
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 04 March 2020
Sec. Behavioral and Psychiatric Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00149
This article is part of the Research Topic Genetic Mechanisms of Biomarkers in Schizophrenia, Bipolar Disorder and Depression View all 18 articles
Background: Schizophrenia risk genes are widely investigated, but a systemic analysis of miRNAs contributing to schizophrenia is lacking.
Methods: Schizophrenia-associated genetic loci profiles were derived from a genome-wide association study (GWAS) from the Schizophrenia Working Group of the Psychiatric Genomics Consortium (PGC) dataset. Experimentally confirmed relationships between miRNAs and their target genes were retrieved from a miRTarBase. A competitive gene set association analysis for miRNA-target regulations was conducted by the Multi-marker Analysis of GenoMic Annotation (MAGMA) and further validated by literature-based functional pathway analysis using Pathway Studio. The association between the targets of three miRNAs and schizophrenia was further validated using a GWAS of antipsychotic treatment responses.
Results: Three novel schizophrenia-risk miRNAs, namely, miR-208b-3p, miR-208a-3p, and miR-494-5p, and their targetomes converged on calcium voltage-gated channel subunit alpha1 C (CACNA1C) and B-cell lymphoma 2 (BCL2), and these are well-known contributors to schizophrenia. Both miR-208a-3p and miR-208b-3p reduced the expression of the RNA-binding protein Quaking (QKI), whose suppression commonly contributes to demyelination of the neurons and to ischemia/reperfusion injury. On the other hand, both QKI and hsa-miR-494-5p were involved in gliomagenesis.
Conclusion: Presented results point at an orchestrating role of miRNAs in the pathophysiology of schizophrenia. The sharing of regulatory networks between schizophrenia and other pathologies may explain higher cardiovascular mortality and lower odds of glioma previously reported in psychiatric patients.
Schizophrenia, a common psychiatric condition characterized by abnormal social behavior and failure to understand reality, affects up to 1% of the human population and causes substantial morbidity and mortality (Barnett, 2018). It is a complex disorder with an estimated heritability of around 80% and an unclear mode of genetic transmission (Hilker et al., 2018). There are many risk genes for schizophrenia, and there is a very small risk attributed to each one (Winchester et al., 2014; Li et al., 2016). In the largest multi-stage genome-wide schizophrenia association study to date, with 34,241 cases, 45,604 controls, and 1,235 affected parent–offspring trios, a total of 128 independent associations spanning 108 conservative loci were identified (Ripke et al., 2014), many of which were consistent with leading pathophysiological hypotheses of schizophrenia development.
It is worth noting that schizophrenia rarely results from the disruption of an individual gene, or even a contiguous chromosomal region. On the contrary, this condition is commonly attributed to the concerted and stable dysregulation of a complex genetic network or a set of networks (Gilman et al., 2012). Because of that, the dysregulation of master regulatory molecules, such as miRNAs, is expected to play a crucial role in the pathogenesis of schizophrenia. Indeed, altered levels of miRNA in the brain, in peripheral blood mononuclear cells, and in serum are found in patients with schizophrenia (Xu et al., 2010; Wang et al., 2014). Consequently, miRNAs that systemically regulate the genes contributing to the risk of schizophrenia may be of particular importance to its pathophysiology.
In this study, we investigated the miRNA-target gene set associated with schizophrenia with a goal of pinpointing potential master miRNA regulators of the gene networks associated with this disorder. To do that, we selected all experimentally confirmed miRNA–target interactions (MTIs) previously collected in a manually curated miRTarBase (Huang et al., 2020), and we then linked them to a schizophrenia-related tissue context through performing a MAGMA analysis (de Leeuw et al., 2015) of confirmed genes rather than miRNAs itself. The finding was validated by PPI network building and an analysis of secondary GWAS datasets concerning differential antipsychotic treatment responses. Our study prioritizes three miRNAs, miR-208a, miR-208b, and miR-494, as potential high-level regulators of schizophrenia phenotypes.
In order to get the most reliable connection, only miRNA–target pairs supported by strong experimental evidence (reporter assay or Western blot) were retrieved from miRTarBase 7.0. (http://mirtarbase.mbc.nctu.edu.tw/php/download.php) (Chou et al., 2017).
A Multi-marker Analysis of GenoMic Annotation (MAGMA) based on a multiple linear principal components regression model was previously designed to analyze the gene set association involved in genome-wide association studies (GWAS) data (de Leeuw et al., 2015). For each miRNA, its target genes were treated as a gene set, and then the competitive MAGMA-based gene-set analysis was utilized to test the association of each gene set using the summary statistics from the PGC2 GWAS (Ripke et al., 2014). The European samples from the 1,000 Genomes data (http://www.1000genomes.org) were used as reference data sets for the summary statistics gene analysis. Potentially confounding effects of gene size and gene density were treated as covariates in a generalized regression model. Multiple comparisons were corrected by a threshold of the false discovery rate (FDR) < 0.05. Then, significantly associated miRNA target sets were validated using the summary result of a GWAS of antipsychotic treatment responses in 2,413 schizophrenia patients (Yu et al., 2018). East Asian samples from the 1,000 Genomes data (http://www.1000genomes.org) were used as reference data sets for the summary statistics gene analysis.
The literature-based pathway analysis has been conducted using Pathway Studio (www.pathwaystudio.com), which allowed us to explore potential functional connections of miRNAs, their targets and schizophrenia by providing high-quality coverage of these connections with evidence extracted from full-text scientific reports.
A total of 8,496 unique miRNA–target pairs were retrieved from miRTarBase, involving 740 miRNAs and 2,853 target genes (Supplementary Table S1). After exclusion of all miRNAs with only one target gene each, a total of 539 miRNAs with two or more targets each were subjected to a gene set association analysis. For each miRNA, its target genes formed a gene set (N = 539). Taken together, all gene sets were comprised of 2,726 unique genes defined in PGC2 genotype data (Supplementary Table S2). The statistics describing miRNA–target gene regulations are shown in Table 1.
Competitive gene set association analysis conducted by MAGMA identified three miRNAs as significantly associated with schizophrenia, namely, hsa-miR-208b-3p (miR-208b) (p = 2.04E-10, FDR = 1.10E-7), hsa-miR-494-5p (miR-494) (p = 1.72E-4, FDR = 0.031), and hsa-miR-208a-3p (miR-208a) (p = 1.75E-4, FDR = 0.031). An analysis of expression for these miRNAs was performed in a comprehensive miRmine dataset (Panwar et al., 2017) that was comprised of 304 high-quality microRNA sequencing experiments. Two of the three miRNAs studied, namely, miR-208b-3p and miR-494-5p, were expressed in various brain tissues at substantial levels. Notably, the expression pattern of miR-208b-3p was restricted to brain and plasma, while miRNA miR-208a-3p was specific to serum, plasma, and placenta. While neither heart nor muscle has been covered by the miRmine dataset, a body of work has demonstrated the importance of miR-208a-3p and miR-208b-3p as myoMiRs, expressed in heart tissues along with their myosin heavy chain encoding genes MYH6 and MYH7, respectively (Siddique et al., 2016).
Table 2 presents a list of target genes regulated by these three miRNAs. Interestingly, all three highlighted miRNAs directly target CDKN1A, pointing to its possible function as a hub gene in the pathology of schizophrenia.
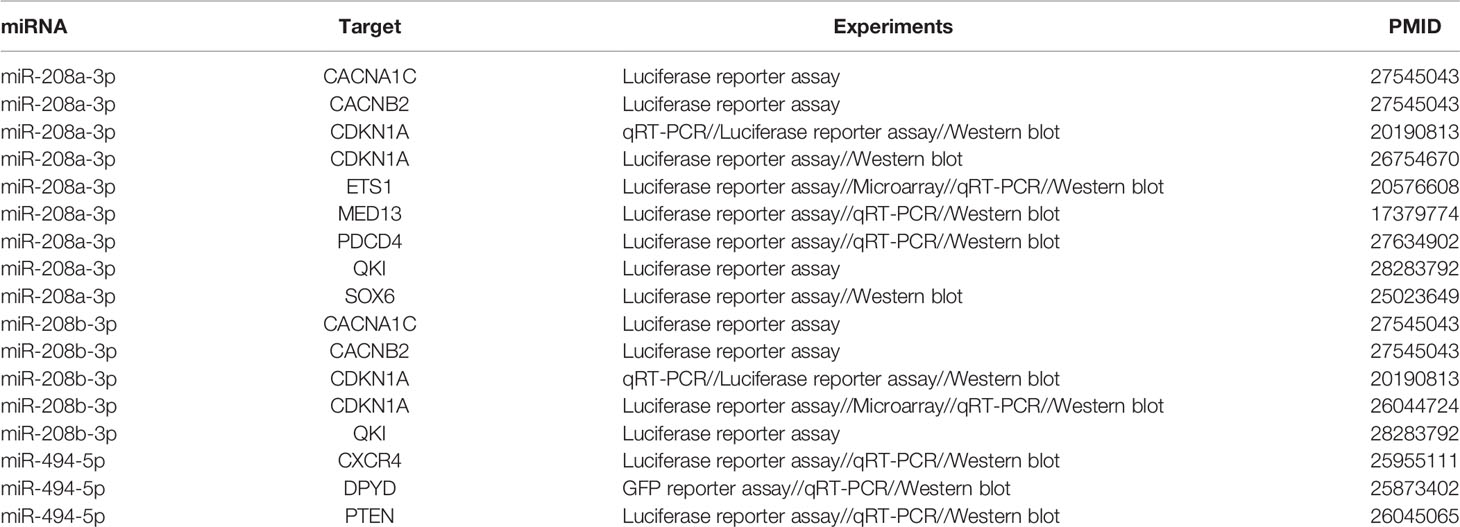
Table 2 Experimentally confirmed target genes of the three miRNAs contributing to the risk of schizophrenia.
A Pathway Studio (www.pathwaystudio.com) analysis provided evidence for multiple functional pathways that link miR-208b-3p (Figure 1A), miR-208a-3p (Figure 1B), and miR-494-5p (Figure 1C) to schizophrenia. The relation types and the reference information are presented in Supplementary Table S3. Notably, all three schizophrenia-implicated networks regulated by miRNA included BCL2, a well-known regulator of apoptosis and mitochondrial dynamics, and a calcium voltage-gated channel subunit alpha1 C (CACNA1C), one of the L-type calcium channels (LTCCs) defining the calcium influx into cells, and these are critical for normal brain development and plasticity (Figure 1).
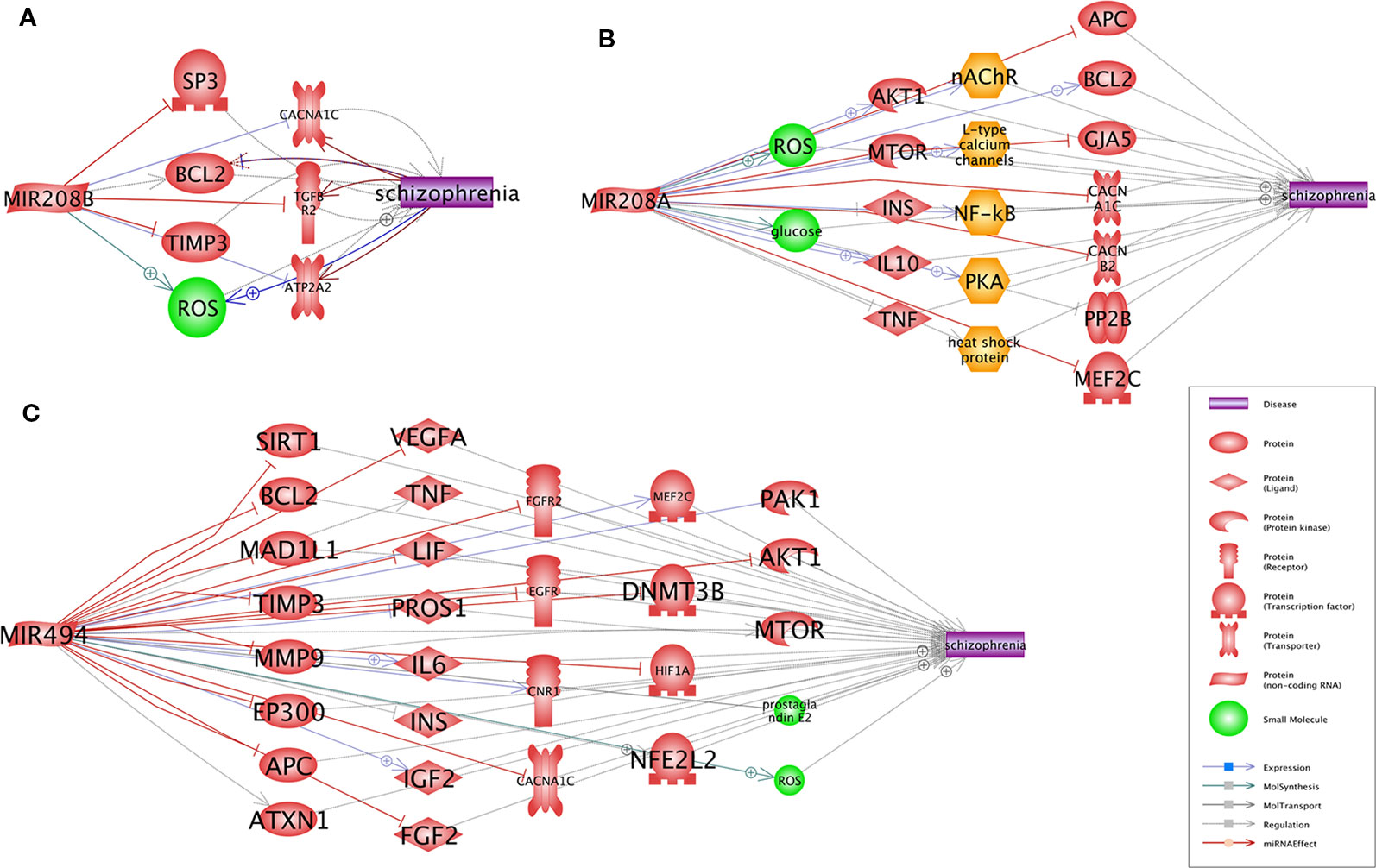
Figure 1 Pathways that link each of three miRNAs, miR-208b (A), miR-208a (B), and miR-494 (C), to schizophrenia. Presented pathways were generated using Pathway Studio (www.pathwaystudio.com) based on known relations mined from existing literature. Each relation has been supported by one or more references summarized in Supplementary Table 3.
A protein–protein interactions (PPIs) analysis was conducted to study the relationship between the target genes of the three miRNAs (miR-208a, miR-208b, and miR-494), as shown in Figure 2. The relation data shown in Figure 2 were acquired from STRING v10.0 (Szklarczyk et al., 2017) and plotted using Cytoscape (Shannon et al., 2003).
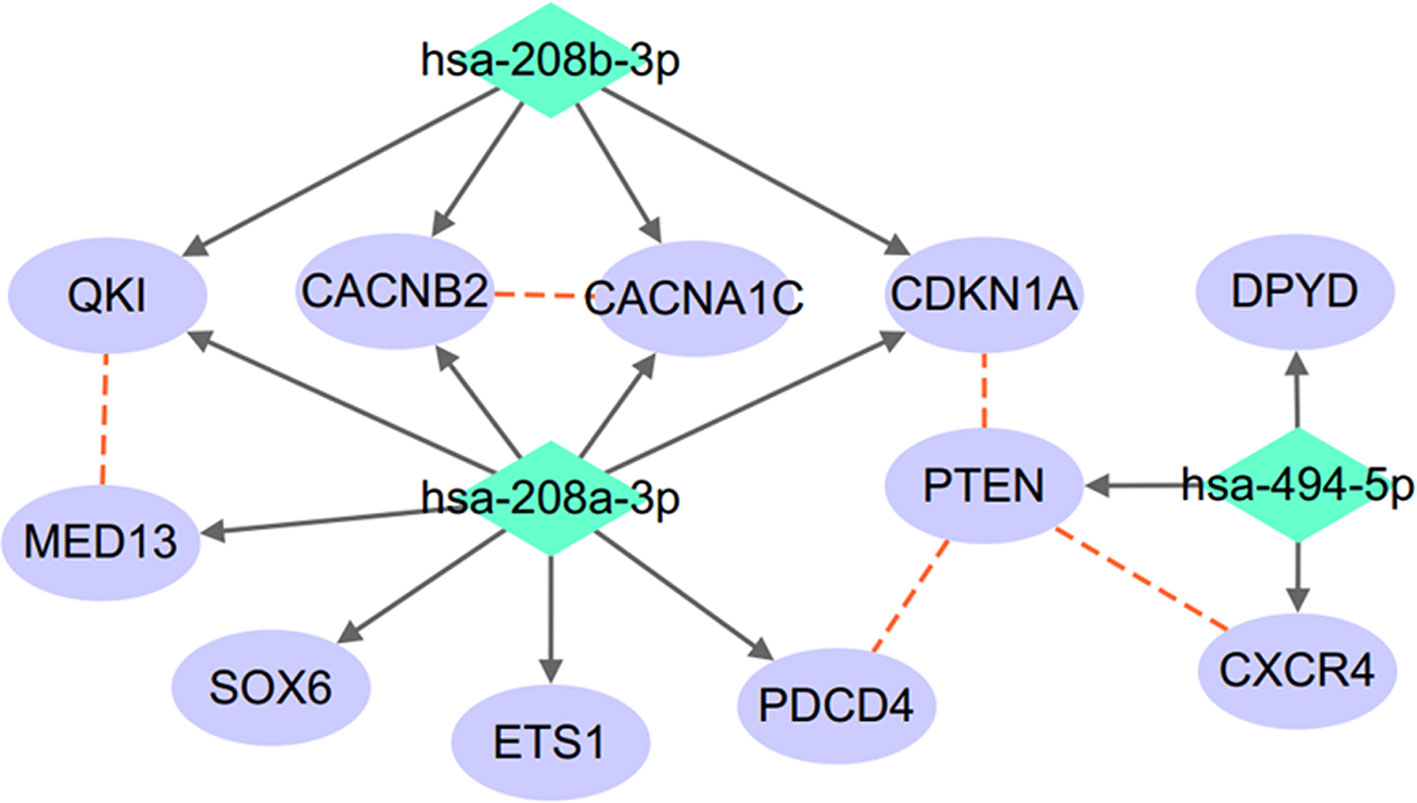
Figure 2 miRNA–target regulatory network connecting miR-208a-3p, miR-208b-3p, and miR-494-5p. Red dashed lines denote protein–protein interactions; solid arrowed lines denote miRNA–target bindings.
As shown in Figure 2, the three miRNAs connect to each other through a complex but relatively compact network through multiple common target-binding proteins. Moreover, many protein components of this network are known to interact with each other, suggesting that this network is not random.
To validate the association of miR-208a-3p, miR-208b-3p, and miR-494-5p and their target sets with schizophrenia, a GWAS of antipsychotic treatment response in 2,413 psychiatric patients (Yu et al., 2018) was mined to detecting enrichment. As shown in Table 3, the gene set regulated by miR-494-5p was associated with the drug treatment response of patients with schizophrenia.
Accumulating evidence suggests that post-transcriptional gene expression regulators, known as microRNAs (miRNAs), play a crucial role in many physiological and pathophysiological processes in human brain. In particular, various areas of the brain and the serum of individuals with schizophrenia were studied for the cellular and extracellular content of miRNA molecules as well as the widespread alterations of their levels reported (Moreau et al., 2011; Santarelli et al., 2011; Banigan et al., 2013). In their typical biomarker discovery design, these and other studies have not aimed at differentiating causal or consequential relationships between the change in the levels of certain miRNA and the development of psychiatric conditions. Nevertheless, the miRNAs encoded by these genes, for example, miR-137 (Kuswanto et al., 2015), were found to harbor the single nucleotide polymorphisms (SNPs) for an increased risk of schizophrenia.
This study highlights three additional miRNAs, hsa-miR-208b-3p, hsa-miR-494-5p, and hsa-miR-208a-3p, as potential contributors to schizophrenia and as the master regulators for the genes previously implicated in this disorder. An analysis of their gene expression showed that these miRNA species were expressed in the brain tissue, the plasma/serum/placenta, or in a combination of these at relatively high levels. Current evidence suggests that the blood–brain barrier does not block the passage of miRNAs between CSF and blood, even if brain-derived miRNAs are somewhat more diluted in blood (Stoicea et al., 2016). While the data on the penetration of miRNA from peripheral tissues to the brain are limited, one can assume that this transfer is highly possible, especially during embryonic development when brain tissue and its compartmentalization is not yet fully formed. Moreover, recent experiments performed in two different rodent models has shown that, in certain conditions, such as during hypoxia, miRNAs actively contribute to an increase in the penetrability of the blood–brain barrier through the inhibition of genes encoding tight junction proteins (Ma et al., 2017; Burek et al., 2019). The role of prenatal and perinatal factors contributing to the risk of schizophrenia was well documented (Kelly and Murray, 2000; Davis et al., 2016). Thereby, one may surmise that the molecular underpinning of this connection may be dependent on plasma miRNAs being carried to the brain in the course of hypoxia or other types of fetal stress, and it may also possibly be dependent on the pathophysiological pairing between miRNAs and mRNAs in non-target tissue.
The accurate identification of miRNA targets remains a formidable challenge. As the output generated by commonly used microRNA–mRNA interaction-predicting software fails to pinpoint experimentally confirmed microRNA-binding regions correctly (Willgoose, 1981; Plotnikova and Skoblov, 2018), we had resorted to limiting our study by investigating only experimentally validated microRNA–mRNA interactions with a subsequent anchoring of them to schizophrenia-related targets by leveraging the data generated over the course of the largest schizophrenia-dissecting GWAS performed to date. Further support for our findings was obtained by the Pathway Studio guided analysis, which allowed us to perform a systems analysis of the molecular pathways engaged by these miRNAs.
Two functional molecules, BCL2 and CACNA1C, were commonly shared between all three miRNA-coordinated “Shortest Path” networks. Notably, both of these molecules were implicated in schizophrenia in numerous previous studies. CACNA1C, which encodes for the Cav1.2 α1 subunit of L-type calcium channels (LTCCs), is one of the best-supported risk loci for schizophrenia and bipolar disorder since it harbors variants with consequences on neural processing and connectivity (Gurung and Prata, 2015; Kabir et al., 2017). For BCL2, the connections to schizophrenia are at the level of cellular processes rather than genetic ones. In the astroglia and the neurons, BCL2 regulates autophagy, which maintains the balance between the synthesis, degradation, and recycling of mitochondria and other cellular components (Aouacheria et al., 2017) as well as prevents apoptosis (Almeida, 2013). The networks we built for schizophrenia risk miRNAs imply the disease-associated deregulation of BCL2/BAX and the resultant enhancement in cell susceptibility to apoptosis, which possibly involves an increase in the production of reactive oxygen species (Wu et al., 2013).
If increases in respective miRNA signals are defined genetically, their observed effects should be systemic rather than brain specific. In this light, it is important to note that the primary fibroblasts collected form antipsychotic-naïve patients with first-episode schizophrenia have greater apoptotic susceptibility, higher caspase-3 activity, and lower BCL2 expression than healthy controls (Gassó et al., 2014). Increased expression of hsa-miR-208b-3p, hsa-miR-494-5p, and hsa-miR-208a-3p may augment susceptibility to schizophrenia by simultaneously conferring susceptibility to apoptosis and altering neural processing and connectivity through the suppression of BCL2 and CACNA1C, respectively.
Importantly, all three schizophrenia-contributing miRNA molecules are far from novel. Cardiomyocyte molecules miR-208a-3p and miR-208b-3p belong to the miR-208 family, which participates in ventricular remodeling (Liu et al., 2016) by promoting myocardial fibrosis (Shyu et al., 2015) and apoptosis of cardiomyocytes (Shannon et al., 2003; Luo et al., 2004; Moreau et al., 2011; Tsai et al., 2013; Huang et al., 2016). Both miR-208a-3p and miR-208b-3p reduce the expression of the RNA-binding protein Quaking, encoded by gene QKI, which inhibits the apoptosis of cardiomyocytes under ischemia/reperfusion condition (de Bruin et al., 2017; Wang F. et al., 2017). Peculiarly, the dysmyelinating mouse mutant shaking (shk), a model of schizophrenia, is a quaking (qk) allele consisting of a 105-nucleotide insertion in the qk regulatory region that decreases the transcription of qk (Chaverneff et al., 2015). Downregulation of the QKI gene was also noted in the brains of schizophrenic patients (Haroutunian et al., 2006). It was hypothesized that deregulation of QKI underlines the defects of oligodendrocyte differentiation and in myelination detected in schizophrenia (Rosenbluth and Bobrowski-Khoury, 2013) as well as in—as described in a separate study—at least some cases of intellectual disability (Darbelli and Richard, 2016). Moreover, in yet another model tissue, auditory nerves, function of both QKI and its protein product substantially decreases in response to noise exposure, leading to demyelination and hearing deficiency (Panganiban et al., 2018). When QKI-regulating molecules of the miR-208 family are overexpressed, their effects are similar to the decrease in the transcription of QKI and should promote the development of the myelination defects. Remarkably, at clinically relevant concentrations of Haloperidol, the expression levels of QKI-encoding mRNA may be restored (Jiang et al., 2009), which would, in turn, alleviate demyelination-related symptoms.
There is no doubt that miR-208-regulated QKI defines the phenotypic plasticity of the vascular smooth muscle cells (van der Veer et al., 2013; Cochrane et al., 2017). These functional pieces of evidence of the involvement of QKI into the development of cardiovascular conditions are also supported by the GWAS, which pointed at QKI as a contributor to coronary heart disease (Dehghan et al., 2016). Patients with schizophrenia are known to have higher mortality rates for all major cardiovascular diagnoses (Wu et al., 2015; Westman et al., 2018). It is tempting to speculate that the connection between these two major disabilities may be, at least in part, explained by the sharing of regulatory networks, particularly ones connecting miRNAs of the miR-208 family and QKI.
Another pathophysiological process characterized by alterations in QKI is the development of gliomas. This gene serves as a tumor suppressor that promotes endolysosome-mediated degradation and suppresses the display of receptors essential for maintaining the self-renewal of neural stem cells outside their niche (Shingu et al., 2017). Consequently, the QKI gene tends to be eliminated in gliomas, either through a complete deletion or through a disruption by translocation (Bandopadhayay et al., 2016). While the roles for upstream regulators of QKI, hsa-miR-208b-3p and hsa-miR-208a-3p, in glioma have not yet been described, another miRNA that affects schizophrenia risk, hsa-miR-494-5p, is a definite glioma suppressor (Li et al., 2015; Zhang et al., 2015; Xu et al., 2018). Importantly, in the case of the latter miRNA, protection against the development of the tumors comes at a cost of elevated susceptibility to neurotoxicity, after exposure to ischemia/reperfusion for example. Notably, knockdown of hsa-miR-494-5p reverses the neurotoxic phenotype in multiple models (Song et al., 2017; Deng et al., 2019; Zhao et al., 2019a; Zhao et al., 2019b). Hsa-miR-494-5p-dependent antagonistic relationships between gliomagenesis and neurotoxicity are intriguing, as they support previously noted decrease in odds of the development of brain tumors in patients with schizophrenia (Grinshpoon et al., 2005; Levav et al., 2007; Wang Y. et al., 2017).
In summary, the presented results point at an orchestrating role of miRNAs in the pathophysiology of schizophrenia. Cellular effects of risk-associated miRNAs, namely, hsa-miR-208b-3p, hsa-miR-494-5p, and hsa-miR-208a-3p, align with the primary etiological hypotheses of schizophrenia and suggest that the three molecules, as well as their target genes, should be investigated for possible pharmacological interventions. The sharing of regulatory networks between schizophrenia and other pathologies may explain higher cardiovascular mortality and lower odds of glioma previously reported in psychiatric patients. Molecular tools for manipulating miRNA activity, including miRNA sponges, are already being developed for cancers (Jung et al., 2015; Fang et al., 2017) and for cardiovascular disease (Bernardo et al., 2018). There is a hope that similarly designed therapeutic interventions may find their utility in the treatment of schizophrenia and other life-long psychiatric illnesses.
The datasets generated for this study can be found in the Supplementary Material.
HC, DL, and FZ developed the study design. FZ, WY, HY, ZZ, HC, and AB analyzed the data. FZ, HC, and AB drafted and then edited the original paper. All authors read and approved the final manuscript.
The study was supported the National Natural Science Foundation of China (No. 81471364) and Primary Research & Development Plan of Jiangsu Province (BE2016630).
Author HC was employed by the company Elsevier Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00149/full#supplementary-material
Supplementary Table S1 | miRNA-target gene pairs retrieved from MiRTarBase.
Supplementary Table S2 | Enrichment of miRNA Target Set in PGC2.
Supplementary Table S3 | References supporting the literature-based pathway linking SCZ and the three miRNAs.
Almeida, A. (2013). Genetic determinants of neuronal vulnerability to apoptosis. Cell Mol. Life Sci. 70 (1), 71–88. doi: 10.1007/s00018-012-1029-y
Aouacheria, A., Baghdiguian, S., Lamb, H. M., Huska, J. D., Pineda, F. J., Hardwick, J. M. (2017). Connecting mitochondrial dynamics and life-or-death events via Bcl-2 family proteins. Neurochem. Int. 109, 141–161. doi: 10.1016/j.neuint.2017.04.009
Bandopadhayay, P., Ramkissoon, L. A., Jain, P., Bergthold, G., Wala, J., Zeid, R., et al. (2016). MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 48 (3), 273–282. doi: 10.1038/ng.3500
Banigan, M. G., Kao, P. F., Kozubek, J. A., Winslow, A. R., Medina, J., Costa, J., et al. (2013). Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PloS One 8, e48814. doi: 10.1371/journal.pone.0048814
Bernardo, B. C., Gregorevic, P., Ritchie, R. H., McMullen, J. R. (2018). Generation of microRNA-34 sponges and tough decoys for the heart: developments and challenges. Front. Pharmacol. 9, 1090. doi: 10.3389/fphar.2018.01090
Burek, M., König, A., Lang, M., Fiedler, J., Oerter, S., Roewer, N., et al. (2019). Hypoxia-Induced MicroRNA-212/132 alter blood-brain barrier integrity through inhibition of tight junction-associated proteins in human and mouse brain microvascular endothelial cells. Transl. Stroke Res. 10 (6), 672–683. doi: 10.1007/s12975-018-0683-2
Chaverneff, F., Mierzwa, A., Weinstock, M., Ketcham, M., Lang, E. J., Rosenbluth, J. (2015). Dysmyelination with preservation of transverse bands in a long-lived allele of the quaking mouse. J. Comp. Neurol. 523 (2), 197–208. doi: 10.1002/cne.23670
Chou, C. H., Shrestha, S., Yang, C. D., Chang, N. W., Lin, Y. L., Liao, K. W., et al. (2017). miRTarBase update 2018: a resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 46 (4), D296–D302. doi: 10.1093/nar/gkx1067
Cochrane, A., Kelaini, S., Tsifaki, M., Bojdo, J., Vilà-González, M., Drehmer, D., et al. (2017). Quaking is a key regulator of endothelial cell differentiation, neovascularization, and angiogenesis. Stem Cells 35 (4), 952–966. doi: 10.1002/stem.2594
Darbelli, L., Richard, S. (2016). Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip. Rev. RNA May7 (3), 399–412. doi: 10.1002/wrna.1344
Davis, J., Eyre, H., Jacka, F. N., Dodd, S., Dean, O., McEwen, S., et al. (2016). A review of vulnerability and risks for schizophrenia: Beyond the two hit hypothesis. Neurosci. Biobehav. Rev. Jun65, 185–194. doi: 10.1016/j.neubiorev.2016.03.017
de Bruin, R. G., Rabelink, T. J., van Zonneveld, A. J., van der Veer, E. P. (2017). Emerging roles for RNA-binding proteins as effectors and regulators of cardiovascular disease. Eur. Heart J. 38 (18), 1380–1388. doi: 10.1093/eurheartj/ehw567
de Leeuw, C. A., Mooij, J. M., Heskes, T., Posthuma, D. (2015). MAGMA: Generalized Gene-Set Analysis of GWAS Data. PloS Comput. Biol. 11 (4), e1004219. doi: 10.1371/journal.pcbi.1004219
Dehghan, A., Bis, J. C., White, C. C., Smith, A. V., Morrison, A. C., Cupples, L. A., et al. (2016). Genome-Wide Association Study for incident myocardial infarction and coronary heart disease in prospective cohort studies: the CHARGE Consortium. PloS One 11 (3), e0144997. doi: 10.1371/journal.pone.0144997
Deng, C., Zhu, J., Yuan, J., Xiang, Y., Dai, L. (2019). Pramipexole inhibits MPP+-Induced neurotoxicity by miR-494-3p/BDNF. Neurochem. Res. 45 (2), 268–277. doi: 10.1007/s11064-019-02910-5
Fang, Y., Zhou, Y., Zhang, Y., He, L., Xue, C., Cao, Y. (2017). Design of miRNA sponges for MDV-1 as a therapeutic strategy against lymphomas. Oncotarget 9 (3), 3842–3852. doi: 10.18632/oncotarget.23379
Gassó, P., Mas, S., Molina, O., Lafuente, A., Bernardo, M., Parellada, E. (2014). Increased susceptibility to apoptosis in cultured fibroblasts from antipsychotic-naïve first-episode schizophrenia patients. J. Psychiatr. Res. 48 (1), 94–101. doi: 10.1016/j.jpsychires.2013.09.017
Gilman, S. R., Chang, J., Xu, B., Bawa, T. S., Gogos, J. A., Karayiorgou, M., et al. (2012). Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat. Neurosci. 15 (12), 1723–1728. doi: 10.1038/nn.3261
Grinshpoon, A., Barchana, M., Ponizovsky, A., Lipshitz, I., Nahon, D., Tal, O., et al. (2005). Cancer in schizophrenia: is the risk higher or lower? Schizophr. Res. 73 (2-3), 333–341. doi: 10.1016/j.schres.2004.06.016
Gurung, R., Prata, D. P. (2015). What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol. Med. 45 (12), 2461–2480. doi: 10.1017/S0033291715000537
Haroutunian, V., Katsel, P., Dracheva, S., Davis, K. L. (2006). The human homolog of the QKI gene affected in the severe dysmyelination “quaking” mouse phenotype: downregulated in multiple brain regions in schizophrenia. Am. J. Psychiatry 163 (10), 1834–1837. doi: 10.1176/ajp.2006.163.10.1834
Hilker, R., Helenius, D., Fagerlund, B., Skytthe, A., Christensen, K., Werge, T. M., et al. (2018). Heritability of schizophrenia and schizophrenia spectrum based on the Nationwide Danish Twin Register. Biol. Psychiatry 83 (6), 492–498. doi: 10.1016/j.biopsych.2017.08.017
Huang, Y., Yang, Y., He, Y., Huang, C., Meng, X., Li, J. (2016). MicroRNA-208a potentiates angiotensin II-triggered cardiac myoblasts apoptosis via inhibiting nemo-like kinase (NLK). Curr. Pharm. Des. 22, 4868–4875. doi: 10.2174/1381612822666160210143047
Huang, H. Y., Lin, Y. C., Li, J., Huang, K. Y., Shrestha, S., Hong, H. C., et al. (2020). miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 48 (D1), D148–D154. doi: 10.1093/nar/gkz896
Jiang, L., Saetre, P., Jazin, E., Carlström, E. L. (2009). Haloperidol changes mRNA expression of a QKI splice variant in human astrocytoma cells. BMC Pharmacol. 9, 6. doi: 10.1186/1471-2210-9-6
Jung, J., Yeom, C., Choi, Y. S., Kim, S., Lee, E., Park, M. J., et al. (2015). Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget 6 (24), 20370–20387. doi: 10.18632/oncotarget.4827
Kabir, Z. D., Martínez-Rivera, A., Rajadhyaksha, A. M. (2017). From Gene to Behavior: L-Type Calcium Channel Mechanisms Underlying Neuropsychiatric Symptoms. Neurotherapeutics 14 (3), 588–613. doi: 10.1007/s13311-017-0532-0
Kelly, J., Murray, R. M. (2000). What risk factors tell us about the causes of schizophrenia and related psychoses. Curr. Psychiatry Rep. 2 (5), 378–385. doi: 10.1007/s11920-000-0019-1
Kuswanto, C. N., Sum, M. Y., Qiu, A., Sitoh, Y. Y., Liu, J., Sim, K. (2015). The impact of genome wide supported microRNA-137 (MIR137) risk variants on frontal and striatal white matter integrity, neurocognitive functioning, and negative symptoms in schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168B (5), 317–326. doi: 10.1002/ajmg.b.32314
Levav, I., Lipshitz, I., Novikov, I., Pugachova, I., Kohn, R., Barchana, M., et al. (2007). Cancer risk among parents and siblings of patients with schizophrenia. Br. J. Psychiatry 190, 156–161. doi: 10.1192/bjp.bp.106.024943
Li, X. T., Wang, H. Z., Wu, Z. W., Yang, T. Q., Zhao, Z. H., Chen, G. L., et al. (2015). miR-494-3p regulates cellular proliferation, invasion, migration, and apoptosis by PTEN/AKT signaling in human glioblastoma cells. Cell Mol. Neurobiol. 35 (5), 679–687. doi: 10.1007/s10571-015-0163-0
Li, Z., Chen, J., Xu, Y., Yi, Q., Ji, W., Wang, P., et al. (2016). Genome-wide analysis of the role of copy number variation in schizophrenia risk in Chinese. Biol. Psychiatry 80 (4), 331–337. doi: 10.1016/j.biopsych.2015.11.012
Liu, X., Meng, H., Jiang, C., Yang, S., Cui, F., Yang, P. (2016). Differential microRNA expression and regulation in the rat model of post-infarction heart failure. PloS One 11, e0160920. doi: 10.1371/journal.pone.0160920
Luo, C., Xu, H., Li, X. M. (2004). Post-stress changes in BDNF and Bcl-2 immunoreactivities in hippocampal neurons: effect of chronic administration of olanzapine. Brain Res. 1025 (1), 194–202. doi: 10.1016/j.brainres.2004.06.089
Ma, Q., Dasgupta, C., Li, Y., Huang, L., Zhang, L. (2017). MicroRNA-210 Suppresses Junction Proteins and Disrupts Blood-Brain Barrier Integrity in Neonatal Rat Hypoxic-Ischemic Brain Injury. Int. J. Mol. Sci. 18 (7), E1356. doi: 10.3390/ijms18071356
Moreau, M. P., Bruse, S. E., David-Rus, R., Buyske, S., Brzustowicz, L. M. (2011). Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry 69, 188–193. doi: 10.1016/j.biopsych.2010.09.039
Panganiban, C. H., Barth, J. L., Darbelli, L., Xing, Y., Zhang, J., Li, H., et al. (2018). Noise-induced dysregulation of Quaking RNA binding proteins contributes to auditory nerve demyelination and hearing loss. J. Neurosci. 38 (10), 2551–2568. doi: 10.1523/JNEUROSCI.2487-17.2018
Panwar, B., Omenn, G. S., Guan, Y. (2017). miRmine: a database of human miRNA expression profiles. Bioinformatics 33 (10), 1554–1560. doi: 10.1093/bioinformatics/btx019
Plotnikova, O. M., Skoblov, M. Y. (2018). Efficiency of the miRNA- mRNA interaction prediction programs. Mol. Biol. (Mosk) 52 (3), 543–554. doi: 10.1134/S0026893318020103
Ripke, S., Neale, B. M., Corvin, A., Walters, J. T., Farh, K. H., Holmans, P. A., et al. (2014). Biological Insights From 108 Schizophrenia-Associated Genetic Loci. Nature 511 (7510), 421–427. doi: 10.1038/nature13595
Rosenbluth, J., Bobrowski-Khoury, N. (2013). Structural bases for central nervous system malfunction in the quaking mouse: dysmyelination in a potential model of schizophrenia. J. Neurosci. Res. 91 (3), 374–381. doi: 10.1002/jnr.23167
Santarelli, D. M., Beveridge, N. J., Tooney, P. A., Cairns, M. J. (2011). Upregulation of Dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry 69, 180–187. doi: 10.1016/j.biopsych.2010.09.030
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498. doi: 10.1101/gr.1239303
Shingu, T., Ho, A. L., Yuan, L., Zhou, X., Dai, C., Zheng, S., et al. (2017). Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat. Genet. 49 (1), 75–86. doi: 10.1038/ng.3711
Shyu, K. G., Wang, B. W., Cheng, W. P., Lo, H. M. (2015). MicroRNA208a increases myocardial endoglin expression and myocardial fibrosis in acute myocardial infarction. Can. J. Cardiol. 31, 679–690. doi: 10.1016/j.cjca.2014.12.026
Siddique, B. S., Kinoshita, S., Wongkarangkana, C., Asakawa, S., Watabe, S. (2016). Evolution and distribution of Teleost myomiRNAs: functionally diversified myomiRs in Teleosts. Mar. Biotechnol. (NY) 18 (3), 436–447. doi: 10.1007/s10126-016-9705-9
Song, S., Lin, F., Zhu, P., Wu, C., Zhao, S., Han, Q., et al. (2017). Extract of Spatholobus suberctus Dunn ameliorates ischemia-induced injury by targeting miR-494. PloS One 12 (9), e0184348. doi: 10.1371/journal.pone.0184348
Stoicea, N., Du, A., Lakis, D. C., Tipton, C., Arias-Morales, C. E., Bergese, S. D. (2016). The MiRNA Journey from Theory to Practice as a CNS Biomarker. Front. Genet. 7, 11. doi: 10.3389/fgene.2016.00011
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45 (Database issue), D362–D3D8. doi: 10.1093/nar/gkw937
Tsai, M. C., Liou, C. W., Lin, T. K., Lin, I. M., Huang, T. L. (2013). Bcl-2 associated with positive symptoms of schizophrenic patients in an acute phase. Psychiatry Res. 210 (3), 735–738. doi: 10.1016/j.psychres.2013.08.032
van der Veer, E. P., de Bruin, R. G., Kraaijeveld, A. O., de Vries, M. R., Bot, I., Pera, T., et al. (2013). Quaking, an RNA-binding protein, is a critical regulator of vascular smooth muscle cell phenotype. Circ. Res. 113 (9), 1065–1075. doi: 10.1161/CIRCRESAHA.113.301302
Wang, J., Wang, Y., Yang, J., Huang, Y. (2014). miRNAs as novel biomarkers of schizophrenia. Exp. Ther. Med. 8 (6), 1671–1676. doi: 10.3892/etm.2014.2014
Wang, F., Yuan, Y., Yang, P., Li, X. (2017). Extracellular vesicles-mediated transfer of miR-208a/b exaggerate hypoxia/reoxygenation injury in cardiomyocytes by reducing QKI expression. Mol. Cell Biochem. 431 (1-2), 187–195. doi: 10.1007/s11010-017-2990-4
Wang, Y., Huang, N., Li, H., Liu, S., Chen, X., Yu, S., et al. (2017). Promoting oligodendroglial-oriented differentiation of glioma stem cell: a repurposing of quetiapine for the treatment of malignant glioma. Oncotarget 8 (23), 37511–37524. doi: 10.18632/oncotarget.16400
Westman, J., Eriksson, S. V., Gissler, M., Hällgren, J., Prieto, M. L., Bobo, W. V., et al. (2018). Increased cardiovascular mortality in people with schizophrenia: a 24-year national register study. Epidemiol. Psychiatr. Sci. 27 (5), 519–527. doi: 10.1017/S2045796017000166
Willgoose, C. E. (1981). Environmental health education for survival. Health Educ. 12 (3), 33–35. doi: 10.1080/00970050.1981.10618153
Winchester, C. L., Pratt, J. A., Morris, B. J. (2014). Risk genes for schizophrenia: translational opportunities for drug discovery. Pharmacol. Ther. 143 (1), 34–50. doi: 10.1016/j.pharmthera.2014.02.003
Wu, J. Q., Kosten, T. R., Zhang, X. Y. (2013). Free radicals, antioxidant defense systems, and schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 46, 200–206. doi: 10.1016/j.pnpbp.2013.02.015
Wu, S. I., Chen, S. C., Liu, S. I., Sun, F. J., Juang, J. J., Lee, H. C., et al. (2015). Relative risk of acute myocardial infarction in people with schizophrenia and bipolar disorder: a population-based cohort study. PLoSOne 10 (8), e0134763. doi: 10.1371/journal.pone.0134763
Xu, Y., Li, F., Zhang, B., Zhang, K., Zhang, F., Huang, X., et al. (2010). MiRNAs and target site screening reveals a pre-miRNA-30e variant associated with schizophrenia. Schizophr. Res. 119 (1), 219–227. doi: 10.1016/j.schres.2010.02.1070
Xu, X. H., Zhang, S. J., Hu, Q. B., Song, X. Y., Pan, W. (2018). Effects of microRNA-494 on proliferation, migration, invasion, and apoptosis of medulloblastoma cells by mediating c-myc through the p38 MAPK signaling pathway. J. Cell Biochem 120 (2), 2594–2606. doi: 10.1002/jcb.27559
Yu, H., Yan, H., Wang, L., Li, J., Tan, L., Deng, W., et al. (2018). Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry 5 (4), 327–338. doi: 10.1016/S2215-0366(18)30049-X
Zhang, L., Niu, T., Huang, Y., Zhu, H., Zhong, W., Lin, J., et al. (2015). Compound 331 selectively induces glioma cell death by upregulating miR-494 and downregulating CDC20. Sci. Rep. 5, 12003. doi: 10.1038/srep12003
Zhao, H., Li, G., Zhang, S., Li, F., Wang, R., Tao, Z., et al. (2019b). Inhibition of histone deacetylase 3 by MiR-494 alleviates neuronal loss and improves neurological recovery in experimental stroke. J. Cereb. Blood Flow Metab. 39 (12), 2392–2405. doi: 10.1177/0271678X19875201
Keywords: miRNA, schizophrenia, miR-208b-3p, miR-208a-3p, miR-494-5p, gliomagenesis, quaking, heart disease
Citation: Cao H, Baranova A, Yue W, Yu H, Zhu Z, Zhang F and Liu D (2020) miRNA-Coordinated Schizophrenia Risk Network Cross-Talk With Cardiovascular Repair and Opposed Gliomagenesis. Front. Genet. 11:149. doi: 10.3389/fgene.2020.00149
Received: 15 October 2019; Accepted: 10 February 2020;
Published: 04 March 2020.
Edited by:
Shaohua Hu, Zhejiang University, ChinaReviewed by:
Owen Murray Rennert, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), United StatesCopyright © 2020 Cao, Baranova, Yue, Yu, Zhu, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuquan Zhang, emhhbmdmcUBuam11LmVkdS5jbg==; Dongbai Liu, ZGxpdTAyQGdvdXNpbmZvLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.