
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 18 July 2019
Sec. RNA
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00626
This article is part of the Research Topic Role of RNA in Molecular Diagnostics of Cancer View all 12 articles
MicroRNAs (miRNAs) are endogenous non-coding small RNA molecules that can be secreted into the circulation and exist in remarkably stable forms. Like intercellular miRNAs, circulating miRNAs participate in numerous regulations of biological process and expressed aberrantly under abnormal or pathological status. The quality and quantity changes of circulating miRNAs are associated with the initiation and progression of cancer and can be easily detected by basic molecular biology techniques. Consequently, considerable effort has been devoted to identify suitable extracellular miRNAs for noninvasive biomarkers in cancer. However, several challenges need to be overcome before the practical application. In this review, we discuss several issues of circulating miRNAs: biological function and basic transport carriers; extracellular cell communication process; roles as reliable cancer biomarkers and usage in targeted cancer therapy; and challenges for clinical application.
MicroRNA (miRNA) was first discovered as the product of the lin-4 gene in Caenorhabditis elegans in 1993 (Lee et al., 1993; Wightman et al., 1993). The small non-coding RNAs (19-22nt) develop post-transcriptional regulation by mRNA cleavage or translation repression, which depended on the complementarity degree of miRNA-mRNA. mRNA cleavage occurs when there is a perfect match, whereas imperfect combination results in gene repression (Bartel, 2004). A large number of studies have confirmed the role of microRNAs in various cancer-associated biological processes, such as proliferation, differentiation, apoptosis, metabolism, invasion, metastasis, and drug resistance. The pathological origin of cancer has also been proven to be directly related to the dysregulation of miRNAs. Moreover, miRNAs are tissue-specific. Different tumors have distinctive miRNA expression profiles. So far, the basic biogenesis and function of the intracellular miRNAs have been reviewed in a number of contexts. On the other hand, the presence of extracellular RNAs in serum/plasma was described first by Bartel (Bartel, 2004; Kibel, 2009) and various miRNAs are proved to exist in a stable cell-free form in body fluids and other extracellular environments, including plasma, serum, urine, saliva, seminal, ascites, amniotic pleural effusions, and cerebrospinal fluid (Weber et al., 2010; Cortez et al., 2011; Husted et al., 2011; Alečković and Kang, 2015; Kibel, 2009). Studies suggested that they are injected to the circulation in different ways. Parts are due to the passive leakage of apoptosis, necrosis, or the environment of inflammation, and parts are secreted actively by exosomes/microvesicles, lipoproteins, and RNA-protein complex (Arroyo et al., 2011; Cheng, 2015). Furthermore, specific miRNAs are selected to pack into exosomes and become one of the most important aspects of the tumor microenvironment, which is the underlying mechanism of tissue/disease specificity of circulating miRNAs. Additionally, circulating miRNAs are correlated with the degree of tumor progression and present differently at different stages of cancer, making them play an important role in cancer immunotherapy. That is to say, the presence of certain type of circulating miRNAs was confirmed essentially in the manifestation, development, invasion, and metastasis of cancer, and the abnormal levels of distinct miRNAs could be observed in every process above (Wang et al., 2018). Although the cancer-related biomarkers, which are widely used clinically, are simple and fast, their disadvantages, like poor early diagnosis and prognostic value, limit their role in targeted therapy and the lack of tissue specificity leads to an urgent need to find novel biomarkers. As a result, circulating miRNAs are becoming candidates of emerging non-invasive cellular and molecular biomarkers of cancer.
The discovery of circulating miRNA is unanticipated, considering that the activity of RNase in plasma and the underlying mechanism came to be known after years of intensive experiments and discussion. In 2008, Patrick and colleagues confirmed that endogenous miRNAs ranging 18 to 24 nt exist in human plasma by cloning, sequencing, and quantification. Storing at room temperature, suffering freeze-thaw, or extreme variations in pH will not lead to a descending of circulating miRNAs (Duttagupta et al., 2011; Kibel, 2009). However, purified plasma (Arroyo et al., 2011) miRNAs and synthetic miRNAs (Tsui et al., 2002; Kibel, 2009) rapidly degraded when cultured with plasma, suggesting that endogenous miRNAs are resisted to RNase because of various modifications. There are two major populations of circulating miRNAs, vesicle-associated and non–vesicle-associated (Figure 1), which reflect the different mechanisms of release. Some tissue-specific miRNAs may release to the circulation in a protein-mediated way and thus presented in protein complex only, such as the liver-specific miR-122 (Chang et al., 2004). However, because releasing vesicles are essential in the process of the maturation and activation of most blood cells, almost all erythrocytes and platelet-related miRNAs are packed into the vesicles in the circulation (Heijnen et al., 1999; Hunter et al., 2010).
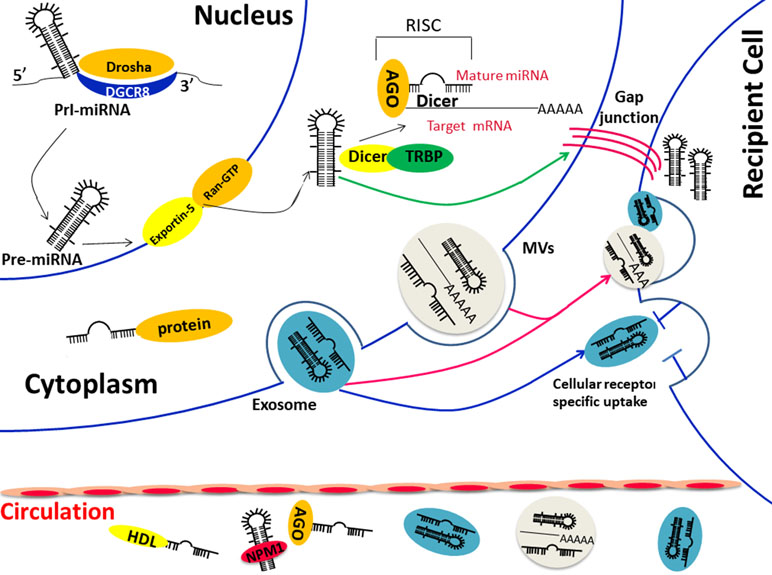
Figure 1 (1) Biological functions and transportation carriers of circulating miRNA; (2) Diverse ways of miRNAs in cell communication: direct fusion and endocytosis of extracellular vesicles (exosomes orMVs) (red arrow) or direct transfer through cell gap junction (green arrow) or the indirect identification of specific surface receptor (blue arrow). Abbreviations: Pre-miRNA, precursor miRNA; Pri-miRNA, primary miRNA; AGO, Argonaute; TRBP, transactivation-responsive RNA-binding protein; DGCR8, DiGeorge Syndrome Critical Region Gene 8; MVBs, multivesicular bodies; NPM1, nucleophosmin 1.
Membrane-bound vesicles, such as exosomes (50–90 nm) and microvesicles (1 μm), comprise one type of extracellular miRNA, which can be detected from vesicles isolated and purified from plasma/serum (Valadi et al., 2007; Taylor and Gercel-Taylor, 2008; Hunter et al., 2010). A study done by a team from America found multiple heterogeneous RNAs via Bioanalyzer from exosomes derived from a mast cell line. Large amounts of small RNA were found while the level of ribosomal RNA was low. Then, more than 120 miRNAs were certified in further analysis by microarray (Valadi et al., 2007). There is no doubt that the formation of the miRNA-vesicle package is confirmed by lots of researches. However, recent studies found that the apoptotic body is also one of the forms of the vesicular carrier, but the miRNA it contains may be a random event that differs from the exosome. In a word, all these results indicated the function of MVBs as transporters for extracellular miRNAs.
On the other hand, other studies confirmed that majority of the circulating miRNAs exist in a non–vesicle-associated form, such as the ribonucleoprotein complex. The copies of miRNAs dropped significantly since proteinase K was added to the plasma, which verifies the hypothesis that miRNAs could be degraded by RNase easily when dissociated from a protease-sensitive complex(10), and these proteins, such as Argonaute2 (Ago), GW182, nucleophosmin1 (NPM1), and high-density lipoprotein (HDL), are confirmed as miRNA carriers in a large number of studies. Ago2, the central protein of miRNA-mediated interference, together with GW182, were verified responsible for the protection and transport of extracellular miRNAs (Wang et al., 2010; Arroyo et al., 2011; Yao et al., 2012; Montani and Bianchi, 2016). The miRNA degradation occurred when Ago2 was isolated from the protein complex (Arroyo et al., 2011) and when GW182 was knocked down (Yao et al., 2012). Another protein that takes part in the protection of external miRNAs is the well-known nucleolar RNA-binding protein, nucleophosmin 1 (NPM1), which is involved in the exporting of RNAs and ribosome (Maggi et al., 2008). Synthetic miR-122 was not degraded by RNase when incubated with NPM1, and the NPM1 was confirmed not only participated in the packaging of miRNAs but also in the process of miRNA exporting (Wang et al., 2010). High-density lipoprotein is widely known as a mediation of excess cellular cholesterol. However, its function is far more complex especially when it involves the transportation and post-transcription effect in the recipient cells of miRNAs (Rothblat and Phillips, 2010; Vickers et al., 2014). As a result, HDL has been used as the delivery of siRNAs in animal models (Kim et al., 2007).
In conclusion, it is just these miRNA transporters that protect circulating miRNAs from RNase in various body fluids. It may be associated with the cell type and tissue specificity, which carrier miRNAs “select” and the carrier of one specific miRNA may be not unique (Arroyo et al., 2011; Li et al., 2012).
Traditional cell–cell communication means gap junctions or cell signal transduction, such as neurotransmitter, hormone, and cytokines, whereas the extracellular miRNA-dependent cell-cell communication is proven to be induced by membrane-derived vesicles recently (Xu et al., 2013). Exosomes were demonstrated to be related to the immune function many years ago, and researchers were surprised to confirm that both mRNA and miRNAs could be packaged to the “magical” vesicles and are exported or released from cells in response to biological stimuli (Cortez et al., 2011). The lipid bilayer is the coat of exosome that reflects the information from the original cell, and the proteins on the surface are important in cell signal pathway (Zhao et al., 2015). Respiratory chain inhibitor rotenone was able to reduce the level of extracellular miRNAs (Wang et al., 2010), and the mRNA and miRNA profiles of exosomes are different from those of the parent cells (Valadi et al., 2007), suggesting that the secretion of exosomes is an ATP-dependent and active-selecting process. On the other hand, the difference between the spectra of intracellular and extracellular miRNAs under the condition of no cytolysis indicated that the cellular control mechanism was involved in the release of extracellular miRNAs(20). In in vitro experiments, many miRNAs were overexpressed in the culture medium after the intervention of serum starvation and proved to be related to the cell cycle arrest, apoptosis, and cell death, suggesting that the biological function of miRNAs may extend outside of the cell and mediate cell–cell communication (Wang et al., 2010). A few selected miRNAs showed a trend of translating from intracellular to extracellular during the first 2 h of serum elimination, which suggested that there is a process of prepackage and storage of miRNAs (Wang et al., 2010). Those encapsulated miRNAs are able to reach the remote area and affect recipient cells, especially various immune cells in the tumor microenvironment, which is important for tumorigenesis (Gong et al., 2012; Lässer, 2012). miRNAs could participate in a directional transfer from T cells to antigen-presenting cells using exosomes as a carrier. Not surprisingly, miRNAs in exosomes are induced by immune cells different from their parent cells, indicating that miRNAs were changed in the process of intercellular communication during immune interactions (Mittelbrunn et al., 2011). Other immune cells, such as dendritic cells (DCs), were proven to transfer signals to neighboring DCs via exosome shuttle miRNAs, and the packed miRNAs were different according to the maturation of the DCs (Montecalvo et al., 2012). That is to say, exosomal miRNAs are one of the complex strategies of immune cell communication. Although the exact underlying mechanism of exosomal miRNAs in cell communication is still unclear now, we have obtained evidence of several hypotheses (Figure 1): 1) direct fusion of vesicles and receptor cell membranes (Mulcahy et al., 2014), 2) active endocytosis or phagocytosis by receptor cells, 3) identification of proteins on the surface of exosomes and receptors of recipient cells (Munich et al., 2012; Corrado et al., 2014), 4) cell gap junction-mediated transfer (Lim et al., 2011; Aucher et al., 2013). The communication between circulating miRNAs and target cells will lead to a series of effects on both physiological and pathological conditions (Kosaka et al., 2010; Pegtel and Kieff, 2010) and the packed miRNAs make an exchange of genetic material additionally (Valadi et al., 2007). Some miRNAs exist in exosomes not derived from their parental cell. These have confirmed that the gene transfer is mediated by exosomes (Valadi et al., 2007; Zhang et al., 2015a). The extracellular miRNAs not only affect the surrounding cells but also the distant tissues, thereby leading to the progression of diseases (Mathivanan et al., 2012).
Adipose tissue contains a different type of circulating exosomal miRNA that can regulate distant cells as a form of adipokine, which is confirmed by the experiment that miRNAs expressed in the brown adipose in one mouse could regulate its target liver reporter in other mouse (Thomou et al., 2017). Additionally, accumulating evidence has shown that circulating miRNAs participate in the invasion and metastasis of cancer via cell communication with recipient cells (Mei et al., 2011). Secreted miRNAs from metastatic cells were transported to endothelial cells and promoted angiogenesis, which are regulated by neutral sphingomyelinase 2 (Kosaka et al., 2013a). All of these studies indicate that (Lee et al., 1993) sending information via extracellular miRNA is another way of intercellular communication (Wightman et al., 1993), miRNA-dependent cell–cell communication is the best explanation for the existence of circulating miRNA, and (Bartel, 2004) circulating miRNAs secreted by cancer cells can trigger tumorigenesis in the recipient cells.
miRNAs are a family of endogenous 19-22nt noncoding RNAs involved in posttranscriptional regulation and participate in various physiological and pathological processes by inhibition of translation or by mRNA cleavage. More than 50% of protein-coding genes are assumed to be the target of miRNAs (Krol et al., 2010). The miRNA expression is frequently dysregulated in cancer, forming tissue-specific expression patterns. In addition to their intracellular biology functions, numerous studies have documented that the dysregulation of extracellular miRNAs is associated with the origin, progression, therapeutic response, and patient survival of the disease since the presence of circulating miRNAs in serum was first described (Wang et al., 2018; Zhang et al., 2018). Mouse prostate cancer xenograft model was used to identify whether the abnormal expressed serum miRNA was tumor-derived in in vivo experiment. Results showed that the levels of miR-620 and miR-629 in the serum were different and can distinguish cancer-bearing mice from controls (Mitchell et al., 2008). There was a surprisingly distinct miRNA expression between endothelial cells cultured with cancer cell lines with control, which means the upregulated miRNAs were induced by tumor cells (Zhuang et al., 2013). At the same time, this study confirmed the hypothesis that stimulated miRNAs were packed into microvesicles and delivered to endothelial cells in follow-up experiments (Skog et al., 2008; Zhuang et al., 2013). On the other hand, the phenomenon that tumor-suppressing miRNAs were amplified, whereas oncogenic miRNAs were reduced, reflected the fact that circulating miRNAs were not the primary products of cancer cells, but the results of global immune response and play an important role in cancer defense and cancer therapy (Aucher et al., 2013). However, another phenomenon is that cancer cells transfer the intercellular tumor-suppressive miRNAs to the extracellular environment, modify tumor microenvironment, and support cancer progression. In other words, extracellular miRNAs can act as both oncomir and suppressor by different stimuli. These studies provided the evidence that miRNAs may derive from tumor cells in response to specific signals, enter the circulation in a stable form as cancer-related molecules, and contribute to early diagnosis, prognosis, and individualizing therapeutic strategies (Table 1 (Gonzales et al., 2011; Silva et al., 2011; Zhou et al., 2011; Bryant et al., 2012; Sun et al., 2012; Valladares-Ayerbes et al., 2012; Kawaguchi et al., 2013; Ng et al., 2013; Nguyen et al., 2013; Takeshita et al., 2013; Tanaka et al., 2013; Wang et al., 2013; Zeng et al., 2013; Enache et al., 2014; Geng et al., 2014; Ogata-Kawata et al., 2014; Zanutto et al., 2014; Zhang et al., 2014; Chen et al., 2014a; Wang et al., 2014; Antolin et al., 2015; Stuckrath et al., 2015; Zhang et al., 2015b; Zhang et al., 2016; Mirzaei H. et al., 2016; Mirzaei H. R. et al., 2016; Zhang et al., 2017; Zhao et al., 2017; Bahrami et al., 2018; Jamali et al., 2018; Shekari et al., 2018; Wang et al., 2018).
The diagnosis of cancer currently suffers from low sensitivity, because many tumors cannot be found at the early stage and delay the treatment until it is too late. MiRNA expression is frequently dysregulated in cancer, forming a particular expression profile and, thus, benefits the early detection of cancer. MiRNAs associated with tumor growth are highly expressed, whereas suppressors are expressed lower. Therefore, these tissue-specific miRNAs are becoming emerging candidates in cancer diagnosis. Selected plasma/serum circulating miRNAs could be used to discriminate various cancer patients from healthy individuals such as breast (Antolin et al., 2015), colorectal (Zanutto et al., 2014), gastric (Zhang et al., 2015b), lung (Zhao et al., 2017), pancreatic (Kawaguchi et al., 2013), and hepatocellular (Mirzaei H. R. et al., 2016) cancers, making them tools for earlier diagnosis. In addition, differential concentrations of miRNAs were detected among different cancer subtypes and differentiation grades in breast cancer; aberrant levels of miRNAs were associated with the HER2 and estrogen receptor status as well, indicating the diagnostic and therapy-selecting potential of circulating miRNAs (Stuckrath et al., 2015). The expression of miR-21 was associated with the clinical stage and molecular subgroup of diffuse large B-cell lymphoma (DLBCL), which means that patients in early stage have a higher concentration of serum miR-21 than those in stage III and IV and patients with different subgroup have an obvious differentiation (Chen et al., 2014b). On the other hand, miRNAs show a remarkable relationship with tumor derivation, which is important in the identification of metastatic tumors with unknown primary origin. A microarray of 48 selected miRNAs could trace and classify 90% primary tumor in metastatic samples (Rosenfeld et al., 2008). The combination of miR-145 and miR-451 could discriminate breast cancer from healthy individuals as well as other types of cancers, including liver cancer, lung cancer, and colorectal cancer, which validated the function of circulating miRNAs in cancer classification (Ng et al., 2013). Furthermore, not only cancers could be distinguished from normal ones but also those who suffered from chronic inflammation were singled out by a distinct miRNA expression pattern. Recent studies suggest that circulating miRNAs are involved in the regulation of inflammation, influence the genetic/epigenetic profile, and capable of predicting the unhealthy incidents (Olivieri et al., 2016). For example, a selected miRNA expression panel could differentiate pancreatic cancer from chronic pancreatitis with relatively high accuracy (Bloomston et al., 2007), whereas it can be used as biomarkers in the identification of hepatitis B virus (HBV) infection and HBV-positive hepatocellular cancer (Li et al., 2010). However, what needs to be pointed out is that the same miRNA can act as either oncogene or suppressor gene, depending on different cancer types (Cortez et al., 2011). MiR-125b could suppress cell proliferation and induce cell cycle arrest in ovarian, thyroid, and oral cancers (Visone et al., 2007; Nam et al., 2008), whereas it functioned oppositely in prostate cancer (Le et al., 2009). As a result, it is important to find out the corresponding abnormally expressed miRNAs in every type of cancers.
On the other hand, the phenomenon illustrated that some of the miRNAs are aberrantly expressed in tumors with an obvious familial aggregation tendency that can be applied to genetic diagnostics. MiR-15 and miR-16 are down-regulated in most of the B-cell chronic lymphocytic leukemia (B-CLL) patients due to the 30-kb region of loss in chromosome 13q14, which is the most frequently deleted genomic region of B-CLL (Calin et al., 2002). Similarly, acute myeloid leukemia patients with chromosomal translocations were proved together with a low level of miR-223 (Fazi et al., 2007). Even single nucleotide polymorphisms (SNPs) in miRNA genes may affect the biogenesis of miRNAs and thus increase the risk of cancer. SNP (rs417309), located in the 3′-UTR of DGCR8, was consistently associated with the possibility of suffering from breast cancer by the mechanism of interrupting the binding of miRNA, whereas an SNP in let-7 complementary sites could increase the risk of non-small cell lung cancer (Chin et al., 2008) as well. These results are also consistent with the studies that genes of miRNAs are most often located at fragile sites and genomic regions associated with cancers (Calin et al., 2004).
A large number of studies have suggested the prognostic and predictive values of cancer-related circulating miRNAs as they participate in the regulation of the development of cancer. In the progression of cancer into a more invasive phenotype, miRNAs change as molecular labels of tumor cells, and the changes can be observed from tumorigenesis throughout the following progression. Therefore, circulating miRNAs are one of the most reliable candidates in disease monitoring. Circulating miR-142-3p correlated with a high risk of recurrence in lung adenocarcinoma patients of early stage (Kaduthanam et al., 2013). The levels of serum miR-155 could reflect the effect of surgery and chemotherapy in breast cancer, whereas the conventional biomarkers, such as carcinoembryonic antigen (CEA) and tissue polypeptide-specific antigen (TPS), were not that sensitive (Sun et al., 2012). Altered circulating miRNAs have also been proven to be bound up with the metastasis of cancer, and miR-141 achieved positive results in a test in prostate cancer patients in terms of identification of micro-metastasis (Gonzales et al., 2011). Decreasing levels of cir-miRNA-126 were related to treatment benefit in metastatic colorectal cancer, as it was proven to be associated with angiogenesis by way of paracrine (Hansen et al., 2015). Similarly, higher levels of circulating miR-122 have a positive correlation with the metastatic recurrence in stage II–III breast cancer patients (Wu et al., 2012). MiR-375 and miR-200b in the serum were significantly upregulated in patients with metastatic prostate cancer compared with patients with localized cancer (Bryant et al., 2012). Some of the other miRNAs that influence the epithelial phenotype of cancer cells were found elevated in the blood of gastric patients and induce invasion and migration (Valladares-Ayerbes et al., 2012). Additionally, circulating miR-214 and miR-373 were related to lymph node metastasis as well (Chen et al., 2013). Responsive miRNAs were observed valuable in therapy monitoring in head and neck squamous cell carcinoma (Summerer et al., 2013).
On the other hand, the changes of circulating miRNAs during chemotherapy and radiotherapy of cancer are well appreciated in many studies. Non-small-cell lung cancer (NSCLC) patients with clinical stage Ib to IIIa often need comprehensive treatment including operation and chemotherapy; a prediction for drug and chemotherapy sensitivity in advance can reduce the unnecessary toxic chemotherapy. A selected serum miRNA panel may serve as a predictor for the purpose of the above and found to be associated with the overall survival of NSCLC (Hu et al., 2010). Serum miR-125 and miR-22 led to the poor response to cisplatin-based and pemetrexed-based chemotherapy separately in NSCLC patients (Cui et al., 2013; Franchina et al., 2014). Serum miR-21 was associated with the relapse-free survival in DLBCL (Lawrie et al., 2008). MiR-150 was sensitive to acute radiation exposure and thus useful for the evaluation of treatment and toxic dose, which is essential for clinical radiation therapy (Jacob et al., 2013). All these studies suggested that circulating miRNAs are promising invasive biomarkers and are considered to be valuable in tumor classification, treatment strategy selection, cancer prognostication, and monitoring.
Currently, more and more studies focus on the biological behavior of circulating miRNAs. It was found that not only mRNAs and proteins that are packed into the MVs or exosomes, but also miRNAs are proved to be existing in the MVs and exosomes abundantly. These exosomes secreted by donor cells containing packed miRNAs could be taken by recipient cells, both in the surrounding and remote area, which is the theoretical basis of miRNA-based targeted cancer therapy via vesicles. However, the delivery method of miRNA is an essential problem to resolve for RNAi therapy. Compared with carriers of targeted therapy, such as viruses, lipid, and polymeric nanoparticles, microvesicles serve as a natural carrier and could avoid attack from the immune system (Cheng, 2015), which is quite important in the persistence of drug intervention. Therefore, MVs and exosomes are used for the delivery of therapeutic RNAi as a more effective strategy in cancer therapy (Kosaka et al., 2013b). In other words, the transfer of synthesis tumor-suppressive miRNAs or antisense of tumor oncogenesis miRNAs into target tumor cells through MVs or exosomes deserves continuing concern. MiR-150 is an immune-related miRNA and participates in the secreting of vascular endothelial growth factor via the regulation of tumor-associated macrophages and plays positive role in tumor growth. Experts transfer antisense miR-150 to MVs and inject “modified” MVs to mice via tail. Results showed that MVs could deliver the antagonucleotide for onco-mirna into tumor efficiently, and thus prevent tumorigenesis (Liu et al., 2013). Using GE11, a kind of peptide that can bind to the epidermal growth factor receptor, miR-let-7a reached the breast cancer tissue of xenograft mice model specifically and inhibited tumor development in vivo successfully (Ohno et al., 2013). MVs derived from human adult liver stem cells containing several anti-miRNAs could inhibit the growth of the tumor (Valentina et al., 2012). Furthermore, anti-mRNAs were also effective in drug resistance. For example, miR-9 was associated with the drug efflux transporter and was found upregulated in temozolomide-resistant glioblastoma multiforme (GBM) cells, whereas mesenchymal stem cells (MSCs)-derived exosomes complete the key biological processes of transferring anti-miR-9 from MSCs to GBM cells and reverse the chemoresistance finally (Munoz et al., 2013). Taken together, tumor-suppressive miRNAs could be delivered to target cancer cells in vivo and can be promising small RNAs for cancer therapy. However, the decomposition by the reticuloendothelial system and the inappropriate immune responses are the major issues of miRNA-relevant cancer therapy before their application.
As we discussed above, circulating miRNAs are becoming potential non-invasive biomarkers for the prediction, prognosis, and therapeutic targets for cancers. Despite their many advantages, there are still challenges to overcome before clinical application.
The fundamental technical constraint to solve is the isolation and purification of samples, as the integrity and purity of RNA are the basic of detection and quantification. Unlike intercellular miRNAs, circulating miRNAs are interference by other components in serum easily and need to be cautious when centrifuged from serum (Cheng et al., 2013). It is necessary to add a step for purification as cell-free miRNAs are modified with exosomes, microvesicles, AGO2/NPM1, and HDL (Lee et al., 1993). Besides, the storage time and conditions also impact the composition of miRNAs; the level of several miRNAs, including miR16, is changed after 24 to 72 h storage either in the situation of 4°C or −20°C (McDonald et al., 2011) and brings another challenge in sample processing. That is to say, different experimental setups and processes all lead to the bias in the final output of miRNAs (Cortez et al., 2011; Schwarzenbach et al., 2014; Lee et al., 2016). Validated and optimized experimental protocols are needed urgently.
Second, the source of samples is also one of the most critical aspects of the ultimate results of circulating miRNAs (Wang et al., 2012). The expression of miRNAs is different between the samples extracted from the serum and plasma even in the same individual. The total RNA concentration is higher in the serum than in the plasma, which may be due to the RNA released from blood cells and platelets (Wang et al., 2012). However, analysis results from studies did not distinguish sample types (serum/plasma) when grouping.
Third, it is still hard to measure circulating miRNAs accurately because of its low concentration and existing form. The major quantifiability detection of circulating miRNAs are qPCR, microarray, and next-generation sequencing (NGS). Quantitative PCR is limited by low throughputs. It is now widely used for the verification of sequencing data. Microarray is influenced by the short length and similar sequence among clusters and families of mature miRNAs. Its requirement of pre-amplification step has a risk of alliterating the actual concentration of circulating miRNAs (Chen et al., 2009). NSG can meet the low concentration of circulating miRNAs due to its low input request and become a preferred method because of its lower cost and higher throughputs. Therefore, it is indispensable to unify the measurement methods and eliminate the deviation.
Additional obstacle lies in the normalization of data, especially the selection of internal control. U6 is widely used for intracellular miRNAs, but it is restricted due to its low expression in body fluids (Singh et al., 2016). Mir-16 was mentioned in many studies, and the inconsistent results also appear in multiple myeloma (Wang and Chen, 2014). Other internal controls, such as UNR6B (Xiang et al., 2014) and miR39 (Wulfken et al., 2011), are not trustworthy yet. The additive artificial non-human miRNAs external control like cel-miR-39 and cel-miR-54 (Zhong et al., 2018) could be a choice to solve this problem. However, it is hard to balance the amount among different samples. On the other hand, because of the expression of extracellular miRNAs in healthy individuals and the variability in acute/chronic inflammation/injury, the identification of a range of negative and cancer-specific “diagnostic” miRNAs is necessary before miRNAs can become clinical test indicators (Zhao et al., 2010; Xiang et al., 2014).
In addition to the technical challenges, the unclear understanding of the function and biology characteristics of circulating miRNAs, such as the secretion and transportation mechanisms, the cell to cell communication, the complicated network between miRNAs and coding gene, and the effects in upstream/downstream pathways are great barriers before clinical transformation (Wang and Chen, 2014). In the past few years, many researchers have devoted to the precise mechanisms of the secretion and uptake of miRNAs. However, it is still not clear if the package of miRNAs is random or specific; the release of cell-free miRNAs is passive or active. Besides, molecules and signals that are involved in the regulation of miRNAs, the precise role of circulating miRNAs in oncogenesis, the great heterogeneity of miRNAs in each type of cancer, different tumor stages, treatment response, and survival are all tasks that require more investigations.
At the same time, there are still challenges in the usage of circulating miRNAs in targeted therapy. Packaged artificial and modified miRNAs in exosomes could increase the stability of miRNAs in vivo (Cortez et al., 2011). However, the restricted tissue specificity and permeability is a big problem. Ligand, antibody, and nanoparticles that carried miRNAs are designed nowadays with improved specificity and decreased immunotoxicity (Chen et al., 2015). Nevertheless, a large number of preclinical studies in animals should be considered to verify their effectiveness.
Since the first discovery of circulating miRNAs, there is a large amount of studies focused on their biological functions and the potential of biomarkers in oncology. As described here, miRNAs in the circulation change as molecular labels of tumor cells throughout the tumorigenesis and development of cancer. Such detectable changes make circulating miRNAs promising non-invasive biomarkers for early cancer diagnosis and predictor of prognosis and cancer treatment. However, several issues including technical and non-technical constraint need to be solved urgently. The further understanding of existing form in circulation and biological function, the deeper exploration of the underlying mechanism of release, transport, and uptake, and the special status in cell communication are all essential before the breakthrough in the application of circulating miRNA-based cancer therapy.
RC and XZ conceived the idea. MC wrote the manuscript. HW and XY revised the manuscript. DZ and YX edited the manuscript.
This work was supported by Program for JLU Science and Technology Innovative Research Team.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alečković, M., Kang, Y. (2015). Regulation of cancer metastasis by cell-free miRNAs. BBA – Rev. Cancer 1855 (1), 24–42. doi: 10.1016/j.bbcan.2014.10.005
Antolin, S., Calvo, L., Blanco-Calvo, M., Santiago, M. P., Lorenzo-Patino, M. J., Haz-Conde, M., et al. (2015). Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer 15, 297. doi: 10.1186/s12885-015-1238-5
Arroyo, J. D., Chevillet, J. R., Kroh, E. M., Ruf, I. K., Prichard, C. C., Gibson, D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 108 (12), 5003–5008. doi: 10.1073/pnas.1019055108
Aucher, A., Rudnicka, D., Davis, D. M. (2013). MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J. Immunol. 191 (12), 6250–6260. doi: 10.4049/jimmunol.1301728
Bahrami, A., Aledavood, A., Anvari, K., Hassanian, S. M., (2018). The prognostic and therapeutic application of microRNAs in breast cancer: tissue and circulating microRNAs. J. Cell. Physiol. 233, 2, 774–786. doi: 10.1002/jcp.25813
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bloomston, M., Frankel, W. L., Petrocca, F., Volinia, S., Alder, H., Hagan, J. P., et al. (2007). MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297 (17), 1901–1908. doi: 10.1001/jama.297.17.1901
Bryant, R. J., Pawlowski, T., Catto, J. W., Marsden, G., Vessella, R. L., Rhees, B., et al. (2012). Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 106 (4), 768–774. doi: 10.1038/bjc.2011.595
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 99 (24), 15524–15529. doi: 10.1073/pnas.242606799
Calin, G. A., Sevignani, C., Dumitru, C. D., Hyslop, T., Noch, E., Yendamuri, S., et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U. S. A. 101 (9), 2999–3004. doi: 10.1073/pnas.0307323101
Chang, J., Nicolas, E., Marks, D., Sander, C., Lerro, A., Buendia, M. A., et al. (2004). miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1 (2), 106–113. doi: 10.4161/rna.1.2.1066
Chen, Q., Ge, X., Zhang, Y., Xia, H., Yuan, D., Tang, Q., et al. (2014a). Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol. Rep. 31 (4), 1863–1870. doi: 10.3892/or.2014.3004
Chen, W., Cai, F., Zhang, B., Barekati, Z., Zhong, X. Y. (2013). The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumor Biol. 34 (1), 455–462. doi: 10.1007/s13277-012-0570-5
Chen, W., Wang, H., Chen, H., Liu, S., Lu, H., Kong, D., et al. (2014b). Clinical significance and detection of microRNA-21 in serum of patients with diffuse large B-cell lymphoma in Chinese population. Eur. J. Haematol. 92 (5), 407–412. doi: 10.1111/ejh.12263
Chen, Y., Gao, D. Y., Huang, L. (2015). In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 81, 128–141. doi: 10.1016/j.addr.2014.05.009
Chen, Y., Gelfond, J. A., McManus, L. M., Shireman, P. K. (2009). Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 10, 407. doi: 10.1186/1471-2164-10-407
Cheng, G. (2015). Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 81, 75–93. doi: 10.1016/j.addr.2014.09.001
Cheng, H. H., Yi, H. S., Kim, Y., Kroh, E. M., Chien, J. W., Eaton, K. D., et al. (2013). Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One 8 (6), e64795. doi: 10.1371/journal.pone.0064795
Chin, L. J., Ratner, E., Leng, S., Zhai, R., Nallur, S., Babar, I., et al. (2008). A SNP in a let-7 microRNA complementary site in the KRAS 3 ‘ untranslated region increases non-small cell lung cancer risk. Cancer Res. 68 (20), 8535–8540. doi: 10.1158/0008-5472.CAN-08-2129
Corrado, C., Raimondo, S., Saieva, L., Flugy, A. M., De, L. G., Alessandro, R. (2014). Exosome-mediated crosstalk between chronic myelogenous leukemia cells and human bone marrow stromal cells triggers an Interleukin 8-dependent survival of leukemia cells. Cancer Lett. 348 (1-2), 71–76. doi: 10.1016/j.canlet.2014.03.009
Cortez, M. A., Buesoramos, C., Ferdin, J., Lopezberestein, G., Sood, A. K., Calin, G. A. (2011). MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 8 (8), 467–477. doi: 10.1038/nrclinonc.2011.76
Cui, E. H., Li, H. J., Hua, F., Wang, B., Mao, W., Feng, X. R., et al. (2013). Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol. Sin. 34 (2), 309–313. doi: 10.1038/aps.2012.125
Duttagupta, R., Jiang, R., Gollub, J., Getts, R. C., Jones, K. W. (2011). Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One 6 (6), e20769. doi: 10.1371/journal.pone.0020769
Enache, L. S., Enache, E. L., Ramiere, C., Diaz, O., Bancu, L., Sin, A., et al. (2014). Circulating RNA molecules as biomarkers in liver disease. Int. J. Mol. Sci. 15 (10), 17644–17666. doi: 10.3390/ijms151017644
Fazi, F., Racanicchi, S., Zardo, G., Starnes, L. M., Mancini, M., Travaglini, L., et al. (2007). Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 12 (5), 457–466. doi: 10.1016/j.ccr.2007.09.020
Franchina, T., Amodeo, V., Bronte, G., Savio, G., Ricciardi, G. R., Picciotto, M., et al. (2014). Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J. Cell Physiol. 229 (1), 97–99. doi: 10.1002/jcp.24422
Geng, Q., Fan, T., Zhang, B., Wang, W., Xu, Y., Hu, H. (2014). Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir. Res. 15, 149. doi: 10.1186/s12931-014-0149-3
Gong, J., Jaiswal, R., Mathys, J. M., Combes, V., Grau, G. E. R., Bebawy, M. (2012). Microparticles and their emerging role in cancer multidrug resistance. Cancer Treat. Rev. 38 (3), 226–234. doi: 10.1016/j.ctrv.2011.06.005
Gonzales, J. C., Fink, L. M., Goodman, O. B., Jr., Symanowski, J. T., Vogelzang, N. J., Ward, D. C. (2011). Comparison of circulating MicroRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin. Genitourin. Cancer 9 (1), 39–45. doi: 10.1016/j.clgc.2011.05.008
Hansen, T. F., Carlsen, A. L., Heegaard, N. H., Sørensen, F. B., Jakobsen, A. (2015). Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br. J. Cancer 112 (4), 624–629. doi: 10.1038/bjc.2014.652
Heijnen, H. F. G., Schiel, A. E., Fijnheer, R., Geuze, H. J., Sixma, J. J. (1999). Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and α-granules. Blood 94 (11), 3791.
Hu, Z., Chen, X., Zhao, Y., Tian, T., Jin, G., Shu, Y., et al. (2010). Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28 (10), 1721–1726. doi: 10.1200/JCO.2009.24.9342
Hunter, M. P., Ismail, N., Zhang, X., Aguda, B. D., Lee, E. J., Yu, L., et al. (2010). Correction: detection of microrna expression in human peripheral blood microvesicles. PLoS One 5 (3), e3694. doi: 10.1371/annotation/b15ca816-7b62-4474-a568-6b60b8959742
Husted, S., Søkilde, R., Rask, L., Cirera, S., Busk, P. K., Eriksen, J., et al. (2011). MicroRNA expression profiles associated with development of drug resistance in Ehrlich ascites tumor cells. Mol. Pharm. 8 (6), 2055–2062. doi: 10.1021/mp200255d
Jacob, N. K., Cooley, J. V., Yee, T. N., Jacob, J., Alder, H., Wickramasinghe, P., et al. (2013). Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS One 8 (2), e57603. doi: 10.1371/journal.pone.0057603
Jamali, L., Tofigh, R., Tutunchi, S., Panahi, G., Borhani, F., Akhavan, S., et al., (2018). Circulating microRNAs as diagnostic and therapeutic biomarkers in gastric and esophageal cancers. J. Cell. Physiol. 233 (11), 8538–8550. doi: 10.1002/jcp.26850
Kaduthanam, S., Gade, S., Meister, M., Brase, J. C., Johannes, M., Dienemann, H., et al. (2013). Serum miR-142-3p is associated with early relapse in operable lung adenocarcinoma patients. Lung Cancer 80 (2), 223–227. doi: 10.1016/j.lungcan.2013.01.013
Kawaguchi, T., Komatsu, S., Ichikawa, D., Morimura, R., Tsujiura, M., Konishi, H., et al. (2013). Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br. J. Cancer 108 (2), 361–369. doi: 10.1038/bjc.2012.546
Kibel, A. S. editor. (2009). Commentary on Circulating microRNAs as stable blood-based markers for cancer detection. Urol. Oncol.-Semin. Ori. 27 (4), 461. doi: 10.1016/j.urolonc.2009.04.001
Kim, S. I., Shin, D., Choi, T. H., Lee, J. C., Cheon, G. J., Kim, K. Y., et al. (2007). Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol. Ther. 15 (6), 1145–1152. doi: 10.1038/sj.mt.6300168
Kosaka, N., Iguchi, H., Hagiwara, K., Yoshioka, Y., Takeshita, F., Ochiya, T. (2013a). Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 288 (15), 10849–10859. doi: 10.1074/jbc.M112.446831
Kosaka, N., Iguchi, H., Yoshioka, Y., Takeshita, F., Matsuki, Y., Ochiya, T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. Commun. Integr. Biol. 285 (5), 17442–17452. doi: 10.1074/jbc.M110.107821
Kosaka, N., Takeshita, F., Yoshioka, Y., Hagiwara, K., Katsuda, T., Ono, M., et al. (2013b). Exosomal tumor-suppressive microRNAs as novel cancer therapy: “exocure” is another choice for cancer treatment. Adv. Drug Deliv. Rev. 65 (3), 376–382. doi: 10.1016/j.addr.2012.07.011
Krol, J., Loedige, I., Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11 (9), 597–610. doi: 10.1038/nrg2843
Lässer, C. (2012). Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin. Biol. Ther. 12 Suppl 1 (6), S189–S197. doi: 10.1517/14712598.2012.680018
Lawrie, C. H., Gal, S., Dunlop, H. M., Pushkaran, B., Liggins, A. P., Pulford, K., et al. (2008). Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 141 (5), 672–675. doi: 10.1111/j.1365-2141.2008.07077.x
Le, M. T. N., Teh, C., Shyhchang, N., Xie, H., Zhou, B., Korzh, V., et al. (2009). MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23 (7), 862–876. doi: 10.1101/gad.1767609
Lee, I., Baxter, D., Min, Y. L., Scherler, K., Kai, W. (2016). The importance of standardization on analyzing circulating RNA. Mol. Diagn. Ther. 21 (3), 259–268. doi: 10.1007/s40291-016-0251-y
Lee, R. C., Feinbaum, R. L., Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 (5), 843. doi: 10.1016/0092-8674(93)90529-Y
Li, L. M., Hu, Z. B., Zhou, Z. X., Chen, X., Liu, F. Y., Zhang, J. F., et al. (2010). Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 70 (23), 9798–9807. doi: 10.1158/0008-5472.CAN-10-1001
Li, L., Zhu, D., Lei, H., Jing, Z., Zhen, B., Xi, C., et al. (2012). Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One 7 (10), e46957. doi: 10.1371/journal.pone.0046957
Lim, P. K., Bliss, S. A., Patel, S. A., Taborga, M., Dave, M. A., Gregory, L. A., et al. (2011). Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 71 (5), 1550–1560. doi: 10.1158/0008-5472.CAN-10-2372
Liu, Y., Zhao, L., Li, D., Yin, Y., Zhang, C. Y., Li, J., et al. (2013). Microvesicle-delivery miR-150 promotes tumorigenesis by up-regulating VEGF, and the neutralization of miR-150 attenuate tumor development. Protein Cell 4 (12), 932–941. doi: 10.1007/s13238-013-3092-z
Maggi, L. B., Kuchenruether, M., Dadey, D. Y. A., Schwope, R. M., Grisendi, S., Townsend, R. R., et al. (2008). Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell. Biol. 28 (23), 7050–7065. doi: 10.1128/MCB.01548-07
Mathivanan, S., Fahner, C. J., Reid, G. E., Simpson, R. J. (2012). ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40 (Database issue), D1241. doi: 10.1093/nar/gkr828
McDonald, J. S., Milosevic, D., Reddi, H. V., Grebe, S. K., Algeciras-Schimnich, A. (2011). Analysis of circulating microRNA: preanalytical and analytical challenges. Clin. Chem. 57 (6), 833–840. doi: 10.1373/clinchem.2010.157198
Mei, Y., Chen, J., Fang, S., Yu, B., Su, F., Ling, L., et al. (2011). Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 10 (1), 117. doi: 10.1186/1476-4598-10-117
Mirzaei, H. R., Sahebkar, A., Mohammadi, M., Yari, R., Salehi, H., Jafari, M. H., et al. (2016). Circulating microRNAs in hepatocellular carcinoma: potential diagnostic and prognostic biomarkers. Curr. Pharm. Des. 22 (34), 5257–5269. doi: 10.2174/1381612822666160303110838
Mirzaei, H., Khataminfar, S., Mohammadparast, S., Sales, S. S., Maftouh, M., Mohammadi, M., et al. (2016). Circulating microRNAs as potential diagnostic biomarkers and therapeutic targets in gastric cancer: current status and future perspectives. Curr. Med. Chem. 23 (36), 4135–4150. doi: 10.2174/0929867323666160818093854
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosovaagadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 105 (30), 10513–10518. doi: 10.1073/pnas.0804549105
Mittelbrunn, M., Gutiérrez-Vázquez, C., Villarroya-Beltri, C., González, S., Sánchez-Cabo, F., González, MÁ, et al. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2 (1), 282. doi: 10.1038/ncomms1285
Montani, F., Bianchi, F. (2016). Circulating Cancer Biomarkers: the Macro-revolution of the Micro-RNA. Ebiomedicine 5 (C), 4–6. doi: 10.1016/j.ebiom.2016.02.038
Montecalvo, A., Larregina, A. T., Shufesky, W. J., Stolz, D. B., Sullivan, M. L., Karlsson, J. M., et al. (2012). Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119 (3), 756–766. doi: 10.1182/blood-2011-02-338004
Mulcahy, L. A., Pink, R. C., Carter, D. R. F. (2014). Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3 (1), 24641. doi: 10.3402/jev.v3.24641
Munich, S., Sobovujanovic, A., Buchser, W. J., Beerstolz, D., Vujanovic, N. L. (2012). Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology 1 (7), 1074–1083. doi: 10.4161/onci.20897
Munoz, J. L., Bliss, S. A., Greco, S. J., Ramkissoon, S. H., Ligon, K. L., Rameshwar, P. (2013). Delivery of functional anti-miR-9 by mesenchymal stem cell–derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol. Ther. Nucleic Acids 2 (10), e126. doi: 10.1038/mtna.2013.60
Nam, E. J., Yoon, H., Sang, W. K., Kim, H., Kim, Y. T., Kim, J. H., et al. (2008). MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 14 (9), 2690–2695. doi: 10.1158/1078-0432.CCR-07-1731
Ng, E. K., Li, R., Shin, V. Y., Jin, H. C., Leung, C. P., Ma, E. S., et al. (2013). Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 8 (1), e53141. doi: 10.1371/journal.pone.0053141
Nguyen, H. C., Xie, W., Yang, M., Hsieh, C. L., Drouin, S., Lee, G. S., et al. (2013). Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate 73 (4), 346–354. doi: 10.1002/pros.22572
Ogata-Kawata, H., Izumiya, M., Kurioka, D., Honma, Y., Yamada, Y., Furuta, K., et al. (2014). Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9 (4), e92921. doi: 10.1371/journal.pone.0092921
Ohno, S. I., Takanashi, M., Sudo, K., Ueda, S., Ishikawa, A., Matsuyama, N., et al. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor MicroRNA to breast cancer cells. Mol. Ther. 21 (1), 185–191. doi: 10.1038/mt.2012.180
Olivieri, F., Capri, M., Bonafè, M., Morsiani, C., Jung, H. J., Spazzafumo, L., et al. (2016). Circulating miRNAs and miRNA shuttles as biomarkers: perspective trajectories of healthy and unhealthy aging. Mech. Ageing Dev. 165 (Pt B), 162–170. doi: 10.1016/j.mad.2016.12.004
Pegtel, D. M., Kieff, E. (2010). Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U. S. A. 107 (14), 6328–6333. doi: 10.1073/pnas.0914843107
Rosenfeld, N., Aharonov, R., Meiri, E., Rosenwald, S., Spector, Y., Zepeniuk, M., et al. (2008). MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 26 (4), 462–469. doi: 10.1038/nbt1392
Rothblat, G. H., Phillips, M. C. (2010). High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr. Opin. Lipidol. 21 (3), 229–238. doi: 10.1097/MOL.0b013e328338472d
Schwarzenbach, H., Nishida, N., Calin, G. A., Pantel, K. (2014). Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 11 (3), 145. doi: 10.1038/nrclinonc.2014.5
Shekari, N., Baradaran, B., Shanehbandi, D., Kazemi, T. (2018). Circulating microRNAs: valuable biomarkers for the diagnosis and prognosis of gastric cancer. Curr. Med. Chem. 25 (6), 698–714. doi: 10.2174/0929867324666171003123425
Silva, J., Garcia, V., Zaballos, A., Provencio, M., Lombardia, L., Almonacid, L., et al. (2011). Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 37 (3), 617–623. doi: 10.1183/09031936.00029610
Singh, R., Ramasubramanian, B., Kanji, S., Chakraborty, A. R., Haque, S. J., Chakravarti, A. (2016). Circulating microRNAs in cancer: hope or hype? Cancer Lett. 381 (1), 113–121. doi: 10.1016/j.canlet.2016.07.002
Skog, J., Wurdinger, T., van Rijn, S., Meijer, D. H., Gainche, L., Sena-Esteves, M., et al. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell. Biol. 10 (12), 1470–1476. doi: 10.1038/ncb1800
Stuckrath, I., Rack, B., Janni, W., Jager, B., Pantel, K., Schwarzenbach, H. (2015). Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget 6 (15), 13387–13401. doi: 10.18632/oncotarget.3874
Summerer, I., Niyazi, M., Unger, K., Pitea, A., Zangen, V., Hess, J., et al. (2013). Changes in circulating microRNAs after radiochemotherapy in head and neck cancer patients. Radiat. Oncol. 8 (1), 296. doi: 10.1186/1748-717X-8-296
Sun, Y., Wang, M., Lin, G., Sun, S., Li, X., Qi, J., et al. (2012). Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One 7 (10), e47003. doi: 10.1371/journal.pone.0047003
Takeshita, N., Hoshino, I., Mori, M., Akutsu, Y., Hanari, N., Yoneyama, Y., et al. (2013). Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br. J. Cancer 108 (3), 644–652. doi: 10.1038/bjc.2013.8
Tanaka, K., Miyata, H., Yamasaki, M., Sugimura, K., Takahashi, T., Kurokawa, Y., et al. (2013). Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann. Surg. Oncol. 20 Suppl 3, S607–S615. doi: 10.1245/s10434-013-3093-4
Taylor, D. D., Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110 (1), 13–21. doi: 10.1016/j.ygyno.2008.04.033
Thomou, T., Mori, M. A., Dreyfuss, J. M., Konishi, M., Sakaguchi, M., Wolfrum, C., et al. (2017). Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542 (7642), 450–455. doi: 10.1038/nature21365
Tsui, N. B., Ng, E. K., Lo, Y. M. (2002). Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin. Chem. 48 (10), 1647–1653.
Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J. J., Lotvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 9 (6), 654–659. doi: 10.1038/ncb1596
Valentina, F., Federica, C., Beatriz, H. M., Claudia, C., Chiara, D. M., Barbara, C., et al. (2012). Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells 30 (9), 1985–1998. doi: 10.1002/stem.1161
Valladares-Ayerbes, M., Reboredo, M., Medina-Villaamil, V., Iglesias-Diaz, P., Lorenzo-Patino, M. J., Haz, M., et al. (2012). Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J. Transl. Med. 10, 186. doi: 10.1186/1479-5876-10-186
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., Remaley, A. T. (2014). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 17 (1), 423–433. doi: 10.1038/ncb3074
Visone, R., Pallante, P., Vecchione, A., Cirombella, R., Ferracin, M., Ferraro, A., et al. (2007). Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene 26 (54), 7590–7595. doi: 10.1038/sj.onc.1210564
Wang, H., Peng, R., Wang, J., Qin, Z., Xue, L. (2018). Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics 10 (1), 59. doi: 10.1186/s13148-018-0492-1
Wang, J., Huang, S. K., Zhao, M., Yang, M., Zhong, J. L., Gu, Y. Y., et al. (2014). Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One 9 (4), e87451. doi: 10.1371/journal.pone.0087451
Wang, K., Yuan, Y., Cho, J. H., McClarty, S., Baxter, D., Galas, D. J. (2012). Comparing the MicroRNA spectrum between serum and plasma. PLoS One 7 (7), e41561. doi: 10.1371/journal.pone.0041561
Wang, K., Zhang, S., Weber, J., Baxter, D., Galas, D. J. (2010). Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 38 (20), 7248–7259. doi: 10.1093/nar/gkq601
Wang, P., Zhuang, L., Zhang, J., Fan, J., Luo, J., Chen, H., et al. (2013). The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol. Oncol. 7 (3), 334–345. doi: 10.1016/j.molonc.2012.10.011
Wang, W. T., Chen, Y. Q. (2014). Circulating miRNAs in cancer: from detection to therapy. J. Hematol. Oncol. 7, 86. doi: 10.1186/s13045-014-0086-0
Weber, J. A., Baxter, D. H., Zhang, S., Huang, D. Y., Huang, K. H., Ming, J. L., et al. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56 (11), 1733. doi: 10.1373/clinchem.2010.147405
Wightman, B., Ha, I., Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 (5), 855–862. doi: 10.1016/0092-8674(93)90530-4
Wu, X., Somlo, G., Yu, Y., Palomares, M. R., Li, A. X., Zhou, W., et al. (2012). De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J. Transl. Med. 10 (1), 42. doi: 10.1186/1479-5876-10-42
Wulfken, L. M., Moritz, R., Ohlmann, C., Holdenrieder, S., Jung, V., Becker, F., et al. (2011). MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One 6 (9), e25787. doi: 10.1371/journal.pone.0025787
Xiang, M., Zeng, Y., Yang, R., Xu, H., Chen, Z., Zhong, J., et al. (2014). U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem. Biophys. Res. Commun. 454 (1), 210–214. doi: 10.1016/j.bbrc.2014.10.064
Xu, L., Yang, B. F., Ai, J. (2013). MicroRNA transport: a new way in cell communication. J. Cell. Physiol. 228 (8), 1713–1719. doi: 10.1002/jcp.24344
Yao, B., Lan, B. L., Chen, Y. C., Chang, L. J., Chan, E. K. L. (2012). Defining a new role of GW182 in maintaining miRNA stability. Embo Rep. 13 (12), 1102–1108. doi: 10.1038/embor.2012.160
Zanutto, S., Pizzamiglio, S., Ghilotti, M., Bertan, C., Ravagnani, F., Perrone, F., et al. (2014). Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br. J. Cancer 110 (4), 1001–1007. doi: 10.1038/bjc.2013.819
Zeng, R. C., Zhang, W., Yan, X. Q., Ye, Z. Q., Chen, E. D., Huang, D. P., et al. (2013). Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med. Oncol. 30 (1), 477. doi: 10.1007/s12032-013-0477-z
Zhang, H., Mao, F., Shen, T., Luo, Q., Ding, Z., Qian, L., et al. (2017). Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol. Lett. 13 (2), 669–676. doi: 10.3892/ol.2016.5462
Zhang, J., Li, S., Li, L., Li, M., Guo, C., Yao, J., et al. (2015a). Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13 (1), 17–24. doi: 10.1016/j.gpb.2015.02.001
Zhang, J., Song, Y., Zhang, C., Zhi, X., Fu, H., Ma, Y., et al. (2015b). Circulating MiR-16-5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics 5 (7), 733–745. doi: 10.7150/thno.10305
Zhang, J., Zhang, K., Bi, M., Jiao, X., Zhang, D., Dong, Q. (2014). Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs 25 (3), 346–352. doi: 10.1097/CAD.0000000000000049
Zhang, K., Wu, X., Wang, J., Lopez, J., Zhou, W., Yang, L., et al. (2016). Circulating miRNA profile in esophageal adenocarcinoma. Am. J. Cancer Res. 6 (11), 2713–2721.
Zhang, L., Zhang, Y., Zhao, Y., Wang, Y., Ding, H., Xue, S., et al. (2018). Circulating miRNAs as biomarkers for early diagnosis of coronary artery disease. Expert Opin. Ther. Pat. 28 (8), 591–601. doi: 10.1080/13543776.2018.1503650
Zhao, H., Shen, J., Medico, L., Wang, D., Ambrosone, C. B., Liu, S. (2010). A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One 5 (10), e13735. doi: 10.1371/journal.pone.0013735
Zhao, L., Liu, W., Xiao, J., Cao, B. (2015). The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 356 (2), 339–346. doi: 10.1016/j.canlet.2014.10.027
Zhao, Y., Song, Y., Yao, L., Song, G., Teng, C. (2017). Circulating microRNAs: promising biomarkers involved in several cancers and other diseases. DNA Cell Biol. 36 (2), 77–94. doi: 10.1089/dna.2016.3426
Zhong, Z., Hou, J., Zhang, Q., Zhong, W., Li, B., Li, C., et al. (2018). Circulating microRNA expression profiling and bioinformatics analysis of dysregulated microRNAs of patients with coronary artery disease. Medicine 97 (27), e11428. doi: 10.1097/md.0000000000011428
Zhou, J., Yu, L., Gao, X., Hu, J., Wang, J., Dai, Z., et al. (2011). Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol. 29 (36), 4781–4788. doi: 10.1200/jco.2011.38.2697
Keywords: miRNAs, circulating, cancer, biomarker, communication, therapy, challenge
Citation: Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R and Zhang X (2019) Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 10:626. doi: 10.3389/fgene.2019.00626
Received: 01 November 2018; Accepted: 17 June 2019;
Published: 18 July 2019.
Edited by:
William Cho, Queen Elizabeth Hospital (QEH), Hong KongReviewed by:
Sumit Arora, University of South Alabama, United StatesCopyright © 2019 Cui, Wang, Yao, Zhang, Xie, Cui, and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranji Cui, Y3VpcmFuamlAamx1LmVkdS5jbg==; Xuewen Zhang, emhhbmcteHVld2VuQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.