting PAX1 methylation. Thus, PAX1 methylation can help identify CIN3+ patients in clinical settings.
LMX1A has been recently reported to be hypermethylated in cervical cancers (Rogeri et al., 2018). LMX1A is a suppressor of tumorigenesis and metastasis of cervical cancer; however, the regulation of LMX1A during tumorigenesis remains to be understood. Lin et al. (2013) have provided two hypotheses for the mechanism of action employed by LMX1A. In conditions of high promoter methylation, Sp1 binds to the promoter of LMX1A and inhibits LMX1A expression. Second, overexpression of EZH2 suppresses LMX1A expression in cancer cells. Thus, Sp1 and EZH2 may work in concert to regulate LMX1A expression (Lin et al., 2013). The sensitivity, specificity, and accuracy of the detection of cervical cancer are 63.3, 35.7, and 89.3%, respectively using LMX1A (Xu et al., 2018). However, another group have used LMX1A methylation to detect CIN3+ in samples with moderate sensitivity (77%) and specificity (88%) (Lai et al., 2010). Thus, whether LMX1A can be used as a biomarker remains to be confirmed.
Upon analyzing various sets of data and cross-referencing, CDH1, CDKN2A, RB1, and TP53 are putative biomarkers; these genes need to be studied for their role in diagnosing cervical cancer (Cardoso et al., 2017). Kim et al. (2018) have determined the methylation status of SIM1 in circulating cell-free DNA and cervical brush specimens by quantitative methylation-specific polymerase chain reaction, indicating that hypermethylated SIM1 is a potential biomarker for cervical cancer. Detecting methylated SIM1 in diagnosing cervical cancer was 38.5 and 100% sensitive and specific, respectively. Since the data is not too promising, it needs improvement. For cervical brush specimens, positive methylated SIM1 was detected in 0, 0, 5.3, 41.2, and 85% of normal, CIN1, CIN2, CIN3, and cancer samples, respectively (Kim et al., 2018). The proportion of methylated SIM1 in cells increases at the CIN3 stage; SIM1 hypermethylation mediates the transition from CIN3 to invasive cervical cancer. Therefore, detecting SIM1 methylation at the CIN3 stage can help prevent the progression of cervical cancer.
Numerous studies have reported the potential of CADM1, MAL, miR-124, SOX1, ERT, and EPB41l3 as biomarkers. DCC was identified to be a crucial candidate as a methylation biomarker during the screening of cervical cancer. DCC is involved in cell cycle control and apoptosis and can be found in several cancers, such as ovarian cancer, etc. However, experiments in the future need to confirm if methylated DCC can serve as a marker during the screening of cervical cancer (Clarke et al., 2017).
Sun et al. (2015) analyzed the methylated promoters of two genes (e.g., DAPK1 and MGMT, MGMT and RARB, and DAPK1 and RARB) using methylation sensitive-high resolution melting analysis to determine if this method enhances the sensitivity and specificity of clinical diagnosis. Methylation of DAPK1 combined with MGMT has a sensitivity and specificity of 43.4 and 68.6%, respectively; this may help develop novel methods to increase the rate of successful diagnosis (Sun et al., 2015).
Vink et al. (2019) reported that combining FAM19A4/miR124-2 methylation with hrHPV testing was more efficient. They assessed cervical cancer globally to find more than 98% cases of FAM19A4/miR124-2 methylation. FAM19A4/miR124-2 methylation can be detected in hrHPV-negative carcinomas; thus, combining these methods complements each other and reduces the rate of missed diagnoses (Leeman et al., 2018; Vink et al., 2019).
Various self-detection methods for cervical cancer have become increasingly popular over the years owing to their convenience of use. Van Ostade et al. (2018) proposed the use of cervical vaginal fluid in screening cervical cancer by self-detection. α-actinin-4 might play a significant role in the detection of cervical cancer from cervical vaginal fluid. Although this method requires significant improvement, it provides an exciting prospect for the early diagnosis of cervical cancer (Van Ostade et al., 2018). Snoek et al. (2019a) have demonstrated that collecting urine is more convenient and comfortable for patients than cervical scrapes and is a promising alternative testing material to screen for cervical cancer. hrHPV testing was conducted from urine sediments, native urine, and cervical scrapes, concluding a near-perfect agreement. The methylation levels of six genes (FAM19A4, GHSR, PHACTR3, PRDM14, SST, and ZIC1) showed differential expression in the urine sediments. The rate of methylation for each marker was at least 97% (Snoek et al., 2019a). Thus, urine-based DNA methylation testing is a novel and promising method that could be used for the diagnosis of cervical cancer.
The biomarkers useful for the prognosis of cervical cancer remain to be elucidated. The treatment of cervical cancer is limited to chemoradiation therapy, surgery, and drug therapy. However, more targets need to be identified for use in the prognosis and treatment of cervical cancer. Table 3 lists the potential for methylation-based therapy against cervical cancer.
Cisplatin-based neoadjuvant chemotherapy (NACT) is performed before surgery depending on the treatment of large or locally advanced squamous cervical cancer and followed by radical hysterectomy. This novel treatment strategy minimizes tumor volume, controls micrometastasis, and enhances the integrity and safety of surgery. However, the resistance and efficacy of NACT before chemotherapy needs to be assessed. Zhu et al. (2017) used immunohistochemistry and bioinformatic analyses to identify IB2 or IIA2 cervical cancer patients; the expression of galectin-1 and integrin α5β1 correlates with cisplatin-based NACT. Among patients with cervical squamous cell carcinoma, the expression of galectin-1 and integrin α5β1 in tumor cells and stromal cells is downregulated after NACT. The expression of galectin-1 and integrin α5β1 is significantly higher in patients with poor NACT efficacy than that of NACT; thus, the downregulation of galectin-1 and integrin α5β1 after chemotherapy is not significant among the patients with poor NACT efficacy. The resistance and prognosis of NACT can be determined by the expression of galectin-1 and integrin α5β1 followed by the selection of a suitable treatment plan for patients (Zhu et al., 2017).
Although chemoradiation therapy is an efficient mode of treatment, it is associated with side effects that result in inefficient prognosis of cervical cancer. Thus, closely related genes were tested to determine if chemoradiation therapy induces epigenetic modifications and regulates gene expression in cervical cancer (Sood et al., 2018). DAPK1 methylation was observed to be decreased with a concomitant increase in the DAPK1 transcripts in the tissue after chemoradiation therapy. DAPK1 has been shown to be associated with the severity of cervical cancer (Wang et al., 2018); DAPK1 upregulation suppresses tumor cell proliferation and improve apoptosis and autophagy (Bhattacharjee et al., 2018; Huang et al., 2018; Ren et al., 2018; Yadav et al., 2018). Thus, chemoradiation has the capacity to increase DAPK1 mRNA levels and reduce the number of tumor cells. This suggests that determining the expression of DAPK1 during chemoradiation therapy will help reduce side effects.
Interestingly, BRCA1 is involved with the same phenotype after chemoradiation therapy (Sood et al., 2018). However, BRCA1 functions differently as compared to DAPK1; BRCA1 is a tumor suppressor (Li et al., 2018a). It predominantly regulates DNA damage repair, transcription regulation, and apoptosis (Maresca et al., 2018; Sun et al., 2018). Therefore, it can serve as novel target during chemoradiation, like DAPK1. Overexpression of BRCA1 is involved in the development of cancer (Li et al., 2018a).
MGMT is another candidate that shows decreased methylation (Ren et al., 2018). MGMT repairs cytotoxic lesions by removing methyl adducts from DNA (Jiapaer et al., 2018; Scicchitano et al., 2018; Yuan et al., 2018), indicating MGMT can be a therapeutic target. Sood et al. (2018) have also tested the potential of ESR1, MYOD1, and MLH1. ESR1 transcript levels increase in the tissue, but decrease for MYOD1 and MLH1. However, it is unclear whether epigenetic modifications of these genes influence the development of cervical cancer after chemoradiation therapy. Moreover, their functions and associated pathways in tumor cells need to be understood in sufficient details.
Hypermethylated promoter of FANCF can be frequently detected in patients with cervical cancer. However, epigallocatechin-3-gallate (EGCG) and -(-)Menthol reverse the hypermethylation in the FANCF promoter. Upon inhibiting M. SssI (DNMT1 analog), these two substances inhibited DNMT1, thereby hypomethylating and reactivating the FANCF promoter and inhibiting cervical cancer progression (Parashar et al., 2017). A similar study reported that as compared to individual treatment, EGCG and eugenol-amarogentin strongly inhibits cellular colony formation and proliferation and induces apoptosis via promoting the hypomethylation of the promoters of LimD1 and P16. Thus, this combination of EGCG and eugenol-amarogentin may be a more efficacious line of therapy (Pal et al., 2018).
STK31 is a new potential therapeutic target in cervical cancer, especially in HPV16/18-positive cervical tumors. STK31 expression can be induced by the HPV16 E7 and E6/E7 oncoproteins by regulating the methylation status of STK31 in the HPV16/18-positive cells, while HPV-negative cervical cancer cells exhibit silencing of STK31. Therefore, inhibiting the tumorigenic activities of STK31 in HPV16/18-positive patients can be used in treating cervical cancer, indicating that STK31 is a crucial target. Moreover, epigenetic drugs increase the efficacy of chemoradiation and reduce chemoresistance. However, not all patients can be administered epigenetics drugs owing to the many restrictions for use in clinical application (Yin et al., 2016).
CDKN2A can also prevent the progression of cervical cancer since patients frequently have hypermethylated CDKN2A. The absence of p16INK4A protein inhibits the expression of HPV E7 oncoprotein and cervical cancer progression. CDKN2A methylation positively correlates with p16INK4A/p14ARF expression in head and neck cancer. There may be a similar correlation in cervical cancer. Therefore, the decrease in methylation of CDKN2A may lead to reduced protein levels of p16INK4A and HPV E7 oncoprotein, thereby inhibiting cervical cancer progression. Thus, CDKN2A is an important therapeutic target (Wijetunga et al., 2016).
Researchers put more focus on using chemical medicine and radiochemotherapy for the treatment of cancers; the function of natural substances, like vegetables or fruits, is usually overlooked. Recently, Kedhari Sundaram et al. (2019) demonstrated that quercetin, a phytochemical from vegetables and fruits, possesses anti-cancer activity by downregulating global DNA methylation and reversing hypermethylation in the TSG promoter, thereby activating TSG expression. However, for clinical application, the bioavailability of quercetin is influenced by factors including source, gender, and form of quercetin. Targeted delivery and liposomal and nanoparticle-based delivery to tumors are methods that improve the bioavailability of compounds and are currently under improvement. Thus, in the future, quercetin is expected to be a safer therapeutic for cervical cancer. Krifa et al. (2013) have demonstrated the anti-cancer effect of the aqueous gall extract (G extract) of Limoniastrum guyonianum and luteolin. These two natural products downregulate UHRF1 and DNMT1 via global DNA hypomethylation and activate p16INK4A-dependent cell cycle checkpoint signaling. Thus, G extract and luteolin inhibit cervical cancer cell proliferation and induce programmed cell death (Krifa et al., 2013). However, they have only used HeLa cells in vitro. The efficacy and association with side effects still remain to be understood.
The Ten-eleven translocation (TET) family oxidize 5-mC to 5-hydroxymethylcytosine (5-hmC) that is further oxidized to 5-formylcytosine and 5-carboxylcytosine. Finally, using thymine-DNA glycosylase (TDG), the carboxyl group from 5-formylcytosine and 5-carboxylcytosine are removed to restore unmethylated cytosine pool (Figure 1; Silva et al., 2017; Jayanthi et al., 2018; Zhu et al., 2018). 5-hmC is an intermediate throughout the process (Li et al., 2016).
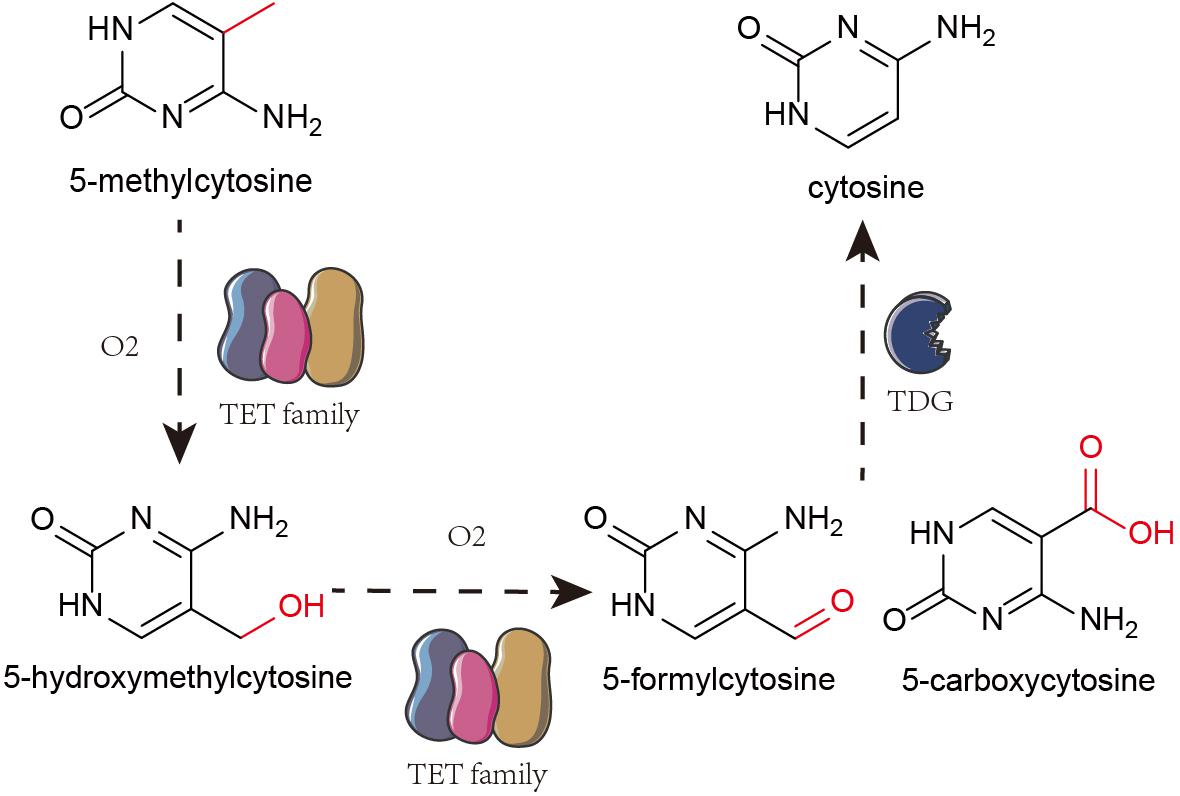
Figure 1. Mechanism of DNA hydroxymethylation. TET family oxidize 5-mc to 5-hmC. And then, TET proteins further oxidize 5-hydroxymethylcytosine (5-hmC) into 5-formylcytosine and 5-carboxylcytosine. Finally, it removes the carboxyl group from 5-formylcytosine and 5-carboxylcytosine by TDG to restore them to unmethylated cytosine.
Compared with the tissues from a normal cervix, the expression of 5-hmC and TET2 decrease in the cervical cancer tissues while the expression of TET1 and TET3 remain unaltered. Thus, the decreased expression of TET2 may decrease 5-hmC (Zhang et al., 2016), suggesting 5-hmC to be a putative prognostic biomarker. Other novel (potential) biomarkers in cervical cancer that undergo hydroxymethylation include ACTG1, SALL3, DNAJA3, SERPINB6, CDC14B, and CALN1 (Wang et al., 2019).
Wu et al. (2019) have demonstrated the downregulation of TET1 and 5-hmC in pancreatic tissues and cell lines. Pancreatic cancer patients with high TET1 levels exhibit longer overall survival than patients with low levels of TET1. Therefore, TET1 can suppress pancreatic tumor development by inhibiting proliferation and metastasis. This is achieved via Wnt signaling where TET1 binds and demethylates the promoter of the secreted frizzled-related protein 2 (SFRP2) to activate SFRP2 transcription (Wu et al., 2019). Interestingly, TET1 can suppress colon cancer, wherein TET1 binds to the promoter of DKK and inhibits Wnt signaling to maintain the hypomethylated state (Neri et al., 2015). Thus, TET1 may be a promising therapeutic in the future.
Glioblastoma melanoma and myeloid cancers are associated with reduced 5-hydroxymethylation. In ∼9% of patients with nasopharyngeal carcinoma, there is a mutation in TET1, TET2, or TET3 (Mohr et al., 2011; Lin et al., 2014; Wille et al., 2015; Salgado et al., 2020). There is a decrease in 5-hmC in melanoma samples as compared to that in nevus samples (Salgado et al., 2020). Wille et al. (2015) have shown that the inhibition of TET protein activity promotes lytic EBV promoter methylation in normal oral keratinocyte cells, thereby increasing Z-mediated lytic reactivation and inhibiting R-mediated lytic reactivation. The BRLF1 promoter contains a CpG-containing Z-binding site. Methylation of this region leads to Z-mediated viral reactivation. However, the presence of 5-hmC in this site in normal oral keratinocyte cells prevents Z-mediated viral reactivation. Thus, the inhibition of TET protein activity and low levels of 5-hmC function in the development of nasopharyngeal carcinoma (Wille et al., 2015).
There is a global decrease in hydroxymethylation in tumors that has elucidated the role of hydroxymethylation in tumor suppression. Li et al. (2019c) demonstrated that, although 5-hmCs were intermediates during oxidation, a proportion of 5-hmCs can be found in the genome. Hydroxymethylated P16 alleles are transcriptionally inactive and hydroxymethylation increases reactivation of P16 in cancer cells (Li et al., 2019b).
Genome-wide DNA hydroxymethylation has been considered a sensitive biomarker for prostate cancer (PCa) detection. Owing to the loss of hydroxymethylation, different cancers maintain low levels of 5-hmC (Feng et al., 2015; Strand et al., 2015; Kamdar et al., 2016; Spans et al., 2016). However, Grelus et al. (2017) have shown that hydroxymethylation markers cannot be single biomarkers for PCa diagnosis. They should be used as a supplement for prostate-specific antigen to diagnose PCa. Although hydroxymethylation can be detected in multiple cancer types, the utility of genomic hydroxymethylation in diagnosing cancer may only be restricted to one type of cancer (Grelus et al., 2017). Thus, whether DNA hydroxymethylation can be used in clinical diagnosis needs to be explored. Even so, we have concluded the function of DNA hydroxymethylation in various kinds of tumors as a reference in Table 4.
Several studies have highlighted the importance of 5-hmC in tumorigenesis. TET proteins are responsible for the presence of mutations or downregulated gene expression in tumorigenesis, thereby reducing the 5-hmC content in malignant cells (Haffner et al., 2011). The catalytic co-substrate α-ketoglutarate may greatly influence TET protein activity. IDH1 and IDH2, that produce α-ketoglutarate, can be present in mutated forms in many different kinds of cancers. Mutated IDH1 and IDH2 are incapable of forming α-ketoglutarate and 2-hydroxyglutarate, thereby inhibiting TET protein activity and stimulating the development of cancer (Kamdar et al., 2016).
Genome-wide loss of 5-hmC does not correspond with the global loss of 5-mC, indicating that the altered levels of 5-hmC in tumorigenesis may involve an independent mechanism (Ficz and Gribben, 2014). However, some studies have demonstrated a correlation between DNA methylation and hydroxymethylation. When the DNA substrate contains 5-hmC, DNMT1 activity is reduced a lot (∼60-fold), thereby diluting the 5-mC pool. Low levels of 5-hmC promote DNMT1 activity (Li et al., 2018b). Using clinical samples, Wang et al. (2019) have demonstrated that 5-hmC negatively correlates with cervical cancer progression and is downregulated upon regulation of 5-mC. Thus, demethylation and hydroxymethylation are inhibited and there is a global accumulation of aberrant DNA hypermethylation in cervical cancer (Figure 2; Wang et al., 2019).
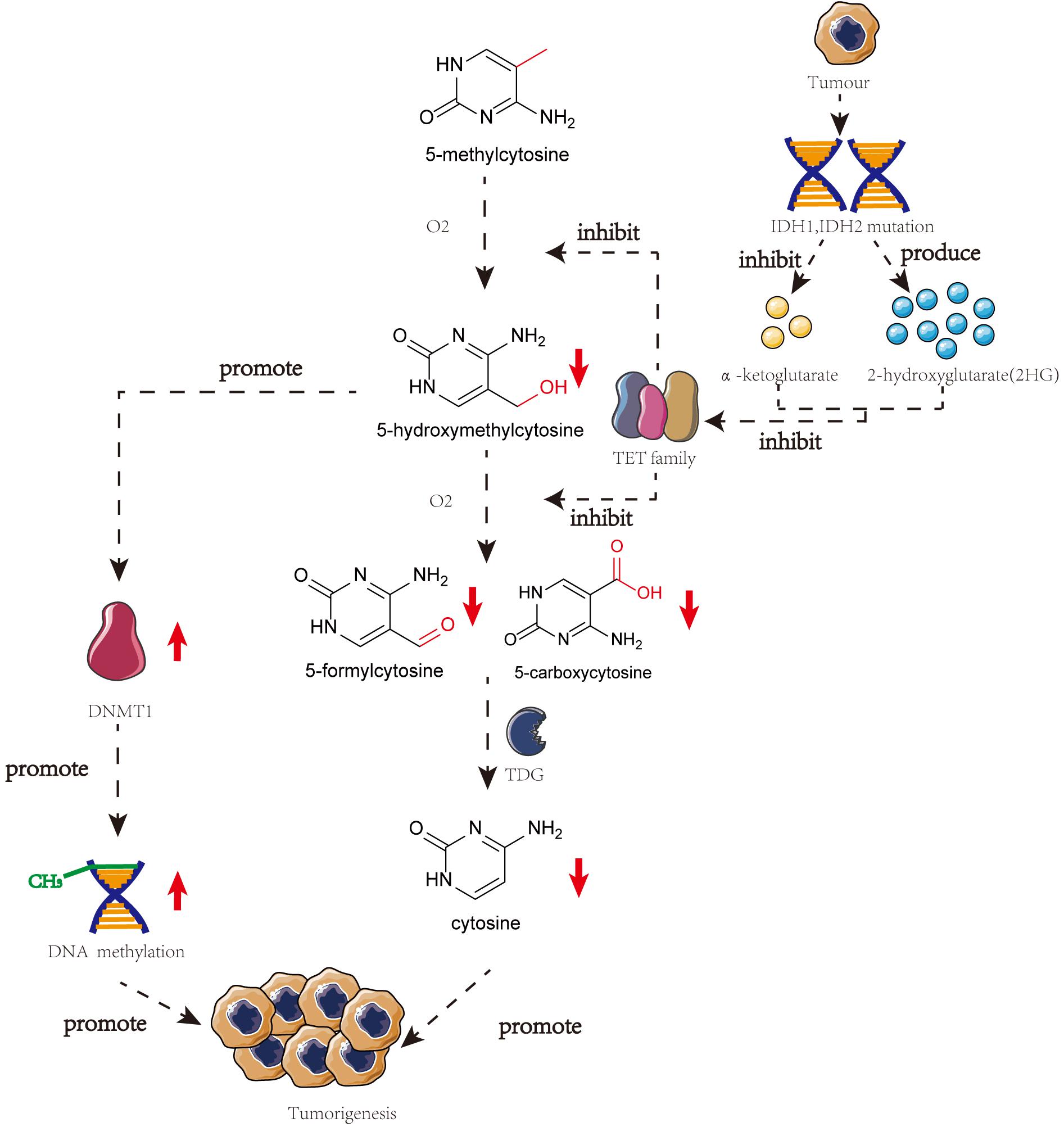
Figure 2. The crosstalk of DNA methylation and DNA hydroxymethylation in the progress of the tumor. In many different kinds of cancers, IDH1, and IDH2 will mutate which can restrain the production of α-ketoglutarate and produce 2HG. Both of these two phenomena will lead to the inhibition of TET family and meanwhile, the low level of 5-hmC can promote the activity of DNMT1. These two pathways will finally result in the tumorigenesis.
The incidence and development of tumors depend on multiple factors, including internal and external factors. Researchers have studied genetic factors in the past. However, there has been an increase in the focus on epigenetics to study gene function and genetic modification in the hope that epigenetics may be the key to developing other novel methods in the early diagnosis or treatment of diseases. In this review, we have summarized the methylation-specific modifications found in cervical cancer-related genes and their clinical application, thereby highlighting the aforementioned importance of epigenetics in cervical cancer. We have also described how DNA hydroxymethylation influences tumorigenesis simultaneously with DNA methylation. These phenomena may provide new methods and strategies for the early prevention and diagnosis, monitoring recurrence, and prognosis and treatment of cervical cancer. However, owing to various restrictions, a large number of samples and detailed research is required for some methods that cannot be used for clinical application yet.
HoZ drafted the manuscript. HeZ and MT revised the manuscript. JH and DW contributed to the acquisition and interpretation of data. TX designed the work. All authors contributed to the manuscript revision, read, and approved the submitted version.
TX was supported by funding from the Department of Science and Technology of Jilin Province (grant numbers 20190101014JH, 20190905004SF, 20180203032YY, and 20170622008JC) and National Key R&D Program of China (grant number 2016YFC1302901).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the Second Hospital of Jilin University for supporting this manuscript.
Bhattacharjee, S., Bose, P., Patel, K., Roy, S. G., Gain, C., Gowda, H., et al. (2018). Transcriptional and epigenetic modulation of autophagy promotes EBV oncoprotein EBNA3C induced B-cell survival. Cell Death Dis. 9:605. doi: 10.1038/s41419-018-0668-9
Cardoso, M. F. S., Castelletti, C. H. M., Lima-Filho, J. L., Martins, D. B. G., and Teixeira, J. A. C. (2017). Putative biomarkers for cervical cancer: SNVs, methylation and expression profiles. Mutat. Res. 773, 161–173. doi: 10.1016/j.mrrev.2017.06.002
Chen, Y., Cui, Z., Xiao, Z., Hu, M., Jiang, C., Lin, Y., et al. (2016). PAX1 and SOX1 methylation as an initial screening method for cervical cancer: a meta-analysis of individual studies in Asians. Ann. Transl. Med. 4:365. doi: 10.21037/atm.2016.09.30
Clarke, M. A., Gradissimo, A., Schiffman, M., Lam, J., Sollecito, C. C., Fetterman, B., et al. (2018). Human papillomavirus DNA methylation as a biomarker for cervical precancer: consistency across 12 genotypes and potential impact on management of HPV-positive women. Clin. Cancer Res. 24, 2194–2202. doi: 10.1158/1078-0432.Ccr-17-3251
Clarke, M. A., Luhn, P., Gage, J. C., Bodelon, C., Dunn, S. T., Walker, J., et al. (2017). Discovery and validation of candidate host DNA methylation markers for detection of cervical precancer and cancer. Int. J. Cancer 141, 701–710. doi: 10.1002/ijc.30781
da Costa, S. C. S., Bonadio, R. C., Gabrielli, F. C. G., Aranha, A. S., Dias Genta, M. L. N., Miranda, V. C., et al. (2019). Neoadjuvant chemotherapy with cisplatin and gemcitabine followed by chemoradiation versus chemoradiation for locally advanced cervical cancer: a randomized phase II trial. J. Clin. Oncol. 37, 3124–3131. doi: 10.1200/jco.19.00674
Del Pino, M., Sierra, A., Marimon, L., Marti Delgado, C., Rodriguez-Trujillo, A., Barnadas, E., et al. (2019). CADM1, MAL, and miR124 promoter methylation as biomarkers of transforming cervical intrapithelial lesions. Int. J. Mol. Sci. 20:2262. doi: 10.3390/ijms20092262
Fang, C., Wang, S. Y., Liou, Y. L., Chen, M. H., Ouyang, W., and Duan, K. M. (2019). The promising role of PAX1 (aliases: HUP48, OFC2) gene methylation in cancer screening. Mol. Genet. Genomic. Med. 7:506. doi: 10.1002/mgg3.506
Feng, J., Wang, Q., Li, G., Zeng, X., Kuang, S., Li, X., et al. (2015). TET1-mediated different transcriptional regulation in prostate cancer. Int. J. Clin. Exp. Med. 8, 203–211.
Ficz, G., and Gribben, J. G. (2014). Loss of 5-hydroxymethylcytosine in cancer: cause or consequence? Genomics 104, 352–357. doi: 10.1016/j.ygeno.2014.08.017
Grelus, A., Nica, D. V., Miklos, I., Belengeanu, V., Ioiart, I., and Popescu, C. (2017). Clinical significance of measuring global hydroxymethylation of white blood cell DNA in prostate cancer: comparison to PSA in a pilot exploratory study. Int. J. Mol. Sci. 18:2465. doi: 10.3390/ijms18112465
Guan, Z., Zhang, J., Wang, J., Wang, H., Zheng, F., Peng, J., et al. (2014). SOX1 down-regulates beta-catenin and reverses malignant phenotype in nasopharyngeal carcinoma. Mol. Cancer 13:257. doi: 10.1186/1476-4598-13-257
Haffner, M. C., Chaux, A., Meeker, A. K., Esopi, D. M., Gerber, J., Pellakuru, L. G., et al. (2011). Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2, 627–637. doi: 10.18632/oncotarget.316
Huang, J., Tan, Z. R., Yu, J., Li, H., Lv, Q. L., Shao, Y. Y., et al. (2017). DNA hypermethylated status and gene expression of PAX1/SOX1 in patients with colorectal carcinoma. Onco. Targets. Ther. 10, 4739–4751. doi: 10.2147/ott.S143389
Huang, Y., Lin, M., Chen, X., Huang, C., Zhang, X., Chen, L., et al. (2018). Evaluation of the prognostic and physiological functions of death associated protein kinase 1 in breast cancer. Oncol. Lett. 15, 8261–8268. doi: 10.3892/ol.2018.8439
Jayanthi, S., Gonzalez, B., McCoy, M. T., Ladenheim, B., Bisagno, V., and Cadet, J. L. (2018). Methamphetamine Induces TET1- and TET3-Dependent DNA Hydroxymethylation of Crh and Avp genes in the rat nucleus accumbens. Mol. Neurobiol. 55, 5154–5166. doi: 10.1007/s12035-017-0750-9
Jiapaer, S., Furuta, T., Tanaka, S., Kitabayashi, T., and Nakada, M. (2018). Potential strategies overcoming the temozolomide resistance for glioblastoma. Neurol. Med. Chir. (Tokyo) 58, 405–421. doi: 10.2176/nmc.ra.2018-0141
Kamdar, S. N., Ho, L. T., Kron, K. J., Isserlin, R., van der Kwast, T., Zlotta, A. R., et al. (2016). Dynamic interplay between locus-specific DNA methylation and hydroxymethylation regulates distinct biological pathways in prostate carcinogenesis. Clin. Epigenetics 8:32. doi: 10.1186/s13148-016-0195-4
Kan, Y. Y., Liou, Y. L., Wang, H. J., Chen, C. Y., Sung, L. C., Chang, C. F., et al. (2014). PAX1 methylation as a potential biomarker for cervical cancer screening. Int. J. Gynecol. Cancer 24, 928–934. doi: 10.1097/igc.0000000000000155
Kedhari Sundaram, M., Hussain, A., Haque, S., Raina, R., and Afroze, N. (2019). Quercetin modifies 5’CpG promoter methylation and reactivates various tumor suppressor genes by modulating epigenetic marks in human cervical cancer cells. J. Cell Biochem. 120, 18357–18369. doi: 10.1002/jcb.29147
Kim, H. J., Kim, C. Y., Jin, J., Bae, M. K., Kim, Y. H., Ju, W., et al. (2018). Aberrant single-minded homolog 1 methylation as a potential biomarker for cervical cancer. Diagn Cytopathol. 46, 15–21. doi: 10.1002/dc.23838
Kong, L. Y., Du, W., Wang, L., Yang, Z., and Zhang, H. S. (2015). PAX1 methylation hallmarks promising accuracy for cervical cancer screening in asians: results from a meta-analysis. Clin. Lab. 61, 1471–1479. doi: 10.7754/clin.lab.2015.150232
Kremer, W. W., Van Zummeren, M., Novianti, P. W., Richter, K. L., Verlaat, W., Snijders, P. J., et al. (2018). Detection of hypermethylated genes as markers for cervical screening in women living with HIV. J. Int. AIDS Soc. 21:e25165. doi: 10.1002/jia2.25165
Krifa, M., Alhosin, M., Muller, C. D., Gies, J. P., Chekir-Ghedira, L., Ghedira, K., et al. (2013). Limoniastrum guyonianum aqueous gall extract induces apoptosis in human cervical cancer cells involving p16 INK4A re-expression related to UHRF1 and DNMT1 down-regulation. J. Exp. Clin. Cancer Res. 32:30. doi: 10.1186/1756-9966-32-30
Lai, H. C., Lin, Y. W., Huang, R. L., Chung, M. T., Wang, H. C., Liao, Y. P., et al. (2010). Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer 116, 4266–4274. doi: 10.1002/cncr.25252
Lai, H. C., Ou, Y. C., Chen, T. C., Huang, H. J., Cheng, Y. M., Chen, C. H., et al. (2014). PAX1/SOX1 DNA methylation and cervical neoplasia detection: a taiwanese gynecologic oncology group (TGOG) study. Cancer Med. 3, 1062–1074. doi: 10.1002/cam4.253
Lee, T. S., Kim, J. W., Kang, G. H., Park, N. H., Song, Y. S., Kang, S. B., et al. (2006). DNA hypomethylation of CAGE promotors in squamous cell carcinoma of uterine cervix. Ann. N. Y. Acad. Sci. 1091, 218–224. doi: 10.1196/annals.1378.068
Leeman, A., Ebisch, R. M. F., Kasius, A., Bosgraaf, R. P., Jenkins, D., van de Sandt, M. M., et al. (2018). Defining hrHPV genotypes in cervical intraepithelial neoplasia by laser capture microdissection supports reflex triage of self-samples using HPV16/18 and FAM19A4/miR124-2 methylation. Gynecol. Oncol. 151, 311–318. doi: 10.1016/j.ygyno.2018.09.006
Lenhart, R., Kirov, S., Desilva, H., Cao, J., Lei, M., Johnston, K., et al. (2015). Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol. Cancer Ther. 14, 2167–2174. doi: 10.1158/1535-7163.Mct-15-0037
Li, A. G., Murphy, E. C., Culhane, A. C., Powell, E., Wang, H., Bronson, R. T., et al. (2018). BRCA1-IRIS promotes human tumor progression through PTEN blockade and HIF-1alpha activation. Proc. Natl. Acad. Sci. U.S.A. 115, E9600–E9609. doi: 10.1073/pnas.1807112115
Li, C., Ke, J., Liu, J., and Su, J. (2019). DNA methylation data-based molecular subtype classification related to the prognosis of patients with cervical cancer. J. Cell Biochem. 121, 2713–2724. doi: 10.1002/jcb.29491
Li, J., Wu, X., Zhou, Y., Lee, M., Guo, L., Han, W., et al. (2018). Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res. 46, 2883–2900. doi: 10.1093/nar/gky063
Li, M., Gao, F., Xia, Y., Tang, Y., Zhao, W., Jin, C., et al. (2016). Filtrating colorectal cancer associated genes by integrated analyses of global DNA methylation and hydroxymethylation in cancer and normal tissue. Sci. Rep. 6:31826. doi: 10.1038/srep31826
Li, P., Gan, Y., Qin, S., Han, X., Cui, C., Liu, Z., et al. (2019). DNA hydroxymethylation increases the susceptibility of reactivation of methylated P16 alleles in cancer cells. Epigenetics 1–14. doi: 10.1080/15592294.2019.1700004
Li, X., Wu, X., Li, Y., Cui, Y., Tian, R., Singh, N., et al. (2019). Promoter hypermethylation of SOX11 promotes the progression of cervical cancer in vitro and in vivo. Oncol. Rep. 41, 2351–2360. doi: 10.3892/or.2019.6993
Lin, D. C., Meng, X., Hazawa, M., Nagata, Y., Varela, A. M., Xu, L., et al. (2014). The genomic landscape of nasopharyngeal carcinoma. Nat. Genet. 46, 866–871. doi: 10.1038/ng.3006
Lin, W. C., Yan, M. D., Yu, P. N., Li, H. J., Kuo, C. C., Hsu, C. L., et al. (2013). The role of Sp1 and EZH2 in the regulation of LMX1A in cervical cancer cells. Biochim. Biophys. Acta 1833, 3206–3217. doi: 10.1016/j.bbamcr.2013.08.020
Liou, Y. L., Zhang, T. L., Yan, T., Yeh, C. T., Kang, Y. N., Cao, L., et al. (2016). Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin. Epigenetics 8:66. doi: 10.1186/s13148-016-0232-3
Mansour, N. M., Bernal, G. M., Wu, L., Crawley, C. D., Cahill, K. E., Voce, D. J., et al. (2015). Decoy receptor DcR1 is induced in a p50/Bcl3-dependent manner and attenuates the efficacy of temozolomide. Cancer Res. 75, 2039–2048. doi: 10.1158/0008-5472.Can-14-2144
Maresca, L., Lodovichi, S., Lorenzoni, A., Cervelli, T., Monaco, R., Spugnesi, L., et al. (2018). Functional interaction between BRCA1 and DNA repair in yeast may uncover a role of RAD50, RAD51, MRE11A, and MSH6 somatic variants in cancer development. Front. Genet. 9:397. doi: 10.3389/fgene.2018.00397
Meng, Y., Liang, H., Hu, J., Liu, S., Hao, X., Wong, M. S. K., et al. (2018). PD-L1 expression correlates with tumor infiltrating lymphocytes and response to neoadjuvant chemotherapy in cervical cancer. J. Cancer 9, 2938–2945. doi: 10.7150/jca.22532
Mersakova, S., Holubekova, V., Grendar, M., Visnovsky, J., Nachajova, M., Kalman, M., et al. (2018). Methylation of CADM1 and MAL together with HPV status in cytological cervical specimens serves an important role in the progression of cervical intraepithelial neoplasia. Oncol. Lett. 16, 7166–7174. doi: 10.3892/ol.2018.9505
Mohr, F., Dohner, K., Buske, C., and Rawat, V. P. (2011). TET genes: new players in DNA demethylation and important determinants for stemness. Exp. Hematol. 39, 272–281. doi: 10.1016/j.exphem.2010.12.004
Neri, F., Dettori, D., Incarnato, D., Krepelova, A., Rapelli, S., Maldotti, M., et al. (2015). TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene 34, 4168–4176. doi: 10.1038/onc.2014.356
Nikolaidis, C., Nena, E., Panagopoulou, M., Balgkouranidou, I., Karaglani, M., Chatzaki, E., et al. (2015). PAX1 methylation as an auxiliary biomarker for cervical cancer screening: a meta-analysis. Cancer Epidemiol. 39, 682–686. doi: 10.1016/j.canep.2015.07.008
Pal, D., Sur, S., Roy, R., Mandal, S., and Kumar Panda, C. (2018). Epigallocatechin gallate in combination with eugenol or amarogentin shows synergistic chemotherapeutic potential in cervical cancer cell line. J. Cell Physiol. 234, 825–836. doi: 10.1002/jcp.26900
Parashar, G., Parashar, N., and Capalash, N. (2017). Menthol induces reversal of promoter hypermethylation and associated up-regulation of the FANCF gene in the SiHa cell line. Asian. Pac. J. Cancer Prev. 18, 1365–1370. doi: 10.22034/apjcp.2017.18.5.1365
Poomipark, N., Flatley, J. E., Hill, M. H., Mangnall, B., Azar, E., Grabowski, P., et al. (2016). Methyl donor status influences DNMT expression and global DNA methylation in cervical cancer cells. Asian. Pac. J. Cancer Prev. 17, 3213–3222.
Ren, X., Li, H., Song, X., Wu, Y., and Liu, Y. (2018). 5-Azacytidine treatment induces demethylation of DAPK1 and MGMT genes and inhibits growth in canine mammary gland tumor cells. Onco. Targets Ther. 11, 2805–2813. doi: 10.2147/ott.S162381
Rogeri, C. D., Silveira, H. C. S., Causin, R. L., Villa, L. L., Stein, M. D., de Carvalho, A. C., et al. (2018). Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol. Oncol. 150, 545–551. doi: 10.1016/j.ygyno.2018.06.014
Rong, G., Zhang, M., Xia, W., Li, D., Miao, J., and Wang, H. (2019). Plasma CADM1 promoter hypermethylation and D-dimer as novel metastasis predictors of cervical cancer. J. Obstet. Gynaecol. Res. 45, 1251–1259. doi: 10.1111/jog.13966
Salgado, C., Oosting, J., Janssen, B., Kumar, R., Gruis, N., and van Doorn, R. (2020). Genome-wide characterization of 5-hydoxymethylcytosine in melanoma reveals major differences with nevus. Genes Chromosomes Cancer 1–9. doi: 10.1002/gcc.22837
Samuelsson, J., Alonso, S., Ruiz-Larroya, T., Cheung, T. H., Wong, Y. F., and Perucho, M. (2011). Frequent somatic demethylation of RAPGEF1/C3G intronic sequences in gastrointestinal and gynecological cancer. Int. J. Oncol. 38, 1575–1577. doi: 10.3892/ijo.2011.972
Sandhu, R., Roll, J. D., Rivenbark, A. G., and Coleman, W. B. (2015). Dysregulation of the epigenome in human breast cancer: contributions of gene-specific DNA hypermethylation to breast cancer pathobiology and targeting the breast cancer methylome for improved therapy. Am. J. Pathol. 185, 282–292. doi: 10.1016/j.ajpath.2014.12.003
Scicchitano, B. M., Sorrentino, S., Proietti, G., Lama, G., Dobrowolny, G., Catizone, A., et al. (2018). Levetiracetam enhances the temozolomide effect on glioblastoma stem cell proliferation and apoptosis. Cancer Cell Int. 18:136. doi: 10.1186/s12935-018-0626-8
Sen, S., Mandal, P., Bhattacharya, A., Kundu, S., Roy Chowdhury, R., Mondal, N. R., et al. (2017). Impact of viral and host DNA methylations on HPV16-related cervical cancer pathogenesis. Tumour. Biol. 39:1010428317699799. doi: 10.1177/1010428317699799
Silva, M. B. D., Melo, A., Costa, L. A., Barroso, H., and Oliveira, N. F. P. (2017). Global and gene-specific DNA methylation and hydroxymethylation in human skin exposed and not exposed to sun radiation. An. Bras. Dermatol. 92, 793–800. doi: 10.1590/abd1806-4841.20175875
Snoek, B. C., Splunter, A. P. V., Bleeker, M. C. G., Ruiten, M. C. V., Heideman, D. A. M., Rurup, W. F., et al. (2019a). Cervical cancer detection by DNA methylation analysis in urine. Sci. Rep. 9:3088. doi: 10.1038/s41598-019-39275-2
Snoek, B. C., Verlaat, W., Babion, I., Novianti, P. W., van de Wiel, M. A., Wilting, S. M., et al. (2019b). Genome-wide microRNA analysis of HPV-positive self-samples yields novel triage markers for early detection of cervical cancer. Int. J. Cancer 144, 372–379. doi: 10.1002/ijc.31855
Sood, S., Patel, F. D., Srinivasan, R., and Dhaliwal, L. K. (2018). Chemoradiation therapy induces in vivo changes in gene promoter methylation & gene transcript expression in patients with invasive cervical cancer. Indian J. Med. Res. 147, 151–157. doi: 10.4103/ijmr.IJMR_1939_16
Spans, L., Van den Broeck, T., Smeets, E., Prekovic, S., Thienpont, B., Lambrechts, D., et al. (2016). Genomic and epigenomic analysis of high-risk prostate cancer reveals changes in hydroxymethylation and TET1. Oncotarget 7, 24326–24338. doi: 10.18632/oncotarget.8220
Strand, S. H., Hoyer, S., Lynnerup, A. S., Haldrup, C., Storebjerg, T. M., Borre, M., et al. (2015). High levels of 5-hydroxymethylcytosine (5hmC) is an adverse predictor of biochemical recurrence after prostatectomy in ERG-negative prostate cancer. Clin. Epigenetics 7:111. doi: 10.1186/s13148-015-0146-5
Su, J., Huang, Y. H., Cui, X., Wang, X., Zhang, X., Lei, Y., et al. (2018). Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 19:108. doi: 10.1186/s13059-018-1492-3
Sun, X. L., Jiang, H., Han, D. X., Fu, Y., Liu, J. B., Gao, Y., et al. (2018). The activated DNA double-strand break repair pathway in cumulus cells from aging patients may be used as a convincing predictor of poor outcomes after in vitro fertilization-embryo transfer treatment. PLoS One 13:e0204524. doi: 10.1371/journal.pone.0204524
Sun, Y., Li, S., Shen, K., Ye, S., Cao, D., and Yang, J. (2015). DAPK1, MGMT and RARB promoter methylation as biomarkers for high-grade cervical lesions. Int. J. Clin. Exp. Pathol. 8, 14939–14945.
Tian, Y., Yuan, Wu, N. Y., Liou, Y. L., Yeh, C. T., Cao, L., et al. (2017). Utility of gene methylation analysis, cytological examination, and HPV-16/18 genotyping in triage of high-risk human papilloma virus-positive women. Oncotarget 8, 62274–62285. doi: 10.18632/oncotarget.19459
Truong, P. K., Lao, T. D., and Le, T. A. H. (2018). Hypermethylation of DcR1 gene-based biomarker in non-invasive cancer screening of vietnamese cervical cancer patients. Iran J. Public Health 47, 350–356.
Vaitkiene, P., Skiriute, D., Skauminas, K., and Tamasauskas, A. (2013). GATA4 and DcR1 methylation in glioblastomas. Diagn Pathol. 8:7. doi: 10.1186/1746-1596-8-7
Van Ostade, X., Dom, M., Tjalma, W., and Van Raemdonck, G. (2018). Candidate biomarkers in the cervical vaginal fluid for the (self-)diagnosis of cervical precancer. Arch. Gynecol. Obstet. 297, 295–311. doi: 10.1007/s00404-017-4587-2
Varghese, V. K., Shukla, V., Kabekkodu, S. P., Pandey, D., and Satyamoorthy, K. (2018). DNA methylation regulated microRNAs in human cervical cancer. Mol. Carcinog. 57, 370–382. doi: 10.1002/mc.22761
Verlaat, W., Snoek, B. C., Heideman, D. A. M., Wilting, S. M., Snijders, P. J. F., Novianti, P. W., et al. (2018a). Identification and validation of a 3-Gene methylation classifier for HPV-based cervical screening on self-samples. Clin. Cancer Res. 24, 3456–3464. doi: 10.1158/1078-0432.Ccr-17-3615
Verlaat, W., Van Leeuwen, R. W., Novianti, P. W., Schuuring, E., Meijer, C., Van Der Zee, A. G. J., et al. (2018b). Host-cell DNA methylation patterns during high-risk HPV-induced carcinogenesis reveal a heterogeneous nature of cervical pre-cancer. Epigenetics 13, 769–778. doi: 10.1080/15592294.2018.1507197
Vink, F. J., Meijer, C., Clifford, G. M., Poljak, M., Ostrbenk, A., Petry, K. U., et al. (2019). FAM19A4/miR124-2 methylation in invasive cervical cancer: a retrospective cross-sectional worldwide study. Int. J. Cancer doi: 10.1002/ijc.32614
Wang, J., Su, Y., Tian, Y., Ding, Y., and Wang, X. (2019). Characterization of DNA hydroxymethylation profile in cervical cancer. Artif. Cells Nanomed. Biotechnol. 47, 2706–2714. doi: 10.1080/21691401.2019.1634578
Wang, X. B., Cui, N. H., Liu, X. N., Ma, J. F., Zhu, Q. H., Guo, S. R., et al. (2018). Identification of DAPK1 promoter hypermethylation as a biomarker for intra-epithelial lesion and cervical cancer: a meta-analysis of published studies. TCGA, and GEO datasets. Front. Genet. 9:258. doi: 10.3389/fgene.2018.00258
Wijetunga, N. A., Belbin, T. J., Burk, R. D., Whitney, K., Abadi, M., Greally, J. M., et al. (2016). Novel epigenetic changes in CDKN2A are associated with progression of cervical intraepithelial neoplasia. Gynecol. Oncol. 142, 566–573. doi: 10.1016/j.ygyno.2016.07.006
Wille, C. K., Nawandar, D. M., Henning, A. N., Ma, S., Oetting, K. M., Lee, D., et al. (2015). 5-hydroxymethylation of the EBV genome regulates the latent to lytic switch. Proc. Natl. Acad. Sci. U.S.A. 112, E7257–E7265. doi: 10.1073/pnas.1513432112
Wu, J., Li, H., Shi, M., Zhu, Y., Ma, Y., Zhong, Y., et al. (2019). TET1-mediated DNA hydroxymethylation activates inhibitors of the Wnt/beta-catenin signaling pathway to suppress EMT in pancreatic tumor cells. J. Exp. Clin. Cancer Res. 38:348. doi: 10.1186/s13046-019-1334-5
Wu, Z., and Zhao, P. (2018). Epigenetic alterations in anesthesia-induced neurotoxicity in the developing brain. Front. Physiol. 9:1024. doi: 10.3389/fphys.2018.01024
Xie, N., Zhou, Y., Sun, Q., and Tang, B. (2018). Novel epigenetic techniques provided by the CRISPR/Cas9 system. Stem. Cells Int. 2018:7834175. doi: 10.1155/2018/7834175
Xu, J., Xu, L., Yang, B., Wang, L., Lin, X., and Tu, H. (2015). Assessing methylation status of PAX1 in cervical scrapings, as a novel diagnostic and predictive biomarker, was closely related to screen cervical cancer. Int. J. Clin. Exp. Pathol. 8, 1674–1681.
Xu, L., Xu, J., Hu, Z., Yang, B., Wang, L., Lin, X., et al. (2018). Quantitative DNA methylation analysis of paired box gene 1 and LIM homeobox transcription factor 1 alpha genes in cervical cancer. Oncol. Lett. 15, 4477–4484. doi: 10.3892/ol.2018.7872
Yadav, P., Masroor, M., Nandi, K., Kaza, R. C. M., Jain, S. K., Khurana, N., et al. (2018). Promoter methylation of BRCA1, DAPK1 and RASSF1A is associated with increased mortality among indian women with breast cancer. Asian. Pac. J. Cancer Prev. 19, 443–448. doi: 10.22034/apjcp.2018.19.2.443
Yeon, M., Byun, J., Kim, H., Kim, M., Jung, H. S., Jeon, D., et al. (2018). CAGE binds to beclin1, regulates autophagic flux and CAGE-derived peptide confers sensitivity to anti-cancer drugs in non-small cell lung cancer cells. Front. Oncol. 8:599. doi: 10.3389/fonc.2018.00599
Yin, F. F., Wang, N., Bi, X. N., Yu, X., Xu, X. H., Wang, Y. L., et al. (2016). Serine/threonine kinases 31(STK31) may be a novel cellular target gene for the HPV16 oncogene E7 with potential as a DNA hypomethylation biomarker in cervical cancer. Virol. J. 13:60. doi: 10.1186/s12985-016-0515-5
Yuan, A. L., Ricks, C. B., Bohm, A. K., Lun, X., Maxwell, L., Safdar, S., et al. (2018). ABT-888 restores sensitivity in temozolomide resistant glioma cells and xenografts. PLoS One 13:e0202860. doi: 10.1371/journal.pone.0202860
Zhang, L. Y., Han, C. S., Li, P. L., and Zhang, X. C. (2016). 5-Hydroxymethylcytosine expression is associated with poor survival in cervical squamous cell carcinoma. Jpn. J. Clin. Oncol. 46, 427–434. doi: 10.1093/jjco/hyw002
Zhou, H., Wang, Y., Lv, Q., Zhang, J., Wang, Q., Gao, F., et al. (2016). Overexpression of ribosomal RNA in the development of human cervical cancer is associated with rDNA promoter hypomethylation. PLoS One 11:e0163340. doi: 10.1371/journal.pone.0163340
Zhu, H., Chen, A., Li, S., Tao, X., Sheng, B., Chetry, M., et al. (2017). Predictive role of galectin-1 and integrin alpha5beta1 in cisplatin-based neoadjuvant chemotherapy of bulky squamous cervical cancer. Biosci. Rep. 37:BSR20170958. doi: 10.1042/bsr20170958
Zhu, Y., Lu, H., Zhang, D., Li, M., Sun, X., Wan, L., et al. (2018). Integrated analyses of multi-omics reveal global patterns of methylation and hydroxymethylation and screen the tumor suppressive roles of HADHB in colorectal cancer. Clin. Epigenetics 10:30. doi: 10.1186/s13148-018-0458-3
Keywords: DNA methylation, DNA hydroxymethylation, cervical cancer, gene, diagnosis, prognosis, treatment
Citation: Zhu H, Zhu H, Tian M, Wang D, He J and Xu T (2020) DNA Methylation and Hydroxymethylation in Cervical Cancer: Diagnosis, Prognosis and Treatment. Front. Genet. 11:347. doi: 10.3389/fgene.2020.00347
Received: 30 December 2019; Accepted: 23 March 2020;
Published: 09 April 2020.
Edited by:
Rui Henrique, Portuguese Oncology Institute, PortugalCopyright © 2020 Zhu, Zhu, Tian, Wang, He and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianmin Xu, eHV0aWFubWluQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.