- Department of Animal Sciences, Reproductive Biotechnology, School of Life Sciences Weihenstephan, Technical University Munich, Freising, Germany
The chicken represents a valuable model for research in the area of immunology, infectious diseases as well as developmental biology. Although it was the first livestock species to have its genome sequenced, there was no reverse genetic technology available to help understanding specific gene functions. Recently, homologous recombination was used to knockout the chicken immunoglobulin genes. Subsequent studies using immunoglobulin knockout birds helped to understand different aspects related to B cell development and antibody production. Furthermore, the latest advances in the field of genome editing including the CRISPR/Cas9 system allowed the introduction of site specific gene modifications in various animal species. Thus, it may provide a powerful tool for the generation of genetically modified chickens carrying resistance for certain pathogens. This was previously demonstrated by targeting the Trp38 region which was shown to be effective in the control of avian leukosis virus in chicken DF-1 cells. Herein we review the current and future prospects of gene editing and how it possibly contributes to the development of resistant chickens against infectious diseases.
Introduction
The chicken represents an important source of protein worldwide and a valuable model for the study of developmental biology in vertebrates (Yasugi and Nakamura, 2000; Speedy, 2003). Chickens are constantly exposed to a plethora of pathogens threatening animal welfare as well as human health (Perdue and Swayne, 2005; Humphrey, 2006). Viral pathogens such as influenza A viruses can be transmitted to humans leading to death (Gao et al., 2013). Furthermore, bacterial agents such as Campylobacter jejuni and Salmonella enteritidis cause food borne illnesses in humans associated with digestive symptoms (Bryan and Doyle, 1995). More recently, using genetically modified chickens as a model for various research areas like developmental biology, immunology, physiology and neurology is gaining importance in the avian research community (Mozdziak and Petitte, 2004; Stern, 2004, 2005). In addition, there is an increasing interest to generate genetically modified chickens resistant to specific pathogens, benefiting from the availability of gene manipulation techniques. This review focuses on the advances made in gene editing in chickens and the future perspectives including the generation of specific-pathogen-resistant birds.
State of the Art
Genetically modified animals have significantly contributed to our understanding of different aspects related to immunity, infectious diseases, neurology, behavior, and developmental biology (Yeh et al., 2002; Lyall et al., 2011; Lalonde et al., 2012; Pinkert, 2014; Park et al., 2017b). While mice were the first animals to be genetically modified (Costantini and Lacy, 1981; Gordon and Ruddle, 1981), pronuclear DNA microinjections allowed the introduction of foreign DNA leading to genetic modifications in livestock including rabbits, sheep and pigs (Hammer et al., 1985). Although this method was used for a long time, it did not allow the induction of targeted gene modifications and had the disadvantage of generating random integrations (Perleberg et al., 2018). The generation of knockout (KO) animals was achieved for the first time by gene targeting in embryonic stem cells (ES) (Evans and Kaufman, 1981; Thomas and Capecchi, 1987). Though the induction of the KO was successful, it had the disadvantage of low efficiency (Thomas and Capecchi, 1987). Due to the absence of true ES lines from farm animals and no solid evidence of germline transmission (Talbot and Blomberg, 2008; Soto and Ross, 2016), stable transfection of sheep somatic cells with human factor IX and neomycin resistance followed by nuclear transfer was the alternative to express foreign DNA in livestock (Schnieke et al., 1997) and afterwards for gene targeting (McCreath et al., 2000). At this time, the generation of KO livestock animals was possible by combining somatic cell nuclear transfer (SNTC) and homologous recombination (Lai et al., 2002; Nottle et al., 2007). The laborious procedure of these methods and the low efficiency for generating targeted KO was improved by homologous recombination (Houdebine, 2002) along with different nucleases (Carlson et al., 2012). The transcription activator-like effector nucleases (TALENs) are composed of series of repeats fused to non-specific FokI-cleavage domains that induce double- stranded DNA breaks upon dimerization (Gaj et al., 2013). More recently the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 system made the process of specific DNA-targeting easier by using single guide RNAs (sgRNAs) (Jinek et al., 2012; Ran et al., 2013; Hsu et al., 2014). CRISPR/Cas9 is an adaptive immune system found in bacteria and archaeal species and uses small-non coding RNAs to guide the Cas9 nuclease to target sites resulting in DNA double-break (Jinek et al., 2012).
In comparison to mammals, difficulties were always associated with the generation of genetically modified chickens due to the complex structure of the chicken zygote (Mozdziak and Petitte, 2004) and the different organization of the chick embryo compared to mammals (Stern, 1990). Over the past 30 years, different research groups paved the way for the generation of genetically modified chickens. Efforts were focused on the stable genomic integration of transgenes and obtaining the highest efficiency of germline transmission. While Pettite and colleagues described the transfer of stage X embryo cells that led to germline transmission, it was not possible to genetically modify these cells and to re-introduce them as germline competent cells into the chicken embryo (Petitte et al., 1990). Although ES were shown to provide a valuable tool for the generation of transgenic mice (Kanatsu-Shinohara et al., 2003), no evidence of germline transmission using chicken ES was reported. Transferred chicken ES cells only contributed to somatic tissue but not to the germline.
The first genetically modified chicken was generated by the insertion of retroviral foreign DNA delivered by avian leukosis virus that was successfully integrated to the germline (Salter et al., 1987). The retroviral vector was injected into the yolk sac near to the developing blastoderm. Since then, various viral vectors have been used to generate genetically modified chickens (Hughes et al., 1986; Bosselman et al., 1989; Salter and Crittenden, 1989; Harvey and Ivarie, 2003; Mozdziak et al., 2003). Drawbacks of viral vectors, such as the replication of deficient viral particles and risks of recombination with wild type viruses, were avoided by plasmid-DNA microinjection into the chicken zygote (Love et al., 1994). The microinjection was done in the germinal disk and led to the generation of transgenic chickens expressing neomycin resistance and a reporter gene lacZ (Love et al., 1994). A total of 5.5% of the generated chicks survived to sexual maturity and later on, one rooster gave 3.4% transmission to his offspring (Love et al., 1994). The germline transmission of integrated transgenes was improved with lentiviral vectors (McGrew et al., 2004). McGrew and colleagues showed the possibility of transduction with lentiviral vectors in G0 birds. Founder cockerels were injected with different plasmids carrying different reporter genes including LacZ and eGFP (McGrew et al., 2004). Lentiviral vectors were injected into the subgerminal cavity of newly laid eggs. Ten of the founder males transmitted 4-45% of the foreign DNA to their offspring (McGrew et al., 2004). Lentiviral vectors offered for the first time the possibility to generate genetically modified chickens with a decent germline transmission efficiency. Nevertheless, the size of the transgene was still limited and precise edits were not possible.
Furthermore, the in ovo injection of the avian retroviral vector RCAS (replication-competent avian sarcoma-leukosis virus with a splice acceptor) carrying enhanced fluorescent protein (eGFP) into unincubated (stage X) blastoderms resulted in stable and widespread expression of eGFP in the embroys. Even though the gonads showed eGFP expression PGCs were eGFP negative indicating viral silencing (Smith et al., 2009).
Like in mammals, chicken primordial germ cells (PGCs) are precursors of gametes and a key element for sperm and oocystes development. At the early hours of embryonic development, PGCs are found in the germinal crescent and migrate afterwards (50–55 h) to the gonads (Kim et al., 2010; Kang et al., 2015) in order to produce sperm and oocystes upon sexual maturity (Fujimoto et al., 1976). The migration of PGCs was found to be greatly influenced by the chemokine stromal cell-derived factor-1 (SDF-1/CXCL12) and its receptor C-X-C chemokine receptor type 4 (CXCR4) (Stebler et al., 2004; Lee et al., 2017c).
The ability to culture PGCs was a milestone in the process of generating transgenic chickens. Genetic modification of PGCs and their subsequent reintroduction into the embryonic vasculature resolved many issues and problems observed with previously established methods. Van de Lavoir and colleagues used BRL or STO feeder cells to cultivate PGCs for up to 217 days. PGCs were shown to retain the germline characteristics by analyzing various germline markers including the chicken vasa homolog (CVH) and were cryoconserved using conventional techniques (Van De Lavoir et al., 2006). PGC-culture was optimized afterwards by Whyte and colleagues that developed feeder and serum free culture conditions that took into consideration the signaling pathways necessary for avian germ cell self-renewal (Whyte et al., 2015). The work of van de Lavoir and colleagues revealed that foreign DNA can be inserted in the genome of PGCs and cells were still restricted to the germline (Van De Lavoir et al., 2006; Leighton et al., 2008). Male PGCs were cultured for a duration between 35 and 110 days during which they were transfected with a construct coding for eGFP and subsequently injected into the vasculature of White Leghorn embryos [stage 13–15 Hamburger and Hamilton (H&H)](Van De Lavoir et al., 2006). Interestingly, the long term culture of PGCs did not influence their ability to colonize the gonads after insertion of foreign DNA, which allowed afterwards the generation of several transgenic chicken lines (Van De Lavoir et al., 2006, 2012; Macdonald et al., 2012). Leighton and colleagues gave new insights about increasing the efficiency of foreign DNA insertion in PGCs mediated by phiC31 integrase that catalyzes site-specific recombination between attB and pseudo attP sites in the chicken genome and increases transgene integration (Leighton et al., 2008).
Lu and colleagues indicated that the piggyBac transposon can be efficiently integrated into the genome of chicken embryo during development via electroporation (Lu et al., 2009). The transfection of PGCs with piggyBac transposon greatly enhanced the integration frequency of foreign DNA into the chicken genome and resulted in the generation of genetically modified chickens (Park and Han, 2012; Glover et al., 2013). In contrast, the injection of piggyBac transposon into the subgerminal cavity of a newly laid egg and subsequent electroporation, resulted in chickens expressing the transgene but no germline transmission was detectable (Liu et al., 2013). At the same time, Tyack and colleagues successfully developed a method for the direct transfection of circulating PGCs using Lipofectamine 2000 in combination with Tol2 transposon and transposase plasmids (Tyack et al., 2013). The plasmid contained the pCAGGS promoter driving the expression of eGFP. Tyack and colleagues found that 5/11 roosters expressed the miniTol DNA in their semen and two of them gave about 1.5% germline transmission (Tyack et al., 2013). This method substantially reduced the time needed for the in vitro isolation and gene manipulation of PGCs; however, it did not increase the germline transmission in G0 (Tyack et al., 2013). In addition, it does not allow clonal selection of PGCs and may result in birds with random integrations of the same transgene. Nevertheless, it is an effective method to produce genetically modified chickens as shown by various publications (Tyack et al., 2013; Lambeth et al., 2016a,b).
The possibility to culture and genetically modify chicken PGCs without losing germline competence made it possible to perform precise gene deletions and integrations in the chicken genome. Specific gene locus KO chickens were generated by Schusser and colleagues via gene targeting by homologous recombination in chicken PGCs (Schusser et al., 2013a, 2016). In the case of targeted immunoglobulin heavy chain J segment, a total of 7 from 27 PGC clones (28%) had a correctly targeted event which reflected a high efficiency comparable to mouse ES cells (Schusser et al., 2013a). Similar efficiency was obtained after targeting the immunoglobulin light chain locus in chicken PGCs. After successful targeting of the immunoglobulin heavy or light chain in chicken PGCs, resulting clones were injected into H&H stage 13–15 embryos in order to generate germline chimeras. Germline transmission rates varied between 0.1 and 48% depending on the used PGC clone (Schusser et al., 2013a, 2016). Resulting homozygous immunoglobulin heavy chain J segment knockout birds showed a depletion of peripheral B cells and antibodies and were the first non-mammalian vertebrates harboring a knockout produced by homologs recombination. In order to perform gene knockouts by homologs recombination in PGCs, isogenic DNA is needed since mismatches in the homology regions are not tolerated (Schusser et al., 2016).
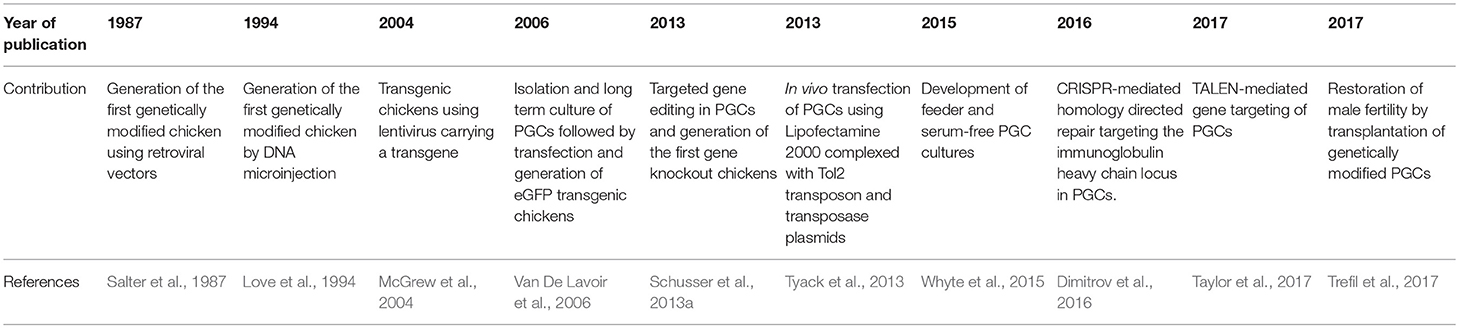
Since PGCs are precursors of sperm, researchers suggested that roosters could be used as recipient for exogenous transfer of genetically-modified PGCs which may improve the germline transmission rate (Trefil et al., 2017). Chicken embryos and adult roosters were chemically or physically sterilized to create a surrogate for external PGC donors (Trefil et al., 2006; Nakamura et al., 2008, 2010; Ghadimi et al., 2017). Nakamura and colleagues partially sterilized chicken embryos by injecting Busulfan into the yolk of fertile eggs before incubation; this led to a significant reduction of endogenous PGCs. Authors demonstrated that the sterilized embryos can be used for exogenous transfer of PGCs resulting in high efficiencies of germline transmission (Nakamura et al., 2008, 2010). Early experiments performed by Trefil and colleagues provided an alternative for chemical sterilization and concluded that repeated gamma irradiation leads to sterilization of roosters (Trefil et al., 2006). Performing injection of donor spermatogonial cells led to reestablishment of male function in 50% of the roosters only 5 weeks after injection (Trefil et al., 2006). Spermatogenesis was restored 4 weeks later in the case of PGC-transplantation compared to spermatogonial cells; however, PGCs exhibited higher efficiency in repopulating the seminiferous epithelium (Trefil et al., 2006, 2017). This was very beneficial in the case of transplantation of genetically modified PGCs into mature roosters after complete irradiation (Trefil et al., 2017). Male fertility was reestablished after the transplantation of GFP- or mCherry-expressing PGCs and resulted in almost 100% germline transmission (Trefil et al., 2017). The prominent advantage of this method is the certainty of the germline transmission and the low number of animals used in the experiment; hence reducing time and costs for testing high number of chimeric roosters. Although using gamma irradiation to sterilize roosters was as efficient as in mice, recent findings in pigs suggested that the knockout of NANOS2, like in mice, results in specific germline ablation with preserved testicular development (Park et al., 2017a); therefore it was suggested that NANOS2 KO pigs may serve as a surrogate for transplantation of donor spermatogonial cells (Park et al., 2017a). Even though the importance of NANOS2 in the transformation of ES into germ cells is well determined, little is known about its function in chickens. The most important steps made in the process of generating genetically modified chickens are summarized in Table 1.
Gene Editing in Avian Cell Lines
The unavailability of fully transgenic chickens for a long time encouraged the development of alternative methods based on in vitro cell culture systems. In vitro studies helped to provide valuable data regarding host susceptibility to specific pathogens and the role of specific genes during host-pathogen interactions. DT-40 cells, an avian leukosis virus induced bursal B- cell lymphoma line, was extensively used to investigate B cell immunology, cell cycle regulation, gene conversion and apoptosis (Uckun et al., 1996; Arakawa et al., 2001; Harris et al., 2002; Arakawa and Buerstedde, 2004). A large number of DT-40 mutants were generated to understand B cell biology and were reviewed elsewhere (Arakawa and Buerstedde, 2004). For instance, studies based on DT-40 cells proved that the activation-induced cytidine deaminase (AID) triggers immunoglobulin gene diversification by gene conversion (Buerstedde et al., 1990; Kim et al., 1990). Furthermore, Szüts and colleagues used mutant DT-40 cells to demonstrate the role of RAD18 in DNA repair and the completion of gene conversion (Szüts et al., 2006). Interestingly, Schusser and colleagues replaced the immunoglobulin light and heavy chain loci in DT-40 cells with human immunoglobulin light and heavy chain loci; this led to the expression of chimeric IgM with human variable regions and chicken constant regions (Schusser et al., 2013b). The later cell line provides a model to study the diversification of the human variable region by gene conversion and somatic hypermutations in chickens. Antigen receptor analysis were performed by deep sequencing confirming that the host machinery in DT-40 cells diversified the integrated human V genes (Leighton et al., 2015).
A different established model for examining gene function in chickens is the Douglas Foster (DF-1) cells, an immortalized chicken fibroblast cell line (Foster, 1998). Recent studies used DF-1 cells to investigate host-pathogen interactions of several avian pathogens with the avian host; this included influenza A viruses, Newcastle disease virus, infectious bursal disease virus and retroviruses (Huang et al., 2003; Lee et al., 2008; Cheng et al., 2015; Hui and Leung, 2015). The overexpression of different avian genes in DF-1 cells helped to examine their role in the innate immunity against viral pathogens (Shao et al., 2014; Cheng et al., 2015; Xu et al., 2015). A well-known tool for the overexpression of various genes is the retroviral vectors derived from the SR-A strain of Rous sarcoma virus (RCAS). The RCAS system is known for its stable transduction in developing chicken embryo and cell culture (Fekete and Cepko, 1993; Bell and Brickell, 1997). Reuter and colleagues used DF-1 cells for the overexpression of the chicken IFN-α and IFN-λ (Reuter et al., 2014). The overexpression of IFN-λ in DF-1 cells did not cause substantial viral resistance against influenza A viruses H1N1, H7N1, and vesicular stomatitis virus (VSV) (Reuter et al., 2014) which suggested that DF-1 cells have weak antiviral activity of IFN-λ (Karpala et al., 2008). This was not the case for IFN-α where the overexpression led to protection against previously mentioned viruses (Reuter et al., 2014). In addition, DF-1 cells were useful to study the function of foreign genes in chicken including intracellular pattern recognition receptor such as the retinoic inducible resistant gene (RIG-I). RIG-I from duck and goose was overexpressed in DF-1 cells and its protective effect against influenza A viruses and infectious bursitis virus (IBDV) was investigated (Barber et al., 2010; Sun et al., 2013; Shao et al., 2014). The overexpression of duck RIG-I in DF-1 cells reduced viral replication and upregulated virus-induced apoptosis following IBVD- and H9N2 influenza virus infections (Shao et al., 2014). Interestingly, the knockdown of the chicken ANP32A, a nuclear protein implicated in mRNA transport and cell death (Reilly et al., 2014), reduced the activity of different avian influenza polymerases in DF-1 cells. This indicated that avian influenza virus polymerases are more adapted to avian ANP32A and proposed this gene as target for antiviral drugs (Long et al., 2016). Furthermore, the overexpression of the chicken GADD45β, a protein associated with cell growth control, apoptotic cell death, and the cellular response to DNA damage (Zazzeroni et al., 2003), helped to limit viral infection which could be used in the future as potential treatment for avian leukosis virus (ALV)-J infections (Zhang et al., 2016).
ALV is one of the most commonly occurring retroviruses in chickens. It induces a variety of neoplastic lesions causing losses in the productivity of affected chicken flocks (Fadly, 2000). Maas and colleagues confirmed that DF-1 cells are much more suitable than primary chicken fibroblasts (CEFs) to study host-pathogen interactions of leukosis viruses with avian cells (Maas et al., 2006). ALV was detected earlier in DF-1 cells and the infection was associated with apparent cytopathogenic effect (CPE) compared to infected-CEFs that had no apparent CPE (Maas et al., 2006). Mutations responsible for the inhibition of ALV subgroup A cell-entry were identified (Klucking et al., 2002) and consisted of four base pairs insertion and one base pair substitution in tumor virus locus A (tva) (Klucking et al., 2002). On the other side, only one base pair substitution in the cysteine-rich domain (CRD) of tvb receptor led to reduced susceptibility of DF-1 cells to infection with ALV subgroup B (Klucking et al., 2002; Reinisová et al., 2008). Interestingly, subgroup J ALV (ALV-J) uses the multimembrane-spanning cell surface protein, the chicken Na+/H+ exchanger type 1 (NHE1), as a receptor. The attachment of the virus to the receptor is crucial to initiate the infection (Barnard et al., 2006). Kučerová and colleagues used mutagenesis to introduce changes in the subgenic fragment of NHE1 (Kučerová et al., 2013); authors described the functional importance of tryptophan reside at position 38 (Trp38) for virus entry (Kučerová et al., 2013).
The rapid development of gene editing tools such as CRISPR/Cas9 rendered cell culture systems much more useful by easily targeting different genes. Precise gene editing of the chicken NHE1 gene using CRISPR/Cas9 system led to resistance of DF-1 cells against ALV-J infection (Lee et al., 2017a). The precise genome editing of NHE1 was performed via homologs directed repair (HDR) that combined CRISPR/Cas9 vectors with single-stranded oligodeoxynucleotide (ssODNs). Authors confirmed previous observations mentioning that mutation in the Trp38 are detrimental for ALV-J infection (Kučerová et al., 2013). On the other side, non-homologous end joining repair (NHEJ) was also established in DF-1 cells. Targeting the tumor virus locus B gene, which serves as entry receptor for ALV subgroup B, resulted in frameshift mutations leading to a KO of the tvb-receptor in DF-1 cells, which conferred resistance against ALV-B (Lee et al., 2017b). Abu-Bonsrah and colleagues targeted a wide range of genes in DF-1 cells such as DROSHA, DICER, MBD3, KIAA1279, CDKN1B, EZH2, HIRA, TYRP1, STMN2, RET, and DGCR, that play a role in embryonic development and pathogenesis of embryonic diseases (Abu-Bonsrah et al., 2016). Efficiency of inducing mutations was analyzed by T7E1 assay. The efficiency ranged between 20 and 65% in DF-1 cells (Abu-Bonsrah et al., 2016). Similar results were obtained after knocking out KIAA1279- and CDKN1B-genes in DT-40 cell-line via electroporation (Abu-Bonsrah et al., 2016). Likewise, Bai and colleagues gave more insights about the efficiency of CRISPR/Cas9 in DF-1 cells by studying gene editing in the presence and the absence of puromycin antibiotic selection (Bai et al., 2016). Three genes including peroxisome proliferator-activated receptor-γ (PPAR-γ), ATP synthase epsilon subunit (ATP5E), and ovalbumin (OVA) were targeted with CRISRP/Cas9 vectors. T7E assay indicated that puromycin selection increased mutation rate in the previously mentioned genes from 0.75, 0.5, and 3.0%, to 60.7, 61.3, and 47.3%, respectively (Bai et al., 2016).
Gene Editing in the Chicken Embryo
The chicken embryo is a well-established model to study developmental processes, gene functions and host-pathogen interactions (Darnell and Schoenwolf, 2000; Chesnutt and Niswander, 2004; Schecterson et al., 2012). Over the last decades, different methods were established to genetically manipulate chicken embryos including electroporation of foreign DNA constructs, transduction with retroviruses and recently the combination of previous known methods with CRISPR/Cas9 system (Gandhi et al., 2017).
For example, Luo and colleagues established a protocol based on ex ovo electroporation of 3.5 days old chicken embryos for the overexpression of Cad7 and eGFP (Luo et al., 2012). This method provided accessibility of different embryonic parts for the electroporation, which are not easily reachable when the embryo is still inside the egg (Luo et al., 2012). Similarly, in ovo electroporation of the embryonic auditory brainstem was previously established (Lu et al., 2017). Plasmids of interest were successfully integrated into the nucleus magnocellularis and nucleus laminaris. Authors indicated the possibility of drug inducible gene expression which was confirmed in the presence of doxycycline (Lu et al., 2017).
A well-established tool for foreign DNA integration is the RCAS-system. Using RCAS in the chicken embryo model indicated that vector proteins and inserted transgenes were mainly detectable in the skin, blood vessels and heart (Sato et al., 2002; Kothlow et al., 2010). Several studies deduced the efficacy of RCAS-system in the case of foreign DNA integration and gene overexpression in chicken embryos (Bell and Brickell, 1997; Sato et al., 2002; Kothlow et al., 2010; Schusser et al., 2011; Reuter et al., 2014). This system is very useful to study the specific function of relevant genes for the innate immunity, particularly during the interaction with influenza A viruses. RCAS vectors expressing various Mx gene isoforms were used for transduction of CEFs. Four days post-transfection, CEFs expressing the retrovirally transduced Mx proteins were injected in the yolk sac of 3 days-old fertilized eggs (Schusser et al., 2011). The overexpression of different Mx isoforms in embryonated eggs did not protect against influenza A virus infection, which was in agreement with the results obtained from chicken fibroblasts (Schusser et al., 2011). In addition, the role of IFN-λ was previously investigated by the generation of mosaic chicken embryos overexpressing chicken IFN-λ (Reuter et al., 2014). Generated embryos exhibited lower viral titers upon challenge with influenza A viruses, NDV Herts-33, or IBV M-41 via the allantoic cavity by at least four log10 units compared to inoculated eggs with empty RCAS vector (Reuter et al., 2014). This clearly demonstrated the protective effect of chicken IFN-λ against different viruses (Reuter et al., 2014). Although the IFN-λ overexpression had detrimental effects at early hours post hatch (Reuter et al., 2014), RCAS system was shown to be successful for maintaining transgene expression after hatch (Kothlow et al., 2010).
A similar system based on gene transfer mediated by lentiviral vectors was described in embryonated eggs (Hen et al., 2012). Usefulness of lentiviral vectors in developmental biology was previously reviewed elsewhere (Stern, 2004). Lentiviral vectors of feline immunodeficiency virus origin were injected into chorioallantoic membrane (CAM) of 11 days old chicken embryos. The injected lentiviral vectors carried yellow fluorescent protein (YFP) or recombinant alpha-melanocyte-stimulating hormone (α-MSH) genes and they were expressed under the cytomegalovirus (CMV) promoter (Hen et al., 2012). High efficiency of transduction was observed in the liver, which implied that this model could be useful for the study of hormones and enzymes.
The application of gene editing technologies via in ovo electroporation of chicken embryos seems to be efficient (Wilson and Stoeckli, 2012). Wilson and Stoeckli used miRNA-based plasmids for knocking down gene expression in the chicken neural tube (Wilson and Stoeckli, 2012). Additionally, Ghandi and colleagues used ex ovo electroporation to knockout Pax7 and Sox10, a key transcription factors in the neural crest, leading to loss of their proteins and transcripts (Gandhi et al., 2017). Overall, collected data indicated that in ovo gene manipulation of the chicken embryo could be used as a model for the study of different embryonic developmental stages (Gandhi et al., 2017; Lu et al., 2017). High targeting efficiency and the simplicity of CRISPR/Cas9 make it now possible to knockout genes in specific tissues/organs of the developing chicken embryo. This allows the study the gene function during development without generating fully gene edited chicken lines.
Generation of Genetically Modified Chickens
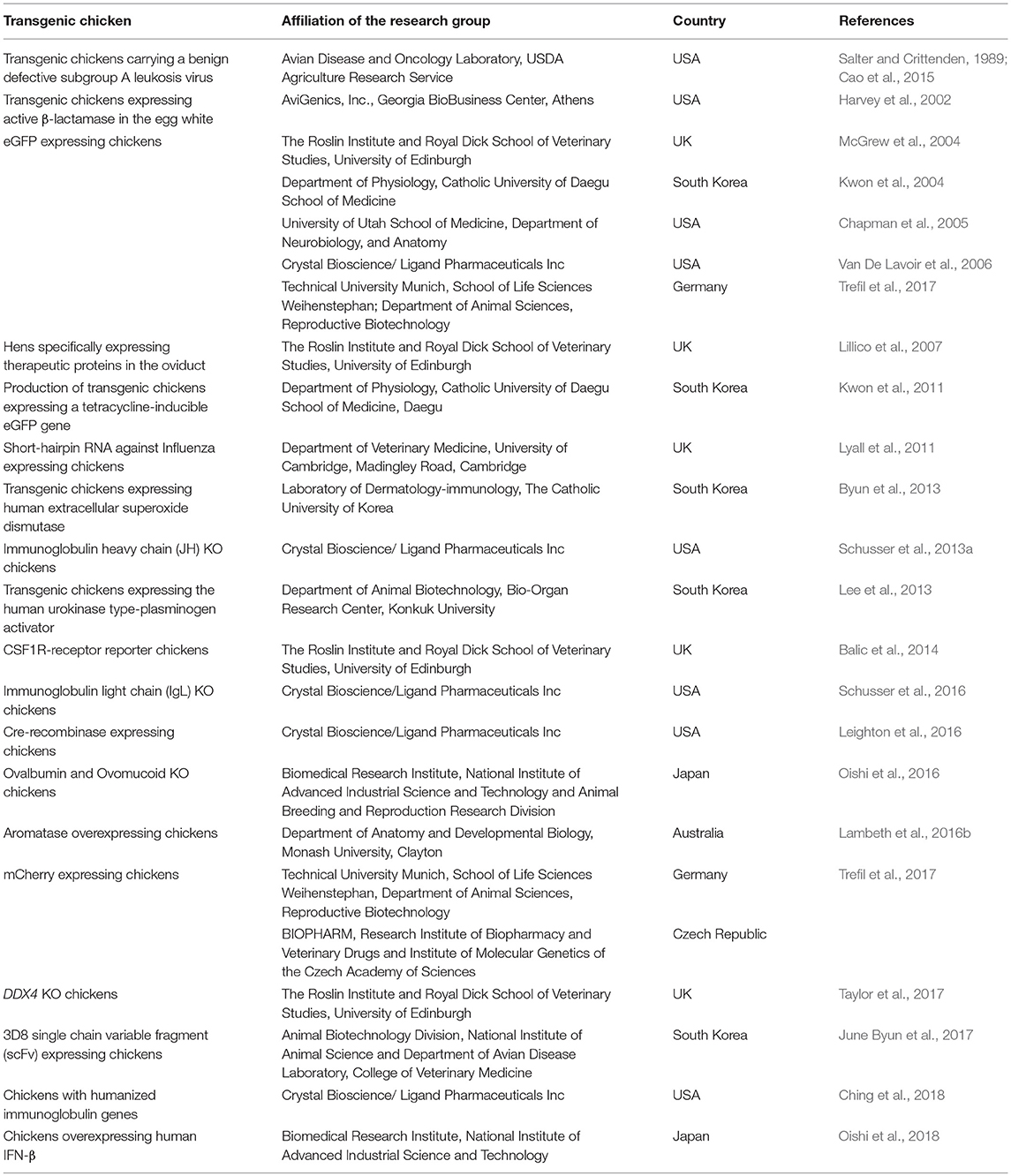
The generation of genetically modified chickens has wide applications in agricultural and biomedical research (Sang, 1994; Ivarie, 2003; Mozdziak and Petitte, 2004). Benefiting from gene editing technologies and germline transmission of PGCs, new knowledge was brought to light about specific gene functions (Schusser et al., 2013a, 2016), resistant for infectious diseases (Lyall et al., 2011) and the possible preservation of endangered species including the Houbara Bastard (Kang et al., 2008; Wernery et al., 2010; Van De Lavoir et al., 2012). Different methods used for gene editing in chickens and the generated chicken lines were stated earlier in this review. In addition, the worldwide availability of genetically modified chicken lines is summarized in Table 2.
Specific gene editing in PGCs was improved using TALEN and CRISPR/Cas9 via HDR (Dimitrov et al., 2016; Oishi et al., 2016; Taylor et al., 2017). Using CRISPR/Cas9, the efficiency of gene targeting was increased remarkably in PGCs (Dimitrov et al., 2016). In order to introduce a loxP site into the immunoglobulin heavy chain locus, Dimitrov and colleagues combined a targeting vector having a total of 2 kb homology arms with CRISPR/Cas9 system targeting the upstream region of the single immunoglobulin heavy chain variable region (VH) in PGCs (Dimitrov et al., 2016). Interestingly, all selected drug resistant PGC clones contained the correct targeting event and the germline transmission rate varied between 0 and 100% depending on the used PGC line (Dimitrov et al., 2016).
Targeting the DDX4 locus, located on the Z chromosome, showed possible role of this gene in the formation of the germ cell lineage (Taylor et al., 2017). Targeted DDX4 KO was achieved with TALEN in combination with a targeting vector. Authors reported a germline transmission rate of 6% from the founder birds (Taylor et al., 2017). G1 female chicks were hemizygous mutant for DDX4, they did not lay eggs and had no yellow or white follicles in the ovaries. Surprisingly this was not the case in DDX4 knockout female mice (Tanaka et al., 2000).
Overall, a significant progress was made in the last decade in producing and using genetically modified chickens to understand developmental biology, immunology, host-pathogen interaction, reproductive biology and physiology. However, efforts to generate resistant chickens for specific pathogens are still at the beginning, probably due to the lack of specific gene targets responsible for acquiring resistance against specific pathogens. This was not the case in other livestock including pigs which were genetically edited to gain resistance against porcine reproductive and respiratory syndrome virus (PRRSV) (Whitworth et al., 2015; Burkard et al., 2017). Using NHEJ, Whitworth and colleagues generated KO pigs with premature stop codon in exon 3 of the viral receptor CD163 (Whitworth et al., 2015). CD163-KO pigs challenged with PRRSV did not exhibit any clinical symptoms, lung pathology, viremia, or antibody response. In addition, Burckard and colleagues generated an exon 7 deletion in CD163 using two sgRNAs to induce the excision of the exon (Burkard et al., 2017). Pigs carrying the mutation were healthy and kept the main biological functions of the protein while macrophages isolated from the CD163 KO animals indicated an inhibition of the viral infection (Burkard et al., 2017).
So far, only few reports are available about the resistance of gene-edited chickens for specific pathogens. Lyall and colleagues generated transgenic chickens expressing short-hairpin RNA intended to function as a decoy that interacts and blocks influenza A virus polymerase (Lyall et al., 2011). Although birds were not resistant to initial infection, viral transmission was prevented (Lyall et al., 2011). A different study demonstrated the possibility to suppress influenza A virus transmission in transgenic birds expressing the 3D8 single chain variable fragment (scFv), a gene that interacts with viral genome leading to suppression of viral shedding (June Byun et al., 2017).
Further Applications in Biomedical Research
The chicken became a very interesting model in biomedical research. Different temporal patterns of bright light were used to study the effect on myopia in chickens. Lan and colleagues found that intermittent episodes of light suppress myopia in chickens more than continuous bright light (Lan et al., 2014). Although the obtained results may not be directly translated into humans (Lan et al., 2014), future applications in optical research seem to be promising. In addition, the chicken was used as a model for xenotransplantation by injection of human stem cells into small induced lesions in the chicken embryo neural tube (Boulland et al., 2010). Authors stated that the reduced immune response during early embryonic development helps to study xenotransplantation without the risk of early immune rejection (Boulland et al., 2010). The chicken was also used as a human multiple myeloma xenograft model (Martowicz et al., 2015); it was suggested that this model may offer novel therapeutic compounds targeting survival and proliferation of multiple myeloma cells. Using the chicken as a bioreactor may greatly benefit human health by providing alternative therapeutic approaches (Zhu et al., 2005). A promising approach using chickens for the production of human antibodies is the replacement of the chicken immunoglobulin variable regions by human V regions and synthetic pseudogene arrays in order to produce affinity matured human antibodies in chickens (Ching et al., 2018). The OmniChicken by Ligand Pharmaceuticals Inc. is a worldwide unique platform to produce human monoclonal antibodies from chickens making use of the phylogenetic difference between mammals and birds. The purification of overexpressed human antibodies from the chicken egg seems also to be a valid application which was reviewed elsewhere (Flemming, 2005). A very recent study conducted by Oishi and colleagues demonstrated the ability of integrating human interferon beta (hIFN-β) into the chicken ovalbumin locus in order to produce hIFN-β in egg white (Oishi et al., 2018). Authors demonstrated the ability of producing foreign proteins in eggs which would have industrial and therapeutic applications.
Future Perspectives
The role of host genes in the susceptibility of chickens to different pathogens was mostly investigated in vitro. Preliminary in vitro investigations provide solid information about the role of these genes prior to the generation of fully gene edited chickens. New technologies including CRISPR/Cas9 make the process of gene editing easy and highly efficient in contrast to the well-established process of homologs recombination. Although gene editing in mammals, particularly mice and pigs, is vastly advanced, gene editing in chickens is entering the golden age. For instance the generation of Cas9-expressing pigs will provide a powerful tool for the study of biological processes (Wang et al., 2017); while this was not done yet in chickens, it seems to be beneficial and may be used in the future to dissect unknown gene functions faster and more easily.
Therapeutic applications using human monoclonal antibodies produced from humanized chickens may be beneficial over in vitro approaches lacking affinity maturation (Ching et al., 2018). In addition, production of antibodies in chicken eggs represents an economic and stress-free method for the production of specific antibodies (Amro et al., 2018). Using chicken eggs to manufacture specific proteins in eggs seems interesting (Lillico et al., 2005; Petitte and Mozdziak, 2007) especially since it may allow improvement of digestibility of sugar complexes in feedstuffs; however, this application may be thwarted by critics that claim the inedibility of the product.
Several advantages are provided by newly invented gene editing technologies including the simplicity of design and application combined with high efficiency (Chira et al., 2017). Understanding the host cell behavior during host-pathogen interactions may help targeting pathogen specific receptors and viral cellular transport (Heaton et al., 2016). The determination of new target genes associated with disease susceptibility should fill the research gap and open the door for new therapeutical approaches. Although the debate about using genetically modified animals in food production will continue to be stimulated, we may obtain new breeds of chickens in the future that are resistant for specific pathogens. We speculate that spending more efforts connecting gene editing technologies with the prevention of infectious diseases will change the way we use to fight pathogens and will probably improve the animal welfare.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This research was supported by grant Schu2446/3-1 to BS from the Deutsche Forschungsgemeinschaft.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abu-Bonsrah, K. D., Zhang, D., and Newgreen, D. F. (2016). CRISPR/Cas9 targets chicken embryonic somatic cells in vitro and in vivo and generates phenotypic abnormalities. Sci. Rep. 6:34524. doi: 10.1038/srep34524
Amro, W. A., Al-Qaisi, W., and Al-Razem, F. (2018). Production and purification of IgY antibodies from chicken egg yolk. J. Genet. Eng. Biotechnol. 16, 99–103. doi: 10.1016/j.jgeb.2017.10.003
Arakawa, H., and Buerstedde, J. M. (2004). Immunoglobulin gene conversion: insights from bursal B cells and the DT40 cell line. Dev. Dyn. 229, 458–464. doi: 10.1002/dvdy.10495
Arakawa, H., Lodygin, D., and Buerstedde, J. M. (2001). Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol 1:7. doi: 10.1186/1472-6750-1-7
Bai, Y., He, L., Li, P., Xu, K., Shao, S., Ren, C., et al. (2016). Efficient genome editing in chicken DF-1 cells using the CRISPR/Cas9 system. G3: Genes Genomes Genet. 6, 917–923. doi: 10.1534/g3.116.027706
Balic, A., Garcia-Morales, C., Vervelde, L., Gilhooley, H., Sherman, A., Garceau, V., et al. (2014). Visualisation of chicken macrophages using transgenic reporter genes: insights into the development of the avian macrophage lineage. Development 141, 3255–3265. doi: 10.1242/dev.105593
Barber, M. R., Aldridge, J. R., Webster, R. G., and Magor, K. E. (2010). Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci.U.S.A. 107, 5913–5918. doi: 10.1073/pnas.1001755107
Barnard, R. J., Elleder, D., and Young, J. A. (2006). Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus–cell membrane fusion. Virology 344, 25–29. doi: 10.1016/j.virol.2005.09.021
Bell, E. J., and Brickell, P. M. (1997). Replication-competent retroviral vectors for expressing genes in avian cells in vitro and in vivo. Mol. Biotechnol. 7, 289–298. doi: 10.1007/BF02740819
Bosselman, R. A., Hsu, R. Y., Boggs, T., Hu, S., Bruszewski, J., Ou, S., et al. (1989). Germline transmission of exogenous genes in the chicken. Science 243, 533–535. doi: 10.1126/science.2536194
Boulland, J.-L., Halasi, G., Kasumacic, N., and Glover, J. C. (2010). Xenotransplantation of human stem cells into the chicken embryo. J. Vis. Exp. 11:2071. doi: 10.3791/2071
Bryan, F. L., and Doyle, M. P. (1995). Health risks and consequences of Salmonella and Campylobacter jejuni in raw poultry. J. Food Prot. 58, 326–344. doi: 10.4315/0362-028X-58.3.326
Buerstedde, J. M., Reynaud, C. A., Humphries, E. H., Olson, W., Ewert, D. L., and Weill, J. C. (1990). Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 9, 921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x
Burkard, C., Lillico, S. G., Reid, E., Jackson, B., Mileham, A. J., Ait-Ali, T., et al. (2017). Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 13:e1006206. doi: 10.1371/journal.ppat.1006206
Byun, S. J., Ji, M.-R., Jang, Y.-J., Hwang, A.-I., Chung, H. K., Kim, J. S., et al. (2013). Human extracellular superoxide dismutase (EC-SOD) expression in transgenic chicken. BMB Rep. 46, 404–409. doi: 10.5483/BMBRep.2013.46.8.251
Cao, W., Mays, J., Kulkarni, G., Dunn, J., Fulton, R. M., and Fadly, A. (2015). Further observations on serotype 2 Marek's disease virus-induced enhancement of spontaneous avian leukosis virus-like bursal lymphomas in ALVA6 transgenic chickens. Avian Pathol. 44, 23–27. doi: 10.1080/03079457.2014.989195
Carlson, D. F., Tan, W., Lillico, S. G., Stverakova, D., Proudfoot, C., Christian, M., et al. (2012). Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. U.S.A. 109, 17382–17387. doi: 10.1073/pnas.1211446109
Chapman, S. C., Lawson, A., Macarthur, W. C., Wiese, R. J., Loechel, R. H., Burgos-Trinidad, M., et al. (2005). Ubiquitous GFP expression in transgenic chickens using a lentiviral vector. Development 132, 935–940. doi: 10.1242/dev.01652
Cheng, Y., Sun, Y., Wang, H., Yan, Y., Ding, C., and Sun, J. (2015). Chicken STING mediates activation of the IFN gene independently of the RIG-I gene. J. Immunol. 195, 3922–3936. doi: 10.4049/jimmunol.1500638
Chesnutt, C., and Niswander, L. (2004). Plasmid-based short-hairpin RNA interference in the chicken embryo. Genesis 39, 73–78. doi: 10.1002/gene.20028
Ching, K. H., Collarini, E. J., Abdiche, Y. N., Bedinger, D., Pedersen, D., Izquierdo, S., et al. (2018). Chickens with humanized immunoglobulin genes generate antibodies with high affinity and broad epitope coverage to conserved targets. MAbs 10, 71–80. doi: 10.1080/19420862.2017.1386825
Chira, S., Gulei, D., Hajitou, A., Zimta, A.-A., Cordelier, P., and Berindan-Neagoe, I. (2017). CRISPR/Cas9: transcending the reality of genome editing. Molecul. Ther. Nucleic Acids 7, 211–222. doi: 10.1016/j.omtn.2017.04.001
Costantini, F., and Lacy, E. (1981). Introduction of a rabbit beta-globin gene into the mouse germ line. Nature 294, 92–94. doi: 10.1038/294092a0
Darnell, D. K., and Schoenwolf, G. C. (2000). The chick embryo as a model system for analyzing mechanisms of development. Dev. Biol. Protocol. 135, 25–29. doi: 10.1385/1-59259-685-1:25
Dimitrov, L., Pedersen, D., Ching, K. H., Yi, H., Collarini, E. J., Izquierdo, S., et al. (2016). Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS ONE 11:e0154303. doi: 10.1371/journal.pone.0154303
Evans, M. J., and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. doi: 10.1038/292154a0
Fadly, A. M. (2000). Isolation and identification of avian leukosis viruses: a review. Avian Pathol. 29, 529–535. doi: 10.1080/03079450020016760
Fekete, D. M., and Cepko, C. L. (1993). Replication-competent retroviral vectors encoding alkaline phosphatase reveal spatial restriction of viral gene expression/transduction in the chick embryo. Mol. Cell. Biol. 13, 2604–2613. doi: 10.1128/MCB.13.4.2604
Flemming, A. (2005). Biotechnology: human antibodies from chicken eggs. Nat. Rev. Drug Discov. 4:884. doi: 10.1038/nrd1883
Foster, D. (1998). Development of a spontaneously immortalized chicken embryo fibroblastic cell line. Virology 248, 305–311.
Fujimoto, T., Ninomiya, T., and Ukeshima, A. (1976). Observations of the primordial germ cells in blood samples from the chick embryo. Dev. Biol. 49, 278–282. doi: 10.1016/0012-1606(76)90273-6
Gaj, T., Gersbach, C. A., and Barbas Iii, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Gandhi, S., Piacentino, M. L., Vieceli, F. M., and Bronner, M. E. (2017). Optimization of CRISPR/Cas9 genome editing for loss-of-function in the early chick embryo. Dev. Biol. 432, 86–97. doi: 10.1016/j.ydbio.2017.08.036
Gao, R., Cao, B., Hu, Y., Feng, Z., Wang, D., Hu, W., et al. (2013). Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897. doi: 10.1056/NEJMoa1304459
Ghadimi, F., Shakeri, M., Zhandi, M., Zaghari, M., Piryaei, A., Moslehifar, P., et al. (2017). Different approaches to establish infertile rooster. Anim. Reprod. Sci. 186, 31–36. doi: 10.1016/j.anireprosci.2017.08.019
Glover, J. D., Taylor, L., Sherman, A., Zeiger-Poli, C., Sang, H. M., and Mcgrew, M. J. (2013). A novel piggyBac transposon inducible expression system identifies a role for AKT signalling in primordial germ cell migration. PLoS ONE 8:e77222. doi: 10.1371/journal.pone.0077222
Gordon, J. W., and Ruddle, F. H. (1981). Integration and stable germ line transmission of genes injected into mouse pronuclei. Science 214, 1244–1246. doi: 10.1126/science.6272397
Hammer, R. E., Pursel, V. G., Rexroad, C. E. Jr, Wall, R. J., Bolt, D. J., Ebert, K. M., et al. (1985). Production of transgenic rabbits, sheep and pigs by microinjection. Nature 315, 680–683. doi: 10.1038/315680a0
Harris, R. S., Sale, J. E., Petersen-Mahrt, S. K., and Neuberger, M. S. (2002). AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12, 435–438. doi: 10.1016/S0960-9822(02)00717-0
Harvey, A. J., and Ivarie, R. (2003). Validating the hen as a bioreactor for the production of exogenous proteins in egg white. Poult. Sci. 82, 927–930. doi: 10.1093/ps/82.6.927
Harvey, A. J., Speksnijder, G., Baugh, L. R., Morris, J. A., and Ivarie, R. (2002). Expression of exogenous protein in the egg white of transgenic chickens. Nat. Biotechnol. 20:396. doi: 10.1038/nbt0402-396
Heaton, N. S., Moshkina, N., Fenouil, R., Gardner, T. J., Aguirre, S., Shah, P. S., et al. (2016). Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity 44, 46–58. doi: 10.1016/j.immuni.2015.12.017
Hen, G., Yosefi, S., Shinder, D., Or, A., Mygdal, S., Condiotti, R., et al. (2012). Gene transfer to chicks using lentiviral vectors administered via the embryonic chorioallantoic membrane. PLoS ONE 7:e36531. doi: 10.1371/journal.pone.0036531
Houdebine, L.-M. (2002). The methods to generate transgenic animals and to control transgene expression. J. Biotechnol. 98, 145–160. doi: 10.1016/S0168-1656(02)00129-3
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Huang, Z., Krishnamurthy, S., Panda, A., and Samal, S. K. (2003). Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 77, 8676–8685. doi: 10.1128/JVI.77.16.8676-8685.2003
Hughes, S. H., Kosik, E., Fadly, A. M., Salter, D. W., and Crittenden, L. B. (1986). Design of retroviral vectors for the insertion of foreign deoxyribonucleic acid sequences into the avian germ line. Poult. Sci. 65, 1459–1467. doi: 10.3382/ps.0651459
Hui, R. K., and Leung, F. C. (2015). Differential expression profile of chicken embryo fibroblast DF-1 cells infected with cell-adapted infectious bursal disease virus. PLoS ONE 10:e0111771. doi: 10.1371/journal.pone.0111771
Humphrey, T. (2006). Are happy chickens safer chickens? Poultry welfare and disease susceptibility. Br. Poult. Sci. 47, 379–391. doi: 10.1080/00071660600829084
Ivarie, R. (2003). Avian transgenesis: progress towards the promise. Trends Biotechnol. 21, 14–19. doi: 10.1016/S0167-7799(02)00009-4
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
June Byun, S., Yuk, S.-S., Jang, Y.-J., Choi, H., Jeon, M.-H., Erdene-Ochir, T., et al. (2017). Transgenic chickens expressing the 3D8 single chain variable fragment protein suppress avian influenza transmission. Sci. Rep. 7:5938. doi: 10.1038/s41598-017-05270-8
Kanatsu-Shinohara, M., Ogonuki, N., Inoue, K., Miki, H., Ogura, A., Toyokuni, S., et al. (2003). Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 69, 612–616. doi: 10.1095/biolreprod.103.017012
Kang, K. S., Lee, H. C., Kim, H. J., Lee, H. G., Kim, Y. M., Lee, H. J., et al. (2015). Spatial and temporal action of chicken primordial germ cells during initial migration. Reproduction 149, 179–187. doi: 10.1530/REP-14-0433
Kang, S. J., Choi, J. W., Kim, S. Y., Park, K. J., Kim, T. M., Lee, Y. M., et al. (2008). Reproduction of wild birds via interspecies germ cell transplantation. Biol. Reprod. 79, 931–937. doi: 10.1095/biolreprod.108.069989
Karpala, A. J., Morris, K. R., Broadway, M. M., Mcwaters, P. G., O'neil, T. E., Goossens, K. E., et al. (2008). Molecular cloning, expression, and characterization of chicken IFN-λ. J. Interferon Cytokine Res. 28, 341–350. doi: 10.1089/jir.2007.0117
Kim, J. N., Park, T. S., Park, S. H., Park, K. J., Kim, T. M., Lee, S. K., et al. (2010). Migration and proliferation of intact and genetically modified primordial germ cells and the generation of a transgenic chicken. Biol. Reprod. 82, 257–262. doi: 10.1095/biolreprod.109.079723
Kim, S., Humphries, E. H., Tjoelker, L., Carlson, L., and Thompson, C. B. (1990). Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol. Cell. Biol. 10, 3224–3231. doi: 10.1128/MCB.10.6.3224
Klucking, S., Adkins, H. B., and Young, J. A. (2002). Resistance to infection by subgroups B, D, and E avian sarcoma and leukosis viruses is explained by a premature stop codon within a resistance allele of the tvb receptor gene. J. Virol. 76, 7918–7921. doi: 10.1128/JVI.76.15.7918-7921.2002
Kothlow, S., Schenk-Weibhauser, K., Ratcliffe, M. J., and Kaspers, B. (2010). Prolonged effect of BAFF on chicken B cell development revealed by RCAS retroviral gene transfer in vivo. Mol. Immunol. 47, 1619–1628. doi: 10.1016/j.molimm.2010.01.011
Kučerová, D., Plachý, J., Reinišová, M., Šenigl, F., Trejbalová, K., Geryk, J., and Hejnar, J. (2013). Nonconserved tryptophan 38 of the cell surface receptor for subgroup J avian leukosis virus discriminates sensitive from resistant avian species. J. Virol. 87, 8399–8407. doi: 10.1128/JVI.03180-12
Kwon, M. S., Koo, B. C., Choi, B. R., Lee, H. T., Kim, Y. H., Ryu, W.-S., et al. (2004). Development of transgenic chickens expressing enhanced green fluorescent protein. Biochem. Biophys. Res. Commun. 320, 442–448. doi: 10.1016/j.bbrc.2004.05.197
Kwon, M. S., Koo, B. C., Roh, J. Y., Kim, M., Kim, J.-H., and Kim, T. (2011). Production of transgenic chickens expressing a tetracycline-inducible GFP gene. Biochem. Biophys. Res. Commun. 410, 890–894. doi: 10.1016/j.bbrc.2011.06.088
Lai, L., Kolber-Simonds, D., Park, K.-W., Cheong, H.-T., Greenstein, J. L., Im, G.-S., et al. (2002). Production of α-1, 3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295, 1089–1092. doi: 10.1126/science.1068228
Lalonde, R., Fukuchi, K., and Strazielle, C. (2012). Neurologic and motor dysfunctions in APP transgenic mice. Rev. Neurosci. 23, 363–379. doi: 10.1515/revneuro-2012-0041
Lambeth, L. S., Morris, K., Ayers, K. L., Wise, T. G., O'neil, T., Wilson, S., et al. (2016a). Overexpression of anti-mullerian hormone disrupts gonadal sex differentiation, blocks sex hormone synthesis, and supports cell autonomous sex development in the chicken. Endocrinology 157, 1258–1275. doi: 10.1210/en.2015-1571
Lambeth, L. S., Morris, K. R., Wise, T. G., Cummins, D. M., O'neil, T. E., Cao, Y., et al. (2016b). Transgenic chickens overexpressing aromatase have high estrogen levels but maintain a predominantly male phenotype. Endocrinology 157, 83–90. doi: 10.1210/en.2015-1697
Lan, W., Feldkaemper, M., and Schaeffel, F. (2014). Intermittent episodes of bright light suppress myopia in the chicken more than continuous bright light. PLoS ONE 9:e110906. doi: 10.1371/journal.pone.0110906
Lee, C.-W., Jung, K., Jadhao, S. J., and Suarez, D. L. (2008). Evaluation of chicken-origin (DF-1) and quail-origin (QT-6) fibroblast cell lines for replication of avian influenza viruses. J. Virol. Methods 153, 22–28. doi: 10.1016/j.jviromet.2008.06.019
Lee, H. J., Lee, K. Y., Jung, K. M., Park, K. J., Lee, K. O., Suh, J. Y., et al. (2017a). Precise gene editing of chicken Na+/H+ exchange type 1 (chNHE1) confers resistance to avian leukosis virus subgroup J (ALV-J). Dev. Comp. Immunol. 77, 340–349. doi: 10.1016/j.dci.2017.09.006
Lee, H. J., Lee, K. Y., Park, Y. H., Choi, H. J., Yao, Y., Nair, V., et al. (2017b). Acquisition of resistance to avian leukosis virus subgroup B through mutations on tvb cysteine-rich domains in DF-1 chicken fibroblasts. Vet. Res. 48:48.
Lee, J. H., Park, J.-W., Kim, S. W., Park, J., and Park, T. S. (2017c). CXC chemokine receptor type 4 (CXCR4) is a key receptor for chicken primordial germ cell migration. J. Reprod. Dev. 63, 555–562. doi: 10.1262/jrd.2017-067
Lee, S. H., Gupta, M. K., Ho, Y. T., Kim, T., and Lee, H. T. (2013). Transgenic chickens expressing human urokinase-type plasminogen activator. Poult. Sci. 92, 2396–2403. doi: 10.3382/ps.2013-03223
Leighton, P. A., Pedersen, D., Ching, K., Collarini, E. J., Izquierdo, S., Jacob, R., et al. (2016). Generation of chickens expressing Cre recombinase. Trans. Res. 25, 609–616. doi: 10.1007/s11248-016-9952-6
Leighton, P. A., Schusser, B., Yi, H., Glanville, J., and Harriman, W. (2015). A diverse repertoire of human immunoglobulin variable genes in a chicken B cell line is generated by both gene conversion and somatic hypermutation. Front. Immunol. 6:126. doi: 10.3389/fimmu.2015.00126
Leighton, P. A., Van De Lavoir, M. C., Diamond, J. H., Xia, C., and Etches, R. J. (2008). Genetic modification of primordial germ cells by gene trapping, gene targeting, and φC31 integrase. Mol. Reprod. Dev. 75, 1163–1175. doi: 10.1002/mrd.20859
Lillico, S. G., Mcgrew, M. J., Sherman, A., and Sang, H. M. (2005). Transgenic chickens as bioreactors for protein-based drugs. Drug Discov. Today 10, 191–196. doi: 10.1016/S1359-6446(04)03317-3
Lillico, S. G., Sherman, A., Mcgrew, M. J., Robertson, C. D., Smith, J., Haslam, C., et al. (2007). Oviduct-specific expression of two therapeutic proteins in transgenic hens. Proc. Natl. Acad. Sci. U.S.A. 104, 1771–1776. doi: 10.1073/pnas.0610401104
Liu, X., Li, N., Hu, X., Zhang, R., Li, Q., Cao, D., et al. (2013). Efficient production of transgenic chickens based on piggyBac. Trans. Res. 22, 417–423. doi: 10.1007/s11248-012-9642-y
Long, J. S., Giotis, E. S., Moncorgé, O., Frise, R., Mistry, B., James, J., et al. (2016). Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 529, 101–104. doi: 10.1038/nature16474
Love, J., Gribbin, C., Mather, C., and Sang, H. (1994). Transgenic birds by DNA microinjection. Biotechnology 12, 60–63.
Lu, T., Cohen, A. L., and Sanchez, J. T. (2017). In Ovo Electroporation in the Chicken Auditory Brainstem. J. Vis. Exp. 9.
Lu, Y., Lin, C., and Wang, X. (2009). PiggyBac transgenic strategies in the developing chicken spinal cord. Nucleic Acids Res. 37:e141. doi: 10.1093/nar/gkp686
Luo, J., Yan, X., Lin, J., and Rolfs, A. (2012). Gene transfer into older chicken embryos by ex ovo electroporation. J. Vis. Exp. 27:4078. doi: 10.3791/4078
Lyall, J., Irvine, R. M., Sherman, A., Mckinley, T. J., Nú-ez, A., Purdie, A., et al. (2011). Suppression of avian influenza transmission in genetically modified chickens. Science 331, 223–226. doi: 10.1126/science.1198020
Maas, R., Van Zoelen, D., Oei, H., and Claassen, I. (2006). Replacement of primary chicken embryonic fibroblasts (CEF) by the DF-1 cell line for detection of avian leucosis viruses. Biologicals 34, 177–181. doi: 10.1016/j.biologicals.2005.09.002
Macdonald, J., Taylor, L., Sherman, A., Kawakami, K., Takahashi, Y., Sang, H. M., et al. (2012). Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proc. Natl. Acad. Sci.U.S.A. 109, E1466–E1472. doi: 10.1073/pnas.1118715109
Martowicz, A., Kern, J., Gunsilius, E., and Untergasser, G. (2015). Establishment of a human multiple myeloma xenograft model in the chicken to study tumor growth, invasion and angiogenesis. J. Vis. Exp. 1:e52665. doi: 10.3791/52665
McCreath, K. J., Howcroft, J., Campbell, K. H., Colman, A., Schnieke, A. E., and Kind, A. J. (2000). Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature 405, 1066–1069. doi: 10.1038/35016604
McGrew, M. J., Sherman, A., Ellard, F. M., Lillico, S. G., Gilhooley, H. J., Kingsman, A. J., et al. (2004). Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 5, 728–733. doi: 10.1038/sj.embor.7400171
Mozdziak, P. E., Borwornpinyo, S., Mccoy, D. W., and Petitte, J. N. (2003). Development of transgenic chickens expressing bacterial beta-galactosidase. Dev. Dyn. 226, 439–445. doi: 10.1002/dvdy.10234
Mozdziak, P. E., and Petitte, J. N. (2004). Status of transgenic chicken models for developmental biology. Dev. Dyn. 229, 414–421. doi: 10.1002/dvdy.10461
Nakamura, Y., Usui, F., Ono, T., Takeda, K., Nirasawa, K., Kagami, H., et al. (2010). Germline replacement by transfer of primordial germ cells into partially sterilized embryos in the chicken. Biol. Reprod. 83, 130–137. doi: 10.1095/biolreprod.110.083923
Nakamura, Y., Yamamoto, Y., Usui, F., Atsumi, Y., Ito, Y., Ono, T., et al. (2008). Increased proportion of donor primordial germ cells in chimeric gonads by sterilisation of recipient embryos using busulfan sustained-release emulsion in chickens. Reprod. Fertility Dev. 20, 900–907. doi: 10.1071/RD08138
Nottle, M. B., Beebe, L. F., Harrison, S. J., Mcilfatrick, S. M., Ashman, R. J., O'connell, P. J., et al. (2007). Production of homozygous α-1, 3-galactosyltransferase knockout pigs by breeding and somatic cell nuclear transfer. Xenotransplantation 14, 339–344. doi: 10.1111/j.1399-3089.2007.00417.x
Oishi, I., Yoshii, K., Miyahara, D., Kagami, H., and Tagami, T. (2016). Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 6:23980. doi: 10.1038/srep23980
Oishi, I., Yoshii, K., Miyahara, D., and Tagami, T. (2018). Efficient production of human interferon beta in the white of eggs from ovalbumin gene–targeted hens. Sci. Rep. 8:10203. doi: 10.1038/s41598-018-28438-2
Park, K. E., Kaucher, A. V., Powell, A., Waqas, M. S., Sandmaier, S. E., Oatley, M. J., et al. (2017a). Generation of germline ablated male pigs by CRISPR/Cas9 editing of the NANOS2 gene. Sci. Rep. 7:40176. doi: 10.1038/srep40176
Park, M. H., Lee, H. J., Lee, H. L., Son, D. J., Ju, J. H., Hyun, B. K., et al. (2017b). Parkin knockout inhibits neuronal development via regulation of proteasomal degradation of p21. Theranostics 7, 2033–2045. doi: 10.7150/thno.19824
Park, T. S., and Han, J. Y. (2012). piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc. Natl. Acad. Sci. U.S.A. 109, 9337–9341. doi: 10.1073/pnas.1203823109
Perdue, M. L., and Swayne, D. E. (2005). Public health risk from avian influenza viruses. Avian Dis. 49, 317–327. doi: 10.1637/7390-060305R.1
Perleberg, C., Kind, A., and Schnieke, A. (2018). Genetically engineered pigs as models for human disease. Dis. Model. Mech. 11:dmm030783. doi: 10.1242/dmm.030783
Petitte, J. N., Clark, M. E., Liu, G., Verrinder Gibbins, A. M., and Etches, R. J. (1990). Production of somatic and germline chimeras in the chicken by transfer of early blastodermal cells. Development 108, 185–189.
Petitte, J. N., and Mozdziak, P. E. (2007). The incredible, edible, and therapeutic egg. Proc. Natl. Acad. Sci. U.S.A. 104, 1739–1740. doi: 10.1073/pnas.0611652104
Pinkert, C. A. (ed.). (2014). “1 - Introduction to Transgenic Animal Technology,” in Transgenic Animal Technology, 3rd Edn, (London: Elsevier), 3–13.
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi: 10.1038/nprot.2013.143
Reilly, P. T., Yu, Y., Hamiche, A., and Wang, L. (2014). Cracking the ANP32 whips: important functions, unequal requirement, and hints at disease implications. Bioessays 36, 1062–1071. doi: 10.1002/bies.201400058
Reinisová, M., Senigl, F., Yin, X., Plachy, J., Geryk, J., Elleder, D., et al. (2008). A single-amino-acid substitution in the TvbS1 receptor results in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroups B and D and resistance to infection by subgroup E in vitro and in vivo. J. Virol. 82, 2097–2105. doi: 10.1128/JVI.02206-07
Reuter, A., Soubies, S., Härtle, S., Schusser, B., Kaspers, B., Staeheli, P., et al. (2014). Antiviral activity of lambda interferon in chickens. J. Virol. 88, 2835–2843. doi: 10.1128/JVI.02764-13
Salter, D. W., and Crittenden, L. B. (1989). Artificial insertion of a dominant gene for resistance to avian leukosis virus into the germ line of the chicken. Theor. Appl. Genet. 77, 457–461. doi: 10.1007/BF00274263
Salter, D. W., Smith, E. J., Hughes, S. H., Wright, S. E., and Crittenden, L. B. (1987). Transgenic chickens: insertion of retroviral genes into the chicken germ line. Virology 157, 236–240. doi: 10.1016/0042-6822(87)90334-5
Sang, H. (1994). Transgenic chickens—methods and potential applications. Trends Biotechnol. 12, 415–420. doi: 10.1016/0167-7799(94)90030-2
Sato, N., Matsuda, K., Sakuma, C., Foster, D. N., Oppenheim, R. W., and Yaginuma, H. (2002). Regulated gene expression in the chicken embryo by using replication-competent retroviral vectors. J. Virol. 76, 1980–1985. doi: 10.1128/JVI.76.4.1980-1985.2002
Schecterson, L. C., Sanchez, J. T., Rubel, E. W., and Bothwell, M. (2012). TrkB downregulation is required for dendrite retraction in developing neurons of chicken nucleus magnocellularis. J. Neurosci. 32, 14000–14009. doi: 10.1523/JNEUROSCI.2274-12.2012
Schnieke, A. E., Kind, A. J., Ritchie, W. A., Mycock, K., Scott, A. R., Ritchie, M., et al. (1997). Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science 278, 2130–2133. doi: 10.1126/science.278.5346.2130
Schusser, B., Collarini, E. J., Pedersen, D., Yi, H., Ching, K., Izquierdo, S., et al. (2016). Expression of heavy chain-only antibodies can support B-cell development in light chain knockout chickens. Eur. J. Immunol. 46, 2137–2148. doi: 10.1002/eji.201546171
Schusser, B., Collarini, E. J., Yi, H., Izquierdo, S. M., Fesler, J., Pedersen, D., et al. (2013a). Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci.U.S.A. 110, 20170–20175. doi: 10.1073/pnas.1317106110
Schusser, B., Reuter, A., Von Der Malsburg, A., Penski, N., Weigend, S., Kaspers, B., et al. (2011). Mx is dispensable for interferon-mediated resistance of chicken cells against influenza A virus. J. Virol. 85, 8307–8315. doi: 10.1128/JVI.00535-11
Schusser, B., Yi, H., Collarini, E. J., Izquierdo, S. M., Harriman, W. D., Etches, R. J., et al. (2013b). Harnessing gene conversion in chicken B cells to create a human antibody sequence repertoire. PLoS ONE 8:e80108. doi: 10.1371/journal.pone.0080108
Shao, Q., Xu, W., Yan, L., Liu, J., Rui, L., Xiao, X., et al. (2014). Function of duck RIG-I in induction of antiviral response against IBDV and avian influenza virus on chicken cells. Virus Res. 191, 184–191. doi: 10.1016/j.virusres.2014.07.028
Smith, C. A., Roeszler, K. N., and Sinclair, A. H. (2009). Robust and ubiquitous GFP expression in a single generation of chicken embryos using the avian retroviral vector, RCASBP. Differentiation 77, 473–482. doi: 10.1016/j.diff.2009.02.001
Soto, D. A., and Ross, P. J. (2016). Pluripotent stem cells and livestock genetic engineering. Transgenic Res. 25, 289–306. doi: 10.1007/s11248-016-9929-5
Speedy, A. W. (2003). Global production and consumption of animal source foods. J. Nutrition 133, 4048S−4053S. doi: 10.1093/jn/133.11.4048S
Stebler, J., Spieler, D., Slanchev, K., Molyneaux, K. A., Richter, U., Cojocaru, V., et al. (2004). Primordial germ cell migration in the chick and mouse embryo: the role of the chemokine SDF-1/CXCL12. Dev. Biol. 272, 351–361. doi: 10.1016/j.ydbio.2004.05.009
Stern, C. D. (1990). The marginal zone and its contribution to the hypoblast and primitive streak of the chick embryo. Development 109, 667–682.
Stern, C. D. (2004). The Chick Embryo–Past, Present and Future as a Model System in Developmental Biology. London: Elsevier.
Stern, C. D. (2005). The chick; a great model system becomes even greater. Dev. Cell 8, 9–17. doi: 10.1016/j.devcel.2004.11.018
Sun, Y., Ding, N., Ding, S. S., Yu, S., Meng, C., Chen, H., et al. (2013). Goose RIG-I functions in innate immunity against Newcastle disease virus infections. Mol. Immunol. 53, 321–327. doi: 10.1016/j.molimm.2012.08.022
Szüts, D., Simpson, L. J., Kabani, S., Yamazoe, M., and Sale, J. E. (2006). Role for RAD18 in homologous recombination in DT40 cells. Mol. Cell. Biol. 26, 8032–8041. doi: 10.1128/MCB.01291-06
Talbot, N. C., and Blomberg, L. A. (2008). The pursuit of ES cell lines of domesticated ungulates. Stem Cell Rev. 4, 235–254. doi: 10.1007/s12015-008-9026-0
Tanaka, S. S., Toyooka, Y., Akasu, R., Katoh-Fukui, Y., Nakahara, Y., Suzuki, R., et al. (2000). The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14, 841–853. doi: 10.1101/gad.14.7.841
Taylor, L., Carlson, D. F., Nandi, S., Sherman, A., Fahrenkrug, S. C., and Mcgrew, M. J. (2017). Efficient TALEN-mediated gene targeting of chicken primordial germ cells. Development 144, 928–934. doi: 10.1242/dev.145367
Thomas, K. R., and Capecchi, M. R. (1987). Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503–512. doi: 10.1016/0092-8674(87)90646-5
Trefil, P., Aumann, D., Koslová, A., Mucksová, J., Benešová, B., Kalina, J., et al. (2017). Male fertility restored by transplanting primordial germ cells into testes: a new way towards efficient transgenesis in chicken. Sci. Rep. 7:14246. doi: 10.1038/s41598-017-14475-w
Trefil, P., Micáková, A., Mucksová, J., Hejnar, J., Poplstein, M., Bakst, M. R., et al. (2006). Restoration of spermatogenesis and male fertility by transplantation of dispersed testicular cells in the chicken. Biol. Reprod. 75, 575–581. doi: 10.1095/biolreprod.105.050278
Tyack, S. G., Jenkins, K. A., O'neil, T. E., Wise, T. G., Morris, K. R., Bruce, M. P., et al. (2013). A new method for producing transgenic birds via direct in vivo transfection of primordial germ cells. Trans. Res. 22, 1257–1264. doi: 10.1007/s11248-013-9727-2
Uckun, F. M., Waddick, K. G., Mahajan, S., Jun, X., Takata, M., Bolen, J., et al. (1996). BTK as a mediator of radiation-induced apoptosis in DT-40 lymphoma B cells. Science 273, 1096–1100. doi: 10.1126/science.273.5278.1096
Van De Lavoir, M.-C., Collarini, E. J., Leighton, P. A., Fesler, J., Lu, D. R., Harriman, W. D., et al. (2012). Interspecific germline transmission of cultured primordial germ cells. PLoS ONE 7:e35664. doi: 10.1371/journal.pone.0035664
Van De Lavoir, M. C., Diamond, J. H., Leighton, P. A., Mather-Love, C., Heyer, B. S., Bradshaw, R., et al. (2006). Germline transmission of genetically modified primordial germ cells. Nature 441, 766–769. doi: 10.1038/nature04831
Wang, K., Jin, Q., Ruan, D., Yang, Y., Liu, Q., Wu, H., et al. (2017). Cre-dependent Cas9-expressing pigs enable efficient in vivo genome editing. Genome Res. 27, 2061–2071. doi: 10.1101/gr.222521.117
Wernery, U., Liu, C., Baskar, V., Guerineche, Z., Khazanehdari, K. A., Saleem, S., et al. (2010). Primordial germ cell-mediated chimera technology produces viable pure-line Houbara bustard offspring: potential for repopulating an endangered species. PLoS ONE 5:e15824. doi: 10.1371/journal.pone.0015824
Whitworth, K. M., Rowland, R. R., Ewen, C. L., Trible, B. R., Kerrigan, M. A., Cino-Ozuna, A. G., et al. (2015). Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22. doi: 10.1038/nbt.3434
Whyte, J., Glover, J. D., Woodcock, M., Brzeszczynska, J., Taylor, L., Sherman, A., et al. (2015). FGF, insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Rep. 5, 1171–1182. doi: 10.1016/j.stemcr.2015.10.008
Wilson, N. H., and Stoeckli, E. T. (2012). In ovo electroporation of miRNA-based plasmids in the developing neural tube and assessment of phenotypes by DiI injection in open-book preparations. J. Vis. Exp. 16:4384. doi: 10.3791/4384
Xu, W., Shao, Q., Zang, Y., Guo, Q., Zhang, Y., and Li, Z. (2015). Pigeon RIG-I function in innate immunity against H9N2 IAV and IBDV. Viruses 7, 4131–4151. doi: 10.3390/v7072813
Yasugi, S., and Nakamura, H. (2000). Gene transfer into chicken embryos as an effective system of analysis in developmental biology. Dev. Growth Differ. 42, 195–197. doi: 10.1046/j.1440-169x.2000.00500.x
Yeh, S., Tsai, M.-Y., Xu, Q., Mu, X.-M., Lardy, H., Huang, K.-E., et al. (2002). Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc. Natl. Acad. Sci.U.S.A. 99, 13498–13503. doi: 10.1073/pnas.212474399
Zazzeroni, F., Papa, S., De Smaele, E., and Franzoso, G. (2003). Cell signalling: cell survival and a Gadd45-factor deficiency: reply. Nature 424, 741–742. doi: 10.1038/424741b
Zhang, X., Yan, Z., Li, X., Lin, W., Dai, Z., Yan, Y., et al. (2016). GADD45β, an anti-tumor gene, inhibits avian leukosis virus subgroup J replication in chickens. Oncotarget 7, 68883–68893. doi: 10.18632/oncotarget.12027
Keywords: chicken, CRISPR/Cas9, transgenic, knockout, Diseases, Immunoglobulins
Citation: Sid H and Schusser B (2018) Applications of Gene Editing in Chickens: A New Era Is on the Horizon. Front. Genet. 9:456. doi: 10.3389/fgene.2018.00456
Received: 29 May 2018; Accepted: 18 September 2018;
Published: 09 October 2018.
Edited by:
Mark S. Fife, Pirbright Institute (BBSRC), United KingdomReviewed by:
Robert Etches, Crystal Bioscience, United StatesJiuzhou Song, University of Maryland, College Park, United States
Copyright © 2018 Sid and Schusser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin Schusser, YmVuamFtaW4uc2NodXNzZXJAdHVtLmRl
 Hicham Sid
Hicham Sid Benjamin Schusser
Benjamin Schusser