- 1Department of Medicine, Imperial College London, London, United Kingdom
- 2Department of Systems Biology, Columbia University Medical Center, New York, NY, United States
- 3Department of Biomedical Informatics, Columbia University Medical Center, New York, NY, United States
- 4CIBER de Diabetes y Enfermedades Metabólicas Asociadas, Barcelona, Spain
- 5Department of Genetics and Development, Columbia University Medical Center, New York, NY, United States
- 6Barbara Davis Center, University of Colorado Anschutz Medical Campus, Aurora, CO, United States
The transcriptional programs of differentiated cells are tightly regulated by interactions between cell type-specific transcription factors and cis-regulatory elements. Long non-coding RNAs (lncRNAs) have emerged as additional regulators of gene transcription. Current evidence indicates that lncRNAs are a very heterogeneous group of molecules. For example, selected lncRNAs have been shown to regulate gene expression in cis or trans, although in most cases the precise underlying molecular mechanisms is unknown. Recent studies have uncovered a large number of lncRNAs that are selectively expressed in pancreatic islet cells, some of which were shown to regulate β cell transcriptional programs. A subset of such islet lncRNAs appears to control the expression of β cell-specific transcription factor (TF) genes by local cis-regulation. In this review, we discuss current knowledge of molecular mechanisms underlying cis-regulatory lncRNAs and discuss challenges involved in using genetic perturbations to define their function. We then discuss known examples of pancreatic islet lncRNAs that appear to exert cis-regulation of TF genes. We propose that cis-regulatory lncRNAs could represent a molecular target for modulation of diabetes-relevant genes.
Introduction
Cell-specific genome regulation in pancreatic islet cells is driven by combinations of transcription factors (TFs) that interact with cis-regulatory elements. Understanding islet-specific transcriptional programs is critically important for strategies to derive β cells for the treatment of type 1 diabetes, as well as for efforts to understand the pathophysiology of monogenic and type 2 diabetes.
In recent years, several lines of evidence have pointed to a potentially important role of long non-coding RNAs (lncRNAs) in the regulation of gene transcription (Rinn and Chang, 2012). LncRNAs are transcripts >200 nucleotides in length that do not encode for proteins. As a result of this broad definition, lncRNAs represent an extremely heterogeneous group of transcripts, a subset of which modulate gene expression through varied mechanisms, including regulation of epigenetic modifications, transcriptional initiation, splicing, mRNA stability and translation (Rinn et al., 2007; Zhao et al., 2008; Tripathi et al., 2010; Carrieri et al., 2012; Kretz et al., 2013; Xing et al., 2014; Hosono et al., 2017; Kopp and Mendell, 2018). This realization poses a need to understand how specific lncRNA subtypes influence regulatory programs.
Several studies have described thousands of lncRNAs expressed in human and mouse pancreatic islets (Ku et al., 2012; Morán et al., 2012; Bramswig et al., 2013; Akerman et al., 2017; Motterle et al., 2017), and have been recently thoroughly reviewed (Pullen and Rutter, 2014; Motterle et al., 2016; Mirza et al., 2017; Singer and Sussel, 2018). The current review focuses on a discrete subset of lncRNAs that have been shown to modulate the transcription of nearby genes, many of which encode for transcription factors. Understanding the cis-regulatory function of certain lncRNAs is likely to provide new insights into genome regulation, and could reveal targets for gene-specific manipulation. However, the analysis of cis-regulatory lncRNAs poses significant experimental challenges. We provide an overview of recent progress in the analysis of cis-regulatory lncRNAs, and discuss obstacles to understand their function. We also discuss specific examples in the recent literature of cis-regulatory pancreatic islet lncRNAs.
Cis-Regulatory lncRNAs
Emerging evidence points to the existence of lncRNAs that regulate nearby genes in cis. In contrast to mRNAs that need to be translated in the cytoplasm to produce functional proteins, lncRNAs can exist in their functional conformation immediately after their transcription, and can thus theoretically exert their function in any cellular compartment, including their site of transcription. Several studies have thus revealed lncRNAs that exert cis-regulation of nearby regulatory elements such as promoters and enhancers (Figure 1A; Wang et al., 2011; Latos et al., 2012; Postepska-Igielska et al., 2015; Yin et al., 2015; Anderson et al., 2016; Kotzin et al., 2016; Luo et al., 2016).
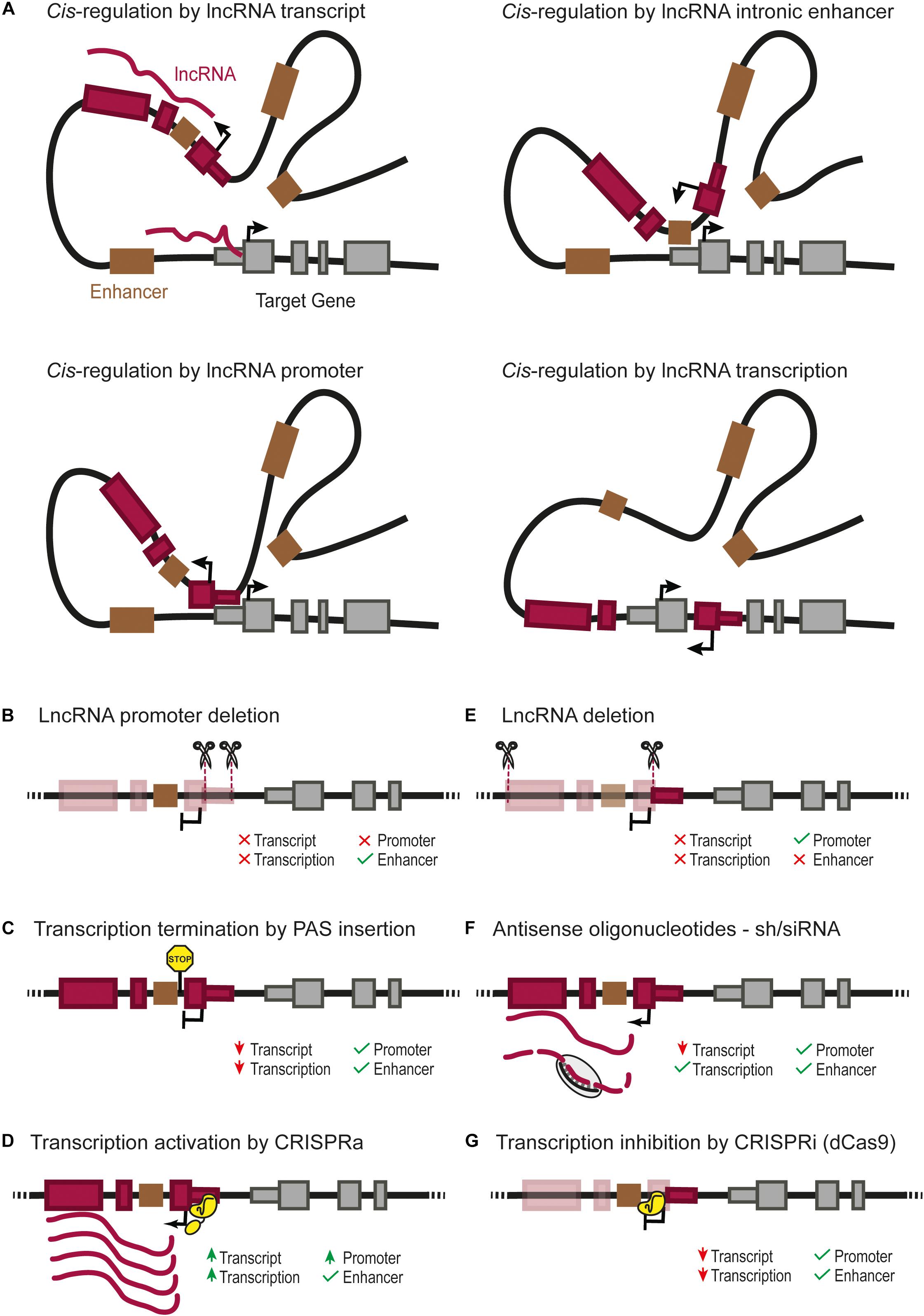
FIGURE 1. (A) Four possible cis-regulatory mechanisms mediated by lncRNA genes. Cis-regulation of nearby genes can be achieved directly by the lncRNA transcript, by cis-regulatory elements overlapping with the lncRNA gene such as an enhancer (Groff et al., 2016), or the lncRNA promoter itself (Engreitz et al., 2016; Paralkar et al., 2016), or by the transcription of the lncRNA (Latos et al., 2012). Different types of genetic perturbation can provide insights into the underlying mechanisms, such as: (B) deletion of the lncRNA promoter, which can affect the transcription of the regulated gene due to the lack of lncRNA transcript, transcription or absence of direct cis-regulation by the lncRNA promoter (Engreitz et al., 2016; Paralkar et al., 2016); (C) insertion of a poly-adenylation signal (PAS) downstream of the transcription start site, which prevents the transcription of the lncRNA through downstream DNA sequences (Sleutels et al., 2002; Ohhata et al., 2007; Yin et al., 2015; Anderson et al., 2016; Engreitz et al., 2016; Paralkar et al., 2016); (D) over-expression of a lncRNA from its endogenous locus can be achieved by CRISPR-activation (CRISPRa) (Gilbert et al., 2014; Konermann et al., 2015; Joung et al., 2017); it should be noted that the CRISPR-activation of the lncRNA promoter can also increase a direct cis-regulatory activity of the lncRNA promoter; (E) deletion of the full lncRNA transcript, which leaves the lncRNA promoter as a potential cis-regulatory element; note that deletion of several exons of the lncRNA will also remove any intronic enhancer (Groff et al., 2016); (F) lncRNA knock-down using short hairpin RNA (siRNA/shRNA) or antisense oligonucleotides, testing the functionality of the lncRNA transcript itself; (G) RNA Polymerase II “roadblock” by CRISPR-dCas9 downstream of the transcriptional start site of the lncRNA (Gilbert et al., 2013; Qi et al., 2013); this selectively blocks the transcription of the lncRNA, helping to discriminate a cis-regulatory function from the transcription/transcript or from genomic cis-regulatory elements.
The precise number of cis-regulatory lncRNAs is unknown. Cell fractionation experiments have revealed that a large portion of lncRNAs is enriched in the nucleus and tightly bound to chromatin (Engreitz et al., 2016; Werner et al., 2017). The expression of chromatin-enriched RNAs has been shown to exhibit higher correlation with their neighboring genes than other lncRNAs, suggesting that many of them could be cis-regulators (Werner et al., 2017). A distinct subset of nuclear lncRNAs are transcribed from active enhancers, and are known as enhancer RNAs (eRNAs). These are often unstable and non-polyadenylated transcripts, and their expression often correlates with the mRNA of the enhancer’s target gene (Kim et al., 2010). In some cases, eRNA transcripts have been shown to regulate the activity of target genes (Bose et al., 2017). However, whether most eRNA transcripts exert true cis-regulatory effects is currently a matter of debate. Furthermore, the distinction between enhancers and promoters is often not unequivocal (Carelli et al., 2018). Thus, although it is useful to classify a subset of lncRNAs as eRNAs, it is possible that lncRNAs that originate from either promoters or enhancers can regulate the expression of nearby genes in cis. The current challenge, therefore, is to define which pairs of lncRNAs and adjacent coding genes show correlated transcription due to true cis-regulatory relationships.
Genetic Approaches to Identify cis-Regulatory lncRNAs
Genetic tools have been used to discriminate cis vs. trans regulation by lncRNAs. Some studies, for example, have used compound heterozygote mice where one chromosome contains an inactive lncRNA allele while the other has a null allele of the coding gene that is regulated by the lncRNA (Dimitrova et al., 2014; Anderson et al., 2016; Kotzin et al., 2016; Herriges et al., 2017). For example, Anderson et al. (2016) showed that homozygous insertion of a premature transcription termination signal to inactivate Upperhand (Uph), a lncRNA expressed upstream of the cardiac-specific TF Hand2, results in ventricular hypoplasia and embryonic lethality, a phenotype similar to the Hand2 gene knockout. While heterozygous mice for the lncRNA or TF are both viable, compound heterozygous mice (Uph+/-;Hand2+/-) display the same phenotype as the Uph-/- mice with a nearly complete absence of Hand2 expression. This result was interpreted as an indication that the inactivation of Uph lncRNA transcription caused decreased expression of Hand2 in cis, thereby suggesting that Uph is a cis-regulator of the cardiac TF Hand2.
The cis-regulatory function of a lncRNA can also be assessed by creating a heterozygous mutation of the lncRNA and then use allelic markers to distinguish the expression of the nearby target gene in the mutated and wild type chromosomes. This can be implemented in hybrid mouse strains that have single nucleotide polymorphisms (SNPs) within the target gene (Engreitz et al., 2016). Heterozygous inactivation of the cis-regulatory lncRNA gene should thus result in allelic imbalance of the target gene, with altered expression of transcripts derived from the same chromosome as the mutations.
The abovementioned approaches can provide genetic evidence that is consistent with cis-regulatory effects of a lncRNA. However, they do not always provide conclusive evidence that the lncRNA transcript is exerting the effect. The distinction between different candidate mechanisms involved can be addressed with complementary genetic approaches that are summarized in Figures 1B–G.
Experimental Challenges to Understand Different Classes of cis-Regulatory lncRNAs
LncRNAs can overlap cis-regulatory elements such as enhancers (Figure 1A). In such cases, the effect of deleting the lncRNA can result from the deletion of one or more enhancers, even if the lncRNA itself has no function, or if there is a cis effect of both the lncRNA and the enhancer (Huarte et al., 2010; Dimitrova et al., 2014; Groff et al., 2016). In certain contexts, a lncRNA and cis-regulatory elements have opposite roles. This is true for Haunt, a lncRNA expressed more than 40 kb upstream of the HOXA cluster (Yin et al., 2015). Deletion and premature transcription termination of Haunt result in opposite effects. Complementary genetic models used in this study showed that the Haunt transcript directly represses HOXA enhancers after exposure to retinoic acid. Thus, cis-regulatory lncRNAs can buffer the expression of their target genes by direct negative regulation of their underlying cis-regulatory elements. The complexity of such studies reveals the peril of using a single perturbation model to study the function of cis-regulatory lncRNAs.
In some cases, the promoter of a lncRNA gene can act as a functional cis-regulatory element for another gene (Figure 1A; Cho et al., 2018). The comparison of promoter deletions vs. premature transcription termination of the lncRNA by insertion of a polyadenylation signal in Lockd or Bendr lncRNA genes showed that the promoter of a lncRNA is capable of exerting cis-regulation of a nearby gene while the transcript itself is not visibly functional (Figures 1B,C; Engreitz et al., 2016; Paralkar et al., 2016). Nevertheless, even in these cases, it is difficult to entirely rule out the function of lncRNA transcripts, since it is still possible that a short exon transcribed before the polyadenylation signal would be the functional part of the lncRNA transcript.
Gain-of-function studies have also provided useful insights into lncRNA functions (Shechner et al., 2015; Joung et al., 2017; Bester et al., 2018). Ectopic expression of lncRNAs, however, also comes with caveats. Cis-regulatory lncRNAs sometimes need to act near their transcription sites, in which case they need to be activated from their endogenous genes, rather than from ectopic artificial transgenes (Dimitrova et al., 2014). CRISPR activation (CRISPRa) could be a powerful tool to accomplish this (Figure 1D; Gilbert et al., 2014; Konermann et al., 2015). However, it remains possible that any downstream effect of activating a lncRNA can be attributed either to the lncRNA transcript itself or to a direct cis-regulatory effect of activating the lncRNA promoter. To this end, it is important to include proper controls such as combining CRISPRa of the lncRNA promoter with the silencing of the lncRNA transcript (Figures 1D–G; Joung et al., 2017). Alternatively, CRISPR/dCas9 technology can be used to artificially tether a lncRNA to a specific locus and test whether the lncRNA function is recapitulated by the exogenous transcript (Shechner et al., 2015). This technique has the power to complement other methods to demonstrate a function intrinsic to the lncRNA transcript, independent from any genomic element.
Several lines of evidence suggest that the cis-regulatory effect of some lncRNAs could result from the act of transcription or transcriptional splicing rather than from the transcript molecule or the promoter activity (Figure 1A; Latos et al., 2012; Engreitz et al., 2016). These mechanisms need to be tested through approaches that specifically disrupt transcription or splicing (Figures 1C,G).
In summary, existing evidence indicates that the cis-regulatory function of lncRNA genes can be attributed to several processes, including an effect of the transcript, its transcription, splicing, or the activity of its promoter. All of these need to be carefully distinguished from overlapping DNA cis-regulatory functions that are unrelated to the lncRNA gene. This warrants a need to investigate cis-regulatory lncRNAs with complementary models that consider these possible genetic mechanisms.
Molecular Mechanisms Underlying cis-Regulatory lncRNAs
Cis-regulation mediated by lncRNA genes can be achieved through various molecular mechanisms, illustrated by several well characterized cases. XIST is an example of a repressive cis-regulatory lncRNA in which the molecular mechanisms have been thoroughly characterized (da Rocha and Heard, 2017). XIST is responsible for random inactivation of the X chromosome. It is known to recruit Polycomb Repressive Complex 2 (PRC2) near the Xist locus on chromosome X, triggering chromosomal condensation (Zhao et al., 2008). Cis-regulatory lncRNAs have also been shown to interact with the members of PRC2 such as the lncRNA Morrbid (Figure 2A), located over 100 kb from Bcl2l11 which binds and represses the promoter of Bcl2l11 in cis (Klattenhoff et al., 2013; Kotzin et al., 2016). Other cis-regulatory lncRNAs appear to promote the formation of heterochromatin at imprinted loci, through binding of the histone methyltransferase G9A (Nagano et al., 2008).
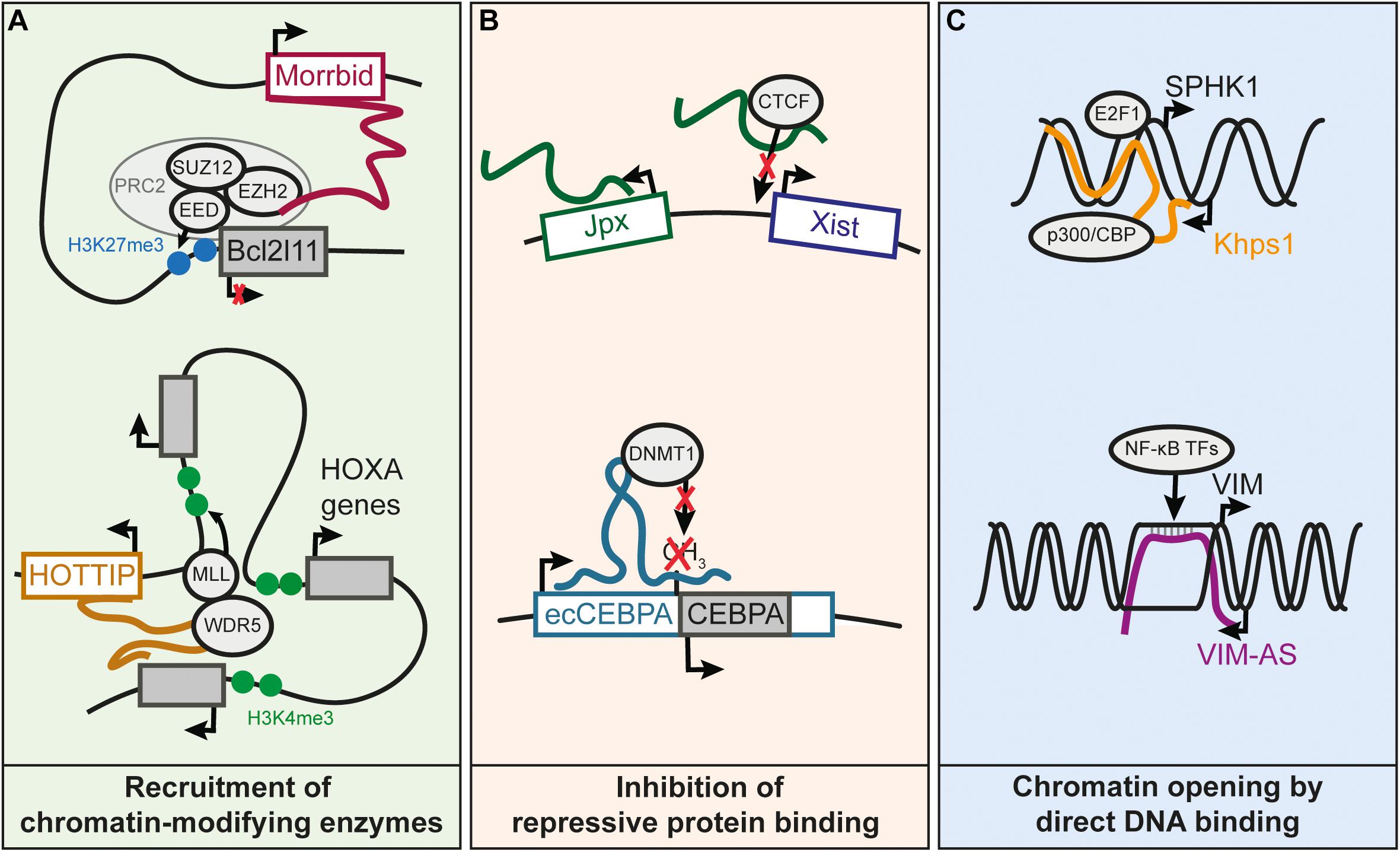
FIGURE 2. Known molecular mechanisms underlying cis-regulation by lncRNAs. (A) Recruitment of chromatin-modifying enzymes by cis-regulatory lncRNAs. Morrbid represses the Bcl2l11 gene by recruiting the PRC2 complex to the Bcl2l11 promoter (Kotzin et al., 2016). HOTTIP recruits the WDR5 subunit of the MLL complex, resulting in the deposition of the H3K4me3 mark and subsequent activation of the HOXA distal genes (Wang et al., 2011). (B) LncRNA prevents binding of an inhibitory protein to a promoter, which results in gene activation. Jpx lncRNA competes with DNA for the binding of CTCF on Xist promoter, and this enables activation of Xist expression (Sun et al., 2013). Transcription of ecCEBPA, a sense-overlapping lncRNA of the CEBPA gene, prevents the silencing of CEBPA by inhibiting DNMT1-dependent DNA methylation of CEBPA promoter (Di Ruscio et al., 2013). (C) Increased chromatin accessibility by DNA binding of lncRNA. The Khps1 lncRNA forms a triple helix with the promoter of SPHK1, which leads to the successive recruitment of p300/CBP and H3K27 acetylation of SPHK1 promoter, facilitating the binding of TFs on the SPHK1 promoter (Postepska-Igielska et al., 2015). VIM-AS lncRNA sustains the expression of the VIM gene by forming an R-loop at the VIM promoter (Boque-Sastre et al., 2015). The R-loop helps keeping the chromatin open, allowing TFs of the NF-kB pathway to bind to the VIM promoter and activate VIM expression.
Several lncRNAs have been shown to recruit transcriptional activating complexes to adjacent genes (Wang et al., 2011; Yang et al., 2014; Bose et al., 2017). The histone lysine-methyltransferase WDR5, a component the MLL complex responsible for H3K4me2/3 deposition, contains an RNA-binding domain that can bind lncRNAs (Yang et al., 2014). For instance, it was reported that activation of the 5′ genes of the HOXA cluster requires WDR5 recruitment by the HOTTIP lncRNA, also encoded in this locus (Wang et al., 2011; Figure 2A). Other lncRNAs have been shown to promote interactions between enhancers and their targets promoters, thereby sustaining transcriptional activity (Luo et al., 2016; Akerman et al., 2017). So far, little is known on the mechanism explaining how some lncRNAs can stabilize or enhance the interaction of a promoter with its enhancers. Recruitment of Mediator subunits is one of the suggested mechanisms (Luo et al., 2016), although further research is needed to understand how lncRNAs could modulate the Mediator complex or any other proteins involved in chromatin structure.
Transcriptional activity can also be maintained by preventing DNA methylation through interference with DNA methyltransferases (Di Ruscio et al., 2013), or by competing with DNA for binding of repressor proteins (Figure 2B; Sun et al., 2013).
Some lncRNAs can function through direct interaction with DNA to create R-loops (i.e., DNA-RNA hybrids through Watson-Crick base pairing) or DNA-RNA triplex structures (Figure 2C; Boque-Sastre et al., 2015; Postepska-Igielska et al., 2015; Li et al., 2016). R-loop formation could help maintain open chromatin at promoters (Boque-Sastre et al., 2015). In contrast to R-loops, the RNA-DNA triplex occurs when RNA interacts with the major groove of the DNA double helix (Li et al., 2016). RNA-DNA triplex interactions have been proposed for lncRNAs regulating their targets in trans or cis. For example, the KHPS1 antisense lncRNA forms a triple helix with the SPHK1 promoter to facilitate the binding of E2F2 that in turn activates both SPHK1 and its antisense lncRNA (Mondal et al., 2015; Postepska-Igielska et al., 2015). In summary, this non-exhaustive list of mechanisms illustrates a wide range of processes through which lncRNAs can be involved in gene cis-regulation.
Co-Regulation of Islet β Cell Programs by TFs and lncRNAs
Human pancreatic islets transcribe well over 1000 lncRNAs, many of which are highly specific to islet cells (Morán et al., 2012). To explore the functionality of these lncRNAs, Akerman et al. (2017) performed a loss-of-function screen of 12 β cell-enriched lncRNAs in a human β cell line named EndoC-βH1 (Ravassard et al., 2011). Transcriptional profiling of cells after lncRNA inhibition revealed several lncRNAs with loss of function phenotypes consistent with a gene regulatory function. Comparison of transcriptional profiles after knock-down of β cell-specific lncRNAs and TFs (PDX1, HNF1A, MAFB, GLIS3, and NKX2-2) showed that genes regulated by β cell TFs are often also regulated by β cell lncRNAs. Moreover, the genes that were regulated by several β cell lncRNAs were in turn associated with clusters of enhancers that are highly bound by cell type-specific TFs (Akerman et al., 2017). Taken together, these results indicated that some cell-specific lncRNAs and TFs regulate common networks. To independently validate this finding, Akerman et al. (2017) performed weighted gene co-expression network analysis (WGCNA) using RNA-seq data from a panel of human islets. This approach uses gene expression correlation across samples to group genes into modules that form part of common transcriptional programs. This revealed specific expression modules that were co-enriched in cell-specific lncRNAs, islet TFs and genes regulated by enhancer clusters (Akerman et al., 2017). Interestingly, knockdown of lncRNAs from these modules led to downregulation of other genes in the same modules, which suggests that the lncRNAs did indeed exert a regulatory function in these cell-specific programs. Taken together, these independent lines of evidence revealed tissue-specific lncRNAs that modulate cell type-specific gene programs.
The mechanisms by which these islet lncRNAs regulate gene expression are still largely unknown. Potential scenarios include the possibility that lncRNAs act in cis or trans. Among the seven lncRNAs that regulate enhancer cluster-associated genes, HI-LNC30, HI-LNC12, HI-LNC78 (also known as TUNAR; (Lin et al., 2014) and HI-LNC80 (also known as OLMALINC; (Mills et al., 2015) had no apparent effect on adjacent genes, arguing against a cis-regulatory mechanism. However, three lncRNAs regulating β cell enriched genes were shown to also control the expression of their neighboring gene. Akerman et al. (2017) showed that knock-down of the PLUTO lncRNA caused decreased expression of the adjacent gene PDX1. Similarly, knockdown of HI-LNC15, the human homolog of βlinc1 (Arnes et al., 2016), caused decreased expression of the adjacent gene NKX2-2. In both cases, knockdown of the lncRNA and the adjacent TF gene resulted in highly correlated changes in differential expression. Thus, it is highly probable that both PLUTO and HI-LNC15 modulate in cis the expression of PDX1 and NKX2-2, respectively, and elicit their regulatory function through this mechanism.
PLUTO Regulates 3D Chromatin Structure at the PDX1 Locus
PDX1 is a key transcriptional regulator of pancreas development and β cell function. It is required for the initial stages of pancreas formation (Jonsson et al., 1994; Stoffers et al., 1997), but is also necessary for the correct function of mature β cells (Ahlgren et al., 1998). PDX1 expression is regulated by a cluster of upstream enhancers (Gerrish et al., 2000; Pasquali et al., 2014). LncRNA PLUTO, which stands for PDX1 Locus Upstream Transcript, is a multiexonic gene that originates approximately 3 kb upstream from PDX1, and is transcribed in the opposite strand through a ∼100 kb region that contains a cluster of active islet enhancers (Akerman et al., 2017). The positive regulatory effect of PLUTO on PDX1 was shown in human β cell lines as well as in human islets, and it can be elicited through short hairpin RNA (shRNA) knock-down or transcription interference of PLUTO with CRISPR/dCas9. Moreover, the genes regulated by PLUTO are essentially the same as the genes regulated by PDX1, suggesting that PLUTO regulates β cell gene expression indirectly through the regulation of PDX1 (Akerman et al., 2017).
Conformation capture experiments indicated that PLUTO promotes interactions of nearby clustered enhancers and the PDX1 promoter. This indicates that PLUTO has a structural function in the regulation of the PDX1 locus (Akerman et al., 2017). Very recently, Amaral et al. (2018) identified a sub-class of lncRNAs that are transcribed from evolutionarily conserved promoters, frequently found in the vicinity of developmental TFs and with an expression correlating with the nearby TF gene (Amaral et al., 2018). Many of these lncRNAs were found to be located near topologically associating domain (TAD) boundaries and overlap CTCF binding sites, pointing to a possible role in genomic 3D organization (Amaral et al., 2018). Thus, PLUTO may be an example of a broad class of lncRNAs that exhibits an analogous function of regulating 3D chromatin structure of genes important for lineage-specific functions.
Interestingly, both PLUTO and PDX1 are down-regulated in islets from donors with type 2 diabetes or impaired glucose tolerance (Akerman et al., 2017). PLUTO lncRNA thus emerges as a cis-regulatory lncRNA that is a candidate player in β cell differentiation, with a potential involvement in diabetes.
Genetic Analysis of βlinc1 Points to a Cell-Specific Differentiation Function
NKX2-2 is another essential transcriptional regulator of islet cell differentiation in the embryo, and β cell function in the adult (Papizan et al., 2011; Gutiérrez et al., 2017). Mice lacking Nkx2-2 fail to form islet α and β cell lineages, and β cell deletion of Nkx2-2 results in the loss of β cell identity. Consistent with the mouse phenotypes, individuals carrying homozygous mutations in NKX2-2 develop neonatal diabetes (Flanagan et al., 2014). The lncRNA βlinc1 (β cell lincRNA1) and its human ortholog HI-LNC15 are each located in gene deserts between Nkx2-2 and Pax1 on chromosome 2 (mice) and chromosome 20 (human), respectively (Arnes et al., 2016). However, unlike Nkx2-2, which is expressed throughout the pancreas and in several different endocrine lineages, βlinc1 expression is highly β cell-specific. Furthermore, deletion of the βlinc1 transcript in mice and siRNA-mediated knockdown of βlinc1 in MIN6 cell lines, resulted in the down-regulation of Nkx2-2 and a large subset of Nkx2-2 β cell targets. ShRNA-mediated knockdown of HI-LNC15 transcript in the human insulin-producing EndoC-βH1 cell line also caused altered expression of genes dysregulated in βlinc1-/- mice, suggesting conservation of βlinc1 transcript function in humans. Interestingly, a disproportionate number of βlinc1-regulated genes are located on chromosome 2, suggesting that similar to PLUTO, βlinc1 may function to regulate gene transcription in cis. Although the extremely low abundance of βlinc1 transcript has impaired further biochemical analysis of its molecular activity, βlinc1 transcript was present in the chromatin fraction and regulated over 70 non-contiguous beta cell-associated genes that are dispersed over a 55 Mb region of chromosome 2, suggesting that the βlinc1 transcript regulates the expression of a set of functionally related β cell genes through a mechanism that remains to be elucidated.
Conclusion
LncRNAs are emerging as important regulators of cell-specific gene expression. In pancreatic islets (and in other cell types), genes encoding for critical developmental proteins such as tissue-specific TFs are often associated with antisense divergent or adjacent intergenic lncRNA (Morán et al., 2012; Luo et al., 2016). This suggests that lncRNAs may regulate many key TFs. Cis-regulation of tissue-specific TFs by lncRNAs is likely to be a widespread mechanism that enables fine-tuning of TF expression. Furthermore, since lncRNAs tend to be much more cell type-restricted than the associated TFs, lncRNAs may confer cell-specific activities to more broadly expressed TFs. For most cell-specific lncRNAs, however, their possible function remains unknown. Functional interrogation of lncRNAs remains extremely challenging, partly due to their low abundance in cells (Wang et al., 2011; Cabili et al., 2015), and because of our limited understanding on how they should be experimentally approached. Nevertheless, future investigations that provide a systematic functional characterization of β cell-specific lncRNAs promise to identify these previously unsuspected regulators of β cell function, and their potential role in monogenic and polygenic forms of diabetes.
Author Contributions
BF-C and AB wrote the first draft of the manuscript. LA, JF, and LS wrote sections of the manuscript.
Funding
Funding for work described here was provided by NIH R01 DK082590, NIH R01 DK DK11140 to LS, Wellcome Trust (WT101033), the Medical Research Council (MR/L02036X/1) to JF, ChroMe Marie Sklodowska Curie Training Network to BF-C and JF (No 675610), Ministerio de Economía y Competitividad (BFU2014-54284-R) and NIHR Imperial Biomedical Research Centre to JF and AB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahlgren, U., Jonsson, J., Jonsson, L., Simu, K., and Edlund, H. (1998). β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes Dev. 12, 1763–1768. doi: 10.1101/gad.12.12.1763
Akerman,İ, Tu, Z., Beucher, A., Rolando, D. M. Y., Sauty-Colace, C., Benazra, M., et al. (2017). Human pancreatic β cell lncRNAs control cell-specific regulatory networks. Cell Metab. 25, 400–411. doi: 10.1016/j.cmet.2016.11.016
Amaral, P. P., Leonardi, T., Han, N., Viré, E., Gascoigne, D. K., Arias-Carrasco, R., et al. (2018). Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 19:32. doi: 10.1186/s13059-018-1405-5
Anderson, K. M., Anderson, D. M., McAnally, J. R., Shelton, J. M., Bassel-Duby, R., and Olson, E. N. (2016). Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436. doi: 10.1038/nature20128
Arnes, L., Akerman,İ, Balderes, D. A., Ferrer, J., and Sussel, L. (2016). βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes Dev. 30, 502–507. doi: 10.1101/gad.273821.115
Bester, A. C., Lee, J. D., Chavez, A., Lee, Y.-R., Nachmani, D., Vora, S., et al. (2018). An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 173, 649.e20–664.e20. doi: 10.1016/j.cell.2018.03.052
Boque-Sastre, R., Soler, M., Oliveira-Mateos, C., Portela, A., Moutinho, C., Sayols, S., et al. (2015). Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 112, 5785–5790. doi: 10.1073/pnas.1421197112
Bose, D. A., Donahue, G., Reinberg, D., Shiekhattar, R., Bonasio, R., and Berger, S. L. (2017). RNA binding to CBP stimulates histone acetylation and transcription. Cell 168, 135.e22–149.e22. doi: 10.1016/j.cell.2016.12.020
Bramswig, N. C., Everett, L. J., Schug, J., Dorrell, C., Liu, C., Luo, Y., et al. (2013). Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J. Clin. Invest. 123, 1275–1284. doi: 10.1172/JCI66514
Cabili, M. N., Dunagin, M. C., McClanahan, P. D., Biaesch, A., Padovan-Merhar, O., Regev, A., et al. (2015). Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16:20. doi: 10.1186/s13059-015-0586-4
Carelli, F. N., Liechti, A., Halbert, J., Warnefors, M., and Kaessmann, H. (2018). Repurposing of promoters and enhancers during mammalian evolution. Nat. Commun. 9:4066. doi: 10.1038/s41467-018-06544-z
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. doi: 10.1038/nature11508
Cho, S. W., Xu, J., Sun, R., Mumbach, M. R., Carter, A. C., Chen, Y. G., et al. (2018). Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398.e22–1412.e22. doi: 10.1016/j.cell.2018.03.068
da Rocha, S. T., and Heard, E. (2017). Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 24, 197–204. doi: 10.1038/nsmb.3370
Di Ruscio, A., Ebralidze, A. K., Benoukraf, T., Amabile, G., Goff, L. A., Terragni, J., et al. (2013). DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503, 371–376. doi: 10.1038/nature12598
Dimitrova, N., Zamudio, J. R., Jong, R. M., Soukup, D., Resnick, R., Sarma, K., et al. (2014). LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell. 54, 777–790. doi: 10.1016/j.molcel.2014.04.025
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. doi: 10.1038/nature20149
Flanagan, S. E., De Franco, E., Lango Allen, H., Zerah, M., Abdul-Rasoul, M. M., Edge, J. A., et al. (2014). Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab. 19, 146–154. doi: 10.1016/j.cmet.2013.11.021
Gerrish, K., Gannon, M., Shih, D., Henderson, E., Stoffel, M., Wright, C. V., et al. (2000). Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J. Biol. Chem. 275, 3485–3492. doi: 10.1074/jbc.275.5.3485
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661. doi: 10.1016/j.cell.2014.09.029
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi: 10.1016/j.cell.2013.06.044
Groff, A. F., Sanchez-Gomez, D. B., Soruco, M. M. L., Gerhardinger, C., Barutcu, A. R., Li, E., et al. (2016). In Vivo characterization of Linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 16, 2178–2186. doi: 10.1016/j.celrep.2016.07.050
Gutiérrez, G. D., Bender, A. S., Cirulli, V., Mastracci, T. L., Kelly, S. M., Tsirigos, A., et al. (2017). Pancreatic β cell identity requires continual repression of non-β cell programs. J. Clin. Invest. 127, 244–259. doi: 10.1172/JCI88017
Herriges, M. J., Tischfield, D. J., Cui, Z., Morley, M. P., Han, Y., Babu, A., et al. (2017). The NANCI–Nkx2.1 gene duplex buffers Nkx2.1 expression to maintain lung development and homeostasis. Genes Dev. 31, 889–903. doi: 10.1101/gad.298018.117
Hosono, Y., Niknafs, Y. S., Prensner, J. R., Iyer, M. K., Dhanasekaran, S. M., Mehra, R., et al. (2017). Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 171, 1559.e20–1572.e20. doi: 10.1016/j.cell.2017.11.040
Huarte, M., Guttman, M., Feldser, D., Garber, M., Koziol, M. J., Kenzelmann-Broz, D., et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419. doi: 10.1016/j.cell.2010.06.040
Jonsson, J., Carlsson, L., Edlund, T., and Edlund, H. (1994). Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371, 606–609. doi: 10.1038/371606a0
Joung, J., Engreitz, J. M., Konermann, S., Abudayyeh, O. O., Verdine, V. K., Aguet, F., et al. (2017). Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548, 343–346. doi: 10.1038/nature23451
Kim, T.-K., Hemberg, M., Gray, J. M., Costa, A. M., Bear, D. M., Wu, J., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. doi: 10.1038/nature09033
Klattenhoff, C. A., Scheuermann, J. C., Surface, L. E., Bradley, R. K., Fields, P. A., Steinhauser, M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. doi: 10.1016/j.cell.2013.01.003
Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. doi: 10.1038/nature14136
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Kotzin, J. J., Spencer, S. P., McCright, S. J., Kumar, D. B. U., Collet, M. A., Mowel, W. K., et al. (2016). The long non-coding RNA morrbid regulates bim and short-lived myeloid cell lifespan. Nature 537, 239–243. doi: 10.1038/nature19346
Kretz, M., Siprashvili, Z., Chu, C., Webster, D. E., Zehnder, A., Qu, K., et al. (2013). Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235. doi: 10.1038/nature11661
Ku, G. M., Kim, H., Vaughn, I. W., Hangauer, M. J., Myung Oh, C., German, M. S., et al. (2012). Research resource: RNA-Seq reveals unique features of the pancreatic β-cell transcriptome. Mol. Endocrinol. 26, 1783–1792. doi: 10.1210/me.2012-1176
Latos, P. A., Pauler, F. M., Koerner, M. V., Şenergin, H. B., Hudson, Q. J., Stocsits, R. R., et al. (2012). Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472. doi: 10.1126/science.1228110
Li, Y., Syed, J., and Sugiyama, H. (2016). RNA-DNA triplex formation by long noncoding RNAs. Cell Chem. Biol. 23, 1325–1333. doi: 10.1016/j.chembiol.2016.09.011
Lin, N., Chang, K.-Y., Li, Z., Gates, K., Rana, Z. A., Dang, J., et al. (2014). An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 53, 1005–1019. doi: 10.1016/j.molcel.2014.01.021
Luo, S., Lu, J. Y., Liu, L., Yin, Y., Chen, C., Han, X., et al. (2016). Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18, 637–652. doi: 10.1016/j.stem.2016.01.024
Mills, J. D., Kavanagh, T., Kim, W. S., Chen, B. J., Waters, P. D., Halliday, G. M., et al. (2015). High expression of long intervening non-coding RNA OLMALINC in the human cortical white matter is associated with regulation of oligodendrocyte maturation. Mol. Brain 8:2. doi: 10.1186/s13041-014-0091-9
Mirza, A. H., Kaur, S., and Pociot, F. (2017). Long non-coding RNAs as novel players in β cell function and type 1 diabetes. Hum. Genomics 11:17. doi: 10.1186/s40246-017-0113-7
Mondal, T., Subhash, S., Vaid, R., Enroth, S., Uday, S., Reinius, B., et al. (2015). MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 6:7743. doi: 10.1038/ncomms8743
Morán, I., Akerman,İ, van de Bunt, M., Xie, R., Benazra, M., Nammo, T., et al. (2012). Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 16, 435–448. doi: 10.1016/j.cmet.2012.08.010
Motterle, A., Gattesco, S., Peyot, M.-L., Esguerra, J. L. S., Gomez-Ruiz, A., Laybutt, D. R., et al. (2017). Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Mol. Metab. 6, 1407–1418. doi: 10.1016/j.molmet.2017.08.005
Motterle, A., Sanchez-Parra, C., and Regazzi, R. (2016). Role of long non-coding RNAs in the determination of β-cell identity. Diabetes Obes. Metab. 18, 41–50. doi: 10.1111/dom.12714
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Feil, R., et al. (2008). The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720. doi: 10.1126/science.1163802
Ohhata, T., Hoki, Y., Sasaki, H., and Sado, T. (2007). Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135, 227–235. doi: 10.1242/dev.008490
Papizan, J. B., Singer, R. A., Tschen, S.-I., Dhawan, S., Friel, J. M., Hipkens, S. B., et al. (2011). Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 25, 2291–2305. doi: 10.1101/gad.173039.111
Paralkar, V. R., Taborda, C. C., Huang, P., Yao, Y., Kossenkov, A. V., Prasad, R., et al. (2016). Unlinking an lncRNA from its associated cis element. Mol. Cell 62, 104–110. doi: 10.1016/j.molcel.2016.02.029
Pasquali, L., Gaulton, K. J., Rodríguez-Seguí, S. A., Mularoni, L., Miguel-Escalada, I., Akerman, Ý, et al. (2014). Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46, 136–143. doi: 10.1038/ng.2870
Postepska-Igielska, A., Giwojna, A., Gasri-Plotnitsky, L., Schmitt, N., Dold, A., Ginsberg, D., et al. (2015). LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 60, 626–636. doi: 10.1016/j.molcel.2015.10.001
Pullen, T. J., and Rutter, G. A. (2014). Roles of lncRNAs in pancreatic beta cell identity and diabetes susceptibility. Front. Genet. 5:193. doi: 10.3389/fgene.2014.00193
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Ravassard, P., Hazhouz, Y., Pechberty, S., Bricout-Neveu, E., Armanet, M., Czernichow, P., et al. (2011). A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 121, 3589–3597. doi: 10.1172/JCI58447
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi: 10.1146/annurev-biochem-051410-092902
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Shechner, D. M., Hacisuleyman, E., Younger, S. T., and Rinn, J. L. (2015). Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 12, 664–670. doi: 10.1038/nmeth.3433
Singer, R. A., and Sussel, L. (2018). Islet long noncoding RNAs: a playbook for discovery and characterization. Diabetes Metab. Res. Rev. 67, 1461–1470. doi: 10.2337/dbi18-0001
Sleutels, F., Zwart, R., and Barlow, D. P. (2002). The non-coding air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. doi: 10.1038/415810a
Stoffers, D. A., Zinkin, N. T., Stanojevic, V., Clarke, W. L., and Habener, J. F. (1997). Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15, 106–110. doi: 10.1038/ng0197-106
Sun, S., del Rosario, B. C., Szanto, A., Ogawa, Y., Jeon, Y., and Lee, J. T. (2013). Jpx RNA activates xist by evicting CTCF. Cell 153, 1537–1551. doi: 10.1016/j.cell.2013.05.028
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Werner, M. S., Sullivan, M. A., Shah, R. N., Nadadur, R. D., Grzybowski, A. T., Galat, V., et al. (2017). Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 24, 596–603. doi: 10.1038/nsmb.3424
Xing, Z., Lin, A., Li, C., Liang, K., Wang, S., Liu, Y., et al. (2014). lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125. doi: 10.1016/j.cell.2014.10.013
Yang, Y. W., Flynn, R. A., Chen, Y., Qu, K., Wan, B., Wang, K. C., et al. (2014). Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 3:e02046. doi: 10.7554/eLife.02046
Yin, Y., Yan, P., Lu, J., Song, G., Zhu, Y., Li, Z., et al. (2015). Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Stem Cell 16, 504–516. doi: 10.1016/j.stem.2015.03.007
Keywords: long non-coding RNAs, transcription factors, pancreatic islets, β cells, cis-regulation
Citation: Font-Cunill B, Arnes L, Ferrer J, Sussel L and Beucher A (2018) Long Non-coding RNAs as Local Regulators of Pancreatic Islet Transcription Factor Genes. Front. Genet. 9:524. doi: 10.3389/fgene.2018.00524
Received: 25 August 2018; Accepted: 18 October 2018;
Published: 06 November 2018.
Edited by:
Jane Mellor, University of Oxford, United KingdomReviewed by:
Kazuhiko Nakabayashi, National Center for Child Health and Development (NCCHD), JapanNejat Dalay, Istanbul University, Turkey
Copyright © 2018 Font-Cunill, Arnes, Ferrer, Sussel and Beucher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anthony Beucher, YWJldWNoZXJAaW1wZXJpYWwuYWMudWs=
 Berta Font-Cunill
Berta Font-Cunill Luis Arnes
Luis Arnes Jorge Ferrer1,4
Jorge Ferrer1,4 Lori Sussel
Lori Sussel Anthony Beucher
Anthony Beucher