- 1Key Laboratory of Cultivation and Protection for Non-Wood Forest Trees, Ministry of Education, Central South University of Forestry and Technology, Changsha, China
- 2Key Lab of Non-wood Forest Products of State Forestry Administration, College of Forestry, Central South University of Forestry and Technology, Changsha, China
- 3Henan Institute of Science and Technology, Xinxiang, China
Tung tree (Vernicia fordii), an economically important woody oil plant, is a monoecious and diclinous species with male and female flowers on the same inflorescence. The extremely low proportion of female flowers leads to low fruit yield in tung orchards. The female flower normally develops along with stamen abortion; otherwise sterile ovules will be produced. However, little knowledge is known about the molecular basis of the female flower development in tung tree. In this study, integrated analyses of morphological and cytological observations, endogenous phytohormone assay and RNA-seq were conducted to understand the molecular mechanism of the female flower development in tung tree. Cytological observation suggested that the abortion of stamens in female flowers (SFFs) belongs to the type of programmed cell death (PCD), which was caused by tapetum degeneration at microspore mother cell stage. A total of 1,366 differentially expressed genes (DEGs) were identified in female flowers by RNA-seq analysis, of which 279 (20.42%) DEGs were significantly enriched in phenylpropanoid biosynthesis, phenylalanine metabolism, flavonoid biosynthesis, starch and sucrose metabolism, and plant hormone signal transduction. Stage-specific transcript identification detected dynamically expressed genes of important transcription regulators in female flowers that may be involved in PCD and floral organ development. Gene expression patterns revealed that 17 anther and pollen development genes and 37 PCD-related genes might be involved in the abortion of SFF. Further analyses of phytohormone levels and co-expression networks suggested that salicylic acid (SA) accumulation could trigger PCD and inhibit the development of SFF in tung tree. This study provides new insights into the role of SA in regulating the abortion of SFF to develop normal female flowers.
Introduction
Flower development attracts great attention as a fascinating topic for studying plant development and evolution. Unisexuality is considered to be an important transition in the evolutionary history of angiosperms (Barrett, 2010). Many flowers become unisexual after floral organs are specified, but during the process of differentiation, carpel or stamen abortion or arrest occurs and organs become non-functional (Sobral et al., 2016). Plant reproduction requires the development of complex structures that interact with each other and that have an inherently limited life span. Thus, programmed cell death (PCD) is involved in shaping the sexual and non-sexual organs of the flower and in their removal once they are no longer needed (Wu and Cheun, 2000). For example, the gynoecium in male flowers is degenerated by PCD just after initiation of floral primordium in Zea mays (Cheng et al., 1983). In Actinidia deliciosa, PCD also induces male sterility in female flowers (Coimbra et al., 2004).
Researchers suggest that PCD has a close relationship with the accumulation of salicylic acid (SA), and SA controls the timing and extent of PCD in the hypersensitive response (HR) (Greenberg, 1997; Alvarez, 2000; Kovacs et al., 2016). As an endogenous signaling molecule, however, SA is suggested in connection with plant flowering. For example, SA induces the expression of FLOWERING LOCUST 2 (FT2) in the regulation of the flowering in Pharbitis nil (Yamada and Takeno, 2014). In Arabidopsis, SA controls early flower development by NON-EXPRESSOR OF PR3 (NPR3) (Shi et al., 2013). Moreover, SA plays an important role in regulating pollen viability and floret fertility in rice (Zhao et al., 2018). SA can also induce mRNA accumulation of Callose synthase 5 (Gsl5) for deposition of callose in pollens (Ostergaard et al., 2002).
Tung tree (Vernicia fordii), native to China, is widely planted in China and other countries for its ornamental purpose and tung oil production (Cui et al., 2018; Li et al., 2018). Tung oil shows strong dryness, strong adhesion, acid and alkali resistance, so it has been widely used in various industrial fields (Cahoon et al., 1999; Chen et al., 2010). In recent years, tung oil has been attracted world-wide attention due to production security, environmental concerns, and negative effect of synthetic chemical coatings on human health (Meininghaus et al., 2000; Tsakas et al., 2011; Wei et al., 2012; Yang et al., 2018). As a monoecious plant species, tung tree produces a low ratio of female to male flowers (approximately 1:27) (McCann, 1942) and functionally abnormal female flowers, which results in low fruit yield in tung orchards. Generally, female tung flowers present bisexual characteristics at early stages, and then switch to unisexual flowers at late stages along with cell death in stamens in female flowers (SFFs) (Mao et al., 2017). However, the reason causing cell death and the transition of tung flowers from bisexuality to unisexuality remains uncertain.
Mao et al. (2017) found that PCD occurred during stamen abortion in female tung flowers, whereas the molecular mechanism is unclear. Toward this end, we conducted integrated analyses of morphological and cytological observations, endogenous phytohormone assay and RNA-seq of male and female flowers. Based on these analyses, we proposed a possible mechanism for female flower development in tung tree.
Materials and Methods
Plant Materials
The flower was collected from 6-year-old tung tree ‘Putaotong’ grown in the experimental area of the Central South University of Forestry and Technology (Qingping Town, Yongshun County, Hunan Province) under natural conditions. The flower was randomly collected every 5 days from March to April, 2017. Male and female flowers at stage 2 in 30 days before flowering (DBF, C1 and X1, Figures 1C,D), stage 4 in 20 DBF (C2 and X2, Figures 1G,H), stage 6 in 10 DBF (C3 and X3, Figures 1K,L) were separately collected from the same plant and used for transcriptome sequencing. Three biological replicates were analyzed for each sample with 100 flowers counted as one replicate. Controlled pollination (‘Putaotong’ × ‘Duiniantong’) was performed in April, 2017. A total of 300 female flowers and 300 bisexual flowers were used to evaluate the fruit setting with three replicates (each 100 flowers).
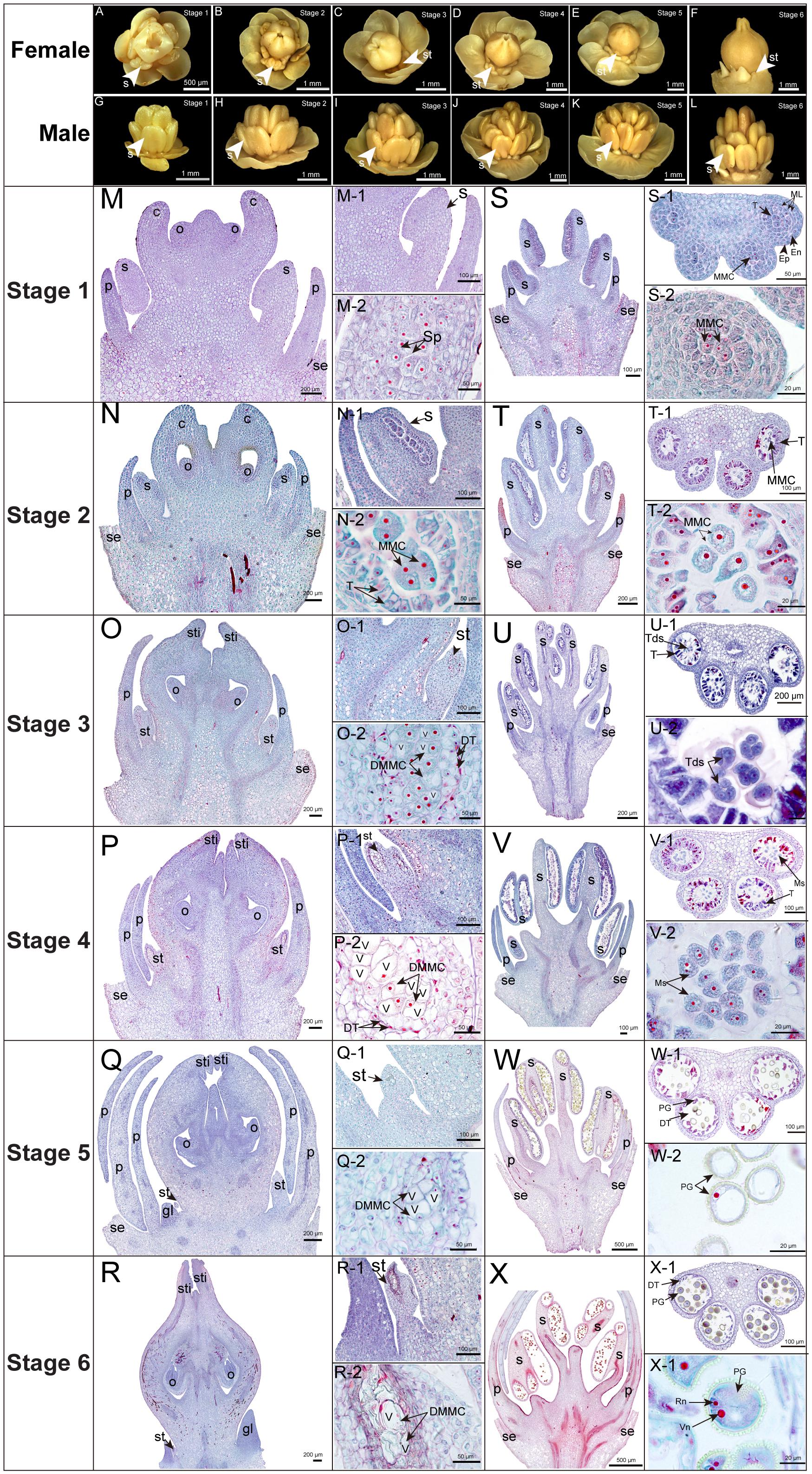
Figure 1. Morphological and cytological observations of male and female flowers. (A–L) Morphological observation. (M–X) Cytological observation. Stages 1–6, 35 DBF (stage 1), 30 DBF (stage 2), 25 DBF (stage 3), 20 DBF (stage 4), 15 DBF (stage 5), 10 DBF (stage 6); se, sepal; p, petal; s, stamen; st, staminode; c, carpel; o, ovule; sti, stigma; gl, gland; Sp, sporogenous cells; MMC, microspore mother cells; Tds, tetrads; Ms, microspore; Pg, pollen grains; Ep, epidermis; En, endothecium; ML, middle layer; T, tapetum; DMMC, degraded microspore mother cells; DT, degraded tapetum; V, vacuole; Vn, vegetative nucleus; Rn, reproduction nucleus.
Morphological and Cytological Observation
Morphology of male and female flowers at six developmental stages was observed by a 3D super depth digital microscope (ZEISS Smartzoom 5, Germany). The samples were prefixed and observed with the paraffin sectioning method described by Liao et al. (2014). The samples were sectioned into 8–10 μm sections using a Leica RM 2265 microtome (Leica Camera AG, Germany), and were stained with Safranin O and Fast Green FCF according to the Sass’s method (Ruzin, 2000). Observations and photomicroscope of the sections were conducted using a microscope (Olympus BX-51, Japan).
RNA Extraction and Sequencing
Total RNAs were isolated using RNAprep pure plant kit DP441 (TIANGEN, China), according to the manufacturer’s protocol. RNA sequencing was performed by Illumina Hiseq 2000 (Illumina, United States). Our reference genome sequences and annotations of tung tree were appliced for the analysis of RNA-seq data. Raw reads of the RNA-seq data are found in NCBI Sequence Read Archive database with the Accession Nos. SRX3843588; SRS3089151; SRS3089154; SRS3089148; SRS3089147; and SRS3089150.
Data Analysis
The detailed processes of de novo assembly and functional annotation were performed as described by Feng et al. (2017) The number of all mapped reads for each gene was counted and normalized into the Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) (Florea et al., 2013). Next, DESeq (Anders and Huber, 2010) was used to identify differentially expressed genes (DEGs), and genes with an adjusted false discovery rate (FDR) ≤ 0.01 and log2FoldChange absolute value of ≥ 1 were marked as significantly different between the two samples. Stage-specific genes were selected as described by Feng et al. (2017). GO and KEGG pathway analyses were performed, and for each category, a two-tailed Fisher’s exact test was employed to test the enrichment of the identified protein against all database proteins. Correction for multiple-hypothesis testing was performed using standard FDR-control methods. The venn diagram, clustering, heatmap, and principal component analysis (PCA) were performed with the venn diagram function, the k-means function, pheatmap, and factoextra package in R software, respectively.
Co-expression Networks Analysis
The transcripts with an average abundance (calculated from three biological replicates) of ≥ 1 (FPKM value) in at least one of the six samples were utilized to construct a co-expression network using the weighted correlation network analysis (WGCNA) R software package (Langfelder and Horvath, 2008). Modules were constructed using the following parameters: maxBlockSize = 10000, power = 18, networkType = “unsigned,” mergeCutHeight = 0.25, minModuleSize = 30. The significant modules in female flowers were determined according to an effective threshold (≥0.85) of the Pearson’s correlation coefficient (PCC) value and a p-value of ≤ 0.05. In order to identify genes which were correlated in expression with those genes involved in anther and pollen development, PCD, and SA, the top 50 genes in each significant module were used for network construction according to correlation degree. The co-expression networks were visualized using the Cytoscape v.3.5.1 program1 (Shannon et al., 2003).
Quantitative Real-Time PCR (qPCR) and Phytohormone Levels Analysis
A PrimeScript RT enzyme with a gDNA eraser (Takara, Japan) was used for cDNA synthesis. QPCR was performed on a Bio-Rad CFX96 Real Time PCR system using SYBR Premix ExTaq II (Takara, Japan). The primers in this step were listed in Supplementary Table S1. Tung Elongation Factor 1-α (EF1α) was used as the internal control (Han et al., 2012). The relative expression levels were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). Phytohormone levels were analyzed using the methods described by Pan et al. (2002).
Statistical Analysis
All experiments were performed with three biological repeats. The data were analyzed by one-way ANOVA procedure in SPSS 22.0 (IBM Corporation, United States). All figures showed the average value of three repeats. All data were expressed as means plus or minus standard deviations (mean ± SD).
Results
Tapetum Degeneration at Microspore Mother Cell Stage Induces PCD and Abortion of SFF
Tung tree is a diclinous species and the functionally normal female flower develops along with stamen abortion in tung tree (Mao et al., 2017). However, the development of bisexual flowers often occurs in inflorescence with abnormal ovaries, leading to lower fruiting ratio (∼5%) than normal female flowers (∼68%) (Supplementary Figure S1). Therefore, we focused on staminode development in female flowers aiming to clarify the process of stamen abortion in tung tree. The development of female flowers could be divided into six important stages (stages 1–6) (Figures 1A–F). Female flowers possessed obvious stamens at stages 1 and 2 which initially degenerated from stage 3 and completely disappeared until stage 6 (Figures 1A–F), whereas no pistil was observed and only intact stamen developed in male flowers across all the development stages (Figures 1G–L).
The SFF showed delayed development in comparison with stamens in male flowers (SMFs). The SFF developed into sporogenous cell stage at 35 DBF (stage 1) (Figure 1M), while the SMF developed into early microspore mother cell (MMC) stage (Figure 1S). When the SFF developed into early MMC stage at 30 DBF (stage 2) (Figure 1N), the SMF developed into later MMC stage (Figure 1T). Interestingly, the SFF stopped growth at early MMC stage with tapetum cells degeneration and large vacuoles in MMC at 25 DBF (stage 3) (Figure 1O), while the SMF developed into tetrads stage (Figure 1U). From 20 to 10 DBF (stages 4 to 6), the SFF shrank at carpel base, and the MMC was mostly occupied by vacuoles with nucleuses in MMC disappearing (Figures 1P–R), while the SMF developed into binuclear pollen stage (Figures 1V–X).
Taken together, the functionally normal female flower developed along with stamen abortion in tung tree. The SFF developed normally at stages 1 and 2, but degenerated from stages 3 to 6. Our morphological and cytological observation further confirmed that the abortion of SFF in tung tree belongs to the type of PCD. The tapetum cell degeneration should contribute to the abortion of SFF, finally leading to PCD in MMC.
DEGs in Male and Female Flowers
In order to elucidate the molecular mechanism responsible for the abortion of SFF in tung tree, the male and female flowers at stage 2 (X1, C1), stage 4 ( X2, C2), and stage 6 (X3, C3) were RNA-sequenced. Based on our tung tree genome sequence, a total of 22,344 genes were identified, of which 18,334 genes were expressed (FPKM value ≥ 1) across six flower samples.
Further, we identified 3,147 DEGs in male flowers (X2_vs_X1, X3_vs_X2) and 1,366 DEGs in female flowers (C2_vs_C1, C3_vs_C2) (Figure 2A). The PCA analysis of the total DEGs, revealed that X1 and C1 had high correlation (Figure 2B), which indicating that the SFF in C1 had normal structure and function as MFF in X1. Besides, among the 1,366 DEGs in female flowers, 531 and 257 genes were significantly up-regulated in C2 and C3, respectively, and 205 and 524 genes were significantly down-regulated in C2 and C3, respectively, as compared to their preceding stage (Figure 2A). Furthermore, in C2_vs_C1 group, there were 356 specific DEGs among up-regulated genes, but 112 among down-regulated genes. In C3_vs_C2 group, 137 specific DEGs were up-regulated and 416 specific DEGs were down-regulated. These results showed that there were more genes highly expressed in C2 than C1 and C3, indicating that a large number of genes might participate in the transition from bisexual to unisexual flowers formation (Figures 2C,D).
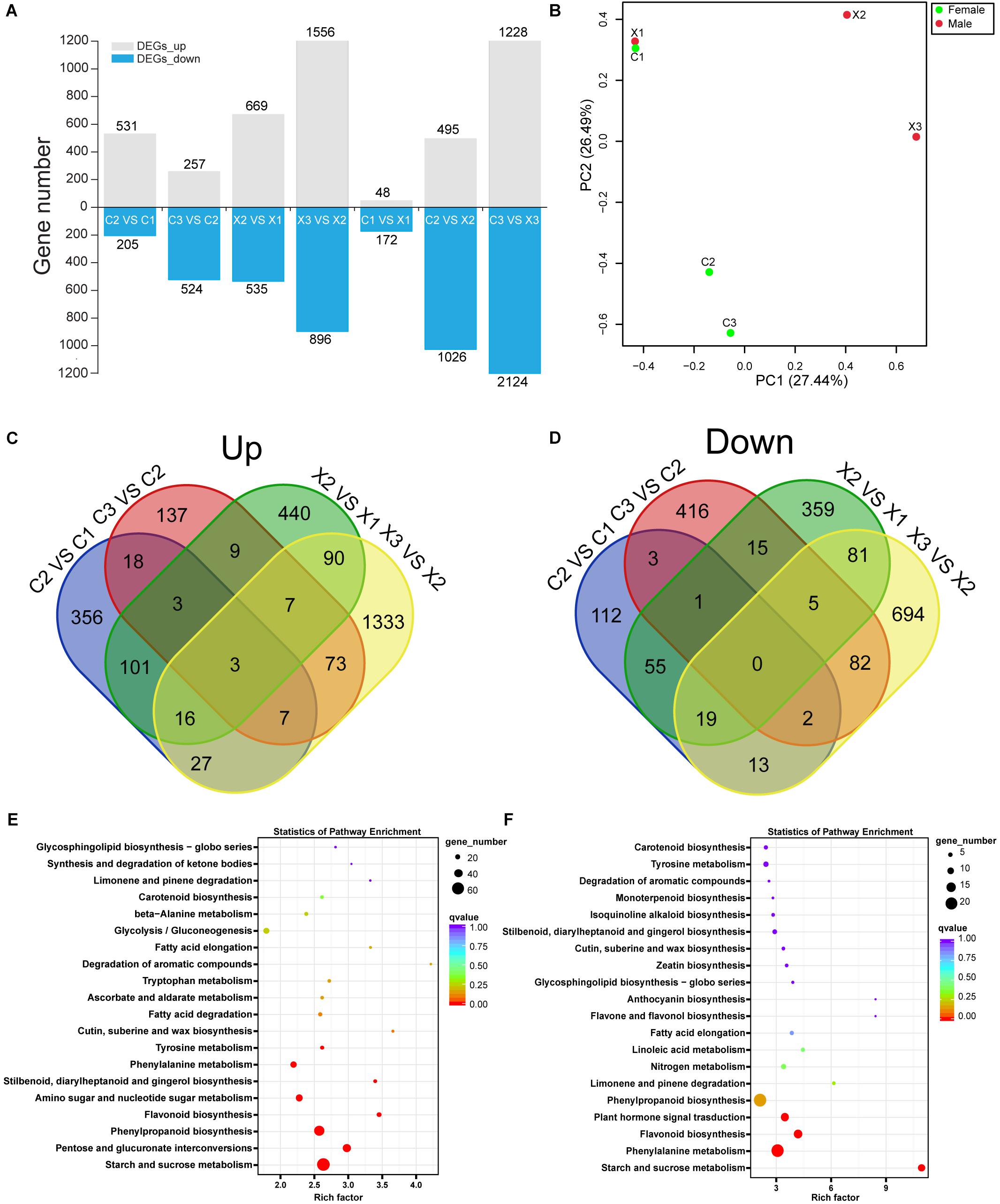
Figure 2. Analysis of DEGs in male and female flowers. (A) Numbers of DEGs in male and female flowers. (B) PCA plot of DEGs in male and female flowers. (C,D) Venn diagrams of the up-regulated and down-regulated DEGs in male and female flowers. (E,F) KEGG enrichment of DEGs in male and female flowers.
KEGG annotation showed that 496 (15.76%) DEGs in male flowers were enriched in 110 KEGG pathways (Supplementary Table S2), of which the starch and sucrose metabolism, phenylpropanoid biosynthesis, pentose and glucuronate interconversions, flavonoid biosynthesis, amino sugar and nucleotide sugar metabolism were significantly enriched pathways (Figure 2E). In contrast, a total of 279 (20.42%) DEGs in female flowers were enriched in 103 KEGG pathways (Supplementary Table S2), of which the phenylpropanoid biosynthesis, phenylalanine metabolism, flavonoid biosynthesis, starch and sucrose metabolism, plant hormone signal transduction were significantly enriched pathways (Figure 2F). It has been reported that sucrose accumulation can change hormonal levels in cut lily flowers and phenylpropanoid can provide the key precursor for SA biosynthesis (Mauchmani and Slusarenko, 1996; Arrom and Munne-Bosch, 2012). In this study, 13 DEGs were found to be significantly enriched in plant hormone signal pathway, indicating that the phytohormones may play an important role in regulating the development of female flowers in tung tree.
Stage-Specific Genes in Male and Female Flowers
In stage 2 (C1, X1), a total of 115 genes were specifically expressed in C1 that were enriched for ribosome biogenesis in eukaryotes (ko03008) and synthesis and degradation of ketone bodies (ko00072) by KEGG enrichment analysis (Figures 3A,G and Supplementary Table S3), including Early flower 4 (ELF4) gene and ELF4-LIKE 4 (EFL4) gene involved in the flowering time and embryoid’s morphogenesis of Arabidopsis, and male fertility genes, like G-type lectin S-receptor-like serine/threonine-protein kinase RLK1, Non-specific lipid-transfer protein 8, and Myb family transcription factor At1g14600 (Boutrot et al., 2006; Xiang et al., 2007; Wan et al., 2008; Adeyemo et al., 2011). A total of 100 stage-specific genes in X1 were enriched for stilbenoid, diarylheptanoid and gingerol biosynthesis (ko00945), phenylpropanoid biosynthesis (ko00940), flavonoid biosynthesis (ko00941), limonene and pinene degradation (ko00903), degradation of aromatic compounds (ko01220), and phenylalanine metabolism (ko00360), including Leucine-rich repeat receptor protein kinase EXS, G-type lectin S-receptor-like serine/threonine-protein kinase RLK1, Phenylalanine ammonia-lyase (PAL), Argonaute 2 (AGO2). These genes are well-known to function in anther or pollen development (Zhao et al., 2002; Xiang et al., 2007; Wan et al., 2008; Feng et al., 2017) (Figure 3B and Supplementary Table S3).
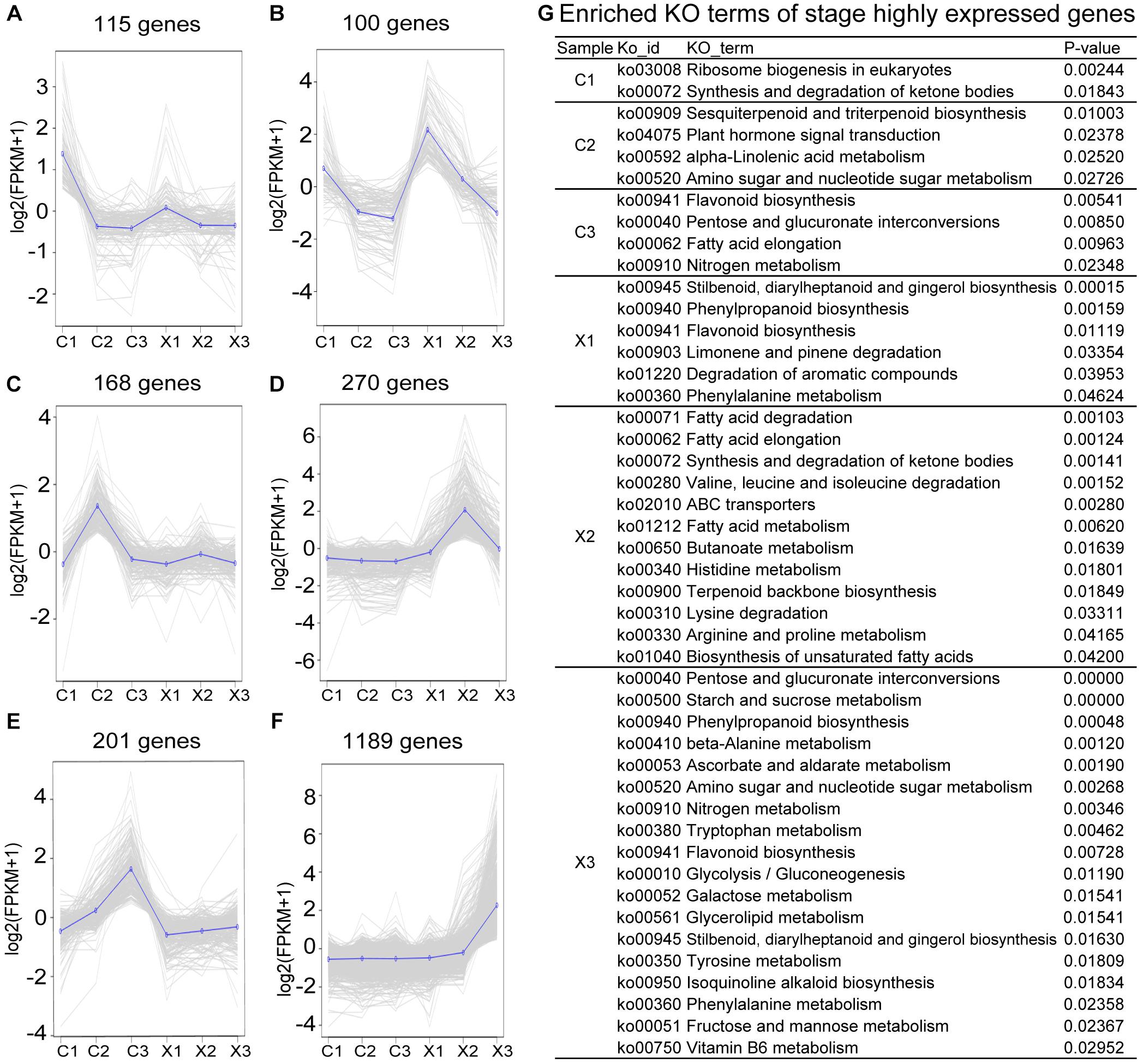
Figure 3. Analysis of stage-specific genes in male and female flowers. (A–F) Expression patterns of specific genes to each developmental stage of female flowers, with the average expression trend highlighted. (G) Enriched KO terms among stage-specific genes (p-values ≤ 0.05).
In stage 4 (C2, X2), SFF began to degenerate in C2, while SMF developed at microspore stage. KEGG enrichment analysis showed that a total of 168 stage-specific genes were mainly enriched in sesquiterpenoid and triterpenoid biosynthesis (ko00909), plant hormone signal transduction (ko04075), alpha-Linolenic acid metabolism (ko00592), Amino sugar and nucleotide sugar metabolism (ko00520), including Vacuolar cation/proton exchanger 3 (CAX3), Arabinogalactan peptide 16 (AGP16), Calcium-binding protein CML 38 (CML38), HVA22-like protein A (HVA22A), and E3 ubiquitin-protein ligase PUB 23 (PUB23). These genes have been reported to function in PCD (Guo and Ho, 2008; Orosa et al., 2017; Sala et al., 2017) (Figures 3C,G and Supplementary Table S3). Moreover, the C2-specific genes included a number of transcription factors such as WRKYs (WRKY18, WRKY46), bHLHs (bHLH96, bHLH3), MYB (MYB108), etc. (Supplementary Table S3), indicating that the transcription factors should play a role in the abortion of SFF. However, there were 270 specific genes in X2 mainly enriched in 12 pathways, including fatty acid degradation (ko00071), fatty acid elongation (ko00062), synthesis and degradation of ketone bodies (ko00072), etc. (Figures 3D,G and Supplementary Table S3).
In stage 6 (C3, X3), a total of 201 specific genes were identified in C3, while 1189 specific genes were in X3 (Figures 3E,F and Supplementary Table S3). KEGG enrichment analysis showed that the C3-specific genes were enriched in flavonoid biosynthesis (ko00941), pentose and glucuronate interconversions (ko00040), fatty acid elongation (ko00062), nitrogen metabolism (ko00910) (Figure 3G). Besides, PCD-related genes including HVA22-like C (HVA22C) and Protein ASPARTIC PROTEASE IN GUARD CELL (ASPG), ovule development controlling gene, Agamous-like 11 (AGL11), and petal development controlling gene, AGL32 were also included in C3 (Supplementary Table S3). In X3, the 1189 specific genes were mainly enriched in 18 pathways, including pentose and glucuronate interconversions (ko00040), starch and sucrose metabolism (ko00500), and phenylpropanoid biosynthesis (ko00940), etc. (Figure 3G).
Identification of Candidate Genes Involved in Anther and Pollen Development in the Abortion of SFF
Cytological observation confirmed that the tapetum cell degeneration may lead to the abortion of SFF in tung tree. A total of 17 key genes were reported to function in anthers and pollens, and they showed two different expression patterns in tung flowers. Pattern 1 included nine genes MYB-related protein Zm38 (MYB35/TDF1), MYB24, ODORANT 1 (MYB103/ODO1), BTB/POZ and TAZ domain-containing protein 3 (TAZ1), BARELY ANY MERISTEM 1 (BAM1), Cytochrome P450 703A2 (CYP703A2), MALE STERILITY (MS1), and Fatty acyl-CoA reductase 2 (MS2) (Zhu et al., 2010; Huang et al., 2017) which were highly expressed in C1, X1, and X2 stages (Figure 4A and Supplementary Table S4). Pattern 2 included eight genes LRR receptor-like serine/threonine-protein kinase RPK 2 (RPK2), SPOROCYTELESS/NOZZLE (SPL/NZZ), ABORTED MICROSPORES (AMS) Somatic embryogenesis receptor-like kinase 1 (SERK1), TAZ1, EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EXS1/EMS1), MYB33 and MYB65 (Millar and Gubler, 2005; Ferguson et al., 2017; Guo et al., 2018), which were highly expressed in C1, X1, X2, and X3 stages (Figure 4A and Supplementary Table S4). QRT-PCR was applied to validate the expression of six genes of anther and pollen at different developmental stages of male and female flowers and generated consistent expression patterns with RNA-Seq data (Figures 4B–G). Together, these 17 key genes in anther and pollen development were suppressed in C2 and C3, and their low expression or silence might result in tapetum and microspore degeneration in tung tree.
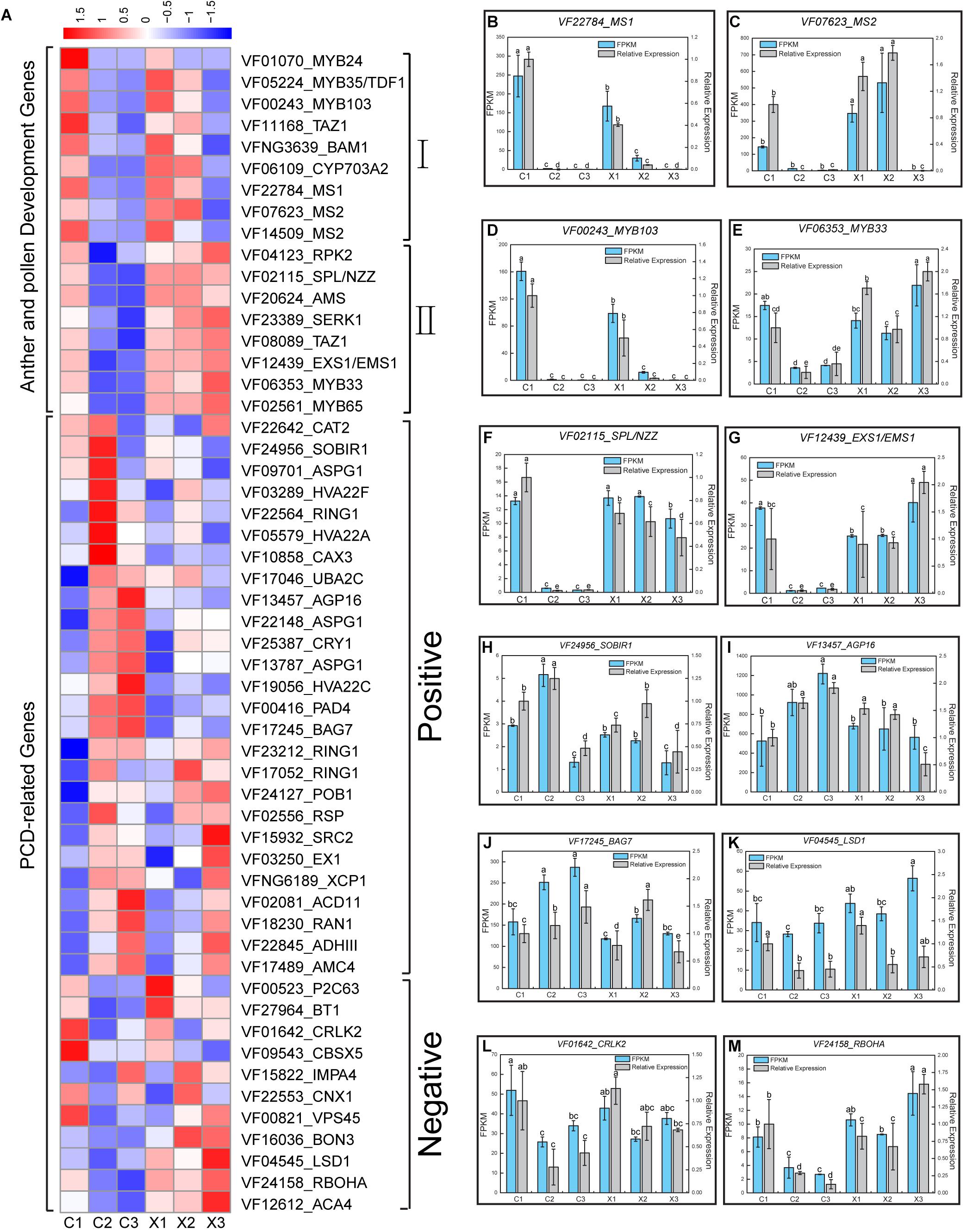
Figure 4. Expression patterns of anther and pollen development genes and PCD-related genes in male and female flowers. (A) Heatmap analysis of the important anther and pollen development genes and PCD-related genes in male and female flowers; I, Genes highly expressed in C1, X1, and X2; II, Genes highly express in C1, X1, X2, and X3. (B–G) Expression levels of MS1, MS2, MYB103, MYB33, SPL/NZZ, and EXS1/EMS1 based on FPKM and qRT-PCR data. (H–M) Expression levels of SORB1, AGP16, BAG7, LSD1, SRLK2, and ROHBA based on FPKM and qRT-PCR data. Error bars indicate SD. Different letters represent significant difference in mRNA levels (p < 0.05).
Identification of Candidate Genes Involved in PCD in the Abortion of SFF
Cytological observation confirmed that the abortion of SFF belongs to PCD type in tung tree. To gain insights into the role of PCD-related genes in the abortion of SFF, the putative 37 key PCD-related genes were identified based on their expression patterns which included 26 positive genes and 11 negative genes (Figure 4A and Supplementary Table S5). Most of the positive genes exhibited high expression levels in C2 and C3, such as Leucine-rich repeat receptor-like serine/threonine/tyrosine-protein kinase (SOBIR1), AGP16, and BAG family molecular chaperone regulator 7 (BAG7) etc. (Figure 4A and Supplementary Table S5). These genes have been reported to behavior in PCD (Gao and Showalter, 1999; Li et al., 2017; Sala et al., 2017). Likewise, negative genes showed low expression levels in C2 and C3 and high expression in C1 such as Lesion Simulating Disease 1 (LSD1), Calcium/calmodulin-regulated receptor-like kinase 2 (CRKL2), Homologies of Respiratory Burst oxidase homolog protein A (RBOHA) and so on (Torres et al., 2002; Yang et al., 2010; Guo et al., 2013) (Figure 4A and Supplementary Table S5). The qRT-PCR analysis of six genes revealed consistent expression patterns with those generated by RNA-seq data (Figures 4H–M). The expression patterns demonstrated the important role of PCD-related genes in the abortion of SFF in tung tree.
Phytohormone Levels in Male and Female Flowers
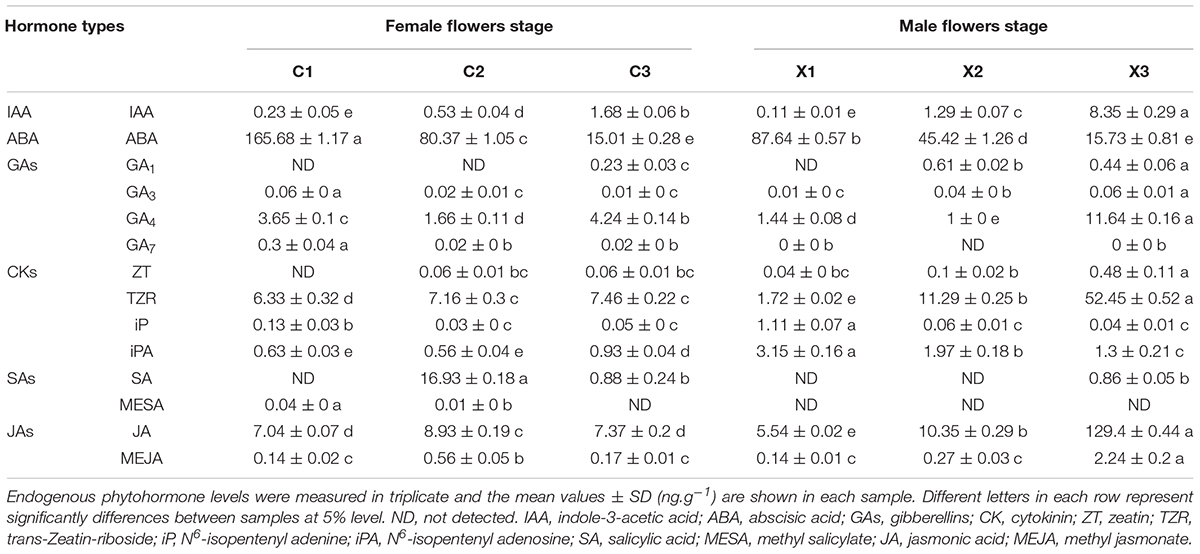
To explore the possible phytohormone regulation in the abortion of SFF, the endogenous levels of auxin, abscisic acid (ABA), gibberellin (GA), jasmonic acid (JA), cytokinin (CK), and SA in tung flowers at different stages were measured (Table 1). The male flower exhibited the same patterns of IAA, ABA, and GA levels with the female flower. The IAA level was up-regulated reaching the highest at stage 3, and it was significantly higher in X3 (8.35 ng/g) than that in C3 (1.68 ng/g) (Table 1). The ABA levels were down-regulated across three stages, and it was significantly higher in C1 (165.68 ng/g) than that in X1 (87.64 ng/g) (Table 1). For GAs, GA4 level was significantly higher than GA1, GA3, and GA7 in all samples. In addition, the GA4 level was the lowest in C2 (1.66 ng/g) and X2 (1.0 ng/g) (Table 1).
The levels of JAs and CKs showed different patterns in comparison with IAA, ABA, and GA (Table 1). JA was significantly higher than MEJA across all samples. It was up-regulated in male flowers with the highest value (129.4 ng/g) in X3 (Table 1). For CKs, the TZR level was significantly higher than Zeatin, IP and IPA. The TZR level was up-regulated in both male and female flowers (Table 1). Notably, SA showed different patterns from other phytohormones. No SA was detected in C1, X1 and X2, and only extremely low levels of SA were detected in C3 (0.88 ng/g) and X3 (0.86 ng/g) (Table 1). In contrast, SA was significantly high in C2 (16.93 ng/g) where the abortion of SFF occurred.
According to the analysis of phytohormone levels, SA is likely to play an important role in the abortion of SFF instead of other phytohormones.
Analysis of SA Synthesis and Signaling Pathway
To uncover the regulatory mechanism of SA involved in the abortion of SFF, we analyzed in detail the pathway of SA synthesis and signaling in tung flowers. Two pathways have been reported to be involved in plant SA synthesis (Chen et al., 2009). One is regulated by phenylalanine ammonia lyase (PAL) and chorismate mutase (CM), and the other by isochorismate synthase (ICS). In Arabidopsis, there are four homologs encoding PAL (PAL1-4), three homologs encoding CM (CM1-3), and two homologs encoding ICS (ICS1/2) (Huang et al., 2010). In tung tree genome, we found three CM members (CM1-3), one PAL and one ICS (ICS2). The three CMs displayed different expression patterns. Generally, FPKM of CM1 was higher than CM2 and CM3 across all samples. CM1 displayed different expression profiles in male and female flowers, and was up-regulated with the development of female flowers (Figure 5A). CM2 was highly expressed in C2 of female flowers, and down-regulated in male flowers (Figure 5A and Supplementary Table S6). CM3 was down-regulated in both male and female flowers (Figure 5A and Supplementary Table S6). PAL was up-regulated in both male and female flowers. ICS2 was highly expressed in C2 of female flowers, and down-regulated in male flowers (Figure 5A and Supplementary Table S6).
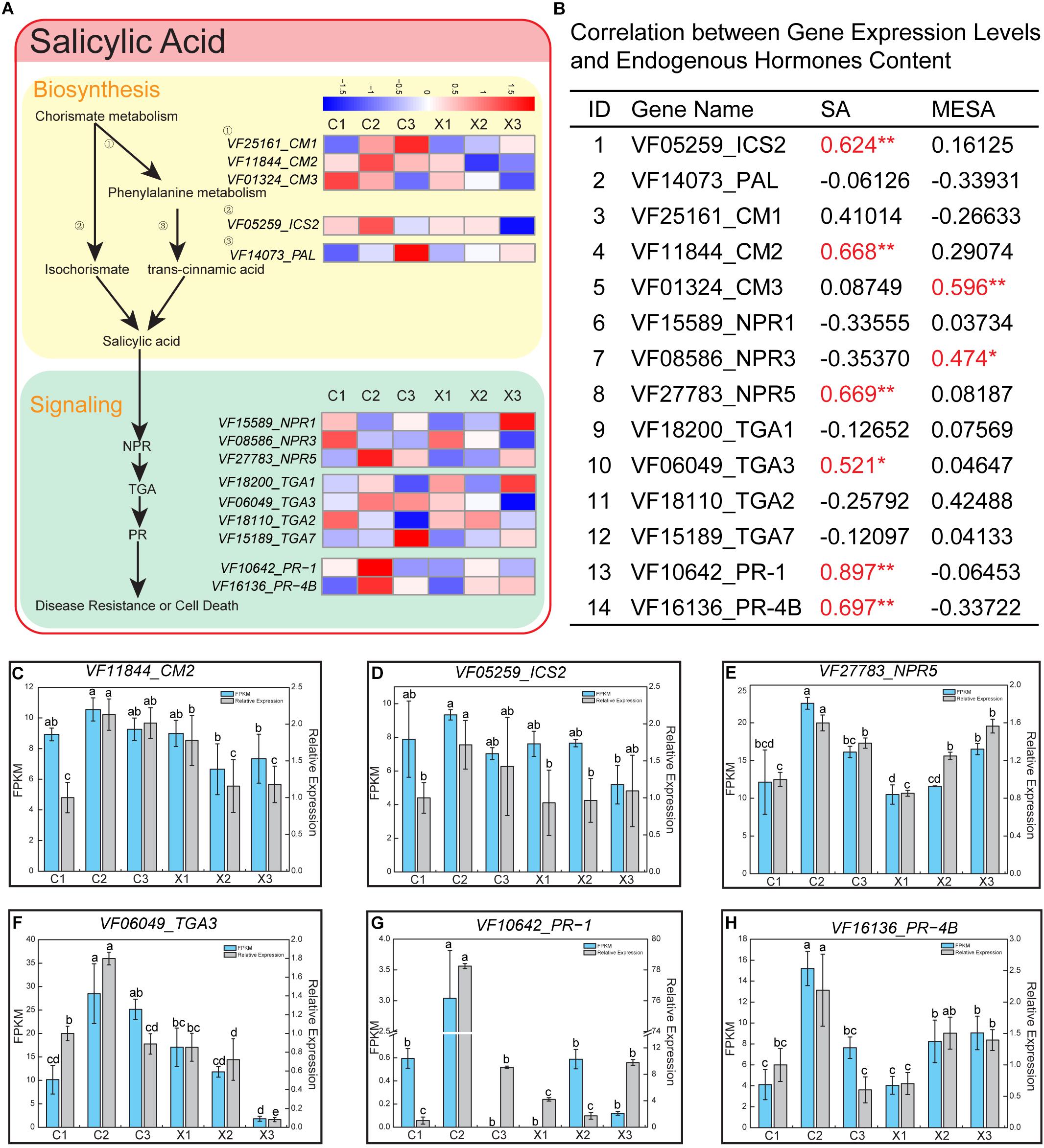
Figure 5. Salicylic acid (SA) pathway in male and female flowers. (A) Representative gene expression profiles of SA biosynthesis and signaling pathway. The color intensity in the heatmap of each gene represents the relative expression levels of these genes at different stages of male and female flowers. (B) Correlation between gene expression levels and endogenous hormones content in male and female flowers. “∗” and “∗∗” after the correlation coefficients represent the correlation between their mRNA levels and endogenous hormone levels significantly different at p < 0.05 and p < 0.01, respectively. (C–H) Expression levels of CM2, ICS2, NPR5, TGA3, PR-1, and PR-4B based on FPKM and qRT-PCR data. Error bars indicate SD. Different letters represent significant difference in mRNA levels (p < 0.05).
In Arabidopsis SA signaling pathway, three receptors are reported, including NON-EXPRESSOR OF PR1 (NPR), transcription factor TGA (TGA), PATHOGENESIS-RELATED (PR) (Kaltdorf and Naseem, 2013). Three homologs of NPR (NPR1/3/5), four TGAs (TGA1-3, TGA7), and two PRs (PR-1, PR-4B) were found in tung tree genome (Figure 5A and Supplementary Table S6). The expression level of NPR3 was higher than NPR1 and NPR5, and down-regulated in male and female flowers of tung tree. NPR1 was down-regulated in female flowers, but up-regulated in male flowers. NPR5 was highly expressed in C2 of female flowers, and up-regulated in male flowers (Figure 5A and Supplementary Table S6). Among the TGA family, TGA3 showed higher expression level than the other three members. It was highly expressed in C2 of female flowers, and down-regulated in male flowers (Figure 5A and Supplementary Table S6). Interestingly, two members of the PR family (PR-1, PR-4B) were highly expressed in C2 of female flowers (Figure 5A and Supplementary Table S6).
Correlation analysis revealed that expression levels of six genes among the selected 14 genes were highly correlated with SA, including ICS2, CM2, NPR5, TGA3, PR-1, and PR-4B, while only two genes (CM3 and NPR3) were significantly correlated with MESA (Figure 5B). Besides, the qRT-PCR analysis of six genes revealed consistent expression patterns with those generated by RNA-seq data (Figures 5C–H). Based on the above analyses, ICS2, CM2, NPR5, TGA3, PR-1, and PR-4B should be the important regulators in the pathways of SA synthesis and signaling and play a role in the abortion of SFF in tung tree.
Co-expression Networks Analysis of Expression Genes Involved in the Abortion of SFF
To gain more insights into the regulatory relationships of anther and pollen development genes, SA- and PCD-related genes in female flowers of tung tree, we performed a WGCNA of transcript expression in male and female flowers (FPKM values ≥ 1). Consequently, 21 co-expression modules were identified for each sample (Figure 6A). An effective PCC threshold of ≥ 0.85 and a p-value of ≤ 0.05 were trained to generate the significant modules. Finally, we found that each sample of female flowers had one significant module, namely C1 with MEblue module (PCC = 0.91, p-value = 0.01), C2 with MEpink module (PCC = 0.88, p-value = 0.02), and C3 with MEbrown module (PCC = 0.85, p-value = 0.03) (Figure 6A).
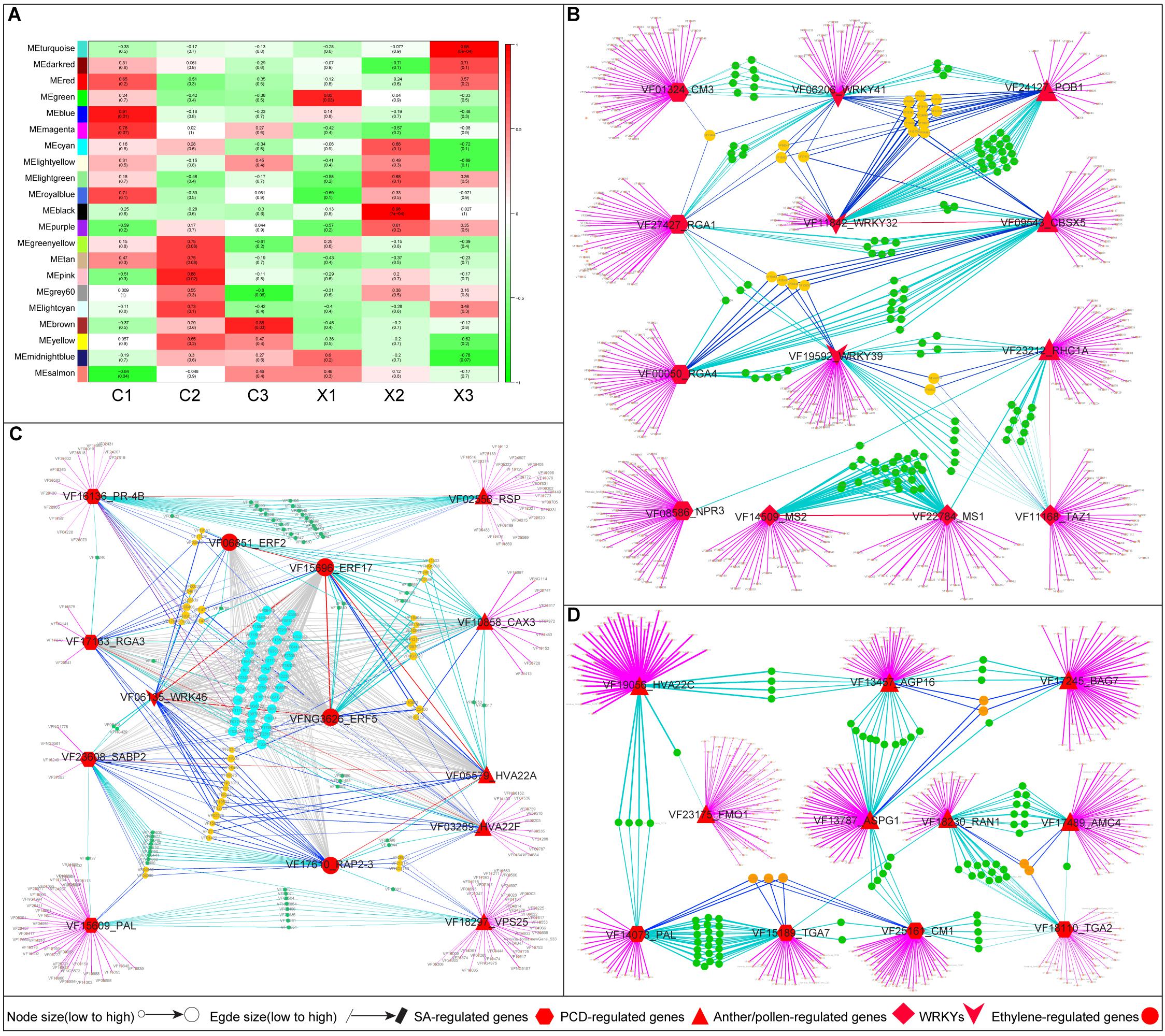
Figure 6. Co-expression networks analysis of genes in different module of female flowers in tung tree. (A) Module trait relationships in six samples. (B–D) Co-expression networks analysis of anther and pollen development genes, SA- and PCD-related genes in MEblue, MEpink, and MEbrown module.
The MEblue module of the co-expression network, representing the development of SFF, contained 3469 genes. Of the 3469 genes, a number of genes were identified as anther and pollen development genes (TAZ1, MS1, MS2), PCD-related genes (RING1, CBSX5, and POB1), and SA-related genes (CM3, NPR3, RGA1, RGA4) (Supplementary Table S7). The TAZ1 (2324 edges), MS1 (2583), MS2 (2297), RING1 (2926), POB1 (3159), CM3 (2512), NPR3 (2534), RGA1 (2990), and RGA4 (3132) had significantly high edge numbers, suggesting their prominent roles in anther and pollen development of female flowers (Figure 6B and Supplementary Table S7). Furthermore, WRKYs were identified as having high connectivity between SA- and PCD-related genes, or SA-related genes and anther and pollen development genes, i.e., WRKY41 (3132 edges), WRKY32 (3178), WRKY39 (2992), suggesting their important biological functions in regulating anther and pollen development of female flowers in tung tree (Figure 6B and Supplementary Table S7).
The MEpink module represented the beginning of the SFF abortion contained 533 genes in which many PCD-, SA- and ethylene-related genes showed high connectivity in the network (Supplementary Table S7), such as (1) the PCD-related genes of VPS25 (354 edges), CAX3 (326), HVA22A (373), and HVA22F (375); (2) the SA-related genes of PAL (408), PR-4B (435), and SABP2 (375), (3) the ethylene-related genes of ethylene-responsive transcription factors (ERF2 (378), ERF5 (379), ERF17 (373), and RAP2-3 (409)) (Figure 6C and Supplementary Table S7). Interestingly, anther and pollen development genes were not found in MEpink module. Furthermore, only one WRKY (WRKY46, 434 edges) was identified as having high connectivity in MEpink module (Figure 6C and Supplementary Table S7). Based on these findings, PCD, SA and ethylene may have critical roles in regulating the abortion of SFF in tung tree.
The MEbrown module, contained 2453 genes and represented the abortion of SFF in tung tree (Supplementary Table S7). Interestingly, only PCD- and SA-related genes were detected in the network, such as AGP16, RAN1, BAG7, FLAVIN-DEPENDENT MONOXYGENASE1 (FMO1), ASGP1, HVA22C, CM1, TGA7, and PAL, etc. (Supplementary Table S7). Furthermore, there were high connectivity among (1) the PCD-related genes, including AGP16 (1872 edges), RAN1 (1509), BAG7 (1925), and FMO1 (1010), (2) the SA-related genes, including CM1 (2114), and PAL (1660) (Figure 6D and Supplementary Table S7). This result suggests that these genes play a key roles in regulating the abortion of SFF in tung tree.
Taken together, the analyses of gene co-expression networks suggested that SA might regulate the anther and pollen development and the PCD, resulting in the abortion of SFF in tung tree.
Discussion
The production of unisexual flowers in flowering plants has evolved more than 1000 times (Renner and Feil, 1993). Unisexual flowering systems promote outbreeding and are considered as driving forces in plant evolution. Although unisexual flowering systems have been studied in A. deliciosa, Quercus sube, and Diospyros kaki (Rocheta et al., 2014; Akagi et al., 2016, 2018), the regulatory mechanisms are unclear.
Anther and Pollen Development Genes Regulate the Abortion of SFF
In tung tree, we found that the SFF was arrested in C2 before the formation of tetrads in microspore meiosis (Figure 1), which was similar with Asparagus officinalis (Caporali et al., 1994). More importantly, the abortion of SFF began with tapetum degeneration. Throughout the abortion process of SFF, the degeneration of tapetum may be a key factor causing the MMC degeneration in SFF. The tapetum plays an important role in pollen grain development which serves as a nutritive tissue, providing metabolites, nutrients, and cell wall precursors for the development of pollen grains, therefore any obstruction of the tapetum development will lead to male sterility in plants (Pacini, 1997; Chen et al., 2018). In tung tree, the tapetum degeneration may result in the abortion of SFF.
The use of classical genetic screens has uncovered a large number of genes involved in anthers development and in the production of viable pollen. In our study, 17 anther and pollen development genes were highly expressed in C1, but lowly expressed or silenced in C2 and C3 (Figure 4). It is worth mentioning that SPL/NZZ, and EXS1/EMS1 have important roles in early anther development and differentiation, while AMS, MS1, MYB33, MYB65, and MYB103 are involved in tapetum and pollen wall development (Pearce et al., 2015). Thus, low expression or silence of anther and pollen development genes, especially the genes of tapetum development, may result in the abortion of SFF in C2 and C3 of female flowers.
PCD Triggers the Abortion of SFF
Programmed cell death is a controlled cellular suicide which plays an important role in plant development. The nuclear DNA degradation, vacuolization, or the activation of specific proteases belongs to specific morphological and biochemical features of PCD (Hautegem et al., 2015). In tung tree, SFF were degenerated when stamens developed into early MMC, and MMC were vacuolize in C2 of female flowers (Figure 1). In Thymelaea and Meliaceae, the gynoecium in male flowers is aborted by PCD at meiosis stage (Caporali et al., 2006; Gouvea et al., 2008). Hence, PCD is essential to many aspects of plant morphogenesis and is a normal component of flower development (Rogers, 2006).
Programmed cell death is controlled by both positive regulators and negative regulators. In this study, positive regulators were highly expressed in C2 or C3 of female flowers, like SOBIR1, ASPG16, and BAG7, which are well-known to positively regulate PCD (Gao and Showalter, 1999; Li et al., 2017; Lu et al., 2017; Sala et al., 2017). Negative regulators showed low expression in C2 and C3, such as RBOHs, which are the source of reactive oxygen species in the oxidative burst and negatively regulate cell death (Torres et al., 2002). These results suggested that PCD-related genes rapidly caused the abortion of SFF in tung tree.
Plant Hormones Are Associated With the Abortion of SFF
A variety of plant hormones are likely to be involved in the regulation of reproductive organ abortion and development pathways in unisexual flowers (Ubeda-Tomas and Bennett, 2010). For example, ethylene increases the percentage of female flowers in Cucumis sativus and Cucumis melo (Boualem et al., 2008; Tao et al., 2018). CK promotes female flowers development in Populus tomentosa, Jatropha curcas, and Castanea henryi (Song et al., 2013; Chen et al., 2014; Fan et al., 2017). In tung tree, contents of GA4, IAA, TZR and JA were significantly higher in X3 of male flowers than C3 of female flowers (Table 1). Only SA was specifically and highly expressed in C2, the initial stage of the abortion of SFF (Table 1). Therefore, the SA accumulation may suppress development of SFF in tung tree.
NPR5, TAG3, PR-1, and PR-4B in SA pathway showed high expression in C2 of female flowers. Interestingly, ICS2 also exhibited the highest expression level in C2 among all samples. In Arabidopsis, isochorismate pathway is the main source of SA accumulation (Wildermuth et al., 2001). Further, the isochorismate pathway also has been shown to be active in tomato and tobacco (Uppalapati et al., 2007). Together, isochorismate pathway may be also the main source of large SA accumulation in C2 of female flowers in tung tree.
SA Mediates the Abortion of SFF by Anther and Pollen Development Genes and PCD-Related Genes
Salicylic acid mediates PCD in plants under biotic stress (Alvarez, 2000). In our study, co-expression networks analysis indicated that anther and pollen development genes, SA- and PCD-related genes showed high connectivity in the network, and that WRKYs were the important connectors between SA-related genes and anther and pollen development genes, and SA- and PCD-related genes (Figure 6). In Arabidopsis, members of WRKYs have interaction with SA-related genes. For example, WRKY58 interacts with NPR1 to regulate SA accumulation in plant defense (Wang et al., 2006). The lower level of SA limits NPR1-NPR3 interaction, enabling NPR1 to restrict the spread of PCD (Fu et al., 2012). SA-dependent signal potentiation loop has been proposed to be negatively regulated by protein LSD1 (Aviv et al., 2002), which also explains the runaway cell death phenotype of the lsd1 mutant. In addition, FMO1 is not only necessary for systemic accumulation of SA and downstream signaling after pathogen infection but also regulates PCD in Arabidopsis (Xu and Brosche, 2014). In rice, SA is suggested to regulate pollen viability and floret fertility (Zhao et al., 2018). Herein, we proposed a possible signaling network where anther and pollen development genes and PCD-related genes work synergistically with SA-related genes to induce the abortion of SFF in tung tree (Figure 6).
Additionally, we constructed a hypothetical model for the development of normal female flowers in tung tree. At the early stage, SFF develops normally in female flowers with low level of SA (Figure 7). With SA accumulation, PCD-related genes are activated and highly expressed, and subsequently anther and pollen development genes, especially tapetum genes are suppressed (Figure 7). Consequently, the abortion of SFF occurs and the normal female flower develops.
Conclusion
We confirmed that the abortion of SFF in tung tree belongs to the PCD type and demonstrated that tapetum degeneration at MMC stage is the major reason causing the abortion of SFF. Furthermore, we constructed a model for the abortion of SFF in tung tree based on integrated analyses of morphological and cytological observations, endogenous phytohormone assay and RNA-seq. During the middle stage in female flowers, SA accumulation triggers PCD activation and arrests anther and pollen development, which ultimately results in the abortion of SFF in tung tree. This study provides valuable information for better understanding the development of female flowers in tung tree.
Author Contributions
LZ, XT, and ML conceived and designed the experiments. ML, WL, XF, and GZ performed the experiments. ML, XF, HL, MS, GZ, YF, and LZ analyzed the data. ML, WL, GZ, XT, HL, YF, and LZ contributed reagents, materials, and analysis tools. ML and LZ wrote the manuscript. All authors discussed results and commented on the manuscript.
Funding
This work was supported by the National Key R&D Program of China (Project No. 2017YFD0600703), the National Natural Science Foundation of China (Project No. 31770720), and the Scientific Innovation Fund for Graduate of Central South University of Forestry and Technology (Project No. CX2017B397).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jin Xu (XISHUANGBANNA TROPICAL BOTANICAL GARDEN, CHINESE ACADEMY OF SCIENCES) for providing 3D super depth digital microscope analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00316/full#supplementary-material
FIGURE S1 | Tung tree flower and fruit. (A,D) Male flower. (B,E) Female flower. (C,F) Bisexual flower. (G,H) Tung fruit of female and bisexual flowers. (I) Fruiting ratio of female and bisexual flowers.
TABLE S1 | Primer of qRT-PCR.
TABLE S2 | KEGG enrichment analysis of DEGs in male and female flowers.
TABLE S3 | Stage-specific genes analysis in male and female flowers.
TABLE S4 | Expression patterns of anther and pollen development genes in male and female flowers.
TABLE S5 | Expression patterns of PCD-related genes in male and female flowers.
TABLE S6 | Salicylic acid pathway in male and female flowers.
TABLE S7 | Co-expression networks analysis of genes in different module of female flowers in Tung Tree.
Footnotes
References
Adeyemo, O. S., Kolmos, E., Tohme, J., Chavariaga, P., Fregene, M., and Davis, S. J. (2011). Identification and characterization of the cassava core-clock gene early flowering 4. Trop. Plant Biol. 4, 117–125. doi: 10.1007/s12042-011-9065-6
Akagi, T., Henry, I. M., Kawai, T., Comai, L., and Tao, R. (2016). Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28, 2905–2915. doi: 10.1105/tpc.16.00532
Akagi, T., Henry, I. M., Ohtani, H., Morimoto, T., Beppu, K., Kataoka, I., et al. (2018). A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in Kiwifruit. Plant Cell 30, 780–795. doi: 10.1105/tpc.17.00787
Alvarez, M. E. (2000). Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol. Biol. 44, 429–442. doi: 10.1023/A:1026561029533
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Arrom, L., and Munne-Bosch, S. (2012). Sucrose accelerates flower opening and delays senescence through a hormonal effect in cut lily flowers. Plant Sci. 188, 41–47. doi: 10.1016/j.plantsci.2012.02.012
Aviv, D., Rustérucci, C., Holt, B., Dietrich, R., Parker, J., and Dangl, J. (2002). Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. 29, 381–391. doi: 10.1046/j.0960-7412.2001.01225.x
Barrett, S. C. H. (2010). Understanding plant reproductive diversity. Philos. T. R. Soc. B. 365, 99–109. doi: 10.1098/rstb.2009.0199
Boualem, A., Fergany, M., Fernandez, R., Troadec, C., Martin, A., Morin, H., et al. (2008). A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321, 836–838. doi: 10.1126/science.1159023
Boutrot, F., Meynard, D., Guiderdoni, E., Joudrier, P., and Gautier, M. F. (2006). The Triticum aestivum non-specific lipid transfer protein (TaLtp) gene family: comparative promoter activity of six TaLtp genes in transgenic rice. Planta 225, 843–862. doi: 10.1007/s00425-006-0397-7
Cahoon, E. B., Carlson, T. J., Ripp, K. G., Schweiger, B. J., Cook, G. A., Hall, S. E., et al. (1999). Biosynthetic origin of conjugated double bonds: production of fatty acid components of high-value drying oils in transgenic soybean embryos. Proc. Natl. Acad. Sci. U.S.A. 96, 12935–12940. doi: 10.1073/pnas.96.22.12935
Caporali, E., Carboni, A., Galli, M. G., Rossi, G., Spada, A., and Longo, G. P. M. (1994). Development of male and female flower in Asparagus officinalis. Search for point of transition from hermaphroditic to unisexual developmental pathway. Sex. Plant Reprod. 7, 239–249. doi: 10.1007/BF00232743
Caporali, E., Roccotiello, E., Cornara, L., Casazza, G., and Minuto, L. (2006). An anatomical study of floral variation in Thymelaea hirsuta(L.) Endl. related to sexual dimorphism. Plant Biosyst. 140, 123–131. doi: 10.1080/11263500600756199
Chen, M. S., Pan, B. Z., Wang, G. J., Ni, J., Niu, L., and Xu, Z. F. (2014). Analysis of the transcriptional responses in inflorescence buds of Jatropha curcas exposed to cytokinin treatment. BMC Plant Biol. 14:318. doi: 10.1186/s12870-014-0318-z
Chen, Y. H., Chen, J. H., Chang, C. Y., and Chang, C. C. (2010). Biodiesel production from tung (Vernicia montana) oil and its blending properties in different fatty acid compositions. Biores. Technol. 101, 9521–9526. doi: 10.1016/j.biortech.2010.06.117
Chen, Z., Zheng, Z., Huang, J., Lai, Z., and Fan, B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal Behav. 4, 493–496. doi: 10.4161/psb.4.6.8392
Chen, Z. S., Liu, X. F., Wang, D. H., Chen, R., Zhang, X. L., Xu, Z. H., et al. (2018). Transcription factor OsTGA10 is a target of the MADS protein OsMADS8 and is required for Tapetum development. Plant Physiol. 176, 819–835. doi: 10.1104/pp.17.01419
Cheng, P. C., Greyson, R. I., and Walden, D. B. (1983). Organ initiation and the development of unisexual flowers in the tassel and ear of zea mays. Am. J. Bot. 70, 450–462. doi: 10.2307/2443252
Coimbra, S., Torrao, L., and Abreu, I. (2004). Programmed cell death induces male sterility in Actinidia deliciosa female flowers. Plant Physiol. Biochem. 42, 537–541. doi: 10.1016/j.plaphy.2004.05.004
Cui, P., Lin, Q., Fang, D., Zhang, L., Li, R., Cheng, J., et al. (2018). Tung tree (Vernicia fordii) genome and transcriptome sequencing reveals coordinate upregulation of fatty acid beta-oxidation and triacylglycerol biosynthesis pathways during eleostearic acid accumulation in seeds. Plant Cell Physiol. 59, 1990–2003. doi: 10.1093/pcp/pcy117
Fan, X. M., Yuan, D. Y., Tian, X. M., Zhu, Z. J., Liu, M. L., and Cao, H. P. (2017). Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in the flowers of chinese chinquapin (Castanea henryi). J. Agri. Food Chem. 65, 10332–10349. doi: 10.1021/acs.jafc.7b03755
Feng, N., Song, G., Guan, J., Chen, K., Jia, M., Huang, D., et al. (2017). Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory genes. Plant Physiol. 174, 1779–1794. doi: 10.1104/pp.17.00310
Ferguson, A. C., Pearce, S., Band, L. R., Yang, C. Y., Ferjentsikova, I., King, J., et al. (2017). Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol. 213, 778–790. doi: 10.1111/nph.14200
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res. 2:188. doi: 10.12688/f1000research.2-188.v2
Fu, Z. Q., Yan, S. P., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gao, M., and Showalter, A. M. (1999). Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J. 19, 321–331. doi: 10.1046/j.1365-313X.1999.00544.x
Gouvea, C. F., Dornelas, M. C., and Rodriguez, A. P. (2008). Floral development in the tribe cedreleae (meliaceae, sub-family swietenioideae): cedrela and toona. Ann. Bot. 101, 39–48. doi: 10.1093/aob/mcm279
Greenberg, J. T. (1997). Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Bio. 48, 525–545. doi: 10.1146/annurev.arplant.48.1.525
Guo, J., Bai, P. F., Yang, Q., Liu, F. R., Wang, X. D., Huang, L. L., et al. (2013). Wheat zinc finger protein TaLSD1, a negative regulator of programmed cell death, is involved in wheat resistance against stripe rust fungus. Plant Physiol. Biochem. 71, 164–172. doi: 10.1016/j.plaphy.2013.07.009
Guo, J. J., Liu, C., Wang, P., Cheng, Q., Sun, L., Yang, W. C., et al. (2018). The aborted microspores (AMS)-like gene is required for anther and microspore development in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 19:1341. doi: 10.3390/ijms19051341
Guo, W. J., and Ho, T. H. (2008). An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol. 147, 1710–1722. doi: 10.1104/pp.108.120238
Han, X., Lu, M., Chen, Y., Zhan, Z., Cui, Q., and Wang, Y. (2012). Selection of reliable reference genes for gene expression studies using real-time pcr in tung tree during seed development. PLoS One 7:e43084. doi: 10.1371/journal.pone.0043084
Hautegem, T. V., Waters, A. J., Goodrich, J., and Nowack, M. K. (2015). Only in dying, life: programmed cell death during plant development. Trends Plant Sci. 20, 102–113. doi: 10.1016/j.tplants.2014.10.003
Huang, H., Gao, H., Liu, B., Qi, T., Tong, J., Xiao, L., et al. (2017). Arabidopsis MYB24 regulates jasmonate-mediated stamen development. Front. Plant Sci. 8:1525. doi: 10.3389/fpls.2017.01525
Huang, J. L., Gu, M., Lai, Z. B., Fan, B. F., Shi, K., Zhou, Y. H., et al. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. doi: 10.1104/pp.110.157370
Kaltdorf, M., and Naseem, M. (2013). How many salicylic acid receptors does a plant cell need? Sci. Signal. 6:jc3. doi: 10.1126/scisignal.2003944
Kovacs, J., Poor, P., Szepesi, A., and Tari, I. (2016). Salicylic acid induced cysteine protease activity during programmed cell death in tomato plants. Acta Biol. Hung. 67, 148–158. doi: 10.1556/018.67.2016.2.3
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Li, W., Liu, M., Zhang, L., Tan, X., Zhang, F., and Wang, Z. (2018). Study of major economic traits in 4 superior families of tung tree. Non Wood For. Res. 36, 29–34. doi: 10.14067/j.cnki.1003-8981.2018.02.005
Li, Y. R., Williams, B., and Dickman, M. (2017). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214, 695–705. doi: 10.1111/nph.14388
Liao, T., Yuan, D. Y., Zou, F., Gao, C., Yang, Y., Zhang, L., et al. (2014). Self-sterility in Camellia oleifera may be due to the prezygotic late-acting self-incompatibility. PloS One 9:e99639. doi: 10.1371/journal.pone.0099639
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Z. G., Xu, J., Li, W. X., Zhang, L., Cui, J. W., He, Q. S., et al. (2017). Transcriptomic analysis reveals mechanisms of sterile and fertile flower differentiation and development in Viburnum macrocephalum f. keteleeri. Front. Plant Sci. 8:261. doi: 10.3389/fpls.2017.00261
Mao, Y. J., Liu, W. B., Chen, X., Xu, Y., Lu, W. L., Hou, J. Y., et al. (2017). Flower development and sex determination between male and female flowers in Vernicia fordii. Front. Plant Sci. 8:1291. doi: 10.3389/fpls.2017.01291
Mauchmani, B., and Slusarenko, A. J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212. doi: 10.1105/tpc.8.2.203
McCann, L. P. (1942). Development of the pistillate flower and structure of the fruit of tung (Aleurites fordii). J. Agr. Res. 65:361.
Meininghaus, R., Gunnarsen, L., and Knudsen, H. N. (2000). Diffusion and sorption of volatile organic compounds in building materials-impact on indoor air quality. Environ. Sci. Technol. 34, 3101–3108. doi: 10.1021/es991291i
Millar, A. A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Orosa, B., He, Q., Mesmar, J., Gilroy, E. M., Mclellan, H., Yang, C., et al. (2017). BTB-BACK domain protein POB1 suppresses immune cell death by targeting ubiquitin E3 ligase PUB17 for degradation. PLoS Genet. 13:e1006540. doi: 10.1371/journal.pgen.1006540
Ostergaard, L., Petersen, M., Mattsson, O., and Mundy, J. (2002). An Arabidopsis callose synthase. Plant Mol. Biol. 49, 559–566. doi: 10.1023/A:1015558231400
Pacini, E. (1997). Tapetum character states analytical keys for tapetum types and activitie. Can. J. Bot. 75, 1448–1459. doi: 10.1139/b97-859
Pan, Q., Han, X., Bai, Y., and Yang, J. (2002). Advances in physiology and ecology studies on stored non-structure carbohydrates in plants. Chin. Bull. Bot. 19, 30–38.
Pearce, S., Ferguson, A., King, J., and Wilson, Z. A. (2015). FlowerNet: a gene expression correlation network for anther and pollen development. Plant Physiol. 167, 1717–1730. doi: 10.1104/pp.114.253807
Renner, S., and Feil, J. P. (1993). Pollinators of tropical dioecious angiosperms. Am. J. Bot. 80, 1100–1107. doi: 10.2307/2445757
Rocheta, M., Sobral, R., Magalhaes, J., Amorim, M. I., Ribeiro, T., Pinheiro, M., et al. (2014). Comparative transcriptomic analysis of male and female flowers of monoecious Quercus suber. Front. Plant Sci. 5:599. doi: 10.3389/fpls.2014.00599
Rogers, H. J. (2006). Programmed cell death in floral organs: how and why do flowers die? Ann. Bot. 97, 309–315. doi: 10.1093/aob/mcj051
Sala, K., Malarz, K., Barlow, P. W., and Kurczynska, E. U. (2017). Distribution of some pectic and arabinogalactan protein epitopes during Solanum lycopersicum (L.) adventitious root development. BMC Plant Biol. 17:25. doi: 10.1186/s12870-016-0949-3
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi, Z., Maximova, S., Liu, Y., Verica, J., and Guiltinan, M. J. (2013). The salicylic acid receptor npr3 is a negative regulator of the transcriptional defense response during early flower development in Arabidopsis. Mol. Plant 6, 802–816. doi: 10.1093/mp/sss091
Sobral, R., Silva, H. G., Morais-Cecilio, L., and Costa, M. M. (2016). The quest for molecular regulation underlying unisexual flower development. Front. Plant Sci. 7:160. doi: 10.3389/fpls.2016.00160
Song, Y. P., Ma, K. F., Ci, D., Chen, Q. Q., Tian, J. X., and Zhang, D. Q. (2013). Sexual dimorphic floral development in dioecious plants revealed by transcriptome, phytohormone, and DNA methylation analysis in Populus tomentosa. Plant Mol. Biol. 83, 559–576. doi: 10.1007/s11103-013-0108-2
Tao, Q., Niu, H., Wang, Z., Zhang, W., Wang, H., Wang, S., et al. (2018). Ethylene responsive factor ERF110 mediates ethylene-regulated transcription of a sex determination-related orthologous gene in two Cucumis species. J. Exp. Bot. 69, 2953–2965. doi: 10.1093/jxb/ery128
Torres, M. A., Dangl, J. L., and Jones, J. D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522. doi: 10.1073/pnas.012452499
Tsakas, M. P., Siskos, A. P., and Siskos, P. A. (2011). “Indoor Air Pollutants and the Impact on Human Health,” in Chemistry, Emission Control, Radioactive Pollution and Indoor Air Quality, ed. M. Nicolas (London: IntechOpen), 447–484.
Ubeda-Tomas, S., and Bennett, M. J. (2010). Plant development: size matters, and it’s all down to hormones. Curr. Biol. 20, R511–R513. doi: 10.1016/j.cub.2010.05.013
Uppalapati, S. R., Ishiga, Y., Wangdi, T., Kunkel, B. N., Anand, A., Mysore, K. S., et al. (2007). The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. MPMI 20, 955–965. doi: 10.1094/MPMI-20-8-0955
Wan, J., Patel, A., Mathieu, M., Kim, S. Y., Xu, D., and Stacey, G. (2008). A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol. Biol. 67, 469–482. doi: 10.1007/s11103-008-9332-6
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
Wei, W. J., Zhang, Y. P., Xiong, J. Y., and Li, M. (2012). A standard reference for chamber testing of material VOC emissions: design principle and performance. Atmos. Environ. 47, 381–388. doi: 10.1016/j.atmosenv.2011.10.051
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 414, 562–565. doi: 10.1038/417571a
Wu, H. M., and Cheun, A. Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44, 267–281. doi: 10.1023/A:1026536324081
Xiang, X., Cao, J. S., Ye, W. Z., Cui, H. M., and Yu, J. N. (2007). Molecular cloning and characterization of BcMYBogu, a novel member of the MYB family involved in OguCMS in Brassica campestris ssp. chinensis. Hereditas 5, 621–628. doi: 10.1360/yc-007-0621
Xu, E. J., and Brosche, M. (2014). Salicylic acid signaling inhibits apoplastic reactive oxygen species signaling. BMC Plant Biol. 14:155. doi: 10.1186/1471-2229-14-155
Yamada, M., and Takeno, K. (2014). Stress and salicylic acid induce the expression of PnFT2 in the regulation of the stress-induced flowering of Pharbitis nil. J. Plant Physiol. 171, 205–212. doi: 10.1016/j.jplph.2013.07.005
Yang, T., Shad Ali, G., Yang, L., Du, L., Reddy, A. S., and Poovaiah, B. W. (2010). Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal Behav. 5, 991–994. doi: 10.1074/jbc.M109.035659
Yang, Y., Liao, Y. W., and Tan, X. F. (2018). Discussion on coordinated development of Vernicia fordii industry and future environment-friendly coatings industry in China. Non Wood For. Res. 36, 188–192. doi: 10.14067/j.cnki.1003-8981.2018.04.030
Zhao, D. Z., Wang, G. F., Speal, B., and Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. doi: 10.1101/gad.997902
Zhao, Q., Zhou, L. J., Liu, J. C., Cao, Z. Z., Du, X. X., Huang, F. D., et al. (2018). Involvement of CAT in the detoxification of HT-induced ROS burst in rice anther and its relation to pollen fertility. Plant Cell Rep. 37, 741–757. doi: 10.1007/s00299-018-2264-y
Keywords: tung tree, salicylic acid, programmed cell death, flower development, unisexual flowers, stamen abortion
Citation: Liu M, Li W, Zhao G, Fan X, Long H, Fan Y, Shi M, Tan X and Zhang L (2019) New Insights of Salicylic Acid Into Stamen Abortion of Female Flowers in Tung Tree (Vernicia fordii). Front. Genet. 10:316. doi: 10.3389/fgene.2019.00316
Received: 27 November 2018; Accepted: 21 March 2019;
Published: 05 April 2019.
Edited by:
Zhixiang Lu, Harvard Medical School, United StatesReviewed by:
Long Gao, University of Pennsylvania, United StatesYi Peng, Northwestern Medicine, United States
Ruijiao Xin, Children’s Hospital of Philadelphia, United States
Copyright © 2019 Liu, Li, Zhao, Fan, Long, Fan, Shi, Tan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Zhang, dHJpd29vZHRpbTkxOEAxMjYuY29t
†These authors have contributed equally to this work
 Meilan Liu
Meilan Liu Wenying Li
Wenying Li Guang Zhao1,2†
Guang Zhao1,2† Lin Zhang
Lin Zhang