
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 13 November 2019
Sec. RNA
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.01112
This article is part of the Research TopicMicroRNA SignalingView all 22 articles
MicroRNAs (miRNAs), belonging to a class of evolutionarily conserved small noncoding RNA of ∼22 nucleotides, are widely involved in skeletal muscle growth and development by regulating gene expression at the post-transcriptional level. While the expression feature and underlying function of miR-216a in mammal skeletal muscle development, especially in cattle, remains to be further elucidated. The aim of this study was to investigate the function and mechanism of miR-216a during bovine primary muscle cells proliferation and differentiation. Herein, we found that the expression level of miR-216a both presented a downward trend during the proliferation and differentiation phases, which suggested that it might have a potential role in the development of bovine skeletal muscle. Functionally, during the cells proliferation phase, overexpression of miR-216a inhibited the expression of proliferation-related genes, reduced the cell proliferation status, and resulted in cells G1 phase arrest. In cells differentiation stages, overexpression of miR-216a suppressed myogenic maker genes mRNA, protein, and myotube formation. Mechanistically, we found that SNIP1 and smad7 were the directly targets of miR-216a in regulating bovine primary muscle cells proliferation and differentiation, respectively. Altogether, these findings suggested that miR-216a functions as a suppressive miRNA in development of bovine primary muscle cells via targeting SNIP1 and smad7.
Skeletal muscle, as an important sports and metabolic organization of animals, accounts for a large proportion of body weight and is directly related to the economic traits such as meat production in cattle. The skeletal muscle development in mammal undergoes two important stages the increase in the number of pre-natal muscle fibers (Russell and Oteruelo, 1981; Hocquette, 2010) and the increase in the volume of muscle fibers after birth (Zhu et al., 2006). Therefore, it is of great value to study the intrinsic mechanism of muscle formation. Skeletal muscle development originates from the embryonic stage, which is derived from the paraxial mesoderm structures in the vertebrate (Christ and Ordahl, 1995). Here, the dorsal somite forms the dermomyotome which can further form the myotome (Tajbakhsh and Buckingham, 2000). Then, the muscle progenitor cells will form and migrate from the myotome. Myotome quickly differentiates into primary myoblasts. Finally, these primary muscle progenitor cells undergo proliferate and differentiate to form skeletal muscle (Buckingham et al., 2003). Orchestral molecules mechanisms are involved in the process of myogenesis, which are mainly regulated by transcription factors likely paired box transcription factors Pax3/Pax7 (Relaix et al., 2006; Lagha et al., 2008; Buckingham and Relaix, 2015), myogenic regulatory factors (MRFs) (Ott et al., 1991), and myocyte enhancer factor 2 family (MEF2) (Edmondson et al., 1994). It’s worth noting that these key transfection factors are also regulated by other multiple regulatory factors, including protein coding gene such as transforming growth factor-beta (TGF-β) (Liu et al., 2001; Gardner et al., 2011) and noncoding RNA likely microRNAs (miRNAs) (Chen et al., 2006).
MiRNAs are a variety of evolutionarily conserved short noncoding RNA. The function and molecular mechanism of miRNAs has been extensively studied in almost all biological processes. The mature sequence of miRNA could complete or partial complementary pair with the 3’ UTR of mRNA, which cause mRNA decay or impede protein translation (Bartel, 2004; Bartel, 2009). MiRNAs, as an integral part of skeletal muscle development, can regulate skeletal muscle proliferation, differentiation, and regeneration by targeting the key factors of the myogenic regulatory network during the different stages of skeletal muscle development process (Chen et al., 2006; Crist et al., 2009; Cheung et al., 2012; Alexander et al., 2013; Diniz and Wang, 2016). Studies have shown that dicer knock-out mice in embryonic skeletal muscle resulted in embryonic lethality, skeletal muscle hypoplasia, myofiber morphogenesis defects, and increased apoptosis (O'Rourke et al., 2007). So far, large numbers of miRNAs have been report to affect skeletal muscle development (Ge and Chen, 2011). Some of them are muscle specific expression miRNAs (myomiRs), including miR-1 (Chen et al., 2006; Sluijter et al., 2010), miR-133 (Chen et al., 2006; Feng et al., 2013), miR-206 (McCarthy, 2008; Dey et al., 2011), etc. miR-1 could promote myogenic cells differentiation by targeting HDAC4 (Chen et al., 2006). miR-133 could promote myoblast proliferation by repressing the expression of SRF (Chen et al., 2006). In the differentiation stage of satellite cells, the expression of miR-206 is upregulated, which can enhance satellite cell differentiation by targeting Pax7 (Dey et al., 2011). Besides that, numerous non-muscle-specific miRNAs (non-myomiRs) also play important role in the regulation of myogenesis (Ju et al., 2015). For example, miR-26a can promote myogenesis by targeting transcription factors Smad1, Smad4, and Ezh2 (enhancer of zeste homologue 2), a negative regulator of myogenesis (Wong and Tellam, 2008; Dey et al., 2012). miR-486 can promote muscle differentiation by activating PI3K/AKT pathway or targets Pax7 directly (Small et al., 2010; Dey et al., 2011). MiR-22 can inhibit proliferation and enhances differentiation of C2C12 myoblasts by targeting TGF-βR1 (Wang et al., 2018b). Although the research of miRNAs is receiving considerable, there are still many miRNAs to be further investigated in skeletal muscle.
miR-216a is a widely expressed miRNA that involved in disease and tumorigenesis by targeting different target genes (Ji et al., 2017; Tao et al., 2017; Wang et al., 2017a). However, little is known about the function of miR-216a in regulation of bovine skeletal muscle development. Therefore, the purpose of current study was to explore the function and mechanism of miR-216a for bovine primary muscle cells proliferation and differentiation. Here, the expression level of miR-216a presented a downward trend both in the proliferation and differentiation phases of bovine skeletal muscle cell. Functional analysis found that miR-216a could inhibit the proliferation and differentiation of bovine skeletal muscle cell by directly targeting SNIP1 and smad7 respectively.
Three months old fetal Qinchuan cattle from Shaanxi Kingbull Livestock Co., Ltd. (Baoji, China) were used in this research. The heart, liver, spleen, lung, kidney, stomach, gut, and leg muscle were collected and kept at -80°C until RNA isolation. Animal care and study protocols were approved by the Animal Care Commission of the College of Veterinary Medicine, Northwest A&F University (permit number: NWAFAC1019).
Bovine skeletal muscle cells are derived from the longissimus muscle or hind limbs of the 3-month-old fetus. The stripped muscle tissue is cut and then digested with collagenase I. The specific separation steps refer to the previous description (Miyake et al., 2012). The growth stage bovine skeletal muscle cells were cultured in DMEM with 20% FBS and 1% penicillin/streptomycin. When inducing skeletal muscle cells differentiation, replace 20% FBS in the medium with 2% horse serum. HEK293T cells (ATCC, USA) were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin. All these cells were cultured at 37°C in a 5% CO2 atmosphere.
The wild-type and mutant-type sequences of smad7 or SNIP1 3’UTR were inserted into the psi-check2 reporter vector, respectively. The mutant type 3’UTR sequences of smad7 or SNIP1 each contained four nucleotide mutations at the miR-216a targeting site. The precursor sequences of miR-216a were cloned into pcDNA3.1 (+) expression vector. The mimic and inhibitor of miR-216a were purchased from Ribobio (Guangzhou, China). The primers used in this study were listed in Supplementary Table 1.
The proliferated bovine primary muscle cells were transfected with miR-216a mimic, inhibitor, and control when the cell density reached 60–70% and collected at 24 h after transfection in 12-well plates. The differentiated bovine primary muscle cells were transfected when the cell density reached 70–80% confluence and 20% FBS medium was changed to 2% horse serum to induce cells differentiation at 24 h after transfection. Cells were collected at three days after induced differentiation. The bovine primary muscle cells used in CCk-8, 5-ethynyl-2′-deoxyuridine (EdU) experiment, and HEK293T cells were cultured in 96-well plates, and they were collected at 24 h after transfection. Thermo transfection reagent was used in this study. The detail procedure of transfection was performed according to the manufacturer’s instructions.
Total RNA of tissues and cells was extracted with TRIzol Reagent (Takara) according to the manufacturer’s instruction. Total RNA was reverse transcribed by using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara). Real-time quantitative PCR (RT-qPCR) was performed with the SYBR Green Kit (Genestar, Beijing, China) on a CFX96 system (Bio-Rad, Hercules, CA, USA) with three biological replicates each time. Relative expression levels of mRNAs and miRNAs were calculated with the 2-∆∆Ct method (Livak and Schmittgen, 2001). U6 and GAPDH gene was used as reference gene for the expression of miRNA and all genes. The primers used in this study were listed in Supplementary Table 1.
Total proteins were extracted from cells by using radio immunoprecipitation assay buffer with 1% PMSF (Solarbio, Beijing, China). The protein concentration was measured by using the BCA Protein Assay Kit (Beyotime, Shanghai, China) and denatured with 5× protein loading buffer at 98°C for 10 min. The specific operation process of WB was described previously (Li et al., 2018). In short, the target proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a methanol-activated polyvinylidene fluoride (PVDF) membrane. The corresponding primary antibody was added to incubate PVDF membrane for overnight at 4°C. At room temperature, the PVDF membrane was re-incubated with the corresponding secondary antibody for 2 h. The protein signal strength was detected using enhanced chemiluminescence reagent. The primary antibodies and secondary antibodies were listed in Supplementary Table 2. The quantified of the protein was performed by the ImageJ program (National Institutes of Health).
For the CCK-8 assay, the bovine primary muscle cells were transfected with the negative control (NC), mimics, and inhibitor when the cell densities were 50–60%. After transfection for 24 h, the culture medium containing 10% CCK-8 reagent was changed and incubated the cells for 2 h. The CCK-8 Reagent Kit (Tiandz, Beijing, China) was performed to measure cell proliferation index at 450 nm by using Microplate Reader (Tecan, Switzerland) and repeated five times for each independent experiment. The Cell-Light EdU DNA Cell Proliferation Kit (Ribobio, Guangzhou, China) was used to measure S phase positive cell according to the manufacturer’s instructions. The EdU positive cells was observed by using fluorescence microscope (AMG EVOS, USA) and each treatment group had three independent replicates.
The muscle cells were seeded into 60 mm diameter petri dish and transfected with the NC, mimics, and inhibitor when the cell densities were 60%. After transfection for 24 h, muscle cells were fixed in PBS containing 70% ethanol, and cell cycle assays were performed with cell cycle staining kit (MultiSciences Biotech Co., Ltd, Hangzhou, China) by using a flow cytometry. The experimental procedure is based on our previous research (Song et al., 2015).
The target genes of miR-216a were predicted with the online software TargetScan (http://www.targetscan.org). The 3’ untranslated region (3’UTR) (wild-type or mutant-type) sequences of the genes SNIP1 and smad7 mRNA was cloned into the psi-Check2 reporter vector, respectively. To confirm their targeting relationship, miR-216a expression plasmid and the psi-Check2 reporter vector (wild or mutant) were co-transfected into HEK293T cells. After transfection for 24 h, the relative luciferase activity was measured by using a Dual-Luciferase® Reporter DLR™Assay System Kit (Promega, USA) according to the manufacturer’s protocols.
Immunofluorescence staining was used to detect the number of MyHC-positive myotubes. After transfection and induction of differentiation for 4 days, the bovine primary muscle cells were fixed with 4% paraformaldehyde in PBS for 20 min. After permeabilized with 0.5% Triton X-100 for 10 min and blocked with 5% BSA at 4°C for 30 min, the cells were incubated with primary antibody-MyHC diluted 1:200 with 5% bovine serum albumin (BSA) in PBS at 4°C overnight. Then the cells were incubated with the corresponding fluorescent secondary antibody [goat anti-mouse immunoglobulin G (IgG) H&L] diluted 1:400 with 5% BSA at 37°C for 2 h. Then cells were stained with 5 mg/ml 4′,6-diamidino-2-phenylindole. The cells were washed three times with PBS for 5 min before each procedure. All images were observed on a fluorescence microscope (DM5000B, Leica, Germany). The antibodies source information was listed in Supplementary Table 2.
Results were presented as mean ± SEM. The data statistical analysis was performed using one-way analysis of variance (ANOVA) or student’s t test. The significance of differences between the groups were considered significant at P <0.05 (* P < 0.05; ** P < 0.01).
To investigate the expression feature of miR-216a in various tissues (heart, liver, spleen, lung, kidney, stomach, gut, and skeletal muscle) during embryonic stage, RT-qPCR analysis data demonstrated that miR-216a was found to be mainly expressed in the heart, but relatively low in skeletal muscle (Figure 1A). During the growth stage (-24, -12, and 0 h) of bovine primary muscle cells, the expression level of miR-216a showed a downward trend (Figure 1B). During the differentiated stage (0, 1, 3, and 5 days) of bovine primary muscle cells, the expression level of miR-216a also showed a downward trend (Figure 1C). Together, the expression characteristics of miR-216a suggested that it might play a negative factor in the proliferation and differentiation of bovine primary muscle cells.
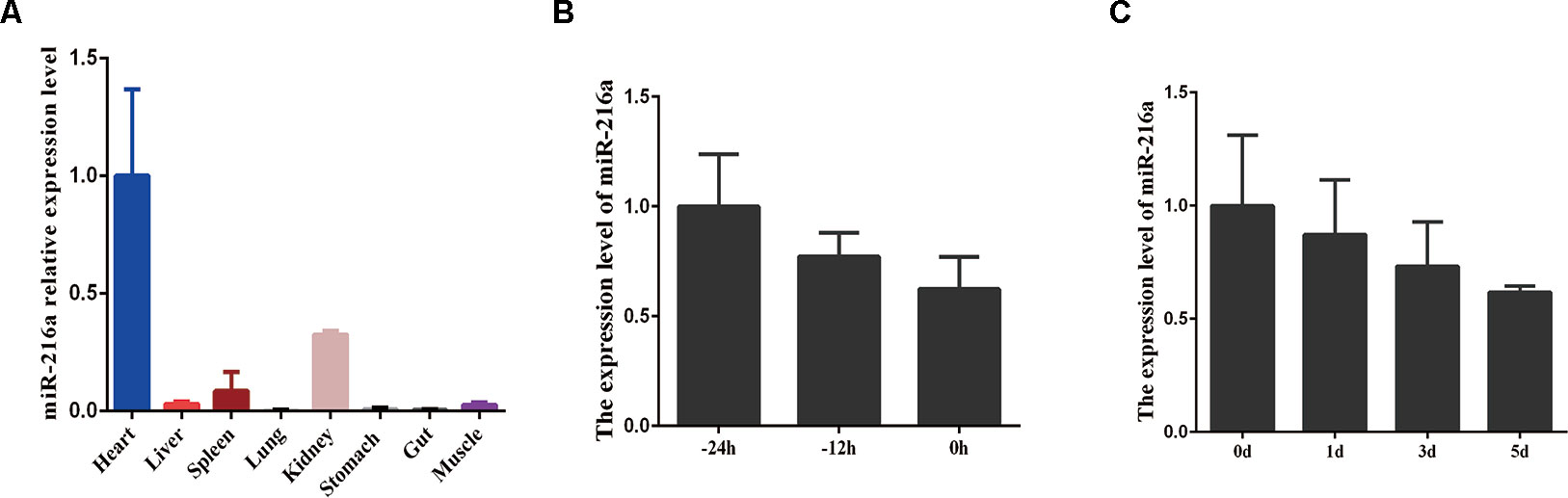
Figure 1 The expression feature of miR-216a in bovine tissues and primary muscle cells. (A) RT-qPCR analysis of relative miR-216a levels in different tissues (heart, liver, spleen, lung, kidneys, stomach, gut, and muscle) of Qinchuan cattle at embryonic stage. (B) RT-qPCR analysis the relative expression level of miR-216a during the growth stage (-24, -12, and 0 h) of bovine primary muscle cells. (C) Real-time quantitative PCR analysis the relative expression level of miR-216a during the differentiation stage (0, 1, 3, and 5 days) of bovine primary muscle cells. Data are presented as the mean ± SEM; n = 3.
To verify the function of miR-216a for bovine primary muscle cells proliferation, miR-216a mimic was transfected into cells to enhance its expression level. After transfection for 24 h, RT-qPCR detection data showed that the relative expression of miR-216a was significantly higher than that of the control group (Figure 2A). At the molecular level, RT-qPCR was used to detect cell proliferation key genes (CyclinD1, PCNA, and CDK2) mRNA expression level. The analysis data demonstrated that the expression of CyclinD1 and CDK2 was significantly lower than that of the control group, but not PCNA (Figure 2B). Western blot analysis showed that the expression level of PCNA and CDK2 protein decreased significantly and the expression level of p53 protein increased significantly (Figures 2C, D). Inhibition of endogenous miR-216a expression was induced by its inhibitor (Figure 2E). Unfortunately, loss of miR-216a did not significantly affect the mRNA levels of the cell proliferation key genes (Figure 2F). But the protein levels of the cell proliferation key genes are significantly affected which consisted with overexpression (Figures 2G, H). At the cellular level, overexpression of miR-216a reduced the number of EdU positive cells, while no significant changes were observed after inhibition of miR-216a (Figure 3A). The results of CCK-8 assay showed that the overexpression of miR-216a resulted in a significant decrease in cell optical density (OD) value, while the cell OD value did not change significantly after interfering with mir-216a (Figure 3B). Detection the cell cycle by flow cytometry showed that overexpression of miR-216a increased the number of cells in G1 phase and decreased the number of cells in S phase, while the inhibition of miR-216a had no significant effect on cell cycle (Figures 3C, D). Together, the above results indicated that miR-216a inhibited the proliferation of bovine primary muscle cells.
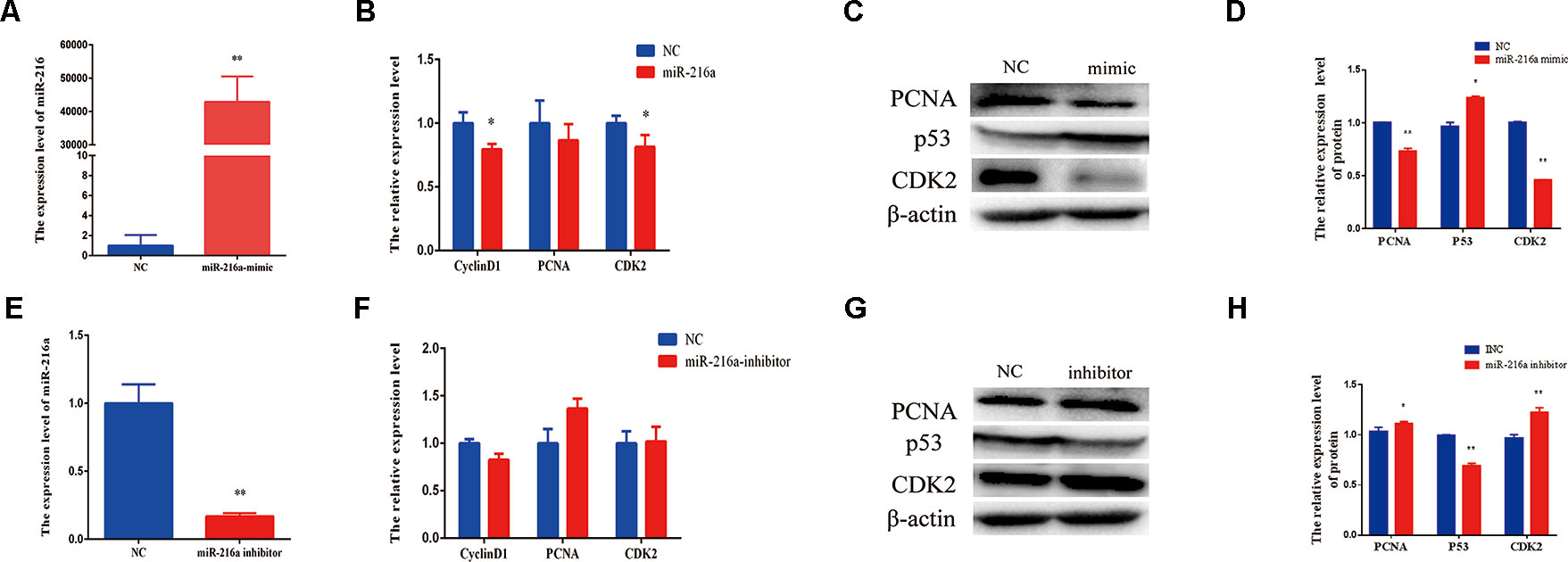
Figure 2 miR-216a regulates bovine primary muscle cells proliferation at the molecular level. (A) The detection of expression efficiency of miR-216a after transfection miR-216a mimic. (B) Gain of miR-216a, cell proliferation key genes (CyclinD1, PCNA, and CDK2) messenger RNA (mRNA) were detected by RT-qPCR. (C) Gain of miR-216a, western blot was used to detect the protein expression level of PCNA, CDK2, and p53. (D) Protein quantitative analysis of PCNA, p53, and CDK2 for (C). (E) The detection of expression level of miR-216a after transfection miR-216a inhibitor. (F) The mRNA expression of cell proliferation genes CyclinD1, PCNA, and CDK2 were detected by real-time quantitative PCR after loss of miR-216a. (G) Western blot was used to detect the protein expression level of PCNA, CDK2, and p53 after loss of miR-216a. (H) Protein quantitative analysis of PCNA, p53, and CDK2 for (G). Data are presented as the mean ± SEM; n = 3; *P < 0.05 and **P < 0.01.
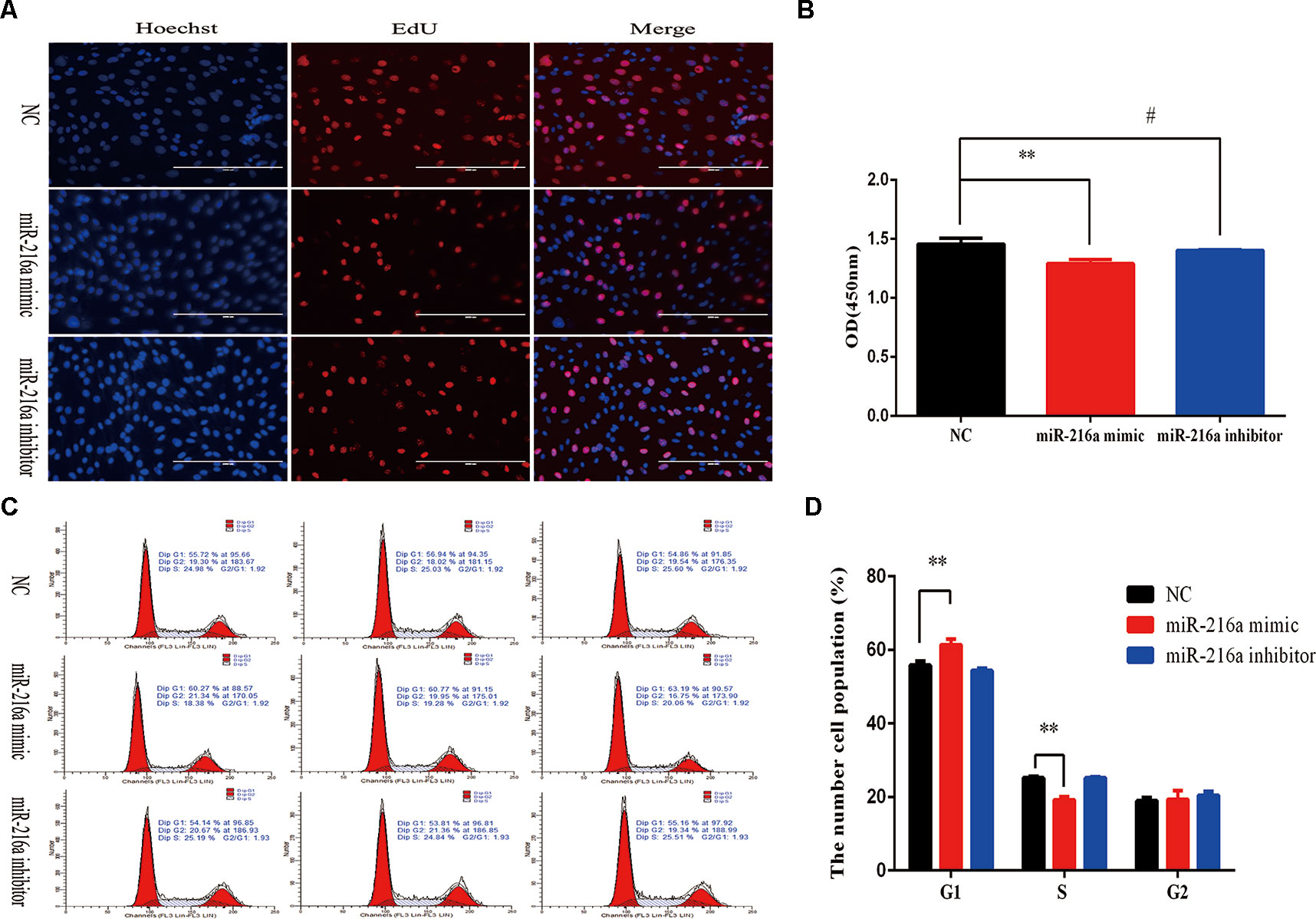
Figure 3 miR-216a regulates bovine primary muscle cells proliferation at the cellular level.(A) Bovine primary muscle cells in the S-phase were stained with 5-ethynyl-2′-deoxyuridine in red, and the cell nuclei were dyed with Hoechst in blue after transfection miR-216a mimic or inhibitor, respectively. (B) During bovine primary muscle cells growth stage, CCK-8 analysis was performed after gain or loss miR-216a. (C) Bovine primary muscle cells phases were analyzed by flow cytometry after transfected with miR-216a mimic or inhibitor, respectively. (D) Statistical results of flow cytometry. Data are presented as the mean ± SEM; n = 3; **P < 0.01 and #P > 0.05.
When the confluence of bovine primary cells reached about 100%, horse serum at a low concentration of 2% was used to induce the differentiation into myotube. Since miR-216a regulates the proliferation of bovine primary muscle cells, it is not clear what role miR-216a plays in the differentiation process. After transfection of miR-216a mimic for 3 days, RT-qPCR was used to detect the mRNA expression level of differentiation marker gene (MyoD1, MyoG, and MyHC). It was found that the mRNA expression levels of MyoD, MyoG, and MyHC were all decreased, and the expression level of MyoG decline was significant (Figure 4A). Western blot results indicated that the protein expression levels of MyoD, MyoG, and MyHC were significantly decreased compared with the control group (Figures 4B, C). Immunofluorescence results showed that overexpression of miR-216a significantly inhibited the formation of myotube (Figure 4D). However, inhibition of miR-216a did not significantly affect the mRNA and protein levels of myoblast differentiation marker genes (Figures 4E–G). In addition, there was no significant difference in myotube formation after inhibition of miR-216a (Figure 4H). All these data demonstrated that overexpression of miR-216a inhibited bovine primary muscle cells differentiation.
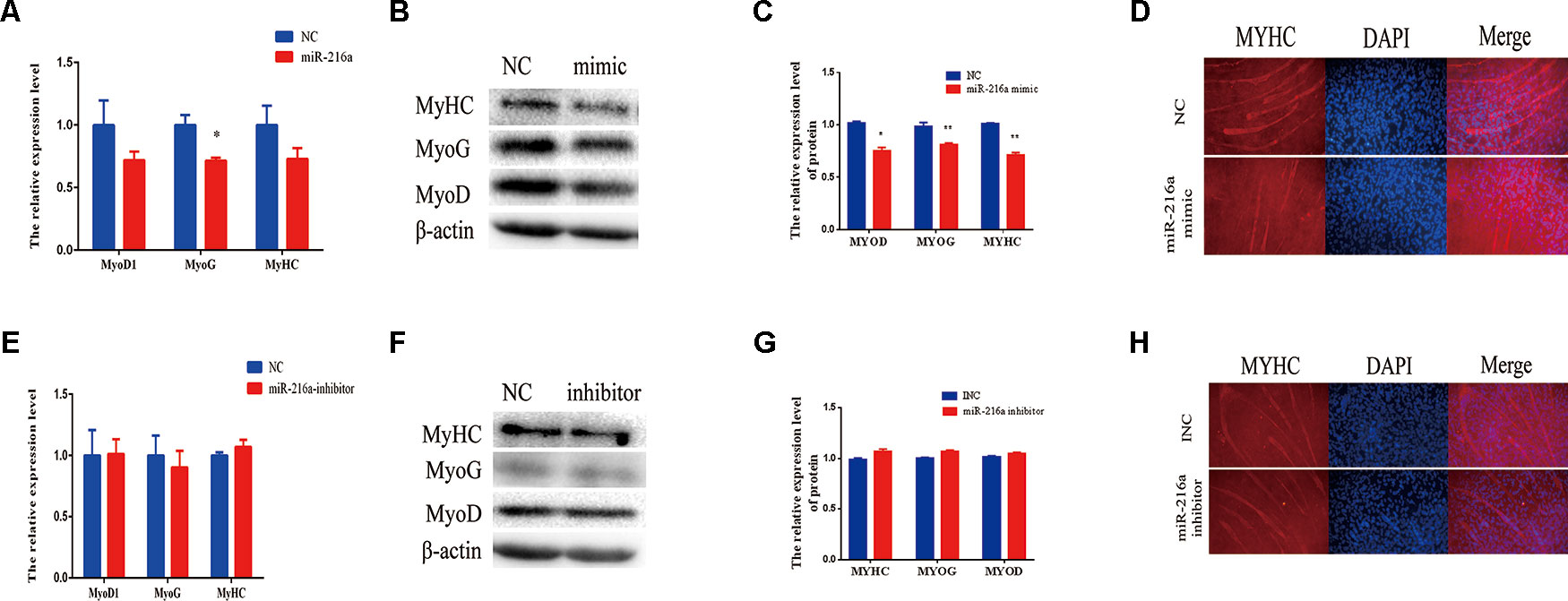
Figure 4 miR-216a regulates bovine primary muscle cells differentiation. (A) RT-qPCR was used to detect the messenger RNA (mRNA) expression of myogenic marker genes MyoD1, MyoG, and MyHC after transfection of miR-216a mimic. (B) Western blot was used to detect the protein expression level of MyoD, MyoG, and MyHC after transfection miR-216a mimic. (C) Protein quantitative analysis of MYHC, MYOG, and MYOD for (B). (D) The myotube formation of bovine primary muscle cells was measured by immunofluorescence assay under the 200 times field of microscope after transfection miR-216a mimic. (E) After loss of miR-216a, the mRNA expression of myogenic marker genes MyoD1, MyoG, and MyHC were detected by RT-qPCR. (F) After loss of miR-216a, the protein level of MyoD, MyoG, and MyHC were detected by western blot. (G) Protein quantitative analysis of MYHC, MYOG, and MYOD for (F). (H) After loss of miR-216a, the myotube formation of bovine primary muscle cells was measured by immunofluorescence assay under the 200 times field of microscope. Data are presented as the mean ± SEM; n = 3; *P < 0.05 and **P < 0.01.
Further, we explored molecular mechanisms underlying the function of miR-216a in impeding bovine primary muscle cells proliferation and differentiation by investigating its target genes. The mature sequence of miR-216a was highly conservative among different species (bovine, mouse, human, and rat) (Figure 5A), which aroused our research interest. To validate the function of miR-216a for bovine muscle cells proliferation and differentiation, predicting and verifying its target genes is essential. Through the online prediction software TargetScan (http://www.Targetscan.org) the seed sequence of miR-216a can target the (3’UTR) of the gene SNIP1 and smad7 mRNA (Figure 5B). After transfection of miR-216a expression plasmid with SNIP1 (wild or mutant) and smad7 (wild or mutant) psi-Check2 reporter vector into HEK293T cells, respectively, the analysis results showed that miR-216a could reduce the luciferase activity of their wild-type reporter vector but has no effect on the mutant reporter vector (Figures 5C, H). During the growth stage of bovine skeletal muscle cells, gain of miR-216a could reduce the mRNA and protein levels of SNIP1 significantly (Figures 5D, E). At the same time, miR-216a inhibitor was transfected into the cells, which had significantly enhanced the mRNA and protein levels of SNIP1 (Figures 5F, G). During the differentiated stage of bovine skeletal muscle cells, gain of miR-216a reduced smad7 mRNA and protein expression level (Figures 5I, J), whereas loss of miR-216a enhanced smad7 mRNA and protein expression level (Figures 5K, L). According to the above results, during the proliferation and differentiation stages of bovine primary muscle cells, miR-216a targeted SNIP1 and smad7, respectively.
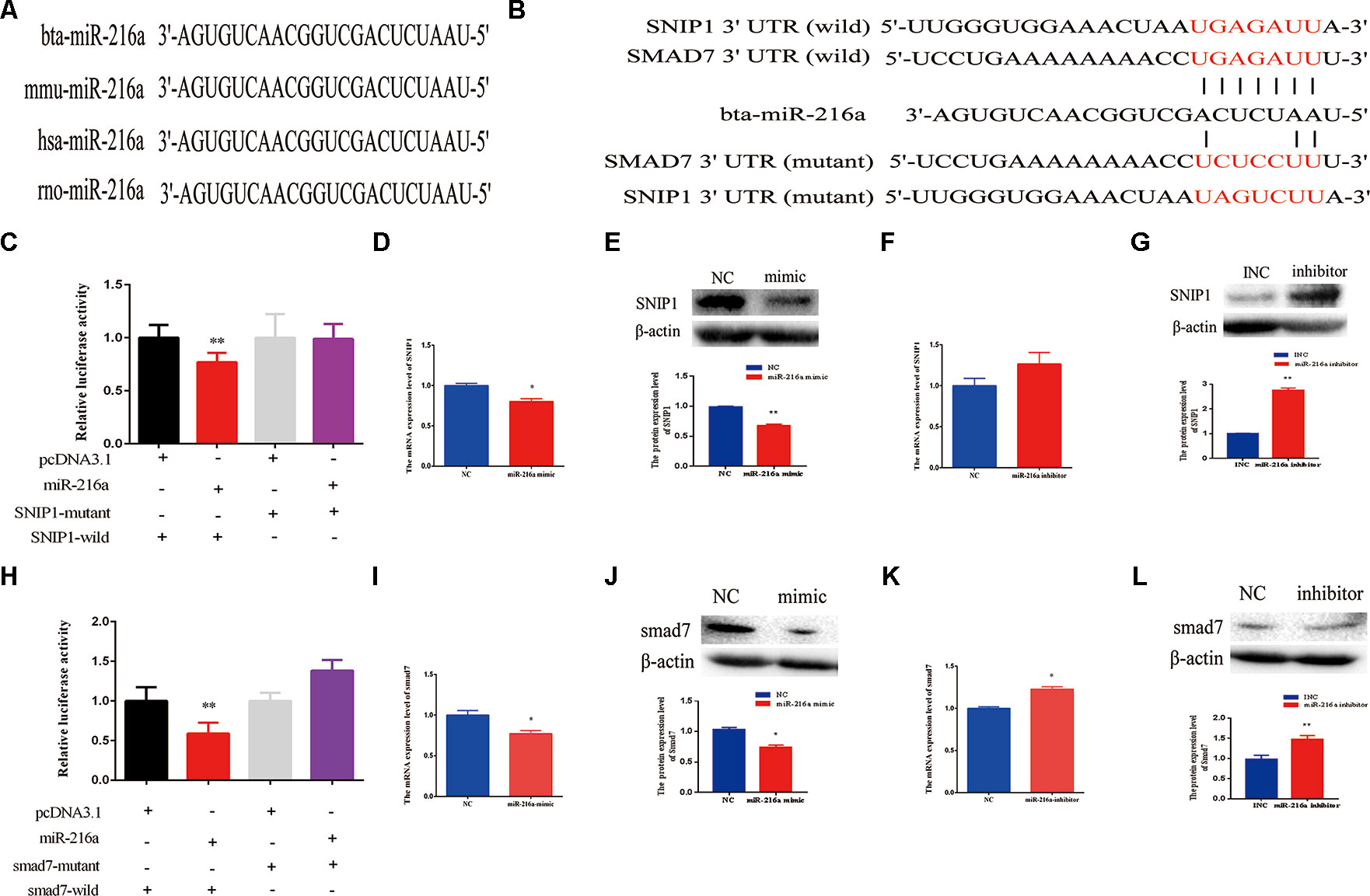
Figure 5 SMAD nuclear interacting protein-1 (SNIP1) and smad7 were the targets of miR-216a. (A) Conservative analysis of the mature sequence of miR-216a among different species (bovine, mouse, human, and rat). (B) The binding sites of miR-216a in the 3′-UTR of SNIP1 and smad7 were predicted by TargetScan software. The sequences in red indicate the target position of the miR-483 seed sequence and certain mutated bases. (C) The relative luciferase activity was detected after transfection of miR-216a expression plasmid with SNIP1 (wild or mutant) psi-Check2 reporter vector into HEK293T cells. (D, E) During the growth stage of bovine primary muscle cells, the messenger RNA (mRNA) and protein expression level of SNIP1 were detected by real-time quantitative PCR (RT-qPCR) and western blot after transfection miR-216a mimic. (F, G) During the growth stage of bovine primary muscle cells, the mRNA and protein expression level of SNIP1 were detected by RT-qPCR and western blot after transfection miR-216a inhibitor. (H) The relative luciferase activity was detected after transfection of miR-216a expression plasmid with smad7 (wild or mutant) psi-Check2 reporter vector into HEK293T cells. (I, J) During the differentiation stage of bovine primary muscle cells, the mRNA and protein expression level of samd7 were detected by RT-qPCR and western blot after transfection miR-216a mimic. (K, L) During the differentiation stage of bovine primary muscle cells, the mRNA and protein expression level of samd7 were detected by RT-qPCR and western blot after transfection miR-216a inhibitor. Data are presented as the mean ± SEM; n = 3; *P < 0.05 and **P < 0.01.
In this study, we found that miR-216a possessed an inhibitory role in the process of proliferation and differentiation of bovine primary muscle cells by targeting SNIP1 and smad7, respectively. Herein, miRNA-216a as a non-muscle-specific miRNA presented a downward trend in the proliferative and differentiation stages of bovine primary muscle cells, which suggested a potential role in the development of bovine skeletal muscle. To evaluate this hypothesis, mimic and inhibitor of miR-216a were used to enhance or block its expression, respectively. The current research found that overexpression of miR-216a can inhibit bovine primary muscle cells proliferation and differentiation. Unfortunately, inhibition of miR-216a expression did not affect the proliferation and differentiation of bovine primary muscle cells. We speculated that this might be caused by the low expression of miR-216a in bovine skeletal muscle or other potential compensation mechanisms. And it has been reported that an individual miRNA might interact with hundreds of mRNA targets while individual mRNAs can be targeted by many different miRNAs (Sassen et al., 2008). It was observed that if one specific miRNA fails, the pathway might be regulated by another miRNA within a complex cross path of miRNA network (Rottiers et al., 2011; Ebert and Sharp, 2012; Lima et al., 2017).
In the current study, we found miR-216a played a significant role in inhibiting skeletal muscle growth and development. According to our prediction and analysis, the mature sequence of miR-216a was found to be highly conserved among different species (bovine, human, mouse, and rat) (Figure 5A). Consistent with the research results, in previous studies, miR-216a also mainly played a suppressive role in disease and tumorigenesis. For instance, miR-216a inhibited tumor cells growth by down-regulating the expression level of Janus kinase 2 (JAK2) in pancreatic cancer (Siliang et al., 2014; Hou et al., 2016). In colorectal cancer, miR-216a can impede tumor cells invasion in vitro and metastasis in vivo by down-regulation of KIAA1199 (Zhang et al., 2017). In gastric cancer, miR-216a restrained tumor cells migration and invasion possibly by targeting JAK2/STAT3-mediated epithelial-mesenchymal transition (EMT) (Tao et al., 2017). In osteosarcoma patients, the expression level of miR-216a was downregulated. Gain of miR-216a can inhibit tumor cells proliferation, migration and invasion in vitro and in vivo via suppressing the expression of CDK14 (Ji et al., 2017). In renal cell carcinoma, miR-216a exerts tumor-suppressing functions by targeting TLR4 (Wang et al., 2018c). Based on the above studies, it’s worth noting that miR-216a has a broad application prospect in cancer therapy. Besides these, in muscle, a certain preliminary study of miR-216a has been reported. For example, miR-216a inhibits proliferation and promotes apoptosis of human airway smooth muscle cells by targeting JAK2 (Yan et al., 2019). And it can also exacerbate TGF-β-induced myofibroblast trans-differentiation via PTEN/AKT signaling (Qu et al., 2019). These results suggest that miR-216a has a potentially important role in muscle development. So, it is important to explore its role in skeletal muscle growth and development.
The current study further confirmed that miR-216a regulated the proliferation and differentiation of bovine primary muscle cells by targeting SNIP1 and smad7. Bioinformatics prediction, dual-luciferase reporter assay, and related verification suggested that SNIP1 and smad7 were the direct targets of miR-216a. In previous studies, SMAD nuclear interacting protein-1 (SNIP1) had been demonstrated function as an oncogene (Fujii et al., 2006), which can partially regulate CyclinD1 to accelerate cell cycle progression through G1 (Roche et al., 2004; Etsu et al., 2010). In a variety of cellular contexts, knockdown of SNIP1 could inhibit the mRNA and protein levels of CyclinD1 (Roche et al., 2004; Bracken et al., 2008). In the current study, overexpression of mir-216a in the growth stage of bovine primary muscle cells significantly inhibited the mRNA and protein levels of SNIP1, and the mRNA expression level of CyclinD1 also decreased at the same stage. Smad7, as a member of the inhibitory Smads (I-Smads) family, can specifically inhibit TGF-β pathway as described above and can also inhibit myostatin signaling by competitive combination with the R-Smads (Xiangyang et al., 2004). It has been reported that TGF-β signal pathway participated in regulating satellite cells (myoblasts) proliferation and differentiation by a complex regulatory network (Greene and Allen, 1991; Hathaway et al., 1991; Liu et al., 2001; Allen and Boxhorn, 2010). It has been reported that Smad7 can promotes and enhances skeletal muscle differentiation not only by blocking smad2/3 transcriptional activity but also by physically interacting with MyoD in vitro experiments (Kollias et al., 2006). Smad7-/- mice have the feature of reducing muscle cells proliferation and differentiation by enhancing smad2/3 signaling, which impedes muscle growth and regeneration (Cohen et al., 2015).
In summary, the current experimental results indicated that miR-216a suppressed bovine primary muscle cells proliferation and differentiation. Bioinformatics analysis, dual luciferase report analysis and experimental verification showed that SNIP1 and smad7 were the targets of miR-216a regulating the proliferation and differentiation of bovine muscle primary cells, respectively. Therefore, the function and mechanism of miR-216a in bovine primary muscle cells growth might provide theoretical basis for elucidating bovine skeletal muscle development in future experiments.
All datasets generated for this study are included in the article/Supplementary Material.
Animal care and study protocols were approved by the Animal Care Commission of the College of Veterinary Medicine, Northwest A&F University (Permit Number: NWAFAC1019).
ZY and CS contributed equally to this work. ZY conducted the analysis and wrote the manuscript. CS designed the study and finalized the manuscript. RJ carried out the part of experiments. YH, XL, CL, and HC provided experimental guidance.
This work was supported by the National Natural Science Foundation of China No.31772574), the Program of National Beef Cattle and Yak Industrial Technology System (CARS-37).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01112/full#supplementary-material
Alexander, M. S., Kawahara, G., Motohashi, N., Casar, J. C., Eisenberg, I., Myers, J. A., et al. (2013). MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ. 20 (9), 1194–1208. doi: 10.1038/cdd.2013.62
Allen, R. E., Boxhorn, L. K. (2010). Regulation of skeletal muscle satellite cell proliferation and differentiation by transforming growth factor-beta, insulin-like growth factor I, and fibroblast growth factor. J. Cell. Physiol. 138 (2), 311–315. doi: 10.1002/jcp.1041380213
Bartel, D. P. (2004). MicroRNAs_ Genomics, Review Biogenesis, Mechanism, and Function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 (2), 215–233. doi: 10.1016/j.cell.2009.01.002
Bracken, C. P., Wall, S. J., Benjamin, B., Panov, K. I., Ajuh, P. M., Perkins, N. D. (2008). Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 68 (18), 7621–7628. doi: 10.1158/0008-5472.CAN-08-1217
Buckingham, M., Bajard, L., Chang, T., Daubas, P., Hadchouel, J., Meilhac, S., et al. (2003). The formation of skeletal muscle: from somite to limb. J. Anat. 202 (1), 59–68. doi: 10.1046/j.1469-7580.2003.00139.x
Buckingham, M., Relaix, F. (2015). PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 44, 115–125. doi: 10.1016/j.semcdb.2015.09.017
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38 (2), 228–233. doi: 10.1038/ng1725
Cheung, T. H., Quach, N. L., Charville, G. W., Liu, L., Park, L., Edalati, A., et al. (2012). Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482 (7386), 524–528. doi: 10.1038/nature10834
Christ, B., Ordahl, C. P. (1995). EARLY STAGES OF CHICK SOMITE DEVELOPMENT. Anat. Embryol. 191 (5), 381–396. doi: 10.1007/BF00304424
Cohen, T. V., Kollias, H. D., Naili, L., Ward, C. W., Wagner, K. R. (2015). Genetic disruption of Smad7 impairs skeletal muscle growth and regeneration. J. Physiol. 593 (11), 2479–2497. doi: 10.1113/JP270201
Crist, C. G., Montarras, D., Pallafacchina, G., Rocancourt, D., Cumano, A., Conway, S. J., et al. (2009). Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. PNAS 106 (32), 13383–13387. doi: 10.1073/pnas.0900210106
Dey, B. K., Gagan, J., Dutta, A. (2011). miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell Biol. 31 (1), 203–214. doi: 10.1128/MCB.01009-10
Dey, B. K., Gagan, J., Yan, Z., Dutta, A. (2012). miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 26 (19), 2180–2191. doi: 10.1101/gad.198085.112
Diniz, G. P., Wang, D. Z. (2016). Regulation of Skeletal Muscle by microRNAs. Compr. Physiol. 6 (3), 1279–1294. doi: 10.1002/cphy.c150041
Ebert, M. S., Sharp, P. A. (2012). Roles for MicroRNAs in Conferring Robustness to Biological Processes. Cell 149 (3), 515–524. doi: 10.1016/j.cell.2012.04.005
Edmondson, D. G., Lyons, G. E., Martin, J. F., Olson, E. N. (1994). Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120, 1251–1263.
Etsu, T., Ayako, T., Masaya, I. (2010). Functions of cyclin D1 as an oncogene and regulation of cyclin D1 expression. Cancer Sci. 98 (5), 629–635. doi: 10.1111/j.1349-7006.2007.00449.x
Feng, Y., Niu, L. L., Wei, W., Zhang, W. Y., Li, X. Y., Cao, J. H., et al. (2013). A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis. 4, e934. doi: 10.1038/cddis.2013.462
Fujii, M., Lyakh, L. A., Bracken, C. P., Fukuoka, J., Hayakawa, M., Tsukiyama, T., et al. (2006). SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol. Cell 24 (5), 771–783. doi: 10.1016/j.molcel.2006.11.006
Gardner, S., Alzhanov, D., Knollman, P., Kuninger, D., Rotwein, P. (2011). TGF-β Inhibits Muscle Differentiation by Blocking Autocrine Signaling Pathways Initiated by IGF-II. Mol. Endocrinol. 25 (1), 128–137. doi: 10.1210/me.2010-0292
Ge, Y., Chen, J. (2011). MicroRNAs in skeletal myogenesis. Cell Cycle 10 (3), 441–448. doi: 10.4161/cc.10.3.14710
Greene, E. A., Allen, R. E. (1991). Growth factor regulation of bovine satellite cell growth in vitro. J. Anim. Sci. 69 (1), 146–152. doi: 10.2527/1991.691146x
Hathaway, M. R., Hembree, J. R., Pampusch, M. S., Dayton, W. R. (1991). Effect of transforming growth factor beta-1 on ovine satellite cell proliferation and fusion. J. Cell. Physiol. 146 (3), 435. doi: 10.1002/jcp.1041460314
Hocquette, J. F. (2010). Endocrine and metabolic regulation of muscle growth and body composition in cattle. Animal 4 (11), 1797–1809. doi: 10.1017/S1751731110001448
Hou, B. H., Jian, Z. X., Cui, P., Li, S. J., Tian, R. Q., Ou, J. R. (2016). miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 589 (17), 2224–2232. doi: 10.1016/j.febslet.2015.06.036
Ji, Q., Xu, X., Li, L., Goodman, S. B., Bi, W., Xu, M., et al. (2017). miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 8 (10), e3103. doi: 10.1038/cddis.2017.499
Ju, H., Yang, Y., Sheng, A., Jiang, X. (2015). Role of microRNAs in skeletal muscle development and rhabdomyosarcoma (review). Mol. Med. Rep. 11 (6), 4019–4024. doi: 10.3892/mmr.2015.3275
Kollias, H. D., Perry, R. L., Miyake, T., Aziz, A., McDermott, J. C. (2006). Smad7 promotes and enhances skeletal muscle differentiation. Mol. Cell Biol. 26 (16), 6248–6260. doi: 10.1128/MCB.00384-06
Lagha, M., Kormish, J. D., Rocancourt, D., Manceau, M., Epstein, J. A., Zaret, K. S., et al. (2008). Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev. 22 (13), 1828–1837. doi: 10.1101/gad.477908
Li, H., Wei, X., Yang, J., Dong, D., Hao, D., Huang, Y., et al. (2018). circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve Its Inhibition of Wnt3a 11, Mol. Ther. Nucleic Acids 11, 272–283. doi: 10.1016/j.omtn.2018.02.012
Lima, T. I., Araujo, H. N., Menezes, E. S., Sponton, C. H., Araujo, M. B., Bomfim, L. H., et al. (2017). Role of microRNAs on the Regulation of Mitochondrial Biogenesis and Insulin Signaling in Skeletal Muscle. J. Cell Physiol. 232 (5), 958–966. doi: 10.1002/jcp.25645
Liu, D., Black, B. L., Derynck, R. (2001). TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 15 (22), 2950–2966. doi: 10.1101/gad.925901
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McCarthy, J. J. (2008). MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta 1779 (11), 682–691. doi: 10.1016/j.bbagrm.2008.03.001
Miyake, M., Takahashi, H., Kitagawa, E., Watanabe, H., Sakurada, T., Aso, H., et al. (2012). AMPK activation by AICAR inhibits myogenic differentiation and myostatin expression in cattle. Cell Tissue Res. 349 (2), 615–623. doi: 10.1007/s00441-012-1422-8
O'Rourke, J. R., Georges, S. A., Seay, H. R., Tapscott, S. J., McManus, M. T., Goldhamer, D. J., et al. (2007). Essential role for Dicer during skeletal muscle development. Dev. Biol. 311 (2), 359–368. doi: 10.1016/j.ydbio.2007.08.032
Ott, M. O., Bober, E., Lyons, G., Arnold, H., Buckingham, M. (1991). Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development 111, 1097–1107.
Qu, C., Liu, X., Ye, T., Wang, L., Liu, S., Zhou, X., et al. (2019). miR216a exacerbates TGFbetainduced myofibroblast transdifferentiation via PTEN/AKT signaling. Mol. Med. Rep. 19 (6), 5345–5352. doi: 10.3892/mmr.2019.10200
Relaix, F., Montarras, D., Zaffran, S., Gayraud-Morel, B., Rocancourt, D., Tajbakhsh, S., et al. (2006). Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 172 (1), 91–102. doi: 10.1083/jcb.200508044
Roche, K. C., Nicola, W., Tom, O. H., Perkins, N. D. (2004). The FHA domain protein SNIP1 is a regulator of the cell cycle and cyclin D1 expression. Oncogene 23 (50), 8185–8195. doi: 10.1038/sj.onc.1208025
Rottiers, V., Najafi-Shoushtari, S. H., Kristo, F. G., Gurumurthy, S., Zhong, L., Li, Y., et al, (2011). MicroRNAs in Metabolism and Metabolic Diseases-miRNA. Cold Spring Harbor Symp. Quant. Biol. 76, 225–233. doi: 10.1101/sqb.2011.76.011049
Russell, R. G., Oteruelo, F. T. (1981). An Ultrastructural Study of the Differentiation of Skeletal Muscle in the Bovine Fetus. Anat. Embryol. 162 (4), 403–417. doi: 10.1007/BF00301866
Sassen, S., Miska, E. A., Caldas, C. (2008). MicroRNA: implications for cancer. Virchows Arch. 452 (1), 1–10. doi: 10.1007/s00428-007-0532-2
Siliang, W., Xiaodong, C., Meiyue, T. (2014). MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol. Rep. 32 (6), 2824–2830. doi: 10.3892/or.2014.3478
Sluijter, J. P., van Mil, A., van Vliet, P., Metz, C. H., Liu, J., Doevendans, P. A., et al. (2010). MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler. Thromb. Vasc. Biol. 30 (4), 859–868. doi: 10.1161/ATVBAHA.109.197434
Small, E. M., O'Rourke, J. R., Moresi, V., Sutherland, L. B., McAnally, J., Gerard, R. D., et al. (2010). Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U. S. A. 107 (9), 4218–4223. doi: 10.1073/pnas.1000300107
Song, C., Wu, G., Xiang, A., Zhang, Q., Li, W., Yang, G., et al. (2015). Over-expression of miR-125a-5p inhibits proliferation in C2C12 myoblasts by targeting E2F3. Acta Biochim. Biophys. Sin. (Shanghai) 47 (4), 244–249. doi: 10.1093/abbs/gmv006
Tajbakhsh, S., Buckingham, M. (2000). The birth of muscle progenitor cells in the mouse: Spatiotemporal considerations. Curr. Topics Dev. Biol. Vol 48 48, 225–268. doi: 10.1016/S0070-2153(08)60758-9
Tao, Y., Yang, S., Wu, Y., Fang, X., Wang, Y., Song, Y., et al. (2017). MicroRNA-216a inhibits the metastasis of gastric cancer cells by targeting JAK2/STAT3-mediated EMT process. Oncotarget 8 (51), 88870–88881. doi: 10.18632/oncotarget.21488
Wang, D., Li, Y., Zhang, C., Li, X., Yu, J. (2017a). MiR-216a-3p inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5. J. Cell. Biochem. 119 (2), 1755–1766. doi: 10.1002/jcb.26336
Wang, H., Zhang, Q., Wang, B., Wu, W., Wei, J., Li, P., et al. (2018b). miR-22 regulates C2C12 myoblast proliferation and differentiation by targeting TGFBR1. Eur. J. Cell Biol. 97 (4), 257–268. doi: 10.1016/j.ejcb.2018.03.006
Wang, W., Zhao, E., Yu, Y., Geng, B., Zhang, W., Li, X. (2018c). MiR-216a exerts tumor-suppressing functions in renal cell carcinoma by targeting TLR4. Am. J. Cancer Res. 8 (3), 476–488.
Wong, C. F., Tellam, R. L. (2008). MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 283 (15), 9836–9843. doi: 10.1074/jbc.M709614200
Xiangyang, Z., Stavros, T., Li-Fang, L., Stotish, R. L. (2004). Myostatin signaling through Smad2, Smad3 and Smad4 is regulated by the inhibitory Smad7 by a negative feedback mechanism. Cytokine 26 (6), 262–272. doi: 10.1016/j.cyto.2004.03.007
Yan, Y. R., Luo, Y., Zhong, M., Shao, L. (2019). MiR-216a inhibits proliferation and promotes apoptosis of human airway smooth muscle cells by targeting JAK2. J. Asthma 56 (9), 938–946. doi: 10.1080/02770903.2018.1509991
Zhang, D., Zhao, L., Shen, Q., Lv, Q., Jin, M., Ma, H., et al. (2017). Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int. J. Cancer 140 (10), 2298–2309. doi: 10.1002/ijc.30656
Keywords: miR-216a, SMAD nuclear interacting protein-1, smad7, skeletal muscle, bovine
Citation: Yang Z, Song C, Jiang R, Huang Y, Lan X, Lei C and Chen H (2019) Micro-Ribonucleic Acid-216a Regulates Bovine Primary Muscle Cells Proliferation and Differentiation via Targeting SMAD Nuclear Interacting Protein-1 and Smad7. Front. Genet. 10:1112. doi: 10.3389/fgene.2019.01112
Received: 09 July 2019; Accepted: 16 October 2019;
Published: 13 November 2019.
Edited by:
Junjie Xiao, Shanghai University, ChinaReviewed by:
Xiaofei Cong, Eastern Virginia Medical School, United StatesCopyright © 2019 Yang, Song, Jiang, Huang, Lan, Lei and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Chen, Y2hlbmhvbmcxMjEyQDI2My5uZXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.