- 1State Key Laboratory of Cell Differentiation and Regulation, Henan Normal University, Xinxiang, China
- 2Henan International Joint Laboratory of Pulmonary Fibrosis, Henan Normal University, Xinxiang, China
- 3Henan Center for Outstanding Overseas Scientists of Pulmonary Fibrosis, Henan Normal University, Xinxiang, China
- 4College of Life Sciences, Henan Normal University, Xinxiang, China
- 5Institute of Biomedical Science, Henan Normal University, Xinxiang, China
- 6Overseas Expertise Introduction Center for Discipline Innovation of Pulmonary Fibrosis (111 Project), Henan Normal University, Xinxiang, China
ZNF143, a human homolog of the transcriptional activator Staf, is a C2H2-type protein consisting of seven zinc finger domains. As a transcription factor (TF), ZNF143 is sequence specifically binding to chromatin and activates the expression of protein-coding and non-coding genes on a genome scale. Although it is ubiquitous expressed, its expression in cancer cells and tissues is usually higher than that in normal cells and tissues. Therefore, abnormal expression of ZNF143 is related to cancer cell survival, proliferation, differentiation, migration, and invasion, suggesting that new small molecules can be designed by targeting ZNF143 as it may be a good potential biomarker and therapeutic target for related cancers. However, the mechanism on how ZNF143 regulates its targeting gene remains unclear. Recently, with the development of chromatin conformation capture (3C) and its derivatives, and high-throughput sequencing technology, new findings have been obtained in the study of ZNF143. Pioneering studies have showed that ZNF143 binds directly to promoters and contributes to chromatin interactions connecting promoters to distal regulatory elements, such as enhancers. Further, it has proved that ZNF143 is involved in CCCTC-binding factor (CTCF) in establishing the conserved chromatin loops by cooperating with cohesin and other partners. These results indicate that ZNF143 is a key loop formation factor. In addition, we report ZNF143 is dynamically bound to chromatin during the cell cycle demonstrated that it is a potential mitotic bookmarking factor. It may be associated with CTCF for mitosis-to-G1 phase transition and chromatin loop re-establishment in early G1 phase. In the future, researchers could further clarify the fine mechanism of ZNF143 in mediating chromatin loops with the help of CUT&RUN (CUT&Tag) and Cut-C technology. Thus, in this review, we summarize the research progress of TF ZNF143 in detail and also predict the potential functions of ZNF143 in cell fate and identity based on our recent discoveries.
Introduction
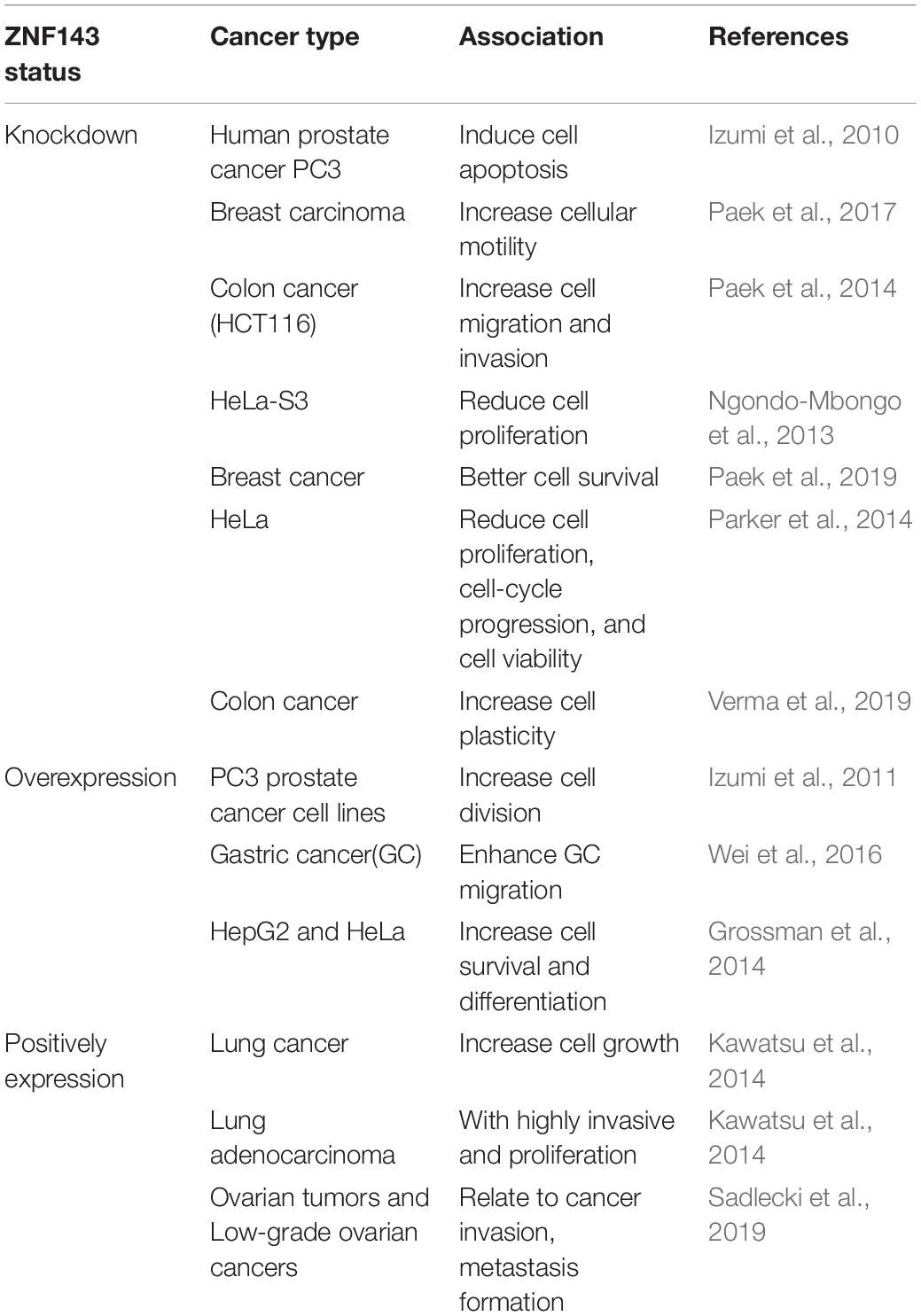
Schuster et al. (1995) found a transcription factor (TF), which can be bound specifically to the promoter of selenocysteine tRNA in Xenopus oocytes and named it Staf (selenocysteine tRNA gene transcription activating factor). In the same year, Tommerup and Vissing (1995) reported zinc finger protein 143 (ZNF143), a human homolog of the transcriptional activator Staf, was located on the human 11th chromosome, 11p15.3–15.4. Subsequently, Adachi et al. (1998) isolated and characterized m-Staf from mouse mammary gland, which is consistent with human ZNF143. ZNF143 is a member of the Kruppel family and is a widely expressed transcriptional activation factor that regulates gene expression associated with cell cycle and DNA replication (Izumi et al., 2010). Therefore, it is widely involved in a variety of cellular and pathogenic processes, such as cell survival, growth, proliferation, etc. (Table 1). However, the molecular mechanism of ZNF143 in regulating gene expression remains elusive.
In recent years, studies have revealed that ZNF143 not only exists in most cancer cells but is also necessary for the normal development of tissues (Izumi et al., 2011; Halbig et al., 2012; Kawatsu et al., 2014; Paek et al., 2014; Wei et al., 2016; Paek et al., 2017). Genome-wide analyses have shown that TF ZNF143 with sequence binding specificity is usually bound to the promoter of its regulatory gene and promotes the formation of chromatin loop by interacting with other chromatin structure and organization factors, such as CCCTC-binding factor (CTCF) and cohesin (Heidari et al., 2014; Bailey et al., 2015; Ye et al., 2016; Yang et al., 2017; Mourad and Cuvier, 2018; Wen et al., 2018). In summary, as a key TF, ZNF143 plays a critical role in chromatin loop formation and gene regulation (Table 2), illustrating great importance in the study of its regulatory mechanism.
The Structural Features of ZNF143
The amino acid sequence of human ZNF143 is highly homologous to both m-Staf and Staf. Among its sequence, 97.1 and 84% residues are identical to those of m-Staf and Staf, respectively (Schuster et al., 1995; Adachi et al., 1998; Myslinski et al., 1998). Structurally, these proteins consist of three regions (A, B, and C) (Figure 1). Analysis of the three regions indicates that the central region B (residues 220–428 in ZNF143 and m-Staf, residues 267–468 in Staf) encompasses seven tandemly repeated zinc fingers of the C2H2 type, is highly basic, while the regions A (residues 1–219 in ZNF143 and m-Staf, residues 1–266 in Staf) encodes four repeated motifs and C (residues 429–626 in ZNF143 and m-Staf, residues 469–600 in Staf) are acidic (Figure 1). The central region of seven zinc fingers domain is the DNA binding domain. Outside of the central domain, N-domain (region A) is the activation domain both for mRNA and snRNA, and the characteristic features of this domain of these three proteins are very simlar. The function of C-domain (region C) is unclear (Myslinski et al., 1998). Strikingly, the four repeated motifs can be observed between residues 39 and 135 in region A of ZNF143/m-Staf (residues 84 and 176 in region A of Staf) (Figure 1). Each repeat motif contains 15 amino acids and the distance between them contains 10–12 amino acids (Schuster et al., 1995).
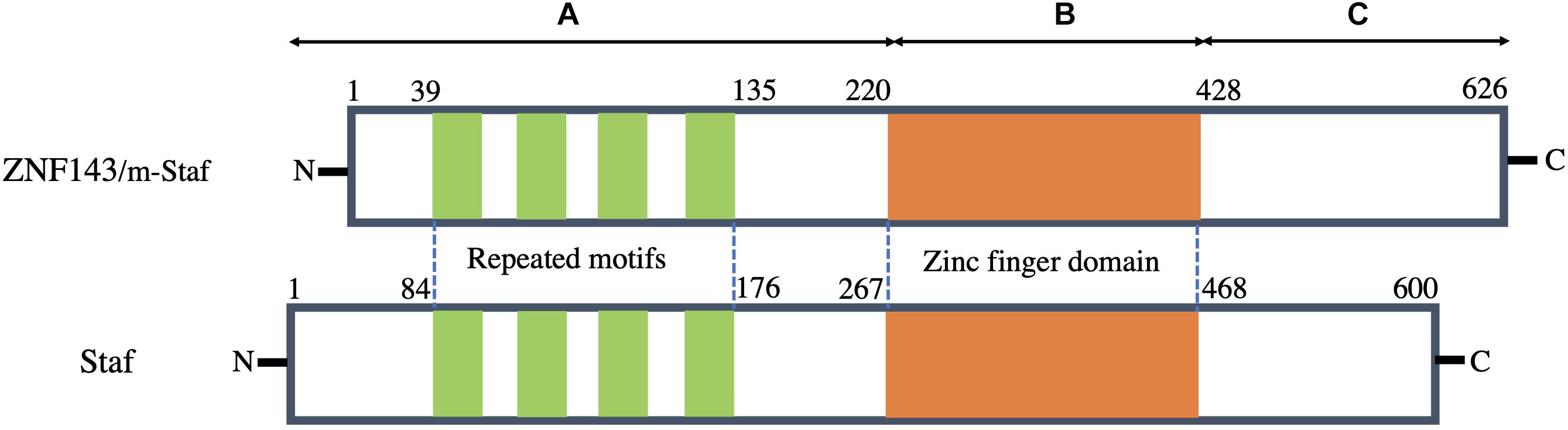
Figure 1. Schematic representation of structural features of human ZNF143 in comparison with m-Staf and Staf. Three regions of these three proteins can be distinguished: A contains mRNA and snRNA activation domains with the presence of the four repeated motifs. B is the central seven zinc finger domain and is therefore a DNA binding domain. C is a region with unknown function.
As a TF, it is noted that the tandemly repeated zinc finger domain (DNA binding domain) and the element of repeated motifs (activation domain) are especially well conserved among these three proteins (Myslinski et al., 1998). It is reported that this TF possesses the capacity to bind over 2000 promoter regions of both mRNA and small nuclear RNA (snRNA) genes (Myslinski et al., 1998, 2006). Recently, Ngondo-Mbongo et al. (2013) have found that ZNF143 has two main DNA binding motifs of high affinity, namely, SBS1(GTTATGGAATTCCCATTATGCACCGCG) and SBS2 (AAACTACAATTCCCATTATGCACCGCG). Both of them are closely related to its specific binding on the chromatin, and thus initiate gene expression and regulation.
The Function of ZNF143
Regulating Cell-Cycle Progression
TF ZNF143 regulates gene expression associated with cell cycle. Many studies utilize knockdown or overexpression methods to evaluate the effect of ZNF143 on cancer cell progression. For example, Izumi et al. (2010) have reported that ZNF143 is associated with cell cycle and cell proliferation, whereas ZNF143 knockdown causes human prostate cancer PC3 cells to stagnate during G2/M and is accompanied with apoptosis. By establishing two forced expression of ZNF143 PC3 cancer cell lines, they found that overexpress genes strongly associated with cell cycle and cell division (Izumi et al., 2011). ZNF143 knockdown induces increased breast cancer motility, which indicates that ZNF143 expression contributes to breast cancer progression (Paek et al., 2017). In addition, low ZNF143 expression exhibits better cell survival through an autophagic process by regulating the p53–Beclin1 axis in breast cancer cells (Paek et al., 2019). ZNF143 is essential and sufficient for Skp2 promoter activity and ZNF143 silencing inhibits cell proliferation; however, ectopic ZNF143 can rescue Skp2 expression (Hernandez-Negrete et al., 2011). Overexpression of ZNF143 enhances transaldolase promoter activity in HepG2 and HeLa cells and ZNF143 plays a key role in controlling cell survival and differentiation (Grossman et al., 2014). Simultaneously, other researchers have reported that THAP11/ZNF143/HCF-1 complex is an indispensable component of the transcriptional regulatory network and disruption of this complex leads to reduced cell proliferation, cell-cycle progression, and cell viability (Parker et al., 2014). Ngondo-Mbongo et al. (2013) have also showed that ZNF143, ICN1, and THAP11 play a pivotal role in modulating cell proliferation of rapidly dividing cells. Myslinski et al. (2007) have found that human BUB1B gene mediates the activity of spindle checkpoints to ensure chromosomal stability and euploidy, requires ZNF143 binding.
Regulating Embryonic Development and Maintaining Stem Cell Identity
As a key TF, ZNF143 has a critical function in regulating embryonic development. Halbig et al. (2012) have found that ZNF143 significantly changes zebrafish embryonic phenotypes. Therefore, ZNF143 is necessary for the normal development of zebrafish embryos. The identification and characterization of paralogous genes is also critical for understanding gene function. In the functional study of ZNF143, Huning and Kunkel (2020) have found that znf143a, a novel paralog of znf143, encodes a strong transcriptional activator protein and performs a similar role in the normal development of zebrafish embryos but expressed at a different level during early development. In mouse embryonic stem (ES) cells, ZNF143 regulates Nanog by regulating the binding of Oct4, and ZNF143 is also critical for maintaining human ES cell identity (Chen et al., 2008).
Potential Drug Design Target
TF ZNF143 is a potential drug design target to treat solid cancers. After cisplatin treatment, the binding activity of ZNF143 and MRP S11 significantly increases. This indicates that ZNF143 is involved in response to DNA damage (Ishiguchi et al., 2004; Torigoe et al., 2005; Wakasugi et al., 2007). P73 promotes ZNF143 binding with cisplatin-modified DNA, indicating that ZNF143 can regulate the transcription of DNA repair genes (Wakasugi et al., 2007). ZNF143 can also mediate cell survival by upregulating glutathione peroxidase (GPX1) activity. Thus, ZNF143 interference can increase drug sensitivity to cisplatin treatment of mitochondrial dysfunction (Lu et al., 2012). GAIP-interacting protein, C-terminus (GIPC) induces ZNF143 expression by participating in IGF-1 signal transduction to regulate reactive oxygen products (Paek and You, 2011). ZNF143 is also involved in the migration and invasion of colon cancer cells through a ZEB1-E- cadherin-linked pathway (Paek et al., 2014). The expression levels of ZNF143 and IL-8 are inversely correlated with three-dimensionally grown spheroids and colon cancer tissues (Verma et al., 2019). ZNF143 is accompanied with an increase in MIB-1 index in patients with lung adenocarcinoma, leading to high cell proliferation activity and poor prognostic treatment (Kawatsu et al., 2014). Wei et al. (2016) have found that ZNF143 expression can enhance the metastasis of gastric cancer cells, indicating that ZNF143 can be a drug target for the treatment of gastric cancer. The reduction in ZNF143 expression eventually leads to the cobaltamine transport protein not effectively transporting cobalamin (Pupavac et al., 2016). The expression patterns of ZNF143 and ZNF281 in serous borderline ovarian tumors (SBOTs) and low-grade epithelial ovarian carcinomas (EOCs) play a key role in cancer invasion, metastasis formation, and chemotherapy resistance (Sadlecki et al., 2019). ZNF143 is an upstream regulator to increase the expression of the RNA binding protein TARBP2 in breast and lung cancers (Fish et al., 2019). Thus, how to effectively design small molecule drugs to target ZNF143 is imminent. Fortunately, Haibara et al. (2017) have found that new small molecules YPC-21661 and YPC-22026 can reduce the expression of their target genes RAD51, PLK1, and Survivin by inhibiting the binding of ZNF143 to their promoters. In the future, it is believed that more and more molecule drugs will be exploited by targeting ZNF143 to treat related cancers.
ZNF143 Regulates Gene Expression and Its Mechanism
ZNF143 Participates in the Regulation of Coding and Non-coding Genes
As an important TF, ZNF143 regulates the expression of various genes. During transcription activation, Schuster et al. (1998) have found that ZNF143 activation domains bound by mRNA and snRNA are different. Myslinski et al. (1998) first have found ZNF143 can activate the transcription from RNA polymerase II TATA box-containing mRNA promoters. For example, Kubota et al. (2000) have reported that ZNF143 is the key TF upregulating the molecular chaperone coding gene Cctα transcription through binding with the two activation elements (CAE1 and CAE2). Mach et al. (2002) have also showed that ZNF143 stimulates transcription of the human interferon regulatory factor-3 (IRF-3) gene by binding to SphI postoctamer homology (SPH) elements in vitro and in transfected cells. ZNF143 plays an important role in the transcription of neuronal nitric-oxide synthase (nNOS) exon 1, the mutation of the binding site of ZNF143 leads to a significant reduction in the activity of this exon (Saur et al., 2002). Barski et al. (2004) use ChIP as well as deletion/mutation analysis reveal that the aldehyde reductase is significantly enhanced by transcription activation after binding to ZNF143. Di Leva et al. (2004) have found that ZNF143, together with CAAT factors, regulates human synaptobrevin-like 1 (SYP-like 1) through binding to the SYBL1 promoter in HeLa cells. Gerard et al. (2007) have reported that ZNF143 binds to the promoter of mitochondrial TF A (Tfam) to regulate transcription initiation and replication of mitochondrial DNA in consistent with Sp1, NRF-1, and NRF-2. ZNF143 binds with the −305/−107 of the BUB1B promoter to regulate BUB1B expression to maintain chromosomal stability and euploidy (Myslinski et al., 2007). Gonzalez et al. (2017) have reported that ZNF143, specifically binds to the 8-bp sequence (CCCAGCAG), ∼100 bases upstream of the C/EBPα transcription start site (TSS), plays an important role in the expression of C/EBPα in myeloid cells.
ZNF143 acts as a transcription-activated factor under the joint action of RNA polymerase III (Schaub et al., 1997). The snRNA and snRNA-type genes require the binding of ZNF143 during transcription, such as human U4C, U6, Y4, 7SK; mouse U6 RNAs and Xenopus U1b1, U2, U5, MRP. However, the binding of ZNF143 to snRNA occurs on a distal sequence element (DSE) (Schaub et al., 1997). By comparing ZNF143 recognition sequence of human U6 snRNA and selenocysteine tRNA, Schaub et al. have found that there are only 47% consistent in sequences. In the seven zinc fingers of ZNF143 recognition sequence, the first zinc finger is necessary for selenocysteine tRNA promoter identification, whereas U6 snRNA is not. The seventh zinc finger is essential for the binding activity of them. The flexibility binding results in differences in transcription activation mechanisms (Schaub et al., 1999a). U6 snRNA transcription activation requires ZNF143–DNA–Oct-1 complex, whereas selenocysteine tRNA requires ZNF143-DNA complex (Schaub et al., 1999b). Schaub et al. (2000) have found that zinc fingers 3–6 are the minimum zinc finger regions.
Self-Regulation of ZNF143
To maintain stable ZNF143 expression at normal levels, the transcription feedback regulation mechanism is the simplest and most direct means. ZNF143 selectively adjusts reverse expression by using a low affinity binding site (TSS2) located downstream of the TSS. When ZNF143 expression is higher than normal, transcripts containing longer 5′-UTR (few translation products) are produced by TSS2 transcription. In addition, when ZNF143 levels are lower than normal, the canonical TSS1 binding site is used to express transcripts containing shorter 5′-UTR (many translation products). This transcriptional auto-regulatory mechanism regulates ZNF143 expression by the conversion of the TSS switch, which plays an important role in cell proliferation and growth (Ngondo and Carbon, 2004). Given that ZNF143 is closely related to many biological processes, its expression must be strictly regulated. Ngondo et al. have found that ZNF143 transcripts have three different lengths of 3′-UTR, with the longer 3′-UTR isoform containing variable polyadenylation sites, miRNA target sites, or AU-rich element (ARE). Thus, it tends to post-transcriptional regulation. The longest 3′-UTR isoform contains an unstabilizing ARE and is targeted by mir-590-3p. These results emphasize that ZNF143 post-transcriptional regulation depends on the long 3′-UTR isoform (Ngondo and Carbon, 2014).
ZNF143 Is a Chromatin-Looping Factor
Myslinski et al. have predicted the whole genome binding sites of ZNF143 through computer simulation (in silico) and biochemical methods. They speculated that at least 2500 ZNF143-binding sites are distributed in 2000 promoter regions throughout the mammalian genome. Further research has found that the presence of ZNF143-binding site alone can initiate the expression of a luciferase reporter gene, suggesting that ZNF143 itself exhibits the ability to recruit the transcription machinery (Myslinski et al., 2006). Recently, Wang et al. have reported the co-localization of RBPJ/Notch1/ZNF143, in which ZNF143 can bind with 40% of the Notch1 sites, and RBPJ shows high promoter binding preference by embedding in the ZNF143 motifs. These results may indicate a dynamic exchange of RBPJ/Notch1 and ZNF143 complexes through competition in the binding sites (Wang et al., 2011). Ngondo-Mbongo et al. (2013) have revealed that ZNF143, THAP11, and Notch1 regulate the common target genes through the mutually exclusive occupation of overlapping binding sites. Michaud et al. (2013) have found that HCF-1 is bound with 5400 CpG island promoters. HCF-1, ZNF143, and THAP11 exhibit co-localization, with HCF-1 in collaboration with ZNF143 and THAP11 plays an important role in the transcriptional regulation of HeLa cells. Parker et al. have found that HCF-1, as a coregulator of the TF E2F proteins, is not directly collected in the promoter region but is mediated by ZNF143 and THAP11. HCF-1/ZNF143/THAP11 as a complex that occupies specific sites of chromatin co-regulates the expression of cell proliferation genes (Parker et al., 2014). However, how DNA sequences guide the THAP11/ZNF143/HCF-1 complex to chromatin remains in dispute. Vinckevicius et al. (2015) have explicitly proposed that ACTACA, as a joint submotif of ZNF143 and THAP11, guides THAP11 and HCF-1 to ZNF143-occupied loci and emphasized the importance of the position, spacing, and direction relative to the ZNF143 core motif.
TF ZNF143 can interact with other transcriptional regulators in mediating chromatin loop formation. Chromatin interactions between promoters and long-region regulatory elements can determine the expression level of a gene (Fraser, 2006; Fraser and Bickmore, 2007). In recent years, with the development of high-throughput sequencing and chromatin conformation capture technologies (3C, chromatin conformation capture; Hi-C, chromatin conformation capture using high throughput sequencing; ChIA-PET, chromatin interaction analysis by paired-end tag sequencing) (Dekker et al., 2002; Fullwood et al., 2009; Lieberman-Aiden et al., 2009), increasing evidence indicates that the interaction between genomic regulatory elements plays an important role in regulating gene expression. Heidari et al. (2014) have discovered that ZNF143 plays an important role in mediated distal chromatin interactions. Bailey et al. (2015) have found that ZNF143, as a novel and key chromatin-looping factor, with sequence specificity dependency at promoters and links the distal regulatory elements together, playing an important role in the establishment of the genomic organization. ZNF143 binds to the PMM2 promoter could establish a functional chromatin loop enabling interaction between the promoter and distal regulatory elements, which allows specific spatiotemporal regulation of PMM2 (Cabezas et al., 2017). ZNF143 knockdown mainly eliminates or destabilizes chromatin loops (Wen et al., 2018). We also found that ZNF143 was involved in the CTCF-mediated chromatin interactions by cooperating with cohesin (Ye et al., 2016). Other researchers have showed that ZNF143 interactes with other regulators are also important for chromatin domain formation. For example, Mourad and Cuvier (2006) have revealed that the formation of 3D chromatin domains is affected by positive driving factors CTCF, cohesin, ZNF143, polycomb proteins, and negative driving factors P300, RXRA, BCL11A, ELK1. CTCF binding sites are not only closely associated with topologically associating domain (TAD) boundaries, but also interact with ZNF143 and Yin Yang (YY)1 (Hong and Kim, 2017).
Conclusion and Prospects
ZNF143 can bind with multi-species, multi-type coding and non-coding genes (Schuster et al., 1995; Schaub et al., 1997; Myslinski et al., 1998). However, ZNF143 binding and co-initiative transcription differs due to the diversity of promoter structures. Although the promoter structure of H1 RNA, the RNA component of the human nuclear RNase P, is similar to that of vertebrate snRNA, H1 RNA’s promoter is distributed within 100 bp of the 5′ flanking sequence and presents a highly compact structure to initiate transcription (Myslinski et al., 2001). ZNF143 binding with U6 found in zebrafish are located upstream of the TATA box and downstream of proximal sequence element (PSE), unlike the U6 of other species (Halbig et al., 2008). The promoter of SCARNA2 is contained within 161 bp upstream of TSS due to its special transcription (different from SCARNA), whereas ZNF143 is the basic regulator (Gerard et al., 2010).
As a general TF, ZNF143 participates in numerous cellular biological activities. Using comparative genomic analysis to identify the distribution of ZNF143 target genes, Myslinski et al. (2006) have found that DNA binding and TFs account for 23%, protein synthesis/degradation/modification account for 21%, and DNA replication/cell cycle/cell growth/differentiation/apoptosis account for 13%. Anno et al. have also found that ZNF143 per se exhibits an inherently bidirectional transcription activity. Thus, ZNF143 has the ability to control the expression of divergent protein–protein and protein–non-coding RNA gene pairs (Anno et al., 2011). ZNF143 is expressed differently in various tissues. It is highly expressed in the lung, ovary and thymus, but weakly expressed in the brain, liver, and kidney (Grossman et al., 2014). ZNF143 is highly expressed in many solid tumors, and it is involved in cisplatin resistance because cisplatin induced ZNF143 binds to cisplatin-modified DNA (Wakasugi et al., 2007; Paek and You, 2011; Lu et al., 2012). Thus, novel small molecules can be designed for ZNF143 to enhance the sensitivity of cisplatin chemotherapy (Haibara et al., 2017). ZNF143 is not only indispensable for the embryonic development of zebrafish but also necessary for ES cell identity and self-renewal capability of ES cell (Chen et al., 2008; Halbig et al., 2012). What is more, histone methylation in the ZNF143 binding sites is usually related to transcription regulation. Yang et al. (2019) have found that both active (H3K4me1, H3K4me3, and H3K27ac) and suppressive (H3K27me3) histone marks can modulate ZNF143 binding, which in turn, regulate gene expression. However, how to develop new and convenient detection systems to study the function of ZNF143 is still a big challenge. Recently, Sathyan et al. (2019) have developed an improved auxin-inducible degron system to study TF function. After rapidly depleting the ZNF143 TF, transcriptional profiling indicates that ZNF143 activates transcription in cis and regulates promoter-proximal paused RNA polymerase density.
CTCF, cohesion, and ZNF143 are three major regulators involved in the establishment and maintenance of long-range chromatin interactions. In mammalian cells, TAD-free analysis indicates that the blocking effects of CTCF, cohesin, and ZNF143 depend on the distance between loci because each protein may participate at different scales of chromatin organization (Mourad and Cuvier, 2018). CTCF and cohesin are the key factors in organizing the mammalian genome to form TADs and loops, and the CTCF loops are formed as a result of cohesin-dependent loop extrusion (Dixon et al., 2012; Nor et al., 2012; Sanborn et al., 2015; Fudenberg et al., 2016; Goloborodko et al., 2016; Busslinger et al., 2017; Nuebler et al., 2018). ZNF143 is not only involved in CTCF/cohesin-mediated chromatin interactions, but also can bind directly to the promoter and connect it to distal regulatory elements (such as enhancer) to form chromatin loops (Heidari et al., 2014; Bailey et al., 2015; Ye et al., 2016). The recurrent C→T conversion at the ZNF143 locus influences the chromatin loop formation and alters distal gene expression in breast cancer (Yang et al., 2018). Lin et al. (2017) have reported a new epigenetic feature called sparse conserved under-methylated CpGs (scUMCs) is involved in cell-specific regulation of long-range chromatin interaction mediated by chromatin-looping factors (CTCF, cohesin, and ZNF143), providing a new direction in the research of the relationship between DNA methylation and chromatin organization. Recent technical developments allow more accurately identify where TFs bind to DNA. Skene et al. have showed that their new in situ methods, such as cleavage under targets and release using nuclease (CUT&RUN) and cleavage under targets and tagmentation (CUT&Tag), will be viewed as a cost-effective and versatile alternative to ChIP because of low backgrounds, which requiring only ∼1/10th the sequencing depth as ChIP (Skene and Henikoff, 2017; Skene et al., 2018; Kaya-Okur et al., 2019; Meers et al., 2019). Based on these methods, Shimbo et al. (2019) have developed cleavage under tethered nuclease for conformational capture (Cut-C) technology to identify chromatin interactions mediated by a protein of interest along with the genome-wide distribution of the target proteins. Thus, using these latest technologies, we may be clearly captured the accuracy of chromatin loops mediated by ZNF143 in a genome-wide scale.
During mitosis, transcription is globally shut down, chromatin condenses, the nuclear envelope is disassembled, and most TFs are stripped off the mitotic chromosomes. How do the new daughter cells faithfully re-establish the cell-type specific transcription program? Recent discoveries that a select set of TFs remain associated with mitotic chromosomes suggest a phenomenon termed mitotic bookmarking (Huang and Wang, 2017). For example, many studies have reported that CTCF is still partially retained in mitotic chromosomes and chromatin structure dynamics during the mitosis-to-G1 phase transition (Burke et al., 2005; Yan et al., 2013; Shen et al., 2015; Teves et al., 2016; Oomen et al., 2019; Palozola et al., 2019; Zhang et al., 2019). Thus, the presence of CTCF during mitosis may function as candidate mitotic bookmarking protein. This mechanism plays a potential and critical role in maintaining cell identity and cell destiny. Meanwhile, ZNF143 can interact with CTCF and mediate the formation of the chromatin loops. We recently discovered that ZNF143 was still partially bound to the chromosome during mitosis and 80% of the retained regions preferentially localized to promoters, supporting that it functioned mainly through promoters (Ye et al., 2020). Thus, the presence of CTCF and ZNF143 during mitosis may be crucial to recruit other regulatory factors to bind to chromosomes and re-establish chromatin loops in early G1 phase (Figure 2). Therefore, further studies on ZNF143 are necessary to help reveal its regulatory mechanism during the cell cycle.
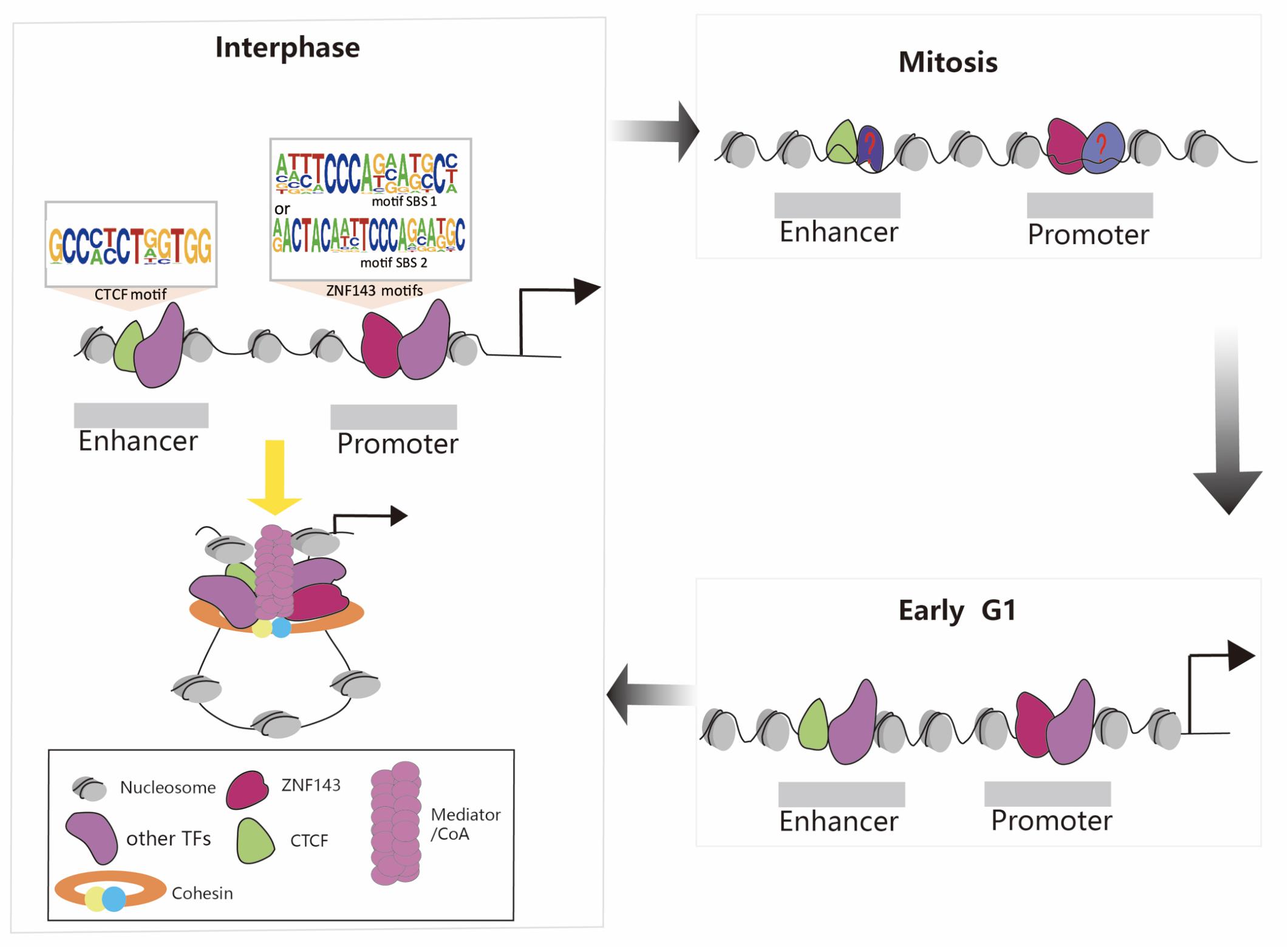
Figure 2. Schematic representation of chromatin loop formation mediated by ZNF143, CTCF, cohesin, and other TFs during the cell cycle. ZNF143 is a potential mitotic bookmarking factor helps to re-establish chromatin loops in early G1.
As a key TF, the role of ZNF143 in cancer progression through transcriptional regulation of genes related to DNA replication and cell cycle (Izumi et al., 2010). Furthermore, Song et al. have showed that miR-590-3p could negatively modulate the expression of ZNF143 via binding to the ZNF143 3′-UTR and ZNF143 can directly activate FAM224A expression through binding to its promoter, forming the A1CF-FAM224A-miR-590-3p-ZNF143 positive feedback loop. This loop plays a critical role in regulating the malignant progression of glioma cells, providing a novel molecular target for glioma therapy (Song et al., 2019). In recent years, with the technology and bioinformatics analysis development, the molecular mechanism of ZNF143-mediated gene transcriptional regulation has been largely exploited. Chromatin looping between promoters and distal regulatory elements depends on DNA binding by ZNF143 and other partners. In the future, how to comprehensively analyze the mechanism of ZNF143 in mediating gene expression of different cell types and discover the novel and potential functions of ZNF143 remains a considerable challenge.
Author Contributions
BY, GaY, and YL drafted the manuscript. CZ, QW, and GuY critically revised the manuscript.
Funding
This work was supported by Postdoctoral Research Grant in Henan Province (001803040), the Key Scientific Research Projects of Henan Higher Education (18A180019).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adachi, K., Saito, H., Tanaka, T., and Oka, T. (1998). Molecular cloning and characterization of the murine staf cDNA encoding a transcription activating factor for the selenocysteine tRNA gene in mouse mammary gland. J. Biol. Chem. 273, 8598–8606. doi: 10.1074/jbc.273.15.8598
Anno, Y. N., Myslinski, E., Ngondo-Mbongo, R. P., Krol, A., Poch, O., Lecompte, O., et al. (2011). Genome-wide evidence for an essential role of the human Staf/ZNF143 transcription factor in bidirectional transcription. Nucleic Acids Res. 39, 3116–3127. doi: 10.1093/nar/gkq1301
Bailey, S. D., Zhang, X., Desai, K., Aid, M., Corradin, O., Cowper-Sal Lari, R., et al. (2015). ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat. Commun. 2:6186.
Barski, O. A., Papusha, V. Z., Kunkel, G. R., and Gabbay, K. H. (2004). Regulation of aldehyde reductase expression by STAF and CHOP. Genomics 83, 119–129. doi: 10.1016/s0888-7543(03)00213-1
Burke, L. J., Zhang, R., Bartkuhn, M., Tiwari, V. K., Tavoosidana, G., Kurukuti, S., et al. (2005). CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 24, 3291–3300. doi: 10.1038/sj.emboj.7600793
Busslinger, G. A., Stocsits, R. R., van der Lelij, P., Axelsson, E., Tedeschi, A., Galjart, N., et al. (2017). Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507. doi: 10.1038/nature22063
Cabezas, O. R., Flanagan, S. E., Stanescu, H., García-Martínez, E., Caswell, R., Lango-Allen, H., et al. (2017). Polycystic kidney disease with hyperinsulinemic hypoglycemia caused by a promoter mutation in phosphomannomutase 2. J. Am. Soc. Nephrol. 28, 2529–2539.
Chen, X., Fang, F., Liou, Y. C., and Ng, H. H. (2008). Zfp143 regulates Nanog through modulation of Oct4 binding. Stem Cells 26, 2759–2767. doi: 10.1634/stemcells.2008-0398
Dekker, J., Rippe, K., Dekker, M., and Kleckner, N. (2002). Capturing chromosome conformation. Science 295, 1306–1311. doi: 10.1126/science.1067799
Di Leva, F., Ferrante, M. I., Demarchi, F., Caravelli, A., Matarazzo, M. R., Giacca, M., et al. (2004). Human synaptobrevin-like 1 gene basal transcription is regulated through the interaction of selenocysteine tRNA gene transcription activating factor-zinc finger 143 factors with evolutionary conserved cis-elements. J. Biol. Chem. 279, 7734–7739. doi: 10.1074/jbc.m308140200
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Fish, L., Navickas, A., Culbertson, B., Xu, Y., Nguyen, H. C. B., Zhang, S., et al. (2019). Nuclear TARBP2 drives oncogenic dysregulation of RNA splicing and decay. Mol. Cell. 75, 967–981.
Fraser, P. (2006). Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 16, 490–495. doi: 10.1016/j.gde.2006.08.002
Fraser, P., and Bickmore, W. (2007). Nuclear organization of the genome and the potential for gene regulation. Nature 447, 413–417. doi: 10.1038/nature05916
Fudenberg, G., Imakaev, M., Lu, C., Goloborodko, A., Abdennur, N., and Mirny, L. A. (2016). Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049. doi: 10.1016/j.celrep.2016.04.085
Fullwood, M. J., Liu, M., Pan, Y. F., Liu, J., Xu, H., Mohamed, Y. B., et al. (2009). An oestrogen-receptor-alpha-bound human chromatin interactome. Nautre 462, 58–64.
Gerard, M. A., Krol, A., and Carbon, P. (2007). Transcription factor hStaf/ZNF143 is required for expression of the human TFAM gene. Gene 401, 145–153. doi: 10.1016/j.gene.2007.07.011
Gerard, M. A., Myslinski, E., Chylak, N., Baudrey, S., Krol, A., and Carbon, P. (2010). The scaRNA2 is produced by an independent transcription unit and its processing is directed by the encoding region. Nucleic Acids Res. 38, 370–381. doi: 10.1093/nar/gkp988
Goloborodko, A., Marko, J. F., and Mirny, L. A. (2016). Chromosome compaction by active loop extrusion. Biophys. J. 110, 2162–2168. doi: 10.1016/j.bpj.2016.02.041
Gonzalez, D., Luyten, A., Bartholdy, B., Zhou, Q., Kardosova, M., Ebralidze, A., et al. (2017). ZNF143 protein is an important regulator of the myeloid transcription factor C/EBPalpha. J. Biol. Chem. 292, 18924–18936. doi: 10.1074/jbc.m117.811109
Grossman, C. E., Qian, Y., Banki, K., and Perl, A. (2014). ZNF143 mediates basal and tissue-specific expression of human transaldolase. J. Biol. Chem. 279, 12190–12205. doi: 10.1074/jbc.m307039200
Haibara, H., Yamazaki, R., Nishiyama, Y., Ono, M., Kobayashi, T., Hokkyo-Itagaki, A., et al. (2017). YPC-21661 and YPC-22026, novel small molecules, inhibit ZNF143 activity in vitro and in vivo. Cancer Sci. 108, 1042–1048. doi: 10.1111/cas.13199
Halbig, K. M., Lekven, A. C., and Kunkel, G. R. (2008). Zebrafish U6 small nuclear RNA gene promoters contain a SPH element in an unusual location. Gene 421, 89–94. doi: 10.1016/j.gene.2008.06.019
Halbig, K. M., Lekven, A. C., and Kunkel, G. R. (2012). The transcriptional activator ZNF143 is essential for normal development in zebrafish. BMC Mol. Biol. 13:3. doi: 10.1186/1471-2199-13-3
Heidari, N., Phanstiel, D. H., He, C., Grubert, F., Jahanbani, F., Kasowski, M., et al. (2014). Genome-wide map of regulatory interactions in the human genome. Genome Res. 24, 1905–1917. doi: 10.1101/gr.176586.114
Hernandez-Negrete, I., Sala-Newby, G. B., Perl, A., Kunkel, G. R., Newby, A. C., and Bond, M. (2011). Adhesion-dependent Skp2 transcription requires selenocysteine tRNA gene transcription-activating factor (STAF). Biochem. J. 436, 133–143. doi: 10.1042/bj20101798
Hong, S., and Kim, D. (2017). Computational characterization of chromatin domain boundary-associated genomic elements. Nucleic Acids Res. 45, 10403–10414. doi: 10.1093/nar/gkx738
Huang, X., and Wang, J. (2017). Mitotic bookmarking: maintaining the stem cell identity during mitosis. Cell Stem Cell 20, 741–742. doi: 10.1016/j.stem.2017.05.002
Huning, L., and Kunkel, G. R. (2020). Two paralogous znf143 genes in zebrafish encode transcriptional activator proteins with similar functions but expressed at different levels during early development. BMC Mol. Cell. Biol. 21:3. doi: 10.1186/s12860-020-0247-7
Ishiguchi, H., Izumi, H., Torigoe, T., Yoshida, Y., Kubota, H., Tsuji, S., et al. (2004). ZNF143 activates gene expression in response to DNA damage and binds to cisplatin-modified DNA. Int. J. Cancer. 111, 900–909. doi: 10.1002/ijc.20358
Izumi, H., Wakasugi, T., Shimajiri, S., Tanimoto, A., Sasaguri, Y., Kashiwagi, E., et al. (2010). Role of ZNF143 in tumor growth through transcriptional regulation of DNA replication and cell-cycle-associated genes. Cancer Sci. 101, 2538–2545. doi: 10.1111/j.1349-7006.2010.01725.x
Izumi, H., Yasuniwa, Y., Akiyama, M., Yamaguchi, T., Kuma, A., Kitamura, N., et al. (2011). Forced expression of ZNF143 restrains cancer cell growth. Cancers 3, 3909–3920. doi: 10.3390/cancers3043909
Kawatsu, Y., Kitada, S., Uramoto, H., Zhi, L., Takeda, T., Kimura, T., et al. (2014). The combination of strong expression of ZNF143 and high MIB-1 labelling index independently predicts shorter disease-specific survival in lung adenocarcinoma. Br. J. Cancer 110, 2583–2592. doi: 10.1038/bjc.2014.202
Kaya-Okur, H. S., Wu, S. J., Codomo, C. A., Pledger, E. S., Bryson, T. D., Henikoff, J. G., et al. (2019). CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10:1930.
Kubota, H., Yokota, S., Yanagi, H., and Yura, T. (2000). Transcriptional regulation of the mouse cytosolic chaperonin subunit gene Ccta/t-complex polypeptide 1 by selenocysteine tRNA gene transcription activating factor family zinc finger proteins. J. Biol. Chem. 275, 28641–28648. doi: 10.1074/jbc.m005009200
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. doi: 10.1126/science.1181369
Lin, X., Su, J., Chen, K., Rodriguez, B., and Li, W. (2017). Sparse conserved under-methylated CpGs are associated with high-order chromatin structure. Genome Biol. 18:163.
Lu, W., Chen, Z., Zhang, H., Wang, Y., Luo, Y., and Huang, P. (2012). ZNF143 transcription factor mediates cell survival through upregulation of the GPX1 activity in the mitochondrial respiratory dysfunction. Cell Death Dis. 3:e422. doi: 10.1038/cddis.2012.156
Mach, C. M., Hargrove, B. W., and Kunkel, G. R. (2002). The Small RNA gene activator protein, SphI postoctamer homology-binding factor/selenocysteine tRNA gene transcription activating factor, stimulates transcription of the human interferon regulatory factor-3 gene. J. Biol. Chem. 277, 4853–4858. doi: 10.1074/jbc.m108308200
Meers, M. P., Bryson, T. D., Henikoff, J. G., and Henikoff, S. (2019). Improved CUT&RUN chromatin profiling tools. eLife 8:e46314.
Michaud, J., Praz, V., James Faresse, N., Jnbaptiste, C. K., Tyagi, S., Schütz, F., et al. (2013). HCFC1 is a common component of active human CpG-island promoters and coincides with ZNF143, THAP11, YY1, and GABP transcription factor occupancy. Genome Res. 23, 907–916. doi: 10.1101/gr.150078.112
Mourad, R., and Cuvier, O. (2006). Computational identification of genomic features that influence 3D chromatin domain formation. PLoS Comput. Biol. 12:e1004908. doi: 10.1371/journal.pcbi.1004908
Mourad, R., and Cuvier, O. (2018). TAD-free analysis of architectural proteins and insulators. Nucleic Acids Res. 46:e27. doi: 10.1093/nar/gkx1246
Myslinski, E., Amé, J. C., Krol, A., and Carbon, P. (2001). An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 29, 2502–2509. doi: 10.1093/nar/29.12.2502
Myslinski, E., Gerard, M. A., Krol, A., and Carbon, P. (2006). A genome scale location analysis of human Staf/ZNF143-binding sites suggests a widespread role for human Staf/ZNF143 in mammalian promoters. J. Biol. Chem. 281, 39953–39962. doi: 10.1074/jbc.m608507200
Myslinski, E., Gerard, M. A., Krol, A., and Carbon, P. (2007). Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res. 35, 3453–3464. doi: 10.1093/nar/gkm239
Myslinski, E., Krol, A., and Carbon, P. (1998). ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J. Biol. Chem. 273, 21998–22006. doi: 10.1074/jbc.273.34.21998
Ngondo, R. P., and Carbon, P. (2004). Transcription factor abundance controlled by an auto-regulatory mechanism involving a transcription start site switch. Nucleic Acids Res. 42, 2171–2184. doi: 10.1093/nar/gkt1136
Ngondo, R. P., and Carbon, P. (2014). ZNF143 is regulated through alternative 3′UTR isoforms. Biochimie 104, 137–146. doi: 10.1016/j.biochi.2014.06.008
Ngondo-Mbongo, R. P., Myslinski, E., Aster, J. C., and Carbon, P. (2013). Modulation of gene expression via overlapping binding sites exerted by ZNF143, Notch1 and THAP11. Nucleic Acids Res. 41, 4000–4014. doi: 10.1093/nar/gkt088
Nor, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nautre 485, 381–385. doi: 10.1038/nature11049
Nuebler, J., Fudenberg, G., Imakaev, M., Abdennur, N., and Mirny, L. A. (2018). Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. U.S.A. 115, E6697–E6706.
Oomen, M. E., Hansen, A. S., Liu, Y., Darzacq, X., and Dekker, J. (2019). CTCF sites display cell cycle-dependent dynamics in factor binding and nucleosome positioning. Genome Res. 29, 236–249. doi: 10.1101/gr.241547.118
Paek, A. R., Lee, C. H., and You, H. J. (2014). A role of zinc-finger protein 143 for cancer cell migration and invasion through ZEB1 and E-cadherin in colon cancer cells. Mol. Carcinog. 53, E161–E168.
Paek, A. R., Mun, J. Y., Hong, K. M., Lee, J., Hong, D. W., and You, H. J. (2017). Zinc finger protein 143 expression is closely related to tumor malignancy via regulating cell motility in breast cancer. BMB Rep. 50, 621–627. doi: 10.5483/bmbrep.2017.50.12.177
Paek, A. R., Mun, J. Y., Jo, M. J., Choi, H., Lee, Y. J., Cheong, H., et al. (2019). The role of ZNF143 in breast cancer cell survival through the NAD(P)H quinone dehydrogenase 1 (-) 53 (-) beclin1 axis under metabolic stress. Cells 8:E296.
Paek, A. R., and You, H. J. (2011). GAIP-interacting protein, C-terminus is involved in the induction of zinc-finger protein 143 in response to insulin-like growth factor-1 in colon cancer cells. Mol. Cells 32, 415–419. doi: 10.1007/s10059-011-0078-7
Palozola, K. C., Lerner, J., and Zaret, K. S. (2019). A changing paradigm of transcriptional memory propagation through mitosis. Nat. Rev. Mol. Cell. Biol. 20, 55–64. doi: 10.1038/s41580-018-0077-z
Parker, J. B., Yin, H., Vinckevicius, A., and Chakravarti, D. (2014). Host cell factor-1 recruitment to E2F-bound and cell-cycle-control genes is mediated by THAP11 and ZNF143. Cell Rep. 9, 967–982. doi: 10.1016/j.celrep.2014.09.051
Pupavac, M., Watkins, D., Petrella, F., Fahiminiya, S., Janer, A., Cheung, W., et al. (2016). Inborn error of cobalamin metabolism associated with the intracellular accumulation of transcobalamin-bound cobalamin and mutations in ZNF143, which codes for a transcriptional activator. Hum. Mutat. 37, 976–982. doi: 10.1002/humu.23037
Sadlecki, P., Grabiec, M., Grzanka, D., Jozwicki, J., Antosik, P., and Walentowicz-Sadlecka, M. (2019). Expression of zinc finger transcription factors (ZNF143 and ZNF281) in serous borderline ovarian tumors and low-grade ovarian cancers. J. Ovarian Res. 12:23.
Sanborn, A. L., Rao, S. S., Huang, S. C., Durand, N. C., Huntley, M. H., Jewett, A. I., et al. (2015). Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E6456–E6465.
Sathyan, K. M., McKenna, B. D., Anderson, W. D., Duarte, F. M., Core, L., and Guertin, M. J. (2019). An improved auxin-inducible degron system preserves native protein levels and enables rapid and specific protein depletion. Genes Dev. 33, 1441–1455. doi: 10.1101/gad.328237.119
Saur, D., Seidler, B., Paehge, H., Schusdziarra, V., and Allescher, H. D. (2002). Complex regulation of human neuronal nitric-oxide synthase exon 1c gene transcription. Essential role of Sp and ZNF family members of transcription factors. J. Biol. Chem. 277, 25798–25814. doi: 10.1074/jbc.m109802200
Schaub, M., Krol, A., and Carbon, P. (1999a). Flexible zinc finger requirement for binding of the transcriptional activator staf to U6 small nuclear RNA and tRNA(Sec) promoters. J. Biol. Chem. 274, 24241–24249. doi: 10.1074/jbc.274.34.24241
Schaub, M., Krol, A., and Carbon, P. (2000). Structural organization of Staf-DNA complexes. Nucleic Acids Res. 28, 2114–2121. doi: 10.1093/nar/28.10.2114
Schaub, M., Myslinski, E., Krol, A., and Carbon, P. (1999b). Maximization of selenocysteine tRNA and U6 small nuclear RNA transcriptional activation achieved by flexible utilization of a Staf zinc finger. J. Biol. Chem. 274, 25042–25050. doi: 10.1074/jbc.274.35.25042
Schaub, M., Myslinski, E., Schuster, C., Krol, A., and Carbon, P. (1997). Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J. 16, 173–181. doi: 10.1093/emboj/16.1.173
Schuster, C., Krol, A., and Carbon, P. (1998). Two distinct domains in Staf to selectively activate small nuclear RNA-type and mRNA promoters. Mol. Cell Biol. 18, 2650–2658. doi: 10.1128/mcb.18.5.2650
Schuster, C., Myslinski, E., Krol, A., and Carbon, P. (1995). Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J. 14, 3777–3787. doi: 10.1002/j.1460-2075.1995.tb00047.x
Shen, W. L., Wang, D., Ye, B. Y., Shi, M. L., Zhang, Y., and Zhao, Z. H. (2015). A possible role of Drosophila CTCF in mitotic bookmarking and maintaining chromatin domains during the cell cycle. Biol. Res. 48:27.
Shimbo, T., Kawamura, M., Wijaya, E., Takaki, E., Kaneda, Y., and Tamai, K. (2019). Cut-C: cleavage under tethered nuclease for conformational capture. BMC Genomics 20:614. doi: 10.1186/s12864-019-5989-2
Skene, P. J., Henikoff, J. G., and Henikoff, S. (2018). Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 13, 1006–1019. doi: 10.1038/nprot.2018.015
Skene, P. J., and Henikoff, S. (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6:e21856.
Song, Y., Shao, L., Xue, Y., Xuelei, R., Xiaobai, L., Chunqing, Y., et al. (2019). Inhibition of the aberrant A1CF-FAM224A- miR-590-3p-ZNF143 positive feedback loop attenuated malignant biological behaviors of glioma cells. J. Exp. Clin. Cancer Res. 38:248.
Teves, S. S., An, L., Hansen, A. S., Xie, L., Darzacq, X., and Tjian, R. (2016). A dynamic mode of mitotic bookmarking by transcription factors. eLife 5:e22280.
Tommerup, N., and Vissing, H. (1995). Isolation and fine mapping of 16 novel human zinc finger-encoding cDNAs identify putative candidate genes for developmental and malignant disorders. Genomics 27, 259–264. doi: 10.1006/geno.1995.1040
Torigoe, T., Izumi, H., Ishiguchi, H., Yoshida, Y., Tanabe, M., Yoshida, T., et al. (2005). Cisplatin resistance and transcription factors. Curr. Med. Chem. Anticancer Agents 5, 15–27.
Verma, V., Paek, A. R., Choi, B. K., Hong, E. K., and You, H. J. (2019). Loss of zinc-finger protein 143 contributes to tumour progression by interleukin-8-CXCR axis in colon cancer. J. Cell. Mol. Med. 23, 4043–4053. doi: 10.1111/jcmm.14290
Vinckevicius, A., Parker, J. B., and Chakravarti, D. (2015). Genomic determinants of THAP11/ZNF143/HCFC1 complex recruitment to chromatin. Mol. Cell. Biol. 35, 4135–4146. doi: 10.1128/mcb.00477-15
Wakasugi, T., Izumi, H., Uchiumi, T., Suzuki, H., Arao, T., Nishio, K., et al. (2007). ZNF143 interacts with p73 and is involved in cisplatin resistance through the transcriptional regulation of DNA repair genes. Oncogene 26, 5194–5203. doi: 10.1038/sj.onc.1210326
Wang, H., Zou, J., Zhao, B., Eric, J., Todd, A., Hoifung, W., et al. (2011). Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14908–14913. doi: 10.1073/pnas.1109023108
Wei, S., Wang, L., Zhang, L., Li, B., Li, Z., Zhang, Q., et al. (2016). ZNF143 enhances metastasis of gastric cancer by promoting the process of EMT through PI3K/AKT signaling pathway. Tumour Biol. 37, 12813–12821. doi: 10.1007/s13277-016-5239-z
Wen, Z., Huang, Z. T., Zhang, R., and Peng, C. (2018). ZNF143 is a regulator of chromatin loop. Cell. Biol. Toxicol. 34, 471–478. doi: 10.1007/s10565-018-9443-z
Yan, J., Enge, M., Whitington, T., Dave, K., Liu, J., Sur, I., et al. (2013). Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813. doi: 10.1016/j.cell.2013.07.034
Yang, J., Wei, X., Tufan, T., Kuscu, C., Unlu, H., Farooq, S., et al. (2018). Recurrent mutations at estrogen receptor binding sites alter chromatin topology and distal gene expression in breast cancer. Genome Biol. 19:190.
Yang, X., Bam, M., Nagarkatti, P. S., and Nagarkatti, M. (2019). Cannabidiol regulates gene expression in encephalitogenic T cells using histone methylation and noncoding rna during experimental autoimmune encephalomyelitis. Sci. Rep. 9:15780.
Yang, Y., Zhang, R. C., Singh, S., and Ma, J. (2017). Exploiting sequence-based features for predicting enhancer- promoter interactions. Bioinformatics 33, i252–i260. doi: 10.1093/bioinformatics/btx257
Ye, B. Y., Shen, W. L., Wang, D., Li, P., Zhang, Z., Shi, M. L., et al. (2016). ZNF143 is involved in CTCF-mediated chromatin interactions by cooperation with cohesin and other partners. Mol. Biol. 50, 496–503.
Ye, B. Y., Shen, W. L., and Zhao, Z. H. (2020). ZNF143 is dynamically bound to a subset of its interphase sites during mitosis. Biochem. Biophys. Res. Commun. 523, 293–298. doi: 10.1016/j.bbrc.2019.12.031
Keywords: transcription factor, ZNF143, biomarker, chromatin organization, loop
Citation: Ye B, Yang G, Li Y, Zhang C, Wang Q and Yu G (2020) ZNF143 in Chromatin Looping and Gene Regulation. Front. Genet. 11:338. doi: 10.3389/fgene.2020.00338
Received: 25 December 2019; Accepted: 20 March 2020;
Published: 07 April 2020.
Edited by:
Huiming Zhang, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Hye Jin You, National Cancer Center, South KoreaDag H. Yasui, University of California, Davis, United States
Copyright © 2020 Ye, Yang, Li, Zhang, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyan Zhang, emN5MTExOXNkY0AxNjMuY29t; Qiwen Wang, d2FuZ3Fpd2VuQGh0dS5lZHUuY24=; Guoying Yu, Z3VveWluZ3l1QGh0dS5lZHUuY24=
†These authors have contributed equally to this work
 Bingyu Ye
Bingyu Ye Ganggang Yang
Ganggang Yang Yuanmeng Li
Yuanmeng Li Chunyan Zhang
Chunyan Zhang Qiwen Wang
Qiwen Wang Guoying Yu1,2,3,4,5,6*
Guoying Yu1,2,3,4,5,6*