
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 29 April 2020
Sec. Epigenomics and Epigenetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00339
This article is part of the Research Topic Exploring the Non-coding Genome as the Next Leap in Genetics View all 10 articles
The transcription factor p63 is an essential regulator of vertebrate ectoderm development, including epidermis, limbs, and craniofacial tissues. Here, we have investigated the evolutionary conservation of p63 binding sites (BSs) between zebrafish and human. First, we have analyzed sequence conservation of p63 BSs by comparing ChIP-seq data from human keratinocytes and zebrafish embryos, observing a very poor conservation. Next, we compared the gene regulatory network orchestrated by p63 in both species and found a high overlap between them, suggesting a high degree of functional conservation during evolution despite sequence divergence and the large evolutionary distance. Finally, we used transgenic reporter assays in zebrafish embryos to functionally validate a set of equivalent p63 BSs from zebrafish and human located close to genes involved in epidermal development. Reporter expression was driven by human and zebrafish BSs to many common tissues related to p63 expression domains. Therefore, we conclude that the gene regulatory network controlled by p63 is highly conserved across vertebrates despite the fact that p63-bound regulatory elements show high divergence.
Mutations affecting the activity of cis-regulatory sequences are thought to be the most prevalent cause of phenotypic divergence in animal evolution (Carroll, 2008; Stern and Orgogozo, 2008). Given that promoter sequences are bound by a collection of highly conserved and widely used transcriptional regulators, they do not seem to be the main driver of cis-regulatory divergence (Brown and Feder, 2005). Indeed, enhancers are usually more variable between species, and they are the type of cis-regulatory element (CRE) thought to be accountable for cis-regulatory divergence (Wray, 2007). Thus, enhancers are potential targets for evolutionary change since they modulate target gene expression in specific tissue contexts and ordinarily exist in groups of redundant elements, making easy the accumulation of genetic modifications by dampening the risk of lethality (Levine, 2010; Wittkopp and Kalay, 2011).
Alterations in CRE sequences will affect cis-regulatory activity through the modification of transcription factor (TF) binding sites (BSs) (Wilson et al., 2008; Zheng et al., 2010) since they are the functional units of cis-regulatory sequences. Possible changes in TF binding are gains or losses of BSs or changes in the affinity of the site (Borneman et al., 2007; Odom et al., 2007; Tuch et al., 2008; Schmidt et al., 2010), although the latter possibility could have a minor contribution since TF binding specificities are highly conserved (Nitta et al., 2015).
p63 is a TF of the p53 family and an essential regulator of ectoderm development, including epidermis, limbs, and craniofacial tissues (Soares and Zhou, 2018). Heterozygous mutations in the TP63 human gene are associated with several hereditary malformations showing ectrodactyly (split hand or foot malformations), ectodermal dysplasia, and orofacial cleft as their principal phenotypes (Celli et al., 1999; Rinne et al., 2006). Null or knockdown animal models exhibit severe limb truncations, as well as absence of skin and derived tissues (Yang et al., 1998; Mills et al., 1999; Bakkers et al., 2002; Lee and Kimelman, 2002; Santos-Pereira et al., 2019).
In this work, we have explored the evolutionary conservation of p63 BSs between zebrafish and human. For this, we have first analyzed the sequence conservation of p63 BSs of human keratinocytes and zebrafish embryos, showing a very poor conservation; however, about one third of the BS-associated genes are orthologous between these two species, suggesting that the p63 gene regulatory network is conserved. Indeed, we validated this functional conservation using transgenic reporter assays in zebrafish and found that diverged enhancers with species-specific p63 binding from both species drove similar expression patterns. Altogether, our data suggest that the gene regulatory network regulated by p63 is conserved across vertebrates and despite the fact that CRE sequences diverged along species evolution.
All experiments involving animals conform to the national and European Community standards for the use of animals in experimentation and were approved by the Ethical Committees from the University Pablo de Olavide, Consejo Superior de Investigaciones Científicas (CSIC), and the Andalusian government.
Wild-type fishes were crossed for 10 min roughly since first eggs laid to obtain synchronous embryos. The eggs were collected in petri dishes with E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 0.1% methylene blue). Wild-type strains for zebrafish were AB/Tübingen (AB/Tu) and were maintained and bred under standard conditions (Westerfield, 2007). Embryo stages were expressed in hours post-fertilization (hpf) as described (Kimmel et al., 1995).
For enhancer cloning, zebrafish and human genomic fragments containing the studied CREs were amplified with iMAX-II DNA Polymerase (Intron Biotechnology) using primers from Supplementary Table S1. The PCR fragments were purified using Isolate II PCR and Gel Kit (BIOLINE), sub-cloned in the pCR8/GW/TOPO vector (Invitrogen) and then transferred, through recombination using Gateway LR technology (Invitrogen), to the enhancer detection vector for zebrafish transgenesis, containing the strong midbrain enhancer z48 and the green fluorescent protein (GFP) reporter gene under the control of the gata2 minimal promoter (Gehrke et al., 2015).
Zebrafish transgenic embryos were generated using the Tol2 method (Kawakami et al., 2004). One-cell-stage embryos were injected with 3–5 nl of a solution containing 30 ng/μl of Tol2 mRNA, 20 ng/μl of phenol:chloroform-purified enhancer detection vector, and 0.05% of phenol red solution. Injected embryos (F0) were selected for GFP expression at 48 hpf, raised to sexual maturity and screened for germline transmission. GFP-expressing F1 embryos were photographed at 24 and 48 hpf stages with a digital CCD camera (MagnaWre, Optronix) mounted on an MZ-12 dissecting scope (Leica). Three independent stable transgenic lines showing similar GFP expression patterns were generated.
ChIP-seq data of human keratinocytes (Kouwenhoven et al., 2010) and ChIPmentation data of 24-hpf zebrafish embryos (Santos-Pereira et al., 2019) were obtained from datasets GSE17611 and GSE123057, respectively. Reads were aligned to the GRCh37/hg19 human genome assembly or the GRCz10/danRer10 zebrafish genome assembly using Bowtie (Langmead et al., 2009). Peaks were called using MACS2 algorithm (Zhang et al., 2008) with a false discovery rate (FDR) < 0.001, and common peaks to all biological replicates, calculated with the Intersect tool from the Bedtools package (Quinlan and Hall, 2010), were considered as high-confidence peaks for each species. The University of California–Sta. Cruz (UCSC) Genome Browser was used to visualize ChIP-seq data (Casper et al., 2018). TF motif enrichment was calculated using the script FindMotifsGenome. pl from Homer software (Heinz et al., 2010) with standard parameters.
For gene assignment to ChIP-seq peaks, zebrafish coordinates were converted to Zv9/danRer7 genome using the Liftover tool of the UCSC Genome Browser (Casper et al., 2018). Both zebrafish and human peaks were assigned to genes using the GREAT tool (Hiller et al., 2013), with the basal plus extension association rule with standard parameters (5 Kb upstream, 1 Kb downstream, 1 Mb maximum extension). Ensembl BioMart tool (Zerbino et al., 2018), release 75 for GRCh37/hg19 and release 91 for GRCz10/danRer10, was used to obtain the orthologous genes in human and zebrafish. The significance of the overlaps between lists of orthologous genes was assessed using the hypergeometric test and the number of total orthologous genes for each species as reference. PANTHER (Mi et al., 2017) was used to calculate Gene Ontology (GO) terms overrepresented. For sequence conservation analyses, genomic coordinates of the p63 BSs in human were converted to zebrafish coordinates, and vice versa, using the Liftover tool from the UCSC Genome Browser (Casper et al., 2018).
p63 is a conserved master regulator of vertebrate ectoderm development. Thus, we wondered whether its BSs were also conserved. To address this question, we took advantage of published ChIP-seq data of p63 in human keratinocytes (Kouwenhoven et al., 2010) and compared them with our ChIP-seq data in zebrafish 24-hpf embryos (Santos-Pereira et al., 2019). Peak calling identified 11,466 high confidence p63 BSs in human keratinocytes and 10,520 in zebrafish embryos. De novo motif discovery analysis identified a p53-like binding sequence that was virtually the same in both species despite their high evolutionary divergence (Figure 1A). These motifs represent indeed incomplete versions of the p63 motif described for human keratinocytes (Kouwenhoven et al., 2010), which is also enriched in the p63 peaks called in both species (Figure 1A).
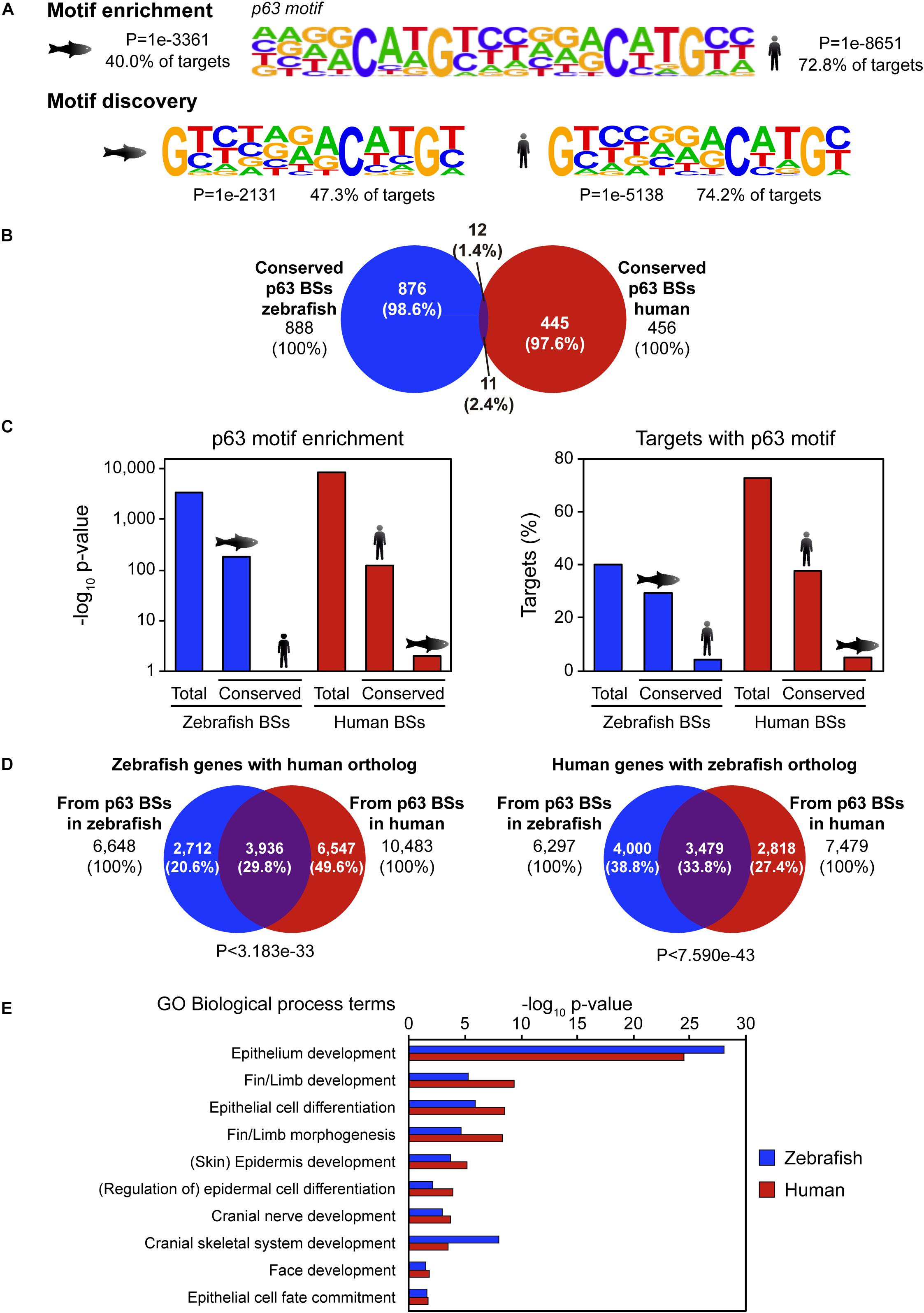
Figure 1. Sequence divergence but functional conservation of p63 binding between zebrafish and human. (A) p63 motif logo found by either motif enrichment (top) or de novo motif discovery (bottom) in p63 ChIP-seq from zebrafish embryos (left) and human keratinocytes (right). Enrichment p-value and percentage of targets with the motif are represented. (B) Venn diagram showing the overlap between p63 binding sites (BSs) in zebrafish and human conserved at the sequence level. (C) p63 motif enrichment (left) and percentage of targets with p63 motif (right) for total and conserved p63 BSs in zebrafish and human. (D) Venn diagrams showing the overlap between zebrafish p63-associated genes with human ortholog (left) or human p63-associated genes with zebrafish ortholog (right) from BSs in both species. P-values for overlap significance using the Hypergeometric test are shown. (E) Gene Ontology (GO) biological process term enrichment for zebrafish and human p63 BSs. Terms related to p63 known functions were selected. Total GO terms are shown in Supplementary Dataset.
We hypothesized that the main CREs involved in p63 regulatory network may be highly conserved at the sequence level. In order to check this possibility, we analyzed sequence conservation between p63 BSs in zebrafish and human and found 888 BSs in zebrafish and 456 BSs in human keratinocytes falling into conserved regions (8.44 and 3.98% of total p63 BSs, respectively; Figure 1B). Next, we analyzed which of those conserved BSs corresponded to p63 BSs in the other species. For this, we intersected the 456 conserved human BSs with the 888 conserved zebrafish p63 BSs, getting as few as 11 human and 12 zebrafish BSs (2.4 and 1.4% of total, respectively; Figure 1B). Although human data comes from cultured keratinocytes and zebrafish samples come from whole embryos, the embryonic epidermis represents a large proportion of the whole embryo, and therefore the results obtained in both systems should be, at some extent, comparable. Taking this into account, these results indicate that most p63 BSs do not have sequence conservation between zebrafish and human.
In order to assess whether the absence of protein binding to conserved BSs in both species was due to a loss of affinity of p63 for these BSs, we analyzed the enrichment of the human and zebrafish p63 motifs (Figure 1A) in these sites. We observed that the p63 motif was highly enriched in the zebrafish conserved BSs, but not in their human orthologous regions. Conversely, the p63 motif was highly enriched in the human conserved BSs, but not in their zebrafish counterparts (Figure 1C). This result indicates that, despite the high degree of conservation of this subset of p63 BSs between the two species, these sequences have diverged enough to lose the p63 binding motif.
Given the absence of sequence conservation of p63 BSs, we wondered whether the gene regulatory network controlled by p63 was conserved even though the CREs’ sequence have diverged between species. For this, we used the GREAT tool to assign putatively regulated genes to the p63 BSs of each species. Thus, the 10,520 p63 BSs in zebrafish were associated with 7,547 genes, while the 11,466 p63 BSs in human were associated with 8,738 genes. In order to compare genes in both species, we selected the BS-associated genes showing at least one ortholog in the other species. This analysis was performed in both directions, i.e., starting either from zebrafish or human BS-associated genes. We found 6,648 zebrafish genes associated with zebrafish p63 BSs that corresponded to 6,297 human orthologs. On the other hand, we found 7,479 human genes associated with keratinocyte p63 BSs that corresponded to 10,483 zebrafish genes (note that the zebrafish genome shows more than one ortholog to many human genes due to the additional whole genome duplication of teleost fishes).
Then, we analyzed the overlap between orthologous genes associated with p63 ChIP-seq data in both species and found that 29.8% of zebrafish genes with human orthologs and 33.8% of human genes with zebrafish orthologs overlapped (P < 3.183e-33 and P < 7.590e-43, respectively) (Figure 1D). In other words, one third of the p63 target genes were conserved between both species. Next, by means of GO term enrichment analyses, we checked whether the biological functions of these conserved genes were the same in both species. These GO analyses showed many biological processes related to p63 functions, among other developmental functions, associated with the conserved genes. These included “skin/epidermis development,” “fin/limb development,” or “face development” (Figure 1E and Supplementary Dataset). However, these p63-related GO terms were absent for the species-specific genes, with the exception of “skin development” for the human orthologs of zebrafish-specific genes (FDR < 1.13e-3). In these species-specific genes, GO terms related to metabolic functions and olfactory perception were among the most enriched ones (Supplementary Dataset). Altogether, our data show that the p63 gene regulatory network is conserved across vertebrates despite the sequence divergence observed at the CRE level.
The observation that p63 BSs have diverged between zebrafish and human at the sequence level, but their associated genetic network is highly conserved, prompted us to seek for non-conserved p63 BSs driving similar expression patterns of orthologous genes in zebrafish and human. For this, we selected a set of six pairs of p63 BSs from both species that were putatively targeting the same epidermal genes and located in equivalent positions in both genomes: upstream of the dlx3b/DLX3, grhl1/GRHL1, and myh9a/MYH9 zebrafish/human genes, and in the introns of st14a/ST14, lama5/LAMA5, and map2k1/MAP2K1. Then, these zebrafish and human sequences were cloned into an enhancer detection vector composed of a gata2 minimal promoter, an enhanced GFP reporter gene, and the strong midbrain enhancer z48 (Gehrke et al., 2015). Thus, after screening for F0 founder individuals, the expression patterns driven by these CREs could be followed by monitoring GFP expression in F1 embryos.
We analyzed the expression driven by these CREs in 24 and 48 hpf zebrafish F1 embryos and found partial to total overlapping tissues for all the CRE couples (Figure 2A). Interestingly, we found that the p63 BSs located upstream to the myh9a/MYH9 genes drove GFP expression to typical p63-expressing tissues in both species, including the skin, the yolk syncytial layer (YSL), and the pectoral fin bud (Figures 2B,C). This pattern is highly consistent with the expression of the zebrafish myh9a gene in epidermis, periderm, and pectoral fin bud (Huang et al., 2013). For other CREs, we found overlapping GFP expression in additional p63-expressing tissues, such as the pharyngeal arches, but also in neural tissue, such as hindbrain and neural tube, sensory organs, such as the eyes and olfactory and otic vesicles, and even in mesodermal lineages, such as the inner cellular mass, pronephros, somites, and notochord for specific cases (Supplementary Figure S1).
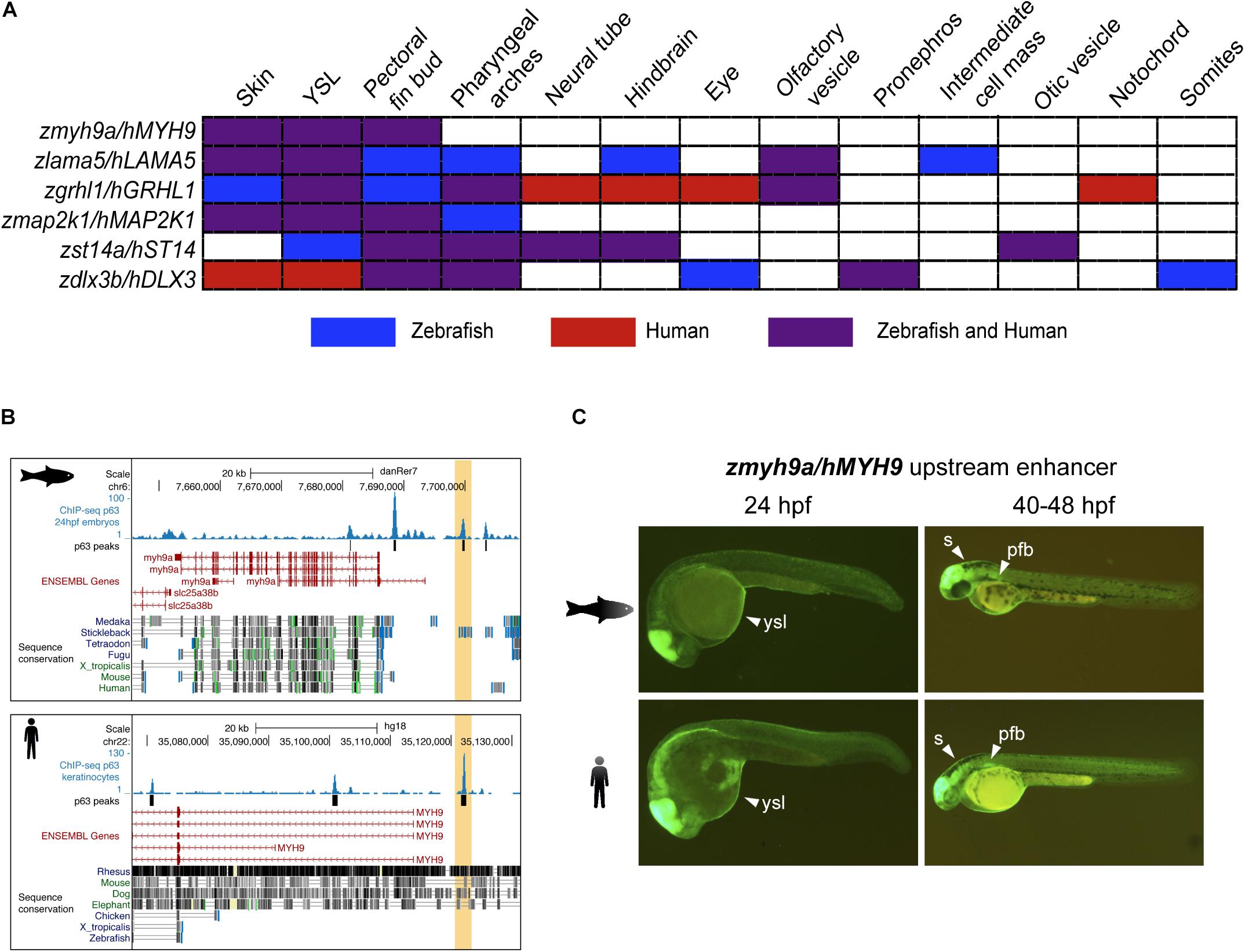
Figure 2. Zebrafish and human p63 binding sites (BSs) drive similar expression patterns. (A) Table showing the overlap in expression patterns driven by cis-regulatory element (CRE) pairs from zebrafish and human, as detected in zebrafish transgenic reporter assays. Note that all BS pairs share at least three expression domains in both species. (B) Genome tracks from zebrafish (top) and human (bottom) showing p63 ChIP-seq signal, p63 peaks, ENSEMBL genes, and vertebrate sequence conservation. The selected p63 BSs for transgenic reporter assays, which are located upstream to the myh9a/MYH9 genes, are highlighted in light orange. (C) Transgenic zebrafish embryos at different stages showing the green fluorescent protein (GFP) expression pattern driven by the enhancers highlighted in (B). Tissues expressing GFP are pointed using white arrowheads. s, skin; ysl, yolk syncytial layer; pfb, pectoral fin bud.
The expression patterns driven by these other p63 BSs were also totally or partially consistent with the expression of the zebrafish genes that are putatively controlled by them. For instance, the lama5 gene has been described to be expressed in the epidermis, apical ectodermal ridges at the pectoral fin buds, lens, midbrain–hindbrain boundary, and pronephros1, consistent with expression driven by the analyzed p63 BSs in skin, YSL, pectoral fin buds, and hindbrain. Also, the gene grhl1 is expressed in the epidermis, periderm, enveloping layer (EVL), olfactory placodes, otic placodes and vesicle, pharyngeal arches, and pronephros (Jänicke et al., 2010), while the analyzed p63 BSs drove reporter expression to the skin, YSL, pectoral fin buds, pharyngeal arches, and olfactory vesicle, as well as the hindbrain, neural tube, eyes, and notochord specifically for the human BS. The gene dlx3b is expressed in the pectoral fin bud, median fin fold, otic vesicle, and pharyngeal arches1, while p63 BSs from both species lead to consistent expression in the pectoral fin buds, pharyngeal arches, and pronephros. For the gene st14a, little overlap is found between the gene expression patterns (Carney et al., 2007) and those driven by the analyzed p63 BSs, although the overlap between BSs from both species is consistent. Finally, no previous information of expression patterns is available for the map2k1 gene.
Altogether, these data suggest that the expression of p63 target genes has been conserved in vertebrates through a rewiring of the enhancers containing p63 BSs.
In this work, we have studied the sequence and functional conservation of genomic BSs of the epidermal TF p63 between human and zebrafish. We observe that most p63 BSs in one species are not conserved at the sequence level in the other species, and that the conserved BSs do not reproduce p63 binding nor p63 motif enrichment in the other species (Figure 1). However, there is a high degree of overlap of the ortholog genes associated with p63 BSs (Figure 1), suggesting conservation of the gene regulatory network across vertebrates despite the large evolutionary distance and the observed sequence divergence. Finally, we have functionally validated this conservation by transgenic reporter assays in zebrafish embryos showing overlapping expression patterns driven by selected BSs from both species associated with common epidermal genes (Figure 2 and Supplementary Figure S1).
Previous studies using transgenesis assays with human and zebrafish developmental enhancers (Fisher et al., 2006; Rada-Iglesias et al., 2011; Taher et al., 2011) or with human and mouse heart enhancers (Blow et al., 2010; May et al., 2012) showed a high degree of functional conservation despite sequence divergence, which is in agreement with our findings. In addition, a computational comparative analysis of p63 BSs in human and mouse keratinocytes found a higher sequence conservation between both species since they diverged more recently, although many of the conserved BSs lost p63 binding and p63 motif enrichment in the other species (Sethi et al., 2017), suggesting an evolutionary rewiring of the p63-associated CREs that is also consistent with our results. In this study, they also found that common p63 BSs putatively control a core network of epithelial genes, while species-specific genes are enriched for metabolic functions (Sethi et al., 2017). Some authors suggest that TF BS turnover is a mechanism that could explain this phenomenon, i.e., the loss of a TF BS can be compensated by the gain of a new BS for the same TF. This can occur directly by changes in the genome sequence involving the lost or gained BSs (Schmidt et al., 2010; Xie et al., 2010) or by insertions of complete cis-regulatory modules by transposable elements (Kunarso et al., 2010; Xie et al., 2010). In any case, we propose that this could be the case of p63 BSs, for which BS losses may have been compensated by BS gains close to the same target genes, leading to divergence of the original sequence due to the absence of selective pressure, but maintaining the control over a common subset of developmental genes.
Publicly available datasets were analyzed in this study. This data can be found here: GSE17611 and GSE123057.
The animal study was reviewed and approved by Comité de Ética de la Universidad Pablo de Olavide (Seville).
JT and JS-P conceived and designed the project. LG-F performed the experiments. LG-F, JS-P, and JT analyzed the data and wrote the manuscript.
This work was supported by a grant from the Spanish Ministry of Economy and Competitiveness to JT (BFU2014-58449-JIN) and an ERC-Advanced grant from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (Grant Agreement No. 740041). JS-P was funded by a Juan de la Cierva-Incorporación fellow from the Spanish Ministry of Economy and Competitiveness (IJCI-2016-29884) and LG-F by a grant from La Marató TV3 (Grant No. 201611).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank J. L. Gómez-Skarmeta for critical reading of the manuscript and the Centro Andaluz de Biología del Desarrollo (CABD) Fish Facility for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00339/full#supplementary-material
Bakkers, J., Hild, M., Kramer, C., Furutani-Seiki, M., and Hammerschmidt, M. (2002). Zebrafish ΔNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell 2, 617–627. doi: 10.1016/S1534-5807(02)00163-166
Blow, M. J., McCulley, D. J., Li, Z., Zhang, T., Akiyama, J. A., Holt, A., et al. (2010). ChIP-seq identification of weakly conserved heart enhancers. Nat. Genet. 42, 806–812. doi: 10.1038/ng.650
Borneman, A. R., Gianoulis, T. A., Zhang, Z. D., Yu, H., Rozowsky, J., Seringhaus, M. R., et al. (2007). Divergence of transcription factor binding sites across related yeast species. Science 317, 815–819. doi: 10.1126/science.1140748
Brown, R. P., and Feder, M. E. (2005). Reverse transcriptional profiling: non-correspondence of transcript level variation and proximal promoter polymorphism. BMC Genomics 6:110. doi: 10.1186/1471-2164-6-110
Carney, T. J., von der Hardt, S., Sonntag, C., Amsterdam, A., Topczewski, J., Hopkins, N., et al. (2007). Inactivation of serine protease Matriptase1a by its inhibitor Hai1 is required for epithelial integrity of the zebrafish epidermis. Development 134, 3461–3471. doi: 10.1242/dev.004556
Carroll, S. B. (2008). Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36. doi: 10.1016/j.cell.2008.06.030
Casper, J., Zweig, A. S., Villarreal, C., Tyner, C., Speir, M. L., Rosenbloom, K. R., et al. (2018). The UCSC genome browser database: 2018 update. Nucleic Acids Res. 46, D762–D769. doi: 10.1093/nar/gkx1020
Celli, J., Duijf, P., Hamel, B. C. J., Bamshad, M., Kramer, B., Smits, A. P. T., et al. (1999). Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99, 143–153. doi: 10.1016/S0092-8674(00)81646-81643
Fisher, S., Grice, E. A., Vinton, R. M., Bessling, S. L., and McCallion, A. S. (2006). Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312, 276–279. doi: 10.1126/science.1124070
Gehrke, A. R., Schneider, I., de la Calle-Mustienes, E., Tena, J. J., Gomez-Marin, C., Chandran, M., et al. (2015). Deep conservation of wrist and digit enhancers in fish. Proc. Natl. Acad. Sci. U.S.A 112, 803–808. doi: 10.1073/pnas.1420208112
Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y. C., Laslo, P., et al. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. doi: 10.1016/j.molcel.2010.05.004
Hiller, M., Agarwal, S., Notwell, J. H., Parikh, R., Guturu, H., Wenger, A. M., et al. (2013). Computational methods to detect conserved non-genic elements in phylogenetically isolated genomes: Application to zebrafish. Nucleic Acids Res. 41:e151. doi: 10.1093/nar/gkt557
Huang, Y., Wang, X., Wang, X., Xu, M., Liu, M., and Liu, D. (2013). Nonmuscle myosin II-B (myh10) expression analysis during zebrafish embryonic development. Gene Expr. Patterns. 13, 265–270. doi: 10.1016/j.gep.2013.04.005
Jänicke, M., Renisch, B., and Hammerschmidt, M. (2010). Zebrafish grainyhead-like1 is a common marker of different non-keratinocyte epidermal cell lineages, which segregate from each other in a Foxi3-dependent manner. Int. J. Dev. Biol. 54, 837–850. doi: 10.1387/ijdb.092877mj
Kawakami, K., Takeda, H., Kawakami, N., Kobayashi, M., Matsuda, N., and Mishina, M. (2004). A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144. doi: 10.1016/j.devcel.2004.06.005
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kouwenhoven, E. N., va Heeringen, S. J., Tena, J. J., Oti, M., Dutilh, B. E., Alonso, M. E., et al. (2010). Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 shfm1 locus. PLoS Genet. 6:e1001065. doi: 10.1371/journal.pgen.1001065
Kunarso, G., Chia, N. Y., Jeyakani, J., Hwang, C., Lu, X., Chan, Y. S., et al. (2010). Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631–634. doi: 10.1038/ng.600
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lee, H., and Kimelman, D. (2002). A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell 2, 607–616. doi: 10.1016/S1534-5807(02)00166-161
Levine, M. (2010). Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763. doi: 10.1016/j.cub.2010.06.070
May, D., Blow, M. J., Kaplan, T., McCulley, D. J., Jensen, B. C., Akiyama, J. A., et al. (2012). Large-scale discovery of enhancers from human heart tissue. Nat. Genet. 44, 89–93. doi: 10.1038/ng.1006
Mi, H., Huang, X., Muruganujan, A., Tang, H., Mills, C., Kang, D., et al. (2017). PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189. doi: 10.1093/nar/gkw1138
Mills, A. A., Zheng, B., Wang, X. J., Vogel, H., Roop, D. R., and Bradley, A. (1999). P63 is a P53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713. doi: 10.1038/19531
Nitta, K. R., Jolma, A., Yin, Y., Morgunova, E., Kivioja, T., Akhtar, J., et al. (2015). Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife 2015:e04837. doi: 10.7554/eLife.04837
Odom, D. T., Dowell, R. D., Jacobsen, E. S., Gordon, W., Danford, T. W., MacIsaac, K. D., et al. (2007). Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 39, 730–732. doi: 10.1038/ng2047
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Rada-Iglesias, A., Bajpai, R., Swigut, T., Brugmann, S. A., Flynn, R. A., and Wysocka, J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–285. doi: 10.1038/nature09692
Rinne, T., Spadoni, E., Kjaer, K. W., Danesino, C., Larizza, D., Kock, M., et al. (2006). Delineation of the ADULT syndrome phenotype due to arginine 298 mutations of the p63 gene. Eur. J. Hum. Genet. 14, 904–910. doi: 10.1038/sj.ejhg.5201640
Santos-Pereira, J. M., Gallardo-Fuentes, L., Neto, A., Acemel, R. D., and Tena, J. J. (2019). Pioneer and repressive functions of p63 during zebrafish embryonic ectoderm specification. Nat. Commun. 10:3049. doi: 10.1038/s41467-019-11121-z
Schmidt, D., Wilson, M. D., Ballester, B., Schwalie, P. C., Brown, G. D., Marshall, A., et al. (2010). Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040. doi: 10.1126/science.1186176
Sethi, I., Gluck, C., Zhou, H., Buck, M. J., and Sinha, S. (2017). Evolutionary re-wiring of p63 and the epigenomic regulatory landscape in keratinocytes and its potential implications on species-specific gene expression and phenotypes. Nucleic Acids Res. 45, 8208–8224. doi: 10.1093/nar/gkx416
Soares, E., and Zhou, H. (2018). Master regulatory role of p63 in epidermal development and disease. Cell. Mol. Life Sci. 75, 1179–1190. doi: 10.1007/s00018-017-2701-z
Stern, D. L., and Orgogozo, V. (2008). The loci of evolution: How predictable is genetic evolution? Evolution 62, 2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x
Taher, L., McGaughey, D. M., Maragh, S., Aneas, I., Bessling, S. L., Miller, W., et al. (2011). Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res. 21, 1139–1149. doi: 10.1101/gr.119016.110
Tuch, B. B., Galgoczy, D. J., Hernday, A. D., Li, H., and Johnson, A. D. (2008). The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. doi: 10.1371/journal.pbio.0060038
Westerfield, M. (2007). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 5th Edn. Eugene: University of Oregon Press.
Wilson, M. D., Barbosa-Morais, N. L., Schmidt, D., Conboy, C. M., Vanes, L., Tybulewicz, V. L. J., et al. (2008). Species-specific transcription in mice carrying human chromosome 21. Science 322, 434–438. doi: 10.1126/science.1160930
Wittkopp, P. J., and Kalay, G. (2011). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Wray, G. A. (2007). The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216. doi: 10.1038/nrg2063
Xie, D., Chen, C. C., Ptaszek, L. M., Xiao, S., Cao, X., Fang, F., et al. (2010). Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 20, 804–815. doi: 10.1101/gr.100594.109
Yang, A., Kaghad, M., Wang, Y., Gillett, E., Fleming, M. D., Dötsch, V., et al. (1998). P63, a P53 Homolog At 3Q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316. doi: 10.1016/S1097-2765(00)80275-80270
Zerbino, D. R., Achuthan, P., Akanni, W., Amode, M. R., Barrell, D., Bhai, J., et al. (2018). Ensembl 2018. Nucleic Acids Res. 46, D754–D761. doi: 10.1093/nar/gkx1098
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. doi: 10.1186/gb-2008-9-9-r137
Keywords: p63, transcription factor binding sites, functional conservation, sequence divergence, human, zebrafish
Citation: Gallardo-Fuentes L, Santos-Pereira JM and Tena JJ (2020) Functional Conservation of Divergent p63-Bound cis-Regulatory Elements. Front. Genet. 11:339. doi: 10.3389/fgene.2020.00339
Received: 22 December 2019; Accepted: 20 March 2020;
Published: 29 April 2020.
Edited by:
Tahsin Stefan Barakat, Erasmus Medical Center, NetherlandsReviewed by:
Jo Huiqing Zhou, Radboud University Nijmegen, NetherlandsCopyright © 2020 Gallardo-Fuentes, Santos-Pereira and Tena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: José M. Santos-Pereira, am1zYW5wZXIxQHVwby5lcw==; Juan J. Tena, amp0ZW5hZ3VAdXBvLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.