- 1Neuroscience and Ophthalmology Research Group, Institute of Inflammation and Ageing, University of Birmingham, Birmingham, United Kingdom
- 2National Institute for Health Research Surgical Reconstruction and Microbiology Research Centre, Queen Elizabeth Hospital Birmingham, Birmingham, United Kingdom
- 3Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana–Champaign, Urbana, IL, United States
Neurodegenerative diseases (NDs) are becoming increasingly prevalent in the world, with an aging population. In the last few decades, due to the devastating nature of these diseases, the research of biomarkers has become crucial to enable adequate treatments and to monitor the progress of disease. Currently, gene mutations, CSF and blood protein markers together with the neuroimaging techniques are the most used diagnostic approaches. However, despite the efforts in the research, conflicting data still exist, highlighting the need to explore new classes of biomarkers, particularly at early stages. Small non-coding RNAs (MicroRNA, Small nuclear RNA, Small nucleolar RNA, tRNA derived small RNA and Piwi-interacting RNA) can be considered a “relatively” new class of molecule that have already proved to be differentially regulated in many NDs, hence they represent a new potential class of biomarkers to be explored. In addition, understanding their involvement in disease development could depict the underlying pathogenesis of particular NDs, so novel treatment methods that act earlier in disease progression can be developed. This review aims to describe the involvement of small non-coding RNAs as biomarkers of NDs and their potential role in future clinical applications.
Introduction
Neurodegenerative diseases (NDs) are classified as a class of disorders affecting the central nervous system and they are characterized by the progressive loss of neuronal tissues. NDs are age-dependent disorders which are increasing internationally, due to the ever increasing elderly population, which is leaving greater numbers of people subjected to the chronic, debilitating nature of these incurable diseases (Heemels, 2016). Currently, the most represented NDs are: Alzheimer’disease (AD) with 5 million people affected in America only, followed by Parkinson’s diseases (PD) with 1 million people; multiple sclerosis (MS) 400,000; Amyotrophoic lateral sclerosis (ALS) 30,000 and Huntington’s disease (HD) with 3,000 incidents (Agrawal and Biswas, 2015).
Some treatments for ND have aimed to reduce the syndrome of NDs; these include L-dopa and deep brain stimulation in PD (Groiss et al., 2009; Nagatsua and Sawadab, 2009). However, very few have aimed to slow or reverse ND development, and those that have been investigated e.g., stem cell therapy (Chung et al., 2002; Rachakonda et al., 2004) highlight the requirement for more research. Late diagnosis leads to strategic treatment being ineffective due to irreversible disease progression (Sheinerman and Umansky, 2013). This has been reported for example, on anti-AD therapies in late-stage clinical trials (including dimebon of Medivation and Pfizer, solanezumab of Eli Lilly and bapineuzumab of Pfizer and Johnson & Johnson). Biomarkers for early diagnosis could prevent or limit disease development through prophylactic or early treatment, which has ignited interest. Currently, the most accurate diagnosis relies on neuropathology, mainly based on autopsy, or in the measurement of cerebrospinal fluid (CSF) proteins, such as tau or Aβ- in AD, which requires invasive procedures. However, blood proteins, such as Aβ1-42 peptide in AD or cytokines for ALS or HD (Agrawal and Biswas, 2015), as well as genetics diagnostics markers such as ApoE isoforms in AD or α-synuclein or Parkin for PD, have also demonstrated potential clinical utility (Agrawal and Biswas, 2015).
Neuroimaging techniques can also help to make the correct diagnosis and monitor the progress of NDs. Magnetic resonance imaging (MRI) is one of the most widely used neuroimaging techniques used for AD (Jack et al., 2011; McKhann et al., 2011) and for dementia with Lewy bodies (DLB) (Ciurleo et al., 2014). Magnetic resonance spectroscopy (MRS) has also showed promise in early diagnosis of PD and traumatic brain injury, measuring metabolic dysfunctions and irreversible neuronal damage (Vagnozzi et al., 2008).
Recently, a new class of circulating RNAs – non-coding RNAs – have been re-evaluated and are being considered as potential biomarkers. After years of the belief that 98% of the genome was “junk” due to its non-coding nature it was realized these genes had biologically functionality. Non-coding genes include introns, pseudogenes, repeat sequences and cis/trans-regulatory elements that function as RNA without translation. Estimations have suggested that 99% of total RNA content is made up of non-coding RNA, with numbers of validated non-coding RNAs (ncRNAs) increasing every year (Palazzo and Lee, 2015).
Currently ncRNAs can be defined by length – small 18–200 nts and long >200nts – or functionality with housekeeping ncRNAs such as ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) or regulatory ncRNAs like microRNAs (miRNAs), small nuclear RNAs (snRNAs), piwi-interacting RNA (piRNAs), tRNA derived small RNAs (tsRNAs) and long non-coding RNAs (lncRNAs) (Dozmorov et al., 2013). Nonetheless, difficulty distinguishing categories persists due to the crossover of properties.
Small non-coding RNAs (sncRNAs) have diverse roles, which in conjunction with other molecules involve gene regulation through either RNA interference, RNA modification or spliceosomal involvement (Table 1). Consequently, during disease progression their expression can alter. MiRNAs are the most studied sncRNA as biomarkers with involvement in various diseases including cancers, aging and neurodegenerative disease (Calin and Croce, 2006; Grasso et al., 2014; Di Pietro et al., 2017). Other sncRNAs have shown promise as biomarkers, with links to neurodegenerative disease (Munoz-Culla et al., 2016). There is the potential for multiple sncRNA biomarkers for neurodegenerative diseases, which if found, could aid diagnosis in a clinical setting while demonstrating the processes underpinning the disease development. In future, this could produce novel therapies to treat neurodegenerative diseases using original methodologies.
In this review, we consider the evolving role of sncRNAs and discuss their involvement in neurodegenerative disease with particular emphasis on their potential as biomarkers.
MicroRNA
MiRNAs are the most studied sncRNA. Their biogenesis commences with the formation of a pri-miRNA made up of two stem-loop structure. A Drosha and DGCR8 complex cleaves the pri-miRNA to form a single stem-loop pre-miRNA. Dicer cleaves the pre-miRNA to create a double stranded miRNA, which is loaded onto Argonaute family of proteins to form the miRISC complex (Figure 1Ai). Accompanied to the miRISC complex, miRNAs regulate gene expression post-transcriptionally through degradation and repression of mRNA sequences by an Argonaute family protein mediated method (Figure 1Aii; O’Brien et al., 2018). A single miRNA can have multiple targets, likewise a target mRNA can be bound to by many different miRNAs, to enable more diverse signaling patterns.
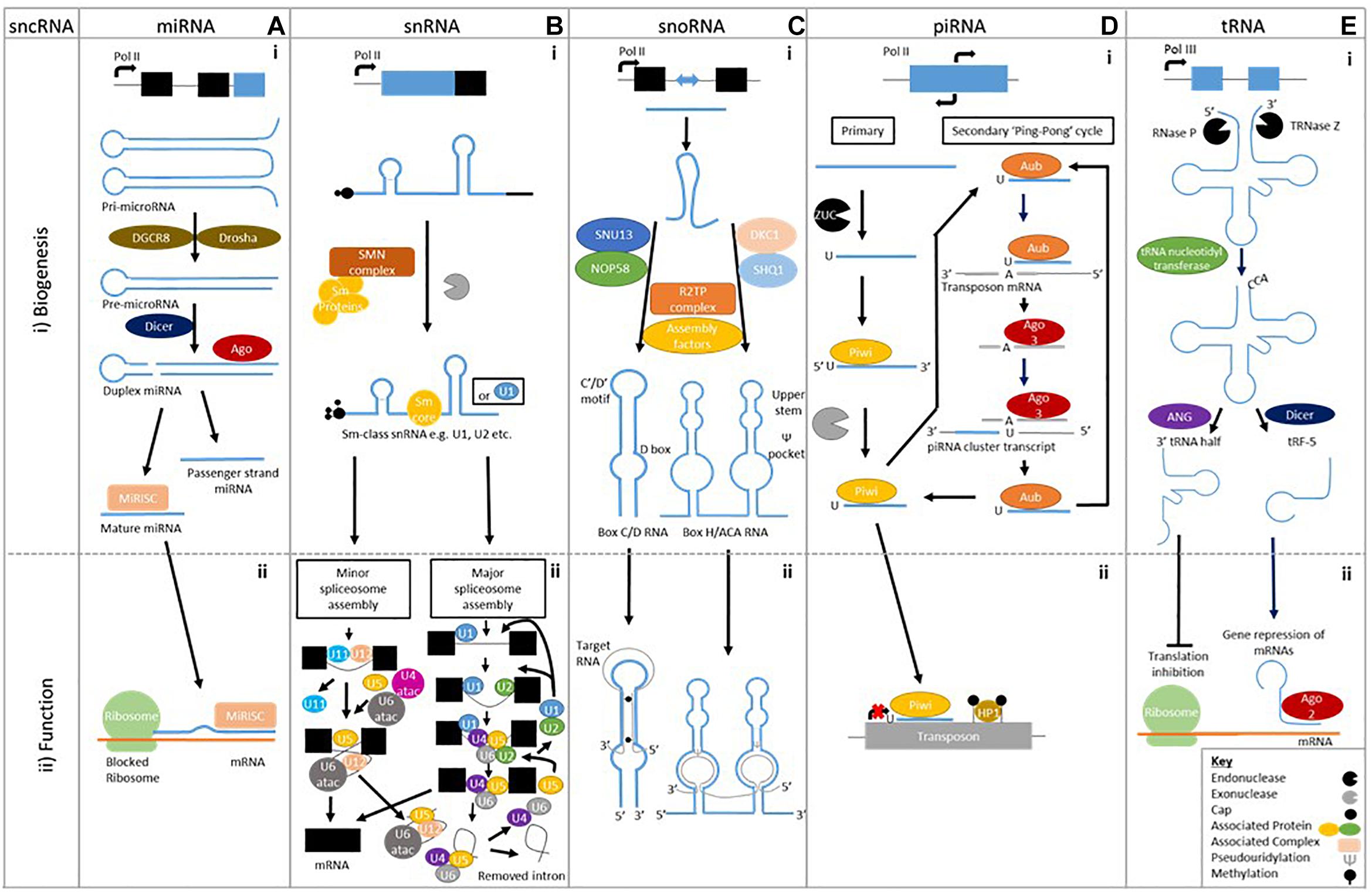
Figure 1. Biogenesis of sncRNAs and an example of their biological function. A (i) MicroRNAs are single stranded ∼22 bp sequences formed from double stranded precursors (ii) that prevent mRNA translation. B (i) Small nuclear RNAs biogenesis is made up of two classes Sm class snRNA and Lsm-class snRNA (Not shown), (ii) which form the major and minor spliceosome. C (i) Small nucleolar RNAs have two different classes formed using different machinery; Box C/D RNA and Box H/ACA RNA, (ii) which cause methylation and pseudouridylation respectively. D (i) Piwi interacting RNAs are formed by either primary alone or by both primary and secondary biogenesis (ii) that prevent transposon translation through methylation. E (i) Transfer RNA cleavage forms transfer RNA derived fragments to be formed, (ii) which can prevent translation or cause gene repression.
MiRNAs show specific signaling in the brain, and were also found differentially expressed in bio-fluids. Although there is no consistent consensus on particular miRNAs or brain area yet, and no specific miRNA overlap between brain tissues and bio-fluids (as reported in Table 2) these findings certainly provide insights in the study of NDs pathogenesis.
MiRNAs are best studied in Alzheimer’s disease (AD), which manifests itself as deposition of neurofibrillary tangles (NFT) and extracellular amyloid-β (Aβ), before neuronal degeneration and clinical symptoms materialize in the form of behavioral changes such as memory issues. NFT, Aβ and neuronal degeneration have been associated with dysregulation of miRNA gene expression, which could emanate from altered Aβ or Tau metabolism. MiRNAs effect Aβ metabolism by interacting with amyloid precursor protein (APP) through direct binding of the 3′untranslated region (3′UTR) to the APP mRNA, indirect inhibition through downregulation of Beta-secretase 1 (BACE1) and ATP-binding cassette transporter (ABCA1) or regulating alternative APP splicing. MiRNAs also affect Tau through regulation of microtubule associated protein tau (MAPT) splicing, affecting tau isoforms 3R and 4R. Direct or indirect binding either modulates phosphorylated Tau-associated protein kinases or influences degradation of phosphorylated tau by binding 3′-UTR BCL2 associated athanogene 2 (BAG2) mRNA (Zhao et al., 2017).
MiRNAs have an established involvement in neurobiological functions and pathogenesis of numerous other neurodegenerative diseases (Serafin et al., 2014; Fransquet and Ryan, 2018; Ricci et al., 2018). Mitochondrial dysfunction caused by miRNA dysregulation leads to oxidative stress, which causes cell death, α-synuclein aggregation and neurodegeneration known to be present in PD (Spano et al., 2015). In ALS, both TAR DNA binding protein (TARDBP) and fused in sarcoma (FUS) are well-established causative genes, which are involved in miRNA processing. TARDBP has specific roles in facilitation of post-transcriptional processing achieved through association directly with miRNA or processing factors such as Dicer (Kawahara and Mieda-Sato, 2012). FUS regulates miRNA-mediated gene silencing through facilitation of the interaction between miRNA, mRNA and RISC components (Zhang et al., 2018). In HD, a miRNA formulation is being trailed as therapeutic agents to alter the aberrant Huntingtin (HTT) protein expression (Aronin and DiFiglia, 2014).
MiRNA involvement in ND development has demonstrated the capability of distinguishing between disease subtypes and shown promise for future stratification. For example in AD, 30 differentially regulated miRNAs found in the brain and blood of AD patients were assigned to different Braak stages, a methodology for classifying AD pathology, with 10 associated with Braak stage III (hsa-mir-107, hsa-mir-26b, hsa-mir-30e, hsa-mir-34a, hsa-mir-485, hsa-mir200c, hsa-mir-210, hsa-mir-146a, hsa-mir-34c, and hsa-mir-125b) (Swarbrick et al., 2019). Likewise in PD, miR-331-5p is differentially expressed in plasma of early onset Parkinson’s disease (EOPD) patients, which was not seen in late onset Parkinson’s disease (LOPD) patients (Cardo et al., 2013; Table 2). Studies comparing between subtypes of NDs are still in the minority and more are required to understand the true capability of miRNA markers in stratification of NDs.
Small Nuclear RNAs
Small nuclear RNAs (snRNAs), the component parts of the spliceosome – responsible for removal of non-coding introns from precursor mRNA – are highly conserved uridine rich sequences with five snRNAs making up its spine; U1, U2, U4, U5, and U6. These snRNAs combine with partner proteins to form the small nuclear ribonucleoprotein (snRNPs) complex, which is essential pre-mRNA splicing to enable production of functional mRNA for protein translation.
Sm-class snRNAs are synthesized by RNA polymerase II and after transcription contain a 7-methylguanosine cap, Sm-protein binding site and 3′ stem-loop. The latter two are recognized by the SMN complex, which recruits a set of Sm proteins to create the Sm-core RNP. Following this, the cap undergoes hypermethylation by trimethylguanosine synthase-1 (TSG1) creating a 2,2,7-trimethylguanosine cap. The 3′ end is then trimmed by an unknown exonuclease before subsequent maturation through modifications (Matera et al., 2007; Figure 1Bi).
Two types of spliceosome “major” and “minor” (0.35% of all introns) can be assembled. Major spliceosome assembly commences by U1 interacting with the 5′ splice site while U2 snRNP binds to the branch point sequence. This leads to the recruitment of the premade U4/U6.U5 tri-snRNP complex, in this state the spliceosome is inactive. After destabilization or release of either U1 or U4, the spliceosome becomes active. The active spliceosome undergoes two phases of catalysis leading to its dissociation – including U2, U5, and U6 that are recycled – when it releases the mRNA, as mRNP (Wahl et al., 2009; Figure 1Bii). The minor spliceosome has divergent and highly conserved 5′ splice site and branch point sequences, which interact with U5 as well as alternative factors U11, U12, and U4atac/U6atac that are functional analog of its major counterpart (Verma et al., 2018; Figure 1Bii). Both spliceosomes show the capability to contribute to the development of neurodegenerative disease, demonstrating snRNA involvement (Bai et al., 2013; Tsuiji et al., 2013; Ratti and Buratti, 2016; Jutzi et al., 2018).
In sporadic and familial AD, U1 snRNP subunits – including U1-70K and U1A – were present in cytoplasmic aggregates, which occurs by the basic-acidic dipeptide (BAD) domain binding to tau in U1-70K (Bishof et al., 2018). Inordinate levels of unspliced RNA also reside, caused by dysregulation of RNA processing. In conjunction with evidence that inhibition of U1 snRNP increases APP, this implicates U1 snRNP dysregulation in the pathogenesis of AD (Bai et al., 2013; Hales et al., 2014a,b). Recent evidence has shown abnormal expression of U1 snRNA can cause premature cleavage of pre-mRNA via polyadenylation (PCPA) at the 3′ poly-A site. This affects splicing and could demonstrate a novel AD causing pathology (Cheng et al., 2017) (Table 3).
U snRNAs are also associated with spinal muscular atrophy (SMA). SMN1 gene dysregulation alters U snRNA levels through its role in U snRNA biosynthesis; nonetheless, the underlying pathology is still unclear (Zhang et al., 2013). Many studies have proposed a reduction in U snRNAs is key to SMA pathology due to their involvement in mRNA processing, with U1 and U11 of particular interest (Gabanella et al., 2007; Zhang et al., 2008). In contrast, U snRNAs can accumulate in the motor neurons of ALS patient spinal cords when compared to control patients, to cause defects showing that U snRNA level can depict disease state, depending of cell type (Tsuiji et al., 2013).
More recently, when considering induced pluripotent stem cell (iPSC) derived motor neurones cultures, a study suggested that an imbalanced ratio of variant U1 to U1 might cause the SMA phenotype rather than an overall reduction in U1 snRNA (Vazquez-Arango et al., 2016). Demonstrating that purely measuring U snRNA level may be an oversimplified measurement and variant U snRNA could indicate the underlying pathophysiology of aberrant spliceosome related neurodegeneration.
Other U snRNAs studied in neurodegenerative disease include U2. A U2 snRNA mutation causes neuron degeneration, through altering pre-mRNA splicing at select splice sites that are associated with alternative pre-mRNA splicing (Jia et al., 2012). In addition, a dipeptide repeat (C90RF72) linked to both ALS and frontotemporal dementia (FTD), interacts and interferes with U2 snRNP. In patient derived cells, this led to mislocalisation but mis-splicing linked to ALS/FTD has yet to be established (Yin et al., 2017).
Mutations found within the gene PRPF4 – which encodes hPrp4 a U4/U6 di-snRNP protein – undertake an important role in the development of retinitis pigmentosa (RP) (Chen et al., 2014). hPrp4 is known to interact with CypH and hPrp3 to regulate the stability of the tri-snRNP, U4/U6.U5. Thus, aberrant splicing could cause RP through direct or indirect mechanisms that have been hypothesized, but not defined.
The minor spliceosome has ND relevance as in ALS, TDP-43 functionality decreases (Colombrita et al., 2012), which reduces the number Gemini of coiled bodies (GEMs). GEMs contribute to U12 snRNA biogenesis, so in spinal motor neurones of ALS patients there was a decrease of U12 snRNA and U11/U12 snRNP, which may disrupts pre-mRNA splicing (Ishihara et al., 2013). Additionally, an ALS mutant (P525L) cannot promote minor intron splicing due to an aberrant FUS gene that routinely binds to U11 snRNP to direct splicing. This leads to mislocalisation of FUS-trapped U11 and U12 snRNAs, which form aggregates in the cytoplasm so incorrect splicing results (Reber et al., 2016). In addition, a cerebral ataxia mutation RNU12 causes minor intron retention in homozygous mutant patients (Elsaid et al., 2017). When combined this demonstrates a likely role for minor intron splicing in motor neurone maintenance.
Small Nucleolar RNAs
Small nucleolar RNAs (SnoRNAs) modify RNA through there conserved motifs, with boxes C/D guiding methylation and H/ACA guiding pseudouridylation, respectively (Ohtani, 2017; Figure 1Cii). Each class of snoRNAs displays a unique secondary structure composed of conserved proteins to form the defined C/D and H/ACA snoRNPs. SnoRNAs mainly target rRNA to modify functionally important regions of the ribosome (Decatur and Fournier, 2002) but other purposes include pre-rRNA endonucleolytic processing (Tollervey and Hurt, 1990), guiding snRNAs such as U6 snRNA (Tycowski et al., 1998) and more recently mRNA guiding (Sharma et al., 2016) or regulation of alternative splicing in pre-mRNAs (Falaleeva et al., 2016).
Box C/D snoRNP biogenesis commences when a protein complex of SNU13 and NOP58 is pre-formed and loaded onto the snoRNA with the help of HSP90/R2TP. This recruits assembly factors and the pre-snoRNPs are transferred to the Cajal bodies where final processing occurs. Box H/ACA RNPs biogenesis starts by SHQ1 and DKC1 combining to prevent to non-specific RNAs binding. SHQ1 is released with the help of the R2TP complex allowing DKC1 to bind H/ACA RNAs at the site of transcription. Numerous assembly factors including NHP2, NOP10, and NAF1 are present during this pre-snoRNP form. When NAF1 – which binds the C-terminal domain of RNA polymerase II to keep H/ACA RNP inactive – is replaced by GAR1, mature and functional H/ACA RNPs are produced. Both forms are transported to the nucleolus to elicit their actions (Massenet et al., 2017; Figure 1Ci).
A study showed differential regulation of two C/D box snoRNAs (e307 and e470) prior to the development of AD in mouse model. After formation of a β-amyloid plaque, this differential expression is no longer present, demonstrating that they could be useful in early diagnosis. No clear evidence of pathogenesis just hypothesized using bioinformatics methods (Gstir et al., 2014) (Table 3).
Despite the fact that autism spectrum disorder (ASD) might not be considered a neurodegenerative disease. Studies have found links in ASD with numerous snoRNA genes found to be differentially expressed using RNA-seq (Wright et al., 2017). Duplication of SNORD115 in mouse chromosome 7 that mirrors human chromosome 15q11-13 – duplication of this is one of the most common chromosomal abnormalities in ASD – has been shown to increase SNORD115 levels and results in abnormal brain development. In addition, SNORD115 (HBII-48 and HBII-52) levels are dysregulated in superior temporal gyrus of human ASD brain samples, which could explain 5-HT changes (Gabriele et al., 2014) and alternative splicing seen in ASD (Voineagu et al., 2011) as HBII-52 may regulate 5-HT2C receptor mRNA levels (Stamova et al., 2015) as well as alternative splicing (Kishore et al., 2010).
Another study demonstrated that maternal alcohol consumption in pregnancy alters the C/D box RNA levels in brain cells during abnormal fetal development. DNA methylation, microRNA and snoRNA levels altered with emphasis on SNORD115 increasing and SNORD116 decreasing (Laufer et al., 2013).
Piwi-Interacting RNA
Piwi-Interacting RNAs (PiRNAs) are a diverse range of small RNAs that are highly enriched in the germline tissues. They interact with PIWI-class Argonaute proteins with sequence bias for only the first 5′ nucleotide to be a Uracil. This diverse population can be mapped back to distinct areas of the genome known as piRNA clusters, which contain highly enriched areas of fragmented dysfunctional transposable element (TE) sequences. These are thought to emanate from the memory of previous TE invasions, and can be utilized to protect against TEs (Toth et al., 2016). In addition, PIWI proteins function at the chromatin level by guiding DNA methylation and deposition of repressive histone marks to silence TE transcription (Le Thomas et al., 2013; Figure 1Dii).
The biogenesis of piRNAs gives rise to two different forms primary and secondary of 26–30 bps in length, stemming from single-stranded precursors (Yan et al., 2011; Mani and Juliano, 2013), which are best studied in Drosophila. Primary piRNAs biogenesis is poorly defined but precursors of around 200 bp stemming nearly entirely from piRNA clusters are cleaved – Zucchini (ZUC) is thought to do this – to enable loading onto a PIWI protein in association with other factors (Figure 1Di). This piRNA-PIWI complex interacts with TEs to prevent insertion through methylation or transcriptional repression, thereby affecting gene expression (Toth et al., 2016).
In Drosophila, secondary piRNAs are formed through a more defined “ping-pong” pathway, which utilizes the primary piRNAs formed from TE fragments present in piRNA clusters loaded onto Aubergine (AUB) to find complementary antisense TE transcripts (Figure 1D). Once found the complementary TE mRNA binds, and is cleaved ten nucleotides along from the 5′ end by AUB, which terminates its function. Additionally it creates a new 5′ end and piRNA precursor, which accompanied by AGO3 is processed into secondary piRNA. The secondary piRNA promotes the development of more cluster-derived piRNAs – it is representative of the sense TE strand – through complementary cluster transcripts to develop a greater repertoire against active TEs (Toth et al., 2016; Figure 1Di).
Originally piRNAs were solely thought to be present in germline cells, more recently they have been found in other areas of the body including blood (Yang et al., 2015), blood plasma (Freedman et al., 2016) and the brain (Roy et al., 2017) as well as interacting with diseases in the liver (Rizzo et al., 2016), cardiovascular system (Loche and Ozanne, 2016) and brain (Roy et al., 2017) demonstrating their roles are far-reaching. In neurodegenerative disease there have been recent studies on PD and AD.
Risk variants APOE (rs2075650) and RNU6-560P (rs10792835 + rs3851179) have been linked with AD through genome-wide association studies (GWAS). These risk variants were significantly correlated with nine (6 APOE and 3 RNU6-560P) different piRNAs, showing regulatory capabilities (Guo X. et al., 2017). PiRNA dysregulation may be integral to the development of AD through aberrant downstream signaling. The link to pathogenesis in AD was clarified in three AD dysregulated piRNAs (piR-38240, piR-34393, and piR-40666) after establishing complementary target genes (CYCS, KPNA6, and RAB11A) through inverse expression correlation (Roy et al., 2017). The target genes were known to regulate AD pathways through oxidative stress induced neurodegeneration, apoptosis and vesicular trafficking of Aβ. This demonstrates a regulatory role for piRNAs in preventing AD and so monitoring dysregulation could allow early diagnosis and implicate a treatment method.
There was a difference found in piRNA expression between PD- and control- patient derived cells. Patient tissue samples showed the same trend, with 70 different piRNAs overlapping between both (Table 3). Two distinct trends come from these piRNAs, up or down regulation (Schulze et al., 2018). In the down-regulated piRNA fraction, those that were short-interspersed nuclear elements (SINE) and long-interspersed nuclear elements (LINE) derived elements in cell lines and LINE in tissues, showed significant enrichment when compared to genome-wide expression (Schulze et al., 2018). This is indicative of an inability to silence SINE and LINE derived elements in PD-derived neurones, which could show a pathogenesis of PD disease.
Transfer RNAs
Transfer RNAs (tRNAs) are the most abundant form of sncRNA, making up 4–10% of all cellular RNAs. Previously thought to be static contributors to gene expression, acting as an adaptor molecule in translation. Recently it has been found that small non-coding tRNAs have unique function that enable wider signaling and dynamic regulation of various functions (Gebetsberger and Polacek, 2013).
Mature tRNA is formed through transcription of precursor tRNA (pre-tRNA) using RNA polymerase III. Endonucleolytic ribonuclease P (RNase P) and ribonuclease Z cleave the transcribed pre-tRNA at the 5′ leader sequence and 3′ polyuracil (poly –U) tail, respectively, before tRNA nucleotidyl transferase adds a 3′CCA tail (Figure 1Ei). Many post-transcriptional modifications will occur during maturation and only tRNAs appropriately processed will leave the nucleus via nuclear receptor-mediated export process, with wrongly processed terminating. The mature tRNAs are between 73–90 nts in length and contain a clover-leaf shaped secondary structure, composing of a D-loop, an anticodon loop, a T-loop, a variable loop and an amino acid acceptor stem (Kirchner and Ignatova, 2015). The mature of pre-tRNA can be cleaved – into specific products unlike previously thought – into two main categories of cleaved tRNAs have been categorized; (1) tRNA-halves, (2) tRNA derived fragments.
tRNA halves are produced by cleavage of the anticodon loop giving rise to two halves; 30–35 nt 5′-tRNA halves and 40–50 nt 3′tRNA halves (Li and Hu, 2012; Figure 1Eii). A subtype of tRNA halves known as tRNA-derived stress-induced RNAs (tiRNAs) are by-products of stress. They induce cleavage by angiogenin (ANG) – a ribonuclease – of mature cytoplasmic tRNAs (Yamasaki et al., 2009).
tRNA derived fragments (tRFs) are produced from either pre-tRNAs or mature tRNAs (Figure 1Eii). Four main types have been established stemming from the fragment location on tRNAs: 5-tRFs, 3-tRFs, 1-tRFs, and 2 tRFs. 5-tRFs – located most abundantly in the nucleus – are generated from cleavage of the D-loop of tRNAs by Dicer, with adenine being present at the 3′ ends. Further subdivision classifies 5-tRFs isoforms into “a” (∼15 nts), “b” (∼22 nts) and “c” (∼30 nts) (Kumar et al., 2015; Lee et al., 2009). 3-tRFs result from cleavage by Dicer, ANG or another member of the Ribonuclease A superfamily of the T-loop, containing a CCA tail sequence (18–22 nts) (Lee et al., 2009; Maraia and Lamichhane, 2011; Kumar et al., 2015). 1-tRFs are formed by the cleavage of the 3′-trailer fragment of pre-tRNAs by either RNaseZ or ELAC2, this usually commences after the 3′-ends of mature tRNA and contains a poly-U 3′-end (Lee et al., 2009; Liao et al., 2010). 2-tRFs, less known about but may be formed from the anticodon loop (Goodarzi et al., 2015).
Numerous neurodegenerative disorders are associated with tRFs. ANG mutants show reduced ribonuclease (RNase) activity and were first implicated in the pathogenesis of amyotrophic lateral sclerosis (ALS) (Greenway et al., 2006). Latterly, a subset of the ALS-associated ANG mutants were observed in Parkinson’s disease (PD) patients (van Es et al., 2011). Recombinant ANG can improve life span and motor function in an ALS [SOD1 (G93A)] mouse model, demonstrating that tRFs may have an important role in motor neuron survival (Kieran et al., 2008) (Table 3).
The link between ANG-induced tiRNAs, cellular stress and neurodevelopment disorders was strengthened with the finding of NSun2 (Blanco et al., 2014). Mutations in the cytosine-5 RNA methyltransferase NSun2 have been shown to cause intellectual disability and a Dubowitz-like syndrome in humans (Abbasi-Moheb et al., 2012; Martinez et al., 2012). NSun2 methylates two different cytosine residues of tRNA. Without NSun2, cytosine-5 RNAs are not methylated, which increases the stress-induced ANG-mediated endonucleolytic cleavage of tRNAs and so 5′-tiRNAs accumulate. Accumulation of these factors leads to cell death in hippocampal and striatal neurons because of translational repression leading to cellular stress. Subsequently, NSun2 knockout mice show reduced neuronal size and impaired formation of synapses, which could explain the impairment of NSun2 gene mutation patients (Blanco et al., 2014).
A mutation in CLP1 gene (R140A) – a RNA kinase involved in tRNA splicing – is present in pontocerebellar hypoplasia (PCH) patients, a heterogeneous group of inherited neurodegenerative disorders characterized by the loss of motor neurons, muscle paralysis, impaired development of various parts of the brain and differential tRNA splicing (Karaca et al., 2014; Schaffer et al., 2014). The role of CLP1 in RNA splicing means the mutant gene has reduced kinase activity and affinity to the tRNA endonuclease complex (TSEN), impairing pre-tRNA cleavage and elevating unspliced pre-tRNAs in patient derived neurons (Schaffer et al., 2014). TSEN cuts the transcript at 3′ intron-extron junctions, so the absence of CLP1 means 5′-unphosphorylated tRF cannot interact with the pre-tRNAtyr 3′-exon and subsequent splicing steps are interrupted (Cassandrini et al., 2010).
N6-threonyl-carbamoyl-adenosine (t6A) is a complex modification of adenosine involved in cytoplasmic tRNA modification. It is located next to the anticodon loop of many tRNAs that decode ANN codons, at position 37 (t6A37). Recently, a biosynthetic defect in the t6A molecule resulting from a mutation in the kinase-associated endopeptidase (KAE1) gene, which is part of the kinase, endopeptidase and other proteins of small size (KEOPS) complex was found in two phenotypically neurodegenerative patients, implicating tRNA modification in neuronal maintenance (Edvardson et al., 2017).
Although, tRNA-derived small non-coding RNAs, have already demonstrated a role in cancer progression (Sun et al., 2018), their role as biomarkers in NDs has not been fully investigated yet.
However, animal studies showed 13 dysregulated tRFs in brain samples of SAMP8 mouse model for AD. In particular, four were upregulated (AS-tDR-011775, AS-tDR-011438, AS-tDR-006835 and AStDR-005058) and 9 down regulated (AS-tDR-013428, AS-tDR-011389, AS-tDR-009392, AS-tDR012690, AS-tDR-010654, AS-tDR-008616, AS-tDR-010789, AS-tDR-011670, and AS-tDR-007919), demonstrating their potential involvement of tRFs in early detection of AD.
Conclusion
The key problem with the ND field is the lack of understanding in the events preceding the development of protein-based markers – such as Tau – currently used to diagnose NDs. By this stage, the diseases become more difficult to treat.
SncRNAs play an important regulatory role in the maintenance of the homeostatic brain. Therefore, changes in their concentration levels can be indicative of mechanistic changes that could precede protein-based markers. One single sncRNA biomarker is unlikely to differentiate between diseases. However, a combination of sncRNA biomarkers could be illustrative of the mechanistic development of NDs to enable early diagnosis, enhanced disease monitoring as well as defining subtle differences between NDs. Consequently, novel treatment methods directly related to their mechanistic underpinning of specific NDs, and potentially other brain related pathologies can be envisaged.
Novel, less-well studied sncRNAs could be integral to understanding the overall disease progression. So new methodologies may be necessary to quantify these changes and allow for future biomarker development.
Author Contributions
CW drafted the manuscript. AB and VDP critically revised the manuscript.
Funding
This study was funded by the National Institute for Health Research (NIHR). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbasi-Moheb, L., Mertel, S., Gonsior, M., Nouri-Vahid, L., Kahrizi, K., Cirak, S., et al. (2012). Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 847–855. doi: 10.1016/j.ajhg.2012.03.021
Absalon, S., Kochanek, D. M., Raghavan, V., and Krichevsky, A. M. (2013). MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 33, 14645–14659. doi: 10.1523/jneurosci.1327-13.2013
Agrawal, M., and Biswas, A. (2015). Molecular diagnostics of neurodegenerative disorders. Front. Mol. Biosci. 2:54. doi: 10.3389/fmolb.2015.00054
Alexandrov, P. N., Dua, P., Hill, J. M., Bhattacharjee, S., Zhao, Y., and Lukiw, W. J. (2012). microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol. 3, 365–373.
Alvarez-Erviti, L., Seow, Y., Schapira, A. H., Rodriguez-Oroz, M. C., Obeso, J. A., and Cooper, J. M. (2013). Influence of microRNA deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in Parkinson’s disease. Cell Death Dis. 4:e545. doi: 10.1038/cddis.2013.73
Aronin, N., and DiFiglia, M. (2014). Huntingtin-lowering strategies in Huntington’s disease: antisense oligonucleotides, small RNAs, and gene editing. Mov. Disord. 29, 1455–1461. doi: 10.1002/mds.26020
Bai, B., Hales, C. M., Chen, P. C., Gozal, Y., Dammer, E. B., Fritz, J. J., et al. (2013). U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 110, 16562–16567. doi: 10.1073/pnas.1310249110
Bai, X., Tang, Y., Yu, M., Wu, L., Liu, F., Ni, J., et al. (2017). Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Sci. Rep. 7:5411. doi: 10.1038/s41598-017-03887-3
Bekris, L. M., Lutz, F., Montine, T. J., Yu, C. E., Tsuang, D., Peskind, E. R., et al. (2013). MicroRNA in Alzheimer’s disease: an exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers 18, 455–466. doi: 10.3109/1354750x.2013.814073
Benigni, M., Ricci, C., Jones, A. R., Giannini, F., Al-Chalabi, A., and Battistini, S. (2016). Identification of miRNAs as potential biomarkers in cerebrospinal fluid from amyotrophic lateral sclerosis patients. Neuromolecular Med. 18, 551–560. doi: 10.1007/s12017-016-8396-8
Bhatnagar, S., Chertkow, H., Schipper, H. M., Yuan, Z., Shetty, V., Jenkins, S., et al. (2014). Increased microRNA-34c abundance in Alzheimer’s disease circulating blood plasma. Front. Mol. Neurosci. 7:2. doi: 10.3389/fnmol.2014.00002
Bishof, I., Dammer, E. B., Duong, D. M., Kundinger, S. R., Gearing, M., Lah, J. J., et al. (2018). RNA-binding proteins with basic-acidic dipeptide (BAD) domains self-assemble and aggregate in Alzheimer’s disease. J. Biol. Chem. 293, 11047–11066. doi: 10.1074/jbc.RA118.001747
Blanco, S., Dietmann, S., Flores, J. V., Hussain, S., Kutter, C., Humphreys, P., et al. (2014). Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039. doi: 10.15252/embj.201489282
Botta-Orfila, T., Morato, X., Compta, Y., Lozano, J. J., Falgas, N., Valldeoriola, F., et al. (2014). Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson’s disease. J. Neurosci. Res. 92, 1071–1077. doi: 10.1002/jnr.23377
Burgos, K., Malenica, I., Metpally, R., Courtright, A., Rakela, B., Beach, T., et al. (2014). Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One 9:e94839. doi: 10.1371/journal.pone.0094839
Butovsky, O., Siddiqui, S., Gabriely, G., Lanser, A. J., Dake, B., Murugaiyan, G., et al. (2012). Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 122, 3063–3087. doi: 10.1172/jci62636
Caggiu, E., Paulus, K., Mameli, G., Arru, G., Sechi, G. P., and Sechi, L. A. (2018). Differential expression of miRNA 155 and miRNA 146a in Parkinson’s disease patients. eNeurologicalSci 13, 1–4. doi: 10.1016/j.ensci.2018.09.002
Calin, G. A., and Croce, C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866. doi: 10.1038/nrc1997
Cardo, L. F., Coto, E., de Mena, L., Ribacoba, R., Moris, G., Menendez, M., et al. (2013). Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J. Neurol. 260, 1420–1422. doi: 10.1007/s00415-013-6900-8
Cardo, L. F., Coto, E., Ribacoba, R., Menendez, M., Moris, G., Suarez, E., et al. (2014). MiRNA profile in the substantia nigra of Parkinson’s disease and healthy subjects. J. Mol. Neurosci. 54, 830–836. doi: 10.1007/s12031-014-0428-y
Cassandrini, D., Biancheri, R., Tessa, A., Di Rocco, M., Di Capua, M., Bruno, C., et al. (2010). Pontocerebellar hypoplasia: clinical, pathologic, and genetic studies. Neurology 75, 1459–1464. doi: 10.1212/WNL.0b013e3181f88173
Chen, X., Liu, Y., Sheng, X., Tam, P. O., Zhao, K., Chen, X., et al. (2014). PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 23, 2926–2939. doi: 10.1093/hmg/ddu005
Cheng, L., Doecke, J. D., Sharples, R. A., Villemagne, V. L., Fowler, C. J., Rembach, A., et al. (2015). Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 20, 1188–1196. doi: 10.1038/mp.2014.127
Cheng, Z., Shang, Y., Gao, S., and Zhang, T. (2017). Overexpression of U1 snRNA induces decrease of U1 spliceosome function associated with Alzheimer’s disease. J. Neurogenet. 31, 337–343. doi: 10.1080/01677063.2017.1395425
Cho, H. J., Liu, G., Jin, S. M., Parisiadou, L., Xie, C., Yu, J., et al. (2013). MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 22, 608–620. doi: 10.1093/hmg/dds470
Chung, S., Sonntag, K. C., Andersson, T., Bjorklund, L. M., Park, J. J., Kim, D. W., et al. (2002). Genetic engineering of mouse embryonic stem cells by Nurr1 enhances differentiation and maturation into dopaminergic neurons. Eur. J. Neurosci. 16, 1829–1838.
Ciurleo, R., Di Lorenzo, G., Bramanti, P., and Marino, S. (2014). Magnetic resonance spectroscopy: an in vivo molecular imaging biomarker for Parkinson’s disease? Biomed Res. Int. 2014:519816. doi: 10.1155/2014/519816
Cogswell, J. P., Ward, J., Taylor, I. A., Waters, M., Shi, Y., Cannon, B., et al. (2008). Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 14, 27–41.
Colombrita, C., Onesto, E., Megiorni, F., Pizzuti, A., Baralle, F. E., Buratti, E., et al. (2012). TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 287, 15635–15647. doi: 10.1074/jbc.M111.333450
de Andrade, H. M., de Albuquerque, M., Avansini, S. H., de, S. R. C., Dogini, D. B., Nucci, A., et al. (2016). MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci. 368, 19–24. doi: 10.1016/j.jns.2016.06.046
De Felice, B., Annunziata, A., Fiorentino, G., Borra, M., Biffali, E., Coppola, C., et al. (2014). miR-338-3p is over-expressed in blood, CFS, serum and spinal cord from sporadic amyotrophic lateral sclerosis patients. Neurogenetics 15, 243–253. doi: 10.1007/s10048-014-0420-2
Decatur, W. A., and Fournier, M. J. (2002). rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351.
Denk, J., Boelmans, K., Siegismund, C., Lassner, D., Arlt, S., and Jahn, H. (2015). MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer’s disease. PLoS One 10:e0126423. doi: 10.1371/journal.pone.0126423
Di Pietro, V., Ragusa, M., Davies, D., Su, Z., Hazeldine, J., Lazzarino, G., et al. (2017). MicroRNAs as novel biomarkers for the diagnosis and prognosis of mild and severe traumatic brain injury. J. Neurotrauma 34, 1948–1956. doi: 10.1089/neu.2016.4857
Diez-Planelles, C., Sanchez-Lozano, P., Crespo, M. C., Gil-Zamorano, J., Ribacoba, R., Gonzalez, N., et al. (2016). Circulating microRNAs in Huntington’s disease: emerging mediators in metabolic impairment. Pharmacol. Res. 108, 102–110. doi: 10.1016/j.phrs.2016.05.005
Ding, H., Huang, Z., Chen, M., Wang, C., Chen, X., Chen, J., et al. (2016). Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Parkinsonism Relat. Disord. 22, 68–73. doi: 10.1016/j.parkreldis.2015.11.014
Dong, H., Li, J., Huang, L., Chen, X., Li, D., Wang, T., et al. (2015). Serum MicroRNA profiles serve as novel biomarkers for the diagnosis of Alzheimer’s disease. Dis. Markers 2015:625659. doi: 10.1155/2015/625659
Dong, H., Wang, C., Lu, S., Yu, C., Huang, L., Feng, W., et al. (2016). A panel of four decreased serum microRNAs as a novel biomarker for early Parkinson’s disease. Biomarkers 21, 129–137. doi: 10.3109/1354750x.2015.1118544
Dozmorov, M. G., Giles, C. B., Koelsch, K. A., and Wren, J. D. (2013). Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinformatics 14(Suppl. 14):S2. doi: 10.1186/1471-2105-14-S14-S2
Edvardson, S., Prunetti, L., Arraf, A., Haas, D., Bacusmo, J. M., Hu, J. F., et al. (2017). tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. 25, 545–551. doi: 10.1038/ejhg.2017.30
Elsaid, M. F., Chalhoub, N., Ben-Omran, T., Kumar, P., Kamel, H., Ibrahim, K., et al. (2017). Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Ann. Neurol. 81, 68–78. doi: 10.1002/ana.24826
Falaleeva, M., Pages, A., Matuszek, Z., Hidmi, S., Agranat-Tamir, L., Korotkov, K., et al. (2016). Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 113, E1625–E1634. doi: 10.1073/pnas.1519292113
Fransquet, P. D., and Ryan, J. (2018). Micro RNA as a potential blood-based epigenetic biomarker for Alzheimer’s disease. Clin. Biochem. 58, 5–14. doi: 10.1016/j.clinbiochem.2018.05.020
Freedman, J. E., Gerstein, M., Mick, E., Rozowsky, J., Levy, D., Kitchen, R., et al. (2016). Diverse human extracellular RNAs are widely detected in human plasma. Nat. Commun. 7:11106. doi: 10.1038/ncomms11106
Freischmidt, A., Muller, K., Ludolph, A. C., and Weishaupt, J. H. (2013). Systemic dysregulation of TDP-43 binding microRNAs in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 1:42. doi: 10.1186/2051-5960-1-42
Freischmidt, A., Muller, K., Zondler, L., Weydt, P., Mayer, B., von Arnim, C. A., et al. (2015). Serum microRNAs in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 36, 2660.e15–2660.e20. doi: 10.1016/j.neurobiolaging.2015.06.003
Freischmidt, A., Muller, K., Zondler, L., Weydt, P., Volk, A. E., Bozic, A. L., et al. (2014). Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain 137, 2938–2950. doi: 10.1093/brain/awu249
Gabanella, F., Butchbach, M. E., Saieva, L., Carissimi, C., Burghes, A. H., and Pellizzoni, L. (2007). Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2:e921. doi: 10.1371/journal.pone.0000921
Gabriele, S., Sacco, R., and Persico, A. M. (2014). Blood serotonin levels in autism spectrum disorder: a systematic review and meta-analysis. Eur. Neuropsychopharmacol. 24, 919–929. doi: 10.1016/j.euroneuro.2014.02.004
Galimberti, D., Villa, C., Fenoglio, C., Serpente, M., Ghezzi, L., Cioffi, S. M., et al. (2014). Circulating miRNAs as potential biomarkers in Alzheimer’s disease. J. Alzheimers Dis. 42, 1261–1267. doi: 10.3233/jad-140756
Gaughwin, P. M., Ciesla, M., Lahiri, N., Tabrizi, S. J., Brundin, P., and Bjorkqvist, M. (2011). Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington’s disease. Hum. Mol. Genet. 20, 2225–2237. doi: 10.1093/hmg/ddr111
Gebetsberger, J., and Polacek, N. (2013). Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 10, 1798–1806. doi: 10.4161/rna.27177
Geekiyanage, H., Jicha, G. A., Nelson, P. T., and Chan, C. (2012). Blood serum miRNA: non-invasive biomarkers for Alzheimer’s disease. Exp. Neurol. 235, 491–496. doi: 10.1016/j.expneurol.2011.11.026
Goodarzi, H., Liu, X., Nguyen, H. C., Zhang, S., Fish, L., and Tavazoie, S. F. (2015). Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802. doi: 10.1016/j.cell.2015.02.053
Grasso, M., Piscopo, P., Confaloni, A., and Denti, M. A. (2014). Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules 19, 6891–6910. doi: 10.3390/molecules19056891
Greenway, M. J., Andersen, P. M., Russ, C., Ennis, S., Cashman, S., Donaghy, C., et al. (2006). ANG mutations segregate with familial and ’sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 38, 411–413. doi: 10.1038/ng1742
Groiss, S. J., Wojtecki, L., Südmeyer, M., and Schnitzler, A. (2009). Review: deep brain stimulation in Parkinson’s disease. Ther. Adv. Neurol. Disord. 2, 20–28. doi: 10.1177/1756285609339382
Gstir, R., Schafferer, S., Scheideler, M., Misslinger, M., Griehl, M., Daschil, N., et al. (2014). Generation of a neuro-specific microarray reveals novel differentially expressed noncoding RNAs in mouse models for neurodegenerative diseases. RNA 20, 1929–1943. doi: 10.1261/rna.047225.114
Gui, Y., Liu, H., Zhang, L., Lv, W., and Hu, X. (2015). Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 6, 37043–37053. doi: 10.18632/oncotarget.6158
Guo, R., Fan, G., Zhang, J., Wu, C., Du, Y., Ye, H., et al. (2017). A 9-microRNA signature in serum serves as a noninvasive biomarker in early diagnosis of Alzheimer’s disease. J. Alzheimers Dis. 60, 1365–1377. doi: 10.3233/jad-170343
Guo, X., Qiu, W., Garcia-Milian, R., Lin, X., Zhang, Y., Cao, Y., et al. (2017). Genome-wide significant, replicated and functional risk variants for Alzheimer’s disease. J. Neural Transm. 124, 1455–1471. doi: 10.1007/s00702-017-1773-0
Hales, C. M., Dammer, E. B., Diner, I., Yi, H., Seyfried, N. T., Gearing, M., et al. (2014a). Aggregates of small nuclear ribonucleic acids (snRNAs) in Alzheimer’s disease. Brain Pathol. 24, 344–351. doi: 10.1111/bpa.12133
Hales, C. M., Seyfried, N. T., Dammer, E. B., Duong, D., Yi, H., Gearing, M., et al. (2014b). U1 small nuclear ribonucleoproteins (snRNPs) aggregate in Alzheimer’s disease due to autosomal dominant genetic mutations and trisomy 21. Mol. Neurodegener. 9:15. doi: 10.1186/1750-1326-9-15
Hara, N., Kikuchi, M., Miyashita, A., Hatsuta, H., Saito, Y., Kasuga, K., et al. (2017). Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 5:10. doi: 10.1186/s40478-017-0414-z
Hebert, S. S., Horre, K., Nicolai, L., Papadopoulou, A. S., Mandemakers, W., Silahtaroglu, A. N., et al. (2008). Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. U.S.A. 105, 6415–6420. doi: 10.1073/pnas.0710263105
Hoss, A. G., Labadorf, A., Beach, T. G., Latourelle, J. C., and Myers, R. H. (2016). microRNA profiles in Parkinson’s disease prefrontal cortex. Front. Aging Neurosci. 8:36. doi: 10.3389/fnagi.2016.00036
Hoss, A. G., Labadorf, A., Latourelle, J. C., Kartha, V. K., Hadzi, T. C., Gusella, J. F., et al. (2015a). miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med. Genomics 8:10. doi: 10.1186/s12920-015-0083-3
Hoss, A. G., Lagomarsino, V. N., Frank, S., Hadzi, T. C., Myers, R. H., and Latourelle, J. C. (2015b). Study of plasma-derived miRNAs mimic differences in Huntington’s disease brain. Mov. Disord. 30, 1961–1964. doi: 10.1002/mds.26457
Ishihara, T., Ariizumi, Y., Shiga, A., Kato, T., Tan, C. F., Sato, T., et al. (2013). Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum. Mol. Genet. 22, 4136–4147. doi: 10.1093/hmg/ddt262
Jack, C. R. Jr., Albert, M. S., Knopman, D. S., McKhann, G. M., Sperling, R. A., Carrillo, M. C., et al. (2011). Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 257–262. doi: 10.1016/j.jalz.2011.03.004
Jia, L. H., and Liu, Y. N. (2016). Downregulated serum miR-223 servers as biomarker in Alzheimer’s disease. Cell Biochem. Funct. 34, 233–237. doi: 10.1002/cbf.3184
Jia, Y., Mu, J. C., and Ackerman, S. L. (2012). Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell 148, 296–308. doi: 10.1016/j.cell.2011.11.057
Jutzi, D., Akinyi, M. V., Mechtersheimer, J., Frilander, M. J., and Ruepp, M.-D. (2018). The emerging role of minor intron splicing in neurological disorders. Cell Stress 2, 40–54. doi: 10.15698/cst2018.03.126
Karaca, E., Weitzer, S., Pehlivan, D., Shiraishi, H., Gogakos, T., Hanada, T., et al. (2014). Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157, 636–650. doi: 10.1016/j.cell.2014.02.058
Kawahara, Y., and Mieda-Sato, A. (2012). TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 3347–3352. doi: 10.1073/pnas.1112427109
Khoo, S. K., Petillo, D., Kang, U. J., Resau, J. H., Berryhill, B., Linder, J., et al. (2012). Plasma-based circulating MicroRNA biomarkers for Parkinson’s disease. J. Parkinsons Dis. 2, 321–331. doi: 10.3233/jpd-012144
Kieran, D., Sebastia, J., Greenway, M. J., King, M. A., Connaughton, D., Concannon, C. G., et al. (2008). Control of motoneuron survival by angiogenin. J. Neurosci. 28, 14056–14061. doi: 10.1523/jneurosci.3399-08.2008
Kiko, T., Nakagawa, K., Tsuduki, T., Furukawa, K., Arai, H., and Miyazawa, T. (2014). MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimers Dis. 39, 253–259. doi: 10.3233/jad-130932
Kirchner, S., and Ignatova, Z. (2015). Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16, 98–112. doi: 10.1038/nrg3861
Kishore, S., Khanna, A., Zhang, Z., Hui, J., Balwierz, P. J., Stefan, M., et al. (2010). The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 19, 1153–1164. doi: 10.1093/hmg/ddp585
Kumar, P., Dezso, Z., MacKenzie, C., Oestreicher, J., Agoulnik, S., Byrne, M., et al. (2013). Circulating miRNA biomarkers for Alzheimer’s disease. PLoS One 8:e69807. doi: 10.1371/journal.pone.0069807
Kumar, P., Mudunuri, S. B., Anaya, J., and Dutta, A. (2015). tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 43, D141–D145. doi: 10.1093/nar/gku1138
Kumar, S., Vijayan, M., and Reddy, P. H. (2017). MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 26, 3808–3822. doi: 10.1093/hmg/ddx267
Laufer, B. I., Mantha, K., Kleiber, M. L., Diehl, E. J., Addison, S. M., and Singh, S. M. (2013). Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Dis. Model. Mech. 6, 977–992. doi: 10.1242/dmm.010975
Le Thomas, A., Rogers, A. K., Webster, A., Marinov, G. K., Liao, S. E., Perkins, E. M., et al. (2013). Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27, 390–399. doi: 10.1101/gad.209841.112
Lee, Y. S., Shibata, Y., Malhotra, A., and Dutta, A. (2009). A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649. doi: 10.1101/gad.1837609
Lehmann, S. M., Kruger, C., Park, B., Derkow, K., Rosenberger, K., Baumgart, J., et al. (2012). An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835. doi: 10.1038/nn.3113
Leidinger, P., Backes, C., Deutscher, S., Schmitt, K., Mueller, S. C., Frese, K., et al. (2013). A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 14:R78. doi: 10.1186/gb-2013-14-7-r78
Li, S., and Hu, G. F. (2012). Emerging role of angiogenin in stress response and cell survival under adverse conditions. J. Cell. Physiol. 227, 2822–2826. doi: 10.1002/jcp.23051
Liao, J. Y., Ma, L. M., Guo, Y. H., Zhang, Y. C., Zhou, H., Shao, P., et al. (2010). Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3’ trailers. PLoS One 5:e10563. doi: 10.1371/journal.pone.0010563
Liao, X. Y., Wang, W. W., Yang, Z. H., Wang, J., Lin, H., Wang, Q. S., et al. (2013). Microarray analysis of transcriptome of medulla identifies potential biomarkers for Parkinson’s disease. Int. J. Genomics 2013:606919. doi: 10.1155/2013/606919
Liguori, M., Nuzziello, N., Introna, A., Consiglio, A., Licciulli, F., D’Errico, E., et al. (2018). Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 11:288. doi: 10.3389/fnmol.2018.00288
Loche, E., and Ozanne, S. E. (2016). Early nutrition, epigenetics, and cardiovascular disease. Curr. Opin. Lipidol. 27, 449–458. doi: 10.1097/mol.0000000000000338
Lugli, G., Cohen, A. M., Bennett, D. A., Shah, R. C., Fields, C. J., Hernandez, A. G., et al. (2015). Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS One 10:e0139233. doi: 10.1371/journal.pone.0139233
Lukiw, W. J. (2007). Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 18, 297–300. doi: 10.1097/WNR.0b013e3280148e8b
Lukiw, W. J., Alexandrov, P. N., Zhao, Y., Hill, J. M., and Bhattacharjee, S. (2012). Spreading of Alzheimer’s disease inflammatory signaling through soluble micro-RNA. Neuroreport 23, 621–626. doi: 10.1097/WNR.0b013e32835542b0
Ma, W., Li, Y., Wang, C., Xu, F., Wang, M., and Liu, Y. (2016). Serum miR-221 serves as a biomarker for Parkinson’s disease. Cell Biochem. Funct. 34, 511–515. doi: 10.1002/cbf.3224
Mani, S. R., and Juliano, C. E. (2013). Untangling the web: the diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 80, 632–664. doi: 10.1002/mrd.22195
Maraia, R. J., and Lamichhane, T. N. (2011). 3’ processing of eukaryotic precursor tRNAs. Wiley Interdiscip. Rev. RNA 2, 362–375. doi: 10.1002/wrna.64
Margis, R., Margis, R., and Rieder, C. R. (2011). Identification of blood microRNAs associated to Parkinsonis disease. J. Biotechnol. 152, 96–101. doi: 10.1016/j.jbiotec.2011.01.023
Martinez, F. J., Lee, J. H., Lee, J. E., Blanco, S., Nickerson, E., Gabriel, S., et al. (2012). Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 49, 380–385. doi: 10.1136/jmedgenet-2011-100686
Martins, M., Rosa, A., Guedes, L. C., Fonseca, B. V., Gotovac, K., Violante, S., et al. (2011). Convergence of miRNA expression profiling, alpha-synuclein interaction and GWAS in Parkinson’s disease. PLoS One 6:e25443. doi: 10.1371/journal.pone.0025443
Massenet, S., Bertrand, E., and Verheggen, C. (2017). Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 14, 680–692. doi: 10.1080/15476286.2016.1243646
Matamala, J. M., Arias-Carrasco, R., Sanchez, C., Uhrig, M., Bargsted, L., Matus, S., et al. (2018). Genome-wide circulating microRNA expression profiling reveals potential biomarkers for amyotrophic lateral sclerosis. Neurobiol. Aging 64, 123–138. doi: 10.1016/j.neurobiolaging.2017.12.020
Matera, A. G., Terns, R. M., and Terns, M. P. (2007). Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220. doi: 10.1038/nrm2124
McKhann, G. M., Knopman, D. S., Chertkow, H., Hyman, B. T., Jack, C. R. Jr., Kawas, C. H., et al. (2011). The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 263–269. doi: 10.1016/j.jalz.2011.03.005
Minones-Moyano, E., Porta, S., Escaramis, G., Rabionet, R., Iraola, S., Kagerbauer, B., et al. (2011). MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 20, 3067–3078. doi: 10.1093/hmg/ddr210
Munoz-Culla, M., Irizar, H., Saenz-Cuesta, M., Castillo-Trivino, T., Osorio-Querejeta, I., Sepulveda, L., et al. (2016). SncRNA (microRNA &snoRNA) opposite expression pattern found in multiple sclerosis relapse and remission is sex dependent. Sci. Rep. 6:20126. doi: 10.1038/srep20126
Nagaraj, S., Laskowska-Kaszub, K., Debski, K. J., Wojsiat, J., Dabrowski, M., Gabryelewicz, T., et al. (2017). Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget 8, 16122–16143. doi: 10.18632/oncotarget.15109
Nagatsua, T., and Sawadab, M. (2009). L-dopa therapy for Parkinson’s disease: past, present, and future. Parkinsonism Relat. Disord. 15(Suppl. 1), S3–S8. doi: 10.1016/s1353-8020(09)70004-5
O’Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9:402. doi: 10.3389/fendo.2018.00402
Ohtani, M. (2017). Transcriptional regulation of snRNAs and its significance for plant development. J. Plant Res. 130, 57–66. doi: 10.1007/s10265-016-0883-3
Packer, A. N., Xing, Y., Harper, S. Q., Jones, L., and Davidson, B. L. (2008). The bifunctional microRNA miR-9/miR-9∗ regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 28, 14341–14346. doi: 10.1523/jneurosci.2390-08.2008
Palazzo, A. F., and Lee, E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6:2. doi: 10.3389/fgene.2015.00002
Rachakonda, V., Pan, T. H., and Le, W. D. (2004). Biomarkers of neurodegenerative disorders: how good are they? Cell Res. 14, 347–358. doi: 10.1038/sj.cr.7290235
Raheja, R., Regev, K., Healy, B. C., Mazzola, M. A., Beynon, V., Von Glehn, F., et al. (2018). Correlating serum microRNAs and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve 58, 261–269. doi: 10.1002/mus.26106
Ratti, A., and Buratti, E. (2016). Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 138(Suppl. 1), 95–111. doi: 10.1111/jnc.13625
Reber, S., Stettler, J., Filosa, G., Colombo, M., Jutzi, D., Lenzken, S. C., et al. (2016). Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 35, 1504–1521. doi: 10.15252/embj.201593791
Reed, E. R., Latourelle, J. C., Bockholt, J. H., Bregu, J., Smock, J., Paulsen, J. S., et al. (2018). MicroRNAs in CSF as prodromal biomarkers for Huntington disease in the PREDICT-HD study. Neurology 90, e264–e272. doi: 10.1212/wnl.0000000000004844
Ricci, C., Marzocchi, C., and Battistini, S. (2018). MicroRNAs as biomarkers in amyotrophic lateral sclerosis. Cells 7:E219. doi: 10.3390/cells7110219
Rizzo, F., Rinaldi, A., Marchese, G., Coviello, E., Sellitto, A., Cordella, A., et al. (2016). Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget 7, 54650–54661. doi: 10.18632/oncotarget.10567
Roy, J., Sarkar, A., Parida, S., Ghosh, Z., and Mallick, B. (2017). Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s disease and their probable role in pathogenesis. Mol. Biosyst. 13, 565–576. doi: 10.1039/c6mb00699j
Sala Frigerio, C., Lau, P., Salta, E., Tournoy, J., Bossers, K., Vandenberghe, R., et al. (2013). Reduced expression of hsa-miR-27a-3p in CSF of patients with Alzheimer disease. Neurology 81, 2103–2106. doi: 10.1212/01.wnl.0000437306.37850.22
Schaffer, A. E., Eggens, V. R., Caglayan, A. O., Reuter, M. S., Scott, E., Coufal, N. G., et al. (2014). CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157, 651–663. doi: 10.1016/j.cell.2014.03.049
Schipper, H. M., Maes, O. C., Chertkow, H. M., and Wang, E. (2007). MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Bio. 1, 263–274.
Schulze, M., Sommer, A., Plotz, S., Farrell, M., Winner, B., Grosch, J., et al. (2018). Sporadic Parkinson’s disease derived neuronal cells show disease-specific mRNA and small RNA signatures with abundant deregulation of piRNAs. Acta Neuropathol. Commun. 6:58. doi: 10.1186/s40478-018-0561-x
Serafin, A., Foco, L., Blankenburg, H., Picard, A., Zanigni, S., Zanon, A., et al. (2014). Identification of a set of endogenous reference genes for miRNA expression studies in Parkinson’s disease blood samples. BMC Res. Notes 7:715. doi: 10.1186/1756-0500-7-715
Serafin, A., Foco, L., Zanigni, S., Blankenburg, H., Picard, A., Zanon, A., et al. (2015). Overexpression of blood microRNAs 103a, 30b, and 29a in L-dopa-treated patients with PD. Neurology 84, 645–653. doi: 10.1212/wnl.0000000000001258
Sharma, E., Sterne-Weiler, T. O., Hanlon, D., and Blencowe, B. J. (2016). Global mapping of human RNA-RNA interactions. Mol. Cell 62, 618–626. doi: 10.1016/j.molcel.2016.04.030
Sheinerman, K. S., Toledo, J. B., Tsivinsky, V. G., Irwin, D., Grossman, M., Weintraub, D., et al. (2017). Circulating brain-enriched microRNAs as novel biomarkers for detection and differentiation of neurodegenerative diseases. Alzheimers Res. Ther. 9:89. doi: 10.1186/s13195-017-0316-0
Sheinerman, K. S., and Umansky, S. R. (2013). Early detection of neurodegenerative diseases: circulating brain-enriched microRNA. Cell Cycle 12, 1–2. doi: 10.4161/cc.23067
Shioya, M., Obayashi, S., Tabunoki, H., Arima, K., Saito, Y., Ishida, T., et al. (2010). Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol. 36, 320–330. doi: 10.1111/j.1365-2990.2010.01076.x
Sorensen, S. S., Nygaard, A. B., and Christensen, T. (2016). miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia - an exploratory study. Transl. Neurodegener. 5:6. doi: 10.1186/s40035-016-0053-5
Soreq, L., Salomonis, N., Bronstein, M., Greenberg, D. S., Israel, Z., Bergman, H., et al. (2013). Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front. Mol. Neurosci. 6:10. doi: 10.3389/fnmol.2013.00010
Spano, M., Signorelli, M., Vitaliani, R., Aguglia, E., and Giometto, B. (2015). The possible involvement of mitochondrial dysfunctions in Lewy body dementia: a systematic review. Funct. Neurol. 30, 151–158.
Stamova, B., Ander, B. P., Barger, N., Sharp, F. R., and Schumann, C. M. (2015). Specific regional and age-related small noncoding RNA expression patterns within superior temporal gyrus of typical human brains are less distinct in autism brains. J. Child Neurol. 30, 1930–1946. doi: 10.1177/0883073815602067
Sun, C., Yang, F., Zhang, Y., Chu, J., Wang, J., Wang, Y., et al. (2018). tRNA-derived fragments as novel predictive biomarkers for trastuzumab-resistant breast cancer. Cell. Physiol. Biochem. 49, 419–431. doi: 10.1159/000492977
Swarbrick, S., Wragg, N., Ghosh, S., and Stolzing, A. (2019). Systematic review of miRNA as biomarkers in Alzheimer’s disease. Mol. Neurobiol. doi: 10.1007/s12035-019-1500-y
Takahashi, I., Hama, Y., Matsushima, M., Hirotani, M., Kano, T., Hohzen, H., et al. (2015). Identification of plasma microRNAs as a biomarker of sporadic Amyotrophic Lateral Sclerosis. Mol. Brain 8:67. doi: 10.1186/s13041-015-0161-7
Tan, L., Yu, J. T., Liu, Q. Y., Tan, M. S., Zhang, W., Hu, N., et al. (2014a). Circulating miR-125b as a biomarker of Alzheimer’s disease. J. Neurol. Sci. 336, 52–56. doi: 10.1016/j.jns.2013.10.002
Tan, L., Yu, J. T., Tan, M. S., Liu, Q. Y., Wang, H. F., Zhang, W., et al. (2014b). Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. J. Alzheimers Dis. 40, 1017–1027. doi: 10.3233/jad-132144
Tasca, E., Pegoraro, V., Merico, A., and Angelini, C. (2016). Circulating microRNAs as biomarkers of muscle differentiation and atrophy in ALS. Clin. Neuropathol. 35, 22–30. doi: 10.5414/np300889
Tatura, R., Kraus, T., Giese, A., Arzberger, T., Buchholz, M., Hoglinger, G., et al. (2016). Parkinson’s disease: SNCA-, PARK2-, and LRRK2- targeting microRNAs elevated in cingulate gyrus. Parkinsonism Relat. Disord. 33, 115–121. doi: 10.1016/j.parkreldis.2016.09.028
Toivonen, J. M., Manzano, R., Olivan, S., Zaragoza, P., Garcia-Redondo, A., and Osta, R. (2014). MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One 9:e89065. doi: 10.1371/journal.pone.0089065
Tollervey, D., and Hurt, E. C. (1990). The role of small nucleolar ribonucleoproteins in ribosome synthesis. Mol. Biol. Rep. 14, 103–106.
Toth, K. F., Pezic, D., Stuwe, E., and Webster, A. (2016). The piRNA pathway guards the germline genome against transposable elements. Adv. Exp. Med. Biol. 886, 51–77. doi: 10.1007/978-94-017-7417-8_4
Tsuiji, H., Iguchi, Y., Furuya, A., Kataoka, A., Hatsuta, H., Atsuta, N., et al. (2013). Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol. Med. 5, 221–234. doi: 10.1002/emmm.201202303
Tycowski, K. T., You, Z. H., Graham, P. J., and Steitz, J. A. (1998). Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell 2, 629–638.
Vagnozzi, R., Signoretti, S., Tavazzi, B., Floris, R., Ludovici, A., Marziali, S., et al. (2008). Temporal window of metabolic brain vulnerability to concussion: a pilot 1H-magnetic resonance spectroscopic study in concussed athletes–part III. Neurosurgery 62, 1286–1295; discussion 1295–1296. doi: 10.1227/01.neu.0000333300.34189.74
Vallelunga, A., Ragusa, M., Di Mauro, S., Iannitti, T., Pilleri, M., Biundo, R., et al. (2014). Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and Multiple System Atrophy. Front. Cell. Neurosci. 8:156. doi: 10.3389/fncel.2014.00156
van Es, M. A., Schelhaas, H. J., van Vught, P. W., Ticozzi, N., Andersen, P. M., Groen, E. J., et al. (2011). Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann. Neurol. 70, 964–973. doi: 10.1002/ana.22611
Vazquez-Arango, P., Vowles, J., Browne, C., Hartfield, E., Fernandes, H. J., Mandefro, B., et al. (2016). Variant U1 snRNAs are implicated in human pluripotent stem cell maintenance and neuromuscular disease. Nucleic Acids Res. 44, 10960–10973. doi: 10.1093/nar/gkw711
Verma, B., Akinyi, M. V., Norppa, A. J., and Frilander, M. J. (2018). Minor spliceosome and disease. Semin. Cell Dev. Biol. 79, 103–112. doi: 10.1016/j.semcdb.2017.09.036
Villa, C., Ridolfi, E., Fenoglio, C., Ghezzi, L., Vimercati, R., Clerici, F., et al. (2013). Expression of the transcription factor Sp1 and its regulatory hsa-miR-29b in peripheral blood mononuclear cells from patients with Alzheimer’s disease. J. Alzheimers Dis. 35, 487–494. doi: 10.3233/jad-122263
Voineagu, I., Wang, X., Johnston, P., Lowe, J. K., Tian, Y., Horvath, S., et al. (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384. doi: 10.1038/nature10110
Wahl, M. C., Will, C. L., and Luhrmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. doi: 10.1016/j.cell.2009.02.009
Wake, C., Labadorf, A., Dumitriu, A., Hoss, A. G., Bregu, J., Albrecht, K. H., et al. (2016). Novel microRNA discovery using small RNA sequencing in post-mortem human brain. BMC Genomics 17:776. doi: 10.1186/s12864-016-3114-3
Waller, R., Goodall, E. F., Milo, M., Cooper-Knock, J., Da Costa, M., Hobson, E., et al. (2017a). Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging 55, 123–131. doi: 10.1016/j.neurobiolaging.2017.03.027
Waller, R., Wyles, M., Heath, P. R., Kazoka, M., Wollff, H., Shaw, P. J., et al. (2017b). Small RNA sequencing of sporadic amyotrophic lateral sclerosis cerebrospinal fluid reveals differentially expressed miRNAs related to neural and glial activity. Front. Neurosci. 11:731. doi: 10.3389/fnins.2017.00731
Wang, W. X., Rajeev, B. W., Stromberg, A. J., Ren, N., Tang, G., Huang, Q., et al. (2008). The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 28, 1213–1223. doi: 10.1523/jneurosci.5065-07.2008
Wright, C., Shin, J. H., Rajpurohit, A., Deep-Soboslay, A., Collado-Torres, L., Brandon, N. J., et al. (2017). Altered expression of histamine signaling genes in autism spectrum disorder. Transl. Psychiatry 7:e1126. doi: 10.1038/tp.2017.87
Wu, Y., Xu, J., Xu, J., Cheng, J., Jiao, D., Zhou, C., et al. (2017). Lower serum levels of miR-29c-3p and miR-19b-3p as biomarkers for Alzheimer’s disease. Tohoku J. Exp. Med. 242, 129–136. doi: 10.1620/tjem.242.129
Yamasaki, S., Ivanov, P., Hu, G. F., and Anderson, P. (2009). Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42. doi: 10.1083/jcb.200811106
Yan, Z., Hu, H. Y., Jiang, X., Maierhofer, V., Neb, E., He, L., et al. (2011). Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39, 6596–6607. doi: 10.1093/nar/gkr298
Yang, X., Cheng, Y., Lu, Q., Wei, J., Yang, H., and Gu, M. (2015). Detection of stably expressed piRNAs in human blood. Int. J. Clin. Exp. Med. 8, 13353–13358.
Yilmaz, S. G., Erdal, M. E., Ozge, A. A., and Sungur, M. A. (2016a). Can peripheral MicroRNA expression data serve as epigenomic (Upstream) biomarkers of Alzheimer’s disease? OMICS 20, 456–461. doi: 10.1089/omi.2016.0099
Yilmaz, S. G., Geyik, S., Neyal, A. M., Soko, N. D., Bozkurt, H., and Dandara, C. (2016b). Hypothesis: do miRNAs targeting the leucine-rich repeat kinase 2 gene (LRRK2) influence Parkinson’s disease susceptibility? OMICS 20, 224–228. doi: 10.1089/omi.2016.0040
Yin, S., Lopez-Gonzalez, R., Kunz, R. C., Gangopadhyay, J., Borufka, C., Gygi, S. P., et al. (2017). Evidence that C9ORF72 dipeptide repeat proteins associate with U2 snRNP to cause Mis-splicing in ALS/FTD Patients. Cell Rep. 19, 2244–2256. doi: 10.1016/j.celrep.2017.05.056
Zeng, Q., Zou, L., Qian, L., Zhou, F., Nie, H., Yu, S., et al. (2017). Expression of microRNA222 in serum of patients with Alzheimer’s disease. Mol. Med. Rep. 16, 5575–5579. doi: 10.3892/mmr.2017.7301
Zhang, T., Wu, Y. C., Mullane, P., Ji, Y. J., Liu, H., He, L., et al. (2018). FUS regulates activity of MicroRNA-mediated gene silencing. Mol. Cell 69, 787–801.e8. doi: 10.1016/j.molcel.2018.02.001
Zhang, Z., Lotti, F., Dittmar, K., Younis, I., Wan, L., Kasim, M., et al. (2008). SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600. doi: 10.1016/j.cell.2008.03.031
Zhang, Z., Pinto, A. M., Wan, L., Wang, W., Berg, M. G., Oliva, I., et al. (2013). Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 110, 19348–19353. doi: 10.1073/pnas.1319280110
Zhao, J., Yue, D., Zhou, Y., Jia, L., Wang, H., Guo, M., et al. (2017). The role of MicroRNAs in Abeta deposition and tau phosphorylation in Alzheimer’s disease. Front. Neurol. 8:342. doi: 10.3389/fneur.2017.00342
Keywords: small non-coding RNAs, microRNAs, neurodegenerative disease, biomarkers, new therapeutic targets
Citation: Watson CN, Belli A and Di Pietro V (2019) Small Non-coding RNAs: New Class of Biomarkers and Potential Therapeutic Targets in Neurodegenerative Disease. Front. Genet. 10:364. doi: 10.3389/fgene.2019.00364
Received: 14 January 2019; Accepted: 05 April 2019;
Published: 26 April 2019.
Edited by:
Yun Zheng, Kunming University of Science and Technology, ChinaReviewed by:
Mauricio Fernando Budini, Universidad de Chile, ChileChao Peng, University of Pennsylvania, United States
Copyright © 2019 Watson, Belli and Di Pietro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Callum N. Watson, Y253NzYzQGJoYW0uYWMudWs=
 Callum N. Watson
Callum N. Watson Antonio Belli
Antonio Belli Valentina Di Pietro
Valentina Di Pietro

