- 1Rice Research Institute and Key Lab for Major Crop Diseases, Sichuan Agricultural University at Wenjiang, Chengdu, China
- 2State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University at Wenjiang, Chengdu, China
MicroRNAs (miRNAs) play essential roles in the regulation of plant growth and defense responses. More and more, miRNA-3ps are reported to act in plant development and immunity. miR156 is a conserved miRNA, and most previous studies focus on its roles in plant growth, development, and yield determinacy. Here, we show that expressing a target mimic of miR156fhl-3p led to enhanced rice blast disease resistance without a yield penalty. miR156fhl-3p was differentially responsive to Magnaporthe oryzae in susceptible and resistant accessions. Transgenic lines expressing a target mimic of miR156fhl-3p (MIM156-3p) exhibited enhanced rice blast disease resistance and increased expression of defense-related genes. MIM156-3p also enhanced the mRNA abundance of SPL14 and WRKY45 by down-regulating miR156-5p and pre-miR156. Moreover, MIM156-3p lines displayed a decreased number of second rachis branches per panicle but enlarged grains, leading to unchanged yield per plant. Consistently, overexpressing miR156h (OX156) led to enhanced susceptibility to M. oryzae and decreased the expression of SPL14 and WRKY45. Our results indicate that miR156fhl-3p mounts a regulatory role on miR156-5p, which subsequently regulates the expression of SPL14 and WRKY45 to improve rice blast disease resistance.
Introduction
MicroRNAs (miRNAs) are a kind of 20- to 24-nucleotide (nt) non-coding RNA molecule that regulate the expression of genes with sequences complementary to the miRNAs (Bartel, 2004; Yu et al., 2017). One miRNA isoform can be transcribed from one or more MIR gene loci (Li et al., 2019). A MIR gene is first transcribed into a pri-miRNA, which is processed by Dicer-like proteins (DCLs) to form pre-miRNA (Rogers and Chen, 2013). The pre-miRNA is further cleaved by DCLs, generating a miRNA-5p/miRNA-3p (previously miRNA/miRNA∗) duplex (Liu et al., 2005). Then, the duplex separates into a mature miRNA arm (-5p or -3p) that is bound by Argonaute (AGO) proteins to form RNA-induced silencing complex (RISC) (Ji et al., 2011; Zhu et al., 2011; Kobayashi and Tomari, 2016). RISC binds to the sequences reversely complementary to the miRNA and triggers DNA methylation, mRNA cleavage, or translational inhibition, respectively (Iwakawa and Tomari, 2015; Yu et al., 2017). The selection of a mature miRNA (-5p or -3p arm) can be different across tissues, developmental stages, and abiotic or biotic stresses (Guo et al., 2014; Hu et al., 2014). Some MIR genes are only processed into miRNA-5p, and some miRNAs are processed into miRNA-5p and miRNA-3p. The miRNA-3p was once considered as a useless by-product of the miRNA biogenesis. Some recent studies showed that a miRNA-5p and a miRNA-3p could target different gene families, each of which is involved in diverse biological processes (Jones-Rhoades et al., 2006; Huang et al., 2019). For example, in Arabidopsis, miR393 can be induced by a well-studied pathogen-associated molecular pattern (PAMP) molecule, flg22, and positively contributes to resistance against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) via miR393-5p-mediated suppression of the expression of Transport Inhibitor Response 1 (TIR1), Auxin signaling F-Box proteins 2 (AFB2), and AFB3 (Navarro et al., 2006). On the other hand, miR393-3p targets MEMB12, a gene encoding a Gogi-localized SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein involved in protein trafficking and regulating plant immune responses (Zhang et al., 2011). However, it is unclear whether the accumulation of a -3p correlates with a -5p.
Rice feeds half of the world population. Rice production is continually threatened by blast disease that is caused by Magnaporthe oryzae (syn. Pyricularia oryzae) (Zhang et al., 2018). In the past decade, more than 70 miRNAs have been identified to be responsive to M. oryzae or its elicitors, and 11 of them have been functionally characterized to play positive or negative roles in rice blast disease resistance (Li et al., 2019; Zhou et al., 2019). For example, miR160 positively regulates rice immunity against M. oryzae via up-regulating defense-related gene expression and hydrogen peroxide accumulation at the infection site (Li et al., 2014). miR167d facilitates M. oryzae infection by repressing the expression of Auxin Responsive Factor 12 (ARF12) (Zhao et al., 2019). miR169 negatively regulates rice immunity by repressing Nuclear Factor Y-A (NF-YA) genes (Wu et al., 2009; Li et al., 2017). miR319 targets the transcription factor Teosinte Branched1/Cycloidea/Proliferating Cell Factor1 (OsTCP21), which manipulates JA synthesis via OsLOX2 and OsLOX5, the key synthetic components of JA, to facilitate the blast fungus infection (Hao et al., 2012; Zhang et al., 2018). miR396 targets Growth Regulating Factor (GRF) genes to balance growth, yield traits, and immunity against M. oryzae (Chandran et al., 2019). Different miR156-3p isoforms are identified to be responsive to the infection of M. oryzae (Li et al., 2014), but their roles are unknown.
miR156 is a conserved and highly abundant miRNA in most plants and acts as a master regulator during the whole growth period (Axtell and Bowman, 2008). In Arabidopsis, miR156 is highly expressed in young seedlings and miR156-5p target SQUAMOSA promoter-binding protein-like transcription factor (SPL) genes to regulate growth and development (Cardon et al., 1999; Wu and Poethig, 2006; Addoquaye et al., 2008; Shikata et al., 2009; Xu et al., 2016). These Arabidopsis SPL genes can be classified into three functionally distinct groups. The first group includes SPL2, SPL9, SPL10, SPL11, SPL13, and SPL15, which regulate juvenile-to-adult vegetative and vegetative-to-reproductive transition. The second group includes SPL3, SPL4, and SPL5, which could promote the floral meristem identity transition. The third group is SPL6, which may be important for physiological processes (Xie et al., 2006; Riese et al., 2007; Preston and Hileman, 2013). In rice, the Osa-miR156 gene family has 12 members that generate three mature miR156-5p isoforms and four mature miR156-3p isoforms. miR156-5p targets 11 rice SPL genes that are involved in the regulation of important agronomic traits and blast disease resistance (Xie et al., 2006). For example, SPL13 positively contributes to rice yield via improving grain size and panicle size (Si et al., 2016). SPL16 positively regulates grain size via binding to GW7, which acts as a critical regulator in rice architecture and grain development (Wang et al., 2012; Wang S. et al., 2015). SPL18 negatively regulates grain number through binding to the promoter of DEP1 that acts as an important regulator of panicle architecture (Huang et al., 2009; Taguchi-Shiobara et al., 2011; Yuan et al., 2019). SPL14 is also known as Ideal Plant Architecture 1 (IPA1), which plays a critical role in rice architecture and resistance to M. oryzae via phosphorylation status and subsequent DNA-binding specificity (Jiao et al., 2010; Wang et al., 2018). Before M. oryzae invasion, SPL14 protein binds to the DEP1 promoter that contains the GTAC motif, to regulate tiller development, leading to plants with fewer unproductive tillers and more grains per panicle, supporting higher yield (Wang et al., 2018). Upon M. oryzae invasion, SPL14 protein is phosphorylated and binds to the WRKY45 promoter that contains TGGGCC motif, leading to enhanced resistance to the blast disease (Wang et al., 2018).
WRKY45 encodes a WRKY transcription factor that binds to W-box or W-like box-type cis-elements in response to pathogens and plays a vital role in plant-pathogen interaction (Li et al., 2004; Eulgem and Somssich, 2007). Overexpression of WRKY45 markedly enhances rice blast resistance, whereas knock out of WRKY45 compromises blast resistance (Shimono et al., 2007). The WRKY45 transcript responds strongly to SA and BTH, but its role is neither downstream nor upstream of NH1, a rice homolog of NPR1. In contrast, a glutathione S-transferase and a cytochrome P450 are found to be downstream of WRKY45. Elevated WRKY45 mRNA level could remarkably enhance resistance to M. oryzae (Shimono et al., 2007; Goto et al., 2015, 2016). However, constitutive WRKY45 overexpression-conferred blast resistance remarkably penalizes yield (Goto et al., 2015). Expressing WRKY45 under the control of pathogen-responsive promoters in combination with a translational enhancer derived from a 5′-untranslated region (UTR) of rice alcohol dehydrogenase (ADH) has strong disease resistance without penalty yield (Goto et al., 2016). Therefore, it is highly anticipated that miR156 plays an essential role in the regulation of rice blast disease resistance via the SPL14-WRKY45 module. However, experimental evidence is lacking, and whether miR156-3p plays roles in blast resistance is unclear.
To investigate the function of miR156-3p in rice blast disease resistance, we constructed the transgenic lines expressing a target mimic of miR156fhl-3p (MIM156-3p) and found that MIM156-3p showed enhanced blast disease resistance. Surprisingly, the MIM156-3p plants exhibited significantly less pre-miR156 and miR156-5p accumulation than control, indicating that blocking miR156fhl-3p interfered with the production of miR156-5p, which in turn led to increase in the expression of SPL14 and WRKY45. In contrast, elevated miR156h expression (OX156) enhanced susceptibility to M. oryzae, which could be due to the suppression of SPL14 and decreased WRKY45 expression. Moreover, MIM156-3p plants displayed an unchanged yield per plant. Together, our data indicate that miR156flh-3p and miR156-5p may be mutually regulated via an unknown mechanism, and miR156 negatively regulates rice blast disease resistance via SPL14 and WRKY45. Our results provide a potential tool to improve blast resistance without yield penalty and a new phenomenon in miRNA synthesis/accumulation.
Materials and Methods
Plant Materials and Growth Conditions
Rice (Oryza sativa) materials used in this study include Lijiangxin Tuan Heigu (LTH) and International Rice Blast Line Pyricularia-Kanto51-m-Tsuyuake (IRBLkm-Ts). LTH is a susceptible accession sensitive to over 1,300 regional isolates of M. oryzae worldwide (Lin et al., 2001). IRBLkm-Ts is a resistant accession, carrying a resistance locus Pi-km (Hiroshi et al., 2000). The rice cultivar Nipponbare was used as wild type (WT) and transgenic background. For germination, rice seeds were immersed in water for 2 d at 37°C in darkness. Then, seedlings were grown in an air-conditioned growth room at 26°C and 70% relative humidity with a 14 h light/10 h dark cycle. Nicotiana benthamiana plants were planted in an air-conditioned growth room at 24°C with a 16 h light/8 h dark cycle. N. benthamiana plants were used for Agrobacterium-infiltration experiments.
Plasmid Construction and Genetic Transformation
The artificial target mimic of miR156fhl-3p was acquired by annealing with primers miR156-mimic-F and miR156-mimic-R (Supplementary Figure S1 and Supplementary Table S1). The fragments were inserted into IPS1 (INDUCED BY PHOSPHATE STARVATION1) to replace the miR399 target site as described previously (Franco-Zorrilla et al., 2007; Wu et al., 2013). Then, the DNA fragments were cloned into BamHI/BglII site of pCAMBIA1300-35S, resulting in overexpressing construct p35S:MIM156-3p. To generate transgenic plants overexpressing miR156h, we amplified the genomic sequences containing 487 bp upstream and 403 bp downstream of the MIR156h gene from Nipponbare genomic DNA with primers miR156h-F and miR156h-R (Supplementary Table S1). The amplified fragments were digested with KpnI/SalI. After purified by DNA Purification Kit (Invitrogen), the fragments were cloned into vector pCAMBIA1300, resulting in the overexpressing construct p35S:miR156h. The plasmids were introduced into the Nipponbare background via Agrobacterium (strain GV3101)-mediated transformation. Positive transformants were screened following a previous report (Li et al., 2014).
Disease Assay and Microscopy Analysis
Three M. oryzae strains, including Zhong10-8-14-GFP (GZ8), 97-27-2, and FJ08-09-1, were used, depending on the availability at the time for inoculation. GZ8 and 97-27-2 were used for punch/spray-inoculation assay, and GZ8 was used for rice sheath inoculation assay in transgenic rice plants in Nipponbare background; FJ08-09-1 was incompatible with resistance accession IRBLkm-Ts and used for spray-inoculation assay in susceptible accession LTH and IRBLkm-Ts. For sporulation, M. oryzae strains were cultured in complete medium and oatmeal and tomato media (OTA) at 28°C with 16 h/8 h light/dark. For disease assays via spray-inoculation, the spores of the indicated strains were collected and adjusted to 1 × 105 spores mL–1 to spray onto three-leaf-seedlings. The inoculated seedlings were incubated in darkness with 100% humidity for 24 h and then put in an air-conditioned growth room (Li et al., 2017). Leaves were collected at 12 and 24 h post-inoculation (hpi) for analysis on the expression of defense-related genes. For disease assays via punch-inoculation, the collected spores (3 × 105 spores mL–1) were drop-inoculated on the punch-wound rice leaves and cultured in buffer containing 0.1% 6-Benzylaminopurine (6-BA). The leaves were incubated in darkness for 24 h, and then continually incubated in 100% humidity and 12 h/12 h light/dark conditions. The disease phenotypes via punch/spray-inoculation were recorded at 5 days post-inoculation (dpi) (Chandran et al., 2019; Zhao et al., 2019). Disease severity was measured by the relative fungal mass that was calculated using the DNA level of MoPot2 against the rice ubiquitin gene via quantitative PCR (Li et al., 2017). For rice-sheath inoculation assays, 7 cm-long leaf sheaths were injected with GZ8 (1 × 105 spores mL–1) spore suspension (Kankanala et al., 2007). The epidermal layer from the inoculated leaf sheaths was analyzed by Laser Scanning Confocal Microscopy (LSCM) (Nikon A1) at 24 and 36 hpi, respectively.
Gene Expression Analyses
Total RNAs were extracted from rice leaves using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. The first-strand cDNA was synthesized by using Primescript RT reagent Kit with gDNA Eraser, following the manufacturer’s instructions [TaKaRa Biotechnology (Dalian) Co., Ltd., Japan]. RT-qPCR was performed by using specific primers and SYBR Green mix (SYBR Green PCR Kit, Bimake) with BIO-RAD C1000TM Thermal Cycler. Rice ubiquitin (UBQ) gene was used as an internal reference for data normalization. The total RNA was reverse-transcribed by using a specific stem-loop RT primer (miR156 stem-loop RT primer) to examine the expression of mature miR156. The RT products were subsequently used as the template for RT-qPCR by using a miR156-specific forward primer and a universal reverse primer. The stem-loop primers were designed as described previously (Chen et al., 2005). The snRNA U6 was used as an internal reference for the detection of miRNAs. RT-qPCR was carried out by using the indicated primers (Supplementary Table S1).
Transient Expression Assay in N. benthamiana
The Agrobacterium strain GV3101 carrying the individual construct in the binary vector pCAMBIA1300 was incubated in LB media containing specific antibiotics (rifampin, 50 μg/mL; kanamycin, 50 μg/mL) at 28°C incubation. The bacteria were collected at 5,000 rpm for 5 min and resuspended in MMA buffer (10 mM MES, 10 mM MgCl2, and 100 μM AS). The Agrobacteria containing the expression constructs were infiltrated into leaves of N. benthamiana for transient expression assay. Leaves were examined for image acquisition by using the Zeiss fluorescence microscope (Zeiss imager A2) between 36 and 72 hpi. Western blot analyses were performed following a previous protocol (Li et al., 2017). Total proteins were extracted from equal amounts of leaves with the protein extraction buffer with 1 × loading buffer (0.05 mg/mL Bromophneil blue; 0.065 M Tris-HCl, pH 6.8; 0.02 g/mL SDS; 0.05 mL/mL 2-mercaptoethanol; 0.1 mL/mL glycerol; 1 × protease inhibitor cocktail EDTA-free, Bimake, b14002). Total proteins were separated by 10% SDS-PAGE and transferred to PVDF membrane (Millipore) by using Trans-Blot Turbo (BIO-RAD). Protein blot was hybridized with the rabbit anti-GFP to determine YFP accumulation, and membranes were stained with Ponceau S as the loading control.
Agronomic Traits Measurement
The rice plants for agronomic traits assay were planted in a paddy field in Chengdu, China (36° N, 103° E), during the rice-growing season from April to September. Five representative plants for each line were sampled for measurement of agronomic traits, including plant height, number of tillers, panicle length, number of primary rachis branches (PBs) per panicle, number of secondary rachis branches (SBs) per panicle, number of seeds per panicle, seed length, seed width, 1,000-grain weight, and grain yield per plant on a SC-G seed-counting and grain-weighing device (Wanshen Ltd., Hangzhou, China). The data were analyzed by a one-way ANOVA followed by post hoc Tukey HSD analysis with significant differences (P < 0.05) by using SPSS statistics software.
Results
miR156fhl-3p Is Responsive to Chitin Treatment and M. oryzae Infection
Rice genome contains twelve MIR156 genes generating three mature miR156-5p isoforms and four miR156-3p isoforms (Supplementary Figure S1A)1. One major miR156-5p isoform was transcribed from Osa-miR156a, Osa-miR156b, Osa-miR156c, Osa-miR156d, Osa-miR156e, Osa-miR156f, Osa-miR156g, Osa-miR156h, Osa-miR156i, and Osa-miR156j (designated miR156-5p); whereas four miR156-3p isoforms were respectively transcribed from Osa-miR156f, Osa-miR156h, and Osa-miR156l (designated miR156fhl-3p), Osa-miR156c and Osa-miR156g (designated miR156cg-3p), Osa-miR156b (miR156b-3p), and Osa-miR156j (miR156j-3p) (Supplementary Figure S1A). In addition, Osa-miR156f and Osa-miR156h generated an identical pre-miRNA (designated pre-miR156fh). In our previous work, we detected the different expression patterns of miRNAs in resistance accession IRBLkm-Ts and susceptible accession LTH following the M. oryzae induction by high-through RNA-Seq assay. The abundance of miR156fhl-3p was remarkably higher than that of miR156b-3p, miR156cg-3p, and miR156j-3p, and all of their abundances were significantly lower in resistance accession IRBLkm-Ts than in susceptible accession LTH (Supplementary Figure S2; Li et al., 2014). Therefore, we focused on miR156fhl-3p in this study. To confirm miR156fhl-3p is responsive to the fungal pathogen, we conducted a time-course analysis of the accumulation pattern of miR156fhl-3p in LTH following chitin treatment, as well as in LTH and IRBLkm-Ts following infection of an M. oryzae strain FJ08-09-1 (Figure 1B). Chitin is a component of the fungal cell wall and can induce immune responses (Kawano and Shimamoto, 2013; Li et al., 2014; Kawasaki et al., 2017). Upon application of chitin, the abundance of miR156fhl-3p was significantly decreased at 1 and 3 h post-treatment (hpt) in comparison with mock treatment (Figure 1A), indicating that miR156fhl-3p is responsive to chitin treatment. In contrast, upon M. oryzae inoculation, the amount of miR156fhl-3p was significantly increased in susceptible accession LTH, and unchanged in IRBLkm-Ts at 12 and 24 hpi (Figure 1C). However, the amount of miR156fhl-3p was significantly less in resistance accession IRBLkm-Ts than that in susceptible accession LTH (Figure 1C). These results suggested that miR156fhl-3p is responsive to chitin and M. oryzae.
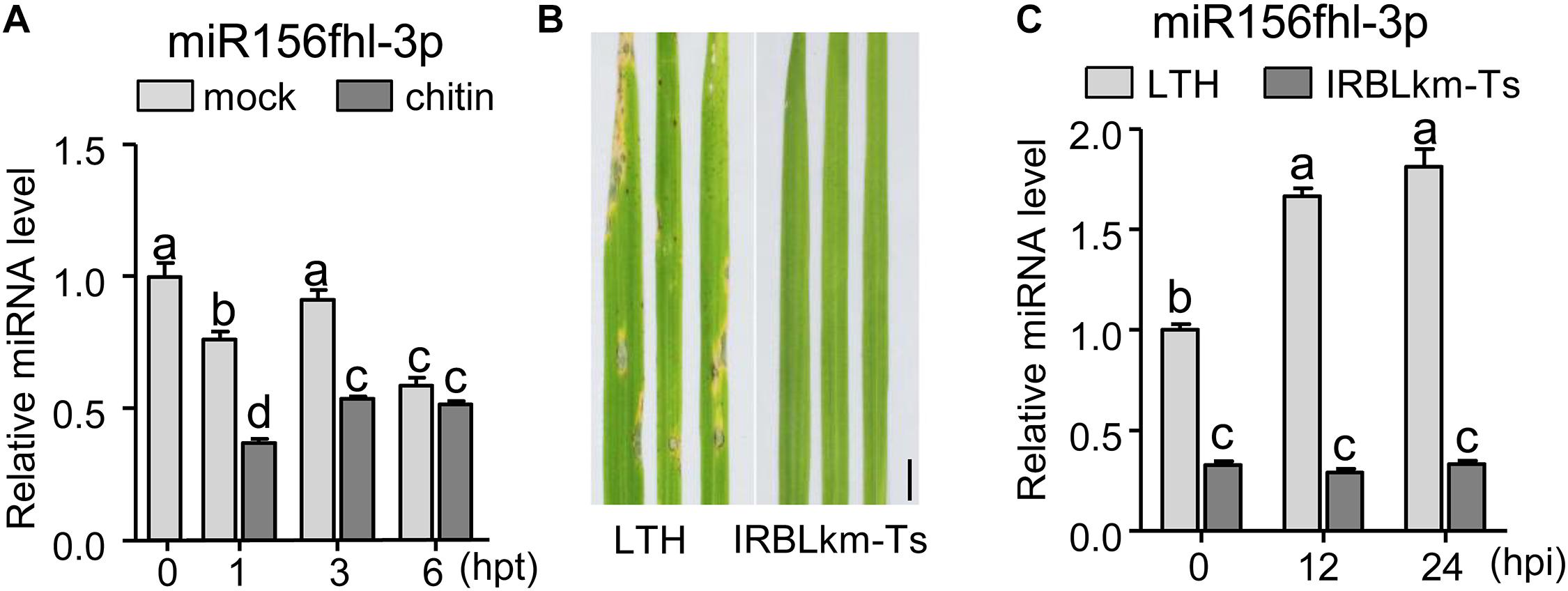
Figure 1. miR156fhl-3p is differentially responsive to chitin and M. oryzae. (A) Expression pattern of miR156fhl-3p upon chitin treatment. (B) Representative leaf sections from LTH and IRBLkm-Ts at 5 dpi upon M. oryzae strain FJ08-09-1. Scale bars, 1 cm. (C) Expression pattern of miR156fhl-3p in LTH and IRBLkm-Ts following M. oryzae inoculation. RNAs were extracted from leaves at the indicated time points. miR156fhl-3p was analyzed using Stem-loop RT-qPCR. SnRNA U6 served as the internal reference. Error bars indicate standard deviation (SD) (n = 3). Different letters above the bars indicate significant differences (P < 0.05) determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis. These experiments were repeated two times with similar results.
Blocking miR156fhl-3p via a Target Mimic Compromises Rice Susceptibility to M. oryzae
To detect the roles of miR156fhl-3p in rice resistance to blast disease, we constructed transgenic lines overexpressing a target mimic of miR156fhl-3p (MIM156-3p), which was complementary to miR156fhl-3p with 3-nt insertion between position 10 and 11 nts and acted as a sponge to hold miR156fhl-3p (Franco-Zorrilla et al., 2007; Supplementary Figure S1B). Out of seven lines, we selected two lines for further investigation, namely MIM156-3p-1 and MIM156-3p-2, which displayed significantly decreased miR156fhl-3p accumulation in comparison with control Nipponbare (WT) plants (Figure 2A). Both transgenic lines showed much smaller disease lesions and less fungal mass than control via punch-inoculation of the M. oryzae strain GZ8 and 97-27-2 (Figures 2B,C). Consistently, both transgenic lines displayed compromised susceptibility with smaller disease lesions and less fungal growth following spray-inoculation with GZ8 and another strain 97-27-2 (Figures 2B,D). These data indicate that blocking miR156fhl-3p compromises rice susceptibility to M. oryzae.
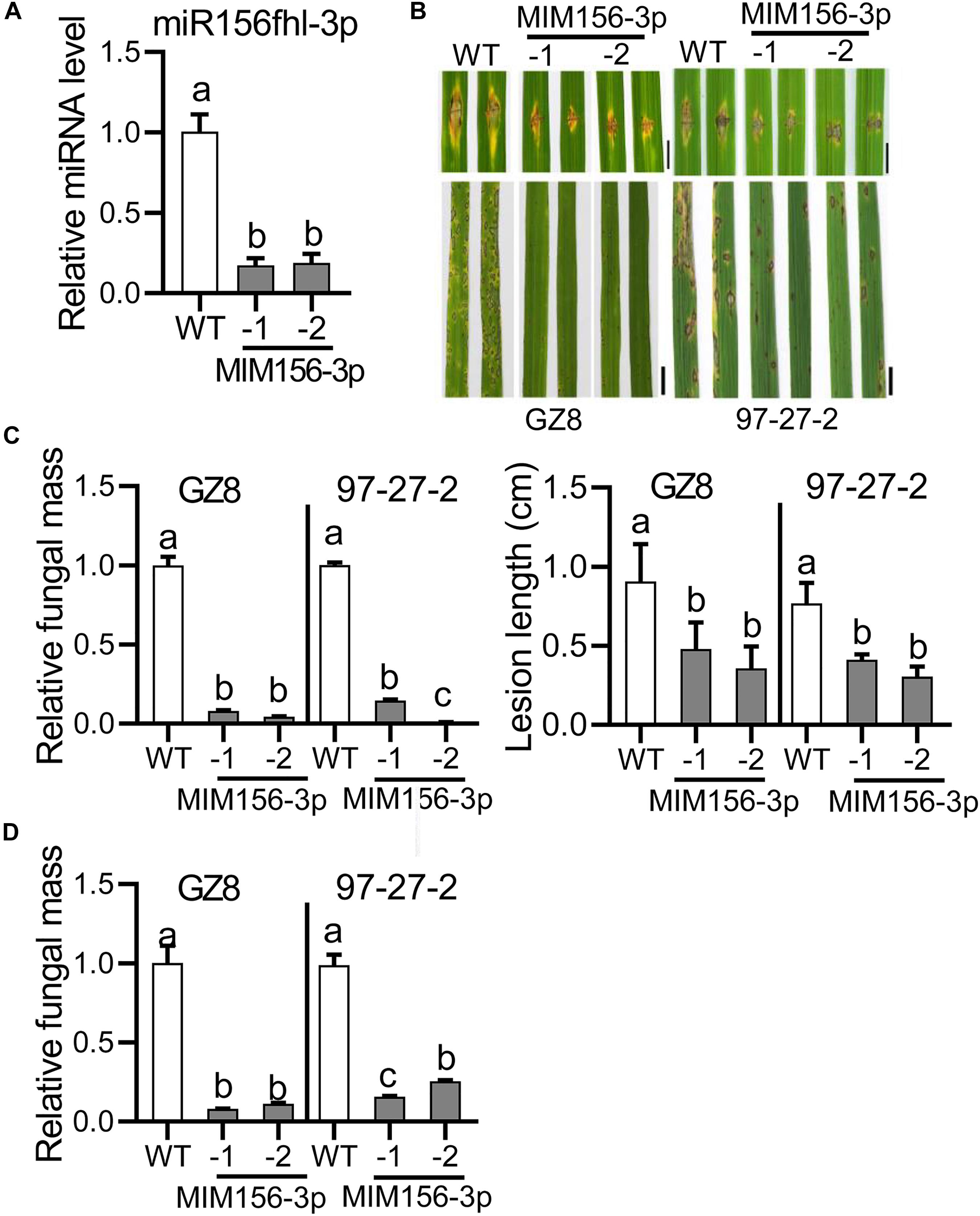
Figure 2. Expression of a target mimic of miR156fhl-3p leads to compromised susceptibility to M. oryzae. (A) Relative amounts of miR156fhl-3p in transgenic lines expressing a target mimic of miR156fhl-3p (MIM156-3p) and WT plants. The amounts of mature miR156fhl-3p were analyzed using stem-loop RT-qPCR. (B) Disease phenotype of control and MIM156-3p lines. Three-leaf-stage seedlings were punch/spray-inoculated with M. oryzae strains GZ8 and 97-27-2. Leaf phenotypes were observed at 5 dpi. Scale bars: Punch-inoculation assays are 5 mm; spray-inoculation assays are 1 cm. (C) Relative fungal mass represented by measuring MoPot2 gene against OsUbiquitin DNA level and average lesion length via punch-inoculation. (D) Relative fungal mass via spray-inoculation. Error bars indicate SD (n = 3). Different letters above the bars indicate significant differences (P < 0.05) determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis. These experiments were repeated two times with similar results.
Expression of a Target Mimic of miR156fhl-3p Delays M. oryzae Infection and Enhances the Induced Expression of Defense-Related Genes
To understand why blocking miR156fhl-3p leads to compromised susceptibility to blast disease, we examined the expression of the two defense-related response marker genes, PR1a and NAC4, upon M. oryzae strain GZ8 infection. The expression of both genes was induced to significantly higher levels by GZ8 in MIM156-3p than that in control at 48 hpi (Figures 3A,B). Then, we compared the infection process in leaf sheath of WT control and MIM156-3p plants with a LSCM after the inoculation of the GFP-tagged strain GZ8. The growth of GZ8 was delayed in MIM156-3p at 24 and 36 hpi in comparison with that in control (Figure 3C). Quantified infection phase assay showed that more than 60% of spores formed the invasive hyphae in local cells at the infection site in WT, whereas only 10% spores formed invasive hyphae in the local cells in MIM156-3p (Figure 3D). At 36 hpi, more than 70% of hyphae infected into the first cell, and 17% of hyphae extend to the neighbor cells in control. In contrast, about 45% of spores formed the appressorium, and 55% spores formed invasive hyphae and infected into the local cells in MIM156-3p (Figure 3D). These observations suggested that blocking miR156fhl-3p delayed the infection process of M. oryzae and promoted the induction of defense-related genes.
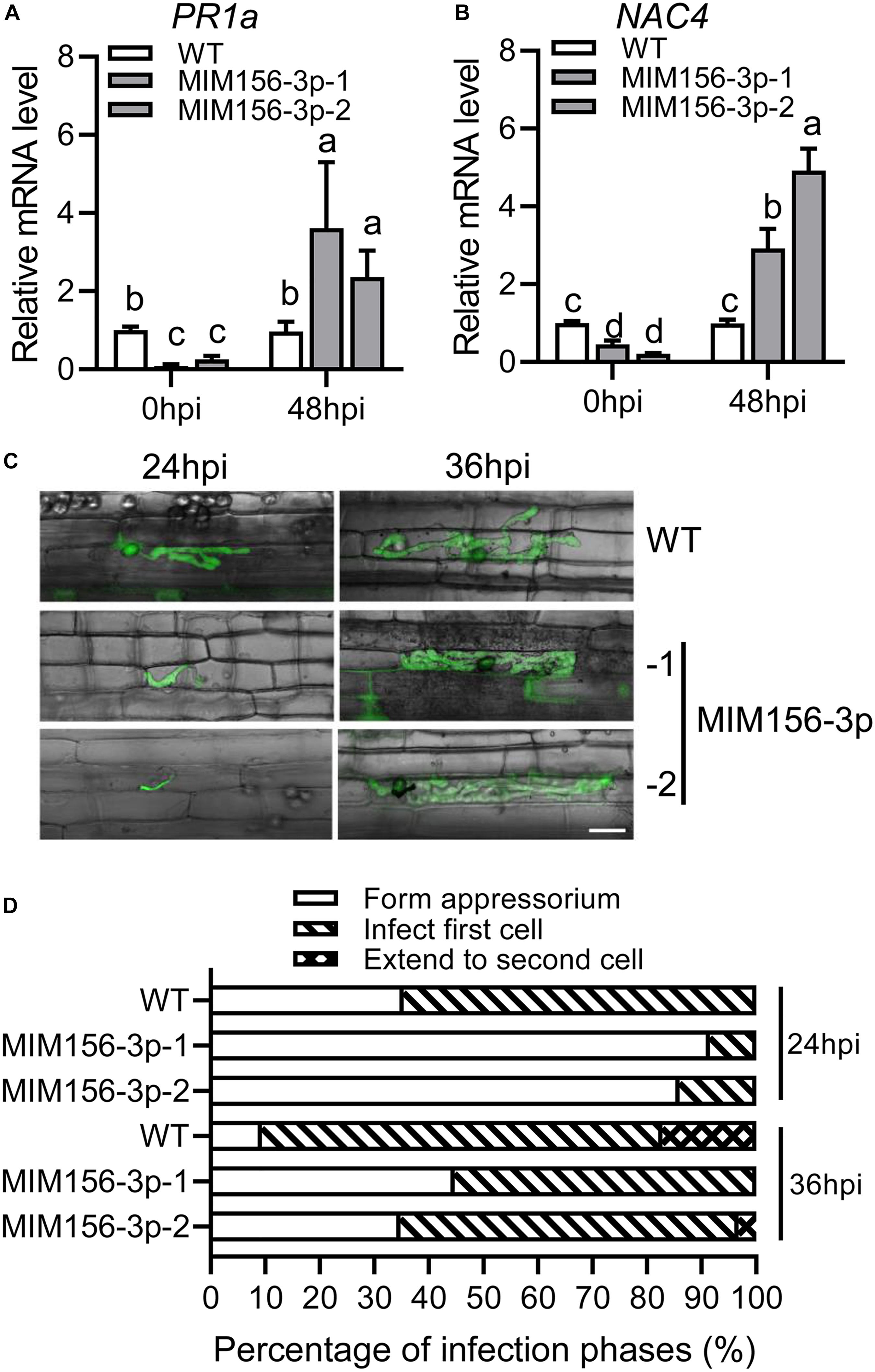
Figure 3. Expression of a target mimic of miR156fhl-3p improves the induction of defense-related genes and delays the M. oryzae infection process. (A,B) Induction of defense-related genes PR1a (A) and NAC4 (B) upon M. oryzae infection in WT and MIM156-3p lines. Error bars indicate SD (n = 3). Different letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis. (C) Representative LSCM images show the development of M. oryzae strain GZ8 at 24 and 36 hpi in sheath cells of WT and MIM156-3p lines. Scale bars, 20 μm. (D) Statistical analysis of M. oryzae growth in (C). More than 200 conidia in each line were analyzed. These experiments were repeated two times with similar results.
Expression of a Target Mimic of miR156fhl-3p Does Not Penalize Agronomic Traits
Activation of blast resistance often penalizes growth and agronomic traits that lead to yield formation (Heil and Baldwin, 2002; Walters and Boyle, 2005; Shimono et al., 2007; Walters and Heil, 2007). To detect whether blocking miR156fhl-3p via target mimicry affected rice growth, we assayed the agronomic traits of MIM156-3p plants. Intriguingly, MIM156-3p showed similar gross plant morphology as WT (Figures 4A,B). Statistical analysis indicated that several agronomic traits were slightly but not significantly decreased in MIM156-3p in comparison with those in WT (Supplementary Figure S3), including plant height, number of tillers per plant, panicle length, and number of primary rachis branches per panicle (No. of PBs). The number of secondary rachis branches per panicle (No. of SBs) was significantly decreased in MIM156-3p plants (Figure 4E), leading to a reduced number of grains per panicle (Figure 4F). In contrast, the seed length (Figures 4C,G) and width (Figures 4D,H) in MIM156-3p were increased, resulting in an increased 1,000-grain weight (Figure 4I). As a result, the grain yield per plant was unchanged in MIM156-3p (Figure 4J) in comparison with that in WT. Altogether, our results indicated that blocking miR156fhl-3p could improve rice blast disease resistance without penalty of yield.
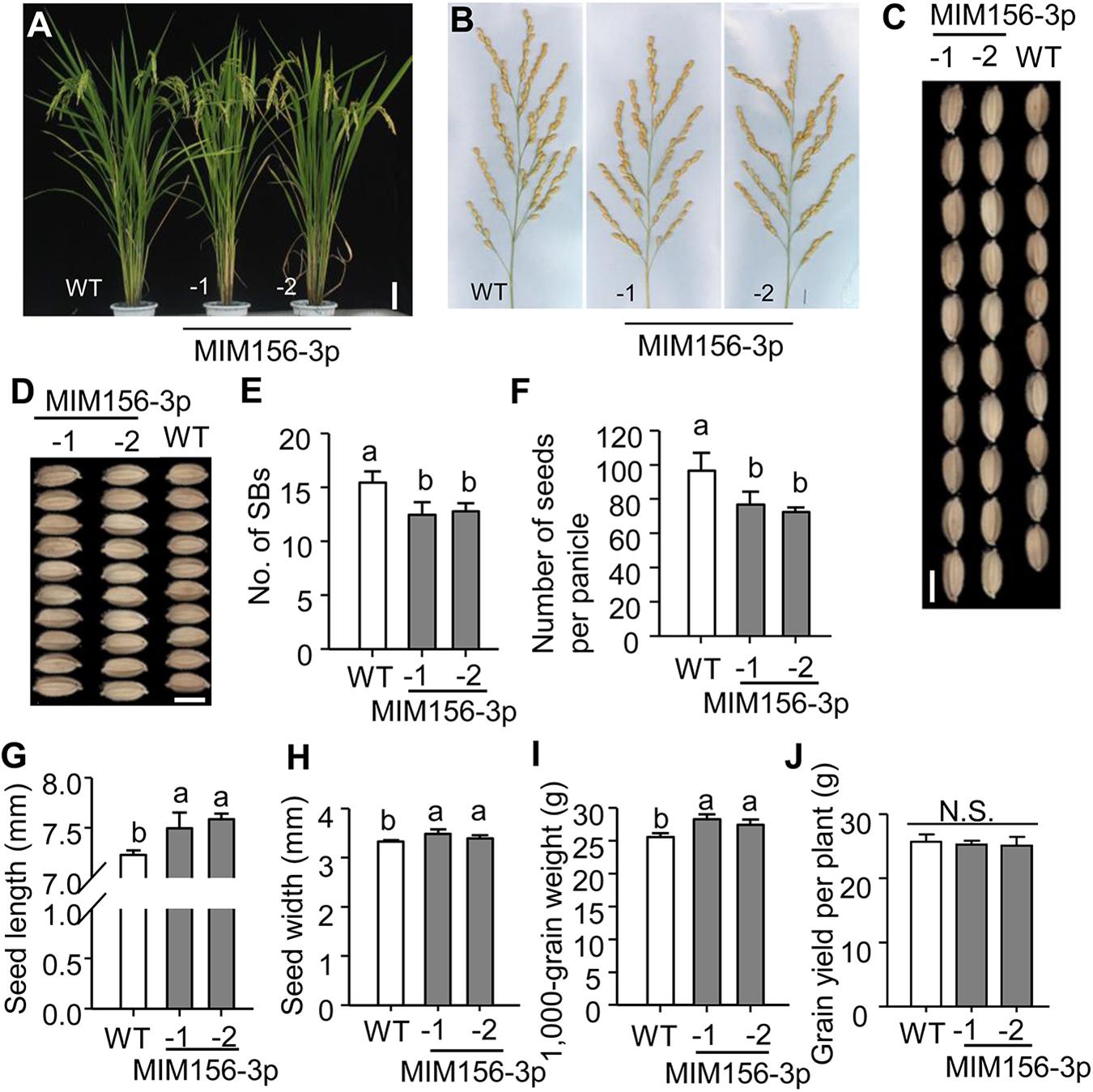
Figure 4. Blocking miR156fhl-3p leads to changed rice agronomic traits without effect on yield. (A) Gross morphology of WT and MIM156-3p lines. Size bar, 10 cm. (B) Panicle morphology of WT and MIM156 lines. Size bar, 1 cm. (C,D) Grain length and width of WT and MIM156-3p. Size bar, 5 mm. (E–J) Statistical analysis of the number of secondary rachis branches per panicle (No. of SBs) (E), the number of seeds per panicle (F), seed length (G), seed width (H), 1,000-grain weight (I), and grain yield per plant (J) of WT and MIM156-3p lines. Error bars indicate standard deviation (n = 5). Different letters above the bars indicate significant differences (P < 0.05) determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis.
Expression of a Target Mimic of miR156fhl-3p Suppresses miR156-5p Accumulation and Increases SPL14 Expression
To uncover how miR156fhl-3p regulated rice resistance against M. oryzae, we first predicted potential target genes of miR156fhl-3p by using psRNATarget2, an online tool for prediction of miRNA targets. We chose two best-matched candidate genes, namely LOC_Os02g17280 and LOC_Os07g37580, for examination (Supplementary Figure S4A). However, we failed to detect changes in their expressions in both MIM156-3p lines (Supplementary Figures S4B,C), indicating that the two candidates were possibly not regulated by miR156fhl-3p at mRNA level. We then tested whether the expression of MIM156-3p affected the accumulation of miR156-5p. Surprisingly, the abundance of mature miR156-5p was significantly decreased in MIM156-3p in comparison with that in WT (Figure 5A). Consistently, the abundance of pre-miR156fh was also significantly decreased in MIM156-3p in comparison with that in WT (Figure 5B). These data indicate that the blocking of miR156fhl-3p by MIM156fhl-3p could suppress the accumulation of miR156-5p. Previously, miR156-5p was reported to regulate the expression of SPL14 (Jiao et al., 2010). The SPL14 protein is phosphorylated during M. oryzae infection, and the phosphorylated SPL14 binds to the promoter of WRKY45 to promote its expression. In turn, WRKY45 positively regulates rice immunity against M. oryzae (Shimono et al., 2007; Wang et al., 2018). We speculated that the expression of SPL14 and WRKY45 should be altered in MIM156-3p lines. As expected and reversely to the decreased accumulation of miR156, the mRNA amounts of SPL14 and WRKY45 were significantly higher in MIM156-3p than in control, and the expression of WRKY45 was further significantly increased compared to that in control at 48 hpi of M. oryzae (Figures 5C,D). These results implied that the mRNA abundance of SPL14 could be regulated by miR156fhl-3p via interference with miR156-5p, thus boosting the expression of WRKY45 to enhance rice blast disease resistance.
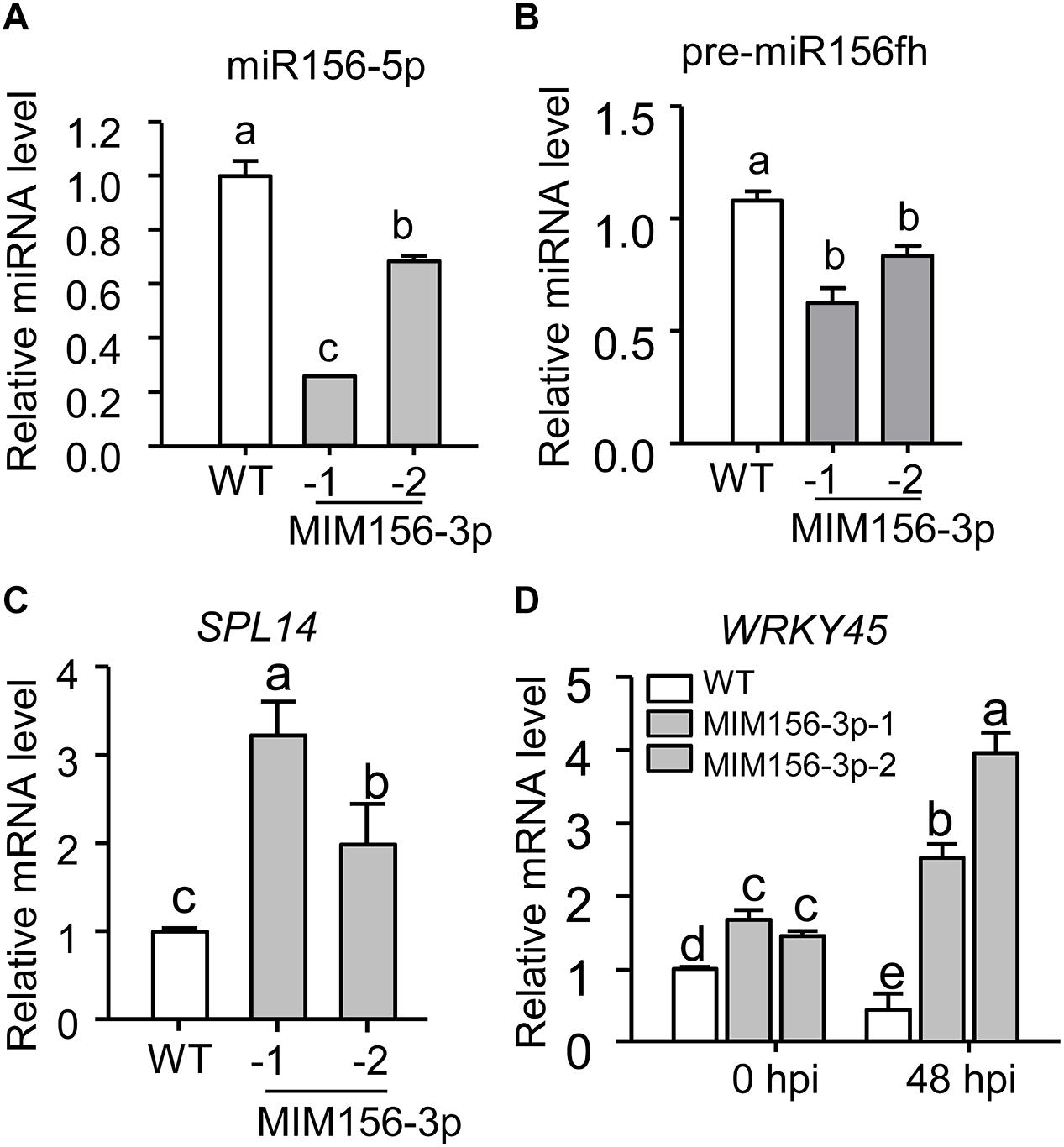
Figure 5. Blocking miR156fhl-3p leads to a decreased abundance of miR156-5p but increased expression of SPL14 and WRKY45. (A,B) Relative amount of mature miR156-5p (A) and pre-miR156fh (B) in WT and MIM156-3p transgenic lines. (C) Relative mRNA amounts of SPL14 in WT and MIM156-3p. (D) Relative mRNA amounts of WRKY45 with or without M. oryzae infection in WT and MIM156-3p at indicated time points. In (A–D), error bars indicate SD (n = 3). Different letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis. These experiments were repeated two times with similar results.
To confirm the interference of miR156fhl-3p with miR156-5p, we established a YFP-based reporter system. In the system, we made a construct expressing a YFP reporter that was fused with the miR156 binding site of SPL14 (SPL14TBS) at the N-terminus (SPL14TBS-YFP) and a construct expressing a mutant target site of SPL14 (SPL14MBS-YFP). SPL14TBS-YFP was separately expressed or co-expressed with miR156 and MIM156-3p in N. benthamiana leaves. Then the YFP reporter was examined by YFP intensity and Western blotting. The YFP intensity and protein accumulation were highly accumulated when SPL14TBS-YFP was expressed alone. In contrast, the YFP intensity and protein accumulation were significantly decreased when co-expressed with miR156 (Figures 6A,B), indicating suppression of miR156 on SPL14TBS-YFP. Intriguingly, the YFP intensity and protein accumulation were recovered when MIM156-3p was added in the co-transient expression (Figures 6A,B). In contrast, the YFP intensity and protein accumulation of SPL14MBS-YFP kept unchanged when co-expressed with miR156 or with both miR156 and MIM156-3p (Figures 6A,B). These data imply that MIM156-3p could release the suppression of SPL14 by miR156 via interference with miR156-5p, leading to activation of SPL14.
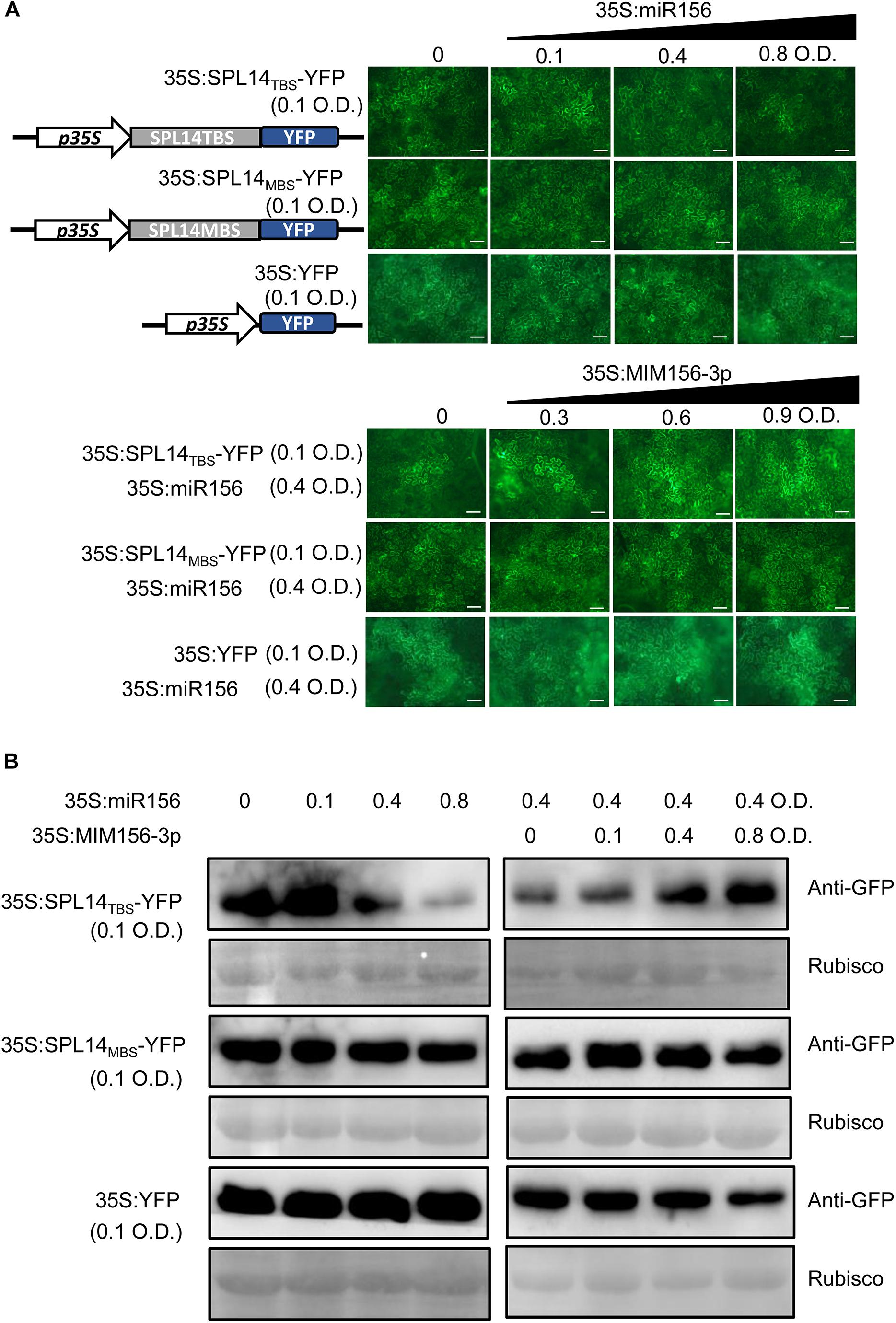
Figure 6. Blocking miR156fhl-3p releases SPL14 from suppression by miR156-5p. (A) Confocal images show the protein levels of SPL14TBS-YFP, SPL14MBS-YFP, and YFP co-expressed with miR156 or with miR156 and MIM156-3p. The indicated SPL14TBS-YFP, SPL14MBS-YFP, and YFP reporter constructs were transiently expressed alone or co-expressed with miR156 and MIM156-3p in N. benthamiana leaves by Agrobacterium-mediated transformation at the indicated optical density (O.D.) concentration. Size bars, 50 μm. (B) Western blotting analysis shows the protein abundance of SPL14TBS-YFP, SPL14MBS-YFP, and YFP. Protein extracts from infiltrated leaves were collected for Western blot analysis using anti-GFP sera and anti-Rubisco sera, respectively. These experiments were repeated two times with similar results.
miR156 Negatively Regulates Rice Blast Resistance
As MIM156-3p plants exhibited enhanced blast resistance via suppressing the accumulation of miR156-5p, we speculated that miR156-5p acts in blast resistance and should be responsive to M. oryzae. To confirm this hypothesis, we first examined the expression pattern of miR156-5p following chitin treatment in LTH and following M. oryzae invasion in LTH and IRBLkm-Ts. We compared the mock and chitin treatment in inducing miR156 expression. The abundance of miR156-5p was increased at 3 and 6 hpt in comparison with mock treatment (Figure 7A), indicating miR156-5p is responsive to chitin. Moreover, miR156-5p was significantly increased in LTH upon M. oryzae infection, whereas it was decreased at 12 and 24 hpi in IRBLkm-Ts (Figure 7B), implying a negative role in pathogen-induced immunity. Then, we constructed transgenic lines overexpressing miR156h (OX156), which formed the same mature sequence as miR156abcdefgij (miR156-5p) (Supplementary Figure S1). OX156 plants showed remarkably decreased plant height (Supplementary Figure S6A) and other agronomic traits (Supplementary Figures S6B–J), as previously reported (Wang L. et al., 2015). We chose two transgenic lines, OX156-1 and OX156-2, for further study (Figure 7C and Supplementary Figure S6A). Both transgenic lines showed enhanced susceptibility to M. oryzae strain GZ8 and 97-27-2, with more significant disease lesions (Figures 7D,F) and fungal mass (Figure 7E) in comparison with control via punch-inoculation. The disease lesions in OX156-1 were even bigger than that in OX156-2 plants, which was consistent with the more accumulation of miR156-5p in OX156-1 than that in OX156-2 (Figures 7C–F). Furthermore, we found that the mRNA amounts of SPL14 and WRKY45 were decreased in OX156 lines (Figures 7G,H), indicating that miR156-5p could facilitate the growth of M. oryzae in a dosage-dependent manner via suppressing the expression of SPL14, which in turn down-regulates WRKY45 expression.
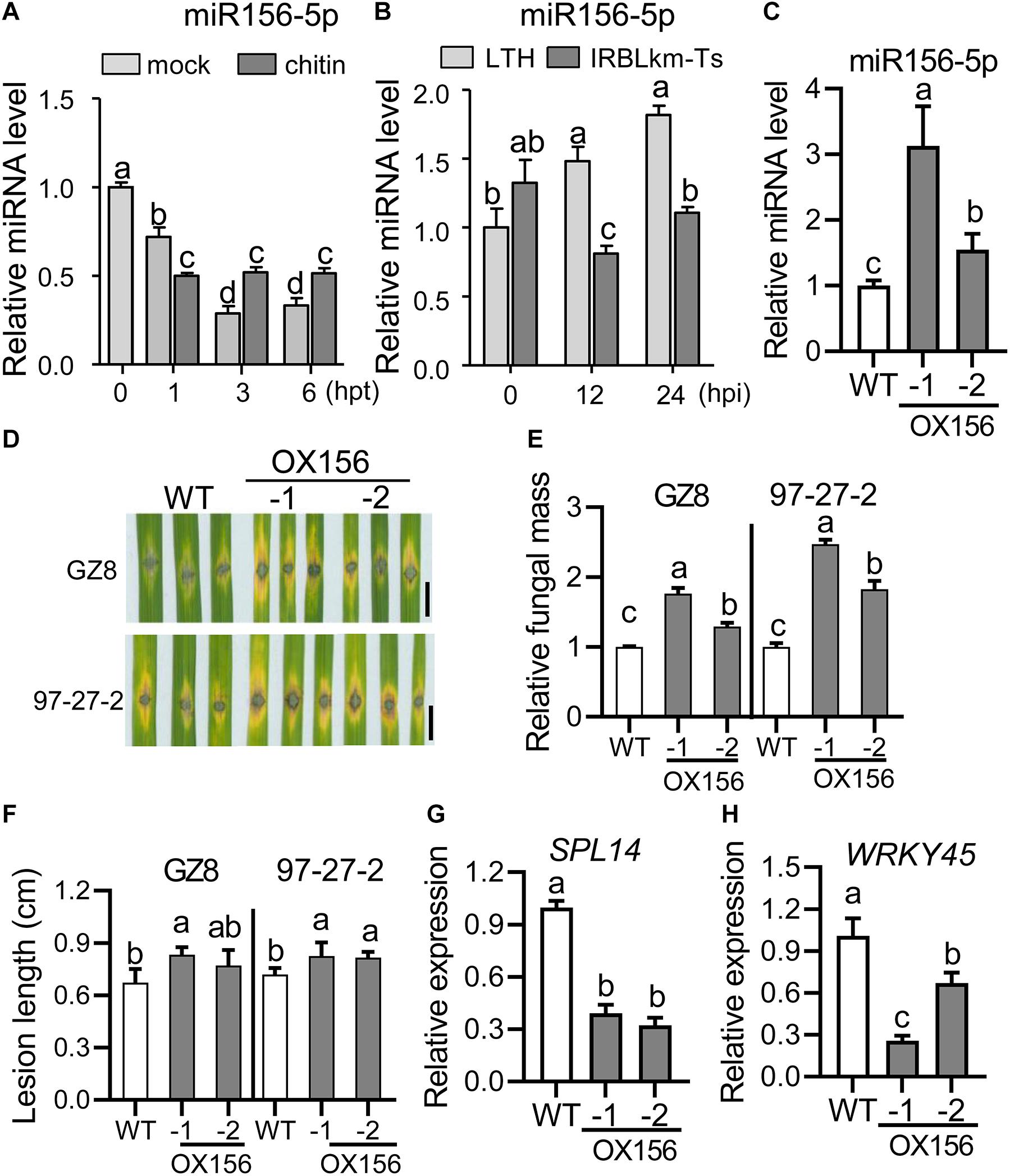
Figure 7. Overexpression of miR156h results in enhanced susceptibility to M. oryzae. (A) Expression pattern of miR156-5p upon chitin treatment. (B) Expression pattern of miR156-5p in LTH and IRBLkm-Ts following M. oryzae inoculation. (C) Relative mRNA amount of miR156-5p in WT and OX156 lines. RNAs were extracted from leaves of three-leaf-stage plants from the indicated lines. The amounts of mature miR156 were analyzed using Stem-loop RT-qPCR. SnRNA U6 served as the internal reference. (D) Disease phenotypes of the indicated lines were observed at 5 dpi of M. oryzae strains GZ8 and 97-27-2. Scale bars, 5 mm. (E) Relative fungal mass represented by measuring MoPot2 gene against OsUbiquitin DNA level. (F) Average lesion length. (G,H) RT-qPCR analysis of relative mRNA amounts of SPL14 (F) and WRKY45 (G) in WT and OX156 lines. For (A–C,E–H), error bars indicate standard deviation (n = 3). Different letters above the bars indicate significant differences (P < 0.05) determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis.
Discussion
Previously, miRNA-3p was marked as miRNA∗ that was considered to be a side-product subjected to decay (Jones-Rhoades et al., 2006; Krol et al., 2010). Later on, miRNA-3p was found to target genes different from those of miRNA-5p and thus performed different fine-tuning functions (Navarro et al., 2006; Zhang et al., 2011). Here, we found that expressing a target mimic of miR156fhl-3p affects the abundance of miR156-5p, which in turn regulates a target gene of miR156-5p to fine-tune the tradeoffs between blast disease resistance and yield traits.
Two techniques provide efficient tools to block the activity of a mature miRNA. One is target mimicry technology that is established based on the modulation of IPS1 on the miR399 activity in Arabidopsis. The other is called short tandem target mimicry (STTM), which exploits two target mimics of a miRNA to increase the efficacy of a target mimic in blocking the function of a miRNA (Yan et al., 2012; Zhang et al., 2017). In our previous study, we found that the abundance of miR156fhl-3p was lower in a resistance accession than in a susceptibility accession; still, its abundance was much more than those of miR156b-3p, miR156cg-3p, and miR156j-3p (Supplementary Figure S2; Li et al., 2014). Here, we exploited target mimicry to explore the function of miR156fhl-3p. Transgenic MIM156-3p plants showed compromised susceptibility to M. oryzae with less fungal mass and shorter lesion length than control (Figures 2B–D). Consistently, defense-related genes, such as PR1a and NAC4, were significantly up-regulated higher in MIM156-3p than in control (Figures 3A,B), and the fungal invasion process was obviously delayed in MIM156-3p plants (Figures 3C,D). Moreover, transgenic MIM156-3p plants also produced a grain yield comparable to that of control, which was built on the orchestration of the number of panicles, panicle size, and grain weight (Figure 4). Therefore, these data indicate that expressing a target mimic of miR156fhl-3p improves blast disease resistance without the yield penalty.
The expression of either a target mimic or an STTM of a miRNA can lead to a decrease in the abundance of the miRNA via an unknown mechanism (Zhang et al., 2017). Here, we found that the abundance of miR156fhl-3p was significantly down-regulated in MIM156-3p lines (Figure 2A). Surprisingly, the abundance of the mature miR156-5p was also significantly down-regulated (Figure 5A), which represents the transcripts from Osa-miR156a, Osa-miR156b, Osa-miR156c, Osa-miR156d, Osa-miR156e, Osa-miR156f, Osa-miR156g, Osa-miR156h, Osa-miR156i, and Osa-miR156j (Supplementary Figure S1). It is well-known that a MIR gene is transcribed into a pri-miRNA that is processed into the precursor of a mature miRNA, namely a pre-miRNA (Rogers and Chen, 2013). A pre-miRNA folds into a hairpin that is cleaved by a DCL protein and exported to the cytoplasm, where the DCL processes it into a miRNA-5p/miRNA-3p duplex (Liu et al., 2005). One or both of the strands of the miRNA-5p/miRNA-3p duplex are incorporated into AGO proteins to mediate the silencing of target genes via sequence complementarity (Zhu et al., 2011; Kobayashi and Tomari, 2016). We also found the down-regulation of pre-miR156fh in MIM156-3p lines (Figure 5B), indicating that the target mimic of miR156fhl-3p may interfere with the expression of miR156 family genes at either the transcription or the procession stage. Previously, the expression of SPL14 in a mutant, i.e., ipa1 (Ideal Plant Architecture1) that carries a mutation at the miR156 target site, was not influenced by the infection of M. oryzae; instead, the phosphorylation of IPA1 was changed during M. oryzae infection, leading to improvement of both blast disease resistance and yield (Wang et al., 2018). We confirmed such interference of a -3p with -5p using an SPL14TBS-YFP-based reporter system, through which we confirmed that the overexpression of a target mimic of miR156fhl-3p interfered with miR156-5p miRNA level, which did release the suppression of miR156-5p on the SPL14TBS-YFP accumulation (Figure 6). Consistently, the expression of both SPL14 and its downstream gene WRKY45 was increased in MIM156-3p (Figures 5C,D). However, we currently could not tell whether the inference of a -3p with a -5p occurs at the transcription or the post-transcription step. Alternatively, the phenotypes observed in MIM156-3p could be due to altered expression of the target genes of miR156fhl-3p. We searched for the target genes of miR156fhl-3p through website prediction. Among these candidate genes, we selected two genes for expression analysis (Supplementary Figure S4A) and found that their expressions were not significantly altered in MIM156-3p (Supplementary Figure S4B). It is unclear whether the target genes of miR156fhl-3p are involved in regulating rice resistance and agronomic traits. Therefore, future work should focus on searching miR156fhl-3p target genes and exploit Northern blotting to examine the abundance of each pri-miR156 and pre-miR156 to figure out which step is impacted by the target mimic of miR156fhl-3p. Also, future research should focus on addressing whether such interference of a -3p with a -5p is specific to MIR156 genes or a common phenomenon among different miRNAs.
MIR156 family genes were classified into two groups based on their functions in regulating rice agronomic straits. Group I includes miR156d, miR156e, miR156f, miR156g, miR156h, miR156i, and miR156j, acting in controlling grain size (Miao et al., 2019); group II includes miR156a, miR156b, miR156c, miR156l, and miR156k, contributing to shoot architecture (Supplementary Figure S5; Miao et al., 2019). Intriguingly, miR156a, miR156b, miR156c, miR156d, miR156e, miR156f, miR156g, miR156h, miR156i, and miR156j are transcribed into the same -5p mature sequences (Supplementary Figure S1). miR156fh is in group I, and miR156l is in group II (Supplementary Figure S5). Transgenic OX156 lines overexpressing miR156h showed dwarfish phenotypes that were similar to previous reports (Supplementary Figure S6; Xie et al., 2006; Dai et al., 2018; Liu et al., 2019). In contrast to MIM156-3p, which exhibited enhanced blast resistance, OX156 showed enhanced susceptibility, which may be due to the down-regulation of SPL14 and WRKY45 (Figure 7). Because miR156h represents the major and the most abundant isoform of miR156, these data indicate that miR156 negatively regulates blast disease resistance via the SPL14-WRKY45 transcription factor module.
Taken together, we propose a model to summarize our discovery (Figure 8). The expression of a miR156fhl-3p target mimic interferes with the accumulation of both miR156fhl-3p and miR156-5p by an unknown mechanism. Previously, miR156-5p was reported to regulate SPL14 expression via mRNA cleavage or transcription suppress (Jiao et al., 2010). Overexpression of SPL14 enhances rice resistance against M. oryzae (Wang et al., 2018). Here, miR156-5p was down-regulated in MIM156-3p plants, leading to the up-regulation of SPL14, which in turn promotes the expression of WRKY45 to enhance resistance against M. oryzae. However, we could not exclude that the target genes of miR156fhl-3p might contribute to rice resistance to M. oryzae and be involved in the regulation of agronomic traits from our current data. In conclusion, our findings demonstrate another layer of regulation on SPL14 expression by miR156 in fine-tuning the tradeoffs between blast disease resistance and yield.
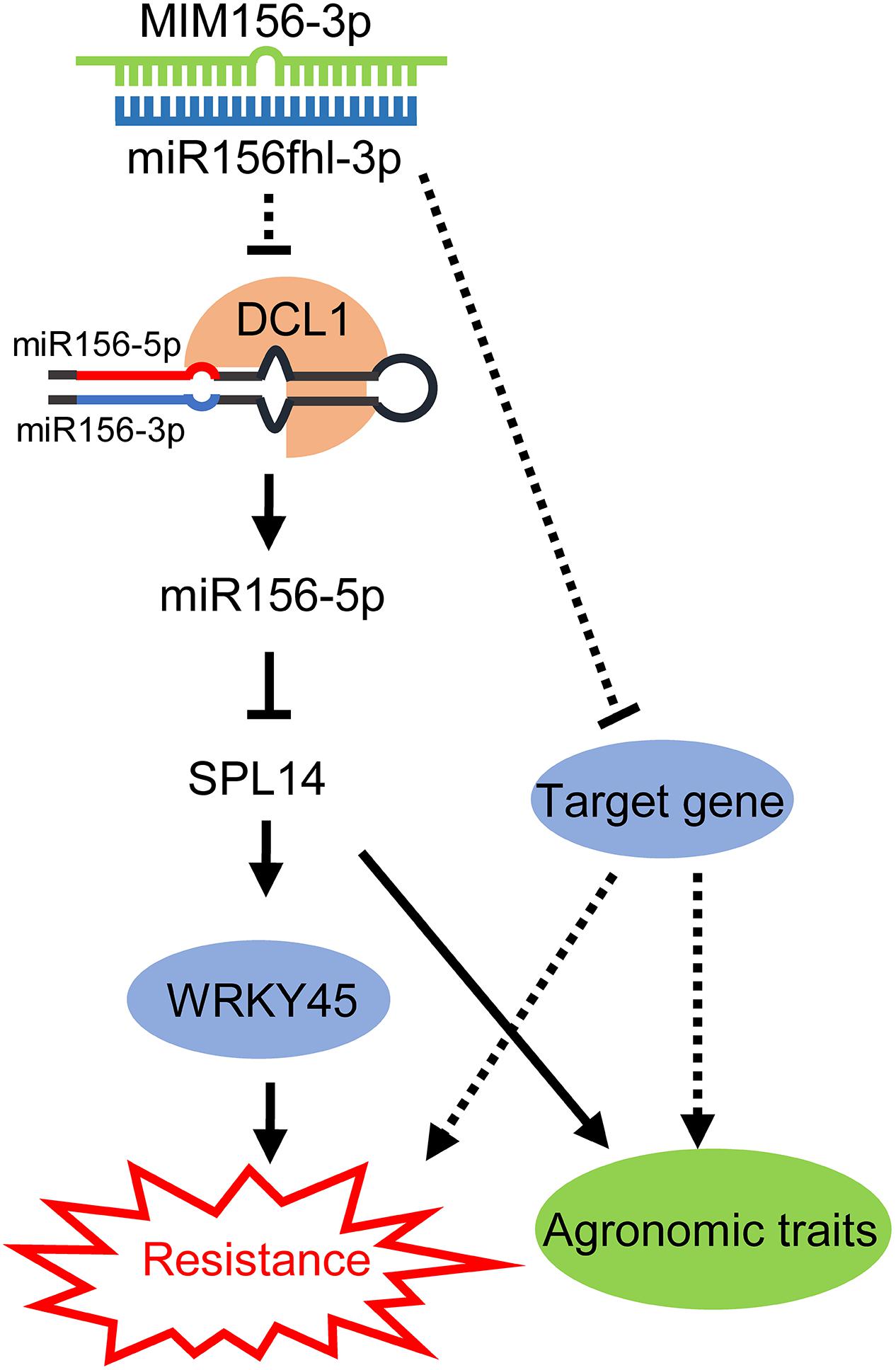
Figure 8. A working model for miR156-3p on regulation of miR156-SPL14 module in resistance and agronomic traits. Overexpression of a target mimic of miR156fhl-3p interferes with miR156fhl-3p, leading to the down-regulation of miR156-5p via an unknown mechanism, which results in the up-regulation of the miR156-5p target gene SPL14. In turn, SPL14 activates WRKY45 expression, leading to enhance rice blast disease resistance without fitness cost. Meanwhile, the unknown target genes of miR156fhl-3p might be involved in regulation of blast disease resistance and agronomic traits.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
W-MW and YL conceived the study. L-LZ, Y-PZ, HW, S-XZ, L-FW, XY, J-FC, X-PL, and X-CM performed the experiment. J-QZ, MP, and HF carried out the field trial. JF, J-WZ, and Y-YH guided the experimental operation. YL, L-LZ, and W-MW contributed to manuscript writing and editing.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31430072 and 31672090 to W-MW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Cai-Lin Lei (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the monogenic resistant lines IRBLkm-Ts.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00327/full#supplementary-material
FIGURE S1 | Schematic diagram of miR156 isoforms and artificial mimicry target of miR156fhl-3p. (A) Sequence alignment of the miR156-5p and miR156-3p isoforms. (B) Alignment of MIM156h-3p with miR156fhl-3p.
FIGURE S2 | RNA-Seq revealed the differential amounts of miR156-3p isoforms following M. oryzae inoculation at indicated time points.
FIGURE S3 | miR156fhl-3p has no significant effect on several agronomic traits. Statistics data of plant height (A), number of tillers (B), panicle length (C) number of primary rachis branches per panicle (No. of PBs) (D), of WT and MIM156-3p lines. Error bars indicate SD (n = 5). Different letters above the bars indicate significant differences (P < 0.05) determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis.
FIGURE S4 | The predicted target genes of miR156fhl-3p are not up-regulated significantly in MIM156. (A) Sequence alignments of miR156fhl-3p and predict target sites of indicated genes. (B,C) RT-PCR analysis of the predicted target genes expression in WT and MIM156-3p plants. Error bars indicate SD (n = 3). Different letters above the bars indicate significant differences (P < 0.05) determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis. These experiments were repeated two times with similar results.
FIGURE S5 | Phylogenetic tree of rice MIR156 gene based on DNA sequences. 100 bp genomic sequences (20 bp upstream sequences corresponding to mature miR156 and 20 bp downstream sequences) were used to construct the phylogenetic tree.
FIGURE S6 | miR156 regulates rice agronomic traits. (A) Gross morphology of WT and transgenic plants over-expressing miR156h (OX156). Scale bars, 10 cm. (B–I) Statistical analysis of plant height (B), number of tillers (C), panicle length (D), number of primary rachis branches per panicle (No. of PBs) (E), number of second rachis branches per panicle (No. of SBs) (F), number of seeds per panicle (G), seed width (H), seed length (I), 1,000-grain weight (J) from the indicated lines. Error bars indicate SD (n = 5). Different letters above the bars indicate significant differences (P < 0.05) determined by one-way ANOVA analysis followed by post hoc Tukey HSD analysis.
TABLE S1 | The primers used in this study.
Footnotes
References
Addoquaye, C., Eshoo, T. W., Bartel, D. P., and Axtell, M. J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. doi: 10.1016/j.cub.2008.04.042
Axtell, M. J., and Bowman, J. L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13, 343–349. doi: 10.1016/j.tplants.2008.03.009
Cardon, G. H., Hohmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. doi: 10.1016/s0378-1119(99)00308-x
Chandran, V., Wang, H., Gao, F., Cao, X.-L., Chen, Y.-P., Li, G.-B., et al. (2019). miR396-OsGRFs module balances growth and rice blast disease-resistance. Front. Plant Sci. 9:1999. doi: 10.3389/fpls.2018.01999
Chen, C., Ridzon, D. A., Broomer, A. J., Zhou, Z., Lee, D. H., Nguyen, J. T., et al. (2005). Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 33, e179. doi: 10.1093/nar/gni178
Dai, Z., Wang, J., Yang, X., Lu, H., Miao, X., and Shi, Z. (2018). Modulation of plant architecture by the miR156f–OsSPL7–OsGH3.8 pathway in rice. J. Exp. Bot. 69, 5117–5130. doi: 10.1093/jxb/ery273
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr.nt Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39:1033. doi: 10.1038/ng2079
Goto, S., Sasakura-Shimoda, F., Suetsugu, M., Selvaraj, M. G., Hayashi, N., Yamazaki, M., et al. (2015). Development of disease-resistant rice by optimized expression of WRKY45. Plant Biotechnol. J. 13, 753–765. doi: 10.1111/pbi.12303
Goto, S., Sasakura-Shimoda, F., Yamazaki, M., Hayashi, N., Suetsugu, M., Ochiai, H., et al. (2016). Development of disease-resistant rice by pathogen-responsive expression of WRKY45. Plant Biotechnol. J. 14, 1127–1138. doi: 10.1111/pbi.12481
Guo, L., Zhang, H., Zhao, Y., Yang, S., and Chen, F. (2014). Selected isomiR expression profiles via arm switching? Gene 533, 149–155. doi: 10.1016/j.gene.2013.09.102
Hao, J., Tu, L., Hu, H., Tan, J., Deng, F., Tang, W., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. doi: 10.1093/jxb/ers278
Heil, M., and Baldwin, I. T. (2002). Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–67. doi: 10.1016/s1360-1385(01)02186-0
Hiroshi, T., Yanoria, M. J. T., Ebron, L. A., Nagao, H., Ikuo, A., Hiroshi, K., et al. (2000). Development of monogenic lines of rice for blast resistance. Breed. Sci. 50, 229–234. doi: 10.1270/jsbbs.50.229
Hu, W., Wang, T., Yue, E., Zheng, S., and Xu, J.-H. (2014). Flexible microRNA arm selection in rice. Biochem. Biophys. Res. Commun. 447, 526–530. doi: 10.1016/j.bbrc.2014.04.036
Huang, C.-Y., Wang, H., Hu, P., Hamby, R., and Jin, H. (2019). Small RNAs–big players in plant-microbe interactions. Cell Host Microbe 26, 173–182. doi: 10.1016/j.chom.2019.07.021
Huang, X., Qian, Q., Liu, Z., Sun, H., He, S., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Iwakawa, H.-O., and Tomari, Y. (2015). The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 25, 651–665. doi: 10.1016/j.tcb.2015.07.011
Ji, L., Liu, X., Yan, J., Wang, W., Yumul, R. E., Kim, Y. J., et al. (2011). ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 7:e1001358. doi: 10.1371/journal.pgen.1001358
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. doi: 10.1105/tpc.106.046300
Kawano, Y., and Shimamoto, K. (2013). Early signaling network in rice PRR-mediated and R-mediated immunity. Curr. Opin. Plant Biol. 16, 496–504. doi: 10.1016/j.pbi.2013.07.004
Kawasaki, T., Yamada, K., Yoshimura, S., and Yamaguchi, K. (2017). Chitin receptor-mediated activation of MAP kinases and ROS production in rice and Arabidopsis. Plant Signal. Behav. 12:e1361076. doi: 10.1080/15592324.2017.1361076
Kobayashi, H., and Tomari, Y. (2016). RISC assembly: coordination between small RNAs and Argonaute proteins. Bioch. Biophys. Acta Gene Regul. Mech. 1859, 71–81. doi: 10.1016/j.bbagrm.2015.08.007
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11:597. doi: 10.1038/nrg2843
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, Y., Jeyakumar, J. M. J., Feng, Q., Zhao, Z.-X., Fan, J., Khaskheli, M. I., et al. (2019). The roles of rice microRNAs in rice-Magnaporthe oryzae interaction. Phytopathol. Res. 1:33. doi: 10.1186/s42483-019-0040-8
Li, Y., Lu, Y.-G., Shi, Y., Wu, L., Xu, Y.-J., Huang, F., et al. (2014). Multiple rice MicroRNAs are involved in immunity against the blast fungus. Plant Physiol. 164, 1077–1092. doi: 10.1104/pp.113.230052
Li, Y., Zhao, S.-L., Li, J.-L., Hu, X.-H., Wang, H., Cao, X.-L., et al. (2017). Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 8:2. doi: 10.3389/fpls.2017.00002
Lin, Z. Z., Jiang, W. W., Wang, J. L., and Lei, C. L. (2001). Research and utilization of universally susceptible property of japonica rice variety lijiangxintuanheigu. Sci. Agric. Sin. 34, 116–117.
Liu, B., Li, P., Li, X., Liu, C., Cao, S., Chu, C., et al. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139, 296–305. doi: 10.1104/pp.105.063420
Liu, M., Shi, Z., Zhang, X., Wang, M., Zhang, L., Zheng, K., et al. (2019). Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat. Plants 5, 389–400. doi: 10.1038/s41477-019-0383-2
Miao, C., Wang, Z., Zhang, L., Yao, J., Hua, K., Liu, X., et al. (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 10, 1–12.
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Sci. 312, 436–439. doi: 10.1126/science.1126088
Preston, J. C., and Hileman, L. (2013). Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4:80. doi: 10.3389/fpls.2013.00080
Riese, M., Höhmann, S., Saedler, H., Münster, T., and Huijser, P. (2007). Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 401, 28–37. doi: 10.1016/j.gene.2007.06.018
Rogers, K., and Chen, X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383–2399. doi: 10.1105/tpc.113.113159
Shikata, M., Koyama, T., Mitsuda, N., and Ohmetakagi, M. (2009). Arabidopsis SBP-Box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50, 2133–2145. doi: 10.1093/pcp/pcp148
Shimono, M., Sugano, S., Nakayama, A., Jiang, C., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Taguchi-Shiobara, F., Kawagoe, Y., Kato, H., Onodera, H., Tagiri, A., Hara, N., et al. (2011). A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets. Breed. Sci. 61, 17–25. doi: 10.1270/jsbbs.61.17
Walters, D., and Boyle, C. (2005). Induced resistance and allocation costs: what is the impact of pathogen challenge? Physiol. Mol. Plant Pathol. 66, 40–44. doi: 10.1016/j.pmpp.2005.04.002
Walters, D., and Heil, M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 71, 3–17. doi: 10.1016/j.pmpp.2007.09.008
Wang, J., Zhou, L., Shi, H., Chern, M., Yu, H., Yi, H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. doi: 10.1126/science.aat7675
Wang, L., Sun, S., Jin, J., Fu, D., Yang, X., Weng, X., et al. (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 15504–15509. doi: 10.1073/pnas.1521949112
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat.re Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. doi: 10.1242/dev.02521
Wu, H.-J., Wang, Z.-M., Wang, M., and Wang, X.-J. (2013). Widespread long noncoding RNAs as endogenous Target Mimics for MicroRNAs in plants. Plant Physiol. 161, 1875–1884. doi: 10.1104/pp.113.215962
Wu, L., Zhang, Q., Zhou, H., Ni, F., Wu, X., and Qi, Y. (2009). Rice microRNA effector complexes and targets. Plant Cell 21, 3421–3435. doi: 10.1105/tpc.109.070938
Xie, K., Wu, C., and Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
Xu, M., Hu, T., Zhao, J., Park, M.-Y., Earley, K. W., Wu, G., et al. (2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 12:e1006263. doi: 10.1371/journal.pgen.1006263
Yan, J., Gu, Y., Jia, X., Kang, W., Pan, S., Tang, X., et al. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24, 415–427. doi: 10.1105/tpc.111.094144
Yu, Y., Jia, T., and Chen, X. (2017). The ‘how’and ‘where’of plant microRNAs. New Phytol. 216, 1002–1017. doi: 10.1111/nph.14834
Yuan, H., Qin, P., Hu, L., Zhan, S., Wang, S., Gao, P., et al. (2019). OsSPL18 controls grain weight and grain number in rice. J. Genet. Genomics 46, 41–51. doi: 10.1016/j.jgg.2019.01.003
Zhang, H., Zhang, J., Yan, J., Gou, F., Mao, Y., Tang, G., et al. (2017). Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. U.S.A. 114, 5277–5282. doi: 10.1073/pnas.1703752114
Zhang, X., Bao, Y., Shan, D., Wang, Z., Song, X., Wang, Z., et al. (2018). Magnaporthe oryzae induces the expression of a MicroRNA to suppress the immune response in rice. Plant Physiol. 177, 352–368. doi: 10.1104/pp.17.01665
Zhang, X., Zhao, H., Gao, S., Wang, W.-C., Katiyar-Agarwal, S., Huang, H.-D., et al. (2011). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393∗-mediated silencing of a Golgi-localized SNARE gene. MEMB12. Mol. Cell 42, 356–366. doi: 10.1016/j.molcel.2011.04.010
Zhao, Z. X., Feng, Q., Cao, X. L., Zhu, Y., Wang, H., Chandran, V., et al. (2019). Osa-miR167d facilitates infection of Magnaporthe oryzae in rice. J. Integrat. Plant Biol. doi: 10.1111/jipb.12816
Zhou, S.-X., Zhu, Y., Wang, L.-F., Zheng, Y.-P., Chen, J.-F., Li, T.-T., et al. (2019). Osa-miR1873 fine-tunes rice immunity against Magnaporthe oryzae and yield traits. J. Integrat. Plant Biol. doi: 10.1111/jipb.12900
Keywords: microRNA, osa-miR156, rice blast disease resistance, Magnaporthe oryzae, SPL14, WRKY45
Citation: Zhang L-L, Li Y, Zheng Y-P, Wang H, Yang X, Chen J-F, Zhou S-X, Wang L-F, Li X-P, Ma X-C, Zhao J-Q, Pu M, Feng H, Fan J, Zhang J-W, Huang Y-Y and Wang W-M (2020) Expressing a Target Mimic of miR156fhl-3p Enhances Rice Blast Disease Resistance Without Yield Penalty by Improving SPL14 Expression. Front. Genet. 11:327. doi: 10.3389/fgene.2020.00327
Received: 03 January 2020; Accepted: 19 March 2020;
Published: 23 April 2020.
Edited by:
Zhu-Qing Shao, Nanjing University, ChinaReviewed by:
Fengming Song, Zhejiang University, ChinaGuo-dong Lu, Fujian Agriculture and Forestry University, China
Copyright © 2020 Zhang, Li, Zheng, Wang, Yang, Chen, Zhou, Wang, Li, Ma, Zhao, Pu, Feng, Fan, Zhang, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Ming Wang, ajMxNndlbm1pbmd3YW5nQHNpY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Ling-Li Zhang
Ling-Li Zhang Yan Li
Yan Li Ya-Ping Zheng1†
Ya-Ping Zheng1† He Wang
He Wang Xuemei Yang
Xuemei Yang Jing Fan
Jing Fan Wen-Ming Wang
Wen-Ming Wang