- 1School of Biomedical Engineering, University of Technology Sydney, Sydney, Australia
- 2School of Software, University of Technology Sydney, Sydney, Australia
- 3The Tumour Bank–CCRU, Kids Research, The Children’s Hospital at Westmead, Sydney, Australia
Pediatric solid tumors are a diverse group of extracranial solid tumors representing approximately 40% of childhood cancers. Pediatric solid tumors are believed to arise as a result of disruptions in the developmental process of precursor cells which lead them to accumulate cancerous phenotypes. In contrast to many adult tumors, pediatric tumors typically feature a low number of genetic mutations in protein-coding genes which could explain the emergence of these phenotypes. It is likely that oncogenesis occurs after a failure at many different levels of regulation. Non-coding RNAs (ncRNAs) comprise a group of functional RNA molecules that lack protein coding potential but are essential in the regulation and maintenance of many epigenetic and post-translational mechanisms. Indeed, research has accumulated a large body of evidence implicating many ncRNAs in the regulation of well-established oncogenic networks. In this review we cover a range of extracranial solid tumors which represent some of the rarer and enigmatic childhood cancers known. We focus on two major classes of ncRNAs, microRNAs and long non-coding RNAs, which are likely to play a key role in the development of these cancers and emphasize their functional contributions and molecular interactions during tumor formation.
Pediatric cancers are often categorized as hematologic, intracranial, or extracranial (Chen et al., 2015). Hematologic cancers include those derived from the blood or blood forming tissues, including bone marrow and the lymph nodes. Intracranial cancers are tumors that develop inside the brain, whereas extracranial solid tumors, often referred to as pediatric solid tumors, arise outside the brain. Collectively, pediatric solid tumors represent approximately 40% of all pediatric cancers and commonly form in the developing sympathetic nervous system (neuroblastoma), retina (retinoblastoma), kidneys (Wilms tumor), liver (hepatoblastoma), bones (osteosarcoma, Ewing sarcoma), or muscles (rhabdomyosarcoma) (Kline and Sevier, 2003; Allen-Rhoades et al., 2018). Solid tumors can originate from cells of any of the three germ layers, the ectoderm, mesoderm, or endoderm, and likely arise due to disruptions in the developmental processes of these precursor cells, leading them to develop cancerous phenotypes (Chen et al., 2015). This contrasts with most adult cancers, which tend to be of epithelial origin and are believed to develop over time due to exposure to toxins and environmental stress. As a result, adult cancers often display a high occurrence of genetic mutations, whereas pediatric solid tumors tend to feature a relatively low number of genetic mutations. This has led to investigations into alternative forms of gene regulation that may contribute to the emergence and development of cancerous cells in pediatric cancers.
Non-coding RNAs (ncRNAs) form a group of functional RNAs lacking protein-coding potential, which play a crucial role in the regulation of gene expression at every level, from epigenetic regulation via methylation and chromatin packaging to post-transcriptional regulation (Cech and Steitz, 2014; Zhao et al., 2016). The most widely studied ncRNAs are the microRNAs (miRNAs), small 20- to 25-nucleotide-long RNAs that play an important role in regulating translation and messenger RNA stability via complementary base pairing (Huang et al., 2011). Other classes of small ncRNAs include small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small transfer RNAs (tRFs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), and small cytoplasmic RNAs (scRNAs). Additionally, long non-coding RNAs (lncRNAs) are a loosely defined group of RNAs normally larger than 200 nucleotides long that lack protein-coding potential and do not fall into any of the other categories but nonetheless play key roles in the regulation of gene expression (Mercer et al., 2009). Increasing data suggest that ncRNAs play a role in regulating all biological processes, and it is no surprise that studies have observed widespread dysregulation of ncRNAs in nearly all forms of cancer (Prensner and Chinnaiyan, 2011; Leichter et al., 2017). Interestingly, dysregulated RNA patterns are often specific to the type of cancer or even subtype and can provide insight into the mechanisms underlying phenotypic differences between tumors or cells within a tumor, such as their aggressiveness or resistance to certain types of treatments (Blenkiron et al., 2007; Li et al., 2014). Additionally, genome-wide association studies have suggested that over 80% of single nucleotide polymorphisms found associated with cancer are outside of coding regions (Carninci et al., 2005; Cheetham et al., 2013). In this review, we will discuss how two major classes of ncRNAs, miRNAs and lncRNAs, may contribute to pediatric solid tumors by participating in the regulation of established oncogenic networks known to drive these cancers.
MicroRNAs and Gene Regulation
Not long after the first human miRNA, let-7, was discovered in 2002 by the Ruvkun lab, miRNAs began to emerge as key participants in tumorigenesis (Pasquinelli et al., 2000). In 2002, two miRNAs were identified as potential tumor suppressors due to their frequent downregulation or deletion in chronic lymphocytic leukemia (Calin et al., 2002). Calin et al. (2004) later showed that many miRNA genes are located close to fragile sites or common breakpoints that frequently occur in cancers, suggesting that their loss of function was a key event in oncogenesis. Since then, oncomiRs—cancer-associated miRNAs—have become a major research focus (Esquela-Kerscher and Slack, 2006). A better understanding of the mechanisms behind miRNA regulation in cancer is invaluable to researchers and clinicians alike, not only to aid in the identification of new drug targets but also for the development of promising RNA-based therapies and their potential use as early detection biomarkers.
miRNAs: Biogenesis and Functions
The life cycle of a miRNA typically begins with its transcription into a primary miRNA (pri-miRNA) by RNA polymerase II (Ha and Kim, 2014). pri-miRNAs share several similarities with messenger RNAs (mRNAs) in that they are 5’ capped, are 3’ polyadenylated, and can be several hundreds or thousands of nucleotides long. In many cases, the pri-miRNA encodes for one miRNA species; however, in humans, a substantial number are polycistronic and encode several different miRNAs together. pri-miRNAs must be processed in the nucleus by the RNAse III enzyme Drosha, which releases shorter ∼65-nucleotide-long precursor RNAs (pre-miRNAs) with a secondary hairpin structure. This hairpin is recognized by the Exportin-5/Ran-GTP transporter, which transports the pre-miRNA from the nucleus to the cytoplasm. In the cytoplasm, the pre-miRNA is further processed by Dicer, another RNAse III enzyme, which cleaves the loop and releases a double-stranded miRNA duplex containing the 5 prime (5p) and 3 prime (3p) sequences. The duplex is then recognized by one of the four human Argonaute proteins, which loads one of the strands and discards the other.
miRNAs carry out their functions by binding to Argonaute and associating with various other proteins to form the RNA induced silencing complex (RISC). As part of this complex, miRNAs serve as guides by binding via complementary base pairing to target sites that are normally found in the 3´-untranslated region (3´UTR) of mRNAs. RISCs can regulate gene expression by direct cleavage of transcripts, transcript destabilization, or blocking translation. In a broader sense, miRNAs play a role in globally “fine-tuning” gene expression and are particularly important in inducing and maintaining differentiated cell states. In cancer, this finely tuned expression is often impaired, enabling gene networks that are normally switched on or off to reverse and begin influencing cellular behavior in a deleterious manner.
miRNAs: Drivers or Passengers in Cancer?
Microarrays and next-generation sequencing technologies enabled global measurements of miRNA expression changes and have revealed miRNA dysregulation to be a hallmark in nearly all cancers. miRNA expression profiles often correlate with cancer subtypes and have been effective at classifying cancer samples for risk stratification (De Preter et al., 2011). However, understanding the contribution of specific miRNAs can prove difficult. miRNAs are predicted to regulate hundreds to thousands of genes; however, their influence may be minor, and often, they must act in concert with other miRNAs. Current miRNA target prediction algorithms are imperfect and do not capture the true range of regulatory targets; therefore, biological validation is still needed (Riffo-Campos et al., 2016). Additionally, opposing behavior is seen with many miRNAs, where the same miRNA may be considered an oncogene in one cancer and a tumor suppressor in another. Because of their integration within complex gene networks, it is often not obvious whether a dysregulated miRNA actively participates in the maintenance of a cancerous state or whether it is simply a bystander. Therefore, it is important to examine how miRNAs participate in oncogenic networks on a functional level in order to properly understand their role.
Transcription factors that play an important role in regulating cell proliferation, migration, and apoptosis are commonly perturbed in pediatric solid tumors. One of the best examples of this is in neuroblastoma, where MYCN amplification is present in approximately 25% of neuroblastoma patients and disproportionally represents high-risk cases (Huang and Weiss, 2013). MYCN upregulation is also observed at a higher frequency in several other pediatric solid tumors including Wilms tumor, rhabdomyosarcoma (Williamson et al., 2005), and retinoblastoma, although generally not to the extent seen in neuroblastoma. Germline inactivation of the Wilms Tumor 1 (WT1) transcription factor has been linked to a genetic predisposition towards Wilms tumor. Several transcription factors, including Twist, Snails, and Zebs, involved in the epithelial-to-mesenchymal transition have also been implicated in the development of osteosarcoma (Yang et al., 2013). miRNAs are often closely tied to transcription factors, either as regulators or as transcriptional targets (Figure 1) (Sin-Chan et al., 2019). One of the earliest studies linking miRNAs to an oncogenic transcription factor was by O’Donnell et al. in 2005 (O’Donnell et al., 2005). In this study, they demonstrated that c-Myc could induce expression of the miR-17∼92 cluster and that several of these miRNAs could in turn regulate E2F1 transcription to control cell proliferation.
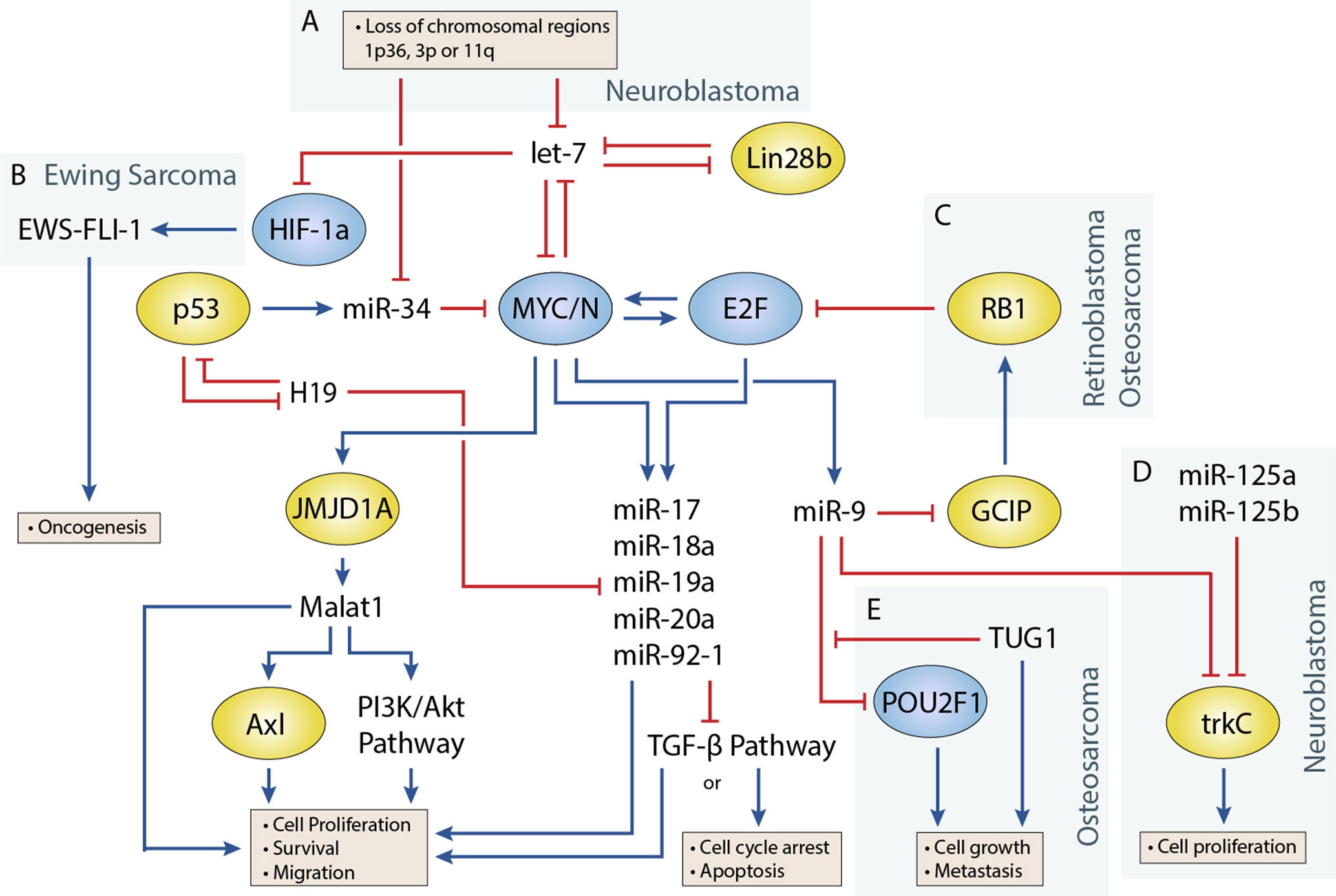
Figure 1 Regulatory circuitry involving non-coding RNAs in various pediatric solid tumors. Shows elements of a key regulatory circuit involving MYC and E2F family transcription factors and many ncRNAs, often dysregulated in pediatric solid tumors. In many cases, recurring dysregulation of specific elements, including miRNAs and lncRNAs, is observed and may represent vulnerabilities in the normal development of specific cell lineages. (A) Loss of chromosomal regions where let-7 and miR-34 miRNAs are localized is frequently observed in neuroblastoma and may represent a key event in the development of many of these cancers. (B) let-7 dysregulation may facilitate overexpression of the oncogenic fusion transcript EWS-FLI-1 in Ewing sarcoma. (C) The RB1 tumor suppressor regulates E2F, and loss of function via mutations can lead to the development of retinoblastoma. In osteosarcoma, miR-9 may be able to act as an oncogenic driver as it is often overexpressed and can downregulate RB1. (D) In neuroblastoma, miR-9 can display tumor-suppressive properties by cooperating with miR-125a and miR-125b to regulate a specific isoform of trkC and suppress cell proliferation (E) The lncRNA TUG1 is suggested to act as a ceRNA against miR-9, which has been shown to display tumor-suppressive properties in some osteosarcoma cell lines.
Disruptions in miRNA Processing
Recent studies have shown that impairments of the miRNA processing machinery are common in Wilms tumor and likely contribute to this disease. For example, a study by Torrezan et al. (2014) found mutations in miRNA processing genes in 33% of tumors, most commonly occurring in the Drosha gene, with other mutations in DICER1, XPO5, DGCR8 and TARBP2. These results are supported by several other studies by Wu et al. (2013), Rakheja et al. (2014), Walz et al. (2015), Wegert et al. (2015), and Gadd et al. (2017). In Rakheja et al.’s study, they further examined the potential consequences of several of these mutations and found that Drosha mutations often led to a loss of RNAse IIIB activity, which prevented processing of pri-miRNAs, leading to a global reduction in mature miRNAs. DICER1 mutations also frequently affected the RNAse IIIB domain; however, this mutation only affected processing of 5p miRNAs from precursors, as DICER1 contains a second RNAse domain for 3p processing. As a result, this mutation led to a shift towards 3p miRNA maturation. These mutations have interesting consequences for global miRNA expression and most likely favor expression of oncogenic miRNAs or reduce expression of miRNAs with tumor-suppressive effects. In line with this, the let-7 family is predominantly 5p-derived, and lower expression of several of its 5p members was found in both Drosha and DICER1 mutants in two of these studies (Rakheja et al., 2014; Walz et al., 2015). Additionally, the miR-200 family was found downregulated in Wilms tumors with mutated miRNA processing genes, which is known to regulate the mesenchymal-to-epithelial transition and has been associated with highly aggressive forms of cancer (Ceppi and Peter, 2014; Walz et al., 2015). The functional role of several oncomiRs has been investigated in detail within the context of pediatric solid tumors and is discussed in the following section.
The miR-17∼92 Cluster is a Downstream Effector of Oncogenic Transcription Factors
The miR-17∼92 cluster is expressed during normal development of the brain, heart, lungs, and immune system (Koralov et al., 2008; Ventura et al., 2008; Bian et al., 2013; Chen et al., 2013) and is known to regulate critical genes involved in cell growth, proliferation, and apoptosis. This cluster is comprised of six different miRNAs that are co-expressed, including miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92-1. Dysregulation of the miR-17∼92 cluster has been shown in several pediatric solid tumors including neuroblastoma, Wilms tumor, retinoblastoma, and osteosarcoma, where a higher expression generally correlates with a poorer prognosis (Chen and Stallings, 2007; Baumhoer et al., 2012; Li et al., 2014). The miR-17∼92 cluster is particularly interesting due to its regulation by the transcription factor MYC and its homologue MYCN, where it seems to act as a mediator for some of MYC/MYCN’s oncogenic effects (Schulte et al., 2008). Other transcription factors known to target the miR-17∼92 cluster include members of the E2F family and STAT3 (Mogilyansky and Rigoutsos, 2013).
Several studies have demonstrated that the miR-17∼92 cluster regulates many downstream components of the transforming growth factor beta (TGF-β) pathway, which is known to participate in a variety of cellular process such as differentiation, proliferation, and immune cell activation. A study by Fontana et al. (2008) demonstrated that in neuroblastoma, miR-17 and miR-20a downregulate the cyclin-dependent kinase inhibitor p21, which is activated by TGF-β. p21 plays a key role in the inhibition of cell cycle progression by blocking the transition from G1 to S phase, and its deregulation leads to uncontrolled cell growth. Additionally, Fontana et al. (2008) showed that miR-17-5p regulated another downstream component of TGF-β, the pro-apoptotic factor Bcl-2 interacting mediator (BIM). Mestdagh et al. (2010) later investigated miR-17∼92 regulation of the TGF-β pathway in more depth and identified miR-17 and miR-20a as regulators of TGFBR2 and miR-18a as a regulator of SMAD2 and SMAD4, both signal transducers for TGF-β receptors. miR-18a and miR-19a have also been shown to repress estrogen receptor (ESR1) expression, and prolonged knockdown of miR-18a induced morphological differentiation of SK-N-BE neuroblastoma cells. Interestingly, the TGF-β pathway interacts with ESR1 signaling via several of the SMADs (Band and Laiho, 2011), suggesting a complex interplay between miR-17∼92 and its targeted pathways necessary for fine-tuning differentiation during neuronal development—a balance that is disrupted when miR-17∼92 is overexpressed. While no studies have investigated in detail the interaction between the miR-17∼92 cluster and TGF-β pathway in Wilms tumor, the TGF-β pathway has been implicated in Wilms tumor development. In contrast with neuroblastoma, the TGF-β pathway appears to function as a promoter of Wilms tumor progression, and TGF-β is highly expressed in primary tumors, even more so in metastatic tumors. This multifaceted behavior of the TGF-β pathway has been shown in other cancers and implies that the pathway’s influence is specific to the tumor it is activated in.
The E2F family of transcription factors serve an important role in cell cycle control as their expression can cause cells to enter the G1 phase to initiate cell division (Chen et al., 2009). Several members, including E2F1, E2F2, and E2F3, all regulate miR-17∼92 expression. In a study by Kort et al. (2008) a member of the E2F family of transcription factors, E2F3, was shown to be exclusively expressed in Wilms tumor and not in other types of kidney tumors. In line with this, they compared expression of the miR-17∼92 miRNAs in Wilms tumor samples to other renal tumor subtypes and found them all to be upregulated. They were also able to show a correlation between E2F3 expression and the stage of Wilms tumor, where it was highest in late-stage metastatic tissues. In retinoblastoma, an early study investigating the miR-17∼92 cluster identified that one of its members, miR-20a, participates in an autoregulatory feedback loop with E2F2 and E2F3 (Sylvestre et al., 2007), as they found both transcription factors are themselves downregulated by miR-20a. The authors suggested that this autoregulation was critical in preventing expression of excessive amounts of E2F transcription factors. Given that MYC/MYCN and E2F have previously been shown to induce each other’s expression, miR-20a appears to play an important role in keeping this positive feedback loop in check (Leone et al., 1997; Strieder and Lutz, 2003). Therefore, it is easy to see how disruption in one or more of these regulatory elements could lead to uncontrolled expression of these proliferative and anti-apoptotic signals.
A later study by Conkrite et al. (2011) investigated miR-17∼92 in retinoblastoma and revealed that this cluster was capable of driving retinoblastoma formation in RB1/p107-deficient mice. RB1 plays a key role in inhibiting cell cycle progression, and germline mutations of this gene can lead to familial retinoblastoma formation (Friend et al., 1986; Classon and Harlow, 2002). RB1’s protein product, pRB, inhibits E2F transcription factors by binding and inactivating them, and its absence enables miR-17∼92–driven tumor formation.
The miR-17∼92 cluster also plays a role in driving tumor progression and metastasis in osteosarcoma (Li et al., 2014). A recent study by Yang et al. (2018) identified QKI2 as a regulatory target of the miR-17∼92 cluster. QKI proteins have previously been shown to inhibit β-catenin and induce differentiation in colon cancer. Yang at el. demonstrated that miR-17∼92 downregulated QKI2, causing upregulation of β-catenin, leading to increased proliferation, invasion, and migration in osteosarcoma (Yang et al., 2018). Additionally, miR-20a has previously been shown to downregulate Fas expression, which is a cell surface marker that interacts with FasL to induce apoptosis in the lungs, where osteosarcoma almost exclusively metastasizes to (Huang et al., 2012).
The miR-17∼92 cluster plays a tumorigenic role in a number of pediatric solid tumors including neuroblastoma, Wilms tumor, retinoblastoma, and osteosarcoma. The use of the miRNA pathway by transcription factors such as the MYC and E2F families enables them to target a wide range of genes and immediately effect gene expression at the post-transcriptional level. Continued research into how miRNAs may operate as oncogenic drivers will likely expand the repertoire of potential drug targets available to us.
Let-7 Dysregulation is a Feature in Many Pediatric Solid Tumors
The let-7 family of miRNAs are among the most well-characterized tumor suppressors due to their frequent downregulation in cancers. In total, there are 12 members of the let-7 family located across eight different chromosomes; however, in most cells, only a selection of these miRNAs will be expressed (Balzeau et al., 2017). let-7 miRNAs are important in regulating the cell cycle and maintaining cells’ differentiated state by targeting a wide range of genes with known roles in cancer biogenesis such as MYC/MYCN, RAS, CDK6, and HMGA2 (Buechner et al., 2011; Wu et al., 2015).
let-7 is regulated by the LIN28 proteins, LIN28A and LIN28B, which mediate uridylation, prevent processing of the let-7 precursor, and are important for maintaining pluripotency in cells (Lehrbach et al., 2009; Balzeau et al., 2017). Both Lin28 genes contain let-7 target sites and participate in a double-negative feedback loop with let-7 (Yin et al., 2017). Overexpression of Lin28 tends to drive cells towards oncogenesis and is a common feature in cancers. In a study by Urbach et al. (2014), Lin28b overexpression was found in approximately 30% of Wilms tumors. Additionally, they found overexpression of Lin28 could induce tumor formation in specific renal intermediates and that restoration of let-7 activity could reverse this effect in mice. Similar examples have been shown in mouse models, where Lin28b overexpression can drive hepatoblastoma and hepatocellular carcinoma in the liver and neuroblastoma in the neural crest (Molenaar et al., 2012; Nguyen et al., 2014). Molenaar et al. (2012) investigated Lin28b in neuroblastoma and demonstrated that Lin28b could enhance MYCN protein levels via let-7 regulation. However, a later study by Powers et al. (2016) showed that Lin28b expression was redundant in certain MYCN-amplified neuroblastoma cells, as overexpression of the MYCN transcript could function as a miRNA sponge for let-7, thereby negating their effect regardless of expression level. Powers et al. showed that most neuroblastomas were characterized by a loss of let-7 with either MYCN overexpression or chromosomal loss of arm 3p or 11q, where several let-7 miRNAs are located (Figure 1A). The authors noted that these events were generally mutually exclusive and suggested that the presence of one event alleviated selective pressure for the other.
A study by Di Fiore et al. (2016) revealed that let-7d could promote and suppress tumor formation within the same system. In this study, they found that let-7d overexpression in osteosarcoma cells reduced several stemness genes, including Lin28b, HMGA2, Oct3/4, and SOX2, and could elicit the mesenchymal-to-epithelial transition with upregulation of the epithelial marker E-cadherin and downregulation of mesenchymal markers N-cadherin and vimentin. However, they also found that let-7d enhanced cell migration and invasion, presumably by acting via the TGF-β pathway, which is known to promote this behavior. let-7d strongly increased versican VI expression, which has previously been shown to activate the TGF-β pathway in osteosarcoma (Li S. et al., 2014).
In Ewing sarcoma, Hameiri-Grossman et al. (2015) found that let-7 downregulated the Ras oncogene, as well as the transcription factor HIF-1a, to reduce EWS-FLI-1 expression (Figure 1B). EWS-FLI-1 is a hybrid transcript that results from a translocation event involving EWS and FLI1, and translocation events such as this are present in nearly all Ewing sarcoma cases and are believed to drive the disease (Delattre et al., 1994).
Loss of Let-7 plays a key role in many pediatric solid tumors as its loss enables expression of transcription factors and other genes that participate in oncogenesis. This has been emphasized in neuroblastoma, where it has been suggested that loss of let-7 function is an essential event in tumor development and positions the miRNA pathway as a central player in pediatric solid tumors.
miR-9 Has Been Shown to Play Oncogenic and Tumor-Suppressive Roles in Different Pediatric Tumors
miR-9 is a highly conserved miRNA involved in several different cellular processes including cell proliferation, differentiation, and migration. Early studies revealed miR-9 to be highly expressed in the brain and play a role both during development and in the adult brain; however, miR-9 has also been associated with many cancers outside the brain, acting as an oncogene or tumor suppressor (Coolen et al., 2013). Mir-9 is upregulated by MYC/MYCN and plays a role in promoting tumor growth and metastasis in several cancers including breast cancer, osteosarcoma, and rhabdomyosarcoma, where it is often overexpressed (Iorio et al., 2005; Luo et al., 2017) However, in other cancers such as neuroblastoma, miR-9’s role is less clear, and studies have argued for oncogenic and tumor suppressor functions (Laneve et al., 2007; Zhi et al., 2014).
The role of miR-9 in osteosarcoma appears to be in promoting cell growth and metastasis (Zhu et al., 2015; Qi et al., 2016). In a study by Zhu et al. (2015), miR-9 knockdown suppressed cell growth and migration of osteosarcoma cells. They were also able to show that miR-9 downregulated RB1 via the Grap2 and cyclin D interacting protein (GCIP), thereby promoting E2F-mediated cell division (Figure 1C). Similar behavior has been observed in the alveolar subtype of rhabdomyosarcoma, where miR-9 contributes to increased cell proliferation and migration (Missiaglia et al., 2017). In this study by Missiaglia et al. (2017), miR-9 was shown to be induced by the PAX3/FOXO1 fusion gene via MYCN, which is specific to this subtype of rhabdomyosarcoma.
In neuroblastoma, miR-9 expression has been shown to be both up- and downregulated in different studies. An early study by Laneve et al. (2007) showed that miR-9 was downregulated in 50% of primary neuroblastoma samples, and follow-up experiments demonstrated that miR-9 could act together with miR-125a and miR-125b to suppress cell proliferation by targeting a truncated isoform of the neurotrophin receptor tropomyosin-related kinase C (trkC) (Figure 1D). However, a later study by Ma et al. (2010) found miR-9 to be a target of MYCN and that miR-9 expression correlated with MYCN and metastatic status in neuroblastoma tumors. In this same study (albeit in breast cancer cells), Ma et al. also demonstrated that miR-9 suppressed E-cadherin to activate β-catenin and promote the epithelial-to-mesenchymal transition. Mir-9 is frequently involved in promoting cell migration; however, its absence has also been shown to produce different responses such as cell cycle arrest or apoptosis in neurons depending on their origin (Bonev et al., 2011). The contradictory behavior seen with studies of miR-9 highlight the diverse roles that individual miRNAs can play, and more comprehensive studies are needed to identify the relevant contextual influences on miRNA behavior.
miR-34 Is a Key Regulator of the Cell Cycle and Drug Resistance in Pediatric Solid Tumors
The miR-34 family has garnered significant interest since its members were discovered to be direct transcriptional targets of the tumor suppressor and transcription factor p53 (Hermeking, 2010). The miR-34 family consists of three miRNAs encoded by two genes, mir-34a and mir-34b/c. All three miRNAs play a key role in regulating apoptosis and the cell cycle by inducing G1 phase arrest. One of the more interesting facts about miR-34a and miR-34b/c is their genomic locations, which are located on chromosomes 1p36 and 11q23, respectively, regions that are frequently lost in pediatric solid tumors (Ruteshouser et al., 2005; Wittmann et al., 2007). In particular, loss of 1p36 occurs in 20–30% of neuroblastoma cases and correlates with MYCN amplification (Caron et al., 1993; Maris et al., 1995), whereas loss of 11q23 in occurs in approximately 40% of cases but almost never occurs with MYCN amplification (Figure 1A) (Guo et al., 1999; Attiyeh et al., 2005). miR-34 members are also regulators of the MYC family, as miR-34a is known to regulate MYCN and miR-34b and mir-34c to regulate c-MYC (Wei et al., 2008).
Studies on mir-34a expression have identified frequent downregulation in neuroblastoma, osteosarcoma, and hepatoblastoma (Jiao et al., 2016). miR-34a is itself considered a tumor suppressor due to its involvement in cell cycle arrest and apoptosis (De Antonellis et al., 2014). In neuroblastoma, Cole et al. (2008) investigated the growth-inhibitory effects of several miRNAs mapping to common chromosomal aberrations by overexpressing them in cell lines. In most cases, overexpression did not lead to a noticeable change in phenotype; however, miR-34a and miR-34c induced significant growth inhibition in cell lines with 1p36 deletion. Growth inhibition and suppression of metastasis by miR-34a have also been shown in osteosarcoma by several studies, where members of key proliferative signal transduction pathways such as c-Met, DUSP1, and Eag1 were identified as regulatory targets (Yan et al., 2012; Wu X. et al., 2013; Gang et al., 2017). The miR-34 family also targets several members of the Notch signaling pathway, which has been linked to both oncogenic and tumor-suppressive roles depending on the cellular context. In osteosarcoma, activation of the Notch pathway is known to contribute to tumor growth, and miR-34a–mediated downregulation of this pathway likely contributes to its tumor-suppressive role. However, in Ewing sarcoma, a recent study investigating miR-34b suggested that it could act as an oncogene, promoting proliferation, migration, and invasion through Notch1 repression (Lu Q. et al., 2018). Prior studies have shown correlations between high mir-34a expression and patient survival, which would indicate a tumor-suppressive role for mir-34a (Nakatani et al., 2012; Marino et al., 2014). It is unclear why miR-34a and miR-34b would display contrasting effects given their shared targets, and further investigation is needed.
Several studies by Pu et al. (2016)and Pu et al. (2017) have suggested that miR-34a may also play a role in promoting multidrug resistance in osteosarcoma. In these studies, they found that miR-34a-5p enhanced multidrug resistance through downregulation of the CD117 and AGTR1 genes in vitro. CD117 is often highly expressed in drug-resistant tumors and is commonly used as a marker for stemness (Adhikari et al., 2010). In contrast, Nakatani et al. found that miR-34a increased chemosensitivity in Ewing sarcoma (Nakatani et al., 2012).
Other miRNAs Involved in Multiple Pediatric Solid Tumors
A substantial number of other miRNAs have been discovered with functional implications in multiple pediatric solid tumors. One such miRNA is miR-125b, which typically exhibits tumor-suppressive properties in cancers such as neuroblastoma, osteosarcoma, and Ewing sarcoma, where it is commonly dysregulated (Laneve et al., 2007; Li J. et al., 2014; Xiao et al., 2019). Previously, it was mentioned that miR-125b participates in a network with miR-125a and miR-9, regulating expression of a truncated trkC isoform to control neuroblastoma growth and differentiation (Laneve et al., 2007; Le et al., 2009). In osteosarcoma, miR-125b was found to regulate STAT3 by downregulating MAP kinase kinase 7 (MKK7), which inactivates STAT3 via dephosphorylation (Xiao et al., 2019). Loss of miR-125b and consequent overexpression of MKK7 led to increased tumor formation and poorer prognosis. In Ewing sarcoma, miR-125b is involved in regulating the PI3K signaling pathway; could inhibit cell proliferation, migration, and invasion; and induce apoptosis through suppression of PIK3CD (Li J et al., 2014). Conversely, in retinoblastoma, miR-125b is overexpressed and has shown oncogenic properties by promoting cell proliferation and migration and inhibiting apoptosis (Bai et al., 2016). Conflicting behavior with miR-125b has been observed in many other cancers, which suggests that its role is highly dependent on cell identity (Sun et al., 2013).
miR-124 has been widely reported to act as a tumor suppressor by inhibiting cell growth and metastasis and acts as a key mediator of differentiation in several pediatric solid tumors (Peng et al., 2014; Feng et al., 2015; Zhao et al., 2017). In neuroblastoma, miR-124a increased the proportion of differentiated cells possessing neurite outgrowths (Le et al., 2009). In retinoblastoma, miR-124 participates in a regulatory network with lncRNAs Malat1 and XIST, which both function as oncogenes by enhancing growth and metastasis through downregulation of miR-124 (Liu S. et al., 2017; Hu et al., 2018). miR-124 itself was shown to target STAT3 to inhibit cell proliferation, migration, and invasion (Liu S. et al., 2016). In Ewing sarcoma, miR-124 expression is suppressed, and expression was found to reduce growth and metastasis via downregulation of mesenchymal genes such as SLUG and cyclin D2 (CCND2) (Li et al., 2017). Finally, in osteosarcoma, retinoblastoma, and Ewing sarcoma, miR-143 has been found to be dysregulated (De Vito et al., 2012; Li S. et al., 2014; Wang et al., 2016; Sun et al., 2018). For example, Li et al. investigated miR-143 function in osteosarcoma and showed that miR-143 participated in the TGF-β pathway by targeting versican, and TGF-β could reduce miR-143 expression to promote cell migration and invasion (Li S. et al., 2014). FOS-like antigen 2 (FOSL2) was also identified as a miR-143 target, which enhanced cell proliferation, migration, and invasion in the absence of miR-143 (Sun et al., 2018). Additional miRNA studies have been listed in Table 1.
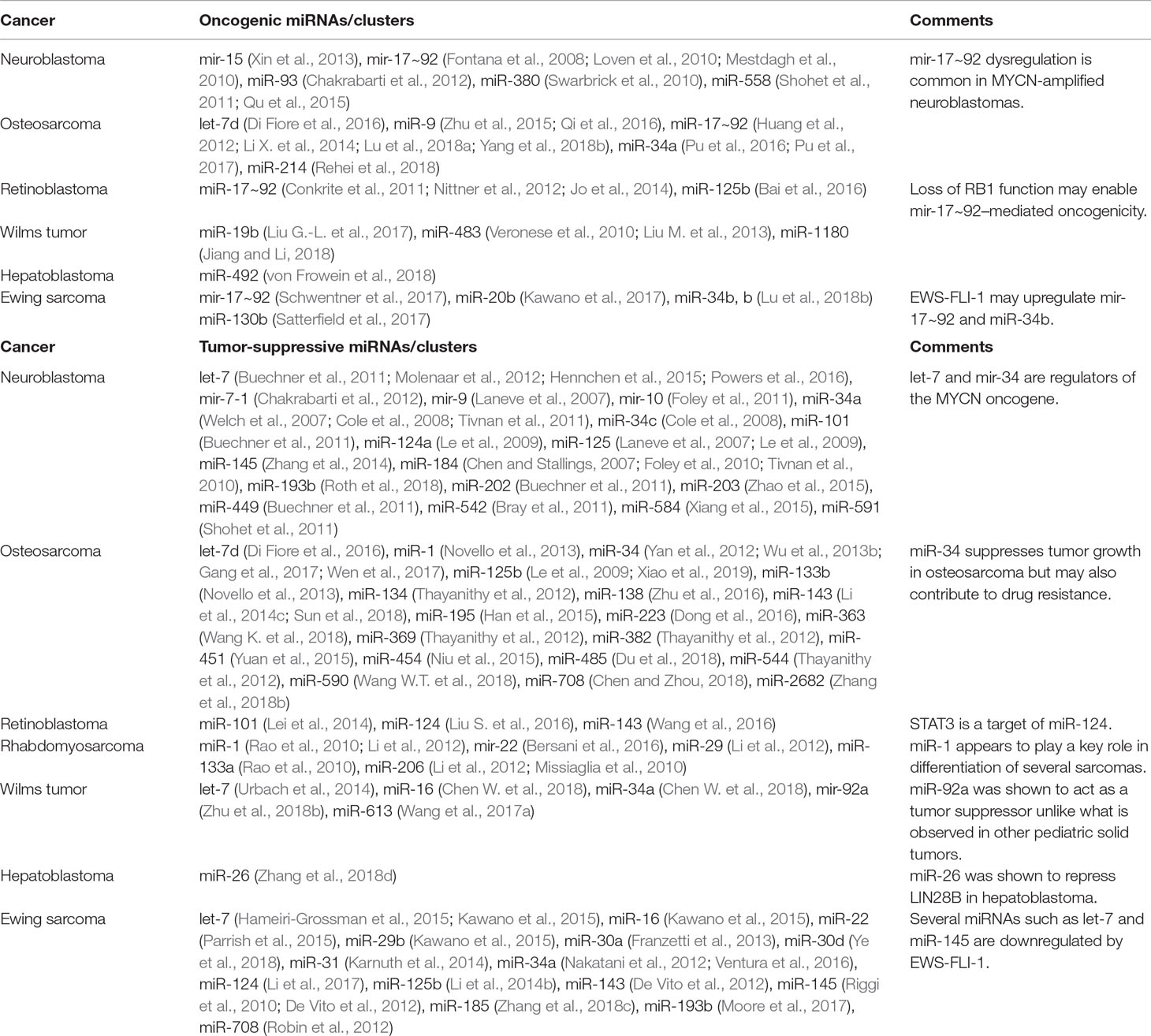
Table 1 miRNAs that have been shown to exhibit oncogenic or tumor-suppressive effects through functional studies in various pediatric solid tumors.
miRNAs Regulate All Aspects of Tumorigenesis
Widespread dysregulation of miRNAs is observed in many pediatric solid tumors, and functional studies have demonstrated that many of these miRNAs can drive or repress oncogenic pathways responsible for cell proliferation, apoptosis, angiogenesis, metastasis, and drug resistance. Importantly, miRNAs such as let-7 and miR-34 play a vital role in pediatric solid tumors by regulating established oncogenic transcription factors such as the MYC and E2F families (Wei et al., 2008; Buechner et al., 2011). Other miRNAs, such as the miR-17∼92 cluster and miR-9, serve as downstream effectors for these transcription factors, although their exact role in tumorigenesis seems to depend on the overall transcriptional landscape (Schulte et al., 2008; Ma et al., 2010). In some cases, viewing miRNAs as oncogenes or tumor suppressors likely represents an oversimplification of their role in cancer, and a better understanding of their participation in oncogenic networks will be needed to clarify their exact contributions.
Long Non-Coding RNAs Regulate Oncogenic Pathways in Pediatric Solid Tumors
For a long time, it was believed that the human genome was mostly comprised of “junk” DNA, despite pervasive transcription of much of the genome outside of protein-coding genes and other known RNAs at the time (Prensner and Chinnaiyan, 2011). Originally thought of as transcriptional noise, lncRNAs have now emerged as functional regulators of nearly all essential cellular processes including growth, differentiation, cell state maintenance, apoptosis, splicing, and epigenetic regulation. The first lncRNA, H19, was discovered in 1990 where an RNA molecule was found spliced and polyadenylated in a manner typical of mRNAs; however, it lacked an open reading frame and was believed to function as an untranslated RNA molecule (Brannan et al., 1990).
Often, lncRNAs participate within protein complexes and can operate as scaffolds, guides, decoys, or allosteric regulators. Many lncRNAs function as epigenetic regulators by interacting with proteins involved in chromatin remodeling and DNA methylation. Frequently, these lncRNAs will be cis-acting and regulate the regions near their transcribed location; however, some are trans-acting. Other lncRNAs function as competing RNAs (ceRNAs), which contain miRNA binding sites in a similar manner to mRNAs in order to compete and reduce the activity of miRNAs.
Several studies have investigated lncRNA expression in pediatric tumors and have successfully identified unique expression profiles in different cancers and tumor subtypes (Mitra et al., 2012; Brunner et al., 2012; Dong et al., 2014; Sahu et al., 2018). For example, Dong et al. (2014) compared hepatoblastoma samples to normal liver tissue in patients and found 2,736 differentially expressed lncRNAs. A study by Pandey et al. (2014) found 24 lncRNAs that could distinguish low- and high-risk neuroblastoma tumors. In a more recent study, Sahu et al. (2018) identified 16 differentially expressed lncRNAs that could be used to predict event-free survival with greater accuracy than other commonly used clinical risk factors. Mechanistic studies into many of these lncRNAs have revealed that they frequently act as an additional layer of regulation within established oncogenic networks involving protein-coding genes and miRNAs. While the field of lncRNAs is still relatively young, many studies have emerged that suggest that lncRNAs are far more integrated into existing gene networks than what has previously been appreciated (Figure 1). In the following section, the roles of some of the better-characterized lncRNAs in pediatric solid tumors will be discussed.
Malat1 Is Induced by MYCN in Neuroblastoma and Competes With Many miRNAs
One of the earliest lncRNAs to be associated with disease was Malat1 (metastasis-associated lung adenocarcinoma transcript 1), which was shown to associate with metastatic tumors in non–small cell lung cancer patients (Ji et al., 2003). Malat1 is abundantly expressed and highly conserved across species, unlike many other lncRNAs, and displays remarkably diverse functions in cellular processes including alternative splicing, nuclear organization, and epigenetic modulation. Studies have suggested an important role for Malat1 in brain development, as it is highly expressed in neurons and its depletion has been shown to affect synapse and dendrite development (Bernard et al., 2010; Chen et al., 2016). However, its importance has been questioned as other studies have found that Malat1-KO mice are viable with no discernable change in phenotype (Nakagawa et al., 2012; Zhang et al., 2012).
In addition to lung cancer, Malat1 is known to contribute to metastasis in other common types of cancer including hepatocellular carcinoma and bladder cancer, with evidence that it acts through induction of the epithelial-to-mesenchymal transition (Ying et al., 2012; Li G. et al., 2014; Yang et al., 2017). The role of Malat1 in several pediatric cancers has also been explored in recent studies. In neuroblastoma, Tee et al. (2014) recently identified a regulatory network involving N-Myc, Malat1, and the histone demethylase JMJD1A. They found that N-Myc upregulated JMJD1A via direct binding of its promoter region and that JMJD1A could demethylate histone H3K9 near the promoter region of Malat1, leading to its upregulation. MYCN-mediated upregulation of Malat1 provides one mechanism in which its amplification can lead to increased metastasis in neuroblastoma patients. Another study by Bi et al. (2017) also demonstrated that Malat1 regulated Axl expression, a transmembrane receptor tyrosine kinase, which is known to activate pathways involved in cell proliferation, survival, and migration. In osteosarcoma, Dong et al. (2015) demonstrated that Malat1 was highly expressed and could activate the PI3K/Akt pathway to promote proliferation and invasion.
Malat1 is known to interact with many miRNAs implicated in cancer. In osteosarcoma, several studies have shown Malat1 can function as a ceRNA for different miRNAs (Wang et al., 2017b; Liu K. et al., 2017b; Sun and Qin, 2018). miR-140-5p is a tumor suppressor that downregulates HDAC4, a histone deacetylase that contributes to tumorigenesis, and competitive binding by Malat1 with miR-140-5p was shown to increase HDAC4 activity (Sun and Qin, 2018). Malat1 was also shown to compete with miR-144-3p binding to ROCK1/ROCK2, promoting proliferation and metastasis (Wang et al., 2017b). In a similar manner, Liu K. et al. (2017) found that Malat1 could regulate cell growth through high-mobility group protein B1 (HMGB1) via ceRNA activity with miR-142-3p and miR-129-5p. Finally, in retinoblastoma, Malat1 downregulated miR-124 activity, leading to activation of the transcription factor SLUG, which is also targeted by miR-124 (Liu S. et al., 2017). SLUG has a known role in the epithelial-to-mesenchymal transition by suppressing E-cadherin via the Wnt/B-catenin pathway (Prasad et al., 2009).
In addition to interactions with miRNAs, Malat1 has also been shown to be processed directly by the Drosha–DGCR8 microprocessor complex through binding sites in the 5’ end of the transcript (MacIas et al., 2012). lncRNAs such as Malat1 cooperate with the miRNA pathway and a number of transcription factors and epigenetic factors to form a complex network responsible for regulating tumorigenesis. The capacity for Malat1 to drive proliferation and metastasis in pediatric solid tumors suggests that dysregulation of any of these regulatory components can be sufficient for the development of cancer and highlights the value of further research into the relatively new field of lncRNAs.
H19: lncRNA Dysregulation via Loss of Imprinting may Contribute to Tumorigenesis
H19 is a paternally imprinted gene that is typically expressed exclusively from the maternal allele. Early reports suggested that H19 functioned as a tumor suppressor capable of inhibiting cell growth (Hao et al., 1993; Zhang et al., 1993; Casola et al., 1997; Fukuzawa et al., 1999). Studies in childhood solid tumors such as hepatoblastoma, Wilms tumor, and embryonic rhabdomyosarcoma supported this idea, as all three cancers often exhibited reduced H19 expression and had frequently lost the maternal 11p15 chromosomal region housing this gene (Fukuzawa et al., 1999). Other studies, in osteosarcoma and retinoblastoma, suggested an oncogenic role for H19, as its upregulation and loss of imprinting were commonly seen (Chan et al., 2014; Li L. et al., 2018). This observation was also seen in many other cancers including breast cancer (Lottin et al., 2002). Recently, the Hedgehog signaling pathway, a regulator of differentiation known to participate in cancer development and metastasis, was shown to induce H19 expression (Chan et al., 2014).
Understanding the exact function of H19 has proved difficult; however, it was known to sit downstream of the insulin growth factor 2 (IGF2) gene, a growth factor known to play a role in tumorigenesis. Early reports suggested interactions between IGF2 and H19, as loss of imprinting of either gene caused biallelic expression of the other gene (Ulaner et al., 2003). Ulaner et al. (2003) proposed a model for H19 and IGF2 involving a CCTF-binding site seated between the two genes, which could facilitate the blocking of IGF2 or transcription of H19 depending on its methylation status. However, this model suggested that H19 may simply serve as a marker for epigenetic disruptions and left open the question of what H19’s actual function is.
More recent studies have demonstrated a role for H19 in epigenetic regulation. H19 binds to several epigenetic regulators including S-adenosylhomocysteine hydrolase (SAHH), methyl-CpG–binding domain protein 1 (MBD1), and enhancer of zeste homolog 2 (EZH2) (Raveh et al., 2015; Zhou et al., 2015). H19 was found to inhibit SAHH, which led to downregulation of DNMT3B-mediated methylation. MBD1 binds methylated DNA and recruits other proteins to mediate transcriptional repression or histone methylation, and H19 was shown to recruit this protein to several genes including IGF2 (Monnier et al., 2013). Finally, EZH2 is a histone methyltransferase that forms part of the Polycomb repressive complex 2 (PRC2) (Sauliere et al., 2006; Zhou et al., 2015).
H19 also plays a role in maintaining cells in an undifferentiated state by associating with the KH-type splicing regulatory protein (KSRP). When multipotent mesenchymal cells were induced, H19 was found to dissociate with KSRP to promote several of its functions including the decay of unstable mRNAs and increasing the expression of specific miRNAs involved in proliferation and differentiation though association with Drosha and Dicer.
H19’s role in cancer has been emphasized by studies highlighting its relationship to the tumor suppressor p53. The H19 locus reciprocally regulates p53, as p53 suppresses H19 transcription and H19 can inactivate p53 by directly interacting with it (Yang et al., 2012). Notably, H19 also encodes for a miRNA in its first exon, miR-675, which suppresses p53 and several other targets including Rb, Igf1r, and several SMAD and cadherin genes. In the absence of functional p53, H19 was shown to promote tumor proliferation and survival under hypoxic conditions. Later studies in colorectal cancer showed that H19 could induce EMT by acting as a ceRNA (Liang et al., 2015). ceRNA function has recently been shown in a retinoblastoma study, targeting the mir-17∼92 cluster (Zhang A. et al., 2018a). In this study, they found that H19 contained seven functional binding sites for mir-17∼92 and was able to sponge mir-17∼92 activity. This led to a de-repression of genes such as p21 and STAT3 targets BCL2, BCL2L1, and BIRC5.
In a review by Raveh et al. (2015) it was proposed that H19 may behave differently in a manner that was dependent on the developmental stage of the cell, which could explain the evidence suggesting both oncogenic and tumor-suppressive roles. Here, the authors found that H19 functioned as a promoter of differentiation during the embryonic period and that absence of H19 at this stage could leave cells vulnerable to forming cancer, thereby seemingly acting as a tumor suppressor. However, in adult cells, where it is not normally expressed, H19 could function as an oncogene by promoting tumor survival and metastasis (Matouk et al., 2015).
TUG1 Regulates Transcription Factors Through Competition With miRNAs in Osteosarcoma
Recent studies have investigated the role of lncRNA TUG1 as a prognostic factor and ceRNA in osteosarcoma. Ma et al. (2016)identified a correlation between upregulation of TUG1 and poor prognosis and metastasis, which was also evident in plasma, and suggested a potential use as a biomarker for patients with osteosarcoma. TUG1 is known to act through ceRNA activity against a number of miRNAs including miR-9, miR-132, miR-144, miR-153, miR-212, and miR-335 (Xie et al., 2016; Cao et al., 2017; Wang et al., 2017a; Li G. et al., 2018; Li H. et al., 2018). These miRNAs are known to regulate pathways involved in proliferation, cell cycle control, migration, and apoptosis. For example, TUG1 was shown to mediate de-repression of the transcription factor POU class 2 homeobox1 (POU2F1) via downregulation of mir-9 (Figure 1E) (Xie et al., 2016). POU2F1 itself participates in various cellular processes including growth, metabolism, stem cell identity, and metastasis (Vázquez-Arreguín and Tantin, 2016). In another example by Cao et al. (2017) they found that TUG1 also regulates migration and the epithelial-to-mesenchymal transition via ceRNA action on miRNA-144-3p. miR-144-3p is a regulator of EZH2, and upregulation of EZH2 induced cell migration through the Wnt/β-catenin pathway (Cao et al., 2017). Studies have also demonstrated direct interactions between TUG1 and the Polycomb repressor complex; however, to our knowledge, this has not been investigated in pediatric solid tumors (Yang et al., 2011).
Other Long Non-Coding RNAs in Pediatric Solid Tumors
In addition to those mentioned above, there are a number of other lncRNAs that have been identified as potential oncogenes or tumor suppressors involved in the pathogenesis of pediatric solid tumors (see Table 2) (Chen et al., 2017; Pandey et al., 2015).
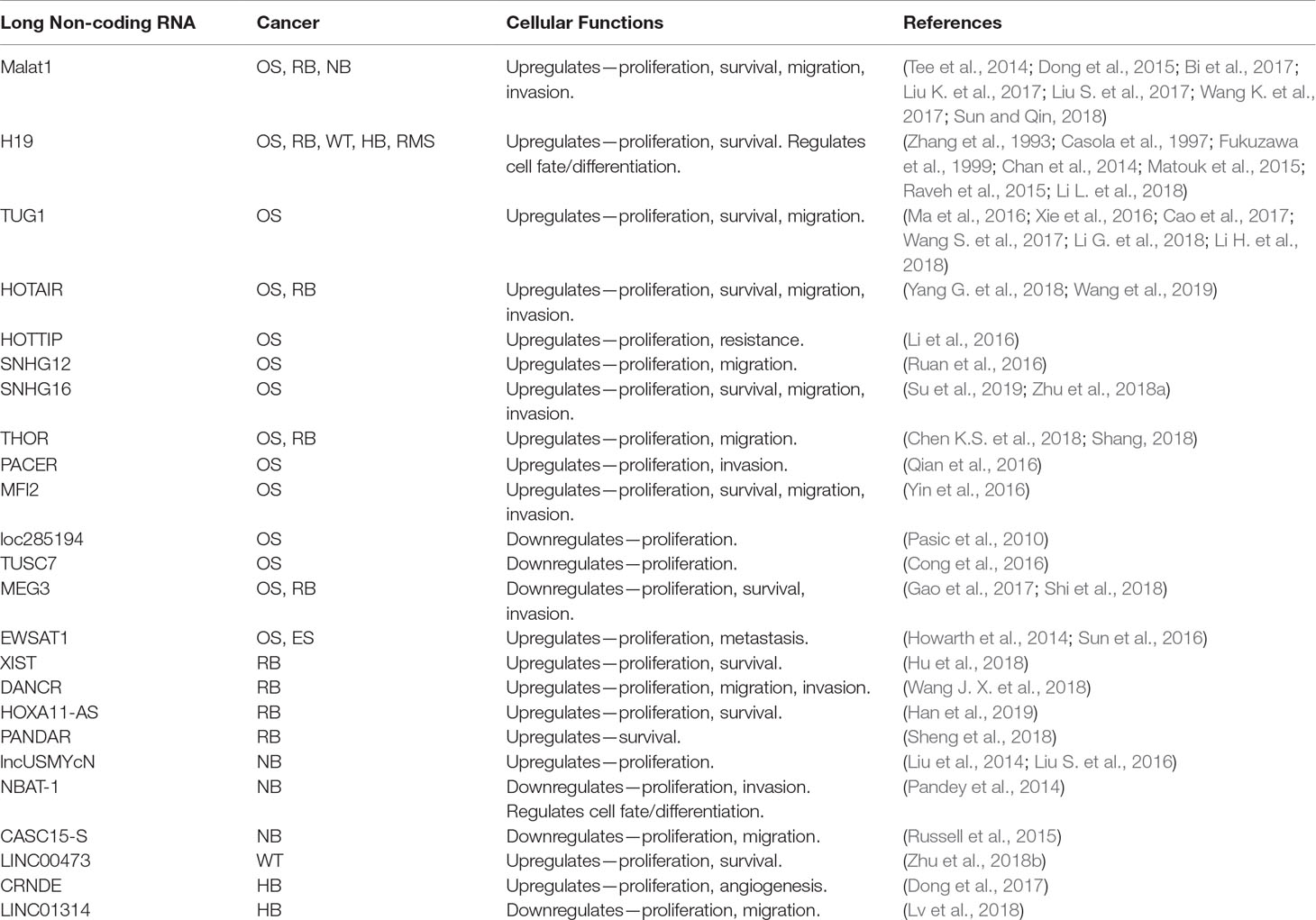
Table 2 lncRNAs that play a role in pediatric solid tumors. OS, osteosarcoma; RB, retinoblastoma; NB, neuroblastoma; WT, Wilms tumor; HB, hepatoblastoma; RMS, rhabdomyosarcoma; ES, Ewing sarcoma.
For example, in osteosarcoma, lncRNAs HOTAIR, SNHG16, SNHG12, THOR, PACER, MFI2, and HOTTIP have all been shown to promote tumor or cell growth (Li et al., 2016; Qian et al., 2016; Ruan et al., 2016; Yin et al., 2016; Chen W. et al., 2018; Su et al., 2019; Wang et al., 2019). HOTAIR is known to play a role in chromatin regulation by acting as a scaffold for PRC2 and lysine-specific histone demethylase 1 (LSD1), and can also act as a ceRNA for miR-217 (Gupta et al., 2010; Tsai et al., 2010; Wang et al., 2019). SNHG16 has been shown to act as a ceRNA for several miRNAs including miR-205 and miR-340 (Zhu C. et al., 2018; Su et al., 2019). Additionally, several lncRNAs are downregulated in osteosarcoma with potential tumor-suppressive activity such as loc285194, MEG3, and TUSC7 (Pasic et al., 2010; Cong et al., 2016; Shi et al., 2018). loc285194 has been identified as a transcriptional target of p53 and can downregulate miR-211 (Liu Q. et al., 2013). In another study, increased p53 expression and a decrease in cell proliferation and invasion were observed when MEG3 was overexpressed (Shi et al., 2018). Furthermore, MEG3 was found to be downregulated by another lncRNA, EWSAT1, which had previously been shown to enhance cell proliferation and metastasis in both osteosarcoma and Ewing sarcoma (Howarth et al., 2014; Sun et al., 2016).
In retinoblastoma, HOTAIR, THOR, and MEG3 appear to have a similar influence as seen in osteosarcoma, where they also acted as oncogenes (HOTAIR and THOR) or tumor suppressors (MEG3) (Gao et al., 2017; Shang, 2018; Yang G. et al., 2018). In the study examining HOTAIR in retinoblastoma, HOTAIR was shown to be engaged in a reciprocal regulatory loop with miR-613 and promoted cell proliferation and activation of the EMT, potentially through upregulation of N-cadherin, vimentin, and α‐SMA (Yang G. et al., 2018). Several lncRNAs have also been found acting as oncogenic ceRNAs including XIST, DANCR, and HOXA11-AS (Hu et al., 2018; Wang J. X. et al., 2018; Han et al., 2019). Finally, PANDAR is upregulated in retinoblastoma and may regulate cell proliferation and apoptosis via the Bcl-2/caspase-3 pathway (Sheng et al., 2018).
A number of studies have also suggested an important role for lncRNAs in neuroblastoma. For example, lncUSMYcN is an lncRNA that is frequently co-amplified alongside MYCN (Liu P. Y. et al., 2016). Liu et al. found that in neuroblastoma, lncUSMycN could upregulate MYCN through transcriptional activation of NCYM (a.k.a. MYCNOS), which codes for a protein that stabilizes MYCN (Suenaga et al., 2014). NCYM RNA has also been suggested to bind to the RNA-binding protein NonO, which is also known to upregulate MYCN expression (Liu et al., 2014; Liu P. Y. et al., 2016). Neuroblastoma associated transcript-1 (NBAT-1) is an epigenetic regulator that interacts with EZH2, and functions as a tumor suppressor due to its important role in neuronal differentiation (Pandey et al., 2014). Loss of NBAT-1 expression was found to increase cell proliferation and invasion (Pandey et al., 2014). Finally, an isoform of lncRNA CASC15, CASC15-S, was also implicated as a key element in neuronal differentiation, and low expression was associated with a poor outcome in patients (Russell et al., 2015).
In Wilms tumor, a study by Zhu et al. identified LINC00473 as an oncogenic lncRNA that is upregulated in unfavorable tumors (Zhu et al., 2018b). LINC00473 was shown to promote tumor growth and metastasis by acting as a ceRNA for the tumor suppressor miR-195 (Zhu et al., 2018b).
A study by Dong et al. identified 1757 upregulated and 979 downregulated lncRNAs comparing hepatoblastoma and normal tissues, suggesting that lncRNAs play a key role in this disease as well (Dong et al., 2014). The lncRNAs Colorectal Neoplasia Differentially Expressed (CRNDE) and LINC01314 have been investigated in more detail in hepatoblastoma (Dong et al., 2017; Lv et al., 2018). CRNDE is known to be frequently upregulated in hepatoblastoma, and knockdown of CRNDE activated the mTOR pathway and inhibited tumor growth and angiogenesis with a corresponding decrease in VEGFA and Ang-2 levels (Dong et al., 2017). LINC01314 was identified as a tumor suppressor, reducing proliferation and migration via downregulation of cell cycle proteins MCM7 and cyclin D1 (Lv et al., 2018).
Concluding Remarks
It is now clear that both miRNAs and lncRNAs form integral parts of the biological networks known to be impaired in pediatric solid tumors. miRNAs such as let-7 and mir-34 are key regulators of many pediatric oncogenes including MYC, MYCN, RAS, and MET (Johnson et al., 2005; Wei et al., 2008; Buechner et al., 2011; Yan et al., 2012). Additionally, ncRNAs such as the miR-17∼92 cluster, mir-9, and Malat1 also serve as downstream effectors of MYC and MYCN (Schulte et al., 2008; Ma et al., 2010; Tee et al., 2014). Many more ncRNAs participate in these and other pathways to form a highly complex regulatory network essential for maintaining an optimal cell state (See Tables 1 and 2). ncRNA dysregulation offers an alternative mechanism to genetic mutations and DNA methylation whereby cell development and differentiation can be disturbed. Despite the relatively rare occurrence of mutations in pediatric solid tumors, copy number variations are common and often occur at regions of the genome that harbor ncRNAs with tumor-suppressive roles (Wei et al., 2008; Powers et al., 2016). Gene expression is often imprecise; however, miRNAs provide a layer of robustness, which helps ensure that biological networks respond appropriately to signals and remain functional despite an ever-increasing cellular disorder (Ebert and Sharp, 2012). lncRNAs, too, play a vital role in maintaining order by forming RNA–protein complexes and serving as ceRNA antagonists against miRNA-mediated repression, although much more work is needed in this field to fully comprehend their range of biological roles. Functional studies have revealed that dysregulation of ncRNAs is capable of driving progenitor cells towards oncogenesis. For example, this has been shown in retinoblastoma, where overexpression of the mir-17∼92 cluster could drive tumor formation in RB/p107-deficient mice (Conkrite et al., 2011).
While genome-wide association studies have revealed that miRNA processing is frequently disrupted in Wilms tumor, this has not been shown to the same extent in other pediatric solid tumors. However, genetic mutations of protein-coding genes are only one way in which disruptions of miRNA processing can be revealed. Most miRNA studies ignore the fact that a high proportion of expressed miRNAs are isoforms (isomiRs). isomiRs originating from the same miRNA gene can possess a great deal of functional variability, with differences in target acquisition or turnover rate that can have a significant impact on overall gene regulation. Studies focusing on isomiR expression will provide an additional layer of resolution to our understanding of miRNA dysregulation.
Recent developments in single-cell technology have revealed heterogeneity in gene expression profiles among individual cells in many cancers such as glioblastoma and neuroblastoma (Patel et al., 2014; Boeva et al., 2017). Such studies suggest that many tumors comprise different cellular subtypes with unique phenotypes such as growth rate, drug resistance, and metastatic potential, which demand a new way of approaching cancer treatments. miRNA expression in pediatric solid tumors may also be heterogenous; however, limitations in single-cell technologies have left this avenue relatively unexplored, and further developments are needed.
So far, ncRNA research has played a key role in advancing our understanding of the mechanisms behind pediatric solid tumor development. Evidence supports an active role for ncRNAs in cancer that extends beyond mere passengers. However, continued research is needed to fully comprehend the molecular events leading to the development of cancer and unlock new possibilities for drug targets and biomarkers, which will ultimately lead to a better outcome for patients afflicted by these diseases.
Author Contributions
CS collected the information and wrote the review. DC and GH provided guidelines, consulted, and edited the manuscript.
Funding
This work was supported by the Australian Research Council DP180100120 project grant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adhikari, A. S., Agarwal, N., Wood, B. M., Porretta, C., Ruiz, B., Pochampally, R. R., et al. (2010). CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 70, 4602–4612. doi: 10.1158/0008-5472.CAN-09-3463
Allen-Rhoades, W., Whittle, S. B., Rainusso, N. (2018). Pediatric solid tumors of infancy: an overview. Pediatr. Rev. 39, 57–67. doi: 10.1542/pir.2017-0057
Attiyeh, E. F., London, W. B., Mossé, Y. P., Wang, Q., Winter, C., Khazi, D., et al. (2005). Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 353, 2243–2253. doi: 10.1056/NEJMoa052399
Bai, S., Tian, B., Li, A., Yao, Q., Zhang, G., Li, F. (2016). MicroRNA-125b promotes tumor growth and suppresses apoptosis by targeting DRAM2 in retinoblastoma. Eye (Lond.) 30, 1630–1638. doi: 10.1038/eye.2016.189
Balzeau, J., Menezes, M. R., Cao, S., Hagan, J. P. (2017). The LIN28/let-7 pathway in cancer. Front. Genet. 8, 1–16. doi: 10.3389/fgene.2017.00031
Band, A. M., Laiho, M. (2011). Crosstalk of TGF-β and estrogen receptor signaling in breast cancer. J. Mammary Gland Biol. Neoplasia 16, 109–115. doi: 10.1007/s10911-011-9203-7
Baumhoer, D., Zillmer, S., Unger, K., Rosemann, M., Atkinson, M. J., Irmler, M., et al. (2012). MicroRNA profiling with correlation to gene expression revealed the oncogenic miR-17-92 cluster to be up-regulated in osteosarcoma. Cancer Genet. 205, 212–219. doi: 10.1016/j.cancergen.2012.03.001
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. doi: 10.1038/emboj.2010.199
Bersani, F., Lingua, M. F., Morena, D., Foglizzo, V., Miretti, S., Lanzetti, L., et al. (2016). Deep sequencing reveals a novel miR-22 regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Res. 76, 6095–6106. doi: 10.1158/0008-5472.CAN-16-0709
Bi, S., Wang, C., Li, Y., Zhang, W., Zhang, J., Lv, Z., et al. (2017). LncRNA-MALAT1–mediated Axl promotes cell invasion and migration in human neuroblastoma. Tumor Biol. 39, 1–7. doi: 10.1177/1010428317699796
Bian, S., Hong, J., Li, Q., Schebelle, L., Pollock, A., Knauss, J. L., et al. (2013). MicroRNA Cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 3, 1398–1406. doi: 10.1016/j.celrep.2013.03.037
Blenkiron, C., Goldstein, L. D., Thorne, N. P., Spiteri, I., Chin, S.-F., Dunning, M. J., et al. (2007). MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 8, R214. doi: 10.1186/gb-2007-8-10-r214
Boeva, V., Louis-Brennetot, C., Peltier, A., Durand, S., Pierre-Eugène, C., Raynal, V., et al. (2017). Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 49, 1408–1413. doi: 10.1038/ng.3921
Bonev, B., Pisco, A., Papalopulu, N. (2011). MicroRNA-9 reveals regional diversity of neural progenitors along the anterior–posterior axis. Dev. Cell. 20, 19–32. doi: 10.1016/j.devcel.2010.11.018
Brannan, C. I., Dees, E. C., Ingram, R. S., Tilghman, S. M. (1990). The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 10, 28–36. doi: 10.1128/MCB.10.1.28
Bray, I., Tivnan, A., Bryan, K., Foley, N. H., Watters, K. M., Tracey, L., et al. (2011). MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma. Cancer Lett. 303, 56–64. doi: 10.1016/j.canlet.2011.01.016
Brunner, A. L., Beck, A. H., Edris, B., Sweeney, R. T., Zhu, S. X., Li, R., et al. (2012). Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 13, R75. doi: 10.1186/gb-2012-13-8-r75
Buechner, J., Tømte, E., Haug, B. H., Henriksen, J. R., Løkke, C., Flægstad, T., et al. (2011). Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer 105, 296–303. doi: 10.1038/bjc.2011.220
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S A. 99, 15524–15529. doi: 10.1073/pnas.242606799
Calin, G. A., Sevignani, C., Dumitru, C. D., Hyslop, T., Noch, E., Yendamuri, S., et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. 101, 2999–3004. doi: 10.1073/pnas.0307323101
Cao, J., Han, X., Qi, X., Jin, X., Li, X. (2017). TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via MIR-144-3p. Int. J. Oncol. 51, 1115–1123. doi: 10.3892/ijo.2017.4110
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). Molecular biology: the transcriptional landscape of the mammalian genome. Science 309, 1559–1563. doi: 10.1126/science.1112014
Caron, H., van Sluis, P., van Hoeve, M., de Kraker, J., Bras, J., Slater, R., et al. (1993). Allelic loss of chromosome 1p36 in neuroblastoma is of preferential maternal origin and correlates with N-myc amplification. Nat. Genet. 4, 187–190. doi: 10.1038/ng0693-187
Casola, S., Pedone, P. V., Cavazzana, A. O., Basso, G., Luksch, R., D'Amore, E. S. G., et al. (1997). Expression and parental imprinting of the H19 gene in human rhabdomyosarcoma. Oncogene 14, 1503–1510. doi: 10.1038/sj.onc.1200956
Cech, T. R., Steitz, J. A. (2014). The noncoding RNA revolution—trashing old rules to forge new ones. Cell 157, 77–94. doi: 10.1016/j.cell.2014.03.008
Ceppi, P., Peter, M. E. (2014). MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene 33, 269–278. doi: 10.1038/onc.2013.55
Chakrabarti, M., Khandkar, M., Banik, N. L., Ray, S. K. (2012). Alterations in expression of specific microRNAs by combination of 4-HPR and EGCG inhibited growth of human malignant neuroblastoma cells. Brain Res. 1454, 1–13. doi: 10.1016/j.brainres.2012.03.017
Chan, L. H., et al. (2014). Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene 33, 4857–4866. doi: 10.1038/onc.2013.433
Cheetham, S. W., Gruhl, F., Mattick, J. S., Dinger, M. E. (2013). Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 108, 2419–2425. doi: 10.1038/bjc.2013.233
Chen, Y., Stallings, R. L. (2007). Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 67, 976–983. doi: 10.1158/0008-5472.CAN-06-3667
Chen, G., Zhou, H. (2018). MiRNA-708/CUL4B axis contributes into cell proliferation and apoptosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 22, 5452–5459. doi: 10.26355/eurrev_201809_15805
Chen, H. Z., Tsai, S. Y., Leone, G. (2009). Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9, 785–797. doi: 10.1038/nrc2696
Chen, J., Huang, Z.-P., Seok, H. Y., Ding, J., Kataoka, M., Zhang, Z., et al. (2013). mir-17–92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 112, 1557–1566. doi: 10.1161/CIRCRESAHA.112.300658
Chen, X., Pappo, A., Dyer, M. A. (2015). Pediatric solid tumor genomics and developmental pliancy. Oncogene 34, 5207–5215. doi: 10.1038/onc.2014.474
Chen, L., Feng, P., Zhu, X., He, S., Duan, J., Zhou, D. (2016). Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J. Cell. Mol. Med. 20, 2102–2110. doi: 10.1111/jcmm.12904
Chen, R., Wang, G., Zheng, Y., Hua, Y., Cai, Z. (2017). Long non-coding RNAs in osteosarcoma. Oncotarget 8, 20462–20475. doi: 10.18632/oncotarget.14726
Chen, K. S., Stroup, E. K., Budhipramono, A., Rakheja, D., Nichols-Vinueza, D., Xu, L., et al. (2018). Mutations in microRNA processing genes in Wilms tumors derepress the IGF2 regulator PLAG1. Genes Dev. 32, 996–1007. doi: 10.1101/gad.313783.118
Chen, W., Chen, M., Xu, Y., Chen, X., Zhou, P., Zhao, X., et al. (2018). Long non-coding RNA THOR promotes human osteosarcoma cell growth in vitro and in vivo. Biochem. Biophys. Res. Commun. 499, 913–919. doi: 10.1016/j.bbrc.2018.04.019
Classon, M., Harlow, E. (2002). The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2, 910–917. doi: 10.1038/nrc950
Cole, K. A., Attiyeh, E. F., Mosse, Y. P., Laquaglia, M. J., Diskin, S. J., Brodeur, G. M., et al. (2008). A Functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 6, 735–742. doi: 10.1158/1541-7786.MCR-07-2102
Cong, M., Li, J., Jing, R., Li, Z. (2016). Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumor Biol. 37, 9441–9450. doi: 10.1007/s13277-015-4414-y
Conkrite, K., Sundby, M., Mukai, S., Michael Thomson, J., Mu, D., Hammond, S. M., et al. (2011). Mir-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 25, 1734–1745. doi: 10.1101/gad.17027411
Coolen, M., Katz, S., Bally-Cuif, L. (2013). miR-9: a versatile regulator of neurogenesis. Front. Cell. Neurosci. 7, 1–11. doi: 10.3389/fncel.2013.00220
De Antonellis, P., Carotenuto, M., Vandenbussche, J., De Vita, G., Ferrucci, V., Medaglia, C., et al. (2014). Early targets of miR-34a in neuroblastoma. Mol. Cell. Proteomics 13, 2114–2131. doi: 10.1074/mcp.M113.035808
De Preter, K., Mestdagh, P., Vermeulen, J., Zeka, F., Naranjo, A., Bray, I., et al. (2011). miRNA expression profiling enables risk stratification in archived and fresh neuroblastoma tumor samples. Clin. Cancer Res. 17, 7684–7692. doi: 10.1158/1078-0432.CCR-11-0610
De Vito, C., Riggi, N., Cornaz, S., Suvà, M.-L., Baumer, K., Provero, P., et al. (2012). A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell 21, 807–821. doi: 10.1016/j.ccr.2012.04.023
Delattre, O., Zucman, J., Melot, T., Garau, X. S., Zucker, J.-M., Lenoir, G. M., et al. (1994). The Ewing family of tumors — a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N. Engl. J. Med. 331, 294–299. doi: 10.1056/NEJM199408043310503
Di Fiore, R., Drago-Ferrante, R., Pentimalli, F., Di Marzo, D., Forte, I. M., Carlisi, D., et al. (2016). Let-7d miRNA shows both antioncogenic and oncogenic functions in osteosarcoma-derived 3AB-OS cancer stem cells. J. Cell. Physiol. 231, 1832–1841. doi: 10.1002/jcp.25291
Dong, R., Jia, D., Xue, P., Cui, X., Li, K., Zheng, S., et al (2014). Genome-wide analysis of long noncoding RNA (lncRNA) expression in hepatoblastoma tissues. PLoS One 9, 1–9. doi: 10.1371/journal.pone.0085599
Dong, Y., Liang, G., Yuan, B., Yang, C., Gao, R., Zhou, X. (2015). MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 36, 1477–1486. doi: 10.1007/s13277-014-2631-4
Dong, J., Liu, Y., Liao, W., Liu, R., Shi, P., Wang, L. (2016). miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J. Bone Oncol. 5, 74–79. doi: 10.1016/j.jbo.2016.05.001
Dong, R., Liu, X.-Q., Zhang, B.-B., Liu, B.-H., Zheng, S., Dong, K.-R. (2017). Long non-coding RNA-CRNDE: a novel regulator of tumor growth and angiogenesis in hepatoblastoma. Oncotarget 8, 42087–42097. doi: 10.18632/oncotarget.14992
Du, K., Zhang, X., Lou, Z., Guo, P., Zhang, F., Wang, B., et al. (2018). MicroRNA485-3p negatively regulates the transcriptional co-repressor CtBP1 to control the oncogenic process in osteosarcoma cells. Int. J. Biol. Sci. 14, 1445–1456. doi: 10.7150/ijbs.26335
Ebert, M. S., Sharp, P. A. (2012). Roles for MicroRNAs in conferring robustness to biological processes. Cell 149, 505–524. doi: 10.1016/j.cell.2012.04.005
Esquela-Kerscher, A., Slack, F. J. (2006). Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. doi: 10.1038/nrc1840
Feng, T., Xu, D., Tu, C., Li, W., Ning, Y., Ding, J., et al. (2015). miR-124 inhibits cell proliferation in breast cancer through downregulation of CDK4. Tumor Biol. 36, 5987–5997. doi: 10.1007/s13277-015-3275-8
Foley, N. H., Bray, I. M., Tivnan, A., Bryan, K., Murphy, D. M., Buckley, P. G., et al. (2010). MicroRNA-184 inhibits neuroblastoma cell survival through targeting the serine/threonine kinase AKT2. Mol. Cancer 9, 83. doi: 10.1186/1476-4598-9-83
Foley, N. H., Bray, I., Watters, K. M., Das, S., Bryan, K., Bernas, T., et al. (2011). MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 18, 1089–1098. doi: 10.1038/cdd.2010.172
Fontana, L., Fiori, M. E., Albini, S., Cifaldi, L., Giovinazzi, S., Forloni, M., et al. (2008). Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One 3, e2236. doi: 10.1371/journal.pone.0002236
Franzetti, G.-A., Laud-Duval, K., Bellanger, D., Stern, M.-H., Sastre-Garau, X., Delattre, O. (2013). MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene 32, 3915–3921. doi: 10.1038/onc.2012.403
Friend, S. H., Bernards, R., Rogelj, S., Weinberg, R. A., Rapaport, J. M., Albert, D. M., et al. (1986). A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323, 643–646. doi: 10.1038/323643a0
Fukuzawa, R., Umezawa, A., Ochi, K., Urano, F., Ikeda, H., Hata, J. I. (1999). High frequency of inactivation of the imprinted H19 gene in ‘sporadic’ hepatoblastoma. Int. J. Cancer 82, 490–497. doi: 10.1002/(SICI)1097-0215(19990812)82:4<490::AID-IJC4>3.0.CO;2-I
Gadd, S., Huff, V., Walz, A. L., Ooms, A. H.A.G., Armstrong, A. E., Gerhard, D. S., et al. (2017). A Children’s Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat. Genet. 49, 1487–1494. doi: 10.1038/ng.3940
Gang, L., Qun, L., Liu, W.-D., Li, Y.-S., Xu, Y.-Z., Yuan, D.-T. (2017). MicroRNA-34a promotes cell cycle arrest and apoptosis and suppresses cell adhesion by targeting DUSP1 in osteosarcoma. Am. J. Transl. Res. 9, 5388–5399.
Gao, Y., Huang, P., Zhang, J. (2017). Hypermethylation of MEG3 promoter correlates with inactivation of MEG3 and poor prognosis in patients with retinoblastoma. J. Transl. Med. 15, 1–10. doi: 10.1186/s12967-017-1372-8
Guo, C., White, P. S., Weiss, M. J., Hogarty, M. D., Thompson, P. M., Stram, D. O., et al. (1999). Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene 18, 4948–4957. doi: 10.1038/sj.onc.1202887
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. doi: 10.1038/nature08975
Ha, M., Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524. doi: 10.1038/nrm3838
Hameiri-Grossman, M., Porat-Klein, A., Yaniv, I., Ash, S., Cohen, I. J., Kodman, Y., et al. (2015). The association between let-7, RAS and HIF-1α in Ewing sarcoma tumor growth. Oncotarget 6, 33834–33848. doi: 10.18632/oncotarget.5616
Han, K., Chen, X., Bian, N., Ma, B., Yang, T., Cai, C., et al. (2015). MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget 6, 8875–8889. doi: 10.18632/oncotarget.3560
Han, N., Zuo, L., Chen, H., Zhang, C., He, P., Yan, H. (2019). Long non-coding RNA homeobox A11 antisense RNA (HOXA11-AS) promotes retinoblastoma progression via sponging miR-506-3p. Onco. Targets Ther. 12, 3509–3517. doi: 10.2147/OTT.S195404
Hao, Y., Crenshaw, T., Moulton, T., Newcomb, E., Tycko, B. (1993). Tumour-suppressor activity of H19 RNA. Nature 365, 764–767. doi: 10.1038/365764a0
Hennchen, M., Stubbusch, J., Abarchan-El Makhfi, I., Kramer, M., Deller, T., Pierre-Eugene, C., et al. (2015). Lin28B and Let-7 in the control of sympathetic neurogenesis and neuroblastoma development. J. Neurosci. 35, 16531–16544. doi: 10.1523/JNEUROSCI.2560-15.2015
Hermeking, H. (2010). The miR-34 family in cancer and apoptosis. Cell Death Differ. 17, 193–199. doi: 10.1038/cdd.2009.56
Howarth, M. M., Simpson, D., Ngok, S. P., Nieves, B., Chen, R., Siprashvili, Z., et al. (2014). Long noncoding RNA EWSAT1-mediated gene repression facilitates Ewing sarcoma oncogenesis. J. Clin. Invest. 124, 5275–5290. doi: 10.1172/JCI72124
Hu, C., Liu, S., Han, M., Wang, Y., Xu, C. (2018). Knockdown of lncRNA XIST inhibits retinoblastoma progression by modulating the miR-124/STAT3 axis. Biomed. Pharmacother. 107, 547–554. doi: 10.1016/j.biopha.2018.08.020
Huang, M., Weiss, W. A. (2013). Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 3, a014415. doi: 10.1101/cshperspect.a014415
Huang, Y., Shen, X. J., Zou, Q., Wang, S. P., Tang, S. M., Zhang, G. Z. (2011). Biological functions of microRNAs: a review. J. Physiol. Biochem. 67, 129–139. doi: 10.1007/s13105-010-0050-6
Huang, G., Nishimoto, K., Zhou, Z., Hughes, D., Kleinerman, E. S. (2012). miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res. 72, 908–916. doi: 10.1158/0008-5472.CAN-11-1460
Iorio, M. V., Ferracin, M., Liu, C. G., Veronese, A., Spizzo, R., Sabbioni, S., et al. (2005). MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070. doi: 10.1158/0008-5472.CAN-05-1783
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041. doi: 10.1038/sj.onc.1206928
Jiang, X., Li, H. (2018). MiR-1180-5p regulates apoptosis of Wilms’ tumor by targeting p73. Onco. Targets. Ther. 11, 823–831. doi: 10.2147/OTT.S148684
Jiao, C., Zhu, A., Jiao, X., Ge, J., Xu, X. (2016). Combined low miR-34s are associated with unfavorable prognosis in children with hepatoblastoma: a Chinese population-based study. J. Pediatr. Surg. 51, 1355–1361. doi: 10.1016/j.jpedsurg.2016.02.091
Jo, D. H., Kim, J. H., Cho, C. S., Cho, Y.-L., Jun, H. O., Yu, Y. S., et al. (2014). STAT3 inhibition suppresses proliferation of retinoblastoma through down-regulation of positive feedback loop of STAT3/miR-17-92 clusters. Oncotarget 5, 11513–11525. doi: 10.18632/oncotarget.2546
Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., et al. (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647. doi: 10.1016/j.cell.2005.01.014
Karnuth, B., Dedy, N., Spieker, T., Lawlor, E. R., Gattenlöhner, S., Ranft, A., et al. (2014). Differentially expressed miRNAs in Ewing sarcoma compared to mesenchymal stem cells: low miR-31 expression with effects on proliferation and invasion. PLoS One 9, e93067. doi: 10.1371/journal.pone.0093067
Kawano, M., Tanaka, K., Itonaga, I., Iwasaki, T., Tsumura, H. (2015). c-Myc represses tumor-suppressive microRNAs, let-7a, miR-16 and miR-29b, and induces cyclin D2–mediated cell proliferation in Ewing’s sarcoma cell line. PLoS One 10, e0138560. doi: 10.1371/journal.pone.0138560
Kawano, M., Tanaka, K., Itonaga, I., Iwasaki, T., Tsumura, H. (2017). MicroRNA-20b promotes cell proliferation via targeting of TGF-β receptor II and upregulates MYC expression in Ewing’s sarcoma cells. Int. J. Oncol. 51, 1842–1850. doi: 10.3892/ijo.2017.4155
Kline, N. E., Sevier, N. (2003). Solid tumors in children. J. Pediatr. Nurs. 18, 96–102. doi: 10.1053/jpdn.2003.12
Koralov, S. B., Muljo, S. A., Galler, G. R., Krek, A., Chakraborty, T., Kanellopoulou, C., et al. (2008). Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132, 860–874. doi: 10.1016/j.cell.2008.02.020
Kort, E. J., Farber, L., Tretiakova, M., Petillo, D., Furge, K. A., Yang, X. J., et al. (2008). The E2F3–oncomir-1 axis is activated in Wilms’ tumor. Cancer Res. 68, 4034–4038. doi: 10.1158/0008-5472.CAN-08-0592
Laneve, P., Di Marcotullio, L., Gioia, U., Fiori, M. E., Ferretti, E., Gulino, A., et al. (2007). The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc. Natl. Acad. Sci. 104, 7957–7962. doi: 10.1073/pnas.0700071104
Le, M. T. N., Xie, H., Zhou, B., Chia, P. H., Rizk, P., Um, M., et al. (2009). MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol. Cell. Biol. 29, 5290–5305. doi: 10.1128/MCB.01694-08
Lehrbach, N. J., Armisen, J., Lightfoot, H. L., Murfitt, K. J., Bugaut, A., Balasubramanian, S., et al. (2009). LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16, 1016–1020. doi: 10.1038/nsmb.1675
Lei, Q., Shen, F., Wu, J., Zhang, W., Wang, J., Zhang, L. (2014). miR-101, downregulated in retinoblastoma, functions as a tumor suppressor in human retinoblastoma cells by targeting EZH2. Oncol. Rep. 32, 261–269. doi: 10.3892/or.2014.3167
Leichter, A. L., Sullivan, M. J., Eccles, M. R., Chatterjee, A. (2017). MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol. Cancer 16, 1–17. doi: 10.1186/s12943-017-0584-0
Leone, G., DeGregori, J., Sears, R., Jakoi, L., Nevins, J. R. (1997). Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 387, 422–426. doi: 10.1038/387422a0
Li, L., Sarver, A. L., Alamgir, S., Subramanian, S. (2012). Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab. Investig. 92, 571–583. doi: 10.1038/labinvest.2012.10
Li, G., Zhang, H., Wan, X., Yang, X., Zhu, C., Wang, A., et al. (2014). Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res. Int. 2014, 1–8. doi: 10.1155/2014/780521
Li, J., You, T., Jing, J. (2014). MiR-125b inhibits cell biological progression of Ewing’s sarcoma by suppressing the PI3K/Akt signalling pathway. Cell Prolif. 47, 152–160. doi: 10.1111/cpr.12093
Li, S., Li, F., Cheng, T. (2014). TGF-β1 promotes osteosarcoma cell migration and invasion through the miR-143–versican pathway. Cell. Physiol. Biochem. 34, 2169–2179. doi: 10.1159/000369660
Li, X., Yang, H., Tian, Q., Liu, Y., Weng, Y. (2014). Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. Neoplasma 61, 453–460. doi: 10.4149/neo_2014_056
Li, Z., Zhao, L., Wang, Q. (2016). Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/β-catenin pathway. Am. J. Transl. Res. 8, 2385–2393.
Li, Y., Shao, G., Zhang, M., Zhu, F., Zhao, B., He, C., et al. (2017). miR-124 represses the mesenchymal features and suppresses metastasis in Ewing sarcoma. Oncotarget 8, 10274–10286. doi: 10.18632/oncotarget.14394
Li, G., Liu, K., Du, X. (2018). Long non-coding RNA TUG1 promotes proliferation and inhibits apoptosis of osteosarcoma cells by sponging miR-132-3p and upregulating SOX4 expression. Yonsei Med. J. 59, 226–235. doi: 10.3349/ymj.2018.59.2.226
Li, H., Tian, G., Tian, F., Shao, L. (2018). Long non-coding RNA TUG1 promotes osteosarcoma cell proliferation and invasion through inhibition of microrna-212-3p expression. Exp. Ther. Med. 16, 779–787. doi: 10.3892/etm.2018.6216
Li, L., Chen, W., Wang, Y., Tang, L., Han, M. (2018). Long non-coding RNA H19 regulates viability and metastasis, and is upregulated in retinoblastoma. Oncol. Lett. 8424–8432. doi: 10.3892/ol.2018.8385
Liang, W.-C., Fu, W.-M., Wong, C.-W., Wang, Y., Wang, W.-M., Hu, G.-X., et al. (2015). The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525. doi: 10.18632/oncotarget.4154
Liu, M., Roth, A., Yu, M., Morris, R., Bersani, F., Rivera, M. N., et al. (2013). The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 27, 2543–2548. doi: 10.1101/gad.224170.113
Liu, Q., Huang, J., Zhou, N., Zhang, Z., Zhang, A., Lu, Z., et al. (2013). LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 41, 4976–4987. doi: 10.1093/nar/gkt182
Liu, P. Y., Erriquez, D., Marshall, G. M., Tee, A. E., Polly, P., Wong, M., et al. (2014). Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J. Natl. Cancer Inst. 106, dju113–dju113. doi: 10.1093/jnci/dju113
Liu, P. Y., Atmadibrata, B., Mondal, S., Tee, A. E., Liu, T. (2016). NCYM is upregulated by lncUSMycN and modulates N-Myc expression. Int. J. Oncol. 49, 2464–2470. doi: 10.3892/ijo.2016.3730
Liu, S., Hu, C., Wang, Y., Shi, G., Li, Y., Wu, H. (2016). miR-124 inhibits proliferation and invasion of human retinoblastoma cells by targeting STAT3. Oncol. Rep. 36, 2398–2404. doi: 10.3892/or.2016.4999
Liu, G.-L., Yang, H.-J., Liu, B., Liu, T. (2017). Effects of microRNA-19b on the proliferation, apoptosis, and migration of Wilms’ tumor cells via the PTEN/PI3K/AKT signaling pathway. J. Cell. Biochem. 118, 3424–3434. doi: 10.1002/jcb.25999
Liu, K., Huang, J., Ni, J., Song, D., Ding, M., Wang, J., et al. (2017). MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142–3p and miR-129–5p. Cell Cycle 16, 578–587. doi: 10.1080/15384101.2017.1288324
Liu, S., Yan, G., Zhang, J., Yu, L. (2017). Knockdown of long noncoding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) inhibits proliferation, migration, and invasion and promoted apoptosis by targeting miR-124 in retinoblastoma. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 26, 581–591. doi: 10.3727/096504017X14953948675403
Lottin, S., Adriaenssens, E., Dupressoir, T., Berteaux, N., Montpellier, C., Coll, J., et al. (2002). Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23, 1885–1895. doi: 10.1093/carcin/23.11.1885
Loven, J., Zinin, N., Wahlstrom, T., Muller, I., Brodin, P., Fredlund, E., et al. (2010). MYCN-regulated microRNAs repress estrogen receptor-α (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc. Natl. Acad. Sci. 107, 1553–1558. doi: 10.1073/pnas.0913517107
Lu, C., Peng, K., Guo, H., Ren, X., Hu, S., Cai, Y., et al. (2018). miR-18a-5p promotes cell invasion and migration of osteosarcoma by directly targeting IRF2. Oncol. Lett. 16, 3150–3156. doi: 10.3892/ol.2018.9032
Lu, Q., Lu, M., Li, D., Zhang, S. (2018). MicroRNA-34b promotes proliferation, migration and invasion of Ewing’s sarcoma cells by downregulating Notch1. Mol. Med. Rep. 18, 3577–3588. doi: 10.3892/mmr.2018.9365
Luo, T., Yi, X., Si, W. (2017). Identification of miRNA and genes involving in osteosarcoma by comprehensive analysis of microRNA and copy number variation data. Oncol. Lett. 14, 5427–5433. doi: 10.3892/ol.2017.6845
Lv, B., Zhang, L., Miao, R., Xiang, X., Dong, S., Lin, T., et al. (2018). Comprehensive analysis and experimental verification of LINC01314 as a tumor suppressor in hepatoblastoma. Biomed. Pharmacother. 98, 783–792. doi: 10.1016/j.biopha.2018.01.013
Ma, L., Young, J., Prabhala, H., Pan, E., Mestdagh, P., Muth, D., et al. (2010). MiR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 12, 247–256. doi: 10.1038/ncb2024
Ma, B., Li, M., Zhang, L., Huang, M., Lei, J.-B., Fu, G.-H., Liu, C.-X., et al. (2016). Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumor Biol. 37, 4445–4455. doi: 10.1007/s13277-015-4301-6
MacIas, S., Plass, M., Stajuda, A., Michlewski, G., Eyras, E., Cáceres, J. F. (2012). DGCR8 HITS-CLIP reveals novel functions for the microprocessor. Nat. Struct. Mol. Biol. 19, 760–766. doi: 10.1038/nsmb.2344
Marino, M. T., Grilli, A., Baricordi, C., Manara, M. C., Ventura, S., Pinca, R. S., et al. (2014). Prognostic significance of miR-34a in Ewing sarcoma is associated with cyclin D1 and ki-67 expression. Ann. Oncol. 25, 2080–2086. doi: 10.1093/annonc/mdu249
Maris, J. M., White, P. S., Beltinger, C. P., Sulman, E. P., Castleberry, R. P., Shuster, J. J., et al. (1995). Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res. 55, 4664–4669.
Matouk, I. J., Halle, D., Raveh, E., Gilon, M., Sorin, V., Hochberg, A. (2015). The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT–MET decision. Oncotarget 7, 3748–3765. doi: 10.18632/oncotarget.6387
Mercer, T. R., Dinger, M. E., Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Mestdagh, P., Boström, A.-K., Impens, F., Fredlund, E., Van Peer, G., De Antonellis, P., et al. (2010). The miR-17-92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol. Cell 40, 762–773. doi: 10.1016/j.molcel.2010.11.038
Missiaglia, E., Shepherd, C. J., Patel, S., Thway, K., Pierron, G., Pritchard-Jones, K., et al. (2010). MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br. J. Cancer 102, 1769–1777. doi: 10.1038/sj.bjc.6605684
Missiaglia, E., Shepherd, C. J., Aladowicz, E., Olmos, D., Selfe, J., Pierron, G., et al. (2017). MicroRNA and gene co-expression networks characterize biological and clinical behavior of rhabdomyosarcomas. Cancer Lett. 385, 251–260. doi: 10.1016/j.canlet.2016.10.011
Mitra, S. A., Mitra, A. P., Triche, T. J. (2012). A central role for long non-coding RNA in cancer. Front. Genet. 3, 1–9. doi: 10.3389/fgene.2012.00017
Mogilyansky, E., Rigoutsos, I. (2013). The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 20, 1603–1614. doi: 10.1038/cdd.2013.125
Molenaar, J. J., Domingo-Fernández, R., Ebus, M. E., Lindner, S., Koster, J., Drabek, K., et al. (2012). LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 44, 1199–1206. doi: 10.1038/ng.2436
Monnier, P., Martinet, C., Pontis, J., Stancheva, I., Ait-Si-Ali, S., Dandolo, L. (2013). H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc. Natl. Acad. Sci. 110, 20693–20698. doi: 10.1073/pnas.1310201110
Moore, C., Parrish, J. K., Jedlicka, P. (2017). MiR-193b, downregulated in Ewing sarcoma, targets the ErbB4 oncogene to inhibit anchorage-independent growth. PLoS One 12, e0178028. doi: 10.1371/journal.pone.0178028
Nakagawa, S., Ip, J. Y., Shioi, G., Tripathi, V., Zong, X., Hirose, T., et al. (2012). Malat1 is not an essential component of nuclear speckles in mice. RNA 18, 1487–1499. doi: 10.1261/rna.033217.112
Nakatani, F., Ferracin, M., Manara, M. C., Ventura, S., Del Monaco, V., Ferrari, S., et al. (2012). miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J. Pathol. 226, 796–805. doi: 10.1002/path.3007
Nguyen, L. H., Robinton, D. A., Seligson, M. T., Wu, L., Li, L., Rakheja, D., et al. (2014). Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 26, 248–261. doi: 10.1016/j.ccr.2014.06.018
Nittner, D., Lambertz, I., Clermont, F., Mestdagh, P., Köhler, C., Nielsen, S. J., et al. (2012). Synthetic lethality between Rb, p53 and Dicer or miR-17-92 in retinal progenitors suppresses retinoblastoma formation. Nat. Cell Biol. 14, 958–965. doi: 10.1038/ncb2556
Niu, G., Li, B., Sun, J., Sun, L. (2015). miR-454 is down-regulated in osteosarcomas and suppresses cell proliferation and invasion by directly targeting c-Met. Cell Prolif. 48, 348–355. doi: 10.1111/cpr.12187
Novello, C., Pazzaglia, L., Cingolani, C., Conti, A., Quattrini, I., Manara, M. C., et al. (2013). MiRNA expression profile in human osteosarcoma: role of miR-1 and miR-133b in proliferation and cell cycle control. Int. J. Oncol. 42, 667–675. doi: 10.3892/ijo.2012.1717
O’Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V., Mendell, J. T. (2005). c-Myc–regulated microRNAs modulate E2F1 expression. Nature 435, 839–843. doi: 10.1038/nature03677
Pandey, G. K., Mitra, S., Subhash, S., Hertwig, F., Kanduri, M., Mishra, K., et al. (2014). The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26, 722–737. doi: 10.1016/j.ccell.2014.09.014
Pandey, G. K., Kanduri, C. (2015). Long noncoding RNAs and neuroblastoma. Oncotarget 6, 18265–18275. doi: 10.18632/oncotarget.4251
Parrish, J. K., Sechler, M., Winn, R. A., Jedlicka, P. (2015). The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in Ewing sarcoma. Oncogene 34, 257–262. doi: 10.1038/onc.2013.541
Pasic, I., Shlien, A., Durbin, A. D., Stavropoulos, D. J., Baskin, B., Ray, P. N., et al. (2010). Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 70, 160–171. doi: 10.1158/0008-5472.CAN-09-1902
Pasquinelli, A. E., Reinhart, B. J., Slack, F., Martindale, M. Q., Kuroda, M. I., Maller, B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. doi: 10.1038/35040556
Patel, A. P., Tirosh, I., Trombetta, J. J., Shalek, A. K., Gillespie, S. M., Wakimoto, H., et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (80-. ) 344, 1396–1401. doi: 10.1126/science.1254257
Peng, X. H., Huang, H. R., Lu, J., Liu, X., Zhao, F. P., Zhang, B., et al. (2014). MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol. Cancer 13, 1–13. doi: 10.1186/1476-4598-13-186
Powers, J. T., Tsanov, K. M., Pearson, D. S., Roels, F., Spina, C. S., Ebright, R., et al. (2016). Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 535, 246–251. doi: 10.1038/nature18632
Prasad, C. P., Rath, G., Mathur, S., Bhatnagar, D., Parshad, R., Ralhan, R. (2009). Expression analysis of E-cadherin, Slug and GSK3beta in invasive ductal carcinoma of breast. BMC Cancer 9, 325. doi: 10.1186/1471-2407-9-325
Prensner, J. R., Chinnaiyan, A. M. (2011). The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407. doi: 10.1158/2159-8290.CD-11-0209
Pu, Y., Zhao, F., Wang, H., Cai, W., Gao, J., Li, Y., et al. (2016). MiR-34a-5p promotes the multi-drug resistance of osteosarcoma by targeting the CD117 gene. Oncotarget 7, 28420–28434. doi: 10.18632/oncotarget.8546
Pu, Y., Zhao, F., Li, Y., Cui, M., Wang, H., Meng, X., et al. (2017). The miR-34a-5p promotes the multi-chemoresistance of osteosarcoma via repression of the AGTR1 gene. BMC Cancer 17, 45. doi: 10.1186/s12885-016-3002-x
Qi, X. J., Wang, J. F., Wang, G. D., Xu, Q., Sun, H. L. (2016). Pivotal role of microRNA-9 in osteosarcoma tumorigenesis and tumor progression. Genet. Mol. Res. 15, 1–10. doi: 10.4238/gmr.15017318
Qian, M., Yang, X., Li, Z., Jiang, C., Song, D., Yan, W., et al. (2016). P50-associated COX-2 extragenic RNA (PACER) overexpression promotes proliferation and metastasis of osteosarcoma cells by activating COX-2 gene. Tumor Biol. 37, 3879–3886. doi: 10.1007/s13277-015-3838-8
Qu, H., Zheng, L., Pu, J., Mei, H., Xiang, X., Zhao, X., et al. (2015). miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum. Mol. Genet. 24, 2539–2551. doi: 10.1093/hmg/ddv018
Rakheja, D., Chen, K. S., Liu, Y., Shukla, A. A., Schmid, V., Chang, T. C., et al. (2014). Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat. Commun. 2, 1–10. doi: 10.1038/ncomms5802
Rao, P. K., Missiaglia, E., Shields, L., Hyde, G., Yuan, B., Shepherd, C. J., et al. (2010). Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 24, 3427–3437. doi: 10.1096/fj.09-150698
Raveh, E., Matouk, I. J., Gilon, M., Hochberg, A. (2015). The H19 long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Mol. Cancer 14, 1–14. doi: 10.1186/s12943-015-0458-2
Rehei, A.-L., Zhang, L., Fu, Y.-X., Mu, W.-B., Yang, D.-S., Liu, Y., et al. (2018). MicroRNA-214 functions as an oncogene in human osteosarcoma by targeting TRAF3. Eur. Rev. Med. Pharmacol. Sci. 22, 5156–5164. doi: 10.26355/eurrev_201808_15711
Riffo-Campos, Á. L., Riquelme, I., Brebi-Mieville, P. (2016). Tools for sequence-based miRNA target prediction: what to choose? Int. J. Mol. Sci. 17, E1987. doi: 10.3390/ijms17121987
Riggi, N., Suvà, M.-L., De Vito, C., Provero, P., Stehle, J.-C., Baumer, K., et al. (2010). EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 24, 916–932. doi: 10.1101/gad.1899710
Robin, T. P., Smith, A., McKinsey, E., Reaves, L., Jedlicka, P., Ford, H. L. (2012). EWS/FLI1 Regulates EYA3 in Ewing sarcoma via modulation of miRNA-708, resulting in increased cell survival and chemoresistance. Mol. Cancer Res. 10, 1098–1108. doi: 10.1158/1541-7786.MCR-12-0086
Roth, S. A., Hald, Ø. H., Fuchs, S., Løkke, C., Mikkola, I., Flægstad, T., et al. (2018). MicroRNA-193b-3p represses neuroblastoma cell growth via downregulation of Cyclin D1, MCL-1 and MYCN. Oncotarget 9, 18160–18179. doi: 10.18632/oncotarget.24793
Ruan, W., Wang, P., Feng, S., Xue, Y., Li, Y. (2016). Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumor Biol. 37, 4065–4073. doi: 10.1007/s13277-015-4256-7
Russell, M. R., Penikis, A., Oldridge, D. A., Alvarez-Dominguez, J. R., McDaniel, L., Diamond, M., et al. (2015). CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 75, 3155–3166. doi: 10.1158/0008-5472.CAN-14-3613
Ruteshouser, E. C., Hendrickson, B. W., Colella, S., Krahe, R., Pinto, L., Huff, V. (2005). Genome-wide loss of heterozygosity analysis of WT1-wild-type and WT1-mutant Wilms tumors. Genes Chromosom. Cancer 43, 172–180. doi: 10.1002/gcc.20169
Sahu, D., Ho, S.-Y., Juan, H.-F., Huang, H.-C. (2018). High-risk, expression-based prognostic long noncoding RNA signature in neuroblastoma. JNCI Cancer Spectr. 2, 1–12. doi: 10.1093/jncics/pky015
Satterfield, L., Shuck, R., Kurenbekova, L., Allen-Rhoades, W., Edwards, D., Huang, S., et al. (2017). miR-130b directly targets ARHGAP1 to drive activation of a metastatic CDC42–PAK1–AP1 positive feedback loop in Ewing sarcoma. Int. J. Cancer 141, 2062–2075. doi: 10.1002/ijc.30909
Sauliere, J., Sureau, A., Expert-Bezancon, A., Marie, J. (2006). The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell. Biol. 26, 8755–8769. doi: 10.1128/MCB.00893-06
Schulte, J. H., Horn, S., Otto, T., Samans, B., Heukamp, L. C., Eilers, U. C., et al. (2008). MYCN regulates oncogenic microRNAs in neuroblastoma. Int. J. Cancer 122, 699–704. doi: 10.1002/ijc.23153
Schwentner, R., Herrero-Martin, D., Kauer, M. O., Mutz, C. N., Katschnig, A. M., Sienski, G., et al. (2017). The role of miR-17-92 in the miRegulatory landscape of Ewing sarcoma. Oncotarget 8, 10980–10993. doi: 10.18632/oncotarget.14091
Shang, Y. (2018). LncRNA THOR acts as a retinoblastoma promoter through enhancing the combination of c-myc mRNA and IGF2BP1 protein. Biomed. Pharmacother. 106, 1243–1249. doi: 10.1016/j.biopha.2018.07.052
Sheng, L., Wu, J., Gong, X., Dong, D., Sun, X. (2018). SP1-induced upregulation of lncRNA PANDAR predicts adverse phenotypes in retinoblastoma and regulates cell growth and apoptosis in vitro and in vivo. Gene 668, 140–145. doi: 10.1016/j.gene.2018.05.065
Shi, Y., Lv, C., Shi, L., Tu, G. (2018). MEG3 inhibits proliferation and invasion and promotes apoptosis of human osteosarcoma cells. Oncol. Lett. 15, 1917–1923. doi: 10.3892/ol.2017.7463
Shohet, J. M., Ghosh, R., Coarfa, C., Ludwig, A., Benham, A. L., Chen, Z., et al. (2011). A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 71, 3841–3851. doi: 10.1158/0008-5472.CAN-10-4391
Sin-Chan, P., Mumal, I., Suwal, T., Ho, B., Fan, X., Singh, I., et al. (2019). A C19MC–LIN28A–MYCN oncogenic circuit driven by hijacked super-enhancers is a distinct therapeutic vulnerability in ETMRs: a lethal brain tumor. Cancer Cell. 36, 51–67. doi: 10.1016/j.ccell.2019.06.002
Strieder, V., Lutz, W. (2003). E2F proteins regulate MYCN expression in neuroblastomas. J. Biol. Chem. 278, 2983–2989. doi: 10.1074/jbc.M207596200
Su, P., Mu, S., Wang, Z. (2019). Long noncoding RNA SNHG16 promotes osteosarcoma cells migration and invasion via sponging miRNA-340. DNA Cell Biol. 38, 170–175. doi: 10.1089/dna.2018.4424
Suenaga, Y., Islam, S. M., Alagu, J., Kaneko, Y., Kato, M., Tanaka, Y., et al. (2014). NCYM, a cis-antisense gene of MYCN, encodes a de novo evolved protein that inhibits GSK3β resulting in the stabilization of MYCN in human neuroblastomas. PLoS Genet. 10, e1003996. doi: 10.1371/journal.pgen.1003996
Sun, Y., Qin, B. (2018). Long noncoding RNA MALAT1 regulates HDAC4-mediated proliferation and apoptosis via decoying of miR-140-5p in osteosarcoma cells. Cancer Med. 7, 4584–4597. doi: 10.1002/cam4.1677
Sun, Y. M., Lin, K. Y., Chen, Y. Q. (2013). Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 6, 1–8. doi: 10.1186/1756-8722-6-6
Sun, L., Yang, C., Xu, J., Feng, Y., Wang, L., Cui, T. (2016). Long noncoding RNA EWSAT1 promotes osteosarcoma cell growth and metastasis through suppression of MEG3 expression. DNA Cell Biol. 35, 812–818. doi: 10.1089/dna.2016.3467
Sun, X., Dai, G., Yu, L., Hu, Q., Chen, J., Guo, W. (2018). MiR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-017-18739-3
Swarbrick, A., Woods, S. L., Shaw, A., Balakrishnan, A., Phua, Y., Nguyen, A., et al. (2010). miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat. Med. 16, 1134–1140. doi: 10.1038/nm.2227
Sylvestre, Y., De Guire, V., Querido, E., Mukhopadhyay, U. K., Bourdeau, V., Major, F., et al. (2007). An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 282, 2135–2143. doi: 10.1074/jbc.M608939200
Tee, A. E., Ling, D., Nelson, C., Atmadibrata, B., Dinger, M. E., Xu, N., et al. (2014). The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget 5, 10000. doi: 10.18632/oncotarget.1785
Thayanithy, V., Sarver, A. L., Kartha, R. V., Li, L., Angstadt, A. Y., Breen, M., et al. (2012). Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone 50, 171–181. doi: 10.1016/j.bone.2011.10.012
Tivnan, A., Foley, N. H., Tracey, L., Davidoff, A. M., Stallings, R. L. (2010). MicroRNA-184-mediated inhibition of tumour growth in an orthotopic murine model of neuroblastoma. Anticancer Res. 30, 4391–4395.
Tivnan, A., Tracey, L., Buckley, P. G., Alcock, L. C., Davidoff, A. M., Stallings, R. L. (2011). MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer 11, 33. doi: 10.1186/1471-2407-11-33
Torrezan, G. T., Ferreira, E. N., Nakahata, A. M., Barros, B. D. F., Castro, M. T. M., Correa, B. R., et al. (2014). Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat. Commun. 5, 4039. doi: 10.1038/ncomms5039
Tsai, M.-C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science (80-. ) 329, 689–693. doi: 10.1126/science.1192002
Ulaner, G. A., Vu, T. H., Li, T., Hu, J. F., Yao, X. M., Yang, Y., et al. (2003). Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum. Mol. Genet. 12, 535–549. doi: 10.1093/hmg/ddg034
Urbach, A., Yermalovich, A., Zhang, J., Spina, C. S., Zhu, H., Perez-Atayde, A. R., et al. (2014). Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 28, 971–982. doi: 10.1101/gad.237149.113
Vázquez-Arreguín, K., Tantin, D. (2016). The Oct1 transcription factor and epithelial malignancies: old protein learns new tricks. Biochim. Biophys. Acta 1859, 792–804. doi: 10.1016/j.bbagrm.2016.02.007
Ventura, A., Young, A. G., Winslow, M. M., Lintault, L., Meissner, A., Erkeland, S. J., et al. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17∼92 family of miRNA clusters. Cell 132, 875–886. doi: 10.1016/j.cell.2008.02.019
Ventura, S., Aryee, D. N., Felicetti, F., De Feo, A., Mancarella, C., Manara, M. C., et al. (2016). CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch–mediated control of NF-κB signaling. Oncogene 35, 3944–3954. doi: 10.1038/onc.2015.463
Veronese, A., Lupini, L., Consiglio, J., Visone, R., Ferracin, M., Fornari, F., et al. (2010). Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 70, 3140–3149. doi: 10.1158/0008-5472.CAN-09-4456
von Frowein, J., Hauck, S. M., Kappler, R., Pagel, P., Fleischmann, K. K., Magg, T., et al. (2018). MiR-492 regulates metastatic properties of hepatoblastoma via CD44. Liver Int. 38, 1280–1291. doi: 10.1111/liv.13687
Walz, A. L., Ooms, A., Gadd, S., Gerhard, D. S., Smith, M. A., Guidry Auvil, J. M., et al. (2015). Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 27, 286–297. doi: 10.1016/j.ccell.2015.01.003
Wang, L.-L., Hu, H.-F., Feng, Y.-Q. (2016). Suppressive effect of microRNA-143 in retinoblastoma. Int. J. Ophthalmol. 9, 1584–1590. doi: 10.18240/ijo.2016.11.08
Wang, H.-F., Zhang, Y.-Y., Zhuang, H.-W., Xu, M. (2017). MicroRNA-613 attenuates the proliferation, migration and invasion of Wilms’ tumor via targeting FRS2. Eur. Rev. Med. Pharmacol. Sci. 21, 3360–3369.
Wang, Y., Zhang, Y., Yang, T., Zhao, W., Wang, N., Li, P., et al. (2017a). Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget 8, 59417–59434. doi: 10.18632/oncotarget.19727
Wang, Y., Yang, T., Zhang, Z., Lu, M., Zhao, W., Zeng, X., et al. (2017b). Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 108, 859–867. doi: 10.1111/cas.13201
Wang, J. X., Yang, Y., Li, K. (2018). Long noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9. J. Cell. Physiol. 233, 6986–6995. doi: 10.1002/jcp.26621
Wang, K., Yan, L., Lu, F. (2018). MiR-363-3p inhibits osteosarcoma cell proliferation and invasion via targeting SOX4. Oncol. Res. 5, 157–163. doi: 10.3727/096504018X15190861873459
Wang, W.-T., Qi, Q., Zhao, P., Li, C.-Y., Yin, X.-Y., Yan, R.-B. (2018). miR-590-3p is a novel microRNA which suppresses osteosarcoma progression by targeting SOX9. Biomed. Pharmacother. 107, 1763–1769. doi: 10.1016/j.biopha.2018.06.124
Wang, B., Qu, X. L., Liu, J., Lu, J., Zhou, Z. Y. (2019). HOTAIR promotes osteosarcoma development by sponging miR-217 and targeting ZEB1. J. Cell. Physiol. 234, 6173–6181. doi: 10.1002/jcp.27394
Wegert, J., Ishaque, N., Vardapour, R., Geörg, C., Gu, Z., Bieg, M., et al. (2015). Mutations in the SIX1/2 Pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 27, 298–311. doi: 10.1016/j.ccell.2015.01.002
Wei, J. S., Song, Y. K., Durinck, S., Chen, Q. R., Cheuk, A. T. C., Tsang, P., et al. (2008). The MYCN oncogene is a direct target of miR-34a. Oncogene 27, 5204–5213. doi: 10.1038/onc.2008.154
Welch, C., Chen, Y., Stallings, R. L. (2007). MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26, 5017–5022. doi: 10.1038/sj.onc.1210293
Wen, J., Zhao, Y. K., Liu, Y., Zhao, J. F. (2017). MicroRNA-34a inhibits tumor invasion and metastasis in osteosarcoma partly by effecting C-IAP2 and Bcl-2. Tumor Biol. 39, 101042831770567. doi: 10.1177/1010428317705761
Williamson, D., Lu, Y. J., Gordon, T., Sciot, R., Kelsey, A., Fisher, C., et al. (2005). Relationship between MYCN copy number and expression in rhabdomyosarcomas and correlation with adverse prognosis in the alveolar subtype. J. Clin. Oncol. 23, 880–888. doi: 10.1200/JCO.2005.11.078
Wittmann, S., Zirn, B., Alkassar, M., Ambros, P., Graf, N., Gessler, M. (2007). Loss of 11q and 16q in Wilms tumors is associated with anaplasia, tumor recurrence, and poor prognosis. Genes Chromosom. Cancer 46, 163–170. doi: 10.1002/gcc.20397
Wu, M. K., Sabbaghian, N., Xu, B., Addidou-Kalucki, S., Bernard, C., Zou, D., et al. (2013). Biallelic DICER1 mutations occur in Wilms tumours. J. Pathol. 230, 154–164. doi: 10.1002/path.4196
Wu, X., Zhong, D., Gao, Q., Zhai, W., Ding, Z., Wu, J. (2013). MicroRNA-34a inhibits human osteosarcoma proliferation by downregulating ether à go-go 1 expression. Int. J. Med. Sci. 10, 676–682. doi: 10.7150/ijms.5528
Wu, L., Nguyen, L. H., Zhou, K., De Soysa, T. Y., Li, L., Miller, J. B., et al. (2015). Precise let-7 expression levels balance organ regeneration against tumor suppression. Elife 4, 1–16. doi: 10.7554/eLife.09431
Xiang, X., Mei, H., Qu, H., Zhao, X., Li, D., Song, H., et al. (2015). miRNA-584-5p exerts tumor suppressive functions in human neuroblastoma through repressing transcription of matrix metalloproteinase 14. Biochim. Biophys. Acta - Mol. Basis Dis. 1852, 1743–1754. doi: 10.1016/j.bbadis.2015.06.002
Xiao, T., Zhou, Y., Li, H., Xiong, L., Wang, J., Wang, Z.‐H., et al. (2019). MiR-125b suppresses the carcinogenesis of osteosarcoma cells via the MAPK–STAT3 pathway. J. Cell. Biochem. 120, 2616–2626. doi: 10.1002/jcb.27568
Xie, C.-H., Cao, Y.-M., Huang, Y., Shi, Q.-W., Guo, J.-H., et al. (2016). Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression. Tumor Biol. 37, 15031–15041. doi: 10.1007/s13277-016-5391-5
Xin, C., Buhe, B., Hongting, L., Chuanmin, Y., Xiwei, H., Hong, Z., et al. (2013). MicroRNA-15a promotes neuroblastoma migration by targeting reversion-inducing cysteine-rich protein with Kazal motifs (RECK) and regulating matrix metalloproteinase-9 expression. FEBS J. 280, 855–866. doi: 10.1111/febs.12074
Yan, K., Gao, J., Yang, T., Ma, Q., Qiu, X., Fan, Q., et al. (2012). MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One. 7, e33778. doi: 10.1371/journal.pone.0033778
Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K. A., Grinstein, J., et al. (2011). ncRNA- and Pc2 methylation–dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. doi: 10.1016/j.cell.2011.08.054
Yang, F., Bi, J., Xue, X., Zheng, L., Zhi, K., Hua, J., et al. (2012). Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 279, 3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x
Yang, G., Yuan, J., Li, K. (2013). EMT transcription factors: implication in osteosarcoma. Med. Oncol. 30, 1–5. doi: 10.1007/s12032-013-0697-2
Yang, L., Li, Y., Wei, Z., Chang, X. (2017). Coexpression network analysis identifies transcriptional modules associated with genomic alterations in neuroblastoma. Biochim. Biophys. Acta - Mol. Basis Dis. 1864, 2341–2348. doi: 10.1016/j.bbadis.2017.12.020
Yang, G., Fu, Y., Lu, X., Wang, M., Dong, H., Li, Q. (2018). LncRNA HOTAIR/miR-613/c-met axis modulated epithelial–mesenchymal transition of retinoblastoma cells. J. Cell. Mol. Med. 22, 5083–5096. doi: 10.1111/jcmm.13796
Yang, H., Peng, Z., Liang, M., Zhang, Y., Wang, Y., Jiang, Y., et al. (2018). The miR-17-92 cluster/QKI2/β-catenin osteosarcoma progression axis promotes 9, 25285–25293. doi: 10.18632/oncotarget.23935
Ye, C., Yu, X., Liu, X., Dai, M., Zhang, B. (2018). miR-30d inhibits cell biological progression of Ewing’s sarcoma by suppressing the MEK/ERK and PI3K/Akt pathways in vitro. Oncol. Lett. 15, 4390–4396. doi: 10.3892/ol.2018.7900
Yin, Z., Ding, H., He, E., Chen, J., Li, M. (2016). Overexpression of long non-coding RNA MFI2 promotes cell proliferation and suppresses apoptosis in human osteosarcoma. Oncol. Rep. 36, 2033–2040. doi: 10.3892/or.2016.5013
Yin, J., Zhao, J., Hu, W., Yang, G., Yu, H., Wang, R., et al. (2017). Disturbance of the let-7/LIN28 double negative feedback loop is associated with radio- and chemo-resistance in non–small cell lung cancer. PLoS One 12, 1–16. doi: 10.1371/journal.pone.0172787
Ying, L., Chen, Q., Wang, Y., Zhou, Z., Huang, Y., Qiu, F. (2012). Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 8, 2289–2294. doi: 10.1039/c2mb25070e
Yuan, J., Lang, J., Liu, C., Zhou, K., Chen, L., Liu, Y. (2015). The expression and function of miRNA-451 in osteosarcoma. Med. Oncol. 32, 324. doi: 10.1007/s12032-014-0324-x
Zhang, Y., Shields, T., Crenshaw, T., Hao, Y., Moulton, T., Tycko, B. (1993). Imprinting of human H19: allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. Am. J. Hum. Genet. 53, 113–124.
Zhang, B., Arun, G., Mao, Y. S., Lazar, Z., Hung, G., Bhattacharjee, G., et al. (2012). The lncRNA malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123. doi: 10.1016/j.celrep.2012.06.003
Zhang, H., Pu, J., Qi, T., Qi, M., Yang, C., Li, S., et al. (2014). MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 alpha. Oncogene 33, 387–397. doi: 10.1038/onc.2012.574
Zhang, A., Shang, W., Nie, Q., Li, T., Li, S. (2018). Long non-coding RNA H19 suppresses retinoblastoma progression via counteracting miR-17-92 cluster. J. Cell. Biochem. 119, 3497–3509. doi: 10.1002/jcb.26521
Zhang, F., Zhu, Y., Fan, G., Hu, S. (2018). MicroRNA-2682-3p inhibits osteosarcoma cell proliferation by targeting CCND2, MMP8 and Myd88. Oncol. Lett. 16, 3359–3364. doi: 10.3892/ol.2018.9029
Zhang, S., Li, D., Jiao, G.-J., Wang, H.-L., Yan, T.-B. (2018). miR-185 suppresses progression of Ewing’s sarcoma via inhibiting the PI3K/AKT and Wnt/β-catenin pathways. Onco. Targets. Ther. 11, 7967–7977. doi: 10.2147/OTT.S167771
Zhang, Y., Zhao, Y., Wu, J., Liangpunsakul, S., Niu, J., Wang, L. (2018). MicroRNA-26-5p functions as a new inhibitor of hepatoblastoma by repressing lin-28 homolog B and aurora kinase a expression. Hepatol. Commun. 2, 861–871. doi: 10.1002/hep4.1185
Zhao, D., Tian, Y., Li, P., Wang, L., Xiao, A., Zhang, M., et al. (2015). MicroRNA-203 inhibits the malignant progression of neuroblastoma by targeting Sam68. Mol. Med. Rep. 12, 5554–5560. doi: 10.3892/mmr.2015.4013
Zhao, Y., Sun, H., Wang, H. (2016). Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 6, 1–6. doi: 10.1186/s13578-016-0109-3
Zhao, Y., Ling, Z., Hao, Y., Pang, X., Han, X., Califano, J. A., et al. (2017). MiR-124 acts as a tumor suppressor by inhibiting the expression of sphingosine kinase 1 and its downstream signaling in head and neck squamous cell carcinoma. Oncotarget 8, 25005–25020. doi: 10.18632/oncotarget.15334
Zhi, F., Wang, R., Wang, Q., Xue, L., Deng, D., Wang, S., et al. (2014). MicroRNAs in neuroblastoma: small-sized players with a large impact. Neurochem. Res. 39, 613–623. doi: 10.1007/s11064-014-1247-9
Zhou, J., Yang, L., Zhong, T., Mueller, M., Men, Y., Zhang, N., et al. (2015). H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 6, 1–13. doi: 10.1038/ncomms10221
Zhu, S. W., Li, J. P., Ma, X. L., Ma, J. X., Yang, Y., Chen, Y., et al. (2015). miR-9 modulates osteosarcoma cell growth by targeting the GCIP tumor suppressor. Asian Pacific J. Cancer Prev. 16, 4509–4513. doi: 10.7314/APJCP.2015.16.11.4509
Zhu, Z., Tang, J., Wang, J., Duan, G., Zhou, L., Zhou, X. (2016). MIR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS One 11, 1–12. doi: 10.1371/journal.pone.0150026
Zhu, C., Cheng, D., Qiu, X., Zhuang, M., Liu, Z. (2018). Long noncoding RNA SNHG16 promotes cell proliferation by sponging microRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell. Physiol. Biochem. 51, 429–440. doi: 10.1159/000495239
Zhu, S., Fu, W., Zhang, L., Fu, K., Hu, J., Jia, W., et al. (2018a). LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif. 51, 1–10. doi: 10.1111/cpr.12416
Keywords: pediatric tumors, miRNA, long noncoding RNA, cancer biology, gene expression
Citation: Smith CM, Catchpoole D and Hutvagner G (2019) Non-Coding RNAs in Pediatric Solid Tumors. Front. Genet. 10:798. doi: 10.3389/fgene.2019.00798
Received: 24 January 2019; Accepted: 30 July 2019;
Published: 20 September 2019.
Edited by:
Yun Zheng, Kunming University of Science and Technology, ChinaReviewed by:
Alessio Naccarati, Italian Institute for Genomic Medicine (IIGM), ItalyZexuan Zhu, Shenzhen University, China
Copyright © 2019 Smith, Catchpoole and Hutvagner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Catchpoole, RGFuaWVsLkNhdGNocG9vbGVAdXRzLmVkdS5hdQ==; Gyorgy Hutvagner, Z3lvcmd5Lmh1dHZhZ25lckB1dHMuZWR1LmF1
 Christopher M. Smith1
Christopher M. Smith1 Gyorgy Hutvagner
Gyorgy Hutvagner