
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 13 August 2019
Sec. Applied Genetic Epidemiology
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00696
This article is part of the Research Topic Host Genetics in Viral Pathogenesis and Controls View all 22 articles
Chronic hepatitis B virus (HBV) infection is still a major health problem worldwide. Recently, a great number of genetic studies based on single nucleotide polymorphisms (SNPs) and genome-wide association studies have been performed to search for host determinants of the development of chronic HBV infection, clinical outcomes, therapeutic efficacy, and responses to hepatitis B vaccines, with a focus on human leukocyte antigens (HLA), cytokine genes, and toll-like receptors. In addition to SNPs, gene insertions/deletions and copy number variants are associated with infection. However, conflicting results have been obtained. In the present review, we summarize the current state of research on host genetic factors and chronic HBV infection, its clinical type, therapies, and hepatitis B vaccine responses and classify published results according to their reliability. The potential roles of host genetic determinants of chronic HBV infection identified in these studies and their clinical significance are discussed. In particular, HLAs were relevant for HBV infection and pathogenesis. Finally, we highlight the need for additional studies with large sample sizes, well-matched study designs, appropriate statistical methods, and validation in multiple populations to improve the treatment of HBV infection.
With 292 million people infected and a global prevalence of 3.9%, hepatitis B virus (HBV) infection is still a major global public health problem (Polaris Observatory Collaborators, 2018). The outcomes of HBV infection are highly diverse, including acute hepatitis, self-limiting recovery, chronic hepatitis, cirrhosis, liver cancer, and liver failure (Figure 1) (European Association For The Study Of The Liver, 2017). In the natural history of HBV infection, there is substantial variation in the course of disease development and clinical outcomes depending on the transmission pattern, timing of infection, sex, immune status, host genetic factors, and underlying diseases in infected individuals. The disease outcome is related to viral, environmental, and host factors (Ganem and Prince, 2004; Rehermann and Bertoletti, 2015). With respect to host factors, in addition to age, gender, alcohol, obesity, diabetes, and renal failure, host gene variants may also affect the clinical course of HBV infection (Laskus et al., 1992; He et al., 2006; Thursz et al., 2011; Yano et al., 2013; Matsuura et al., 2016). Since the implementation of the Human Genome Project, great progress has been made in genetic and disease-related research, providing a basis for advances in precision medicine. Host genetic polymorphisms mainly include single nucleotide polymorphisms (SNPs), insertions/deletions, and copy number variations (CNVs). Owing to technological limitations, current research mainly focuses on the relationships between SNPs and susceptibility to diseases.
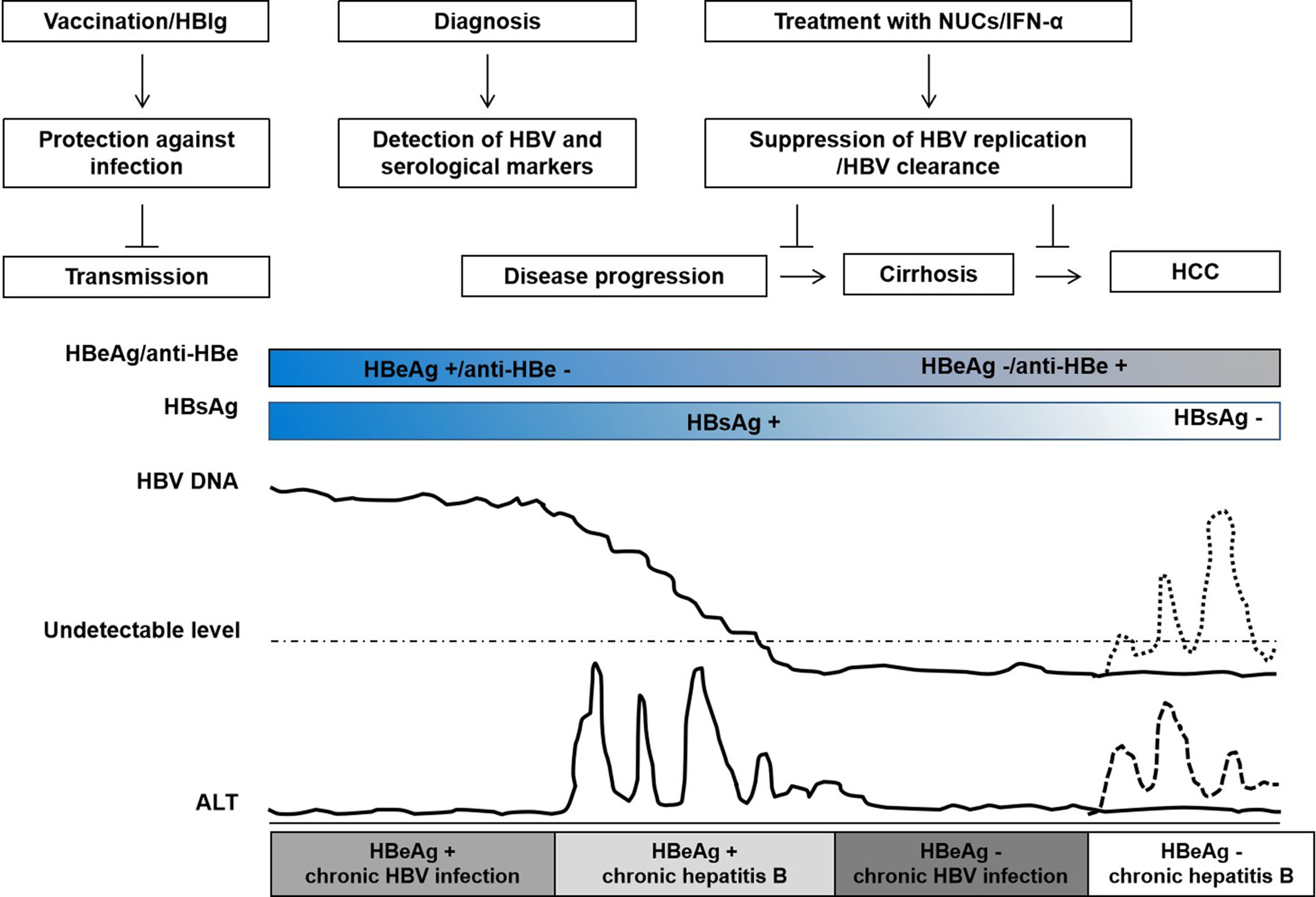
Figure 1 Natural history of chronic hepatitis B virus (HBV) infection, interventions, and clinical outcomes. The natural history of HBV infection is typically classified into four clinical states: HBeAg+ chronic HBV infection, HBeAg+ chronic hepatitis, HBeAg– chronic HBV infection, and HBeAg− chronic hepatitis according to the guideline of European Association for the Study of the Liver (EASL). Patients with chronic HBV infection do not necessarily experience all states. HBV vaccine and/or HBIg injection may prevent HBV transmission; during HBeAg+ chronic hepatitis, treatment with nucleos(t)ide analogs (NUCs) or interferon-alpha (IFN-α) may suppress HBV replication, slow disease progression, and reduce the risk of cirrhosis and hepatocellular carcinoma; HBV reactivation may occur during HBeAg– chronic HBV infection in patients with HBsAg and/or anti-HBc positive after chemotherapy or immunosuppression. The course of HBV infection and disease progression are variable, indicating that host genetic determinants play an important role in HBV infection.
Two approaches are frequently used to screen susceptibility genes for HBV-related diseases and outcomes. First, genome-wide association study (GWAS) have become a common tool for genetic association research focused on complex diseases (Hardy and Singleton, 2009), including HBV infection. In the first stage of GWAS, patient samples are typically used to screen a large number of candidate SNP loci using TaqMan probes, SNPstream genotyping technology, SNaPshot genotyping, SNP chips, and other methods, with several rounds of validation to finally identify SNPs. Second, the traditional approach is the selection of candidate genes predicted to play a role in HBV-related diseases or treatment responses based on theory and previous analyses, followed by verification by comparisons of SNP genotype frequencies in patient and control groups.
Many clinical relevant aspects of HBV prevention, infection, and treatment are potentially related to host genetics (Kamatani et al., 2009; Mbarek et al., 2011; Brouwer et al., 2014; Chang et al., 2014; Hosaka et al., 2015; Jiang et al., 2015; Jiang et al., 2016a; Brouwer et al., 2019). Hepatitis B vaccines are widely used and effective for the prevention of new HBV infections. However, the overall success rate of hepatitis B vaccination is about 90% (Bruce et al., 2016; Hu et al., 2018). The outcome of HBV infection can vary dramatically, depending on both host and viral factors. A major part of patients do not experience clinically relevant symptoms during the acute infection phase while others have acute illness with symptoms that last several weeks. Without any intervention, about 95% of intrauterine or perinatal HBV infections but only about 5% in adults develop into chronic infections (Stevens et al., 1975; Wright and Lau, 1993; Petrova and Kamburov, 2010). HBV persistence is a major risk for developing chronic hepatitis B, cirrhosis, hepatocellular carcinoma, or acute liver failure (Wright and Lau, 1993; Zhang et al., 2013a). A small number of patients develop occult HBV infection (OBI) without detectable serum HBsAg but HBV DNA in the serum or liver (Torbenson and Thomas, 2002; Raimondo et al., 2008; Raimondo et al., 2019; Seed et al., 2019). Interferon-alpha (IFN-α) and nucleos(t)ide analogs (NUCs) have been approved and are widely used for the treatment of chronic hepatitis B (European Association For The Study Of The Liver, 2017). Regimens such as antiviral treatment with tenofovir and entecavir provide results in viral suppression in around 95% of patients, have limited associated resistance, and prevent liver disease progression, cirrhosis, and HCC. However, these treatments do not regularly achieve clinical cure or HBsAg clearance and the incidence of cirrhosis or HCC is still significantly higher in treated patients than those without HBV infection (Figure 1). Many studies have shown that host genetic polymorphisms may influence HBV infection, including hepatitis B vaccine responses, chronic HBV infection (CHI), intrauterine transmission (IT), OBI, liver cirrhosis (LC), hepatocellular carcinoma (HCC), liver transplantation (LT), and the antiviral efficacy of IFNs and NUCs (Kamatani et al., 2009; Davila et al., 2010; Li et al., 2012b; Chang et al., 2014). These studies have demonstrated that host genetics play an important role in HBV infection and pathogenesis (Figure 2). Yet, many contradictory or ambiguous findings have been reported.
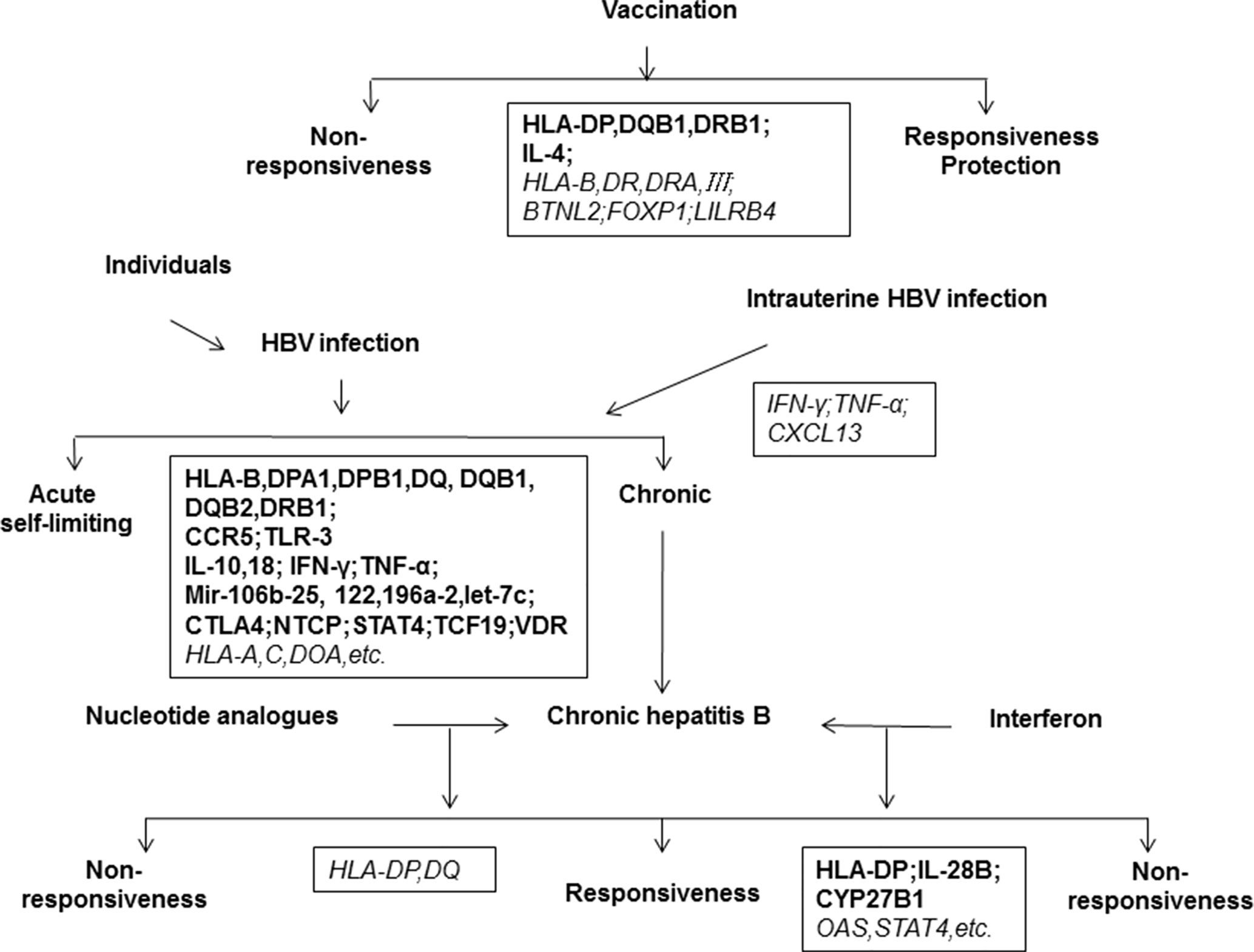
Figure 2 Host genetic determinants of HBV infection. The outcomes of HBV infection are highly diverse, including acute hepatitis, self-limiting recovery, chronic hepatitis, cirrhosis, liver cancer, and liver failure. The disease outcome is related to viral, environmental, and host factors. This map shows important host genes that may be associated with clinical outcomes of HBV infection or Hepatitis B vaccination. Black boxes show the relevant genes identified, in which the bold letter indicates a high degree of correlation and the italic indicates a possible correlation to clinical features of HBV infection.
In this review, we summarize the current state of research on the role of host genetic determinants of CHI, its clinical type, therapeutic responses, and responses to hepatitis B vaccines (Tables S2–7). More than 500 publications varying in quality have focused on this topic. Thus, we assessed the reliability of these studies according to the sample size, type of technology, and analysis methods. Note that for great many genes related to HBI or vaccine efficacy, only a single study was available. For intrauterine infections, OBI, drug efficacy, and other factors, all published papers were included owing to the small sample size. We also considered whether there are valid contradictions among studies. We classified studies into three categories: very relevant (I), possibly relevant (II), and unable to confirm (III) (Table 1; Table S1), as a guide for readers. The topic of host genetics and end-stage liver diseases, including HCC, is reviewed by An et al. (An et al., 2018) in this special issue and therefore is not included in our review.

Table 1 Classification of host genetic determinants associated with hepatitis B virus (HBV)-related liver diseases and responses to HBV vaccines.
Early epidemiological data showed that chronic HBV infection is very frequently established in infants born to HBV carrier mothers or in young children (Stevens et al., 1975; Li et al., 2015) and shows a strong gender disparity with significantly higher susceptibility of male patients (Lee et al., 1999; Su et al., 2007; Liaw et al., 2009; Wang et al., 2015). Adequate host immune responses to HBV are essential for sustained viral control (Bertoletti and Gehring, 2006). HBV control is associated with vigorous, multi-specific T cell responses in the host. In chimpanzees, T cell depletion prevents HBV clearance (Thimme et al., 2003; Asabe et al., 2009; Hoogeveen et al., 2018), demonstrating the essential role of cellular immunity in HBV control. Chronic HBV infection is associated with impaired immune responses (Guidotti et al., 2015; Lebosse et al., 2017). Thus, great efforts have been made to identify associations between host immunogenetics and chronic HBV infection. A number of host genetic variants, such as mutations in human leukocyte antigens (HLAs), cytokine and chemokine genes, toll-like receptor (TLRs), microRNAs, vitamin D–related genes, and HBV receptor sodium taurocholate cotransporting polypeptide (NTCP), have been found to influence the outcome of HBV infection (Table 2; Tables S1, S2). The SNPs loci of genes very relevant or possibly relevant with chronic HBV infection are listed in Table 2. Other SNPs loci of genes that are still unable to confirm are listed in Tables S1 and S2.
HLA is the human major histocompatibility complex (MHC) and a central element of antiviral immune defense (Shiina et al., 2009). HLA genes are classified as class I (HLA-A, B, C, E, F, and G) and II (HLA-DP, DQ, DR, DM, and DO) (Koller et al., 1989; van Lith et al., 2010). When a pathogen enters the body, it is engulfed by antigen-presenting cells (APCs) and the pathogen proteins are digested into small pieces and loaded onto HLA antigens. They are then displayed by APCs to T cells, which produce a variety of effector molecules to eliminate the pathogen. Therefore, HLAs can affect the outcome of infectious diseases (Singh et al., 2007; Goulder and Walker, 2012; Wang et al., 2016). Genetic polymorphisms, especially in HLA class II genes, are significantly associated with HBV infection and pathogenesis according to many studies (Kamatani et al., 2009; Zhang et al., 2014b; He et al., 2015; Wasityastuti et al., 2016; Trinks et al., 2017). Kamatani et al. (Kamatani et al., 2009) performed a GWAS of chronic HBV infection using 786 cases and 2,201 HBsAg seronegative controls in the Japanese population, followed by a replication study of three additional Japanese and Thai cohorts consisting of 1,300 cases and 2,100 controls. HLA-DPA1 rs3077 and HLA-DPB1 rs9277535 were significantly associated with chronic HBV infection in the Asian population (combined OR [95% CI] = 0.56 [0.51–0.61], P = 2.31 × 10–38; OR [95% CI] = 0.57 [0.52–0.62], P = 6.34 × 10–39, respectively). They also identified four associated haplotypes: HLA-DPA1*0202-DPB1*0501 and HLA-DPA1*0202-DPB1*0301 were associated with susceptibility to chronic hepatitis B and HLA-DPA1*0103-DPB1*0402 and HLA-DPA1*0103-DPB1*0401 showed a protective effect. Several GWAS of Asian populations have also shown that SNPs in HLA regions, such as HLA-DP (rs9277535, rs3077, rs9366816, and rs9277542), HLA-DQ (rs2856718, rs7453920, rs9276370, rs7756516, and rs7453920), HLA-DPB1 (positions 84–87), HLA-C (Leu-15, rs3130542, and rs2853953), HLA-DRB1*13, HLA-J (rs400488), and HLA-DOA (rs378352), are associated with chronic HBV infection (Mbarek et al., 2011; Nishida et al., 2012; Hu et al., 2013; Kim et al., 2013; Chang et al., 2014; Jiang et al., 2015; Zhu et al., 2016b). Many studies have provided evidence based on case–control samples and conventional PCR-based detection methods that HLA molecules may be crucial determinants of the outcomes of HBV infection in Asian, European, African-American, Saudi Arabian, and Caucasian populations (Thomas et al., 2012; Al-Qahtani et al., 2014; Trinks et al., 2017). However, some studies have reported contradictory results. A study of the Chinese Zhuang population found no association between HLA-DPA1 rs3077 and chronic HBV infection (Wang et al., 2011). Akgöllü et al. (Akgöllü et al., 2017) suggested that HLA-DP rs3077 is not associated with HBV infection in the Turkish population. Vermehren et al. (Vermehren et al., 2012) failed to confirm an association of HLA-DPB1 rs9277535 with hepatitis B in Caucasians. A comprehensive meta-analysis suggested that HLA-DP/DQ (rs3077, rs9277535, rs9275572, rs9275319, rs2856718, and rs7453920) is associated with susceptibility to HBV or the clearance of HBV infection (Zhang et al., 2013e; Yu et al., 2015; Xu et al., 2017b; Xu et al., 2018). A meta-analysis by Huang et al. (Huang et al., 2016) suggested that HLA-DQB1*0201, DQB1*0301, and DQB1*0502 are associated with an increased risk of CHB (chronic hepatitis B), while HLA-DQB1*0303 and DQB1*0604 are associated with a decreased risk of CHB.
Another meta-analysis indicated that HLA B*07 and B*58 protect against chronic HBV infection (Seshasubramanian et al., 2018). Recent studies have shown that non-classical HLA-class I molecules, including HLA-E may also be related to hepatitis B virus infection (Zidi et al., 2016). Available data are consistent with the fact that cellular immunity is a major determinant for HBV control. However, it is not clear how the identified genetic variation influences immune functions in the host and specific immune responses to HBV. These critical questions need to be answered in future studies.
Cytokines are key antiviral and immunomodulatory molecules produced by immune cells and certain non-immune cells; they are involved in host defense against HBV infection and pathogenesis (Koziel, 1999; Shin et al., 2016). The antiviral and immunomodulatory functions of a number of cytokines, like type I and II IFNs, TNF-α, IL-10, and IL-21, play essential roles in the direct suppression of HBV replication in hepatocytes (Phillips et al., 2010), mediating the antiviral functions of T cells (Freeman et al., 2012), and the modulation of adaptive immune responses to HBV (Lin and Young, 2014; Gehring and Protzer, 2019). IFN-γ and TNF-α are major antiviral mediators of specific CD8+T cells and are required for HBV control in primary HBV infections (Phillips et al., 2010; Bertoletti and Ferrari, 2013). IL-10 may contribute to negative regulation of host immune responses and thereby plays a role in viral persistence (Saraiva and O’Garra, 2010). IL-18 is a cytokine that belongs to the IL-1 superfamily and is produced by macrophages and other cells (Mavragani and Moutsopoulos, 2014). Recently, IL-21 has drawn attention in the field of HBV research owing to its potential association with viral control via follicular helper T cell functions (Johnson and Jameson, 2009). A series of studies have identified associations between genes encoding cytokines (including IL-1, IL-4, IL-6, IL-10/IL-10RB, IL-12/IL-12B, IL-18, IL-27, IL-28B, IFN-γ, TNF-α, and TGF-β) and chronic HBV infection (Cheong et al., 2006; Du et al., 2006; Liu et al., 2006; Chen et al., 2010; Fletcher et al., 2011; Ma et al., 2014; Saxena et al., 2014; Talaat et al., 2014; He et al., 2015; Karataylı et al., 2015; Karra et al., 2015; Eskandari et al., 2017b). Clinical researches have revealed correlations between IL-18 -607C/A and -137G/C (rs1946519 and rs187238) polymorphisms and the risk of HBV infection (Karra et al., 2015). The study of a Indian population by Karra et al. involving 271 patients with hepatitis B–related liver diseases, including 109 with spontaneous recovery, 162 patients with persistent HBV infection, and 280 healthy volunteers, indicated that the −607A allele in the promoter region of the IL-18 gene may protect against HBV infection (healthy controls vs. cases, OR [95% CI] = 0.711 [0.559–0.904], P = 0.005). The AA genotype was associated with spontaneous clearance (spontaneous recovery vs. persistent HBV infection, OR [95% CI] = 2.765 [1.582–4.832], P = 0.0002), while the CC genotype was associated with HBV infection (persistent HBV infection vs. spontaneous recovery, OR [95% CI] = 0.318 [0.151–0.67], P = 0.001). Moreover, the -137C allele (cases vs. healthy controls, OR [95% CI] = 1.355 [1.037–1.771], P = 0.025) and the GC genotype (cases vs. healthy controls, OR [95% CI] = 1.558 [1.11–2.185], P = 0.01) were associated with persistent HBV infections. SNPs (−1,082, −592) within the IL-10 gene have been reported to be associated with susceptibility or the clearance of HBV infection (Cheong et al., 2006; Talaat et al., 2014). Further studies are needed to determine the functions of these cytokines in HBV infection and thereby to explain these conflicting results.
IL-28B SNPs are major determinants of HCV clearance and the efficacy of IFN therapy (Ge et al., 2009; Prokunina-Olsson et al., 2013). This locus encodes a cytokine that is distantly related to type I interferons and the IL-10 family and is induced by viral infection (Sheppard et al., 2003). A systematic review by Lee et al. (Lee et al., 2014) including 4,028 patients with chronic hepatitis B and 2,327 spontaneously recovered controls from 11 case–control studies indicated that there is no significant association between IL28B SNPs (rs12979860, rs12980275, and rs8099917) and spontaneous HBV clearance. These findings are not surprising, as the mechanisms underlying HBV control may differ substantially from those underlying HCV infection.
Interferon gamma (IFN-γ) is a type II interferon and is critical for innate and adaptive immunity against viral, bacterial, and protozoal infections (Schoenborn and Wilson, 2007). Cellular responses to IFN-γ are activated by its interaction with a heterodimeric receptor consisting of IFN-γ receptor 1 (IFN-γR1) and IFN-γ receptor 2 (IFN-γR2) (Robek et al., 2004). Previous studies have reported that several polymorphisms in IFN-γ, IFN-γR1, and IFN-γR2 are associated with the natural history of HBV infection (Liu et al., 2006; Khanizadeh et al., 2012). In a study by Liu et al. (Liu et al., 2006) including 181 patients with HBV infection and 272 gender, age-matched healthy controls, the A allele frequency of IFN-γ +874 (OR [95% CI] = 2.25 [1.69–2.99], P < 0.0001), and the AG haplotype (+874A and +2109G) (P < 0.0001) were found to significantly influence susceptibility to HBV infection in the Chinese population. A meta-analysis by Sun et al. (Sun et al., 2015) suggested that the IFN-γ +874T/A polymorphism contributes to an increased risk of HBV-related diseases, especially in Asians.
Chemokines are a family of small cytokines found in all vertebrates, some viruses, and some bacteria; they guide cells of the innate and adaptive immune systems. Some chemokines are involved in the control of immune cells during immune surveillance, directing lymphocytes to lymph nodes, to screen for pathogen invasion via interactions with APCs residing in these tissues (Kehrl, 2006; Mélik-Parsadaniantz and Rostène, 2008).
To date, studies of cytokine and chemokine genes have evaluated limited numbers of SNPs with small sample sizes, and rather inconsistent results have been obtained. Therefore, future large-scale and multi-center studies are needed to establish the relationship between the genetic control of cytokine- and chemokine-related functions and chronic HBV infection, given the importance of cytokines in anti-HBV immune responses.
TLRs play key roles as pattern recognition receptors; they activate the innate immune system (Chang, 2010; Kondo et al., 2011) and are required for efficient host defense against viral infection. Deficiencies in TLR or TLR-related signaling greatly reduce host-specific T cell responses to HBV and may contribute to HBV persistence (Ma et al., 2017). Thus, genetic variation in TLRs is expected to impact the susceptibility to chronic HBV infection (Al-Qahtani et al., 2012a; He et al., 2015; Huang et al., 2015; Zhu et al., 2017). Multiple SNPs in TLR genes (TLR3 rs1879026, rs3775290, and rs3775291, TLR7 rs179010, and TLR9 rs352140) have been examined for their association with the risk of HBV infection (He et al., 2015; Huang et al., 2015; Zhu et al., 2017). SNPs in TLR3 may be associated with an increased risk of chronic HBV infection. Al-Qahtani et al. (Al-Qahtani et al., 2012a) investigated SNPs in the TLR3 gene in Saudi Arabian patients chronically infected with HBV, including 707 patients and 600 uninfected controls. Only TLR3 rs1879026 (OR [95% CI] = 0.809 [0.655–0.999], P = 0.0480) and the GCGA haplotype (rs1879026, rs5743313, rs5743314, and rs5743315) (P = 0.0339) potentially contribute to the risk of HBV infection. Huang et al. (Huang et al., 2015) indicated that the TT genotype of TLR3 rs3775290 is closely correlated with a decreased risk of CHB. A meta-analysis by Geng et al. (Geng et al., 2016) demonstrated a significant effect of TLR3 rs3775291 and HBV-related diseases.
MicroRNAs (miRNAs) are a group of endogenous, highly conserved, small noncoding RNAs that modulate various cellular processes and play key roles in host–virus interactions and the pathogenesis of viral diseases (Qureshi et al., 2014). HBV replication is also regulated by miRNAs via their cellular targets (Zhang et al., 2011a; Zhang et al., 2013d; Lin et al., 2017). Hundreds of SNPs in miRNAs have been reported. Recent studies have shown that some miRNAs may play a role in anti-HBV defense (Su et al., 2011). In addition, miRNA polymorphisms are related to hepatitis B infection (Bae et al., 2012; Cheong et al., 2013; Al-Qahtani et al., 2017). A study of a Saudi Arabian population including 1,352 HBV-infected patients and 600 uninfected healthy individuals found that SNPs in different microRNA genes, including miR-149 (rs2292832), miR-146a (rs2910164), miR-196a-2 (rs11614913), and miR-30a (rs1358379), were associated with hepatitis B infection, while other SNPs in microRNA genes, including miR-423 (rs6505162), miR-492 (rs2289030), miR-146a (rs2910164), miR-196a-2 (rs11614913), and miR-30a (rs1358379) were associated with HBV clearance (Al-Qahtani et al., 2017). A meta-analysis (Zhou et al., 2015) has shown that the carriers of miR-196a-2*T (rs11614913), miR-122*del (rs3783553), miR-106b-25*A (rs999885), and miR-let-7c*del (rs6147150) alleles in the Asian population have an increased risk of chronic HBV infection.
Cytotoxic T-lymphocyte-associated protein 4 (CTLA4) is expressed by T lymphocytes and plays a key role as a negative regulator of T-cell responses (Buchbinder and Hodi, 2015). Several studies have evaluated the association between CTLA4 polymorphisms and chronic HBV infection. In particular, the + 49A/G polymorphism (rs231775) of CTLA4 may influence susceptibility to HBV infection in the Chinese population (Chen et al., 2010). Mohammad et al. (Mohammad et al., 2006) found that CTLA-4 -318 polymorphisms (rs5742909), but not +49 and −1,172 polymorphisms (rs733618) are significantly associated with susceptibility to chronic HBV infection in the Iranian population (OR = 0.49, 95% = 0.206–1.162, P = 0.012). Another study also showed that the CTLA4 –318C > T, but not +49G > A, is associated with chronic HBV infection (Schott et al., 2007). A meta-analysis (Xu et al., 2013) suggested that the A allele of the CTLA4 +49 polymorphism is significantly associated with an increased risk of persistent HBV infection, whereas the G allele may influence viral clearance. Another meta-analysis also suggested that the CTLA-4 +49A/G polymorphism is significantly correlated with persistent HBV infection in the Asian population (Huang et al., 2013).
NTCP encoded by the solute carrier family 10 member 1 (SLC10A1) gene is mainly expressed in hepatocytes and a functional receptor for HBV and hepatitis D virus (Hagenbuch and Meier, 1994; Yan et al., 2012a). NTCP rs2296651 (S267F) has been found to be related to HBV susceptibility in the Asian population (Hu et al., 2016; Yang et al., 2016a; Nfor et al., 2018; Wu et al., 2018a). In a larger cohort of Taiwanese patients, including 3,801 with chronic HBV infection and 3,801 matched HBsAg seronegative controls, the AA genotype of the S267F variant (rs2296651) was associated with resistance to chronic HBV infection (OR = 0.13, 95% CI = 0.05–0.34, P < 0.001), but there were no significant associations with serological outcomes, including HBV DNA detectability and HBeAg and HBsAg seroclearance (Hu et al., 2016). However, the S267F variant is absent in the Moroccan population (Ezzikouri et al., 2017). A large cohort study did not confirm the association between the common and rare alleles or CNVs in the SLC10A1 gene with the risk of persistent HBV infection in a population from Southern China (Zhang et al., 2017). A meta-analysis including 14,591 chronically HBV-infected patients and 12,396 healthy controls suggested that the A allele and GA genotypes of rs2296651 are inversely correlated with chronic HBV infection. They also reported that NTCP rs4646287, rs7154439, and rs4646296 show no significant correlation with HBV infection (Wang et al., 2017a).
The signal transducer and activator of transcription (STAT) protein family mediates many aspects of cellular immunity, proliferation, apoptosis, and differentiation (Miklossy et al., 2013). STAT proteins are involved in the development and function of the immune system and play a role in maintaining immune tolerance (Singh et al., 2009). In HBV infection, polymorphisms in STAT4 are related to the clinical outcome of HBV infection (Lu et al., 2015b; Jiang et al., 2016b). A case–control study including 1,610 Chinese patients with chronic HBV infection and 1,423 uninfected control subjects showed that the STAT4 SNPs rs7574865, rs10168266, rs11889341, and rs8179673 are significantly associated with the risk of HBV infection and inversely related to HBV clearance. They also found that the CTCTT haplotype, formed by the SNPs rs8179673, rs7574865, rs4274624, rs11889341, and rs10168266, is related to HBV infection susceptibility and clearance in the Chinese Han population. A meta-analysis including 8,944 chronically HBV-infected patients, 8,451 healthy individuals, and 2,081 subjects with HBV clearance demonstrated that STAT4 rs7574865 is significantly associated with HBV infection (OR [95% CI] = 1.14 [1.07–1.21]; P = 3.8 × 10−5) and clearance (OR [95% CI] = 1.20 [1.07–1.35], P = 0.002) (Jiang et al., 2016b). Lu et al. (Lu et al., 2015b) showed that STAT4 minor alleles (rs7574865, rs7582694, rs11889341, and rs8179673) are associated with spontaneous HBV clearance. Another meta-analysis by Liao et al. (Liao et al., 2014) did not confirm the correlation between STAT4 rs7574865 and HBV susceptibility or clearance. Thus, the association of STAT4 polymorphisms with HBV infection requires further verification.
Signal transducer and activator of transcription 3 (STAT3) and nuclear factor-kappaB (NF-κB) pathways may play a significant role in chronic HBV infection. A Chinese Han population study showed that STAT3 rs1053004 and rs1053005 polymorphisms and haplotypes formed by rs1053004 and rs1053005 might contribute to the susceptibility to chronic HBV infection (Li et al., 2018). Another Chinese study also found that the SNPs rs2233406 (CT versus CC) and rs3138053 (AG or AG+GG versus AA) in the promoter region of NFKBIA were significantly associated with decreased risk of HBV persistence, especially in genotype B HBV-infected subjects (Zhang et al., 2014a). Unfortunately, related research is still not sufficient and mainly focused on HCC. Their role of HBV infection needs to be better defined by future research.
Using a GWAS approach, several loci that are strongly associated with HBV clearance or susceptibility to chronic HBV infection have been identified, including TCF19 (Kim et al., 2013; Jiang et al., 2015), UBE2L3 (Hu et al., 2013; Jiang et al., 2015), CFB (Jiang et al., 2015), NOTCH4 (Jiang et al., 2015), CD40 (Jiang et al., 2015), EHMT2 (Kim et al., 2013; Jiang et al., 2015; Shin et al., 2019), and INTS10 (Li et al., 2016).
Vitamin D has the ability to activate the innate immune system and dampen the adaptive immune system (Hewison, 2011). A vitamin D deficiency is associated with an increased risk or severity of viral infections (Beard et al., 2011). Vitamin D has both anti-inflammatory and anti-microbial effects and may play an active role in HBV infection (Bellamy et al., 1999; Beard et al., 2011; Gao et al., 2017). Vitamin D–related genes include 1-α-hydroxylase (CYP27B1), cytochrome P450 family 2 subfamily R polypeptide 1 (CYP2R1), vitamin D–binding protein (GC), VDR, 7-dehydrocholesterol reductase (DHCR7), sterol 27-hydroxylase (CYP27A1), and cytochrome P450 family 24 subfamily A member 1 (CYP24A1). Vitamin D–related genes have recently been found to play an important role in HBV susceptibility (Chatzidaki et al., 2012; Gao et al., 2017). The majority of these studies have focused on four SNPs in VDR: ApaI (rs7975232), TaqI (rs731236), FokI (rs10735810), and BsmI (rs1544410). Chatzidaki et al. (Chatzidaki et al., 2012) found that the joint haplotype of the VDR ApaI α allele and TaqI T allele is related to the outcome of HBV infection in children in the context of mother-to-child transmission (RR [95% CI] = 1.74 [0.97–3.13], P = 0.049). A meta-analysis (He et al., 2018) has reported that the genotypes FF, Ff, and allele F of FokI in VDR increase the risk of HBV infection. However, no associations were found between VDR ApaI and BsmI polymorphisms and HBV infection.
There is a marked difference in HBV infection rates between populations in East/Southeast Asia and Western world. For instance, the prevalence of HBsAg was as high as 10% in the Chinese population prior to the initiation of the HBV vaccination program but less than 1% in the Caucasian population (Fattovich et al., 2008; Yin et al., 2010). The reason for this difference is not completely clear. The role of host genetic determinants is also not well-established. Li et al. (Li et al., 2015) summarized multiple studies and found that some genetic loci with rare alleles include rs3138053 (NFKBIA), rs2856718, rs7453920, and rs9275319 (HLA-DQ), and rs9277378, rs2395309, rs2301220, and rs9277341 (HLA-DP) are significantly associated with decreased risks of development of chronic HBV infection. These loci are different between the Chinese Han and European populations, indicating that the Chinese Han patients may be inherently more prone to develop chronic infection once exposed to HBV, whereas Europeans tend to recover from HBV infection spontaneously (Li et al., 2015). A study including Chinese Han and Uyghur populations reported that there are some differences in HLA-DP/DQ polymorphisms that affect susceptibility and resistance to HBV infection (Xiang et al., 2016). A meta-analysis by Geng et al. (2016) demonstrated that TNF-α-238 increases the risk in the European population but not in an Asian population in all genetic models. Therefore, the genetic determinants may contribute the different rates of chronic HBV infection in Asian and European populations. However, future global multicenter research is needed to precisely define such host genetic determinants.
HBV infection often occurs in families. If a mother is chronically HBV infected, infants have an extremely high chance of establishing a chronic HBV infection in the absence of medical interventions (Stevens et al., 1975; Chen et al., 2018; Dionne-Odom et al., 2018). HBV exposure during infancy or early childhood is more prone to the development of chronic infection and HBV infection-related diseases (Lai and Yuen, 2007). Studies have shown that spontaneous HBsAg clearance is most likely to occur in patients born to mothers of non-HBsAg carriers (Chiu et al., 2014). Globally, a total of 42.1% of infants naturally born to HBsAg carrier mothers but only 2.9% of infants with HBV passive–active immunoprophylaxis acquired HBV infection perinatally (Li et al., 2015). The rate of vertical HBV infection had stayed constant (approximately 0.3%) until immunoprophylaxis of mother-to-infant transmission was implemented in 1986 in Japan. In contrast, horizontal HBV transmission decreased from 1.43 to 0.10% in men and from 0.95 to 0.03% in women over years. The decrease of horizontal transmission of HBV may be due to many factors, including improved socioeconomic environments, advanced medical maneuvers and equipment, and careful vaccination procedures (Sato et al., 2014). However, the current studies on host genetic and chronic HBV infection did not separate vertical from horizontal transmission, nor did they stratify the age at the time of infection. The role of environmental exposure including early exposure of HBV in the childhood when immune function is immature, and their interaction with the genetic predisposition is still not clarified.
In studies of associations between host genetic determinants and chronic HBV infection to date, reported OR values for significant associations typically range from 1 to 2. Thus, these determinants have much less influence than the HBV infection status of the mother or the age of the HBV infection. Nevertheless, some genetic variants, like mutations in NTCP, may play a decisive role in HBV infection. NTCP variation could prevent the binding of the HBV virion to receptors on hepatocytes and therefore limit viral infection (Yan et al., 2012a). Such findings may provide mechanistic insights into the viral life cycle and new targets for drug development. Many genetic determinants (HLA, TLR, IL) may modulate host innate and adaptive immunity and significantly influence the course and outcome HBV infection (Kamatani et al., 2009; Mbarek et al., 2011; Kim et al., 2013; Jiang et al., 2015; Geng et al., 2016) (Table 2; Table S2). It will be useful to identify the functional relevance of these genetic polymorphisms in cellular signaling and other processes, with the goal of identifying new targets. Many factors contribute to the development of chronic HBV infection, such as the mode of transmission (vertical or horizontal transmission), age at the time of infection, the presence of underlying diseases involving other organs, and the use of immune inhibitors (Pawlowska et al., 2016; Chien et al., 2019). The majority of studies do not including subgroup analyses of these factors, which may have a significant impact on the reliability of the results. Furthermore, there are substantial variations among studies from different countries and regions due to differences in the study populations, genetic background, and polymorphisms. Therefore, it may be necessary to conduct stratified analyses of HBV infection time, ethnicity, host immune status, and mode of transmission to further determine the impact of HLA-DP, DQ, and DR on chronic hepatitis B or clearance. Nevertheless, the available information suggests that genetic variation in host immune-related genes has substantial contribution to the outcome of HBV infection. Future research is necessary to uncover the mechanisms underlying these associations.
Mother-to-child transmission is the major route of HBV infection in developing countries (Stevens et al., 1975; Shao et al., 2007; Li et al., 2015). Intrauterine transmission is the main explanation for mother-to-child transmission, since the implementation of effective measures against HBV infection, such as antiviral treatment, vaccination, and the application of hepatitis B immunoglobulins (Gu et al., 2004; Brown et al., 2016; Ren et al., 2016). Therefore, blocking intrauterine HBV transmission is an important part of the eradication of HBV infection in the general population (Peng et al., 2019). Unfortunately, the incidence of intrauterine HBV infection remains high (Han et al., 2011). The exact mechanism underlying intrauterine infection with HBV has not been fully elucidated (Xu et al., 2008). Previous studies have shown that genetic factors could play an important role in determining the outcome of HBV intrauterine infection (Yu et al., 2006; Xu et al., 2008; Gao et al., 2015b; Liu et al., 2018d). Based on twin and family study, host genetic factors are critical for the development of chronic HBV infection (Lin et al., 1989). Up to date, SNPs in the host genes IFN-γ, TNF-α, CXCL13, PDCD1, TLR3, and TLR9 were found to be relevant for intrauterine transmission (Yu et al., 2006; Gao et al., 2015b; Wan et al., 2016; Liu et al., 2018c; Gao et al., 2015b) (Table S3).
A number of host genes may play important roles in HBV intrauterine infection (Table 3, Tables S1, S3). Yu et al. (Yu et al., 2006) showed that a SNP (+874) in IFN-γ is associated with HBV intrauterine infection (both P < 0.05). Wan et al. (Wan et al., 2016) investigated associations between candidate genes (SLC10A1, HLA-DP, HLA-C, CXCR5, CXCL13, TLR-3, TLR-4, TLR-9, and UBE2L3) and HBV intrauterine infection in a sample of 44 neonates with HBV intrauterine infection and 662 neonatal controls. They demonstrated that only the rs355687 CT genotype of CXCL13 was associated with susceptibility to HBV intrauterine infection (OR [95% CI] = 0.25 [0.08–0.82], P = 0.022).
Table 3 Host genetic factors associated with intrauterine hepatitis B infection or response to interferon-alpha (IFN-α) or nucleos(t)ide analog (NUC).
A case–control study of the Chinese population including 69 HBsAg-positive mother–newborn pairs with intrauterine infection and 138 mother–newborn pairs without intrauterine infection assessed the involvement of the LTβR/APOBEC3B signaling pathway and PD-1/PD-L1 signaling pathway genes, including LTA, LTBR, TNFSF14, PDCD1, APOBEC3B, CD274, CD40, and CD40LG, in HBV intrauterine transmission. They found that the maternal rs2227981 TT genotype of the PDCD1 gene is associated with a decreased risk of intrauterine HBV infection (OR 0.11, 95% CI = 0.01–0.95, P = 0.045). There was no significant correlation between the remaining genes and the risk of intrauterine HBV infection (Liu et al., 2018c). Only a few studies have evaluated genetic susceptibility to intrauterine transmission. It is often difficult to distinguish whether a child acquired an HBV infection by the intrauterine or intra-natal route. HBV infection by intrauterine transmission is mainly identified by a blood test at birth. However, this approach may result in false positives due to maternal blood contamination during birth, especially when sensitive PCR methods for HBV DNA detection are used. Moreover, the sample sizes of existing studies are not large enough; thus, the reliability of studies could be questioned. Further research is needed to improve the diagnostic accuracy of intrauterine HBV infection and explore the relationship between HBV intrauterine infection and susceptibility genes as well as other risk factors associated with HBV intrauterine infection.
There is no effective intervention available to prevent intrauterine HBV infection. Neither antiviral treatment during the late phase of pregnancy nor perinatal vaccination/hyperimmunoglobulin application changed the rate of intrauterine HBV infection (Pan et al., 2012a; Brown et al., 2016). Therefore, the identification of relevant genetic determinants could be very helpful.
OBI is defined by serum HBV DNA positivity but negative serum HBsAg detection (Torbenson and Thomas, 2002; Raimondo et al., 2008; Raimondo et al., 2019; Seed et al., 2019). If blood from patients with OBI is transfused to other persons, post-transfusion hepatitis may occur (Seo et al., 2011). OBI may also be associated with hepatitis B vaccination failure, vertical HBV transmission, organ transplant failure, cirrhosis, and liver cancer (Raimondo et al., 2013).
The mechanisms underlying OBI have yet to be elucidated and may be related to HBV mutations in the pre-S/S genomic region or antigen–antibody cycle immune complex formation (Zhang et al., 2011b; Samal et al., 2012). Some investigators have suggested that host genetic factors, especially immune-related factors including HLAs, IL10, CXCL12, and VDR, may play a role in the occurrence of OBI (Arababadi et al., 2010; Hassanshahi et al., 2010; Mardian et al., 2017; Wang et al., 2017b) (Tables S1, S4). Wang et al. (Wang et al., 2017b) performed a case–control study with 107 patients with OBI and 280 healthy individuals to assess associations of SNPs in HLA loci, including HLA-A, -B, -C, -DRB1, and -DQB1, in Chinese patients. They found that HLA-B*44:03 (OR = 2.146, 95% CI = 1.070–4.306, P = 0.028), C*07:01 (OR = 4.693, 95% CI = 1.822–12.086, P = 0.000), DQB1*02:02 (OR = 1.919, 95% CI = 1.188–3.101, P = 0.007), and DRB1*07:01 (OR = 2.012, 95% CI = 1.303–3.107, P = 0.001) were strongly associated with susceptibility to OBI, while DRB1*08:03 (OR = 0.395, 95% CI = 0.152–1.027, P = 0.049), DRB1*15:01 (OR = 0.495, 95% CI = 0.261–0.940, P = 0.029), and DQB1*06:02 (OR = 0.500, 95% CI = 0.249–1.005, P = 0.048) alleles were more prevalent in the healthy control group. Mardian et al. (Mardian et al., 2017) found a significant association between the minor allele “T” of HLA-DP (rs3077) and the TGA haplotype (rs3077–rs3135021–rs9277535) and OBI in Indonesian blood donors. Therefore, HLA polymorphisms are an important factor in OBI infection and additional studies are needed to validate these findings. Iranian researchers have shown that the genotypes and alleles of IL-10 (-592) (Ahmadabadi et al., 2012), the +801 region of CXCL12 (Hassanshahi et al., 2010), and the T/T allele of exon 9 of VDR (Arababadi et al., 2010) may be associated with OBI.
Owing to the low frequency of OBI, the accumulation of cases is difficult and time-consuming. Thus, the number of cases included in studies is always small (generally less than 100 cases). In addition, the occurrence of OBI is also related to HBV mutations (Samal et al., 2012). OBI diagnostic criteria are not uniform among studies. Both viral and host factors need to be considered in future analyses. Patients with OBI are positive for HBV DNA but negative for HBsAg. Thus, there could be specific mechanisms for the control of HBsAg production without affecting HBV DNA synthesis. Recently, Lin et al. found that autophagy and related genes (e.g., Rab7) may determine HBsAg production (Lin et al., 2017; Lin et al., 2019), but further studies are needed to evaluate this.
IFN-α and NUCs have been approved and are widely used for the treatment of chronic hepatitis B and slow down liver disease progression, cirrhosis, and HCC (Lee and Keeffe, 2011; European Association For The Study Of The Liver, 2017). However, NUCs and IFN-α alone or combined treatment are rare to achieve functional cure (Reijnders et al., 2010; Zoulim and Locarnini, 2012; Terrault et al., 2016; Revill et al., 2019). Extensive studies have identified host genetic genes, alanine aminotransferase (ALT) levels, HBV genotype, and HBV DNA, anti-HBc, and HBsAg levels as important predictors of treatment efficacy (Brouwer et al., 2014; Fan et al., 2016b; Zhu et al., 2016a). Accumulating evidence suggests that host genetics play an important role in the patient response to IFN-α or NUCs (Table 3; Tables S1, S5, S6).
IFN-α is a cytokine with a broad-spectrum effect against viruses and tumors. It is able to inhibit cell growth and exerts immunomodulatory effects. IFN-α exerts antiviral activity mainly via the Janus-activated kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway and interferon-stimulating genes (ISGs) (Levy and Darnell, 2002). IFN-α binds to type I IFN receptor (IFN-αR1) on the surface of target cells and induces the synthesis of antiviral proteins, such as MxA, 2’,5’-oligoadenylate synthetase (OAS), protein kinase (PKR), and adenosine deaminases acting on RNA (ADAR) (Samuel, 2001). In addition, IFN-α upregulates HLAs and thereby modulates adaptive immunity. Thus, host genetic polymorphisms, particularly those associated with IFN signaling pathways, may potentially alter the responsiveness to IFN-α therapy. Many studies have evaluated the relationship between the outcome of IFN-α treatment for CHB and host genetics and identified the potential relevance of HLAs, STAT4, vitamin D–related genes, and some ISGs (Tseng et al., 2011; Brouwer et al., 2014; Wu et al., 2014; Jiang et al., 2016a; Thanapirom et al., 2017; Brouwer et al., 2019) (Table S5).
The first GWAS published to date did not find any candidate gene associated with the efficacy of IFN-α treatment. This may be explained by an insufficient sample size, with only 43 patients (Chang et al., 2014). Recently, Brouwer et al. (Brouwer et al., 2019) published a GWAS on the efficacy prediction of PEG-IFN in the treatment of CHB, including 1,058 patients from 21 centers from Europe, Asia, and North America. A SNP rs78900671 GC allele (TRAPPC9, COL22A1) had been identified to have the strongest association with response to PEG-IFN treatment (P = 6.43×10-7). Another SNP PRELID2 rs371991 (P = 3.44×10-6) was associated with the primary response to PEG-IFN treatment in HBeAg-positive patients while G3BP2 rs3821977 (P = 2.46×10-6) was associated with response in HBeAg-negative patients. Yet the correlation between these SNP loci and the efficacy of interferon treatment needs confirmation by future studies.
HLA molecules play an important role in the recognition of viral proteins and regulation of adaptive immunity. Adaptive immunity is upregulated by IFN-α treatment and essential for sustained responses and HBV control (Bengsch and Thimme, 2018; Ma et al., 2018b). Thus, HLAs may be an important genetic marker for the prediction of IFN-α efficacy. Many studies have analyzed the relationship between HLA genes and the IFN-α response (Tseng et al., 2011; Zhu et al., 2011; Brouwer et al., 2014; Tangkijvanich et al., 2016). A study of a Taiwan population by Tseng et al. (Tseng et al., 2011) including 115 HBeAg-positive patients with CHB who received peginterferon (PEG-IFN) therapy showed that the HLA-DPA1 rs3077 GG genotype (OR [95% CI] = 3.49 [1.12–10.84], P = 0.031) is associated with HBeAg seroconversion at 6 months of therapy. Brouwer et al. (Brouwer et al., 2014) evaluated 262 Caucasian patients with chronic HBV infection who were treated with PEG-IFN for 1 year and found that the HLA-DPB1 (rs9277535) G-allele and haplotype block GG combining HLA-DPB1 (rs9277535) and HLA-DPA1 (rs3077) are strongly associated with virological (HBV DNA <2,000 IU/ml at 6 months post-treatment) and serological responses (HBeAg seroconversion combined with HBV DNA <2,000 IU/ml at 6 months post-treatment) to PEG-IFN therapy. Zhu et al. (Zhu et al., 2011) reported that the HLA-DQB1*0303 or DRB1*08 alleles and the *1101-*4601-*0102 (HLA-A, B, C) or *0302-*0303-*09 (HLA-DQA1, DQB1, DRB1) haplotypes are associated with the efficacy of IFN therapy in the Chinese population. However, some studies have failed to observe a relationship between HLA variants and the response to IFN-α- or PEG IFN-based treatment (Limothai et al., 2016).
IFN-λ3 is encoded by the IL-28B gene, which regulates the immune system by activating the JAK-STAT signaling pathway, thereby exerting an antiviral effect (Liu et al., 2012a). Recent GWAS has revealed that IL28B is associated with the efficacy of anti-HCV treatment (Ge et al., 2009). In a large international collaborative study of 205 HBeAg-positive patients with CHB who received PEG-IFN-α-2a or PEG-IFN-α-2b, IL28B rs12979860 and rs12980275 were independent predictors of HBeAg seroconversion in response to PEG-IFN (Sonneveld et al., 2012). Boglione et al. (Boglione et al., 2014) examined the effect of the IL28B gene polymorphisms rs12979860, rs8099917, and rs12980275 on the response to PEG-IFN in 190 chronically HBV-infected, HBeAg-negative patients and showed that rs12979860 CC, rs8099917 TT, and rs12980275 AA are significantly associated with HBV-DNA < 2,000 IU/ml post-treatment, while rs12979860 CC and rs12980275 AA are significantly associated with qHBsAg > 1 log during therapy. Many other research teams worldwide have indicated that IL-28B is involved in the efficacy of IFN-α therapy (Lampertico et al., 2013; Wu et al., 2015a; Cusato et al., 2017). However, variable and inconclusive results have been obtained (Cheng et al., 2014; Tangkijvanich et al., 2016). A study in Taiwan showed that IL28B rs12979860 is not associated with the virological response to PEG-IFN alpha-2b with or without entecavir in patients with HBeAg-negative CHB (Tangkijvanich et al., 2016). A Chinese study also suggested that IL28B (rs12979860 and rs8099917) has no significant association with the response to IFN-α or PEG IFN (Cheng et al., 2014). To determine whether IL28B polymorphisms play a role in the response to interferons, further large-scale cohort studies of homogenous patient populations are necessary.
Vitamin D has important immunomodulatory effects on innate and adaptive immune responses (Hewison, 2011). Vitamin D enhances the IFN-α-mediated activation of the JAK-STAT pathway, increases the expression levels of antiviral proteins, and improves the overall antiviral ability (Lange et al., 2014). Vitamin D–related genes, including VDR (Cusato et al., 2017), CYP27B1 (Cusato et al., 2017), CYP24A1 (Cusato et al., 2017), CYP2R1 (Thanapirom et al., 2017), DHCR7 (Thanapirom et al., 2017), and DBP/GC (Cusato et al., 2017; Thanapirom et al., 2017), have been evaluated for potential associations with the outcome of IFN-α therapy in patients with CHB. Several studies in China, Thailand, and Italy have shown that polymorphisms in the rs4646536 and rs10877012 loci of CYP27B1 may be related to the efficacy of IFN-α in patients with CHB; OR values reported in studies of Italy (Cusato et al., 2017), Thailand (Limothai et al., 2017), and Italy (Boglione et al., 2015) were 2.87, 3.13, and 3.13 for the rs4646536 locus (all P < 0.01); and the OR values for studies of Italy (Cusato et al., 2017) was 3.13 for the rs10877012 locus (both P < 0.001). Therefore, the TT (rs4646536) and GG (rs10877012) genotypes may be predictors of sustained HBeAg seroconversion in HBeAg-positive patients treated with IFN-α. Cusato et al. (Cusato et al., 2017) evaluated SNPs in vitamin D pathway genes in HBeAg-negative patients with CHB treated with PEG-IFN-α, suggesting that VDR (rs7975232 [ApaI], rs11568820 [Cdx2], rs10735810 [FokI]), CYP27B1 (rs4646536 [+2838], rs10877012 [−1260]), VDBP (rs7041), and CYP24A1 (rs2248359) are correlated with the clinical outcomes of PEG-IFN-α treatment. Thanapirom et al. (Thanapirom et al., 2017) showed that the CYP2R1 (rs12794714) TT genotype is a predictor of sustained HBeAg seroconversion in HBeAg-positive patients treated with Peg-IFN-α (OR [95% CI] = 4.53 [1.51–13.61], P = 0.01), whereas SNPs in DHCR7 (rs12785878), CYP27B1 (rs10877012), CYP2R1 (rs2060793), GC (rs4588, rs7041, rs222020, and rs2282679), and VDR (FokI, BsmI, Tru9I, ApaI, TaqI) are not related to HBeAg seroconversion. Several studies have investigated the role of other SNPs in vitamin D pathway genes in predicting the outcome of IFN-α therapy, but their predictive value remains controversial (Thanapirom et al., 2017). Differences among studies could be explained by several factors, such as differences in study design, sample size, ethnicity, and genotyping methods.
SNPs in JAK-STAT pathway genes and ISGs may contribute to the antiviral effects of IFN-based therapy (Kong et al., 2007; Ren et al., 2011; Wu et al., 2014; Jiang et al., 2016a). OAS3, MxA-88, and STAT4 genes in the IFN signaling pathway may have predictive value for the efficacy of interferons, but further studies are needed to verify this.
NUCs (e.g., lamivudine [LAM], adefovir [ADV], telbivudine [LdT], entecavir [ETV], and tenofovir [TDF]) inhibit HBV replication mainly by inhibiting HBV DNA polymerase. They bind competitively to HBV DNA polymerase and are incorporated into the DNA chain, resulting in the termination of DNA synthesis and inhibition of HBV replication (Lok et al., 2007; European Association For The Study Of The Liver, 2017). As the direct effect of NUCs on HBV does not require host functions and may not be directly related to host genetics, few studies have evaluated the correlation between host gene SNPs and drug efficacy, and these have generally yielded negative results (Table 3; Tables S1, S6).
There were significant differences in the patterns of HBsAg decline, and seroclearance during LAM therapy in Japanese patients with HBeAg-positive CHB was associated with HLA-DPA1 (rs3077), HLA-DPB1 (rs9277535), and A alleles at rs3077 and rs9277535 (Hosaka et al., 2015). A GWAS (Chang et al., 2014) with small sample sizes (LAM group, n = 119; ETV group, n = 64) of male Taiwanese individuals revealed that the TT genotype of rs9276370 (HLA-DQA2) is highly associated with a non-sustained response in the LAM group (OR [95% CI] = 5.41 [1.73–16.95], P = 0.0037), but not in the ETV group (OR [95% CI] = 0.68 [0.16–2.86], P = 0.5954). Zhang et al. (Zhang et al., 2014b) reported that the HLA-DQ (rs9275572) A allele is associated with viral and biochemical responses to LAM treatment in Chinese patients.
Using a small sample size (n = 76), our team evaluated the virological responses associated with the ESR1 PvuII T/C genotype in nucleoside-naive patients with chronic hepatitis B treated with ETV after 48 and 96 weeks of treatment; we did not detect associations between XbaI polymorphisms and virological response (Zhang et al., 2013c). Another study also found that CTLA4 (rs231775) and HLA-DPA1 (rs3077) are associated with predictors of relapse and outcomes after discontinuing TDF and ETV therapy in Taiwan patients with CHB (Su et al., 2018). However, the sample sizes of these studies are insufficient.
The main issues with studies of the efficacy of IFN/NUCs are as follows. 1) ALT, HBsAg, HBeAg, anti-HBc, HBV DNA, and HBV genotype may affect the efficacy of IFN/NUCs, and it is difficult to control for these confounding factors. 2) Some studies do not distinguish between HBeAg-positive or -negative patients, resulting in the inconsistent use of drugs as well as differences in treatment duration. 3) The criteria for determining efficacy and time points for evaluations are often inconsistent. 4) Many studies include small sample sizes and a lack of secondary verification. 5) Different and even contradictory results have been reported. Despite studies showing that several SNP sites in IL28B are closely related to the efficacy of IFN-α for the treatment of HCV infection, it is difficult to confirm that any host genetic determinant is significantly related to the efficacy of IFN-α or NUCs in chronic HBV infection without resolving these issues. It is necessary to clarify whether host genetic factors determine the efficacy of antiviral therapies. Currently, available antiviral treatments based on IFN-α/NUCs are not effective to clear HBV covalently closed circular DNA (cccDNA). Information about the involvement of host factors in antiviral activity may be helpful to optimize treatment approaches.
Attempts to improve the response by administering two different NUCs or a combination of NUCs and IFN-α failed to increase the rate of “functional cure.” New drug candidates targeting different steps of HBV life cycle including entry, cccDNA formation, viral transcription, capsid assembly, and secretion of viral envelope proteins were developed and partly tested in clinical trials (Qazi et al., 2018; Tu and Urban, 2018; Yang and Lu, 2018; Yang et al., 2019). Immunotherapeutic approaches to enhance antiviral immunity using IL-15, TLR agonists, retinoic acid inducible gene I (RIG-I) agonist, stimulator of IFN genes (STING) agonist or based on engineered T cells are also under investigation (Ma et al., 2015; Zhang and Lu, 2015; Guo et al., 2017; Koh et al., 2018). A new drug targeting cIAPs has being now tested in a clinical trial phase I (Liu et al., 2018a). Checkpoint inhibitors like anti-PD1 are able to stimulate host adaptive immunity and considered as potential drug candidates to treat chronic HBV infection (Liu et al., 2014a; Liu et al., 2018b). Such innovative drugs, Along with current available therapies, will offer the prospect of a markedly improved response to treatments and an increased rate of functional cure (Ko et al., 2015; Blank et al., 2016; Di Scala et al., 2016; Zhu et al., 2019). Those new drug candidates, especially for immunotherapies, may exert the antiviral actions in dependence on host genetic background. Some genetic determinants found in the previous studies like SNPs in HLA loci may reappear again in such studies. Their efficacy in anti-HBV treatment related to human genetics remains to be a topic for future studies.
Hepatitis B vaccines are widely used for the prevention of new HBV infections and are the most effective way to prevent HBV transmission (Kew, 1995; Ni et al., 2012; Bruce et al., 2016). The overall success rate of vaccination is about 90%. Several factors, such as age, gender, body mass index (BMI), immunosuppression, vaccine escape mutations, and premature birth, are associated with a failure to respond to hepatitis B vaccination (Zuckerman, 2006; Kang et al., 2014; Yang et al., 2016b). A twin study indicated that the immune response to hepatitis B vaccination has a high heritability (77%) (Newport et al., 2004). Yan et al. investigate the overall contribution of genetic and environmental effects on poor response to HBV vaccination in Chinese infants. They found that the HBV vaccine response in infants is dominantly determined by genetic effect by 91%, compared with perinatal environmental factors (Yan et al., 2013). Genetic factors may have important roles in the immune response to hepatitis B vaccination, and many studies have focused on the identification of these genes including HLAs and IL4 (Png et al., 2011; Cui et al., 2013; Li et al., 2013b; Nishida et al., 2018) (Table 4; Tables S1, S7).
A recent GWAS has shown that genetic variants in HLA loci are strongly associated with chronic HBV infection. Difference in host immune responses to HBV antigens are largely explained by HLA alleles (Ovsyannikova et al., 2004). Associations of SNPs in HLA regions, including -A, -B, -DR, and -DQ loci, with the response to hepatitis B vaccines have been identified (Davila et al., 2010; Png et al., 2011; Yoon et al., 2014; Roh et al., 2016). A GWAS of an Indonesian population including 3,614 hepatitis B vaccine recipients suggested that HLA loci, including HLA-DR (rs3135363) (P = 6.53 × 10−22; OR = 1.53, 95% CI = 1.35–1.74), HLA-DP (rs9277535) (P = 2.91 ×10−12; OR = 0.72, 95% CI = 0.63–0.81), and HLA class III (rs9267665) (P = 1.24 × 10−17; OR = 2.05, CI = 1.64–2.57), are strongly associated with the response to hepatitis B vaccination (Png et al., 2011). Other GWAS have found that HLA-DRB1 (rs477515, rs28366298, and rs13204672), HLA-DP (rs7770370), and multiple HLA-DRB1-DQB1 haplotypes are associated with the response to hepatitis B vaccines (Davila et al., 2010; Wu et al., 2015b). Although similar results have been obtained in several additional studies (Yoon et al., 2014; Roh et al., 2016; Sakai et al., 2017), other studies do not support these associations, likely owing to the sample sizes, statistical methods, or the use of different vaccines (Okada et al., 2017; Xu et al., 2017a). A meta-analysis (Li et al., 2013b) has shown that HLA class II genotypes (HLA-DRB1*01, DRB1*1301, DRB1*15, HLA-DQB1*05, DQB1*0501, DQB1*06, DQB1*0602) are associated with the antibody response to the hepatitis B vaccine, while the opposite results were found for DRB1*03 (DRB1*0301), DRB1*04, DRB1*07, DRB1*1302, and DQB1*02. Taken together, HLA is an important marker for the response to the hepatitis B vaccine; multicenter studies are needed for verification and to determine the mechanism underlying this association.
Cytokines play key roles in regulating immune responses and may modulate responses to HBV vaccines (Harber et al., 2000; Howe et al., 2005). Among these cytokine genes, IL-4 induces the differentiation of naive helper T cells to Th2 cells and is a key regulator of humoral immune responses. Genetic polymorphisms in IL-4 have been reported to be associated with the response to the hepatitis B vaccine in many studies (Chen et al., 2011; Wang et al., 2012), such as IL-4 (rs2070874, rs2243250, rs2227282, rs2243248, rs2227284) (Chen et al., 2011; Wang et al., 2012), IL-4RA (rs1805015) (Chen et al., 2011), IL-12B (rs17860508) (Pan et al., 2012b), and IL-13 (rs1295686) (Chen et al., 2011) that are associated with responses to the hepatitis B vaccine. In a meta-analysis of eight published studies, Cui et al. (Cui et al., 2013) evaluated associations of IL4 genetic polymorphisms, including rs2243250 (C > T), rs2070874 (C > T), rs2227284 (A > C), rs2227282 (C > G), and rs2243248 (G > T), with the response to the hepatitis B vaccine. The analysis suggested that the T allele of rs2243250, the T allele of rs2070874, and the C allele of rs2227284 in IL4 may be useful biomarkers for predicting favorable responses to the hepatitis B vaccine, especially in the Asian population. Relatively few studies have examined other cytokine genes.
Other studies have suggested that polymorphisms in other host genes (BTNL2, FOXP1, LILRB4, TLR2, VDR, etc.) may be related to responses to hepatitis B vaccination (Davila et al., 2010; Chen et al., 2011; Grzegorzewska et al., 2014), but further validation of these results is required.
The overall success rate of vaccination is about 90% (Zuckerman, 2006), and the factors that affect the success of vaccination mainly include 1) the type of vaccine; 2) the dose, mode, and frequency of inoculation; 3) the vaccination schedule; 4) ethnic differences; 5) the age at vaccination; 6) the presence of severe heart, liver, and kidney disease; 7) OBI status; and 8) assessment criteria for successful vaccination. When analyzing the effect of SNPs on the success of vaccination, it is important to match patients with respect to the main factors or perform appropriate statistical analyses to account for these factors. However, at present, correlation analyses are relatively simple. In addition, most subjects are under 20 years of age, and age is related to the rate of antibody production in response to the hepatitis B vaccine. These factors can lead to differences and contradictory results among studies. Further in-depth analyses of host genes may be helpful for understanding the mechanisms underlying the response to hepatitis B vaccination and for exploring new approaches for the development of more effective hepatitis B vaccines. Furthermore, studies should evaluate whether so-called non-responders, who do not produce anti-HBs antibodies after vaccination, may develop chronic HBV infection. It would be useful to determine the genetic factors leading to non-responsiveness to HBV vaccines.
A large number of studies have reported host genetic factors that are associated with chronic HBV infection, clinical type, therapeutic responses, or responses to hepatitis B vaccines (Figure 2). Host innate and adaptive immunity represent the major determinants of the outcome of HBV infection. The genetic diversity of many genes related to innate and adaptive immunity have been found to be relevant for hepatitis B vaccination, HBV infection, and antiviral therapy (Figure 3).
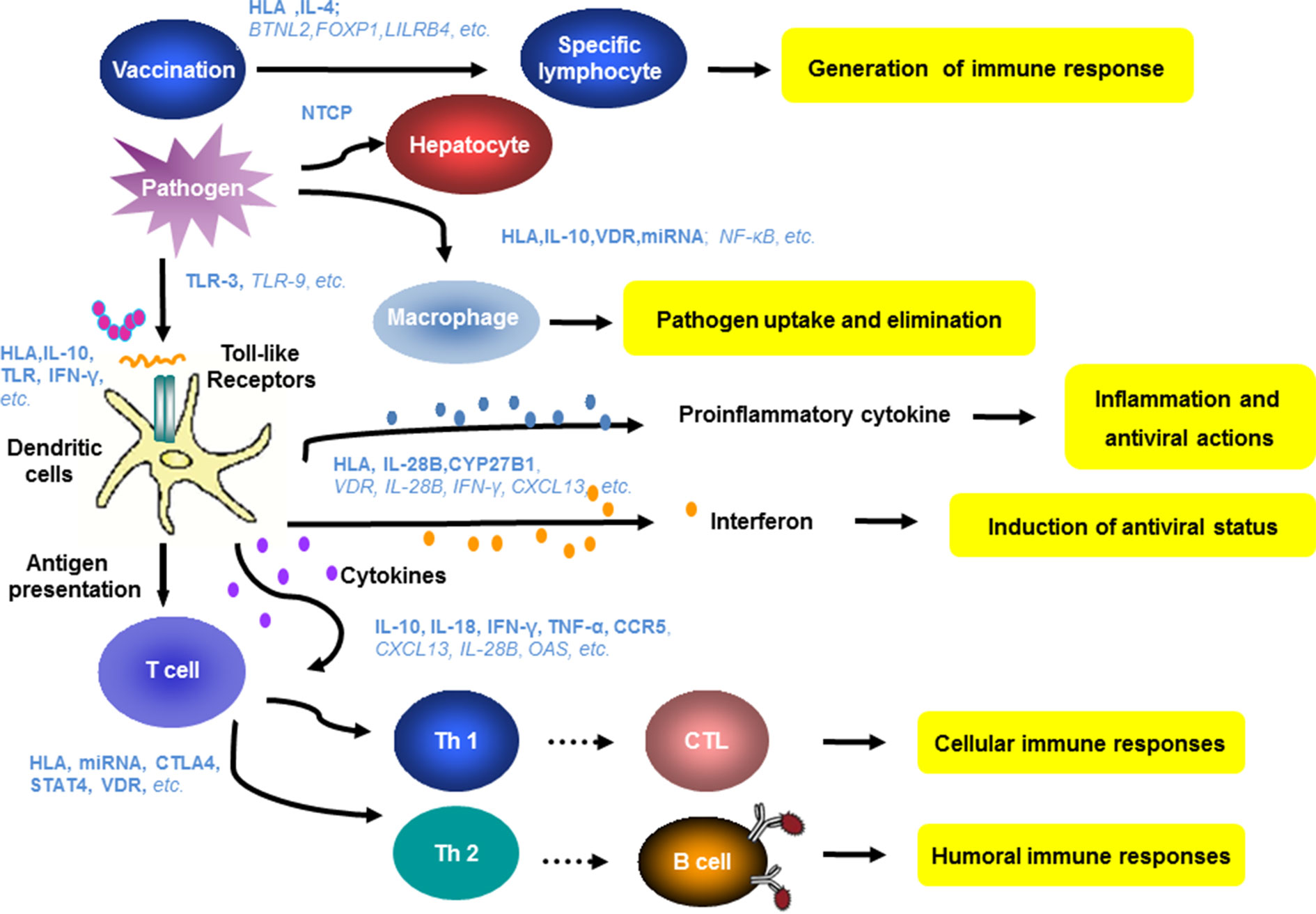
Figure 3 Host genes and immune mechanisms involved in HBV infection. Host innate and adaptive immunity represent the major determinants of the outcome of HBV infection. The genetic diversity of many genes related to innate and adaptive immunity have been found to be relevant for hepatitis B vaccination, HBV infection, and antiviral therapy. Blue letters show the relevant genes identified in the previous studies. The bold and italic letters indicate a high degree or potential correlation of the indicated host factors to clinical features of HBV infection.
HBV vaccines induce specific B cell responses, leading to the production of protective antibodies. The genetic polymorphism in many genes like HLAs and IL-4 has an impact on antigen presentation or cytokine production and thereby on antibody production, resulting in different degrees of responsiveness to vaccination (Png et al., 2011; Wang et al., 2012; Cui et al., 2013; Li et al., 2013b; Akcay et al., 2018; Nishida et al., 2018). The polymorphism in NTCP may determine the process of HBV entry (Yan et al., 2012a; Hu et al., 2016; Yang et al., 2016a; Nfor et al., 2018; Wu et al., 2018a). Many host factors may influence the innate and adaptive immunity including activation and phagocytosis of macrophages, antigen presentation by APCs, and priming and functionality of HBV-specific T cells, influencing HBV clearance and persistence (Bertoletti and Ferrari, 2016; Akcay et al., 2018; Lang et al., 2019; Rehermann and Thimme, 2019). It is well recognized that the recognition and presentation of HBV antigens by DCs to specific T cells initiate specific T cell antibody responses (Schurich et al., 2011; Das et al., 2012; Guidotti et al., 2015; Gehring and Protzer, 2019; Rehermann and Thimme, 2019; Zhu et al., 2019). IFNs and proinflammatory cytokines also contribute to HBV pathogenesis and control (Maini and Gehring, 2016; Hong and Bertoletti, 2017; Lang et al., 2019; Zhu et al., 2019). The genetic polymorphisms in relevant human genes could directly or indirectly determine these immune functions (Figure 3). Owing to the complexity of the genetic basis for infection characteristics, it is unlikely that clinical parameters can be attributed to variation in a single genetic factor. Furthermore, contradictory results are not unexpected. As age and mother-to-child transmission of HBV infection have great impacts (Li et al., 2015), host genetic factors are usually not the only determinants for chronic HBV infection, HBV intrauterine infection, its clinical outcome, or antiviral therapies.
Owing to the complexity of the pathogenesis of HBV, future studies should apply strict criteria for patient selection and for the selection of matched controls according to various factors, like age, route of transmission, time of infection, race, and host immune status, to identify relevant host factors. It is also necessary to standardize the diagnosis of HBV-related diseases. Results should be verified using different ethnic groups and confirmed by future controlled studies with large sample sizes as well as meta-analyses. Importantly, understanding the biological functions of candidate genetic markers is essential to gain mechanistic insight into HBV-related diseases.
At the moment, genetic markers associated with host responses to HBV infection/vaccines and HBV-related diseases could be classified into three categories according to the quality of studies. Some markers, like human HLAs, are clearly important determinants of HBV pathogenesis and the response to HBV vaccines. However, the precise functions of these loci in HBV infection are unclear. For example, the immunological effects of genetic variation in HLAs are not obvious, though these mutations are likely to contribute to HBV-specific immunity. Future research should consider interactions between genetic factors and other factors, including viral genotypes/subtypes, population characteristics, other genes, and environmental factors (Table 1; Table S1). Additional candidate genes that may play a role in the pathogenesis of HBV require confirmation by large-scale, multi-center validation studies. In these future studies, it will be important to stratify populations, match samples, and choose appropriate control groups to improve the reliability of results (Table 1; Table S1). Additional candidate genes with unconfirmed relevance owing to small samples sizes and studies with different designs should be evaluated by future controlled studies with suitable study subjects and meta-analyses (Table 1; Table S1).
Recently, our understanding in HBV infection and pathogenesis is fast growing. The knowledge about the molecular and physiological mechanisms of HBV pathogenesis will help us to improve future study design and develop better criteria for patient recruitment. New diagnostic methods like quantitative HBsAg and anti-HBc assays may provide more information to select suitable patient cohorts. The development of genomic and proteomic analysis tools will also enable us to define novel parameters—for example, at the level of single cells or protein chemistry, thereby opening new ways to define genetic determinants for HBV pathogenesis.
ZZ and CW reviewed the literature. ZZ, CW, ZL, and GZ wrote the manuscript. JL and ML participated in the coordination of the study and manuscript modification. ML conceived the project. All authors contributed, read, and approved the manuscript.
This work was supported by the Natural Science Foundation of Anhui Province (Grant no. 1608085MH162).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00696/full#supplementary-material
Ahmadabadi, B. N., Hassanshahi, G., Arababadi, M. K., Leanza, C., Kennedy, D. (2012). The IL-10 promoter polymorphism at position -592 is correlated with susceptibility to occult HBV infection. Inflammation 35, 818–821. doi: 10.1007/s10753-011-9381-x
Akgöllü, E., Bilgin, R., Akkız, H., Ülger, Y., Kaya, B. Y., Karaoğullarından, Ü., et al. (2017). Association between chronic hepatitis B virus infection and HLA-DP gene polymorphisms in the Turkish population. Virus Res. 232, 6–12. doi: 10.1016/j.virusres.2017.01.017
Al-Qahtani, A., Al-Ahdal, M., Abdo, A., Sanai, F., Al-Anazi, M., Khalaf, N., et al. (2012a). Toll-like receptor 3 polymorphism and its association with hepatitis B virus infection in Saudi Arabian patients. J. Med. Virol. 84, 1353–1359. doi: 10.1002/jmv.23271
Al-Qahtani, A., Al-Anazi, M., Viswan, N. A., Khalaf, N., Abdo, A. A., Sanai, F. M., et al. (2012b). Role of single nucleotide polymorphisms of KIF1B gene in HBV-associated viral hepatitis. PLoS One 7, e45128. doi: 10.1371/journal.pone.0045128
Al-Qahtani, A. A., Al-Anazi, M. R., Abdo, A. A., Sanai, F. M., Al-Hamoudi, W., Alswat, K. A., et al. (2014). Association between HLA variations and chronic hepatitis B virus infection in Saudi Arabian patients. PLoS One 9, e80445. doi: 10.1371/journal.pone.0080445
Al-Qahtani, A. A., Al-Anazi, M. R., Nazir, N., Wani, K., Abdo, A. A., Sanai, F. M., et al. (2017). Association of single nucleotide polymorphisms in microRNAs with susceptibility to hepatitis B virus infection and HBV-related liver complications: a study in a Saudi Arabian population. J. Viral Hepat. 24, 1132–1142. doi: 10.1111/jvh.12749
Akcay, I. M., Katrinli, S., Ozdil, K., Doganay, G. D., Doganay, L. (2018). Host genetic factors affecting hepatitis B infection outcomes: insights from genome-wide association studies. World J. Gastroenterol. 24, 3347–3360. doi: 10.3748/wjg.v24.i30.3347
An, P., Xu, J., Yu, Y., Winkler, C. A. (2018). Host and viral genetic variation in HBV-related hepatocellular carcinoma. Front. Genet. 9, 261. doi: 10.3389/fgene.2018.00261
Arababadi, M. K., Pourfathollah, A. A., Jafarzadeh, A., Hassanshahi, G., Rezvani, M. E. (2010). Association of exon 9 but not intron 8 VDR polymorphisms with occult HBV infection in south-eastern Iranian patients. J. Gastroenterol. Hepatol. 25, 90–93. doi: 10.1111/j.1440-1746.2009.05950.x
Asabe, S., Wieland, S. F., Chattopadhyay, P. K., Roederer, M., Engle, R. E., Purcell, R. H., et al. (2009). The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J. Virol. 83, 9652–9662. doi: 10.1128/JVI.00867-09
Bae, J. S., Kim, J., Pasaje, C. F. A., Cheong, H. S., Lee, T. H., Koh, I. S., et al. (2012). Association study of genetic variations in microRNAs with the risk of hepatitis B-related liver diseases. Dig. Liver Dis. 44, 849–854. doi: 10.1016/j.dld.2012.04.021
Beard, J. A., Bearden, A., Striker, R. (2011). Vitamin D and the anti-viral state. J. Clin. Virol. 50, 194–200. doi: 10.1016/j.jcv.2010.12.006
Bellamy, R., Ruwende, C., Corrah, T., K., P. W. J. M., Thursz, M., Whittle, H. C., et al. (1999). Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J. Infect. Dis. 179, 721–724. doi: 10.1086/314614
Bengsch, B., Thimme, R. (2018). For whom the interferons toll – TLR7 mediated boosting of innate and adaptive immunity against chronic HBV infection. J. Hepatol. 68, 883–886. doi: 10.1016/j.jhep.2018.01.025
Bertoletti, A., Ferrari, C. (2013). Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Postgrad. Med. J. 89, 294–304. doi: 10.1136/postgradmedj-2011-301073rep
Bertoletti, A., Ferrari, C. (2016). Adaptive immunity in HBV infection. J. Hepatol. 64, S71–S83. doi: 10.1016/j.jhep.2016.01.026
Bertoletti, A., Gehring, A. J. (2006). The immune response during hepatitis B virus infection. J. Gen. Virol. 87, 1439–1449. doi: 10.1099/vir.0.81920-0
Blank, A., Markert, C., Hohmann, N., Carls, A., Mikus, G., Lehr, T., et al. (2016). First-in-human application of the novel hepatitis B and hepatitis D virus entry inhibitor myrcludex B. J. Hepatol. 65, 483–489. doi: 10.1016/j.jhep.2016.04.013
Boglione, L., Cusato, J., Allegra, S., Esposito, I., Patti, F., Cariti, G., et al. (2014). Role of IL28-B polymorphisms in the treatment of chronic hepatitis B HBeAg-negative patients with peginterferon. Antiviral Res. 102, 35–43. doi: 10.1016/j.antiviral.2013.11.014
Boglione, L., Cusato, J., De Nicolò, A., Cariti, G., Di Perri, G., D’Avolio, A. (2015). Role of CYP27B1+2838 promoter polymorphism in the treatment of chronic hepatitis B HBeAg negative with PEG-interferon. J. Viral Hepat. 22, 318–327. doi: 10.1111/jvh.12288
Brouwer, W. P., Chan, H. L., Lampertico, P., Hou, J., Tangkijvanich, P., Reesink, H. W., et al. (2019). Genome wide association study identifies genetic variants associated with early and sustained response to (Peg)interferon in chronic hepatitis B patients: the GIANT-B study. Clin. Infect. Dis. doi: 10.1093/cid/ciz084
Brouwer, W. P., Sonneveld, M. J., Tabak, F., Simon, K., Cakaloglu, Y., Akarca, U. S., et al. (2014). Polymorphisms of HLA-DP are associated with response to peginterferon in Caucasian patients with chronic hepatitis B. Aliment Pharmacol. Ther. 40, 811–818. doi: 10.1111/apt.12910
Brown, R. S., McMahon, B. J., Lok, A. S., Wong, J. B., Ahmed, A. T., Mouchli, M. A., et al. (2016). Antiviral therapy in chronic hepatitis B viral infection during pregnancy: a systematic review and meta-analysis. Hepatology 63, 319–333. doi: 10.1002/hep.28302
Bruce, M. G., Bruden, D., Hurlburt, D., Zanis, C., Thompson, G., Rea, L., et al. (2016). Antibody levels and protection after hepatitis B vaccine: results of a 30-year follow-up study and response to a booster dose. J. Infect. Dis. 214, 16–22. doi: 10.1093/infdis/jiv748
Buchbinder, E., Hodi, F. S. (2015). Cytotoxic T lymphocyte antigen-4 and immune checkpoint blockade. J. Clin. Invest. 125, 3377–3383. doi: 10.1172/JCI80012
Cao, B., Liu, X., Hou, F., Li, W., Han, Z., Zhang, Q., et al. (2009). The haplotype of the MxA gene promoter is associated with hepatitis B virus infection in a Chinese population. Liver Int. 29, 1383–1388. doi: 10.1111/j.1478-3231.2009.02053.x
Chang, Z. L. (2010). Important aspects of Toll-like receptors, ligands and their signaling pathways. Inflamm. Res. 59, 791–808. doi: 10.1007/s00011-010-0208-2
Chang, S. W., Fann, C. S., Su, W. H., Wang, Y. C., Weng, C. C., Yu, C. J., et al. (2014). A genome-wide association study on chronic HBV infection and its clinical progression in male Han-Taiwanese. PLoS One 9, e99724. doi: 10.1371/journal.pone.0099724
Chatzidaki, V., Choumerianou, D., Dimitriou, H., Kouroumalis, E., Galanakis, E. (2012). Genetic variants associated with susceptibility to mother-to-child transmission of hepatitis B virus. Eur. J. Gastroenterol. Hepatol. 24, 1185–1190. doi: 10.1097/MEG.0b013e328356440f
Chen, D., Zeng, Y., Zhou, J., Yang, L., Jiang, S., Huang, J., et al. (2010). Association of candidate susceptible loci with chronic infection with hepatitis B virus in a Chinese population. J. Med. Virol. 82, 371–378. doi: 10.1002/jmv.21716
Chen, H. L., Zha, M. L., Cai, J. Y., Qin, G. (2018). Maternal viral load and hepatitis B virus mother-to-child transmission risk: a systematic review and meta-analysis. Hepatol. Res. 48, 788–801. doi: 10.1111/hepr.13072
Chen, J., Liang, Z., Lu, F., Fang, X., Liu, S., Zeng, Y., et al. (2011). Toll-like receptors and cytokines/cytokine receptors polymorphisms associate with non-response to hepatitis B vaccine. Vaccine 29, 706–711. doi: 10.1016/j.vaccine.2010.11.023
Cheng, L., Sun, X., Tan, S., Tan, W., Dan, Y., Zhou, Y., et al. (2014). Effect of HLA-DP and IL28B gene polymorphisms on response to interferon treatment in hepatitis B e-antigen seropositive chronic hepatitis B patients. Hepatol. Res. 44, 1000–1007. doi: 10.1111/hepr.12284
Cheong, H. S., Lee, J. H., Yu, S. J., Yoon, J. H., Lee, H. S., Cheong, J. Y., et al. (2015). Association of VARS2-SFTA2 polymorphisms with the risk of chronic hepatitis B in a Korean population. Liver Int. 35, 1934–1940. doi: 10.1111/liv.12740
Cheong, J. Y., Cho, S. W., Hwang, I. L., Yoon, S. K., Lee, J. H., Park, C. S., et al. (2006). Association between chronic hepatitis B virus infection and interleukin-10, tumor necrosis factor-alpha gene promoter polymorphisms. J. Gastroenterol. Hepatol. 21, 1163–1169. doi: 10.1111/j.1440-1746.2006.04304.x
Cheong, J. Y., Shin, H. D., Kim, Y. J., Cho, S. W. (2013). Association of polymorphism in MicroRNA 219-1 with clearance of hepatitis B virus infection. J. Med. Virol. 85, 808–814. doi: 10.1002/jmv.23551
Chien, R., Kao, J., Peng, C., Chen, C., Liu, C., Huang, Y., et al. (2019). Taiwan consensus statement on the management of chronic hepatitis B. J. Formos. Med. Assoc. 118, 7–38. doi: 10.1016/j.jfma.2018.11.008
Chiu, Y. C., Liao, S. F., Wu, J. F., Lin, C. Y., Lee, W. C., Chen, H. L., et al. (2014). Factors affecting the natural decay of hepatitis B surface antigen in children with chronic hepatitis B virus infection during long-term follow-up. J. Pediatr. 165, 767–772. doi: 10.1016/j.jpeds.2014.06.059
Cui, W., Sun, C., Deng, B., Liu, P. (2013). Association of polymorphisms in the interleukin-4 gene with response to hepatitis B vaccine and susceptibility to hepatitis B virus infection: a meta-analysis. Gene 525, 35–40. doi: 10.1016/j.gene.2013.04.065
Cusato, J., Boglione, L., De Nicolò, A., Imbornone, R., Cardellino, C. S., Ghisetti, V., et al. (2017). Association of vitamin D pathway SNPs and clinical response to interferon in a cohort of HBeAg-negative patients. Pharmacogenomics 18, 651–661. doi: 10.2217/pgs-2016-0041
Das, A., Ellis, G., Pallant, C., Lopes, A. R., Khanna, P., Peppa, D., et al. (2012). IL-10-producing regulatory B cells in the pathogenesis of chronic hepatitis B virus infection. J. Immunol. 189, 3925–3935. doi: 10.4049/jimmunol.1103139
Davila, S., Froeling, F. E. M., Tan, A., Bonnard, C., Boland, G. J., Snippe, H., et al. (2010). New genetic associations detected in a host response study to hepatitis B vaccine. Genes Immun. 11, 232–238. doi: 10.1038/gene.2010.1
Deng, G., Zhou, G., Zhai, Y., Li, S., Li, X., Li, Y., et al. (2004). Association of estrogen receptor alpha polymorphisms with susceptibility to chronic hepatitis B virus infection. Hepatology 40, 318–326. doi: 10.1002/hep.20318
Di Scala, M., Otano, I., Gil-Fariña, I., Vanrell, L., Hommel, M., Olagüe, C., et al. (2016). Complementary Effects of Interleukin-15 and alpha interferon induce immunity in hepatitis B virus transgenic mice. J. Virol. 90, 8563–8574. doi: 10.1128/JVI.01030-16
Dionne-Odom, J., Njei, B., Tita, A. T. N. (2018). Elimination of vertical transmission of hepatitis B in Africa: a review of available tools and new opportunities. Clin. Ther. 40, 1255–1267. doi: 10.1016/j.clinthera.2018.05.016
Du, T., Guo, X. H., Zhu, X. L., Li, J. H., Lu, L. P., Gao, J. R., et al. (2006). Association of TNF-alpha promoter polymorphisms with the outcomes of hepatitis B virus infection in Chinese Han population. J Viral Hepat. 13, 618-624. doi: 10.1111/j.1365-2893.2006.00731.x
Eskandari, E., Metanat, M., Pahlevani, E., Nakhzari-Khodakheir, T. (2017b). Association between TGFβ1 polymorphisms and chronic hepatitis B infection in an Iranian population. Rev. Soc. Bras. Med. Trop. 50, 301–308. doi: 10.1590/0037-8682-0266-2016
European Association For The Study Of The Liver (2017). EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 67, 370–398. doi: 10.1016/j.jhep.2017.03.021
Ezzikouri, S., Chihab, H., Elhabazi, A., Wakrim, L., Benjelloun, S. (2017). Lack of Ser267Phe variant of sodium taurocholate cotransporting polypeptide among Moroccans regardless of hepatitis B virus infection status. BMC Infect. Dis. 17, 99. doi: 10.1186/s12879-017-2214-2
Fan, J. H., Hou, S. H., Qing-Ling, L., Hu, J., Peng, H., Guo, J. J. (2016a). Association of HLA-DQ and IFNL4 polymorphisms with susceptibility to hepatitis B virus infection and clearance. Ann. Hepatol. 15, 532–539. doi: 10.5604/16652681.1202946
Fan, R., Sun, J., Yuan, Q., Xie, Q., Bai, X., Ning, Q., et al. (2016b). Baseline quantitative hepatitis B core antibody titre alone strongly predicts HBeAg seroconversion across chronic hepatitis B patients treated with peginterferon or nucleos(t)ide analogues. Gut 65, 313–320. doi: 10.1136/gutjnl-2014-308546
Fattovich, G., Bortolotti, F., Donato, F. (2008). Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J. Hepatol. 48, 335–352. doi: 10.1016/j.jhep.2007.11.011
Fletcher, G. J., Samuel, P., Christdas, J., Gnanamony, M., Ismail, A. M., Anantharam, R., et al. (2011). Association of HLA and TNF polymorphisms with the outcome of HBV infection in the South Indian population. Genes Immun. 12, 552–558. doi: 10.1038/gene.2011.32
Freeman, B. E., Hammarlund, E., Raue, H. P., Slifka, M. K. (2012). Regulation of innate CD8+ T-cell activation mediated by cytokines. Proc. Natl. Acad. Sci. U.S.A. 109, 9971–9976. doi: 10.1073/pnas.1203543109
Ganem, D., Prince, A. M. (2004). Hepatitis B virus infection–natural history and clinical consequences. N. Engl. J. Med. 350, 1118–1129. doi: 10.1056/NEJMra031087
Gao, W., Wang, R., Wang, X., Wu, H., Wang, Y., Lu, X., et al. (2017). Vitamin D serum levels and receptor genetic polymorphisms are associated with hepatitis B virus and HIV infections and IFN-λ levels. Biomark. Med. 11, 733–740. doi: 10.2217/bmm-2017-0022
Gao, Y., Guo, J., Zhang, F., Guo, Z., Zhang, L. R., Wang, T., et al. (2015b). Evaluation of neonatal Toll-like receptors 3 (c.1377C/T) and 9 (G2848A) gene polymorphisms in HBV intrauterine transmission susceptibility. Epidemiol. Infect. 143, 1868–1875. doi: 10.1017/S0950268814002921
Ge, D., Fellay, J., Thompson, A. J., Simon, J. S., Shianna, K. V., Urban, T. J., et al. (2009). Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401. doi: 10.1038/nature08309
Gehring, A. J., Protzer, U. (2019). Targeting innate and adaptive immune responses to cure chronic HBV infection. Gastroenterology 156, 325–337. doi: 10.1053/j.gastro.2018.10.032
Geng, P., Song, L., An, H., Huang, J., Li, S., Zeng, X. (2016). Toll-like receptor 3 is associated with the risk of HCV infection and HBV-related diseases. Medicine (Baltimore) 95, e2302. doi: 10.1097/MD.0000000000002302
Goulder, P. J., Walker, B. D. (2012). HIV and HLA class I: an evolving relationship. Immunity 37, 426–440. doi: 10.1016/j.immuni.2012.09.005
Grzegorzewska, A. E., Jod Owska, E. B., Mostowska, A., Sowi Ska, A., Jagodzi Ski, P. P. (2014). Single nucleotide polymorphisms of vitamin D binding protein, vitamin D receptor and retinoid X receptor alpha genes and response to hepatitis B vaccination in renal replacement therapy patients. Expert Rev. Vaccines 13, 1395–1403. doi: 10.1586/14760584.2014.962521
Gu, S. Q., Zhu, Q. R., Yu, H., Fei, L. E., Dong, Z. Q., Pu, D. P. (2004). Relationship between genetic polymorphism of tumor necrosis factor-alpha and susceptibility to intrauterine HBV infection. Zhonghua Gan Zang Bing Za Zhi 12, 538–539.
Guidotti, L. G., Isogawa, M., Chisari, F. V. (2015). Host–virus interactions in hepatitis B virus infection. Curr. Opin. Immunol. 36, 61–66. doi: 10.1016/j.coi.2015.06.016
Guo, F., Tang, L., Shu, S., Sehgal, M., Sheraz, M., Liu, B., et al. (2017). Activation of stimulator of interferon genes in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob. Agents Chemother. 61, e00771-17. doi: 10.1128/AAC.00771-17
Guo, X., Yang, G., Yuan, J., Ruan, P., Zhang, M., Chen, X., et al. (2013). Genetic variation in interleukin 28B and response to antiviral therapy in patients with dual chronic infection with hepatitis B and C viruses. PLoS One 8, e77911. doi: 10.1371/journal.pone.0077911
Guo, X., Zhang, Y., Li, J., Ma, J., Wei, Z., Tan, W., et al. (2011). Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology 53, 422–428. doi: 10.1002/hep.24048
Hagenbuch, B., Meier, P. J. (1994). Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J. Clin. Invest. 93, 1326–1331. doi: 10.1172/JCI117091
Han, G., Cao, M., Zhao, W., Jiang, H., Wang, C., Bai, S., et al. (2011). A prospective and open-label study for the efficacy and safety of telbivudine in pregnancy for the prevention of perinatal transmission of hepatitis B virus infection. J. Hepatol. 55, 1215–1221. doi: 10.1016/j.jhep.2011.02.032
Harber, M., Sundstedt, A., Wraith, D. (2000). The role of cytokines in immunological tolerance: potential for therapy. Expert Rev. Mol. Med. 2, 1–20. doi: 10.1017/S1462399400002143
Hardy, J., Singleton, A. (2009). Genomewide association studies and human disease. N. Engl. J. Med. 360, 1759–1768. doi: 10.1056/NEJMra0808700
Hassanshahi, G., Arababadi, M. K., Khoramdelazad, H., Yaghini, N., Zarandi, E. R. (2010). Assessment of CXCL12 (SDF-1α) polymorphisms and its serum level in posttransfusion occult HBV-infected patients in Southeastern Iran. Arch. Med. Res. 41, 338–342. doi: 10.1016/j.arcmed.2010.07.001
He, D., Tao, S., Guo, S., Li, M., Wu, J., Huang, H., et al. (2015). Interaction of TLR-IFN and HLA polymorphisms on susceptibility of chronic HBV infection in Southwest Han Chinese. Liver Int. 35, 1941–1949. doi: 10.1111/liv.12756
He, Q., Huang, Y., Zhang, L., Yan, Y., Liu, J., Song, X., et al. (2018). Association between vitamin D receptor polymorphisms and hepatitis B virus infection susceptibility: a meta-analysis study. Gene 645, 105–112. doi: 10.1016/j.gene.2017.12.027
He, Y. L., Zhao, Y. R., Zhang, S. L., Lin, S. M. (2006). Host susceptibility to persistent hepatitis B virus infection. World J. Gastroenterol. 12, 4788–4793. doi: 10.3748/wjg.v12.i30.4788
Hewison, M. (2011). Vitamin D and innate and adaptive immunity. Vitam. Horm. 86, 23–62. doi: 10.1016/B978-0-12-386960-9.00002-2
Hoan, N. X., Van Tong, H., Giang, D. P., Cuong, B. K., Toan, N. L., Wedemeyer, H., et al. (2017). SOCS3 genetic variants and promoter hypermethylation in patients with chronic hepatitis B. Oncotarget 8, 17127–17139. doi: 10.18632/oncotarget.15083
Hong, M., Bertoletti, A. (2017). Tolerance and immunity to pathogens in early life: insights from HBV infection. Semin. Immunopathol. 39, 643–652. doi: 10.1007/s00281-017-0641-1
Hong, M., Sandalova, E., Low, D., Gehring, A. J., Fieni, S., Amadei, B., et al. (2015). Trained immunity in newborn infants of HBV-infected mothers. Nat. Commun. 6, 6588. doi: 10.1038/ncomms7588
Hoogeveen, R. C., Robidoux, M. P., Schwarz, T., Heydmann, L., Cheney, J. A., Kvistad, D., et al. (2018). Phenotype and function of HBV-specific T cells is determined by the targeted epitope in addition to the stage of infection. Gut. 68, 893-904. doi: 10.1136/gutjnl-2018-316644
Hosaka, T., Suzuki, F., Kobayashi, M., Fukushima, T., Kawamura, Y., Sezaki, H., et al. (2015). HLA-DP genes polymorphisms associate with hepatitis B surface antigen kinetics and seroclearance during nucleot(s)ide analogue therapy. Liver Int. 35, 1290–1302. doi: 10.1111/liv.12652
Howe, R. C., Dhiman, N., Ovsyannikova, I. G., Poland, G. A. (2005). Induction of CD4 T cell proliferation and in vitro Th1-like cytokine responses to measles virus. Clin. Exp. Immunol. 140, 333–342. doi: 10.1111/j.1365-2249.2005.02766.x
Hu, L., Zhai, X., Liu, J., Chu, M., Pan, S., Jiang, J., et al. (2012). Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology. 55, 1426-1431. doi: 10.1002/hep.24799
Hu, H., Liu, J., Lin, Y., Luo, W., Chu, Y., Chang, C., et al. (2016). The rs2296651 (S267F) variant on NTCP (SLC10A1) is inversely associated with chronic hepatitis B and progression to cirrhosis and hepatocellular carcinoma in patients with chronic hepatitis B. Gut 65, 1514–1521. doi: 10.1136/gutjnl-2015-310686
Hu, Y., Chen, Y., Wang, Y., Liang, H. (2018). Hepatitis B vaccination among 1999–2017 birth cohorts in Zhejiang Province: the determinants associated with infant coverage. Int. J. Environ. Res. Public Health 15, E2915. doi: 10.3390/ijerph15122915
Hu, Z., Liu, Y., Zhai, X., Dai, J., Jin, G., Wang, L., et al. (2013). New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat. Genet. 45, 1499–1503. doi: 10.1038/ng.2809
Hu, Z., Yang, J., Xiong, G., Shi, H., Yuan, Y., Fan, L., et al. (2014). HLA-DPB1 variant effect on hepatitis B virus clearance and liver cirrhosis development among Southwest Chinese population. Hepat. Mon. 14, e19747. doi: 10.5812/hepatmon.19747
Huang, J. M., Xiong, L. S., Wang, J., Liu, Y. F., Zhu, Q. R., Lei, J., et al. (2016). Association between the HLA-DQB1 polymorphisms and the susceptibility of chronic hepatitis B: a comprehensive meta-analysis. Biomed. Rep. 4, 557–566. doi: 10.3892/br.2016.632
Huang, R., Hao, Y., Fan, Y., Yang, C., Wu, K., Cao, S., et al. (2013). Association between cytotoxic T-lymphocyte-associated antigen 4 + 49A/G polymorphism and persistent hepatitis B virus infection in the Asian population: evidence from the current studies. Genet. Test. Mol. Biomarkers 17, 601–606. doi: 10.1089/gtmb.2013.0069
Huang, X., Li, H., Wang, J., Huang, C., Lu, Y., Qin, X., et al. (2015). Genetic polymorphisms in Toll-like receptor 3 gene are associated with the risk of hepatitis B virus-related liver diseases in a Chinese population. Gene 569, 218–224. doi: 10.1016/j.gene.2015.05.054
Jiang, D., Wu, X., Qian, J., Ma, X., Yang, J., Li, Z., et al. (2016a). Genetic variation in STAT4 predicts response to interferon-α therapy for hepatitis B e antigen-positive chronic hepatitis B. Hepatology 63, 1102–1111. doi: 10.1002/hep.28423
Jiang, D. K., Ma, X. P., Yu, H., Cao, G., Ding, D. L., Chen, H., et al. (2015). Genetic variants in five novel loci including CFB and CD40 predispose to chronic hepatitis B. Hepatology 62, 118–128. doi: 10.1002/hep.27794
Jiang, X., Su, K., Tao, J., Fan, R., Xu, Y., Han, H., et al. (2016b). Association of STAT4 polymorphisms with hepatitis B virus infection and clearance in Chinese Han population. Amino Acids 48, 2589–2598. doi: 10.1007/s00726-016-2283-3
Johnson, L. D., Jameson, S. C. (2009). A chronic need for IL-21. Science 324, 1525–1526. doi: 10.1126/science.1176487
Kamatani, Y., Wattanapokayakit, S., Ochi, H., Kawaguchi, T., Takahashi, A., Hosono, N., et al. (2009). A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 41, 591–595. doi: 10.1038/ng.348
Kang, W., Ding, Z., Shen, L., Zhao, Z., Huang, G., Zhang, J., et al. (2014). Risk factors associated with immunoprophylaxis failure against mother to child transmission of hepatitis B virus and hepatitis B vaccination status in Yunnan province, China. Vaccine 32, 3362–3366. doi: 10.1016/j.vaccine.2014.04.045
Karataylı, S. C., Bozdayı, M., Karataylı, E., Öztürk, T., Husseini, A. A., Albayrak, R., et al. (2015). Interleukin-28 gene polymorphisms may contribute to HBsAg persistence and the development of HBeAg-negative chronic hepatitis B. Liver Int. 35, 846–853. doi: 10.1111/liv.12595
Karra, V. K., Gumma, P. K., Chowdhury, S. J., Ruttala, R., Polipalli, S. K., Chakravarti, A., et al. (2015). IL-18 polymorphisms in hepatitis B virus related liver disease. Cytokine 73, 277–282. doi: 10.1016/j.cyto.2015.02.015
Kehrl, J. H. (2006). Chemoattractant receptor signaling and the control of lymphocyte migration. Immunol. Res. 34, 211–227. doi: 10.1385/IR:34:3:211
Kew, M. C. (1995). Protective efficacy of hepatitis B vaccination. Lancet 345, 1065–1066. doi: 10.1016/S0140-6736(95)90813-7
Khanizadeh, S., Ravanshad, M., Mohebbi, S. R., Naghoosi, H., Tahaei, M. E., Mousavi Nasab, S. D., et al. (2012). Polymorphisms within the promoter region of the gamma interferon (IFN-γ) receptor1 gene are associated with the susceptibility to chronic HBV infection in an Iranian population. Hepat. Mon. 12, e7283. doi: 10.5812/hepatmon.7283
Kim, L. H., Cheong, H. S., Namgoong, S., Kim, J. O., Kim, J., Park, B. L., et al. (2015). Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci of STAT4 and HLA-DQ in a Korean population. Infect. Genet. Evol. 33, 72–76. doi: 10.1016/j.meegid.2015.04.013
Kim, Y. J., Kim, H. Y., Kim, J. S., Lee, J. H., Yoon, J. H., Kim, C. Y., et al. (2009). Putative association of transforming growth factor-α polymorphisms with clearance of hepatitis B virus and occurrence of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. J. Viral Hepat. 17, 518–526. doi: 10.1111/j.1365-2893.2009.01205.x
Kim, Y. J., Kim, H. Y., Lee, J., Yu, S. J., Yoon, J., Lee, H., et al. (2013). A genome-wide association study identified new variants associated with the risk of chronic hepatitis B. Hum. Mol. Genet. 22, 4233–4238. doi: 10.1093/hmg/ddt266
Kim, Y. J., Lee, H. S., Yoon, J. H., Kim, C. Y., Park, M. H., Kim, L. H., et al. (2003). Association of TNF-alpha promoter polymorphisms with the clearance of hepatitis B virus infection. Hum. Mol. Genet. 12, 2541–2546. doi: 10.1093/hmg/ddg262
Ko, C., Park, W. J., Park, S., Kim, S., Windisch, M. P., Ryu, W. S. (2015). The FDA-approved drug irbesartan inhibits HBV-infection in HepG2 cells stably expressing sodium taurocholate co-transporting polypeptide. Antivir. Ther. 20, 835–842. doi: 10.3851/IMP2965
Koh, S., Kah, J., Tham, C. Y. L., Yang, N., Ceccarello, E., Chia, A., et al. (2018). Nonlytic lymphocytes engineered to express virus-specific T-cell receptors limit HBV infection by activating APOBEC3. Gastroenterology 155, 180–193. doi: 10.1053/j.gastro.2018.03.027
Koller, B. H., Geraghty, D. E., DeMars, R., Duvick, L., Rich, S. S., Orr, H. T. (1989). Chromosomal organization of the human major histocompatibility complex class I gene family. J. Exp. Med. 169, 469–480. doi: 10.1084/jem.169.2.469
Kondo, Y., Ueno, Y., Shimosegawa, T. (2011). Toll-like receptors signaling contributes to immunopathogenesis of HBV infection. Gastroenterol. Res. Pract. 2011, 810939. doi: 10.1155/2011/810939
Kong, X., Zhang, X., Gong, Q., Gao, J., Zhang, S., Wang, L., et al. (2007). MxA induction may predict sustained virologic responses of chronic hepatitis B patients with IFN-alpha treatment. J. Interferon Cytokine Res. 27, 809–818. doi: 10.1089/jir.2006.0163
Koziel, M. J. (1999). Cytokines in viral hepatitis. Semin. Liver Dis. 19, 157–169. doi: 10.1055/s-2007-1007107
Lai, C. L., Yuen, M. F. (2007). The natural history of chronic hepatitis B. J. Viral Hepat. 14Suppl 1, 6–10. doi: 10.1111/j.1365-2893.2007.00909.x
Lampertico, P., Viganò, M., Cheroni, C., Facchetti, F., Invernizzi, F., Valveri, V., et al. (2013). IL28B polymorphisms predict interferon-related hepatitis B surface antigen seroclearance in genotype D hepatitis B e antigen–negative patients with chronic hepatitis B. Hepatology 57, 890–896. doi: 10.1002/hep.25749
Lang, J., Neumann-Haefelin, C., Thimme, R. (2019). Immunological cure of HBV infection. Hepatol. Int. 13, 113–124. doi: 10.1007/s12072-018-9912-8
Lange, C. M., Gouttenoire, J., Duong, F. H. T., Morikawa, K., Heim, M. H., Moradpour, D. (2014). Vitamin D receptor and Jak-STAT signaling crosstalk results in calcitriol-mediated increase of hepatocellular response to IFN-α. J. Immunol. 192, 6037–6044. doi: 10.4049/jimmunol.1302296
Laskus, T., Radkowski, M., Lupa, E., Horban, A., Cianciara, J., Slusarczyk, J. (1992). Prevalence of markers of hepatitis viruses in out-patient alcoholics. J. Hepatol. 15, 174–178. doi: 10.1016/0168-8278(92)90032-K
Lebosse, F., Testoni, B., Fresquet, J., Facchetti, F., Galmozzi, E., Fournier, M., et al. (2017). Intrahepatic innate immune response pathways are downregulated in untreated chronic hepatitis B. J. Hepatol. 66, 897–909. doi: 10.1016/j.jhep.2016.12.024
Lee, M., Keeffe, E. B. (2011). Hepatitis B: modern end points of treatment and the specter of viral resistance. Gastroenterol. Clin. North Am. 40, 495–505. doi: 10.1016/j.gtc.2011.06.004
Lee, C. M., Lu, S. N., Changchien, C. S., Yeh, C. T., Hsu, T. T., Tang, J. H., et al. (1999). Age, gender, and local geographic variations of viral etiology of hepatocellular carcinoma in a hyperendemic area for hepatitis B virus infection. Cancer 86, 1143–1150. doi: 10.1002/(SICI)1097-0142(19991001)86:7<1143::AID-CNCR7>3.0.CO;2-Z
Lee, D. H., Lee, J. H., Kim, Y. J., Park, N. H., Cho, Y., Lee, Y. B., et al. (2014). Relationship between polymorphisms near the IL28B gene and spontaneous HBsAg seroclearance: a systematic review and meta-analysis. J. Viral Hepat. 21, 163–170. doi: 10.1111/jvh.12193
Levy, D. E., Darnell, J. E. (2002). Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662. doi: 10.1038/nrm909
Li, J., Yang, D., He, Y., Wang, M., Wen, Z., Liu, L., et al. (2011a). Associations of HLA-DP variants with hepatitis B virus infection in southern and northern Han Chinese populations: a multicenter case-control study. PLoS One 6, e24221. doi: 10.1371/journal.pone.0024221
Li, M., Li, F., Li, N., Sang, J., Fan, X., Deng, H., et al. (2018). Association of polymorphism rs1053005 in STAT3 with chronic hepatitis B virus infection in Han Chinese population. BMC Med. Genet. 19, 52. doi: 10.1186/s12881-018-0569-x
Li, N., Zhu, Q., Li, Z., Han, Q., Chen, J., Lv, Y., et al. (2013a). IL21 and IL21R polymorphisms and their interactive effects on serum IL-21 and IgE levels in patients with chronic hepatitis B virus infection. Hum. Immunol. 74, 567–573. doi: 10.1016/j.humimm.2013.01.005
Li, S., Deng, Y., Chen, Z. P., Huang, S., Liao, X. C., Lin, L. W., et al. (2011b). Genetic polymorphism of interleukin-16 influences susceptibility to HBV-related hepatocellular carcinoma in a Chinese population. Infect. Genet. Evol. 11, 2083–2088. doi: 10.1016/j.meegid.2011.09.025
Li, S., Qian, J., Yang, Y., Zhao, W., Dai, J., Bei, J. X., et al. (2012b). GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 8, e1002791. doi: 10.1371/journal.pgen.1002791
Li, Y., Si, L., Zhai, Y., Hu, Y., Hu, Z., Bei, J., et al. (2016). Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat. Commun. 7, 11664. doi: 10.1038/ncomms11664
Li, Z., Hou, X., Cao, G. (2015). Is mother-to-infant transmission the most important factor for persistent HBV infection? Emerg. Microbes Infect. 4, e30. doi: 10.1038/emi.2015.30
Li, Z., Nie, J., Li, J., Zhuang, H. (2013b). The effect of HLA on immunological response to hepatitis B vaccine in healthy people: a meta-analysis. Vaccine 31, 4355–4361. doi: 10.1016/j.vaccine.2013.06.108
Liao, Y., Cai, B., Li, Y., Chen, J., Tao, C., Huang, H., et al. (2014). Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One 9, e111677. doi: 10.1371/journal.pone.0111677
Liaw, Y. F., Jen, C. L., You, S. L., Chen, T. H., Lu, S. N., Yeh, S. H., et al. (2009). Hepatitis B virus DNA levels and outcomes in chronic hepatitis B. Hepatology 49, S72–S84. doi: 10.1002/hep.22884
Limothai, U., Chuaypen, N., Khlaiphuengsin, A., Chittmittraprap, S., Poovorawan, Y., Tangkijvanich, P. (2017). Association of vitamin-D-related genetic variations and treatment response to pegylated interferon in patients with chronic hepatitis B. Antivir. Ther. 22, 681–688. doi: 10.3851/IMP3154
Limothai, U., Chuaypen, N., Khlaiphuengsin, A., Posuwan, N., Wasitthankasem, R., Poovorawan, Y., et al. (2016). Association of interferon-gamma inducible protein 10 polymorphism with treatment response to pegylated interferon in HBeAg-positive chronic hepatitis B. Antivir. Ther. 21, 97–106. doi: 10.3851/IMP2992
Lin, F. C., Young, H. A. (2014). Interferons: success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 25, 369–376. doi: 10.1016/j.cytogfr.2014.07.015
Lin, T. M., Chen, C. J., Wu, M. M., Yang, C. S., Chen, J. S., Lin, C. C., et al. (1989). Hepatitis B virus markers in Chinese twins. Anticancer Res. 9, 737–741.
Lin, Y., Deng, W., Pang, J., Kemper, T., Hu, J., Yin, J., et al. (2017). The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell. Microbiol. 19, e12709. doi: 10.1111/cmi.12709
Lin, Y., Wu, C., Wang, X., Kemper, T., Squire, A., Gunzer, M., et al. (2019). Hepatitis B virus is degraded by autophagosome-lysosome fusion mediated by Rab7 and related components. Protein Cell 10, 60–66. doi: 10.1007/s13238-018-0555-2
Liu, H., Hou, J., Zhang, X. (2018a). Targeting cIAPs, a new option for functional cure of chronic hepatitis B infection? Virol. Sin. 33, 459–461. doi: 10.1007/s12250-018-0062-x
Liu, J., Zhang, E., Ma, Z., Wu, W., Kosinska, A., Zhang, X., et al. (2014a). Enhancing virus-specific immunity in vivo by combining therapeutic vaccination and PD-L1 blockade in chronic hepadnaviral infection. PLoS Pathog. 10, e1003856. doi: 10.1371/journal.ppat.1003856
Liu, J., Pan, W., Yang, D. (2018b). The era of immune checkpoint therapy: from cancer to viral infection-a mini comment on the 2018 medicine nobel prize. Virol. Sin. 33, 467–471. doi: 10.1007/s12250-018-0077-3
Liu, M., Cao, B., Zhang, H., Dai, Y., Liu, X., Xu, C. (2006). Association of interferon-gamma gene haplotype in the Chinese population with hepatitis B virus infection. Immunogenetics 58, 859–864. doi: 10.1007/s00251-006-0161-y
Liu, M. Q., Zhou, D. J., Wang, X., Zhou, W., Ye, L., Li, J. L., et al. (2012a). IFN-lambda3 inhibits HIV infection of macrophages through the JAK-STAT pathway. PLoS One 7, e35902. doi: 10.1371/journal.pone.0035902
Liu, T., Wan, Z., Peng, S., Wang, Y., Chen, H., Li, X., et al. (2018c). Genetic variations in LTA gene and PDCD1 gene and intrauterine infection of hepatitis B virus: a case–control study in China. Amino Acids 50, 877–883. doi: 10.1007/s00726-018-2568-9
Liu, Y., Song, C., Ni, H., Jiao, W., Gan, W., Dong, X., et al. (2018d). UBE2L3, a susceptibility gene that plays oncogenic role in hepatitis B-related hepatocellular carcinoma. J. Viral Hepat. 25, 1363–1371. doi: 10.1111/jvh.12963
Liu, Y., Xie, K., Wen, J., Deng, M., Li, J., Hu, Z. (2014b). A genetic variant in microRNA-122 regulatory region confers risk for chronic hepatitis B virus infection and hepatocellular carcinoma in Han Chinese. J. Med. Virol. 86, 1669–1674. doi: 10.1002/jmv.23996
Liu, Y., Zhang, Y., Wen, J., Liu, L., Zhai, X., Liu, J., et al. (2012b). A genetic variant in the promoter region of miR-106b-25 cluster and risk of HBV infection and hepatocellular carcinoma. PLoS One 7, e32230. doi: 10.1371/journal.pone.0032230
Lok, A. S., Zoulim, F., Locarnini, S., Bartholomeusz, A., Ghany, M. G., Pawlotsky, J., et al. (2007). Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology 46, 254–265. doi: 10.1002/hep.21698
Lu, Y., Zhu, Y., Peng, J., Wang, X., Wang, F., Sun, Z. (2015b). STAT4 genetic polymorphisms association with spontaneous clearance of hepatitis B virus infection. Immunol. Res. 62, 146–152. doi: 10.1007/s12026-015-8645-1
Ma, N., Zhang, X., Yang, L., Zhou, J., Liu, W., Gao, X., et al. (2018a). Role of Functional IFNL4, IFNLR1, IFNA, IFNAR2 polymorphisms in Hepatitis B virus-related liver disease in Han Chinese population. J. Viral Hepat. 25, 306–313. doi: 10.1111/jvh.12817
Ma, N., Zhang, X., Yu, F., Gao, P., Fan, Q., Liu, L., et al. (2014). Role of IFN-ks, IFN-ks related genes and the DEPDC5 gene in hepatitis B virus-related liver disease. J. Viral Hepat. 21, e29–e38. doi: 10.1111/jvh.12235
Ma, Z., Cao, Q., Xiong, Y., Zhang, E., Lu, M. (2018b). Interaction between hepatitis B virus and toll-like receptors: current status and potential therapeutic use for chronic hepatitis B. Vaccines (Basel) 6, 6. doi: 10.3390/vaccines6010006
Ma, Z., Liu, J., Wu, W., Zhang, E., Zhang, X., Li, Q., et al. (2017). The IL-1R/TLR signaling pathway is essential for efficient CD8+ T-cell responses against hepatitis B virus in the hydrodynamic injection mouse model. Cell. Mol. Immunol. 14, 997–1008. doi: 10.1038/cmi.2017.43
Ma, Z., Zhang, E., Yang, D., Lu, M. (2015). Contribution of Toll-like receptors to the control of hepatitis B virus infection by initiating antiviral innate responses and promoting specific adaptive immune responses. Cell. Mol. Immunol. 12, 273–282. doi: 10.1038/cmi.2014.112
Maini, M. K., Gehring, A. J. (2016). The role of innate immunity in the immunopathology and treatment of HBV infection. J. Hepatol. 64, S60–S70. doi: 10.1016/j.jhep.2016.01.028
Mardian, Y., Yano, Y., Wasityastuti, W., Ratnasari, N., Liang, Y., Putri, W. A., et al. (2017). Genetic polymorphisms of HLA-DP and isolated anti-HBc are important subsets of occult hepatitis B infection in Indonesian blood donors: a case-control study. Virol. J. 14, 201. doi: 10.1186/s12985-017-0865-7
Matsuura, K., Isogawa, M., Tanaka, Y. (2016). Host genetic variants influencing the clinical course of Hepatitis B virus infection. J. Med. Virol. 88, 371–379. doi: 10.1002/jmv.24350
Mavragani, C. P., Moutsopoulos, H. M. (2014). Sjögren’s syndrome. Annu. Rev. Pathol. 9, 273–285. doi: 10.1146/annurev-pathol-012513-104728
Mbarek, H., Ochi, H., Urabe, Y., Kumar, V., Kubo, M., Hosono, N., et al. (2011). A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum. Mol. Genet. 20, 3884–3892. doi: 10.1093/hmg/ddr301
Mélik-Parsadaniantz, S., Rostène, W. (2008). Chemokines and neuromodulation. J. Neuroimmunol. 198, 62–68. doi: 10.1016/j.jneuroim.2008.04.022
Miao, F., Sun, H., Pan, N., Xu, J., Qiu, J., Shen, Y., et al. (2013). Association of human leukocyte antigen class I polymorphism with spontaneous clearance of hepatitis B surface antigen in Qidong Han population. Clin. Dev. Immunol. 2013, 145725. doi: 10.1155/2013/145725
Miklossy, G., Hilliard, T. S., Turkson, J. (2013). Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov. 12, 611–629. doi: 10.1038/nrd4088
Mohammad, A. A., Hajilooi, M., Ranjbar, M., Fallahian, F., Mousavi, S. (2006). Cytotoxic T-lymphocyte antigen 4 gene polymorphisms and susceptibility to chronic hepatitis B. World J. Gastroenterol. 12, 630–635. doi: 10.3748/wjg.v12.i4.630
Moudi, B., Heidari, Z., Mahmoudzadeh-Sagheb, H., Hashemi, M. (2016). Gene polymorphisms of macrophage migration inhibitory factor affect susceptibility to chronic hepatitis B virus infection in an Iranian cohort. Microbiol. Immunol. 60, 390–396. doi: 10.1111/1348-0421.12382
Namgoong, S., Shin, J., Cheong, H. S., Kim, L. H., Kim, J. O., Seo, J. Y., et al. (2018). Genetic association of complement component 2 variants with chronic hepatitis B in a Korean population. Liver Int. 38, 1576–1582. doi: 10.1111/liv.13675
Newport, M. J., Goetghebuer, T., Weiss, H. A., Whittle, H., Siegrist, C., Marchant, A., et al. (2004). Genetic regulation of immune responses to vaccines in early life. Genes Immun. 5, 122–129. doi: 10.1038/sj.gene.6364051
Nfor, O. N., Wu, M. F., Debnath, T., Lee, C. T., Lee, W., Liu, W. H., et al. (2018). Hepatitis B virus infection in Taiwan: the role of NTCP rs2296651 variant in relation to sex. J. Viral Hepat. 25, 1116–1120. doi: 10.1111/jvh.12912
Ni, Y. H., Chang, M. H., Wu, J. F., Hsu, H. Y., Chen, H. L., Chen, D. S. (2012). Minimization of hepatitis B infection by a 25-year universal vaccination program. J. Hepatol. 57, 730–735. doi: 10.1016/j.jhep.2012.05.021
Nishida, N., Ohashi, J., Sugiyama, M., Tsuchiura, T., Yamamoto, K., Hino, K., et al. (2015). Effects of HLA-DPB1 genotypes on chronic hepatitis B infection in Japanese individuals. Tissue Antigens 86, 406–412. doi: 10.1111/tan.12684
Nishida, N., Sawai, H., Kashiwase, K., Minami, M., Sugiyama, M., Seto, W., et al. (2014). New susceptibility and resistance HLA-DP alleles to HBV-related diseases identified by a trans-ethnic association study in Asia. PLoS One 9, e86449. doi: 10.1371/journal.pone.0086449
Nishida, N., Sawai, H., Matsuura, K., Sugiyama, M., Ahn, S. H., Park, J. Y., et al. (2012). Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS One 7, e39175. doi: 10.1371/journal.pone.0039175
Nishida, N., Sugiyama, M., Sawai, H., Nishina, S., Sakai, A., Ohashi, J., et al. (2018). Key HLA-DRB1-DQB1 haplotypes and role of the BTNL2 gene for response to a hepatitis B vaccine. Hepatology 68, 848–858. doi: 10.1002/hep.29876
Okada, Y., Uno, N., Sato, S., Mori, S., Sasaki, D., Kaku, N., et al. (2017). Strong influence of human leukocyte antigen-DP variants on response to hepatitis B vaccine in a Japanese population. Vaccine 35, 5662–5665. doi: 10.1016/j.vaccine.2017.08.045
Ovsyannikova, I. G., Jacobson, R. M., Vierkant, R. A., Shane Pankratz, V., Jacobsen, S. J., Poland, G. A. (2004). Associations between human leukocyte antigen (HLA) alleles and very high levels of measles antibody following vaccination. Vaccine 22, 1914–1920. doi: 10.1016/j.vaccine.2003.11.016
Pan, C. Q., Duan, Z. P., Bhamidimarri, K. R., Zou, H. B., Liang, X. F., Li, J., et al. (2012a). An algorithm for risk assessment and intervention of mother to child transmission of hepatitis B virus. Clin. Gastroenterol. Hepatol. 10, 452–459. doi: 10.1016/j.cgh.2011.10.041
Pan, L., Zhang, L., Zhang, W., Wu, X., Li, Y., Yan, B., et al. (2014). A genome-wide association study identifies polymorphisms in the HLA-DR region associated with non-response to hepatitis B vaccination in Chinese Han populations. Hum. Mol. Genet. 23, 2210–2219. doi: 10.1093/hmg/ddt586
Pan, L., Zhang, W., Liang, Z., Wu, X., Zhu, X., Li, J., et al. (2012b). Association between polymorphisms of the cytokine and cytokine receptor genes and immune response to hepatitis B vaccination in a Chinese Han population. J. Med. Virol. 84, 26–33. doi: 10.1002/jmv.22251
Park, B. L., Kim, Y. J., Cheong, H. S., Kim, L. H., Choi, Y. H., Lee, H. S., et al. (2006). Association of common promoter polymorphisms of MCP1 with hepatitis B virus clearance. Exp. Mol. Med. 38, 694–702. doi: 10.1038/emm.2006.82
Pawlowska, M., Pniewska, A., Pilarczyk, M., Kozielewicz, D., Domagalski, K. (2016). Prophylaxis of vertical HBV infection. Expert Opin. Drug Saf. 15, 1361–1368. doi: 10.1080/14740338.2016.1211106
Peng, S., Wan, Z., Liu, T., Zhu, H., Du, Y. (2019). Incidence and risk factors of intrauterine transmission among pregnant women with chronic hepatitis B virus infection. J. Clin. Gastroenterol. 53, 51–57. doi: 10.1097/MCG.0000000000001001
Petrova, M., Kamburov, V. (2010). Breastfeeding and chronic HBV infection: clinical and social implications. World J. Gastroenterol. 16, 5042–5046. doi: 10.3748/wjg.v16.i40.5042
Phillips, S., Chokshi, S., Riva, A., Evans, A., Williams, R., Naoumov, N. V. (2010). CD8+ T cell control of hepatitis B virus replication: direct comparison between cytolytic and noncytolytic functions. J. Immunol. 184, 287–295. doi: 10.4049/jimmunol.0902761
Png, E., Thalamuthu, A., Ong, R. T. H., Snippe, H., Boland, G. J., Seielstad, M. (2011). A genome-wide association study of hepatitis B vaccine response in an Indonesian population reveals multiple independent risk variants in the HLA region. Hum. Mol. Genet. 20, 3893–3898. doi: 10.1093/hmg/ddr302
Polaris Observatory Collaborators (2018). Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol. Hepatol. 3, 383–403. doi: 10.1016/S2468-1253(18)30056-6
Prokunina-Olsson, L., Muchmore, B., Tang, W., Pfeiffer, R. M., Park, H., Dickensheets, H., et al. (2013). A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45, 164–171. doi: 10.1038/ng.2521
Qazi, S., Schlicksup, C. J., Rittichier, J., VanNieuwenhze, M. S., Zlotnick, A. (2018). An assembly-activating site in the hepatitis B virus capsid protein can also trigger disassembly. ACS Chem. Biol. 13, 2114–2120. doi: 10.1021/acschembio.8b00283
Qureshi, A., Thakur, N., Monga, I., Thakur, A., Kumar, M. (2014). VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets. Database (Oxford) 2014, 1–10. doi: 10.1093/database/bau103
Raimondo, G., Allain, J. P., Brunetto, M. R., Buendia, M. A., Chen, D. S., Colombo, M., et al. (2008). Statements from the Taormina expert meeting on occult hepatitis B virus infection. J. Hepatol. 49, 652–657. doi: 10.1016/j.jhep.2008.07.014
Raimondo, G., Caccamo, G., Filomia, R., Pollicino, T. (2013). Occult HBV infection. Semin. Immunopathol. 35, 39–52. doi: 10.1007/s00281-012-0327-7
Raimondo, G., Locarnini, S., Pollicino, T., Levrero, M., Zoulim, F., Lok, A. S., et al. (2019). Update of the statements on biology and clinical impact of occult hepatitis b virus infection. J. Hepatol. doi: 10.1016/j.jhep.2019.03.034
Rebbani, K., Ababou, M., Nadifi, S., Kandil, M., Marchio, A., Pineau, P., et al. (2017). Myxovirus resistance 1 gene polymorphisms and outcomes of viral hepatitis B and C infections in Moroccan patients. J. Med. Virol. 89, 647–652. doi: 10.1002/jmv.24642
Rehermann, B., Bertoletti, A. (2015). Immunological aspects of antiviral therapy of chronic hepatitis B virus and hepatitis C virus infections. Hepatology 61, 712–721. doi: 10.1002/hep.27323
Rehermann, B., Thimme, R. (2019). Insights from antiviral therapy into immune responses to hepatitis B and C virus infection. Gastroenterology 156, 369–383. doi: 10.1053/j.gastro.2018.08.061
Reijnders, J. G., Perquin, M. J., Zhang, N., Hansen, B. E., Janssen, H. L. (2010). Nucleos(t)ide analogues only induce temporary hepatitis B e antigen seroconversion in most patients with chronic hepatitis B. Gastroenterology 139, 491–498. doi: 10.1053/j.gastro.2010.03.059
Ren, S., Yu, H., Zhang, H., Liu, Y., Huang, Y., Ma, L., et al. (2011). Polymorphisms of interferon-inducible genes OAS associated with interferon-α treatment response in chronic HBV infection. Antiviral Res. 89, 232–237. doi: 10.1016/j.antiviral.2011.01.006
Ren, Y., Guo, Y., Feng, L., Li, T., Du, Y. (2016). Controversy and strategies exploration in blocking mother-to-child transmission of hepatitis B. Int. Rev. Immunol. 35, 249–259. doi: 10.3109/08830185.2015.1096934
Revill, P. A., Chisari, F. V., Block, J. M., Dandri, M., Gehring, A. J., Guo, H., et al. (2019). A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. doi: 10.1016/S2468-1253(19)30119-0
Robek, M. D., Boyd, B. S., Wieland, S. F., Chisari, F. V. (2004). Signal transduction pathways that inhibit hepatitis B virus replication. Proc. Natl. Acad. Sci. U.S.A. 101, 1743–1747. doi: 10.1073/pnas.0308340100
Roh, E. Y., Yoon, J. H., In, J. W., Lee, N., Shin, S., Song, E. Y. (2016). Association of HLA-DP variants with the responsiveness to Hepatitis B virus vaccination in Korean Infants. Vaccine 34, 2602–2607. doi: 10.1016/j.vaccine.2016.03.090
Romporn, S., Hirankarn, N., Tangkijvanich, P., Kimkong, I. (2013). Association of IFNAR2 and IL10RB genes in chronic hepatitis B virus infection. Tissue Antigens 82, 21–25. doi: 10.1111/tan.12133
Sakai, A., Noguchi, E., Fukushima, T., Tagawa, M., Iwabuchi, A., Kita, M., et al. (2017). Identification of amino acids in antigen-binding site of class II HLA proteins independently associated with hepatitis B vaccine response. Vaccine 35, 703–710. doi: 10.1016/j.vaccine.2016.08.068
Samal, J., Kandpal, M., Vivekanandan, P. (2012). Molecular mechanisms underlying occult hepatitis B virus infection. Clin. Microbiol. Rev. 25, 142–163. doi: 10.1128/CMR.00018-11
Samuel, C. E. (2001). Antiviral actions of interferons. Clin. Microbiol. Rev. 14, 778–809. doi: 10.1128/CMR.14.4.778-809.2001
Saraiva, M., O’Garra, A. (2010). The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181. doi: 10.1038/nri2711
Sato, T., Do, S. H., Asao, T., Akita, T., Katayama, K., Tatara, K., et al. (2014). Estimating numbers of persons with persistent hepatitis B virus infection transmitted vertically and horizontally in the birth cohort during 1950-1985 in Japan. Hepatol. Res. 44, E181–E188. doi: 10.1111/hepr.12288
Saxena, R., Chawla, Y. K., Verma, I., Kaur, J. (2014). Effect of IL-12B, IL-2, TGF-β1, and IL-4 polymorphism and expression on hepatitis B progression. J. Interferon Cytokine Res. 34, 117–128. doi: 10.1089/jir.2013.0043
Schoenborn, J. R., Wilson, C. B. (2007). Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 96, 41–101. doi: 10.1016/S0065-2776(07)96002-2
Schott, E., Witt, H., Pascu, M., van Boemmel, F., Weich, V., Bergk, A., et al. (2007). Association of CTLA4 single nucleotide polymorphisms with viral but not autoimmune liver disease. Eur. J. Gastroenterol. Hepatol. 19, 947–951. doi: 10.1097/MEG.0b013e3282efa240
Schurich, A., Khanna, P., Lopes, A. R., Han, K. J., Peppa, D., Micco, L., et al. (2011). Role of the coinhibitory receptor cytotoxic T lymphocyte antigen-4 on apoptosis-Prone CD8 T cells in persistent hepatitis B virus infection. Hepatology 53, 1494–1503. doi: 10.1002/hep.24249
Seed, C. R., Allain, J. P., Lozano, M., Laperche, S., Gallian, P., Gross, S., et al. (2019). International forum on occult hepatitis B infection and transfusion safety. Vox Sang. 114, 397–406. doi: 10.1111/vox.12744
Seo, D. H., Whang, D. H., Song, E. Y., Kim, H. S., Park, Q. (2011). Prevalence of antibodies to hepatitis B core antigen and occult hepatitis B virus infections in Korean blood donors. Transfusion 51, 1840–1846. doi: 10.1111/j.1537-2995.2010.03056.x
Seshasubramanian, V., Soundararajan, G., Ramasamy, P. (2018). Human leukocyte antigen A, B and hepatitis B infection outcome: a meta-analysis. Infect. Genet. Evol. 66, 392–398. doi: 10.1016/j.meegid.2017.07.027
Shao, Z., Xu, D., Xu, J., Li, J., Yan, Y., Men, K., et al. (2007). Maternal hepatitis B virus (HBV) DNA positivity and sexual intercourse are associated with HBV intrauterine transmission in China: a prospective case-control study. J. Gastroenterol. Hepatol. 22, 165–170. doi: 10.1111/j.1440-1746.2006.04462.x
Sheppard, P., Kindsvogel, W., Xu, W., Henderson, K., Schlutsmeyer, S., Whitmore, T. E., et al. (2003). IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4, 63–68. doi: 10.1038/ni873
Shi, K. Q., Cai, X. H., Xiao, D. D., Wu, S. J., Peng, M. M., Lin, X. F., et al. (2012). Tumour necrosis factor-α-857T allele reduces the risk of hepatitis B virus infection in an Asian population. J. Viral Hepat. 19, e66–e72. doi: 10.1111/j.1365-2893.2011.01540.x
Shiina, T., Hosomichi, K., Inoko, H., Kulski, J. K. (2009). The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 54, 15–39. doi: 10.1038/jhg.2008.5
Shin, E. C., Sung, P. S., Park, S. H. (2016). Immune responses and immunopathology in acute and chronic viral hepatitis. Nat. Rev. Immunol. 16, 509–523. doi: 10.1038/nri.2016.69
Shin, H. D., Park, B. L., Cheong, H. S., Yoon, J. H., Kim, Y. J., Lee, H. S. (2007). SPP1 polymorphisms associated with HBV clearance and HCC occurrence. Int. J. Epidemiol. 36, 1001–1008. doi: 10.1093/ije/dym093
Shin, J., Cheong, H. S., Kim, J. Y., Lee, J., Yu, S. J., Yoon, J., et al. (2019). Identification of additional EHMT2 variant associated with the risk of chronic hepatitis B by GWAS follow-up study. Genes Immun. 20, 1–9. doi: 10.1038/s41435-017-0004-x
Shin, J. G., Cheong, H. S., Lee, K., Ju, B. G., Lee, J. H., Yu, S. J., et al. (2017). Identification of novel OCT4 genetic variant associated with the risk of chronic hepatitis B in a Korean population. Liver Int. 37, 354–361. doi: 10.1111/liv.13245
Singh, R., Kaul, R., Kaul, A., Khan, K. (2007). A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J. Gastroenterol. 13, 1770–1787. doi: 10.3748/wjg.v13.i12.1770
Singh, R. K., Dai, Y., Staudinger, J. L., Muma, N. A. (2009). Activation of the JAK-STAT pathway is necessary for desensitization of 5-HT2A receptor-stimulated phospholipase C signalling by olanzapine, clozapine and MDL 100907. Int. J. Neuropsychopharmacol. 12, 651–665. doi: 10.1017/S1461145708009590
Sonneveld, M. J., Wong, V. W. S., Woltman, A. M., Wong, G. L. H., Cakaloglu, Y., Zeuzem, S., et al. (2012). Polymorphisms near IL28B and serologic response to peginterferon in HBeAg-positive patients with chronic hepatitis B. Gastroenterology 142, 513–520.e1. doi: 10.1053/j.gastro.2011.11.025
Stevens, C. E., Beasley, R. P., Tsui, J., Lee, W. C. (1975). Vertical transmission of hepatitis B antigen in Taiwan. N. Engl. J. Med. 292, 771–774. doi: 10.1056/NEJM197504102921503
Su, C., Hou, Z., Zhang, C., Tian, Z., Zhang, J. (2011). Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol. J. 8, 354. doi: 10.1186/1743-422X-8-354
Su, F. H., Chen, J. D., Cheng, S. H., Lin, C. H., Liu, Y. H., Chu, F. Y. (2007). Seroprevalence of hepatitis-B infection amongst Taiwanese university students 18 years following the commencement of a national hepatitis-B vaccination program. J. Med. Virol. 79, 138–143. doi: 10.1002/jmv.20771
Su, T., Yang, H., Tseng, T., Liou, J., Liu, C., Chen, C., et al. (2018). Distinct relapse rates and risk predictors after discontinuing tenofovir and entecavir therapy. J. Infect. Dis. 217, 1193–1201. doi: 10.1093/infdis/jix690
Sun, Y., Lu, Y., Li, T., Xie, L., Deng, Y., Li, S., et al. (2015). Interferon gamma +874T/A polymorphism increases the risk of hepatitis virus-related diseases: evidence from a meta-analysis. PLOS ONE 10, e0121168. doi: 10.1371/journal.pone.0121168
Suneetha, P. V., Sarin, S. K., Goyal, A., Kumar, G. T., Shukla, D. K., Hissar, S. (2006). Association between vitamin D receptor, CCR5, TNF-α and TNF-β gene polymorphisms and HBV infection and severity of liver disease. J. Hepatol. 44, 856–863. doi: 10.1016/j.jhep.2006.01.028
Talaat, R. M., Dondeti, M. F., El-Shenawy, S. Z., Khamiss, O. A. (2014). Association between IL-10 gene promoter polymorphism and hepatitis B viral infection in an Egyptian population. Biochem. Genet. 52, 387–402. doi: 10.1007/s10528-014-9655-8
Tangkijvanich, P., Chittmittraprap, S., Poovorawan, K., Limothai, U., Khlaiphuengsin, A., Chuaypen, N., et al. (2016). A randomized clinical trial of peginterferon alpha-2b with or without entecavir in patients with HBeAg-negative chronic hepatitis B: role of host and viral factors associated with treatment response. J. Viral Hepat. 23, 427–438. doi: 10.1111/jvh.12467
Terrault, N. A., Bzowej, N. H., Chang, K. M., Hwang, J. P., Jonas, M. M., Murad, M. H., et al. (2016). AASLD guidelines for treatment of chronic hepatitis B. Hepatology 63, 261–283. doi: 10.1002/hep.28156
Thanapirom, K., Suksawatamnuay, S., Sukeepaisarnjareon, W., Tanwandee, T., Charatcharoenwitthaya, P., Thongsawat, S., et al. (2017). Genetic variation in the vitamin D pathway CYP2R1 gene predicts sustained HBeAg seroconversion in chronic hepatitis B patients treated with pegylated interferon: a multicenter study. PLoS One 12, e0173263. doi: 10.1371/journal.pone.0173263
Thimme, R., Wieland, S., Steiger, C., Ghrayeb, J., Reimann, K. A., Purcell, R. H., et al. (2003). CD8+ T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 77, 68–76. doi: 10.1128/JVI.77.1.68-76.2003
Thio, C. L., Mosbruger, T. L., Kaslow, R. A., Karp, C. L., Strathdee, S. A., Vlahov, D., et al. (2004). Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. J. Virol. 78, 11258–11262. doi: 10.1128/JVI.78.20.11258-11262.2004
Thomas, R., Thio, C. L., Apps, R., Qi, Y., Gao, X., Marti, D., et al. (2012). A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J. Virol. 86, 6979–6985. doi: 10.1128/JVI.00406-12
Thursz, M., Yee, L., Khakoo, S. (2011). Understanding the host genetics of chronic hepatitis B and C. Semin. Liver Dis. 31, 115–127. doi: 10.1055/s-0031-1276642
Torbenson, M., Thomas, D. L. (2002). Occult hepatitis B. Lancet Infect. Dis. 2, 479–486. doi: 10.1016/S1473-3099(02)00345-6
Trinks, J., Nishida, N., Hulaniuk, M. L., Caputo, M., Tsuchiura, T., Marciano, S., et al. (2017). Role of HLA-DP and HLA-DQ on the clearance of hepatitis B virus and the risk of chronic infection in a multiethnic population. Liver Int. 37, 1476–1487. doi: 10.1111/liv.13405
Tseng, T., Yu, M., Liu, C., Lin, C., Huang, Y., Hsu, C., et al. (2011). Effect of host and viral factors on hepatitis B e antigen-positive chronic hepatitis B patients receiving pegylated interferon-α-2a therapy. Antivir. Ther. 16, 629–637. doi: 10.3851/IMP1841
Tu, T., Urban, S. (2018). Virus entry and its inhibition to prevent and treat hepatitis B and hepatitis D virus infections. Curr. Opin. Virol. 30, 68–79. doi: 10.1016/j.coviro.2018.04.004
van Lith, M., Ewen-Smith, R. M., Benham, A. M. (2010). HLA-DP, HLA-DQ, and HLA-DR have different requirements for invariant chain and HLA-DM. J. Biol. Chem. 285, 40800–40808. doi: 10.1074/jbc.M110.148155
Vermehren, J., Lotsch, J., Susser, S., Wicker, S., Berger, A., Zeuzem, S., et al. (2012). A common HLA-DPA1 variant is associated with hepatitis B virus infection but fails to distinguish active from inactive Caucasian carriers. PLoS One 7, e32605. doi: 10.1371/journal.pone.0032605
Wan, Z., Lin, X., Li, T., Zhou, A., Yang, M., Hu, D., et al. (2016). Genetic variant in CXCL13 gene is associated with susceptibility to intrauterine infection of hepatitis B virus. Sci. Rep. 6, 26465. doi: 10.1038/srep26465
Wang, L., Wu, X., Zhang, W., Zhu, D., Wang, Y., Li, Y., et al. (2011). Evaluation of genetic susceptibility loci for chronic hepatitis B in Chinese: two independent case-control studies. PLoS One 6, e17608. doi: 10.1371/journal.pone.0017608
Wang, L., Zou, Z., Wang, K. (2016). Clinical relevance of HLA gene variants in HBV infection. J. Immunol. Res. 2016, 9069375. doi: 10.1155/2016/9069375
Wang, P., Mo, R., Lai, R., Xu, Y., Lu, J., Zhao, G., et al. (2017a). Genetic variations of NTCP are associated with susceptibility to HBV infection and related hepatocellular carcinoma. Oncotarget 8, 105407–105424. doi: 10.18632/oncotarget.22211
Wang, S. H., Chen, P. J., Yeh, S. H. (2015). Gender disparity in chronic hepatitis B: mechanisms of sex hormones. J. Gastroenterol. Hepatol. 30, 1237–1245. doi: 10.1111/jgh.12934
Wang, T., Shen, C., Chen, L., Liu, S., Ji, Y. (2017b). Association of human leukocyte antigen polymorphisms with occult hepatitis B virus infection in a Shaanxi Han population. J. Gene Med. 19, e2987. doi: 10.1002/jgm.2987
Wang, Y., Xu, P., Zhu, D., Zhang, S., Bi, Y., Hu, Y., et al. (2012). Association of polymorphisms of cytokine and TLR-2 genes with long-term immunity to hepatitis B in children vaccinated early in life. Vaccine 30, 5708–5713. doi: 10.1016/j.vaccine.2012.07.010
Wasityastuti, W., Yano, Y., Ratnasari, N., Triyono, T., Triwikatmani, C., Indrarti, F., et al. (2016). Protective effects of HLA-DPA1/DPB1 variants against hepatitis B virus infection in an Indonesian population. Infect. Genet. Evol. 41, 177–184. doi: 10.1016/j.meegid.2016.03.034
Wen, J., Liu, Y., Liu, J., Liu, L., Song, C., Han, J., et al. (2015). Expression quantitative trait loci in long non-coding RNA ZNRD1-AS1 influence both HBV infection and hepatocellular carcinoma development. Mol. Carcinog. 54, 1275–1282. doi: 10.1002/mc.22200
Wong, D. K., Watanabe, T., Tanaka, Y., Seto, W. K., Lee, C. K., Fung, J., et al. (2013). Role of HLA-DP polymorphisms on chronicity and disease activity of hepatitis B infection in Southern Chinese. PLoS One 8, e66920. doi: 10.1371/journal.pone.0066920
Wright, T. L., Lau, J. Y. (1993). Clinical aspects of hepatitis B virus infection. Lancet 342, 1340–1344. doi: 10.1016/0140-6736(93)92250-W
Wu, H., Zhao, G., Qian, F., Liu, K., Xie, J., Zhou, H., et al. (2015a). Association of IL28B polymorphisms with peginterferon treatment response in Chinese Han patients with HBeAg-positive chronic hepatitis B. Liver Int. 35, 473–481. doi: 10.1111/liv.12491
Wu, T., Chen, C., Lai, S., Lin, H. H., Chu, C., Wang, L. (2015b). SNP rs7770370 in HLA-DPB1 loci as a major genetic determinant of response to booster hepatitis B vaccination: results of a genome-wide association study. J. Gastroenterol. Hepatol. 30, 891–899. doi: 10.1111/jgh.12845
Wu, W., Zeng, Y., Lin, J., Wu, Y., Chen, T., Xun, Z., et al. (2018a). Genetic variants in NTCP exon gene are associated with HBV infection status in a Chinese Han population. Hepatol. Res. 48, 364–372. doi: 10.1111/hepr.13007
Wu, X., Shi, W., Wu, J., Zhu, X., Chen, K., Zheng, S., et al. (2014). A functional polymorphism in ADAR1 gene affects HBsAg seroclearance both spontaneously and interferon induced. Liver Int. 34, 1560–1565. doi: 10.1111/liv.12444
Wu, X., Zhu, X., Zhu, S., Li, J., Ma, J., Li, Z., et al. (2009). A pharmacogenetic study of polymorphisms in interferon pathway genes and response to interferon-alpha treatment in chronic hepatitis B patients. Antiviral Res. 83, 252–256. doi: 10.1016/j.antiviral.2009.06.003
Xiang, X., Guo, Y., Yang, L., Ge, Q., Mijit, S., Xu, F. (2016). Association of human leukocyte antigen DP/DQ gene polymorphisms with chronic hepatitis B in Chinese Han and Uygur populations. Infect. Genet. Evol. 43, 407–411. doi: 10.1016/j.meegid.2016.06.022
Xu, B., Zhu, D., Bi, Y., Wang, Y., Hu, Y., Zhou, Y. (2017a). Minimal association of alleles of human leukocyte antigen class II gene and long-term antibody response to hepatitis B vaccine vaccinated during infancy. Vaccine 35, 2457–2462. doi: 10.1016/j.vaccine.2017.03.021
Xu, H., Zhao, M., He, J., Chen, Z. (2013). Association between cytotoxic T-lymphocyte associated protein 4 gene +49 A/G polymorphism and chronic infection with hepatitis B virus: a meta-analysis. J. Int. Med. Res. 41, 559–567. doi: 10.1177/0300060513483387
Xu, T., Sun, M., Wang, H. (2017b). Relationship between HLA-DQ gene polymorphism and hepatitis B virus infection. Biomed. Res. Int. 2017, 9679843. doi: 10.1155/2017/9679843
Xu, T., Zhu, A., Sun, M., Lv, J., Qian, Z., Wang, X., et al. (2018). Quantitative assessment of HLA-DQ gene polymorphisms with the development of hepatitis B virus infection, clearance, liver cirrhosis, and hepatocellular carcinoma. Oncotarget 9, 96–109. doi: 10.18632/oncotarget.22941
Xu, Y. Y., Yu, J. Y., Zhong, Y. W., Song, H. B., Liu, H. H., Jia, L. L., et al. (2008). Association between the frequency of class II HLA antigens and the susceptibility to intrauterine infection of hepatitis B virus. Int J Biol Sci 4, 111–115. doi: 10.7150/ijbs.4.111
Yan, H., Zhong, G., Xu, G., He, W., Jing, Z., Gao, Z., et al. (2012a). Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1, e00049. doi: 10.7554/eLife.00049
Yan, K., Cai, W., Cao, F., Sun, H., Chen, S., Xu, R., et al. (2013). Genetic effects have a dominant role on poor responses to infant vaccination to hepatitis B virus. J. Hum. Genet. 58, 293–297. doi: 10.1038/jhg.2013.18
Yan, Z., Tan, S., Dan, Y., Sun, X., Deng, G., Wang, Y. (2012b). Relationship between HLA-DP gene polymorphisms and clearance of chronic hepatitis B virus infections: case–control study and meta-analysis. Infect. Genet. Evol. 12, 1222–1228. doi: 10.1016/j.meegid.2012.03.026
Yang, C., Pan, L., Zhang, L., Wu, X., Zhu, X., Yan, B., et al. (2014). BTNL2 associated with the immune response to hepatitis B vaccination in a Chinese Han population. J. Med. Virol. 86, 1105–1112. doi: 10.1002/jmv.23934
Yang, J., Yang, Y., Xia, M., Wang, L., Zhou, W., Yang, Y., et al. (2016a). A genetic variant of the NTCP gene is associated with HBV infection status in a Chinese population. BMC Cancer 16, 211. doi: 10.1186/s12885-016-2257-6
Yang, L., Lu, M. (2018). Small molecule inhibitors of hepatitis B virus nucleocapsid assembly: a new approach to treat chronic HBV infection. Curr. Med. Chem. 25, 802–813. doi: 10.2174/0929867324666170704121800
Yang, L., Liu, F., Tong, X., Hoffmann, D., Zuo, J., Lu, M. (2019). Treatment of chronic hepatitis B virus infection using small molecule modulators of nucleocapsid assembly: recent advances and perspectives. ACS Infect. Dis. 5, 713–724. doi: 10.1021/acsinfecdis.8b00337
Yang, S., Tian, G., Cui, Y., Ding, C., Deng, M., Yu, C., et al. (2016b). Factors influencing immunologic response to hepatitis B vaccine in adults. Sci. Rep. 6, 27251. doi: 10.1038/srep27251
Yano, Y., Seo, Y., Azuma, T., Hayashi, Y. (2013). Hepatitis B virus and host factors. Hepatobiliary Surg. Nutr. 2, 121–123. doi: 10.3978/j.issn.2304-3881.2012.10.10
Yin, J., Zhang, H., He, Y., Xie, J., Liu, S., Chang, W., et al. (2010). Distribution and hepatocellular carcinoma-related viral properties of hepatitis B virus genotypes in Mainland China: a community-based study. Cancer Epidemiol. Biomarkers Prev. 19, 777–786. doi: 10.1158/1055-9965.EPI-09-1001
Yoon, J. H., Shin, S., In, J. W., Chang, J. Y., Song, E. Y., Roh, E. Y. (2014). Association of HLA alleles with the responsiveness to hepatitis B virus vaccination in Korean infants. Vaccine 32, 5638–5644. doi: 10.1016/j.vaccine.2014.08.007
Yu, H., Zhu, Q. R., Gu, S. Q., Fei, L. E. (2006). Relationship between IFN-gamma gene polymorphism and susceptibility to intrauterine HBV infection. World J. Gastroenterol. 12, 2928–2931. doi: 10.3748/wjg.v12.i18.2928
Yu, L., Cheng, Y., Cheng, M., Yao, Y., Zhang, Q., Zhao, X., et al. (2015). Quantitative assessment of common genetic variations in HLA-DP with hepatitis B virus infection, clearance and hepatocellular carcinoma development. Sci. Rep. 5, 14933. doi: 10.1038/srep14933
Yu, S. J., Kim, J. W., Lee, J. H., Yoon, J. H., Lee, H. S., Cheong, J. Y., et al. (2014). Association of a microRNA-323b polymorphism with the persistence of hepatitis B virus infection by the enhancement of viral replication. J. Viral Hepat. 21, 853–859. doi: 10.1111/jvh.12215
Zhang, E., Lu, M. (2015). Toll-like receptor (TLR)-mediated innate immune responses in the control of hepatitis B virus (HBV) infection. Med. Microbiol. Immunol. 204, 11–20. doi: 10.1007/s00430-014-0370-1
Zhang, C., Zhong, Y., Guo, L. (2013a). Strategies to prevent hepatitis B virus infection in China: immunization, screening, and standard medical practices. Biosci. Trends 7, 7–12. doi: 10.5582/bst.2013.v7.1.7
Zhang, Q., Ji, X. W., Hou, X. M., Lu, F. M., Du, Y., Yin, J. H., et al. (2014a). Effect of functional nuclear factor-kappaB genetic polymorphisms on hepatitis B virus persistence and their interactions with viral mutations on the risk of hepatocellular carcinoma. Ann. Oncol. 25, 2413–2419. doi: 10.1093/annonc/mdu451
Zhang, Q., Yin, J., Zhang, Y., Deng, Y., Ji, X., Du, Y., et al. (2013b). HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. J. Virol. 87, 12176–12186. doi: 10.1128/JVI.02073-13
Zhang, T. T., Ye, J., Xia, S. L., Zhang, Y. F., Su, Q., Zhang, Z. H., et al. (2013c). Polymorphism of estrogen receptor alpha (ESR1) is associated with virological response to entecavir (ETV) in nucleoside-naïve adult patients with chronic hepatitis B. Infection 41, 371–378. doi: 10.1007/s15010-012-0320-z
Zhang, X., Hou, J., Lu, M. (2013d). Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front. Genet. 4, 202. doi: 10.3389/fgene.2013.00202
Zhang, X., Jia, J., Dong, J., Yu, F., Ma, N., Li, M., et al. (2014b). HLA-DQ polymorphisms with HBV infection: different outcomes upon infection and prognosis to lamivudine therapy. J. Viral Hepat. 21, 491–498. doi: 10.1111/jvh.12159
Zhang, X., Ni, X., Jia, J., Dong, J., Yu, F., Ma, N., et al. (2013e). Association of the rs3077 and rs9277535 polymorphisms in HLA-DP with hepatitis B virus infection and spontaneous clearance: a meta-analysis. Scand. J. Gastroenterol. 48, 736–744. doi: 10.3109/00365521.2013.787643
Zhang, X., Zhang, E., Ma, Z., Pei, R., Jiang, M., Schlaak, J. F., et al. (2011a). Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology 53, 1476–1485. doi: 10.1002/hep.24195
Zhang, Y., Li, Y., Wu, M., Cao, P., Liu, X., Ren, Q., et al. (2017). Comprehensive assessment showed no associations of variants at the SLC10A1 locus with susceptibility to persistent HBV infection among Southern Chinese. Sci. Rep. 7, 46490. doi: 10.1038/srep46490
Zhang, Z. H., Li, L., Zhao, X. P., Glebe, D., Bremer, C. M., Zhang, Z. M., et al. (2011b). Elimination of hepatitis B virus surface antigen and appearance of neutralizing antibodies in chronically infected patients without viral clearance. J. Viral Hepat. 18, 424–433. doi: 10.1111/j.1365-2893.2010.01322.x
Zhao, Q., Peng, L., Huang, W., Li, Q., Pei, Y., Yuan, P., et al. (2012). Rare inborn errors associated with chronic hepatitis B virus infection. Hepatology 56, 1661–1670. doi: 10.1002/hep.25850
Zheng, M. H., Xiao, D. D., Lin, X. F., Wu, S. J., Peng, M. M., Yu, X. Y., et al. (2012). The tumour necrosis factor-α-238A allele increases the risk of chronic HBV infection in European populations. J. Viral Hepat. 19, e11–e17. doi: 10.1111/j.1365-2893.2011.01491.x
Zhou, G. Q., Meng, H., Wang, J. R., Sun, F. X., Wang, X. J., Wang, R. B., et al. (2015). Functional polymorphisms in microRNA gene and hepatitis B risk among Asian population: a meta-analysis. Genet. Mol. Res. 14, 4767–4777. doi: 10.4238/2015.May.11.9
Zhu, H., Wang, C., Zhang, Y., Wei, S., Li, X., Zhang, Z. (2016a). Prediction model for sustained hepatitis B e antigen seroconversion to peginterferon alfa-2a in chronic hepatitis B. J. Gastroenterol. Hepatol. 31, 1963–1970. doi: 10.1111/jgh.13414
Zhu, J., Zhang, T., Cao, L., Li, A., Zheng, K., Zhang, N., et al. (2017). Toll like receptor7 polymorphisms in relation to disease susceptibility and progression in Chinese patients with chronic HBV infection. Sci. Rep. 7, 12417. doi: 10.1038/s41598-017-12698-5
Zhu, M., Dai, J., Wang, C., Wang, Y., Qin, N., Ma, H., et al. (2016b). Fine mapping the MHC region identified four independent variants modifying susceptibility to chronic hepatitis B in Han Chinese. Hum. Mol. Genet. 25, 1225–1232. doi: 10.1093/hmg/ddw003
Zhu, W., Liu, H., Zhang, X. (2019). Toward curative immunomodulation strategies for chronic hepatitis B virus infection. ACS Infect. Dis. 5, 703–712. doi: 10.1021/acsinfecdis.8b00297
Zhu, X., Du, T., Wu, X., Guo, X., Niu, N., Pan, L., et al. (2011). Human leukocyte antigen class I and class II genes polymorphisms might be associated with interferon α therapy efficiency of chronic hepatitis B. Antiviral Res. 89, 189–192. doi: 10.1016/j.antiviral.2011.01.001
Zidi, I., Laaribi, A. B., Bortolotti, D., Belhadj, M., Mehri, A., Yahia, H. B., et al. (2016). HLA-E polymorphism and soluble HLA-E plasma levels in chronic hepatitis B patients. HLA 87, 153–159. doi: 10.1111/tan.12767
Zoulim, F., Locarnini, S. (2012). Management of treatment failure in chronic hepatitis B. J. Hepatol. 56 Suppl 1, S112–S122. doi: 10.1016/S0168-8278(12)60012-9
Keywords: hepatitis B virus, genetic determinants, human leukocyte antigen, susceptibility gene, single nucleotide polymorphism, genome-wide association study
Citation: Zhang Z, Wang C, Liu Z, Zou G, Li J and Lu M (2019) Host Genetic Determinants of Hepatitis B Virus Infection. Front. Genet. 10:696. doi: 10.3389/fgene.2019.00696
Received: 07 November 2018; Accepted: 03 July 2019;
Published: 13 August 2019.
Edited by:
Cheryl Ann Winkler, Frederick National Laboratory for Cancer Research (NIH), United StatesReviewed by:
Michael Scheurer, Baylor College of Medicine, United StatesCopyright © 2019 Zhang, Wang, Liu, Zou, Li and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengji Lu, bWVuZ2ppLmx1QHVuaS1kdWUuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.