
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 24 September 2018
Sec. Livestock Genomics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00386
 ) × Megalobrama amblycephala (
) × Megalobrama amblycephala ( )
) Liu Cao1,2†
Liu Cao1,2† QinBo Qin1,2†
QinBo Qin1,2† Qiong Xiao1,2†
Qiong Xiao1,2† HongTing Yin2†
HongTing Yin2† Jin Wen2†
Jin Wen2† QiWen Liu1,2
QiWen Liu1,2 Xu Huang1,2
Xu Huang1,2 YangYang Huo1,2
YangYang Huo1,2 Min Tao1,2
Min Tao1,2 Chun Zhang1,2
Chun Zhang1,2 Kaikun Luo1,2
Kaikun Luo1,2 ShaoJun Liu1,2*
ShaoJun Liu1,2*Nucleolar dominance is related to the expression of 45S rRNA genes inherited from one progenitor due to the silencing of the other progenitor’s rRNA genes. To investigate nucleolar dominance associated with tetraploidization, we analyzed the changes regarding the genetic traits and expression of 45S rRNA genes in tetraploidy hybrid lineage including F1 allotetraploids (4n = 148) and F2 autotetraploids (4n = 200) derived from the distant hybridization of Carassius auratus red var. (2n = 100) ( ) × Megalobrama amblycephala (2n = 48) (
) × Megalobrama amblycephala (2n = 48) ( ). Results showed that nucleolar dominance from the females was established in F1 hybrids and it was inherited in F2 hybrids, suggesting that tetraploidization can lead to rapid establishment of nucleolar dominance in the hybrid origin’s tetraploid lineage. These results extend the knowledge of nucleolar dominance in polyploidy hybrid animals, which are of significance for the evolution of hybrids in vertebrates.
). Results showed that nucleolar dominance from the females was established in F1 hybrids and it was inherited in F2 hybrids, suggesting that tetraploidization can lead to rapid establishment of nucleolar dominance in the hybrid origin’s tetraploid lineage. These results extend the knowledge of nucleolar dominance in polyploidy hybrid animals, which are of significance for the evolution of hybrids in vertebrates.
Polyploidy species have played a major role in the evolution and the adaptation of eukaryotes in both animals (Muller, 1925; Peer et al., 2009) and plants (Levy and Feldman, 2002; Soltis et al., 2009; Jiao et al., 2011). Hybridization is one of the primary mechanisms for the origin of species leading to the formation of polyploids (Otto and Whitton, 2000; Liu, 2010). In the newly formed allopolyploids, the genomes changes react in some genomic reorganizations and modifications of parental genomes. It is assumed that these genomic changes facilitate the establishment and success of the newly formed polyploids (Song et al., 1995; Ozkan et al., 2001; Kashkush et al., 2002; Raskina et al., 2002). In previous studies, fertile allotetraploids (abbreviated as F1) (AABB, 4n = 148) were successfully obtained in the first generation derived from the distant hybridization of Carassius auratus red var. (RCC) (RR, 2n = 100) ( ) × Megalobrama amblycephala (BSB) (BB, 2n = 48) (
) × Megalobrama amblycephala (BSB) (BB, 2n = 48) ( ) as a result of chromosome doubling of diploid hybrid embryos (AB, 2n = 74) (Liu et al., 2007). Fertile autotetraploids (abbreviated as F2) (AAAA, 4n = 200) were obtained in the second generation through crossing autodiploid sperm and autodiploid ova produced by abnormal chromosome behavior during meiosis of F1 hybrids (Qin et al., 2014). In contrast with the F1 hybrids, the F2 hybrids have increasing the fertility and reach sexual maturity at 1 year old.
) as a result of chromosome doubling of diploid hybrid embryos (AB, 2n = 74) (Liu et al., 2007). Fertile autotetraploids (abbreviated as F2) (AAAA, 4n = 200) were obtained in the second generation through crossing autodiploid sperm and autodiploid ova produced by abnormal chromosome behavior during meiosis of F1 hybrids (Qin et al., 2014). In contrast with the F1 hybrids, the F2 hybrids have increasing the fertility and reach sexual maturity at 1 year old.
By self-mating, F2 hybrids can generate the next generation of the autotetraploids. Until now, a stable autotetraploid line (F2 – F13; 4n = 200) is formed which can provides an excellent materials to investigate the mechanisms that drive diploidization in autotetraploids and is useful in the production of the sterile triploids (Qin et al., 2014). This hybrid origin’s tetraploid lineage is an attractive model to elucidate genomic changes associated with tetraploidization.
Nucleoli, the sites of 45S rRNA gene transcription (the 5.8S, 18S, and 28S rRNA) and ribosome assembly, form at the chromosome loci where tandemly repeated 45S rRNA genes are actively transcribed (Reeder, 1985; Comai, 2000). In a number of interspecific hybrids, nucleolar dominance describes expression of 45S rRNA genes inherited from one progenitor while silencing of the other progenitor’s rRNA genes (Tucker et al., 2010). About 70 years of research has contributed to our current understanding of nucleolar dominance, through studies based on plants, invertebrates, frogs, flies, fish, and mammals (Mcstay, 2006; Preuss and Pikaard, 2007). The transcriptional dominance of the ribosomal genes of one species over the ribosomal genes of another species was first described in some interspecific hybrids of Crepis (Honjo and Reeder, 1973), and was then confirmed by the studies of other hybrids or allopolyploid species (Lacadena et al., 1984; Reeder, 1985; Gustafson et al., 1988). At present, the knowledge of understanding the nucleolar dominance in polyploid animals is still limited, because the formation of distant polyploid hybrid lineages in animals is challenging in practice.
The genetics traits and expression of 45S rRNA genes have been studied by detecting single nucleotide polymorphism (SNP) sites in the 18S rRNA gene (Flowers and Burton, 2006; Xiao et al., 2016). Nucleotide sequence variation in the internal transcribed spacer (ITS) region of the 45S rRNA genes has been the candidate of choice for species identification (Jobes and Thien, 1997; Hajibabaei et al., 2007; Son et al., 2009) and phylogenomic analyses (Yonemori et al., 2002; Liu et al., 2008). Here, we analyzed the genetic traits and expression of 45S rRNA genes in RCC, BSB, F1 and F2 hybrids. The results showed that nucleolar dominance from the females was established in F1 hybrids and it was inherited in F2 hybrids, suggesting that tetraploidization can lead to rapid establishment of nucleolar dominance in the hybrid origin’s tetraploid lineage. These results extend the knowledge of nucleolar dominance in polyploidy hybrid animals, which are of significance for the evolution of hybrids in vertebrates.
The allotetraploids (F1, AABB, 4n = 148) were obtained in the first generation of the distant hybridization of Carassius auratus red var. (RCC, AA, 2n = 100  ) × Megalobrama amblycephala (BSB, BB, 2n = 48
) × Megalobrama amblycephala (BSB, BB, 2n = 48 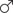 ) as a result of chromosome doubling of diploid hybrid embryos (AB, 2n = 74) (Liu et al., 2007). The autotetraploids (F2, AAAA, 4n = 200) were obtained in the second generation through crossing autodiploid sperm and autodiploid ova produced by abnormal chromosome behavior during meiosis of F1 hybrids (Qin et al., 2014). All samples were reared and bred at the Engineering Research Center of Polyploid Fish Breeding and Reproduction of the State Education Ministry, China, located at Hunan Normal University.
) as a result of chromosome doubling of diploid hybrid embryos (AB, 2n = 74) (Liu et al., 2007). The autotetraploids (F2, AAAA, 4n = 200) were obtained in the second generation through crossing autodiploid sperm and autodiploid ova produced by abnormal chromosome behavior during meiosis of F1 hybrids (Qin et al., 2014). All samples were reared and bred at the Engineering Research Center of Polyploid Fish Breeding and Reproduction of the State Education Ministry, China, located at Hunan Normal University.
To determine ploidy, a 5S gene probe was made from RCC genome and amplified by PCR using the primers 5′-TTCGAAAAGAGAGAATAATCTA-3′ and 5′-AACTCGTCTAAACC CGAACTA-3′ (Qin et al., 2010). The fluorescence in situ hybridization (FISH) probes were produced by Dig-11-dUTP labeling (using a nick translation kit; Roche, Germany) of purified PCR products. FISH was performed according to He et al. (2012). Two hundred metaphase chromosome spreads from 10 individuals were analyzed for each type of fish (RCC, BSB, F1 and F2 hybrids). Preparations were examined under an inverted microscope (CW4000, Leica, Germany), with a confocal imaging system (LCS SP2, Leica). Captured images were colored and superimposed in Adobe Photoshop CS6.
The ITS sequences from different genomes were amplified using ITS primers (5′-AGTCGTAACAAGGTTTCCGTAGGTG-3′; 3′-TTATGGCCGTGCTCTGGCTAT-5′), with DNA and cDNA as templates. This pair of primers was designed based on the 45S sequence of common carp (GenBank, Accession No. JN628435.1). Unique cleavage sites for restriction enzymes (NotI, DraI, StuI; New England Biolabs) were located in ITS sequences from different genomes using a sequence alignment program (GeneTool) (Beisvag et al., 2006). Three individuals for each type of fish (RCC, BSB, F1 and F2 hybrids) were used to validate these restriction enzymes sites.
A pair of primers reported by Singh et al. (2009) (18S, 5′-TTGGTGACTCTCGATAACCTCGGGC-3′; 18S, 5′-CCTTGTTACGACTTTTACTTCCTC-3′) was used to amplify the 18S rDNA fragment. Genomic DNA and total RNA were simultaneously isolated from single adult fish. Thirty individuals of each fish (including RCC, BSB, F1 and F2 hybrids) were selected at random. DNA was extracted with a universal genomic DNA extraction kit (TaKaRa). The liver tissue of different individuals was used to isolate total RNA using a Total RNA Isolation System (Omega Bio-Tek, Norcross, GA, United States). RNA was treated with gDNA Eraser (TaKaRa) and first-strand cDNA was synthesized with Superscript III (Invitrogen) using an 18S rRNA gene-specific primer (5′-CCTTGTTACGACTTTTACTTCCTC-3′). Then the 18S F/P primer pair was used to amplify the cDNA fragment of 18S rDNA by PCR. For all samples, PCR controls were used to detect the contamination of RNA with genomic DNA. Sequences were analyzed with BioEdit (Hall, 1999) and Clustal W (Thompson et al., 1994). To examine the SNP sites, we designed three pairs of primers according to 100 upstream and downstream nucleotides in the SNP sites (Supplementary Table S1). Except the forward primer in position 41 was designed according to 200 upstream nucleotides.
The 5S rDNA probe (477 bp, GenBank: GQ485557) hybridized with the metaphase chromosomes of RCC, BSB, F1 and F2 hybrids. Hybridization of the 5S rDNA probe yielded eight 5S gene loci in RCC (Figure 1A and Table 1), but none in BSB (Figure 1B and Table 1); thus, RCC and BSB-derived chromosomes could be discriminated by the 5S rDNA probe. In the F1 hybrids, eight RCC-derived 5S rDNA gene loci were detected in the metaphase chromosomes (Figure 1C and Table 1). Sixteen RCC-derived 5S rDNA gene loci were detected in the metaphase chromosomes of the F2 hybrids (Figure 1D and Table 1).
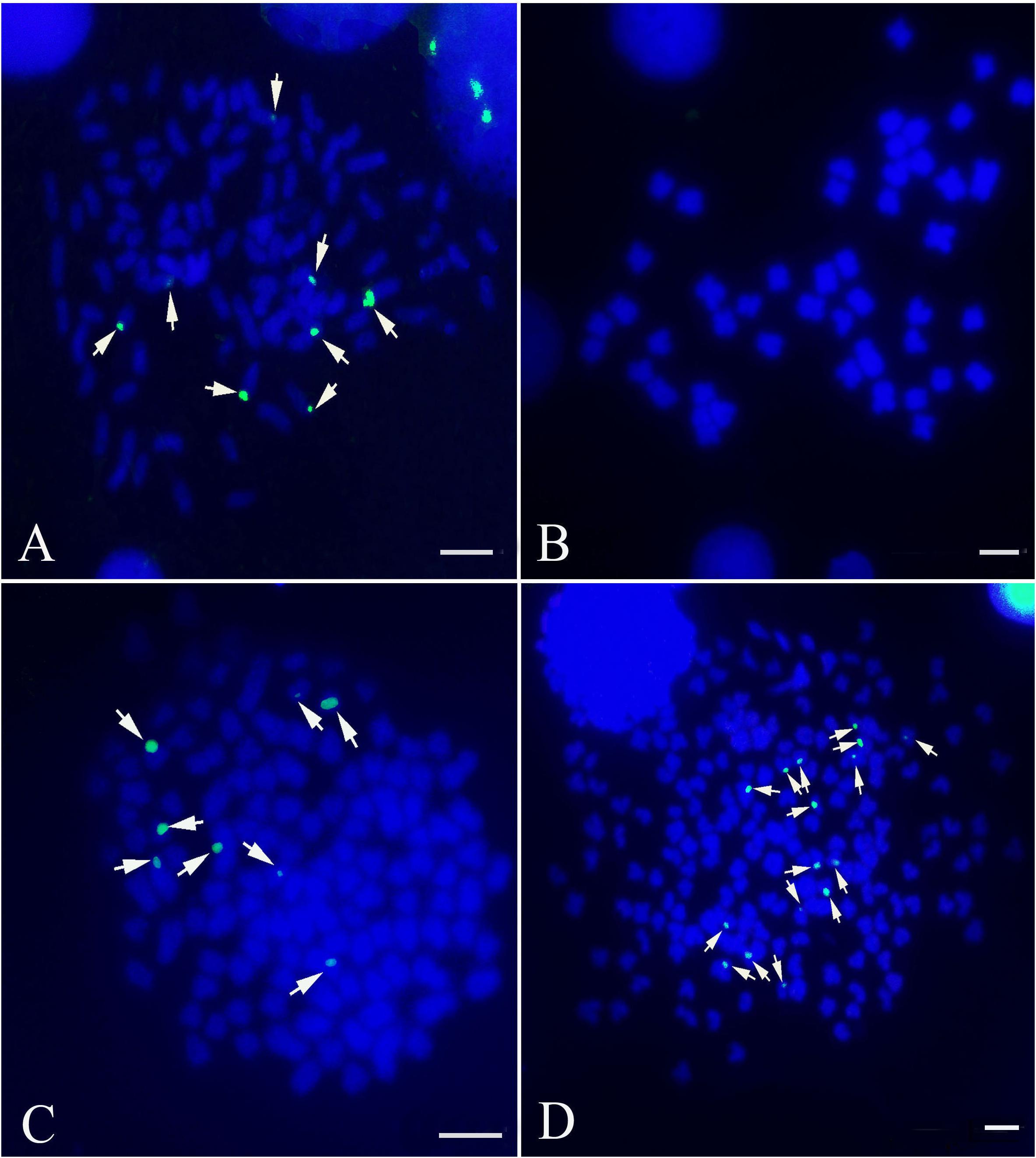
FIGURE 1. Examination of hybridizing signals by the 5S rDNA probe. The white arrows indicate the 5S rDNA gene loci. The eight 5S gene loci were found in RCC (A), but none in BSB (B). The eight 5S gene loci in F1 (C) and sixteen 5S gene loci in F2 (D) are shown. Bars (A–D): 3 μm. RCC, Carassius auratus red var.; BSB, Megalobrama amblycephala; F1, allotetraploids; F2, autotetraploids.
Internal transcribed spacer sequence polymorphisms allow distinct nucleolar organizing regions (NORs) to be identified via specific restriction enzyme sites in parental individuals and polyploid hybrid progeny (Figure 2). Lengths of DNA fragments (including partial 18S rDNA, complete ITS regions, and partial 28S rDNA sequences) were amplified from the BSB (1296 bp) (Accession No. MG830472) and RCC (1283 bp) (Accession No. MG830471). In the F1 and F2 hybrids, the length of the ITS region was 1283 bp (Figure 2A). The NotI restriction enzyme could digest the ITS region into two smaller bands in the RCC (904 and 379 bp), and in the F1 (904 and 379 bp) and F2 (904 and 379 bp) hybrids, but not in the BSB (Figure 2B). The DraI restriction site was used because it was found in the ITS region of BSB (1039 bp), but not in the ITS region of RCC, or of the F1 or F2 hybrids (Figure 2C). The StuI restriction site could be found in the ITS region of both BSB (658 bp) and RCC (926 bp). In the F1 and F2 hybrids only a proportion of the individuals carried the StuI restriction site, although where it was present was in the same position as in the RCC (Figure 2D). (Note: Three samples showed the same results.)
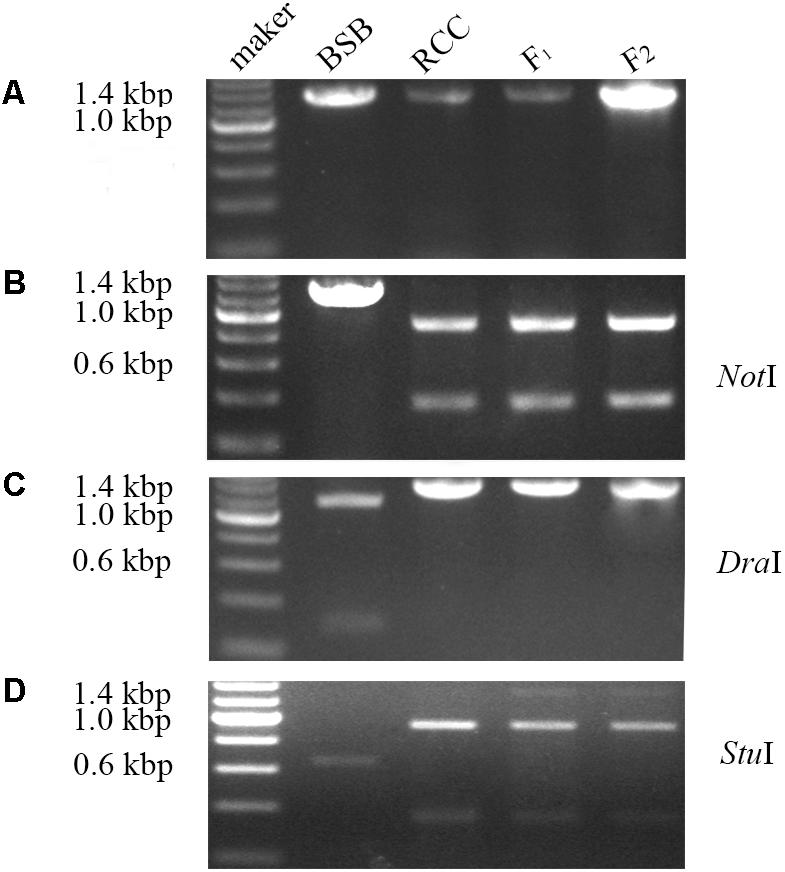
FIGURE 2. Analysis of the inheritance pattern of complete ITS region in distant hybrid lineage of RCC × BSB. (A) The complete DNA sequence of ITS from distant hybrid lineage of RCC × BSB without digestion by enzyme. (B) The digested product by NotI from distant hybrid lineage of RCC × BSB. (C) The digested product by DraI from distant hybrid lineage of RCC × BSB. (D) The digested product by StuI from distant hybrid lineage of RCC × BSB.
Expression of the complete ITS sequence was amplified in BSB (1296 bp), RCC (1283 bp), and the F1 (1283 bp) and F2 (1283 bp) hybrids (Figure 3A). A NotI restriction site was found in the cDNA fragment of the ITS region in RCC (904 bp), but not in BSB. Both the F1 and F2 hybrids carried a restriction site in the same position as RCC, although in some of the F2 hybrids the position of this restriction site varied (Figure 3B). A DraI restriction site was found in the cDNA fragment of the ITS region in BSB (1039 bp), but not in RCC, or the F1 and F2 hybrids (Figure 3C). A StuI restriction site was found in the cDNA fragment of the ITS regions of both BSB (658 bp), RCC (926 bp), the F1 (926 bp) and F2 (926 bp) hybrids (Figure 3D). (Note: Three samples showed the same results.)
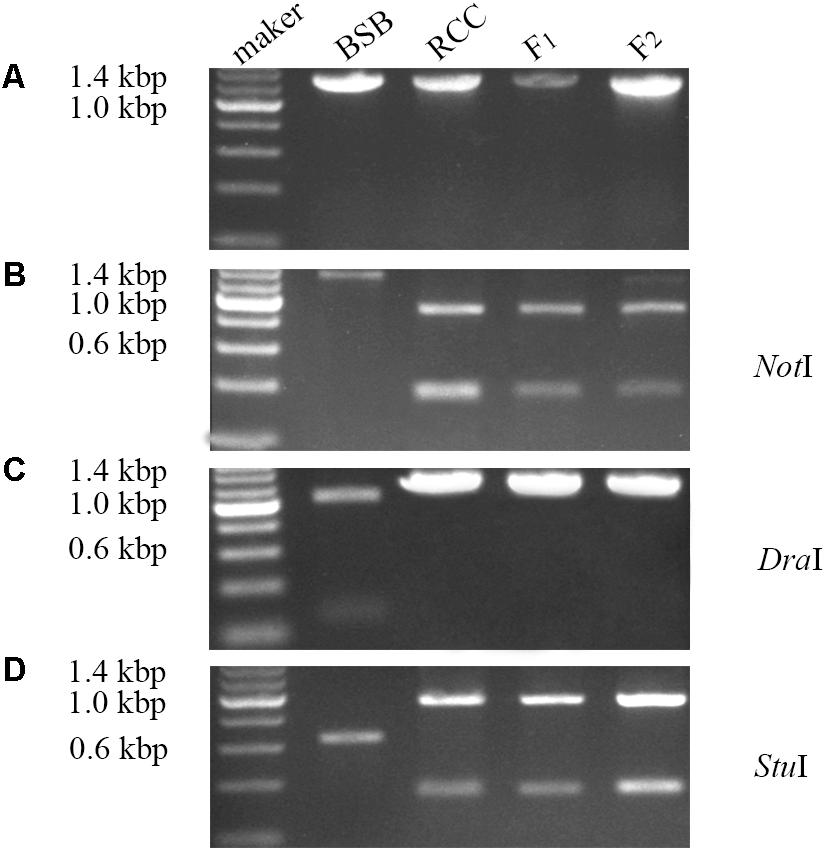
FIGURE 3. Analysis of the expression pattern of complete ITS region in distant hybrid lineage of RCC × BSB. (A) The complete cDNA sequence of ITS from distant hybrid lineage of RCC × BSB without digestion by enzyme. (B) The digested product by NotI from distant hybrid lineage of RCC × BSB. (C) The digested product by DraI from distant hybrid lineage of RCC × BSB. (D) The digested product by StuI from distant hybrid lineage of RCC × BSB.
We examined the intra- and interspecific sequence divergence for 60 sequences from 30 RCC individuals and 60 sequences from 30 BSB individuals. The sequence of the 18S rRNA gene in RCC was 1558 bp (Accession No. MG830470). In BSB, the sequence of the 18S rRNA gene was homogeneous with the sequence provided by Xiao et al. (2016) (Accession No. AB860215), except for position 12 (indicated with asterisk in Supplementary Figure S1). The similarity between 18S rRNA gene fragments amplified from RCC and BSB genome was 97.00% (Table 2 and Supplementary Figure S1).
Thirty samples each of fish (RCC, BSB, F1 and F2 hybrids) were selected. According to genotypes of 18S rRNA gene, the inheritance pattern in hybrids fell into two classes. In pattern 1, the DNA sequence of the 18S rRNA gene was consistent with that of RCC. In pattern 2, the DNA sequence of the 18S rRNA gene had variations in four positions compared with that of RCC: position 41 (T→C), 486 (A→G), 1124 (T→C), and 1157 (T→C) (Figure 4 and Supplementary Figure S1). The proportion of F1 hybrids belonging to pattern 1 was 76.67% and the proportion belonging to pattern 2 was 23.33% (Table 3). The proportion of F2 hybrids belonging to pattern 1 was 73.33% and the proportion belonging to pattern 2 was 26.67% (Table 4). The four positions 41, 486, 1124, and 1157 in the F1 and F2 hybrids were examined by primers (Supplementary Table S1 and Supplementary File S1). The cDNAs of the 18S rRNA gene in all the F1 and F2 hybrids were consistent with that of RCC, whether they belonged to pattern 1 or 2 (Figure 4).
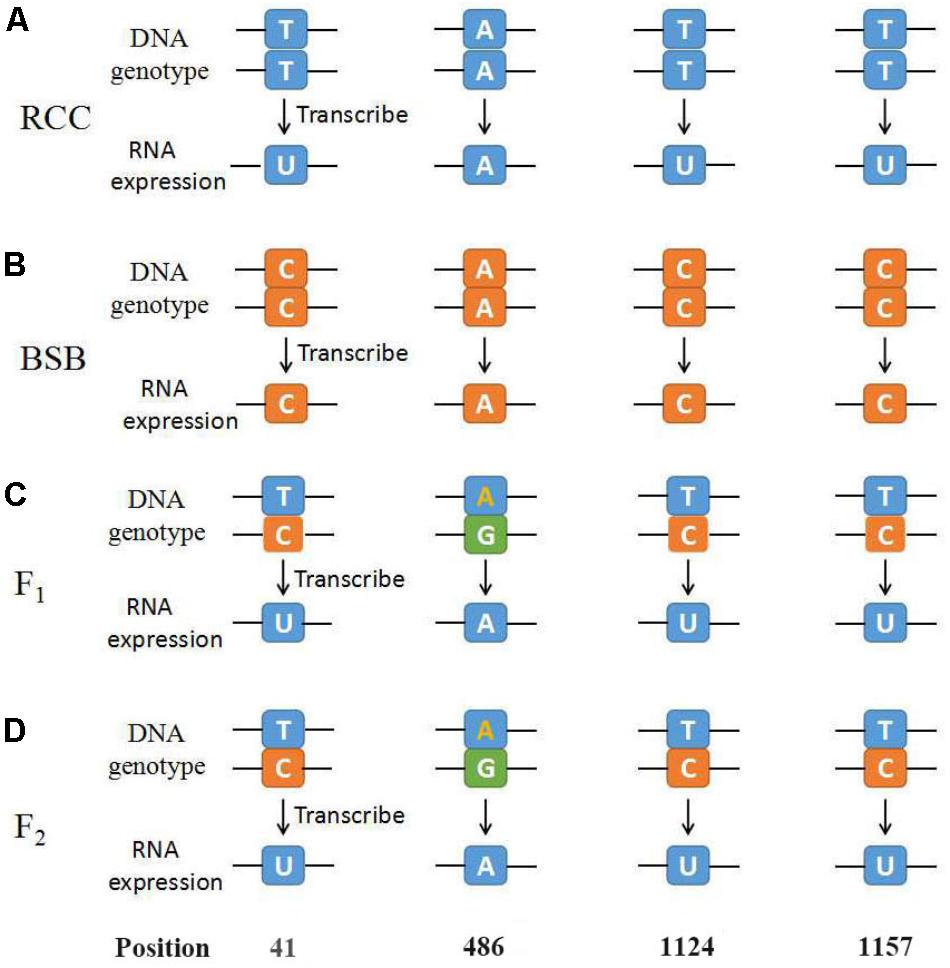
FIGURE 4. The diagrammatic drawing of genotype and expression of 45S rRNA gene at position 41, 486, 1124, and 1157. (A) The genotype and expression of 45S rRNA gene in RCC. (B) The genotype and expression of 45S rRNA gene in BSB. (C) The genotype and expression of 45S rRNA gene in F1 hybrids of RCC × BSB. (D) The genotype and expression of 45S rRNA gene in F2 hybrids of RCC × BSB. For every pattern of each kind of fish, one sample was used to present.
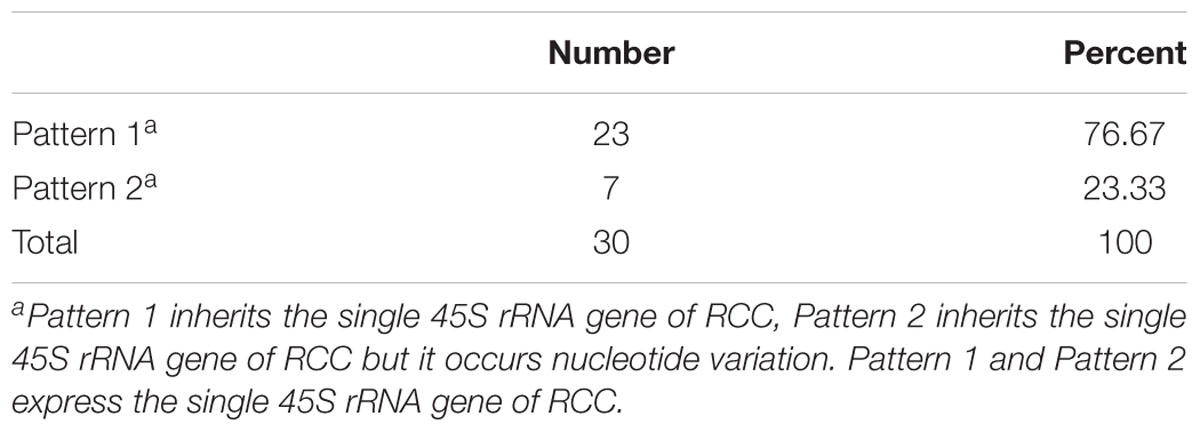
TABLE 3. The frequency distribution of the 2 different genetic expression patterns of 45S rRNA gene in the F1 hybrids.
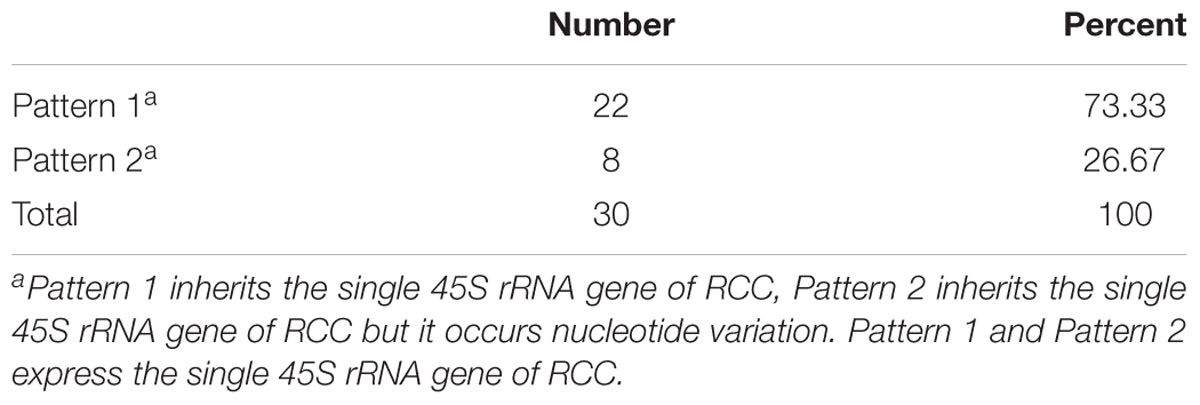
TABLE 4. The frequency distribution of the two different genetic expression patterns of 45S rRNA gene in the F2 hybrids.
The 45S rRNA gene is a classic locus for the study of both genomic structure and expression levels in polyploids, as part of a study of the more general topic of nucleolar dominance. The knowledge of understanding the nucleolar dominance in hybrid origin’s fish and polyploidy livestock is scarce. The limited data indicate that nucleolar dominance have established in the red vizcacha rat (Gallardo et al., 2006). However, the nucleolar dominance established in a hybrids fish lineage is not realized until the second generation (Xiao et al., 2016). In the present study, we observed that in the F1 hybrids of RCC × BSB, the inherited and expressed 45S rRNA genes were derived from the females, RCC. Thus, preferential expression evolved rapidly in the initial generations. The F1 hybrids can successfully survive in the first generation of RCC × BSB, but diploids hybrids cannot, probably because of the incompatibility of paternal genome (Qin et al., 2015). Polyploidization-associated genomic changes have been found in F1 hybrids (Qin et al., 2016). Thus, we speculate that tetraploidization facilitate rapid establishment of nucleolar dominance in F1 hybrids. As reported, genetics changes in the hybrids may have some relationship with the unique nucleolar dominance patterns in the hybrid lineage (Xiao et al., 2016). In Xenopus hybrids, nucleolar dominance appears to be associated with a major non-Mendelian reduction in the number of 45S rRNA gene copies rather than a specific pattern of their expression. The loss of rRNA gene copies in F1 hybrids was non-random with respect to the parental species, with the transcriptionally dominant variant preferentially removed from hybrid zygotes (Katarzyna et al., 2015). The “epigenetic landscape” created by cytosine methylation and various histone modification also can play an important role in selective silencing of rRNA genes (Chen and Pikaard, 1997; Olga et al., 2007). The specific molecular basis for choosing 45S rRNA genes of RCC to express in F1 hybrids still needs further exploration.
Four single nucleotide variations were observed in the DNA sequences of 18S rRNA gene of F1 hybrids. Incomplete homogenization of 18S ribosomal DNA sequences have been reported in some studies (Mentewab et al., 2011; Chelomina et al., 2016). Polyploidy and hybridization are the most often discussed reasons for nucleotide variability in the rDNA sequences (Muir et al., 2001; Liu et al., 2003). Thus, tetraploidization and distant hybridization may be responsible for the appearance of this nucleotide variation. Exosome mediated quality control of misfolded pre-rRNAs following their polyadenylation (Kadaba et al., 2006; Slomovic et al., 2006; Chekanova et al., 2007) as well as the ‘non-functional rRNA decay’ leading to decreased stability of the mature rRNA contained in fully assembled ribosomes and ribosomal subunits (Lariviere et al., 2006). These mechanisms can explain that cDNAs of the 18S rRNA gene in all the F1 and F2 hybrids were consistent with that of RCC, whether they belonged to pattern 1 or 2. It was further confirmed by the digestion products of the restriction enzyme in the complete ITS region. In addition, we found an unrecognized band due to the NotI enzyme in the cDNA of the complete ITS of F2 hybrids. Because the 5.8S region in all parents and hybrids was conservation, the variant restriction enzyme site existed in ITS region. The F2 hybrids were produced through the elimination of BSB genetic material and an autopolyploidization process. It is reasonable to assume that these genomic changes cause variation of the cDNA of the ITS region in the F2 hybrids.
Nucleolar dominance from the females was established in the F1 hybrids and it was inherited in the F2 hybrids, suggesting that tetraploidization can lead to rapid establishment of nucleolar dominance in the hybrid origin’s tetraploid lineage. These results extend the knowledge of nucleolar dominance in polyploidy hybrid animals, which are of significance for the evolution of hybrids in vertebrates.
All the fish were cultured in ponds at the Protection Station of Polyploid Fish, Hunan Normal University. According to the Care and Use of Agricultural Animals in Agricultural Research and Teaching, fish treatments were performed. These were approved through the Science and Technology Bureau of China. Approval from the Department of Wildlife Administration was not required for the experiments conducted in this study. Before dissection, fish were deeply anesthetized with 100 mg/L MS-222 (Sigma-Aldrich).
SL contributed to the conception and designed the study. LC, QQ, QX, HY, and JW carried out the experimental work and participated in drafting the manuscript. QL, XH, and YH analyzed the sequences. MT, CZ, and KL participated in interpretation and discussion of the results. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31430088 and 31730098), the earmarked fund for China Agriculture Research System (Grant No. CARS-45), the Hunan Provincial Natural Science and Technology Major Project (Grant No. 2017NK1031), the Key Research and Development Program of Hunan Province (Grants No. 2018NK2072), the Natural Science Foundation of Hunan Province for Distinguished Young Scholars (Grant No. 2017JJ1022), the Major Program of the Educational Commission of Hunan Province (Grant No. 17A133), and the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (Grant No. 20134486).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to sincerely thank many researchers who help to complete this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00386/full#supplementary-material
FIGURE S1 | Sequence alignment of 18S rDNA of pattern 2 in RCC, BSB, F1 and F2 hybrids. Variable sites are shaded; asterisks mark position 12.
TABLE S1 | The primers are designed to examine the position 41, 486, 1124, and 1157 in F1 and F2 hybrids.
FILE S1 | (A) The sequencing peak figures of the PCR products amplified from genomic of 18S rRNA gene. Only the position 41 and 2 upstream and downstream nucleotides are showed. (B) The sequencing peak figures of the PCR products amplified from genomic of 18S rRNA gene. Only the position 486 and 2 upstream and downstream nucleotides are showed. (C) The sequencing peak figures of the PCR products amplified from genomic of 18S rRNA gene. Only the position 1124 and 2 upstream and downstream nucleotides are showed. (D) The sequencing peak figures of the PCR products amplified from genomic of 18S rRNA gene. Only the position 1157 and 2 upstream and downstream nucleotides are showed.
Beisvag, V., Jünge, F. K., Bergum, H., Jølsum, L., Lydersen, S., Günther, C. C., et al. (2006). GeneTools – application for functional annotation and statistical hypothesis testing. BMC Bioinformatics 7:470. doi: 10.1186/1471-2105-7-470
Chekanova, J. A., Gregory, B. D., Reverdatto, S. V., Chen, H., Kumar, R., Hooker, T., et al. (2007). Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131, 1340–1353. doi: 10.1016/j.cell.2007.10.056
Chelomina, G. N., Rozhkovan, K. V., Voronova, A. N., Burundukova, O. L., Muzarok, T. I., and Zhuravlev, Y. N. (2016). Variation in the number of nucleoli and incomplete homogenization of 18S ribosomal DNA sequences in leaf cells of the cultivated Oriental ginseng (Panax ginseng Meyer). J. Ginseng Res. 40, 176–184. doi: 10.1016/j.jgr.2015.07.005
Chen, Z. J., and Pikaard, C. S. (1997). Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11, 2124–2136.
Comai, L. (2000). Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43, 387–399.
Flowers, J. M., and Burton, R. S. (2006). Ribosomal RNA gene silencing in interpopulation hybrids of Tigriopus californicus: nucleolar dominance in the absence of intergenic spacer subrepeats. Genetics 173, 1479–1486. doi: 10.1534/genetics.106.058719
Gallardo, M. H., González, C. A., and Cebrián, I. (2006). Molecular cytogenetics and allotetraploidy in the red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae). Genomics. 88, 214–221. doi: 10.1016/j.ygeno.2006.02.010
Gustafson, J. P., Dera, A. R., and Petrovic, S. (1988). Expression of modified rye ribosomal RNA genes in wheat. Proc. Natl. Acad. Sci. U.S.A. 85, 3943–3945.
Hajibabaei, M., Singer, G. A. C., Hebert, P. D. N., and Hickey, D. A. (2007). DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 23, 167–172. doi: 10.1016/j.tig.2007.02.001
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
He, W. G., Qin, Q. B., Liu, S. J., Li, T. L., Wang, L., Xiao, J., et al. (2012). Organization and variation analysis of 5S rDNA in different ploidy-level hybrids of red crucian carp × topmouth culter. PLoS One 7:e38976. doi: 10.1371/journal.pone.0038976
Honjo, T., and Reeder, R. H. (1973). Preferential transcription of Xenopus laevis ribosomal RNA in interspecies hybrids between Xenopus laevis and Xenopus mulleri. J. Mol. Biol 80, 217–228. doi: 10.1016/0022-2836(73)90168-X
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Jobes, D. V., and Thien, L. B. (1997). A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) Sequences. Plant Mol. Biol. Rep. 15, 326–334.
Kadaba, S., Wang, X., and Anderson, J. T. (2006). Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12, 508–521. doi: 10.1261/rna.2305406
Kashkush, K., Feldman, M., and Levy, A. A. (2002). Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160, 1651–1659.
Katarzyna, M., Sebastian, M., Young Bun, K., Gracielaet, S., Jung Hun, O., Lin, K., et al. (2015). Nucleolar dominance and maternal control of 45S rDNA expression. Proc. Biol. Sci. 282:20152201. doi: 10.1098/rspb.2015.2201
Lacadena, J. R., Cermeño, M. C., Orellana, J., and Santos, J. L. (1984). Evidence for wheat-rye nucleolar competition (amphiplasty) in triticale by silver-staining procedure. Theor. Appl. Genet. 67, 207–213. doi: 10.1007/BF00317037
Lariviere, F. J., Cole, S. E., Ferullo, D. J., and Moore, M. J. (2006). A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell. 24, 619–626.
Levy, A. A., and Feldman, M. (2002). The impact of polyploidy on grass genome evolution. Plant Physiol. 130, 1587–1593. doi: 10.1104/pp.015727
Liu, Q., Ge, S., Tang, H., Zhang, X., Zhu, G., and Lu, B. R. (2008). Phylogenetic relationships in Elymus (Poaceae: Triticeae) based on the nuclear ribosomal internal transcribed spacer and chloroplast trnL-F sequences. New Phytol. 170, 278–289. doi: 10.1111/j.1469-8137.2006.01665.x
Liu, S., Qin, Q., Xiao, J., Lu, W., Shen, J., Li, W., et al. (2007). The formation of the polyploid hybrids from different subfamily fish crossings and its evolutionary significance. Genetics 176:1023. doi: 10.1534/genetics.107.071373
Liu, S. J. (2010). Distant hybridization leads to different ploidy fishes. Sci. China Life Sci. 53, 416–425. doi: 10.1007/s11427-010-0057-9
Liu, Z. L., Zhang, D., Wang, X. Q., Ma, X. F., and Wang, X. R. (2003). Intragenomic and interspecific 5S rDNA sequence variation in five Asian pines. Am.J. Bot. 90, 17–24. doi: 10.3732/ajb.90.1.17
Mentewab, A. B., Jacobsen, M. J., and Flowers, R. A. (2011). Incomplete homogenization of 18 S ribosomal DNA coding regions in Arabidopsis thaliana. BMC Res. Notes 4:93. doi: 10.1186/1756-0500-4-93
Muir, G., Fleming, C. C., and Schlötterer, C. (2001). Three divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Mol. Biol. Evol. 18, 112–119. doi: 10.1093/oxfordjournals.molbev.a003785
Muller, H. J. (1925). Why polyploidy is rarer in animals than in plants. Am. Nat. 59, 346–353. doi: 10.1111/j.1095-8312.2004.00332.x
Olga, P., Lawrence, R. J., Manuela, S., Sasha, P., Pedro, C. N., Keith, E., et al. (2007). Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS One 2:e1157. doi: 10.1371/journal.pone.0001157
Otto, S. P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. doi: 10.1146/annurev.genet.34.1.401
Ozkan, H., Levy, A. A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13, 1735–1747.
Peer, Y. V. D., Maere, S., and Meyer, A. (2009). The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10:725. doi: 10.1038/nrg2600
Preuss, S., and Pikaard, C. S. (2007). rRNA gene silencing and nucleolar dominance: Insights into a chromosome-scale epigenetic on/off switch. Biochim. Biophys. Acta. 1769, 383–392. doi: 10.1016/j.bbaexp.2007.02.005
Qin, Q., He, W., Liu, S., Wang, J., Xiao, J., and Liu, Y. (2010). Analysis of 5S rDNA organization and variation in polyploid hybrids from crosses of different fish subfamilies. J. Exp. Zool. B Mol. Dev. Evol. 314, 403–411. doi: 10.1002/jez.b.21346
Qin, Q., Lai, Z., Cao, L., Xiao, Q., Wang, Y., and Liu, S. (2016). Rapid genomic changes in allopolyploids of Carassius auratus red var. ( )× Megalobrama amblycephala (
)× Megalobrama amblycephala ( ). Sci Rep 6, 34417. doi: 10.1038/srep34417
). Sci Rep 6, 34417. doi: 10.1038/srep34417
Qin, Q., Wang, J., Dai, J., Wang, Y., and Liu, S. (2015). Induced All-Female Autotriploidy in the Allotetraploids of Carassius auratus red var. ( ) × Megalobrama amblycephala (
) × Megalobrama amblycephala ( ). Marine Biotechnol. 17, 1–9. doi: 10.1007/s10126-015-9647-7
). Marine Biotechnol. 17, 1–9. doi: 10.1007/s10126-015-9647-7
Qin, Q., Wang, Y., Wang, J., Dai, J., Xiao, J., Hu, F., et al. (2014). The Autotetraploid Fish Derived from hybridization of Carassius auratus red var. (Female) × Megalobrama amblycephala (Male). Biol. Reprod. 91:93. doi: 10.1095/biolreprod.114.122283
Raskina, O., Belyayev, A., and Nevo, E. (2002). Repetitive DNas of wild emmer wheat (Triticum dicoccoides) and their relation to S-genome species: molecular cytogenetic analysis. Genome 45, 391–401. doi: 10.1139/g01-142
Reeder, R. H. (1985). Mechanisms of nucleolar dominance in animals and plants. J. Cell Biol. 101:2013–2016.
Singh, M., Kumar, R., Nagpure, N. S., Kushwaha, B., Gond, I., and Lakra, W. S. (2009). Chromosomal localization of 18S and 5S rDNA using FISH in the genus Tor (Pisces, Cyprinidae). Genetica 137, 245–252. doi: 10.1007/s10709-009-9367-x
Slomovic, S., Laufer, D., Geiger, D., and Schuster, G. (2006). Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 34, 2966–2975. doi: 10.1093/nar/gkl357
Soltis, D. E., Albert, V. A., Leebensmack, J., Bell, C. D., Paterson, A. H., Zheng, C., et al. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348. doi: 10.3732/ajb.0800079
Son, S. W., Kim, J. H., Kim, K. J., and Park, S. J. (2009). Molecular evidence for the hybridity of Ilex wandoensis and the phylogenetic study of Korean Ilex based on ITS sequence data. Genes & Genomics. 31, 53–63.
Song, K., Lu, P., Tang, K., and Osborn, T. C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. U.S.A. 92, 7719–7723.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment throughsequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680.
Tucker, S., Vitins, A., and Pikaard, C. S. (2010). Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 22, 351–356. doi: 10.1016/j.ceb.2010.03.009
Xiao, J., Hu, F., Luo, K., Li, W., and Liu, S. (2016). Unique nucleolar dominance patterns in distant hybrid lineage derived from Megalobrama Amblycephala × Culter Alburnus. BMC Genet 17:150. doi: 10.1186/s12863-016-0457-3
Keywords: nucleolar dominance, distant hybridization, polyploid hybrid progeny, 45S rRNA gene, tetraploidization
Citation: Cao L, Qin Q, Xiao Q, Yin H, Wen J, Liu Q, Huang X, Huo Y, Tao M, Zhang C, Luo K and Liu S (2018) Nucleolar Dominance in a Tetraploidy Hybrid Lineage Derived From Carassius auratus red var. ( ) × Megalobrama amblycephala (
) × Megalobrama amblycephala ( ). Front. Genet. 9:386. doi: 10.3389/fgene.2018.00386
). Front. Genet. 9:386. doi: 10.3389/fgene.2018.00386
Received: 12 June 2018; Accepted: 27 August 2018;
Published: 24 September 2018.
Edited by:
Peng Xu, Xiamen University, ChinaReviewed by:
Chenghui Wang, Shanghai Ocean University, ChinaCopyright © 2018 Cao, Qin, Xiao, Yin, Wen, Liu, Huang, Huo, Tao, Zhang, Luo and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: ShaoJun Liu, bHNqQGh1bm51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.