
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet. , 12 February 2019
Sec. RNA
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00103
This article is part of the Research Topic Current Advances in the Research of RNA Regulatory Enzymes View all 12 articles
RNA-splicing endonuclease (EndA) cleaves out introns from archaeal and eukaryotic precursor (pre)-tRNA and is essential for tRNA maturation. In archaeal EndA, the molecular mechanisms underlying complex assembly, substrate recognition, and catalysis have been well understood. Recently, certain studies have reported novel findings including the identification of new subunit types in archaeal EndA structures, providing insights into the mechanism underlying broad substrate specificity. Further, metagenomics analyses have enabled the acquisition of numerous DNA sequences of EndAs and intron-containing pre-tRNAs from various species, providing information regarding the co-evolution of substrate specificity of archaeal EndAs and tRNA genetic diversity, and the evolutionary pathway of archaeal and eukaryotic EndAs. Although the complex structure of the heterothermic form of eukaryotic EndAs is unknown, previous reports regarding their functions indicated that mutations in human EndA cause neurological disorders including pontocerebellar hypoplasia and progressive microcephaly, and yeast EndA significantly cleaves mitochondria-localized mRNA encoding cytochrome b mRNA processing 1 (Cpb1) for mRNA maturation. This mini-review summarizes the aforementioned results, discusses their implications, and offers my personal opinion regarding future directions for the analysis of the structure and function of EndAs.
Transfer RNAs (tRNAs) play a fundamental role as adapter molecules for mRNA translation. Maturation events in tRNAs, including removals of the 5′-leader, 3′-trailer, and intron sequences, modification, and addition of 3′-CCA sequences and amino acids are essential for protein synthesis. During tRNA maturation, tRNA splicing is one of the most significant processes in intron splicing and ligation of the two halves of exons in the precursor (pre)-tRNA. Pre-tRNA introns are either auto-catalytically or enzymatically cleaved out in the three domains of life. Group I introns found in pre-tRNA in some bacteria and higher eukaryotic plastids are auto-catalytically cleaved out with an external guanosine-5′-triphosphate (GTP) (Xu et al., 1990; Haugen et al., 2005). By contrast, the introns in cytoplasmic eukaryotic and archaeal pre-tRNAs are enzymatically cleaved out by an RNA-splicing endonuclease (EndA) (Abelson et al., 1998) and the two halves of the exon are subsequently ligated by a tRNA ligase (Phizicky et al., 1986; Westaway et al., 1988; Englert et al., 2011; Popow et al., 2011; Tanaka et al., 2011). Eukaryotic EndA has been extensively identified and characterized in yeast, xenopus, and human. The yeast and human isoform comprise four distinct subunits, referred to as either Sen2, Sen15, Sen34, and Sen54 or αβγσ (Rauhut et al., 1990; Trotta et al., 1997, 2006; Paushkin et al., 2004), although the complete structure of the heterothermic form of eukaryotic EndAs remains unknown. The intron cleavage mechanism of eukaryotic EndAs has been demonstrated owing to early advancements by Dr. John Abelson’s and Dr. Glauco Tocchini-Valentini’s groups (Reyes and Abelson, 1988; Baldi et al., 1992; Bufardeci et al., 1993). Furthermore, archaeal EndAs are classified into three types [α4, α′2, (αβ)2] in accordance with the subunit components (Tocchini-Valentini et al., 2005b) until the 𝜀2 type of archaeal End is newly identified and characterized (Fujishima et al., 2011; Hirata et al., 2012). Currently, four types of EndAs are found in archaea. The general mechanism underlying the recognition and cleavage of pre-tRNA by archaeal EndA was previously reported by Dr. John Abelson’s and Dr. Hong Li’s groups (Li et al., 1998; Li and Abelson, 2000; Xue et al., 2006). Eukaryotic EndA follows a similar mechanism, implicating an evolutionary association between archaeal and eukaryotic EndAs. Furthermore, Calvin and Li (2008) reported the molecular mechanisms underlying complex assembly, substrate recognition, and catalysis in archaeal EndA. Their review article still provides robust evidence regarding the mechanisms underlying substrate recognition and intron-cleavage by archaeal EndAs. This mini-review is focused on recent advancements regarding the structure, function, and evolution of archaeal and eukaryotic EndAs and additionally provides a perspective for future studies on the structure and function of EndAs.
Information regarding the four types of archaeal EndA structures, i.e., α4, α′2, (αβ)2, and 𝜀2, has been obtained from extensive crystallographic studies (Table 1), whereas only the structure of one subunit (Sen15) of eukaryotic EndA has been determined by nuclear magnetic resonance (NMR) spectroscopy (Song and Markley, 2007). Initially, Dr. John Abelson’s group determined the X-ray structure of the homotetrameric form (α4) of archaeal EndA in Methanocaldococcus jannaschii (Li et al., 1998) and of the homodimeric form (α′2) in Archaeoglobus fulgidus (Li and Abelson, 2000). The α′2 type of EndA has also been determined in Thermoplasma acidophilum by another group (Kim et al., 2007). The overall structures of two types are suggestive of a rectangular parallelepiped conformation (Figures 1A,B). Briefly, the N-terminal domain of one α subunit in α4 type of archaeal EndA consists of three α helices and a mixed antiparallel/parallel β sheet, and the C-terminal domain comprises two α helices and a central four-stranded mixed β sheet. Homotetramer formation is achieved by two significant interactions: interaction between two β–β strands at the domain interface between two α subunits and interaction between a negatively charged L10 loop of the α subunit with a positively charged pocket of the opposing α subunit. The interactions are conserved in the four types of archaeal EndAs. The α subunit of α′2 type of EndA is considered the fusion protein of two α subunits of α4 type of EndA because of the evolutionary association between the α4 and α′2 types, based on their sequence similarity, and the two α subunits are connected by a linker from the C-terminal domain of α subunit to the N-terminal domain of another subunit. X-ray structures of (αβ)2 type of archaeal EndAs have been reported in Nanoarchaeum equitans (Mitchell et al., 2009), Pyrobaculum aerophilum (Yoshinari et al., 2009), Aeropyrum pernix (Hirata et al., 2011; Okuda et al., 2011), and Methanopyrus kandleri (Kaneta et al., 2018). The (αβ)2 type EndA comprises two α catalytic subunits and two β structural subunits, and the four subunits are assembled into a heterotetramer (αβ)2 through the aforementioned interactions. The overall structures are very similar to those of the α4 and α′2 types of EndAs, although the structure of P. aerophilum EndA is more compact than that of other EndAs because of the absence of the N-terminal domain of structural β subunit. Furthermore, a new type of 𝜀2 EndA was identified and characterized in Candidatus Micrarchaeum acidiphilum (ARMAN-2) (Fujishima et al., 2011; Hirata et al., 2012), which is deeply branched within Euryarchaeota. ARMAN-2 EndA forms an 𝜀2 homodimer through evolutionarily conserved interactions in the other three types of archaeal EndAs. The 𝜀 protomer is very unique and is separated into three units (αN, α, and βC) fused by two distinct linkers, although the overall shape of ARMAN-2 𝜀2 EndA is similar to that of the other three types of archaeal EndAs. Structure-based sequence analysis suggests that all four types of archaeal EndAs evolved from a common ancestor.
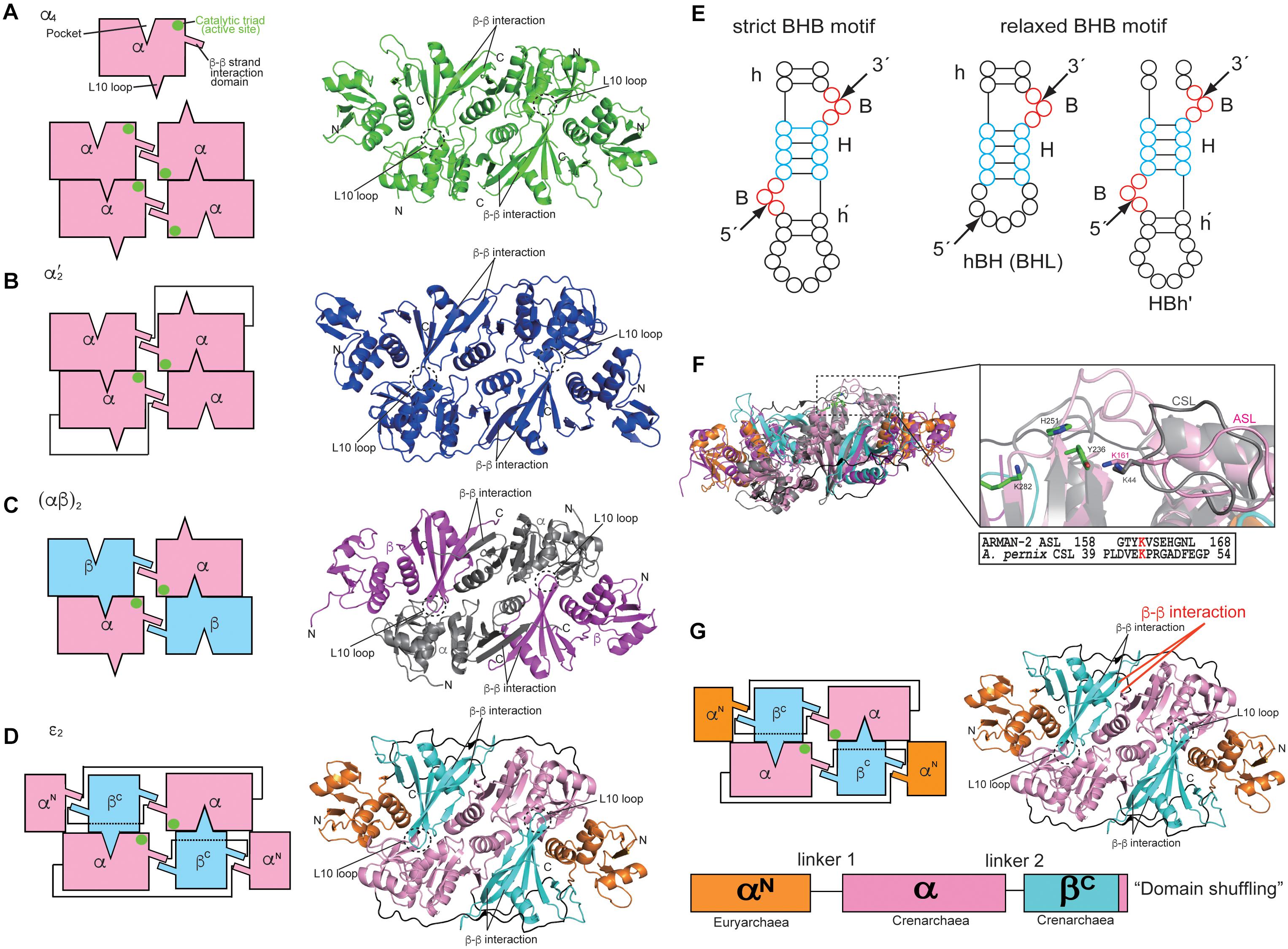
Figure 1. Structures and characteristics of four types of archaeal RNA-splicing endonucleases (EndAs): (A) α4 type Methanocaldococcus jannaschii EndA; (B) α′2 type Archaeoglobus fulgidus EndA; (C) (αβ)2 type Aeropyrum pernix EndA; (D) 𝜀2 type ARMAN-2 EndA. Interactions among the subunits are represented by cartoon models on the left side. The β–β interaction responsible for inter/intra-unit formation, the L10 loop and pocket responsible for dimer/tetramer formation are highlighted. The catalytic triads are marked by green circles. The right panels show the ribbon models of EndAs. (E) Left, strict BHB motif; Right, relaxed BHB motifs (hBH and HBh′). (F) Comparison of the ARMAN-2-specific loop (ASL) in the ARMAN-2 EndA and Crenarchaea-specific loop (CSL) in the Aeropyrum pernix EndA Left: superimposed structures of ARMAN-2 EndA and Aeropyrum pernix EndA. Ribbon diagram of the ARMAN-2 EndA and Aeropyrum pernix EndA are represented by colors similar to those in (C,D). Right: close-up view of the structure of the ASL region (pink) of ARMAN-2 EndA superimposed on the structure of the CSL region (gray) of Aeropyrum pernix EndA. The catalytic triad comprised three catalytic residues (Y236, H251, and K282), shown by a stick model (green). Structure-based sequence alignment is shown at the bottom of the superimposed structures. The conserved K161 in ASL and K44 in CSL are highlighted in red. (G) Gene recombination of three units in the 𝜀 protomer of ARMAN-2 EndA. Interactions among units are represented by cartoon models on the left side. The panels on the right side show the ribbon models of EndAs. The β–β interactions responsible for inter/intra-unit formation are altered for gene recombination (red). These figures are illustrated with some modifications using previous figures (Hirata et al., 2012) and reproduced with permission based on the copyright policy from Oxford University Press.
Three catalytic residues (tyrosine, histidine, and lysine) are conserved in the four types of EndAs, and each subunit assembly of the archaeal EndAs leads to the formation of two intron cleavage sites at the active site (Figures 1A–D, green circle). Similarly, two sets of the two substrate recognition residues [two arginines in α subunit of α4 and α′2 types or arginine and tryptophan residues in α subunit of (αβ)2 and 𝜀2 types] are positioned at a similar location adjacent to the three catalytic residues. Thus, each multimeric conformation of archaeal EndAs is essential for catalysis and substrate tRNA recognition. In eukaryotes, yeast EndA is a heterotetramer (αβγσ) comprising two catalytic (Sen2 and Sen34) and two accessory (Sen15 and Sen54) subunits identified on the basis of homology with their human counterparts (Trotta et al., 1997). The Sen2 and Sen34 share homology with the α subunit of archaeal EndAs and employ the catalytic residues (histidine, tyrosine, and lysine) identical to their archaeal counterparts. Therefore, eukaryotic and archaeal EndAs are presumed to employ a molecular mechanism of cleavage similar to that of ribonuclease A, using the three catalytic residues (Raines, 1998; Calvin and Li, 2008). The complex structure of the heterodimer in eukaryotic EndA is unknown, although the NMR structure of human Sen15 is known (Table 1). The structural arrangement of human Sen15 is similar to that of the C-terminal domain of the α subunit in M. jannaschii α4 EndA. Together, these findings implicate an evolutionary relationship between the eukaryotic and archaeal isoforms of EndA.
Initial studies on the substrate specificity of archaeal EndAs were conducted by Dr. Charles Daniels’ and Roger Garrett’s groups (Kjems and Garrett, 1988; Thompson and Daniels, 1988; Palmer et al., 1992; Kleman-Leyer et al., 1997; Lykke-Andersen and Garrett, 1997; Lykke-Andersen et al., 1997a,b). Archaeal EndAs are known to recognize a bulge-helix-bulge (BHB) motif (Figure 1E), which comprises two bulges (3 nt) separated by one helix (4 nt) located at the exon–intron boundary of pre-tRNAs (Marck and Grosjean, 2003). The canonical BHB motif is frequently present in the anticodon loop between position 37 and 38 (37/38) of archaeal pre-tRNA; however, in some cases, this motif is present in pre-mRNA and pre-rRNA for their maturation (Kjems and Garrett, 1991; Yoshinari et al., 2006). In contrast with this canonical BHB motif, two types of relaxed BHB motifs, non-canonical introns (hBH and HBh′), are present in pre-tRNAs (Figure 1E). The relaxed BHB motifs of hBH and HBh′ disrupt either 5′ or 3′ bulges in the canonical BHB motif. One of the bulges is often absent to form a relaxed bulge-helix-loop (BHL). Furthermore, the unique features of disrupted tRNA genes include multiple (two or three) intron-containing tRNAs (Sugahara et al., 2008; Tocchini-Valentini et al., 2009), split and tri-split tRNAs, wherein tRNA fragments are encoded by two or three genes (Randau et al., 2005; Fujishima et al., 2009), and permuted tRNAs, wherein the sequences of 5′ and 3′ halves of tRNA genes are inverted (Chan et al., 2011). Remarkably, the canonical and relaxed BHB motifs are located not only at the anticodon loop position 37/38 but also at the D-loop, T-loop, and acceptor-stem of archaeal pre-tRNAs (Marck and Grosjean, 2003; Yoshihisa, 2014). Although introns with canonical and relaxed BHB motifs are distributed at the various positions in pre-tRNA, archaeal EndAs actually recognize and cleave introns. However, only two types, i.e., (αβ)2 and 𝜀2 EndAs, can efficiently eliminate introns with relaxed BHB motifs, thereby displaying broad substrate specificity in the EndAs. Eukaryotic EndA recognizes and eliminates introns with a canonical BHB motif from archaeal pre-tRNA, although in most eukaryotic pre-tRNAs, the introns are located at the anticodon loop 37/38 and includes the BHL motif. To eliminate introns with a BHL motif, eukaryotic EndA requires a mature domain of pre-tRNA, wherein the interaction between the D- and T-loops yields a unique structure, the so-called “elbow” (Reyes and Abelson, 1988; Calvin and Li, 2008). The α′2 type of archaeal EndA from Archaeoglobus fulgidus can eliminate introns with the BHL motif at position 37/38 in the case of full-length pre-tRNA (Tocchini-Valentini et al., 2005a).
The (αβ)2 and 𝜀2 EndAs have broad substrate specificity, which can efficiently cleave not only the introns with canonical BHB motif but also those with a relaxed BHB motif. The molecular mechanism underlying the broad substrate specificity of (αβ)2 EndA is unknown. To clarify the mechanism, structural and biochemical analyses of the (αβ)2 type of EndA from hyperthermophilic crenarchaeon Aeropyrum pernix was performed (Hirata et al., 2011). At the time, (αβ)2-type EndAs were reported exclusively in crenarchaea and nanoarchaea, except for euryarchaeon Methanopyrus kandleri (Marck and Grosjean, 2003). Our studies on A. pernix EndA reported a Crenarchaea-specific loop (CSL), which was conserved in crenarchaeal EndAs and located adjacent to the active site (Figure 1F). Furthermore, insertion of CSL in A. fulgidus α′2 EndA conferred A. pernix EndA with broad substrate specificity, which originally had narrow substrate specificity. In the A. pernix EndA with a CSL insert, an alanine-substituted mutant of the conserved Lys residue of CSL disrupted the broad substrate specificity. Together, these findings suggest that the Lys residue of CSL plays a significant role as an RNA binding site and is responsible for the broad substrate specificity in the (αβ)2 of crenarchaeal EndAs. Similarly, the 𝜀2 type of ARMAN-2 EndA possesses an ARMAN-2 specific loop (ASL), which confers broad substrate specificity, and the Lys residue of ASL functions as the RNA recognition site. Although the ASL conformation in ARMAN-2 EndA is markedly similar to that of CSL in A. pernix EndA, there are no obvious sequence similarities between the ASL and CSL, except for the conserved Lys residue, which functions as the substrate recognition site. Together, these findings indicate that the ASL was acquired by a distinctly independent evolutionary pathway toward the CSL (i.e., “convergent evolution”). However, it is still unknown why each Lys residue conserved in the CSL and ASL is required for intron cleavage, despite the presence of three catalytic residues in the EndAs. However, M. kandleri EndA was identified as the (αβ)2 type lacking specific loops such as the ASL and CSL (Kaneta et al., 2018). While M. kandleri EndA slightly cleaves introns with a relaxed BHB motif in M. kandleri pre-tRNAGlu (UUC), it could not eliminate introns from a mini-helix RNA with a BHL motif. Therefore, the M. kandleri EndA is considered to be of the (αβ)2 type with constrained substrate specificity.
The α4 type of archaeal EndA, which encodes a single catalytic α subunit, is proposed to be the prototype of the EndAs (Tocchini-Valentini et al., 2005b), and the subsequent subfunctionalization of gene duplication and fusion has yielded the other three types [α′2, (αβ)2 and 𝜀2]. Intriguingly, 𝜀2-type ARMAN-2 EndA appears to have undergone a genetic recombination of the three subunits, euryarchaeal α subunit, crenarchaeal α subunit, and crenarchaeal β subunit (Hirata et al., 2012), comprising three units (αN-α-βC) of the 𝜀 protomer (Figure 1G). Each unit is clearly divided into a domain structure, thus providing a good example of the so-called “domain shuffling” occurring naturally. Moreover, the C-terminal subdomain of the crenarchaeal β subunit may have been incorporated into the terminus of the crenarchaeal α subunit, which may have primarily led to changes in the structural location of β–β interaction responsible for subunit assembly.
The sequence of archaeal α subunit is locally conserved in the two catalytic subunits (Sen2 and Sen34) of the heterotetrameric form (αβγδ) of eukaryotic EndA with approximately 50 amino acid residues. Therefore, eukaryotic EndA is considered to have evolved from the archaeal (αβ)2 EndA with the acquisition of new subunits (γ and δ). Remarkably, the primitive eukaryotic red alga Cyanidioschyzon merolae harbors many disrupted tRNA genes with a relaxed BHB motif as employed in Archaea (Soma et al., 2007, 2013; Soma, 2014). The C. merolae EndA is expected to comprise three subunits [cmSen2p, cmSen34p, and cmSen54p (αβγ)] for processing these pre-tRNAs; however, it does not contain the ASL and CSL. Thus, heterotrimer form of C. merolae EndA might be an intermediate in the evolutionary transition between the heterotetramer of archaeal EndA to heterotetramer of eukaryotic EndA. Furthermore, recent bioinformatics analysis has reported that archaeal species with specific loops such as the ASL and CSL in EndAs clearly represent a trend of increased intron-containing tRNA genes with BHB and relaxed BHB motifs, suggesting coevolution of tRNA gene diversity and broad substrate specificity (Kaneta et al., 2018). These findings further update the previous concept of co-evolution (Tocchini-Valentini et al., 2005b; Fujishima and Kanai, 2014).
Vertebrate and Saccharomyces cerevisiae EndAs are localized in the nucleus (Paushkin et al., 2004) and on the mitochondrial outer membrane (Yoshihisa et al., 2003, 2007), respectively. A recent study reported that S. cerevisiae EndA cleaves the mitochondria-localized mRNA encoding Cbp1 (cytochrome b mrNA processing 1) and this cleavage requires a predicted stem-loop structure of the endonucleolytic cleavage-inducible sequence of Cpb1 with synergistic effects of other factors (Tsuboi et al., 2015). These significant findings provide evidence regarding the biological role of mitochondrial-localized S. cerevisiae EndA and suggest that the EndA has broad substrate specificity owing to specific recognition of the predicted stem-loop structure without the BHB motif. Furthermore, the human EndA complex (TSen2, TSen15, TSen34, and TSen54) reportedly cleaves introns from pre-tRNAs, and the TSen2 subunit is involved in pre-mRNA′3 end formation (Paushkin et al., 2004). These reports further expand the possibility that the substrates of EndA are non-coding RNAs involved in the regulation of gene expression. To confirm the possibility, crosslinking RNA-EndA complex using UV irradiation combined with immunoprecipitation and RNA sequencing could be a useful method to identify the non-coding RNAs as the substrate of EndA. More importantly, recessive mutations in the genes of three subunits (TSen2, TSen34, and TSen54) cause pontocerebellar hypoplasia (PCH) types 2A-C, 4, and 5 (Budde et al., 2008; Namavar et al., 2011a,b; Bierhals et al., 2013; Maraş-Genç et al., 2015). PCH2 is reportedly involved in progressive cerebral atrophy and microcephaly, dyskinesia, seizures and early childhood mortality. Furthermore, a recent study reported that three homozygous TSEN15 cause a milder version of the PCH2-related pathology (Breuss et al., 2016). Hence, appropriate EndA function is required for brain development in humans. However, the mechanism underlying the pathogenesis of PCH which is caused by human EndA mutations remains unclear because its complex structure is yet unknown.
The mechanism underlying the recognition and cleavage of RNA introns by EndAs is known; however, they have gained increasing interest, since the evolutionary pathway from archaeal to eukaryotic EndA and the mechanism underlying the broad substrate specificity of archaeal and eukaryotic EndA warrant further investigation. The conserved Lys residue in CSL and ASL of the (αβ)2 and 𝜀2 types of archaeal EndAs might function as the catalytic and RNA recognition residue. Eukaryotic EndAs probably possess broad substrate specificity, similar to the archaeal (αβ)2- and 𝜀2- type EndAs, whereas the mechanism underlying the broad substrate specificity may vary between the eukaryotic and archaeal EndAs. Further structural analysis is required to elucidate the detailed mechanism underlying broad substrate specificity by archaeal and eukaryotic EndAs. In particular, the structural information of human EndA may be useful for drug design that improves the inadequate EndA function, which causes the developmental retardation of human brain described above.
The author confirms being the sole contributor of this work and has approved it for publication.
This work was supported by JSPS KAKENHI grant number JP18K06088 (to AH).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abelson, J., Trotta, C., and Li, H. (1998). tRNA splicing. J. Biol. Chem. 273, 12685–12688. doi: 10.1074/jbc.273.21.12685
Baldi, M. I., Mattoccia, E., Bufardeci, E., Fabbri, S., and Tocchini-Valentini, G. P. (1992). Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science 255, 1404–1408. doi: 10.1126/science.1542788
Bierhals, T., Korenke, G. C., Uyanik, G., and Kutsche, K. (2013). Pontocerebellar hypoplasia type 2 and TSEN2: review of the literature and two novel mutations. Eur. J. Med. Genet. 56, 325–330. doi: 10.1016/j.ejmg.2013.03.009
Breuss, M. W., Sultan, T., James, K. N., Rosti, R. O., Scott, E., Musaev, D., et al. (2016). Autosomal-recessive mutations in the tRNA splicing endonuclease subunit TSEN15 cause pontocerebellar hypoplasia and progressive microcephaly. Am. J. Hum. Genet. 99, 228–235. doi: 10.1016/j.ajhg.2016.08.009
Budde, B., Namavar, Y., Barth, P., Poll-The, B., Nürnberg, G., Becker, C., et al. (2008). tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 40, 1113–1118. doi: 10.1038/ng.204
Bufardeci, E., Fabbri, S., Baldi, M. I., Mattoccia, E., and Tocchini-Valentini, G. P. (1993). In vitro genetic analysis of the structural features of the pre-tRNA required for determination of the 3’ splice site in the intron excision reaction. EMBO J. 12, 4697–4704. doi: 10.1002/j.1460-2075.1993.tb06158.x
Calvin, K., and Li, H. (2008). RNA-splicing endonuclease structure and function. Cell. Mol. Life Sci. 65, 1176–1185. doi: 10.1007/s00018-008-7393-y
Chan, P. P., Cozen, A. E., and Lowe, T. M. (2011). Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 12:R38. doi: 10.1186/gb-2011-12-4-r38
Englert, M., Sheppard, K., Aslanian, A., Yates, J. R., and Söll, D. (2011). Archaeal 3’-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295. doi: 10.1073/pnas.1018307108
Fujishima, K., and Kanai, A. (2014). tRNA gene diversity in the three domains of life. Front. Genet. 5:142. doi: 10.3389/fgene.2014.00142
Fujishima, K., Sugahara, J., Kikuta, K., Hirano, R., Sato, A., Tomita, M., et al. (2009). Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl. Acad. Sci. U.S.A. 106, 2683–2687. doi: 10.1073/pnas.0808246106
Fujishima, K., Sugahara, J., Miller, C. S., Baker, B. J., Di Giulio, M., Takesue, K., et al. (2011). A novel three-unit tRNA splicing endonuclease found in ultrasmall Archaea possesses broad substrate specificity. Nucleic Acids Res. 39, 9695–9704. doi: 10.1093/nar/gkr692
Haugen, P., Simon, D. M., and Bhattacharya, D. (2005). The natural history of group I introns. Trends Genet. 21, 111–119. doi: 10.1016/j.tig.2004.12.007
Hirata, A., Fujishima, K., Yamagami, R., Kawamura, T., Banfield, J. F., Kanai, A., et al. (2012). X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity. Nucleic Acids Res. 40, 10554–10566. doi: 10.1093/nar/gks826
Hirata, A., Kitajima, T., and Hori, H. (2011). Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop. Nucleic Acids Res. 39, 9376–9389. doi: 10.1093/nar/gkr615
Kaneta, A., Fujishima, K., Morikazu, W., Hori, H., and Hirata, A. (2018). The RNA-splicing endonuclease from the euryarchaeaon Methanopyrus kandleri is a heterotetramer with constrained substrate specificity. Nucleic Acids Res. 46, 1958–1972. doi: 10.1093/nar/gky003
Kim, Y., Mizutani, K., Rhee, K., Nam, K., Lee, W., Lee, E., et al. (2007). Structural and mutational analysis of tRNA intron-splicing endonuclease from Thermoplasma acidophilum DSM 1728: catalytic mechanism of tRNA intron-splicing endonucleases. J. Bacteriol. 189, 8339–8346. doi: 10.1128/JB.00713-07
Kjems, J., and Garrett, R. A. (1988). Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell 54, 693–703. doi: 10.1016/S0092-8674(88)80014-X
Kjems, J., and Garrett, R. A. (1991). Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc. Natl. Acad. Sci. U.S.A. 88, 439–443. doi: 10.1073/pnas.88.2.439
Kleman-Leyer, K., Armbruster, D., and Daniels, C. (1997). Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell 89, 839–847. doi: 10.1016/S0092-8674(00)80269-X
Li, H., and Abelson, J. (2000). Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol. 302, 639–648. doi: 10.1006/jmbi.2000.3941
Li, H., Trotta, C., and Abelson, J. (1998). Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280, 279–284. doi: 10.1126/science.280.5361.279
Lykke-Andersen, J., Aagaard, C., Semionenkov, M., and Garrett, R. (1997a). Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem. Sci. 22, 326–331. doi: 10.1016/S0968-0004(97)01113-4
Lykke-Andersen, J., Garrett, R., and Kjems, J. (1997b). Mapping metal ions at the catalytic centres of two intron-encoded endonucleases. EMBO J. 16, 3272–3281. doi: 10.1093/emboj/16.11.3272
Lykke-Andersen, J., and Garrett, R. (1997). RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J. 16, 6290–6300. doi: 10.1093/emboj/16.20.6290
Maraş-Genç, H., Uyur-Yalçin, E., Rosti, R., Gleeson, J. G., and Kara, B. (2015). TSEN54 gene-related pontocerebellar hypoplasia type 2 presenting with exaggerated startle response: report of two cases in a family. Turk. J. Pediatr. 57, 286–289.
Marck, C., and Grosjean, H. (2003). Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA 9, 1516–1531. doi: 10.1261/rna.5132503
Mitchell, M., Xue, S., Erdman, R., Randau, L., Söll, D., and Li, H. (2009). Crystal structure and assembly of the functional Nanoarchaeum equitans tRNA splicing endonuclease. Nucleic Acids Res. 37, 5793–5802. doi: 10.1093/nar/gkp537
Namavar, Y., Barth, P. G., Kasher, P. R., van Ruissen, F., Brockmann, K., Bernert, G., et al. (2011a). Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain 134, 143–156. doi: 10.1093/brain/awq287
Namavar, Y., Chitayat, D., Barth, P. G., van Ruissen, F., de Wissel, M. B., Poll-The, B. T., et al. (2011b). TSEN54 mutations cause pontocerebellar hypoplasia type 5. Eur. J. Hum. Genet. 19, 724–726. doi: 10.1038/ejhg.2011.8
Okuda, M., Shiba, T., Inaoka, D. K., Kita, K., Kurisu, G., Mineki, S., et al. (2011). A conserved lysine residue in the crenarchaea-specific loop is importantfor the crenarchaeal splicing endonuclease activity. J. Mol. Biol. 405, 92–104. doi: 10.1016/j.jmb.2010.10.050
Palmer, J. R., Baltrus, T., Reeve, J. N., and Danielsm, C. J. (1992). Transfer RNA genes from the hyperthermophilic Archaeon, Methanopyrus kandleri. Biochim. Biophys. Acta 1132, 315–318. doi: 10.1016/0167-4781(92)90168-Y
Paushkin, S., Patel, M., Furia, B., Peltz, S., and Trotta, C. (2004). Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3’ end formation. Cell 117, 311–321. doi: 10.1016/S0092-8674(04)00342-3
Phizicky, E. M., Schwartz, R. C., and Abelson, J. (1986). Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J. Biol. Chem. 261, 2978–2986.
Popow, J., Englert, M., Weitzer, S., Schleiffer, A., Mierzwa, B., Mechtler, K., et al. (2011). HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764. doi: 10.1126/science.1197847
Randau, L., Münch, R., Hohn, M., Jahn, D., and Söll, D. (2005). Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5’- and 3’-halves. Nature 433, 537–541. doi: 10.1038/nature03233
Rauhut, R., Green, P., and Abelson, J. (1990). Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. J. Biol. Chem. 265, 18180–18184.
Reyes, V., and Abelson, J. (1988). Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55, 719–730. doi: 10.1016/0092-8674(88)90230-9
Soma, A. (2014). Circularly permuted tRNA genes: their expression and implications for their physiological relevance and development. Front. Genet. 5:63. doi: 10.3389/fgene.2014.00063
Soma, A., Onodera, A., Sugahara, J., Kanai, A., Yachie, N., Tomita, M., et al. (2007). Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 318, 450–453. doi: 10.1126/science.1145718
Soma, A., Sugahara, J., Onodera, A., Yachie, N., Kanai, A., Watanabe, S., et al. (2013). Identification of highly-disrupted tRNA genes in nuclear genome of the red alga, Cyanidioschyzon merolae 10D. Sci. Rep. 3:2321. doi: 10.1038/srep02321
Song, J., and Markley, J. L. (2007). Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold. J. Mol. Biol. 366, 155–164. doi: 10.1016/j.jmb.2006.11.024
Sugahara, J., Kikuta, K., Fujishima, K., Yachie, N., Tomita, M., and Kanai, A. (2008). Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 25, 2709–2716. doi: 10.1093/molbev/msn216
Tanaka, N., Meineke, B., and Shuman, S. (2011). RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 286, 30253–30257. doi: 10.1074/jbc.C111.274597
Thompson, L. D., and Daniels, C. J. (1988). A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J. Biol. Chem. 263, 17951–17959.
Tocchini-Valentini, G. D., Fruscoloni, P., and Tocchini-Valentini, G. P. (2005a). Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl. Acad. Sci. U.S.A. 102, 15418–15422. doi: 10.1073/pnas.0506750102
Tocchini-Valentini, G. D., Fruscoloni, P., and Tocchini-Valentini, G. P. (2005b). Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc. Natl. Acad. Sci. U.S.A. 102, 8933–8938. doi: 10.1073/pnas.0502350102
Tocchini-Valentini, G. D., Fruscoloni, P., and Tocchini-Valentini, G. P. (2009). Processing of multiple-intron-containing pre-tRNA. Proc. Natl. Acad. Sci. U.S.A. 106, 20246–20251. doi: 10.1073/pnas.0911658106
Trotta, C., Miao, F., Arn, E., Stevens, S., Ho, C., Rauhut, R., et al. (1997). The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89, 849–858. doi: 10.1016/S0092-8674(00)80270-6
Trotta, C., Paushkin, S., Patel, M., Li, H., and Peltz, S. (2006). Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441, 375–377. doi: 10.1038/nature04741
Tsuboi, T., Yamazaki, R., Nobuta, R., Ikeuchi, K., Makino, S., Ohtaki, A., et al. (2015). The tRNA splicing endonuclease complex cleaves the mitochondria-localized CBP1 mRNA. J. Biol. Chem. 290, 16021–16030. doi: 10.1074/jbc.M114.634592
Westaway, S. K., Phizicky, E. M., and Abelson, J. (1988). Structure and function of the yeast tRNA ligase gene. J. Biol. Chem. 263, 3171–3176.
Xu, M., Kathe, S., Goodrich-Blair, H., Nierzwicki-Bauer, S., and Shub, D. (1990). Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science 250, 1566–1570. doi: 10.1126/science.2125747
Xue, S., Calvin, K., and Li, H. (2006). RNA recognition and cleavage by a splicing endonuclease. Science 312, 906–910. doi: 10.1126/science.1126629
Yoshihisa, T. (2014). Handling tRNA introns, archaeal way and eukaryotic way. Front. Genet. 5:213. doi: 10.3389/fgene.2014.00213
Yoshihisa, T., Ohshima, C., Yunoki-Esaki, K., and Endo, T. (2007). Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 12, 285–297. doi: 10.1111/j.1365-2443.2007.01056.x
Yoshihisa, T., Yunoki-Esaki, K., Ohshima, C., Tanaka, N., and Endo, T. (2003). Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell 14, 3266–3279. doi: 10.1091/mbc.e02-11-0757
Yoshinari, S., Itoh, T., Hallam, S., DeLong, E., Yokobori, S., Yamagishi, A., et al. (2006). Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease. Biochem. Biophys. Res. Commun. 346, 1024–1032. doi: 10.1016/j.bbrc.2006.06.011
Keywords: RNA-splicing endonuclease, intron-containing tRNA, broad substrate specificity, co-evolution of protein and RNA, archaea and eukaryote
Citation: Hirata A (2019) Recent Insights Into the Structure, Function, and Evolution of the RNA-Splicing Endonucleases. Front. Genet. 10:103. doi: 10.3389/fgene.2019.00103
Received: 19 November 2018; Accepted: 30 January 2019;
Published: 12 February 2019.
Edited by:
Tohru Yoshihisa, University of Hyogo, JapanReviewed by:
Naoki Shigi, National Institute of Advanced Industrial Science and Technology (AIST), JapanCopyright © 2019 Hirata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akira Hirata, aGlyYXRhLmFraXJhLm1nQGVoaW1lLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.