- 1Mental Health Center, Shantou University Medical College, Shantou, China
- 2Shantou Central Hospital, Affiliated Shantou Hospital of Sun Yat-sen University, Shantou, China
- 3Laboratory of Genomic and Precision Medicine, Wuxi School of Medicine, Jiangnan University, Wuxi, China
- 4Shenzhen Kang Ning Hospital, Shenzhen, China
- 5Beijing Genomics Institute – Shenzhen, Shenzhen, China
Schizophrenia is a complex psychiatric disorder with high genetic heterogeneity, however, the contribution of rare mutations to the disease etiology remains to be further elucidated. We herein performed exome sequencing in a Han Chinese schizophrenia family and identified a missense mutation (c.6724C>T, p.R2242C) in the teneurin transmembrane protein 4 (TENM4) gene in the SCZD2 locus, a region previously linked to schizophrenia at 11q14-21. The mutation was confirmed to co-segregate with the schizophrenia phenotype in the family. Subsequent investigation of TENM4 exons 31, 32, and 33 adjacent to the p.R2242C mutation revealed two additional missense mutations in 120 sporadic schizophrenic patients. Residues mutated in these mutations, which are predicted to be deleterious to protein function, were highly conserved among vertebrates. These rare mutations were not detected in 1000 Genomes, NHLBI Exome Sequencing Project databases, or our in-house 1136 non-schizophrenic control exomes. Analysis of RNA-Seq data showed that TENM4 is expressed in the brain with high abundance and specificity. In line with the important role of TENM4 in central nervous system development, our findings suggested that increased rare variants in TENM4 could be associated with schizophrenia, and thus TENM4 could be a novel candidate gene for schizophrenia in the SCZD2 locus.
Introduction
Schizophrenia (OMIM 181500) is a complex psychiatric disorder, and is also a public health problem affecting approximately 1% of the world population, leading to reduced life expectancy by an average of 20–25 years (Tiihonen et al., 2009). Family and twin studies have demonstrated a strong genetic component in schizophrenia with heritability estimated to be 60–80% (Sullivan et al., 2003; Shih et al., 2004).
High heritability in the disease underlines substantial effects from genetic variants. The frequency of these of alleles ranges from common to extremely rare. Common variants associated with schizophrenia have been widely studied by candidate gene association studies (Stefansson et al., 2002; Chen et al., 2009a, 2) and genome-wide association studies (GWASs) (Bergen and Petryshen, 2012). For example, a recent multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls identify 128 independent associations spanning 108 loci that meet genome-wide significance (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). It is estimated that half to a third of the genetic risk of schizophrenia is indexed by common alleles in GWAS.
Apart from common variants with small effect, recent studies have shown that rare variants or mutations with large effect may also help to understand remaining component of psychiatric etiology unexplained by common variants (Pulver et al., 1994;Jacquet et al., 2002; Ament et al., 2015). For example, a mutation in the Neuronal PAS domain protein 3 (NPAS3) gene segregates with mental illness in a family affected by schizophrenia and major depression (Yu et al., 2014, 3). Recent exome sequencing studies have advanced understanding of rare variants and mutations in schizophrenia (Bustamante et al., 2017; Singh et al., 2017; John et al., 2019). Studies in parent-proband trios have revealed de novo mutations with high genetic heterogeneity in schizophrenia (Xu et al., 2011, 2012). Moreover, analysis of exomes from 2536 schizophrenia cases and 2543 controls has emphasized burden raised from extremely rare (less than 1 in 10,000), disruptive mutations in the patients (Purcell et al., 2014). However, the contribution of rare mutations to schizophrenia remains to be further elucidated. In spite of a number of genetic loci identified by linkage studies in schizophrenia,1 only a few genes have been mapped in these loci, such as PRODH (SCZD4) (Jacquet et al., 2002), DISC1 (SCZD9) (Debono et al., 2012, 1), SHANK3 (SCZD15) (Gauthier et al., 2010), NRXN1 (SCZD17) (Rujescu et al., 2009), and SLC1A1 (SCZD18) (Myles-Worsley et al., 2013, 1). Most of these linkage loci have not yet been defined molecularly, which might harbor unknown genes or mutations with large effects in disease risk determination that remain to be identified.
In the current study, by using exome sequencing we identified a novel rare mutation in the teneurin transmembrane protein 4 gene (TENM4, also named ODZ4), which cosegregated with schizophrenia in a Han Chinese family. Moreover, additional rare TENM4 mutations were found in a cohort of 120 unrelated sporadic schizophrenic patients. All mutation was not detected in 1441 non-schizophrenic control exomes. TENM4 is located within the schizophrenia disorder 2 (SCZD2) locus at 11q14-21, in which the underlying gene has not been identified by far. The study thus demonstrated increased TENM4 mutation burden in schizophrenia and suggested that TENM4 could probably be a candidate gene for schizophrenia in the SCZD2 locus.
Materials and Methods
Sample Collection and Clinical Examination
This study has been approved by the Ethics Committee of the Mental Health Center of Shantou University Medical College and was performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and all subsequent revisions. All participants in the paper gave their informed consent for their participation, and the publication of clinical data and indirectly identifiable information prior to their inclusion in the study. The family with schizophrenia was recruited at the Mental Health Center of Medical College of Shantou University, Shantou, China (Figure 1). Clinical diagnoses in the proband (III-2) and family members were based on a Chinese version of the Structured Clinical Interview for DSM-IV-TR Axis I Disorder-Patient Edition (SCID-I/P) (First et al., 1997) criteria derived from a standard interview and from a case-note review by two trained psychiatrists. Four affected family members (II-2, III-2, III-3, and III-4) were interviewed with Social Disability Screening Schedule (SDSS) (World Health Organization, 1988) to assess the clinical-specific features and social abilities. Clinical global impression-severity of illness (CGI-SI) (Programs and Guy et al., 1976) was used to assess the disease severity (Table 1). All of them were diagnosed as paranoid schizophrenia and had social or occupational dysfunction and continuous signs of the disturbance for more than 15 years. Four individuals affected with treatment-resistant schizophrenia were enrolled. The two unaffected family members were confirmed to have no sign of mental illness.
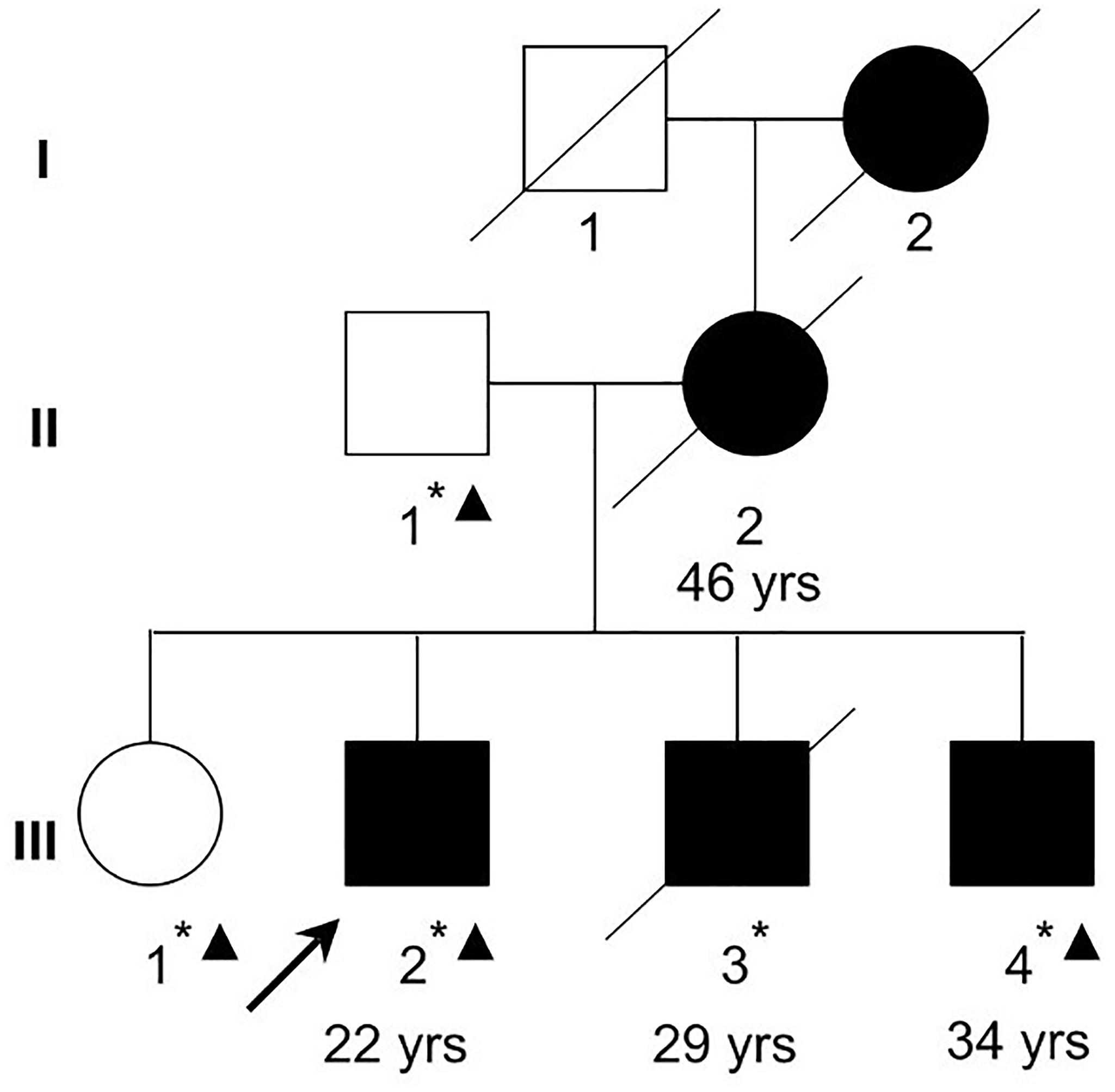
Figure 1. A Chinese Han family with schizophrenia. Filled squares and circles denote affected males and females, respectively. Normal individual is shown as empty symbols. All family members in the second and third generations were examined. The ages at diagnosis for the examined affected family members are shown below the symbols. Whole-exome sequencing was performed in two affected (III-2 and III-4) and two unaffected (II-1 and III-1) family members. Asterisks denote individuals with blood samples and DNA collected. Triangles denote individuals (II-1, III-1, III-2, and III-4) whose DNA were used in exome sequencing in the current study.

Table 1. Demographic information and clinical features of four affected family members from schizophrenia family enrolled in the exome sequencing study.
A cohort of 120 unrelated sporadic schizophrenic patients was recruited using the same criteria as described above. All of them were diagnosed as schizophrenia, and their main manifestations included auditory hallucination, persecutory delusion, and social or occupational dysfunction.
A group of 205 unrelated non-schizophrenia individuals in Sanger sequencing validation had no history of psychotic diseases following clinical interview FH-RDC criteria (Endicott, 1978).
Peripheral blood was collected from all participants except for II-2 whose blood and DNA was not available due to her death before blood sample collection for the current study. Her genotypes and mutation status was then inferred from her children and spouse’s genotypes. Genomic DNA was extracted by using the QIAmp Blood kit (Qiagen, Hilden, Germany).
Exome Capture and Sequencing
Genomic DNA (3 μg) from two affected (III-2 and III-4) and two unaffected members (II-1 and III-1) was used to perform exome sequencing by Axeq Technologies (Rockville, MD, United States). Whole exome was captured by SeqCap EZ Human Exome Library v3.0 (Roche NimbleGen, Madison, WI, United States) and sequenced on an Illumina HiSeq 2000 (Illumina, Hayward, CA, United States) with a paired-end 100 bp length configuration.
Read Mapping and Variant Detection
The reads were mapped against UCSC hg19 Human Reference Genome2 by using BWA3. The single nucleotide variations (SNVs) and Indels were detected by SAMTOOLS4, and annotated using ANNOVAR (Wang et al., 2010) previously known and reported variants were identified and filtered using dbSNP 1355 and 1000 Genomes project6 data. Functional impact of variants was predicted by PROVEAN, SIFT, and PolyPhen.
Sanger Sequencing
Genomic sequence of TENM4 was obtained from the NCBI reference sequence database7. Primers designed accordingly by Primer 3 were summarized in Supplementary Table S5. Polymerase chain reaction (PCR) amplification was performed using the GeneAmp PCR System 9700 (ABI, Foster City, CA, United States) in a 25-μl mixture containing 1.5 mM MgCl2, 0.2 mM of each dNTP (Sangon, Shanghai, China), 1 U Taq DNA polymerase (Invitrogen, Carlsbad, CA, United States), 0.2 μM primers, and 20 ng of genomic DNA. Sanger sequencing was performed using the BigDye Terminator Cycle Sequencing v3.1 kit (ABI, Foster City, CA, United States) and the 3130xl Genetic Analyzer (ABI, Foster City, CA, United States) following the protocol suggested by the manufacturer. Sequence alignment and analysis of variations were performed using the NovoSNP program8.
Non-schizophrenic Control Exomes
Variants were screened against exomes from Chinese Han individuals without schizophrenia to remove common variants. The set of control exome data were consolidated from several previous exome sequencing studies performed by our group. Various exome capture chips were used for this dataset, such as Roche NimbleGen SeqCap EZ Human Exome Library v3.0, Agilent SureSelect All Human Exon v4.0 kit, or Ilumina TruSeq Exome Enrichment kit. Sequencing was performed in Illumina Hiseq 2000. The data quality criteria and filtering process followed the same standard as that for the family.
Statistical Analysis
Difference in scores of clinical assessment was analyzed using Student’s t-test. Association of SNVs with schizophrenia was performed using Fisher’s exact test.
RNA-Seq of Human Tissues
Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values of RNA-seq of the brain, cerebellum, heart, kidney, liver, and testis from human, macaque, mouse, and opossum were obtained from the Baseline Atlas of Gene Expression Altas website8 (Keane et al., 2011). Relative expression level of TENM4 mRNA was calculated as the original expression normalized by the highest expression among the six tissues in each species.
Results
Clinical Data of the Schizophrenia Family
A Chinese Han family of five family members with three generations affected by schizophrenia was recruited from our longitudinal follow-up, and latest interview was done in 2004 for II-2 and III-3, and 2011 for III-2 and III-4, respectively (Figure 1, Table 1). All affected family members received the interviews were all diagnosed as schizophrenia (Table 1). The affected patients all had disease-onset after adulthood (age of onset later than 22 years old), and manifested auditory hallucination, persecutory delusion, and were easy to be offended or irritated and aggressive in the acute stage. They developed gradually into difficulty in concentration, and blunting affect, leading to social isolation and work disability for multiple episodes.
Exome Sequencing Data of the Family
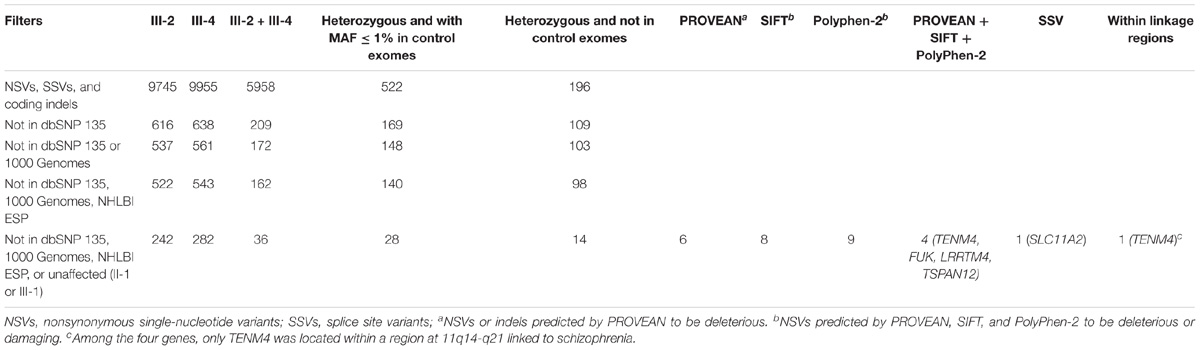
In order to identify the underlying genetic predisposition in the family, exome sequencing was performed in two affected (III-2 and III-4) and two unaffected (II-1 and III-1) family members. The original exome sequencing data from the four family members were summarized in Supplementary Table S1. The exome sequencing achieved 96.2% mean coverage and 62.5× mean depth of target region, which allowed high quality of variant calling (Supplementary Table S2). The variants called from the exome data were analyzed using a step-by-step filtering method (Table 2). The variants were firstly filtered to remove all noncoding and synonymous variants, and keep only nonsynonymous single-nucleotide variants (NSVs), splice site variant (SSV), and coding indels. The selected variants were then filtered against databases including dbSNP 135, 1000 Genomes, the NHLBI Exome Sequencing Project, and non-psychiatric exomes from the Exome Aggregation Consortium (ExAC) to remove common and known variants in the public databases. Given the pattern of dominant inheritance in the family, only heterozygous variants shared by the two affected (III-2 and III-4) but not found in the two unaffected (II-1 and III-1) were kept for subsequent analysis. In order to look for rare variants, the remaining variants were then filtered against exomes of non-schizophrenic controls. Twenty-eight remaining variants had a MAF less than 1% in the control exomes (Table 2), none of which was in PRODH, DISC1, SHANK3, NRXN1, or SLC1A1. These variants were then evaluated for impact on protein function to by three bioinformatic programs PROVEAN, SIFT, and PolyPhen-2. Higher priority was given to four NSVs and one SSV that were predicted to be deleterious and were not detected in control exomes (Table 2). These five variants were then analyzed with prior knowledge of gene function as shown in Supplementary Table S4. SLC11A2 is a known disease-causing gene for autosomal recessive anemia (OMIM #206100) (Iolascon et al., 2006). Likewise TSPAN12 has been characterized as a disease-causing gene for exudative vitreoretinopathy (OMIM #613310) (Nikopoulos et al., 2010). These two genes were known disease-causing genes of distinct phenotypes from schizophrenia. A recently study on autozygosity showed lack of an apparent phenotype in individuals with loss of function variants of FUK (Alsalem et al., 2013). Therefore, these three genes were excluded from further study. TENM4 is located within a region that was previously linked to schizophrenia (SCZD2, OMIM %603342). LRRTM4 has highly selective expression in the brain and can mediate excitatory synapse development on dentate gyrus granule cells (Siddiqui et al., 2013). Therefore, TENM4 and LRRTM4 were then included for subsequent validation experiment.
Cosegregation With Schizophrenia in the Family
Sanger sequencing results showed that the LRRTM4 variant was not found in one affected family member III-3, and thus were excluded from further analysis due to lack of co-segregation with the disease phenotype. The c.6724C>T variant in exon 32 of TENM4 was heterozygous in the four affected (III-2, III-3, and III-4 by sequencing and II-2 by inferring) (Figure 2A and Supplementary Figure S1), and were not detected in the two unaffected (II-1 and III-1). These results showed that the c.6724C>T variant cosegregated with the schizophrenia phenotype in the schizophrenia family. The variant was not detected in any of another group of 205 unrelated non-schizophrenic controls by Sanger sequencing, in consistent to its absence in the public databases and our in-house control exomes as described above.
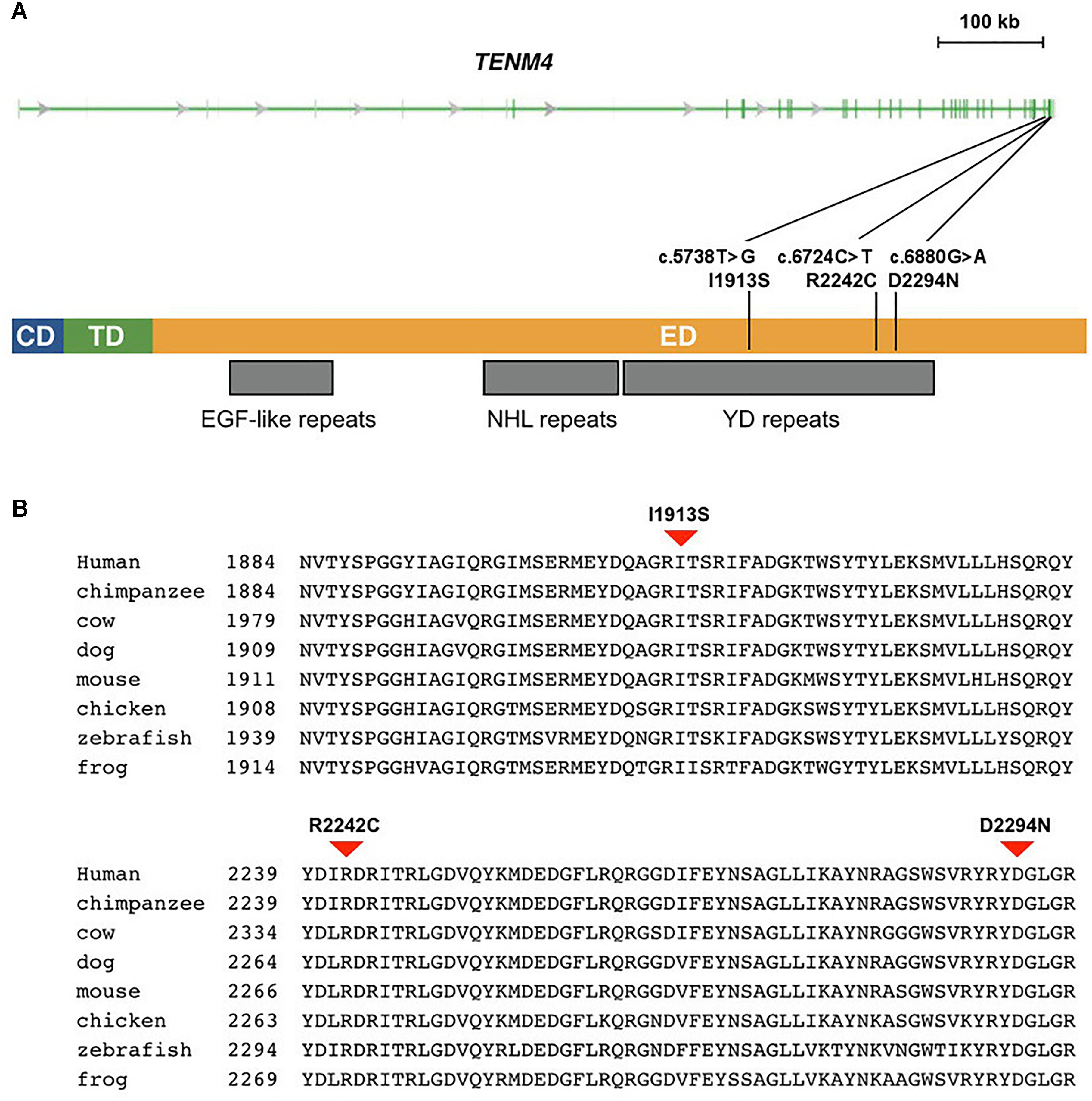
Figure 2. Missense mutations identified in schizophrenia patients in TENM4 at 11q14.1. (A) The genomic features of the TENM4 gene and the domains in the encoded protein. TENM4 contains a cellular domain (CD), a transmembrane domain (TD), and a large extracellular domain (ED) containing EGF-like, NHL repeats, and YD repeats (indicated underneath the protein). The three mutations detected in schizophrenic patients are shown. (B) Sequence alignment of TENM4 orthologs in vertebrates. The residues (I1913, R2242, and D2294) mutated in the three mutations were conserved among vertebrates from zebrafish through human. The three mutations were predicted to be deleterious to the protein function.
TENM4 Mutations in Unrelated Sporadic Schizophrenic Patients
As TENM4 could possible serve as mutation hotspots, we further investigate TENM4 exons 31, 32, and 33 which were adjacent to c.6724C>T in 120 unrelated sporadic schizophrenic patients using Sanger sequencing (Supplementary Table S5). Two additional missense alterations, c.5738T>G (p.I1913S) in exon 31 and c.6880G>A (p.D2294N) in exon 32 were detected (Table 3), both of which were predicted to be deleterious to protein function by PROVEAN, SIFT, and PolyPhen-2. The variant p.D2294N was found in a single heterozygous individual (MAF = 8.3 × 10−6) in the ExAC database but not found in our controls. And p.I1913S was not detected in the public databases, control exomes, or in the Sanger sequencing results of 205 unrelated controls. A rare synonymous variant (p.K1922K) was also significantly associated with schizophrenia (p < 0.05). In contrast, only one missense variant with a MAF less than 0.04% (1 in 2882) in the YD repeats was detected in the same exons in controls, which was predicted to be benign, and was not significantly different between patients and controls (p = 1). Further comparison of clinical features did not find evident difference between patients with and without either of p.I1913S and p.D2294N.
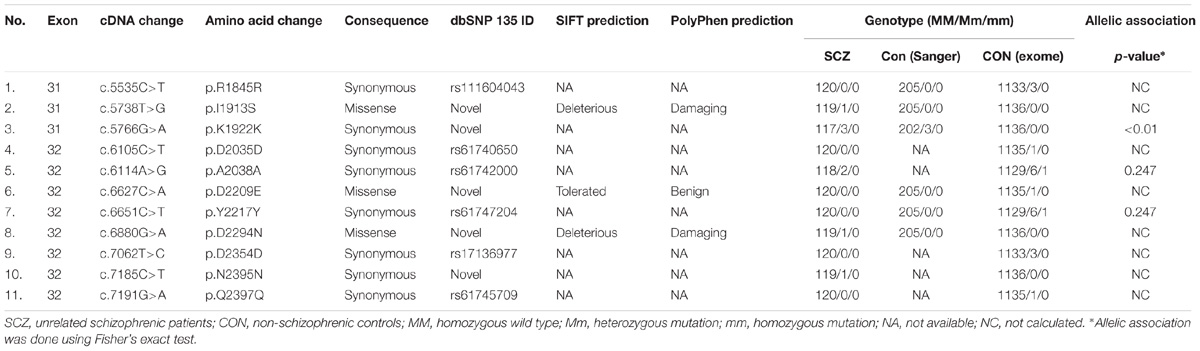
Table 3. Additional variants in unrelated schizophrenic patients and controls in TENM4 exons adjacent to the mutation found in the family.
TENM4 Residues Mutated in Schizophrenia
The human TENM4 protein sequence was aligned to its orthologs in other vertebrates. The alignment showed that the TENM4 residues mutated in schizophrenic patients were evolutionarily conserved (Figure 2). They were all located within a region that consisted of 23 tyrosine/aspartic acid (YD) repeats in the extracellular domain of the transmembrane TENM4 protein.
To evaluate burden of rare variants in the gene, all coding variants of TENM4 were examined in the exomes of the family and non-schizophrenia controls. The data were summarized in Supplementary Table S6. In total, 39 variants were detected in the gene from the exomes including the novel mutation pR2242C exclusively found in the affected family members. We analyzed the distribution of variants predicted to be deleterious in control exomes. Three novel rare variants that were predicted to damaging were exclusively found in control exomes, including a missense variant (p.R325W) in the teneurin N-terminus, a missense variant (p.E1098G) between the EGF-like and NHL repeats, and one stop-gained variant (p.Q2735X) in the last exon close to the C-terminus. However, by carefully looking into the positions of novel and known variants predicted to be deleterious in control exomes, none of them was found to be within the YD repeats. The three control individuals carrying these variants had no symptom of schizophrenia. Moreover, either control exomes or the two unaffected did not have any variant predicted to be deleterious in the YD repeats. In contrast, novel, rare deleterious variants in the YD repeat region were exclusively observed in affected family members and unrelated patients. These findings might suggest increased burden of rare deleterious variants within the YD repeats of TENM4 was associated with schizophrenia (variant burden test p < 0.001).
Expression Specificity of TENM4 in the Brain
We then analyzed RNA-seq data of six human tissues including brain, cerebrum, heart, liver, kidney, and testis, which were retrieved from Gene Expression Atlas. The results showed that TENM4 was highly expressed in human brain tissues (Supplementary Figure S2). In addition, the gene exhibited higher specificity to the brain compared to other tissues. Moreover, highest expression level of TENM4 in the brain among six different tissues was observed in mouse, opossum, and monkey, suggesting that the high specificity of TENM4 expression in the brain was evolutionarily conserved in mammals.
Discussion
In the current study, we conducted exome sequencing in a three-generation family affected by schizophrenia, and identified a novel candidate gene TENM4 in a linkage locus SCZD2 at 11q14-21. One missense mutation cosegregated with the schizophrenia phenotype, and two more missense mutations were detected in unrelated sporadic schizophrenic patients. These findings hence implicated a potential role of TENM4 in etiology of schizophrenia.
The SCZD2 locus at 11q14-21 has not yet been defined molecularly for any disease gene for schizophrenia (Yamada et al., 2004). A balanced t(1;11)(q42.1;q14.3) translocation was first reported to co-segregate with schizophrenia (Millar et al., 2000). The 11q14-21 region has been linked to mental illnesses, including schizophrenia and bipolar disorder (Fanous et al., 2012). A recent study has further demonstrated that polymorphisms in the SCZD2 locus are associated with schizophrenia in Scottish population (Debono et al., 2012), emphasizing the possibility that the SCZD2 region may harbor genetic variants contributing to risk of schizophrenia. By showing co-segregation with schizophrenia in the family, our findings thus suggested that TENM4 could be a candidate gene in the SCZD2 locus.
TENM4 is located at 11q14.1 and contains 34 exons, which encode a large protein composed of 2769-amino acid residues. The molecular function of the gene has not yet been well characterized by far. It probably plays as a type II transmembrane protein, and functions as a cellular signal transducer. TENM4 is a member of the TENEURIN/ODD OZ (TEN/ODZ) protein family, which is first identified in Drosophila (Baumgartner and Chiquet-Ehrismann, 1993; Baumgartner et al., 1994), and contains many members with orthologs identified in mammals, vertebrates, insects, and nematodes (Minet and Chiquet-Ehrismann, 2000). This family of transmembrane proteins shares a common TENEURIN N-terminus intracellular domain, 5–8 EGF-like repeats, and more than 20 tyrosine/aspartic acid (YD) repeats. TENM4 were highly expressed in mammalian brain (Zhou et al., 2003). TENM4 mutations in mice can be lethal, and the mutants exhibit anomalies in gastrulation (Lossie et al., 2005). It is noted that missense mutations in TENM4 have recently been reported to cause autosomal-dominant essential tremor (Hor et al., 2015). TENM4 rare variants are also found to be associated with bipolar disorder (Ament et al., 2015). Furthermore, a role of Tenm4 has been implicated in a rodent model of schizophrenia (Neary et al., 2017). These studies taken together were in line with a substantial role of TENM4 in mammalian central nervous system development and functions.
In the current study, a rare missense mutation (c.6724C>T; p.R2242C) in TENM4 showed cosegregation with schizophrenia in a three-generation family. In addition, two more rare mutations were identified in unrelated sporadic schizophrenic patients (Table 3), suggesting the exon surrounding p.R2242C in TENM4 could be a potential mutation hotspot for schizophrenia. Such findings thus pointed to potential association of TENM4 with schizophrenia. It was noted that more TENM4 missense were observed in these schizophrenic patients than in controls. It could be due to higher clinical homogeneity in our patients. All of the unrelated schizophrenic patients in the current study were diagnosed as paranoid schizophrenia. It should also be noted that the local population in the current study could be more genetically homogeneous as reported in a recent study (Chen et al., 2009b). The absence of rare TENM4 mutations in non-schizophrenic controls was confirmed by using Sanger sequencing in 205 controls plus exome sequencing in a larger control cohort, which should ensure that the extreme rareness of these mutations was unlikely due to technical bias if any. Among the three TENM4 mutations found in schizophrenia patients, p.D2294N was found in a heterozygous individual (MAF = 8.3 × 10−6) in the ExAC database indicated that these deleterious TENM4 mutations might exist in the general population, yet with an extremely low frequency. We further checked loss-of-function variants in TENM4 in the ExAC database. In total six loss-of-function variants were found for the canonical TENM4 isoform, each of which was detected in a single heterozygous individual (MAF = 8.3 × 10−6), suggesting that loss-of-function variants in TENM4 were extremely rare. For comparison, there were 27 loss-of-function variants found in DISC1 in the ExAC database, with the highest MAF = 0.0005. Therefore, the function of TENM4 protein could be relatively conserved and was unlikely to be tolerant to loss-of-function mutations.
The TENM4 protein contains 1 teneurin N-terminal domain, 8 EGF-like repeats, 5 NHL (NCL-1, HT2A, and Lin-41) repeats, and 23 YD repeats. Our findings suggested that distribution of variants predicted to be deleterious was unlikely random. The mutations detected in schizophrenia patients in the current study were located within a YD repeat containing region in the extracellular domain of TENM4 protein, which was essential for extracellular protein–protein interaction and signal transduction for transmembrane proteins. In contrast, only a benign missense variant with low MAF, and no deleterious variant were found in the YD repeat region among non-schizophrenia control exomes. A premature termination codon variant was identified in the last exon close to the C-terminus in one of our control exomes. However, it was not within the YD repeat. Encoding 98.7% of full-length protein, the variant was unlikely to trigger nonsense-mediated decay of mRNA to affect the protein function (Cirulli et al., 2011). Such findings suggested that increased burden of rare deleterious variants in YD repeats of TENM4 was probably associated with schizophrenia. The YD repeat sequences contain two tandem copies of a 21-residue extracellular repeat named for a YD dipeptide, the most strongly conserved motif of the repeat (Minet et al., 1999). These repeats appear in general to be involved in binding carbohydrate, and their function has not been understood clearly. In human, YD repeats are found in all four of TENM proteins. YD repeats are found in all four of TENM proteins. Prior studies suggest that these conserved repeats might be involved in neuron development. Minet et al. (1999) reported that culture substrate coated with the YD repeat region of chicken teneurin-1 supported neuron outgrowth from dorsal root ganglion explants. Therefore, defects in YD repeats might contribute to development of psychiatric diseases due to its role in neuron development. And our results thus warranted further studies of the YD repeats and TENM4 in molecular mechanism of schizophrenia.
Although TENM4 has not been clinically linked to schizophrenia previously, recent studies have already implicated its possible role in mental illness and cognition. In an epigenetic study, the Tenm4 gene has recently been implicated in a rodent model of schizophrenia (Neary et al., 2017). In addition, TENM4 is associated with bipolar disorder in a recent genome-wide association study with a large sample size (Sklar et al., 2011). And genetic studies have shown prominent genetic overlap between schizophrenia and bipolar disorder. In a large population-based study in Sweden and Demark (Lichtenstein et al., 2009), risk of bipolar disorder was associated with a family history of schizophrenia. Moreover, a recent study has reported that rare variants in TENM4 might contribute to etiology of bipolar disorder (Ament et al., 2015). Moreover, in a study on genetic correlation on brain magnetic resonance imaging and cognitive tests, TENM4 has been ranked among the top 25 loci with Boston Naming test score via generalized estimating equations, suggesting its link to cognition function33. Association of cognitive impairment with schizophrenia has been implicated in previous studies (O’Carroll, 2000). Although our study subjects have not been assessed for cognition function directly in our sample collection, our records showed that all affected in the family received education and had been employed, suggesting they were unlikely to have cognitive impairment before disease onset. After disease onset, as the disease progressed, they gradually exhibited cognitive impairments with marked negative symptoms including disorganized thinking, lack of judgment and insight, social dysfunction, and loss of ability to work. Likewise, all of the unrelated schizophrenia patients had evident hallucination and delusion during the acute phase. As the disease progressed, cognitive impairments were observed in these patients including poor attention, active social avoidance, lack of judgment and insight, poor rapport, and disturbance of volition. The rare deleterious TENM4 variants found in schizophrenia patients might not affect cognition directly. Therefore, our data suggested association of TENM4 with schizophrenia, while its association with cognitive impairment needs to be investigated in further study. Nevertheless, taken together with these previous studies, our findings were in line with a substantial role of TENM4 in central nervous system development and diseases.
Schizophrenia is of high genetic heterogeneity, and genetic variants with different effects can contribute to the disease risk. In addition to common variants with relatively small effects identified by both GWAS and candidate gene studies, the role of rare variants or mutations with large effects has been emphasized in psychiatric disorders such as autism (Chaste et al., 2014), especially by exome sequencing studies (Cukier et al., 2014; Toma et al., 2014). Schizophrenia-associated rare mutations have been depicted in recent exome sequencing studies (Purcell et al., 2014). However, most of these variants need to be further confirmed, and for most of them validation still remain a big challenge due to their low frequency (<1%) and uncertain penetrance, especially for schizophrenia. The mapping of rare mutations can be enhanced by analyzing rare variants cosegregated with disease phenotypes in high-density families of schizophrenia, and by integrating prior knowledge of linkage loci. The association between TENM4 mutations with schizophrenia in the current study was not contradictory to previous literature regarding architecture of schizophrenia genetic risk. First, to the best of our knowledge, mutations with large risk to schizophrenia have been reported in at least five genes, DISC1, NRXN1, PRODH, SHANK3, and SLC1A1 according to records in OMIM (Supplementary Table S3). Second, the observation of TENM4 mutations in some of our schizophrenic patients does not rule out possible contribution of other genetic factors such as polymorphisms with small effects or mutations with large effects to the disease in the same patients.
In the current study, by using exome sequencing, we demonstrated cosegregation of a novel missense mutation in TENM4 with schizophrenia in a Chinese family. With additional rare TENM4 mutations significantly associated with schizophrenia, our findings suggested that TENM4 were a candidate gene for the disease in the SCZD2 locus at 11q14-21. Actual specific functional effects of these TENM4 rare variants were thus warranted in further study.
Additional Information
Accession Codes
Whole-exome sequence data for the schizophrenia family has been deposited in GenBank Sequence Read Archive (SRA) under the accession code SPR172436.
URLs
Online Mendelian Inheritance in Man (OMIM), http://omim.org/; PROVEAN, http://provean.jcvi.org/; SIFT, http://sift.jcvi.org/; PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/; Gene Expression Atlas, http://www.ebi.ac.uk/gxa/; ExAC, http://exac.broadinstitute.org/.
Author Contributions
C-BX and X-HZ contributed equally to this work. X-HH and J-HC conceived the project and planned the experiments. C-BX, X-HZ, YW, Q-LC, C-RW, J-TH, H-SZ, and W-HX clinically characterized the cases and collected blood samples. C-BX, Z-HX, and JZ performed the validation experiments. J-HC, XY, S-SY, Z-YP, and H-MY analyzed and interpreted the experiment data. H-MY provided the control exome data. C-BX and X-HH interpreted the clinical data. All authors contributed to the final manuscript.
Funding
This study was supported in part by grants from the National Natural Science Foundation of China (Nos. 31671311, 81371033, 81000397, and 81573510), the National first-class discipline program of Light Industry Technology and Engineering (LITE2018-14), the National “Tenth Five-Year” Science and Technology Program (2002BA711A08), the Project of Guangdong Province Medical Research Foundation (A2012409), Natural Science Foundation of Guangdong Province, China (No. S2013010015618), Science and Technology Planning Project of Guangdong Province, China (No. 2010B031600130), the “Six Talent Peak” Plan of Jiangsu Province (No. SWYY-127), the Innovative and Entrepreneurial Talents of Jiangsu Province, the Program for High-Level Entrepreneurial and Innovative Talents Introduction of Jiangsu Province, Guangdong High-Level Personnel of Special Support Program, Yangfan Plan of Talents Recruitment Grant, and Fundamental Research Funds for the Central Universities (JUSRP51712B).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the participating patients, institutions, and medical staff, without whose contribution this work would not have been possible.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00725/full#supplementary-material
Footnotes
- ^ https://www.ebi.ac.uk/arrayexpress/experiments/E-GEOD-30352/
- ^ http://genome.ucsc.edu/
- ^ http://bio-bwa.sourceforge.net/
- ^ http://samtools.sourceforge.net/
- ^ http://www.ncbi.nlm.nih.gov/snp/
- ^ http://www.1000genomes.org
- ^ http://www.ncbi.nlm.nih.gov/refseq/
- ^ http://www.molgen.ua.ac.be/bioinfo/novosnp/
References
Alsalem, A. B., Halees, A. S., Anazi, S., Alshamekh, S., and Alkuraya, F. S. (2013). Autozygome sequencing expands the horizon of human knockout research and provides novel insights into human phenotypic variation. PLoS Genet. 9:e1004030. doi: 10.1371/journal.pgen.1004030
Ament, S. A., Szelinger, S., Glusman, G., Ashworth, J., Hou, L., Akula, N., et al. (2015). Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc. Natl. Acad. Sci. U.S.A. 112, 3576–3581. doi: 10.1073/pnas.1424958112
Baumgartner, S., and Chiquet-Ehrismann, R. (1993). Tena, a Drosophila gene related to tenascin, shows selective transcript localization. Mech. Dev. 40, 165–176. doi: 10.1016/0925-4773(93)90074-8
Baumgartner, S., Martin, D., Hagios, C., and Chiquet-Ehrismann, R. (1994). Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 13, 3728–3740. doi: 10.1002/j.1460-2075.1994.tb06682.x
Bergen, S. E., and Petryshen, T. L. (2012). Genome-wide association studies (GWAS) of schizophrenia: does bigger lead to better results? Curr. Opin. Psychiatry 25, 76–82. doi: 10.1097/YCO.0b013e32835035dd
Bustamante, M. L., Herrera, L., Gaspar, P. A., Nieto, R., Maturana, A., Villar, M. J., et al. (2017). Shifting the focus toward rare variants in schizophrenia to close the gap from genotype to phenotype. Am. J. Med. Genet. B Neuropsychiatr. Genet. 174, 663–670. doi: 10.1002/ajmg.b.32550
Chaste, P., Sanders, S. J., Mohan, K. N., Klei, L., Song, Y., Murtha, M. T., et al. (2014). Modest impact on risk for autism spectrum disorder of rare copy number variants at 15q11.2, specifically breakpoints 1 to 2. Autism Res. 7, 355–362. doi: 10.1002/aur.1378
Chen, J., Tsang, S.-Y., Zhao, C.-Y., Pun, F. W., Yu, Z., Mei, L., et al. (2009a). GABRB2 in schizophrenia and bipolar disorder: disease association, gene expression and clinical correlations. Biochem. Soc. Trans. 37, 1415–1418. doi: 10.1042/BST0371415
Chen, J., Zheng, H., Bei, J.-X., Sun, L., Jia, W., Li, T., et al. (2009b). Genetic structure of the han chinese population revealed by genome-wide SNP variation. Am. J. Hum. Genet. 85, 775–785. doi: 10.1016/j.ajhg.2009.10.016
Cirulli, E. T., Heinzen, E. L., Dietrich, F. S., Shianna, K. V., Singh, A., Maia, J. M., et al. (2011). A whole-genome analysis of premature termination codons. Genomics 98, 337–342. doi: 10.1016/j.ygeno.2011.07.001
Cukier, H. N., Dueker, N. D., Slifer, S. H., Lee, J. M., Whitehead, P. L., Lalanne, E., et al. (2014). Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol. Autism 5:1. doi: 10.1186/2040-2392-5-1
Debono, R., Topless, R., Markie, D., Black, M. A., and Merriman, T. R. (2012). Analysis of the DISC1 translocation partner (11q14.3) in genetic risk of schizophrenia. Genes Brain Behav. 11, 859–863. doi: 10.1111/j.1601-183X.2012.00832.x
Endicott, J. (1978). Family History: Research Diagnostic Criteria: (FH-RDC). New York, NY: New York State Psychiatric Institute.
Fanous, A. H., Middleton, F. A., Gentile, K., Amdur, R. L., Maher, B. S., Zhao, Z., et al. (2012). Genetic overlap of schizophrenia and bipolar disorder in a high-density linkage survey in the Portuguese Island population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 159B, 383–391. doi: 10.1002/ajmg.b.32041
First, M. B., Spitzer, R. L., Gibbon, M., and Williams, J. B. W. (1997). User’s Guide for the Structured Clinical Interview for Dsm-IV Axis I Disorders: Scid-1 Clinician Version, 1 Edn. Washington, DC: American Psychiatric Publisher.
Gauthier, J., Champagne, N., Lafrenière, R. G., Xiong, L., Spiegelman, D., Brustein, E., et al. (2010). De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 107, 7863–7868. doi: 10.1073/pnas.0906232107
Guy, W. (National) Institute of Mental Health (U.S.). Psychopharmacology Research Branch., Early Clinical Drug Evaluation Program. (1976). ECDEU Assessment Manual for Psychopharmacology. (Rev. 1976). Rockville, MD: U.S. Dept. of Health, Education, and Welfare, Public Health Service, Alcohol, Drug Abuse, and Mental Health Administration, National Institute of Mental Health, Psychopharmacology Research Branch, Division of Extramural Research Programs.
Hor, H., Francescatto, L., Bartesaghi, L., Ortega-Cubero, S., Kousi, M., Lorenzo-Betancor, O., et al. (2015). Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor. Hum. Mol. Genet. 24, 5677–5686. doi: 10.1093/hmg/ddv281
Iolascon, A., d’Apolito, M., Servedio, V., Cimmino, F., Piga, A., and Camaschella, C. (2006). Microcytic anemia and hepatic iron overload in a child with compound heterozygous mutations in DMT1 (SCL11A2). Blood 107, 349–354. doi: 10.1182/blood-2005-06-2477
Jacquet, H., Raux, G., Thibaut, F., Hecketsweiler, B., Houy, E., Demilly, C., et al. (2002). PRODH mutations and hyperprolinemia in a subset of schizophrenic patients. Hum. Mol. Genet. 11, 2243–2249. doi: 10.1093/hmg/11.19.2243
John, J., Sharma, A., Kukshal, P., Bhatia, T., Nimgaonkar, V. L., Deshpande, S. N., et al. (2019). Rare variants in tissue inhibitor of metalloproteinase 2 as a risk factor for schizophrenia: evidence from familial and cohort analysis. Schizophr. Bull. 45, 256–263. doi: 10.1093/schbul/sbx196
Keane, T. M., Goodstadt, L., Danecek, P., White, M. A., Wong, K., Yalcin, B., et al. (2011). Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294. doi: 10.1038/nature10413
Lichtenstein, P., Yip, B. H., Björk, C., Pawitan, Y., Cannon, T. D., Sullivan, P. F., et al. (2009). Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 373, 234–239. doi: 10.1016/S0140-6736(09)60072-6
Lossie, A. C., Nakamura, H., Thomas, S. E., and Justice, M. J. (2005). Mutation of l7Rn3 shows that Odz4 is required for mouse gastrulation. Genetics 169, 285–299. doi: 10.1534/genetics.104.034967
Millar, J. K., Wilson-Annan, J. C., Anderson, S., Christie, S., Taylor, M. S., Semple, C. A. M., et al. (2000). Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423. doi: 10.1093/hmg/9.9.1415
Minet, A. D., and Chiquet-Ehrismann, R. (2000). Phylogenetic analysis of teneurin genes and comparison to the rearrangement hot spot elements of E. coli. Gene 257, 87–97. doi: 10.1016/S0378-1119(00)00388-7
Minet, A. D., Rubin, B. P., Tucker, R. P., Baumgartner, S., and Chiquet-Ehrismann, R. (1999). Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. J. Cell Sci. 112, 2019–2032.
Myles-Worsley, M., Tiobech, J., Browning, S. R., Korn, J., Goodman, S., Gentile, K., et al. (2013). Deletion at the SLC1A1 glutamate transporter gene co-segregates with schizophrenia and bipolar schizoaffective disorder in a 5-generation family. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 162B, 87–95. doi: 10.1002/ajmg.b.32125
Neary, J. L., Perez, S. M., Peterson, K., Lodge, D. J., and Carless, M. A. (2017). Comparative analysis of MBD-seq and MeDIP-seq and estimation of gene expression changes in a rodent model of schizophrenia. Genomics 109, 204–213. doi: 10.1016/j.ygeno.2017.03.004
Nikopoulos, K., Gilissen, C., Hoischen, A., van Nouhuys, C., Boonstra, F. N., Arts, P., et al. (2010). Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am. J. Hum. Genet. 86, 240–247. doi: 10.1016/j.ajhg.2009.12.016
O’Carroll, R. (2000). Cognitive impairment in schizophrenia. Adv. Psychiatr. Treat. 6, 161–168. doi: 10.1192/apt.6.3.161
Pulver, A. E., Karayiorgou, M., Wolyniec, P. S., Lasseter, V. K., Kasch, L., Nestadt, G., et al. (1994). Sequential strategy to identify a susceptibility gene for schizophrenia: report of potential linkage on chromosome 22q12-q13.1: part 1. Am. J. Med. Genet. 54, 36–43. doi: 10.1002/ajmg.1320540108
Purcell, S. M., Moran, J. L., Fromer, M., Ruderfer, D., Solovieff, N., Roussos, P., et al. (2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190. doi: 10.1038/nature12975
Rujescu, D., Ingason, A., Cichon, S., Pietiläinen, O. P. H., Barnes, M. R., Toulopoulou, T., et al. (2009). Disruption of the neurexin 1 gene is associated with schizophrenia. Hum. Mol. Genet. 18, 988–996. doi: 10.1093/hmg/ddn351
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. doi: 10.1038/nature13595
Shih, R. A., Belmonte, P. L., and Zandi, P. P. (2004). A review of the evidence from family, twin and adoption studies for a genetic contribution to adult psychiatric disorders. Int. Rev. Psychiatry 16, 260–283. doi: 10.1080/09540260400014401
Siddiqui, T. J., Tari, P. K., Connor, S. A., Zhang, P., Dobie, F. A., She, K., et al. (2013). An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron 79, 680–695. doi: 10.1016/j.neuron.2013.06.029
Singh, T., Walters, J. T. R., Johnstone, M., Curtis, D., Suvisaari, J., Torniainen, M., et al. (2017). The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet. 49, 1167–1173. doi: 10.1038/ng.3903
Sklar, P., Ripke, S., Scott, L. J., Andreassen, O. A., Cichon, S., Craddock, N., et al. (2011). Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983. doi: 10.1038/ng.943
Stefansson, H., Sigurdsson, E., Steinthorsdottir, V., Bjornsdottir, S., Sigmundsson, T., Ghosh, S., et al. (2002). Neuregulin 1 and susceptibility to schizophrenia. Am. J. Hum. Genet. 71, 877–892. doi: 10.1086/342734
Sullivan, P. F., Kendler, K. S., and Neale, M. C. (2003). Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192. doi: 10.1001/archpsyc.60.12.1187
Tiihonen, J., Lönnqvist, J., Wahlbeck, K., Klaukka, T., Niskanen, L., Tanskanen, A., et al. (2009). 11-year follow-up of mortality in patients with schizophrenia: a population-based cohort study (FIN11 study). Lancet 374, 620–627. doi: 10.1016/S0140-6736(09)60742-X
Toma, C., Torrico, B., Hervás, A., Valdés-Mas, R., Tristán-Noguero, A., Padillo, V., et al. (2014). Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol. Psychiatry 19, 784–790. doi: 10.1038/mp.2013.106
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
World Health Organization (1988). WHO Psychiatric Disability Assessment Schedule (WHO/DAS): With a Guide to Its Use. Geneva: World Health Organization.
Xu, B., Ionita-Laza, I., Roos, J. L., Boone, B., Woodrick, S., Sun, Y., et al. (2012). De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 44, 1365–1369. doi: 10.1038/ng.2446
Xu, B., Roos, J. L., Dexheimer, P., Boone, B., Plummer, B., Levy, S., et al. (2011). Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 43, 864–868. doi: 10.1038/ng.902
Yamada, K., Iwayama-Shigeno, Y., Yoshida, Y., Toyota, T., Itokawa, M., Hattori, E., et al. (2004). Family-based association study of schizophrenia with 444 markers and analysis of a new susceptibility locus mapped to 11q13.3. Am. J. Med. Genet. B Neuropsychiatr. Genet. 127B, 11–19. doi: 10.1002/ajmg.b.20166
Yu, L., Arbez, N., Nucifora, L. G., Sell, G. L., DeLisi, L. E., Ross, C. A., et al. (2014). A mutation in NPAS3 segregates with mental illness in a small family. Mol. Psychiatry 19, 7–8. doi: 10.1038/mp.2012.192
Keywords: association, co-segregation, schizophrenia, exome analysis, rare mutation
Citation: Xue C-B, Xu Z-H, Zhu J, Wu Y, Zhuang X-H, Chen Q-L, Wu C-R, Hu J-T, Zhou H-S, Xie W-H, Yi X, Yu S-S, Peng Z-Y, Yang H-M, Hong X-H and Chen J-H (2019) Exome Sequencing Identifies TENM4 as a Novel Candidate Gene for Schizophrenia in the SCZD2 Locus at 11q14-21. Front. Genet. 9:725. doi: 10.3389/fgene.2018.00725
Received: 10 August 2018; Accepted: 22 December 2018;
Published: 28 January 2019.
Edited by:
Cunyou Zhao, Southern Medical University, ChinaReviewed by:
Xingyin Liu, Nanjing Medical University, ChinaXiao Dong, Albert Einstein College of Medicine, United States
Mariza De Andrade, Mayo Clinic, United States
Copyright © 2019 Xue, Xu, Zhu, Wu, Zhuang, Chen, Wu, Hu, Zhou, Xie, Yi, Yu, Peng, Yang, Hong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Hong Hong, aG9uZ3hpYW9ob25nQDIxY24uY29t Jian-Huan Chen, Y2poX2Jpb0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Chao-Biao Xue
Chao-Biao Xue Zhou-Heng Xu3†
Zhou-Heng Xu3† Jian-Huan Chen
Jian-Huan Chen