
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 17 April 2020
Sec. RNA
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00262
This article is part of the Research Topic Microbial Regulation of Translation View all 10 articles
Type II Toxin–antitoxin (TA) modules are bacterial operons that encode a toxic protein and its antidote, which form a self-regulating genetic system. Antitoxins put a halter on toxins in many ways that distinguish different types of TA modules. In type II TA modules, toxin and antitoxin are proteins that form a complex which physically sequesters the toxin, thereby preventing its toxic activity. Type II toxins inhibit various cellular processes, however, the translation process appears to be their favorite target and nearly every step of this complex process is inhibited by type II toxins. The structural features, enzymatic activities and target specificities of the different toxin families are discussed. Finally, this review emphasizes that the structural folds presented by these toxins are not restricted to type II TA toxins or to one particular cellular target, and discusses why so many of them evolved to target translation as well as the recent developments regarding the role(s) of these systems in bacterial physiology and evolution.
Bacterial toxin-antitoxin (TA) systems are generally composed of a toxic protein and its inhibitor. These small modules were originally discovered on plasmids in the 1980s where they were found to promote plasmid maintenance in growing bacterial populations (Karoui et al., 1983; Ogura and Hiraga, 1983; Jaffe et al., 1985; Gerdes et al., 1986; Hiraga et al., 1986). TA modules are classified into different types depending on the nature and mode of action of the antitoxins, as the toxins are always proteins. Antitoxins are either small RNAs that block translation of the toxin mRNA (type I) (Gerdes and Wagner, 2007) or sequesters the toxic protein (type III) (Fineran et al., 2009; Blower et al., 2011) or proteins that inhibit the activity of the toxin through direct interaction (type II) (Tam and Kline, 1989; Kamada et al., 2003; Takagi et al., 2005) or antagonize the toxic activity on the target, without any direct interaction with the toxins (type IV) (Brown and Shaw, 2003). This review will focus on type II systems. These elements are not only found in plasmids but also in other types of mobile genetic elements (such as phages and ICEs) as well as in chromosomes (see e.g., Anantharaman and Aravind, 2003; Pandey and Gerdes, 2005; Guglielmini and Van Melderen, 2011; Leplae et al., 2011; Ramisetty et al., 2016; Coray et al., 2017). While the roles of TAs, when located in mobile genetic elements, are reminiscent to that on plasmids, i.e., maintenance (Szekeres et al., 2007; Wozniak and Waldor, 2009; Huguet et al., 2016), the roles of chromosomally-encoded systems remains a largely debated topic in the field. These systems have been involved in the adaptation to adverse conditions and are considered to be stress response modules (Hayes and Van Melderen, 2011; Page and Peti, 2016; Harms et al., 2018), with a mainstream model proposing that TA systems are essential effectors of persistence to antibiotics (Gerdes and Maisonneuve, 2012). However, seminal papers supporting this hypothesis are now being retracted (Maisonneuve et al., 2018a, b; Germain et al., 2019) and contradictory data are being published (Harms et al., 2017; Shan et al., 2017; Goormaghtigh et al., 2018a; Pontes and Groisman, 2019). The involvement of TA systems in the persistence phenomenon was based on the observation that successive deletions of 10 TA systems in Escherichia coli lead to a gradual decrease of persistence frequency in the presence of lethal doses of ampicillin or ciprofloxacin (Maisonneuve et al., 2011). In subsequent studies, time-lapse microscopy experiments with E. coli strains containing fluorescent reporters revealed that cells that are able to recover from an ampicillin treatment (namely persister cells) are those in which the ppGpp level is high and TA systems are activated (Maisonneuve et al., 2013). Furthermore, the same group proposed that the HipBA system is the major regulator of persistence as the HipA toxin, by phosphorylating glutamyl-tRNA synthetase (see below), will trigger ppGpp production and the activation of the 10 other TA systems (Germain et al., 2015). We, along with other researchers, have identified major problems both with the E. coli strains and fluorescent reporters that were used in these studies (Harms et al., 2017; Goormaghtigh et al., 2018a). First, the strain deleted for the 10 TA systems is lysogenized with several copies of the Phi80 phages (Harms et al., 2017; Goormaghtigh et al., 2018a). This explains why that strain presents a lower persistence frequency to ciprofloxacin. Indeed, activation of the SOS response by fluoroquinolones will lead to lambdoid prophage activation and a strong decrease in viability. Second, the fluorescent reporters used to monitor ppGpp levels and TA activation are likely to not be functional, either forming protein aggregates or not being more fluorescent than the fluorescence background of control strains without the reporter (Goormaghtigh et al., 2018a). In an effort to solve the issue of TAs and persistence, we, along with other researchers, constructed a strain in which the 10 TA systems are deleted, devoid of any phage contaminants, and showed that this strain presents the same level of persistence to ampicillin or ofloxacin as the wild-type strain (Harms et al., 2017; Goormaghtigh et al., 2018a). Moreover, newly designed reporters monitoring the activation of the yefM-yoeB TA system allowed us to show that there is no correlation between persister cells and the activation of this TA system (Goormaghtigh et al., 2018a). Moreover, we recently showed that another TA system, MqsrA (see below), that was thought to be a global regulator involved in stress responses and biofilm formation, does not appear to play any significant role neither in oxidative or bile stresses nor in macrocolony formation (Fraikin et al., 2019). Therefore, our data strongly argue against the idea that TA systems are pivotal elements of antibiotic persistence. This basically leaves the principal questions in the field open (for a recent review see Fraikin et al., 2020). Conditions in which TA systems are activated and what the outcomes of such activations are, still remain undetermined.
Type II toxins are very diverse in their molecular mode of action, however, almost all the families described to date comprise toxins that target protein synthesis (Harms et al., 2018). Several major mechanisms of translation inhibition by type II toxins can be distinguished: RNA hydrolysis of (i) solvent exposed RNAs, (ii) mRNAs in ribosomes, (iii) rRNAs, (iv) tRNAs or interference with the tRNA machinery, where in addition to the above mentioned hydrolysis of tRNAs, toxicity is exhibited by the modification of tRNA cargo (v) and the inactivation of enzymes that service the tRNAs (vi) i.e., phosphorylation of aminoacyl-tRNA synthetases that charge the tRNAs or EF-Tu which delivers the tRNAs to the ribosome (Figure 1). Most type II toxin activities lead to the general inhibition of protein synthesis and the subsequent inhibition of growth. In this review, we follow this classification to discuss the specificity and evolution of the different families of toxins from TA systems. We discuss the consequences of toxin-mediated translation inhibition on cell physiology and phenotypes and raise questions about their biological functions in light of recent discoveries.
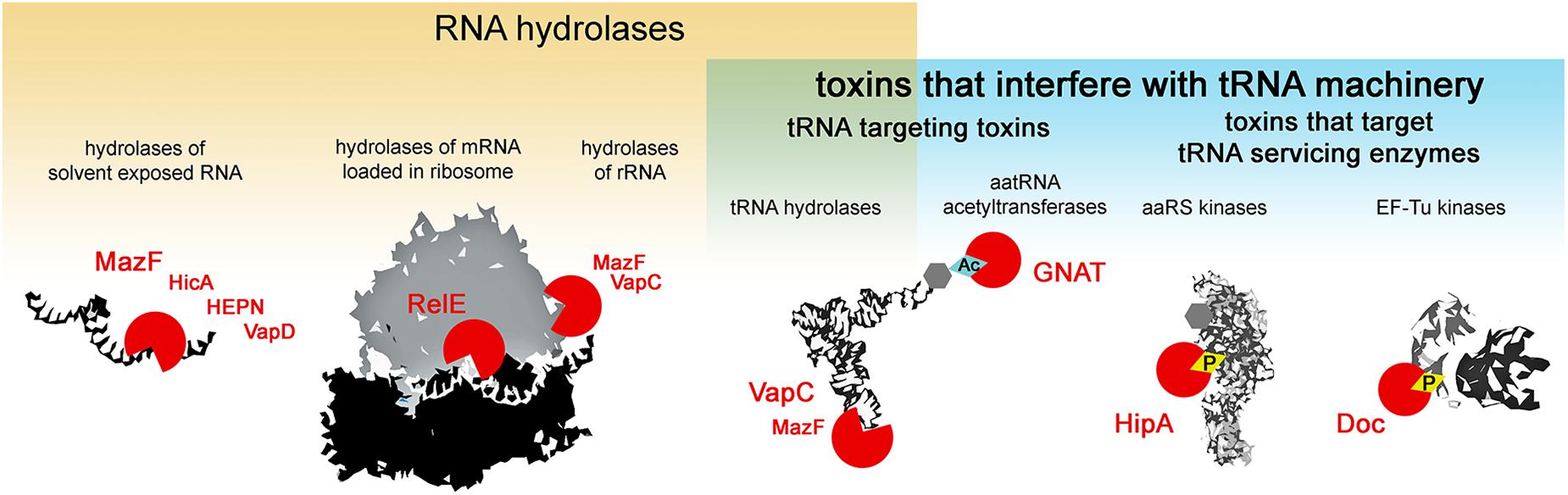
Figure 1. Activities of type II TA toxins. Cellular targets of the toxins are depicted in black and white; toxins are depicted as red circles – open circles for toxins hydrolyzing chemical bonds, circles with diamond for toxins transferring chemical groups on the targets. Ac stands for acetylation, and P for phosphorylation.
MazF toxins are RNA endonucleases that exhibit cleavage specificity to sequences spanning from three to seven bases (Figure 2). Most of the MazF toxins prefer U upstream of cleavage (at position −1) and AC downstream of cleavage (at positions +1 and +2, respectively) with less stringency outside of the two to four main recognized bases (Figure 2). The E. coli MazF cleaves in the coding as well as untranslated regions of mRNAs, independent from the reading frame, and in rRNA precursors (Mets et al., 2017; Culviner and Laub, 2018; Mets et al., 2019). Slight changes in sequence specificity of different MazF enzymes from different bacteria, correspond well to their amino acid sequence similarity (Figure 2). A particular member of the MazF family, the MazF-mt9 toxin from Mycobacterium tuberculosis, cleaves a tRNA substrate (Schifano et al., 2016), similar to the VapC toxins that will be discussed later in this review.
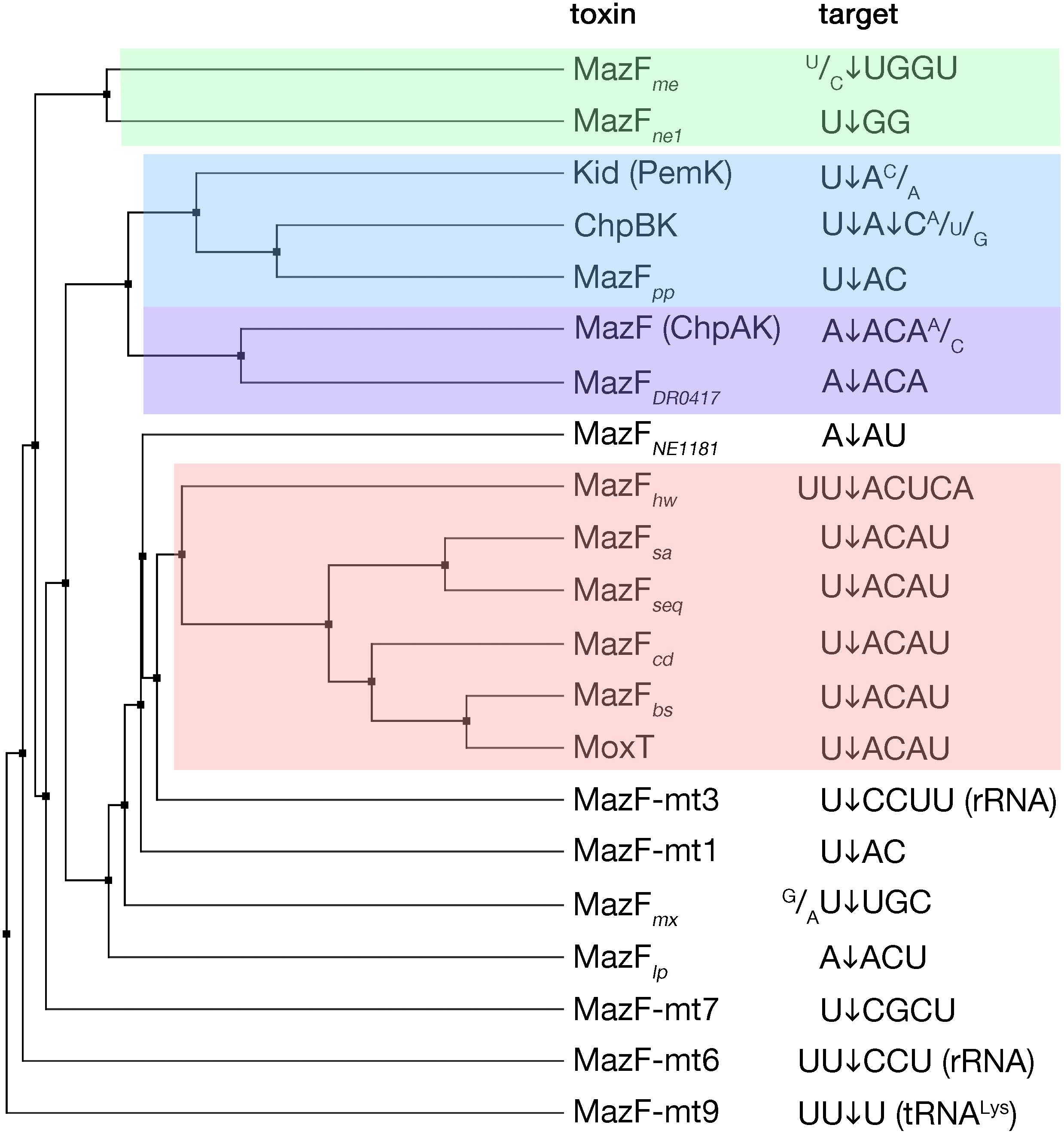
Figure 2. MazF toxins and their specificity of RNA cleavage. MazF protein sequences were aligned and an average distance tree was build (BLOSUM 62, JalView) (Waterhouse et al., 2009). Substrate specificity is indicated (right). The cleavage position of the substrate is indicated by an arrow. Subsets of toxins sharing similar target sequences are boxed with different colors. Protein identifiers and cleavage specificity were taken from: E. coli MazF (ChpAK) (NP_417262.1) (Culviner and Laub, 2018; Mets et al., 2019), ChpBK (NP_418646.1) (Zhang et al., 2005), Kid (PemK) (YP_003937673.1) (Munoz-Gomez et al., 2004; Zhang et al., 2004); Bacillus subtilis MazFbs (AOR96854.1) (Park et al., 2011); Bacillus anthracis MoxT (NP_842807.1) (Verma and Bhatnagar, 2014); Staphylococcus aureus MazFsa (BBJ19047.1) (Zhu et al., 2009); Staphylococcus equorum MazFseq (AFV93478.1) (Schuster et al., 2013); Clostridium difficile MazFcd (YP_001089981.1) (Rothenbacher et al., 2012); Legionella pneumophila MazFlp (CCD10720.1) (Shaku et al., 2018); Deinococcus radiodurans MazFDR0417 (AAF09995.1) (Miyamoto et al., 2017); Haloquadratum walsbyi MazFhw (WP_048066888.1) (Yamaguchi et al., 2012); Pseudomonas putida MazFpp (NP_742932.1) (Miyamoto et al., 2016a); Myxococcus xanthus MazFmx (SDX28280.1) (Nariya and Inouye, 2008); Methanohalobium evestigatum MazFme (WP_013195679.1); Nitrosomonas europaea MazFne1 (WP_011111532.1) (Miyamoto et al., 2018), MazFNE1181 (CAD85092.1) (Miyamoto et al., 2016b); Mycobacterium tuberculosis MazF-mt1 (NP_217317.1) (Zhu et al., 2006), MazF-mt3 (NP_216507.1) (Zhu et al., 2008; Schifano et al., 2014), MazF-mt6 (NP_215618.1) (Schifano et al., 2013), MazF-mt7 (NP_216011.1) (Zhu et al., 2008), MazF-mt9 (YP_004837055.2) (Barth et al., 2019).
The structures of many MazF toxins alone or in complex with an mRNA substrate or with their cognate antitoxins have been solved (Hargreaves et al., 2002; Kamada et al., 2003; Simanshu et al., 2013; Zorzini et al., 2014, 2016; Ahn et al., 2017; Chen et al., 2017; Hoffer et al., 2017). The MazF monomer consists of 2 beta-sheets composed of antiparallel beta-strands linked by three or four small alpha-helices (Figure 3). The residues constituting the active site are located on the β1-β2 and β3-β4 linkers (Kamada et al., 2003; Simanshu et al., 2013; Zorzini et al., 2016). Although MazF toxins possess what is known as the SH3-barrel-fold, they are not related to other members of this fold (Anantharaman and Aravind, 2003). Two MazF subunits form a dimer with an extensive dimeric interface. A concave positively charged groove at the interface between the two subunits of the MazF dimer binds RNA in an extended alignment with bases facing upwards toward the groove (Simanshu et al., 2013; Zorzini et al., 2016). Structural studies have shown that a MazF dimer binds to one mRNA molecule and covers at least seven bases (Simanshu et al., 2013; Zorzini et al., 2016), explaining the extent of possible recognition and necessity of an unstructured RNA substrate, i.e., solvent exposed bases. MazF cleavage leaves 2′, 3′-cyclic phosphate at the 3′-end and 5′-OH group at the 5′-end of the cleavage site, which are used as a signature to study the cleavage products generated by the in vivo overexpression of MazF toxins (Schifano et al., 2014; Mets et al., 2017).
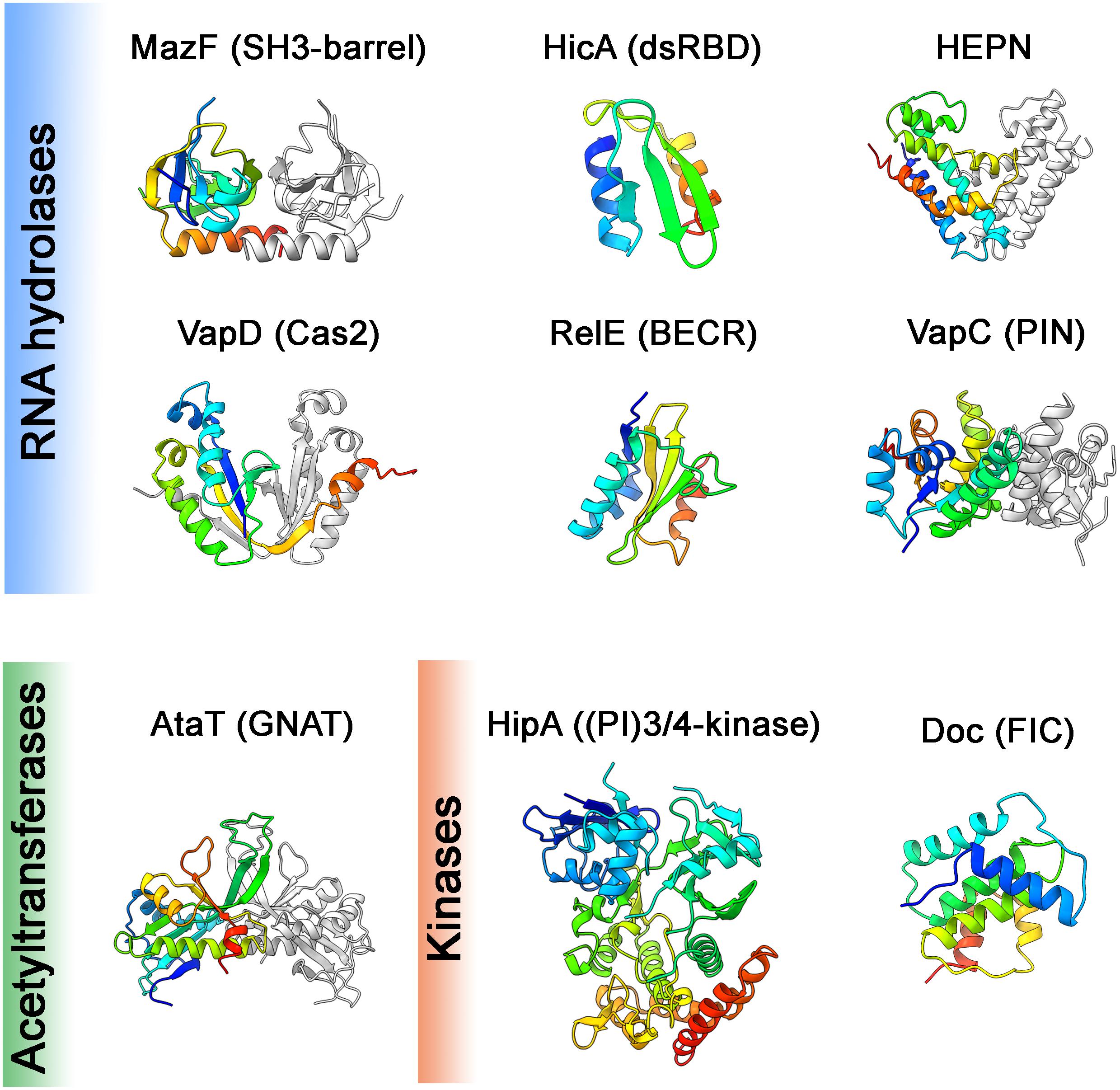
Figure 3. Toxin structures. Structures of the toxins from different families representing different structural folds (noted in brackets) are shown. These domains are common in proteins of the defense and offense systems (HEPN, Cas2, BECR, FIC), RNA processing (dsRBD, PIN), eukaryotic cell signaling (SH3-barrel, (PI)3/4-kinase) or various other functions (GNAT). Structures are colored in rainbow from N-terminus (blue) to C-terminus (red), in cases of dimers the second monomer is in gray. Structures were visualized using ChimeraX, PDB codes were following: E. coli MazF:3nfc; E. coli HicAB:6hpb (Manav et al., 2019); S. oneidensis SO_3166-SO3165 (HEPN): 5yep (Jia et al., 2018); H. pylori HP0315 (VapD): 3ui3 (Kwon et al., 2012); S. flexneri VapCD7A: 5ecw (Xu et al., 2016); E. coli RelER81A, R83A: 2kc9 (Li et al., 2009); E. coli HipAS150A: 3tpb (Schumacher et al., 2012); E. coli AtaTY144F: 6gtp (Jurenas et al., 2019); prophage P1 Phd-Doc: 3k33 (Garcia-Pino et al., 2010). In the cases of structures in complexes with antitoxin, the coordinates of antitoxin were deleted.
In addition to cleaving mRNAs, the E. coli MazF toxin was shown to cleave the 16S rRNA (Vesper et al., 2011). It was proposed that rRNA-cleavage generates specialized ribosomes able to translate specific pools of mRNAs constituting a regulon that is required to cope with various stresses (Vesper et al., 2011; Sauert et al., 2016). The model relies on data showing that E. coli MazF cleaves the 16S rRNA in the decoding center before ACA sequences at nucleotide positions 1396 and 1500 (↓1396ACA and ↓1500ACA) within the 30S ribosomal subunit. It was speculated that MazF-mediated cleavage removes the anti-SD sequence, thereby generating specialized ribosomes that are able to translate specialized MazF-processed leaderless mRNAs that also lack SD (Vesper et al., 2011; Sauert et al., 2016). However, these particular 16S rRNA bases are paired in the 30S subunits, making the MazF-dependent cleavage very unlikely. It was later shown that indeed, MazF cleaves the rRNAs in their precursor states, before the 30S subunit biogenesis, and at multiple sites (Mets et al., 2017; Culviner and Laub, 2018). This is in agreement with structural data showing that MazF binds to single-stranded RNA (ssRNA) and interacts with the bases (Simanshu et al., 2013; Zorzini et al., 2016). In fact, fragmentation of rRNAs as well as mRNAs including those coding for ribosomal proteins leads to the accumulation of aberrant ribosomal subunits and generates irregular particles with fragmented rRNA upon MazF expression (Mets et al., 2017). Accordingly, the synthesis of the vast majority of cellular proteins drop in response to MazF cleavage, without enrichment of any specific functional protein groups. Proteomic studies upon MazF expression did not find any ‘death’ or ‘survival’ proteins (Mets et al., 2019), associated to the proposed programmed cell death pathway (Amitai et al., 2009).
The TA-mediated programmed cell death (PCD) theory proposed that some TA systems, in particular MazEF, serve as built-in suicide modules (Engelberg-Kulka et al., 2004). Multiple different and unrelated stressing conditions, such as amino acids starvation and elevated ppGpp (guanosine tetraphosphate) concentrations, antibiotic treatments, high temperature, H2O2 treatment or phage infections were proposed to trigger MazF-dependent PCD. All these conditions would lead to the inhibition of mazEF expression. Since the MazE antitoxin is unstable and degraded by ATP-dependent proteases, this would liberate MazF and provoke the death of a large subpopulation of cells (Engelberg-Kulka et al., 2004). Within the frame of this pathway, it has been suggested that MazF itself is induced by a ‘quorum sensing’ peptide called extracellular death factor (EDF). This pentapeptide was identified to have the NNWNN sequence (Kolodkin-Gal et al., 2007) and is thought to amplify the endoribonucleolytic activity of MazF and other homologs and prevent the interaction of MazF toxins with their cognate antitoxins (Belitsky et al., 2011). However, alongside the PCD theory, the involvement of the EDF peptide in MazF-mediated PCD is questionable. First, in the original paper that describes the activity of the EDF peptide, the authors observed a drastic decrease in bacterial viability upon the addition of 2.5 ng/ml (4 nM) of EDF to growing E. coli cultures. This effect was also observed in MazF-deleted strains although at higher concentrations, starting from 200 ng/ml (300 nM) (Kolodkin-Gal et al., 2007). In subsequent publications, the authors showed that in vitro MazF ribonucleolytic activity was enhanced by only 26% by the addition of 1.5 μM of EDF (Belitsky et al., 2011). Further addition of up to 7.5 μM of EDF increased MazF activity to a maximum of 57% (Belitsky et al., 2011). Such a high EDF concentration, corresponding to a 300-fold molar excess, was used to show the disruption of the MazE-MazF complex in vitro. Assuming that EDF is produced in vivo under physiological conditions, it is very unlikely that such high concentrations will be reached. Moreover, in vivo, viability of bacteria is highly affected at EDF concentrations that are by several logs lower (4 nM versus 7.5 μM). The same group later discovered more EDF-like peptides (6 aa or longer) in Bacillus subtilis and Pseudomonas aeruginosa, however all of these peptides were only active in combination with rifampicin (Kolodkin-Gal et al., 2007; Kumar et al., 2013). Although in the original studies, rifampicin was used to induce MazF-mediated PCD (Kolodkin-Gal et al., 2007; Kumar et al., 2013), other studies did not find any MazF-related effect of rifampicin, nor any MazF-mediated PCD (Tsilibaris et al., 2007; Ramisetty et al., 2016). Thus, the cellular functions of MazF remain an open question.
Probably, the most intriguing case is that of M. tuberculosis which encodes 10 MazF toxins among its large TA arsenal (Sala et al., 2014). Out of those, the MazF-mt3 and MazF-mt6 toxins cleave mainly mRNAs at the U↓CCUU and UU↓CCU sequences, respectively. In addition, MazF-mt3 cleaves the anti-SD sequence of the 16S rRNA and both the MazF-mt3 and MazF-mt6 cleave the 23S rRNA loop 70 (L70) opening to the ribosomal A site (Schifano et al., 2013, 2014; Hoffer et al., 2017). It has been shown that MazF-mt6 can cleave the 23S rRNA in mature 50S subunit, albeit with 30% efficiency as compared to free RNA fragment coding for 23S L70 sequence (Hoffer et al., 2017). This indicates that like other MazF toxins, MazF-mt6 cleaves the solvent exposed target sequence. Alternatively, cleavage might be possible in rRNA precursors as described for E. coli MazF (Mets et al., 2017; Culviner and Laub, 2018). Another MazF toxin from M. tuberculosis, MazF-mt9, cleaves the tRNALys43–UUU in its anticodon sequence (Schifano et al., 2016; Barth et al., 2019). Since tRNALys19–CUU does not compensate for tRNALys43–UUU, ribosomes stall at the AAA codon in MazF-mt9 overexpression conditions (Barth et al., 2019). Ribosome stalled transcripts are further cleaved by specific RNAses such as RNAse J and are therefore eliminated (Barth et al., 2019). Since the AAA codon is rare in the GC-rich M. tuberculosis (5.3/1000), the authors speculate that expression of MazF-mt9 generates a specific proteome consisting of the proteins whose genes are poor in AAA Lys codons (Barth et al., 2019). However, it remains unclear whether genes devoid of the AAA codon would be sufficient to generate a functional proteome.
Interestingly, MazF toxins share high structural similarity to the F plasmid CcdB toxin (Loris et al., 1999; Hargreaves et al., 2002; Gogos et al., 2003; Kamada et al., 2003) that binds to the GyrA subunits of DNA-gyrase and induces double-strand breaks and the SOS response (Bernard and Couturier, 1992; Bernard et al., 1993; Dao-Thi et al., 2005). Despite their structural similarity, the two toxins bind their substrates using different sites on the toxin dimer interface – CcdB binds GyrA via the α4 helices, while, MazF recognizes RNA using the β1-β2, β3-β4, β4-β5 loops and a short α1 helix (Dao-Thi et al., 2005; Zorzini et al., 2016). On the other hand, the activity of MazF and CcdB toxins is regulated by their cognate antitoxins in a common way, which further supports the hypothesis of a common ancestor (Zorzini et al., 2016).
HicA toxin and its cognate HicB antitoxin owe their gene names to a genetic locus linked to the pilus gene cluster (hif contiguous) in Haemophilus influenzae (Mhlanga-Mutangadura et al., 1998). HicA toxins possess small ∼50 amino acid double-stranded RNA binding domains (dsRBD). HicA folds into a three-stranded antiparallel beta-sheet flanked by two alpha helices that reside on one side of the sheet (Figure 3) (Makarova et al., 2006; Butt et al., 2014). The positively charged surface is predicted to bind RNA and the catalytic histidine residue located in the β2 strand is required for RNase activity (Bibi-Triki et al., 2014; Butt et al., 2014; Kim et al., 2018). The E. coli HicA toxin cleaves mRNAs and tmRNA in vivo independently of translation and no consensus of cleavage was reported (Jorgensen et al., 2009). Yersinia pestis HicA3 was shown to degrade in vitro transcribed mRNAs (Bibi-Triki et al., 2014) and Sinorhizobium meliloti HicA degrades purified rRNA (Thomet et al., 2019). However, there is not enough data available to be able to conclude what the precise targets of HicA toxins are in vivo.
Outside of the TA context, it has been shown that the dsRBD domain binds double-stranded RNA where the α1 helix interacts with the minor groove and the α2 helix with the major groove of RNA molecules (Ryter and Schultz, 1998). All the HicA toxins studied to date hydrolyze RNA in addition to binding (Jorgensen et al., 2009; Bibi-Triki et al., 2014; Thomet et al., 2019), however there are no details on the molecular substrate binding and hydrolysis.
The typical antitoxin partner HicB protein comprises DNA-binding domain fused to a degenerated RNAse H-fold (Makarova et al., 2006). These two domains (dsRBD and RNAse H) are also found in the architecture of eukaryotic RNA interference (RNAi) machinery (Makarova et al., 2006).
The HEPN (Higher Eukaryotes and Prokaryotes Nucleotide-binding domain) superfamily contains proteins that have all-alpha helical catalytic domains. Outside of the TA context, HEPN-domain proteins typically have metal-independent endonuclease activities, although some only bind RNA without degrading it (Anantharaman et al., 2013). HEPN domains are shared between TA and prokaryotic defense systems, such as abortive infection modules, restriction-modification systems and CRISPR-Cas systems, as well as eukaryotic antiviral, antitransposon systems and rRNA processing enzymes (Anantharaman et al., 2013). In the TA context, HEPN is frequently found in association to MNT (minimal nucleotidyltransferase) domain proteins. Shewanella oneidensis SO_3166 toxin possesses a HEPN domain and its cognate antitoxin SO_3165 an MNT domain (Yao et al., 2015). The SO_3166 toxin cleaves mRNA, but not rRNA or tRNA in vitro, however, the sequence specificity has not been determined. The toxin has a typical all-alpha helical HEPN fold and conserved Arg-(4-6X)-His motif, and forms a dimer with a potential composite active site in a central cleft that could accommodate RNA substrate (Figure 3) (Jia et al., 2018).
RnlA (or RNase LS)-like toxins that exhibit RNAse activity (Otsuka et al., 2007) also contain a catalytic domain belonging to the HEPN superfamily (Anantharaman et al., 2013). RNA cleavage by RnlA was shown to be dependent on translation (Otsuka and Yonesaki, 2012), more specifically on translation termination (Yamanishi and Yonesaki, 2005). RnlA induces sequence non-specific mRNA cleavage more frequently occurring 3′ to pyrimidines (Kai and Yonesaki, 2002). Homologous toxin LsoA shares the same cleavage pattern, however, cleavage sites are not identical (Otsuka and Yonesaki, 2012). The RnlA and LsoA toxins were shown to be part of functional TA systems with their cognate antitoxins RnlB and LsoB, respectively (Koga et al., 2011; Otsuka and Yonesaki, 2012). Interestingly, it was shown that the RnlA and LsoA toxins have a common antitoxin, Dmd, encoded by the T4 phage (Otsuka and Yonesaki, 2012). This antitoxin does not share any sequence similarity with the homologous but not interchangeable RnlB and LsoB antitoxins. T4 phages devoid of the Dmd-encoding gene are unable to propagate on E. coli, opening the possibility that the RnlAB and LsoAB systems act as defense mechanisms (Kai and Yonesaki, 2002; Otsuka and Yonesaki, 2012). Later it was shown that the activity of the RnlA and LsoA toxins is enhanced by RNAse HI (Naka et al., 2014) – an RNase responsible for RNA cleavage in DNA-RNA duplexes and removing the RNA primer in DNA replication (Miller et al., 1973; Itoh and Tomizawa, 1980). RNase H-fold domains are often fused to HEPN domains in larger architectures, suggesting that other RNase LS family proteins could also target RNA in DNA–RNA duplexes (Anantharaman et al., 2013). RnlA toxins in addition to the HEPN domain (also described as DBD) contain two additional domains, NTD and NRD that share similar topology of 4–5 stranded antiparallel beta-sheets with two alpha helices. It was shown that the DBD domain is responsible for dimerization of the toxin, its toxicity, and it is neutralized by both the RnlB and Dmd antitoxins (Wei et al., 2013). The functions of the NTD and NRD domains still need to be investigated.
The vapD gene was first detected in the chromosome of the anaerobic bacterium Dichelobacter nodosus. The genes of this locus, prevalent in virulent strains, were designated as vapABCD (Katz et al., 1992). The vapBC locus was later shown to encode a type II TA system (Daines et al., 2007); the same was demonstrated for vapD and a small upstream encoded ORF designated vapX (Daines et al., 2004). Later it was found that the VapD toxin from Helicobacter pylori displays a ferredoxin-like fold and therefore is structurally related to CRISPR-associated protein Cas2 (Figure 3). VapD functions as an endoribonuclease and cleaves mRNA preferentially before A or G nucleotides (Kwon et al., 2012). The presence of solitary vapD toxin in the Neisseria gonorrhoeae pEP5289 plasmid was suggested to be a factor restricting plasmids’ host range. A small cryptic neisserial plasmid pJD1, however, contains full VapXD. When the VapX antitoxin from pJD1 is expressed in E. coli, it increases the conjugation rate of pEP5289. This suggests that solitary VapD could limit the host range of pEP5289-like plasmids to the ones that contain the pJD1 plasmid carrying an intact VapXD module, and is therefore able to neutralize incoming VapD (Pachulec and van der Does, 2010). Although it has been proposed that vapXD locus as well as vapBC locus contributes to the virulence of H. influenzae (Ren et al., 2012), no molecular mechanism has been demonstrated yet. VapXD satisfies the definition of the type II TA module and comprises the toxin that likely shares its origins with the Cas2 protein, however, virtually nothing else is known about its molecular mechanism or its physiological function.
Almost all RelE toxins cleave mRNAs in the A site of the ribosome, between the second and third position of the codon. RelE family toxins described to date share as low as 11–20% sequence identity but retain the conserved fold which is similar to ribosome independent endoribonucleases T1, Sa2, and U2 (Neubauer et al., 2009; Schureck et al., 2015). The well-studied RelE-like toxins (E. coli RelE, YoeB, YafQ, and Proteus vulgaris HigB) possess different active sites, have different preferences for targeted mRNA codons, and differ in their ability to associate with 30S and/or 70S ribosomes (Neubauer et al., 2009; Feng et al., 2013; Maehigashi et al., 2015; Schureck et al., 2016b; Pavelich et al., 2019). Analysis of the cleavage specificity of RelE homologs from a wide range of bacterial species or isolates shows that the cleavage specificity is not strict (Goeders et al., 2013). The specificity likely originates from subtle differences in the association with the ribosome, rather than recognition of specific mRNA bases. RelE toxins belong to the large superfamily of BECR-fold proteins (barnase-EndoU-colicin E5/D-RelE fold) that is found in different polymorphic toxin systems (Zhang et al., 2012).
RelE contains an antiparallel beta-sheet (usually 4-stranded) flanked by two to four surface-exposed alpha helices enriched with positively charged residues (Figure 3) (Neubauer et al., 2009; Schureck et al., 2016b). The positively charged residues that decorate the alpha helices and mediate interaction with the negatively charged 16S rRNA backbone are thought to be a unique feature of the ribosome-dependent RelE family of endoribonucleases (Maehigashi et al., 2015; Schureck et al., 2016b). Despite the overall structural similarity, the residues that comprise the active sites of several well-studied RelE toxins are different, as well as the ribosome conformation induced by these toxins (Neubauer et al., 2009; Feng et al., 2013; Maehigashi et al., 2015; Schureck et al., 2016b). Inside the ribosomal A site, RelE toxins reorient and activate the mRNA for 2′-OH-induced hydrolysis. Although the ribosome is not directly involved in catalysis, it is required to achieve the correct orientation of the mRNA for the cleavage reaction (Neubauer et al., 2009). Generally, RelE toxins induce strong reorganization of the mRNAs at the ribosomal A site and cause the hydrolysis between the second and third position of the codon in the A site (Neubauer et al., 2009; Feng et al., 2013; Goeders et al., 2013; Maehigashi et al., 2015; Schureck et al., 2016b). Toxins pull the mRNA out of its typical tRNA bound state. In presence of the toxin, all three A site nucleotides are shifted by more than 7 Å (Neubauer et al., 2009; Schureck et al., 2016b). Conserved residues of the toxin orient the first two A site codon bases for hydrolysis, while the third base is usually oriented with the aid of 16s rRNA (Neubauer et al., 2009; Schureck et al., 2016b). Different RelE family toxins interact with all three nucleotides of the cleaved codon in different ways, leading to subtle specificities for nucleobases at each position (Neubauer et al., 2009; Feng et al., 2013; Maehigashi et al., 2015; Schureck et al., 2016b).
The E. coli RelE toxin extensively relies on the ribosome for both mRNA binding and cleavage, as it has lost conserved histidine and glutamate residues used for RNA cleavage in ribosome independent RNases like RNAse T1 (Heinemann and Saenger, 1983; Takagi et al., 2005; Neubauer et al., 2009). RelE instead uses conserved basic residues both for interaction and catalysis (Neubauer et al., 2009). Almost 1/5 of the residues of RelE are basic and provide a large potential for an interaction with negatively charged RNA. In particular, a large accumulation of these residues is observed on the three helices that interact with the 16S rRNA. Other RelE toxins, such as HigB, YoeB, or YafQ do not rely on basic residues and instead function more like the RNase T1, through conserved histidine and glutamate (or histidine or tyrosine) residues for catalysis (Kamada and Hanaoka, 2005; Maehigashi et al., 2015; Schureck et al., 2016b). In addition, the RelE toxin leaves 2′-3′-cyclic phosphate at the new 3′ end (Neubauer et al., 2009), while others (YefM, YoeB, HigB) further hydrolyze it to a 3′-phosphate product (Feng et al., 2013; Maehigashi et al., 2015; Schureck et al., 2016b), like RNase T1 (Heinemann and Saenger, 1983).
Most of the RelE toxins display virtually no codon specificity and cleave between the second and third positions of the codon with the only conserved preference of purines at the third position (Goeders et al., 2013). However, several RelE family toxins were reported to be selective for the AAA lysine codon. This codon is preferred by the E. coli YafQ, Rhodopseudomonas palustris RelERpa, Nostoc sp. RelENsp, Sinorhizobium meliloti RelESme and Treponema denticola RelETde (Prysak et al., 2009; Goeders et al., 2013). P. vulgaris HigB cleaves A-rich codons (Hurley and Woychik, 2009; Schureck et al., 2016b), and was shown to cleave the 30S bound mRNA, indicating that it can interfere with the initiation step of translation after IF1 dissociation (Schureck et al., 2016a). The AAA and other A-rich codons are the most frequent at the beginning of the ORFs in E. coli (Sato et al., 2001). Several studies have shown that the AT-rich content at the 5′ of the ORF likely reduces the secondary structures and has been shown to serve as a translation ramp for efficient protein expression in E. coli (Goodman et al., 2013; Verma et al., 2019). Moreover, ribosome profiling indicates that ribosomes spend a lot of time at the beginning of the transcripts (Oh et al., 2011), which might provide more time for these toxins to access their substrates. Therefore, at least a subset of RelE toxins could be considered as translation initiation inhibitors.
Several RelE toxins were shown to cleave at different sites with respect to the codon. For example, E. coli YhaV and Mycobacterium avium RelE cleave mRNA preferably between the codons (i.e., after the third base of the codon) (Goeders et al., 2013; Choi et al., 2017). However, the molecular mechanisms of substrate recognition are not yet described for these toxins. The YafO toxin was shown to be a ribosome-dependent endoribonuclease that cleaves mRNAs 11–13 bases downstream of the initiation codon (Zhang et al., 2009; Christensen-Dalsgaard et al., 2010). Such cleavage event was speculated to be located near the mRNA entrance tunnel rather than at the A site as determined for the RelE-like toxins (Schureck et al., 2016a). Although initially YafO was considered a RelE-like toxin (Christensen-Dalsgaard et al., 2010), the lack of structural information about this toxin or its interaction with ribosome lead to YafO being classified as a separate family (Leplae et al., 2011). Several RelE-family toxins, such as E. coli MqsR (YgiU), Brucella abortus BrnT and H. pylori HP0894 are suggested to cleave RNA in a ribosome-independent manner (Brown et al., 2009; Christensen-Dalsgaard et al., 2010; Han et al., 2011; Heaton et al., 2012) and could therefore be functionally closest to ribosome independent RNases T1, Sa2, and U2 found outside of the TA context. MqsR is a RelE-fold toxin that possesses an additional beta strand (Brown et al., 2009; Christensen-Dalsgaard et al., 2010). MqsR preferentially cleaves the 5′-G↓CN-3′ triplet (where N is preferentially U, but C or A are tolerated) in mRNA or rRNA precursors (Yamaguchi et al., 2009; Mets et al., 2017). The H. pylori HP0894 toxin prefers purines upstream of cleavage and its major cleavage activity was observed between first and second base at termination codons UAA and UAG (Han et al., 2011), therefore its independence from the ribosome is questionable. In fact, E. coli RelE was also suggested to inhibit translation termination, as in addition to CAG sense codon, it cleaves UAG or UAA stop codons between the second and third nucleotide and subsequently prevents class 1 release factors from binding the ribosome (Pedersen et al., 2003). It remains unclear whether RelE toxins targeting translation initiation would be able to compete for the A site with initiation factors, and those acting during elongation with tRNA. To date, only the affinity of YafQ toxin to the 70S assembled complex has been reported (∼360 nM) and is comparable to those of the above-mentioned factors (Maehigashi et al., 2015). It remains unclear why toxins rely on the ribosome for activity as simply blocking the translation cleavage of free mRNA would both be sufficient and efficient. It has been suggested that this dependence may indicate a specialized mechanism that would allow a response to stresses (Schureck et al., 2016a). On the other hand, selectivity for specific ribosomal complexes, such as initiating ribosomes, might be an efficient way to inhibit translation and is likely to give the same effect of reducing the global translation rate as ribosome-independent but more sequence-specific mRNA cleavage.
Bioinformatic searches and structural comparisons have detected the relationship between RelE-family toxins and those similar to ParE encoded by RK2 plasmid (Anantharaman and Aravind, 2003; Sterckx et al., 2016). ParE-family toxins act at the level of DNA replication by poisoning DNA-gyrase or by currently unidentified mechanisms (Jiang et al., 2002; Hallez et al., 2010; Yuan et al., 2010; Sterckx et al., 2016). Some residues are highly conserved across the RelE/ParE superfamily (Anantharaman and Aravind, 2003) and their three-dimensional structure is strikingly similar (Dalton and Crosson, 2010; Sterckx et al., 2016). The major differences between the mRNAses and replication inhibitors are the extended N-terminal alpha helices as well as the absence of the C-terminal helix observed in ParE toxins (Dalton and Crosson, 2010; Sterckx et al., 2016). ParE toxins are also devoid of the major catalytic residues used by RelE for mRNA cleavage in the ribosome (Neubauer et al., 2009; Dalton and Crosson, 2010). Evidence of the evolutionary relationships between RelE-like and ParE-like toxins, is the conserved principle of the binding of their cognate antitoxins to conserved hydrophobic motifs on the toxins, although different families of antitoxins can associate with both ParE and RelE-like toxins (Dalton and Crosson, 2010; Leplae et al., 2011; Sterckx et al., 2016).
The VapC toxins are characterized by the presence of a PIN domain that presents a structural similarity with the classical Rossmann-fold associated with the binding of nucleotides and nucleotide-based cofactors (Rao and Rossmann, 1973; Matelska et al., 2017; Senissar et al., 2017). The PIN domain, although originally owing its name to the type IV pili protein PilT (PilT N-terminal like nucleases), is generally found in proteins that present various endonuclease functions such as tRNA and rRNA maturation, nonsense mediated mRNA decay, and DNA replication and repair in all domains of life (Matelska et al., 2017; Senissar et al., 2017). In the PIN motif, alternating beta strands and alpha helices (α/β/α sandwich) fold into a central five stranded parallel beta-sheet decorated with alpha helices on both sides (Figure 3). All VapC toxins, although presenting low sequence similarity, share conserved acidic residues (D, E, D, D/N) that are distant in the primary amino acid sequence, but cluster together in the protein to form an active site that coordinates divalent-cations, such as Mg2+ and Mn2+ that are required for cleavage of single-stranded RNA (Fatica et al., 2004; Daines et al., 2007; Das et al., 2014; Matelska et al., 2017; Senissar et al., 2017). VapC toxins cleave the 3′-O-P bond of single stranded RNA to produce 3′-hydroxyl and 5′-phosphate cleavage products (McKenzie et al., 2012). So far, VapC toxins targeting the initiation tRNAfMet, different elongation tRNAs, and the sarcin-ricin loop (SRL) of the 23 S rRNA have been identified (Figure 4) (Winther and Gerdes, 2011; Winther et al., 2013, 2016). All the VapC toxins targeting tRNAs cleave in the anticodon stem-loop (ASL), either in the anticodon sequence itself or at the 3′-side of the anticodon before the stem structure (Figure 4). The M. tuberculosis VapC-mt20 cleaves the 23S rRNA SRL loop in the helix 95 that structurally mimics the ASL of tRNAs (Winther et al., 2013), therefore all VapC toxins tested so far cleave ASL-like structures. Unlike other ribonuclease toxins (namely MazF and RelE – see above), VapC toxins appear to recognize both RNA sequence and structure (Winther et al., 2013; Cruz et al., 2015; Walling and Butler, 2018; Cintron et al., 2019). Ribonucleotide modifications in the ASL region might be important for governing the specificity of VapC toxins (Winther et al., 2016; Cintron et al., 2019). Strikingly, VapC-mt20 requires the presence of the ribosome for 23S rRNA cleavage and does not cleave isolated SRL rRNA fragments (Winther et al., 2013). Since the helix 95 is exposed to the solvent at the surface of the ribosome (Yusupov et al., 2001), it is accessible for cleavage, however no information of VapC-mt20 interactions with ribosomes is currently available (Winther et al., 2013).
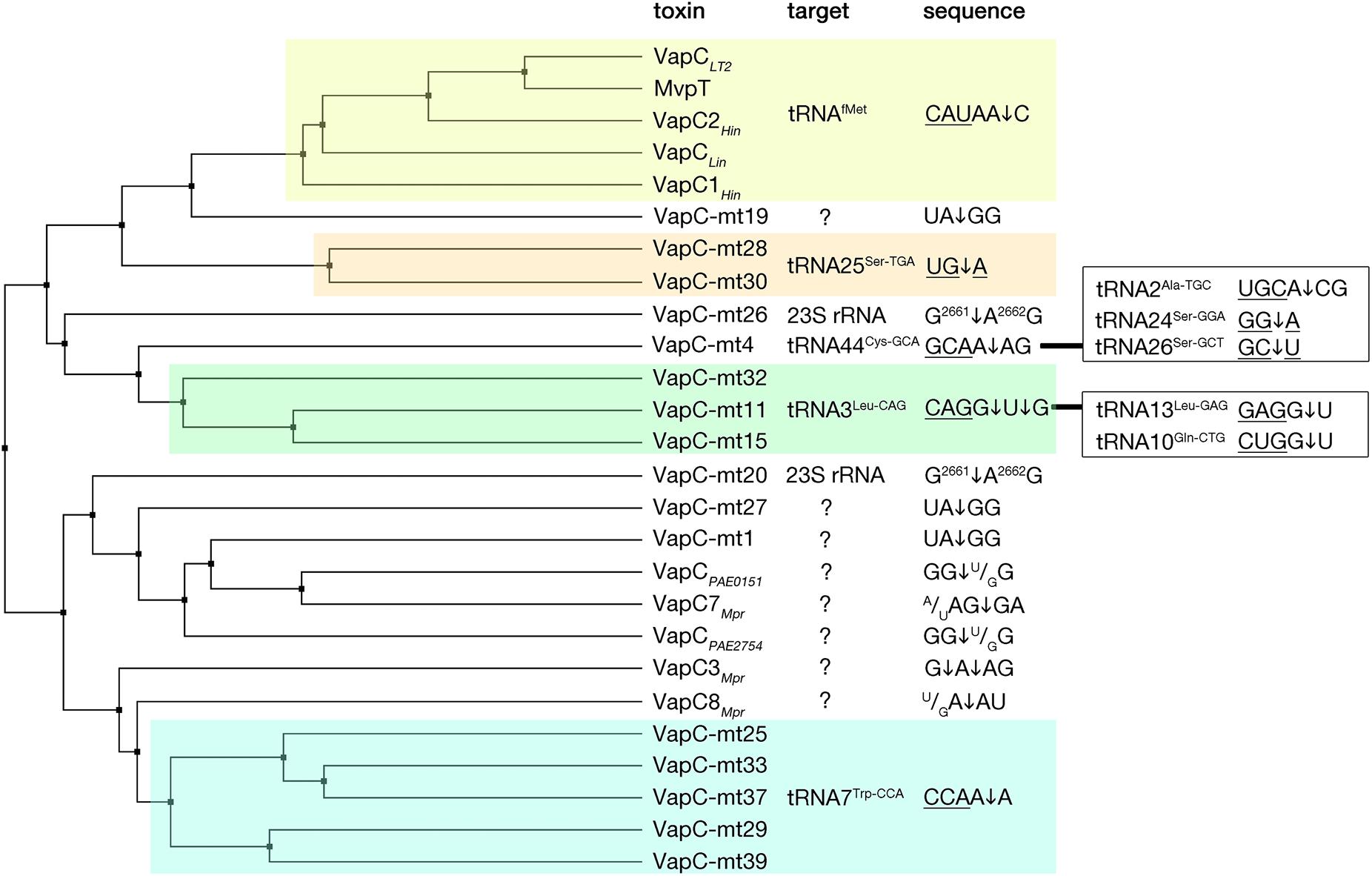
Figure 4. VapC toxins and their specificity of RNA cleavage. VapC protein sequences were aligned and an average distance tree was build (BLOSUM 62, JalView) (Waterhouse et al., 2009). Substrate specificity is indicated (right). Toxin cleavage position is marked by an arrow, tRNA anticodon sequence is underlined, in the case of rRNA numbers of nucleotide positions are indicated in superscript. Additional targets and sequences are marked in the boxes on the right. Subsets of toxins sharing similar target sequences are boxed with different colors. Protein identifiers and cleavage specificity were taken from: Haemophilus influenzae VapC1Hin (NP_438487.1), VapC2Hin (NP_439108.1) (Walling and Butler, 2018); Salmonella enterica VapCLT2 (NP_461950.1), Shigella flexneri MvpT (AMN61047.1) (Winther and Gerdes, 2011); Leptospira interrogans VapCLin (AAS71220.1) (Lopes et al., 2014); Mycobacterium tuberculosis VapC-mt4 (NP_215109.1) (Cruz et al., 2015; Winther et al., 2016), VapC-mt11 (NP_216077.1) (Winther et al., 2016; Cintron et al., 2019), VapC-mt15(NP_216526.1), VapC-mt25 (NP_214791.1), VapC-Mt28 (NP_215123.1), VapC-mt29 (NP_215131.1), VapC-mt30(NP_215138.1), VapC-mt32(NP_215630.1), VapC-mt33 (NP_215758.1), VapC-mt37 (NP_216619.1), VapC-mt39 (NP_217046.1) (Winther et al., 2016), VapC-mt20 (NP_217065.1), VapC-mt26 (NP_215096.1) (Winther et al., 2013, 2016), VapC-mt1 (NP_214579.1) (McKenzie et al., 2012; Sharrock et al., 2018), VapC-mt19 (NP_217064.1), VapC-mt27 (NP_215112.1) (Sharrock et al., 2018), Pyrobaculum aerophilum VapCPAE2754 (WP_011008882.1), VapCPAE0151 (WP_011007068.1) (McKenzie et al., 2012); Metallosphaera prunae VapC3Mpr (WP_012020824.1), VapC7Mpr (WP_012021162.1), VapC8Mpr (WP_012021192.1) (Mukherjee et al., 2017).
While some VapC toxins cleave tRNAs that service highly abundant codons, others cleave tRNAs that service rare codons, however it is unlikely that they could be compensated by other tRNAs (Cruz et al., 2015; Winther et al., 2016; Cintron et al., 2019). Therefore, it is most likely that the overexpression of VapC toxins leads to global inhibition of translation. Breaks in the ASL will interfere with the aminoacylation of those tRNAs that are recognized through their ASL by aminoacyl-tRNA synthetases (McClain et al., 1998), while charged tRNAs cleaved in ASL are unlikely to generate productive interaction with the A site of the ribosome. It has been proposed that tRNA halves produced by VapC toxins could have further functions in the stress response (Cruz et al., 2015), however the lifetime and possible interactions of VapC cleavage products remain unexplored.
Cleavage of tRNAfMet reported for Salmonella, Shigella, and several other VapC toxins (Winther and Gerdes, 2011) (Figure 4) will lead to inhibition of the initiation – a rate limiting step of protein synthesis (Laursen et al., 2005). It has been shown that expression of VapCLT2, which leads to cleavage of initiator tRNAfMet, would boost the translation start from elongation codons (Winther and Gerdes, 2011). However, translation of only one model mRNA was followed (Winther and Gerdes, 2011), therefore it is difficult to conclude whether induction of VapC would lead to synthesis of specific proteins initiating with other codons and whether it would lead to a sufficient, functional, and specialized proteome.
Several VapC toxins were reported to cleave the SRL loop of 23S rRNA (Winther et al., 2013; Winther et al., 2016) (Figure 4) that owes its name to two toxins – the α-sarcin that cleaves it at the identical position as VapC-mt20 and the ricin that removes the adenine base in the SRL (Olmo et al., 2001; Grela et al., 2019). The SRL loop is located in the GTPase associated center of the large ribosomal subunit where translational GTPases are recruited. The SRL loop is required for docking and stimulation of GTP hydrolysis of all translation GTPases (Munishkin and Wool, 1997; Rodnina et al., 1999), therefore its cleavage leads to overall inhibition of protein synthesis in prokaryotes as demonstrated for VapC, as well as in eukaryotes in case of α-sarcin (Olmo et al., 2001; Winther et al., 2013).
Numerous structures of VapCs show that these toxins are dimers and that their cognate antitoxins neutralize the toxic activity by interfering with the binding of the metal ions (Miallau et al., 2009; Dienemann et al., 2011; Min et al., 2012). However, there is still no structural information regarding their interaction with the RNA substrates, making it difficult to predict details of target recognition and catalysis. It has been demonstrated that by comparing divergent VapCs that recognize particular tRNA or rRNA substrates, they can be grouped in sub-families sharing amino acid sequence similarity (Figure 4). Predicted specificity of new VapCs within these families was confirmed (Winther et al., 2016). Interestingly, VapC toxins can be encoded abundantly in one genome, with an outstanding example of M. tuberculosis encoding as much as 50 VapC toxins (Ramage et al., 2009; Sala et al., 2014). Despite the high abundance and sequence/structure similarities, these TAs appear to not cross-talk, at least for those that are functional and that have been tested – their toxins are neutralized only by their cognate antitoxins – although in some cases this insulation could be alleviated by a single mutation (Ramage et al., 2009; Walling and Butler, 2018). Moreover, mycobacterial VapC toxins do not overlap functionally, since many of them target different tRNAs or the SRL of 23S rRNA (Figure 4) (Winther et al., 2016). Nevertheless, the question of the functional benefit, if any, of having large arrays of VapCs, remains open.
GNAT-fold acetyltransferase toxins identified so far, including TacT, AtaT, and ItaT, acetylate aminocylated-tRNAs on the amino group of the cargo amino acids (Cheverton et al., 2016; Jurenas et al., 2017a; Wilcox et al., 2018). For convenience, we will refer to this family as AtaTs for Aminoacyl-tRNA-acetylating Toxins. GNAT-fold comprises a central beta-sheet composed of six to seven strands surrounded by four alpha helices (Figure 3). The alpha helix α3 located between the β4 and β5 strands encodes the signature motif R/Q-X-X-G-X-A/G, also referred to as ‘P-loop,’ which binds the pyrophosphates of acetyl-Coenzyme A which is used as a substrate for the transfer of the acetyl group (Neuwald and Landsman, 1997). The GNAT (general control non-repressible 5 (GCN5)-related N-acetyltransferases) family of proteins comprises more than 300,000 enzymes that acetylate various substrates from small metabolites to proteins and tRNAs (Ikeuchi et al., 2008; Salah Ud-Din et al., 2016). The GNAT-fold toxins from type II TA systems described to date form a distinct monophyletic group of GNAT acetyltransferases (Wilcox et al., 2018). Despite being related, these toxins have diverged to target different species of tRNAs charged with their respective amino acids (Jurenas et al., 2017b; Wilcox et al., 2018). The AtaT toxin encoded by the E. coli O157:H7 strain specifically acetylates the initiator tRNA Met-tRNAfMet. Acetylation of the Met loaded on the Met-tRNAfMet impairs its interaction with the initiation factor 2 (IF2) and precludes the formation of the 30S translation initiation complex (Jurenas et al., 2017a). Formylation of the initiator fMet-tRNAfMet is essential for normal growth (Shah et al., 2019) and is impaired by the Met acetylation, resulting in a strong growth inhibition and generating a dead-end product acMet-tRNAfMet. In vivo expression of AtaT manifests in accumulation of ribosome assembly intermediates, reflecting a strong inhibition of translation initiation (Jurenas et al., 2017a). Interestingly, AtaT is able to discriminate between the initiator Met-tRNAfMet and the elongator Met-tRNAMet in vitro (Jurenas et al., 2017a) although the molecular basis of this specificity has not yet been determined. The E. coli HS strain encoded-ItaT toxin acetylates the elongator Ile-tRNAIle. This leads to the inhibition of translation elongation at Ile codons. It has been shown that tRNAs charged with N-blocked amino acid cannot form ternary complex EF-Tu:GTP:tRNA (Janiak et al., 1990), therefore acetylated elongator tRNAs would not be delivered to the ribosome. TacTs from Salmonella have been shown to have more relaxed specificities and target several elongator tRNAs. However, some specificity still occurs as different TacTs have slightly different preferences for subsets of elongation aa-tRNAs. For example, the TacT and TacT2 toxins mostly target the Gly-tRNAGly, while TacT3 prefers Ile or Leu charged tRNAs (Rycroft et al., 2018). TacTs were suggested to play an important role for persistence of Salmonella in macrophages (Helaine et al., 2014; Cheverton et al., 2016; Rycroft et al., 2018). However, recent data did not find any involvement of TacT or other type II TA systems from Salmonella in persistence (Claudi et al., 2014; Pontes and Groisman, 2019). On the other hand, like other TAs, many AtaRT-like TA systems are associated to mobile genetic elements, such as plasmids, transposons and integrons and could be involved in their maintenance (Iqbal et al., 2015; McVicker and Tang, 2016; Jurenas et al., 2017b).
HipA toxins phosphorylate aminoacyl-tRNA synthetases on conserved serines located in their ATP-binding sites, therefore leading to their inactivation (Kaspy et al., 2013; Vang Nielsen et al., 2019). Since phosphorylated aa-tRNA-synthetases cannot charge their respective tRNAs, ribosomes stall at the generated hungry codons. Consequently, an increase of ppGpp concentration is observed, associated to the activation of RelA, the effect known as the stringent response (Wendrich et al., 2002; Germain et al., 2013; Vang Nielsen et al., 2019). HipA toxins are serine-threonine kinases that belong to the phosphatidylinositol (PI) 3/4–kinase superfamily (Correia et al., 2006). Their C-terminal domain is all-alpha helical and has a similar fold to human CDK2/cyclin A kinase (Figure 3) (Schumacher et al., 2009). The C-terminal domain comprises the active site with a conserved aspartate, an ATP-binding ‘P-loop motif’ and a Mg2+ binding aspartate (Correia et al., 2006). The N-terminus is an α/β globular domain specific to the HipA toxins (Figure 3) (Correia et al., 2006; Schumacher et al., 2009). Interestingly, it can be encoded as a separate protein, as seen in a three-component TA system HipBST from E. coli O27. The HipT toxin exhibits sequence similarity with the C-terminal kinase region of the HipA toxin and also targets specific tRNA synthetase (see below). The gene located upstream of the hipT gene, hipS, encodes a small protein (∼100 amino acids) corresponding to the N-terminal domain of HipA which is able to counteract the toxic activity of HipT (Vang Nielsen et al., 2019). The first gene of the operon, hipB, encodes a protein that enhances the ability of HipS to counteract HipT. Indeed, it has been proposed, that in addition to HipB antitoxin, the N-terminal domain of HipA (similar to the HipS protein) could be involved in regulation of HipA toxin activity through dimerization that blocks the active site (Schumacher et al., 2015).
The E. coli K12 HipA toxin phosphorylates the Ser 239 located in the ATP-binding site of the glutamyl-tRNA synthetase (GltX) (Germain et al., 2013; Kaspy et al., 2013). The homologous E. coli O127 HipT toxin phosphorylates tryptophanyl-tRNA synthetase (TrpS) at the conserved Ser 197 (corresponding to Ser 239 in the glutamyl-tRNA synthetase) (Vang Nielsen et al., 2019). The Ser 239 of GltX is located in a conserved flexible loop (characteristic to type I aa-tRNA-synthetases). Conformational changes upon tRNAGlu binding make this loop more exposed (Sekine et al., 2003). It was shown that HipA only phosphorylates the tRNAGlu-bound HipA (Germain et al., 2013). The conserved motif of GltX that is phosphorylated by HipA is required for ATP binding. Thus, phosphorylation of GltX likely precludes the binding of ATP at the first step of aminoacylation reaction (Sekine et al., 2003; Germain et al., 2013). Meanwhile, the HipT toxin is able to phosphorylate the TrpS tryptophanyl-tRNA synthetase independently of tRNATrp binding (Vang Nielsen et al., 2019). In fact, GltX activates glutamate to glutamyl adenylate only in presence of cognate tRNA, while TrpS can activate tryptophan to tryptophanyl-adenylate without binding tRNATrp (Giege and Springer, 2016). These differences likely correspond to the conformation and accessibility of the loop in the ATP-binding site of the targeted aminoacyl-tRNA synthetases.
It has been shown that HipA inactivates itself by auto-phosphorylation (Correia et al., 2006). Typically, kinases auto-phosphorylate on the solvent exposed activation loops, HipA instead auto-phosphorylates at Ser150 located in its ATP-binding site (P-loop motif) located in the core of the protein (Schumacher et al., 2012). Autophosphorylation on Ser150 leads to conformational changes of the P-loop motif which then hinders binding of ATP (Correia et al., 2006; Schumacher et al., 2012). Likely due to the flexibility of the P-loop, autophosphorylation is an intermolecular event and therefore is likely to happen when amounts of free HipA increase, thereby providing an auto-regulation and reported as a possibility to revive from HipA-induced growth inhibition (Korch and Hill, 2006; Schumacher et al., 2012). HipT also auto-phosphorylates at Ser57 and Ser59, which are adjacent to the P-loop motif of the kinase, and which corresponds to the position of autophosphorylation of HipA (Vang Nielsen et al., 2019). The HipA toxin from Shewanella oneidensis was also shown to auto-phosphorylate at a similar position and it was proposed that this modification is important for complex formation with the cognate HipB antitoxin and its further binding to DNA as well as the stability of this complex (Wen et al., 2014). The autophosphorylation most likely regulates the activity and the expression of the HipA toxins.
The discovery and the name of the E. coli K12 HipA toxin is related to the isolation of the hyper-the HipA7 persistent mutant (Moyed and Bertrand, 1983). The HipA7 strain shows a 100 to 1000-fold increase in persistence (Korch et al., 2003). It was shown that the hipA7 allele codes for the mutations G22S and D291A in the HipA protein, which was later described to encode the toxin HipA from the hipBA TA system (Black et al., 1991; Korch et al., 2003). Recently, a proteomics study suggested that HipA phosphorylates multiple targets in addition to the principal target GltX, notably the ribosomal protein L11 (RplK) and other proteins involved in translation, transcription, and replication (Semanjski et al., 2018). This is in agreement with previous reports showing that HipA overexpression inhibits protein, RNA and DNA synthesis in vivo (Korch and Hill, 2006). However, no phosphorylation resulting from endogenous HipA encoded on the chromosome was observed, since it is repressed by HipB (Semanjski et al., 2018). Furthermore, the HipA7 strain showed phosphorylation of GltX and to a lesser extent of the phage shock protein PspA (Semanjski et al., 2018). This is in agreement with previous observations that the G22S mutation in the N-terminal domain of HipA7 likely results in compromised dimerization and failure to form a HipA:HipB:operator-DNA complex required for neutralization of HipA and transcription repression of the hipBA locus (Schumacher et al., 2015). Induction of HipA7 however showed more phosphorylation targets (Semanjski et al., 2018), therefore indicating that they might be physiologically non-relevant and result from a high expression of the protein. Therefore, GltX is likely to be the main target of E. coli K-12 HipA toxin. HipA7 also showed less auto-phosphorylation than HipA, indicating that the mutant either has less activity or compromised folding or stability, as induction of chaperons was reported upon overexpression of HipA7 (Semanjski et al., 2018). Accordingly, it has been previously reported that the HipA7 does not have a strong inhibitory effect on protein synthesis (Korch and Hill, 2006). In conclusion, the hipA7 allele likely results in smaller effective HipA7 concentrations as compared to the wild-type system and in overall reduced activity of phosphorylation, however the HipA7 toxin is more likely to be released from the HipBA7 complex. It was later proposed that the HipBA system, together with 10 other TA systems in which the toxins are RNases, are the central effectors of antibiotic persistence in E. coli (Maisonneuve et al., 2018a, b; Germain et al., 2019). However, it was later demonstrated that these TA systems are not involved in persistence (see above) (Ramisetty et al., 2016; Harms et al., 2017; Shan et al., 2017; Goormaghtigh et al., 2018a) and the idea that ppGpp is required for persistence has been challenged as well (Bhaskar et al., 2018; Pontes and Groisman, 2019).
Another family of proteins that are YjjJ-like belong to the same PI 3/4-kinase superfamily as HipA and possesses similarities in the catalytic domain (Correia et al., 2006; Maeda et al., 2017). YjjJ although encoded without cognate antitoxin was shown to be toxic, but strikingly it could be neutralized by the HipB antitoxin. YjjJ comprises a DNA-binding motif in its N-terminus that is not present in HipA-like toxins. YjjJ appears to have different cellular targets as it does not inhibit macromolecule synthesis and may affect cell division (Maeda et al., 2017).
Doc toxin is a Fic-fold protein that catalyzes phosphorylation of the elongation factor EF-Tu (Castro-Roa et al., 2013). Fic-fold proteins typically perform NMPylation (AMPylation, GMPylation, or UMPylation) as a post-translational modification on proteins using a phosphate-containing compound, usually ATP (Garcia-Pino et al., 2014; Veyron et al., 2018). In contrast, Doc toxin catalyzes the transfer of the phosphor moiety of ATP, instead of transferring the AMP, and is therefore a kinase (Castro-Roa et al., 2013). Doc phosphorylates EF-Tu at the conserved threonine Thr283 which leads to translation arrest (Castro-Roa et al., 2013). The binding site of Doc on EF-Tu likely overlaps with the tRNA binding site since ternary complex formation prevents Doc binding. In agreement with that, Doc preferentially phosphorylates the GDP-bound state of EF-Tu (Castro-Roa et al., 2013). Phosphorylation of the Thr382 located on the loop of the beta-barrel domain III of EF-Tu locks it in an unfavorable open conformation typical of GDP-bound EF-Tu (Talavera et al., 2018). Conformational dynamics of EF-Tu are the essence of its function and GTP hydrolysis has a major effect on aa-tRNA binding and interaction with the ribosome. Once locked in an open state, EF-Tu exhibits decreased affinity for aa-tRNA to a similar extent as the affinity of GDP-bound EF-Tu (Talavera et al., 2018) and is not compatible with translation (Castro-Roa et al., 2013). Fic domain toxins that perform AMPylation have also been reported to constitute type II TA modules (Harms et al., 2015). FicT toxins target TopoIV and Gyrase, and block their ATPase activity (Harms et al., 2015). Fic and Doc domain families, together sometimes referred to as Fido proteins, have conserved the alpha-helical core arranged in the bundle with two additional alpha helices perpendicular to the bundle (Figure 3) (Kinch et al., 2009). Although active site geometry is conserved, a single substitution in the active site motifs for Doc in comparison to Fic (K73 in Doc while G114 in Fic) leads to inverted orientation of ATP and therefore the transfer of γ-phosphate (Castro-Roa et al., 2013).
In contrast to some TA toxins, like RnlA that targets invading phages, Doc is itself encoded by a phage and provides stability to its lysogenic state. The Doc toxin is encoded by the P1 phage that is maintained as a plasmid during its lysogenic cycle. Doc was named after its impact on the remarkable stability of lysogens due to elimination of cells that have lost P1 (death on curing) (Lehnherr et al., 1993).
Protein synthesis is one of the most complex processes in the cell. Translation involves the step by step assembly of ribosomes, coordinated movements of translation machinery at every addition of a new amino acid into the nascent polypeptide chain and well-organized termination, leading to the release of a newly synthesized protein as well as the recycling of ribosomes and translation factors (Arenz and Wilson, 2016a). The complexity of this process provides a multitude of intervention possibilities that have been explored by antibiotics, bacteriocins and secreted toxins (Zhang et al., 2012; Arenz and Wilson, 2016b; Kumariya et al., 2019). Toxins that are part of type II TA systems target translation in a multitude of ways – from destroying the transcripts before or during translation, to affecting ribosome biogenesis or interrupting the charging of tRNAs or the delivery of amino acids into growing polypeptide chains. Targeting translation allows not only choices, but also room for specialization – potential targets include a great number of tRNAs, tRNA synthetases, translation factors, and the ribosome itself (rRNAs and ribosomal proteins). Specialization of toxins portrayed in this review is already seen in almost all the TA toxin families that we know of to date. MazF toxins have diverged to target different mRNA and precursor rRNA sequences – although the majority of them cleave downstream of U and upstream of ACA nucleobases, some of them prefer an A downstream of the cleavage site and C or G upstream (Figure 2). Furthermore, the recently described MazF-mt9 is specialized for a particular tRNALys43–UUU species (Barth et al., 2019). VapCs also exhibit specificity for a multitude of different tRNAs or even a tRNA stem-loop structure-mimicking the 23S SRL (Winther et al., 2016) (Figure 4). The AtaT-like toxins that target charged tRNAs are also specific to different tRNAs and even though their toxicity relies on the acetylation of the cargo amino acid charged on its cognate tRNA, these toxins most likely recognize both the amino acids and the tRNA sequence (Jurenas et al., 2017a; Rycroft et al., 2018; Wilcox et al., 2018). Likewise, different HipA toxins have been recently demonstrated to phosphorylate different aminoacyl-tRNA synthetases (Germain et al., 2013; Kaspy et al., 2013; Vang Nielsen et al., 2019). RelE family toxins seem to recognize the mRNA pre-loaded ribosome rather than the particular mRNA sequence. Although some RelE toxins show some preference toward certain nucleobases at certain positions of a codon, the specificity comes not only from the toxin itself, but also from the conformation of ribosomes induced by the binding of these toxins (Neubauer et al., 2009; Feng et al., 2013; Maehigashi et al., 2015; Schureck et al., 2016b; Pavelich et al., 2019). Consequently, the specialization of RelEs involves the evolution of the interactions with ribosomes that in turn direct the substrate recognition.
In this review, we have discussed a number of toxin families that inhibit translation at different steps. Not surprisingly, some of these families have potential evolutionary links with proteins involved in the translation maintenance. Toxins targeting RNAs share folds with RNA metabolism associated proteins, in particular those used in RNA maturation and processing (dsRBD, HEPN and PIN-like domains) (Anantharaman and Aravind, 2006; Makarova et al., 2006; Anantharaman et al., 2013). These protein folds are not limited to type II TA toxins, they also take part in other defense and offense systems in prokaryotes as well as in eukaryotes; namely, dsRBD, HEPN, ferredoxin-like, PIN, FIC, and BECR domains can be detected in RNA interference, antitransposon and antiviral systems (Makarova et al., 2006; Kwon et al., 2012; Zhang et al., 2012; Anantharaman et al., 2013). The MazF endoribonucleases comprise a distinct SH3-like barrel-fold rather than a typical nucleic acid-binding domain. The classical SH3 domain is common in eukaryotic cell-to-cell communication and signal transduction proteins, such as signaling kinases, but is less evident in prokaryotes. It has been speculated that bacterial SH3-domain proteins act in eukaryotic cell invasion by corrupting cell signaling (Whisstock and Lesk, 1999). MazF however belongs to the group of proteins that possess domains that structurally resemble SH3, but have diverse functions and enzymatic activities (Whisstock and Lesk, 1999). Therefore, it is not clear whether the SH3 structural motif observed in MazF has a true evolutionary link with other SH3-domain proteins. Similarly, HipA toxins comprise a fold similar to (PI)3/4-kinases found in eukaryotes. These eukaryotic kinases produce 3′ phosphoinositide lipids that bind and activate proteins and therefore participate in signaling cascades (Lempiainen and Halazonetis, 2009). However, a certain class of eukaryotic PI3K family proteins are also Ser/Thr kinases (like HipA toxins) that respond to DNA damage (ATM and DNA-PKcs), nutrient stress (mTOR) or are involved in nonsense-mediated mRNA decay (SMG-1) or transcription regulation (TRRAP) (Lempiainen and Halazonetis, 2009). Whether these eukaryotic proteins have real evolutionary links with HipA toxins remains unclear.
GNAT-fold acetyltransferases are among the most abundant protein folds, however all GNAT-fold TA toxins analyzed to date target the amino group of the amino acid charged on their respective tRNAs (Cheverton et al., 2016; Jurenas et al., 2017a; Wilcox et al., 2018). GNAT-fold acetyltransferases are known to target a wide variety of substrates (Salah Ud-Din et al., 2016). A certain class of GNAT enzymes also acetylate the alpha-amino group of amino acids, however in the context of proteins, i.e., the N-terminal amino acid of peptides after methionine removal. In eukaryotes, this modification is co-translational and affects the majority of proteins, while in bacteria it is limited to specific cases of several ribosomal proteins (Vetting et al., 2008; Favrot et al., 2016). As for GNATs that interact with tRNA, TmcA – an enzyme implicated in translation fidelity – acetylates the wobble cytidine in the anticodon to prevent its misreading (Ikeuchi et al., 2008). Another family of GNATs – the Fem enzymes – use charged tRNAs as substrates for the synthesis of peptidoglycan precursors (Dare and Ibba, 2012). Fic/Doc toxins and secreted toxic effectors generally target eukaryotic and prokaryotic GTPases involved in protein signaling, translation or replication, however, new Fic targets, such as chaperons are emerging (Veyron et al., 2018). Interestingly, three TA toxin folds have also evolved to target topoisomerases (MazF/CcdB, RelE/ParE, Doc/Fic). It is not clear what the link is between translation machinery and topoisomerases. However, topoisomerases are probably the second favorite target of TA toxins. DNA damage caused by these toxins induces a SOS response and DNA repair and could favor rearrangements of genetic material providing higher chances for TAs to relocate. However, it is not clear why the mechanism of choice is blocking topoisomerases. It is worth noting that the translation machinery and topoisomerases are also among the most common targets of antibiotics. Lastly, novel enzymatic activities and targets of TA toxins have been increasingly reported, for example ADP-rybosyltransferase-fold toxins were shown to act by NAD+ phosphorolysis and its depletion, or by DNA ribosylation (Jankevicius et al., 2016; Freire et al., 2019). These are the first examples of ADP-ribosyltransferase toxins (ART) involved in TA systems, however, many examples of secreted ART toxins are known and predicted, those also involve translation inhibitors, such as the diphtheria toxin (Zhang et al., 2012).
The role(s) of TA systems in bacterial physiology and evolution is a long-standing debate (Magnuson, 2007; Tsilibaris et al., 2007; Van Melderen and Saavedra De Bast, 2009; Van Melderen, 2010; Ramisetty et al., 2016; Harms et al., 2017; Culviner and Laub, 2018; Goormaghtigh et al., 2018a, b; Holden and Errington, 2018; Kaldalu et al., 2019; Mets et al., 2019; Pontes and Groisman, 2019; Wade and Laub, 2019; Fraikin et al., 2020). Since their discovery on plasmids in the 1980’s and on chromosomes almost 20 years later, the TA field has been going through waves of hypothesis ranging from replicon maintenance, programmed cell death, stress response, generation of specialized ribosomes, persistence to antibiotics, to phage abortive infection mechanisms. The mainstream hypothesis for the last 10 years was the central role played by type II TA systems in persistence to antibiotics. The hypothesis is that TA systems would be induced in persister subpopulations, thereby stopping their growth and allowing these cells to tolerate the presence of antibiotics. While this hypothesis prevailed for several years, contradicting data accumulated and eventually lead to the retraction of the main papers thereby questioning the involvement of TA systems in drug tolerance (Tsilibaris et al., 2007; Conlon et al., 2016; Ramisetty et al., 2016; Harms et al., 2017; Shan et al., 2017; Goormaghtigh et al., 2018a, b; Kaldalu and Tenson, 2019; Pontes and Groisman, 2019). Although TA systems are occasionally found upregulated under stress conditions in transcriptomic data (Keren et al., 2004), this could be a natural consequence of de-repression of TA loci due to instability and degradation of the antitoxins. Since TA systems present selfish behavior, it is tempting to compare them to viral elements, and to look at TAs from the perspective of genes, and not of the organism. In this review we have provided a detailed view of the specialization of toxins sharing the same fold. Such a specialization seems to be the general trend for type II TA toxins and is seen in virtually all families that have at least several studied examples. If such systems would be part of a stress-response system, however, one would expect the selection and conservation of the ‘best’ mode of action. On the contrary, the reservoir and activities of TA systems from different strains is highly variable and reminds the variety seen in offense and defense systems that are used for competition between species where innovations are beneficial. Among the driving forces for the evolution of different substrate specificities could be the tight neutralization of each toxin by its cognate antitoxin. Co-evolution of different antitoxins and different toxins relies on their vast contacts, necessary for neutralization and transcriptional autoregulation. This dependency should in addition allow for a faster evolution and selection of changes. Further, the competition between an incoming near-identical TA system likely provides selective pressure. The incoming TAs, if identical, would be neutralized by an existing copy of the TA system and such an ‘anti-addiction’ module therefore would prevent stable establishment of identical TAs (Van Melderen and Saavedra De Bast, 2009). Indeed, it has been shown that in some cases the different TAs in the same organism are only insulated by 1 amino acid difference (Walling and Butler, 2018), indicating that a strong selection might apply on TAs to avoid cross-talks. Lastly, the high abundance of TA systems on mobile genetic elements supports the idea that TA systems are primarily elements involved in intergenomic conflicts – inheritance of existing, defense against incoming and offense or spread of new genetic material.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
DJ was supported by a post-doctoral fellowship from the ‘Fondation pour la Recherche Médicale’ and EMBO non-stipendiary long-term fellowship. Work in LV’s lab was supported by the Fonds National de la Recherche Scientifique (FNRS, T.0147.15F PDR and J.0061.16F CDR), the Fonds d’Encouragement à la Recherche ULB (FER-ULB), the Interuniversity Attraction Poles Program initiated by the Belgian Science Policy Office (MicroDev), the Fonds Jean Brachet, and the Fondation Van Buuren.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ahn, D. H., Lee, K. Y., Lee, S. J., Park, S. J., Yoon, H. J., Kim, S. J., et al. (2017). Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor. J Biol Chem 292, 18832–18847. doi: 10.1074/jbc.M117.807974
Amitai, S., Kolodkin-Gal, I., Hananya-Meltabashi, M., Sacher, A., and Engelberg-Kulka, H. (2009). Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet 5:e1000390. doi: 10.1371/journal.pgen.1000390
Anantharaman, V., and Aravind, L. (2003). New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol 4, R81. doi: 10.1186/gb-2003-4-12-r81
Anantharaman, V., and Aravind, L. (2006). The NYN domains: novel predicted RNAses with a PIN domain-like fold. RNA Biol 3, 18–27. doi: 10.4161/rna.3.1.2548
Anantharaman, V., Makarova, K. S., Burroughs, A. M., Koonin, E. V., and Aravind, L. (2013). Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct 8, 15. doi: 10.1186/1745-6150-8-15
Arenz, S., and Wilson, D. N. (2016a). Bacterial Protein Synthesis as a Target for Antibiotic Inhibition. Cold Spring Harb Perspect Med 6, a025361. doi: 10.1101/cshperspect.a025361
Arenz, S., and Wilson, D. N. (2016b). Blast from the Past: Reassessing Forgotten Translation Inhibitors, Antibiotic Selectivity, and Resistance Mechanisms to Aid Drug Development. Mol Cell 61, 3–14. doi: 10.1016/j.molcel.2015.10.019
Barth, V. C., Zeng, J. M., Vvedenskaya, I. O., Ouyang, M., Husson, R. N., and Woychik, N. A. (2019). Toxin-mediated ribosome stalling reprograms the Mycobacterium tuberculosis proteome. Nat Commun 10, 3035. doi: 10.1038/s41467-019-10869-8
Belitsky, M., Avshalom, H., Erental, A., Yelin, I., Kumar, S., London, N., et al. (2011). The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol Cell 41, 625–635. doi: 10.1016/j.molcel.2011.02.023
Bernard, P., and Couturier, M. (1992). Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol 226, 735–745. doi: 10.1016/0022-2836(92)90629-x
Bernard, P., Kezdy, K. E., Van Melderen, L., Steyaert, J., Wyns, L., Pato, M. L., et al. (1993). The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J Mol Biol 234, 534–541. doi: 10.1006/jmbi.1993.1609
Bhaskar, A., De Piano, C., Gelman, E., McKinney, J. D., and Dhar, N. (2018). Elucidating the role of (p)ppGpp in mycobacterial persistence against antibiotics. IUBMB Life 70, 836–844. doi: 10.1002/iub.1888
Bibi-Triki, S., Li de la Sierra-Gallay, I., Lazar, N., Leroy, A., Van Tilbeurgh, H., Sebbane, F., et al. (2014). Functional and structural analysis of HicA3-HicB3, a novel toxin-antitoxin system of Yersinia pestis. J Bacteriol 196, 3712–3723. doi: 10.1128/JB.01932-14
Black, D. S., Kelly, A. J., Mardis, M. J., and Moyed, H. S. (1991). Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol 173, 5732–5739. doi: 10.1128/jb.173.18.5732-5739.1991
Blower, T. R., Pei, X. Y., Short, F. L., Fineran, P. C., Humphreys, D. P., Luisi, B. F., et al. (2011). A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat Struct Mol Biol 18, 185–190. doi: 10.1038/nsmb.1981
Brown, B. L., Grigoriu, S., Kim, Y., Arruda, J. M., Davenport, A., Wood, T. K., et al. (2009). Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog 5:e1000706. doi: 10.1371/journal.ppat.1000706
Brown, J. M., and Shaw, K. J. (2003). A novel family of Escherichia coli toxin-antitoxin gene pairs. J Bacteriol 185, 6600–6608. doi: 10.1128/jb.185.22.6600-6608.2003
Butt, A., Higman, V. A., Williams, C., Crump, M. P., Hemsley, C. M., Harmer, N., et al. (2014). The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem J 459, 333–344. doi: 10.1042/BJ20140073
Castro-Roa, D., Garcia-Pino, A., De Gieter, S., van Nuland, N. A. J., Loris, R., and Zenkin, N. (2013). The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol 9, 811–817. doi: 10.1038/nchembio.1364
Chen, R., Tu, J., Liu, Z., Meng, F., Ma, P., Ding, Z., et al. (2017). Structure of the MazF-mt9 toxin, a tRNA-specific endonuclease from Mycobacterium tuberculosis. Biochem Biophys Res Commun 486, 804–810. doi: 10.1016/j.bbrc.2017.03.132
Cheverton, A. M., Gollan, B., Przydacz, M., Wong, C. T., Mylona, A., Hare, S. A., et al. (2016). A Salmonella Toxin Promotes Persister Formation through Acetylation of tRNA. Mol Cell 63, 86–96. doi: 10.1016/j.molcel.2016.05.002
Choi, W., Yamaguchi, Y., Lee, J. W., Jang, K. M., Inouye, M., Kim, S. G., et al. (2017). Translation-dependent mRNA cleavage by YhaV in Escherichia coli. FEBS Lett 591, 1853–1861. doi: 10.1002/1873-3468.12705
Christensen-Dalsgaard, M., Jorgensen, M. G., and Gerdes, K. (2010). Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol 75, 333–348. doi: 10.1111/j.1365-2958.2009.06969.x
Cintron, M., Zeng, J. M., Barth, V. C., Cruz, J. W., Husson, R. N., and Woychik, N. A. (2019). Accurate target identification for Mycobacterium tuberculosis endoribonuclease toxins requires expression in their native host. Sci Rep 9, 5949. doi: 10.1038/s41598-019-41548-9
Claudi, B., Sprote, P., Chirkova, A., Personnic, N., Zankl, J., Schurmann, N., et al. (2014). Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158, 722–733. doi: 10.1016/j.cell.2014.06.045
Conlon, B. P., Rowe, S. E., Gandt, A. B., Nuxoll, A. S., Donegan, N. P., Zalis, E. A., et al. (2016). Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1, 16051. doi: 10.1038/nmicrobiol.2016.51
Coray, D. S., Wheeler, N. E., Heinemann, J. A., and Gardner, P. P. (2017). Why so narrow: Distribution of anti-sense regulated, type I toxin-antitoxin systems compared with type II and type III systems. RNA Biol 14, 275–280. doi: 10.1080/15476286.2016.1272747
Correia, F. F., D’Onofrio, A., Rejtar, T., Li, L., Karger, B. L., Makarova, K., et al. (2006). Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J Bacteriol 188, 8360–8367. doi: 10.1128/JB.01237-06
Cruz, J. W., Sharp, J. D., Hoffer, E. D., Maehigashi, T., Vvedenskaya, I. O., Konkimalla, A., et al. (2015). Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat Commun 6, 7480. doi: 10.1038/ncomms8480
Culviner, P. H., and Laub, M. T. (2018). Global Analysis of the E. coli Toxin MazF Reveals Widespread Cleavage of mRNA and the Inhibition of rRNA Maturation and Ribosome Biogenesis. Mol Cell 70, 868.e–880.e. doi: 10.1016/j.molcel.2018.04.026
Daines, D. A., Jarisch, J., and Smith, A. L. (2004). Identification and characterization of a nontypeable Haemophilus influenzae putative toxin-antitoxin locus. BMC Microbiol 4:30. doi: 10.1186/1471-2180-4-30
Daines, D. A., Wu, M. H., and Yuan, S. Y. (2007). VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol 189, 5041–5048. doi: 10.1128/JB.00290-07
Dalton, K. M., and Crosson, S. (2010). A conserved mode of protein recognition and binding in a ParD-ParE toxin-antitoxin complex. Biochemistry 49, 2205–2215. doi: 10.1021/bi902133s
Dao-Thi, M. H., Van Melderen, L., De Genst, E., Afif, H., Buts, L., Wyns, L., et al. (2005). Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol 348, 1091–1102. doi: 10.1016/j.jmb.2005.03.049
Dare, K., and Ibba, M. (2012). Roles of tRNA in cell wall biosynthesis. Wiley Interdiscip Rev RNA 3, 247–264. doi: 10.1002/wrna.1108
Das, U., Pogenberg, V., Subhramanyam, U. K., Wilmanns, M., Gourinath, S., and Srinivasan, A. (2014). Crystal structure of the VapBC-15 complex from Mycobacterium tuberculosis reveals a two-metal ion dependent PIN-domain ribonuclease and a variable mode of toxin-antitoxin assembly. J Struct Biol 188, 249–258. doi: 10.1016/j.jsb.2014.10.002
Dienemann, C., Boggild, A., Winther, K. S., Gerdes, K., and Brodersen, D. E. (2011). Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J Mol Biol 414, 713–722. doi: 10.1016/j.jmb.2011.10.024
Engelberg-Kulka, H., Sat, B., Reches, M., Amitai, S., and Hazan, R. (2004). Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol 12, 66–71. doi: 10.1016/j.tim.2003.12.008
Fatica, A., Tollervey, D., and Dlakic, M. (2004). PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA 10, 1698–1701. doi: 10.1261/rna.7123504
Favrot, L., Blanchard, J. S., and Vergnolle, O. (2016). Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation. Biochemistry 55, 989–1002. doi: 10.1021/acs.biochem.5b01269
Feng, S., Chen, Y., Kamada, K., Wang, H., Tang, K., Wang, M., et al. (2013). YoeB-ribosome structure: a canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res 41, 9549–9556. doi: 10.1093/nar/gkt742
Fineran, P. C., Blower, T. R., Foulds, I. J., Humphreys, D. P., Lilley, K. S., and Salmond, G. P. (2009). The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A 106, 894–899. doi: 10.1073/pnas.0808832106
Fraikin, N., Goormaghtigh, F., and Van Melderen, L. (2020). Type II toxin-antitoxin systems: evolution and revolutions. J Bacteriol 202, doi: 10.1128/JB.00763-19 ∗∗query
Fraikin, N., Rousseau, C. J., Goeders, N., and Van Melderen, L. (2019). Reassessing the Role of the Type II MqsRA Toxin-Antitoxin System in Stress Response and Biofilm Formation: mqsA Is Transcriptionally Uncoupled from mqsR. mBio 10, e2678–e2619. doi: 10.1128/mBio.02678-19
Freire, D. M., Gutierrez, C., Garza-Garcia, A., Grabowska, A. D., Sala, A. J., Ariyachaokun, K., et al. (2019). An NAD(+) Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death. Mol Cell 73, 1282.e–1291.e. doi: 10.1016/j.molcel.2019.01.028
Garcia-Pino, A., Balasubramanian, S., Wyns, L., Gazit, E., De Greve, H., Magnuson, R. D., et al. (2010). Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 142, 101–111. doi: 10.1016/j.cell.2010.05.039
Garcia-Pino, A., Zenkin, N., and Loris, R. (2014). The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem Sci 39, 121–129. doi: 10.1016/j.tibs.2014.01.001
Gerdes, K., and Maisonneuve, E. (2012). Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol 66, 103–123. doi: 10.1146/annurev-micro-092611-150159
Gerdes, K., Rasmussen, P. B., and Molin, S. (1986). Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A 83, 3116–3120. doi: 10.1073/pnas.83.10.3116
Gerdes, K., and Wagner, E. G. (2007). RNA antitoxins. Curr Opin Microbiol 10, 117–124. doi: 10.1016/j.mib.2007.03.003
Germain, E., Castro-Roa, D., Zenkin, N., and Gerdes, K. (2013). Molecular mechanism of bacterial persistence by HipA. Mol Cell 52, 248–254. doi: 10.1016/j.molcel.2013.08.045
Germain, E., Roghanian, M., Gerdes, K., and Maisonneuve, E. (2015). Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 112, 5171–5176. doi: 10.1073/pnas.1423536112
Germain, E., Roghanian, M., Gerdes, K., and Maisonneuve, E. (2019). Retraction for Germain et al., Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 116, 11077. doi: 10.1073/pnas.1906160116
Giege, R., and Springer, M. (2016). Aminoacyl-tRNA Synthetases in the Bacterial World. EcoSal Plus 7, doi: 10.1128/ecosalplus.ESP-0002-2016 ∗∗query∗
Goeders, N., Dreze, P. L., and Van Melderen, L. (2013). Relaxed cleavage specificity within the RelE toxin family. J Bacteriol 195, 2541–2549. doi: 10.1128/JB.02266-12
Gogos, A., Mu, H., Bahna, F., Gomez, C. A., and Shapiro, L. (2003). Crystal structure of YdcE protein from Bacillus subtilis. Proteins 53, 320–322. doi: 10.1002/prot.10457
Goodman, D. B., Church, G. M., and Kosuri, S. (2013). Causes and effects of N-terminal codon bias in bacterial genes. Science 342, 475–479. doi: 10.1126/science.1241934
Goormaghtigh, F., Fraikin, N., Putrins, M., Hallaert, T., Hauryliuk, V., Garcia-Pino, A., et al. (2018a). Reassessing the Role of Type II Toxin-Antitoxin Systems in Formation of Escherichia coli Type II Persister Cells. MBio 9, e640–e618. doi: 10.1128/mBio.00640-18
Goormaghtigh, F., Fraikin, N., Putrins, M., Hauryliuk, V., Garcia-Pino, A., Udekwu, K., et al. (2018b). Reply to Holden and Errington, “Type II Toxin-Antitoxin Systems and Persister Cells”. MBio 9, e1838–e1818. doi: 10.1128/mBio.01838-18
Grela, P., Szajwaj, M., Horbowicz-Drozdzal, P., and Tchorzewski, M. (2019). How Ricin Damages the Ribosome. Toxins (Basel) 11, 241. doi: 10.3390/toxins11050241
Guglielmini, J., and Van Melderen, L. (2011). Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mob Genet Elements 1, 283–290. doi: 10.4161/mge.18477
Hallez, R., Geeraerts, D., Sterckx, Y., Mine, N., Loris, R., and Van Melderen, L. (2010). New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol Microbiol 76, 719–732. doi: 10.1111/j.1365-2958.2010.07129.x
Han, K. D., Matsuura, A., Ahn, H. C., Kwon, A. R., Min, Y. H., Park, H. J., et al. (2011). Functional identification of toxin-antitoxin molecules from Helicobacter pylori 26695 and structural elucidation of the molecular interactions. J Biol Chem 286, 4842–4853. doi: 10.1074/jbc.M109.097840
Hargreaves, D., Santos-Sierra, S., Giraldo, R., Sabariegos-Jareno, R., de la Cueva-Mendez, G., Boelens, R., et al. (2002). Structural and functional analysis of the kid toxin protein from E. coli plasmid R1. Structure 10, 1425–1433.
Harms, A., Brodersen, D. E., Mitarai, N., and Gerdes, K. (2018). Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol Cell 70, 768–784. doi: 10.1016/j.molcel.2018.01.003
Harms, A., Fino, C., Sorensen, M. A., Semsey, S., and Gerdes, K. (2017). Prophages and Growth Dynamics Confound Experimental Results with Antibiotic-Tolerant Persister Cells. MBio 8, e1964–e1917. doi: 10.1128/mBio.01964-17
Harms, A., Stanger, F. V., Scheu, P. D., de Jong, I. G., Goepfert, A., Glatter, T., et al. (2015). Adenylylation of Gyrase and Topo IV by FicT Toxins Disrupts Bacterial DNA Topology. Cell Rep 12, 1497–1507. doi: 10.1016/j.celrep.2015.07.056
Hayes, F., and Van Melderen, L. (2011). Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol 46, 386–408. doi: 10.3109/10409238.2011.600437
Heaton, B. E., Herrou, J., Blackwell, A. E., Wysocki, V. H., and Crosson, S. (2012). Molecular structure and function of the novel BrnT/BrnA toxin-antitoxin system of Brucella abortus. J Biol Chem 287, 12098–12110. doi: 10.1074/jbc.M111.332163
Heinemann, U., and Saenger, W. (1983). Crystallographic study of mechanism of ribonuclease T1-catalysed specific RNA hydrolysis. J Biomol Struct Dyn 1, 523–538. doi: 10.1080/07391102.1983.10507459
Helaine, S., Cheverton, A. M., Watson, K. G., Faure, L. M., Matthews, S. A., and Holden, D. W. (2014). Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208. doi: 10.1126/science.1244705
Hiraga, S., Jaffe, A., Ogura, T., Mori, H., and Takahashi, H. (1986). F plasmid ccd mechanism in Escherichia coli. J Bacteriol 166, 100–104. doi: 10.1128/jb.166.1.100-104.1986
Hoffer, E. D., Miles, S. J., and Dunham, C. M. (2017). The structure and function of Mycobacterium tuberculosis MazF-mt6 toxin provide insights into conserved features of MazF endonucleases. J Biol Chem 292, 7718–7726. doi: 10.1074/jbc.M117.779306
Holden, D. W., and Errington, J. (2018). Type II Toxin-Antitoxin Systems and Persister Cells. MBio 9, e1574–e1518. doi: 10.1128/mBio.01574-18
Huguet, K. T., Gonnet, M., Doublet, B., and Cloeckaert, A. (2016). A toxin antitoxin system promotes the maintenance of the IncA/C-mobilizable Salmonella Genomic Island 1. Sci Rep 6, 32285. doi: 10.1038/srep32285
Hurley, J. M., and Woychik, N. A. (2009). Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem 284, 18605–18613. doi: 10.1074/jbc.M109.008763
Ikeuchi, Y., Kitahara, K., and Suzuki, T. (2008). The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J 27, 2194–2203. doi: 10.1038/emboj.2008.154
Iqbal, N., Guerout, A. M., Krin, E., Le Roux, F., and Mazel, D. (2015). Comprehensive Functional Analysis of the 18 Vibrio cholerae N16961 Toxin-Antitoxin Systems Substantiates Their Role in Stabilizing the Superintegron. J Bacteriol 197, 2150–2159. doi: 10.1128/JB.00108-15
Itoh, T., and Tomizawa, J. (1980). Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A 77, 2450–2454. doi: 10.1073/pnas.77.5.2450
Jaffe, A., Ogura, T., and Hiraga, S. (1985). Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol 163, 841–849.
Janiak, F., Dell, V. A., Abrahamson, J. K., Watson, B. S., Miller, D. L., and Johnson, A. E. (1990). Fluorescence characterization of the interaction of various transfer RNA species with elongation factor Tu.GTP: evidence for a new functional role for elongation factor Tu in protein biosynthesis. Biochemistry 29, 4268–4277. doi: 10.1021/bi00470a002
Jankevicius, G., Ariza, A., Ahel, M., and Ahel, I. (2016). The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA. Mol Cell 64, 1109–1116. doi: 10.1016/j.molcel.2016.11.014
Jia, X., Yao, J., Gao, Z., Liu, G., Dong, Y. H., Wang, X., et al. (2018). Structure-function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN-MNT toxin-antitoxin system. J Biol Chem 293, 6812–6823. doi: 10.1074/jbc.RA118.002421
Jiang, Y., Pogliano, J., Helinski, D. R., and Konieczny, I. (2002). ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 44, 971–979. doi: 10.1046/j.1365-2958.2002.02921.x
Jorgensen, M. G., Pandey, D. P., Jaskolska, M., and Gerdes, K. (2009). HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol 191, 1191–1199. doi: 10.1128/JB.01013-08
Jurenas, D., Chatterjee, S., Konijnenberg, A., Sobott, F., Droogmans, L., Garcia-Pino, A., et al. (2017a). AtaT blocks translation initiation by N-acetylation of the initiator tRNA(fMet). Nat Chem Biol 13, 640–646. doi: 10.1038/nchembio.2346
Jurenas, D., Garcia-Pino, A., and Van Melderen, L. (2017b). Novel toxins from type II toxin-antitoxin systems with acetyltransferase activity. Plasmid 93, 30–35. doi: 10.1016/j.plasmid.2017.08.005
Jurenas, D., Van Melderen, L., and Garcia-Pino, A. (2019). Mechanism of regulation and neutralization of the AtaR-AtaT toxin-antitoxin system. Nat Chem Biol 15, 285–294. doi: 10.1038/s41589-018-0216-z
Kai, T., and Yonesaki, T. (2002). Multiple mechanisms for degradation of bacteriophage T4 soc mRNA. Genetics 160, 5–12.
Kaldalu, N., Maivali, U., Hauryliuk, V., and Tenson, T. (2019). Reanalysis of Proteomics Results Fails To Detect MazF-Mediated Stress Proteins. MBio 10, e949–e919. doi: 10.1128/mBio.00949-19
Kaldalu, N., and Tenson, T. (2019). Slow growth causes bacterial persistence. Sci Signal 12, eaay1167. doi: 10.1126/scisignal.aay1167
Kamada, K., and Hanaoka, F. (2005). Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell 19, 497–509. doi: 10.1016/j.molcel.2005.07.004
Kamada, K., Hanaoka, F., and Burley, S. K. (2003). Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell 11, 875–884.
Karoui, H., Bex, F., Dreze, P., and Couturier, M. (1983). Ham22, a mini-F mutation which is lethal to host cell and promotes recA-dependent induction of lambdoid prophage. EMBO J 2, 1863–1868.
Kaspy, I., Rotem, E., Weiss, N., Ronin, I., Balaban, N. Q., and Glaser, G. (2013). HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun 4, 3001. doi: 10.1038/ncomms4001
Katz, M. E., Strugnell, R. A., and Rood, J. I. (1992). Molecular characterization of a genomic region associated with virulence in Dichelobacter nodosus. Infect Immun 60, 4586–4592.
Keren, I., Shah, D., Spoering, A., Kaldalu, N., and Lewis, K. (2004). Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186, 8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004
Kim, D. H., Kang, S. M., Park, S. J., Jin, C., Yoon, H. J., and Lee, B. J. (2018). Functional insights into the Streptococcus pneumoniae HicBA toxin-antitoxin system based on a structural study. Nucleic Acids Res 46, 6371–6386. doi: 10.1093/nar/gky469
Kinch, L. N., Yarbrough, M. L., Orth, K., and Grishin, N. V. (2009). Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One 4:e5818. doi: 10.1371/journal.pone.0005818
Koga, M., Otsuka, Y., Lemire, S., and Yonesaki, T. (2011). Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 187, 123–130. doi: 10.1534/genetics.110.121798
Kolodkin-Gal, I., Hazan, R., Gaathon, A., Carmeli, S., and Engelberg-Kulka, H. (2007). A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318, 652–655. doi: 10.1126/science.1147248
Korch, S. B., Henderson, T. A., and Hill, T. M. (2003). Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol 50, 1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x
Korch, S. B., and Hill, T. M. (2006). Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J Bacteriol 188, 3826–3836. doi: 10.1128/JB.01740-05
Kumar, S., Kolodkin-Gal, I., and Engelberg-Kulka, H. (2013). Novel quorum-sensing peptides mediating interspecies bacterial cell death. MBio 4, 1–12. doi: 10.1128/mBio.00314-13
Kumariya, R., Garsa, A. K., Rajput, Y. S., Sood, S. K., Akhtar, N., and Patel, S. (2019). Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb Pathog 128, 171–177. doi: 10.1016/j.micpath.2019.01.002
Kwon, A. R., Kim, J. H., Park, S. J., Lee, K. Y., Min, Y. H., Im, H., et al. (2012). Structural and biochemical characterization of HP0315 from Helicobacter pylori as a VapD protein with an endoribonuclease activity. Nucleic Acids Res 40, 4216–4228. doi: 10.1093/nar/gkr1305
Laursen, B. S., Sorensen, H. P., Mortensen, K. K., and Sperling-Petersen, H. U. (2005). Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69, 101–123. doi: 10.1128/MMBR.69.1.101-123.2005
Lehnherr, H., Maguin, E., Jafri, S., and Yarmolinsky, M. B. (1993). Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol 233, 414–428. doi: 10.1006/jmbi.1993.1521
Lempiainen, H., and Halazonetis, T. D. (2009). Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J 28, 3067–3073. doi: 10.1038/emboj.2009.281
Leplae, R., Geeraerts, D., Hallez, R., Guglielmini, J., Dreze, P., and Van Melderen, L. (2011). Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 39, 5513–5525. doi: 10.1093/nar/gkr131
Li, G. Y., Zhang, Y., Inouye, M., and Ikura, M. (2009). Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J Biol Chem 284, 14628–14636. doi: 10.1074/jbc.M809656200
Lopes, A. P., Lopes, L. M., Fraga, T. R., Chura-Chambi, R. M., Sanson, A. L., Cheng, E., et al. (2014). VapC from the leptospiral VapBC toxin-antitoxin module displays ribonuclease activity on the initiator tRNA. PLoS One 9:e101678. doi: 10.1371/journal.pone.0101678
Loris, R., Dao-Thi, M. H., Bahassi, E. M., Van Melderen, L., Poortmans, F., Liddington, R., et al. (1999). Crystal structure of CcdB, a topoisomerase poison from E. coli. J Mol Biol 285, 1667–1677. doi: 10.1006/jmbi.1998.2395
Maeda, Y., Lin, C. Y., Ishida, Y., Inouye, M., Yamaguchi, Y., and Phadtare, S. (2017). Characterization of YjjJ toxin of Escherichia coli. FEMS Microbiol Lett 364, fnx086. doi: 10.1093/femsle/fnx086
Maehigashi, T., Ruangprasert, A., Miles, S. J., and Dunham, C. M. (2015). Molecular basis of ribosome recognition and mRNA hydrolysis by the E. coli YafQ toxin. Nucleic Acids Res 43, 8002–8012. doi: 10.1093/nar/gkv791
Magnuson, R. D. (2007). Hypothetical functions of toxin-antitoxin systems. J Bacteriol 189, 6089–6092. doi: 10.1128/JB.00958-07
Maisonneuve, E., Castro-Camargo, M., and Gerdes, K. (2013). (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150. doi: 10.1016/j.cell.2013.07.048
Maisonneuve, E., Castro-Camargo, M., and Gerdes, K. (2018a). (p)ppGpp Controls Bacterial Persistence by Stochastic Induction of Toxin-Antitoxin Activity. Cell 172, 1135. doi: 10.1016/j.cell.2018.02.023
Maisonneuve, E., Shakespeare, L. J., Jorgensen, M. G., and Gerdes, K. (2011). Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108, 13206–13211. doi: 10.1073/pnas.1100186108
Maisonneuve, E., Shakespeare, L. J., Jorgensen, M. G., and Gerdes, K. (2018b). Retraction for Maisonneuve et al., Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 115, E2901. doi: 10.1073/pnas.1803278115
Makarova, K. S., Grishin, N. V., and Koonin, E. V. (2006). The HicAB cassette, a putative novel, RNA-targeting toxin-antitoxin system in archaea and bacteria. Bioinformatics 22, 2581–2584. doi: 10.1093/bioinformatics/btl418
Manav, M. C., Turnbull, K. J., Jurenas, D., Garcia-Pino, A., Gerdes, K., and Brodersen, D. E. (2019). The E. coli HicB Antitoxin Contains a Structurally Stable Helix-Turn-Helix DNA Binding Domain. Structure 27, 1675.e–1685.e. doi: 10.1016/j.str.2019.08.008
Matelska, D., Steczkiewicz, K., and Ginalski, K. (2017). Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res 45, 6995–7020. doi: 10.1093/nar/gkx494
McClain, W. H., Schneider, J., Bhattacharya, S., and Gabriel, K. (1998). The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc Natl Acad Sci U S A 95, 460–465. doi: 10.1073/pnas.95.2.460
McKenzie, J. L., Duyvestyn, J. M., Smith, T., Bendak, K., Mackay, J., Cursons, R., et al. (2012). Determination of ribonuclease sequence-specificity using Pentaprobes and mass spectrometry. RNA 18, 1267–1278. doi: 10.1261/rna.031229.111
McVicker, G., and Tang, C. M. (2016). Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nat Microbiol 2, 16204. doi: 10.1038/nmicrobiol.2016.204
Mets, T., Kasvandik, S., Saarma, M., Maivali, U., Tenson, T., and Kaldalu, N. (2019). Fragmentation of Escherichia coli mRNA by MazF and MqsR. Biochimie 156, 79–91. doi: 10.1016/j.biochi.2018.10.004
Mets, T., Lippus, M., Schryer, D., Liiv, A., Kasari, V., Paier, A., et al. (2017). Toxins MazF and MqsR cleave Escherichia coli rRNA precursors at multiple sites. RNA Biol 14, 124–135. doi: 10.1080/15476286.2016.1259784
Mhlanga-Mutangadura, T., Morlin, G., Smith, A. L., Eisenstark, A., and Golomb, M. (1998). Evolution of the major pilus gene cluster of Haemophilus influenzae. J Bacteriol 180, 4693–4703.
Miallau, L., Faller, M., Chiang, J., Arbing, M., Guo, F., Cascio, D., et al. (2009). Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J Biol Chem 284, 276–283. doi: 10.1074/jbc.M805061200
Miller, H. I., Riggs, A. D., and Gill, G. N. (1973). Ribonuclease H (hybrid) in Escherichia coli. Identification and characterization. J Biol Chem 248, 2621–2624.
Min, A. B., Miallau, L., Sawaya, M. R., Habel, J., Cascio, D., and Eisenberg, D. (2012). The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg(2)(+) ion in the active site and a putative RNA-binding site. Protein Sci 21, 1754–1767. doi: 10.1002/pro.2161
Miyamoto, T., Kato, Y., Sekiguchi, Y., Tsuneda, S., and Noda, N. (2016a). Characterization of MazF-Mediated Sequence-Specific RNA Cleavage in Pseudomonas putida Using Massive Parallel Sequencing. PLoS One 11:e0149494. doi: 10.1371/journal.pone.0149494
Miyamoto, T., Ota, Y., Yokota, A., Suyama, T., Tsuneda, S., and Noda, N. (2017). Characterization of a Deinococcus radiodurans MazF: A UACA-specific RNA endoribonuclease. Microbiologyopen 6, e00501. doi: 10.1002/mbo3.501
Miyamoto, T., Yokota, A., Ota, Y., Tsuruga, M., Aoi, R., Tsuneda, S., et al. (2018). Nitrosomonas europaea MazF Specifically Recognises the UGG Motif and Promotes Selective RNA Degradation. Front Microbiol 9:2386. doi: 10.3389/fmicb.2018.02386
Miyamoto, T., Yokota, A., Tsuneda, S., and Noda, N. (2016b). AAU-Specific RNA Cleavage Mediated by MazF Toxin Endoribonuclease Conserved in Nitrosomonas europaea. Toxins (Basel) 8, 174. doi: 10.3390/toxins8060174
Moyed, H. S., and Bertrand, K. P. (1983). hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155, 768–775.
Mukherjee, A., Wheaton, G. H., Counts, J. A., Ijeomah, B., Desai, J., and Kelly, R. M. (2017). VapC toxins drive cellular dormancy under uranium stress for the extreme thermoacidophile Metallosphaera prunae. Environ Microbiol 19, 2831–2842. doi: 10.1111/1462-2920.13808
Munishkin, A., and Wool, I. G. (1997). The ribosome-in-pieces: binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA. Proc Natl Acad Sci U S A 94, 12280–12284. doi: 10.1073/pnas.94.23.12280
Munoz-Gomez, A. J., Santos-Sierra, S., Berzal-Herranz, A., Lemonnier, M., and Diaz-Orejas, R. (2004). Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett 567, 316–320. doi: 10.1016/j.febslet.2004.05.005
Naka, K., Koga, M., Yonesaki, T., and Otsuka, Y. (2014). RNase HI stimulates the activity of RnlA toxin in Escherichia coli. Mol Microbiol 91, 596–605. doi: 10.1111/mmi.12479
Nariya, H., and Inouye, M. (2008). MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132, 55–66. doi: 10.1016/j.cell.2007.11.044
Neubauer, C., Gao, Y. G., Andersen, K. R., Dunham, C. M., Kelley, A. C., Hentschel, J., et al. (2009). The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095. doi: 10.1016/j.cell.2009.11.015
Neuwald, A. F., and Landsman, D. (1997). GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci 22, 154–155.
Ogura, T., and Hiraga, S. (1983). Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell 32, 351–360. doi: 10.1016/0092-8674(83)90454-3
Oh, E., Becker, A. H., Sandikci, A., Huber, D., Chaba, R., Gloge, F., et al. (2011). Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308. doi: 10.1016/j.cell.2011.10.044
Olmo, N., Turnay, J., Gonzalez de Buitrago, G., Lopez, de Silanes, I., Gavilanes, J. G., et al. (2001). Cytotoxic mechanism of the ribotoxin alpha-sarcin. Induction of cell death via apoptosis. Eur J Biochem 268, 2113–2123. doi: 10.1046/j.1432-1327.2001.02086.x
Otsuka, Y., Koga, M., Iwamoto, A., and Yonesaki, T. (2007). A role of RnlA in the RNase LS activity from Escherichia coli. Genes Genet Syst 82, 291–299.
Otsuka, Y., and Yonesaki, T. (2012). Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol Microbiol 83, 669–681. doi: 10.1111/j.1365-2958.2012.07975.x
Pachulec, E., and van der Does, C. (2010). Conjugative plasmids of Neisseria gonorrhoeae. PLoS One 5:e9962. doi: 10.1371/journal.pone.0009962
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol 12, 208–214. doi: 10.1038/nchembio.2044
Pandey, D. P., and Gerdes, K. (2005). Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33, 966–976. doi: 10.1093/nar/gki201
Park, J. H., Yamaguchi, Y., and Inouye, M. (2011). Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett 585, 2526–2532. doi: 10.1016/j.febslet.2011.07.008
Pavelich, I. J., Maehigashi, T., Hoffer, E. D., Ruangprasert, A., Miles, S. J., and Dunham, C. M. (2019). Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability. Nucleic Acids Res 47, 10400–10413. doi: 10.1093/nar/gkz760
Pedersen, K., Zavialov, A. V., Pavlov, M. Y., Elf, J., Gerdes, K., and Ehrenberg, M. (2003). The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140. doi: 10.1016/s0092-8674(02)01248-5
Pontes, M. H., and Groisman, E. A. (2019). Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci Signal 12, eaax3938. doi: 10.1126/scisignal.aax3938
Prysak, M. H., Mozdzierz, C. J., Cook, A. M., Zhu, L., Zhang, Y., Inouye, M., et al. (2009). Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol 71, 1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x
Ramage, H. R., Connolly, L. E., and Cox, J. S. (2009). Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767
Ramisetty, B. C., Ghosh, D., Roy Chowdhury, M., and Santhosh, R. S. (2016). What Is the Link between Stringent Response, Endoribonuclease Encoding Type II Toxin-Antitoxin Systems and Persistence? Front Microbiol 7:1882. doi: 10.3389/fmicb.2016.01882
Rao, S. T., and Rossmann, M. G. (1973). Comparison of super-secondary structures in proteins. J Mol Biol 76, 241–256. doi: 10.1016/0022-2836(73)90388-4
Ren, D., Walker, A. N., and Daines, D. A. (2012). Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol 12:263. doi: 10.1186/1471-2180-12-263
Rodnina, M. V., Savelsbergh, A., and Wintermeyer, W. (1999). Dynamics of translation on the ribosome: molecular mechanics of translocation. FEMS Microbiol Rev 23, 317–333. doi: 10.1111/j.1574-6976.1999.tb00402.x
Rothenbacher, F. P., Suzuki, M., Hurley, J. M., Montville, T. J., Kirn, T. J., Ouyang, M., et al. (2012). Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J Bacteriol 194, 3464–3474. doi: 10.1128/JB.00217-12
Rycroft, J. A., Gollan, B., Grabe, G. J., Hall, A., Cheverton, A. M., Larrouy-Maumus, G., et al. (2018). Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat Commun 9, 1993. doi: 10.1038/s41467-018-04472-6
Ryter, J. M., and Schultz, S. C. (1998). Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J 17, 7505–7513. doi: 10.1093/emboj/17.24.7505
Sala, A., Bordes, P., and Genevaux, P. (2014). Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 6, 1002–1020. doi: 10.3390/toxins6031002
Salah Ud-Din, A. I., Tikhomirova, A., and Roujeinikova, A. (2016). Structure and Functional Diversity of GCN5-Related N-Acetyltransferases (GNAT). Int J Mol Sci 17, 1018. doi: 10.3390/ijms17071018
Sato, T., Terabe, M., Watanabe, H., Gojobori, T., Hori-Takemoto, C., and Miura, K. (2001). Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J Biochem 129, 851–860. doi: 10.1093/oxfordjournals.jbchem.a002929
Sauert, M., Wolfinger, M. T., Vesper, O., Muller, C., Byrgazov, K., and Moll, I. (2016). The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res 44, 6660–6675. doi: 10.1093/nar/gkw115
Schifano, J. M., Cruz, J. W., Vvedenskaya, I. O., Edifor, R., Ouyang, M., Husson, R. N., et al. (2016). tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res 44, 1256–1270. doi: 10.1093/nar/gkv1370
Schifano, J. M., Edifor, R., Sharp, J. D., Ouyang, M., Konkimalla, A., Husson, R. N., et al. (2013). Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci U S A 110, 8501–8506. doi: 10.1073/pnas.1222031110
Schifano, J. M., Vvedenskaya, I. O., Knoblauch, J. G., Ouyang, M., Nickels, B. E., and Woychik, N. A. (2014). An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat Commun 5, 3538. doi: 10.1038/ncomms4538
Schumacher, M. A., Balani, P., Min, J., Chinnam, N. B., Hansen, S., Vulic, M., et al. (2015). HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524, 59–64. doi: 10.1038/nature14662
Schumacher, M. A., Min, J., Link, T. M., Guan, Z., Xu, W., Ahn, Y. H., et al. (2012). Role of unusual P loop ejection and autophosphorylation in HipA-mediated persistence and multidrug tolerance. Cell Rep 2, 518–525. doi: 10.1016/j.celrep.2012.08.013
Schumacher, M. A., Piro, K. M., Xu, W., Hansen, S., Lewis, K., and Brennan, R. G. (2009). Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323, 396–401. doi: 10.1126/science.1163806
Schureck, M. A., Dunkle, J. A., Maehigashi, T., Miles, S. J., and Dunham, C. M. (2015). Defining the mRNA recognition signature of a bacterial toxin protein. Proc Natl Acad Sci U S A 112, 13862–13867. doi: 10.1073/pnas.1512959112
Schureck, M. A., Maehigashi, T., Miles, S. J., Marquez, J., and Dunham, C. M. (2016a). mRNA bound to the 30S subunit is a HigB toxin substrate. RNA 22, 1261–1270. doi: 10.1261/rna.056218.116
Schureck, M. A., Repack, A., Miles, S. J., Marquez, J., and Dunham, C. M. (2016b). Mechanism of endonuclease cleavage by the HigB toxin. Nucleic Acids Res 44, 7944–7953. doi: 10.1093/nar/gkw598
Schuster, C. F., Park, J. H., Prax, M., Herbig, A., Nieselt, K., Rosenstein, R., et al. (2013). Characterization of a mazEF toxin-antitoxin homologue from Staphylococcus equorum. J Bacteriol 195, 115–125. doi: 10.1128/JB.00400-12
Sekine, S., Nureki, O., Dubois, D. Y., Bernier, S., Chenevert, R., Lapointe, J., et al. (2003). ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO J 22, 676–688. doi: 10.1093/emboj/cdg053
Semanjski, M., Germain, E., Bratl, K., Kiessling, A., Gerdes, K., and Macek, B. (2018). The kinases HipA and HipA7 phosphorylate different substrate pools in Escherichia coli to promote multidrug tolerance. Sci Signal 11, eaat5750. doi: 10.1126/scisignal.aat5750
Senissar, M., Manav, M. C., and Brodersen, D. E. (2017). Structural conservation of the PIN domain active site across all domains of life. Protein Sci 26, 1474–1492. doi: 10.1002/pro.3193
Shah, R. A., Varada, R., Sah, S., Shetty, S., Lahry, K., Singh, S., et al. (2019). Rapid formylation of the cellular initiator tRNA population makes a crucial contribution to its exclusive participation at the step of initiation. Nucleic Acids Res 47, 1908–1919. doi: 10.1093/nar/gky1310
Shaku, M., Park, J. H., Inouye, M., and Yamaguchi, Y. (2018). Identification of MazF Homologue in Legionella pneumophila Which Cleaves RNA at the AACU Sequence. J Mol Microbiol Biotechnol 28, 269–280. doi: 10.1159/000497146
Shan, Y., Brown Gandt, A., Rowe, S. E., Deisinger, J. P., Conlon, B. P., and Lewis, K. (2017). ATP-Dependent Persister Formation in Escherichia coli. MBio 8, e2267–e2216. doi: 10.1128/mBio.02267-16
Sharrock, A., Ruthe, A., Andrews, E. S. V., Arcus, V. A., and Hicks, J. L. (2018). VapC proteins from Mycobacterium tuberculosis share ribonuclease sequence specificity but differ in regulation and toxicity. PLoS One 13:e0203412. doi: 10.1371/journal.pone.0203412
Simanshu, D. K., Yamaguchi, Y., Park, J. H., Inouye, M., and Patel, D. J. (2013). Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol Cell 52, 447–458. doi: 10.1016/j.molcel.2013.09.006
Sterckx, Y. G., Jove, T., Shkumatov, A. V., Garcia-Pino, A., Geerts, L., De Kerpel, M., et al. (2016). A unique hetero-hexadecameric architecture displayed by the Escherichia coli O157 PaaA2-ParE2 antitoxin-toxin complex. J Mol Biol 428, 1589–1603. doi: 10.1016/j.jmb.2016.03.007
Szekeres, S., Dauti, M., Wilde, C., Mazel, D., and Rowe-Magnus, D. A. (2007). Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol Microbiol 63, 1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x
Takagi, H., Kakuta, Y., Okada, T., Yao, M., Tanaka, I., and Kimura, M. (2005). Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol 12, 327–331. doi: 10.1038/nsmb911
Talavera, A., Hendrix, J., Versees, W., Jurenas, D., Van Nerom, K., Vandenberk, N., et al. (2018). Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors. Sci Adv 4, eaa9714. doi: 10.1126/sciadv.aap9714
Tam, J. E., and Kline, B. C. (1989). The F plasmid ccd autorepressor is a complex of CcdA and CcdB proteins. Mol Gen Genet 219, 26–32. doi: 10.1007/bf00261153
Thomet, M., Trautwetter, A., Ermel, G., and Blanco, C. (2019). Characterization of HicAB toxin-antitoxin module of Sinorhizobium meliloti. BMC Microbiol 19:10. doi: 10.1186/s12866-018-1382-6
Tsilibaris, V., Maenhaut-Michel, G., Mine, N., and Van Melderen, L. (2007). What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol 189, 6101–6108. doi: 10.1128/JB.00527-07
Van Melderen, L. (2010). Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol 13, 781–785. doi: 10.1016/j.mib.2010.10.006
Van Melderen, L., and Saavedra De Bast, M. (2009). Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet 5:e1000437. doi: 10.1371/journal.pgen.1000437
Vang Nielsen, S., Turnbull, K. J., Roghanian, M., Baerentsen, R., Semanjski, M., Brodersen, D. E., et al. (2019). Serine-Threonine Kinases Encoded by Split hipA Homologs Inhibit Tryptophanyl-tRNA Synthetase. MBio 10, e1138–e1119. doi: 10.1128/mBio.01138-19
Verma, M., Choi, J., Cottrell, K. A., Lavagnino, Z., Thomas, E. N., Pavlovic-Djuranovic, S., et al. (2019). A short translational ramp determines the efficiency of protein synthesis. Nat Commun 10, 5774. doi: 10.1038/s41467-019-13810-1
Verma, S., and Bhatnagar, R. (2014). MoxT toxin of Bacillus anthracis exhibits sequence specific ribonuclease activity. Biochem Biophys Res Commun 450, 998–1004. doi: 10.1016/j.bbrc.2014.06.092
Vesper, O., Amitai, S., Belitsky, M., Byrgazov, K., Kaberdina, A. C., Engelberg-Kulka, H., et al. (2011). Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147, 147–157. doi: 10.1016/j.cell.2011.07.047
Vetting, M. W., Bareich, D. C., Yu, M., and Blanchard, J. S. (2008). Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci 17, 1781–1790. doi: 10.1110/ps.035899.108
Veyron, S., Peyroche, G., and Cherfils, J. (2018). FIC proteins: from bacteria to humans and back again. Pathog Dis 76, fty012. doi: 10.1093/femspd/fty012
Wade, J. T., and Laub, M. T. (2019). Concerns about “Stress-Induced MazF-Mediated Proteins in Escherichia coli”. MBio 10, e825–e819. doi: 10.1128/mBio.00825-19
Walling, L. R., and Butler, J. S. (2018). Homologous VapC Toxins Inhibit Translation and Cell Growth by Sequence-Specific Cleavage of tRNA(fMet). J Bacteriol 200, e582–e517. doi: 10.1128/JB.00582-17
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wei, Y., Gao, Z. Q., Otsuka, Y., Naka, K., Yonesaki, T., Zhang, H., et al. (2013). Structure-function studies of Escherichia coli RnlA reveal a novel toxin structure involved in bacteriophage resistance. Mol Microbiol 90, 956–965. doi: 10.1111/mmi.12409
Wen, Y., Behiels, E., Felix, J., Elegheert, J., Vergauwen, B., Devreese, B., et al. (2014). The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence. Nucleic Acids Res 42, 10134–10147. doi: 10.1093/nar/gku665
Wendrich, T. M., Blaha, G., Wilson, D. N., Marahiel, M. A., and Nierhaus, K. H. (2002). Dissection of the mechanism for the stringent factor RelA. Mol Cell 10, 779–788.
Whisstock, J. C., and Lesk, A. M. (1999). SH3 domains in prokaryotes. Trends Biochem Sci 24, 132–133. doi: 10.1016/s0968-0004(99)01366-3
Wilcox, B., Osterman, I., Serebryakova, M., Lukyanov, D., Komarova, E., Gollan, B., et al. (2018). Escherichia coli ItaT is a type II toxin that inhibits translation by acetylating isoleucyl-tRNAIle. Nucleic Acids Res 46, 7873–7885. doi: 10.1093/nar/gky560
Winther, K., Tree, J. J., Tollervey, D., and Gerdes, K. (2016). VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res 44, 9860–9871. doi: 10.1093/nar/gkw781
Winther, K. S., Brodersen, D. E., Brown, A. K., and Gerdes, K. (2013). VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat Commun 4, 2796. doi: 10.1038/ncomms3796
Winther, K. S., and Gerdes, K. (2011). Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A 108, 7403–7407. doi: 10.1073/pnas.1019587108
Wozniak, R. A., and Waldor, M. K. (2009). A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet 5:e1000439. doi: 10.1371/journal.pgen.1000439
Xu, K., Dedic, E., and Brodersen, D. E. (2016). Structural analysis of the active site architecture of the VapC toxin from Shigella flexneri. Proteins 84, 892–899. doi: 10.1002/prot.25002
Yamaguchi, Y., Nariya, H., Park, J. H., and Inouye, M. (2012). Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat Commun 3, 607. doi: 10.1038/ncomms1621
Yamaguchi, Y., Park, J. H., and Inouye, M. (2009). MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem 284, 28746–28753. doi: 10.1074/jbc.M109.032904
Yamanishi, H., and Yonesaki, T. (2005). RNA cleavage linked with ribosomal action. Genetics 171, 419–425. doi: 10.1534/genetics.105.042515
Yao, J., Guo, Y., Zeng, Z., Liu, X., Shi, F., and Wang, X. (2015). Identification and characterization of a HEPN-MNT family type II toxin-antitoxin in Shewanella oneidensis. Microb Biotechnol 8, 961–973. doi: 10.1111/1751-7915.12294
Yuan, J., Sterckx, Y., Mitchenall, L. A., Maxwell, A., Loris, R., and Waldor, M. K. (2010). Vibrio cholerae ParE2 poisons DNA gyrase via a mechanism distinct from other gyrase inhibitors. J Biol Chem 285, 40397–40408. doi: 10.1074/jbc.M110.138776
Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H., et al. (2001). Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896. doi: 10.1126/science.1060089
Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M., and Aravind, L. (2012). Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7, 18. doi: 10.1186/1745-6150-7-18
Zhang, J., Zhang, Y., Zhu, L., Suzuki, M., and Inouye, M. (2004). Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem 279, 20678–20684. doi: 10.1074/jbc.M314284200
Zhang, Y., Yamaguchi, Y., and Inouye, M. (2009). Characterization of YafO, an Escherichia coli toxin. J Biol Chem 284, 25522–25531. doi: 10.1074/jbc.M109.036624
Zhang, Y., Zhu, L., Zhang, J., and Inouye, M. (2005). Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem 280, 26080–26088. doi: 10.1074/jbc.M502050200
Zhu, L., Inoue, K., Yoshizumi, S., Kobayashi, H., Zhang, Y., Ouyang, M., et al. (2009). Staphylococcus aureus MazF specifically cleaves a pentad sequence. J Bacteriol 191, 3248–3255. doi: 10.1128/JB.01815-08
Zhu, L., Phadtare, S., Nariya, H., Ouyang, M., Husson, R. N., and Inouye, M. (2008). The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol 69, 559–569. doi: 10.1111/j.1365-2958.2008.06284.x
Zhu, L., Zhang, Y., Teh, J. S., Zhang, J., Connell, N., Rubin, H., et al. (2006). Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem 281, 18638–18643. doi: 10.1074/jbc.M512693200
Zorzini, V., Buts, L., Sleutel, M., Garcia-Pino, A., Talavera, A., Haesaerts, S., et al. (2014). Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics. Nucleic Acids Res 42, 6709–6725. doi: 10.1093/nar/gku266
Keywords: toxins, translation, persistence, programmed cell death, mobile genetic elements
Citation: Jurėnas D and Van Melderen L (2020) The Variety in the Common Theme of Translation Inhibition by Type II Toxin–Antitoxin Systems. Front. Genet. 11:262. doi: 10.3389/fgene.2020.00262
Received: 15 October 2019; Accepted: 05 March 2020;
Published: 17 April 2020.
Edited by:
Michael Ibba, The Ohio State University, United StatesReviewed by:
Christine M. Dunham, Emory University, United StatesCopyright © 2020 Jurėnas and Van Melderen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laurence Van Melderen, bHZtZWxkZXJAdWxiLmFjLmJl; bGF1cmVuY2UudmFuLm1lbGRlcmVuQHVsYi5hYy5iZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.