- 1Department of Anesthesia, McGill University, Montreal, QC, Canada
- 2School of Behavioral and Brain Sciences, The University of Texas at Dallas, Richardson, TX, United States
- 3Center for Advanced Pain Studies, The University of Texas at Dallas, Richardson, TX, United States
- 4Alan Edwards Centre for Research on Pain, McGill University, Montreal, QC, Canada
Translational control of gene expression has emerged as a key mechanism in regulating different forms of long-lasting neuronal plasticity. Maladaptive plastic reorganization of peripheral and spinal nociceptive circuits underlies many chronic pain states and relies on new gene expression. Accordingly, downregulation of mRNA translation in primary afferents and spinal dorsal horn neurons inhibits tissue injury-induced sensitization of nociceptive pathways, supporting a central role for translation dysregulation in the development of persistent pain. Translation is primarily regulated at the initiation stage via the coordinated activity of translation initiation factors. The mRNA cap-binding protein, eukaryotic translation initiation factor 4E (eIF4E), is involved in the recruitment of the ribosome to the mRNA cap structure, playing a central role in the regulation of translation initiation. eIF4E integrates inputs from the mTOR and ERK signaling pathways, both of which are activated in numerous painful conditions to regulate the translation of a subset of mRNAs. Many of these mRNAs are involved in the control of cell growth, proliferation, and neuroplasticity. However, the full repertoire of eIF4E-dependent mRNAs in the nervous system and their translation regulatory mechanisms remain largely unknown. In this review, we summarize the current evidence for the role of eIF4E-dependent translational control in the sensitization of pain circuits and present pharmacological approaches to target these mechanisms. Understanding eIF4E-dependent translational control mechanisms and their roles in aberrant plasticity of nociceptive circuits might reveal novel therapeutic targets to treat persistent pain states.
Introduction
Chronic pain is a debilitating condition affecting more than 20 percent of the population worldwide (Steglitz et al., 2012; de Souza et al., 2017). Chronic pain is most commonly triggered by tissue inflammation or nerve injury, which can be caused by metabolic diseases (diabetes), autoimmune diseases, viral infection (herpes zoster), cancer, chemotherapy drugs (e.g., platinums, taxanes, epothilones, and vinca alkaloids), and nerve entrapment or blunt trauma. Chronic pain, however, can also appear without any recognizable trigger such as in fibromyalgia, migraine, irritable bowel syndrome, and interstitial cystitis.
In most cases, the pain is a result of increased sensitivity of peripheral or central nociceptive circuits to stimulation, causing painful sensation in response to a normally innocuous stimulus. The increase in sensitivity, also referred as sensitization, is mediated by a combination of mechanisms taking place at several levels along the pain pathway including primary sensory neurons, spinal cord, and higher brain areas (Todd, 2010; Yekkirala et al., 2017).
Long-lasting increases in the sensitivity and responsiveness of pain circuits is ultimately accompanied by changes in gene expression, which support biochemical and structural alterations in neuronal and non-neuronal cells involved in pain processing. Gene expression is a multi-step process that is tightly regulated at different levels. Regulation of the rate by which mRNA is translated into protein is called translational control (Sonenberg and Hinnebusch, 2009; Robichaud et al., 2018). Translational control has a strong impact on the abundance of proteins in the cell, and its dysregulation contributes to many pathologies in the nervous system including developmental abnormalities, metabolic dysregulation, autism spectrum disorder (ASD), and epilepsy (Buffington et al., 2014; Tahmasebi et al., 2018). Tissue injury, metabolic diseases, and certain drugs (e.g., anticancer and opioids) cause an upregulation of mRNA translation in pain-processing tissues such as dorsal root ganglion (DRG) and dorsal horn of the spinal cord (Melemedjian and Khoutorsky, 2015; Khoutorsky and Price, 2018). Inhibition of translational control signaling in these tissues reduces the sensitization of nociceptive circuits and alleviates pain, demonstrating a central role of translational upregulation in the development of persistent pain (Price et al., 2007; Jimenez-Diaz et al., 2008; Asante et al., 2009; Geranton et al., 2009; Price and Geranton, 2009; Melemedjian et al., 2010; Xu et al., 2011; Bogen et al., 2012; Ferrari et al., 2013; Obara and Hunt, 2014). The rate of mRNA translation is controlled via several mechanisms (Costa-Mattioli et al., 2009; Robichaud et al., 2018). The recruitment of the ribosome to the mRNA is a central step in translation initiation and the major site for regulation. A key mechanism to regulate this process is controlling the activity of the eukaryotic translation initiation factor 4E (eIF4E), which binds a mRNA “cap” structure (a 7-methylguanosine linked to the first nucleotide at the 5′ end of all nuclear transcribed eukaryotic mRNAs) and initiates ribosome recruitment (Altmann et al., 1985; Sonenberg and Hinnebusch, 2009). In this review, we focus on the regulation of eIF4E-dependent mRNA translation initiation in nociceptive plasticity, highlighting a central role of this mechanism in the development of chronic pain.
Translational Control Mechanisms
The process of translation can be divided into three phases: initiation, elongation, and termination. Most of the regulation of translation occurs at the initiation step (Sonenberg and Hinnebusch, 2009; Merrick and Pavitt, 2018). Initiation is regulated by a large number of translation initiation factors, which mediate the recruitment of the ribosome to the mRNA, followed by scanning of the 5′ untranslated region (5′ UTR) of the mRNA for the presence of an AUG start codon. A critical step in this process is the binding of eIF4E to the mRNA cap. Following binding to the cap, eIF4E binds a mRNA helicase, eIF4A, and a large scaffolding protein, eIF4G, to form a tri-subunit complex named eIF4F (Figure 1). eIF4F facilitates the recruitment of the 43S preinitiation complex (PIC) to the mRNA. The PIC is composed of a small 40S ribosomal subunit, translation factors eIF1, eIF1A, and eIF3, and a ternary complex (eIF2: GTP bound to initiator, Met-tRNAiMet). Recruitment of the PIC is followed by scanning of the mRNA 5′ UTR and joining of a large ribosomal subunit (60S), upon encountering a start codon, to form an 80S ribosome that is competent to proceed to the elongation phase of translation. Importantly, the helicase activity of eIF4F (mediated by eIF4A) is required for unwinding the mRNAs 5′ UTR secondary structure to allow the scanning process and translation to proceed (Parsyan et al., 2011).
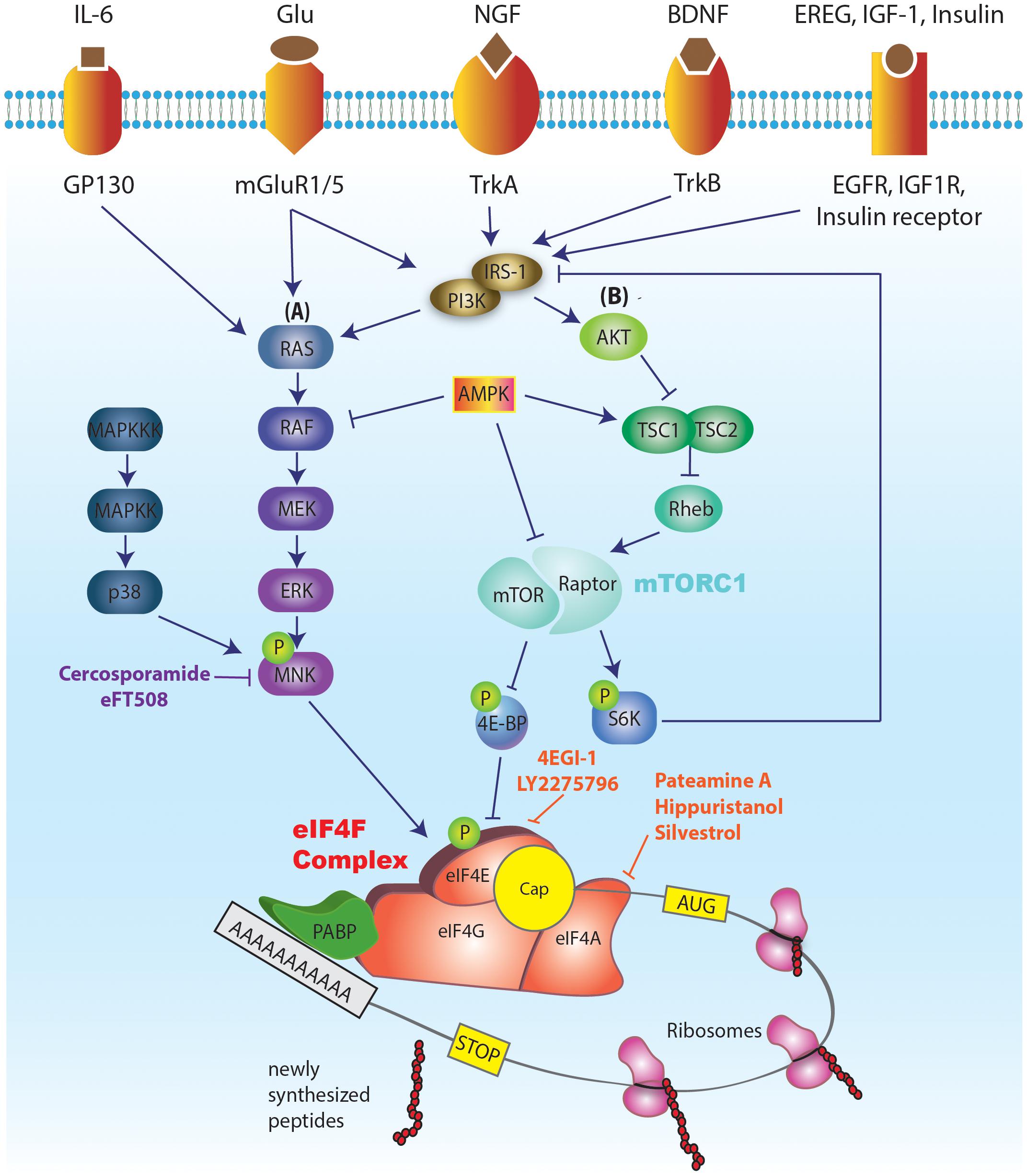
FIGURE 1. Schematic illustration of the major signaling pathways regulating eIF4E activity and translation initiation. The cap binding ability of eIF4E makes it a central regulator of translation. A critical step in the translation initiation process is the binding of eIF4E to the mRNA cap. eIF4E mediates the formation of the eIF4F complex on the mRNA cap structure (a 7mGp bound to the first nucleotide). eIF4F complex, in addition to eIF4E, consists of eIF4G (scaffolding protein) and eIF4A (helicase). Successful formation of eIF4F complex on the mRNA cap further promotes the recruitment of the pre-initiation complex (PIC), followed by 5′ UTR scanning to reach the start codon AUG and joining of 60S ribosomal subunit. This event marks the completion of translation initiation. eIF4E is a downstream effector of both mTORC1 (via 4E-BP-dependent repression) and ERK (via eIF4E phosphorylation by MNK 1/2). The activities of mTORC1 and ERK signaling pathways are in turn modulated by a multitude of external [tyrosine receptor kinase A (trkA) and trkB, receptors from the insulin receptor family (IR, IGF1R, EGFR), and metabotropic glutamate and NMDA receptors] and internal cues [status of cellular energy (via AMPK), oxygen levels (via activation of AMPK and REDD1; Regulated in DNA damage and development 1), and DNA damage (via the induction of p53 target genes)]. Various inhibitors of cap dependent translation initiation have been identified. 4EGI-1 inhibits eIF4E’s interaction with eIF4G, thus inhibiting the formation of eIF4F complex. Cercosporamide blocks MNK phosphorylation, which in turn prevents phosphorylation of eIF4E. Inhibitors of eIF4A have also been identified which function by either blocking its helicase activity (hippuristanol) or by preventing its participation in the eIF4F complex (pateamine A, and silvestrol).
Other major mechanisms involved in the regulation of translation initiation include regulation of ternary complex availability [via phosphorylation of the alpha subunit of the eukaryotic initiation factor 2 (eIF2α)] (Trinh and Klann, 2013); regulation of the length of mRNA poly(A) tail which promotes translation and protects mRNA from degradation (Gray et al., 2000; Kahvejian et al., 2001; Derry et al., 2006); and finally translation initiation via a cap-independent mechanism (mediated by internal ribosome entry site, IRES) (Pelletier and Sonenberg, 1988; Macejak and Sarnow, 1991; Leppek et al., 2018). Since the expression levels of eIF4E are the lowest among all translation initiation factors, the formation of the eIF4F complex and correspondingly, translation initiation are the rate-limiting steps for translation under most circumstances.
eIF4E is a Central Regulator of Cap-Dependent Translation
Eukaryotic translation initiation factor 4E activity is tightly regulated via two mechanisms. Translational repressor 4E-binding protein (4E-BP) binds eIF4E and prevents its association with eIF4G, and thus precludes the formation of the eIF4F complex (Gingras et al., 1999; Peter et al., 2015). In mammals, there are three 4E-BP isoforms – 4E-BP1, 4E-BP2, and 4E-BP3, which have similar functions but exhibit differences in tissue distribution. The binding of 4E-BP to eIF4E depends on the 4E-BP phosphorylation state. Upon phosphorylation by the mechanistic target of rapamycin complex 1 (mTORC1), the affinity of 4E-BP to eIF4E is reduced, leading to its dissociation from eIF4E and allowing the formation of eIF4F complex at the mRNA cap. This promotes the recruitment of 43S PIC to the mRNA and stimulation of translation (Figure 1).
Eukaryotic translation initiation factor 4E activity is required for translation initiation of all capped mRNAs. Complete loss of eIF4E, as in eIF4E−/− mice is not compatible with life and leads to embryos death before embryonic day 6.5 (Truitt et al., 2015). Partial loss of eIF4E does not have a strong impact on general translation, mostly because it induces a compensatory degradation of hypophosphorylated 4E-BP1 (Yanagiya et al., 2012). Even though all nuclear transcribed eukaryotic mRNAs have a cap, not all cellular mRNAs are equally sensitive to eIF4E activity. The translation of “eIF4E-sensitive mRNAs” is preferentially stimulated by increased eIF4E activity. For example, housekeeping mRNAs such as GAPDH and β-actin are less sensitive to eIF4E as compared to mRNAs involved in cell growth, proliferation, and immune responses [e.g., c-MYC, cyclins, BCL-2, MCL1, osteopontin, survivin, vascular endothelial growth factor (VEGF), fibroblast growth factors (FGF), and matrix metalloproteinase 9 (MMP-9)] (Rousseau et al., 1996; Sonenberg and Gingras, 1998; Bhat et al., 2015; Chu and Pelletier, 2018). The mRNA features rendering eIF4E-sensitivity have been typically associated with 5′ UTRs enriched with high-complexity secondary structures (Pelletier and Sonenberg, 1985; Sonenberg and Gingras, 1998). It has been demonstrated that a long 5′ UTR favors the formation of stable secondary structures, and that the proximity of these structures to the cap obstructs eIF4F complex formation. On the other hand, hairpin structures with a greater free energy, located further away from the cap, restrict 5′ UTR scanning (the progression of the PIC toward the start codon) (Kozak, 1989; Pickering and Willis, 2005). However, translation of a subset of mRNAs without long 5′ UTR can still be sensitive to eIF4E, indicating that other 5′ UTR signatures may also render this sensitivity (Leppek et al., 2018). Potential mechanisms include the presence of 5′ terminal oligopyrimidine tracts (5′TOPs) (Thoreen et al., 2012) and cis-regulatory elements (Wolfe et al., 2014; Truitt et al., 2015; Hinnebusch et al., 2016; Truitt and Ruggero, 2016; Leppek et al., 2018) at the 5′ UTR. For example, a Cytosine-rich 15-nucleotide motif, termed Cytosine Enriched Regulator of Translation (CERT), was shown to be responsible for conferring eIF4E sensitivity under oncogenic transformation and oxidative stress (Truitt et al., 2015).
Although most studies have attributed the elevated translation of mRNAs with highly structured 5′ UTRs to the cap-binding ability of eIF4E and it being the limiting component of the eIF4F complex, other studies did not find that the cap-binding ability completely explained eIF4E function and explored further mechanisms of eIF4E-mediated translation regulation. This led to the identification of an additional function of eIF4E – stimulation of eIF4A helicase activity, which is independent of its cap-binding ability (Feoktistova et al., 2013). Feoktistova et al. (2013) showed that the eIF4E binding site on eIF4G has an autoinhibitory function. Binding of eIF4E to eIF4G counteracts this autoinhibition, and in turn enables eIF4G to stimulate eIF4A activity (rate of duplex unwinding). They show that this function of eIF4E is independent of its cap-binding activity, suggesting that eIF4E can stimulate translation by two distinct mechanisms (Feoktistova et al., 2013).
In addition to regulation by mTORC1/4E-BP, eIF4E activity is also controlled via phosphorylation of its sole phosphorylation site, Ser 209, by mitogen activated protein kinase [MAPK]-interacting protein kinases (MNKs) 1 and 2, downstream of the extracellular-signal-regulated kinase (ERK) and the p38 MAPK signaling cascades (Figure 1; Pyronnet et al., 1999; Waskiewicz et al., 1999). The phosphorylation of eIF4E is associated with altered translation of a subset of mRNAs, although the mechanisms underlying the effect of this phosphorylation event on translational efficiency and transcript-specificity remain elusive.
Since eIF4E is a downstream effector of both mTORC1 (via 4E-BP-dependent repression) and ERK (via eIF4E phosphorylation), its activity can be modulated by a multitude of external and internal cues that activate these central cellular signaling pathways. Numerous membrane receptors activate mTORC1 and ERK signaling in neurons including tyrosine receptor kinase A (trkA) and trkB, receptors from the insulin receptor family (IR, IGF1R, EGFR), and metabotropic glutamate and NMDA receptors. In addition to the extracellular cues, these pathways integrate intracellular signals conveying information on the status of cellular energy (via AMPK), oxygen levels [via activation of AMPK and REDD1 (Regulated in DNA damage and development 1)], and DNA damage (via the induction of p53 target genes) (Saxton and Sabatini, 2017; Figure 1).
eIF4E in Regulation of Peripheral Nociceptive Plasticity
Tissue injury induces profound changes in the phenotype of sensory neurons, increasing their excitability and changing the connectivity within peripheral tissues and spinal cord. These alterations are driven by pro-inflammatory molecules released from injured tissues, such as neurotrophin nerve growth factor (NGF) and cytokine interleukin 6 (IL-6), as well as by neuronal activity evoked by direct injury to the nerve. ERK and mTORC1, two central intracellular pathways, are stimulated by tissue inflammation and nerve injury, diabetes, cancer, and drug-induced neuropathies (Melemedjian and Khoutorsky, 2015; Khoutorsky and Price, 2018). In addition to the phosphorylation-mediated activation of mTOR, downstream of PI3K/AKT pathway, a recent study showed that nerve injury stimulates local axonal mTOR mRNA translation (Terenzio et al., 2018). Translation profiling of DRG tissue from mice subjected to nerve injury showed that ERK is a key regulatory hub controlling both transcriptional and translation gene expression networks (Uttam et al., 2018).
Inhibition of ERK and mTORC1 signaling alleviates the development of pain hypersensitivity in a variety of pain models (Ji et al., 2009; Chen et al., 2018; Khoutorsky and Price, 2018). Since ERK and mTORC1 pathways converge on eIF4E to control the rate of cap-dependent translation, it was suggested that eIF4E might play a central role in the sensitization of pain circuits via regulating the translation of specific mRNAs. The physiological significance of eIF4E phosphorylation was studied using mice lacking eIF4E phosphorylation (knock-in mutation of serine209 to alanine, eIF4ES209A). These mice display greatly reduced mechanical and thermal hypersensitivity in response to intraplantar administration of IL-6, NGF, and carrageenan, as well as diminished hyperalgesic priming (Moy et al., 2017). Moreover, the increase in excitability of eIF4ES209A primary sensory neurons in response to IL-6 and NGF was reduced as compared to wild-type (WT) controls. These findings were recapitulated in MNK1/2 knockout mice, which also lack eIF4E phosphorylation. In the nerve injury model of neuropathic pain, spared nerve injury (SNI), the development of mechanical and cold hypersensitivity was reduced in both eIF4ES209A and MNK1/2 knockout mice. Notably, local intraplantar inhibition of MNK with cercosporamide reduced mechanical hypersensitivity in response to NGF and alleviated hyperalgesic priming (Moy et al., 2017). These findings support the notion that the stimulation of eIF4E phosphorylation is imperative for the phenotypic changes of sensory neurons, promoting the hyperalgesic state and contributing to the development of chronic pain, and that this likely occurs independently of effects on inflammation (Moy et al., 2018b). Experiments with local administration of cercosporamide also indicate that pro-inflammatory mediators- or tissue injury-induced phosphorylation of eIF4E mediates sensitization of sensory neurons via local mRNA translation.
The advances in translational profiling techniques have provided important insights into the potential mechanisms by which eIF4E phosphorylation regulates neuronal functions. In the brain, eIF4E phosphorylation controls the translation of mRNAs involved in inflammatory responses such as IκBα, a repressor of the transcription factor NF-κB that regulates the expression of the cytokine tumor necrosis factor (TNFα) (Aguilar-Valles et al., 2018). Genome-wide translational profiling of the brain from eIF4ES209A mice revealed that eIF4E phosphorylation controls translation of mRNAs involved in inflammation (IL-2 and TNFα), organization of the extracellular matrix (Prg2, Mmp9, Adamts16, Acan), and the serotonin pathway (Slc6a4) (Amorim et al., 2018).
In the DRG, phosphorylation of eIF4E stimulates translation of brain derived neurotropic factor (Bdnf) mRNA. eIF4ES209A mice show reduced protein levels of BDNF under baseline conditions and fail to translate Bdnf mRNA to protein in response to pro-inflammatory cytokines despite an increase in Bdnf mRNA levels (Moy et al., 2018a). BDNF is a key molecule mediating pain plasticity (Obata and Noguchi, 2006) and identification of MNK/eIF4E signaling as a central regulator of Bdnf translation has important therapeutic implications (Moy et al., 2018a). Cell-type specific translational profiling of nociceptors [using translating ribosome affinity purification (TRAP) approach] (Heiman et al., 2014) in a mouse model of chemotherapy-induced neuropathic pain revealed that MNK-eIF4E signaling controls translation of RagA mRNA, a key regulator of mTORC1 (Megat et al., 2018). This finding suggests crosstalk between ERK/MNK/eIF4E and mTORC1 signaling pathways in promoting pain hypersensitivity in chemotherapy-induced neuropathies.
In addition to phosphorylation, eIF4E in primary sensory neurons is also regulated via mTORC1/4E-BP. IL-6 and NGF activate mTORC1, which promotes 4E-BP1 phosphorylation, increased eIF4F complex formation and nascent protein synthesis in cultured sensory neurons (Melemedjian et al., 2010). Intraplantar administration of IL-6 or NGF induced mechanical allodynia, which is blocked by subcutaneous administration of the mTORC1 inhibitor rapamycin, as well as by 4EGI-1, an inhibitor of eIF4F complex formation that disrupts eIF4E and eIF4G interaction. Intraplantar 4EGI-1 also blocked the establishment of the sensitization state in a hyperalgesic priming model in response to IL-6 and NGF injection (Asiedu et al., 2011).
These findings support a model that local activation of mTORC1 stimulates eIF4F complex formation, promoting pain hypersensitivity via axonal mRNA translation. 4E-BP1 is a major isoform involved in regulation of nociception, whereas in the brain 4E-BP2 is the dominant isoform. 4E-BP1 is highly expressed in nociceptors and mice lacking 4E-BP1, but not 4E-BP2, exhibit enhanced mechanical hypersensitivity. Notably, eif4ebp1 knockout mice show no alterations in thermal sensitivity, suggesting a mechanical-specific effect of eIF4E activation via 4E-BP-dependent mechanisms (Khoutorsky et al., 2015).
A second major downstream effector of mTORC1, p70S6 ribosomal kinase (S6K1 and S6K2) may not play as significant a role in the regulation of nociceptive sensitization. Mice lacking S6K1/2 do exhibit increased mechanical pain sensitivity, but normal thermal thresholds, and an inhibitor of S6K1/2 recapitulates this phenotype (Melemedjian et al., 2013). This finding seems paradoxical; however, further analysis revealed that loss of S6K1/2 function engages a feedback loop that stimulates enhanced ERK phosphorylation, driving mechanical sensitization (Melemedjian et al., 2013). Therefore, it is tempting to speculate that most of the pain inhibitory effects of mTORC1 inhibition are mediated via the suppression of 4E-BP1/eIF4E-dependent protein synthesis. The role of other translation-independent outputs of mTORC1, such as regulation of autophagy, lipogenesis, and mitochondrial function, remain unknown.
eIF4E in Regulation of Spinal Plasticity
The spinal cord integrates peripheral somatosensory inputs to generate, after processing, an output that is conveyed to the brain where the perception of pain ultimately arises. Peripheral injury, disease, and certain drugs can cause an increase in the gain of spinal nociceptive circuits, resulting in disproportional amplification of somatosensory inputs, and therefore increased pain. These maladaptive plastic changes in the spinal cord, frequently referred to as central sensitization, significantly contribute to the development of pathological pain states. Central sensitization leads to a lowered threshold for the induction of pain (allodynia), an increase in the responsiveness to noxious stimuli (hyperalgesia), and an enlargement of the receptive field, resulting in pain sensation from non-injured areas (secondary hyperalgesia).
Long-lasting spinal plasticity critically relies on new protein synthesis to allow alterations in the cellular proteome, and consequently, sensitization of the pro-nociceptive circuits. Numerous studies have demonstrated the activation of ERK and mTORC1 signaling in the spinal cord following peripheral tissue injury, cancer, and opioid treatment (Geranton et al., 2009; Ji et al., 2009; Norsted Gregory et al., 2010; Xu et al., 2011, 2014; Shih et al., 2012; Jiang et al., 2013; Liang et al., 2013; Zhang et al., 2013). Intrathecal delivery of pharmacological inhibitors targeting these pathways efficiently alleviates pathological pain without affecting the baseline mechanical and thermal sensitivity (Ji et al., 2009; Melemedjian and Khoutorsky, 2015; Martin et al., 2017). There is evidence that the beneficial effect of mTORC1 inhibition on pain in the spinal cord is largely mediated via mTORC1/4E-BP1-dependent regulation of eIF4E activity. Pain hypersensitivity produced by intrathecal injection of epiregulin (EREG), an endogenous agonist of the epidermal growth factor receptor (EGFR) upstream of mTORC1, is blocked by intrathecal injection of 4EGI-1 (Martin et al., 2017). Moreover, specific deletion of 4E-BP1 in the dorsal horn of the spinal cord causes mechanical hypersensitivity (Khoutorsky et al., 2015). Mice lacking 4E-BP1 show increased excitatory and inhibitory synaptic transmission in lamina II neurons as well as enhanced potentiation of spinal excitatory field potentials following sciatic nerve stimulation. Taken together, these results indicate that enhanced eIF4F complex formation in the spinal cord promotes spinal plasticity and contributes to the development of central sensitization.
Therapeutic Approaches to Target eIF4E-Dependent Mechanisms to Alleviate Pain
Several lines of evidence suggest that targeting eIF4E is a potentially promising therapeutic strategy to inhibit aberrant pain plasticity. First, due to low expression levels, eIF4E’s activity is a rate-limiting factor for translation initiation and a central node of regulation. eIF4E integrates signals from two major signaling pathways, ERK and mTORC1, both of which have important functions in the development of pain. Second, eIF4E does not strongly affect general translation, but mainly regulates the translation of a subset of mRNAs involved in cell growth, proliferation, immune responses, and neuronal plasticity. Mice with partial reduction of eIF4E protein levels, such as eIF4E heterozygous mice (Truitt et al., 2015) or mice expressing short hairpin RNA against eIF4E (Lin et al., 2012) show no developmental abnormalities or changes in survival rate or body weight. Third, whereas acute inhibition of mTORC1 is effective in alleviating pain, long-term mTORC1 inhibition leads to the hyperactivation of ERK via a mTORC1-S6K1-IRS1 negative feedback loop (Veilleux et al., 2010; Melemedjian et al., 2013). Since ERK is a well-known sensitizer of neurons involved in pain transmission, both in the periphery and the spinal cord, chronic mTORC1 inhibition leads to mechanical hypersensitivity and pain. Thus, long-term treatment with compounds targeting mTORC1 is unlikely to be clinically applicable. Conversely, chronic inhibition of eIF4E does not activate these compensatory mechanisms. Mice lacking eIF4E phosphorylation do not exhibit alterations in pain sensation at baseline, but show reduced nociceptive plasticity in response to pro-inflammatory and nerve injury stimuli (Moy et al., 2017). Finally, compelling preclinical studies have demonstrated beneficial effects of pharmacologically targeting eIF4E in alleviating persistent pain using 4EGI-1, an inhibitor of eIF4 complex formation or cercosporamide, an inhibitor of MNK. Efforts to develop and test new translation inhibitors are fuelled by their potential use for treatment of cancer (Stumpf and Ruggero, 2011), malaria (Baragana et al., 2015), and bacterial infection (Bhat et al., 2015). Here, we overview the existing and newly developed pharmacological approaches to target eIF4E-dependent translation.
MNK Inhibitors
CGP57380 and cercosporamide are two small molecule inhibitors targeting MNK1 and MNK2 (Bhat et al., 2015). Cercosporamide, extracted from the fungus Cercosporidium henningsii, is an antifungal agent and a phytotoxin. It has antiproliferative and proapoptotic activities in cancer cells in preclinical animal models of lung and colon carcinomas (Konicek et al., 2011). It readily crosses the blood-brain barrier (BBB) and efficiently reduces p-eIF4E in the brain after peripheral administration (Gkogkas et al., 2013). However, both CGP57380 and cercosporamide have been shown to exhibit off-target effects (Bain et al., 2007; Bhat et al., 2015). More specific MNK inhibitors have been recently developed. eFT508 is a new generation Mnk1/2 inhibitor, which is potent, selective and orally bioavailable (Dreas et al., 2017). Its efficacy has been assessed in preclinical models of diffuse large B-cell lymphoma, and it causes a dose dependent decrease in eIF4E-phosphorylation (Reich et al., 2018). eFT508 is now in phase II clinical trial for the treatment of colorectal cancer. A recent study showed that eFT508 efficiently reduces eIF4E phosphorylation in DRG without affecting other major signaling pathways (ERK, 4E-BP, and AKT) and general translation (Megat et al., 2018). eFT508 also alleviated paclitaxel-induced mechanical and thermal sensitivity, supporting its further testing in other chronic pain conditions. BAY 1143269 is another potent, and selective orally administered MNK1 inhibitor (Santag et al., 2017). Additional MNK inhibitors include: 5-(2-(phenylamino)pyrimidin-4-yl)thiazol-2(3H)-one derivatives (Diab et al., 2014), resorcylic acid lactone analogs (Xu et al., 2013), and retinoic acid metabolism blocking agents (RAMBAs) (Ramalingam et al., 2014). These compounds need to be better characterized in both in vitro and in vivo studies.
Inhibitors of eIF4F Complex
Three inhibitors disrupting eIF4G:eIF4E interaction have been described: 4EGI-1 (Moerke et al., 2007), 4E1RCat, and 4E2RCat (Cencic et al., 2011). 4EGI-1 is a small molecule, which binds eIF4E at the site distal to the eIF4G-binding epitope, causing localized conformational changes and dissociation of eIF4G from eIF4E (Papadopoulos et al., 2014). 4EGI-1 also impairs mitochondrial functions (Yang et al., 2015). 4EGI-1 has been used in studies examining the role of eIF4F complex in memory (Hoeffer et al., 2011) and autism (Gkogkas et al., 2013; Santini et al., 2013), where it was delivered directly to the brain (intracerebroventricular injection) as it does not readily penetrate the BBB. Rigidified analogs of 4EGI-1 have been developed, showing improved potency in inhibition of eIF4E/eIF4G interaction (Mahalingam et al., 2014).
4E1RCat, and 4E2RCat block the interaction of eIF4E with both eIF4G and 4E-BP1, and thereby prevent the eIF4F complex formation (Cencic et al., 2011). These compounds have not been used yet in the nervous system in vivo. Antisense oligonucleotide (ASO) targeting eIF4E (LY2275796) with improved tissue stability and nuclease resistance has been developed (Graff et al., 2007). Since eIF4E is overexpressed in many human cancers (by ∼3- to 10-fold) (Bhat et al., 2015), LY2275796 has been tested as an anti-cancer treatment. Administration of LY2275796 to patients resulted in a reduction of eIF4E mRNA and protein levels in tumor cells but caused dose-dependent toxicity (Hong et al., 2011). The antiviral drug ribavirin has been proposed to mimic the mRNA “cap” to inhibit eIF4E/mRNA interaction (Kentsis et al., 2004). This notion was later disputed, and ribavirin’s biological effects were attributed to translation-independent activities (Westman et al., 2005; Yan et al., 2005).
eIF4A Inhibitors
eIF4A helicase activity is critically required for the eIF4F complex formation and unwinding of the 5′ UTR to allow scanning to occur. Therefore, targeting eIF4A might be an additional approach to inhibit eIF4F-dependent translation initiation, particularly for mRNAs with highly structured 5′ UTRs. Pateamine A, hippuristanol, and recoglate family members [e.g., silvestrol and Rocaglamide A (RocA)] are the commonly known inhibitors of eIF4A, out of which only pateamine A is known to cause irreversible inhibition (Pelletier et al., 2015). Hippuristanol is a member of the polyoxygenated steroids family, and it blocks the helicase activity of eIF4A by binding to the C-terminal of eIF4A and imposing allosteric hindrance, thus preventing eIF4A to bind RNA (Sun et al., 2014). On the other hand, pateamine A increases the sequence non-specific RNA-binding activity of free eIF4A, thus preventing eIF4A from participating in the formation of eIF4 complex (Bordeleau et al., 2006; Cencic et al., 2009). Out of these eIF4A inhibitors, silvestrol has been most widely assessed in in vivo preclinical cancer models, owing to its high potency, bioavailability, and relatively low toxicity (Raynaud et al., 2007). Recently, RocA was identified as a sequence-selective inhibitor of translation which acts by stabilizing eIF4A binding on polyurine sequences, thus impeding 43S scanning and leading to upstream premature translation initiation (Iwasaki et al., 2016). The anticancer potential of rocaglates has been widely examined, however, the mechanisms underlying their cytotoxic and anti-proliferative effects have been studied only recently (Becker et al., 2016). The role of eIF4A inhibitors in pain has yet to be examined.
Conclusion
A central role of eIF4E-dependent translational control in mediating maladaptive nociceptive plasticity provides an opportunity to develop new therapeutics to prevent the development of the hypersensitivity state or even reverse established pain states by weakening ongoing activity-dependent plasticity. Existing compounds targeting eIF4E (cercosporamide and 4EGI-1) lack specificity and have poor solubility and BBB permeability (4EGI-1). Therefore, validation of other existing inhibitors for in vivo applications and development of more specific and efficacious inhibitors are required. Another important research direction is uncovering cell type-specific translational landscapes (for example using TRAP) in different pain conditions. This work might reveal mRNAs whose aberrant translation drives the pain phenotype and allow targeting these transcripts or the encoded proteins to reverse the hypersensitivity. It is, however, conceivable that a complex pattern of translation drives the hypersensitivity, involving a combinatory effect of several translationally activated and repressed mRNAs. In this scenario, targeting upstream regulatory mechanisms, such as formation of eIF4F complex, might be a more feasible therapeutic approach. Combination of diverse inhibition strategies could be beneficial to achieve long-lasting effects on pain without triggering compensatory mechanisms.
In summary, a growing recognition of the importance of the eIF4E-dependent translational control in regulation of cellular functions in general and neuronal plasticity in particular, have substantially accelerated studies in the field of pain and advanced our knowledge of how eIF4E-dependent translational dysregulation causes maladaptive plasticity and contributes to the sensitization of the pain pathway. Identification of new molecular targets and pharmacological compounds to target these mechanisms might constitute a basis for next-generation pain therapeutics.
Author Contributions
All authors participated in writing the manuscript.
Funding
This work was supported by QPRN grant (AK) and NIH grant R01NS065926 (TP).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aguilar-Valles, A., Haji, N., De Gregorio, D., Matta-Camacho, E., Eslamizade, M. J., Popic, J., et al. (2018). Translational control of depression-like behavior via phosphorylation of eukaryotic translation initiation factor 4E. Nat. Commun. 9, 2459. doi: 10.1038/s41467-018-04883-5
Altmann, M., Edery, I., Sonenberg, N., and Trachsel, H. (1985). Purification and characterization of protein synthesis initiation factor eIF-4E from the yeast Saccharomyces cerevisiae. Biochemistry 24, 6085–6089. doi: 10.1021/bi00343a009
Amorim, I. S., Kedia, S., Kouloulia, S., Simbriger, K., Gantois, I., Jafarnejad, S. M., et al. (2018). Loss of eIF4E phosphorylation engenders depression-like behaviors via selective mRNA translation. J. Neurosci. 38, 2118–2133. doi: 10.1523/JNEUROSCI.2673-17.2018
Asante, C. O., Wallace, V. C., and Dickenson, A. H. (2009). Formalin-induced behavioural hypersensitivity and neuronal hyperexcitability are mediated by rapid protein synthesis at the spinal level. Mol. Pain 5:27. doi: 10.1186/1744-8069-5-27
Asiedu, M. N., Tillu, D. V., Melemedjian, O. K., Shy, A., Sanoja, R., Bodell, B., et al. (2011). Spinal protein kinase M zeta underlies the maintenance mechanism of persistent nociceptive sensitization. J. Neurosci. 31, 6646–6653. doi: 10.1523/JNEUROSCI.6286-10.2011
Bain, J., Plater, L., Elliott, M., Shpiro, N., Hastie, C. J., McLauchlan, H., et al. (2007). The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315. doi: 10.1042/BJ20070797
Baragana, B., Hallyburton, I., Lee, M. C., Norcross, N. R., Grimaldi, R., Otto, T. D., et al. (2015). A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315–320. doi: 10.1038/nature14451
Becker, M. S., Müller, P. M., Bajorat, J., Schroeder, A., Giaisi, M., Amin, E., et al. (2016). The anticancer phytochemical rocaglamide inhibits Rho GTPase activity and cancer cell migration. Oncotarget 7, 51908–51921. doi: 10.18632/oncotarget.10188
Bhat, M., Robichaud, N., Hulea, L., Sonenberg, N., Pelletier, J., and Topisirovic, I. (2015). Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 14, 261–278. doi: 10.1038/nrd4505
Bogen, O., Alessandri-Haber, N., Chu, C., Gear, R. W., and Levine, J. D. (2012). Generation of a pain memory in the primary afferent nociceptor triggered by PKCepsilon activation of CPEB. J. Neurosci. 32, 2018–2026. doi: 10.1523/JNEUROSCI.5138-11.2012
Bordeleau, M. E., Cencic, R., Lindqvist, L., Oberer, M., Northcote, P., Wagner, G., et al. (2006). RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol. 13, 1287–1295. doi: 10.1016/j.chembiol.2006.10.005
Buffington, S. A., Huang, W., and Costa-Mattioli, M. (2014). Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 37, 17–38. doi: 10.1146/annurev-neuro-071013-014100
Cencic, R., Carrier, M., Galicia-Vazquez, G., Bordeleau, M. E., Sukarieh, R., Bourdeau, A., et al. (2009). Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One 4:e5223. doi: 10.1371/journal.pone.0005223
Cencic, R., Hall, D. R., Robert, F., Du, Y., Min, J., Li, L., et al. (2011). Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc. Natl. Acad. Sci. U.S.A. 108, 1046–1051. doi: 10.1073/pnas.1011477108
Chen, G., Luo, X., Qadri, M. Y., Berta, T., and Ji, R. R. (2018). Sex-dependent glial signaling in pathological pain: distinct roles of spinal microglia and astrocytes. Neurosci. Bull. 34, 98–108. doi: 10.1007/s12264-017-0145-y
Chu, J., and Pelletier, J. (2018). Therapeutic opportunities in eukaryotic translation. Cold Spring Harb. Perspect. Biol. 10:a032995. doi: 10.1101/cshperspect.a032995
Costa-Mattioli, M., Sossin, W. S., Klann, E., and Sonenberg, N. (2009). Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26. doi: 10.1016/j.neuron.2008.10.055
de Souza, J. B., Grossmann, E., Perissinotti, D. M. N., de Oliveira Junior, J. O., da Fonseca, P. R. B., and Posso, I. P. (2017). Prevalence of chronic pain, treatments, perception, and interference on life activities: Brazilian population-based survey. Pain Res. Manag. 2017:4643830. doi: 10.1155/2017/4643830
Derry, M. C., Yanagiya, A., Martineau, Y., and Sonenberg, N. (2006). Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb. Symp. Quant. Biol. 71, 537–543. doi: 10.1101/sqb.2006.71.061
Diab, S., Teo, T., Kumarasiri, M., Li, P., Yu, M., Lam, F., et al. (2014). Discovery of 5-(2-(phenylamino)pyrimidin-4-yl)thiazol-2(3H)-one derivatives as potent Mnk2 inhibitors: synthesis, SAR analysis and biological evaluation. ChemMedChem 9, 962–972. doi: 10.1002/cmdc.201300552
Dreas, A., Mikulski, M., Milik, M., Fabritius, C.-H., Brzozka, K., and Rzymski, T. (2017). Mitogen-activated protein kinase (MAPK) interacting kinases 1 and 2 (MNK1 and MNK2) as targets for cancer therapy: recent progress in the development of MNK inhibitors. Curr. Med. Chem. 24, 3025–3053. doi: 10.2174/0929867324666170203123427
Feoktistova, K., Tuvshintogs, E., Do, A., and Fraser, C. S. (2013). Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl. Acad. Sci. U.S.A. 110, 13339–13344. doi: 10.1073/pnas.1303781110
Ferrari, L. F., Bogen, O., Chu, C., and Levine, J. D. (2013). Peripheral administration of translation inhibitors reverses increased hyperalgesia in a model of chronic pain in the rat. J. Pain 14, 731–738. doi: 10.1016/j.jpain.2013.01.779
Geranton, S. M., Jimenez-Diaz, L., Torsney, C., Tochiki, K. K., Stuart, S. A., Leith, J. L., et al. (2009). A rapamycin-sensitive signaling pathway is essential for the full expression of persistent pain states. J. Neurosci. 29, 15017–15027. doi: 10.1523/JNEUROSCI.3451-09.2009
Gingras, A.-C., Gygi, S. P., Raught, B., Polakiewicz, R. D., Abraham, R. T., Hoekstra, M. F., et al. (1999). Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13, 1422–1437. doi: 10.1101/gad.13.11.1422
Gkogkas, C. G., Khoutorsky, A., Ran, I., Rampakakis, E., Nevarko, T., Weatherill, D. B., et al. (2013). Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377. doi: 10.1038/nature11628
Graff, J. R., Konicek, B. W., Vincent, T. M., Lynch, R. L., Monteith, D., Weir, S. N., et al. (2007). Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 117, 2638–2648. doi: 10.1172/JCI32044
Gray, N. K., Coller, J. M., Dickson, K. S., and Wickens, M. (2000). Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19, 4723–4733. doi: 10.1093/emboj/19.17.4723
Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P., and Heintz, N. (2014). Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9, 1282–1291. doi: 10.1038/nprot.2014.085
Hinnebusch, A. G., Ivanov, I. P., and Sonenberg, N. (2016). Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. doi: 10.1126/science.aad9868
Hoeffer, C. A., Cowansage, K. K., Arnold, E. C., Banko, J. L., Moerke, N. J., Rodriguez, R., et al. (2011). Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc. Natl. Acad. Sci. U.S.A. 108, 3383–3388. doi: 10.1073/pnas.1013063108
Hong, D. S., Kurzrock, R., Oh, Y., Wheler, J., Naing, A., Brail, L., et al. (2011). A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin. Cancer Res. 17, 6582–6591. doi: 10.1158/1078-0432.CCR-11-0430
Iwasaki, S., Floor, S. N., and Ingolia, N. T. (2016). Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534, 558–561. doi: 10.1038/nature17978
Ji, R. R., Gereau, R. W. T., Malcangio, M., and Strichartz, G. R. (2009). MAP kinase and pain. Brain Res. Rev. 60, 135–148. doi: 10.1016/j.brainresrev.2008.12.011
Jiang, F., Pang, X. Y., Niu, Q. S., Hua, L. M., Cheng, M., and Ji, Y. H. (2013). Activation of mammalian target of rapamycin mediates rat pain-related responses induced by BmK I, a sodium channel-specific modulator. Mol. Pain 9:50. doi: 10.1186/1744-8069-9-50
Jimenez-Diaz, L., Geranton, S. M., Passmore, G. M., Leith, J. L., Fisher, A. S., Berliocchi, L., et al. (2008). Local translation in primary afferent fibers regulates nociception. PLoS One 3:e1961. doi: 10.1371/journal.pone.0001961
Kahvejian, A., Roy, G., and Sonenberg, N. (2001). The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 66, 293–300. doi: 10.1101/sqb.2001.66.293
Kentsis, A., Topisirovic, I., Culjkovic, B., Shao, L., and Borden, K. L. (2004). Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc. Natl. Acad. Sci. U.S.A. 101, 18105–18110. doi: 10.1073/pnas.0406927102
Khoutorsky, A., Bonin, R. P., Sorge, R. E., Gkogkas, C. G., Pawlowski, S. A., Jafarnejad, S. M., et al. (2015). Translational control of nociception via 4E-binding protein 1. eLife 4:e12002. doi: 10.7554/eLife.12002
Khoutorsky, A., and Price, T. J. (2018). Translational control mechanisms in persistent pain. Trends Neurosci. 41, 100–114. doi: 10.1016/j.tins.2017.11.006
Konicek, B. W., Stephens, J. R., McNulty, A. M., Robichaud, N., Peery, R. B., Dumstorf, C. A., et al. (2011). Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 71, 1849–1857. doi: 10.1158/0008-5472.CAN-10-3298
Kozak, M. (1989). Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 9, 5134–5142. doi: 10.1128/MCB.9.11.5134
Leppek, K., Das, R., and Barna, M. (2018). Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174. doi: 10.1038/nrm.2017.103
Liang, L., Tao, B., Fan, L., Yaster, M., Zhang, Y., and Tao, Y. X. (2013). mTOR and its downstream pathway are activated in the dorsal root ganglion and spinal cord after peripheral inflammation, but not after nerve injury. Brain Res. 1513, 17–25. doi: 10.1016/j.brainres.2013.04.003
Lin, C. J., Nasr, Z., Premsrirut, P. K., Porco, J. A. Jr., Hippo, Y., Lowe, S. W., et al. (2012). Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 1, 325–333. doi: 10.1016/j.celrep.2012.02.010
Macejak, D. G., and Sarnow, P. (1991). Internal initiation of translation mediated by the 5’ leader of a cellular mRNA. Nature 353, 90–94. doi: 10.1038/353090a0
Mahalingam, P., Takrouri, K., Chen, T., Sahoo, R., Papadopoulos, E., Chen, L., et al. (2014). Synthesis of rigidified eIF4E/eIF4G inhibitor-1 (4EGI-1) mimetic and their in vitro characterization as inhibitors of protein-protein interaction. J. Med. Chem. 57, 5094–5111. doi: 10.1021/jm401733v
Martin, L. J., Smith, S. B., Khoutorsky, A., Magnussen, C. A., Samoshkin, A., Sorge, R. E., et al. (2017). Epiregulin and EGFR interactions are involved in pain processing. J. Clin. Invest. 127, 3353–3366. doi: 10.1172/JCI87406
Megat, S., Ray, P., Moy, J., Lou, T.-F., Barragan-Iglesias, P., Li, Y., et al. (2018). Nociceptor translational profiling reveals the RagA-mTORC1 network as a critical generator of neuropathic pain. bioRxiv [Preprint]. doi: 10.1101/336784
Melemedjian, O. K., Asiedu, M. N., Tillu, D. V., Peebles, K. A., Yan, J., Ertz, N., et al. (2010). IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J. Neurosci. 30, 15113–15123. doi: 10.1523/JNEUROSCI.3947-10.2010
Melemedjian, O. K., and Khoutorsky, A. (2015). Translational control of chronic pain. Prog. Mol. Biol. Transl. Sci. 131, 185–213. doi: 10.1016/bs.pmbts.2014.11.006
Melemedjian, O. K., Khoutorsky, A., Sorge, R. E., Yan, J., Asiedu, M. N., Valdez, A., et al. (2013). mTORC1 inhibition induces pain via IRS-1-dependent feedback activation of ERK. Pain 154, 1080–1091. doi: 10.1016/j.pain.2013.03.021
Merrick, W. C., and Pavitt, G. D. (2018). Protein synthesis initiation in eukaryotic cells. Cold Spring Harb. Perspect. Biol. doi: 10.1101/cshperspect.a033092 [Epub ahead of print].
Moerke, N. J., Aktas, H., Chen, H., Cantel, S., Reibarkh, M. Y., Fahmy, A., et al. (2007). Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128, 257–267. doi: 10.1016/j.cell.2006.11.046
Moy, J. K., Khoutorsky, A., Asiedu, M. N., Black, B. J., Kuhn, J. L., Barragan-Iglesias, P., et al. (2017). The MNK-eIF4E signaling axis contributes to injury-induced nociceptive plasticity and the development of chronic pain. J. Neurosci. 37, 7481–7499. doi: 10.1523/JNEUROSCI.0220-17.2017
Moy, J. K., Khoutorsky, A., Asiedu, M. N., Dussor, G., and Price, T. J. (2018a). eIF4E phosphorylation influences Bdnf mRNA translation in mouse dorsal root ganglion neurons. Front. Cell. Neurosci. 12:29. doi: 10.3389/fncel.2018.00029
Moy, J. K., Kuhn, J. L., Szabo-Pardi, T. A., Pradhan, G., and Price, T. J. (2018b). eIF4E phosphorylation regulates ongoing pain, independently of inflammation, and hyperalgesic priming in the mouse CFA model. Neurobiol. Pain 4, 45–50. doi: 10.1016/j.ynpai.2018.03.001
Norsted Gregory, E., Codeluppi, S., Gregory, J. A., Steinauer, J., and Svensson, C. I. (2010). Mammalian target of rapamycin in spinal cord neurons mediates hypersensitivity induced by peripheral inflammation. Neuroscience 169, 1392–1402. doi: 10.1016/j.neuroscience.2010.05.067
Obara, I., and Hunt, S. P. (2014). Axonal protein synthesis and the regulation of primary afferent function. Dev. Neurobiol. 74, 269–278. doi: 10.1002/dneu.22133
Obata, K., and Noguchi, K. (2006). BDNF in sensory neurons and chronic pain. Neurosci. Res. 55, 1–10. doi: 10.1016/j.neures.2006.01.005
Papadopoulos, E., Jenni, S., Kabha, E., Takrouri, K. J., Yi, T., Salvi, N., et al. (2014). Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc. Natl. Acad. Sci. U.S.A. 111, E3187–E3195. doi: 10.1073/pnas.1410250111
Parsyan, A., Svitkin, Y., Shahbazian, D., Gkogkas, C., Lasko, P., Merrick, W. C., et al. (2011). mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12, 235–245. doi: 10.1038/nrm3083
Pelletier, J., Graff, J., Ruggero, D., and Sonenberg, N. (2015). Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 75, 250–263. doi: 10.1158/0008-5472.CAN-14-2789
Pelletier, J., and Sonenberg, N. (1985). Insertion mutagenesis to increase secondary structure within the 5’ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40, 515–526. doi: 10.1016/0092-8674(85)90200-4
Pelletier, J., and Sonenberg, N. (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325. doi: 10.1038/334320a0
Peter, D., Igreja, C., Weber, R., Wohlbold, L., Weiler, C., Ebertsch, L., et al. (2015). Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell 57, 1074–1087. doi: 10.1016/j.molcel.2015.01.017
Pickering, B. M., and Willis, A. E. (2005). The implications of structured 5 ’ untranslated regions on translation and disease. Semin. Cell Dev. Biol. 16, 39–47. doi: 10.1016/j.semcdb.2004.11.006
Price, T. J., and Geranton, S. M. (2009). Translating nociceptor sensitivity: the role of axonal protein synthesis in nociceptor physiology. Eur. J. Neurosci. 29, 2253–2263. doi: 10.1111/j.1460-9568.2009.06786.x
Price, T. J., Rashid, M. H., Millecamps, M., Sanoja, R., Entrena, J. M., and Cervero, F. (2007). Decreased nociceptive sensitization in mice lacking the fragile X mental retardation protein: role of mGluR1/5 and mTOR. J. Neurosci. 27, 13958–13967. doi: 10.1523/JNEUROSCI.4383-07.2007
Pyronnet, S., Imataka, H., Gingras, A. C., Fukunaga, R., Hunter, T., and Sonenberg, N. (1999). Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18, 270–279. doi: 10.1093/emboj/18.1.270
Ramalingam, S., Gediya, L., Kwegyir-Afful, A. K., Ramamurthy, V. P., Purushottamachar, P., Mbatia, H., et al. (2014). First MNKs degrading agents block phosphorylation of eIF4E, induce apoptosis, inhibit cell growth, migration and invasion in triple negative and Her2-overexpressing breast cancer cell lines. Oncotarget 5, 530–543. doi: 10.18632/oncotarget.1528
Raynaud, F. I., Eccles, S., Clarke, P. A., Hayes, A., Nutley, B., Alix, S., et al. (2007). Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 67, 5840–5850. doi: 10.1158/0008-5472.CAN-06-4615
Reich, S. H., Sprengeler, P. A., Chiang, G. G., Appleman, J. R., Chen, J., Clarine, J., et al. (2018). Structure-based design of pyridone-aminal eFT508 targeting dysregulated translation by selective mitogen-activated protein kinase interacting kinases 1 and 2 (MNK1/2) inhibition. J. Med. Chem. 61, 3516–3540. doi: 10.1021/acs.jmedchem.7b01795
Robichaud, N., Sonenberg, N., Ruggero, D., and Schneider, R. J. (2018). Translational control in cancer. Cold Spring Harb. Perspect. Biol. 10, 254–266. doi: 10.1101/cshperspect.a032896
Rousseau, D., Kaspar, R., Rosenwald, I., Gehrke, L., and Sonenberg, N. (1996). Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. U.S.A. 93, 1065–1070. doi: 10.1073/pnas.93.3.1065
Santag, S., Siegel, F., Wengner, A. M., Lange, C., Bomer, U., Eis, K., et al. (2017). BAY 1143269, a novel MNK1 inhibitor, targets oncogenic protein expression and shows potent anti-tumor activity. Cancer Lett. 390, 21–29. doi: 10.1016/j.canlet.2016.12.029
Santini, E., Huynh, T. N., MacAskill, A. F., Carter, A. G., Pierre, P., Ruggero, D., et al. (2013). Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493, 411–415. doi: 10.1038/nature11782
Saxton, R. A., and Sabatini, D. M. (2017). mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. doi: 10.1016/j.cell.2017.02.004
Shih, M. H., Kao, S. C., Wang, W., Yaster, M., and Tao, Y. X. (2012). Spinal cord NMDA receptor-mediated activation of mammalian target of rapamycin is required for the development and maintenance of bone cancer-induced pain hypersensitivities in rats. J. Pain 13, 338–349. doi: 10.1016/j.jpain.2011.12.006
Sonenberg, N., and Gingras, A.-C. (1998). The mRNA 5’ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10, 268–275. doi: 10.1016/S0955-0674(98)80150-6
Sonenberg, N., and Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. doi: 10.1016/j.cell.2009.01.042
Steglitz, J., Buscemi, J., and Ferguson, M. J. (2012). The future of pain research, education, and treatment: a summary of the IOM report Relieving pain in America: a blueprint for transforming prevention, care, education, and research. Transl. Behav. Med. 2, 6–8. doi: 10.1007/s13142-012-0110-2
Stumpf, C. R., and Ruggero, D. (2011). The cancerous translation apparatus. Curr. Opin. Genet. Dev. 21, 474–483. doi: 10.1016/j.gde.2011.03.007
Sun, Y., Atas, E., Lindqvist, L. M., Sonenberg, N., Pelletier, J., and Meller, A. (2014). Single-molecule kinetics of the eukaryotic initiation factor 4AI upon RNA unwinding. Structure 22, 941–948. doi: 10.1016/j.str.2014.04.014
Tahmasebi, S., Khoutorsky, A., Mathews, M. B., and Sonenberg, N. (2018). Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. doi: 10.1038/s41580-018-0034-x [Epub ahead of print].
Terenzio, M., Koley, S., Samra, N., Rishal, I., Zhao, Q., Sahoo, P. K., et al. (2018). Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416–1421. doi: 10.1126/science.aan1053
Thoreen, C. C., Chantranupong, L., Keys, H. R., Wang, T., Gray, N. S., and Sabatini, D. M. (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. doi: 10.1038/nature11083
Todd, A. J. (2010). Neuronal circuitry for pain processing in the dorsal horn. Nat. Rev. Neurosci. 11, 823–836. doi: 10.1038/nrn2947
Trinh, M. A., and Klann, E. (2013). Translational control by eIF2alpha kinases in long-lasting synaptic plasticity and long-term memory. Neurobiol. Learn. Mem. 105, 93–99. doi: 10.1016/j.nlm.2013.04.013
Truitt, M. L., Conn, C. S., Shi, Z., Pang, X., Tokuyasu, T., Coady, A. M., et al. (2015). Differential requirements for eIF4E dose in normal development and cancer. Cell 162, 59–71. doi: 10.1016/j.cell.2015.05.049
Truitt, M. L., and Ruggero, D. (2016). New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 16, 288–304. doi: 10.1038/nrc.2016.27
Uttam, S., Wong, C., Amorim, I. S., Jafarnejad, S. M., Tansley, S. N., Yang, J., et al. (2018). Translational profiling of dorsal root ganglia and spinal cord in a mouse model of neuropathic pain. Neurobiol. Pain 4, 35–44. doi: 10.1016/j.ynpai.2018.04.001
Veilleux, A., Houde, V. P., Bellmann, K., and Marette, A. (2010). Chronic inhibition of the mTORC1/S6K1 pathway increases insulin-induced PI3K activity but inhibits Akt2 and glucose transport stimulation in 3T3-L1 adipocytes. Mol. Endocrinol. 24, 766–778. doi: 10.1210/me.2009-0328
Waskiewicz, A. J., Johnson, J. C., Penn, B., Mahalingam, M., Kimball, S. R., and Cooper, J. A. (1999). Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19, 1871–1880. doi: 10.1128/MCB.19.3.1871
Westman, B., Beeren, L., Grudzien, E., Stepinski, J., Worch, R., Zuberek, J., et al. (2005). The antiviral drug ribavirin does not mimic the 7-methylguanosine moiety of the mRNA cap structure in vitro. RNA 11, 1505–1513. doi: 10.1261/rna.2132505
Wolfe, A. L., Singh, K., Zhong, Y., Drewe, P., Rajasekhar, V. K., Sanghvi, V. R., et al. (2014). RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513, 65–70. doi: 10.1038/nature13485
Xu, J., Chen, A., Joy, J., Xavier, V. J., Ong, E. H., Hill, J., et al. (2013). Rational design of resorcylic acid lactone analogues as covalent MNK1/2 kinase inhibitors by tuning the reactivity of an enamide Michael acceptor. ChemMedChem 8, 1483–1494. doi: 10.1002/cmdc.201300231
Xu, J. T., Zhao, J. Y., Zhao, X., Ligons, D., Tiwari, V., Atianjoh, F. E., et al. (2014). Opioid receptor-triggered spinal mTORC1 activation contributes to morphine tolerance and hyperalgesia. J. Clin. Invest. 124, 592–603. doi: 10.1172/JCI70236
Xu, Q., Fitzsimmons, B., Steinauer, J., O’Neill, A., Newton, A. C., Hua, X. Y., et al. (2011). Spinal phosphinositide 3-kinase-Akt-mammalian target of rapamycin signaling cascades in inflammation-induced hyperalgesia. J. Neurosci. 31, 2113–2124. doi: 10.1523/JNEUROSCI.2139-10.2011
Yan, Y., Svitkin, Y., Lee, J. M., Bisaillon, M., and Pelletier, J. (2005). Ribavirin is not a functional mimic of the 7-methyl guanosine mRNA cap. RNA 11, 1238–1244. doi: 10.1261/rna.2930805
Yanagiya, A., Suyama, E., Adachi, H., Svitkin, Y. V., Aza-Blanc, P., Imataka, H., et al. (2012). Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell. 46, 847–858. doi: 10.1016/j.molcel.2012.04.004
Yang, X., Dong, Q. F., Li, L. W., Huo, J. L., Li, P. Q., Fei, Z., et al. (2015). The cap-translation inhibitor 4EGI-1 induces mitochondrial dysfunction via regulation of mitochondrial dynamic proteins in human glioma U251 cells. Neurochem. Int. 90, 98–106. doi: 10.1016/j.neuint.2015.07.019
Yekkirala, A. S., Roberson, D. P., Bean, B. P., and Woolf, C. J. (2017). Breaking barriers to novel analgesic drug development. Nat. Rev. Drug Discov. 16:810. doi: 10.1038/nrd.2017.202
Keywords: eIF4E, mRNA translation, persistent pain, sensitization, treatment
Citation: Uttam S, Wong C, Price TJ and Khoutorsky A (2018) eIF4E-Dependent Translational Control: A Central Mechanism for Regulation of Pain Plasticity. Front. Genet. 9:470. doi: 10.3389/fgene.2018.00470
Received: 07 August 2018; Accepted: 24 September 2018;
Published: 24 October 2018.
Edited by:
Maritza Jaramillo, University of Quebec, CanadaCopyright © 2018 Uttam, Wong, Price and Khoutorsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arkady Khoutorsky, YXJrYWR5Lmtob3V0b3Jza3lAbWNnaWxsLmNh
 Sonali Uttam
Sonali Uttam Calvin Wong
Calvin Wong Theodore J. Price
Theodore J. Price Arkady Khoutorsky
Arkady Khoutorsky