- Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, PA, United States
Although the genetic code is degenerate, synonymous codons for the same amino acid are not translated equally. Codon-specific translation is important for controlling gene expression and determining the proteome of a cell. At the molecular level, codon-specific translation is regulated by post-transcriptional epigenetic modifications of tRNA primarily at the wobble position 34 and at position 37 on the 3′-side of the anticodon. Modifications at these positions determine the quality of codon-anticodon pairing and the speed of translation on the ribosome. Different modifications operate in distinct mechanisms of codon-specific translation, generating a diversity of regulation that is previously unanticipated. Here we summarize recent work that demonstrates codon-specific translation mediated by the m1G37 methylation of tRNA at CCC and CCU codons for proline, an amino acid that has unique features in translation.
Introduction
The redundancy of the genetic code offers an opportunity for fine-tuning of protein synthesis, virtually at every codon position. Although the sequence of codons in an mRNA provides the template for translation into amino acid building blocks of the protein, synonymous codons are not translated equally. Codon bias, which is defined by the frequencies of usage among synonymous codons, is a specific feature unique to each genome and each gene and can impact the fitness of each organism (Plotkin and Kudla, 2011). The choice of synonymous codons determines how these codons are differentially translated at the molecular level. Translation is controlled by tRNA species with the anticodon that is cognate to each codon. The quality of a codon-anticodon pairing interaction is determined not only by the level of the tRNA available for the codon (Ikemura, 1982; Komar, 2009; Nedialkova and Leidel, 2015), but also and perhaps more importantly by post-transcriptional modifications to the tRNA anticodon, either at nucleobases or backbones (Takai and Yokoyama, 2003; Armengod et al., 2014; Grosjean and Westhof, 2016; Agris et al., 2018). These modifications are catalyzed by distinct enzymes that bear high relevance to human health and disease (Sarin and Leidel, 2014). Many of these enzymes are dedicated to modifications of the anticodon at position 34 (the wobble position) and at position 37 on the 3′-side of the anticodon. It is the nature of post-transcriptional modifications at these anticodon-associated positions that exert differential pairing with synonymous codons, resulting in differential translation among these codons. The mechanism of codon-specific translation is therefore the underlying basis that determines how codon bias impacts the fitness of each organism. It is an emerging new concept that directly influences the expression of the proteome at the cell level.
The m1G37-Methylation in tRna
While there are more than 100 post-transcriptional modifications in tRNA databases, the majority by themselves are not required for cell survival or growth. One exception is the N1-methylation of G37 on the 3′-side of the tRNA anticodon, generating m1G37 (Figure 1A), which as a single methylated nucleobase is not only essential for life but is also conserved in evolution present in all three domains of life (Bjork et al., 2001). In the bacterial domain, the biosynthesis of m1G37 is catalyzed by the tRNA methyl transferase TrmD (Hou et al., 2017), whereas in the eukaryotic and archaeal domains, it is catalyzed by Trm5 (Bjork et al., 2001). While both TrmD and Trm5 perform the same methyl transfer reaction, using S-adenosyl methionine (AdoMet) as the methyl donor, they are fundamentally different in structure, where TrmD is a member of the SpoU-TrmD family (Anantharaman et al., 2002; Ahn et al., 2003; Elkins et al., 2003; Ito et al., 2015; Hori, 2017) and Trm5 is a member of the Rossmann-fold family (Goto-Ito et al., 2008, 2009). TrmD and Trm5 also differ in virtually all aspects of the reaction mechanism (Christian et al., 2004, 2006, 2010a,b, 2013, 2016; Christian and Hou, 2007; Lahoud et al., 2011; Sakaguchi et al., 2012, 2014). Due to the dependence on m1G37 for cell survival, TrmD is required for growth in several bacterial species, including Escherichia coli and Salmonella (Gamper et al., 2015a), and Trm5 is required for growth in the single-cell eukaryote yeast Saccharomyces cerevisiae (Bjork et al., 2001), where it provides the important role of preventing mis-charging of tRNA (Perret et al., 1990). Additionally, the Trm5-dependent synthesis of m1G37 is present in both the cytosolic and mitochondrial compartments (Lee et al., 2007) and it is the initiation event that leads to further modifications to m1I37 (Brule et al., 2004) and to wyosine and derivatives in the cytosol (Urbonavicius et al., 2014). The molecular basis for how m1G37-tRNA is essential for cell survival is largely elucidated in E. coli and Salmonella (Gamper et al., 2015a).
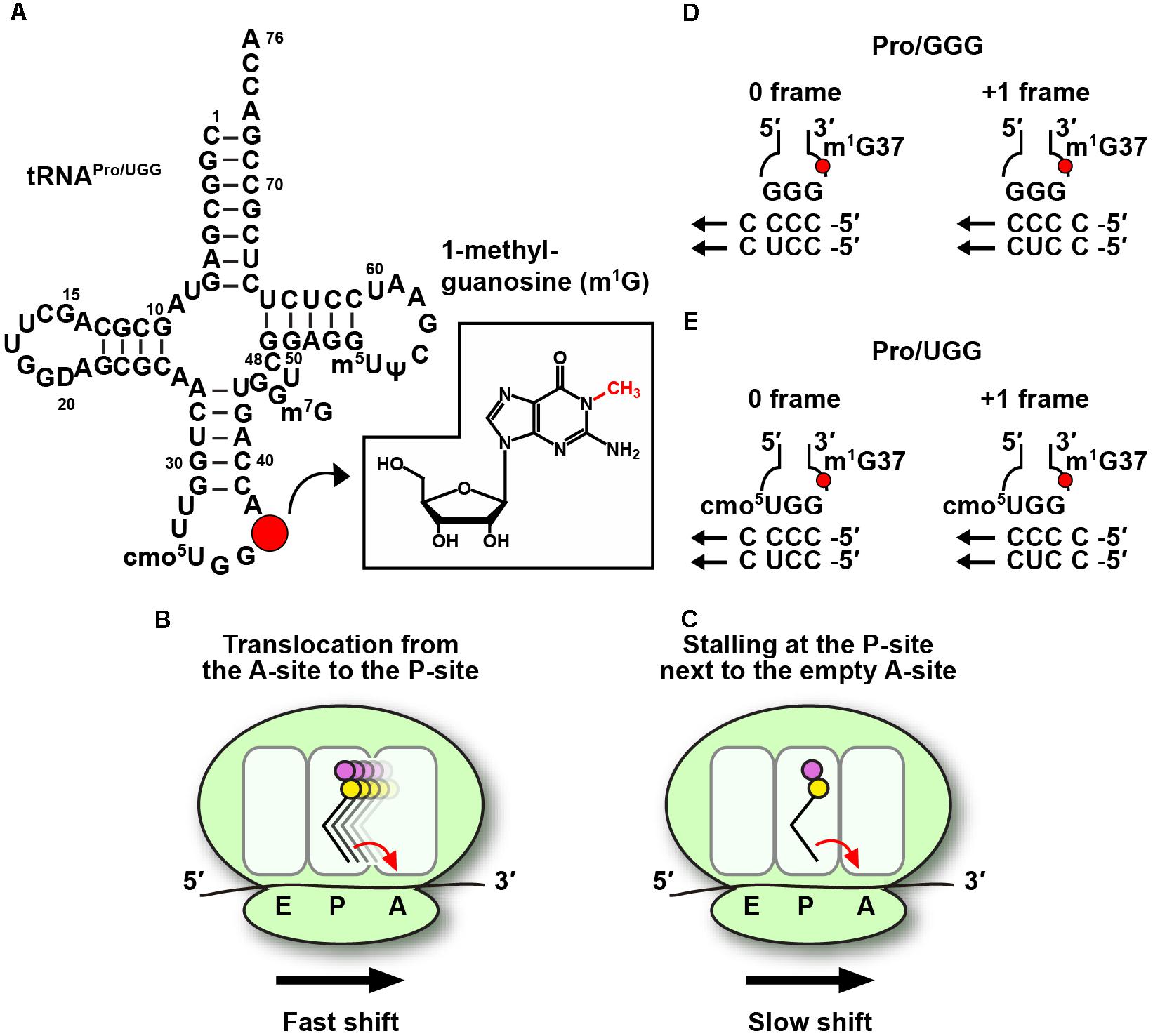
Figure 1. Codon-specific translation by m1G37-tRNA. (A) The sequence and cloverleaf structure of Escherichia coli UGG isoacceptor of tRNAPro (tRNAPro/UGG) is shown with m1G37 indicated by a red circle and displayed in the chemical structure in the inset. (B) Lack of m1G37 promotes the tRNA to make +1-frameshifts in a fast mechanism during tRNA translocation from the A- to the P-site on the ribosome, and also (C) in a much slower mechanism during tRNA stalling on the P-site next to an empty A-site. The fast and slow mechanisms are marked in the figure for panels B,C, respectively. The arrow in each panel indicates the direction of the ribosome translocation in the 5′-to-3′ direction on the mRNA. The propensity of (D) the GGG isoacceptor and (E) the UGG isoacceptor of E. coli tRNAPro to make the +1-frameshift is based on similar base pairing stability with cognate codons CCC and CCU. The UGG isoacceptor contains the cmo5U34 modified base at the wobble position, which allows pairing with all four nucleobases.
Maintenance of Protein Synthesis Reading Frame by m1G37-tRna
In bacteria, all three tRNAPro species, which read CCN codons, contain m1G37. Elimination of m1G37 by inactivation of TrmD leads to accumulation of ribosomal +1-frameshifts (+1-shifts), particularly at sites where mRNA sequences contain successive Cs in a row (Bjork et al., 1989). Unlike ribosomal mis-coding errors, which replace a correct amino acid with an incorrect one but maintain the protein synthesis reading frame, +1-shifts move the ribosome to the next nucleotide in the 5′ to 3′ direction of the mRNA, resulting in loss of the reading frame, premature termination of protein synthesis, and ultimately cell death. The ability of m1G37-tRNA to suppress ribosomal +1-shifts is an important finding that has activated in-depth mechanistic studies with high significance. mRNA sequences such as the pyrimidine-ending CCC and CCU codons for proline (Pro), followed by another pyrimidine, resulting in CC[C/U]-[C/U] sequence motifs, are particularly prone to +1-shifts (Yourno and Tanemura, 1970) and are often found within the first 15 codons of protein-coding genes (Gamper et al., 2015a), where ribosomal translation is highly sensitive to attenuation (Chen and Inouye, 1990). Additionally, some of the +1-shift prone sequences are directly adjacent to the start codon (Gamper et al., 2015a), at the second codon position of the reading frame, where ribosomal translation is in the unique stage of transitioning from the initiation to the elongation phase.
The maintenance of protein synthesis reading frame in normal cellular conditions is achieved with unexpectedly high fidelity. Despite the dynamics of each ribosome that successively move tRNA-mRNA complexes from the A- to P- to E-site, and despite the rapid rate of protein synthesis at 10–20 amino acids per second, +1-shifts typically occur in less than one per 30,000 amino acids (Jorgensen and Kurland, 1990), at least 10-fold lower relative to other types of translational errors. In a genetic reporter system developed in E. coli, m1G37-tRNA was found to have the strongest suppression power when the shift-prone CCC-C sequence was placed next to the start codon (Gamper et al., 2015a). Lack of m1G37 in tRNAPro at this second-codon position increased +1-shifts by almost 10-fold in the reporter system, higher than increases of +1-shifts at any other position throughout the reporter gene. An in vitro assay was then developed using reconstituted E. coli ribosomes, with the goal to assess the rate of +1-shifts relative to the rate of peptide bond formation (Gamper et al., 2015a). Both fast and slow mechanisms were uncovered (Gamper et al., 2015a). The fast mechanism occurs on a timescale comparable to that of peptide bond formation. It takes place when tRNAPro, carrying the first peptide bond made at the A-site, is moving into the P-site by a process known as translocation (Figure 1B). Thus, due to the ability of a +1-shift to compete with peptide bond formation on a similar timescale, should this process happen, it would compromise the accuracy of the reading frame. The slow mechanism occurs on a timescale 100-fold slower relative to peptide bond formation (Gamper et al., 2015a). It takes place when tRNAPro, carrying the first peptide bond, is sitting at the P-site next to an empty A-site (Figure 1C), which is usually the case during nutrient starvation that depletes charged aminoacyl-tRNAs. The model that has emerged from these mechanistic studies is that the ribosome is most prone to +1-shifts after it completes the first peptide bond formation and is moving tRNAPro from the A- to the P-site. This is also the transitioning point of the ribosome from the initiation phase (where the first peptide bond is made) to the elongation phase (where the second peptide bond will be made when the third codon enters the ribosome A-site). The finding that m1G37 has the strongest suppression of +1-shifts at the second codon position indicates its pivotal role in maintaining the reading frame during the transition point of the ribosome. Should m1G37 be eliminated leading to ribosomal +1-shifts at the transition point, this would generate a prematurely terminated peptide in a non-productive and energetically costly translational error.
Codon-Specific Translation by m1G37-tRna
Of the four CCN codons for Pro, CC[C/U] codons are most dependent on the presence of m1G37 (Jorgensen and Kurland, 1990) and the translation of these codons is noticeably slowed down upon deficiency of the methylation (Bjork and Nilsson, 2003). The codon-anticodon base-pairing interaction for CC[C/U] codons was thought to involve quadruplet base-pairing as implicated in previous studies (Hohsaka et al., 2001; Taki et al., 2002). However, there is no structural evidence of quadruplet pairing at the ribosomal A-site (Maehigashi et al., 2014) and the isolation of suppressor mutations of +1-shifts that are not in tRNA sequence suggests that quadruplet pairing is unlikely (Qian et al., 1998; Farabaugh, 2000). More recent work favors a model of tRNA slippage by a triplet codon-anticodon pairing interaction (Qian et al., 1998), which is supported by a detailed kinetic analysis (Gamper et al., 2015a). In the triplet slippage model, the +1-shift-prone sequences CC[C/U]-[C/U] are each read by two isoacceptors of tRNAPro: the GGG isoacceptor exploits wobble pairing without additional modifications, whereas the UGG isoacceptor requires the 5-carboxy-methoxy (cmo5) modification to U34 at the wobble position (Nasvall et al., 2004). In all cases, the codon-anticodon pairing interaction in the unshifted 0-frame and in the shifted +1-frame is similar (Figures 1D,E), indicating minimum energetic penalty for the tRNA to shift. The role of m1G37 in suppressing the shift is to re-organize the structure of the anticodon loop to stabilize the pairing in the 0-frame (Maehigashi et al., 2014). A differential effect of m1G37 in suppressing the shift between the GGG and UGG isoacceptors is that the methylation by itself is insufficient in the GGG isoacceptor and requires the assistance of the elongation factor EF-P (Gamper et al., 2015a), whereas it is dominant in the UGG isoacceptor (Gamper et al., 2015a,b). In contrast, the dependence on m1G37 for reading-frame accuracy is much reduced with the purine-ending CC[G/A] codons for Pro, which are read by two isoacceptors of tRNAPro: the CGG isoacceptor for the CCG codon and the UGG isoacceptor for both. Notably, the codon-anticodon pairing interaction in the +1-frame is highly unfavorable relative to the 0-frame, indicating that CC[G/A] codons are intrinsically more stable with pairing of the anticodon and are less likely to shift and less sensitive to m1G37 deficiency. This comparison illustrates the notion that synonymous codons have different requirements for pairing with the anticodon, and that such differences are the underlying basis of codon-specific translation.
The discovery that m1G37 has differential control of the translation of Pro codons is intriguing. Pro is a unique amino acid that contains an α-imino, rather than an α-amino group. It is the slowest substrate relative to all other amino acids for peptide bond formation, both as the acceptor or the donor of making a peptide bond by the ribosome (Pavlov et al., 2009). The aminoacyl group of Pro when charged to tRNAPro is also the least stable compared to others and it has the lowest affinity to the elongation factor EF-Tu (Peacock et al., 2014). Additionally, Pro is the only amino acid that can enable peptide backbones to make turns and change direction, so its position and distribution in a protein sequence directly impact on folding of the protein, particularly if trans-membrane domains are involved (Yohannan et al., 2004). Also, human disease-associated mutations are frequently found at Pro positions (Partridge et al., 2004). These considerations suggest that m1G37-tRNA has the ability to regulate a diverse process of cellular activities via differential translation of Pro codons at specific positions during gene expression.
Besides the universal association of m1G37 with all isoacceptors of tRNAPro, the CCG isoacceptor of tRNAArg for reading the Arg CGG codon, and the GAG isoacceptor of tRNALeu for reading the Leu CU[C/U] codons, also contain m1G37. The mechanism for why these isoacceptors carry m1G37 has not been investigated. It does not appear to involve suppression of +1-shifts, based on analysis of the codon-anticodon pairing interaction, but may involve release of ribosomes from stalling at the specific codons. Notably, the m1G37-containing CCG tRNAArg is only one of the several isoacceptors of the Arg family, whereas other members of the family do not need m1G37. Similarly, while m1G37 is present in the GAG, CAG, and UAG isoacceptors of the Leu family, it is absent from the UAA isoacceptor. This provides additional evidence for codon-specific translation that is dependent on the presence of post-transcriptional modifications in the anticodon for pairing with a codon.
Codon-Specific Translation in Mg2+ Homeostasis
An example of codon-specific translation mediated by m1G37 is the maintenance of Mg2+ homeostasis in Gram-negative bacteria. Mg2+ is the most abundant divalent cation in all living cells and is maintained at mM concentrations. For Salmonella enterica serovar Typhimurium (hereafter Salmonella), the etiologic agent of human gastroenteritis, the infection of the human gut into the metal-scarce macrophage compartment, requires Mg2+ transport into cells for survival and virulence of the pathogen (Papp-Wallace and Maguire, 2008a; Groisman et al., 2013). This transport is activated upon expression of the major Mg2+ transporter gene mgtA and is regulated at two levels: the transcriptional activation of the structural gene and the ribosomal translational control of the 5′-leader ORF ahead of the structural gene (Figure 2A). First, the initial transcription activation is by the membrane-bound two-component PhoPQ system, in which sensing of low external Mg2+ by PhoQ promotes phosphorylation of PhoP, which activates transcription of mgtA and many virulence genes (Papp-Wallace and Maguire, 2008a; Groisman et al., 2013). Second, the subsequent transcriptional regulation of mgtA is determined by the speed of ribosomal translation of the 5′-leader ORF of the mgtA mRNA. Rapid translation of this ORF exposes the Rho-utilization (rut) sequence, resulting in attenuation of transcription before the mgtA gene (Cromie et al., 2006). In contrast, slow or stalled translation of the ORF induces a structural change in the 5′-leader mRNA that places the rut sequence inaccessible to Rho-dependent termination, thus allowing transcription through mgtA (Park et al., 2010; Hollands et al., 2014; Figure 2B). This translation-dependent attenuation of transcription is common to regulation of expression of amino-acid biosynthesis genes (Merino and Yanofsky, 2005). In addition, Salmonella has a second inducible Mg2+ transporter gene mgtB expressed from the virulence operon mgtCBR under a similar control of transcriptional attenuation that is determined by the speed of ribosomal translation of its 5′-leader ORF (Papp-Wallace and Maguire, 2008a; Groisman et al., 2013). While Salmonella has a third and constitutively expressed Mg2+ transporter gene corA (Papp-Wallace and Maguire, 2008b), it is the inducible expression of mgtA and mgtB that maintains Mg2+ at virtually constant levels. While the external Mg2+ level can change by 5 orders of magnitude, the internal level varies by less than fivefold (Papp-Wallace and Maguire, 2008b). Without this Mg2+ homeostasis, Salmonella cannot survive in host cells. Thus, the translation-dependent attenuation of transcription of mgtA is the major determinant of Mg2+ homeostasis for Salmonella.
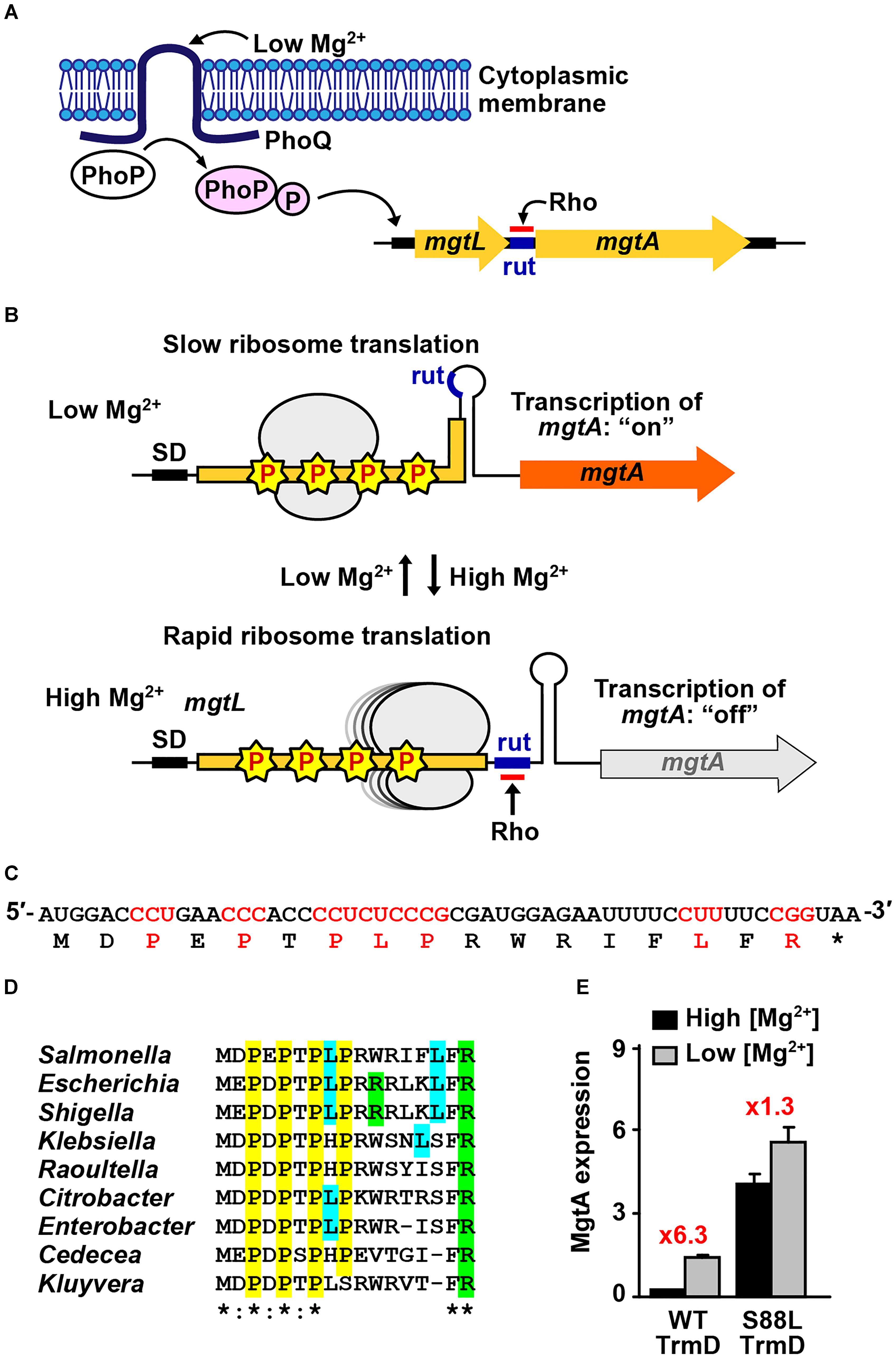
Figure 2. Codon-specific translation in Mg2+ homeostasis. (A) Mg2+ homeostasis in Salmonella is maintained by the membrane-bound two-component system PhoPQ sensing of the external low Mg2+, which activates transcription of the major transporter gene mgtA. Transcription of mgtA is determined by ribosomal translation of the 5′-leader ORF, which contains several m1G37-dependent Pro codons. (B) Low levels of Mg2+ slow down ribosomal translation due to stalling at m1G37-dependent codons, resulting in a structure of the 5′-leader ORF that places rut in a stem-loop region inaccessible to Rho, thus allowing transcription through mgtA, whereas high levels of Mg2+ enable rapid ribosomal translation of the 5′-leader ORF, which exposes the rut sequence ahead of the mgtA structure gene and attenuates transcription. (C) The codon sequence of the 5′-leader ORF is shown, where m1G37-dependent codons are highlighted. Asterisk “∗” indicates a termination codon. (D) The m1G37-dependent codons in the 5′-leader ORF are highly conserved across different species of Gram-negative bacteria. Asterisk “∗” indicates complete conservation among all the sequences, whereas a colon “:” indicates conservation between those with strongly similar properties. (E) Salmonella cells expressing the native trmD show a robust response of activation of transcription of mgtA from high to low Mg2+ (6.3-fold), whereas cells expressing a mutant trmD show a diminished response (1.3-fold), consistent with codon-specific translation at m1G37-dependent codons in the 5′-leader ORF. Data are obtained from published work (Gall et al., 2016).
Analysis of the 5′-leader ORF of mgtA shows a strong bias for codons that are dependent on m1G37-tRNA for translation (Park et al., 2010). These include Pro codons CC[C/U] at positions 3, 5, and 7, Leu codons CU[C/U] at positions 8 and 15, and Arg codon CGG at position 17 (Figure 2C). These m1G37-dependent codons are highly conserved across diverse Gram-negative bacteria (Figure 2D), indicating significance under evolutionary pressure. The codon conservation raises the possibility of codon-specific translation, where the speed of ribosomal translation of these codons is controlled by the presence of m1G37. A strong support for this possibility is that TrmD, the bacterial methyl transferase that synthesizes m1G37, is strictly dependent on Mg2+ for catalytic activity (Sakaguchi et al., 2014). While the requirement for Mg2+ for several tRNA methyl transferases is known (Hurwitz et al., 1964; Kumagai et al., 1982), the requirement in TrmD is strictly at the transition state of the catalytic mechanism (Sakaguchi et al., 2014), which is an unexpected finding that links the synthesis of m1G37 to cellular concentrations of the metal ion.
In a genetic reporter system, where mgtA was fused to lacZ, the transcription of mgtA was monitored in Salmonella cells grown in media containing high or low Mg2+ (1.6 vs. 0.016 mM) (Gall et al., 2016). It was found that cells expressing the native trmD showed more than a sixfold activation of transcription upon switching from high to low Mg2+ media, whereas cells expressing a mutant trmD showed less than a twofold activation (Figure 2E). The mutant trmD harbors a mutation (S88L) near the AdoMet binding site (Masuda et al., 2013), which prevents the enzyme from binding to the methyl donor and from performing the Mg2+-dependent methyl transfer. The reported observation supports a model of codon-specific translation in the 5′-leader ORF. For cells expressing the native trmD, the level of Mg2+ modulates the level of TrmD-dependent m1G37-tRNA synthesis, which in turn modulates the speed of ribosomal translation of m1G37-dependent codons in the 5′-leader ORF. At high Mg2+, TrmD is active and the abundantly synthesized m1G37-tRNA facilitates ribosomal translation through the 5′-leader ORF, thus attenuating the transcription of mgtA. At low Mg2+, by contrast, TrmD is inactive, m1G37-tRNA synthesis is reduced, and ribosomal translation of m1G37-dependent codons is stalled, thus activating transcription through the mgtA gene and producing a robust response. This response to changes of Mg2+ concentrations is reduced in cells expressing the mutant trmD, consistent with the observed lower level of activation of mgtA transcription.
Perspective
Codon bias has the ability to regulate protein expression by controlling the efficiency or accuracy of protein synthesis. Codon bias can be executed by post-transcriptional modifications of the tRNA anticodons that determine the quality of pairing interaction with the cognate codons at local positions. While it is well documented that altering the codon usage synonymously can alter the expression levels of the manipulated genes (Kudla et al., 2009; Navon and Pilpel, 2011; Goodman et al., 2013; Zhou et al., 2013), much less is known how the alteration is correlated with post-transcriptional modifications of the tRNA anticodons in response to changes of the codon usage. While we present one example in the regulation of bacterial Mg2+ homeostasis by the m1G37 modification of tRNA (Gall et al., 2016), there are increasing studies demonstrating the ability of post-transcriptional modifications in the anticodon region to alter protein expression. Examples include cmo5U34 (Chionh et al., 2016), mcm5U34 (Begley et al., 2007), and Q34 (Tuorto et al., 2018) to the wobble position, and t6A37 on the 3′-side of the anticodon (Lin H. et al., 2018), all of which are induced in response to stress, indicating the ability to reprogram the proteome during cellular adaptation to stress. More broadly, even modifications that are outside of the anticodon but are important for tRNA stability can regulate protein expression by altering the abundance of a specific tRNA, thus impacting on the progress of disease. Examples include m1A58 in the T loop (Richter et al., 2018) and m7G46 in the V loop (Lin S. et al., 2018). The diversity of post-transcriptional modifications of tRNA is a key feature of the importance of tRNA biology. We are only at the tip of the iceberg at the forefront of exciting new discoveries.
Author Contributions
Y-MH prepared the manuscript. IM prepared the figures. HG contributed to discussion. All authors made a substantial intellectual contribution to the work and approved its presentation.
Funding
This work was supported by NIH grants GM108972 and GM114343 to Y-MH and a Japanese JSPS overseas postdoctoral fellowship to IM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agris, P. F., Eruysal, E. R., Narendran, A., Vare, V. Y. P., Vangaveti, S., and Ranganathan, S. V. (2018). Celebrating wobble decoding: half a century and still much is new. RNA Biol. 15, 537–553. doi: 10.1080/15476286.2017.1356562
Ahn, H. J., Kim, H. W., Yoon, H. J., Lee, B. I., Suh, S. W., and Yang, J. K. (2003). Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J. 22, 2593–2603. doi: 10.1093/emboj/cdg269
Anantharaman, V., Koonin, E. V., and Aravind, L. (2002). SPOUT. a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4, 71–75.
Armengod, M. E., Meseguer, S., Villarroya, M., Prado, S., Moukadiri, I., Ruiz-Partida, et al. (2014). Modification of the wobble uridine in bacterial and mitochondrial tRNAs reading NNA/NNG triplets of 2-codon boxes. RNA Biol. 11, 1495–1507. doi: 10.4161/15476286.2014.992269
Begley, U., Dyavaiah, M., Patil, A., Rooney, J. P., DiRenzo, D., Young, C. M., et al. (2007). Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28, 860–870. doi: 10.1016/j.molcel.2007.09.021
Bjork, G. R., Jacobsson, K., Nilsson, K., Johansson, M. J., Bystrom, A. S., and Persson, O. P. (2001). A primordial tRNA modification required for the evolution of life? EMBO J. 20, 231–239. doi: 10.1093/emboj/20.1.231
Bjork, G. R., and Nilsson, K. (2003). 1-methylguanosine-deficient tRNA of Salmonella enterica serovar Typhimurium affects thiamine metabolism. J. Bacteriol. 185, 750–759. doi: 10.1128/JB.185.3.750-759.2003
Bjork, G. R., Wikstrom, P. M., and Bystrom, A. S. (1989). Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244, 986–989. doi: 10.1126/science.2471265
Brule, H., Elliott, M., Redlak, M., Zehner, Z. E., and Holmes, W. M. (2004). Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43, 9243–9255. doi: 10.1021/bi049671q
Chen, G. F., and Inouye, M. (1990). Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 18, 1465–1473. doi: 10.1093/nar/18.6.1465
Chionh, Y. H., McBee, M., Babu, I. R., Hia, F., Lin, W., Zhao, W., et al. (2016). tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7:13302. doi: 10.1038/ncomms13302
Christian, T., Evilia, C., and Hou, Y. M. (2006). Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline. Biochemistry 45, 7463–7473. doi: 10.1021/bi0602314
Christian, T., Evilia, C., Williams, S., and Hou, Y. M. (2004). Distinct origins of tRNA(m1G37) methyltransferase. J. Mol. Biol. 339, 707–719. doi: 10.1016/j.jmb.2004.04.025
Christian, T., Gamper, H., and Hou, Y. M. (2013). Conservation of structure and mechanism by Trm5 enzymes. RNA 19, 1192–1199. doi: 10.1261/rna.039503.113
Christian, T., and Hou, Y. M. (2007). Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J. Mol. Biol. 373, 623–632. doi: 10.1016/j.jmb.2007.08.010
Christian, T., Lahoud, G., Liu, C., Hoffmann, K., Perona, J. J., and Hou, Y. M. (2010a). Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA 16, 2484–2492. doi: 10.1261/rna.2376210
Christian, T., Lahoud, G., Liu, C., and Hou, Y. M. (2010b). Control of catalytic cycle by a pair of analogous tRNA modification enzymes. J. Mol. Biol. 400, 204–217. doi: 10.1016/j.jmb.2010.05.003
Christian, T., Sakaguchi, R., Perlinska, A. P., Lahoud, G., Ito, T., Taylor, E. A., et al. (2016). Methyl transfer by substrate signaling from a knotted protein fold. Nat. Struct. Mol. Biol. 23, 941–948. doi: 10.1038/nsmb.3282
Cromie, M. J., Shi, Y., Latifi, T., and Groisman, E. A. (2006). An RNA sensor for intracellular Mg(2+). Cell 125, 71–84. doi: 10.1016/j.cell.2006.01.043
Elkins, P. A., Watts, J. M., Zalacain, M., van Thiel, A., Vitazka, P. R., Redlak, M., et al. (2003). Insights into catalysis by a knotted TrmD tRNA methyltransferase. J. Mol. Biol. 333, 931–949. doi: 10.1016/j.jmb.2003.09.011
Farabaugh, P. J. (2000). Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 64, 131–170. doi: 10.1016/S0079-6603(00)64004-7
Gall, A. R., Datsenko, K. A., Figueroa-Bossi, N., Bossi, L., Masuda, I., Hou, Y. M., et al. (2016). Mg2+ regulates transcription of mgtA in Salmonella Typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc. Natl. Acad. Sci. U.S.A. 113, 15096–15101. doi: 10.1073/pnas.1612268113
Gamper, H. B., Masuda, I., Frenkel-Morgenstern, M., and Hou, Y. M. (2015a). Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNNat. Nat. Commun. 6:7226. doi: 10.1038/ncomms8226
Gamper, H. B., Masuda, I., Frenkel-Morgenstern, M., and Hou, Y. M. (2015b). The UGG isoacceptor of tRNAPro is naturally prone to frameshifts. Int. J. Mol. Sci. 16, 14866–14883. doi: 10.3390/ijms160714866
Goodman, D. B., Church, G. M., and Kosuri, S. (2013). Causes and effects of N-terminal codon bias in bacterial genes. Science 342, 475–479. doi: 10.1126/science.1241934
Goto-Ito, S., Ito, T., Ishii, R., Muto, Y., Bessho, Y., and Yokoyama, S. (2008). Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins 72, 1274–1289. doi: 10.1002/prot.22019
Goto-Ito, S., Ito, T., Kuratani, M., Bessho, Y., and Yokoyama, S. (2009). Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat. Struct. Mol. Biol. 16, 1109–1115. doi: 10.1038/nsmb.1653
Groisman, E. A., Hollands, K., Kriner, M. A., Lee, E. J., Park, S. Y., and Pontes, M. H. (2013). Bacterial Mg2+ homeostasis, transport, and virulence. Annu. Rev. Genet. 47, 625–646. doi: 10.1146/annurev-genet-051313-051025
Grosjean, H., and Westhof, E. (2016). An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 44, 8020–8040. doi: 10.1093/nar/gkw608
Hohsaka, T., Ashizuka, Y., Taira, H., Murakami, H., and Sisido, M. (2001). Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry 40, 11060–11064. doi: 10.1021/bi0108204
Hollands, K., Sevostiyanova, A., and Groisman, E. A. (2014). Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc. Natl. Acad. Sci. U.S.A. 111, E1999–E2007. doi: 10.1073/pnas.1319193111
Hori, H. (2017). Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA. Biomolecules 7:E23. doi: 10.3390/biom7010023
Hou, Y. M., Matsubara, R., Takase, R., Masuda, I., and Sulkowska, J. I. (2017). TrmD A Methyl Transferase for tRNA Methylation With m(1)G37. Enzymes 41, 89–115. doi: 10.1016/bs.enz.2017.03.003
Hurwitz, J., Gold, M., and Anders, M. (1964). The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. iv. the properties of the soluble ribonucleic acid-methylating enzymes. J. Biol. Chem. 239, 3474–3482.
Ikemura, T. (1982). Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J. Mol. Biol. 158, 573–597. doi: 10.1016/0022-2836(82)90250-9
Ito, T., Masuda, I., Yoshida, K., Goto-Ito, S., Sekine, S., Suh, S. W., et al. (2015). Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmProc. Proc. Natl. Acad. Sci. U.S.A. 112, E4197–E4205. doi: 10.1073/pnas.1422981112
Jorgensen, F., and Kurland, C. G. (1990). Processivity errors of gene expression in Escherichia coli. J. Mol. Biol. 215, 511–521. doi: 10.1016/S0022-2836(05)80164-0
Komar, A. A. (2009). A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 34, 16–24. doi: 10.1016/j.tibs.2008.10.002
Kudla, G., Murray, A. W., Tollervey, D., and Plotkin, J. B. (2009). Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258. doi: 10.1126/science.1170160
Kumagai, I., Watanabe, K., and Oshima, T. (1982). A thermostable tRNA (guanosine-2′)-methyltransferase from Thermus thermophilus HB27 and the effect of ribose methylation on the conformational stability of tRNA. J. Biol. Chem. 257, 7388–7395.
Lahoud, G., Goto-Ito, S., Yoshida, K., Ito, T., Yokoyama, S., and Hou, Y. M. (2011). Differentiating analogous tRNA methyltransferases by fragments of the methyl donor. RNA 17, 1236–1246. doi: 10.1261/rna.2706011
Lee, C., Kramer, G., Graham, D. E., and Appling, D. R. (2007). Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J. Biol. Chem. 282, 27744–27753. doi: 10.1074/jbc.M704572200
Lin, H., Miyauchi, K., Harada, T., Okita, R., Takeshita, E., Komaki, H., et al. (2018). CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 9:1875. doi: 10.1038/s41467-018-04250-4
Lin, S., Liu, Q., Lelyveld, V. S., Choe, J., Szostak, J. W., and Gregory, R. I. (2018). Mettl1/Wdr4-Mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255e5. doi: 10.1016/j.molcel.2018.06.001
Maehigashi, T., Dunkle, J. A., Miles, S. J., and Dunham, C. M. (2014). Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc. Natl. Acad. Sci. U.S.A. 111, 12740–12745. doi: 10.1073/pnas.1409436111
Masuda, I., Sakaguchi, R., Liu, C., Gamper, H., and Hou, Y. M. (2013). The temperature sensitivity of a mutation in the essential tRNA modification enzyme tRNA methyltransferase D (TrmD). J. Biol. Chem. 288, 28987–28996. doi: 10.1074/jbc.M113.485797
Merino, E., and Yanofsky, C. (2005). Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 21, 260–264. doi: 10.1016/j.tig.2005.03.002
Nasvall, S. J., Chen, P., and Bjork, G. R. (2004). The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA 10, 1662–1673. doi: 10.1261/rna.7106404
Navon, S., and Pilpel, Y. (2011). The role of codon selection in regulation of translation efficiency deduced from synthetic libraries. Genome Biol. 12:R12. doi: 10.1186/gb-2011-12-2-r12
Nedialkova, D. D., and Leidel, S. A. (2015). Optimization of codon translation rates via trna modifications maintains proteome integrity. Cell 161, 1606–1618. doi: 10.1016/j.cell.2015.05.022
Papp-Wallace, K. M., and Maguire, M. E. (2008a). Magnesium transport and magnesium homeostasis. EcoSal Plus 3, doi: 10.1128/ecosalplus.5.4.4.2
Papp-Wallace, K. M., and Maguire, M. E. (2008b). Regulation of CorA Mg2+ channel function affects the virulence of Salmonella enterica serovar typhimurium. J. Bacteriol. 190, 6509–6516. doi: 10.1128/JB.00144-08
Park, S. Y., Cromie, M. J., Lee, E. J., and Groisman, E. A. (2010). A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell 142, 737–748. doi: 10.1016/j.cell.2010.07.046
Partridge, A. W., Therien, A. G., and Deber, C. M. (2004). Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins 54, 648–656. doi: 10.1002/prot.10611
Pavlov, M. Y., Watts, R. E., Tan, Z., Cornish, V. W., Ehrenberg, M., and Forster, A. C. (2009). Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 106, 50–54. doi: 10.1073/pnas.0809211106
Peacock, J. R., Walvoord, R. R., Chang, A. Y., Kozlowski, M. C., Gamper, H., and Hou, Y. M. (2014). Amino acid-dependent stability of the acyl linkage in aminoacyl-tRNRNA. RNA 20, 758–764. doi: 10.1261/rna.044123.113
Perret, V., Garcia, A., Grosjean, H., Ebel, J. P., Florentz, C., and Giege, R. (1990). Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature 344, 787–789. doi: 10.1038/344787a0
Plotkin, J. B., and Kudla, G. (2011). Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42. doi: 10.1038/nrg2899
Qian, Q., Li, J. N., Zhao, H., Hagervall, T. G., Farabaugh, P. J., and Bjork, G. R. (1998). A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell 1, 471–482. doi: 10.1016/S1097-2765(00)80048-9
Richter, U., Evans, M. E., Clark, W. C., Marttinen, P., Shoubridge, E. A., Suomalainen, A., et al. (2018). RNA modification landscape of the human mitochondrial tRNA(Lys) regulates protein synthesis. Nat. Commun. 9:3966. doi: 10.1038/s41467-018-06471-z
Sakaguchi, R., Giessing, A., Dai, Q., Lahoud, G., Liutkeviciute, Z., Klimasauskas, S., et al. (2012). Recognition of guanosine by dissimilar tRNA methyltransferases. RNA 18, 1687–1701. doi: 10.1261/rna.032029.111
Sakaguchi, R., Lahoud, G., Christian, T., Gamper, H., and Hou, Y. M. (2014). A divalent metal ion-dependent N(1)-methyl transfer to G37-tRNChem. Chem. Biol. 21, 1351–1360. doi: 10.1016/j.chembiol.2014.07.023
Sarin, L. P., and Leidel, S. A. (2014). Modify or die?–RNA modification defects in metazoans. RNA Biol. 11, 1555–1567. doi: 10.4161/15476286.2014.992279
Takai, K., and Yokoyama, S. (2003). Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 31, 6383–6391. doi: 10.1093/nar/gkg839
Taki, M., Hohsaka, T., Murakami, H., Taira, K., Sisido, M., Hohsaka, T., et al. (2002). Position-specific incorporation of a fluorophore-quencher pair into a single streptavidin through orthogonal four-base codon/anticodon pairs. J. Am. Chem. Soc. 124, 14586–14590. doi: 10.1021/ja017714+
Tuorto, F., Legrand, C., Cirzi, C., Federico, G., Liebers, R., Muller, M., et al. (2018). Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 37:e99777. doi: 10.15252/embj.201899777
Urbonavicius, J., Meskys, R., and Grosjean, H. (2014). Biosynthesis of wyosine derivatives in tRNA(Phe) of Archaea: role of a remarkable bifunctional tRNA(Phe):m1G/imG2 methyltransferase. RNA 20, 747–753. doi: 10.1261/rna.043315.113
Yohannan, S., Yang, D., Faham, S., Boulting, G., Whitelegge, J., and Bowie, J. U. (2004). Proline substitutions are not easily accommodated in a membrane protein. J. Mol. Biol. 341, 1–6. doi: 10.1016/j.jmb.2004.06.025
Yourno, J., and Tanemura, S. (1970). Restoration of in-phase translation by an unlinked suppressor of a frameshift mutation in Salmonella typhimurium. Nature 225, 422–426. doi: 10.1038/225422a0
Keywords: synonymous codons, methyl transferases TrmD and Trm5, codon-anticodon pairing interaction, ribosomal +1-frameshifts and stalling, protein synthesis
Citation: Hou Y-M, Masuda I and Gamper H (2019) Codon-Specific Translation by m1G37 Methylation of tRNA. Front. Genet. 9:713. doi: 10.3389/fgene.2018.00713
Received: 21 November 2018; Accepted: 20 December 2018;
Published: 10 January 2019.
Edited by:
Akio Kanai, Keio University, JapanCopyright © 2019 Hou, Masuda and Gamper. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya-Ming Hou, eWEtbWluZy5ob3VAamVmZmVyc29uLmVkdQ==
 Ya-Ming Hou
Ya-Ming Hou Isao Masuda
Isao Masuda Howard Gamper
Howard Gamper