
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 24 April 2019
Sec. Nutrigenomics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00352
This article is part of the Research TopicRegulation and Manipulation of Nutrient-Controlling Genes in CropsView all 6 articles
Malting quality is an important determinant of the value of barley grain used in malting and brewing. With recent sequencing and assembling of the barley genome, an increasing number of quantitative trait loci (QTLs) and genes related to malting quality have been identified and cloned, which lays a good molecular genetic basis for barley quality improvement. In this review, we describe the following indicators of malting quality: malt extract (ME), diastatic power (DP), kolbach index (KI), wort viscosity (VIS), free amino nitrogen (FAN) content, soluble protein (SP) content, wort β-glucan (WBG) content, and protein content (PC), and have list related QTLs/genes with high phenotypic variation in multiple populations or environments. Meanwhile, the correlations among the quality parameters and parts of significant indicators suitable for improvement are discussed based on nutrient composition and content required for high-quality malt, which will provide reference for molecular marker-assisted selection (MAS) of malting quality in barley.
Barley (Hordeum vulgare) is the fourth largest cereal crop in the world, widely used for livestock feed, food, and industrial utilization (Bond et al., 2015; FAOSTAT1). In industrial applications, barley is processed into malt and mainly used for brewing and distilling, in which malting quality is an important factor in determining the quality of the manufactured products (Kochevenko et al., 2018). About 30% of the barley produced globally is used for malting, thus breeding barley varieties with high-quality malt for processing is an important goal (Bond et al., 2015; Walker and Panozzo, 2016; Kochevenko et al., 2018).
In barley endosperm, the nutrients such as starch and protein are stored and directly determine the barely quality (Bamforth, 2003; Jamar et al., 2011). During seed germination, the gibberellic acid (GA), released by the embryo, induces a large number of hydrolases in aleurone layer and begin to degrade the endosperm cell wall (Zentella et al., 2002; Bamforth, 2017). Then, many hydrolases enter into the endosperm cells and start to degrade proteins, starches, and lipids. In the procession, the conversion of nutrients has always continued, and low molecular weight sugars, amino acids, fatty acids, and enzymes are formed, which provide substances for subsequent fermentation. Among them, the amount and quality of the converted substances determine the malting quality (Autio et al., 2001; Georg-Kraemer et al., 2001; Bamforth, 2009).
In order to provide a genetic basis for the breeding of high-quality barley varieties, we focus on the known major quantitative trait loci (QTLs) related to malting quality and systematically analyze the association between the quality traits and the improvement prospects in this paper.
Malting quality is mainly determined by malt extract (ME), diastatic power (DP), viscosity (VIS), wort β-glucan (WBG) content, kolbach index (KI), free amino nitrogen (FAN) content, soluble protein (SP) content, and grain protein (GP) (Cu et al., 2016). ME, the ratio of extracted malt soluble matter to dry malt weight, is directly related to malt production, with a higher ME being required for better malting quality (Sarkar et al., 2008). DP represents the ability to hydrolyze starch to simple sugars during barley germination, which is directly proportional to the yield and quality of beer brewing (Henson and Duke, 2007). KI, also known as malt protein solubility, is an indispensable parameter of malting quality. Viscosity reflects the solubility and filtration speed of the malt wort, and low VIS value is an important indicator of high-quality malt. β-Glucan is the main component of the endosperm cell wall, and high concentration of WBG will hinder the hydrolysis process in malt. FAN content contains amino acids and small peptides from protein degradation by protease, and is the only nitrogen source for yeast growth. In addition, the protein content (PC) in the grain is also a significant factor influencing malting quality. All of these traits in combination affect the malting quality.
Besides the genetic factors, malting quality is also affected by environmental conditions (Qi et al., 2005; Chen et al., 2006), and there are complex inhibitory relationships among malting quality traits. For instance, the GP in the grains is negatively correlated with ME and positively correlated with DP (Eagles et al., 1995; Mather et al., 1997). Therefore, it is difficult to directly locate and clone the malt quality-related genes. However, as an effective strategy, QTL analysis had been widely used in identification and localization of QTLs in different crops, but large genome size and high homology limited polymorphic DNA markers development for the establishment of the genetic map in barley. Excitingly, with the completion of the assembly of the barley genome, many QTLs regulating malting quality traits have been located in smaller intervals using various genetic markers, and some essential genes have been cloned. Up to now, More than 200 malting quality QTLs/genes have been reported (Wei et al., 2009), but only a small number of QTLs/genes have been successfully applied in molecular breeding (Han et al., 1997; Igartua et al., 2000; Rae et al., 2007; Li et al., 2010; Xu et al., 2018). One reason may be the population size, thus the linkage of unfavorable genetic traits will reduce the accuracy of QTL screening in small populations (Zhang et al., 2012; Cu et al., 2016). On the other hand, the QTLs accounting for lower phenotypic variation are easily affected by environmental factors, which are inconvenient for breeding selection.
Malt extract includes the soluble matters produced by malt itself and the enzymatic hydrolysis during saccharification and fermentation, which reflects the degree of malt dissolution and the amount of enzyme formation in the malting process. ME is quantitative trait controlled by multiple genes, which also vary among different varieties. At present, a number of QTLs related to ME with high variances have been identified and located on chromosomes 1, 2, 4, 5, and 7 (Table 1).
Using a double haploid (DH) population, Wang J. et al. (2015) identified two QTLs, in which the QMe.NaTx-2H was mapped in the 24–35 cM region of chromosome 2 (2H) and explained 48.4% of the total phenotypic variance. A cell wall hydrolytic enzyme, endo-1,4-xylanase A (MLOC_60943.2), was found near the marker GBM1121, which was closely linked to QMe.NaTx-2H. The main function of this gene is to degrade the endosperm cell wall and facilitate other substances more easily digestible in the cells, so it is suspected to be the target gene regulating ME. Another QTL, detected on 1H, contributed little to phenotypic variance and was not identified in other environments. Laidò et al. (2009) detected a QTL Qme1.1 on 1H, which was positioned at 60.3 cM and explained 21.1% of the phenotypic variance. Using different population, a ME-associate QTL, with extremely high phenotypic variance, was also detected in this region, which verifies that there is a major QTL for ME in the interval (Panozzo et al., 2007; Laidò et al., 2009). Matthies et al. (2014) analyzed 174 European barley cultivars by genome-wide association analysis (GWAS) and also identified a major QTL near Qme1.1. They found that this QTL was not only related to ME but also regulated VIS. Elía et al. (2010) and Wang J. et al. (2015) mapped two QTLs on short and long arm of 2H, respectively, both of which exhibited high phenotypic variances. Singh et al. (2017) cloned a major QTL thaumatin-like protein 8 (TLP8) near the telomeric region of 4H, which acts on β-glucan through redox reaction, thereby affecting the ME. In addition, two closely spaced QTLs were also detected on 4H, explaining approximately 8–13% and 4–10% of the variances, respectively (Walker et al., 2013). On 5H, two QTLs for ME were identified, and one accounted for 35.7–53.6% of the variance (Elía et al., 2010; Zhou et al., 2012).
Diastatic power is a critical parameter of malting quality. In general, a higher DP is required for better malting quality and a higher ME (Henson and Duke, 2007). The DP of barley malt represents the collective activity of four starch-degrading enzymes, namely α-amylase, β-amylase, limit dextrinase, and α-glucosidase (Gibson et al., 1995; Walker and Panozzo, 2016). The conversion of starch to fermentable products in the endosperm is primarily catalyzed by α-amylase, followed by β-amylase, limit dextrinase, and α-glucosidase (Bamforth, 2009). During this process, DP is significantly positively correlated with amylase activity, which can be determined by measuring the activity of amylase. Therefore, the identification of QTLs/genes related to amylase activity and its application in breeding is one effective means of improving malting quality.
Diastatic power, not highly influenced by environmental conditions, is mainly determined by genetic factors and easier to improve. In recent years, multiple major DP-related QTLs have been mapped or cloned using positional cloning and comparative genomics method (Table 1). Eight major QTLs influencing α-amylase activity have been listed, and two of which, encoding α-amylase1 and α-amylase2, have been cloned (Elía et al., 2010; GrainGenes 3.0). By SNP-based maps, von Korff et al. (2008) identified three QTLs and each accounted for about 10% of phenotypic variances. Two major QTLs for α-amylase were located on 5H, and explained 25.6% and 12.4% of the phenotypic variances, respectively (Mohammadi et al., 2015; Zhou et al., 2016). Cu et al. (2016) mapped three QTLs that significantly increased DP on 1H and 4H. The QTL, located on 1H at 50.56 cM, accounted for 15% of the variance and could critically increase α-amylase activity. Another two QTLs, which significantly increased the β-amylase activity, were identified on 4H and contributed to 11.89% and 30.74–49.66% of the phenotypic variances, respectively. The QTL linked to bPb-9820, which encodes β-amylase 1, has been cloned and can be used for improving malting quality (Cu et al., 2016). Another cloned β-amylase gene is β-amylase 2, which was located at 64.68 cM on 2H (Elía et al., 2010). QTL qBAM3, mapped on 3H between markers bPb-4564 and bPb-3634, was also linked to β-amylase activity and explained 12.81% of the phenotypic variance (Wei et al., 2009).
α-Glucosidase is an essential enzyme in the starch degradation pathway, and four α-glucosidase genes have been cloned. A gene immediately related to DP was found at 4.7 cM on 1H by GWAS. Aglu3, encoding an α-glucosidase, was found in this interval and contributes to the conversion of gelatinized starch and glucan to sugars; however, the genetic effect of Aglu3 is only 1.57% (Pauli et al., 2015). HvAGL197,α-glucosidase-related gene, is involved in the conversion of maltose to glucose instead of starch degradation (Stanley et al., 2011). Aglu2 and Aglu5, another two α-glucosidase-related genes, were located on 3H and 2H (Szűcs et al., 2009; Pauli et al., 2015). Two major QTLs controlling limit dextrinase activity were positioned on 5H and 7H, respectively. The QTL qLD5 was located on 5H at 174–178.4 cM, explaining 19.4% of the phenotypic variance (Wei et al., 2009). The last QTL was positioned at 58.7 cM on 7H and accounted for 31.0% of the phenotypic variance, which is significantly higher compared to the other two QTLs (Wang X. et al., 2015).
Kolbach index is typically measured as the ratio of soluble nitrogen to total nitrogen in the wort. In brewing applications, the degree of protein degradation in barley malt will have distinct effects on yeast growth and wort filtration. When the degree of protein degradation is low, the corresponding enzyme activity is also reduced, resulting in lower ME, protein turbidity, and wort filtration difficulty. When decomposition is high, the corresponding KI is also increased and the normal proportion of protein components is compromised, resulting in accelerated yeast aging and thin beer taste. Therefore, the KI of elite malt should be controlled between 41 and 48% (Molina-Cano et al., 1997).
Although KI is affected by environmental conditions, the genetic background of different varieties is also significant. Using genetic populations, QTLs contributing to high variances were identified on 1–7H under multiple environmental conditions (Elía et al., 2010; Zhou et al., 2012; Islamovic et al., 2014; Wang J. et al., 2015; Kochevenko et al., 2018) (Table 1). Among them, the QTL on 5H was positioned at an interval of 141–150 cM and accounted for 56.8–77.5% of the variance, which is much higher than that on other chromosomes (Wang J. et al., 2015). Secondly, QTL, on 6H at 57.20 cM, explained 23.40% of the phenotypic variance (Islamovic et al., 2014). These two major QTLs have great potential in breeding applications. However, no KI-related genes have thus far been cloned.
The main components of the cell wall of barley endosperm are non-starch polysaccharides, arabinoxylan, and β-glucan, which can form highly viscous solutions, reducing the leaching rate of ME, filtration rate, and the finished beer quality (Jamar et al., 2011). So, low VIS in malt is necessary for high malting quality. Although β-glucan is a primary component in cell wall, it is not the only parameter controlling VIS. At present, a number of QTLs regulating VIS have been identified (Table 1). Laidò et al. (2009) mapped two major QTLs, loci Qvis1.1 and Qvis5.1, which were located on 1H at 60.3 cM and 5H at 137.5 cM, and explained as high as 39.5% and 24.3% of the variances, respectively. von Korff et al. (2008) also detected a QTL on 1H at 68 cM closely to Qvis1.1. Kochevenko et al. (2018) positioned a QTL QVIS-3 on 3H (63.66 cM) explaining 13.82% of the phenotypic variance, and another QTL with slight phenotypic variance was found at the top of 4H. Wang J. et al. (2015) discovered locus QVi.NaTx-7H at 20.79–27.87 cM on 7H, contributing to 14.5% of the phenotypic variance. In addition, locus QVi.NaTx-1H, contributing to 17.8% of the variance, was detected at 61.15 cM on 1H, which may be the same QTL as Qvis1.1 (Wang J. et al., 2015).
β-Glucan is mainly distributed in the aleurone layer and endosperm cell wall, accounting for 75% of the endosperm cell wall composition (Jamar et al., 2011). In the process of barley malt production and beer brewing, the incomplete degradation of endosperm cell wall will cause excessive WBG, which would influence the expansion of hydrolase and protease into the malt cells and decrease the extract content in the wort (Bamforth, 2003; Li et al., 2010; Bamforth, 2017). Meanwhile, excessive residual β-glucan in the malt will lead to an increase of VIS, which is not conducive to the filtration of wort and beer, and results in reduced beer quality (Vis and Lorenz, 1998; Bamforth, 2003). Therefore, reducing the VIS and improving the filterability of beer is significant for breeders and brewers.
Although WBG is affected by both genotypic and environmental factors, the genetic background is more significant (Jamar et al., 2011), and multiple QTLs for WBG have been identified using genetic populations (Table 1). Panozzo et al. (2007) located a QTL on 1H at 59 cM, accounting for 36.0–52.8% of the variance. Cu et al. (2016) located a QTL on 1H at 50.56 cM that explained 13.92% of the variance. Mohammadi et al. (2015) also detected a QTL on 1H at 58 cM using six-row spring breeding lines, which resulted in high reduction of WBG. In fact, the genetic distances of the three QTLs are relatively close, suggesting that they may be the same QTL. In addition, a genetic locus affecting WBG was positioned on 2H at 65.6 cM, which overlapped with the QTL interval (Adh8-ABGOI9) controlling grain β-glucan content. It is speculated that these two loci constitute the same QTL and can control both wort and grain β-glucan content (Han et al., 1995; Panozzo et al., 2007). Seventy-seven lines were evaluated for malting quality and selected for WBG analysis, and a QTL was mapped on 5H at 187 cM that can significantly reduce WBG (Mohammadi et al., 2015). Using the DH population from the Japanese barley variety “Mikamo Golden” and the North American variety “Harrington,” Zhou et al. (2012) identified a QTL on 6H at 35–55 cM and explained 17.2% of the variance. Kim et al. (2011) detected two QTLs on 7H using F5-derived lines, and accounted for 44.4% and 31.6–37.6% of the variances, respectively. Several genes have been cloned in barley grains by comparative genomics method, such as the CslF gene cluster on 2H and HvCslF6 on 7H, which are involved in the synthesis of β-glucan (Han et al., 1995; Burton et al., 2006; Taketa et al., 2011). Loss of function of CslF genes can decrease the β-glucan content significantly.
Malt β-glucanase plays a critical role in the degradation of β-glucan during malting process (McCleary and Shameer, 1987; Han et al., 1995). Two β-glucanase genes, Glb 1 and Glb 2, were cloned from barley malt and located on 5H and 1H, explaining 12.3% and 10.8% of the variances, respectively (Han et al., 1995; GrainGenes 3.02). In addition, Han et al. (1995) detected a QTL on 4H at 79.5 cM that explained 12.6% of the variance. Panozzo et al. (2007) detected two major QTLs, one was detected on 1H at 50.9 cM, and the other was mapped on 4H at 22.9 cM. The major QTLs on 4H were detected under a variety of conditions, which indicated that the effect of environment on these QTLs was relatively low.
Free amino nitrogen is the only nitrogen source for yeast cell growth and reproduction, and its content in the wort plays a decisive role in yeast growth, synthesis, and metabolite changes (Stewart et al., 2013). FAN not only provides nutrition for yeast during beer fermentation, but also constitutes the flavor substance of beer. Thus, FAN is a significant indicator of beer quality. Although high PC in the grains can increase the FAN content, it also leads to a decrease of ME (Qi et al., 2005). Therefore, FAN in the wort is generally maintained at 180–220 mg/L.
In the genetic analysis of FAN (Table 1), Panozzo et al. (2007) detected four major QTLs, of which two were from the same DH lines linked to the Bmag0211 and Bmag0345 loci on 1H, which contributed to as high as 64.0% and 39–60% of the phenotypic variances, respectively. In another DH population, Panozzo et al. (2007) also identified two loci on 3H and 5H, with a lower variance compared with the first DH population. Cu et al. (2016) mapped two major QTLs, and one was located at 50.56 cM on 1H, near the Bmag0211 interval, another one was narrowed to an interval of 66.16–73.26 cM on 7H (Cu et al., 2016). Islamovic et al. (2014) positioned a major QTL on 7H at 34 cM, which was overlapped with QAn.NaTx-7H, a QTL located by Wang J. et al. (2015). The QTL on this location explained about 15% of the phenotypic variance at two different isolation populations.
Soluble protein content in the wort is a parameter for evaluating wort quality. It affects the nutritional composition, flavors, foam, and abiotic stability of beer in the brewing process.
A number of QTLs for SP have been identified using different genetic populations, and the QTLs with higher contribution rates were detected on 1, 3, 5, and 7H (Table 1). Matthies et al. (2014) analyzed 174 European barley cultivars by GWAS and identified two QTLs, located on 1H at 94.9 cM, and 5H at 184.4 cM, respectively. A QTL was detected on 1H at 50.56 cM under different environmental conditions, and the QTL linkage marker 009-148 was not only linked to the traits controlling SP, but also related to FAN and KI. It is speculated that this region should be a critical site for malting quality (Cu et al., 2016). In addition, three major QTLs, contributing to 27.31%, 33.6%, and 29% of the phenotypic variances, were identified on 3H, 5H, and 7H, respectively (Mather et al., 1997; Zhou et al., 2016; Kochevenko et al., 2018). These reported QTLs contribute high variances and can be used for marker-assisted selection (MAS) improving the SP content of the malt.
Protein is one of the main components of malt products. In the malting process, excessive GP will reduce ME, increase wort VIS, and decrease beer stability. At the same time, proteolysis can provide the only nitrogen source for yeast growth and various hydrolytic enzymes for starch degradation, and thus the PC of high-quality barley is generally 9–12% (Mather et al., 1997).
Protein content is extremely susceptible to environmental factors, and up to now, only a few QTLs with high contribution rates for malt protein content (MPC) and grain protein content (GPC) have been reported (Mather et al., 1997) (Table 1). Using a DH population, only qPC2.1 and qPC7.1, associated with MPC, were identified on 2H and 7H, and explained 14.72% and 12.02% of the variances, respectively (Li et al., 2005). Although there is no significant difference between the total protein content in the barley grains and in the malt, the proportion of protein components is altered following germination (Celus et al., 2006), and more QTLs for GPC are detected (Table 1). Laidò et al. (2009) located two QTLs, qPC1.1 and qPC6.1, on 1H and 6H that explained 29.1% and 18.7% of the variance, respectively. Using two DH populations, Elía et al. (2010) mapped seven QTLs on 1, 2, 3, 5, and 7H, explaining 13.4% to 62.3% of the variance. However, only three were identified in more than two environmental conditions, and a QTL at 82 cM on 2H was detected in all the planting conditions, explaining an average of 54% of the variance (Elía et al., 2010).
The genetic elements controlling malting quality traits interact with each other to form a regulatory network that determines the malting quality (Figure 1). Among them, GPC has a great influence on the hydrolase activity, malt saccharification, beer fermentation and the biostability of the finished beer (Bond et al., 2015). In addition, GPC is also an essential factor regulating the leaching rate of ME. These two indicators are negatively correlated, and a high GPC can lead to reduced ME in the malt (Qi et al., 2005; Mohammadi et al., 2015). During the hydrolysis of malt starch, GPC can provide an abundance of enzymes for starch degradation, including α-amylase and β-amylase (Mather et al., 1997; Elía et al., 2010), and thus GPC is positively correlated with DP, but negatively correlated with KI. During germination, proteins in the grains are decomposed into amino acids by protease, which can provide a nitrogen source for the growth of beer yeast, but the correlation between GPC and FAN is low (Stewart et al., 2013; Pauli et al., 2015; Cu et al., 2016).
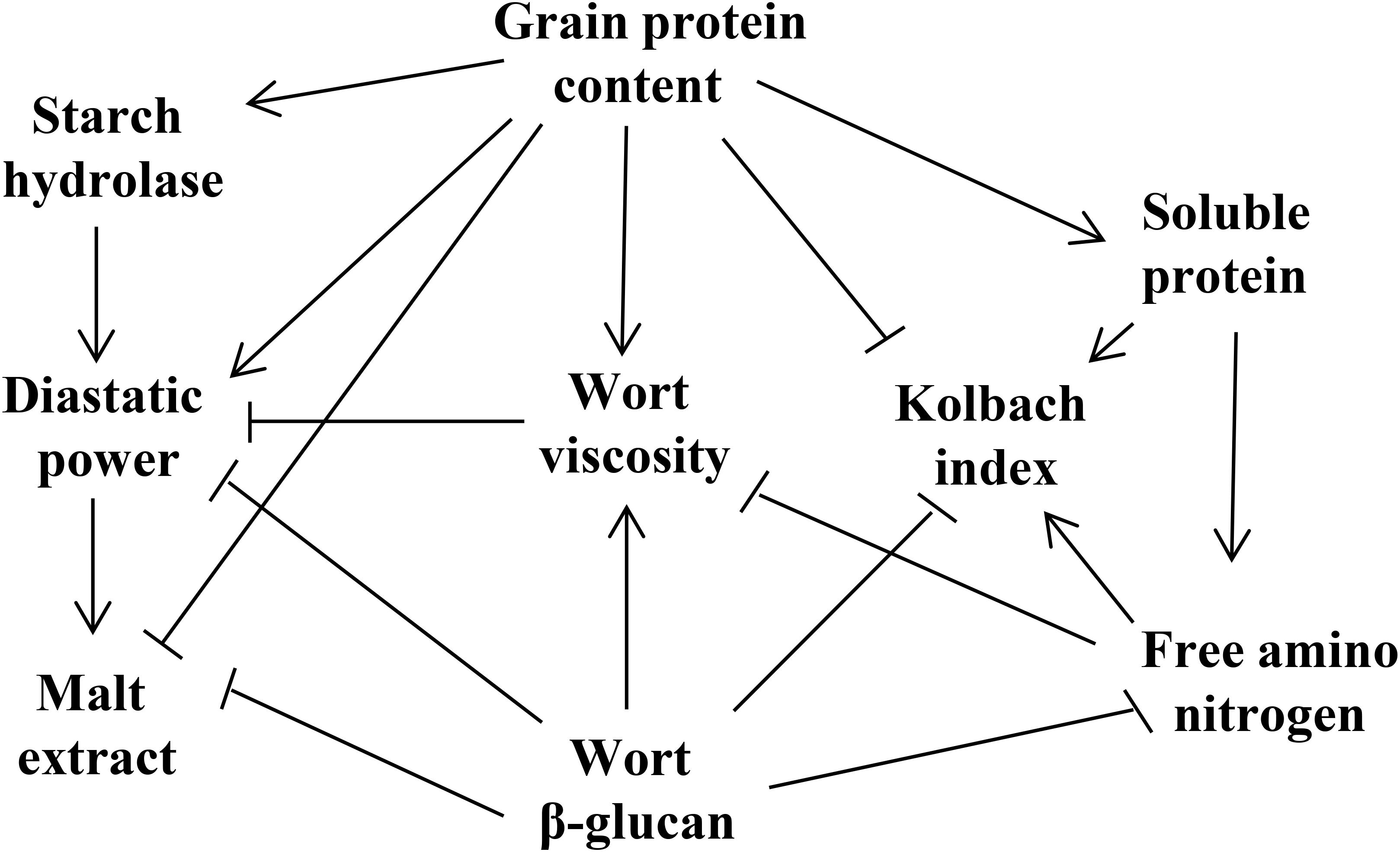
Figure 1. The correlation of malting quality traits. Arrows indicate positive interactions between parameters, and terminally barred lines indicate negative interactions.
Soluble protein, derived from GP, produces various amino acids and small molecular peptides following degradation. Therefore, SP is significantly positively interacted with GPC, FAN, and KI (Cu et al., 2016). FAN is mainly used in the subsequent beer fermentation process. Although FAN is negatively interacted with VIS and WBG, there is no direct relation with the two traits. Thus, it is supposed that FAN may indirectly influence VIS and WBG by altering the PC. KI was found to be negatively interacted with GPC and VIS, and positively correlated with SP and FAN with a high correlation coefficient (Wang J. et al., 2015; Cu et al., 2016).
Wort β-glucan is negatively interacted with most malting quality traits (Pauli et al., 2015). Excessive WBG is the residue of the incomplete degradation of cell wall, which may affect the expansion of various hydrolases in the germinated grains and reduce the ME content. Additionally, WBG is also significantly negatively correlated with FAN, KI, and SP, and positively interacted with GPC (Pauli et al., 2015). High-quality malt requires lower wort VIS, and excessive residues of β-glucan will increase wort VIS and lead to filtration issues (Bamforth, 2003). Thus, WBG is significantly positively correlated with VIS, both of which are unfavorable traits for improving malting quality. In addition, a high VIS will result in increased beer turbidity (Wei et al., 2009).
Diastatic power, representation of the starch hydrolase activity, is positively correlated with starch hydrolases (Gibson et al., 1995; Mohammadi et al., 2015). In addition, DP is positively interacted with GPC, FAN, and ME, but significantly negatively correlated with WBG and VIS (Emebiri et al., 2004; Pauli et al., 2015).
Malting quality in barley is a comprehensive reflection of many indicators, which are easily influenced by environmental conditions. However, there are also differences among varieties. For example, Molina-Cano et al. (1997) detected ME, wort VIS, KI, and DP in numerous varieties and found that there were highly significant differences among the four quality traits, of which VIS and ME were particularly correlated with the genotype. High-quality malt requires high ME and DP, optimal PC, and low WBG. However, it is difficult to improve multiple traits simultaneously in breeding because of a longer breeding cycles, so improving single or several traits is necessary during the improvement process.
In this review, we listed the QTLs/genes identified in recent years and located them on seven chromosomes (Table 1 and Figure 2), and found that most QTLs/genes controlling malting quality were mainly positioned in the intervals of 1H, 4H, 5H, and 7H, which indicates that malting quality traits with high phenotypic variances may prefer to cluster in these regions. Though improvement of a few traits was considered as the first choice in the past, substitution of a large interval containing elite traits in barley will be also feasible in the future, which can achieve the breeding programs in short term.
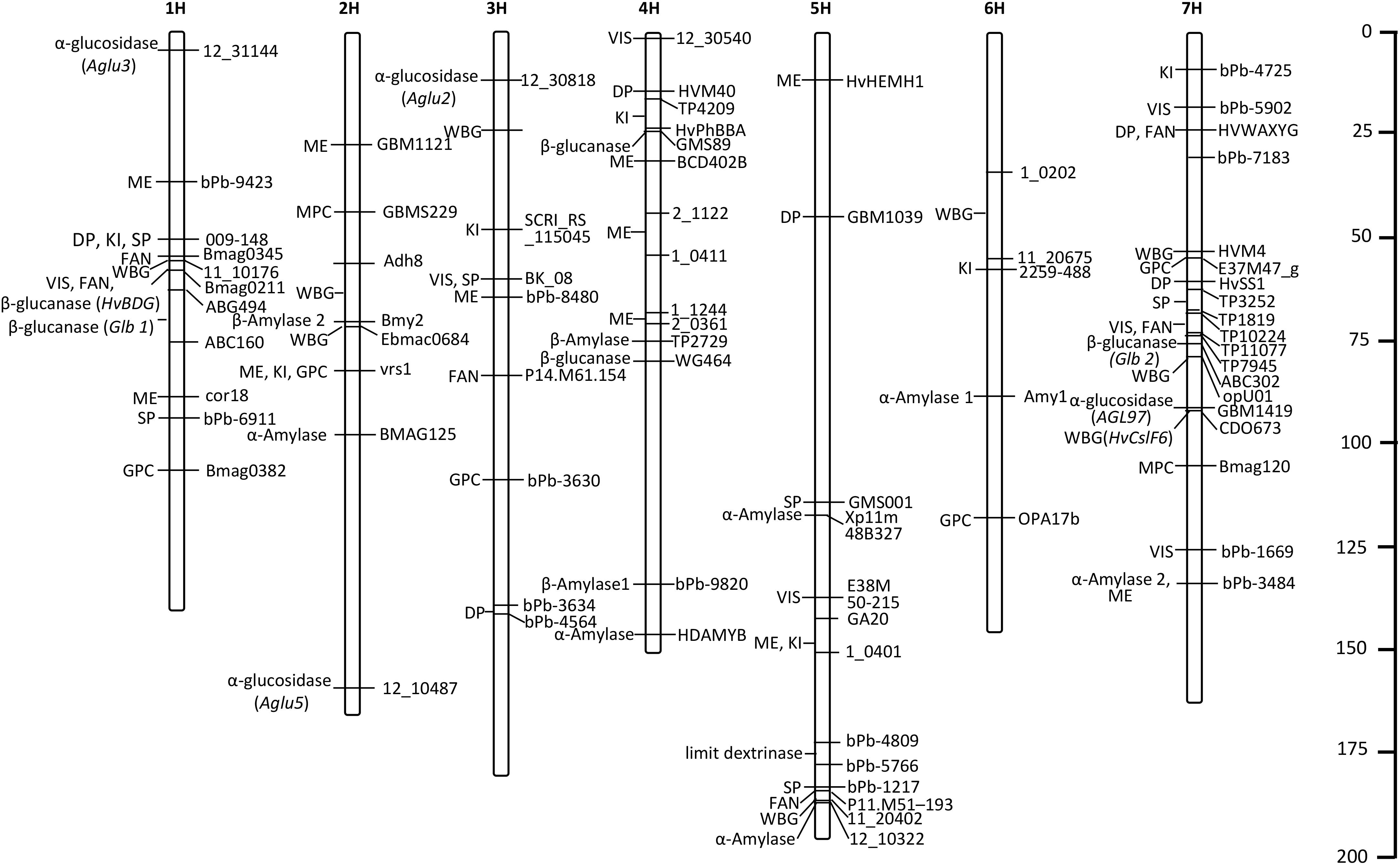
Figure 2. The distributions of QTLs/genes accounting for high phenotypic variances. ME, malt extract; DP, diastatic power; VIS, wort viscosity; KI, kolbach index; FAN, free amino nitrogen; SP, soluble protein; WBG, wort β-glucan; MPC, malt protein content; GPC, grain protein content.
Previous studies have provided valuable experience for traits improvement. ME mainly influenced by the genetic background, is the most significant element determining beer yield in the malting process, thus it is a quantitative trait that is extremely easy to improve (Sarkar et al., 2008). DP is also proportional to the yield and quality of beer brewing (Henson and Duke, 2007), and the correlation coefficient between DP and the three main amylases, α-amylase, β-amylase, and limit dextrinase, was as high as 0.79 (Gibson et al., 1995). Among these, β-amylase is the most important enzyme affecting DP, followed by α-amylase (Clancy et al., 2003; Henson and Duke, 2007). Therefore, the applications of these two hydrolases can significantly improve the DP level. During malting, GPC is not only altered by its own genetic basis, but also more susceptible to environment. It is necessary to coordinate the relationship between GPC and malting quality through genetic improvement or cultivation (Mather et al., 1997). WBG is negatively correlated with most barley quality traits, and reducing its content is also favorable. The cultivation of barley varieties with low BG content or high β-glucanase activity will facilitate to reduce the β-glucan content in the wort.
In our review, more than 60 QTLs/genes were listed, in which 15 genes have been cloned, and others were located in intervals between genetic markers. In molecular breeding, genes with high heritability will be better than identified QTLs because selection targets are more accurate and clear. Secondly, QTLs explaining high variances will also be chosen for high-quality breeding, because these QTLs will not be seriously influenced by the environment or other traits. Therefore, DP and WBG must be better choices for the improvement of malting quality, then VIS should be considered according to Table 1. In addition, the proportion of malting quality parameters in barley breeding is different, due to the standards set by different countries or regions. Breeders and scientists in different countries should adjust their strategies to improve malting quality according to their own national industrial standards and purposes (European Brewery Convention, 1998; Laidò et al., 2009; Punda, 2009).
The corresponding phenotypes of malting quality traits in barley are affected by both genetic and environmental elements, and positive and negative interactions among these traits are also present, which increase the difficulty for breeding application. Barley, a diploid cereal crop, has a large genome size of 5.1 Gb and highly repetitive DNA composition (Mascher et al., 2017), which also adds interference from multiple micro-effective homologous genes. The use of conventional breeding techniques to improve malting quality will face a series of difficulties, such as low selection efficiency and longer selection cycles, and it is difficult to aggregate multiple quantitative trait genes. With the discovery of regulatory mechanisms of major malting quality traits, combining MAS with conventional techniques to construct economical and efficient molecular breeding technology systems has become an important research direction in malt barley breeding.
Marker-assisted selection is convenient for breeding of malting barley and have been applied successfully. For example, Xu et al. (2018) transferred a thermostable β-amylase from wild barley into a commercial variety, and identified several elite lines with high DP. In the barley MAS procedures, elite barley varieties without serious defects and donor parent containing favorable alleles should be selected for the improvement of malting quality. Then, polymorphic markers closely linked to the alleles needs to be designed for the identification of hybrid plants. Hybridization and backcross must be carried out for transferring alleles from donor parents to elite varieties, and MAS will be implemented for selection of superior alleles in the segregating population.
In this review, we analyze the main malting quality indicators, ME, DP, VIS, KOL, FAN, GPC, SP, and WBG, and their correlation with each other. In addition, we also find that starch hydrolase, β-glucan, and PC have greatest impact on malting quality and play important roles in the regulatory network. Although the target traits can be selected quickly by MAS, but it also may be difficult to improve the traits controlled by QTLs with a low genetic contribution. Meanwhile, these traits are easily affected by environmental factors, which may weaken the improvement effect. Here, we list the major QTLs/genes that regulate the eight indicators (Table 1 and Figure 2), particularly those major loci that contribute to large phenotypic variances and have been detected in multiple populations or environmental conditions, which may provide more genetic information to breeders for facilitating the targeted improvement of malting quality.
DX and YF conceived the work and wrote the manuscript. YF and XZ prepared the figures and tables.
This study was funded by the National Natural Science Foundation of China (Grant No. 31401316) to XZ, State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (Grant No. 2010DS700124-KF1913) and National Key Technology Research and Development Program (Grant No. 2015BAD01B02) to DX.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript. We would also like to thank Professor Longbiao Guo and Mr. David Randall for improving English language.
Andriotis, V. M., Saalbach, G., Waugh, R., Field, R. A., and Smith, A. M. (2016). The maltase involved in starch metabolism in barley endosperm is encoded by a single gene. PLoS One 11:e0151642. doi: 10.1371/journal.pone.0151642
Autio, K., Simoinen, T., Suortti, T., Salmenkallio-Marttila, M., Lassila, K., and Wilhelmson, A. (2001). Structural and enzymic changes in germinated barley and rye. J. Inst. Brew. 107, 19–25. doi: 10.1002/j.2050-0416.2001.tb00075.x
Bamforth, C. W. (2003). Barley and malt starch in brewing: a general review. Tech. Q. Master Brew. Assoc. Am. 40, 89–97.
Bamforth, C. W. (2009). Current perspectives on the role of enzymes in brewing. J. Cereal Sci. 50, 353–357. doi: 10.1016/j.jcs.2009.03.001
Bamforth, C. W. (2017). Progress in brewing science and beer production. Annu. Rev. Chem. Biomol. Eng. 8, 161–176. doi: 10.1146/annurev-chembioeng-060816-101450.
Bond, J., Capehart, T., Allen, E., and Kim, G. (2015). Boutique Brews, Barley, and the Balance Sheet: Changes in Malt Barley Industrial Use Require an Updated Forecasting Approach. Washington, DC: Economic Research Division, United Stated Department of Agriculture, 18–23.
Burton, R. A., Wilson, S. M., Hrmova, M., Harvey, A. J., Shirley, N. J., Medhurst, A., et al. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311, 1940–1942. doi: 10.1126/science.1122975
Celus, I., Brijs, K., and Delcour, J. A. (2006). The effects of malting and mashing on barley protein extractability. J. Cereal Sci. 44, 203–211. doi: 10.1016/j.jcs.2006.06.003
Chen, J., Dai, F., Wei, K., and Zhang, G. P. (2006). Relationship between malt qualities and β-amylase activity and protein content as affected by timing of nitrogen fertilizer application. J. Zhejiang Univ. Sci. B 7, 79–84. doi: 10.1631/jzus.2006.B0079
Clancy, J. A., Han, F., and Ullrich, S. E. (2003). Comparative mapping of beta-amylase activity loci among three barley crosses. Crop Sci. 43, 1043–1052. doi: 10.2135/cropsci2003.1043
Cu, S. T., March, T. J., Stewart, S., Degner, S., Coventry, S., Box, A., et al. (2016). Genetic analysis of grain and malt quality in an elite barley population. Mol. Breed. 36:129. doi: 10.1007/s11032-016-0554-z
Eagles, H. A., Bedggood, A. G., Panozzo, J. F., and Martin, P. J. (1995). Cultivar and environmental effects on malting quality in barley. Aust. J. Agric. Res. 46, 831–844. doi: 10.1071/AR9950831
Elía, M., Swanston, J. S., Moralejo, M., Casas, A., Pérez-Vendrell, A. M., Ciudad, F. J., et al. (2010). A model of the genetic differences in malting quality between European and North American barley cultivars based on a QTL study of the cross Triumph × Morex. Plant Breed. 129, 280–290. doi: 10.1111/j.1439-0523.2009.01694.x
Emebiri, L. C., Moody, D. B., Panozzo, J. F., and Read, B. J. (2004). Mapping of QTL for malting quality attributes in barley based on a cross of parents with low grain protein concentration. Field Crops Res. 87, 195–205. doi: 10.1016/j.fcr.2003.11.002
European Brewery Convention (1998). ‘Analytica EBC’. Method 4.5. Congress Method. Nurnberg: Fachverlag Hans Carl.
Gao, W., Clancy, J. A., Han, F., Jones, B. L., Budde, A., Wesenberg, D. M., et al. (2004). Fine mapping of a malting-quality QTL complex near the chromosome 4H S telomere in barley. Theor. Appl. Genet. 109, 750–760. doi: 10.1007/s00122-004-1688-7
Georg-Kraemer, J. E., Mundstock, E. C., and Cavalli-Molina, S. (2001). Developmental expression of amylases during barley malting. J. Cereal Sci. 33, 279–288. doi: 10.1016/j.plaphy.2010.09.019
Gibson, T. S., Solah, V., Holmes, M. R. G., and Taylor, H. R. (1995). Diastatic power in malted barley: contributions of malt parameters to its development and the potential of barley grain beta-amylase to predict malt diastatic power. J. Inst. Brew. 101, 277–280. doi: 10.1002/j.2050-0416.1995.tb00867.x
Han, F., Romagosa, I., Ullrich, S. E., Jones, B. L., Hayes, P. M., and Wesenberg, D. M. (1997). Molecular marker-assisted selection for malting quality traits in barley. Mol. Breed. 3, 427–437. doi: 10.1023/A:1009608312385
Han, F., Ullrich, S. E., Chirat, S., Menteur, S., Jesrin, L., Sarrafi, A., et al. (1995). Mapping of β-glucan content and β-glucanase activity loci in barley grain and malt. Theor. Appl. Genet. 91, 921–927. doi: 10.1007/BF00223901
Henson, C. A., and Duke, S. H. (2007). Osmolyte concentration as an indicator of malt quality. J. Am. Soc. Brew. Chem. 65, 59–62. doi: 10.1094/ASBCJ-2007-0112-01
Igartua, E., Edney, M., Rossnagel, B. G., Spaner, D., Legge, W. G., Scoles, G. J., et al. (2000). Marker-based selection of QTL affecting grain and malt quality in two-row barley. Crop Sci. 40, 1426–1433. doi: 10.2135/cropsci2000.4051426x
Islamovic, E., Obert, D. E., Budde, A. D., Schmitt, M., Brunick, R., Kilian, A., et al. (2014). Quantitative trait loci of barley malting quality trait components in the Stellar/01Ab8219 mapping population. Mol. Breed. 34, 59–73. doi: 10.1007/s11032-014-0017-3
Jamar, C., du Jardin, P., and Fauconnier, M. L. (2011). Cell wall polysaccharides hydrolysis of malting barley (Hordeum vulgare L.): a review. Biotechnol. Agron. Soc. Environ. 15, 301–313.
Kim, H. S., Park, K. G., Baek, S. B., and Kim, J. G. (2011). Inheritance of (1-3)(1-4)-Beta-D-Glucan content in barley (Hordeum vulgare L.). J. Crop Sci. Biotechnol. 14, 239–245. doi: 10.1007/s12892-011-0045-z
Kochevenko, A., Jiang, Y., Seiler, C., Surdonja, K., Kollers, S., Reif, J. C., et al. (2018). Identification of QTL hot spots for malting quality in two elite breeding lines with distinct tolerance to abiotic stress. BMC Plant Biol. 18:106. doi: 10.1186/s12870-018-1323-4
Laidò, G., Barabaschi, D., Tondelli, A., Gianinetti, A., Stanca, A. M., Li Destri Nicosia, O., et al. (2009). QTL alleles from a winter feed type can improve malting quality in barley. Plant Breed. 128, 598–605. doi: 10.1111/j.1439-0523.2009.01636.x
Li, C., Cakir, M., and Lance, R. (2010). “Genetic improvement of malting quality through conventional breeding and marker-assisted selection,” in Genetics and Improvement of Barley Malt Quality, eds G. Zhang and C. Li (Berlin: Springer), 260–292.
Li, J. Z., Huang, X. Q., Heinrichs, F., Ganal, M. W., and Röder, M. S. (2005). Analysis of QTLs for yield, yield components, and malting quality in a BC3-DH population of spring barley. Theor. Appl. Genet. 110, 356–363. doi: 10.1007/s00122-004-1847-x
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Mather, D. E., Tinker, N. A., LaBerge, D. E., Edney, M., Jones, B. L., Rossnagel, B. G., et al. (1997). Regions of the genome that affect grain and malt quality in a North American two-row barley cross. Crop Sci. 37, 544–554. doi: 10.2135/cropsci1997.0011183X003700020039x
Matthies, I. E., Malosetti, M., Röder, M. S., and van Eeuwijk, F. (2014). Genome-wide association mapping for kernel and malting quality traits using historical European barley records. PLoS One 9:e110046. doi: 10.1371/journal.pone.0110046
McCleary, M. V., and Shameer, I. (1987). Assay of malt-β-glucanase using azobarley glucan: an improved precipitant. J. Inst. Brew. 93, 87–90. doi: 10.1002/j.2050-0416.1987.tb04481.x
Mohammadi, M., Blake, T. K., Budde, A. D., Chao, S., Hayes, P. M., Horsley, R. D., et al. (2015). A genome-wide association study of malting quality across eight US barley breeding programs. Theor. Appl. Genet. 128, 705–721. doi: 10.1007/s00122-015-2465-5
Molina-Cano, J., Francesch, M., Perez-Vendrell, A. M., Ramo, T., Voltas, J., and Brufau, J. (1997). Genetic and environmental variation in malting and feed quality of barley. J. Cereal Sci. 25, 37–47. doi: 10.1007/s00122-015-2481-5
Panozzo, J. F., Eckermann, P. J., Mather, D. E., Moody, D. B., Black, C. K., Collins, H. M., et al. (2007). QTL analysis of malting quality traits in two barley populations. Aust. J. Agric. Res. 58, 858–866. doi: 10.1071/AR06203
Pauli, D., Brown-Guedira, G., and Blake, T. K. (2015). Identification of malting quality QTLs in advanced generation breeding germplasm. J. Am. Soc. Brew. Chem. 73, 29–40. doi: 10.1094/ASBCJ-2015-0129-01
Qi, J. C., Chen, J. X., Wang, J. M., Wu, F. B., Cao, L. P., and Zhang, G. P. (2005). Protein and hordein fraction content in barley seeds as affected by sowing date and their relations to malting quality. J. Zhejiang Univ. Sci. B 6, 1069–1075. doi: 10.1631/jzus.2005.B1069
Rae, S., Macaulay, M., Ramsay, L., Leigh, F., Matthews, D., O’Sullivan, D., et al. (2007). Molecular barley breeding. Euphytica 158, 295–303. doi: 10.1007/s10681-006-9166-8
Sarkar, B., Verma, R. P., and Mishra, B. (2008). Association of important melting traits in barley. Indian J. Agric. Sci. 78, 853–857.
Singh, S., Tripathi, R. K., Lemaux, P. G., Buchanan, B. B., and Singh, J. (2017). Redox-dependent interaction between thaumatin-like protein and β-glucan influences malting quality of barley. Proc. Natl. Acad. Sci. U.S.A. 114, 7725–7730. doi: 10.1073/pnas.1701824114
Stanley, D., Rejzek, M., Naested, H., Smedley, M., Otero, S., Fahy, B., et al. (2011). The role of α-glucosidase in germinating barley grains. Plant Physiol. 155, 932–943. doi: 10.1104/pp.110.168328
Stewart, G. G., Hill, A., and Lekkas, C. (2013). Wort FAN – its characteristics and importance during fermentation. J. Am. Soc. Brew. Chem. 71, 179–185. doi: 10.1094/ASBCJ-2013-0921-01
Szűcs, P., Blake, V. C., Bhat, P. R., Chao, S., Close, T. J., Cuesta-Marcos, A., et al. (2009). An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome 2, 134–140. doi: 10.3835/plantgenome2008.01.0005
Taketa, S., Yuo, T., Tonooka, T., Tsumuraya, Y., Inagaki, Y., Haruyama, N., et al. (2011). Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1, 3; 1, 4)-β-D-glucan biosynthesis. J. Exp. Bot. 63, 381–392. doi: 10.1093/jxb/err285
Vis, R. B., and Lorenz, K. (1998). Malting and brewing with a high β-glucan barley. LWT - Food Sci. Technol. 31, 20–26. doi: 10.1006/fstl.1997.0285
von Korff, M., Wang, H., Léon, J., and Pillen, K. (2008). AB-QTL analysis in spring barley: III. Identification of exotic alleles for the improvement of malting quality in spring barley (H. vulgare ssp. spontaneum). Mol. Breed. 21, 81–93. doi: 10.1007/s11032-007-9110-1
Walker, C. K., Ford, R., Muñoz-Amatriaín, M., and Panozzo, J. F. (2013). The detection of QTLs associated with endosperm hardness, grain density, malting quality and plant development traits in barley using rapid phenotyping tools. Theor. Appl. Genet. 126, 2533–2551. doi: 10.1007/s00122-013-2153-2
Walker, C. K., and Panozzo, J. F. (2016). Genetic characterization, expression and association of quality traits and grain texture in barley (Hordeum vulgare L.). Euphytica 212, 1–15. doi: 10.1007/s10681-016-1767-2
Wang, J., Yang, J., Zhang, Q., Zhu, J., Jia, Q., Hua, W., et al. (2015). Mapping a major QTL for malt extract of barley from a cross between TX9425 × Naso Nijo. Theor. Appl. Genet. 128, 943–952. doi: 10.1007/s00122-015-2481-5
Wang, X., Zhang, X., Cai, S., Ye, L., Zhou, M., Chen, Z., et al. (2015). Genetic diversity and QTL mapping of thermostability of limit dextrinase in barley. J. Agric. Food Chem. 63, 3778–3783. doi: 10.1021/acs.jafc.5b00190
Wei, K., Xue, D. W., Huang, Y. Z., Jin, X. L., Wu, F. B., and Zhang, G. P. (2009). Genetic mapping of quantitative trait loci associated with beta-amylase and limit dextrinase activities and beta-glucan and protein fraction contents in barley. J. Zhejiang Univ. Sci. B 10, 839–846. doi: 10.1631/jzus.B0920135
Xu, Y., Zhang, X. Q., Harasymow, S., Westcott, S., Zhang, W., and Li, C. (2018). Molecular marker-assisted backcrossing breeding: an example to transfer a thermostable β-amylase gene from wild barley. Mol. Breed. 38:63. doi: 10.1007/s11032-018-0828-8
Zentella, R., Yamauchi, D., and Ho, T.-H. D. (2002). Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14, 2289–2301. doi: 10.1105/tpc.003376
Zhang, G. P., Li, C. D., and Liu, X. (2012). Advance in Barley Sciences: Proceedings of 11th International Barley Genetics Symposium. Dordrecht: Springer, 37–46.
Zhou, G., Panozzo, J., Zhang, X. Q., Cakir, M., Harasymow, S., and Li, C. (2016). QTL mapping reveals genetic architectures of malting quality between Australian and Canadian malting barley (Hordeum vulgare L.). Mol. Breed. 36:70. doi: 10.1007/s11032-016-0492-9
Keywords: malting quality, quantitative trait loci, phenotypic variance, correlation, marker-assisted selection, barley
Citation: Fang Y, Zhang X and Xue D (2019) Genetic Analysis and Molecular Breeding Applications of Malting Quality QTLs in Barley. Front. Genet. 10:352. doi: 10.3389/fgene.2019.00352
Received: 05 December 2018; Accepted: 02 April 2019;
Published: 24 April 2019.
Edited by:
Shuijin Hua, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Meixue Zhou, University of Tasmania, AustraliaCopyright © 2019 Fang, Zhang and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawei Xue, ZHd4dWVAaHpudS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.