- 1Faculty of Agronomy, S. Seifullin Kazakh AgroTechnical University, Astana, Kazakhstan
- 2Biological Sciences, College of Science and Engineering, Flinders University, Bedford Park, SA, Australia
- 3School of Agriculture, Food and Wine, University of Adelaide, Adelaide, SA, Australia
- 4Wheat Initiative, Julius-Kühn-Institute, Berlin, Germany
Intracellular vesicle trafficking genes, Rab, encoding small GTP binding proteins, have been well studied in medical research, but there is little information concerning these proteins in plants. Some sub-families of the Rab genes have not yet been characterized in plants, such as RabC – otherwise known as Rab18 in yeast and animals. Our study aimed to identify all CaRab gene sequences in chickpea (Cicer arietinum L.) using bioinformatics approaches, with a particular focus on the CaRabC gene sub-family since it featured in an SNP database. Five isoforms of the CaRabC gene were identified and studied: CaRabC-1a, -1b, -1c, -2a and -2a∗. Six accessions of both Desi and Kabuli ecotypes, selected from field trials, were tested for tolerance to abiotic stresses, including salinity, drought and rapid dehydration and compared to plant growth under control conditions. Expression analysis of total and individual CaRabC isoforms in leaves of control plants revealed a very high level of expression, with the greatest contribution made by CaRabC-1c. Salinity stress (150 mM NaCl, 12 days in soil) caused a 2-3-fold increased expression of total CaRabC compared to controls, with the highest expression in isoforms CaRabC-1c, -2a∗ and -1a. Significantly decreased expression of all five isoforms of CaRabC was observed under drought (12 days withheld water) compared to controls. In contrast, both total CaRabC and the CaRabC-1a isoform showed very high expression (up-to eight-fold) in detached leaves over 6 h of dehydration. The results suggest that the CaRabC gene is involved in plant growth and response to abiotic stresses. It was highly expressed in leaves of non-stressed plants and was down-regulated after drought, but salinity and rapid dehydration caused up-regulation to high and very high levels, respectively. The isoforms of CaRabC were differentially expressed, with the highest levels recorded for CaRabC-1c in controls and under salinity stress, and for CaRabC-1a – in rapidly dehydrated leaves. Genotypic variation in CaRabC-1a, comprising eleven SNPs, was found through sequencing of the local chickpea cultivar Yubileiny and germplasm ICC7255 in comparison to the two fully sequenced reference accessions, ICC4958 and Frontier. Amplifluor-like markers based on one of the identified SNPs in CaRabC-1a were designed and successfully used for genotyping chickpea germplasm.
Introduction
Plant genomes include a superfamily of genes that encode small GTP-binding proteins (Guanosine triphosphatases) that are classified into four groups: Arf, Rab, Ran and Rho; and an additional Ras-GTP gene group is found only in yeast and animals (Ma, 2007). Small GTP-binding proteins were first described in medical research, where the term “Ras” stemmed from their association with rat sarcoma (Chang et al., 1982; Bishop, 1985; Chavrier et al., 1990). The remaining three-letter names are not related in structure or function to the genes but rather refer to their product or some other feature (Coffin et al., 1981). Small GTP-binding proteins are known to be involved in a diverse range of activities in eukaryotes that are vital for growth, development and repair; from cytoskeletal organization, vacuolar storage and signaling, to modulation of gene expression (Takai et al., 2001). The mechanism for the regulation of GTP-binding proteins is conserved in all organisms and involves cycling between active (GTP-bound) and inactive (GDP-bound) forms, so they are often described as “molecular switches” that are turned “on” or “off” via the hydrolysis of GTP (Marshall, 1993). Activation requires the dissociation of GDP, which can be either stimulated by a regulatory factor named GEP (GDP/GTP Exchange Protein) or inhibited by GDI (GDP Dissociation Inhibitor; Takai et al., 2001; Liu et al., 2015; Martín-Davison et al., 2017).
Rab proteins, encoded by Rab-GTP genes, are normally prenylated at their carboxyl terminus. The hydrophobic prenyl-groups facilitate attachment to membranes and are therefore integral to the biological role performed by Rab proteins in vesicle trafficking via endocytic and exocytic pathways between the endoplasmic reticulum, Golgi membrane network, endosome, plasma membrane and all intracellular membranes (Alory and Balch, 2003). Rab proteins are highly conserved across kingdoms, from yeast to animals and plants (Haubruck et al., 1987; Marcote et al., 2000), but are most often present as a small family of highly similar genes. They are divided into either nine (Ma, 2007) or 18 clades (Agarwal et al., 2009) based on their structure, with only eight clades represented in plants. Historically, different nomenclatures were adopted for identification of Rab genes in plants compared to animals. For example, in plants, eight capitalized letters from A to H were used in the names of Rab genes, while the numbers 1 to 11 were applied in human, animal and yeast research. In the absence of a universal system of nomenclature for Rab genes and their proteins, a list of all known genes and their respective identifiers for both nomenclatures is given later in the text.
The genes for Rab GTP-binding proteins should not be confused with the similarly named Dehydrin genes in plants, which are also known as RAB, meaning “Responsive to ABA” (Abscisic acid). Dehydrins encode proteins belonging to the large but very different group of Late embryogenesis abundant proteins, LEA (Hundertmark and Hincha, 2008). For example, AtRAB18 (or AtRab18) was described and studied in Arabidopsis thaliana in response to various abiotic stresses and ABA treatment (Lång and Palva, 1992; Rushton et al., 2012; Hernández-Sánchez et al., 2017). Despite the identical name, this gene is neither structurally nor functionally related to the Rab-GTP genes, and care must be taken to clearly distinguish between the two. The mixing of these two different types of genes is unfortunately apparent in recent publications. For example, Jiang et al. (2017) studied the correctly designated TaRab18 (=TaRabC1) gene in response to stripe rust in bread wheat, but this gene was incorrectly compared with RAB18 (Responsive to ABA) in Arabidopsis, rice and maize. As a result, the Authors wrongly cited work by Lång and Palva (1992) and others on the Dehydrin AtRab18 to support their findings on the sensitivity of TaRab18 (=TaRabC1) to ABA.
In plants, Rab-GTP genes are reportedly involved in multiple physiological processes (Borg et al., 1997; Rehman and Sansebastiano, 2014; He et al., 2018; Lawson et al., 2018) and are often highly expressed in response to biotic and abiotic stresses (Marcote et al., 2000; Stenmark and Olkkonen, 2001; Zerial and McBride, 2001; Rutherford and Moore, 2002; Ma, 2007; Woollard and Moore, 2008; Agarwal et al., 2009). However, despite the numerous links, little is known about the precise molecular mechanisms underlying their involvement in plant stress responses.
One of the first studies to report a link between Rab protein and abiotic stress was a report by O’Mahony and Oliver (1999) who found increased transcript levels of the Rab2 gene (otherwise known as RabB) in the desiccation-tolerant grass Sporobolus stapfianus in response to dehydration, but decreased transcript levels after rehydration. This suggested the involvement of SsRab2 in both the short-term response and later recovery from desiccation. SsRab2 was found to share 90% similarity to Rab2 proteins found in rice, maize, Arabidopsis, Lotus japonicus and soybean (O’Mahony and Oliver, 1999). Since that time, links to various stresses have been established for genes encoding Rab proteins in numerous plants, and especially in species with high abiotic stress-tolerance such as Lilium formolongi – LfRabB (Howlader et al., 2017), poplar – PtRabE1b (Zhang et al., 2018), and Mesembryanthemum crystallinum – McRab5b (=McRabF) (Bolte et al., 2000). Interestingly, many plant species were studied for RabG genes and their corresponding proteins including the halophyte species, Aeluropus lagopoides – AlRab7 (=AlRabG) (Rajan et al., 2015) and food grain crop, Pennisetum glaucum – PgRab7 (=PgRabG) (Agarwal et al., 2008), as well as more stress susceptible crops such as rice, Oryza sativa – OsRab7 (=OsRabG) (Nahm et al., 2003) and peanut, Arachis hypogaea – AhRabG (Sui et al., 2017), and the model species A. thaliana – AtRab7 (=AtRabG) (Mazel et al., 2004). A comprehensive analysis of all MpRab genes was reported for the liverwort, Marchantia polymorpha (Minamino et al., 2018).
Rab transcripts are often found to show different responses to abiotic stresses. For example, in rice, dehydration triggered a strong increase in OsRab7 (=OsRabG) transcript after 4 h and then a decrease after 10 h. However, no significant changes were found in response to cold or salinity stress (Nahm et al., 2003). Similarly, in the halophytic grass A. lagopoides, AlRab7 (=AlRabG) was upregulated by dehydration, but salinity stress caused no significant increase in transcript levels (Rajan et al., 2015). In another halophyte, M. crystallinum, expression of McRab5b (=McRabF) was higher after 2 h and continued to rise over 3 days of very strong salt stress (400 mM NaCl), but wilting or osmotic stress triggered no change in expression (Bolte et al., 2000). These differences obviously reflect various roles of the intracellular membrane system to abiotic stresses and may provide the key to uncovering the precise molecular mechanisms underlying differential plant susceptibility or tolerance to an environmental stress.
A number of studies have used a transgenic approach to shed light on the mechanisms explaining the link between Rab proteins and plant stress and to explore how Rab proteins could play a role in the breeding of more stress-tolerant crops. For example, Mazel et al. (2004) constitutively overexpressed AtRabG3e (=AtRab7) in Arabidopsis. The transgenic plants accumulated more sodium in vacuoles and showed greater tolerance to salinity and osmotic stress. Evidence was also found for increased endocytosis in roots and leaves and entry of Reactive oxygen species into the cell to trigger signaling and subsequent activation of stress tolerance mechanisms (Mazel et al., 2004; Baral et al., 2015). AhRabG, OsRab7 (=OsRabG) and OsRab11 (=OsRabA) were also overexpressed in transgenic peanut and rice, respectively, producing plants that showed relatively higher salinity tolerance compared to wild-type plants (Peng et al., 2014; Sui et al., 2017; Chen and Heo, 2018). In transgenic peanut plants, of 132 genes differentially expressed, most were identified as transcription factors (TF) relating to salinity tolerance (Sui et al., 2017).
The aim of this study was to identify and analyze a possible candidate gene involved in the tolerance to drought, salinity and rapid dehydration in chickpea, C. arietinum, a species that is becoming increasingly popular as a cash crop in agricultural areas with the requirements for moderate tolerance to high temperatures, drought and salinity stress during the growing season. A candidate gene CaRabC1, belonging to the family of Rab-GTP genes, was identified from an SNP database using bioinformatic and molecular genetic analyses. Currently, the only report concerning chickpea Rab-GTP genes was published by Muñoz et al. (2001), who identified a Rab-specific GDI in chickpea seedlings showing 96% homology to MsRab11f (=MsRabG), a GDI in Medicago truncatula (Yaneva and Niehaus, 2005). Our study therefore represents the first report of the Rab-GTP family of genes in C. arietinum. We present the results of bioinformatic analyses of the identified genes and tests conducted to assess the expression of all isoforms of the CaRabC gene family in response to salinity, drought and rapid dehydration in selected chickpea genotypes. Amplifluor-like markers based on one of the identified SNPs in CaRabC-1a were used for genotyping of chickpea germplasm.
Materials and Methods
Plant Material
A germplasm collection comprising 250 chickpea (C. arietinum L) samples from the ICRISAT Reference set plus local accessions were tested over 3 years in field trials in Northern and Central Kazakhstan. Six accessions were selected during field trials for further molecular analyses, as listed in Table 1. The first accession, cv. Yubileiny, originated from Krasnokutskaya Breeding Station, in the Saratov region (Russia), and is used as a Standard for local field trials with chickpea accessions. The remaining five chickpea lines were selected from the original 230 collected in the ICRISAT Reference set, to represent diverse gene-pool sources.

Table 1. List and short description of six selected chickpea germplasm accessions used for molecular analyses.
Identification of the Gene of Interest Using Bioinformatics and Molecular Phylogenetic Comparative Analysis
Bioinformatics and systems biology methods were applied in this study to identify a target gene or “Gene of Interest” (GoI) with a potential role in tolerance to abiotic stresses in chickpea. Initially, the SNP database for C. arietinum L.1 was used to search and select one suitable SNP with a short fragment of sequence for further study. The full-length nucleotide sequence of the GoI and its corresponding polypeptide sequence was retrieved from the same database and used for both BLASTN and BLASTP in NCBI and in GenomeNet Database Resources, hosted by Kyoto University, Japan2. All chickpea gene sequences with KEGG and NCBI identification and the encoded proteins were downloaded from GenomeNet and NCBI databases, while chromosome locations were checked using LIS, Legume Information System database3. The A. thaliana genes displaying the highest level of similarity to each GoI within the gene family were identified using alignments from the same database.
Multiple sequence alignments of nucleotide sequences for the Rab family of genes were conducted in CLUSTALW using GenomeNet Database Resources4, while CLC Main Workbench software5 was used for protein amino acid sequence alignment.
The molecular dendrogram was constructed using BLASTP at GenomeNet Database Resources (See footnote 2) with the function of ETE3 v3.0.0b32 (Huerta-Cepas et al., 2016) and MAFFT v6.861b applied using the default options (Katoh and Standley, 2013). The FastTree v2.1.8 program with default parameters was used for phylogenetic tree preparation (Price et al., 2009).
Abiotic Stress Treatments: Salinity, Drought and Rapid Dehydration
Three experiments applying abiotic stress treatments (salinity, slow drought and rapid dehydration) were carried out in chickpea for RT-qPCR analyses using the same conditions as described earlier in our publication for wheat (Zotova et al., 2018). The size of containers used, number of plants, soil type and growth conditions were all as described and no artificial inoculation of rhizobium was applied.
For salt stress, twenty-four uniform seedlings in each of six accessions were grown for one month in two separate containers. On “Day 0,” three plants from each accession (three biological replicates) were randomly selected from each container, before the salt stress was applied. The two youngest fully developed leaves were collected from each selected plant into a 10-ml plastic tube and immediately frozen in liquid nitrogen and stored at –80°C until RNA extraction. Subsequently, 200 ml of 150 mM NaCl was applied to the container, covering the entire soil surface but avoiding any direct contact with the plants. The NaCl treatments were applied four times, on every third day following Day 0 (over 12 days in total) in treatment containers, while the same volume of tap-water without NaCl was used under the same schedule in the control containers. No solution was lost through drainage from any container. No supplementary CaCl2 was added despite the recommended requirements in experiments with hydroponics. This is because the soil mix used contained sufficient available calcium and no symptoms of Ca deficiency were apparent in the treated plants. After 12 days, as on Day 0, the two youngest fully developed leaves were collected from each of three plants both in salt treatments and controls. Leaf samples were immediately frozen in liquid nitrogen and stored at –80°C for RNA extraction.
Experiments with slowly droughted plants and rapid dehydration of detached leaves were carried out using exactly the same schedule as described in Experiments 1 and 2 in our previous paper on wheat (Zotova et al., 2018).
RNA Extraction, cDNA Synthesis and qPCR Analysis
Frozen leaf samples were ground as described below for DNA extraction. TRIzol-like reagent was used for RNA extraction following the protocol described by Shavrukov et al. (2013) and all other steps for RNA extraction and cDNA synthesis were as described previously (Zotova et al., 2018). The procedures included DNase treatment (Qiagen, Germany), and the use of a MoMLV Reverse Transcriptase kit (Biolabmix, Novosibirsk, Russia). All cDNA samples were checked for quality control using PCR and yielded bands of the expected size on agarose gels.
Diluted (1:2) cDNA samples were used for qPCR analyses using either a QuantStudio-7 Real-Time PCR instrument (Thermo Fisher Scientific, United States) at S. Seifullin Kazakh AgroTechnical University, Astana, Kazakhstan, or Real-Time qPCR system, Model CFX96 (BioRad, Gladesville, NSW, Australia) at Flinders University, Australia. The qPCR protocol was similar in both instruments as published earlier (Shavrukov et al., 2016), wherein the total volume of 10 μl q-PCR reactions included either 5 μl of 2xBiomaster HS-qPCR SYBR Blue (Biolabmix, Novosibirsk, Russia) for experiments in Kazakhstan or 5 μl of 2xKAPA SYBR FAST (KAPA Biosystems, United States) for experiments in Australia, 4 μl of diluted cDNA, and 1 μl of two gene-specific primers (3 μM of each primer) (Supplementary material 1). Expression data for the target genes were calculated relative to the average expression of the two reference genes: CAC, Clathrin adaptor complexes, medium subunit (Reddy et al., 2016) and GAPDH, Glyceraldehyde-3-phosphate dehydrogenase (Garg et al., 2010) (Supplementary material 1). At least three biological and two technical replicates were used in each qPCR experiment.
DNA Extraction, Sequencing and SNP Identification
Plants were grown in control (non-stressed) conditions in containers with soil as described above. Five uniform one month-old individual plants were selected from each accession and five leaves were collected and bulked for leaf samples. Leaf samples frozen in liquid nitrogen were ground in 10-ml tubes with two 9-mm stainless ball bearings using a Vortex mixer. DNA was extracted from the bulked leaf samples with phenol-chloroform as described in our earlier papers (Shavrukov et al., 2016; Zotova et al., 2018). One microliter of DNA was checked on a 0.8% agarose gel to assess quality, and concentration was measured by Nano-Drop (Thermo Fisher Scientific, United States).
To identify SNPs in the GoI and compare them with annotated accessions in databases, primers were designed in exon regions flanking introns (Supplementary material 1). PCR was performed in 60 μl volume reactions containing 8 μl of template DNA adjusted to 20 ng/μl, and with the following components in the final concentrations listed: 1xPCR Buffer, 2.2 mM MgCl2, 0.2 mM each of dNTPs, 0.25 μM of each primer and 4.0 units of Taq-DNA polymerase in each reaction (Maxima, Thermo Fisher Scientific, United States). PCR was conducted on a SimpliAmp Thermal Cycler (Thermo Fisher Scientific, United States), using a program recommended by the Taq-polymerase manufacturer, with the following steps: initial denaturation, 95°C, 4 min; 35 cycles of 95°C for 20 s, 55°C for 20 s, 72°C for 1 min, and final extension, 72°C for 5 min. Single bands of the expected size were confirmed after visualization of 5 μl of the PCR product in 1% agarose gel. The remaining PCR reaction volume (55 μl) was purified using FavorPrep PCR Purification kit (Favorgene Biotec Corp., Taiwan) following the Manufacturer’s protocol. The concentrations of purified PCR products were measured using NanoDrop (Thermo Fisher Scientific, United States) and later used as a template (100 ng) in a sequencing reaction with the Beckman Coulter Sequencing kit, following the Manufacturer’s protocol. Sanger sequencing and analysis of results were performed on a Beckman Coulter Genetic Analysis System, Model CEQ 8000 (Beckman Coulter, United States) following the Manufacturer’s protocol and software at S. Seifullin Kazakh AgroTechnical University, Astana (Kazakhstan). The identified SNPs were used to design allele-specific primers that were applied in Amplifluor-like SNP analysis. Two fully sequenced chickpea accessions, ICC4958 of the Desi ecotype, and Frontier of the Kabuli ecotype, were used as the reference genomes6.
SNP Amplifluor Analysis
Amplifluor-like SNP analysis was carried out using a QuantStudio-7 Real-Time PCR instrument (Thermo Fisher Scientific, United States) as described previously (Jatayev et al., 2017; Zotova et al., 2018) with the following modifications: Each reaction contained 3 μl of template DNA adjusted to 20 ng/μl, 5 μl of Hot-Start 2xBioMaster (MH020-400, Biolabmix, Novosibirsk, Russia7) with all other components as recommended by the manufacturers, including MgCl2 (2.0 mM). One microliter of a mixture of two fluorescently labeled Universal probes was added (0.25 μM each) and 1 μl of allele-specific primer mix (0.15 μM of each of two forward primers and 0.78 μM of the common reverse primer). Four microliter of Low ROX (Thermo Fisher Scientific, United States) was added as a passive reference label to the Master-mix as prescribed for the qPCR instrument. Assays were performed in 96-well microplates. Sequences of the Universal probes and primers as well as the sizes of amplicons are presented in Supplementary material 1.
PCR was conducted using a program adjusted from those published earlier (Jatayev et al., 2017; Zotova et al., 2018): initial denaturation, 95°C, 2 min; 14 “doubled” cycles of 95°C for 10 s, 60°C for 10 s, 72°C for 20 s, 95°C for 10 s, 55°C for 20 s and 72°C for 50 s; with recording of allele-specific fluorescence after each cycle. Genotyping by SNP calling was determined automatically by the instrument software, but each SNP result was also checked manually using amplification curves and final allele discrimination. Experiments were repeated twice over different days, where two technical replicates confirmed the confidence of SNP calls.
Statistical Analysis
IBM SPSS Statistical software was applied to calculate means, standard errors, and to estimate the probabilities for significance using ANOVA tests.
Results
Bioinformatics and Comparative Phylogenetic Analysis
During the initial screening of SNP No. 2103, rs853191221 [C. arietinum] within the chickpea SNP database (Supplementary material 2), NCBI BLAST analysis revealed the closest nucleotide accession to be XM_012715175.1, encoding a Ras-related protein in C. arietinum with the corresponding RabC1-like gene (LOC101496214, transcript variant X2, mRNA). We designated this gene as the isoform CaRabC-1a.
To identify the full list of all members of CaRab genes in chickpea, bioinformatics approaches were used to search and analyze annotated sequences and whole genome sequences available in public databases using comparisons to the reference genome of A. thaliana. As a result, eight sub-families of CaRab gene were identified, with 54 isoforms. The corresponding accession IDs for the genes and proteins, as well as references to Arabidopsis genes with the highest level of similarity are shown in Table 2.
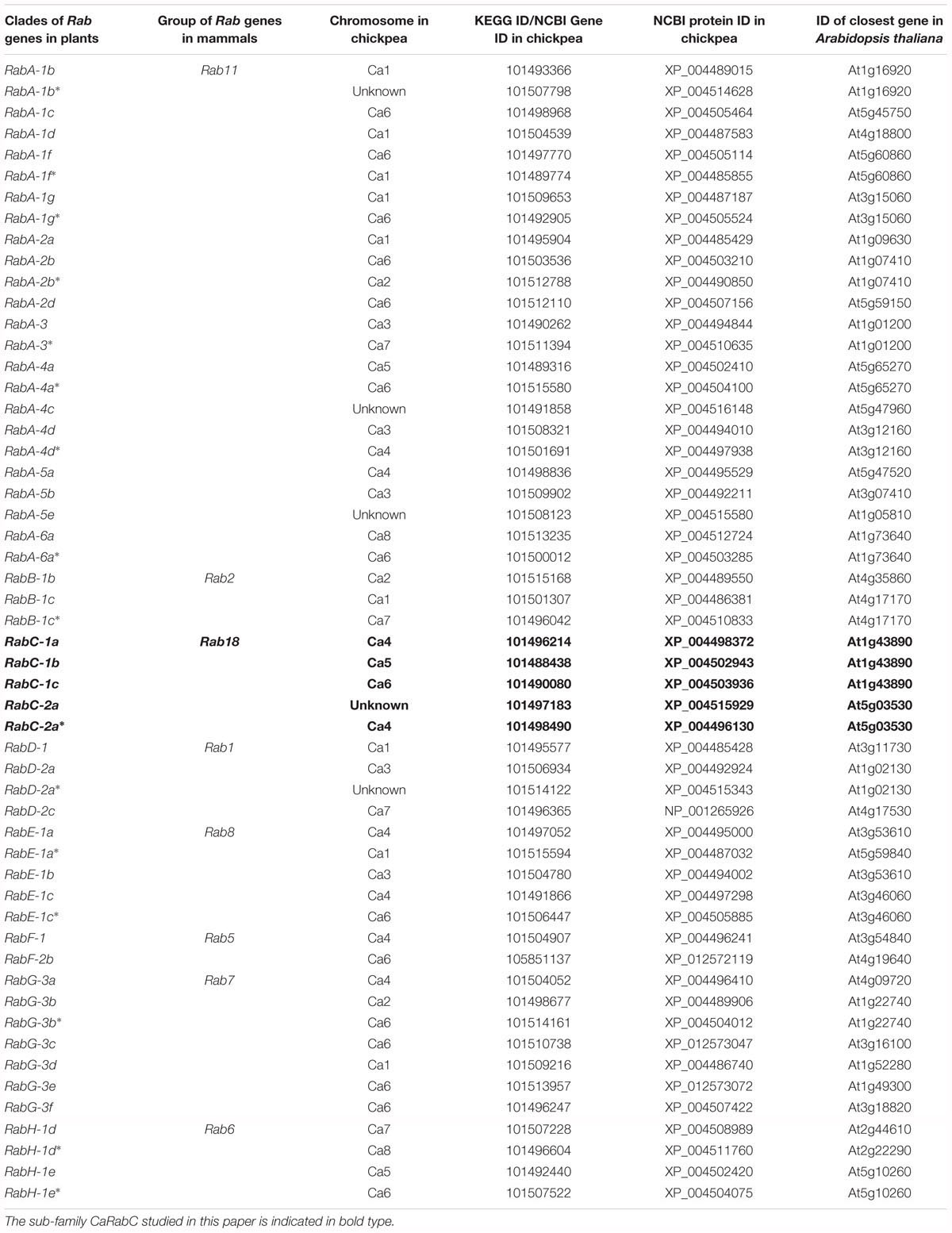
Table 2. The eight identified sub-families (RabA – RabH) of the chickpea CaRab, with 54 isoforms and their corresponding accession ID listed for genes and proteins as well as reference to closest genes in Arabidopsis.
The sequences of all 54 isoforms of CaRab genes identified in chickpea were used to construct a phylogenetic tree (Figure 1). Eight distinct clades were identified in the molecular dendrogram, and the letter corresponding to each sub-family name is used to distinguish the corresponding clade. The biggest and most diverse was Clade A, the CaRabA gene sub-family while Clades B and F contained only two accessions each. Clades D, G, H and F are molecularly similar, but most distanced from other sub-families of the CaRab gene. The sub-family CaRabC contained five isoforms with the closest sub-families being CaRabD and CaRabE (Figure 1).
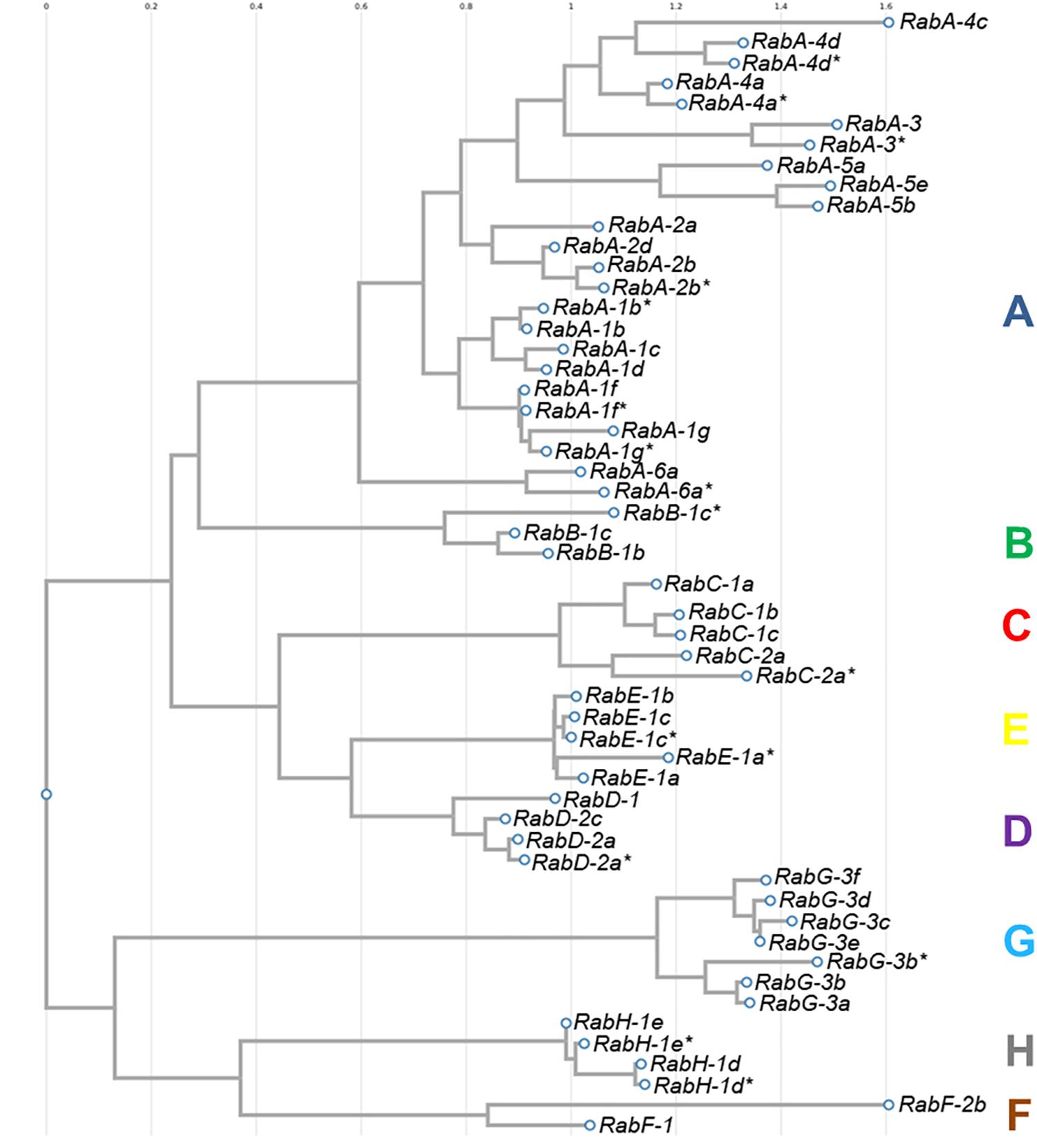
Figure 1. Phylogenetic tree of 54 CaRab gene isoforms identified in chickpea using the FastTree v2.1.8 program. Clades are designated by colored letters identical to the sub-family CaRab gene name.
Protein sequence analysis of five isoforms from sub-family CaRabC (Figure 2) showed distinct separation of CaRabC-1 from CaRabC-2. The closest molecular similarity was found between CaRabC-1b and CaRabC-1c with the next greatest similarity shared with CaRabC-1a, while CaRabC-2a and CaRabC-2a∗ were the most diverged from all others (Figure 2).
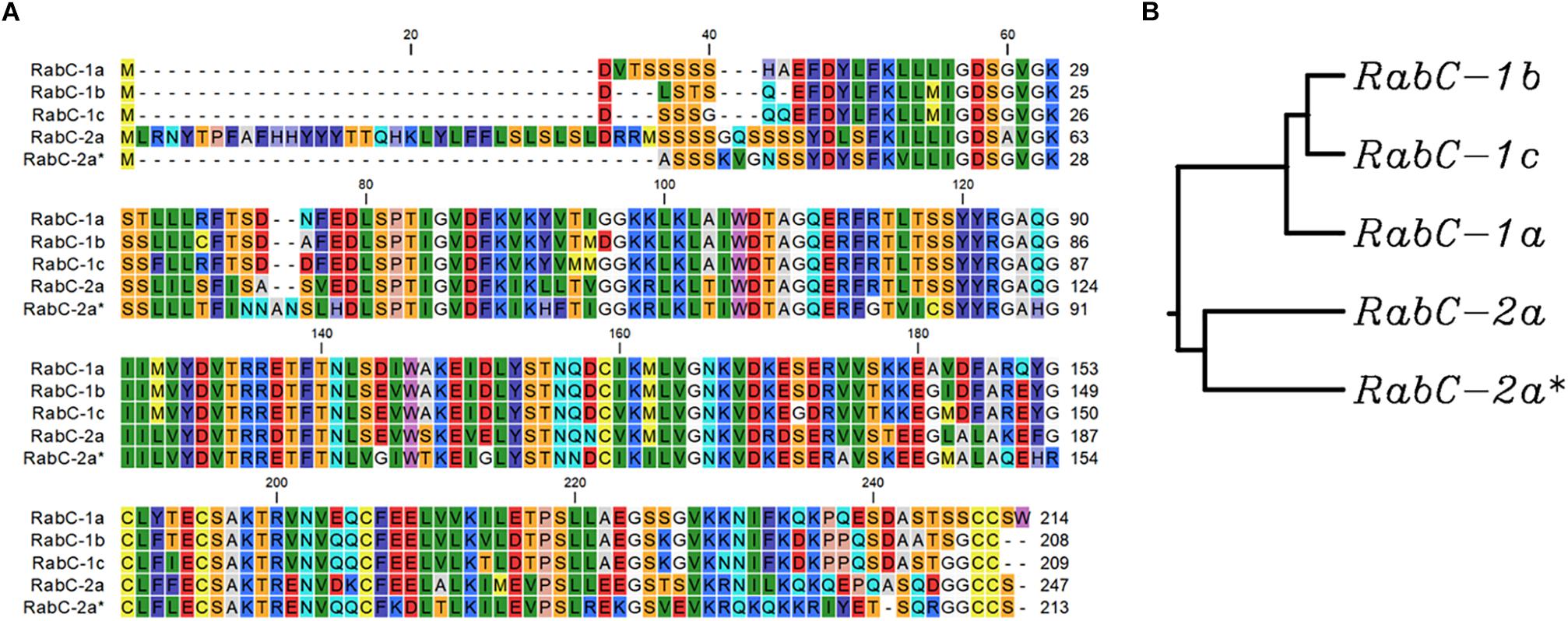
Figure 2. A comparison of amino acid sequences (A), and Rooted UPGMA phylogenetic tree with branch length (B) of the five isoforms of CaRabC proteins identified in chickpea. Multiple sequence alignment conducted presented using CLC Main Workbench software.
RT-qPCR and Gene Expression Analysis
Primers for RT-qPCR analysis were designed based on the alignment and comparison of CDS sequences of five identified CaRabC isoforms listed in Table 2. To estimate the total expression level of all five CaRabC genes combined, common primers with degenerative nucleotides were designed based on the longest consensus regions in the alignments. In addition, 3’-ends of gene-specific primers were designed for specific SNPs to maximize the specificity of qPCR analysis for each of the five isoforms of CaRabC gene (Supplementary material 3).
Initially, the expression level of CaRabC gene was determined in control plants grown under favorable conditions for all isoforms combined, as well as for each of them separately (Figure 3A). All six studied chickpea accessions, 3 Kabuli and 3 Desi (dark green and dark blue, respectively, in Figure 3A), showed a very high level of total CaRabC gene expression, ranging from 11.2 to 18.4 relative expression units, with non-significant differences among the six studied genotypes. The expression level of a single isoform of CaRabC-1c had maximal (63–88%) contribution in the CaRabC gene expression in total. Two isoforms, CaRabC-2a and -2a∗, both showed very similar levels of 1.9–2.5 expression units. A level of around 1 expression unit was observed in the isoform CaRabC-1a, similar to the average level for the two reference genes used in this study. An extremely low level of expression (approximately 10-fold lower than both reference genes) was shown for the last isoform CaRabC-1b (Figure 3A).
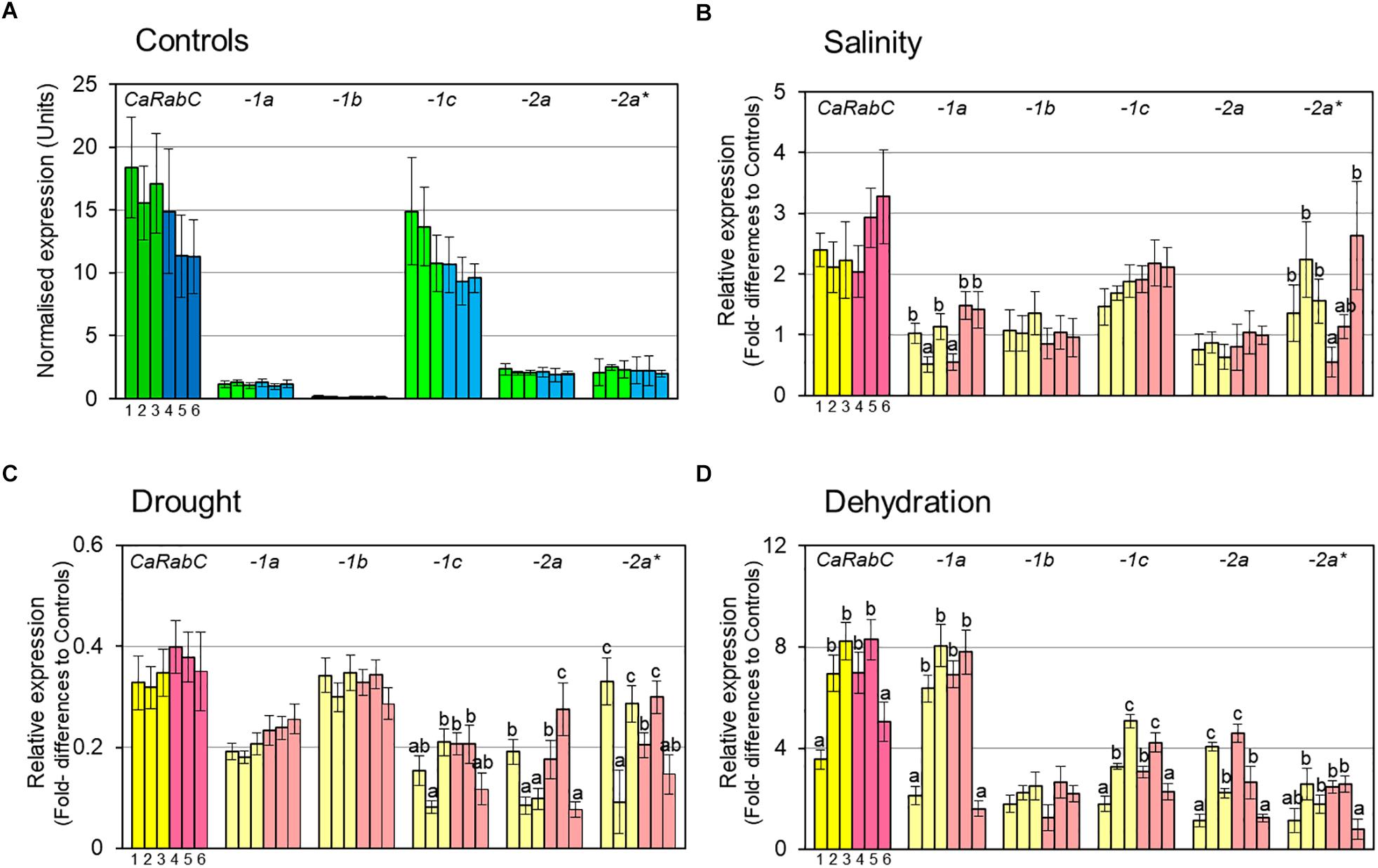
Figure 3. RT-qPCR analysis of CaRabC gene family expression in chickpea leaves: (A) In favorable, non-stressed conditions (Controls) for 3 Kabuli and 3 Desi (dark green and dark blue, respectively); and the relative gene expression compared to Controls under: (B) Gradual salt stress application, 150 mM NaCl, 7 days; (C) Slowly developing drought in pots with soil, 12 days; and (D) Rapid dehydration of detached leaves, 6 h, room temperature. All isoforms of CaRabC gene combined (darker colors) and the five separate isoforms (lighter colors) of the CaRabC-1a, -1b, -1c, -2a and -2a∗ (for corresponding gene family) were analyzed separately. Each set contained six chickpea accessions, including three Kabuli ecotypes, shown in yellow (1, Yubileiny; 2, ICC-7255; and 3, ICC-4841), and three Desi ecotypes, shown in pink (4, ICC-1392; 5, ICC-4918; and 6, ICC-12726). Data were normalized using an average for two reference genes, calculated with ANOVA, and are presented as means for three biological and two technical replicates ± SE, shown as error bars. Significant differences (at least for P > 0.95) for each gene isoform and within each set of chickpea accessions are shown by different letters according to ANOVA tests.
For salinity stress (Figure 3B), a high level of expression of the total CaRabC gene family was observed with 2-3.3-fold higher expression relative to Controls, but no significant differences were found within each set of six studied accessions due to relatively wide variability between replicates. In all studied genotypes, the isoform CaRabC-1c made the highest contribution to the gene expression (around 1.5–2-fold above the Controls). Only two accessions, No. 2 (ICC-7255, Kabuli) and No. 6 (ICC-12726, Desi), showed a higher level of CaRabC-2a∗ isoform expression (2.2- and 2.6-fold, respectively) but these data were quite variable. Significant genetic variation was found for expression levels of CaRabC-1a and CaRabC-2a∗. Expression levels of two isoforms, CaRabC-1b and CaRabC-2a, did not differ from Controls (Figure 3B).
A different expression pattern for the CaRabC gene family was found for the drought experiment, where total expression was down-regulated by 0.3–0.4-fold compared to Controls (Figure 3C). The highest contribution to gene expression was made by the isoform CaRabC-1b. There was no significant genetic variation for CaRabC-1a and CaRabC-1b among the studied germplasm while the other three isoforms were quite variable (Figure 3C).
In contrast, rapid dehydration of detached leaves resulted in an up-to 8-fold increase of expression for the total CaRabC gene family expression, as well as isoform CaRabC-1a, compared to controls (Figure 3D). With the exception of CaRab1b, significant genetic variation was observed among the studied chickpea accessions for all other isoform expression profiles.
Amplicon Sequencing Showed an SNP in the Candidate Gene CaRabC-1a
The initial SNP discovered was annotated at position 516 from the start-codon in the identified CDS, LOC101496214, based on the reverse-compliment order in the SNP-containing fragment. The full nucleotide sequence of the accession and position of this initial SNP is presented in Supplementary material 2.
To check for the presence/absence of the initial SNP in the studied chickpea accessions, several pairs of primers were designed flanking the SNP. The most successful primer pair, F5&R5, amplified a fragment of 1148 bp. A fragment of the alignment showing polymorphic amplicons from the germplasm sequences compared to two fully sequenced reference chickpea accessions (ICC4958, Desi ecotype and Frontier, Kabuli ecotype) in CaRabC-1a is presented in Figure 4. The sequencing of the amplified fragments revealed the presence of 11 new SNPs in two chickpea accessions, Yubileiny and ICC7255, both Kabuli ecotypes (Table 1), compared to the two reference accessions. All 11 identified SNPs recorded high scores, and clear nucleotide peaks at the SNP positions were assessed manually. Interestingly, the initial SNP recorded in the database was monomorphic among the two reference accessions and two genotypes sequenced in our study.
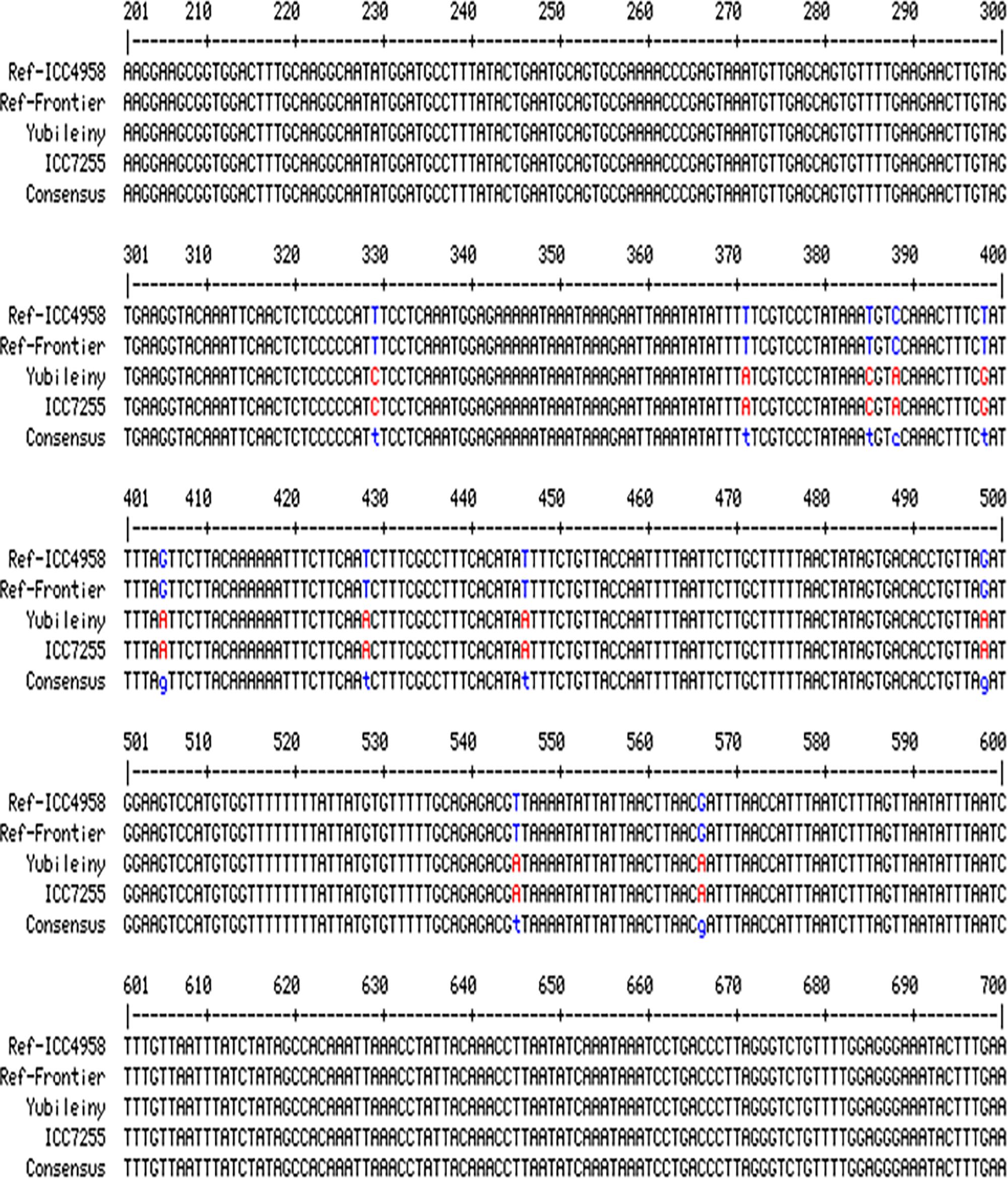
Figure 4. Partial alignments of CaRabC-1a (LOC101496214) amplicons. The alignment was produced with primers F5&R5 from the studied chickpea germplasm, Yubileiny and ICC7255, compared to two fully sequenced reference accessions, ICC4958 and Frontier. The studied amplicons were located on Chromosome 4, position 27,819,854–27,818,706 (Reverse order), in the reference accession ICC4958 (Desi ecotype), while the amplicon position in the second reference accession Frontier (Kabuli ecotype), was at 38,617,439–38,616,292 (Reverse order), also on Chromosome 4. Eleven identified SNPs are shown in red for the two studied accessions against those indicated in blue for the two reference accessions.
SNP Screening in CaRabC-1a Using Amplifluor-Like Markers
Allele-specific primers, KATU-C22-F&R, were designed for one of the selected SNPs from the 11 identified in the studied fragment of isoform CaRabC-1a to use with Amplifluor-like genotyping analysis. Details on the design of primers and positions of the studied SNPs are presented in Supplementary material 4. The example in Figure 5 shows allele discrimination using Amplifluor-like SNP marker KATU-C22, where allele 1 (FAM) has been identified in chickpea accessions with SNP genotypes similar to reference accessions ICC4958 and Frontier but allele 2 (VIC) was found in germplasm similar to Yubileiny and ICC7255 (Figure 5).
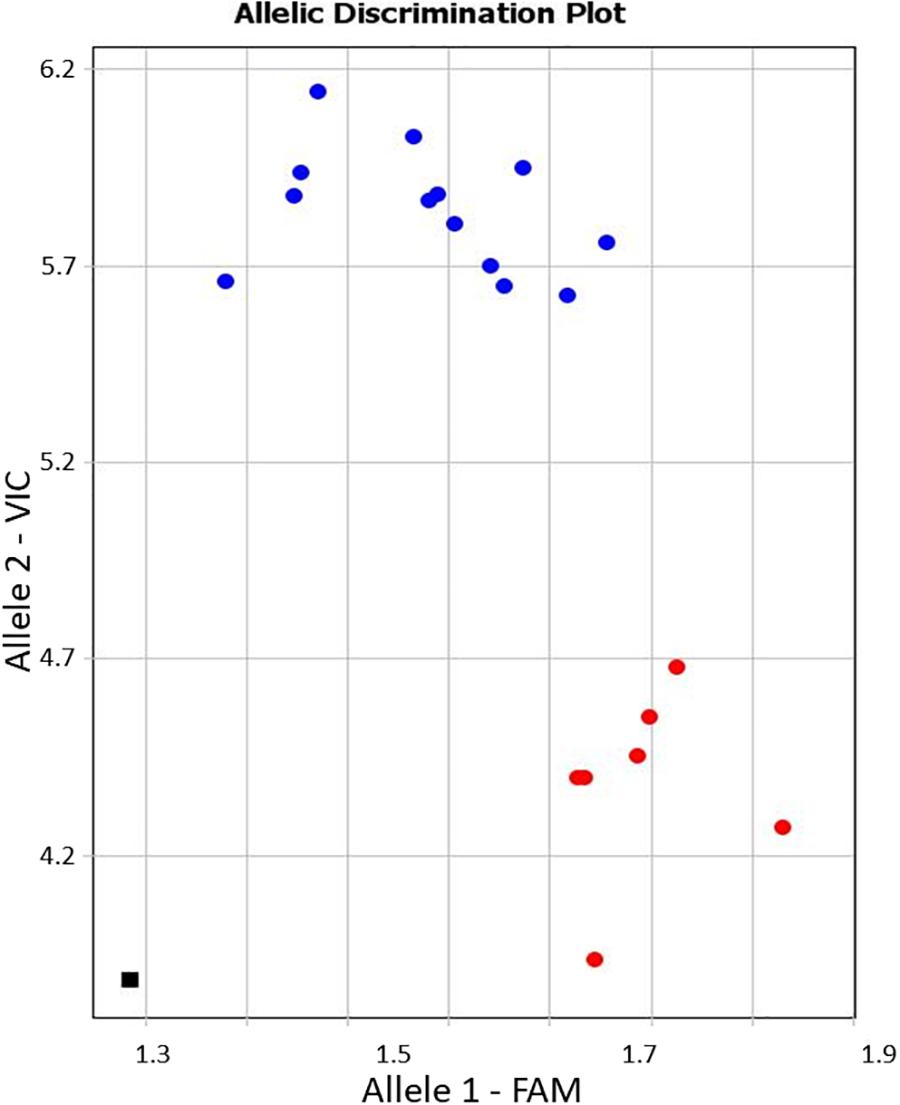
Figure 5. An example of allele discrimination in the chickpea germplasm collection using the Amplifluor-like SNP marker KATU-C22. X- and Y-axes show Relative amplification units, ΔRn, for FAM and VIC fluorescence signals, respectively. Red dots represent homozygote (aa) genotypes with allele 1 (FAM), and blue dots represent homozygote (bb) genotypes for allele 2 (VIC) identified with automatic SNP calling. The black square shows the no template control (NTC) using water instead of template DNA.
Discussion
Rab-GTP proteins are well known in oncology studies in human and animals, but in plants there is increasing evidence that they play a central role in the tolerance to abiotic and biotic stresses. Nevertheless, it appears that the mechanism of membrane trafficking with which they are associated is similar in cells of both humans and plants. Most Rab genes of the eight clades represented in the molecular phylogenetic tree in plants, have similar corresponding groups of genes in human and other animal genomes. A greater or lesser diversity of isoforms for each clade of Rab genes just reflects the differing outcomes of evolution in the plant and animal kingdoms.
In plants, the most studied groups of Rab genes are from Clades G and H, where multiple vacuolar trafficking pathway components were demonstrated (Vernoud et al., 2003; Peng et al., 2014; Uemura and Ueda, 2014; Brillada and Rojas-Pierce, 2017). These types of Rab genes encode proteins that have been associated with a response to salinity and osmotic stresses, and are thought to associate with pre-vacuolar vesicles. Thus, Rab proteins may enhance relocation of Na+ ions to the vacuole, after they reach a toxic level in the cytoplasm of cells. Whilst there has been less attention placed on other groups of Rab genes, including the diverse Clade A with its many isoforms and the non-diverse Clade B with only two gene members, there is practically nothing known about Clade C of Rab in plants (Vernoud et al., 2003; Jha et al., 2014; Rehman and Sansebastiano, 2014; Lawson et al., 2018). Despite the strong similarity between A. thaliana and C. arietinum, our bioinformatic results show significant differences in the number of Rab isoforms in most clades.
In the work described here, 54 isoforms of CaRab genes were identified in chickpea, indicating an evolutionary reorganization when compared to A. thaliana, where 57 AtRab isoforms have been identified (Vernoud et al., 2003). Clade C in the chickpea dendrogram has not been previously identified, described or studied, and contains the five isoforms: CaRabC-1a, -1b, -1c, -2a and -2a∗. The first three isoforms show similarity to AtRabC-1 (At1g43890, Table 1) while the latter two isoforms in chickpea were similar to another single isoform AtRabC-2a (At5g03530). The isoform AtRabC-2b (At3g09910), listed in a comprehensive analysis of the Rab genes in A. thaliana (Vernoud et al., 2003), has no ortholog in the C. arietinum genome. To avoid any misunderstanding with the classification of CaRabC-2a and -2a∗ isoforms, we have used an asterisk instead of another letter, to indicate its very similar polypeptide structure.
Following the bioinformatics study, the expression analyses of total CaRabC for all five isoforms revealed high levels of expression of the gene family in leaves of non-stressed young chickpea plants compared to two reference genes (Figure 3A). More importantly, a single isoform, CaRabC-1c, made the major contribution to the gene expression, indicating a very active role of this isoform in chickpea plant development under non-stressed conditions. In the absence of other reports comparing expression of individual and combined (bulk) isoforms of Rab genes in plants, our conclusions await further verification and discussion.
Under salt stress, the dominance of the CaRabC-1c isoform in expression profiles was not as pronounced as under control conditions and was more comparable to other isoforms in some of the studied chickpea accessions, particularly CaRabC-1a and CaRabC-2a∗. Therefore, at least three isoforms of CaRabC were salinity-responsive and the two latter ones were strongly genotype-dependent (Figure 3B).
An unexpected result was found in the comparison of CaRabC gene expression in response to slowly progressing drought of whole plants and rapid dehydration of detached leaves. Only a few reports have described expression of different genes in parallel experiments with drought and dehydration. For example, a peroxisomal isoform of APX, Ascorbate peroxidase, was down-regulated under strong drought but up-regulated in desiccated leaves in a cultivar of cowpea, Vigna unguiculata (D’Arcy-Lameta et al., 2006). Similar results were reported for two genes associated with loss of water during slow drought progression compared to rapid dehydration of barley leaves: HvMT2, a metallothionein-like protein, and 2HvLHCB, Chlorophyll a-b binding protein of LHCII type III (Gürel et al., 2016). Therefore, there are examples of genes related to drought and dehydration that can be down- and up-regulated, in several plant species. However, our results show for the first time that all isoforms of CaRabC were strongly down-regulated under the slowly developing drought, but very strongly up-regulated in rapidly dehydrated leaves (Figures 3C,D).
Amplifluor-like SNP markers and other molecular markers are very helpful in identifying genetic polymorphisms in diverse germplasm accessions. In the current study, the molecular marker KATU-C22 was useful for genotyping one isoform CaRabC-1a (Figure 5). This allows for tracking of the different variants of this gene and the possibility of linking variants with an associated phenotype. Additional markers are now needed for all other isoforms of CaRabC and other GoI, but this will require further investment in sequencing in the future. It also may be worth looking for SNPs in the upstream promoter regions of the gene family, since this could explain the variation in expression between the genotypes.
CaRabC is just one sub-family from a large CaRab gene family involved in controlling cell membrane trafficking, and like the other Rab genes investigated to date (reviewed in Flowers et al., 2018), it is responsive and potentially associated with the adaptation of plants to abiotic stresses. For comparison, in the bacteria Salmonella, the Rab18 protein (related to RabC in plants) is actively involved in endocytosis and is localized in the early endocytic compartment of cells (Hashim et al., 2000). In plants, there is increasing evidence for the role of endocytosis under salinity and osmotic stress (Martín-Davison et al., 2017). The implications of increased endocytosis during these stresses would be a reduction in total plasma-membrane area, thereby limiting water loss from the cell through a decrease in the number of aquaporins. Additionally, it may represent a mechanism to obtain Na+ ions directly from outside the cell for accumulation in the vacuole, thus keeping the cytoplasmic level of Na+ low (Baral et al., 2015). In future work, we hope to explore the role of CaRabC on endocytosis and Na+ compartmentalization. There has been very little work published to date concerning endocytosis and extended drought. The different responses shown in the changes in expression observed in this study between salinity and dehydration (both components of osmotic stress), is intriguing and probably indicative of the underlying biological role of RabC proteins themselves.
Further research is required in several selected chickpea accessions to assess tolerance to salinity, drought and rapid dehydration. This would allow us to explore possible associations between sequence variants and levels of stress tolerance. The genotype-dependent role of each isoform of CaRabC as well as other genes from the gene family will be studied, and we plan to carry out these experiments in the near future. These new experiments should elaborate on the mechanism and clarify the suggested roles of these proteins in cell polarization and recycling to the plasma membrane, as suggested by Vernoud et al. (2003) and Rutherford and Moore (2002), respectively. Hopefully, our study of CaRabC extends the knowledge of Rab gene family structure and function in plants.
Author Contributions
GK conducted the experiments with chickpea germplasm and the genotyping with Amplifluor-like SNP analysis. AK and SJ supervised the experiments and interpreted the results. AsZ conducted the experiments with plant stresses and sampling. AyZ carried out sequencing. AT worked with plants in the field trial. BA coordinated the experiments in the field. SL analyzed gene sequences in databases and wrote the corresponding sections. CS analyzed the results, and revised and edited the manuscript. CJ analyzed the qRT-PCR data and revised the corresponding section. KS coordinated the qRT-PCR study and revised other sections. PL supervised the study and revised the final version of the manuscript. YS coordinated all experiments and wrote the first version of the manuscript.
Funding
This study was supported by the Ministry of Education and Science (Kazakhstan), Research Program BR05236500 (SJ).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the staff and students of S. Seifullin Kazakh AgroTechnical University, Astana (Kazakhstan) and Flinders University, SA (Australia) for their support in this research and help with critical comments to the manuscript. The results of this study were presented at the International Conference ‘Bioinformatics and Computational Biology’, August 2018, Novosibirsk, Russia. The authors acknowledge the Organizing Committee for their support in the presentation and publication of this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00040/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/snp
- ^ https://www.genome.jp/tools/blast
- ^ https://legumeinfo.org/organism/Cicer/arietinum
- ^ https://www.genome.jp/tools-bin/clustalw
- ^ https://www.qiagenbioinformatics.com/products/clc-main-workbench
- ^ http://www.cicer.info/databases.php
- ^ http://biolabmix.ru/en/products
References
Agarwal, P., Reddy, M. K., Sopory, S. K., and Agarwal, P. K. (2009). Plant Rabs: characterization, functional diversity, and role in stress tolerance. Plant Mol. Biol. Rep. 27, 417–430. doi: 10.1007/s11105-009-0100-9
Agarwal, P. K., Agarwal, P., Jain, P., Jha, B., Reddy, M. K., and Sopory, S. K. (2008). Constitutive overexpression of a stress-inducible small GTP-binding protein PgRab7 from Pennisetum glaucum enhances abiotic stress tolerance in transgenic tobacco. Plant Cell Rep. 27, 105–115. doi: 10.1007/s00299-007-0446-0
Alory, C., and Balch, W. E. (2003). Molecular evolution of the Rab-escort-protein/guanine-nucleotide-dissociation-inhibitor superfamily. Mol. Biol. Cell 14, 3857–3867. doi: 10.1091/E03-04-0227
Baral, A., Shruthi, K. S., and Mathew, M. K. (2015). Vesicular trafficking and salinity responses in plants. IUBMB Life 67, 677–686. doi: 10.1002/iub.1425
Bolte, S., Schiene, K., and Dietz, K. J. (2000). Characterization of a small GTP-binding protein of the Rab5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol. Biol. 42, 923–936. doi: 10.1023/A:1006449715236
Borg, S., Brandstrup, B., Jensen, T. J., and Poulsen, C. (1997). Identification of new protein species among 33 different small GTP-binding proteins encoded by cDNAs from Lotus japonicus, and expression of corresponding mRNAs in developing root nodules. Plant J. 11, 237–250. doi: 10.1046/j.1365-313X.1997.11020237.x
Brillada, C., and Rojas-Pierce, M. (2017). Vacuolar trafficking and biogenesis: a maturation in the field. Curr. Opin. Plant Biol. 40, 77–81. doi: 10.1016/j.pbi.2017.08.005
Chang, E. H., Gonda, M. A., Ellis, R. W., Scolnick, E. M., and Lowy, D. R. (1982). Human genome contains four genes homologous to transforming genes of harvey and kirsten murine sarcoma viruses (molecular cloning/restriction endonuclease mapping/heteroduplex analysis/intervening sequences/gene family). Proc. Natl. Acad. Sci. U.S.A. 79, 4848–4852. doi: 10.1073/pnas.79.16.4848
Chavrier, P., Parton, R. G., Hauri, H. P., Simons, K., and Zerial, M. (1990). Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62, 317–329. doi: 10.1016/0092-8674(90)90369-P
Chen, C., and Heo, J. B. (2018). Overexpression of constitutively active OsRab11 in plants enhances tolerance to high salinity levels. J. Plant Biol. 61, 169–176. doi: 10.1007/s12374-018-0048-0
Coffin, J. M., Varmus, H. E., Bishop, J. M., Essex, M., Hardy, W. D., Martin, J. G. S., et al. (1981). Proposal for naming host cell-derived inserts in retrovirus genomes. J. Virol. 40, 953–957.
D’Arcy-Lameta, A., Ferrari-Iliou, R., Contour-Ansel, D., Pham-Thi, A. T., and Zuily-Fodil, Y. (2006). Isolation and characterization of four ascorbate peroxidase cDNAs responsive to water deficit in cowpea leaves. Ann. Bot. 97, 133–140. doi: 10.1093/aob/mcj010
Flowers, T. J., Glenn, E. P., and Volkov, V. (2018). Could vesicular transport of Na+ and Cl- be a feature of salt tolerance in halophytes? Ann. Bot. doi: 10.1093/aob/mcy164 [Epub ahead of print].
Garg, R., Sahoo, A., Tyagi, A. K., and Jain, M. (2010). Validation of internal control genes for quantitate gene expression studies in chickpea (Cicer arietinum L.). Biochem. Biophys. Res. Commun. 396, 283–288. doi: 10.1016/j.bbrc.2010.04.079
Gürel, F., Öztürk, N. Z., Yörük, E., Uçarlı, C., and Poyraz, N. (2016). Comparison of expression patterns of selected drought-responsive genes in barley (Hordeum vulgare L.) under shock-dehydration and slow drought treatments. Plant Growth Regul. 80, 183–193. doi: 10.1007/s10725-016-0156-0
Hashim, S., Mukherjee, K., Raje, M., Basu, S. K., and Mukhopadhyay, A. (2000). Live Salmonella modulate expression of Rab proteins to persist in a specialized compartment and escape transport to lysosomes. J. Biol. Chem. 275, 16281–16288. doi: 10.1074/jbc.275.21.16281
Haubruck, H., Disela, C., Wagner, P., and Gallwitz, D. (1987). The ras-related ypt protein is an ubiquitous eukaryotic protein: isolation and sequence analysis of mouse cDNA clones highly homologous to the yeast YPTJ gene. EMBO J. 6, 4049–4053. doi: 10.1002/j.1460-2075.1987.tb02750.x
He, M., Lan, M., Zhang, B., Zhou, Y., Wang, Y., Zhu, L. A., et al. (2018). Rab-H1b is essential for trafficking of cellulose synthase and for hypocotyl growth in Arabidopsis thaliana. J. Integr. Plant Biol. 60, 1051–1069. doi: 10.1111/jipb.12694
Hernández-Sánchez, I. E., Maruri-López, I., Graether, S. P., and Jiménez-Bremont, J. F. (2017). In vivo evidence for homo- and heterodimeric interactions of Arabidopsis thaliana dehydrins AtCOR47, AtERD10, and AtRAB18. Sci. Rep. 7:17036. doi: 10.1038/s41598-017-15986-2
Howlader, J., Park, J. I., Robin, A. H. K., Sumi, K. R., and Nou, I. S. (2017). Identification, characterization and expression profiling of stress-related genes in easter lily (Lilium formolongi). Genes 8:172. doi: 10.3390/genes8070172
Huerta-Cepas, J., Serra, F., and Bork, P. (2016). ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Hundertmark, M., and Hincha, D. K. (2008). LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118. doi: 10.1186/1471-2164-9-118
Jatayev, S., Kurishbayev, A., Zotova, L., Khasanova, G., Serikbay, D., Zhubatkanov, A., et al. (2017). Advantages of amplifluor-like SNP markers over KASP in plant genotyping. BMC Plant Biol. 17:254. doi: 10.1186/s12870-017-1197-x
Jha, Y., Sablok, G., Subbarao, N., Sudhakar, R., Fazil, M. H. U. T., Subramanian, R. B., et al. (2014). Bacterial-induced expression of RAB18 protein in Oryza sativa salinity stress and insights into molecular interaction with GTP ligand. J. Mol. Recognit. 27, 521–527. doi: 10.1002/jmr.2371
Jiang, Z., Wang, H., Zhang, G., Zhao, R., Bie, T., Zhang, R., et al. (2017). Characterization of a small GTP-binding protein gene TaRab18 from wheat involved in the stripe rust resistance. Plant Physiol. Biochem. 113, 40–50. doi: 10.1016/j.plaphy.2017.01.025
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lång, V., and Palva, T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20, 951–962. doi: 10.1007/BF00027165
Lawson, T., Mayes, S., Lycett, G. W., and Chin, C. F. (2018). Plant Rabs and the role in fruit ripening. Biotechnol. Genet. Eng. Rev. 34, 181–197. doi: 10.1080/02648725.2018.1482092
Liu, Z., Luo, C., Li, L., Dong, L., and Can, V. (2015). Isolation, characterization and expression analysis of the GDP dissociation inhibitor protein gene MiRab-GDI from Mangifera indica L. Sci. Hortic. 185, 14–21. doi: 10.1016/j.scienta.2015.01.008
Ma, Q. H. (2007). Small GTP-binding proteins and their functions in plants. J. Plant Growth Regul. 26, 369–388. doi: 10.1007/s00344-007-9022-7
Marcote, M. J., Gu, F., Gruenberg, J., and Aniento, F. (2000). Membrane transport in the endocytic pathway: animal versus plant cells. Protoplasma 210, 123–132. doi: 10.1007/BF01276852
Marshall, C. J. (1993). Protein prenylation: a mediator of protein-protein interactions. Science 259, 1865–1866. doi: 10.1126/science.8456312
Martín-Davison, A. S., Pérez-Díaz, R., Soto, F., Madrid-Espinoza, J., González-Villanueva, E., Pizarro, L., et al. (2017). Involvement of SchRabGDI1 from Solanum chilense in endocytic trafficking and tolerance to salt stress. Plant Sci. 263, 1–11. doi: 10.1016/j.plantsci.2017.06.007
Mazel, A., Leshem, Y., Tiwari, B. S., and Levine, A. (2004). Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol. 134, 118–128. doi: 10.1104/pp.103.025379
Minamino, N., Kanazawa, T., Era, A., Ebine, K., Nakano, A., and Ueda, T. (2018). RAB GTPases in the basal land plant Marchantia polymorpha. Plant Cell Physiol. 59, 845–856. doi: 10.1093/pcp/pcy027
Muñoz, F. J., Esteban, R., Labrador, E., and Dopico, B. (2001). Expression of a novel chickpea Rab-GDI cDNA mainly in seedlings. Plant Physiol. Biochem. 39, 363–366. doi: 10.1016/S0981-9428(01)01249-9
Nahm, M. Y., Kim, S. W., Yun, D., Lee, S. Y., Cho, M. J., and Bahk, J. D. (2003). Molecular and biochemical analyses of OsRab7, a rice Rab7 homolog. Plant Cell Physiol. 44, 1341–1349. doi: 10.1093/pcp/pcg163
O’Mahony, P. J., and Oliver, M. J. (1999). Characterization of a desiccation-responsive small GTP-binding protein (Rab2) from the desiccation-tolerant grass Sporobolus stapfianus. Plant Mol. Biol. 39, 809–821. doi: 10.1023/A:1006183431854
Peng, X., Ding, X., Chang, T., Wang, Z., Liu, R., Zeng, X., et al. (2014). Overexpression of a vesicle trafficking gene, OsRab7, enhances salt tolerance in rice. Sci. World J. 214:483526. doi: 10.1155/2014/483526
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Rajan, N., Agarwal, P., Patel, K., Sanadhya, P., Khedia, J., and Agarwal, P. K. (2015). Molecular characterization and identification of target protein of an important vesicle trafficking gene AlRab7 from a salt excreting halophyte Aeluropus lagopoides. DNA Cell Biol. 34, 83–91. doi: 10.1089/dna.2014.2592
Reddy, D. S., Bhatnagar-Mathur, P., Reddy, P. S., Cindhuri, K. S., Ganesh, A. S., and Sharma, K. K. (2016). Identification and validation of reference genes and their impact on normalized gene expression studies across cultivated and wild Cicer species. PLoS One 11:e0148451. doi: 10.1371/journal.pone.0148451
Rehman, R. U., and Sansebastiano, D. G. P. (2014). “Plant Rab GTPases in membrane trafficking and signalling,” in Plant Signaling: Understanding the Molecular Crosstalk, eds K. R. Hakeem, R. U. Rehman, and I. Tahir (New Delhi: Springer), 51–73. doi: 10.1007/978-81-322-1542-4-3
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Rutherford, S., and Moore, I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5, 518–528. doi: 10.1016/S1369-5266(02)00307-2
Shavrukov, Y., Bovill, J., Afzal, I., Hayes, J. E., Roy, S. J., Tester, M., et al. (2013). HVP10 encoding V-PPase is a prime candidate for the barley HvNax3 sodium exclusion gene: evidence from fine mapping and expression analysis. Planta 237, 1111–1122. doi: 10.1007/s00425-012-1827-3
Shavrukov, Y., Zhumalin, A., Serikbay, D., Botayeva, M., Otemisova, A., Absattarova, A., et al. (2016). Expression level of the DREB2-type gene, identified with Amplifluor SNP markers, correlates with performance, and tolerance to dehydration in bread wheat cultivars from Northern Kazakhstan. Front. Plant Sci. 7:1736. doi: 10.3389/fpls.2016.01736
Stenmark, H., and Olkkonen, V. M. (2001). The rab GTPase family. Genome Biol. 2:3007. doi: 10.1186/gb-2001-2-5-reviews3007
Sui, J. M., Li, G., Chen, G. X., Yu, M. Y., Ding, S. T., Wang, J. S., et al. (2017). Digital expression analysis of the genes associated with salinity resistance after overexpression of a stress-responsive small GTP-binding RabG protein in peanut. Genet. Mol. Res. 16:gmr16019432. doi: 10.4238/gmr16019432
Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153–208. doi: 10.1152/physrev.2001.81.1.153
Uemura, T., and Ueda, T. (2014). Plant vacuolar trafficking driven by RAB and SNARE proteins. Curr. Opin. Plant Biol. 22, 116–121. doi: 10.1016/j.pbi.2014.10.002
Vernoud, V., Horton, A. C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191–1208. doi: 10.1104/pp.013052
Woollard, A. A. D., and Moore, I. (2008). The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 11, 610–619. doi: 10.1016/j.pbi.2008.09.010
Yaneva, I., and Niehaus, K. (2005). Molecular cloning and characterisation of a Rab-binding GDP-dissociation inhibitor from Medicago truncatula. Plant Physiol. Biochem. 43, 203–212. doi: 10.1016/j.plaphy.2005.01.019
Zerial, M., and McBride, H. (2001). RAB proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–119. doi: 10.1038/35052055
Zhang, J., Li, Y., Liu, B., Wang, L., Zhang, L., Hu, J., et al. (2018). Characterization of the Populus Rab family genes and the function of PtRabE1b in salt tolerance. BMC Plant Biol. 18:124. doi: 10.1186/s12870-018-1342-1
Zotova, L., Kurishbayev, A., Jatayev, S., Khassanova, G., Zhubatkanov, A., Serikbay, D., et al. (2018). Genes encoding transcription factors TaDREB5 and TaNFYC-A7 are differentially expressed in leaves of bread wheat in response to drought, dehydration and ABA. Front. Plant Sci. 9:1441. doi: 10.3389/fpls.2018.01441
Keywords: abiotic stresses, Amplifluor-like SNP markers, bioinformatics, CaRab gene, differential gene expression, gene isoforms
Citation: Khassanova G, Kurishbayev A, Jatayev S, Zhubatkanov A, Zhumalin A, Turbekova A, Amantaev B, Lopato S, Schramm C, Jenkins C, Soole K, Langridge P and Shavrukov Y (2019) Intracellular Vesicle Trafficking Genes, RabC-GTP, Are Highly Expressed Under Salinity and Rapid Dehydration but Down-Regulated by Drought in Leaves of Chickpea (Cicer arietinum L.). Front. Genet. 10:40. doi: 10.3389/fgene.2019.00040
Received: 25 October 2018; Accepted: 18 January 2019;
Published: 07 February 2019.
Edited by:
Yuriy L. Orlov, Russian Academy of Sciences, RussiaReviewed by:
Awais Rasheed, International Maize and Wheat Improvement Center, MexicoMehar Hasan Asif, National Botanical Research Institute (CSIR), India
Copyright © 2019 Khassanova, Kurishbayev, Jatayev, Zhubatkanov, Zhumalin, Turbekova, Amantaev, Lopato, Schramm, Jenkins, Soole, Langridge and Shavrukov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuri Shavrukov, eXVyaS5zaGF2cnVrb3ZAZmxpbmRlcnMuZWR1LmF1
 Gulmira Khassanova1
Gulmira Khassanova1 Satyvaldy Jatayev
Satyvaldy Jatayev Aybek Zhumalin
Aybek Zhumalin Sergiy Lopato
Sergiy Lopato Carly Schramm
Carly Schramm Colin Jenkins
Colin Jenkins Kathleen Soole
Kathleen Soole Yuri Shavrukov
Yuri Shavrukov